Abstract
Background
Neuroinflammation involving the central nervous system (CNS), such as depression, is associated with a significantly increased risk of cancer and cancer-specific mortality due to breast cancer. It is of great significance to learn about the regulatory process of CNS in breast cancer progression.
Methods
We established a depressive MMTV-PyVT mouse model. The expression levels of neurotransmitters in the serum of depression animal models were assessed by enzyme-linked immunosorbent assay (ELISA). Changes of the microglia cells in the mice’s brains were evaluated by immunofluorescence and reverse transcription-polymerase chain reaction (RT-PCR). Breast cancer progression was assessed by immunohistochemistry (IHC) analysis. To further investigate the mechanism by which ant-depressant drugs disrupt breast cancer progression, protein sequencing and network pharmacology were applied to identify related targets. Furthermore, we used conditioned medium from BV-2 microglia to culture breast cancer cells and treated the cells with quercetin at different concentrations; cell viability was assessed by the MTT assay.
Results
Our results show a possible regulatory target between neuroinflammation in the CNS and development of breast cancer, along with the reversal effect of quercetin on breast cancer progression.
Conclusions
Chronic stress may be an indicator of breast cancer and that quercetin could be an effective treatment for breast cancer patients with chronic stress.
Keywords: Neuroinflammation, breast cancer, quercetin, depression, microglia, network pharmacology
Introduction
Breast cancer is the most common clinical malignancy in women worldwide (1,2), with China accounting for 12.2% of new breast cancer cases and 9.6% of deaths worldwide each year (3,4). The neurotoxic effects of anti-cancer drugs have been confirmed in clinical and pre-clinical research (5). Breast cancer patients are prone to presenting with age-related cognitive deficits and/or cognitive decline during chemotherapy (6). Depression is a common psychological complication of breast cancer (7,8), which has profound effects on neural structure and function (9,10), and the presence of chronic stress has a major impact on depression (11,12). The incidence of depression in every generation worldwide is increasing (13,14). Continuous psychological stress often induces breast cancer patients into a state of chronic stress (15,16). Traditional Chinese medicine has been proven to be effective in the prevention and treatment of various psychiatric diseases, including depression (17-19).
Xiaoyao pill (State Medical Permit No. Z41021831), a classical Chinese medicine formula, has been widely used in mental disorders for thousands of years. It has been demonstrated to ameliorate depressive-like behaviors in rats with chronic restraint stress (CRS) (20) and to impart anti-depressive effects in mouse models of chronic unpredictable mild stress-induced depression (21). However, the pharmacological mechanism of antidepressant drugs in the promotion of breast cancer remains unclear.
Previous studies on breast cancer seldom focus on the relation between central nervous system (CNS) and breast cancer. Our study tried to figure out the main target genes between CNS and breast cancer for the further researches on the treatment on breast cancer patients with chronic stress. We therefore established a CRS MMTV-PyVT mouse model (Figure 1A) to identify the active compounds and related potential targets of the antidepressant drug and to elucidate the effect of this antidepressant drug in breast cancer invasion and metastasis. We present the following article in accordance with the ARRIVE reporting checklist (available at http://dx.doi.org/10.21037/atm-21-2558).
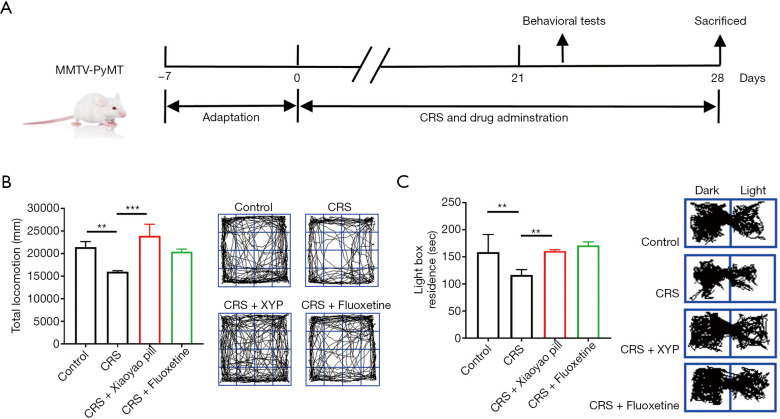
Figure 1.
Effect of CRS on depressive-like behaviors in MMTV-PyVT mice. CRS robustly induced notable depressive-like behaviors in MMTV-PyMT mice, which was be relieved by the antidepressant Xiaoyao pill. (A) Schematic timeline of the experimental procedures. Antidepressant-like effects of antidepressant Xiaoyao pill on MMTV-PyMT mice in open-field (B) and light/dark box (C) tests. **, P<0.01; ***, P<0.001. CRS, chronic restraint stress; SEM, standard error of mean; XYP, Xiaoyao pill.
Methods
Animals
Five-week-old female FVB/N-Tg (MMTV-PyVT) 634 Mul/J mice weighing about 16–18 g were acquired from Shanghai Model Organisms Center, Inc. (Shanghai, China). The experimental scheme was approved by the experimental animal ethics committee of Guangdong Provincial Hospital of Chinese Medicine (Reference No. 2019048). Animal welfare and experimental procedures were performed in compliance with the Guide for the Care and Use of Laboratory Animals (National Institutes of Health, the United States). A protocol was prepared before the study without registration.
After 7 days of acclimatization, the mice were randomly assigned to the control group (n=10) or the CRS-treated group (n=10); CRS treatment (restrained stress 4 hours/day) was maintained for 3 weeks until the mice exhibited a depression-like behavior phenotype. After the last behavioral examination, the mice were sacrificed after anesthesia, and serum samples as well as hippocampus and breast cancer tissues were collected.
Cell culture
Breast cancer cell lines MCF-7, T47D, HCC1937, HCC1806, MDA-MB-231, and SKB-R3, along with BV-2 microglia cells were purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA), cultured in Dulbecco’s modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS), and incubated in the incubator (WCI-180, Wiggens, Germany) with a humid atmosphere of 37 °C and 5% CO2. All cell culture reagents were obtained from Thermo Fisher Scientific (Waltham, MA, USA).
Reagents and preparation
The Xiaoyao pill, which was obtained from Guangdong Provincial Hospital of Chinese Medicine, was dissolved in normal saline to yield a suspension. According to the conversion coefficient between adult and mice, the dosage of the Xiaoyao pill for mice was 6.1 g/kg. The mice were treated with Xiaoyao pill by intragastric administration once a day. Fluoxetine hydrochloride was purchased from Guangdong Provincial Hospital of Chinese Medicine and dissolved in normal saline to yield a suspension. The mice received fluoxetine via intragastric administration at a dose of 1.8 mg/kg once a day. Quercetin (CAS No. 117-39-5, purity ≥95%) was obtained from Sigma-Aldrich (St. Louis, MO, USA). Stock solutions of 100 mM concentration were prepared in dimethyl sulfoxide (DMSO) and stored at –20 °C. Quercetin was diluted as needed in the experimental medium.
Behavioral tests
All the behavioral tests were performed at the end of a 3-week period of CRS exposure, and included the open-field test (OFT) and light/dark box test (LBT). Behavioral performances were recorded using a video camera. All the tests were recorded in an animal behavior analysis system (Shanghai Jiliang Software Technology Co., Ltd., China). OFT was performed on day 32 to measure locomotor activity. The locomotor activity of the mice in an open-field area (40 cm × 40 cm × 40 cm) was recorded for 10 minutes after drug administration. LBT consisted of 2 compartments with a lid divided by a connecting door (3.5 cm × 3.5 cm). The light compartment (24 cm × 20 cm × 20 cm) was illuminated by a single 60 W (300 lx) fluorescent lamp that was positioned 30 cm above the compartment. The dark compartment (12 cm × 20 cm × 20 cm) was entirely black. Each mouse was individually placed in the light compartment facing the door and monitored for 5 minutes. The number of transfers between the two compartments and the time spent in the white compartment was recorded by a video motion tracking system. Between trials, the compartments were cleaned with 75% ethanol to remove odor cues.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
In this study, we used PrimeScriptTM RT Master Mix (Takara Bio, Shiga, Japan) for the reverse transcription (RT) reaction according to the manufacturer’s instructions (2 μL 5× PrimeScript RT Master Mix, total RNA, RNase-free H2O up to 10 mL). RT was conducted at 37 °C for 15 minutes, and reverse transcriptase inactivation was conducted at 85 °C for 5 seconds. The obtained RT reaction solution was used to prepare a PCR reaction solution for real-time PCR reaction. Subsequently, we mixed 10 mL of TB Green® Premix Ex TaqTM II (Takara Bio, Shiga, Japan), 0.8 mL of the PCR forward primer (10 mM), 0.8 mL of the PCR reverse primer (10 mM), 0.4 mL of ROX reference dye or Dye II (50×) ×2, 2 mL of the RT reaction solution [complement DNA (cDNA) solution] ×3, and 6 mL of sterilized distilled water (dH2O) in a total volume of 20 mL. PCR was conducted under the following conditions: predenaturation at 95 °C for 30 seconds, followed by 40 cycles of denaturation at 95 °C for 5 seconds, and annealing for 1 minute at 60 °C and 72 °C for 30 seconds. The assays were performed 3 times for each sample, and the mean number of genomes was calculated. Finally, a 60–95 °C melting curve was generated.
Immunohistochemistry (IHC)
IHC assay was used to detect the content of Laminin A and Ki67. Paraffin sections were separated in xylene and rehydrated in an ethanol gradient (absolute ethanol, 95% ethanol, 70% ethanol, and 50% ethanol). After the antigen was extracted in 10 mM citric acid buffer, the tissue sections were incubated in 3% H2O2 for 10 minutes, and sealed at room temperature for 1 hour. The tissue sections were then incubated overnight with primary antibodies. Sections were then washed with Tris-Buffered Saline + Tween 20 and incubated with SignalStain Boost IHC Detection Reagent [Cell Signaling Technology (CST), Danvers, MA, USA] for 30 minutes at room temperature. After that, the sections were stained with a SignalStain DAB Substrate Kit (CST) and observed under a FluoView FV1000 confocal microscope (Olympus, Shinjuku, Tokyo, Japan). All experiments were performed in triplicate.
Proteomics analysis
Proteins meeting the screening criteria of differential expression exhibited ploidy greater than 1.2-fold (up- and downregulated), and those with P values (t-test) less than 0.05 were considered differentially expressed proteins. Analytical methods included significant difference analysis, Gene Ontology (GO) annotation and enrichment analysis, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway annotation and enrichment analysis, and cluster analysis.
Enzyme-linked immunosorbent assay (ELISA)
The sandwich ELISA method was used to measure the levels of 5-hydroxytryptamine, adrenaline, noradrenaline, and dopamine in the mouse serum samples using kits from Omnimabs (Omnimabs, Alhambra, CA, USA). The ELISA kit used purified antibodies immobilized onto the surface of the microtiter well plates. The samples, including standards, were added into antibody-coated microtiter wells via a pipette. Following incubation and washing of the wells, biotinylated antibody and streptavidin combined with horseradish peroxidase (HRP) were added, and the plate was incubated again. A complex was formed between the antibody and the antigen enzyme-linked antibody. After another wash, a tetramethylbenzidine (TMB) substrate solution was added to the well plates. Ammonium sulfate (NH4)2SO4 was used to stop the reaction, and colorimetric changes were assessed. Color changes were evaluated at a wavelength of 450 nm using a microplate reader. The optical density (OD) values were directly proportional to the protein concentration. A standard curve was used to calculate protein concentrations. All experiments were performed in triplicate.
Immunofluorescence
Mouse hippocampal tissues were fixed on glass coverslips and treated as indicated. Upon fixation/permeabilization with cold 100% methanol for 5 minutes at room temperature, the cells were washed with phosphate-buffered saline (PBS) twice and blocked in TBS buffer (50 mM Tris-Cl, 150 mM NaCl, pH 7.5) containing 0.1% Tween-20, 2% bovine serum albumin (BSA), and 0.1% NaN3 (Sigma-Aldrich) for 1 hour at room temperature. The primary rabbit anti-Iba1 antibody (CST, #17198) diluted 1:200 in blocking solution was then added and incubated with cells at 4 °C overnight. After three washes in PBS, the cells were incubated with Alexa Fluor 488 conjugated anti-rabbit secondary antibody (Thermo Fisher Scientific) for 2 hours at room temperature. Coverslips were washed 4 times with PBS and mounted on SuperFrost Plus slides with Vectashield anti-fade mounting medium with DAPI (Vector Labs, Burlingame, CA, USA). Images of the cells were captured using a confocal microscope.
Network construction and analyses
Active compounds and related target collection
All compounds contained in the Xiaoyao pill were queried in the Traditional Chinese Medicine Systems Pharmacology Database and Analysis Platform (TCMSP) (http://lsp.nwsuaf.edu.cn/). Oral bioavailability (OB) greater than 30% and drug-likeness (DL) greater than 0.18 were used as criteria in screening for potentially active compounds. All the targets of the active compounds in Xiaoyao pill were retrieved from TCMSP. The targets were then submitted to the UniProt database (http://www.UniProt.org/) to obtain their gene names.
Collecting gene targets related to diseases
The gene targets associated with “breast cancer”, “depression”, “anxiety”, and “immune” were obtained from two databases: GeneCards (https://www.genecards.org/) and Online Mendelian Inheritance in Man (OMIM) (https://www.omim.org/). After removal of the duplicate targets, the overlapping target genes related to “breast cancer”, “depression or anxiety”, “immune”, and the differential genes of previous animal experiment were collected as candidate targets.
Creating an active compound-gene target network
An active compound-gene target-disease network was constructed and displayed using Cytoscape 3.7.2 (http://cytoscape.org/). The network can identify target genes that associate active compounds in Xiaoyao pill. The Search Tool for the Retrieval of Interacting Genes/Proteins (STRING; http://www.string-db.org/) database provides experimental and predicted information on the interactions of proteins. In addition, candidate targets were input into STRING to acquire relevant information on protein interactions with the species limited to “Homo sapiens” and a combined score >0.4. A protein-protein interaction (PPI) network was created. The top 30 hub proteins were visualized using the R programming language (The R Foundation for Statistical Computing). GO functional enrichment and KEGG pathway enrichment analyses were performed using the online functional annotation and enrichment tool Metascape (https://metascape.org/). The downloaded results were sorted based on enrichment score, count values, and P values, and the results were then submitted to a bioinformatics online tool (http://www.bioinformatics.com.cn) that allows data visualization. Previous proteomic results showed 23 differentially expressed proteins, which were used to overlap with the candidate targets. One intersectional target was viewed as the key target in this research. First, the 3D structures of the ligand molecules were obtained from the TCMSP database and stored as a Mol2 file. Second, the protein structures were downloaded from the RCSB Protein Data Bank (http://www.rcsb.org/) and saved as PDB files. Third, after preparing the files of ligand molecules and the target protein through Pymol and Autodock Tools, Autodock Vina 1.1.2 (http://vina.scripps.edu/) was used to analyze the binding properties of the ligands of the protein. Finally, after docking, one ligand with the lowest affinity score for the protein was selected for further analysis.
MTT assay
The supernatant culture medium of BV-2 was collected in advance [after soaking in 1 μg/L lipopolysaccharide (LPS) for 24 hours; it was collected after replacing the conventional culture medium for 24 hours] and mixed with the conventional culture medium in a 1:1 ratio as an intervention method. MCF-7, T47D, HCC1937, HCC1806, MDA-MB-231, and SKB-R3 cells were cultured in 1 μg/L LPS-stimulated supernatant of BV-2 microglia (mixed with fresh medium in a 1:1 ratio) for 24 hours. The T47D and MCF-7 cells were soaked in BV-2 supernatant nutrient solution for 24 hours, subjected to quercetin (20, 40, 80, 160 nmol/L) and processed for 24 hours, and treated with quercetin or BV-2 supernatant for 24 hours separately. Cell viability was assessed using the MTT assay.
Various breast cancer cells at the logarithmic phase were seeded at a density of 5×103 per well of a 96-well microtiter plate containing 100 μL of culture medium. After the different processing methods were completed (the specific conditions are described below) for each group, the cells were incubated at 37 °C and 5% CO2 in an incubator for 24 hours, and then 10 μL of MTT solution (5 mg/mL) was added to each well and incubated for 4 hours. After 100 μL of DMSO was added to each well, the absorbance was measured using a microplate reader at a wavelength of 570 nm.
Statistical analysis
All results are represented as mean ± standard error of mean (SEM). Comparisons were made by one-way analysis of variance followed by Tukey’s test. Statistical analysis was performed by GraphPad Prism V.9.0.0 [121] (San Diego, California, USA). Values of P<0.05 were considered statistically significant. All analyses were performed with three independent experiments.
Results
Effect of CRS on depressive-like behaviors in MMTV-PyVT mice
Compared with the controls, mice with CRS showed a significant decrease in the total locomotion distance, and the depressive-like behavior was alleviated by the Xiaoyao pill. The mice treated with the Xiaoyao pill and fluoxetine showed substantial improvement in OFT indices compared with CRS-induced mice (Figure 1B). CRS induced a decrease in residence time for mice in the light box, which was also reversed by the Xiaoyao pill (Figure 1C). Moreover, the therapeutic effect of the Xiaoyao pill was more significant than that of the fluoxetine.
Effect of CRS on neurotransmitter and microglia cells in MMTV-PyVT mice
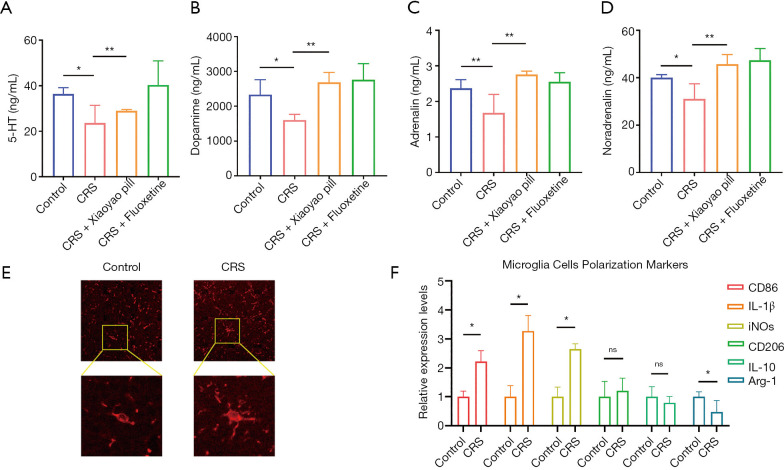
The levels of 5-hydroxytryptamine (Figure 2A), dopamine (Figure 2B), adrenaline (Figure 2C), and noradrenaline (Figure 2D) in the serum of the CRS group mice decreased, and this downregulation was suppressed by the Xiaoyao pill.
Figure 2.
Effect of CRS on neurotransmitter and microglia cells in MMTV-PyVT mice. Effect of CRS on neurotransmitter in serum was reversed by antidepressant Xiaoyao pill (A,B,C,D). Immunofluorescence detection of microglia marker IBA1 (E) (200× above, 400× below). Relative expression levels of M1 polarization markers (CD86, IL-1β, iNOS) and M2 polarization markers (CD206, IL-10, Arg-1) in microglia cells after CRS treatment (F). *, P<0.05; **, P<0.01. CRS, chronic restraint stress; CD86, cluster of differentiation 86; IL-1β, interleukin 1 beta; iNOS, inducible nitric oxide synthase; SEM, standard error of mean.
CRS induced the polarization of microglia cells in the brain of MMTV-PyVT mice (Figure 2E). However, the relative expression levels of M1 polarization markers [cluster of differentiation 86 (CD86), interleukin 1 beta (IL-1β), and inducible nitric oxide synthase (iNOS)] in microglia cells increased, while the relative expression levels of M2 polarization markers (CD206, IL-10, and Arg-1) decreased (Figure 2F), indicating that microglia cells were mostly polarized to the M1 type.
Effect of CRS on breast cancer progression in MMTV-PyVT mice
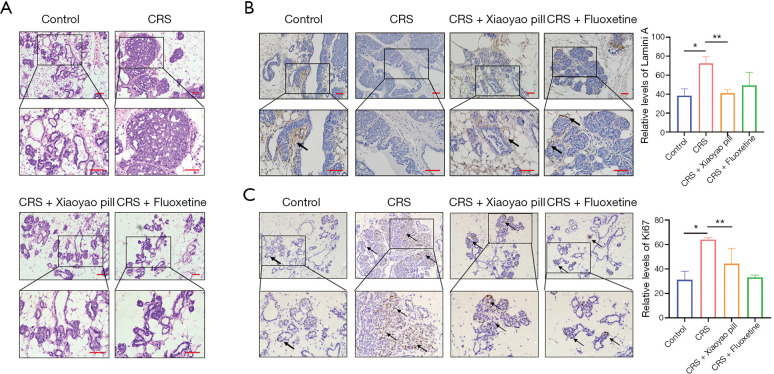
In the CRS group, breast ducts and acini were completely filled by tumor cells, and atypia was obvious. Hematoxylin and eosin (HE) staining also confirmed this stage. However, there were numerous ducts in the control and treatment groups (including in the CRS + Xiaoyao pill group and the CRS + fluoxetine group). A few lobules were observed in dysplastic cells that were attached to the ducts. HE staining showed that these were in the early stage of breast cancer and breast hyperplasia (Figure 3A). Laminin A staining showed that the CRS group was in the early stage of breast cancer, and the other groups were in the adenoma stage (Figure 3B). Ki67 staining showed that there were more proliferating cells in the CRS group than in the control and treatment groups (Figure 3C).
Figure 3.
Effect of CRS on promotion of breast cancer in MMTV-PyVT mice, which was postponed by antidepressant Xiaoyao pill. Breast tumor HE staining (A), Laminin A IHC staining (B) (arrows stand for Laminin A), and Ki67 IHC staining (C) (arrows stand for Ki67). (200× above, 400× below) *, P<0.05; **, P<0.01. CRS, chronic restraint stress; HE, hematoxylin and eosin; IHC, immunohistochemistry.
Protein sequencing of differentially expressed proteins
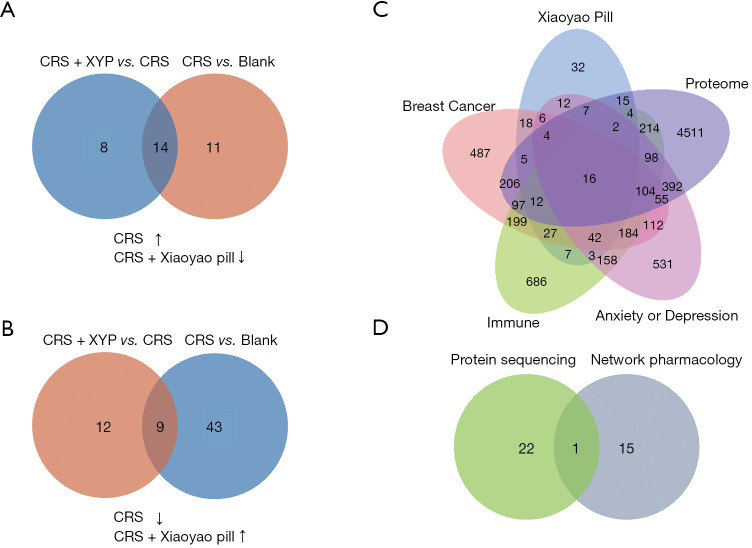
According to the result of protein sequencing, 25 proteins were upregulated after CRS stimulation, of which 14 were downregulated by the Xiaoyao pill (Figure 4A, Tables 1,2). A further 52 proteins were downregulated after CRS stimulation, and 9 were upregulated by the Xiaoyao pill (Figure 4B, Tables 3,4). Based on TCMSP, 120 active compounds in the Xiaoyao pill were identified (Table 5), and 212 targets of active compounds were collected. A total of 1,547 target genes related to breast cancer were collected from GeneCards and OMIM databases. In addition, 1,726 target genes related to depression or anxiety and 1,853 target genes related to immunity were retrieved from these databases. Furthermore, previous animal experiments identified 5,742 differentially expressed genes based on the results of proteomics. After merging 5 kinds of gene targets, 16 overlapping targets were identified as candidate targets (Figure 4C, Table 6). Based on key target screening, STAT1 was identified at the intersectional target (Figure 4D).
Figure 4.
Protein sequencing and network pharmacology for target proteins. (A,B) Venn diagram of the proteins upregulated and downregulated after CRS, as well as by the Xiaoyao pill after CRS. (C) Venn diagram of the targets for active compounds of the Xiaoyao pill, breast cancer, immune, animal experiment, and depression or anxiety. (D) Venn diagram of the intersectional target of protein sequencing and network pharmacology. CRS, chronic restraint stress; XYP, Xiaoyao pill.
Table 1. Proteins upregulated after CRS (CRS vs. blank).
| Accession | Gene name/description | CRS/blank | P value |
|---|---|---|---|
| Q64339 | Isg15 | 2.0682 | 0.0025 |
| Q64282 | Ifit1 | 1.8564 | 0.0023 |
| P01867 | Igh-3 | 1.7271 | 0.0059 |
| P42225 | Stat1 | 1.6328 | 0.0004 |
| P01807 | Ig heavy chain V region X44 | 1.5950 | 0.0165 |
| P01635 | Ig kappa chain V-V region K2 (Fragment) | 1.4875 | 0.0018 |
| P01837 | Igkc | 1.4394 | 0.0211 |
| Q60590 | Orm1 | 1.4287 | 0.0034 |
| Q07797 | Lgals3bp | 1.3669 | 0.0163 |
| P06327 | Gm5629 | 1.3592 | 0.0023 |
| Q61107 | Gbp4 | 1.3358 | 0.0016 |
| Q9D8B3 | Chmp4b | 1.3301 | 0.0011 |
| Q91X72 | Hpx | 1.3109 | 0.0258 |
| Q03963 | Eif2ak2 | 1.2984 | 0.0003 |
| Q9Z0E6 | Gbp2 | 1.2746 | 0.0108 |
| P01865 | Igh-1a | 1.2709 | 0.0012 |
| P10854 | H2bc14 | 1.2568 | 0.0400 |
| Q6Q899 | Ddx58 | 1.2532 | 0.0319 |
| Q64112 | Ifit2 | 1.2506 | 0.0018 |
| P68369 | Tuba1a | 1.2302 | 0.0326 |
| Q60766 | Irgm1 | 1.2302 | 0.0086 |
| Q9CQ10 | Chmp3 | 1.2266 | 0.0004 |
| Q6PHZ2 | Camk2d | 1.2244 | 0.0228 |
| P14824 | Anxa6 | 1.2156 | 0.0249 |
| Q19LI2 | A1bg | 1.2044 | 0.0174 |
CRS, chronic restraint stress.
Table 2. Proteins downregulated by the Xiaoyao pill after CRS (CRS + Xiaoyao pill vs. CRS).
| Accession | Gene name/description | CRS + Xiaoyao pill/CRS | P value |
|---|---|---|---|
| Q6PDM2 | Srsf1 | 0.8329 | 0.0025 |
| Q8R081 | Hnrnpl | 0.8303 | 0.0288 |
| Q60590 | Orm1 | 0.8257 | 0.0193 |
| P01899 | H2-D1 | 0.8226 | 0.0177 |
| O70370 | Ctss | 0.8203 | 0.0004 |
| Q6ZWQ7 | Spcs3 | 0.8195 | 0.0218 |
| Q9Z204 | Hnrnpc | 0.8176 | 0.0110 |
| P62806 | H4c1 | 0.8052 | 0.0468 |
| Q9Z0E6 | Gbp2 | 0.7987 | 0.0141 |
| Q6Q899 | Ddx58 | 0.7976 | 0.0336 |
| P10854 | H2bc14 | 0.7942 | 0.0146 |
| Q60766 | Irgm1 | 0.7787 | 0.0053 |
| P01635 | Ig kappa chain V-V region K2 (Fragment) | 0.7760 | 0.0110 |
| Q9ESB3 | Hrg | 0.7732 | 0.0122 |
| P68369 | Tuba1a | 0.7719 | 0.0071 |
| Q03963 | Eif2ak2 | 0.7640 | 0.0021 |
| Q61107 | Gbp4 | 0.7524 | 0.0028 |
| P01867 | Igh-3 | 0.7265 | 0.0265 |
| Q07797 | Lgals3bp | 0.7012 | 0.0083 |
| P42225 | Stat1 | 0.6271 | 0.0008 |
| Q64282 | Ifit1 | 0.5579 | 0.0024 |
| Q64339 | Isg15 | 0.5098 | 0.0033 |
CRS, chronic restraint stress.
Table 3. Proteins downregulated after CRS (CRS vs. blank).
| Accession | Gene name/description | CRS/blank | P value |
|---|---|---|---|
| Q8K0T7 | Unc13c | 0.8329 | 0.0044 |
| Q62210 | Birc2 | 0.8305 | 0.0498 |
| Q6A152 | Cyp4x1 | 0.8290 | 0.0064 |
| Q9WVF8 | Tusc2 | 0.8274 | 0.0011 |
| Q99K30 | Eps8l2 | 0.8270 | 0.0138 |
| P50608 | Fmod | 0.8265 | 0.0400 |
| P03995 | Gfap | 0.8244 | 0.0045 |
| Q9R049 | Amfr | 0.8229 | 0.0132 |
| P39429 | Traf2 | 0.8222 | 0.0064 |
| Q8BFP9 | Pdk1 | 0.8214 | 0.0078 |
| Q99N89 | Mrpl43 | 0.8207 | 0.0464 |
| Q8BH98 | Mboat1 | 0.8205 | 0.0006 |
| Q9JKL4 | Ndufaf3 | 0.8204 | 0.0048 |
| P20152 | Vim | 0.8193 | 0.0123 |
| Q8VCF1 | Cant1 | 0.8130 | 0.0058 |
| Q9ERD6 | Ralgps2 | 0.8128 | 0.0088 |
| Q8K0D2 | Habp2 | 0.8119 | 0.0203 |
| O55026 | Entpd2 | 0.8084 | 0.0103 |
| Q61216 | Mre11 | 0.8072 | 0.0270 |
| P46735 | Myo1b | 0.8036 | 0.0084 |
| P98191 | Cds1 | 0.8017 | 0.0062 |
| Q8VED5 | Krt79 | 0.4590 | 0.0372 |
| Q8BGZ7 | Krt75 | 0.4537 | 0.0265 |
| Q03311 | Bche | 0.4468 | 0.0000 |
| Q99LC9 | Pex6 | 0.3942 | 0.0334 |
| Q8BQM8 | Eml5 | 0.3845 | 0.0320 |
| Q7TNG8 | Ldhd | 0.7991 | 0.0159 |
| Q8N7N5 | Dcaf8 | 0.7974 | 0.0002 |
| Q8BKH7 | Mapkap1 | 0.7955 | 0.0215 |
| Q91WR3 | Ascc2 | 0.7802 | 0.0074 |
| Q3UZA1 | Rcsd1 | 0.7801 | 0.0440 |
| Q9JJ26 | Mefv | 0.7797 | 0.0034 |
| Q8VE96 | Slc35f6 | 0.7783 | 0.0102 |
| Q9R0X0 | Med20 | 0.7679 | 0.0404 |
| Q6TEK5 | Vkorc1l1 | 0.7649 | 0.0009 |
| Q6P6M7 | Sepsecs | 0.7497 | 0.0352 |
| Q9Z1R4 | D17h6s53e | 0.7488 | 0.0226 |
| O89110 | Casp8 | 0.7428 | 0.0120 |
| P70302 | Stim1 | 0.7387 | 0.0005 |
| Q9WUP4 | Srd5a3 | 0.7223 | 0.0288 |
| Q99N93 | Mrpl16 | 0.6986 | 0.0107 |
| Q80X60 | Efcab3 | 0.6936 | 0.0092 |
| A2APT9 | Klhdc7a | 0.6843 | 0.0233 |
| P04104 | Krt1 | 0.6677 | 0.0341 |
| Q9R0M0 | Celsr2 | 0.6605 | 0.0004 |
| Q8BZS9 | Dhx32 | 0.6522 | 0.0322 |
| Q3UV17 | Krt76 | 0.6159 | 0.0437 |
| Q922U2 | Krt5 | 0.6096 | 0.0480 |
| Q6IFX2 | Krt42 | 0.5902 | 0.0192 |
| Q61781 | Krt14 | 0.5115 | 0.0245 |
| Q9D845 | Tex9 | 0.5061 | 0.0073 |
| Q3U595 | UPF0545 protein C22orf39 homolog | 0.5006 | 0.0246 |
CRS, chronic restraint stress.
Table 4. Proteins upregulated by the Xiaoyao pill after CRS (CRS + Xiaoyao pill vs. CRS).
| Accession | Gene name/description | CRS + Xiaoyao pill/CRS | P value |
|---|---|---|---|
| Q9CYN2 | Spcs2 | 1.6329 | 0.0478 |
| Q80X60 | Efcab3 | 1.6057 | 0.0063 |
| Q9JJ26 | Mefv | 1.4960 | 0.0006 |
| Q8BH98 | Mboat1 | 1.4269 | 0.0000 |
| Q3UNA4 | Nxt2 | 1.3960 | 0.0005 |
| Q62210 | Birc2 | 1.3787 | 0.0122 |
| Q8VCF1 | Cant1 | 1.3094 | 0.0442 |
| P50608 | Fmod | 1.2951 | 0.0265 |
| Q8VE96 | Slc35f6 | 1.2775 | 0.0023 |
| P35441 | Thbs1 | 1.2617 | 0.0285 |
| O55026 | Entpd2 | 1.2583 | 0.0004 |
| Q80YR2 | Fam160b2 | 1.2482 | 0.0159 |
| P56382 | Atp5f1e | 1.2359 | 0.0448 |
| P63141 | Kcna2 | 1.2320 | 0.0130 |
| P51830 | Adcy9 | 1.2311 | 0.0291 |
| Q62234 | Myom1 | 1.2224 | 0.0266 |
| Q91WR3 | Ascc2 | 1.2196 | 0.0225 |
| Q80Z38 | Shank2 | 1.2179 | 0.0143 |
| Q9D6J4 | Necab3 | 1.2039 | 0.0381 |
| P62878 | Rbx1 | 1.2028 | 0.0296 |
| P03975 | Iap | 1.2002 | 0.0173 |
CRS, chronic restraint stress.
Table 5. A total of 120 active compounds of the Xiaoyao pill were selected from TCMSP.
| No. | Mol ID | Molecule name | Herbs | OB | DL |
|---|---|---|---|---|---|
| 1 | MOL001918 | Paeoniflorgenone | Bai Shao | 87.59 | 0.37 |
| 2 | MOL001919 | (3S,5R,8R,9R,10S,14S)-3,17-dihydroxy-4,4,8,10,14-pentamethyl-2,3,5,6,7,9-hexahydro-1H-cyclopenta[a]phenanthrene-15,16-dione | Bai Shao | 43.56 | 0.53 |
| 3 | MOL001924 | Paeoniflorin | Bai Shao | 53.87 | 0.79 |
| 4 | MOL000492 | (+)-Catechin | Bai Shao | 54.83 | 0.24 |
| 5 | MOL000022 | 14-acetyl-12-senecioyl-2E,8Z,10E-atractylentriol | Bai Zhu | 63.37 | 0.3 |
| 6 | MOL000033 | (3S,8S,9S,10R,13R,14S,17R)-10,13-dimethyl-17-[(2R,5S)-5-propan-2-yloctan-2-yl]-2,3,4,7,8,9,11,12,14,15,16,17-dodecahydro-1H-cyclopenta[a]phenanthren-3-ol | Bai Zhu | 36.23 | 0.78 |
| 7 | MOL000049 | 3β-acetoxyatractylone | Bai Zhu | 54.07 | 0.22 |
| 8 | MOL000072 | 8β-ethoxy atractylenolide III | Bai Zhu | 35.95 | 0.21 |
| 9 | MOL001689 | Acacetin | Bo He | 34.97 | 0.24 |
| 10 | MOL002881 | Diosmetin | Bo He | 31.14 | 0.27 |
| 11 | MOL000471 | Aloe-emodin | Bo He | 83.38 | 0.24 |
| 12 | MOL005190 | Eriodictyol | Bo He | 71.79 | 0.24 |
| 13 | MOL005573 | Genkwanin | Bo He | 37.13 | 0.24 |
| 14 | MOL000006 | Luteolin | Bo He | 36.16 | 0.25 |
| 15 | MOL001645 | Linoleyl acetate | Chai Hu | 42.1 | 0.2 |
| 16 | MOL004598 | 3,5,6,7-tetramethoxy-2-(3,4,5-trimethoxyphenyl)chromone | Chai Hu | 31.97 | 0.59 |
| 17 | MOL004609 | Areapillin | Chai Hu | 48.96 | 0.41 |
| 18 | MOL013187 | Cubebin | Chai Hu | 57.13 | 0.64 |
| 19 | MOL004624 | Longikaurin A | Chai Hu | 47.72 | 0.53 |
| 20 | MOL004653 | (+)-Anomalin | Chai Hu | 46.06 | 0.66 |
| 21 | MOL004718 | A-spinasterol | Chai Hu | 42.98 | 0.76 |
| 22 | MOL000490 | Petunidin | Chai Hu | 30.05 | 0.31 |
| 23 | MOL000273 | (2R)-2-[(3S,5R,10S,13R,14R,16R,17R)-3,16-dihydroxy-4,4,10,13,14-pentamethyl-2,3,5,6,12,15,16,17-octahydro-1H-cyclopenta[a]phenanthren-17-yl]-6-methylhept-5-enoic acid | Fu ling | 30.93 | 0.81 |
| 24 | MOL000275 | Trametenolic acid | Fu ling | 38.71 | 0.8 |
| 25 | MOL000279 | Cerevisterol | Fu ling | 37.96 | 0.77 |
| 26 | MOL000282 | Ergosta-7,22E-dien-3beta-ol | Fu ling | 43.51 | 0.72 |
| 27 | MOL000283 | Ergosterol peroxide | Fu ling | 40.36 | 0.81 |
| 28 | MOL000296 | Hederagenin | Fu ling | 36.91 | 0.75 |
| 29 | MOL001484 | Inermine | Gan Cao | 75.18 | 0.54 |
| 30 | MOL001792 | Dfv | Gan Cao | 32.76 | 0.18 |
| 31 | MOL002311 | Glycyrol | Gan Cao | 90.78 | 0.67 |
| 32 | MOL000239 | Jaranol | Gan Cao | 50.83 | 0.29 |
| 33 | MOL002565 | Medicarpin | Gan Cao | 49.22 | 0.34 |
| 34 | MOL003656 | Lupiwighteone | Gan Cao | 51.64 | 0.37 |
| 35 | MOL003896 | 7-Methoxy-2-methyl isoflavone | Gan Cao | 42.56 | 0.2 |
| 36 | MOL000392 | Formononetin | Gan Cao | 69.67 | 0.21 |
| 37 | MOL000417 | Calycosin | Gan Cao | 47.75 | 0.24 |
| 38 | MOL004805 | (2S)-2-[4-hydroxy-3-(3-methylbut-2-enyl)phenyl]-8,8-dimethyl-2,3-dihydropyrano[2,3-f]chromen-4-one | Gan Cao | 31.79 | 0.72 |
| 39 | MOL004806 | Euchrenone | Gan Cao | 30.29 | 0.57 |
| 40 | MOL004808 | Glyasperin B | Gan Cao | 65.22 | 0.44 |
| 41 | MOL004810 | Glyasperin F | Gan Cao | 75.84 | 0.54 |
| 42 | MOL004811 | Glyasperin C | Gan Cao | 45.56 | 0.4 |
| 43 | MOL004814 | Isotrifoliol | Gan Cao | 31.94 | 0.42 |
| 44 | MOL004815 | (E)-1-(2,4-dihydroxyphenyl)-3-(2,2-dimethylchromen-6-yl)prop-2-en-1-one | Gan Cao | 39.62 | 0.35 |
| 45 | MOL004820 | Kanzonols W | Gan Cao | 50.48 | 0.52 |
| 46 | MOL004824 | (2S)-6-(2,4-dihydroxyphenyl)-2-(2-hydroxypropan-2-yl)-4-methoxy-2,3-dihydrofuro[3,2-g]chromen-7-one | Gan Cao | 60.25 | 0.63 |
| 47 | MOL004827 | Semilicoisoflavone B | Gan Cao | 48.78 | 0.55 |
| 48 | MOL004828 | Glepidotin A | Gan Cao | 44.72 | 0.35 |
| 49 | MOL004829 | Glepidotin B | Gan Cao | 64.46 | 0.34 |
| 50 | MOL004833 | Phaseolinisoflavan | Gan Cao | 32.01 | 0.45 |
| 51 | MOL004835 | Glypallichalcone | Gan Cao | 61.6 | 0.19 |
| 52 | MOL004838 | 8-(6-hydroxy-2-benzofuranyl)-2,2-dimethyl-5-chromenol | Gan Cao | 58.44 | 0.38 |
| 53 | MOL004841 | Licochalcone B | Gan Cao | 76.76 | 0.19 |
| 54 | MOL004848 | Licochalcone G | Gan Cao | 49.25 | 0.32 |
| 55 | MOL004849 | 3-(2,4-dihydroxyphenyl)-8-(1,1-dimethylprop-2-enyl)-7-hydroxy-5-methoxy-coumarin | Gan Cao | 59.62 | 0.43 |
| 56 | MOL004855 | Licoricone | Gan Cao | 63.58 | 0.47 |
| 57 | MOL004856 | Gancaonin A | Gan Cao | 51.08 | 0.4 |
| 58 | MOL004857 | Gancaonin B | Gan Cao | 48.79 | 0.45 |
| 59 | MOL004863 | 3-(3,4-dihydroxyphenyl)-5,7-dihydroxy-8-(3-methylbut-2-enyl)chromone | Gan Cao | 66.37 | 0.41 |
| 60 | MOL004864 | 5,7-dihydroxy-3-(4-methoxyphenyl)-8-(3-methylbut-2-enyl)chromone | Gan Cao | 30.49 | 0.41 |
| 61 | MOL004866 | 2-(3,4-dihydroxyphenyl)-5,7-dihydroxy-6-(3-methylbut-2-enyl)chromone | Gan Cao | 44.15 | 0.41 |
| 62 | MOL004879 | Glycyrin | Gan Cao | 52.61 | 0.47 |
| 63 | MOL004882 | Licocoumarone | Gan Cao | 33.21 | 0.36 |
| 64 | MOL004883 | Licoisoflavone | Gan Cao | 41.61 | 0.42 |
| 65 | MOL004884 | Licoisoflavone B | Gan Cao | 38.93 | 0.55 |
| 66 | MOL004885 | Licoisoflavanone | Gan Cao | 52.47 | 0.54 |
| 67 | MOL004891 | Shinpterocarpin | Gan Cao | 80.3 | 0.73 |
| 68 | MOL004898 | (E)-3-[3,4-dihydroxy-5-(3-methylbut-2-enyl)phenyl]-1-(2,4-dihydroxyphenyl)prop-2-en-1-one | Gan Cao | 46.27 | 0.31 |
| 69 | MOL004903 | Liquiritin | Gan Cao | 65.69 | 0.74 |
| 70 | MOL004904 | Licopyranocoumarin | Gan Cao | 80.36 | 0.65 |
| 71 | MOL004907 | Glyzaglabrin | Gan Cao | 61.07 | 0.35 |
| 72 | MOL004908 | Glabridin | Gan Cao | 53.25 | 0.47 |
| 73 | MOL004910 | Glabranin | Gan Cao | 52.9 | 0.31 |
| 74 | MOL004911 | Glabrene | Gan Cao | 46.27 | 0.44 |
| 75 | MOL004912 | Glabrone | Gan Cao | 52.51 | 0.5 |
| 76 | MOL004913 | 1,3-dihydroxy-9-methoxy-6-benzofurano[3,2-c]chromenone | Gan Cao | 48.14 | 0.43 |
| 77 | MOL004914 | 1,3-dihydroxy-8,9-dimethoxy-6-benzofurano[3,2-c]chromenone | Gan Cao | 62.9 | 0.53 |
| 78 | MOL004915 | Eurycarpin A | Gan Cao | 43.28 | 0.37 |
| 79 | MOL004924 | (–)-Medicocarpin | Gan Cao | 40.99 | 0.95 |
| 80 | MOL004935 | Sigmoidin-B | Gan Cao | 34.88 | 0.41 |
| 81 | MOL004941 | (2R)-7-hydroxy-2-(4-hydroxyphenyl)chroman-4-one | Gan Cao | 71.12 | 0.18 |
| 82 | MOL004945 | (2S)-7-hydroxy-2-(4-hydroxyphenyl)-8-(3-methylbut-2-enyl)chroman-4-one | Gan Cao | 36.57 | 0.32 |
| 83 | MOL004948 | Isoglycyrol | Gan Cao | 44.7 | 0.84 |
| 84 | MOL004949 | Isolicoflavonol | Gan Cao | 45.17 | 0.42 |
| 85 | MOL004957 | Hmo | Gan Cao | 38.37 | 0.21 |
| 86 | MOL004959 | 1-methoxyphaseollidin | Gan Cao | 69.98 | 0.64 |
| 87 | MOL004961 | Quercetin der. | Gan Cao | 46.45 | 0.33 |
| 88 | MOL004966 | 3’-hydroxy-4’-o-methylglabridin | Gan Cao | 43.71 | 0.57 |
| 89 | MOL000497 | Licochalcone a | Gan Cao | 40.79 | 0.29 |
| 90 | MOL004974 | 3’-methoxyglabridin | Gan Cao | 46.16 | 0.57 |
| 91 | MOL004978 | 2-[(3R)-8,8-dimethyl-3,4-dihydro-2H-pyrano[6,5-f]chromen-3-yl]-5-methoxyphenol | Gan Cao | 36.21 | 0.52 |
| 92 | MOL004980 | Inflacoumarin A | Gan Cao | 39.71 | 0.33 |
| 93 | MOL004985 | Icos-5-enoic acid | Gan Cao | 30.7 | 0.2 |
| 94 | MOL004988 | Kanzonol F | Gan Cao | 32.47 | 0.89 |
| 95 | MOL004989 | 6-prenylated eriodictyol | Gan Cao | 39.22 | 0.41 |
| 96 | MOL004990 | 7,2’,4’-trihydroxy-5-methoxy-3-arylcoumarin | Gan Cao | 83.71 | 0.27 |
| 97 | MOL004991 | 7-Acetoxy-2-methylisoflavone | Gan Cao | 38.92 | 0.26 |
| 98 | MOL004993 | 8-prenylated eriodictyol | Gan Cao | 53.79 | 0.4 |
| 99 | MOL004996 | Gadelaidic acid | Gan Cao | 30.7 | 0.2 |
| 100 | MOL000500 | Vestitol | Gan Cao | 74.66 | 0.21 |
| 101 | MOL005000 | Gancaonin G | Gan Cao | 60.44 | 0.39 |
| 102 | MOL005001 | Gancaonin H | Gan Cao | 50.1 | 0.78 |
| 103 | MOL005003 | Licoagrocarpin | Gan Cao | 58.81 | 0.58 |
| 104 | MOL005007 | Glyasperins M | Gan Cao | 72.67 | 0.59 |
| 105 | MOL005008 | Glycyrrhiza flavonol A | Gan Cao | 41.28 | 0.6 |
| 106 | MOL005012 | Licoagroisoflavone | Gan Cao | 57.28 | 0.49 |
| 107 | MOL005016 | Odoratin | Gan Cao | 49.95 | 0.3 |
| 108 | MOL005017 | Phaseol | Gan Cao | 78.77 | 0.58 |
| 109 | MOL005018 | Xambioona | Gan Cao | 54.85 | 0.87 |
| 110 | MOL005020 | Dehydroglyasperins C | Gan Cao | 53.82 | 0.37 |
| 111 | MOL006129 | 6-methylgingediacetate2 | Sheng Jiang | 48.73 | 0.32 |
| 112 | MOL001771 | Poriferast-5-en-3beta-ol | Sheng Jiang | 36.91 | 0.75 |
| 113 | MOL000211 | Mairin | Bai Shao, Gan Cao | 55.38 | 0.78 |
| 114 | MOL000359 | Sitosterol | Bai Shao, Gan Cao, Bo He | 36.91 | 0.75 |
| 115 | MOL000422 | Kaempferol | Chai Hu, Bai Shao, Gan Cao | 41.88 | 0.24 |
| 116 | MOL000449 | Stigmasterol | Chai Hu, Dang Gui, Sheng Jiang | 43.83 | 0.76 |
| 117 | MOL000354 | Isorhamnetin | Chai Hu, Gan Cao | 49.6 | 0.31 |
| 118 | MOL000098 | Quercetin | Chai Hu, Gan Cao | 46.43 | 0.28 |
| 119 | MOL000358 | Beta-sitosterol | Dang Gui, Bai Shao, Sheng Jiang | 36.91 | 0.75 |
| 120 | MOL004328 | Naringenin | Gan Cao, Bo He | 59.29 | 0.21 |
CRS, chronic restraint stress; TCMSP, Traditional Chinese Medicine Systems Pharmacology Database and Analysis Platform; OB, oral bioavailability; DL, drug-likeness.
Table 6. The 16 overlapping gene considered as candidate targets.
| No. | Gene symbol | Protein name |
|---|---|---|
| 1 | CASP3 | Caspase-3 |
| 2 | AKT1 | RAC-alpha serine/threonine-protein kinase |
| 3 | STAT1 | Signal transducer and activator of transcription 1-alpha/beta |
| 4 | VCAM1 | Vascular cell adhesion protein 1 |
| 5 | CAT | Catalase |
| 6 | MAPK14 | Mitogen-activated protein kinase 14 |
| 7 | GSK3B | Glycogen synthase kinase-3 beta |
| 8 | EGFR | Epidermal growth factor receptor |
| 9 | MAPK1 | Mitogen-activated protein kinase 1 |
| 10 | RAF1 | RAF proto-oncogene serine/threonine-protein kinase |
| 11 | GJA1 | Gap junction alpha-1 protein |
| 12 | NOS3 | Nitric oxide synthase, endothelial |
| 13 | CTSD | Cathepsin D |
| 14 | MAPK3 | Mitogen-activated protein kinase 3 |
| 15 | APOB | Apolipoprotein B-100 |
| 16 | STAT3 | Signal transducer and activator of transcription 3 |
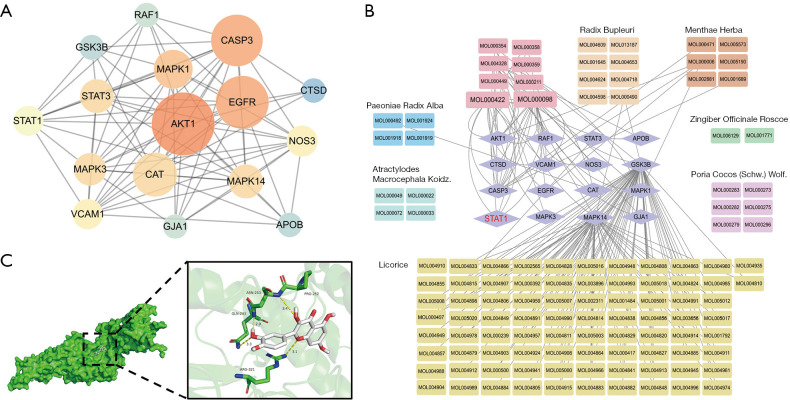
Based on 16 candidate targets, a PPI network was constructed by importing the gene ID of the candidate targets to the STRING database (Figure 5A). The results indicated that the mechanisms of the Xiaoyao pill in breast cancer progression in relation to depression or anxiety were associated with responses to LPS (Figure 5B). STAT1 was mapped to the compounds for molecular docking. According to the affinity score of molecular docking, STAT1 and the active compound quercetin were selected for further investigation (Figure 5C).
Figure 5.
Active compounds in the Xiaoyao pill and candidate targets. (A) PPI network. (B) Active herb compounds-target genes network. Light purple diamond: target genes; brownish-green rectangle: herbs; others rectangle: active compounds. (C) The result of molecular docking. The optimal docking pose and ligand-residue interaction between STAT1 protein model (green) and positive ligand quercetin (white). PPI, protein-protein interaction.
Effect of polarized microglia cells on breast cancer cell proliferation
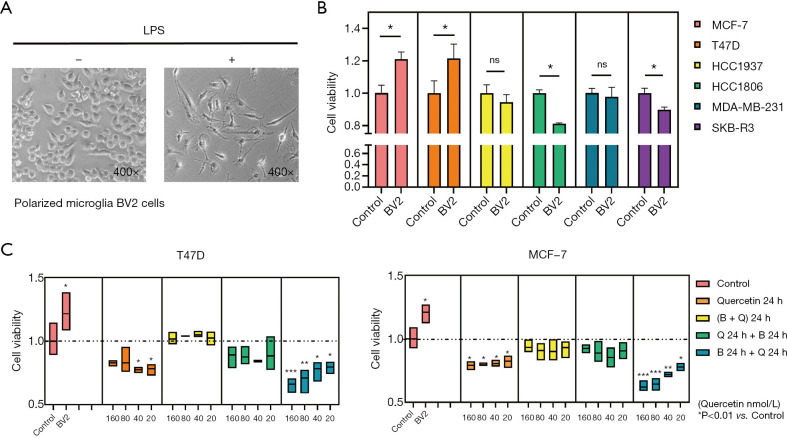
The supernatant culture medium of BV-2 microglia activated by LPS (Figure 6A) was used in coculturing breast cancer cells, and changes in cell viability of different breast cancer cells were assessed. It was found that the BV-2 supernatant promoted the proliferation of MCF-7 and T47D cells, but not HCC1937, HCC1806, MDA-MB-231, and SKB-R3 cells (Figure 6B). Based on this, the effect of quercetin on MCF-7 and T47D cells was subsequently verified (Figure 6C). The results suggested that quercetin could reverse the proliferative effect of BV-2 supernatant on breast cancer cells, thereby inhibiting the proliferation of MCF-7 and T47D cells.
Figure 6.
Effect of polarized microglia cells on proliferation in breast cancer cells. (A) Morphology of BV-2 microglia before and after 1 µg/L LPS stimulation (400×). (B) Cell viability of MCF-7, T47D, HCC1937, HCC1806, MDA-MB-231, and SKB-R3 cells after stimulation by the supernatant of BV-2 microglia. (C) Cell viability of T47D and MCF-7 cells after stimulation by the supernatant of BV-2 microglia and quercetin. *, P<0.05; **, P<0.01; ***, P<0.001 vs. control group. LPS, lipopolysaccharide; B, BV2 supernatant; Q, quercetin.
Discussion
Compared to acute stress, chronic stress leads to the continuous release of neuroendocrine hormones (22-24), resulting in pathological changes (25-27). Chronic stress can increase the risk for the onset and progression of tumors because of its effects on the neuroendocrine system (28-30). Our study indicated that CRS robustly induced notable depressive-like behaviors in MMTV-PyMT mice, which could be relieved by the antidepressant Xiaoyao pill. Furthermore, CRS promoted the occurrence of breast cancer in MMTV-PyMT mice, which was also disrupted by the antidepressant drug.
Depression is associated with low levels of neurotransmitters in serum, including 5-hydroxytryptamine, dopamine, adrenaline, and noradrenaline (31,32). Recent studies have shown that damage to the normal structure and function of microglia is due to strong inflammatory activation or decline, and chronic unpredictable stress exposure may lead to depression and related disorders in neuroplasticity and neurogenesis. Therefore, some forms of depression can be considered as a microglial disease (33). Antidepressant therapies have been shown to inhibit inflammation and microglial M1-polarization (34). Our results showed that CRS induced microglial M1-polarization in MMTV-PyMT mice. In addition, CRS decreased the levels of neurotransmitters in serum, and both the Xiaoyao pill group and the fluoxetine group had significantly higher levels of these neurotransmitters in serum. The effect of antidepressant drugs on depression involves multiple targets and pathways. Network pharmacology provides an in-depth way for researching the complex mechanisms of how antidepressant drugs can be used in treating depression and in disrupting breast cancer progression. The results of enrichment analysis indicate that the mechanisms of the antidepressant drug in breast cancer progression in relation to depression or anxiety were associated with responses to LPS. In addition, STAT1 was the intersectional target that was mapped to the compounds by molecular docking. It was reported that STAT1 drives M1 microglia activation and neuroinflammation (35) and that autophagy regulates microglial polarization via the STAT1/6 pathway (36). According to the affinity score of molecular docking, STAT1 and the active compound quercetin were selected for further investigation.
Quercetin can cross the blood-brain barrier and has been shown to play a neuroprotective role through effective anti-inflammatory and antioxidant effects in neurological diseases (37,38). Furthermore, quercetin can act as an adjuvant therapy drug in breast cancer treatment (39). Previous study suggested that quercetin exerts an antimanic-like effect at doses that do not impair spontaneous locomotor activity (40). And it was reported that quercetin modulates inflammatory cytokines expression during microglia interactions (41).
Neuroinflammation is defined as a cascade of cell damage and biochemical reactions occurring in the CNS (42,43), which includes the activation of microglia cells (44) and macrophages (45). In this study, we examined the expression of the type M1 microglia surface marker CD86; soluble mediators IL-1β, IL-10, and iNOS; type M2 microglia surface marker CD206; and Arg-1 messenger RNA (mRNA) in the brain of mice exposed to CRS stimulation. The results indicated that CD86 and IL-1β cell damage and biochemical reactions occur in the CNS, including the activation of microglia cells and macrophages.
We stimulated the BV-2 microglial cell line with LPS in vitro to simulate microglia cell polarization. We collected its supernatant culture medium for conditional coculture with breast cancer cells to explore the polarizing effect of microglial cells on breast cancer cell proliferation. We examined 6 types of breast cancer cells, among which MCF-7 and T47D cells were sensitive to the BV-2 supernatant; however, the BV-2 supernatant did not promote the proliferation of HCC1937, HCC1806, MDA-MB-231, and SKB-R3 cells. This difference may be attributable to different phenotypes of various breast cancer cell lines. MCF-7 and T47D cells are both estrogen receptor-positive (ER+) type breast cancer cells; therefore, quercetin may have a specific inhibitory effect on ER+ type breast cancer cells. However, the identification of breast cancer cells that are sensitive to quercetin requires further research.
Our results showed that CRS promotes an earlier occurrence of breast cancer in MMTV-PyMT mice and increases levels of these neurotransmitters in the serum of mice. These findings strongly suggest that the CNS can directly regulate breast cancer progression and that quercetin could be the most active compound in this reversal effect on the CNS during breast cancer progression. Our results further demonstrate that chronic stress may be an indicator of breast cancer and that quercetin could be an effective treatment for breast cancer patients with chronic stress.
Supplementary
The article’s supplementary files as
Acknowledgments
We would like to thank LetPub (www.letpub.com) for their English-language assistance in the preparation of this manuscript.
Funding: This work was supported by grants from the National Natural Science Foundation of China (No. 81904206 and 81974571); Guangdong Natural Science Foundation (No. 2017A030313719); Foundation Project of Guangzhou University of Chinese Medicine (No. XKP2019002); Key Project at Central Government Level: The Ability Establishment of Sustainable Use for Valuable Chinese Medicine Resources (No. 2060302); Zhuhai City Flexible Introduction of High-Level Health Professor Project.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The experimental scheme was approved by the experimental animal ethics committee of Guangdong Provincial Hospital of Chinese Medicine (Reference No. 2019048). Animal welfare and experimental procedures were performed in compliance with the Guide for the Care and Use of Laboratory Animals (National Institutes of Health, the United States). A protocol was prepared before the study without registration.
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at http://dx.doi.org/10.21037/atm-21-2558
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-21-2558
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-21-2558). The authors have no conflicts of interest to declare.
(English Language Editor: J. Gray)
References
- 1.DeSantis CE, Ma J, Gaudet MM, et al. Breast cancer statistics, 2019. CA Cancer J Clin 2019;69:438-51. 10.3322/caac.21583 [DOI] [PubMed] [Google Scholar]
- 2.Britt KL, Cuzick J, Phillips KA. Key steps for effective breast cancer prevention. Nat Rev Cancer 2020;20:417-36. 10.1038/s41568-020-0266-x [DOI] [PubMed] [Google Scholar]
- 3.Fan L, Strasser-Weippl K, Li JJ, et al. Breast cancer in China. Lancet Oncol 2014;15:e279-89. 10.1016/S1470-2045(13)70567-9 [DOI] [PubMed] [Google Scholar]
- 4.Huang Z, Wen W, Zheng Y, et al. Breast cancer incidence and mortality: trends over 40 years among women in Shanghai, China. Ann Oncol 2016;27:1129-34. 10.1093/annonc/mdw069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Winocur G, Johnston I, Castel H. Chemotherapy and cognition: International cognition and cancer task force recommendations for harmonising preclinical research. Cancer Treat Rev 2018;69:72-83. 10.1016/j.ctrv.2018.05.017 [DOI] [PubMed] [Google Scholar]
- 6.Lange M, Heutte N, Rigal O, et al. Decline in cognitive function in older adults with early-stage breast cancer after adjuvant treatment. Oncologist 2016;21:1337-48. 10.1634/theoncologist.2016-0014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carreira H, Williams R, Müller M, et al. Associations between breast cancer survivorship and adverse mental health outcomes: a systematic review. J Natl Cancer Inst 2018;110:1311-27. Erratum in: J Natl Cancer Inst 2020;112:118. 10.1093/jnci/djy177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beutel ME, Weißflog G, Leuteritz K, et al. Efficacy of short-term psychodynamic psychotherapy (STPP) with depressed breast cancer patients: results of a randomized controlled multicenter trial. Ann Oncol 2014;25:378-84. 10.1093/annonc/mdt526 [DOI] [PubMed] [Google Scholar]
- 9.Scarpa JR, Fatma M, Loh YE, et al. Shared transcriptional signatures in major depressive disorder and mouse chronic stress models. Biol Psychiatry 2020;88:159-68. 10.1016/j.biopsych.2019.12.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lawson RP, Nord CL, Seymour B, et al. Disrupted habenula function in major depression. Mol Psychiatry 2017;22:202-8. 10.1038/mp.2016.81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sheline YI, Liston C, McEwen BS. Parsing the hippocampus in depression: chronic stress, hippocampal volume, and major depressive disorder. Biol Psychiatry 2019;85:436-8. 10.1016/j.biopsych.2019.01.011 [DOI] [PubMed] [Google Scholar]
- 12.Ho YC, Lin TB, Hsieh MC, et al. Periaqueductal gray glutamatergic transmission governs chronic stress-induced depression. Neuropsychopharmacology 2018;43:302-12. 10.1038/npp.2017.199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schramm E, Klein DN, Elsaesser M, et al. Review of dysthymia and persistent depressive disorder: history, correlates, and clinical implications. Lancet Psychiatry 2020;7:801-12. 10.1016/S2215-0366(20)30099-7 [DOI] [PubMed] [Google Scholar]
- 14.Wang YH, Li JQ, Shi JF, et al. Depression and anxiety in relation to cancer incidence and mortality: a systematic review and meta-analysis of cohort studies. Mol Psychiatry 2020;25:1487-99. 10.1038/s41380-019-0595-x [DOI] [PubMed] [Google Scholar]
- 15.Le CP, Nowell CJ, Kim-Fuchs C, et al. Chronic stress in mice remodels lymph vasculature to promote tumour cell dissemination. Nat Commun 2016;7:10634. 10.1038/ncomms10634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abrahams HJG, Gielissen MFM, Verhagen CAHHVM, et al. The relationship of fatigue in breast cancer survivors with quality of life and factors to address in psychological interventions: a systematic review. Clin Psychol Rev 2018;63:1-11. 10.1016/j.cpr.2018.05.004 [DOI] [PubMed] [Google Scholar]
- 17.Chen VC, Yang YH, Yu Kuo T, et al. Methylphenidate and the risk of burn injury among children with attention-deficit/hyperactivity disorder. Epidemiol Psychiatr Sci 2020;29:e146. 10.1017/S2045796020000608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang J, He H, Qiao Y, et al. Priming of microglia with IFN-γ impairs adult hippocampal neurogenesis and leads to depression-like behaviors and cognitive defects. Glia 2020;68:2674-92. 10.1002/glia.23878 [DOI] [PubMed] [Google Scholar]
- 19.Su KP. Are we all the same? The critical role of translational brain, behavior, and immunity research in East Asia. Brain Behav Immun 2019;82:1-2. 10.1016/j.bbi.2019.07.011 [DOI] [PubMed] [Google Scholar]
- 20.Wang J, Li X, He S, et al. Regulation of the kynurenine metabolism pathway by Xiaoyao San and the underlying effect in the hippocampus of the depressed rat. J Ethnopharmacol 2018;214:13-21. 10.1016/j.jep.2017.11.037 [DOI] [PubMed] [Google Scholar]
- 21.Gao L, Huang P, Dong Z, et al. Modified Xiaoyaosan (MXYS) exerts anti-depressive effects by rectifying the brain blood oxygen level-dependent fMRI signals and improving hippocampal neurogenesis in mice. Front Pharmacol 2018;9:1098. Erratum in: Front Pharmacol 2018;9:1420. 10.3389/fphar.2018.01098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lightman SL, Birnie MT, Conway-Campbell BL. Dynamics of ACTH and cortisol secretion and implications for disease. Endocr Rev 2020;41:470-90. 10.1210/endrev/bnaa002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hill MN, Kumar SA, Filipski SB, et al. Disruption of fatty acid amide hydrolase activity prevents the effects of chronic stress on anxiety and amygdalar microstructure. Mol Psychiatry 2013;18:1125-35. 10.1038/mp.2012.90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lotan A, Lifschytz T, Wolf G, et al. Differential effects of chronic stress in young-adult and old female mice: cognitive-behavioral manifestations and neurobiological correlates. Mol Psychiatry 2018;23:1432-45. 10.1038/mp.2017.237 [DOI] [PubMed] [Google Scholar]
- 25.Eagle AL, Manning CE, Williams ES, et al. Circuit-specific hippocampal ΔFosB underlies resilience to stress-induced social avoidance. Nat Commun 2020;11:4484. 10.1038/s41467-020-17825-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xiao Q, Zhou X, Wei P, et al. A new GABAergic somatostatin projection from the BNST onto accumbal parvalbumin neurons controls anxiety. Mol Psychiatry 2020. [Epub ahead of print]. doi: . 10.1038/s41380-020-0816-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oliveira TG, Chan RB, Bravo FV, et al. The impact of chronic stress on the rat brain lipidome. Mol Psychiatry 2016;21:80-8. 10.1038/mp.2015.14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wagner KV, Marinescu D, Hartmann J, et al. Differences in FKBP51 regulation following chronic social defeat stress correlate with individual stress sensitivity: influence of paroxetine treatment. Neuropsychopharmacology 2012;37:2797-808. 10.1038/npp.2012.150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu J, Qu L, Wan C, et al. A novel β2-AR/YB-1/β-catenin axis mediates chronic stress-associated metastasis in hepatocellular carcinoma. Oncogenesis 2020;9:84. 10.1038/s41389-020-00268-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu W, Sun M, Zhang HP, et al. Prolactin mediates psychological stress-induced dysfunction of regulatory T cells to facilitate intestinal inflammation. Gut 2014;63:1883-92. Erratum in: Gut 2020;69:e7. 10.1136/gutjnl-2013-306083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ambrase A, Lewis CA, Barth C, et al. Influence of ovarian hormones on value-based decision-making systems: Contribution to sexual dimorphisms in mental disorders. Front Neuroendocrinol 2021;60:100873. 10.1016/j.yfrne.2020.100873 [DOI] [PubMed] [Google Scholar]
- 32.Zhao T, Chen Y, Sun Z, et al. Prenatal sevoflurane exposure causes neuronal excitatory/inhibitory imbalance in the prefrontal cortex and neurofunctional abnormality in rats. Neurobiol Dis 2020;146:105121. 10.1016/j.nbd.2020.105121 [DOI] [PubMed] [Google Scholar]
- 33.Yirmiya R, Rimmerman N, Reshef R. Depression as a microglial disease. Trends Neurosci 2015;38:637-58. 10.1016/j.tins.2015.08.001 [DOI] [PubMed] [Google Scholar]
- 34.Kalkman HO, Feuerbach D. Antidepressant therapies inhibit inflammation and microglial M1-polarization. Pharmacol Ther 2016;163:82-93. 10.1016/j.pharmthera.2016.04.001 [DOI] [PubMed] [Google Scholar]
- 35.Butturini E, Boriero D, Carcereri de Prati A, et al. STAT1 drives M1 microglia activation and neuroinflammation under hypoxia. Arch Biochem Biophys 2019;669:22-30. 10.1016/j.abb.2019.05.011 [DOI] [PubMed] [Google Scholar]
- 36.Qin C, Liu Q, Hu ZW, et al. Microglial TLR4-dependent autophagy induces ischemic white matter damage via STAT1/6 pathway. Theranostics 2018;8:5434-51. Erratum in: Theranostics 2020;10:8818-20. 10.7150/thno.27882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.de Oliveira MR, Nabavi SM, Braidy N, et al. Quercetin and the mitochondria: a mechanistic view. Biotechnol Adv 2016;34:532-49. 10.1016/j.biotechadv.2015.12.014 [DOI] [PubMed] [Google Scholar]
- 38.Chakraborty J, Singh R, Dutta D, et al. Quercetin improves behavioral deficiencies, restores astrocytes and microglia, and reduces serotonin metabolism in 3-nitropropionic acid-induced rat model of Huntington's Disease. CNS Neurosci Ther 2014;20:10-9. 10.1111/cns.12189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu Z, Zhao D, Zheng X, et al. Quercetin exerts bidirectional regulation effects on the efficacy of tamoxifen in estrogen receptor-positive breast cancer therapy: an in vitro study. Environ Toxicol 2020;35:1179-93. 10.1002/tox.22983 [DOI] [PubMed] [Google Scholar]
- 40.Kanazawa LKS, Vecchia DD, Wendler EM, et al. Quercetin reduces manic-like behavior and brain oxidative stress induced by paradoxical sleep deprivation in mice. Free Radic Biol Med 2016;99:79-86. 10.1016/j.freeradbiomed.2016.07.027 [DOI] [PubMed] [Google Scholar]
- 41.da Silva AB, Cerqueira Coelho PL, das Neves Oliveira M, et al. The flavonoid rutin and its aglycone quercetin modulate the microglia inflammatory profile improving antiglioma activity. Brain Behav Immun 2020;85:170-85. 10.1016/j.bbi.2019.05.003 [DOI] [PubMed] [Google Scholar]
- 42.Gilhus NE, Deuschl G. Neuroinflammation - a common thread in neurological disorders. Nat Rev Neurol 2019;15:429-30. 10.1038/s41582-019-0227-8 [DOI] [PubMed] [Google Scholar]
- 43.Yuan J, Amin P, Ofengeim D. Necroptosis and RIPK1-mediated neuroinflammation in CNS diseases. Nat Rev Neurosci 2019;20:19-33. 10.1038/s41583-018-0093-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reverchon F, de Concini V, Larrigaldie V, et al. Hippocampal interleukin-33 mediates neuroinflammation-induced cognitive impairments. J Neuroinflammation 2020;17:268. 10.1186/s12974-020-01939-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liaw K, Sharma R, Sharma A, et al. Systemic dendrimer delivery of triptolide to tumor-associated macrophages improves anti-tumor efficacy and reduces systemic toxicity in glioblastoma. J Control Release 2021;329:434-44. 10.1016/j.jconrel.2020.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as