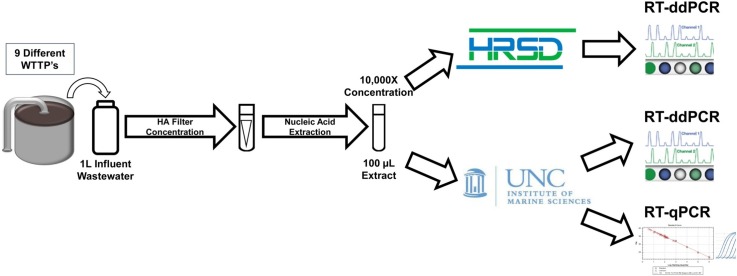
Graphical abstract
Keywords: SARS-CoV-2, Wastewater, Molecular methods, PCR
Abstract
Throughout the COVID-19 global pandemic there has been significant interest and investment in using Wastewater-Based Epidemiology (WBE) for surveillance of viral pathogen presence and infections at the community level. There has been a push for widescale implementation of standardized protocols to quantify viral loads in a range of wastewater systems. To address concerns regarding sensitivity, limits of quantification, and large-scale reproducibility, a comparison of two similar workflows using RT-qPCR and RT-ddPCR was conducted. Sixty raw wastewater influent samples were acquired from nine distinct wastewater treatment plants (WWTP’s) served by the Hampton Roads Sanitation District (HRSD, Virginia Beach, Virginia) over a 6-month period beginning March 9th, 2020. Common reagents, controls, master mixes and nucleic acid extracts were shared between two individual processing groups based out of HRSD and the UNC Chapel Hill Institute of Marine Sciences (IMS, Morehead City, North Carolina). Samples were analyzed in parallel using One-Step RT-qPCR and One-Step RT-ddPCR with Nucleocapsid Protein 2 (N2) specific primers and probe. Influent SARS-CoV-2 N2 concentrations steadily increased over time spanning a range from non-detectable to 2.13E + 05 copies/L. Systematic dilution of the extracts indicated that inhibitory components in the wastewater matrices did not significantly impede the detection of a positive N2 signal for either workflow. The RT-ddPCR workflow had a greater analytical sensitivity with a lower Limit of Detection (LOD) at 0.066 copies/μl of template compared to RT-qPCR with a calculated LOD of 12.0 copies/μL of template. Interlaboratory comparisons using non-parametric correlation analysis demonstrated that there was a strong, significant, positive correlation between split extracts when employing RT-ddPCR for analysis with a ρ value of 0.86.
1. Introduction
The etiological agent of COVID-19, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), is responsible for the current pandemic. SARS-CoV-2 is a member of the family Coronaviridae which are enveloped, single-stranded, positive-sense RNA viruses with pathogenic potential for humans and various animals (Zeng et al., 2020; Huang et al., 2020). Transmission is primarily through respiratory droplets resulting in the development of COVID-19 disease (Meselson, 2020). Additionally, numerous studies have demonstrated that SARS-CoV-2 viral RNA is detectable in stool from patients that exhibit symptoms as well as in stool of asymptomatic carriers (Wu et al., 2020; Xiao et al., 2020; Tang et al., 2020). It has been shown that individuals infected with COVID-19 can continuously shed SARS-CoV-2 for up to as many as 20 days following initial contraction at variable rates dependent on the constitution of the infected individual with viral concentrations decreasing over time throughout the duration of the infection (Bai et al., 2020; Xu et al., 2020; Cevik et al., 2021). As viral material continues to be shed in feces and saliva throughout the course of an infection, it enters wastewater systems that deliver sewage to wastewater treatment plants (WWTPs) responsible for treatment and removal of pollutants from industrial wastewater (Lodder and De Roda Husman, 2020; Ahmed et al., 2020a). The combination of SARS-CoV-2 viral concentrations and concurrent wastewater treatment plant influent flow measurements can be used to quantify the viral load in a municipal wastewater system, thereby providing a metric of the prevalence of infection in the community (Randazzo et al., 2020; Westhaus et al., 2021; Kitajima et al., 2020). The wastewater-based epidemiology (WBE) approach has proven effective in previous surveillance efforts for the closely related SARS-CoV in wastewater from hospitals in China as well as the poliovirus eradication program piloted by the World Health Organization in 1988 (Asghar et al., 2014). As the pandemic of COVID-19 persists, the utility of WBE continues to be recognized as a complementary monitoring tool by acting as an early warning signal for outbreaks of COVID-19 infections (Ahmed et al., 2020a, 2020b; Ahmed et al., 2021).
Currently, there are numerous molecular workflows utilized to quantify SARS-CoV-2 RNA in wastewater matrices (Kitajima et al., 2020). Among these molecular workflows, RT-qPCR and RT-ddPCR have become the dominant platforms for the quantification of SARS-CoV-2 RNA for WBE purposes and have been described in numerous works e.g. Ahmed et al., 2020a, 2020b; Lu et al., 2020; Medema et al., 2020; Wu et al., 2020; Ahmed et al., 2021; and Feng et al., 2021. However, the standardization across platforms has been challenging as there are advantages and disadvantages with each workflow regarding rapidity of result acquisition, cost, reagent availability, user-friendliness, equipment, and sensitivity (Cao et al., 2013; Hayden et al., 2013; Falzone et al., 2020; Vasudevan et al., 2021). However, some of the reported studies do not present the range of necessary quality control evaluations, and others do not apply the approaches across a range of expected SARS-CoV-2 concentrations. As a result, there are inconsistencies amongst reports that inform management, public policy and decision-making entities. Due to this lack of standardization, there is a growing demand to determine if a particular workflow is best suited for SARS-CoV-2 specific WBE surveillance and whether there is a limitation in reproducibility that would restrict wide-scale application (Kitajima et al., 2020; Sims and Kasprzyk-Hordern, 2020). This is particularly pertinent with the current circulation of mutation-based strains and the need to track these and subsequent important strains (Baric, 2020; Public Health, 2021) Here, we present a comparison between two molecular workflows using RT-qPCR and RT-ddPCR for quantification of SARS-CoV-2 and evaluate the impact of inhibition on quantification of the SARS-CoV-2 target. The study was designed to quantify sensitivity and reproducibility for the quantification of SARS-CoV-2 RNA in untreated wastewater influent over time, with samples collected across WWTP types from nine separate WWTP’s in southeast Virginia.
2. Materials and methods
2.1. Sample sites and collection
Hampton Roads Sanitation District (HRSD) serves a population of 1.7 million people across 18 cities in the Commonwealth of Virginia with 9 major and 7 minor facilities. A combination of grab and 24-Hour flow-weighted raw wastewater influent samples were collected from the nine major WWTPs. A total of 60 influent samples were aseptically collected from March 9th, 2020 to September 6th, 2020 with the use of Teledyne ISCO 3710 portable samplers paced to take 150 mL aliquots at variable intervals dependent on the individual treatment plant flow rates. Grab and composited samples were collected between the hours of 8 and 11 AM and transported on wet ice to HRSD in less than six hours for immediate filtration.
2.2. Wastewater influent concentration
Prior to wastewater concentration, the pH of the wastewater samples was adjusted to 3.5 with 10 M HCl solution followed by inoculation with MgCl2 x 6 H2O (to a final concentration of 25 mM). Magnetic stir bars were added to the samples and a magnetic stir plate was used to agitate until thoroughly mixed. 50 mL of sample was then filtered to dryness through a 47 mm dia., 0.45 μm Mixed Cellulose Ester (HA, HAWP04700, Millipore Corp., Bedford, MA) filter using vacuum filtration manifolds. Using sterilized forceps, HA filters were transferred to a microcentrifuge tube and immediately analyzed or stored at −80 °C for no more than 1 week.
2.3. Viral RNA extraction
From each sampling event, viral RNA was extracted using the AllPrep PowerViral DNA/RNA Kit according to the manufacturer’s instructions (cat no. 28,000–50, QIAGEN, Germantown, MD) and each round of nucleic acid extraction included a negative extraction control (NEC) as defined in Section 2.7. In brief, HA filters were transferred to a 0.1 mm Glass PowerBead Tube followed by an addition of 600 μL of Precipitation Solution 1 and 6 μL of β-mercaptoethanol. Additionally, each sample was spiked with 10 μL of Zoetis Calf Guard Cattle Vaccine (BCoV, ValleyVet Supply, Marysville, KS) with a predetermined concentration (1.0E6 copies) to act as a total process control. Bead tubes were secured and disrupted using a Mini-Bead-Beater-96 (cat no. 1001, BioSpec, Bartlesville, OK) for 2 min at maximum speed. Upon completion of bead beating, samples were centrifuged at a speed of 13,000 x g for 1 min. The supernatant was subsequently processed according to the manufacturer’s instructions followed by an elution with 100 μL of RNase free H2O and the resulting extracts were stored at −80 °C until they could be shared between both processing teams.
2.4. SARS-CoV-2 specific quantitative RT-qPCR
Purified RNA was analyzed with the Reliance One-Step Multiplex Supermix (cat no. 12010220, Bio-Rad, Hercules, CA) and the CFX96 Real-Time System with a C1000 Touch Thermal Cycler (Bio-Rad). Primer pairs specific for the Nucleocapsid Protein 2 (N2) and BCoV were used in equimolar ratios at 1.0 μM per reaction and fluorescent probes were used at a concentration of 0.1 μM (Biosearch Technologies, Petaluma, CA; Table 1 ). Reaction mixtures contained 5 μL of RNA template and each reaction mixture reached a total volume of 20 μL. Each 96 well plate (cat no. HSP9655, Bio-Rad, Hercules, CA) included a negative extraction control, negative reverse transcription control, and a no template control. Plates were transferred to the thermal cycler and reverse transcription was initiated at 50 °C for 10 min followed by DNA polymerase activation and template denaturation which were performed at 95 °C for 10 min. After initial denaturation was complete, there were 40 cycles of denaturation for 3 s at 95 °C and annealing/extension for 30 s at 55 °C. All PCR runs were analyzed using Bio-Rad CFX Maestro Software 1.1 (Version 4.1.2433.1219). Unknown samples were run in triplicate for the N2 target and the BCoV target. 6-point Standard curves were generated using Twist Synthetic SARS-CoV-2 RNA Control 3 (cat no. 102860, TwistBioscience, South San Francisco, CA) at a stock concentration of 1.8E + 5 copies/μL. Control RNA was serially diluted 1:10 in AE Buffer (cat no. 19077, QIAGEN, Germantown, MD) and run in triplicate on each 96-well plate with concentrations ranging from 9.00E + 5 copies/reaction to 9.00E + 0 copies/reaction (Supplemental Fig. 1). Ct values were determined using CFX Maestro Software regression analysis and samples with copy numbers below the calculated limit of detection (LOD) were deemed non-detectable.
Table 1.
Primer and Probe Information.
| Target Gene | Primer and Probes | Sequence 5′-3′ | Reference |
|---|---|---|---|
| N2 Nucleocapsid Protein | nCoV N2 Forward | TTACAAACATTGGCCGCAAA | Lu et al. (2020). US CDC Real-Time Reverse Transcription PCR Panel for Detection of Severe Acute Respiratory Syndrome Coronavirus 2. Emerg Infect Dis, 26(8). |
| nCoV N2 Reverse | GCGCGACATTCCGAAGAA | ||
| nCoV N2 FAM Probe | FAM-ACAATTTGCCCCCAGCGCTTCAG-BHQ-1 | ||
| Bovine Coronavirus | BCoV Forward | CTGGAAGTTGGTGGAGTT | Decaro et al. (2008). "Detection of bovine coronavirus using a TaqMan-based real-time RT-PCR assay." Journal of Virological Methods 151(2): 167−171. |
| BCoV Reverse | ATTATCGGCCTAACATACATC | ||
| BCoV HEX Probe | HEX-CCTTCATATCTATACACATCAAGTTGTT-BHQ-1 |
2.5. SARS-CoV-2 specific RT-ddPCR
Purified RNA was analyzed using Bio-Rad’s One-Step RT-ddPCR Advanced Kit for Probes and a QX200 Droplet Reader equipped with Bio-Rad QuantaSoft Software (Version 1.74.0917). The same primer pairs specific for N2 and BCoV that were used for RT-qPCR were also employed for RT-ddPCR analysis and were used in equimolar ratios at 0.9 μM per reaction whereas fluorescent probes were used at a concentration of 0.25 μM (Table 1). Reaction mixtures contained 5 μL of RNA template and, with all reagent components, each reaction mixture reached a total volume of 25 μL. Reaction mixtures were prepared in a pre-PCR room to reduce the possibility of reagent contamination. 20 μL of reaction mixture was used to generate droplets using a Bio-Rad Droplet Generator according to the manufacturer’s instructions and the resulting emulsion was transferred to a new 96-well plate (cat no. 951020389, Eppendorf, Enfield, CT) to undergo PCR amplification. Plates were transferred to C1000 Touch Thermal Cyclers (Bio-Rad) and underwent reverse transcription for 1 h at 50 °C. Polymerase activation and template denaturation were performed at 95 °C for 10 min. After initial denaturation was complete, there were 40 cycles of denaturation for 30 s at 95 °C and annealing/extension for 1 min at 55 °C. Enzyme deactivation was then performed at 98 °C for 10 min. Plates were then held at 25 °C for one minute. Once PCR amplification was completed, the plates were then transferred to the QX200 Droplet Reader (Bio-Rad) for analysis. Threshold values were manually set one standard deviation above the baseline (Cao et al., 2015; Baker et al., 2018). For analysis, replicate sample wells were merged, and samples were considered non-detectable if there were fewer than 3 positive partitions, and were deemed non-quantifiable if the average number of partitions was < 10,000 accepted droplets and if concentrations were below the calculated LOD (Deprez et al., 2016).
2.6. Limits of detection
In order for wastewater surveillance to be an effective strategy for understanding community prevalence of SARS-CoV-2, the LOD according to workflow and platform should be determined. In this study, the LOD was interpreted as a metric of sensitivity. For RT-ddPCR, the Limit of Blank (LOB) was determined using eight technical replicates of eight negative matrix samples derived from influent collected at multiple WWTPs throughout eastern North Carolina. The LOB was calculated as the mean of all sixty-four replicates and the LOD was then calculated as two standard deviations beyond the defined LOB (Hayden et al., 2013). For RT-qPCR the LOD was calculated by analyzing serial dilutions of a synthetic RNA standard with 18 technical replicates at 6 orders of magnitude. The LOD was defined as the concentration at which ≤ 60% of technical replicates were detectable (Gonzalez et al., 2020).
2.7. Data analysis
The agreement for the detection of SARS-CoV-2 RNA in wastewater between the two molecular workflows was determined using Cohen’s Kappa Coefficient and percent agreement statistics (McHugh, 2012; Obermeier et al., 2016). The Kappa value was interpreted as follows: ĸ values ≤ 0 are indicative of no agreement, 0.01–0.20 as none to slight, 0.21–0.40 as fair, 0.41– 0.60 as moderate, 0.61–0.80 as substantial, and 0.81–1.00 as near perfect agreement (McHugh, 2012). Shapiro-Wilks tests were performed to assess the normality of the data distribution where ρ values < 0.05 were considered deviant from a normal distribution (Royston, 1982). Quantitative data were log transformed to reduce observed kurtosis and non-parametric Spearman rank correlation was performed to interpret relationships between variables (Akoglu, 2018). Correlation coefficient values were interpreted as follows: ρ values < 0.3 are indicative of a poor relationship, 0.30 – 0.50 as fair, 0.6 – 0.8 as moderately strong, and > 0.8 as very strong (Akoglu, 2018). Calculated Spearman rank correlation values with a p value < 0.05 were considered statistically significant. All plots were generated using R Statistical Computing Software version 4.0.1 (R Core Team, 2020) and the ggpubr package (Kassambara, 2018).
2.8. Quality control and data evaluation
To rule out contamination of samples and efficacy of each workflow, positive and negative controls were used for the separate processing steps consistent with the MIQE Guidelines (Bustin et al., 2009; Huggett et al., 2013, 2020). The following were implemented for each assay performed using both RT-qPCR and RT-ddPCR:
-
a
Positive Extraction Process Control: A known quantity of BCoV was introduced to each sample following influent concentration and immediately preceding extraction.
-
b
Negative Extraction Processing Control (NEC): A sterile filter was extracted using the same extraction kit as unknown samples.
-
c
Negative Reverse Transcription Control (NRT): Prior to PCR analysis, an aliquot of Supermix was held at 95 °C for 1 min to inactivate the reverse transcriptase enzyme and 5 μL of the heat treated Supermix was incorporated into a reaction mixture.
-
d
No Template Control (NTC): Buffer AE (QIAGEN) containing no target analyte was used within a reaction mixture with all required components including primers, probes, and mastermix.
-
e
Inhibition Control: Nucleic acid extract was diluted in Buffer AE (QIAGEN) at a 1:2 dilution as well as a 1:5 dilution to reduce the concentration of inhibitory substances that may be present in wastewater matrices.
Extraction recoveries derived from BCoV concentrations were calculated using the following equation:
Calculated recoveries fell within a range between 2.55% and 86.9% for RT-ddPCR and between 6.92% and 99.3% for RT-qPCR.
For RT-qPCR, samples were considered to be positive (detectable) if amplification/positive signal was observed in two of the three technical replicates and concentrations were above the calculated LOD. Samples were considered quantifiable if all replicates were positive with a Ct deviation <1 Ct (Staley et al., 2012). For RT-ddPCR, samples were considered to be positive (detectable) if there were at least three positive partitions following the merging of three wells and if sample concentrations were above the calculated LOD and were considered quantifiable if the average total number of partitions was >10,000. Based on these criteria, all samples that were detectable were also quantifiable and negative controls (NEC’s, NRT’s, and NTC’s) were non-detectable for both RT-qPCR and RT-ddPCR.
2.9. Assessing inhibition
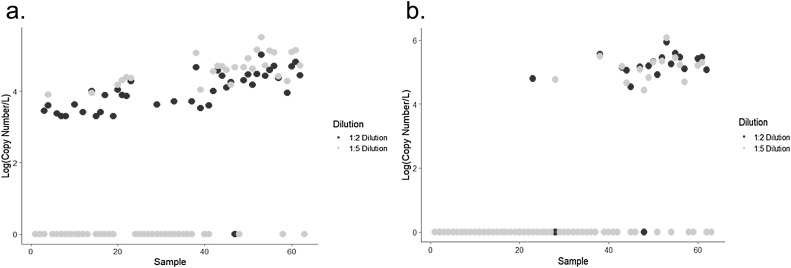
Wastewater as a matrix is known to be fraught with inhibitory high molecular weight compounds such as humic acids, polysaccharides and proteins that impede PCR and molecular detection (Schlindwein et al., 2009). Therefore, after thawing, all sample extracts were diluted using AE buffer as previously described in Section 2.3 with the aim of decreasing the concentration of potential inhibitors. For both RT-qPCR and RT-ddPCR, several samples did change from a positive signal to non-detectable when diluted further from 1:2 to 1:5. There was moderate agreement between samples diluted at a 1:2 dilution versus a 1:5 dilution for detecting a positive signal regardless of which workflow was employed with ĸ values of 0.54 and 0.60 respectively (Fig. 1 ). Therefore, moving forward, all comparisons were made using samples that were diluted 1:2 unless indicated otherwise.
Fig. 1.
Effect of Dilution on Detection of SARS-CoV-2 RNA in Wastewater: Extracts had been diluted at both a 1:2 (black dots) and 1:5 dilution (grey dots) in AE buffer prior to reverse transcription and PCR amplification and samples were analyzed using both (a) RT-ddPCR and (b) RT-qPCR by the processing group based out of IMS.
3. Results
3.1. Intra-laboratory comparison between RT-ddPCR and RT-qPCR
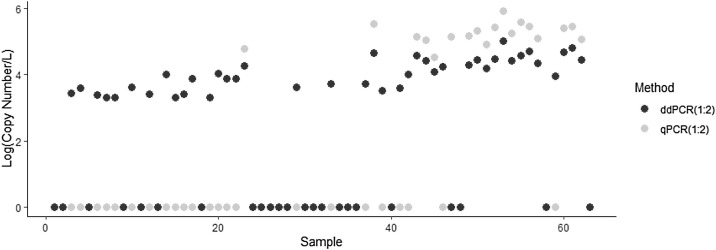
During the sampling window, raw wastewater influent was collected from 9 separate WWTP’s between March 9th, 2020 and September 6th, 2020. SARS-CoV-2 N2 concentrations consistently increased over time and ranged from non-detectable to 3.2E + 05 copies/L for RT-ddPCR and from non-detectable to 1.16E + 06 for RT-qPCR. The workflow for RT-ddPCR led to a lower N2-specific classical LOD of 0.066 copies/μL of template whereas the LOD for RT-qPCR analysis averaged 12.0 copies/μL of template. For RT-ddPCR, of the 63 unknown samples at both a 1:2 and 1:5 dilution, a total of 33 were considered non-detectable according to the previously defined criterium (Section 2.8) in that there were fewer than 3 positive partitions following the merging of wells. For the workflow of RT-qPCR, 14 of the unknown samples at both a 1:2 and 1:5 dilution yielded a positive signal but were below the calculated LOD and so were considered non-detectable. For the first half of the sampling collection period, only RT-ddPCR had the capability to identify a positive signal for N2 (Fig. 2 ). Samples from the same extract that had been analyzed using both platforms revealed that RT-qPCR analysis consistently identifies higher N2 concentrations than RT-ddPCR. All calculated concentrations were within 1 log of each other for samples that were positive using both molecular workflows and percent difference did not exceed 20.0%. Between the two workflows there was a positive agreement of 95.0%, negative agreement of 67.7% and an overall percent agreement of 73.8%. Spearman rank correlation analysis provided a ρ value of 0.717 which is indicative of moderately strong agreement.
Fig. 2.
Direct Comparison of Workflow Performance and Sensitivity: Sample extracts that had been diluted at a 1:2 dilution in AE buffer were analyzed in parallel using RT-ddPCR (black dots) and RT-qPCR (grey dots) by the processing group at IMS. N2 concentrations are presented as log transformed copy numbers per liter of influent filtered. A qualitative comparison between workflows resulted in a ĸ value of 0.314 with overall an overall agreement of 73.8%. Spearman rank correlation analysis resulted in a ρ value of 0.717.
3.2. WWTP specific comparison
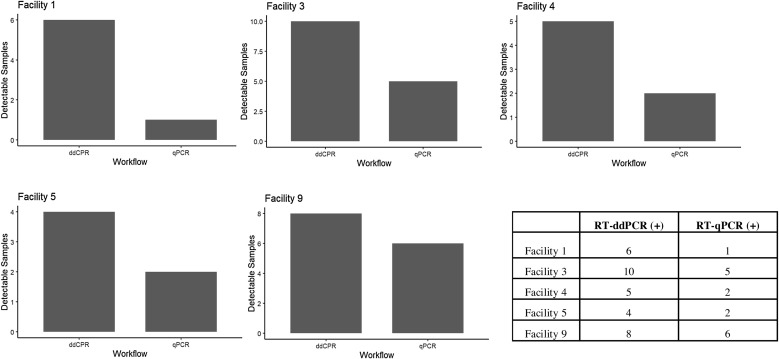
As each individual WWTP serves a different community with unique inputs, it was of interest to assess the variation in SARS-CoV-2 quantification according to treatment plant. This is of interest because matrix composition between individual WWTP’s could preferentially favor one workflow over another. To address this, samples from different WWTP’s supporting variable catchment populations and sizes throughout the collection window were used to determine whether the origin of a sample would impact the efficacy of a particular molecular platform (Table 2 ). Five WWTP’s with a large enough sample size (n > 7) were used to determine if there was plant dependent variability in platform performance. For each of the five WWTP’s, at a 1:2 dilution, the RT-ddPCR workflow was able to consistently identify a positive signal with a greater frequency than RT-qPCR (Fig. 3 ). Due to the superior analytical sensitivity of RT-ddPCR based upon the calculated LOD, this is particularly evident early on within the collection window when there was a lower anticipated viral load. Four of the five facilities had ĸ values < 0.15 suggesting slight or no agreement between workflow analyses. Only facility four had a ĸ value > 0.15 (ĸ = 0.33; moderate agreement).
Table 2.
Facility Information.
| Catchment Population | Collection Events | |
|---|---|---|
| Facility 1 | 343,016 | 10 |
| Facility 2 | 69,059 | 6 |
| Facility 3 | 78,322 | 14 |
| Facility 4 | 192,347 | 8 |
| Facility 5 | 141,543 | 7 |
| Facility 6 | 118,497 | 1 |
| Facility 7 | 197,608 | 2 |
| Facility 8 | 187,832 | 2 |
| Facility 9 | 99,112 | 8 |
| Total: 60 |
Fig. 3.
RT-ddPCR is the more Sensitive Workflow Regardless of WWTP Origin: Samples originating from the same WWTP were analyzed using the two previously described molecular workflows of RT-ddPCR and RT-qPCR. For all WWTPs with > than 7 collection events, RT-ddPCR was able to outperform RT-qPCR with respect to the detection of SARS-CoV-2 RNA.
3.3. Interlaboratory comparison using RT- ddPCR
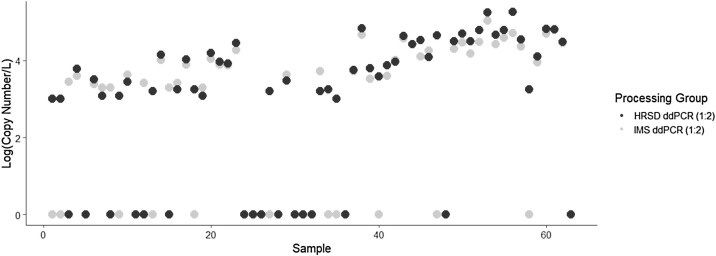
For widespread surveillance efforts, reproducibility is critical for consistency in reporting. To ensure that RT-ddPCR is an effective platform for large-scale implementation, split extracts samples were analyzed in parallel using RT-ddPCR by both processing teams from HRSD and IMS (Fig. 4 ). Correlation analysis resulted in a ρ value of 0.86, indicative a very strong positive relationship between quantification of N2 signal in raw wastewater influent by both teams. For all samples that detected a positive N2 signal by both HRSD and IMS (n = 39), 76.3% of the samples were < 0.25 log different from one another, 13.2% diverged by < 0.5 log and 10.5% of samples N2 concentrations that were different by > 0.5 log. There was 95% positive percent agreement, 67.7% negative percent agreement, and 73.8% overall percent agreement and fair agreement based on kappa analysis with a ĸ value of 0.31.
Fig. 4.
Interlaboratory Assessment of Reproducibility for RT-ddPCR Workflow: Nucleic acid extracts that had been shared between both HRSD (black dots) and IMS (grey dots) were both analyzed using the RT-ddPCR workflow in parallel. There was fair agreement between both groups based on kappa coefficient analysis with a ĸ value of 0.31 and a 95% positive percent positive agreement with a 73.8% overall percent agreement. Spearman correlation analysis provided a ρ of 0.86 which is evidence of a strong relationship between results produced by both groups.
4. Discussion
The use of WBE relies on the accurate quantification of viral pathogen targets in order to develop and implement public health decision making practices for disease mitigation. There are three main suggested and highlighted purposes of WBE for COVID-19 related targets, 1.) evaluation of increasing and decreasing trends in overall load of SARS-CoV-2 in any given municipality, 2.) quantification of SARS-CoV-2 in municipalities to estimate the prevalence of infected individuals in a community and 3.) uncovering emergence and or disappearance of SARS-CoV-2 from wastewater in specific environments such as dormitories, corporate environments, and schools (Yong, 2020; Gibas et al., 2021). These suggested uses of WBE rely on development of molecular workflows that are fully quantitative and are bolstered by strong quality control, are consistent with recommended quality assurance practices, and exhibit reliable reproducibility.
As the pandemic has continued, WBE is widely recognized as a powerful tool for surveillance purposes (Ahmed et al., 2020b). One of the primary concerns in optimizing surveillance efforts is the overall sensitivity of workflows that are being utilized for WBE (Foladori et al., 2020). In this comparative study, a LOD of 0.066 copies/μL of template and 12.0 copies/μL of template for RT-ddPCR and RT-qPCR respectively, was achieved for both molecular workflows. In these particular workflows, the LODs for each platform equate to 1.32E + 2 copies/L and 2.4E + 4 copies/L respectively. These are an improvement from what has previously been reported in the literature (Gonzalez et al., 2020; Arnaout et al., 2020) and with the LOD for RT-ddPCR being more than tenfold lower than the alternative workflow of RT-qPCR, RT-ddPCR offers a significant advantage by allowing for earlier detection of SARS-COV-2 signal in the wastewater. While RT-qPCR can offer advantages in speed, we also found that using the workflows presented here, that RT-ddPCR was capable of delivering results on the same business day. Using RT-ddPCR, this study was able to capture the onset and early development of the pandemic as clinical cases of COVID-19 began to rise in a region covering nearly 1.5 million individuals as early as the end of March (03/25/2020). Conversely, using RT-qPCR, the first detection of SARS-CoV-2 RNA was seen as late as June (06/02/2020).
To investigate the loss of target signal through processing and analysis steps inherent to the workflow, recovery was assessed using an attenuated bovine coronavirus. A known concentration of BCoV was spiked into each sample following concentration and immediately before extraction as described in Section 2.3. There was substantial variability between samples with no observed relationship between calculated recoveries and extraction batch or date of collection. Though not statistically significant, the average recovery calculated based on BCoV concentrations was lower in RT-ddPCR than when using RT-qPCR. This reduction in recovery may be attributed to the additional step of droplet generation that is required for analysis using RT-ddPCR as, prior to PCR, droplets are fragile, and the integrity of the droplets can be easily compromised (Anderson and Maldarelli, 2018). It is also imperative to note that, in this study, all samples had > 10,000 accepted droplets per replicate. Regardless of lower recovery, the superior sensitivity of RT-ddPCR allowed for the capture of low concentration targets that was otherwise missed by RT-qPCR.
Inhibition was also addressed to account for impeded quantification resulting from complex matrix composition. By diluting extracts, concentrated enzymatic inhibitors that are also present in raw wastewater influent pose a lesser risk with respect to compromised quantification (Cao et al., 2012). Investigation of the potential role inhibition plays in molecular analysis revealed that strategic dilution of extracts did not significantly improve the quantification for the N2 target. Correlation statistics indicated that there was good agreement between dilutions for both molecular workflows and it was seen for multiple samples that diluting samples too much completely removed the viral signal, especially for RT-ddPCR. This data suggests that dilution is not critical for quantification of N2 in wastewater matrices. This ultimately improves workflow efficiency in that a step in the processing of samples is removed which reduces potential error. Regardless, it is still critical to consider and include exhaustive quality control measures in all molecular workflows for accurate reporting (Ahmed et al., 2020b)
Previous work has revealed that there are observable and significant differences in the performance of molecular methods dependent on the origin of wastewater sample acquisition (Reemtsma et al., 2010; Loos et al., 2013). In this study, RT-ddPCR was superior with respect to analytical sensitivity and viral quantification regardless of the WWTP from which the sample originated. This may be derived from the low sample size and overall low concentrations of the target for the duration of the study and so future work would involve a larger sample size. Though this study was unable to identify a facility to facility difference between workflow performance, it is still an important aspect of WBE methodology that needs to be taken in to account and should be considered for ongoing WBE efforts.
Reproducibility is also a significant concern for consistency and reliability of data acquisition for widespread surveillance efforts. Previous studies have indicated that poor reproducibility is far too common as there are many variable aspects associated with quantitative molecular tests (Hayden et al., 2013; Ahmed et al., 2021). As such, this study aimed to determine whether RT-ddPCR could alleviate such concerns regarding reproducibility. Upon direct comparison between quantitative results from both processing, there was a strong correlation between viral concentrations in untreated wastewater influent making RT-ddPCR a highly reproducible workflow and thus well suited for widescale WBE surveillance efforts. Though RT-ddPCR displayed a greater analytical sensitivity, RT-qPCR offers the advantage of working within a wider dynamic range and has a relatively rapid turnaround time from sample collection to reporting output (Taylor et al., 2017). As such, the application of RT-qPCR can and should still be considered for WBE.
The findings from this study are relevant to a wide range of locations, including new assessments conducted in Charlotte, NC and across the country using similar workflows (Gibas et al., 2021; Ahmed et al., 2021). Unfortunately, to capture more of the population, there is much to be done on WBE applications, including improvements and optimization of approaches for package treatment plants, septic systems, and individual treatment or living facilities. However, this study is valuable at a time when vaccine distribution is increasing and, at some point, sensitivity and quantification of viral pathogens using reproducible and optimized approaches are paramount. With the global report of clinical cases decreasing and as restrictions set forth by governing bodies begin to loosen, fully quantitative results from WWTP’s will be desired to identify regions of continued vulnerability at the community level. Sensitivity will also be particularly pertinent as the COVID-19 pandemic continues and mutant variants are rising in prominence requiring early action to mitigate spread of these new strains (Baric, 2020; Orive et al., 2020).
5. Conclusion
This study illustrates that in using WBE for the surveillance of SARS-CoV-2, RT-ddPCR for routine quantification may offer significant advantages over RT-qPCR. RT-ddPCR demonstrated resistance to inhibitors in a wastewater matrix and proved to be highly reproducible between two processing groups. In parallel workflows, RT-ddPCR offered substantially greater analytical sensitivity when compared to RT-qPCR, yielding fewer false negative results. As such, RT-ddPCR has greater potential as an early warning system given that viral quantification was possible at an earlier timepoint. Regardless of which workflow is employed, thorough quality control measures should be implemented to maximize the effectiveness of WBE approaches in the future.
CRediT authorship contribution statement
Mark Ciesielski: Conceptualization, Methodology, Formal analysis, Investigation, Data curation, Writing - original draft, Writing - review & editing, Visualization. Denene Blackwood: Methodology, Validation, Formal analysis, Investigation, Writing - original draft, Writing - review & editing, Project administration. Thomas Clerkin: Validation, Formal analysis, Investigation, Writing - original draft, Visualization. Raul Gonzalez: Conceptualization, Methodology, Validation, Formal analysis, Resources, Writing - original draft, Project administration, Funding acquisition. Hannah Thompson: Validation, Investigation, Writing - original draft. Allison Larson: Validation, Investigation, Writing - original draft. Rachel Noble: Conceptualization, Methodology, Validation, Formal analysis, Resources, Data curation, Writing - original draft, Writing - review & editing, Supervision, Project administration, Funding acquisition.
Declaration of Competing Interest
The authors report no declarations of interest.
Acknowledgements
Initial funding was received by R.T. Noble for a Pilot COVID-19 Wastewater Surveillance Project from the North Carolina Policy Collaboratory. Additional follow-on support was received for the project entitled “Tracking SARS-CoV-2 in the Wastewater Across a Range of North Carolina Municipalities” which was also funded by the North Carolina Policy Collaboratory.
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.jviromet.2021.114230.
Appendix A. Supplementary data
The following is Supplementary data to this article:
References
- Ahmed W., Angel N., Edson J., Bibby K., Bivins A., O’Brien J.W., et al. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: a proof of concept for the wastewater surveillance of COVID-19 in the community. Sci. Total Environ. 2020;728:138764. doi: 10.1016/j.scitotenv.2020.138764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Bivins A., Bertsch P.M., Bibby K., Choi P.M., Farkas K., et al. Surveillance of SARS-CoV-2 RNA in wastewater: methods optimisation and quality control are crucial for generating reliable public health information. Curr. Opin. Environ. Sci. Health. 2020;17:82–93. doi: 10.1016/j.coesh.2020.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Simpson S., Bertsch P., Bibby K., Bivins A., Blackall L., et al. MDPI AG; 2021. Minimizing Errors in RT-PCR Detection and Quantification of SARS-CoV-2 RNA for Wastewater Surveillance. Retrieved from. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akoglu H. User’s guide to correlation coefficients. Turk. J. Emerg. Med. 2018;18(3):91–93. doi: 10.1016/j.tjem.2018.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson E.M., Maldarelli F. Quantification of HIV DNA using droplet digital PCR techniques. Curr. Protoc. Microbiol. 2018;51(1):e62. doi: 10.1002/cpmc.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnaout R., Lee R.A., Lee G.R., Callahan C., Yen C.F., Smith K.P., et al. bioRxiv. 2020. SARS-CoV2 testing: the limit of detection matters. [DOI] [Google Scholar]
- Asghar H., Diop O.M., Weldegebriel G., Malik F., Shetty S., El Bassioni L., et al. Environmental surveillance for polioviruses in the global polio eradication initiative. J. Infect. Dis. 2014;210(suppl 1):S294–S303. doi: 10.1093/infdis/jiu384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai Y., Yao L., Wei T., Tian F., Jin D.-Y., Chen L., Wang M. Presumed asymptomatic carrier transmission of COVID-19. JAMA. 2020;323(14):1406. doi: 10.1001/jama.2020.2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker C.S., Steel D., Nieukirk S., Klinck H. Environmental DNA (eDNA) from the wake of the whales: droplet digital PCR for detection and species identification. Front. Mar. Sci. 2018;5 doi: 10.3389/fmars.2018.00133. [DOI] [Google Scholar]
- Baric R.S. Emergence of a highly fit SARS-CoV-2 variant. N. Engl. J. Med. 2020;383(27):2684–2686. doi: 10.1056/nejmcibr2032888. [DOI] [PubMed] [Google Scholar]
- Bustin S.A., Benes V., Garson J.A., Hellemans J., Huggett J., Kubista M., et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009;55(4):611–6622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- Cao Y., Griffith J.F., Dorevitch S., Weisberg S.B. Effectiveness of qPCR permutations, internal controls and dilution as means for minimizing the impact of inhibition while measuring Enterococcus in environmental waters. J. Appl. Microbiol. 2012;113(1):66–75. doi: 10.1111/j.1365-2672.2012.05305.x. [DOI] [PubMed] [Google Scholar]
- Cao Y., Sivaganesan M., Kinzelman J., Blackwood A.D., Noble R.T., Haugland R.A., et al. Effect of platform, reference material, and quantification model on enumeration of Enterococcus by quantitative PCR methods. Water Res. 2013;47(1):233–241.. doi: 10.1016/j.watres.2012.09.056. [DOI] [PubMed] [Google Scholar]
- Cao Y., Raith M.R., Griffith J.F. Droplet digital PCR for simultaneous quantification of general and human-associated fecal indicators for water quality assessment. Water Res. 2015;70:337–349. doi: 10.1016/j.watres.2014.12.008. [DOI] [PubMed] [Google Scholar]
- Cevik M., Tate M., Lloyd O., Maraolo A.E., Schafers J., Ho A. SARS-CoV-2, SARS-CoV, and MERS-CoV viral load dynamics, duration of viral shedding, and infectiousness: a systematic review and meta-analysis. Lancet Microbe. 2021;2(1):e13–e22. doi: 10.1016/S2666-5247(20)30172-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decaro N., Elia G., Campolo M., Desario C., Mari V., Radogna A., et al. Detection of bovine coronavirus using a TaqMan-based real-time RT-PCR assay. J. Virol. Methods. 2008;151(2):167–171. doi: 10.1016/j.jviromet.2008.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deprez L., Corbisier P., Kortekaas A.-M., Mazoua S., Beaz Hidalgo R., Trapmann S., Emons H. Validation of a digital PCR method for quantification of DNA copy number concentrations by using a certified reference material. Biomol. Detect. Quantif. 2016;9:29–39. doi: 10.1016/j.bdq.2016.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falzone L., Musso N., Gattuso G., Bongiorno D., Palermo C., Scalia G., et al. Sensitivity assessment of droplet digital PCR for SARS-CoV-2 detection. Int. J. Mol. Med. 2020;46(3):957–9964.. doi: 10.3892/ijmm.2020.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S., Roguet A., McClary-Gutierrez J.S., Newton R.J., Kloczko N., Meiman J.G., McLellan S.L. Cold Spring Harbor Laboratory; 2021. Evaluation of Sampling Frequency and Normalization of SARS-CoV-2 Wastewater Concentrations for Capturing COVID-19 Burdens in the Community. Retrieved from. [DOI] [Google Scholar]
- Foladori P., Cutrupi F., Segata N., Manara S., Pinto F., Malpei F., et al. SARS-CoV-2 from faeces to wastewater treatment: what do we know? A review. Sci. Total Environ. 2020;743:140444. doi: 10.1016/j.scitotenv.2020.140444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibas C., Lambirth K., Mittal N., Juel M.A.I., Barua V.B., Brazell L.R., et al. medRxiv. 2021. Implementing building-level SARS-CoV-2 wastewater surveillance on a university campus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez R., Curtis K., Bivins A., Bibby K., Weir M.H., Yetka K., et al. COVID-19 surveillance in Southeastern Virginia using wastewater-based epidemiology. Water Res. 2020;186:116296. doi: 10.1016/j.watres.2020.116296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayden R.T., Gu Z., Ingersoll J., Abdul-Ali D., Shi L., Pounds S., Caliendo A.M. Comparison of droplet digital PCR to real-time PCR for quantitative detection of cytomegalovirus. J. Clin. Microbiol. 2013;51(2):540–546. doi: 10.1128/jcm.02620-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi: 10.1016/s0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huggett J.F., Foy C.A., Benes V., Emslie K., Garson J.A., Haynes R., et al. The digital MIQE guidelines: minimum information for publication of quantitative digital PCR experiments. Clin. Chem. 2013;59(6):892–902. doi: 10.1373/clinchem.2013.206375. [DOI] [PubMed] [Google Scholar]
- Huggett J.F., Whale A.S., De Spiegelaere W., Trypsteen W., Nour A.A., Bae Y.-K., et al. The digital MIQE guidelines update: minimum information for publication of quantitative digital PCR experiments for 2020. Clin. Chem. 2020;66(8):1012–1029. doi: 10.1093/clinchem/hvaa125. [DOI] [PubMed] [Google Scholar]
- Kassambara A. 2018. Ggpubr:’ ggplot2’ Based Publication Ready Plots. R Package Version 0.2.https://CRAN.R-project.org/package=ggpubr [Google Scholar]
- Kitajima M., Ahmed W., Bibby K., Carducci A., Gerba C.P., Hamilton K.A., et al. SARS-CoV-2 in wastewater: state of the knowledge and research needs. Sci. Total Environ. 2020;739:139076. doi: 10.1016/j.scitotenv.2020.139076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodder W., De Roda Husman A.M. SARS-CoV-2 in wastewater: potential health risk, but also data source. Lancet Gastroenterol. Hepatol. 2020;5(6):533–534. doi: 10.1016/s2468-1253(20)30087-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loos R., Carvalho R., Antonio D.C., Comero S., Locoro G., Tavazzi S., et al. EU-wide monitoring survey on emerging polar organic contaminants in wastewater treatment plant effluents. Water Res. 2013;47(17):6475–6487. doi: 10.1016/j.watres.2013.08.024. [DOI] [PubMed] [Google Scholar]
- Lu X., Wang L., Sakthivel S.K., Whitaker B., Murray J., Kamili S., et al. US CDC real-time reverse transcription PCR panel for detection of severe acute respiratory syndrome coronavirus 2. Emerg. Infect. Dis. 2020;26(8) doi: 10.3201/eid2608.201246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHugh M.L. Interrater reliability: the kappa statistic. Biochem. Med. (Zagreb) 2012;22(3):276–282. https://www.ncbi.nlm.nih.gov/pubmed/23092060 Retrieved from. [PMC free article] [PubMed] [Google Scholar]
- Medema G., Heijnen L., Elsinga G., Italiaander R., Brouwer A. Cold Spring Harbor Laboratory; 2020. Presence of SARS-Coronavirus-2 in Sewage. Retrieved from. [DOI] [PubMed] [Google Scholar]
- Meselson M. Droplets and aerosols in the transmission of SARS-CoV-2. N. Engl. J. Med. 2020;382(21) doi: 10.1056/nejmc2009324. 2063-2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obermeier P., Muehlhans S., Hoppe C., Karsch K., Tief F., Seeber L., et al. Enabling precision medicine with digital case classification at the point-of-care. EBioMedicine. 2016;4:191–196. doi: 10.1016/j.ebiom.2016.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orive G., Lertxundi U., Barcelo D. Early SARS-CoV-2 outbreak detection by sewage-based epidemiology. Sci. Total Environ. 2020;732:139298. doi: 10.1016/j.scitotenv.2020.139298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Public Health England . 2021. Investigation of novel SARS-CoV-2 variant: Variant of Concern 202012/01. Technical Brief 3 (Publication No. GW-1856)https://www.gov.uk/government/publications/phe-investigation-of-novel-sars-cov-2-variant-of-concern-20201201-technical-briefing-3-6-january-2021 [Google Scholar]
- R Core Team . R Foundation for Statistical Computing; Vienna, Austria: 2020. R: A Language and Environment for Statistical Computing. URL https://www.R-project.org/ [Google Scholar]
- Randazzo Walter, Cuevas-Ferrando Enric, Sanjuán Rafael, Domingo-Calap Pilar, Sánchez Gloria. Metropolitan wastewater analysis for COVID-19 epidemiological surveillance. medRxiv. 2020 doi: 10.1016/j.ijheh.2020.113621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reemtsma T., Miehe U., Duennbier U., Jekel M. Polar pollutants in municipal wastewater and the water cycle: occurrence and removal of benzotriazoles. Water Res. 2010;44(2):596–604. doi: 10.1016/j.watres.2009.07.016. [DOI] [PubMed] [Google Scholar]
- Royston J.P. An extension of shapiro and Wilk’s W test for normality to large samples. Appl. Stat. 1982;31(2):115. doi: 10.2307/2347973. [DOI] [Google Scholar]
- Schlindwein A., Simões C., Barardi C. Comparative study of two extraction methods for enteric virus recovery from sewage sludge by molecular methods. Memórias do Instituto Oswaldo Cruz. 2009;104(4):576–579. doi: 10.1590/s0074-02762009000400007. [DOI] [PubMed] [Google Scholar]
- Sims N., Kasprzyk-Hordern B. Future perspectives of wastewater-based epidemiology: monitoring infectious disease spread and resistance to the community level. Environ. Int. 2020;139:105689. doi: 10.1016/j.envint.2020.105689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staley C., Gordon K.V., Schoen M.E., Harwood V.J. Performance of two quantitative PCR methods for microbial source tracking of human sewage and implications for microbial risk assessment in recreational waters. Appl. Environ. Microbiol. 2012;78(20):7317–7326. doi: 10.1128/aem.01430-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang A., Tong Z.-D., Wang H.-L., Dai Y.-X., Li K.-F., Liu J.-N., et al. Detection of novel coronavirus by RT-PCR in stool specimen from asymptomatic child, China. Emerg. Infect. Dis. 2020;26(6):1337–1339. doi: 10.3201/eid2606.200301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor S.C., Laperriere G., Germain H. Droplet Digital PCR versus qPCR for gene expression analysis with low abundant targets: from variable nonsense to publication quality data. Sci. Rep. 2017;7(1):2409. doi: 10.1038/s41598-017-02217-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasudevan H.N., Xu P., Servellita V., Miller S., Liu L., Gopez A., et al. Digital droplet PCR accurately quantifies SARS-CoV-2 viral load from crude lysate without nucleic acid purification. Sci. Rep. 2021;11(1) doi: 10.1038/s41598-020-80715-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westhaus S., Weber F.-A., Schiwy S., Linnemann V., Brinkmann M., Widera M., et al. Detection of SARS-CoV-2 in raw and treated wastewater in Germany – suitability for COVID-19 surveillance and potential transmission risks. Sci. Total Environ. 2021;751:141750. doi: 10.1016/j.scitotenv.2020.141750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Xiao A., Zhang J., Moniz K., Endo N., Armas F., et al. medRxiv. 2020. SARS-CoV-2 titers in wastewater foreshadow dynamics and clinical presentation of new COVID-19 cases. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao F., Tang M., Zheng X., Liu Y., Li X., Shan H. Evidence for gastrointestinal infection of SARS-CoV-2. Gastroenterology. 2020;158(6):1831–1833. doi: 10.1053/j.gastro.2020.02.055. e1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., Li X., Zhu B., Liang H., Fang C., Gong Y., et al. Characteristics of pediatric SARS-CoV-2 infection and potential evidence for persistent fecal viral shedding. Nat. Med. 2020;26(4):502–505. doi: 10.1038/s41591-020-0817-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yong Clement. The Straits Times; 2020. NEA Monitoring Wastewater in Bid to Give Dorms the Virus All-Clear.https://www.straitstimes.com/singapore/nea-monitoring-wastewater-in-bid-to-give-dorms-the-virus-all-clear [Google Scholar]
- Zeng W., Liu G., Ma H., Zhao D., Yang Y., Liu M., et al. Biochemical characterization of SARS-CoV-2 nucleocapsid protein. Biochem. Biophys. Res. Commun. 2020;527(3):618–623. doi: 10.1016/j.bbrc.2020.04.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.