Abstract
Multiparametric assays for risk stratification are widely used in the management of both node negative and node positive hormone receptor positive invasive breast cancer. Recent data from multiple sources suggests that different tests may provide different risk estimates at the individual patient level. The TEAM pathology study consists of 3284 postmenopausal ER+ve breast cancers treated with endocrine therapy Using genes comprising the following multi-parametric tests OncotypeDx®, Prosigna™ and MammaPrint® signatures were trained to recapitulate true assay results. Patients were then classified into risk groups and survival assessed. Whilst likelihood χ2 ratios suggested limited value for combining tests, Kaplan–Meier and LogRank tests within risk groups suggested combinations of tests provided statistically significant stratification of potential clinical value. Paradoxically whilst Prosigna-trained results stratified Oncotype-trained subgroups across low and intermediate risk categories, only intermediate risk Prosigna-trained cases were further stratified by Oncotype-trained results. Both Oncotype-trained and Prosigna-trained results further stratified MammaPrint-trained low risk cases, and MammaPrint-trained results also stratified Oncotype-trained low and intermediate risk groups but not Prosigna-trained results. Comparisons between existing multiparametric tests are challenging, and evidence on discordance between tests in risk stratification presents further dilemmas. Detailed analysis of the TEAM pathology study suggests a complex inter-relationship between test results in the same patient cohorts which requires careful evaluation regarding test utility. Further prognostic improvement appears both desirable and achievable.
Subject terms: Prognostic markers, Breast cancer
Introduction
Multi-parametric molecular tests are central to the treatment management of early breast cancer and their use is incorporated into most major guidelines1 as a pre-requisite for the staging of breast cancer patients, to direct prognostication and to select patients for chemotherapy treatment2,3. Two major challenges related to their use need to be addressed. Firstly, reports highlighting disagreements between tests are disquieting for physicians, health care providers, and patients alike4 since they raise the question “have I recommended/received the right test?” Secondly, the lack of consistency at an individual patient level between different tests suggests additional prognostic information may result from novel tests. Recent results from the MINDACT and TAILORx studies validate the utility of tests to direct chemotherapy use in node-negative patients2,5,6, which may be extended as new evidence emerges from retrospective3 or prospective studies7,8. In this context an error in assigning appropriate risk classifications would have significant impact on patient treatment and outcomes. Additionally, given recent evidence documenting the long-term risk of relapse for ER+ve breast cancer and the increasing use of extended endocrine therapy9 the selection of the appropriate test to detect recurrence risk over extended time periods is also critical.
Reports of disagreements between tests, based on in silico analyses of existing expression array data, were frequently attributed to methodological challenges and incomplete gene coverage10–14. However, recently direct comparisons, where tests were performed exactly to vendor protocols, demonstrate marked disagreement in risk categorization and subtyping of individual tumors between widely used multiparameter assays4. Furthermore, comparisons between tests in clinical trials derived cohorts provide consistent evidence that combining test results generally improves prognostic value15,16. These results may reflect the relatively modest performance of individual multiparametric tests17.
To date, no direct comparison between different multiparameter assays in a large patient cohort with associated follow-up provides robust information on the impact of discrepant test results for patients. We developed a method to compare signatures using a combined quantitative mRNA array covering key molecular signatures17, trained against the results of the same signatures measured by original methodology18. We analyzed >3000 samples from the TEAM pathology cohort19 using “trained” signatures to demonstrate the impact of disagreements between tests on patient outcome in the context of a recent clinical trial cohort.
Results
Comparing signature-trained risk scores—Likelihood ratios
We compared the ability of trained signatures to predict DMFS10 using the likelihood ratio χ2(LRχ2) based on the Cox models as a measure of the overall prognostic information provided by each model. We illustrated the performance of each “trained” test using Kaplan–Meier survival curves and estimated Hazard ratios as described above (see Fig. 1). We calculated the change in LRχ2 values(ΔLRχ2) between the reclassified and single signature models to assess prognostic improvement of reclassification with a second signature versus the single signature using existing trinary and binary (Table 1) cut points as outlined above.
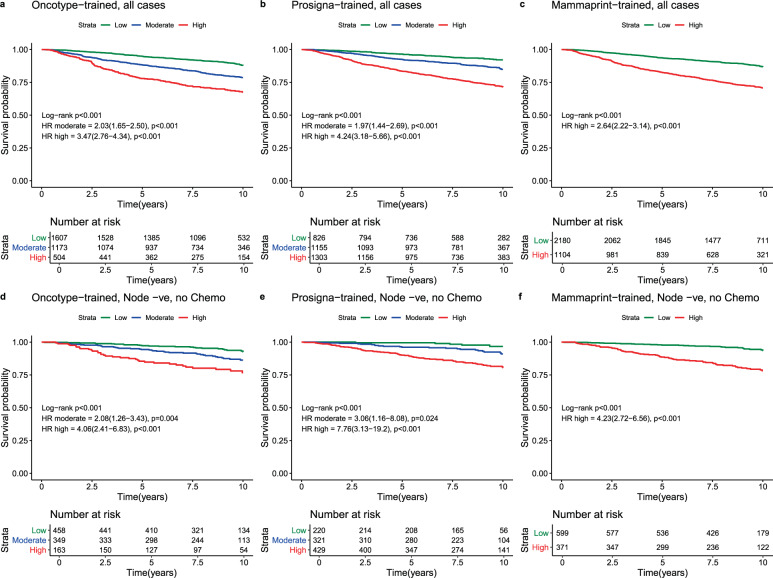
Fig. 1. Test performance in ER+ve, HER2-ve breast cancer from the TEAM cohort.
Kaplan–Meier survival curves with Log-rank Hazard ratios for cases of ER+ve, HER2−ve breast cancer from the entire TEAM cohort for Oncotype-trained (Panel a), Prosigna-trained (Panel b), and Mammaprint-trained results (Panel c) and for ER+ve, HER2−ve Node negative breast cancers treated without chemotherapy from the TEAM cohort for Oncotype-trained (Panel d), Prosigna-trained (Panel e), and Mammaprint-trained results (Panel 5). Log-Rank P values for each test are in brackets. Within each panel low (green), moderate (blue) and high (red) risk survival curves are plotted with LogRank Hazard ratios for high risk and intermediate risk (Oncotype-trained and Prosigna-trained only) calculated against low risk cases in each sub-group. 95% Confidence intervals for LogRank Hazard ratios are in brackets. For each group the number at risk (Low, moderate, high) are presented under the X axis.
Table 1.
Likelihood χ2 ratios by test and cohort.
| ER+/HER2− (N = 3284) | ||||||
|---|---|---|---|---|---|---|
| Trinary classification | Binary classification | |||||
| df | LRχ2 | p-value | df | LRχ2 | p-value | |
| Univariate models | ||||||
| Oncotype | 2 | 118.0 | <0.0001 | 1 | 109.87 | <0.0001 |
| Prosigna | 2 | 146.9 | <0.0001 | 1 | 127.31 | <0.0001 |
| Mammaprint | 1 | 119.5 | <0.0001 | 1 | 119.45 | <0.0001 |
| Bivariate models | ||||||
| Oncotype + Prosigna | 4 | 177.9 | <0.0001 | 2 | 164.47 | <0.0001 |
| Oncotype + Mammaprint | 3 | 145.7 | <0.0001 | 2 | 143.34 | <0.0001 |
| Prosigna + Mammaprint | 3 | 168.8 | <0.0001 | 2 | 155.11 | <0.0001 |
| Bivariate vs. univariate | ||||||
| Oncotype + Prosigna vs. Oncotype | 2 | 59.97 | <0.0001 | 1 | 54.60 | <0.0001 |
| Oncotype + Mammaprint vs. Oncotype | 1 | 27.78 | <0.0001 | 1 | 33.48 | <0.0001 |
| Prosigna + Oncotype vs. Prosigna | 2 | 31.02 | <0.0001 | 1 | 37.16 | <0.0001 |
| Prosigna + Mammaprint vs. Prosigna | 1 | 21.89 | <0.0001 | 1 | 27.80 | <0.0001 |
| MammaPrint + Oncotype vs. Mammaprint | 2 | 26.28 | <0.0001 | 1 | 23.89 | <0.0001 |
| Mammaprint + Prosigna vs. Mammaprint | 2 | 49.34 | <0.0001 | 1 | 35.65 | <0.0001 |
LRχ2 = likelihood ratio chi-squared value, all models run exiting at 10 years. Likelihood χ2 ratios(LRχ2) for univariate(single test) or bivariate(two tests in sequence) derived using 10-year distant metastasis free survival as end point, ER+/HER2+ve cases = all ER+ve/HER2−ve cases (irrespective of nodal status and chemotherapy), ΔLRχ2 = change in Likelihood χ2 ratio when two tests are used sequentially. Trinary classification: results using results from Oncotype-Dx trained and Prosigna-trained tests categorized as low, intermediate, and high risk, binary classification: results using dichotomous results for all tests, see text for cut-points, ΔLRχ2 = change in LRχ2 for comparison of 2 tests versus a single test.
In ER+/HER2− cases (n = 3284), the Prosigna-trained signature provided greater prognostic information compared to Oncotype-trained and MammaPrint-trained signatures(LRχ2 = 146.9 vs. 118.0 and 119.5, respectively; Table 1). In bivariate models (combining 2 tests) the greatest LRχ2 was observed with Oncotype-trained and Prosigna-trained results (Table 1). Comparing bivariate and univariate results combining Oncotype-trained and Prosigna-trained results increased the LRχ2 to a far greater extent versus Oncotype-trained results (ΔLRχ2 = 60.0) than versus Prosigna-trained (ΔLRχ2 = 31.0) results. Similarly, when combining tests with Mammaprint-trained results adding Prosigna-trained results showed a greater increase in LRχ2 (ΔLRχ2 = 49.3) than did combining Mammaprint-trained results with Oncotype-trained results (ΔLRχ2 = 26.3). Adding Mammaprint-trained results to either Oncotype-trained or Prosigna-trained results to, versus either test produced the smallest improvements in the LRχ2 (Table 1). Nonetheless, all test combinations outperformed single tests to a highly statistically significant degree (p < 0.0001; Table 1).
When test results for Oncotype-trained and Prosigna-trained results were dichotomized, there were less marked differences in univariate models between these tests and Mammaprint-trianed results (Table 1). Again the largest increase in LRχ2 was observed when comparing combined Oncotype-trained and Prosigna-trained classification versus Oncotype-trained alone. All other bivariate models outperformed univariate models to a lesser, but still statistically significant, degree (p < 0.0001; Table 1).
Analysis of test performance by outcome in reclassified patients
We analyzed agreement between tests by investigating the extent to which re-classifying results for individual patients by performing tests in sequence affected predicted outcome. Example, we estimated the effects of performing a Prosigna-trained test on tumors previously classified as intermediate risk by the Oncotype-trained test.
Entire ER+ve/HER2−ve population
Oncotype-trained
Of 3284 ER+ve/HER2−ve breast cancers with results for the Oncotype-trained risk classification, 48.9% were classified low risk (DMFS10 = 87.9%), 35.8% intermediate risk (DMFS10 = 78.6%) and 15.3% high risk (DMFS10 = 67.5%) (Table 2; Figs. 1a, 2).
Table 2.
Oncotype-trained results stratified by other test results, trinary classification.
| Oncotype-trained first | |||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Oncotype-trained low risk | Oncotype-trained intermediate risk | Oncotype-trained high risk | |||||||||||||||||||||||||
| HR (95% CI) | DMFS (95% CI) | P* (N) | HR (95% CI) | DMFS (95% CI) | P (N) | HR (95% CI) | DMFS (95% CI) | P (N) | |||||||||||||||||||
| All cases | REF |
87.9 (85.8–89.6) |
<0.001 (1607) |
2.03 (1.65–2.50) |
78.6 (75.9–81.1) |
<0.001 (1173) |
3.47 (2.76–4.34) |
67.5 (62.8–71.7) |
<0.001 (504) |
||||||||||||||||||
| N−Ch− | REF |
92.5 (88.8–95.0) |
<0.001 (458) |
2.08 (1.26–3.43) |
86.3 (81.6–89.9) |
0.004 (349) |
4.06 (2.41–6.83) |
76.7 (68.4–83.0) |
<0.001 (163) |
||||||||||||||||||
| N+Ch− | REF |
86.4 (83.1–89.1) |
<0.001 (683) |
2.03 (1.48–2.78) |
77.0 (72.0–81.2) |
<0.001 (403) |
4.64 (3.32–6.47) |
55.1 (46.4–63.0) |
<0.001 (161) |
||||||||||||||||||
| Ch+ | REF |
85.4 (81.2–88.8) |
<0.001 (463) |
2.03 (1.45–2.82) |
73.8 (68.8–78.2) |
<0.001 (418) |
2.48 (1.68–3.65) |
70.8 (63.0–77.2) |
<0.001 (179) |
||||||||||||||||||
| Prosigna-trained low | Prosigna trained Int | Prosigna trained high | Prosigna-trained low | Prosigna trained Int | Prosigna-trained high | Prosigna-trained low | Prosigna-trained Int | Prosigna-trained high | |||||||||||||||||||
| HR | DMFS | P* | HR | DMFS | P | HR | DMFS | P | HR | DMFS | P* | HR | DMFS | P | HR | DMFS | P | HR | DMFS | P* | HR | DMFS | P | HR | DMFS | P | |
| All cases | REF |
92.2 (89.5–94.2) |
<0.001 (643) |
1.39 (0.94–2.06) |
88.5 (85.3–91.0) |
0.099 (685) |
3.19 (2.12–4.82) |
75.4 (68.3–81.1) |
<0.001 (279) |
REF |
91.5 (85.2–95.2) |
<0.001 (174) |
2.41 (1.30–4.48) |
81.4 (76.5–85.4) |
0.005 (381) |
3.70 (2.05–6.67) |
73.3 (69.2–77.0) |
<0.001 (618) |
REF | 100 |
0.114 (9) |
NA |
72.6 (61.0–81.3) |
NA (89) |
NA |
65.7 (60.4–70.5) |
NA (406) |
| N−Ch− | REF |
97.1 (92.5–98.9) |
0.011 (174) |
2.52 (0.80–7.92) |
92.4 (86.3–95.8) |
0.114 (189) |
5.08 (1.59–16.2) |
83.8 (70.1–91.6) |
0.006 (95) |
REF |
94.1 (65.0–99.1) |
0.090 (43) |
4.09 (0.52–31.9) |
87.5 (77.3–93.4) |
0.180 (106) |
6.09 (0.83–44.8) |
83.7 (77.1–88.5) |
0.076 (200) |
REF | 100 |
0.058 (3) |
NA |
94.7 (68.1–99.2) |
NA (26) |
NA |
72.8 (63.4–80.2) |
NA (134) |
| N+Ch− | REF |
89.3 (84.4–92.7) |
<0.001 (263) |
0.93 (0.53–1.62) |
89.1 (84.1–92.5) |
0.795 (309) |
2.88 (1.63–5.07) |
70.3 (58.1–79.6) |
<0.001 (111) |
REF |
94.3 (83.4–98.1) |
<0.001 (54) |
2.81 (0.83–9.41) |
81.8 (73.1–87.9) |
0.094 (142) |
5.68 (1.78–18.2) |
69.0 (61.4–75.4) |
0.003 (207) |
REF | 100 |
0.148 (1) |
NA |
74.5 (51.7–87.7) |
NA (25) |
NA |
51.1 (41.5–59.8) |
NA (135) |
| Ch+ | REF |
91.9 (86.5–95.2) |
<0.001 (205) |
1.98 (1.03–3.80) |
83.4 (76.0–88.7) |
0.041 (185) |
3.76 (1.83–7.70) |
71.7 (57.0–82.1) |
<0.001 (73) |
REF |
87.9 (76.9–93.9) |
0.007 (77) |
2.26 (1.03–4.94) |
76.2 (67.4–83.0) |
0.041 (133) |
3.03 (1.45–6.33) |
67.4 (59.7–74.0) |
0.003 (208) |
REF | 100 |
0.108 (5) |
NA |
56.4 (37.0–71.9) |
NA (38) |
NA |
73.8 (65.1–80.7) |
NA (136) |
| Mammaprint-trained low | Mammaprint trained high | Mammaprint-trained low | Mammaprint trained high | Mammaprint-trained low | Mammaprint trained high | ||||||||||||||||||||||
| HR | DMFS | P* | HR | DMFS | P | HR | DMFS | P* | HR | DMFS | P | HR | DMFS | P* | HR | DMFS | P | ||||||||||
| All cases | REF |
89.1 (87.1–90.8) |
<0.001 (1483) |
2.79 (1.83–4.25) |
72.1 (60.9–80.6) |
<0.001 (124) |
REF |
83.2 (79.6–86.1) |
<0.001 (645) |
1.70 (1.30–2.23) |
73.2 (68.8–77.2) |
<0.001 (528) |
REF |
70.4 (53.7–82.1) |
0.465 (52) |
1.24 (0.70–2.18) |
67.1 (62.2–71.6) |
0.466 (452) |
|||||||||
| N−Ch− | REF |
93.8 (90.0–96.2) |
0.002 (407) |
3.57 (1.49–8.55) |
80.8 (62.8–90.7) |
0.004 (51) |
REF |
92.2 (85.6–95.8) |
0.003 (174) |
2.87 (1.40–5.88) |
80.8 (73.4–86.4) |
0.004 (175) |
REF | 100 |
0.034 (18) |
NA |
74.0 (65.0–81.0) |
NA (145) |
|||||||||
| N+Ch− | REF |
87.7 (84.4–90.3) |
<0.001 (639) |
2.94 (1.55–5.59) |
67.6 (48.0–81.2) |
0.001 (44) |
REF |
84.2 (78.2–88.7) |
<0.001 (237) |
2.30 (1.47–3.60) |
66.9 (58.3–74.1) |
<0.001 (166) |
REF |
54.2 (28.0–74.5) |
0.945 (20) |
1.03 (0.49–2.15) |
55.3 (46.0–63.6) |
0.945 (141) |
|||||||||
| Ch+ | REF |
86.8 (82.6–90.0) |
0.002 (434) |
3.03 (1.43–6.41) |
64.2 (37.6–81.8) |
0.004 (29) |
REF |
75.8 (69.1–81.2) |
0.318 (233) |
1.23 (0.82–1.83) |
71.2 (63.0–77.9) |
0.319 (185) |
REF |
61.1 (29.8–81.9) |
0.537 (14) |
0.75 (0.30–1.89) |
71.9 (63.8–78.4) |
0.538 (165) |
|||||||||
HR = hazard ratio, 95% CI = 95% confidence interval, P* = p value of log-rank test to compare survival distributions, REF = reference group, P = p value of Wald test for comparison versus reference (low risk) group, DMFS = distant metastasis free survival at 10 years (see text), (N) = number of cases in subgroups, All cases = all ER+ve/HER2-ve cases, N−Ch− = Node negative cases treated without chemotherapy, N+Ch− = Node positive cases treated without chemotherapy, Ch+ = cases treated with chemotherapy (node negative and node positive combined), Int = intermediate, P*:p-value of log-rank test to compare survival distributions (global statistical significance of the model).P: p-value of Wald-test to evaluate whether the hazard ratio is 1 (statistical significance of each individual coefficient).
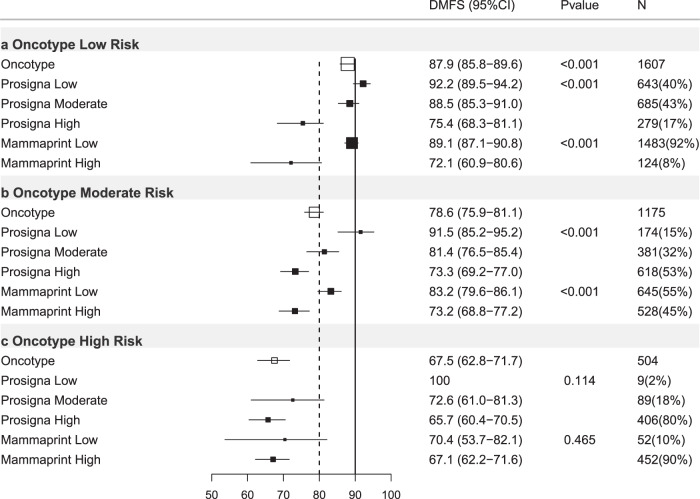
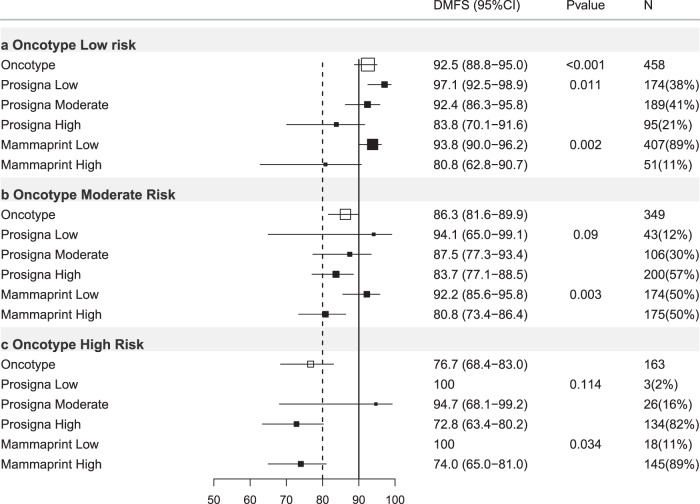
Fig. 2. Forest plot of Oncotype-trained test results re-stratified by other tests, all ER+ve/HER2−ve cases.
DMFS10 = distant metastasis free survival at 10 years post diagnosis. (95% CI) = 95% confidence interval, P value = p value, N = number of cases in each subgroup, % = percentage of cases within each risk strata. X axis = percent distant metastasis free survival. Open boxes represent primary test DMFS10 by risk group. Solid boxes represent sub-stratification by secondary tests with 95% confidence intervals (bars). Top panel (a) oncotype-trained low risk cases stratified by prosigna-trained and Mammaprint-trained results. Middle panel (b) oncotype-trained moderate risk group. Bottom panel (c) oncotype-trained high risk group.
Oncotype-trained stratified by Prosigna-trained
When Oncotype-trained results were further stratified by Prosigna-trained results a significant proportion (56.5%) of cases changed risk category (Supplementary Table 2). In Oncotype-trained low-risk cases, 279 (17.4%) were re-classified as high risk by Prosigna-trained results and 9 Oncotype-trained high-risk cases (1.8%) were re-classified as low risk by Prosigna-trained results. Oncotype-trained low risk/Prosigna-trained high-risk cases exhibited a significantly reduced DMFS10 (75.4%) relative to cases low risk by both signatures (HR = 3.19; 95%CI 2.12–4.82; p < 0.001; Table 2; Fig. 2). For Oncotype-trained intermediate-risk cases, 174 (14.8%) were classified as Prosigna-trained low risk with a DMFS10 = 91.5% (p < 0.001; Table 2; Fig. 2), and 618 (52.6%) were classified as Prosigna-trained high risk (DMFS10 = 73.3%; Table 2; Fig. 2). Few Oncotype-trained high-risk tumors were low risk by Prosigna-trained scores and no events were observed in these cases.
Oncotype-trained stratified by MammaPrint-trained
124 Oncotype-trained low-risk cases (8%) were high risk by MammaPrint-trained (DMFS10 = 72.1%; Table 2; Fig. 2; p < 0.001). 52 Oncotype-trained high-risk cases (10%) were low risk by MammaPrint-trained (DMFS10 = 70.4%; Table 2; Fig. 2; p = 0.465). Finally 528 (45%) Oncotype-trained intermediate-risk cases were MammaPrint-trained high risk(DMFS10 = 73.2%; Table 2; Fig. 2; p < 0.001).
Prosigna-trained results
Of 3284 ER+ve/HER2−ve cases with results for Prosigna-trained risk available 25.2% were low risk (DMFS10 = 92.1%, 95%CI 89.8–94.0%), 35.2% intermediate risk (DMFS10 = 84.9%, 95%CI 82.3–87.1%) and 39.7% high risk (DMFS10 = 71.4%, 95%CI 68.6–74.1%; Table 3; Figs. 1b, 3).
Table 3.
Prosigna-trained results stratified by other test results, trinary classification.
| Prosigna-trained first | |||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Prosigna-trained low risk | Prosigna-trained intermediate risk | Prosigna-trained high risk | |||||||||||||||||||||||||
| HR (95% CI) | DMFS (95% CI) | P* (N) | HR (95% CI) | DMFS (95% CI) | P (N) | HR (95% CI) | DMFS (95% CI) | P (N) | |||||||||||||||||||
| All cases | REF |
92.1 (89.8–94.0) |
<0.001 (826) |
1.97 (1.44–2.69) |
84.9 (82.3–87.1) |
<0.001 (1155) |
4.24 (3.18–5.66) |
71.4 (68.6–74.1) |
<0.001 (1303) |
||||||||||||||||||
| N−Ch− | REF |
96.7 (92.0–98.7) |
<0.001 (220) |
3.06 (1.16–8.08) |
91.0 (86.4–94.1) |
0.024 (321) |
7.76 (3.13–19.2) |
80.5 (75.7–84.3) |
<0.001 (429) |
||||||||||||||||||
| N+Ch− | REF |
90.1 (85.8–93.2) |
<0.001 (318) |
1.35 (0.85–2.15) |
86.2 (82.1–89.4) |
0.204 (476) |
4.43 (2.93–6.69) |
63.9 (58.6–68.7) |
<0.001 (453) |
||||||||||||||||||
| Ch+ | REF |
90.9 (86.3–94.0) |
<0.001 (287) |
2.64 (1.63–4.27) |
77.8 (72.5–82.2) |
<0.001 (356) |
3.74 (2.37–5.92) |
70.4 (65.2–74.9) |
<0.001 (417) |
||||||||||||||||||
| Oncotype-trained Low | Oncotype-trained Int | Oncotype-trained high | Oncotype-trained low | Oncotype-trained Int | Oncotype-trained high | Oncotype-trained low | Oncotype-trained Int | Oncotype-trained high | |||||||||||||||||||
| HR | DMFS | P* | HR | DMFS | P | HR | DMFS | P | HR | DMFS | P* | HR | DMFS | P | HR | DMFS | P | HR | DMFS | P* | HR | DMFS | P | HR | DMFS | P | |
| All cases | REF |
92.2 (89.5–94.2) |
0.737 (643) |
1.07 (0.56–2.03) |
91.5 (85.2–95.2) |
0.835 (174) |
NA | 100 |
NA (9) |
REF |
88.5 (85.3–91.0) |
<0.001 (685) |
1.86 (1.31-2.66) |
81.4 (76.5-85.4) |
0.001 (381) |
3.08 (1.89-5.01) |
72.6 (61.0-81.3) |
<0.001 (89) |
REF |
75.4 (68.3–81.1) |
<0.001 (279) |
1.27 (0.92–1.75) |
73.3 (69.2–77.0) |
0.148 (618) |
1.78 (1.28–2.47) |
65.7 (60.4–70.5) |
0.001 (406) |
| N−Ch− | REF |
97.1 (92.5–98.9) |
0.955 (174) |
1.03 (0.12–9.26) |
94.1 (65.0–99.1) |
0.976 (43) |
NA | 100 |
NA (3) |
REF |
92.4 (86.3-95.8) |
0.395 (189) |
1.69 (0.72-3.97) |
87.5 (77.3-93.4) |
0.232 (106) |
0.69 (0.09-5.34) |
94.7 (68.1-99.2) |
0.721 (26) |
REF |
83.8 (70.1-91.6) |
0.012 (95) |
1.26 (0.61-2.59) |
83.7 (77.1-91.6) |
0.535 (200) |
2.35 (1.15-4.78) |
72.8 (63.4-80.2) |
0.019 (134) |
| N+Ch− | REF |
89.3 (84.4–92.7) |
0.701 (263) |
0.62 (0.19–2.07) |
94.3 (83.4–98.1) |
0.441 (54) |
NA | 100 |
NA (1) |
REF |
89.1 (84.1-92.5) |
0.008 (309) |
1.93 (1.09-3.43) |
81.8 (73.1-87.9) |
0.025 (142) |
3.21 (1.32-7.80) |
74.5 (51.7–87.7) |
0.010 (25) |
REF |
70.3 (58.1–79.6) |
<0.001 (111) |
1.29 (0.80–2.09) |
69.0 (61.4–75.4) |
0.291 (207) |
2.27 (1.41–3.65) |
51.1 (41.5–59.8) |
0.001 (135) |
| Ch+ | REF |
91.9 (86.5–95.2) |
0.552 (205) |
1.51 (0.63–3.60) |
87.9 (76.9–93.9) |
0.353 (77) |
NA | 100 |
NA (5) |
REF |
83.4 (76.0-88.7) |
<0.001 (185) |
1.72 (1.01-2.94) |
76.2 (67.4-83.0) |
0.046 (133) |
3.47 (1.83-6.58) |
56.4 (37.0–71.9) |
<0.001 (38) |
REF |
71.7 (57.0–82.1) |
0.644 (73) |
1.24 (0.71–2.15) |
67.4 (59.7–74.0) |
0.445 (208) |
1.06 (0.58–1.92) |
73.8 (65.1–80.7) |
0.854 (136) |
| Mammaprint-trained low | Mammaprint trained high | Mammaprint-trained low | Mammaprint-trained high | Mammaprint-trained low | Mammaprint-trained high | ||||||||||||||||||||||
| HR | DMFS | P* | HR | DMFS | P | HR | DMFS | P* | HR | DMFS | P | HR | DMFS | P* | HR | DMFS | P | ||||||||||
| All cases | REF |
92.2 (89.8–94.0) |
0.753 (814) |
1.37 (0.19–9.92) |
90.0 (47.3–98.5) |
0.754 (12) |
REF |
86.1 (83.3–88.5) |
0.005 (944) |
1.69 (1.16–2.44) |
79.4 (72.6–84.7) |
0.006 (211) |
REF |
78.1 (73.0–82.3) |
<0.001 (422) |
1.64 (1.26–2.12) |
68.4 (64.9–71.7) |
<0.001 (881) |
|||||||||
| N−Ch− | REF |
96.6 (91.8–98.6) |
0.763 (217) |
NA | 100 |
NA (3) |
REF |
93.1 (88.1–96.0) |
0.031 (249) |
2.47 (1.06–5.79) |
83.8 (70.2–91.5) |
0.037 (72) |
REF |
89.6 (80.6–94.5) |
0.002 (133) |
2.78 (1.43–5.44) |
76.5 (70.6–81.4) |
0.003 (296) |
|||||||||
| N+Ch− | REF |
90.0 (85.7–93.1) |
0.601 (315) |
NA | 100 |
NA (3) |
REF |
86.6 (82.2–90.0) |
0.371 (415) |
1.38 (0.68–2.84) |
83.6 (70.7–91.1) |
0.373 (61) |
REF |
75.7 (66.8–82.5) |
<0.001 (166) |
2.09 (1.41–3.10) |
57.4 (50.8–63.5) |
<0.001 (287) |
|||||||||
| Ch+ | REF |
91.1 (86.5–94.2) |
0.280 (281) |
2.88 (0.39–21.4) |
75.0 (12.8–96.1) |
0.302 (6) |
REF |
79.3 (73.3–84.2) |
0.066 (278) |
1.62 (0.96-2.73) |
72.7 (60.8-81.5) |
0.069 (78) |
REF |
69.0 (58.8-77.2) |
0.948 (122) |
0.99 (0.65-1.49) |
70.9 (64.7-76.2) |
0.948 (295) |
|||||||||
HR = Hazard ratio. 95%CI = 95% confidence interval. P* = p value of log-rank test to compare survival distributions. REF = reference group. P = p value of Wald test for comparison versus reference (low risk) group. DMFS = distant metastasis free survival at 10 years (see text). (N) = number of cases in subgroups. All cases = all ER+ve/HER2-ve cases. N−Ch− = Node negative cases treated without chemotherapy. N+Ch− = Node positive cases treated without chemotherapy. Ch+= cases treated with chemotherapy (node negative and node positive combined). Int = intermediate.
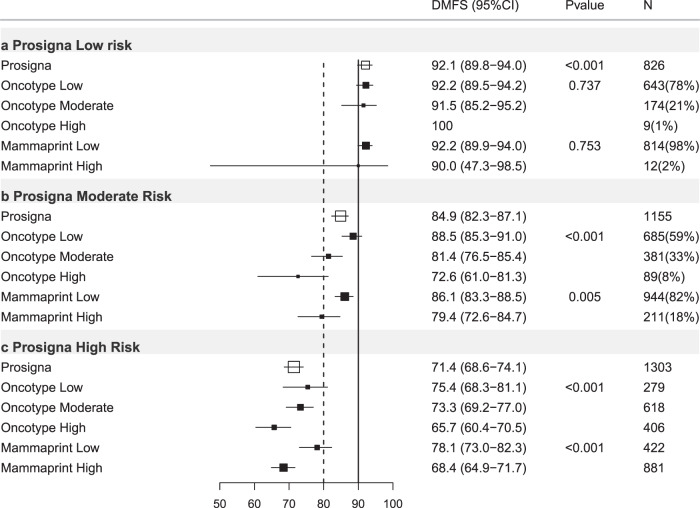
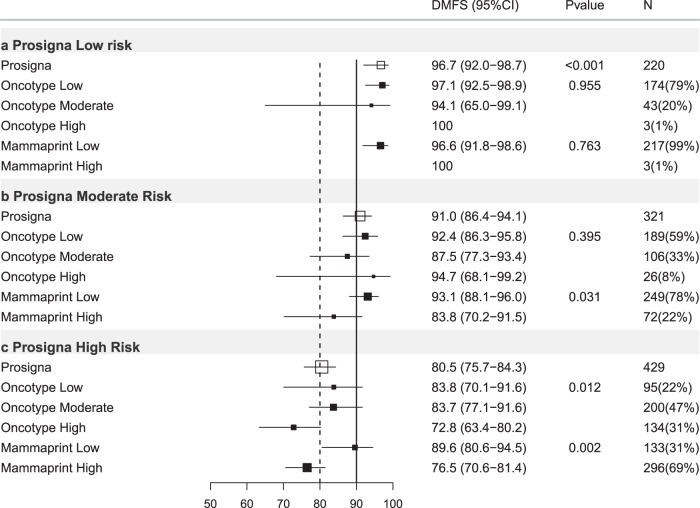
Fig. 3. Forest plot of Prosigna-trained test results re-stratified by other tests, all ER+ve/HER2-ve cases.
DMFS10 = distant metastasis free survival at 10 years post diagnosis, (95% CI) = 95% confidence interval, P = p value, N = number of cases in each subgroup, % = percentage of cases within each risk strata, X axis = percent distant metastasis free survival. Open boxes represent primary test DMFS10 by risk group. Solid boxes represent sub-stratification by secondary tests with 95% confidence intervals (bars). Top panel (a) prosigna-trained low risk cases stratified by Oncotype-trained and Mammaprint-trained results. Middle panel (b) prosigna-trained moderate-risk group. Bottom panel (c) prosigna-trained high risk group.
Prosigna-trained results stratified by Oncotype-trained results
In Prosigna-trained low-risk cases there were no significant differences in outcome across Oncotype-trained risk groups, all Prosigna trained low-risk cases experienced DMFS10 > 90% (Table 3; Fig. 3a). Similarly all Prosigna-trained high risk cases experienced a DMFS10 ≤ 80%; those that were also Oncotype-DX-trained high risk experienced significantly poorer outcome (DMFS10 = 65.7% 95%CI 60.4–70.5%, p < 0.001) than low or intermediate risk by Oncotype-trained (Table 3; Fig. 3c). Of 1155 Prosigna-trained intermediate-risk cases, 685 (59%) were classified low risk by the Oncotype-trained test (DMFS10 = 88.5%; p < 0.001), 89 cases (8%) were Oncotype-trained high risk (DMFS10 = 72.6%; p < 0.001, Table 3; Fig. 3b).
Prosigna-trained stratified by MammaPrint-trained
Excluding Prosigna-trained intermediate-risk cases the majority of results (79.7%) remained in the same risk category (Supplementary Table 2). No stratification of Prosigna-trained low-risk cases occurred using MammaPrint-trained results (Table 3; Fig. 3a). All Prosigna-trained high-risk cases had DMFS10 < 80%, 32% were MammaPrint-trained low risk (Table 3; Fig. 3c). For Prosigna-trained intermediate-risk cases 18% were MammaPrint-trained high risk (DMFS10 = 79.4%; p = 0.005; Table 3, Fig. 3b).
MammaPrint-trained
Of 3284 ER+ve/HER2−ve breast cancers with MammaPrint-Trained risk classification, 66.3% were low risk (DMFS10 = 86.9%) and 33.7% high risk (DMFS10 = 70.7%; Table 4, Figs. 1c, 4).
Table 4.
Mammaprint-trained results stratified by other test results, trinary classification.
| Mammaprint-trained first | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mammaprint-trained low risk | Mammaprint-trained high risk | |||||||||||||||||
| HR (95% CI) | DMFS (95% CI) | P* (N) | HR (95% CI) | DMFS (95% CI) | P (N) | |||||||||||||
| All cases | REF |
86.9 (85.1–88.4) |
<0.001 (2180) |
2.64 (2.22–3.14) |
70.7 (67.6–73.6) |
<0.001 (1104) |
||||||||||||
| N-Ch− | REF |
93.5 (90.5–95.6) |
<0.001 (599) |
4.23 (2.72–6.56) |
78.2 (73.0–82.5) |
<0.001 (371) |
||||||||||||
| N+Ch− | REF |
85.9 (83.1–88.3) |
<0.001 (896) |
3.34 (2.56–4.36) |
62.4 (56.5–67.7) |
<0.001 (351) |
||||||||||||
| Ch+ | REF |
82.3 (78.8–85.3) |
<0.001 (681) |
1.86 (1.41–2.46) |
71.2 (65.8–75.9) |
<0.001 (379) |
||||||||||||
| Oncotype-trained low | Oncotype-trained Int | Oncotype-trained high | Oncotype-trained low | Oncotype-trained Int | Oncotype-trained high | |||||||||||||
| HR | DMFS | P* | HR | DMFS | P | HR | DMFS | P | HR | DMFS | P* | HR | DMFS | P | HR | DMFS | P | |
| All cases | REF |
89.1 (87.1–90.8) |
<0.001 (1483) |
1.74 (1.33–2.28) |
83.2 (79.6–86.1) |
<0.001 (645) |
3.22 (1.82–5.69) |
70.4 (53.7–82.1) |
<0.001 (52) |
REF |
72.1 (60.9–80.6) |
0.031 (124) |
1.10 (0.72–1.68) |
73.2 (68.8–77.2) |
0.668 (528) |
1.47 (0.97–2.24) |
67.1 (62.2–71.6) |
0.072 (452) |
| N−Ch− | REF |
93.8 (90.0–96.2) |
0.485 (407) |
1.35 (0.62–2.92) |
92.2 (85.6–95.8) |
0.452 (174) |
NA | 100 |
NA (18) |
REF |
80.8 (62.8–90.7) |
0.196 (51) |
1.12 (0.49–2.56) |
80.8 (73.2–86.4) |
0.784 (175) |
1.68 (0.74–3.80) |
74.0 (65.0–81.0) |
0.212 (145) |
| N+Ch− | REF |
87.7 (84.4–90.3) |
<0.001 (639) |
1.49 (0.98–2.29) |
84.2 (78.2–88.7) |
0.064 (237) |
5.14 (2.46–10.7) |
54.2 (28.0–74.5) |
<0.001 (20) |
REF |
67.6 (48.0–81.2) |
0.047 (44) |
1.22 (0.63–2.35) |
66.9 (58.3–74.1) |
0.552 (166) |
1.83 (0.96–3.49) |
55.3 (46.0–63.6) |
0.067 (141) |
| Ch+ | REF |
86.8 (82.6–90.0) |
<0.001 (434) |
2.05 (1.38–3.06) |
75.8 (69.1–81.2) |
<0.001 (233) |
3.52 (1.40–8.86) |
61.1 (29.8–81.9) |
0.007 (14) |
REF |
64.2 (37.6–81.8) |
0.903 (29) |
0.85 (0.40–1.81) |
71.2 (63.0–77.9) |
0.676 (185) |
0.90 (0.42–1.92) |
71.9 (63.8–78.4) |
0.792 (165) |
| Prosigna-trained low | Prosigna-trained Int | Prosigna-trained high | Prosigna-trained low | Prosigna-trained Int | Prosigna-trained high | |||||||||||||
| HR | DMFS | P* | HR | DMFS | P | HR | DMFS | P | HR | DMFS | P* | HR | DMFS | P | HR | DMFS | P | |
| All cases | REF |
92.2 (89.8–94.0) |
<0.001 (814) |
1.77 (1.27–2.46) |
86.1 (83.3–88.5) |
0.001 (944) |
3.01 (2.11–4.28) |
78.1 (73.0–82.3) |
<0.001 (422) |
REF |
90.0 (47.3–98.5) |
0.006 (12) |
2.17 (0.30–15.8) |
79.4 (72.6–84.7) |
0.446 (211) |
3.57 (0.50–25.5) |
68.4 (64.9–71.7) |
0.204 (881) |
| N−Ch− | REF |
96.6 (91.8–98.6) |
0.069 (217) |
2.24 (0.80–6.28) |
93.1 (88.1–96.0) |
0.126 (249) |
3.37 (1.15–9.86) |
89.6 (80.6-94.5) |
0.027 (133) |
REF | 100 |
0.222 (3) |
NA |
83.8 (70.2-91.5) |
NA (72) |
NA |
76.5 (70.6–81.4) |
NA (296) |
| N+Ch− | REF |
90.0 (85.7–93.1) |
<0.001 (315) |
1.28 (0.79–2.06) |
86.6 (82.2–90.0) |
0.315 (415) |
2.65 (1.59–4.42) |
75.7 (66.8–82.5) |
<0.001 (166) |
REF | 100 |
0.002 (3) |
NA |
83.6 (70.7–91.1) |
NA (61) |
NA |
57.4 (50.8–63.5) |
NA (287) |
| Ch+ | REF |
91.1 (86.5–94.2) |
<0.001 (281) |
2.42 (1.45–4.03) |
79.3 (73.3–84.2) |
0.001 (278) |
3.91 (2.26–6.79) |
69.0 (58.8–77.2) |
<0.001 (122) |
REF |
75.0 (12.8–96.1) |
0.951 (6) |
1.38 (0.19–10.3) |
72.7 (60.8–81.5) |
0.752 (78) |
1.37 (0.19–9.83) |
70.9 (64.7–76.2) |
0.756 (295) |
HR = hazard ratio, 95%CI = 95% confidence interval. P* = p value of log-rank test to compare survival distributions. REF = reference group. P = p value of Wald test for comparison versus reference (low risk) group. DMFS = distant metastasis free survival at 10 years (see text). (N) = number of cases in subgroups. All cases = all ER+ve/HER2-ve cases. N−Ch− = node negative cases treated without chemotherapy. N+Ch− = Node positive cases treated without chemotherapy. Ch+ = cases treated with chemotherapy (node negative and node positive combined). Int = intermediate.
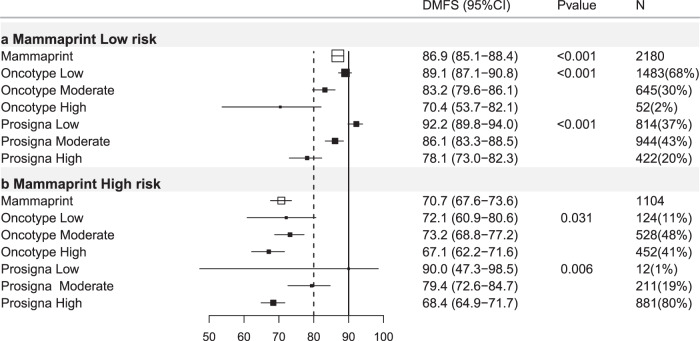
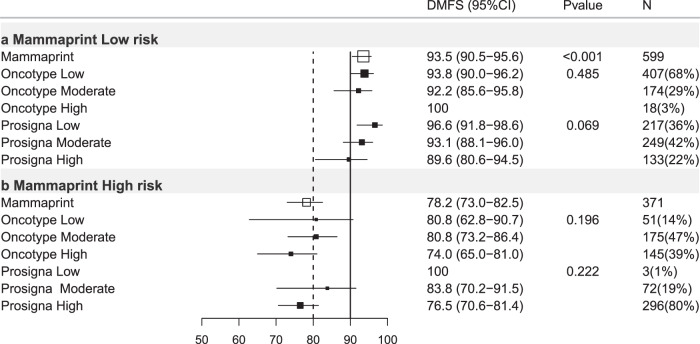
Fig. 4. Forest plot of Mammaprint-trained test results re-stratified by other tests, all ER+ve/HER2-ve cases.
DMFS10 = distant metastasis free survival at 10 years post diagnosis. (95% CI) = 95% confidence interval, P = p value, N = number of cases in each subgroup, % = percentage of cases within each risk strata, X axis = percent distant metastasis free survival. Open boxes represent primary test DMFS10 by risk group. Solid boxes represent sub-stratification by secondary tests with 95% confidence intervals (bars). Top panel (a) Mammaprint-trained low-risk cases stratified by Oncotype-trained and Prosigna-trained results. Bottom panel (b) Mammaprint-trained high-risk group.
MammaPrint-trained stratified by Oncotype-trained
Of 2180 MammaPrint-trained low-risk cases, 68% were low risk by Oncotype-trained results (DMFS10 = 89.1%; Table 4; Fig. 4a). Mammaprint-trained low risk Oncotype-trained intermediate-risk cases (30%) exhibited DMFS10 = 83.2% (Table 4, p < 0.001) and Oncotype-trained high-risk cases exhibited DMFS10 = 70.4% (Table 4, p < 0.001; Fig. 4a). In MammaPrint-trained high-risk cases DMFS10 ranged from 73.2–67.3 across Oncotype-trained-subgroups and there were marked differences in outcome across Oncotype-trained categories (Table 4, Fig. 4b).
MammaPrint-Trained results stratified by Prosigna-trained results
In MammaPrint-trained low-risk cases 20% were Prosigna-trained high risk (DMFS10 = 78.1%; Table 4, p < 0.001) and 43% intermediate risk (DMFS10 = 86.1% Table 4; p < 0.001, Fig. 4a). Amongst MammaPrint-trained high-risk cases, only a small (n = 12) subgroup of Mammaprint-trained high, Prosigna trained low results exhibited DMFS10 = 90% (p = 0.006, Fig. 4b).
Sub-group analysis ER+ve/HER2-ve, Node-ve patients not treated with chemotherapy
Oncotype-trained
Of 970 cases in this subgroup, 47.2% were Oncotype-trained low (DMFS10 = 92.5%), 36.0% intermediate (DMFS10 = 86.3%) and 16.8% high risk (DMFS10 = 76.7%, Table 2; Figs. 1d; 5) respectively.
Fig. 5. Forest plot of Oncotype-trained test results re-stratified by other tests, Node-ve ER+ve/HER2-ve cases treated without chemotherapy.
DMFS10 = distant metastasis free survival at 10 years post diagnosis. (95% CI) = 95% confidence interval, P = p value, N = number of cases in each subgroup, % = percentage of cases within each risk strata. X axis = percent distant metastasis free survival. Open boxes represent primary test DMFS10 by risk group. Solid boxes represent sub-stratification by secondary tests with 95% confidence intervals (bars). Top panel (a) Oncotype-trained low-risk cases stratified by Prosigna-trained and Mammaprint-trained results. Middle panel (b) Oncotype-trained moderate risk group. Bottom panel (c) Oncotype-trained high-risk group.
Oncotype-trained results stratified by Prosigna-trained results
When Oncotype-trained results were stratified by Prosigna-trained results, 57.3% changed risk category (Supplementary Table 3). In Oncotype Dx-trained low risk 95 cases (21%) were Prosigna-trained high risk with DMFS10 = 83.8% (p = 0.006, Table 2; Fig. 5). In Oncotype-trained intermediate-risk cases 12% were Prosigna-trained low risk (DMFS10 = 94.1%; Table 2, p = 0.090; Fig. 5). The 57% of Oncotype-trained intermediate-risk cases classified as Prosigna-trained high risk exhibited DMFS10 = 83.7% (Table 2; p = 0.076, Fig. 5). Only three Oncotype-trained high-risk cases were Prosigna-trained low risk no events were observed in these cases.
Oncotype-trained stratified by MammaPrint-trained
11% of Oncotype-trained low-risk cases were MammaPrint-trained high risk (DMFS10 = 80.8%, p = 0.004; Table 2, Fig. 5a). In Oncotype-trained intermediate-risk patients 50% were MammaPrint-trained low risk(DMFS10 = 92.2%, p = 0.002; Table 2, Fig. 5b). In Oncotype Dx-trained high-risk cases 11% were MammaPrint-trained low risk, no events were observed in these 18 cases (Table 2, Fig. 5c). MammaPrint-trained scores identified 37.5% of Oncotype-trained cases (intermediate or high) as low risk (DMFS10 > 90%).
Prosigna-trained stratified by Oncotype-trained
Neither Prosigna-trained low nor moderate risk cases showed statistically significant sub-stratification for outcome by Oncotype-trained risk scores (Table 3, Fig. 6a, b). Within Prosigna-trained high-risk cases 22% were Oncotype-trained low risk, however, DMFS10 for this group was 83.8% (Table 3, Fig. 6c).
Fig. 6. Forest plot of Prosigna-trained test results re-stratified by other tests, Node-ve ER+ve/HER2-ve cases treated without chemotherapy.
DMFS10 = distant metastasis free survival at 10 years post diagnosis. (95% CI) = 95% confidence interval, P = p value, N = number of cases in each subgroup, % = percentage of cases within each risk strata, X axis = percent distant metastasis free survival. Open boxes represent primary test DMFS10 by risk group. Solid boxes represent sub-stratification by secondary tests with 95% confidence intervals (bars). Top panel (a) Prosigna-trained low-risk cases stratified by Oncotype-trained and Mammaprint-trained results. Middle panel (b) Prosigna-trained moderate risk group. Bottom panel (c) Prosigna-trained high risk group.
Prosigna-trained stratified by MammaPrint-trained
No impact of MammaPrint-trained scores was observed in the Prosigna-trained low-risk group (Table 3, Fig. 6a), with only three discordant results. For both moderate and high risk Prosigna-trained results a group of MammaPrint-trained low-risk cases were identified (DMFS10 = 93.1% and 89.6%, respectively, Table 3; Fig. 6b, c).
MammaPrint-trained results
No impact of Oncotype-trained on Mammaprint-trained scores was observed (Fig. 7; Table 4). In Mammaprint trained low-risk cases 22% were categorized as Prosigna-trained high risk, with a modest reduction in DMFS10 = 89.6% (p = 0.027, Table 4).
Fig. 7. Forest plot of Mammaprint-trained test results re-stratified by other tests, Node-ve ER+ve/HER2−ve cases treated without chemotherapy.
DMFS10 = Distant metastasis free survival at 10 years post diagnosis. (95% CI) = 95% confidence interval, P = p value, N = number of cases in each subgroup, % = percentage of cases within each risk strata, X axis = percent distant metastasis free survival. Open boxes represent primary test DMFS10 by risk group. Solid boxes represent sub-stratification by secondary tests with 95% confidence intervals (bars). Top panel (a) Mammaprint-trained low-risk cases stratified by Oncotype-trained and Prosigna-trained results. Bottom panel (b) Mammaprint-trained high-risk group.
Discussion
Our analysis of 3284 ER+ve/HER2−ve cases using trained signatures demonstrates that the Prosigna-trained signature provides potentially more prognostic information than either the Oncotype-trained or MammaPrint-trained signatures (Table 1). This result is consistent with results in the smaller TransATAC cohort20 using original vendor methodology.
Critical to our study is the close correlation between the computationally derived “signature trained” scores and true results as shown by us previously18. For ROR-PT results the correlation coefficient between “trained” and true assay results was 0.93, comparing true to “trained” results showed 90% of cases within the same risk category (low, intermediate, high—see ref. 18). Similarly for “Oncotype-Dx trained” results the correlation coefficient between true and “trained” results was 0.87 with 75% of results giving the same risk category (see ref. 18) and only 1% of cases disagreeing by more than 1 risk category. For Mammaprint trained results, which were calculated only as categorical high versus low risk groups, over 90% of cases were classified in the same risk group by “trained” and true results18. Full details of these results are reported elsewhere18.
We also show when two trained tests are combined the overall amount of information is always greater than a single test alone. In this study, adding stratification by Prosigna-trained results to Oncotype-trained results provided the greatest LRχ2, and the improvement was greater for this combined model versus Oncotype-trained results alone than for Prosigna-trained results alone. Collectively these results suggest that, in this study, Prosigna-trained results, either alone or combined with other test results, provide potentially greater prognostic information. However, most critically, all test combinations (where two tests were used for patient stratification) outperformed models with only one test to a highly statistically significant degree. This both confirms earlier reports20 and suggests that differences between tests reflect quantitative and qualitative differences in the degree of prognostic information collected. This conclusion is supported by recent comparisons by the ATAC group, showing the impact of different signaling modules in ER+ve/HER2−ve cases21 across different signatures. The conclusion from this work is that different tests capture different aspects of prognostic drivers and therefore that future improvements in prognostic testing remain achievable.
Critically, we dissected the effect of applying a second test to risk-stratified subgroups defined by the initial result; e.g. we examined the effect of applying the Prosigna-trained signature to the “intermediate risk” group identified by the Oncotype-trained signature etc. When combining tests, Prosigna-trained results added value to both Oncotype-trained and MammaPrint-trained results (Table 1). The improved prognostic impact of Prosigna-trained results applied across all ER+ve/HER2−ve cases after Oncotype-trained results was reflected by Prosigna-trained results sub-stratifying patients across both low and intermediate risk Oncotype trained groups (Fig. 2a, b). Even within the node negative ER+ve/HER2−ve population not treated with chemotherapy (Table 2; Fig. 5a, b) Oncotype-trained low and intermediate-risk groups were also further stratified by Prosigna-trained results and 20.7% of Oncotype-trained low-risk cases were identified as high risk by Prosigna-trained results, with DMFS10 of 83.8%, which is important as results from prospective trials suggest these cases may benefit from chemotherapy2,6. This difference was more striking when Oncotype-trained results were dichotomized using cut-points applied in the Tailor-X trial. In ER+HER2−ve, node negative patients treated without chemotherapy 17–24% of cases with Oncotype-trained results ≥25 were low risk (DMFS10 > 90%) when stratified by Mammaprint-trained or Prosigna-trained results respectively (Supplementary Table 4; Supplementary Fig. 2). Conversely 18–30% of Oncotype-trained low risk cases (<25) were high risk when stratified by Mammaprint-trained or Prosigna-trained results and exhibited DMFS < 90% (Supplementary Table 4; Supplementary Fig. 2)
Conversely, only in Prosigna-trained intermediate risk cases did Oncotype-trained results provide additional stratification by risk (Fig. 3; Table 3). However this stratification was not observed in the sub-group of node negative cases treated without chemotherapy (Fig. 6). No stratification of Prosigna-trained low or high risk cases was observed using either Oncotype-trained or Mammaprint trained results (Fig. 3; Table 3). When using dichotomized risk scores for Prosigna-trained ER+ve/HER2−ve node-negative cases treated without chemotherapy no further stratification using dichotomized Oncotype-trained results was seen (Supplementary Table 5; Supplementary Fig. 5) and all Prosigna-high risk cases exhibited DMFS10 < 85% regardless of dichotomized Oncotype-trained results (Supplementary Table 5; Supplementary Fig. 5). These results are illustrative of and highlight the potential clinical impact of disagreements between tests at an individual patient level previously demonstrated in the OPTIMA-prelim cohort4.
A number of conclusions that can be drawn from our analyses. Firstly that, as with previous analyses20 there is additional prognostic value to be gained from combining multiple molecular tests in the research setting. The corollary is that no single existing assay captures the sum of prognostic information available at the transcriptomic level. This confirms earlier findings22 that improvements in prognostic assays remain possible. Such improvements may, however, require integration of additional molecular features beyond transcriptomics23,24. Secondly, there was evidence, albeit from sub-group analyses, that the known interaction between clinical risk, treatment, and molecular risk profiling may differ depending on the test chosen. If taken at face value, this might provide support for the use of different testing strategies in different patient risk strata.
Our analysis has some potentially important limitations. In particular we have used a computational approach to generate test scores for the different tests described herein. At an individual tumor level, the trained score may not be identical to the equivalent generated using original methodology. We trained our signatures in an independent cohort using the same signatures measured using original methodology18, achieving extremely high correlations with commercial test results. Additionally, the broad agreement between our analysis with the(more limited) analysis of Sestak et al. 20 using original methodology and a slightly different statistical approach is highly reassuring.
Additionally, although our cohort is exclusively postmenopausal ER-positive, 30% of cases were treated with adjuvant chemotherapy. All patients in the TEAM trial were postmenopausal, with a median age of 64 years, results presented here may not be representative of the premenopausal population. We included chemotherapy-treated patients to maximize the power of our main analysis. However, the conclusions of our analysis performed on the node-negative subgroup who were not chemotherapy-treated are broadly similar to those in the analysis of the entire cohort, suggesting that these findings are robust both in this clinically critical node negative sub-group and indeed across all patients in the TEAM cohort.
The goal of our study was to provide robust information on the impact of discordant risk classification by different molecular prognostic signatures in postmenopausal, ER+ve early breast cancer. Existing evidence highlights discordance between tests4,25, which is reiterated here. There is clear evidence that adding clinical information to test results provides additional prognostic information15,26–29, which is supported by sub-group analyses performed here, and that information provided by any individual assay is relatively modest17. To date comparisons between tests have been limited either by relatively small sample sizes or by a lack of evidence that signatures extracted from global expression data reflect actual test performance and can therefore inform patients and clinicians on the impact of discordant test results on outcome in the real-world setting. This study provides data on a large clinical trial cohort (the TEAM trial) using test signatures trained in a second cohort (OPTIMA-prelim4) to match actual commercial test performance.
In summary, our study provides novel evidence for the potential clinical impact of discordant molecular test results in a large population. Further improvements in test performance are potentially within reach and would be of benefit to patients. Evidence presented here suggests the differences in test performance are more nuanced than previously reported and that careful consideration to test selection, in the context of treatment and clinical risk may be appropriate.
Methods
Study design
Our primary analyses explored the impact of signature-trained prognostic scores, categorized in accordance with published cut-points for each assay, for patients with centrally confirmed estrogen receptor positive (ER+ve) HER2 negative (HER2−ve) disease30–32. HER2 positive (HER2+ve) cases were excluded since during recruitment of the TEAM trial HER2 targeted therapies were not used in this setting. We performed a secondary analysis using dichotomized scores for Oncotype Dx and Prosigna to reflect the results of the TailorX study. We also report a complete cohort analysis, including HER2+ve cases (see Supplementary Information), since no assay used was trained on samples treated with HER2-targeted therapies. Supplementary analyses further sub-divide patient groups into node negative cases treated with endocrine therapy (but not chemotherapy), node positive cases treated with endocrine therapy (but not chemotherapy) and cases treated with chemotherapy and endocrine therapy (both node negative and node positive, supplementary methods, data and figures).
Patient samples
Patient samples were derived from the Tamoxifen Exemestane Adjuvant Multicenter (TEAM) Trial pathology study (Supplementary Table 1; NCT00279448/NCT0032126/NCT0036270, NTR267, UMIN C000000057)19,33 and included only hormone receptor positive, post-menopausal cancers. Patients provided informed consent and this study was approved by the University of Toronto REB (protocol number 29021).
RNA profiling using NanoString
Profiling of all samples was performed using mRNA previously extracted and analyzed using a custom NanoString codeset as described previously22. Five 4 μm formalin-fixed paraffin-embedded (FFPE) sections per case were deparaffinised, tumor areas were macro-dissected and RNA extracted using the Ambion® Recoverall™ Total Nucleic Acid Isolation Kit-RNA extraction protocol (Life TechnologiesTM, ON, Canada). RNA aliquots were quantified using a Nanodrop-8000 spectrophometer (Delaware, USA). All 3825 RNAs extracted from the TEAM pathology cohort were successfully assayed. Probes for each gene were designed and synthesized at NanoString® Technologies (Seattle, WA, USA); and 250 ng of RNA for each sample were hybridized, processed and analyzed using the NanoString® nCounter® Analysis System, according to NanoString® Technologies protocols.
Signature-trained Risk Stratification Scores from candidate assays
We compared two different approaches to the generation of simulated risk scores18, and selected a training and validation approach using results obtained from the OPTIMA prelim study4 to fit risk stratification scores generated for this study to those derived from the relevant commercial assay. For all tests, we used the suffix-trained to discriminate the computationally derived assays scores from the commercially derived scores, e.g. Oncotype-trained vs. Oncotype-DX™.
Methods for cross comparisons between Tests
Results were available for 3811 subjects. Cases were grouped into the pre-defined risk categories for each test as follows: Oncotype DX—low risk < 18, intermediate risk 18–31 (supplementary methods), high risk ≥ 31; Prosigna-ROR-PT—low risk < 41, intermediate risk 41–60, high risk ≥ 613,20,34; MammaPrint—low risk and high risk18. We also performed a dichotomized risk analysis for Oncotype Dx using low/intermediate risk 0–25 and high risk > 25, in line with the TailorX study2, and for Prosigna RT using low/intermediate risk < 61 and high risk ≥ 61. Grouped analyses were performed as follows: (1) ER+/HER2−ve (n = 3284); and (2) hormone-receptor positive (HR+) regardless of HER2 status (n = 3811). Subjects were considered HR+ve if ER and/or progesterone receptor (PR) was reported as positive33. Differences in distant metastasis free survival (DMFS; i.e. time to first distant recurrence or death, excluding ipsilateral breast cancer recurrences but including distant metastasis, contralateral breast cancer and death from breast cancer) were evaluated using the Kaplan–Meier method with test equality of survivor functions assessed by log-rank and graphs with risk tables generated. 10-year survival function with 95% confidence intervals (95%CI) were calculated as DMFS10. Hazard ratios (HRs) were calculated using Cox proportional hazards regression models, with appropriate adjustments to obtain HRs for each risk level, with low risk set as reference. To assess the prognostic information of each signature, we evaluated the likelihood ratio χ2 (LRχ2) statistics based on the Cox models, and the difference in LRχ2(ΔLRχ2) was calculated to assess prognostic improvement. All analyses were performed using Stata 14.2 (StataCorp, College Station, TX) and R 4.0.2. Reported p-values were two-sided with p < 0.05 considered statistically significant.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Acknowledgements
Research at the Ontario Institute for Cancer Research is supported by the Government of Ontario. P.C.B. was supported by Genome Canada, by CIHR New Investigator Award and by a Terry Fox Research Institute New Investigator Award. R.C.S. was supported by the National Institute for Health Research University College London Hospitals Biomedical Research Centre. The funder had no role in the analysis or reporting of results.
Author contributions
J.M.S.B., J.B., R.C.S., D.R. contributed to the conception and design of the work, the acquisition, analysis, and interpretation of data, and were in involved in drafting, critical review and approval of the final submitted version. They have agreed to be personally accountable for their contributions. E.K., K.X., G.R.P., C.Q.Y., and P.C.B. contributed the acquisition, analysis, and interpretation of data, and were in involved in critical review and approval of the final submitted version. They have agreed to be personally accountable for their contributions. T.P., E.M., A.H., J.A.D., C.M., L.D., C.S. and C.J.H.v.d.V. contributed the acquisition and interpretation of data, and were in involved in critical review and approval of the final submitted version. They have agreed to be personally accountable for their contributions. All authors contributed to the conception or design of the work or to the acquisition, analysis or interpretation of data. All authors were involved in the drafting and/or critical review.
Data availability
The data generated and analyzed during this study are described in the following data record: 10.6084/m9.figshare.1461711335. The data generated and analyzed as part of this study take the form of 3811 individual Nanostring data files (one per sample). These data represent part of a clinical trial and were used under license for the current study, therefore restrictions apply to their availability. The data are housed in institutional storage at The Ontario Institute for Cancer Research (OICR) and are not publicly available, but can be made available upon request subject to approval from the TEAM steering committee and after appropriate data sharing agreements have been completed. Requests for data access should be directed to the senior author (J.M.S.B.).
Code availability
The codes that support these findings are subject to patent applications and restrictions related to licenses. Codes are available from the author J.M.S.B. upon reasonable request and with the permission of the Ontario Institute for Cancer Research (OICR).
Competing interests
J.M.S.B. has received consultancy or honoraria from Insight Genetics Inc., BioNTechAG, Biothernostics Inc., RNA Diagnsotics Inc., oncoXchange, NanoString Technologies Inc, and research funding from ThermoFisher Scientific, Genoptics, Agendia, NanoString Technologies Inc., Biotheranostics Inc. J.B. has received honoraria from ThermoFisher Scientific. G.P. has received consulting fees from Merck, Astra-Zeneca, Profound Medical, outside of submitted work; Honorarium for DSMB membership from Takeda outside of submitted work. A.H. has received honoraria from MedConcept GmbH, Med Update GmbH, Pfizer, Roche Pharma AG, Streamed up GmbH, Tesaro Bio Germany GmbH and serves on Advisory Boards for PharmaMar, Roche Pharma AG, Tesaro Bio Germany GmbH. C.M. has received consultancy from Genomic Heath. All other authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: John M.S. Bartlett, Jane Bayani, Elizabeth Kornaga.
Supplementary information
The online version contains supplementary material available at 10.1038/s41523-021-00297-7.
References
- 1.Vieira, A. F. & Schmitt, F. An update on breast cancer multigene prognostic tests—emergent clinical biomarkers. Front. Med.5, 10.3389/fmed.2018.00248 (2018). [DOI] [PMC free article] [PubMed]
- 2.Sparano JA, et al. Adjuvant chemotherapy guided by a 21-gene expression assay in breast cancer. N. Engl. J. Med. 2018;379:111–121. doi: 10.1056/NEJMoa1804710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sestak I, et al. Prediction of chemotherapy benefit by EndoPredict in patients with breast cancer who received adjuvant endocrine therapy plus chemotherapy or endocrine therapy alone. Breast Cancer Res. Treat. 2019;176:377–386. doi: 10.1007/s10549-019-05226-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartlett JM, et al. Comparing breast cancer multiparameter tests in the OPTIMA Prelim trial: no test is more equal than the others. J. Natl Cancer Inst. 2016;108:djw050. doi: 10.1093/jnci/djw050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cardoso F, et al. 70-Gene signature as an aid to treatment decisions in early-stage breast cancer. N. Engl. J. Med. 2016;375:717–729. doi: 10.1056/NEJMoa1602253. [DOI] [PubMed] [Google Scholar]
- 6.Sparano JA, et al. Clinical and genomic risk to guide the use of adjuvant therapy for breast cancer. N. Engl. J. Med. 2019;380:2395–2405. doi: 10.1056/NEJMoa1904819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bartlett J, et al. Selecting breast cancer patients for chemotherapy: the opening of the UK OPTIMA trial. Clin. Oncol. (R. Coll. Radiol.) 2013;25:109–116. doi: 10.1016/j.clon.2012.10.005. [DOI] [PubMed] [Google Scholar]
- 8.Ramsey SD, et al. Integrating comparative effectiveness design elements and endpoints into a phase III, randomized clinical trial (SWOG S1007) evaluating oncotypeDX-guided management for women with breast cancer involving lymph nodes. Contemp. Clin. Trials. 2013;34:1–9. doi: 10.1016/j.cct.2012.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pan H, et al. 20-Year risks of breast-cancer recurrence after stopping endocrine therapy at 5 years. N. Engl. J. Med. 2017;377:1836–1846. doi: 10.1056/NEJMoa1701830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Prat A, Ellis MJ, Perou CM. Practical implications of gene-expression-based assays for breast oncologists. Nat. Rev. Clin. Oncol. 2012;9:48–57. doi: 10.1038/nrclinonc.2011.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fan C, et al. Concordance among gene-expression-based predictors for breast cancer. N. Engl. J. Med. 2006;355:560–569. doi: 10.1056/NEJMoa052933. [DOI] [PubMed] [Google Scholar]
- 12.Kelly CM, et al. Agreement in risk prediction between the 21-gene recurrence score assay (Oncotype DX(R)) and the PAM50 breast cancer intrinsic Classifier in early-stage estrogen receptor-positive breast cancer. Oncologist. 2012;17:492–498. doi: 10.1634/theoncologist.2012-0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mackay A, et al. Microarray-based class discovery for molecular classification of breast cancer: analysis of interobserver agreement. J. Natl Cancer Inst. 2011;103:662–673. doi: 10.1093/jnci/djr071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weigelt B, et al. Breast cancer molecular profiling with single sample predictors: a retrospective analysis. Lancet Oncol. 2010;11:339–349. doi: 10.1016/S1470-2045(10)70008-5. [DOI] [PubMed] [Google Scholar]
- 15.Dowsett M, et al. Comparison of PAM50 risk of recurrence score with oncotype DX and IHC4 for predicting risk of distant recurrence after endocrine therapy. J. Clin. Oncol. 2013;31:2783–2790. doi: 10.1200/JCO.2012.46.1558. [DOI] [PubMed] [Google Scholar]
- 16.Sgroi DC, et al. Prediction of late distant recurrence in patients with oestrogen-receptor-positive breast cancer: a prospective comparison of the breast-cancer index (BCI) assay, 21-gene recurrence score, and IHC4 in the TransATAC study population. Lancet Oncol. 2013;14:1067–1076. doi: 10.1016/S1470-2045(13)70387-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bayani J, et al. Molecular stratification of early breast cancer identifies drug targets to drive stratified medicine. npj Breast Cancer. 2017;3:3. doi: 10.1038/s41523-016-0003-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bartlett JMS, et al. Computational approaches to support comparative analysis of multiparametric tests: modelling versus Training. PLoS ONE. 2020;15:e0238593–e0238593. doi: 10.1371/journal.pone.0238593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van de Velde CJH, et al. Adjuvant tamoxifen and exemestane in early breast cancer (TEAM): a randomised phase 3 trial. Lancet. 2011;377:321–331. doi: 10.1016/S0140-6736(10)62312-4. [DOI] [PubMed] [Google Scholar]
- 20.Sestak I, et al. Comparison of the performance of 6 prognostic signatures for estrogen receptor–positive breast cancer: a secondary analysis of a randomized clinical trialprognostic signatures for estrogen receptor–positive breast cancerprognostic signatures for estrogen receptor–positive breast cancer. JAMA Oncol. 2018;4:545–553. doi: 10.1001/jamaoncol.2017.5524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Buus R, et al. Molecular drivers of oncotype DX, Prosigna, EndoPredict, and the Breast Cancer Index: a TransATAC study. J. Clin. Oncol. 2020;20:00853. doi: 10.1200/JCO.20.00853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bayani J, et al. Molecular stratification of early breast cancer identifies drug targets to drive stratified medicine. npj Breast Cancer. 2017;3:3. doi: 10.1038/s41523-016-0003-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bayani J, et al. Identification of distinct prognostic groups: implications for patient selection to targeted therapies among anti-endocrine therapy-resistant early breast cancers. JCO Precis. Oncol. 2019;3:1–13. doi: 10.1200/PO.18.00373. [DOI] [PubMed] [Google Scholar]
- 24.Pereira B, et al. The somatic mutation profiles of 2,433 breast cancers refines their genomic and transcriptomic landscapes. Nat. Commun. 2016;7:11479. doi: 10.1038/ncomms11479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vallon-Christersson J, et al. Cross comparison and prognostic assessment of breast cancer multigene signatures in a large population-based contemporary clinical series. Sci. Rep. 2019;9:12184. doi: 10.1038/s41598-019-48570-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cuzick J, et al. Prognostic value of a combined ER, PgR, Ki67, HER2 immunohistochemical (IHC4) score and comparison with the GHI recurrence score - results from TransATAC. Cancer Res. 2009;69:503S–503S. [Google Scholar]
- 27.Dowsett M, et al. Prediction of risk of distant recurrence using the 21-gene recurrence score in node-negative and node-positive postmenopausal patients with breast cancer treated with anastrozole or tamoxifen: a TransATAC study. J. Clin. Oncol. 2010;28:1829–1834. doi: 10.1200/JCO.2009.24.4798. [DOI] [PubMed] [Google Scholar]
- 28.Cuzick J, et al. Prognostic value of a combined estrogen receptor, progesterone receptor, Ki-67, and human epidermal growth factor receptor 2 immunohistochemical score and comparison with the genomic health recurrence score in early breast cancer. J. Clin. Oncol. 2011;29:4273–4278. doi: 10.1200/JCO.2010.31.2835. [DOI] [PubMed] [Google Scholar]
- 29.Sestak I, et al. Factors predicting late recurrence for estrogen receptor-positive breast cancer. J. Natl Cancer Inst. 2013;105:1504–1511. doi: 10.1093/jnci/djt244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bartlett JM, Rea D, Rimm DL. Quantification of hormone receptors to guide adjuvant therapy choice in early breast cancer: better methods required for improved utility. J. Clin. Oncol. 2011;29:3715–3716. doi: 10.1200/JCO.2011.37.3704. [DOI] [PubMed] [Google Scholar]
- 31.Bartlett JM, et al. Mammostrat as an immunohistochemical multigene assay for prediction of early relapse risk in the tamoxifen versus exemestane adjuvant multicenter trial pathology study. J. Clin. Oncol. 2012;30:4477–4484. doi: 10.1200/JCO.2012.42.8896. [DOI] [PubMed] [Google Scholar]
- 32.Bartlett JM, et al. Do type 1 receptor tyrosine kinases inform treatment choice? A prospectively planned analysis of the TEAM trial. Br. J. Cancer. 2013;109:2453–2461. doi: 10.1038/bjc.2013.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bartlett JMS, et al. Estrogen receptor and progesterone receptor as predictive biomarkers of response to endocrine therapy: a prospectively powered pathology study in the tamoxifen and exemestane adjuvant multinational trial. J. Clin. Oncol. 2011;29:1531–1538. doi: 10.1200/JCO.2010.30.3677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sestak I, et al. Abstract P5-06-05: discordant classification and outcomes between Prosigna and Oncotype Dx Recurrence Score for ER-positive, HER2-negative, node-negative breast cancer. Cancer Res. 2020;80:P5-06-05. [Google Scholar]
- 35.Bartlett, J. M. et al. Metadata Record for the Article: Comparative Survival Analysis of Multiparametric Tests—when Molecular Tests Disagree—A TEAM Pathology Study10.6084/m9.figshare.14617113 (2021). [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data generated and analyzed during this study are described in the following data record: 10.6084/m9.figshare.1461711335. The data generated and analyzed as part of this study take the form of 3811 individual Nanostring data files (one per sample). These data represent part of a clinical trial and were used under license for the current study, therefore restrictions apply to their availability. The data are housed in institutional storage at The Ontario Institute for Cancer Research (OICR) and are not publicly available, but can be made available upon request subject to approval from the TEAM steering committee and after appropriate data sharing agreements have been completed. Requests for data access should be directed to the senior author (J.M.S.B.).
The codes that support these findings are subject to patent applications and restrictions related to licenses. Codes are available from the author J.M.S.B. upon reasonable request and with the permission of the Ontario Institute for Cancer Research (OICR).