Abstract
The molecular features of hepatitis B virus (HBV) infection, eradication, and pathogenesis are poorly understood, partly due to the lack of an adequate animal model that faithfully reproduces the course of infection. Although Tupaia belangeri were previously recognized as HBV-susceptible animals, the course of infection in adult tupaias remains obscure. Herein, we performed a longitudinal study and demonstrated that adult tupaias were efficiently infected (90% infection rate) with 108 copies of the HBV genome. HBV replicated vigorously, produced high levels of covalently closed circular DNA (cccDNA) in hepatocytes, and released hepatitis B surface antigen (HBsAg), hepatitis Be antigen (HBeAg), and HBV DNA into the serum at day 9 post-inoculation (p.i.), which then decreased on day 15 p.i. The kinetics were consistent with the expression of liver HBsAg and HBeAg, as determined with immunohistochemistry. The viral products in serum at day 9 and 15 p.i. represented de novo synthesized viral products, as treatment with a viral entry inhibitor completely abolished these products from the serum. Viral clearance and serological conversion occurred at day 21 p.i. and were accompanied by elevated alanine transaminase (ALT) levels and liver pathology, such as inflammatory infiltration and hepatocyte ballooning degeneration. Although ALT levels eventually returned to normal levels by day 42 p.i., the liver pathology persisted until at least day 120 p.i. The HBV infection process in tupaia, therefore, exhibits features similar to that of human acute HBV infection, including viral replication, viral eradication, ALT elevation, and liver pathology. Thus, adopting the tupaia model to study host-HBV interactions presents an important advance which could facilitate further investigation and understanding of human HBV infection, especially for features like cccDNA that current small-animal models cannot effectively model.
Keywords: acute infection, hepatitis B virus, pathogenesis, small animal model, viral replication
Hepatitis B virus (HBV) infection is a major public health problem worldwide; approximately 350 million people are chronically infected with HBV at present, and 1 million annual deaths are due to HBV-related liver cirrhosis and hepatocellular carcinoma [2, 28]. Currently, available treatments for HBV, including interferon and nucleotide/nucleoside analogs [27], may reduce viral load in the host but are rarely able to cure HBV infection. One major contributing factor to this inability to find a cure is an incomplete understanding of the molecular and clinical features of HBV infection, including viral replication, eradication, and pathogenesis, which is partially due to the absence of convenient animal models [29, 30, 49] that accurately model human disease pathogenesis.
HBV has a narrow host range and it infects humans and chimpanzees through natural means only. Chimpanzees are the most well-known HBV-susceptible animal species [3, 15, 45]; however, the utilization of chimpanzees as an animal model is severely restricted because of their limited availability and high costs. Animal models based on HBV-related hepadnaviruses, such as woodchuck, ground squirrel, and duck HBV, have provided important insights into factors involved in the establishment of viral infection, viral persistence, and hepatocarcinogenesis [9, 19, 22, 23, 33, 40]. However, HBV-specific life cycle steps, such as viral entry and host immune responses, cannot be addressed in these animal models.
The development of HBV-transgenic mice has also provided important insights regarding hepatocellular biology and pathology. However, although HBV-transgenic mice can produce infectious HBV, the full viral life cycle, including uptake and elimination of the virus by the host, cannot be fully investigated in this model system because mice are not naturally susceptible to HBV infection. In order to generate virus particles in these mice, the virus has to be produced from viral genome copies that have been integrated into the host chromosomes rather than the natural covalently closed circular DNA (cccDNA) template that HBV normally uses in its natural hosts [5, 16]. In recent years, artificial HBV-permissive animal models have been developed by transplanting primary human or Tupaia belangeri (tree shrew) hepatocytes into immunodeficient mice [7, 8, 35]; however, a lack of host immune responses to HBV infection renders this model ineffective for studying the pathogenesis underlying HBV infection since the host immune response to HBV is the most crucial factor in HBV disease [6, 17].
The northern tree shrew, tupaia, is evolutionarily related to primates and can be chronically infected with HBV, leading to observable histopathological changes in the liver [38, 43]. Currently, the tupaia is the only non-primate susceptible to human HBV [38]. Tupaia are susceptible to chronic infections, relatively close to those in humans. However, in contrast with chimpanzees, tree shrews develop HBV-associated HCC. The susceptibility of tupaia hepatocytes to infection in vitro and in vivo revealed that the tree shrew has the potential to be an immunocompetent HBV infection model [20, 39, 47]. However, the lack of tupaia-specific experimental reagents such as antibodies against Tupaia belangeri have limited its application as an effective model for HBV infection study. Moreover, a normal course of human HBV infection in adult tupaias has not yet been assessed. Here, we characterized the full course of HBV infection in adult tupaias in a 42-day longitudinal study period. In doing so, we assessed the tupaia host response according to the parameters normally associated with human acute HBV infection, including changes in serum alanine transaminase (ALT) levels, serological response, and the development of liver pathology. The results from the current study provide evidence to support establishing tupaias as a suitable animal model for acute HBV infection in order to further investigate the molecular mechanisms underlying HBV, the pathogenesis of HBV-related liver disease, and various features of the host immune response to HBV.
MATERIALS AND METHODS
Animals
Adult hybrid tupaias more than 60 days old were obtained from the Kunming Institute of Zoology, Chinese Academy of Sciences (Kunming, China). They were bred and maintained under optimal conditions, including 26–28°C (room temperature) and daily 12-hr light/dark cycles. All animal experiments were approved by the Committee on Laboratory Animal Care of the Third Military Medical University (Chongqing, China).
In vivo infection of tupaias
The HBV-containing human serum used to infect tupaias was obtained from chronic hepatitis B (CHB) patients under the approval of the local ethics committee and with patient consent. Fourteen tupaias were inoculated intraperitoneally (i.p.) with 1 ml of HBV DNA-positive human serum containing 108 copies of the HBV genome 2 days after receiving i.p. triamcinolone steroid treatment (5 mg/kg body weight) as described previously [42]. Eleven tupaias in the control group were injected with phosphate buffered saline (PBS). Additional detailed procedures are provided in the Supplementary Data.
Infection inhibition by an inhibitory peptide for viral entry
To distinguish de novo synthesized viral products from those carried over from the original i.p. HBV inoculation, HBV infection was blocked by hepalatide (a peptide derived from the HBV-encoded Pre-S1) as previously described [14, 37]. Additional detailed procedures are provided in the Supplementary Data.
Detection of HBV antigens, antibodies, HBV DNA, and ALT in serum
HBV-derived surface antigen (HBsAg) and e antigen (HBeAg) as well as antibodies against HBsAg (HBsAb), HBeAg (HBeAb), and the core antigen (HBcAg) (HBcAb) in the serum were quantified using the commercially available electrochemiluminescence immunoassay (ECLIA) kits. For antibody measurements, concentrations greater than 10 IU/l for HBsAb and cut-off indices (COIs) of less than 1 for HBeAb and HBcAb were considered to be positive results. Serum HBV DNA titers were determined with a real-time PCR Fluorescence Quantification Kit (Shanghai Fosun Biology High Technology Ltd., Shanghai, China). Serum ALT was measured using an automated clinical analyzer (Automatic Analyzer 7060, Hitachi, Tokyo, Japan).
HBV titer assay
HBV titer was determined by measuring HBV DNA with a real-time PCR-fluorescence quantification kit (Shanghai Fosun Biology High Technology Ltd.) with quantification range from 103 to 108 copies/ml. Samples were diluted with HBV negative human serum if HBV titer was higher than 108 copies/ml.
Histological analysis
Liver biopsy samples were fixed in neutral buffered formalin followed by paraffin embedding. The sections were stained with hematoxylin and eosin (H&E) for pathological analysis. For immunohistochemistry, a mouse monoclonal antibody against HBsAg and a rabbit polyclonal antibody against HBcAg (Maixin Bio, Fuzhou, China) were used according to the manufacturer’s instructions.
Detection of intrahepatic HBV cccDNA
Protein-free DNA was extracted from the homogenized tupaia liver biopsy samples as previously described [45]. cccDNA in the isolated DNA samples was detected with rolling circle amplification (RCA) combined with Southern blot analysis as previously described [32]. cccDNA was further quantitatively analyzed with real-time PCR using a commercially available kit (Shanghai Fosun Biology High Technology Ltd.). Additional detailed procedures are provided in the Supplementary Data.
Statistical analysis
The statistical significance between serum ALT levels before HBV inoculation and on day 21 post-infection (p.i.) was determined with a two-tailed paired-data t-test using SPSS 16.0 software (SPSS Inc., Chicago, IL, USA). Values of P less than 0.05 were considered to be statistically significant.
RESULTS
Adult tupaias displayed vigorous enhancement of serum HBV and viral proteins after intraperitoneal inoculation of HBV-containing serum from CHB patients
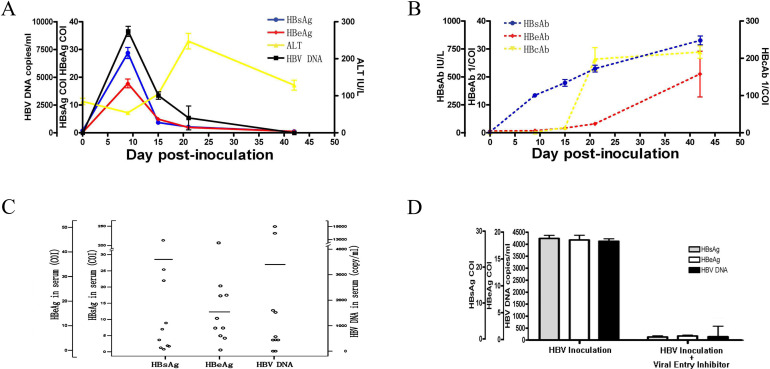
In order to characterize the HBV life cycle in adult tupaia hosts, we infected tupaias with human HBV in serum obtained from CHB patients. Similar to the successful infection strategy used in previous reports [42], adult tupaias were first transiently immunosuppressed with the corticosteroid triamcinolone and then injected with HBV-containing serum from CHB patients (108 copies of the HBV genome). HBsAg, HBeAg, and HBV DNA titers reached their peak at day 9, declined at days 15 and 21, and became undetectable by day 42. (Fig. 1A); these levels remained relatively consistent with each other throughout the HBV infection process. These data indicated that HBV inoculation in adult tupaias elicited an obvious transient course of infection that was similar to the acute self-limited HBV infection observed in humans.
Fig. 1.
The kinetics of hepatitis B virus (HBV) viral products, anti-HBV antibodies, and alanine aminotransferase (ALT) in tupaia serum. Tupaias were inoculated i.p. with 1 ml of HBV DNA-positive human serum containing 108 copies of the HBV genome 2 days after receiving i.p. triamcinolone steroid treatment (5 mg/kg body weight). (A) Analysis of serum HBsAg and HBeAg levels with electrochemiluminescence immunoassay (ECLIA); HBV DNA using real-time PCR; and ALT using a clinical biochemical analyzer. HBsAg and HBeAg levels higher than the cut-off index (COI) of 1 were considered positive. The reference normal ALT level was identified as 56.82 IU/l based on the average ALT levels from 50 healthy tupaias. All values are shown as mean ± SD from 10 tupaias. (B) Analysis of serum HBsAb, HBeAb, and HBcAb with ECLIA. Concentrations of HBsAb higher than 10 IU/l were considered positive. HBeAb and HbcAb values are presented as 1/COI and were considered positive if 1/COI was greater than 1. All values are shown as mean ± SD from 10 tupaias. (C) Individual serum HBsAg, HBeAg, and HBV DNA levels on day 9 p.i. among 10 HBV-inoculated tupaias. (D) In vivo HBV infection in tupaias was blocked by a peptide derived from Pre-S1. An HBV entry inhibitor was administered to 10 HBV-inoculated tupaias. HBsAg, HBeAg, and HBV DNA levels in the serum on day 9 p.i. were compared between HBV-inoculated animals and HBV-inoculated animals that also received an HBV entry inhibitor. All values are shown as mean ± SD from 10 tupaias.
In the human population, this transient course of HBV infection is heterogeneous among individuals. Similarly, HBV infection also displayed heterogeneity among adult tupaias as illustrated by the variation in serum levels of HBsAg, HBeAg, and HBV DNA titers at day 9 p.i. (Fig. 1C). More specifically, serum samples from 9/10 tupaias were positive for HBsAg, HBeAg, and HBV DNA, indicating an overall typical infection rate of 90%; 1/10 animals remained negative throughout the observation period (Supplementary Table 1). Among the 9 infected animals, 2 (#3 and #7) experienced a “super infection” with high levels of serum HBsAg, HBeAg, and HBV DNA on day 9 p.i.; however, the majority of infected tupaias underwent a routine infection course with moderate HBsAg, HBeAg, and HBV DNA levels.
Serum HBV and viral proteins were synthesized de novo in tupaia
To verify that the viral antigens and DNA detected in the serum of the inoculated animals were synthesized de novo and to exclude any contribution from the original HBV inoculum, 2 mg/kg hepalatide, an HBV entry inhibitor derived from the Pre-S113-59 polypeptide of HBV genotype C with an N-terminal myristoylation, similar to Myrcludex B derived from Pre-S12-48 of the HBV genotype D [18, 32], was administered to a separate group of 10 HBV-inoculated tupaias. In our previous study, we showed that hepalatide could bind to NTCP, a functional receptor for HBV infection [46]. Here, we confirmed that hepalatide bound to NTCP (Supplementary Fig. 1A–C) and efficiently inhibited HBV infection of primary tupaia hepatocytes by preventing the binding of virus to NTCP at a 50% inhibitory concentration (IC50) of 1.6 nM (Supplementary Fig. 1D and 1E). Administering hepalatide to tupaia in vivo almost completely abolished the presence of serum HBsAg, HBeAg, and HBV DNA on day 9 p.i. (Fig. 1D), indicating that these viral products were de novo synthesized by HBV replication in tupaia hepatocytes.
High serum ALT levels were maintained even after HBV clearance
As a marker of hepatocyte damage, serum ALT levels were analyzed longitudinally in 10 HBV-inoculated tupaias. ALT levels slightly increased during the initial decrease in the cycling viral HBsAg, HBeAg, and HBV DNA products, and its peak occurred at day 21 p.i. after the observed significant reduction in viral products (Fig. 1A). Indeed, serum ALT levels were significantly elevated in HBV-inoculated adult tupaias on day 21 p.i. as compared to baseline levels (P<0.05). ALT levels returned to normal by day 42 p.i., coinciding with the elimination of HBsAg, HBeAg, and HBV DNA from the serum, a phenomenon consistent with acute HBV infections in chimpanzees and humans [17, 44]. Therefore, all HBV-inoculated tupaias exhibited ALT elevation on day 21 p.i., including tupaia #3, which had ALT concentrations of 11 IU/ml at baseline and 31 IU/ml on day 21 p.i. (Supplementary Table 3).
Seroconversion occurred after viral elimination in tupaia
To further determine the host response to HBV infection in tupaia, we evaluated the generation of antibodies against major HBV proteins (HBsAb, HBeAb, and HBcAb) with ECLIA. In contrast to the various viral replication patterns, the serologic response observed in the tupaias was typically uniform, with HBsAb appearing during antigenemia, and HBeAb and HBcAb arising after antigenemia had already ceased (Fig. 1B). Specifically, the presence of HBsAg and HBsAb overlapped between days 9 and 15 p.i., with HBsAb levels peaking at day 42 p.i. upon clearance of circulating HBsAg and the development of HBsAb, HBeAb, and HBcAb. Seroconversion occurred in all 10 HBV-inoculated tupaias after viral clearance. Taken together, the serological profile of the tupaias in the present study was consistent with a previous report on HBV in vivo infections in tupaias [42] and very similar to acute self-limited HBV infection in humans [44].
HBV infection induced acute inflammation in the liver and long-lasting pathological changes to hepatocytes
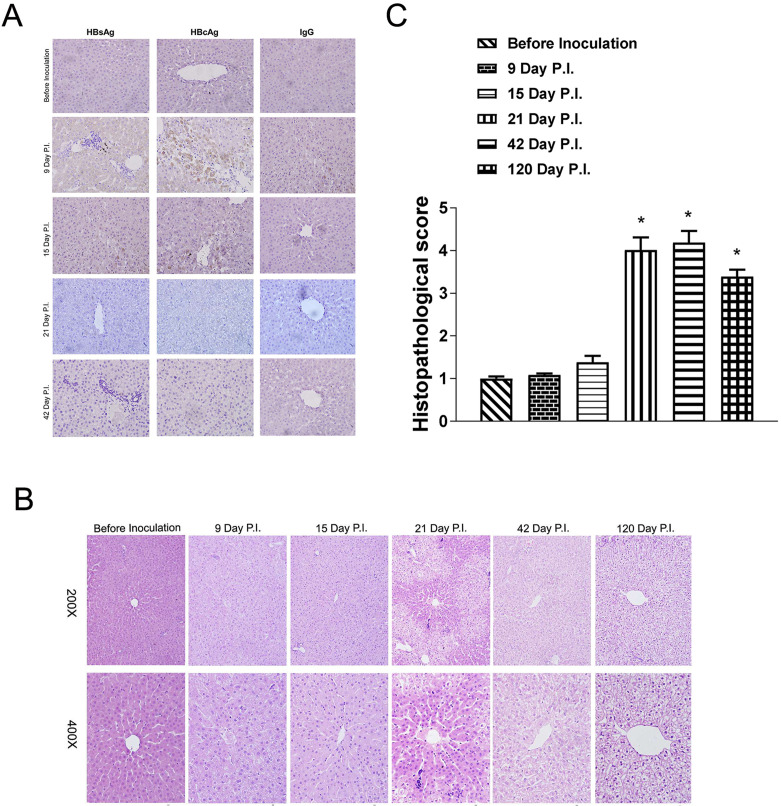
The increased serum ALT levels that occurred along with HBV clearance suggested that HBV infection in tupaia hepatocytes induced liver injury. To formally evaluate whether liver injury occurred in HBV-infected tupaia, liver biopsy samples from a subset of 4 HBV-inoculated tupaias were obtained longitudinally and stained with H&E for histopathological assessment. Figure 2A shows the biopsy samples obtained over time from 1 representative tupaia. Only weak staining was observed in the cytoplasm of perilobular hepatocytes on days 9 and 15 p.i., and no significant lesions in the liver were observed until day 21 p.i. However, coincident with the elevated ALT activity observed on day 21 p.i., mild acute hepatitis was noted in all 4 tupaias with features that included scattered mononuclear cells throughout the lobules and occasional focal infiltration. The inflammatory exudate spread through the slightly enlarged portal tracts, and these infiltrates expanded into portal areas. The portal tract walls were thickened with activated sinusoidal cells and infiltrated by mononuclear cells. Occasionally, lymphoid cells accumulated and formed follicles within the tissue. Perilobular hepatocellular variations were predominantly pronounced in the periportal regions at this time point, leaving the centrilobular hepatocytes nearly normal. The pathological changes in the hepatocytes were characterized by vacuolization, swelling, and pale-staining cytoplasm, although the lobular architecture remained essentially intact. In addition, apoptotic hepatocytes identified by their rounded, deeply eosinophilic cytoplasm and deeply basophilic nuclear remnants were rare, whereas binucleate and multinucleate hepatocytes were present. By 6 weeks p.i., the hepatic inflammation had subsided with minimal portal infiltrates. In contrast, hepatocellular variation extended throughout all parts of the lobules. This included the centrilobular zone, which was pervaded by hydropic degeneration of the hepatocytes despite the complete normalization of serum ALT levels and clearance of HBsAg on day 42 p.i. Unexpectedly, long-lasting hepatocellular variation persisted without significant hepatic inflammatory infiltration in a final biopsy specimen obtained from a tupaia that survived to 120 days p.i (Fig. 2B and 2C).
Fig. 2.
Detection of viral antigens in tupaia liver biopsy samples using immunohistochemistry. (A) Tupaia livers were biopsied 4 days before hepatitis B virus (HBV) inoculation and on days 9, 15, 21, and 42 p.i. Liver samples were sectioned and stained with HBsAg and HBcAg antibodies. The images represent sequential biopsy sections from a single HBV-inoculated tupaia. (B and C) A series of liver biopsy samples were obtained 4 days before HBV inoculation and on days 9, 15, 21, 42, and 120 p.i., and then stained with hematoxylin eosin. The images represent sequential biopsy sections from a single HBV-inoculated tupaia.
HBV antigen expression in hepatocytes indicated vigorous HBV replication in the tupaia liver
To further confirm successful HBV infection in adult tupaias, liver biopsy samples were obtained by surgery from a subgroup of 4 tupaias that received the same HBV inoculation as the main group of 10 tupaias in the abovementioned serology analysis, and HBsAg and HBcAg expression was detected with immunohistochemistry. In contrast to a previous report showing that viral antigens were detected in only a few tupaia hepatocytes [44], HBsAg and HBcAg expression was detected in nearly all hepatocytes on day 9 p.i. (Fig. 3A and 3B), consistent with studies on acute HBV infection in chimpanzees [6, 17]. HBcAg was mainly expressed in the cytoplasm and occasionally in the nuclei of some tupaia hepatocytes at this time point. Viral protein expression decreased on days 15 and 21 p.i. and was cleared by day 42 p.i., which paralleled the observed changes in these viral proteins within the serum at the same time points. Taken together, these data indicated that vigorous viral replication occurred in the livers of HBV-inoculated tupaias [24].
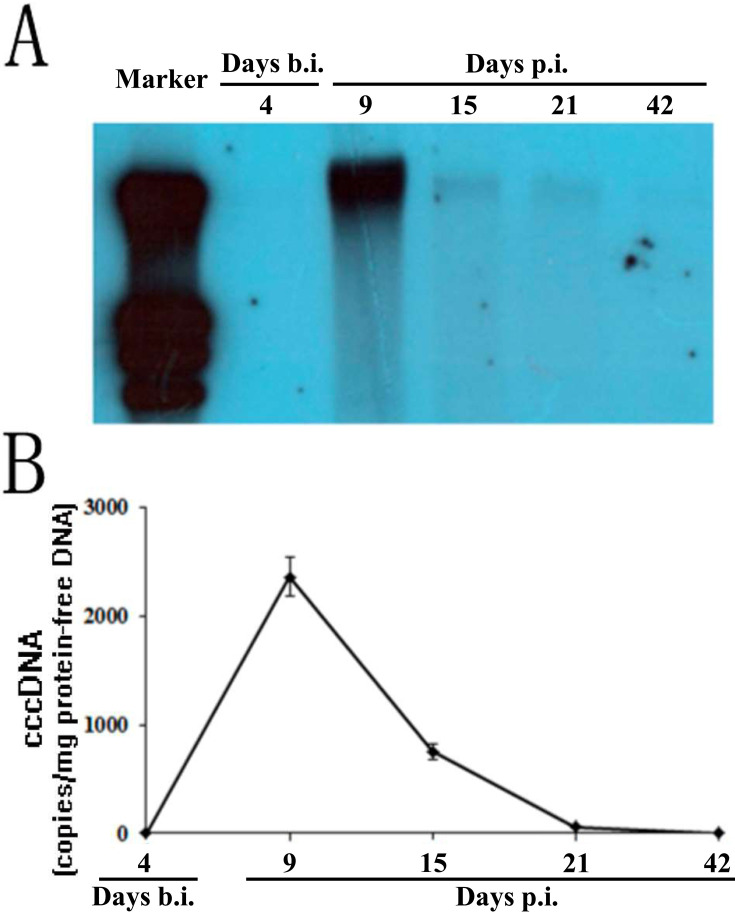
Fig. 3.
Detection of cccDNA in tupaia liver biopsy samples. Tupaias were biopsied 4 days before hepatitis B virus inoculation and on days 9, 15, 21, and 42 p.i., and protein-free DNA was extracted from the biopsy samples. (A) The protein-free DNA was subjected to rolling circle amplification (RCA), and the reaction product was linearized by digestion with the restriction enzyme SpeI followed by Southern blotting with the digoxin-labeled HBV DNA-specific probe. (B) cccDNA in protein-free DNA extracted from liver biopsy samples was quantified with quantitative PCR. cccDNA levels in the liver biopsy are displayed as the copy number per microgram of DNA. The figures represent the blotting signal and cccDNA quantification of longitudinal liver tissue from a single HBV-inoculated tupaia. All values are shown as mean ± SD from 3 independent experiments run in duplicate.
cccDNA formation in hepatocytes confirmed successful HBV infection in tupaias
To definitively confirm that a full course of HBV infection occurred in the tupaia liver after virus inoculation, we looked for the formation of cccDNA, which could only be generated when filling in the gaps within HBV DNA after successful entry into hepatocytes and served as the crucial template for HBV replication. Using Southern blotting, the RCA signal was evaluated over time in liver tissue from 1 tupaia animal using a digoxin-labeled HBV DNA-specific probe, and the results indicated that cccDNA appeared on day 9 p.i., decreased by days 15 and 21 p.i., and almost completely disappeared by day 42 p.i. (Fig. 3A and 3B).
To further evaluate cccDNA formation in hepatocytes, we next quantitatively analyzed cccDNA with real-time PCR using a commercially available kit with primers selective for amplifying cccDNA. In this real-time PCR system, cccDNA was specifically amplified, and false negative reactions were avoided in the presence of HBV DNA at no more than 107 copies/ml. cccDNA was undetectable before HBV inoculation and peaked on day 9 p.i. with 2345 copies/µg protein-free DNA (Fig. 3B and Supplementary Table 2). Consistent with the RCA results, cccDNA levels declined on days 15 and 21 p.i., and finally became undetectable on day 42 p.i. Collectively, cccDNA analysis with both RCA and real-time PCR revealed that HBV inoculation resulted in successful infection of tupaia hepatocytes in vivo.
DISCUSSION
HBeAg is considered a marker of HBV replication and infectivity [11]. Although tupaias were found to be susceptible to HBV infection [44], the present study thoroughly demonstrated a full understanding of HBV infection kinetics. We found that serum from CHB patients containing 108 HBV genome copies was sufficient to infect adult tupaias. Highly variable responses to HBV infection occurred among the individual tupaias, and we speculated that this variability might be due to the species not being inbred. Despite this variability, however, the subsequent course of events during HBV infection in tupaias was typical in terms of the dynamic changes in serum viral antigen, HBV DNA, and ALT levels as well as in viral protein expression, cccDNA, and liver histopathology.
After HBV infected the hepatocytes, HBV cccDNA accumulated in the cells for later use as the replication template, and HBV produced its core antigen (HBcAg) in the hepatocyte nuclei and cytoplasm as well as its HBsAg in the cytoplasm. Antigenemia caused by HBsAg and HBeAg release then persisted along with the viremia caused by the progeny virions, both peaking at day 9 p.i. Following the viral replication peak at day 9 p.i., the infection transitioned into the elimination period beginning around day 15 p.i. During this elimination period, the levels of circulating viral components fell sharply, closely correlating to intrahepatic virus levels before the onset of liver injury. In terms of liver injury, the maximal hepatic damage occurred after HBV clearance at day 21 p.i., possibly as a result of the functional oscillation of specific T cells that express various regulatory factors at different stages of the infection [21].
In the tupaia model, rapid and massive decreases in HBV levels occurred before the onset of liver injury and lymphocyte infiltration, similar to what was observed in the chimpanzee and woodchuck animal models [17, 23, 44]. These consistent results among the different animal models suggest that noncytopathic antiviral mechanisms contribute to viral clearance during acute viral hepatitis, likely by purging viral cccDNA from the infected cells [15]. HBV-specific CD8+ T lymphocytes are effectors for viral clearance and abolish HBV replication in the liver of transgenic mice by a noncytopathic mechanism induced by inflammatory cytokines [24, 26]. Our data showed that the initial reduction in cccDNA levels preceded the onset of ALT elevation and intrahepatic inflammation at day 15 p.i. in HBV-infected tupaias. This finding suggests a noncytopathic-mediated control of hepatocellular cccDNA contents during acute viral hepatitis, which might provide clues to understanding the mechanism involved in cccDNA elimination.
The tupaia model serves as an in vivo model to refine therapeutics that target the cccDNA pathway. For example, Interferon (IFN)-γ or lymphotoxin-β has been shown to degrade cccDNA in the nucleus through activating nuclear deaminases [31], and IFN-γ may also epigenetically regulate and inhibit the cccDNA minichromosome [4]. Another study showed suppression of IFN-β in the tupaia infection model of HBV [41]. Moreover, the development of phenomena on cccDNA, such as CpG methylation [48], observed in patient samples or in hepatocyte/hepatocarcinoma cell lines could be monitored in vivo in real time in the tupaia model after infection.
In association with the peak in serum ALT on day 21 p.i., dramatic changes in liver morphology occurred. Furthermore, the cytopathological variation in liver cells extended to the centrilobular zone despite the normalization of serum ALT levels on day 42 p.i. Damage to the liver cells was sustained, and liver cells still had not recovered even by day 120 p.i. We speculate that the decreased ALT levels observed on day 42 p.i. may have been due to a reduction in granzyme B-expressing specific T cells and the persistence of other inflammatory cells, such as CD69+ or PD-1+ T cells, at day 120 p.i. [21].
The immune response acts like a double-edged sword for the host in terms of protection against pathogens vs. the generation of immune-mediated pathology. The inherent capacity of the immune system to efficiently restrict HBV during acute infection provides a strong rationale for immunotherapeutic treatments for chronic HBV infection. Since the morphologic changes within the HBV-infected tupaia liver are very similar to those observed in HBV-infected humans [1, 10, 36], the tupaia animal model therefore presents a convenient and valuable approach in which to gain further insight into the complex biology of the immune response. In particular, the tupaia animal model may be the most valuable for further investigating the intrahepatocellular pathogenesis mechanisms underlying HBV-induced liver injury.
The tree shrew model contains a complete and natural process from HBV infection to elimination, making it an ideal model for studying the life history of HBV. However, the tree shrew is not a routine experimental animal, and is lack of genetic uniformity and relevant reagents of interest, such as antibodies, immune cell markers, etc. With the availability of high-quality transcriptomics of the entire tupaia genome [12, 13, 34], such issues may be solved in the future.
Collectively, our data depicted the landscape of viral replication, immune response, and liver pathology in a tree shrew model of HBV infection, providing evidence for studying HBV infection and new therapies for hepatitis B.
COMPETING INTERESTS
The authors declare that they have no competing interests.
Supplementary
Acknowledgments
This work was supported by National Natural Science Foundation of China (No. 81170396), Natural Science Foundation of Chongqing (No. cstc2011jjA10056), National Science and Technology key Project of China (No. 2011ZX09102-009-08) and Program for Changjiang Scholars and Innovative Research Team in University.
REFERENCES
- 1.Abe H., Beninger P. R., Ikejiri N., Setoyama H., Sata M., Tanikawa K.1982. Light microscopic findings of liver biopsy specimens from patients with hepatitis type A and comparison with type B. Gastroenterology 82: 938–947. doi: 10.1016/S0016-5085(82)80259-X [DOI] [PubMed] [Google Scholar]
- 2.Alter M. J.2003. Epidemiology and prevention of hepatitis B. Semin. Liver Dis. 23: 39–46. doi: 10.1055/s-2003-37583 [DOI] [PubMed] [Google Scholar]
- 3.Asabe S., Wieland S. F., Chattopadhyay P. K., Roederer M., Engle R. E., Purcell R. H., Chisari F. V.2009. The size of the viral inoculum contributes to the outcome of hepatitis B virus infection. J. Virol. 83: 9652–9662. doi: 10.1128/JVI.00867-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Belloni L., Allweiss L., Guerrieri F., Pediconi N., Volz T., Pollicino T., Petersen J., Raimondo G., Dandri M., Levrero M.2012. IFN-α inhibits HBV transcription and replication in cell culture and in humanized mice by targeting the epigenetic regulation of the nuclear cccDNA minichromosome. J. Clin. Invest. 122: 529–537. doi: 10.1172/JCI58847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chisari F. V.1996. Hepatitis B virus transgenic mice: models of viral immunobiology and pathogenesis. Curr. Top. Microbiol. Immunol. 206: 149–173. [DOI] [PubMed] [Google Scholar]
- 6.Chisari F. V., Isogawa M., Wieland S. F.2010. Pathogenesis of hepatitis B virus infection. Pathol. Biol. (Paris) 58: 258–266. doi: 10.1016/j.patbio.2009.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dandri M., Burda M. R., Zuckerman D. M., Wursthorn K., Matschl U., Pollok J. M., Rogiers X., Gocht A., Köck J., Blum H. E., von Weizsäcker F., Petersen J.2005. Chronic infection with hepatitis B viruses and antiviral drug evaluation in uPA mice after liver repopulation with tupaia hepatocytes. J. Hepatol. 42: 54–60. doi: 10.1016/j.jhep.2004.09.021 [DOI] [PubMed] [Google Scholar]
- 8.Dandri M., Burda M. R., Török E., Pollok J. M., Iwanska A., Sommer G., Rogiers X., Rogler C. E., Gupta S., Will H., Greten H., Petersen J.2001. Repopulation of mouse liver with human hepatocytes and in vivo infection with hepatitis B virus. Hepatology 33: 981–988. doi: 10.1053/jhep.2001.23314 [DOI] [PubMed] [Google Scholar]
- 9.Delmas J., Schorr O., Jamard C., Gibbs C., Trépo C., Hantz O., Zoulim F.2002. Inhibitory effect of adefovir on viral DNA synthesis and covalently closed circular DNA formation in duck hepatitis B virus-infected hepatocytes in vivo and in vitro. Antimicrob. Agents Chemother. 46: 425–433. doi: 10.1128/AAC.46.2.425-433.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Desmet V. J.1988. Liver lesions in hepatitis B viral infection. Yale J. Biol. Med. 61: 61–83. [PMC free article] [PubMed] [Google Scholar]
- 11.Elgouhari H. M., Abu-Rajab Tamimi T. I., Carey W. D.2008. Hepatitis B virus infection: understanding its epidemiology, course, and diagnosis. Cleve. Clin. J. Med. 75: 881–889. doi: 10.3949/ccjm.75a.07019 [DOI] [PubMed] [Google Scholar]
- 12.Fan Y., Huang Z. Y., Cao C. C., Chen C. S., Chen Y. X., Fan D. D., He J., Hou H. L., Hu L., Hu X. T., Jiang X. T., Lai R., Lang Y. S., Liang B., Liao S. G., Mu D., Ma Y. Y., Niu Y. Y., Sun X. Q., Xia J. Q., Xiao J., Xiong Z. Q., Xu L., Yang L., Zhang Y., Zhao W., Zhao X. D., Zheng Y. T., Zhou J. M., Zhu Y. B., Zhang G. J., Wang J., Yao Y. G.2013. Genome of the Chinese tree shrew. Nat. Commun. 4: 1426. doi: 10.1038/ncomms2416 [DOI] [PubMed] [Google Scholar]
- 13.Fan Y. M., Hernesniemi J., Oksala N., Levula M., Raitoharju E., Collings A., Hutri-Kähönen N., Juonala M., Marniemi J., Lyytikäinen L. P., Seppälä I., Mennander A., Tarkka M., Kangas A. J., Soininen P., Salenius J. P., Klopp N., Illig T., Laitinen T., Ala-Korpela M., Laaksonen R., Viikari J., Kähönen M., Raitakari O. T., Lehtimäki T.2014. Upstream Transcription Factor 1 (USF1) allelic variants regulate lipoprotein metabolism in women and USF1 expression in atherosclerotic plaque. Sci. Rep. 4: 4650. doi: 10.1038/srep04650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gripon P., Cannie I., Urban S.2005. Efficient inhibition of hepatitis B virus infection by acylated peptides derived from the large viral surface protein. J. Virol. 79: 1613–1622. doi: 10.1128/JVI.79.3.1613-1622.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guidotti L. G., Rochford R., Chung J., Shapiro M., Purcell R., Chisari F. V.1999. Viral clearance without destruction of infected cells during acute HBV infection. Science 284: 825–829. doi: 10.1126/science.284.5415.825 [DOI] [PubMed] [Google Scholar]
- 16.Guidotti L. G., Ishikawa T., Hobbs M. V., Matzke B., Schreiber R., Chisari F. V.1996. Intracellular inactivation of the hepatitis B virus by cytotoxic T lymphocytes. Immunity 4: 25–36. doi: 10.1016/S1074-7613(00)80295-2 [DOI] [PubMed] [Google Scholar]
- 17.Guidotti L. G., Chisari F. V.2006. Immunobiology and pathogenesis of viral hepatitis. Annu. Rev. Pathol. 1: 23–61. doi: 10.1146/annurev.pathol.1.110304.100230 [DOI] [PubMed] [Google Scholar]
- 18.Guidotti L. G., Matzke B., Schaller H., Chisari F. V.1995. High-level hepatitis B virus replication in transgenic mice. J. Virol. 69: 6158–6169. doi: 10.1128/JVI.69.10.6158-6169.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guo J. T., Zhou H., Liu C., Aldrich C., Saputelli J., Whitaker T., Barrasa M. I., Mason W. S., Seeger C.2000. Apoptosis and regeneration of hepatocytes during recovery from transient hepadnavirus infections. J. Virol. 74: 1495–1505. doi: 10.1128/JVI.74.3.1495-1505.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20Guo W. N., Zhu B., Ai L., Yang D. L., Wang B. J.2018. Animal models for the study of hepatitis B virus infection. Zool. Res. 39: 25–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Isogawa M., Furuichi Y., Chisari F. V.2005. Oscillating CD8+ T cell effector functions after antigen recognition in the liver. Immunity 23: 53–63. doi: 10.1016/j.immuni.2005.05.005 [DOI] [PubMed] [Google Scholar]
- 22.Jilbert A. R., Wu T. T., England J. M., Hall P. M., Carp N. Z., O’Connell A. P., Mason W. S.1992. Rapid resolution of duck hepatitis B virus infections occurs after massive hepatocellular involvement. J. Virol. 66: 1377–1388. doi: 10.1128/JVI.66.3.1377-1388.1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kajino K., Jilbert A. R., Saputelli J., Aldrich C. E., Cullen J., Mason W. S.1994. Woodchuck hepatitis virus infections: very rapid recovery after a prolonged viremia and infection of virtually every hepatocyte. J. Virol. 68: 5792–5803. doi: 10.1128/JVI.68.9.5792-5803.1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kakimi K., Guidotti L. G., Koezuka Y., Chisari F. V.2000. Natural killer T cell activation inhibits hepatitis B virus replication in vivo. J. Exp. Med. 192: 921–930. doi: 10.1084/jem.192.7.921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim T. H., Cho E. Y., Oh H. J., Choi C. S., Kim J. W., Moon H. B., Kim H. C.2006. The degrees of hepatocyte cytoplasmic expression of hepatitis B core antigen correlate with histologic activity of liver disease in the young patients with chronic hepatitis B infection. J. Korean Med. Sci. 21: 279–283. doi: 10.3346/jkms.2006.21.2.279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kimura K., Kakimi K., Wieland S., Guidotti L. G., Chisari F. V.2002. Activated intrahepatic antigen-presenting cells inhibit hepatitis B virus replication in the liver of transgenic mice. J. Immunol. 169: 5188–5195. doi: 10.4049/jimmunol.169.9.5188 [DOI] [PubMed] [Google Scholar]
- 27.Lai C. L., Yuen M. F.2008. Chronic hepatitis B-new goals, new treatment. N. Engl. J. Med. 359: 2488–2491. doi: 10.1056/NEJMe0808185 [DOI] [PubMed] [Google Scholar]
- 28.Lavanchy D.2004. Hepatitis B virus epidemiology, disease burden, treatment, and current and emerging prevention and control measures. J. Viral Hepat. 11: 97–107. doi: 10.1046/j.1365-2893.2003.00487.x [DOI] [PubMed] [Google Scholar]
- 29.Lok A. S., McMahon B. J.2009. Chronic hepatitis B: update 2009. Hepatology 50: 661–662. doi: 10.1002/hep.23190 [DOI] [PubMed] [Google Scholar]
- 30.Lok A. S., McMahon B. J.2007. Chronic hepatitis B. Hepatology 45: 507–539. doi: 10.1002/hep.21513 [DOI] [PubMed] [Google Scholar]
- 31.Lucifora J., Xia Y., Reisinger F., Zhang K., Stadler D., Cheng X., Sprinzl M. F., Koppensteiner H., Makowska Z., Volz T., Remouchamps C., Chou W. M., Thasler W. E., Hüser N., Durantel D., Liang T. J., Münk C., Heim M. H., Browning J. L., Dejardin E., Dandri M., Schindler M., Heikenwalder M., Protzer U.2014. Specific and nonhepatotoxic degradation of nuclear hepatitis B virus cccDNA. Science 343: 1221–1228. doi: 10.1126/science.1243462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Margeridon S., Carrouée-Durantel S., Chemin I., Barraud L., Zoulim F., Trépo C., Kay A.2008. Rolling circle amplification, a powerful tool for genetic and functional studies of complete hepatitis B virus genomes from low-level infections and for directly probing covalently closed circular DNA. Antimicrob. Agents Chemother. 52: 3068–3073. doi: 10.1128/AAC.01318-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marion P. L., Salazar F. H., Winters M. A., Colonno R. J.2002. Potent efficacy of entecavir (BMS-200475) in a duck model of hepatitis B virus replication. Antimicrob. Agents Chemother. 46: 82–88. doi: 10.1128/AAC.46.1.82-88.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ogasawara T., Oogane M., Al-Mahdawi M., Tsunoda M., Ando Y.2019. Effect of second-order magnetic anisotropy on nonlinearity of conductance in CoFeB/MgO/CoFeB magnetic tunnel junction for magnetic sensor devices. Sci. Rep. 9: 17018. doi: 10.1038/s41598-019-53439-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ohashi K., Marion P. L., Nakai H., Meuse L., Cullen J. M., Bordier B. B., Schwall R., Greenberg H. B., Glenn J. S., Kay M. A.2000. Sustained survival of human hepatocytes in mice: A model for in vivo infection with human hepatitis B and hepatitis delta viruses. Nat. Med. 6: 327–331. doi: 10.1038/73187 [DOI] [PubMed] [Google Scholar]
- 36.Okuno T., Sano A., Deguchi T., Katsuma Y., Ogasawara T., Okanoue T., Takino T.1984. Pathology of acute hepatitis A in humans. Comparison with acute hepatitis B. Am. J. Clin. Pathol. 81: 162–169. doi: 10.1093/ajcp/81.2.162 [DOI] [PubMed] [Google Scholar]
- 37.Petersen J., Dandri M., Mier W., Lütgehetmann M., Volz T., von Weizsäcker F., Haberkorn U., Fischer L., Pollok J. M., Erbes B., Seitz S., Urban S.2008. Prevention of hepatitis B virus infection in vivo by entry inhibitors derived from the large envelope protein. Nat. Biotechnol. 26: 335–341. doi: 10.1038/nbt1389 [DOI] [PubMed] [Google Scholar]
- 38.Ruan P., Yang C., Su J., Cao J., Ou C., Luo C., Tang Y., Wang Q., Yang F., Shi J., Lu X., Zhu L., Qin H., Sun W., Lao Y., Li Y.2013. Histopathological changes in the liver of tree shrew (Tupaia belangeri chinensis) persistently infected with hepatitis B virus. Virol. J. 10: 333. doi: 10.1186/1743-422X-10-333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sanada T., Tsukiyama-Kohara K., Yamamoto N., Ezzikouri S., Benjelloun S., Murakami S., Tanaka Y., Tateno C., Kohara M.2016. Property of hepatitis B virus replication in Tupaia belangeri hepatocytes. Biochem. Biophys. Res. Commun. 469: 229–235. [DOI] [PubMed] [Google Scholar]
- 40.Severini A., Liu X. Y., Wilson J. S., Tyrrell D. L.1995. Mechanism of inhibition of duck hepatitis B virus polymerase by (-)-beta-L-2′,3′-dideoxy-3′-thiacytidine. Antimicrob. Agents Chemother. 39: 1430–1435. doi: 10.1128/AAC.39.7.1430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shayestehpour M., Moghim S., Salimi V., Jalilvand S., Yavarian J., Romani B., Mokhtari-Azad T.2017. Targeting human breast cancer cells by an oncolytic adenovirus using microRNA-targeting strategy. Virus Res. 240: 207–214. doi: 10.1016/j.virusres.2017.08.016 [DOI] [PubMed] [Google Scholar]
- 42.Walter E., Keist R., Niederöst B., Pult I., Blum H. E.1996. Hepatitis B virus infection of tupaia hepatocytes in vitro and in vivo. Hepatology 24: 1–5. [DOI] [PubMed] [Google Scholar]
- 43.Wang Q., Schwarzenberger P., Yang F., Zhang J., Su J., Yang C., Cao J., Ou C., Liang L., Shi J., Yang F., Wang D., Wang J., Wang X., Ruan P., Li Y.2012. Experimental chronic hepatitis B infection of neonatal tree shrews (Tupaia belangeri chinensis): a model to study molecular causes for susceptibility and disease progression to chronic hepatitis in humans. Virol. J. 9: 170. doi: 10.1186/1743-422X-9-170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Webster G. J., Reignat S., Maini M. K., Whalley S. A., Ogg G. S., King A., Brown D., Amlot P. L., Williams R., Vergani D., Dusheiko G. M., Bertoletti A.2000. Incubation phase of acute hepatitis B in man: dynamic of cellular immune mechanisms. Hepatology 32: 1117–1124. doi: 10.1053/jhep.2000.19324 [DOI] [PubMed] [Google Scholar]
- 45.Wieland S. F., Spangenberg H. C., Thimme R., Purcell R. H., Chisari F. V.2004. Expansion and contraction of the hepatitis B virus transcriptional template in infected chimpanzees. Proc. Natl. Acad. Sci. USA 101: 2129–2134. doi: 10.1073/pnas.0308478100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yan H., Zhong G., Xu G., He W., Jing Z., Gao Z., Huang Y., Qi Y., Peng B., Wang H., Fu L., Song M., Chen P., Gao W., Ren B., Sun Y., Cai T., Feng X., Sui J., Li W.2012. Sodium taurocholate cotransporting polypeptide is a functional receptor for human hepatitis B and D virus. eLife 1: e00049. doi: 10.7554/eLife.00049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang C., Ruan P., Ou C., Su J., Cao J., Luo C., Tang Y., Wang Q., Qin H., Sun W., Li Y.2015. Chronic hepatitis B virus infection and occurrence of hepatocellular carcinoma in tree shrews (Tupaia belangeri chinensis). Virol. J. 12: 26. doi: 10.1186/s12985-015-0256-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang Y., Zhang S., Ma K., Fu B., Anand M.2014. Woody species diversity in forest plantations in a mountainous region of Beijing, China: effects of sampling scale and species selection. PLoS One 9: e115038. doi: 10.1371/journal.pone.0115038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zoulim F., Berthillon P., Guerhier F. L., Seigneres B., Germon S., Pichoud C., Cheng Y. C., Trepo C.2002. Animal models for the study of HBV infection and the evaluation of new anti-HBV strategies. J. Gastroenterol. Hepatol. 17 Suppl: S460–S463. doi: 10.1046/j.1440-1746.17.s4.10.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.