Abstract
Loss of function (LOF) in IL11RA infers IL11 signaling as important for fertility, fibrosis, inflammation and incompletely penetrant craniosynostosis. The impact of LOF in IL11 has not been characterized. We generated IL11 knockout (Il11−/−) mice that are born in expected ratios and have normal hematological profiles. Lung fibroblasts from Il11−/− mice are resistant to pro-fibrotic stimulation with TGFβ1. Following bleomycin-induced lung injury, Il11−/− mice are protected from pulmonary fibrosis and exhibit lesser ERK, STAT3 and NF-kB activation, reduced Il1b, Timp1, Ccl2 and diminished IL6 expression, both at baseline and after injury: placing Il11 activity upstream of IL6 in this model. Il11−/− female mice are infertile. Unlike Il11ra1−/− mice, Il11−/− mice do not have craniosynostosis, have normal long bone mass and reduced body weights. These data further establish the role of IL11 signaling in lung fibrosis while suggesting that bone development abnormalities can be associated with mutation of IL11RA but not IL11, which may have implications for therapeutic targeting of IL11 signaling.
Subject terms: Extracellular signalling molecules, Interleukins
Introduction
Interleukin 11 (IL11) was originally described as a factor important for hematopoiesis, notably platelet production, but more recently found to drive fibro-inflammatory disorders1. IL11 is a member of the IL6 family of cytokines, which share the gp130 coreceptor, but while IL6 has been studied in very great detail with an armamentarium of genetic tools to dissect its function, IL11 remains poorly characterised1, 2.
It is apparent from the published literature that the majority of our understanding of the biology associated with loss-of-function (LOF) in IL11 signaling is inferred from studies of IL11RA mutant humans or mice1. The field of lL11 biology has lacked a mouse genetic model specific for IL11 LOF, which represents a gap in our understanding. This is important as, in the case of the family member IL6, there are both similarities and differences between effects of LOF in the IL6 cytokine as compared to LOF in its cognate alpha chain receptor (IL6RA)3, 4. As such, it is possible that the phenotype of IL11RA LOF may not map precisely to IL11 function. Furthermore, studies of IL11RA1 LOF have been conducted in a single mouse strain and there are additional genes close by in the targeted locus, which is a potential shortcoming.
Based on the genetic studies of IL11RA mutants, IL11 signaling is thought important for a number of phenotypes. Il11ra1-deleted female mice are infertile5 and mutation in Il11ra1 in the mouse is associated with incompletely penetrant snout displacement and tooth abnormalities: a craniosynostosis-like phenotype6. Several human studies have identified individuals with IL11RA mutations who have mild features of craniosynostosis, joint laxity, scoliosis and delayed tooth eruption, although ascertainment bias has been suggested6–8. Unlike mice, female humans with mutations in IL11RA appear fertile in some studies9.
Here, we report the generation of mice with germline deletion of Il11 that we characterise at baseline and in the context of pro-fibrotic stimulation in vitro and in vivo. We also report similarities and differences between the phenotypes of Il11−/− and Il11ra1−/− mice through direct experimentation and in comparison with the published Il11ra1−/− literature (reviewed in Ref.1).
Results
Generation and anatomical characterization of Il11 knockout mice
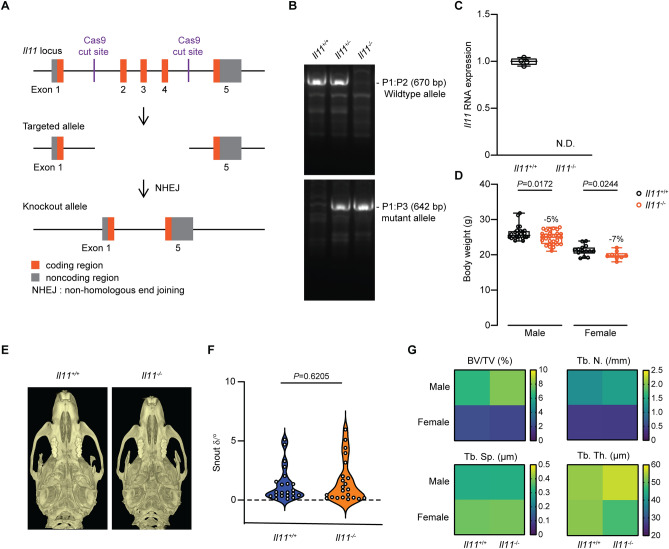
Three separate transcripts of mouse Il11 (ENSMUSG00000004371) have been annotated and using Crispr/Cas9, we deleted Exon 2–4 of the longest transcript (ENSMUST00000094892.11: Il11-201). This deletion causes a reading frame shift after the first two amino acids of IL11 resulting in a mutant 62 amino acids peptide that does not align to any known peptide sequences resulting in the inactivation of all known transcripts (Fig. 1A).
Figure 1.
Generation and anatomical characterization of Il11 knockout mice. (A) Schematic design of Crispr/Cas9 mediated deletion of exons 2 to 4 of the mouse Il11 locus on a C57BL/6J background. (B) Representative genotyping of Il11-deficient or wild-type mice, showing a wild-type band (670 bp) and mutant band (642 bp). (C) RT-qPCR of Il11 expression in whole lung tissue from wild-type and Il11−/− mice (n = 4). N.D., not detected. (D) Body weight of 10–12 week old wild-type (male n = 26; female n = 14) and Il11−/− mice (male n = 29; female n = 9). (E) Representative µCT images showing the ventral view of skulls from wild-type and Il11−/− mice. (F) The degree of deviation from linear snout growth (δ/°) was determined in Il11−/− as compared to wild-type mice (Il11+/+ n = 23 and Il11−/− n = 22). (G) Heatmaps showing the quantification of trabecular bone parameters of Il11−/− and wild-type mice (12 weeks of age); n = 5–7 mice per genotype per gender and quantitative data are plotted in Supplementary Fig. S1. BV/TV, Trabecular bone volume per total volume, Tb.Th., trabecular thickness, Tb. N., trabecular number and Tb. Sp., trabecular separation. Data shown in C, D are: centre line, median value; box edges, 25th and 75th percentiles; whiskers, minimum and maximum values and data in F are shown as a violin plot. P values were determined by Student’s t-test.
Il11 knockout mice (Il11−/−) were generated on a C57BL/6J background and genotypes were determined by sequencing and PCR (Fig. 1B). To address whether the mutant alleles resulted in the loss of Il11 RNA expression, we isolated total RNA from whole lung tissue from Il11−/− mice and did not observe any detectable expression of Il11 RNA by RT-qPCR in Il11−/− mutants (Fig. 1C). We observed a slight (5–7% lower) but statistically significant reduction in body weights of male and female Il11−/− mice (10–12 weeks old) as compared to age and gender matched wild-type controls (Fig. 1D), which has not been reported in Il11ra1−/− mice of a similar age by us or others.
It is reported that approximately 40–50% of Il11ra1−/− mice display a craniosynostosis-like phenotype10. We assessed the skulls of adult mice (> 12 weeks of age) in Il11−/− mice (n = 22) and littermate control wild-type mice (n = 23) and compared data with an analysis of Il11ra1−/− mice (n = 12) maintained locally, also on a C57BL/6J background. Similar to what was reported previously, we observed snout deformities in 42% of Il11ra1−/− mice (5 out of 12 mice) (Supplementary Fig. S1A)10. However, we did not observe significant differences in the proportions of Il11−/− mice with snout deformities as compared to littermate controls (P = 0.6) (Fig. 1E and Supplementary Fig. S1A). In separate blinded analyses by experts in the field using microcomputed tomography (µCT) analysis, it was confirmed that there was no significant difference in snout phenotypes in Il11−/− mice as compared to wild-type mice, unlike snouts of Il11ra1−/− mice that were distorted and served as a positive control (Fig. 1F and Supplementary Fig. S1B).
Previous studies of Il11ra1−/− mice have shown that IL11RA1-deletion results in a high bone mass phenotype11. We characterized the bone phenotype of Il11−/− mice by µCT analysis of the distal femora and showed that trabecular bone parameters were similar in Il11−/− and wild-type mice (Fig. 1G and Supplementary Fig. S1C–F). In gross anatomy studies, the indexed organ-to-body weight ratios of the heart, lung, liver, kidney, spleen and pancreas were comparable in Il11−/− and wild-type mice (Supplementary Fig. S2).
Female Il11 knockout mice are infertile
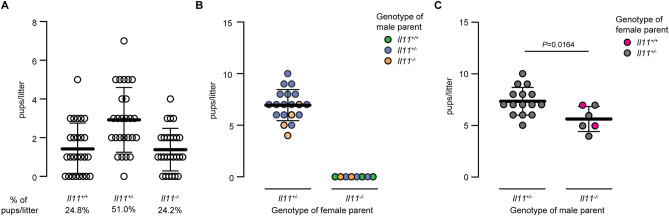
Deletion of Il11ra1 in mice leads to female infertility due to defective embryo implantation, whereas Il11ra1−/− males are fertile5, 12, 13. We found that intercrosses of heterozygotes Il11+/− mice gave rise to viable and apparently normal homozygous (Il11−/−) mutant mice in the expected Mendelian ratios (Fig. 2A). We determined whether maternal Il11 expression is required for fertility by mating homozygous (Il11−/−) female mice with male mice of variable Il11 genotype (Il11+/+, Il11+/− or Il11−/−) and found that female mice deficient for Il11 never had a detectable pregnancy nor gave birth to offspring (Fig. 2B), which mirrors the infertility phenotype of homozygous Il11ra1−/− female mice5. Crossing homozygous (Il11−/−) male mice with either wild-type (Il11+/+) or heterozygous (Il11+/−) female mice resulted in viable offspring of expected Mendelian ratios. However, litter sizes derived from Il11−/− male mice were significantly smaller as compared to intercrosses of heterozygotes (Fig. 2C).
Figure 2.
Il11 knockout female mice are infertile. (A) Genotype distribution of pups per litter in heterozygous breeding (Il11+/− X Il11+/−; n = 26 litters). (B) Litter size based on parents’ genotype. (C) Litter size based on the breeding of heterozygous Il11+/− or homozygous Il11−/− male mice with wild-type or heterozygous Il11+/− female mice. Data shown as mean ± SD. P value was determined by Student’s t test.
Blood hematology and chemistry profiles are normal in Il11 knockout mice
We evaluated the hematological profile of adult Il11−/− mice (10–14 weeks of age) and observed that null mice had normal peripheral red and white blood cell counts as well as normal platelet counts and volumes as compared to wild-type mice (Table 1). Likewise, we profiled serum chemistry and observed normal levels of serum markers of liver function (albumin, alanine aminotransferase, total bilirubin), kidney function (blood urea nitrogen, sodium and potassium) and bone turnover (alkaline phosphatase, calcium and phosphate) in Il11−/− mice (Table 1). These data indicate that Il11−/− mice have ordinary blood hematological and chemistry profiles under normal physiological conditions.
Table 1.
Hematology and serum metabolic profiles of Il11 knockout mice.
| Blood counts | Il11+/+ (n = 5) | Il11−/− (n = 7) | Sig.a |
|---|---|---|---|
| WBC (109/L) | 6.74 ± 2.60 | 6.30 ± 1.60 | NS |
| Lymphocytes (109/L) | 5.00 ± 1.83 | 4.84 ± 1.67 | NS |
| Monocytes (109/L) | 0.23 ± 0.16 | 0.31 ± 0.31 | NS |
| Neutrophils (109/L) | 1.52 ± 1.65 | 1.15 ± 0.98 | NS |
| RBC (1012/L) | 10.18 ± 2.46 | 9.02 ± 0.21 | NS |
| HGB (g/dL) | 11.98 ± 2.23 | 13.52 ± 0.43 | NS |
| HCT (%) | 45.42 ± 12.16 | 40.66 ± 1.16 | NS |
| Platelets (109/L) | 521 ± 358 | 387 ± 75 | NS |
| Mean Platelet Volume (fl) | 6.12 ± 0.16 | 6.15 ± 0.26 | NS |
| Serum metabolic markers | Il11+/+ (n = 5) | Il11−/− (n = 6) | Sig.a |
|---|---|---|---|
| Albumin (g/dL) | 3.34 ± 0.32 | 3.23 ± 0.30 | NS |
| Alkaline Phosphatase (U/L) | 89.20 ± 7.33 | 98.83 ± 21.15 | NS |
| Alanine Aminotransferase (U/L) | 35.00 ± 20.10 | 59.17 ± 27.69 | NS |
| Amylase (U/L) | 730 ± 142 | 696 ± 91 | NS |
| Total Bilirubin (mg/dL) | 0.32 ± 0.04 | 0.32 ± 0.04 | NS |
| Blood Urea Nitrogen (mg/dL) | 20.00 ± 5.39 | 19.67 ± 3.56 | NS |
| Calcium (mg/dL) | 9.62 ± 0.16 | 9.67 ± 0.55 | NS |
| Phosphate (mg/dL) | 10.88 ± 2.20 | 10.57 ± 0.96 | NS |
| Glucose (mg/dL) | 328 ± 108 | 389 ± 91.91 | NS |
| Sodium (mmol/L) | 147 ± 1.87 | 143 ± 3.92 | NS |
| Potassium (mmol/L) | 5.24 ± 0.44 | 5.43 ± 0.67 | NS |
| Total Protein (g/dL) | 4.34 ± 0.17 | 4.10 ± 0.13 | NS |
| Globulin (g/dL) | 0.98 ± 0.19 | 0.83 ± 0.23 | NS |
Data shown as mean ± SD.
WBC white blood cells, RBC red blood cells, HGB hemoglobin count, HCT hematocrit, NS non-significant.
aSignificance was determined using Bonferroni post-hoc test.
IL11 is required for myofibroblast differentiation
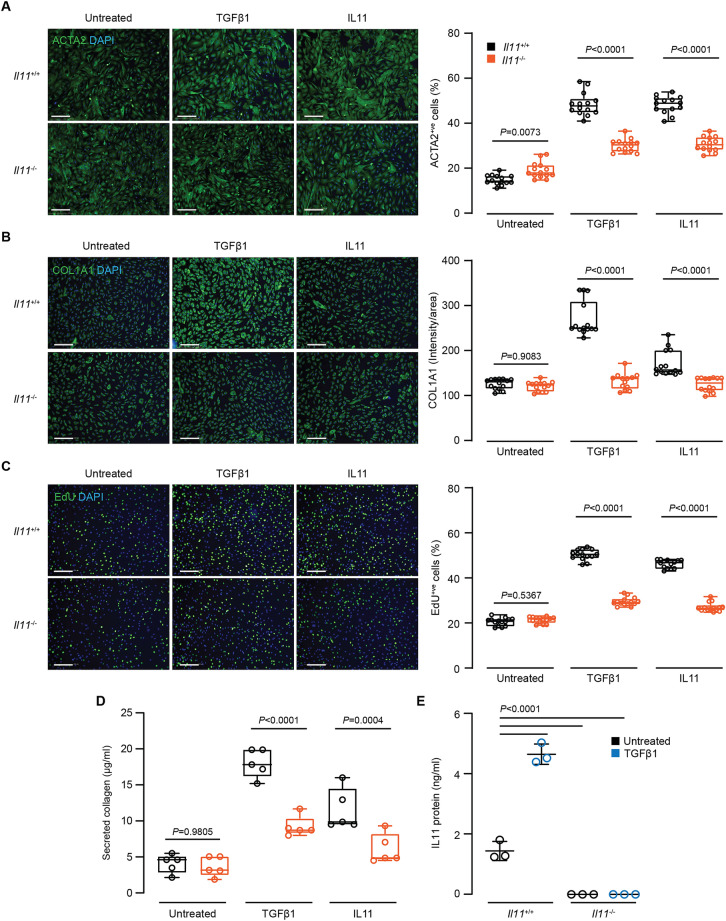
We found previously that TGFβ1-induced myofibroblast transdifferentiation is impaired in Il11ra1−/− lung fibroblasts14. To examine whether the loss of the endogenous IL11 autocrine feed-forward loop similarly perturbed fibroblast activation, we stimulated lung fibroblasts from Il11−/− mice with recombinant mouse TGFβ1 or IL11 (5 ng/ml; 24 h) and monitored fibroblast activation using automated high-throughput immunofluorescence imaging and Sirius red-based quantification of secreted collagen. In keeping with the data from Il11ra1-deleted fibroblasts, the differentiation of Il11−/− fibroblasts into ACTA2+ve and COL1A1 expressing myofibroblasts following TGFβ1 stimulation was significantly diminished (Fig. 3A,B). Cell proliferation (as determined by EdU+ve staining) and secreted collagen levels into the culture supernatant were also significantly reduced in Il11−/− fibroblasts following TGFβ1 stimulation (Fig. 3C,D).
Figure 3.
Reduced activation of primary lung fibroblasts from Il11 knockout mice. Automated immunofluorescence quantification of (A) ACTA2+ve cells, (B) COL1A1 expression (intensity/area) and (C) EdU+ve cells in TGFβ1- or IL11-treated lung fibroblasts from Il11−/− or wild-type mice (5 ng/ml, 24 h). One representative dataset from two independent biological experiments is shown (14 measurements per condition per experiment). Scale bars: 200 µm. (D) Secreted collagen concentrations in supernatant of cells treated as described in (A,B) were quantified (n = 5). (E) Secreted IL11 in culture supernatant from TGFβ1-treated lung fibroblasts from Il11−/− or wild-type mice (5 ng/ml, 24 h; n = 3). Data in (A–D) are shown as: centre line, median value; box edges, 25th and 75th percentiles; whiskers, minimum and maximum values; and shown as mean ± SD in (E). P values in (A–D) were determined by ANOVA (Sidak’s test) and by ANOVA (Tukey’s test) in (E).
We next addressed whether disruption of the IL11 locus prevented IL11 protein expression by performing ELISA on culture supernatants and found that IL11 protein was not expressed by Il11−/− lung fibroblasts at baseline or after TGFβ1 stimulation (Fig. 3E). Interestingly, recombinant mouse IL11 (5 ng/ml) did not fully restore pro-fibrotic phenotypes in Il11−/− fibroblasts (Fig. 3A–D). We determined the expression of IL11RA in Il11−/− lung fibroblasts by immunofluorescence staining and detected reduced levels of IL11RA expression in IL11-deleted cells (Supplementary Fig. S3), which might contribute towards a diminished response to exogenous IL11 stimulation, as may the absence of an IL11 feed-forward autocrine loop.
Bleomycin-induced pulmonary fibrosis is attenuated in Il11 knockout mice
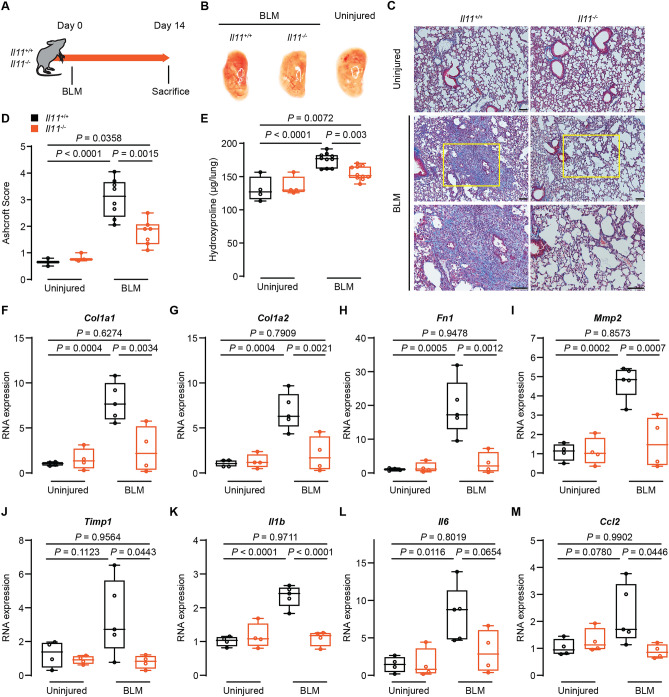
IL11 expression is elevated in the mouse lung after bleomycin (BLM)-induced injury and BLM-induced lung fibrosis is attenuated in Il11ra1−/− mice15. To determine if the genetic deletion of Il11 provided similar protection, we challenged Il11−/− mice with BLM. In preliminary experiments, we found that Il11−/− mice had a higher mortality rate compared to Il11ra1−/− mice, with the same dose/protocol we used previously15. We therefore lowered the dose of BLM and chose to evaluate the mice lungs at an earlier time point (14 days post-BLM) when fibrosis is equally established (Fig. 4A).
Figure 4.
Bleomycin-induced pulmonary fibrosis is attenuated in Il11 knockout mice. (A) Schematic showing the induction of lung fibrosis in Il11−/− and wild-type mice. A single dose of bleomycin (BLM) was administered oropharyngeally and the mice were sacrificed 14 days post-BLM. (B) Representative gross lung anatomy of Il11−/− and wild-type mice 14 days post-BLM. (C) Masson’s trichrome staining of lung sections, (D) histology assessment of fibrosis and (E) total lung hydroxyproline content of Il11−/− and wild-type mice 14 days post-BLM. Scale bars: 100 µm. (Il11+/+ uninjured n = 3–4; Il11+/+ + BLM n = 7–10; Il11−/− uninjued n = 3–4; Il11−/− + BLM n = 7–10). Relative RNA expression of (F) Col1a1, (G) Col1a2, (H) Fn1, (I) Mmp2, (J) Timp1, (K) Il1b, (L) Il6 and (M) Ccl2 in lung lysates from Il11−/− and wild-type mice 14 days post-BLM (n = 4). Data shown as: centre line, median value; box edges, 25th and 75th percentiles; whiskers, minimum and maximum values. P values were determined by ANOVA (Tukey’s test).
By gross morphology analysis, we observed reduced macroscopic lung damage in Il11−/− mice than that seen in wild-type mice (Fig. 4B). As compared to wild-type mice, following BLM injury Il11−/− mice gained more weight (Day 0–14), had reduced pulmonary infiltrates and alveolar septal thickening whereas indexed lung weights were unchanged (Supplementary Fig. S4). Consistent with these effects, blinded histological analysis of Masson’s trichrome stained lung sections showed that Il11−/− mice had reduced parenchymal disruption and fibrosis (Fig. 4C,D). These changes were associated with significantly lower total lung hydroxyproline (collagen) content in Il11−/− mice (Fig. 4E).
Evaluation of fibrotic gene expression in lung lysates in a subset of representative lung samples showed reduced RNA levels of extracellular matrix and protease genes such as Col1a1, Col1a2, Fn1, Mmp2 and Timp1 in BLM-challenged Il11−/− mice as compared to wild-type mice (Fig. 4F–J). Reduced expression of several inflammatory response genes (such as Il1b, Il6 and Ccl2) were also seen in the lungs of Il11−/− mice following BLM injury (Fig. 4K–M).
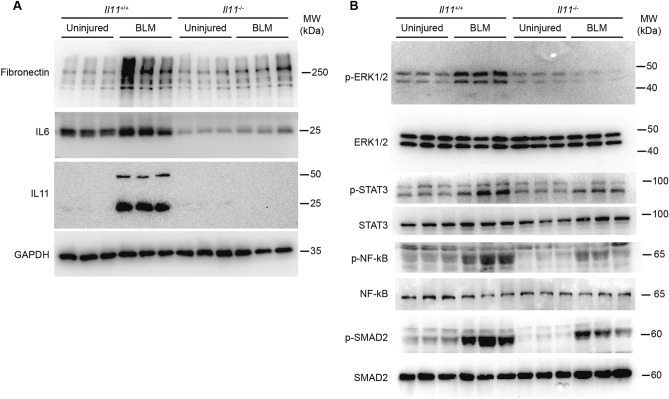
Western blot analysis in a subset of representative lung samples showed that IL11 protein expression was strongly upregulated in the lungs of BLM-injured wild-type mice and was not expressed at all in the lungs of Il11−/− mice at all, as expected (Fig. 5A). Furthermore, in BLM-treated Il11−/− mice, lung protection was accompanied by reduced pulmonary fibronectin and IL6 protein expression (Fig. 5A) and reduced activation of multiple signaling pathways implicated in lung fibro-inflammation including ERK, STAT3, NF-kB and SMAD2 (Fig. 5B). Notably, IL6 levels in Il11−/− mice were lower than wild type control levels even at baseline, in the absence of lung injury. These data show that Il11−/− mice are protected from BLM-induced lung fibrosis and inflammation, similar to Il11ra1−/− mice15, 16.
Figure 5.
Bleomycin-induced pulmonary fibronectin, IL6, IL11 expression and ERK, STAT3, NF-kB and SMAD2 phosphorylation in Il11 knockout and wild type mice. Western blots of (A) fibronectin, IL6 and IL11 and (B) phosphorylated and total protein levels ERK1/2, STAT3, NF-kB and SMAD2 in lung homogenates of Il11−/− and wild-type mice 14 days post-BLM (n = 3 for each genotype/condition).
Discussion
Here we provide a phenotypic description of mice with germline deletion of Il11 and explore how this relates to the phenotypes of Il11ra1-null mice, published previously. We show that IL11 signaling in Il11−/− mice is important for qualitative fibrotic phenotypes in fibroblasts in vitro and lung fibrosis in vivo. These data, taken together with recent studies of Il11ra1−/− mice and the use of anti-IL11 or anti-IL11RA antibodies in mouse models of fibrosis14, 15, 17, firmly establish IL11-mediated ERK signaling as of central importance for fibrosis.
Il11ra1−/− mice have normal hematopoiesis at baseline and after bone marrow or hemolytic stress12 and long term administration of neutralizing antibodies against IL11 or IL11RA have no effect on blood counts in mice17. In agreement with mouse data, there is no description of hematological abnormalities in patients with IL11RA1 mutations18. While IL11 is still considered by some to have a role in platelet homeostasis, the data shown here in Il11−/− mice provide a further line of evidence that IL11 is unrelated to physiological hematopoiesis, although we did not study bone marrow stress.
It was surprising to observe differences in skull morphology and bone phenotypes between Il11−/− and that published, and replicated by us, for Il11ra1−/− genotypes, both maintained on C57BL/6J backgrounds. This said, it was recently shown that Ccl27 expression may be disrupted at the Il11ra1 locus in Il11ra1−/− mice19 although this is unrelated to the craniosynostosis phenotypes seen in humans with bi-allelic IL11RA LOF mutations. The fact that Il11−/− mice do not have snout deformities implies that this is unrelated to the loss of IL11 signaling per se. In a recent study, a biallelic non-synonymous variant in IL6ST (gp130), which results in a loss of IL11-signaling, conferred a craniosynostosis phenotype, which replicated in a mouse model, although again this phenotype is mediated at the membrane receptor level20. However, female mutant mice in the study were fertile and also exhibited reduced LIF signaling. Intriguingly, while LOF mutations in IL11 are common in the general population they have not been associated with craniosynostosis despite large scale sequencing projects8 whereas IL11RA mutations are widely reported6, 7.
IL11 signaling has been reported to play a role in skeletal remodeling by promoting the differentiation and function of osteoblasts and osteoclasts21–23. Intriguingly, Il11−/− mice did not exhibit a high bone mass phenotype, or other bone abnormalities, as observed previously in Il11ra1−/− mice11. This suggests further that the LOF in IL11 itself does not affect bone homeostasis and provides a safety signal if long-term anti-IL11 therapy was ever considered as a therapeutic intervention. While adult Il11−/− mice were found to have lower body weights than their littermate controls, this was not apparent in adult Il11ra1−/− mice which had comparable body weights to their corresponding wild-type controls. This was not evaluated further here and requires additional study.
There are parallels between the variant phenotypes between Il11ra1- and Il11-deficient mice and those seen for Il6- and Il6ra-deficient mice. For instance, physiologically important immune phenotypes associated with loss of canonical IL6-mediated JAK/STAT signaling are seen with both Il6- or Il6ra genotypes. However, dissimilar ERK-mediated wound healing phenotypes are only seen in Il6ra mutants, which are dominant over Il6 LOF effects (i.e. IL6-independent)4. Furthermore, Il6 or Il6ra mutant mice also have different responses to experimental models of colitis3.
A possible explanation for the difference between IL6 family member alpha chain receptor versus ligand mutants could relate to reduced/no interaction of mutant alpha chains with the gp130. Thus, biallelic LOF in one alpha chain (e.g. IL6R) might reduce its competition for the shared gp130 receptor and potentiate the activity of another gp130-binding alpha chain (e.g. IL11RA). This premise could account for the increased ERK signaling seen in IL6RA mutant mice, perhaps reflecting increased IL11-driven ERK signaling4. This concept is also suggested further by human genetics: autosomal recessive mutations in gp130 are associated with craniosynostosis, whereas dominant negative (autosomal dominant) variation is not24, 25.
There is variation in reported fertility phenotypes associated with LOF mutations in the IL11 pathway in mice and humans (Table 2). Female mice lacking Il11RA1 are infertile due to defective decidualization in response to embryo implantation5, 13. In contrast, women with homozygous IL11RA variants appear able to reproduce9, which could be explained by species differences in decidualization26. In this study, we found that Il11−/− female mice are infertile, consistent with a recent report27. Interestingly, we observed a reduction in litter sizes from Il11−/− male mice, suggesting additional male-related effects of IL11 on fertility, which warrants further study.
Table 2.
Fertility reported with genetic loss-of-function in IL11 signaling in mice and humans.
| Gene | Mutation/variant | Gender effects | References |
|---|---|---|---|
| Human | |||
| IL11RA | Exon 4 donor splice site (c.479 + 6T > G) | Females: fertile | 9 |
| Mouse | |||
| Il6st | Gp130 p.R279Q (selective IL11 signaling deficiency) |
Females: homozygous mutants have reduced litter size Males: homozygous mutants have reduced litter size |
20 |
| Il11ra1 | Deletion of Exons 8–13 |
Females: homozygous mutants are infertile due to defective decidulization Males: normal fertility |
5, 12 |
| Il11ra1 | Hypomorphic mutations |
Females: fertility of homozygous mutants are severely impaired Males: normal fertility |
13 |
| Il11 | Crispr/Cas9 frameshift in Exon 3 |
Females: homozygous mutants are infertile (data not shown) Males: not reported |
27 |
| Il11 | Crispr/Cas9 deletion of Exons 2–4. Frameshift from 2nd amino acid (aa), mutant 62 aa protein |
Females: homozygous mutants are infertile Males: homozygous mutants have reduced litter size |
This study |
While IL11 is increasingly recognized as important for tissue fibrosis, more recent data has shown a role for IL11 signaling in inflammatory fibroblasts and stromal immunity in the lung and colon16, 28, 29. Fitting with this, we found that Il11−/− mice were protected from BLM-induced lung inflammation with lower Il1b, Il6 and ccl2 mRNA levels, diminished immune cell NF-kB and STAT3 activation and reduced pulmonary infiltrates. Notably, at the protein level, IL6 was not only not upregulated in the injured lung of Il11−/− mice and IL6 levels were also lower in Il11−/− mouse lungs at baseline. We point out that, for reasons outlined above, we do not attempt to compare the magnitude of protection against lung damage following BLM challenge between the Il11−/− mice used here and previous data from Il11ra1−/− mice15. Instead, we wish to emphasize the shared qualitative protection against lung fibro-inflammation between Il11−/− and Il11ra1−/− genotypes.
In conclusion, loss of IL11 signaling due to mutation in either Il11 or Il11RA1 is protective against lung fibrosis that relates to reduced autocrine IL11 activity in myofibroblasts14, 30. However, the craniosynostosis and bone phenotypes seen in Il11RA1 deficient mice are not seen in Il11−/− mice, suggesting that developmental, STAT3-related bone effects are not due to defective IL11 signaling per se and instead, specific to Il11RA1 LOF. This is in agreement with a recent study that, apart from the fertility issue we describe here, reported no other phenotype in Il11−/− mice27. This is important, as therapeutic targeting of IL11 signaling for fibrotic lung and liver diseases is being considered and it may be that targeting the ligand has potential advantages over the receptor. To facilitate further study of IL11, the Il11−/− mice described in this manuscript have been made available to the scientific community at the Jackson Laboratories repository.
Materials and methods
Animal studies
All experimental procedures were approved and conducted in accordance with guidelines set by the Institutional Animal Care and Use Committee at SingHealth (Singapore) and the SingHealth Institutional Biosafety Committee and with the recommendations in the Guidelines on the Care and Use of Animals for Scientific Purposes of the National Advisory Committee for Laboratory Animal Research (NACLAR) and with the Animal Research: Reporting of In Vivo Experiments (ARRIVE) guidelines. Animals were maintained in a specific pathogen-free environment and given ad libitum access to food and water.
Generation of Il11 knockout mice
Crispr/Cas9 technique was used to knock out the IL11 gene (ENSMUST00000094892.11: Il11-201 transcript). Specific single guide RNA (sgRNA) sequences with recognition sites on introns 1 and 4 along with Cas9 were microinjected into fertilized C57BL/6J zygotes, and subsequently transferred into pseudopregnant mice (Shanghai Model Organisms, Centre, Inc). It is predicted that this deletion would cause a shift in the reading frame from the splicing of coding sequences present within exons 1 and 5, resulting in the generation of a mutant peptide (62 amino acids in length) that does not align to that of known proteins. This effectively results in the inactivation of the gene. Successfully generated F0 mice were identified by PCR and sequencing and further backcrossed to wild type C57BL/6J mice and maintained on this background. Wild-type allele was identified by a 670 bp PCR product using genotyping primer sequences as follows: P1: 5′-CGGGGGCGGACGGGAGACG-3′ and P2: 5′-CCAGGAGGGATCGGGTTAGGAGAA-3′. Whereas, mutant (knockout) alleles were identified by a second PCR reaction using primers P1 and P3: 5′-CAGCTAGGGACGACACTTGAGAT-3′.
Bleomycin model of lung injury
The bleomycin model of lung fibrosis was performed as previously described15. Briefly, female mice (8–10 weeks of age) were anesthetized by isoflurane inhalation and subsequently administered a single dose of bleomycin (Sigma-Aldrich) oropharyngeally at 0.5 mg/kg body weight in a volume of saline not exceeding 50 µl per mouse. Uninjured animals received equal volumes of saline as sham controls. Mice were sacrificed 14 days post-bleomycin administration and lungs were collected for downstream analysis.
Analysis of craniosynostosis-like snout phenotype and trabecular bone mass analysis
Il11ra1−/− mice on a C57BL/6J background were originally described in Ref.12 and obtained from The Jackson’s laboratory. For the analysis of skull phenotypes of Il11−/−, Il11ra1−/− and wild-type mice, the heads of both male and female adult mice (> 12 weeks of age) were dissected and cleaned by removing the soft tissue surrounding the skull. The bones were fixed in 10% neutral buffered formalin. For the analysis of trabecular bone mass, the hindlimbs of 12-week old mice were dissected and tibiae and femora bones were fixed in 10% neutral buffered formalin. Micro-computed tomography of the skulls and femora of l11−/−, Il11ra1−/− and wild-type mice was performed using the Skyscan 1276 system and analysed using NRecon (version 1.6.9.8), Dataviewer (version 1.4.4) and CT analyzer (version 1.11.8.0) as previously described31. For the analysis of skull phenotypes, classification was performed by scoring for the presence (craniosynostosis-like phenotype) or absence (no phenotype) of twisted snouts by two independent investigators blinded to genotypes, and concordant scores were obtained between the investigators. Deviation from linear nasal bone growth was determined by assessing the angle between the tip of the snout bone and the sagittal suture at the base of the frontal bone by ImageJ software analysis (v1.8)32.
Hematologic analysis
Blood was collected by cardiac puncture from anesthetized male and female adult mice (10–14 weeks of age). Differential red and white cell counts, hematocrit, hemoglobin and platelet counts were determined using the VetScan HM5 hematology analyzer (Abraxis Inc.). Blood chemistry profiles from male and female mice were determined using the VetScan VS2 system, partnered with the VetScan comprehensive diagnostic profile discs (Abraxis Inc.).
Reagents
Recombinant proteins: Mouse TGFβ1 (7666-MB, R&D Systems), mouse IL11 (rmIL11, UniProtKB: P47873, GenScript). Antibodies: anti-smooth muscle actin (ab7817, abcam), anti-Collagen I (ab34710, abcam), anti-IL11RA (ab125015, abcam), goat anti-mouse Alexa Flour 488-conjugated secondary antibody (ab150113, abcam), goat anti-rabbit Alexa Flour 488-conjugated secondary antibody (ab150077, abcam), DAPI (D1306, Thermo Fisher Scientific). Primary antibodies for Western blots include: anti-Fibronectin antibody (ab2413, Abcam); anti-IL6 (12912, Cell Signaling); anti-p-ERK1/2 (4370, Cell Signaling), anti-ERK1/2s (4695, Cell Signaling), anti-p-SMAD2 (ab216482, Abcam), anti-p-NF-kB (ab28856, Abcam), anti-p-STAT3 (4113, Cell Signaling), anti-SMAD2 (5339, Cell Signaling), anti-NF-kB (8242, Cell Signaling), anti-STAT3 (4904, Cell Signaling) and anti-GAPDH (2118, Cell Signaling). Monoclonal anti-IL11 antibody (X203), generated in our previous study15, was used to detect IL11 protein expression in tissue lysates. Secondary antibodies for Western blots include: anti-rabbit HRP (7074, Cell Signaling) or anti-mouse HRP (7076, Cell Signaling).
Primary mouse lung fibroblasts cultures
Primary mouse lung fibroblast were isolated from 8 to 12 weeks old Il11−/− and wild-type mice as previously described15. Tissues were minced, digested for 30 min with mild agitation at 37 °C in DMEM (11995-065, Gibco) containing 1% penicillin/streptomycin (P/S, 15140-122, Gibco) and 0.14 Wunsch U ml−1 Liberase (5401119001, Roche). Cells were subsequently cultured in complete DMEM supplemented with 10% fetal bovine serum (10500, Hyclone), 1% P/S, in a humidified atmosphere at 37 °C and 5% CO2. Fresh medium was renewed every 2–3 days. Fibroblasts were allowed to explant from the digested tissues and enriched via negative selection with magnetic beads against mouse CD45 (leukocytes), CD31 (endothelial) and CD326 (epithelial) using a QuadroMACS separator (Miltenyi Biotec) according to the manufacturer’s protocol. All experiments were carried out at low cell passage (< P3) and cells were cultured in serum-free media for 16 h prior to stimulation.
Operetta high-content imaging and analysis
Immunofluorescence imaging and quantification of fibroblast activation were performed on the Operetta High Content Imaging System (PerkinElmer) as previously described14, 15. Briefly, lung fibroblasts were seeded in 96-well CellCarrier black plates (PerkinElmer) and following experimental conditions, the cells were fixed in 4% paraformaldehyde (Thermo Fisher Scientific) and permeabilized with 0.1% Triton X-100 in phosphate-buffered saline (PBS). EdU-Alexa Fluor 488 was incorporated using Click-iT EdU Labelling kit (C10350, Thermo Fisher Scientific) according to manufacturer’s protocol. The cells were then incubated with primary antibodies (anti-ACTA2 or anti-COL1A1) and visualized using anti-mouse or anti-rabbit Alexa Flour 488-conjugated secondary antibodies. Plates were scanned and images were collected with the Operetta high-content imaging system (PerkinElmer). Each treatment condition was run in duplicate wells, and 14 fixed fields were imaged and analysed per condition. The percentage of activated myofibroblasts (ACTA2+ve cells) and proliferating cells (EdU+ve cells) was quantified using the Harmony software version 3.5.2 (PerkinElmer). Quantification of COL1A1 immunostaining was performed using the Columbus software (version 2.7.2, PerkinElmer), and fluorescence intensity was normalized to cell area.
Lung histology analysis
Freshly dissected left lungs from each Il11−/− and wild-type mouse in the bleomycin experiment were fixed in 10% formalin overnight, dehydrated and embedded in paraffin and sectioned for Haematoxylin and Eosin or Masson’s trichrome staining as described previously15. Histological analysis for fibrosis was performed blinded to genotype and treatment exposure according to Ashcroft scoring method33.
Quantification of collagen content in culture supernatant and lung tissue
Detection of soluble collagen in the supernatant of lung fibroblasts cultures was performed as previously described15. Briefly, the cell culture supernatant was concentrated using a Polyethylene glycol concentrating solution (90626, Chondrex) and collagen content was quantified using a Sirius red collagen detection kit (9062, Chondrex), according to the manufacturer’s protocol. Total lung hydroxyproline content of the right caudal lobes of each mouse mouse in the bleomycin experiment were was measured as previously described15, using the Quickzyme Total Collagen assay kit (Quickzyme Biosciences).
ELISA
Detection of secreted IL11 into the supernatant of lung fibroblast cultures was performed using the mouse IL-11 DuoSet ELISA kit according to manufacturers’ instructions.
RT-qPCR
Total RNA was extracted from snap-frozen mouse right lung tissues using Trizol reagent (Invitrogen) followed by RNeasy column (Qiagen) purification and cDNA was prepared using an iScript cDNA synthesis kit (Biorad) following manufacturer's instructions. Quantitative RT–PCR gene expression analysis was performed with QuantiFast SYBR Green PCR kit (Qiagen) using a StepOnePlus (Applied Biosystem). Relative expression data were normalized to Gapdh mRNA expression using the 2−ΔΔCt method. Primers sequences used for are as follows: Il11 5′-AATTCCCAGCTGACGGAGATCACA-3′ and 5′-TCTACTCGAAGCCTTGTCAGCACA-3′; Col1a1 5′-GGGGCAAGACAGTCATCGAA-3′ and 5′-GTCCGAATTCCTGGTCTGGG-3′; Col1a2 5′-AGGATTGGTCAGAGCAGTGT-3′ and 5′-TCCACAACAGGTGTCAGGGT-3′; Fn1 5′-CACCCGTGAAGAATGAAGA-3′ and 5′-GGCAGGAGATTTGTTAGGA-3′; Mmp2 5′-ACAAGTGGTCCGCGTAAAGT-3′ and 5′-AAACAAGGCTTCATGGGGGC-3′; Timp1 5′-GGGCTAAATTCATGGGTTCC-3′ and 5′-CTGGGACTTGTGGGCATATC-3′; Ccl2 5′-GAAGGAATGGGTCCAGACAT-3′ and 5′-ACGGGTCAACTTCACATTCA-3′; Il6 5′-CTCTGGGAAATCGTGGAAAT-3′ and 5′-CCAGTTTGGTAGCATCCATC-3′; Il1b 5′-CACAGCAGCACATCAACAAG-3′ and 5′-GTGCTCATGTCCTCATCCTG-3′; Gapdh 5′-CTGGAAAGCTGTGGCGTGAT-3 and 5′-GACGGACACATTGGGGGTAG-3′.
Western blot analysis
Total proteins were extracted from snap-frozen mouse right lung tissues using RIPA lysis buffer (Thermo Fisher Scientific) and separated by SDS-PAGE, transferred to a PVDF membrane (Biorad), and incubated overnight with the appropriate primary antibodies. Blots were visualized using the ECL detection system (Pierce) with the appropriate secondary antibodies.
Statistical analysis
All statistical analyses were performed using Graphpad Prism (version 8). Statistical analyses were performed using two-sided Student’s t-test, or ANOVA as indicated in the figure legends. For comparisons between multiple treatment groups, P values were corrected for multiple hypothesis testing using Sidak’s test, Tukey’s test or Bonferonni’s post-hoc test. P values < 0.05 were considered statistically significant.
Supplementary Information
Acknowledgements
We would like to acknowledge B. L. George and N. Ko for their support. This research is supported by the National Medical Research Council (NMRC), Singapore STaR awards (NMRC/STaR/0029/2017), MOH‐CIRG18nov‐0002, Goh Foundation, Tanoto Foundation to S.A.C. A.A.W. is supported by NMRC/OFYIRG/0053/2017. S.S is supported by NMRC/OFYIRG18nov-0014.
Author contributions
B.N., A.A.W. and S.A.C. designed the study. B.N., A.A.W., S.V., J.D., S.L., S.G.S. and J.T. performed in vitro and in vivo experiments. N.E.M., E.C.W. and N.A.S. conducted and analysed microCT of mice skulls and bones. B.N., A.A.W., S.V. and S.P.C. analyzed the data. B.N., A.A.W., S.S. and S.A.C. prepared and wrote the manuscript with input from the other co-authors.
Data availability
All data generated and analysed in the current study are presented in the manuscript or available from the corresponding author upon request.
Competing interests
S.A.C. and S.S. are co-inventors of the patent applications (WO2017103108, WO2017103108 A2, WO 2018/109174 A2, WO 2018/109170 A2) for “Treatment of fibrosis”. A.A.W., S. S. and S.A.C. are co-inventors of the patent applications (GB1900811.9, GB 1902419.9, GB1906597.8) for “Treatment of hepatotoxicity, nephrotoxicity, and metabolic diseases''. S. S., S. A.C. and B.N. are co-inventors of the patent application (WO/2019/073057) for “Treatment of SMC mediated disease”. S.A.C. and S.S. are co-founders and shareholders of Enleofen Bio Pte. Ltd., a company that made anti-IL11 therapeutics, which were acquired for further development by Boehringer Ingelheim. All other co-authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Benjamin Ng and Anissa A. Widjaja.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-93623-9.
References
- 1.Cook SA, Schafer S. Hiding in plain sight: Interleukin-11 emerges as a master regulator of fibrosis, tissue integrity, and stromal inflammation. Annu. Rev. Med. 2020;71:263–276. doi: 10.1146/annurev-med-041818-011649. [DOI] [PubMed] [Google Scholar]
- 2.Widjaja AA, Chothani SP, Cook SA. Different roles of interleukin 6 and interleukin 11 in the liver: Implications for therapy. Hum. Vaccin. Immunother. 2020;16:2357–2362. doi: 10.1080/21645515.2020.1761203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sommer J, et al. Interleukin-6, but not the interleukin-6 receptor plays a role in recovery from dextran sodium sulfate-induced colitis. Int. J. Mol. Med. 2014;34:651–660. doi: 10.3892/ijmm.2014.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McFarland-Mancini MM, et al. Differences in wound healing in mice with deficiency of IL-6 versus IL-6 receptor. J. Immunol. 2010;184:7219–7228. doi: 10.4049/jimmunol.0901929. [DOI] [PubMed] [Google Scholar]
- 5.Robb L, et al. Infertility in female mice lacking the receptor for interleukin 11 is due to a defective uterine response to implantation. Nat. Med. 1998;4:303–308. doi: 10.1038/nm0398-303. [DOI] [PubMed] [Google Scholar]
- 6.Nieminen P, et al. Inactivation of IL11 signaling causes craniosynostosis, delayed tooth eruption, and supernumerary teeth. Am. J. Hum. Genet. 2011;89:67–81. doi: 10.1016/j.ajhg.2011.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Keupp K, et al. Mutations in the interleukin receptor IL11RA cause autosomal recessive Crouzon-like craniosynostosis. Mol. Genet. Genom. Med. 2013;1:223–237. doi: 10.1002/mgg3.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miller KA, et al. Diagnostic value of exome and whole genome sequencing in craniosynostosis. J. Med. Genet. 2017;54:260–268. doi: 10.1136/jmedgenet-2016-104215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Keupp K, et al. Mutations in the interleukin receptor IL 11 RA cause autosomal recessive Crouzon-like craniosynostosis. Mol. Genet. Genom. Med. 2013;1:223–237. doi: 10.1002/mgg3.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Agthe M, et al. Mutations in craniosynostosis patients cause defective interleukin-11 receptor maturation and drive craniosynostosis-like disease in mice. Cell Rep. 2018;25:10–18.e5. doi: 10.1016/j.celrep.2018.09.005. [DOI] [PubMed] [Google Scholar]
- 11.Sims NA, et al. Interleukin-11 receptor signaling is required for normal bone remodeling. J. Bone Miner. Res. 2005;20:1093–1102. doi: 10.1359/JBMR.050209. [DOI] [PubMed] [Google Scholar]
- 12.Nandurkar HH, et al. Adult mice with targeted mutation of the interleukin-11 receptor (IL11Ra) display normal hematopoiesis. Blood. 1997;90:2148–2159. doi: 10.1182/blood.V90.6.2148. [DOI] [PubMed] [Google Scholar]
- 13.Bilinski P, Roopenian D, Gossler A. Maternal IL-11Rα function is required for normal decidua and fetoplacental development in mice. Genes Dev. 1998;12:2234–2243. doi: 10.1101/gad.12.14.2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schafer S, et al. IL-11 is a crucial determinant of cardiovascular fibrosis. Nature. 2017;552:110–115. doi: 10.1038/nature24676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ng B, et al. Interleukin-11 is a therapeutic target in idiopathic pulmonary fibrosis. Sci. Transl. Med. 2019;11:eaaw1237. doi: 10.1126/scitranslmed.aaw1237. [DOI] [PubMed] [Google Scholar]
- 16.Ng B, et al. Fibroblast-specific IL11 signaling drives chronic inflammation in murine fibrotic lung disease. FASEB J. 2020 doi: 10.1096/fj.202001045RR. [DOI] [PubMed] [Google Scholar]
- 17.Widjaja AA, et al. Inhibiting interleukin 11 signaling reduces hepatocyte death and liver fibrosis, inflammation, and steatosis in mouse models of non-alcoholic steatohepatitis. Gastroenterology. 2019 doi: 10.1053/j.gastro.2019.05.002. [DOI] [PubMed] [Google Scholar]
- 18.Brischoux-Boucher E, et al. IL11RA-related Crouzon-like autosomal recessive craniosynostosis in 10 new patients: Resemblances and differences. Clin. Genet. 2018;94:373–380. doi: 10.1111/cge.13409. [DOI] [PubMed] [Google Scholar]
- 19.Widjaja AA, et al. Redefining IL11 as a regeneration-limiting hepatotoxin and therapeutic target in acetaminophen-induced liver injury. Sci. Transl. Med. 2021;13:eaba8146. doi: 10.1126/scitranslmed.aba8146. [DOI] [PubMed] [Google Scholar]
- 20.Schwerd T, et al. A variant in IL6ST with a selective IL-11 signaling defect in human and mouse. Bone Res. 2020;8:1–12. doi: 10.1038/s41413-020-0098-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Takeuchi Y, et al. Interleukin-11 as a stimulatory factor for bone formation prevents bone loss with advancing age in mice. J. Biol. Chem. 2002;277:49011–49018. doi: 10.1074/jbc.M207804200. [DOI] [PubMed] [Google Scholar]
- 22.Kudo O, et al. Interleukin-6 and interleukin-11 support human osteoclast formation by a RANKL-independent mechanism. Bone. 2003;32:1–7. doi: 10.1016/S8756-3282(02)00915-8. [DOI] [PubMed] [Google Scholar]
- 23.Matsumoto T, Kuriwaka-Kido R, Kondo T, Endo I, Kido S. Regulation of osteoblast differentiation by interleukin-11 via AP-1 and Smad signaling. Endocr. J. 2012;59:91–101. doi: 10.1507/endocrj.EJ11-0219. [DOI] [PubMed] [Google Scholar]
- 24.Schwerd T, et al. A biallelic mutation in IL6ST encoding the GP130 co-receptor causes immunodeficiency and craniosynostosis. J. Exp. Med. 2017;214:2547–2562. doi: 10.1084/jem.20161810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Béziat, V. et al. Dominant-negative mutations in human IL6ST underlie hyper-IgE syndrome. J. Exp. Med.217, e20191804 (2020). 10.1084/jem.20191804 [DOI] [PMC free article] [PubMed]
- 26.Dimitriadis E, Salamonsen LA, Robb L. Expression of interleukin-11 during the human menstrual cycle: Coincidence with stromal cell decidualization and relationship to leukaemia inhibitory factor and prolactin. Mol. Hum. Reprod. 2000;6:907–914. doi: 10.1093/molehr/6.10.907. [DOI] [PubMed] [Google Scholar]
- 27.Deguchi Y, et al. Generation of and characterization of anti-IL-11 antibodies using newly established Il11-deficient mice. Biochem. Biophys. Res. Commun. 2018;505:453–459. doi: 10.1016/j.bbrc.2018.09.128. [DOI] [PubMed] [Google Scholar]
- 28.Smillie CS, et al. Intra- and inter-cellular rewiring of the human colon during ulcerative colitis. Cell. 2019;178:714–730.e22. doi: 10.1016/j.cell.2019.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ng B, Cook SA, Schafer S. Interleukin-11 signaling underlies fibrosis, parenchymal dysfunction, and chronic inflammation of the airway. Exp. Mol. Med. 2020;52:1871–1878. doi: 10.1038/s12276-020-00531-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dong J, et al. Hepatocyte-specific IL11 cis-signaling drives lipotoxicity and underlies the transition from NAFLD to NASH. Nat. Commun. 2021;12:66. doi: 10.1038/s41467-020-20303-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vrahnas C, et al. Increased autophagy in EphrinB2-deficient osteocytes is associated with elevated secondary mineralization and brittle bone. Nat. Commun. 2019;10:3436. doi: 10.1038/s41467-019-11373-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rueden CT, et al. Image J2: ImageJ for the next generation of scientific image data. BMC Bioinform. 2017;18:529. doi: 10.1186/s12859-017-1934-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ashcroft T, Simpson JM, Timbrell V. Simple method of estimating severity of pulmonary fibrosis on a numerical scale. J. Clin. Pathol. 1988;41:467–470. doi: 10.1136/jcp.41.4.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated and analysed in the current study are presented in the manuscript or available from the corresponding author upon request.