Abstract
Background & aims
Nonalcoholic fatty liver disease (NAFLD) has multifactorial origin. Genetic and environmental factors lead to the biology of this complex disorder. In this study, we screened parents of cases with NAFLD and compared them with parents of cases without NAFLD to see its familial aggregation and the role of patatin-like phospholipase domain containing 3 (PNPLA3).
Method
It was a cross-sectional study. Parents of probands with NAFLD and without NAFLD were screened with abdominal sonography, anthropometry, blood tests, transient elastography, and PNPLA3 polymorphism.
Results
We had enrolled 303 individuals: 51 probands with NAFLD, 50 probands without NAFLD, and their 202 parents. Parents of the NAFLD group had significantly higher metabolic risk factors as compared with parents of the non-NAFLD group. They had a significantly higher rate of fatty liver (P = 0.0001), mean serum aspartate aminotransferase levels (P = 0.011), mean serum alanine aminotransferase levels (P = 0.001),raised fasting and postprandial blood sugar levels, lower mean platelets (P = 0.033) and serum albumin levels (P = 0.005), and higher mean liver stiffness (P = 0.001) on transient elastography.
Frequency of PNPLA3 polymorphism within NAFLD group was higher compared to the non-NAFLD group (mutant GG-13.3 vs 3.3%). Similarly, parents of NAFLD group had mutant GG in 15 % versus 5% in parents of non-NAFLD group, (P = 0.105, odds ratio 6), though it was not statistically significant but may be relevant. In this study, offsprings of parents with nonalcoholic steatohepatitis were likely to have GG homozygous allele. A NAFLD16 score based on parent's parameters was calculated to predict the probability of NAFLD occurrence in an overweight obese individual.
Conclusion
Screening of parents of individuals with NAFLD will help in the identification of undiagnosed NAFLD cases and other metabolic risk factors among them as there is a familial aggregation of NAFLD. One can predict the occurrence of NAFLD in the next generation using the NAFLD16 score.
Keywords: familial aggregation of NAFLD, NAFLD, NAFLD16 score, transient elastography, PNPLA3
Abbreviations: ALT, Alanine Aminotransferase; APRI, AST/Platelet Ratio Index; AST, Aspartate Aminotransferase; BMI, Body Mass Index; FBS, Fasting Blood Sugar; FIB-4, Fibrosis-4 Index; HDL, High-Density Lipoprotein; HOMA IR, Homeostatic Model Assessment of Insulin Resistance; HWE, Hardy-Weinberg Equilibrium; I148M, isoleucine to methionine; IAAT, Intra-Adipose Tissue Thickness; LSM, Liver Stiffness Measurement; NAFLD, Nonalcoholic Fatty Liver Disease; NASH, Nonalcoholic Steatohepatitis; PLBS, Postprandial Blood Sugar; PNPLA3, Patatin-like Phospholipase Domain Containing 3; SNP, single-nucleotide polymorphism; TE, Transient Elastography; USG, Ultrasonography; WHR, Waist-Hip Ratio
Introduction
Nonalcoholic fatty liver disease (NAFLD) at present is the most common cause of chronic liver disease among adults.1 It can progress from nonalcoholic fatty liver (NAFL) to nonalcoholic steatohepatitis (NASH) and even to cirrhosis and hepatocellular carcinoma. The growing epidemic of obesity and metabolic syndrome has increased the prevalence and impact of NAFLD. In the United States, the prevalence of NAFLD has been reported to be from 10% to 46%, and most of the biopsy-based studies have reported the prevalence of NASH as 3–5%.2 The prevalence of NAFLD in India varies from 9% to 35%.3 Obesity is a strong risk factor but by itself is not sufficient to produce NAFLD.4 Hence, there is a complex multiphysiological phenomenon that leads to NAFLD in an individual. Recent literature on the genetics of NAFLD in search of missing heritability of NAFLD suggests that there may be a role of mitochondrial genetics, microribonucleic acids, long noncoding RNAs, epigenetic factors, single-nucleotide polymorphisms (SNPs) such as patatin-like phospholipase domain containing 3 (PNPLA3), transmembrane 6 superfamily member 2, apolipoprotein C3, transcription factor 7-like 2, and microsomal triglyceride transfer protein in its pathogenesis.5 Four genome-wide association studies have been conducted in population and have reported PNPLA3, SAMM50, PARVB, and GATAD2A genes which are significantly associated with NAFLD.6 Among all these, PNPLA3 has been shown to be maximally affecting NAFLD outcomes.
Recent literature review demonstrates a strong association between an SNP in PNPLA3 gene rs738409 at I148M (causing an isoleucine-to-methionine substitution at position 148) and severity of NAFLD. PNPLA3 gene is located on human chromosome 22q13. PNPLA3 plays a role in hepatic triglyceride hydrolysis and hepatic fat content by coding for a protein ‘adiponutrin’. The lipase activity against triglycerides and acylglycerol transacetylase activity are exhibited by PNPLA3, and its expression is responsible for energy mobilization and the storage of lipid droplets. Its allelic variant results in a change from isoleucine to methionine (I148M) that hinders its lipase activity, thereby increasing the risk of fatty liver and its severity.7 Interestingly, the at-risk PNPLA3 rs738409 GG genotype is found in 13–19% of the general population in Asian studies.8 Carriage of this PNPLA3 rs738409 SNP is associated with a greater risk of not only progressive steatohepatitis and fibrosis but also of hepatocellular carcinoma (HCC). Screening of at-risk siblings and offspring with PNPLA3 gene polymorphism can be a possible intervention to have a favorable outcome at an early (presymptomatic) stage.
Familial clustering of NAFLD and PNPLA3 polymorphism has been infrequently reported from India.9
In the present study, we have screened parents of patients with NAFLD and overweight individuals without NAFLD to find out lineage inheritance of NAFLD. Parents were screened for metabolic parameters with anthropometry, ultrasound, transient elastography (TE), and PNPLA3 gene polymorphism. This cross-sectional study showed multifactorial etiology and the role of PNPLA3 gene polymorphism in NAFLD inheritance.
Methods
This observational cross-sectional study was carried out in the gastroenterology department of a tertiary care center in a metropolitan city. The study approval was obtained from the institutional ethics committee. All patients and their parents were enrolled after taking informed and genetic consent for the PNPLA3 gene polymorphism study. The duration of the study was from 1st January 2019 to 30th June 2019. All procedures were in accordance with the ethical standards of the Helsinki declaration.
Inclusion criteria
-
1)
Age of more than 12 years.
-
2)
NAFLD group definition: included proband with fatty liver on abdominal ultrasonography (USG) and having Body Mass Index (BMI) of > 23 kg/m2seen in the gastroenterology out-patient department (OPD) or ward and their parents were enrolled.
-
3)
Without NAFLD group definition: included proband not showing fatty liver on abdominal USG but having BMI of >23 kg/m2 seen in the gastroenterology OPD or ward and their parents were enrolled. Both groups were matched for age, sex, BMI, and environmental factors.
Exclusion criteria
The following were the exclusion criteria for the study:
-
1)
Individuals or his/her parents with other causes of chronic liver disease such as alcoholic liver disease, hepatitis B/C, Wilson disease, and autoimmune hepatitis were excluded from the study.
-
2)
Individuals or his/her parents with daily alcohol consumption >20 g/day for men and >10 g/day for women were excluded from the study.
-
3)
Individuals with history of chronic intake of a hepatotoxic drug were excluded from the study.
-
4)
Individuals who were pregnant were excluded from the study
-
5)
Individuals with an abnormal thyroid profile were excluded from the study.
-
6)
Individuals with history of acute hepatitis in the last 6 months were excluded from the study.
-
7)
Individuals with chronic diseases (HIV, celiac disease) were excluded from the study.
Methodology
In both groups (individuals and their parents), detailed history regarding dietary habits (includes history on the number of days an individual eating vegetarian or nonvegetarian food, soft drinks, and fast food per week), diabetes mellitus, hypertension, alcohol intake was obtained. Waist circumference, hip circumference, and waist-hip ratio (WHR; men: 0.9, women: 0.8) were calculated for obesity. Participants were classified based upon BMI as normal weight (BMI: 18.5–22.9 kg/m2), overweight (BMI: 23.0–24.9 kg/m2), and obese (BMI: ≥ 25.0 kg/m2) according to the Indian standard.10
Subjects were instructed to fast overnight for 12 h before blood investigations. Blood investigations performed in both groups were following: hemoglobin, platelet levels, liver function tests, lipid profile (serum cholestrol and triglycerides), fasting blood sugar (FBS), postprandial blood sugar (PLBS) hepatitis B surface antigen (HBsAg), Anti-HCV, and PNPLA3 I148M polymorphism (rs738409 SNP). Autoimmune hepatitis and Wilson disease were ruled out with antinuclear antibody, antismooth muscle antibody, anti-liver-kidney muscle antibody, serum IgG levels, and serum ceruloplasmin levels. USG-guided measurement of intra abdominal adipose tissue thickness and TE using a ECHOSENS fibroscan 402 machine for liver stiffness measurement (LSM) were also carried out. Fatty liver on abdominal USG and alanine aminotransferase (ALT) >40 U/L was defined as NASH, based on the clinical scenario without any liver biopsy.
PNPLA3 gene polymorphism
There are 3 types of polymorphisms: wild (CC), heterozygous (CG), and homozygous (GG). Genomic DNA was isolated from whole blood using the QIAmp Blood DNA Kit (Qiagen). Genotyping for the PNPLA3 gene (rs738409) was performed using predesigned TaqMan probes (Assay Id C_7241_10, Applied BioSystems) for allelic discrimination as per the protocol on Quant studio 3 Real-Time PCR System (Applied BioSystems). The success rate reported by this assay was 100%.
We could do genotyping for PNPLA3 in only 180 individuals (30 probands with NAFLD, 30 probands without NAFLD, 60 parents of patients with NAFLD, 60 parents of individuals without NAFLD) due to financial constraints.
Ultrasound grading of diffuse hepatic steatosis
Grade I
Diffusely increased hepatic echogenicity but periportal and diaphragmatic echogenicity is still appreciable.
Grade II
Diffusely increased hepatic echogenicity obscuring periportal echogenicity but diaphragmatic echogenicity is still appreciable.
Grade III
Diffusely increased hepatic echogenicity obscuring periportal and diaphragmatic echogenicity.11
Various scores such as Fibrosis-4 Index,12 NAFLD fibrosis score,13 AST/Platelet Ratio Index (APRI),14 and BARD15 score were calculated.
Statistical analysis
The sample size was calculated with the help of previous literature statistics. According to a study which found that fatty liver among parents of probands with fatty liver was 78%, based on that comparison of percentages (two groups), alpha = 0.05, beta = 0.2, power = 80, and percentage n = 8 × (p1q1 + p2q2)/(p1-p2)2, q1 = 1−p1, q2 = 1−p2, where p1 = 78, p2 = 35, q1 = 22, q2 = 65 gives n = 21.6, which was the calculated sample size of probands.16 But according to normal distribution and to account attrition loss, we decided to take a sample size of 51 (NAFLD) and 50 (non-NAFLD) probands in each group and their respective parents (202). Thus, a total of 303 individuals were enrolled. We enrolled more number of individuals than estimated to increase the precision of the study.
Sensitivity is equivalent to p here; p is taken as 35 based on an Indian study showing the prevalence of NAFLD in India as 9–35%.3 To compare various qualitative and quantitative data, chi-square (or Fisher's exact) and Student t-test tests were used, respectively. Different parameters affecting the familial clustering of NAFLD were analyzed using univariate and multivariate analysis. SPSS, version 22, was used for data analysis. Probability of Y (outcome, e.g. NAFLD in an individual) was predicted by P(Y) = 1/1 + e − (b0 + b1X1 + b2X2.) where P(Y) is the probability of Y occurring, e is the base of natural logarithms, X1/X2/X3, and so on are predictor variables, and b0/b1/b2, and so on are beta coefficients. The sample size was calculated with the formula method using previous literature statistics.3 Allele frequency of PNPLA3 I148M was tested for Hardy-Weinberg equilibrium (HWE) applying the Fisher exact test or chi-square test.
Results
A total of 303 individuals (51 with NAFLD and 50 without NAFLD) and their parents (202) were enrolled.
Description of parents (n = 202)
Clinical features
On history review, parents of the NAFLD group were found to be significantly symptomatic than parents of the non-NAFLD group (31.4% vs 12%, P = 0.001). Among 32 symptomatic parents of the NAFLD group, 30 had vague right upper abdominal discomfort, whereas in the without NAFLD group, 8 had vague right upper abdominal discomfort.
Metabolic risk factors
Parents of the NAFLD group had significantly higher metabolic risk factors: 42.2% had diabetes mellitus and 23.5% had hypertension. Fatty liver was seen more frequently in the parents of the NAFLD proband as compared with the non-NAFLD proband (73.5% vs 30%). Incidence of obesity and its indicators such as acanthosis nigricans, BMI, and WHR were also found significantly higher in parents of the NAFLD group compared with the non-NAFLD group (Table 1). When analyzed, whether single or both the parents of an individual are having any metabolic risk factors, we found that there were more individuals among the NAFLD group whose both parents were having metabolic risk factors.
Table 1.
Comparison of Dietary, Clinical, and Laboratory Features of NAFLD Versus without NAFLD Groups and Their Parents.
| Proband |
Parent of |
|||||
|---|---|---|---|---|---|---|
| NAFLD |
Non-NAFLD |
Proband with NAFLD |
Proband without NAFLD |
|||
| N = 51 | N = 50 | P value | N = 102 | N = 100 | P value | |
| Age | 30.16 ± 10.97 | 29.42 ± 10.73 | 0.74 | 54.06 (10.88) | 51.86 (10.67) | 0.15 |
| Presentation: | ||||||
| Asymptomatic | 35 (68.6%) | 45 (90%) | 0.01∗ | 70 (68.6%) | 88 (88%) | 0.01∗ |
| Symptomatic | 16 (31.4%) | 5 (10%) | 32 (31.4%) | 12 (12%) | ||
| Diet history | ||||||
| Nonvegetarian consumption in days/week | 1.72 ± 1.3 | 1.07 ± 0.58 | 0.12 | 1.27 ± 0.91 | 1.28 ± 0.54 | 0.013∗ |
| Fast food consumption in days/week | 1.62 ± 1.1 | 0.57 ± 0.2 | 0.9 | 0.58 ± 0.29 | 0.43 ± 0.1 | 0.06 |
| Soft drink consumption days/week | 0.78 ± 0.43 | 0.54 ± 0.22 | 0.16 | 0.29 ± 0.1 | 0.14 ± 0.02 | 0.15 |
| Exercise in days/week | 1.37 ± 0.53 | 1.47 ± 0.6 | 0.58 | 2 ± 0.72 | 0.95 ± 0.2 | 0.053 |
| Acanthosis nigricans | 7 (13.7%) | 3 (6%) | 0.19 | 17 (16.7%) | 5 (5%) | 0.02∗ |
| Hypertension | 1 (1.96%) | 1 (2%) | 0.99 | 24 (23.5%) | 10 (10%) | 0.01∗ |
| Diabetes mellitus | 5 (9.8%) | 5 (10%) | 0.97 | 43 (42.2%) | 16 (16%) | 0.01∗ |
| Smoking | 6 (11.8%) | 5 (10%) | 0.77 | 32 (31.4%) | 16 (16%) | 0.01 |
| BMI (kg/m2) | 28.02 ± 6.43 | 26.64 ± 7.88 | 0.34 | 27.43 ± 5.12 | 25.37 ± 5.49 | 0.01∗ |
| Obesity | 30 (58.8%) | 30 (60%) | 0.99 | 68 (67.3%) | 48 (48%) | 0.03∗ |
| Waist-hip ratio | ||||||
| (a) Child | 0.92 ± 0.06 | 0.91 ± 0.05 | 0.36 | |||
| b) Father | 0.92 ± 0.05 | 0.95 ± 0.05 | 0.01∗ | |||
| c) Mother | 0.91 ± 0.06 | 0.88 ± 0.06 | 0.04∗ | |||
| Platelet/μl | 145951.63 ± 102943.20 | 160322.99 ± 117213.78 | 0.64 | 150594.36 ± 132902.78 | 241839.62 ± 108176.33 | 0.04∗ |
| FBS (mg/dl) | 95.04 ± 14.88 | 98.14 ± 24.87 | 0.45 | 122.06 ± 49.34 | 102.5 ± 28.8 | 0.01∗ |
| PLBS (mg/dl) | 130.7 ± 55.41 | 129.2 ± 44.01 | 0.88 | 161.13 ± 62.5 | 140.5 ± 48.1 | 0.01∗ |
| AST (U/L) | 28.35 ± 12.3 | 24.58 ± 8.85 | 0.08 | 29.15 ± 15.28 | 24.18 ± 12.07 | 0.02∗ |
| AST >40 U/L | 6 (11.7%) | 3 (6%) | 0.51 | 18 (17.85%) | 8 (8%) | 0.04∗ |
| ALT (U/L) | 32.43 ± 13.94 | 30.12 ± 16.97 | 0.46 | 36.76 ± 19.85 | 26.35 ± 13.12 | 0.0001∗ |
| ALT >40 U/L | 13 (25.5%) | 8 (20%) | 0.35 | 40 (39.2%) | 17 (17%) | 0.01∗ |
| Serum albumin (g/dl) | 3.76 ± 0.36 | 3.92 ± 0.35 | 0.04∗ | 3.71 ± 0.38 | 3.86 ± 0.41 | 0.01∗ |
| Cholesterol (mg/dl) | 155.6 ± 43.98 | 150.6 ± 53.28 | 0.61 | 170.08 ± 47.12 | 161.6 ± 49.23 | 0.22 |
| Triglyceride (mg/dl) | 144.7 ± 83.23 | 140.8 ± 64.07 | 0.79 | 154.11 ± 55.82 | 144.5 ± 58.78 | 0.22 |
FBS = fasting blood sugar, PLBS = postprandial blood sugar, AST = Aspartate transaminase, ALT = alanine transaminase, BMI = body mass index, NAFLD = nonalcoholic fatty liver disease.
∗Significant P value <0.05.
Biochemical parameters
Among the laboratory parameters, there were significantly higher mean serum aspartate aminotransferase (AST) levels (29.15 ± 15.28 vs 24.18 ± 12.07, P = 0.011), higher mean serum ALT levels (36.76 ± 19.85 vs 26.35 ± 13.12, P = 0.0001), higher mean fasting and postprandial blood sugar levels, lower mean platelets (150,594.36 ± 132902.78 vs 241839.62 ± 108176.33, P = 0.033), and lower mean serum albumin levels (3.71 ± 0.38 vs 3.86 ± 0.41, P = 0.005) found in parents of the proband with NAFLD.
Transient elastography
TE showed higher mean LSM values (7.4 ± 3.97 vs 5.46 ± 2.51, P = 0.001) in parents of the proband with NAFLD (Table 2). The number of parents having LSM >7.9 kPa (F2 fibrosis) was found in among 41 (40.19%) parents of NAFLD proband and in only 17 (17%) parents of the non NAFLD proband (P = 0.025) group. The number of parents having LSM >15 kPa was 5 (4.9%) among parents of probands with NAFLD, whereas none in other group (P = 0.025).
Table 2.
Comparison of Fatty Liver, Liver Fibrosis and Various Scores Among NAFLD Versus without NAFLD Groups and Their Parents.
| Proband |
Parents of |
|||||
|---|---|---|---|---|---|---|
| NAFLD |
Non-NAFLD |
Proband with NAFLD |
Proband without NAFLD |
|||
| N = 51 | N = 50 | P value | N = 102 | N = 100 | P value | |
| Fatty liver on USG | 100% | 0% | 75 (73.5%) | 30 (30%) | 0.01∗ | |
| IAAT (cm) | 2.46 ± 0.95 | 2.26 ± 0.57 | 0.21 | 2.28 ± 0.91 | 2.45 ± 1.05 | 0.22 |
| Liver stiffness (kPa) | 6.65 ± 3.12 | 5.38 ± 2.88 | 0.07 | 7.4 ± 3.97 | 5.46 ± 2.51 | 0.01∗ |
| FIB4 score | 0.8 ± 0.7 | 0.57 ± 0.33 | 0.22 | 1.35 ± 1.27 | 1.11 ± 0.97 | 0.14 |
| NAFLD fibrosis score | −2.72 ± 1.49 | −2.55 ± 1.75 | 0.61 | −1.27 ± 1.71 | −2.22 ± 1.51 | 0.01∗ |
| BARD score | 1.63 ± 1.06 | 1.3 ± 1.02 | 0.12 | 1.81 ± 1.25 | 1.82 ± 0.99 | 0.97 |
| APRI Score | 0.32 ± 0.27 | 0.25 ± 0.13 | 0.13 | 0.36 ± 0.35 | 0.27 ± 0.26 | 0.03∗ |
IAAT = intraabdominal adipose tissue thickness, USG = ultrasonography, NAFLD fibrosis score = nonalcoholic fatty liver disease fibrosis score, BARD score = body mass index aspartate transaminase/alanine transaminase diabetes mellitus, APRI = aspartate transaminase to platelet ratio, NAFLD = nonalcoholic fatty liver diseases.
∗Significant P value <0.05.
NAFLD risk scores
The NAFLD fibrosis score and APRI score were significantly higher among parents of probands with NAFLD than among parents of probands without NAFLD (−1.27 ± (−1.71) vs −2.22 ± (−1.51), P = 0.0001) (Table 2). Among them, parents having the NAFLD fibrosis score >0.676 were 13 of 102 vs 5 of 100, with P = 0.033, respectively.
PNPLA3 polymorphisms
Among the parents of the NAFLD group, frequency of the PNPLA3 gene was 9 (15%) and 18 (30%), respectively, for GG and CG allele (Table 3), which was not statistically significant as compared with the non-NAFLD group. But the odds ratio found was 6, showing an association of PNPLA3 gene with familial inheritance of NAFLD (Table 4).
Table 3.
PNPLA3 Distribution Among NAFLD and without NAFLD Groups and Their Parents.
| PNPLA3 |
Probands |
Parents of |
||||
|---|---|---|---|---|---|---|
| NAFLD |
Non-NAFLD |
Proband with NAFLD |
Probands without NAFLD |
|||
| N = 30 | N = 30 | P value | N = 60 | N = 60 | P value | |
| Wild (CC) | 16 (53.3%) | 16 (53.3%) | 1 | 33 (55%) | 31 (51.7%) | 0.76 |
| Heterozygous (CG) | 10 (33.3%) | 13 (43.3%) | 0.42 | 18 (30%) | 26 (43.3%) | 0.28 |
| Homozygous (GG) | 4 (13.3%) | 1 (3.3%) | 0.16 | 9 (15%) | 3 (5%) | 0.2 |
| Allele frequency | ||||||
| C | 0.70 | 0.75 | 0.70 | 0.73 | ||
| G | 0.30 | 0.25 | 0.30 | 0.27 | ||
PNPLA3 = patatin-like phospholipase domain containing 3, NAFLD = nonalcoholic fatty liver disease.
Table 4.
PNPLA3 Polymorphism in the NAFLD Group.
| Proband PNPLA3 Genotype |
|||||
|---|---|---|---|---|---|
| Mutant (CG/GG) | Wild (CC) | P value | Odds ratio | ||
| Parents PNPLA3 |
Mutant (CG/GG) | 12 | 8 | 0.057 | 6∗ |
| Wild (CC) | 2 | 8 | |||
PNPLA3 = patatin-like phospholipase domain containing 3, NAFLD = nonalcoholic fatty liver disease.
∗Significant P value <0.05.
Description of probands (n = 101)
A total of 101 individuals (51 with NAFLD and 50 without NAFLD) were enrolled.
PNPLA3 polymorphisms
In probands with NAFLD, PNPLA3 gene polymorphism was seen in 14 individuals, where 4 (13.3%) had homozygous GG genotype and 10 (33.3%) had heterozygous CG genotype. All allele frequency followed the HWE. Probands of parents with NASH were found to have homozygous GG PNPLA3 polymorphism.
NAFLD predictive equation (NAFLD 16 score)
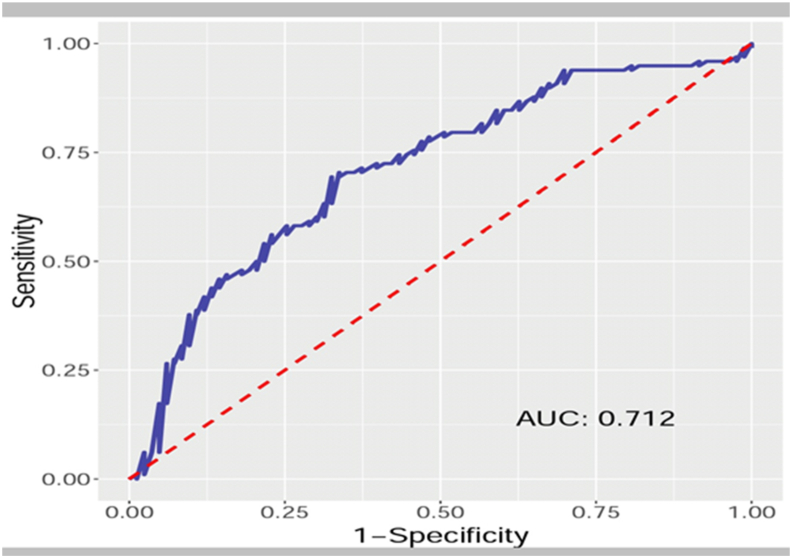
Based on the logistic regression model, prediction of NAFLD in an individual depending upon the parent's parameters was derived by the following equation: P[Y] = 1/1+e− ["25.36 + 0.19 X Hypertensiony+0.21 X DM y ± 1.1 X acanthosis nigricansy+0.38 X Weight±0.33 X Height±0.86 X BMI±0.34 X Waist Circumference+ 0.29 X Hip Circumference+35.81 X Waist-Hip Ratio X Platelet X AST+0.02 X ALT±1.1 X Albumin±0.01 X Cholesterol±0.01 X Triglyceride± 0.28 X Fatty Livery+1.46 X USG Liver Grade1+3.02 X USG Liver Grade2+3.05 X USG Liver Grade 3 + 0.02 X Liver Stiffness±0.83 X PNPLA3 Heterozygous+0.72 X PNPLA3 Homozygous± 0.19 X PNPLA3 Wild where ‘History of Hypertension, diabetes mellitus, acanthosis nigricans, fatty liver in any parent’ is y = 1 or y = 0 when present or absent, respectively. This NAFLD 16 score leads us to predict in an overweight/obese individual the probability of developing NAFLD with the sensitivity of 71.1%, the accuracy of 67.4%, and specificity of 64.3% (Figure 1) (Table 5).
Figure 1.
Receiver operating characteristic (ROC) curve for NAFLD prediction in an individual based on parent's parameters. NAFLD = nonalcoholic fatty liver disease.
Table 5.
Univariate and Multivariate Analysis of NAFLD Group parents.
| Serial no. | Parameters | Odds ratio (univariate), P value | Odds ratio (multivariate), P value |
|---|---|---|---|
| 1. | Hypertension | 0.36 (0.16–0.78, P = 0.01∗) | 0.83 (0.25–2.68, P = 0.75) |
| 2. | Diabetes mellitus | 0.26 (0.13–0.50, P < 0.01∗) | 0.81 (0.24–2.67, P = 0.73) |
| 3. | Acanthosis Nigricans |
0.26 (0.08–0.70, P = 0.01∗) | 3.00 (0.55–17.01, P = 0.21) |
| 4. | Weight (kg) | 0.97 (0.95–0.99, P = 0.01∗) | 0.68 (0.50–0.87, P = 0.01∗) |
| 5. | Height (cm) | 1.01 (0.98–1.03, P = 0.68) | 1.40 (1.12–1.82, P = 0.01∗) |
| 6. | Waist circumference (cm) | 0.97 (0.95–1.00, P = 0.02∗) | 1.41 (0.75–2.75, P = 0.29) |
| 7. | Hip circumference (cm) | 0.98 (0.95–1.00, P = 0.10) | 0.75 (0.41–1.31, P = 0.32) |
| 8. | Platelet/μl | 1.00 (1.00–1.00, P = 0.04∗) | 1.00 (1.00–1.00, P = 0.34) |
| 9. | AST (U/L) | 0.97 (0.95–0.99, P = 0.01∗) | 1.00 (0.96–1.03, P = 0.97) |
| 10. | ALT (U/L) | 0.96 (0.94–0.98, P < 0.01∗) | 0.98 (0.94–1.01, P = 0.12) |
| 11. | Serum albumin (g/dl) | 2.84 (1.38–6.09, P = 0.01∗) | 3.02 (0.98–9.88, P = 0.06) |
| 12. | Cholesterol (mg/dl) | 1.00 (0.99–1.00, P = 0.21) | 1.01 (1.00–1.02, P = 0.11) |
| 13. | Triglyceride (mg/dl) | 1.00 (0.99–1.00, P = 0.22) | 1.01 (1.00–1.02, P = 0.14) |
| 14. | Fatty liver on USG | 0.15 (0.08–0.28, P < 0.01∗) | 1.33 (0.10–34.90, P = 0.84) |
| 15. | Liver stiffness on Fibroscan (kPa) | 0.85 (0.76–0.94, P = 0.01∗) | 0.98 (0.85–1.12, P = 0.75) |
| 16. | PNPLA3 genotype | ||
| 1. Wild (CC) | 0.99 (0.51–1.90, P = 0.97) | 1.20 (0.37–4.04, P = 0.76) | |
| 2. Heterozygous (CG) | 1.52 (0.73–3.21, P = 0.270) | 2.28 (0.70–7.75, P = 0.17) | |
| 3. Homozygous (GG) | 0.35 (0.07–1.27, P = 0.12) | 0.49 (0.07–2.94, P = 0.44) | |
BMI = body mass index, AST = aspartate transaminase, ALT = alanine transaminase, PNPLA3 = patatin-like phospholipase domain containing 3, USG = ultrasonography, NAFLD = nonalcoholic fatty liver disease.
∗Significant P value <0.05.
Discussion
The concept of heritability is based on a mathematical calculation, which involves three phenotype variance sources: genetics (G), environment (E), and individual. G represents the fraction of variation between individuals in a population that is due to their genetic background. Unfortunately, knowledge of the G × E interaction in the biology of NAFLD and NASH remains largely unexplored. We aimed to establish the hierarchal association of PNPLA3 polymorphic gene in NAFLD. Data on familial aggregation and heritability of NAFLD are scanty in the literature. Patients with a positive family history of NAFLD have an increased risk for an early and more severe form of disease.16 Struben et al17 have reported that NASH and cryptogenic cirrhosis may coexist in families speculating that these findings may represent inherited susceptibility to fatty liver injury. In our study, we found fatty liver was seen in 75 (73.5%) parents of the probands with NAFLD as compared with only 30 (30%) parents of probands without NAFLD. Similarly, one prospective study which used MR spectroscopy among siblings and parents of probands with fatty liver detected fatty liver in 59% and 78%, respectively, compared with 17% and 37% among relatives of probands without fatty liver.16 Siddiqui et al18 found that the prevalence of the disease in relatives is nearly 3 times higher than that of the general population if the patient had early decompensated NAFLD-related cirrhosis.
We found that one of the parents had NASH in 36.3% of cases among the NAFLD proband group. This association was higher as compared to a similar retrospective cohort study, Willner et al19 showed that 18% of patients with NAFLD had one or both parents affected with NASH. This quite higher prevalence rate could be attributed to environmental, ethnic, or racial factors.
Parents of probands with NAFLD had a significantly higher rate of metabolic risk factors such as hypertension (32%), diabetes (50.9%), dyslipidemia (38%), acanthosis nigricans (17%), raised BMI (27.4 ± 5.12), WHR (96.26 ± 12.39), and elevated serum ALT and AST levels (P = 0.001). Few studies also found a similar association of metabolic factors in parents of children with NAFLD.16, 20 Bhadoria et al reported that a family history of at least one metabolic trait was seen in more than two-thirds of the cirrhotic cases, that is, 68.8% (779/1133).21 They reported prevalence of diabetes as 52.5%, hypertension 46.2%, dyslipidemia 6.7%, coronary artery disease 21.2%, and obesity 53.3% among first-degree relatives of patients with NAFLD-related cirrhosis, suggesting a positive family history of metabolic risk factors to be corroborative with early and severe NAFLD cirrhosis.21
We observed that PNPLA3 homozygous (GG) gene polymorphism was seen more frequently in parents of the NAFLD group (n = 9, 15%) than in parents of the without NAFLD group (n = 3, 5%). This shows that the presence of mutant PNPLA3 polymorphism has 6 times increased risk in the predisposition of NAFLD (Table 3). A recent Indian study carried out in overweight/obese children with NAFLD showed similar results, where PNPLA3 polymorphism homozygous/GG and heterozygous/CG was seen in 24 (34.8%) and 23 (33.3%) children, respectively.9
Recently, a meta-analysis from Dai G et al22 showed that PNPLA3 rs738409 polymorphism has a strong association with fatty liver and histological injury. In addition, G allele carriers were more frequently associated with NASH and liver fibrosis.5,22,23 In our study, PNPLA3 homozygous polymorphism (GG) has not shown a statistically significant association, and this can be due to incomplete penetrance, variable expression, multifactorial causation of NAFLD, and smaller sample size. To the best of our knowledge, Mendelian inheritance for PNPLA3 alleles has not yet been described. We also found that individuals with any parent having NASH are more likely to get homozygous/GG allele leading to early and more severe disease.
TE showed higher mean LSM values (7.4 ± 3.97 vs 5.46 ± 2.51, P = 0.001) in parents of the NAFLD group. The number of parents having LSM >7.9 kPa was 41 (40.19%) vs only 17 (17%) among parents of both groups (P = 0.025). Similar to a study carried out, Siddiqui et al18,24 studied caregivers of patients with NAFLD-related cirrhosis and found the median LSM value of 6.7 kPa (4.4–9.7) with the distribution of fibrosis stage 2, 3, and 4 as 28%, 28%, and 14%, respectively. Liver cirrhosis was found in 5 (4.9%) parents of the NAFLD group, while none of the parents from the non-NAFLD group had cirrhosis. This suggests the importance of noninvasive tools for screening and assessment of the severity of NAFLD.25
We have compared various scores such as the NAFLD fibrosis score, APRI, and FIB 4 score, of which the NAFLD fibrosis score and APRI score were found to be significantly higher among parents of the NAFLD group. We also found that NAFLD fibrosis score >0.676 which is equivalent to F3– F4 fibrosis was seen in 12.7% of parents of the NAFLD group while it was seen in only 5% of parents of the non-NAFLD group (P = 0.033).13
A literature review showed that the ClinLipMet Score given by Zhou et al based on AST, fasting insulin, PNPLA3 rs738409 genotype, and plasma metabolites such as glutamate, isoleucine, glycine and so on demonstrated good accuracy for the diagnosis of NASH (AUC approximately 0.87).26
We have developed an equation (NAFLD 16 Score) that can help in predicting NAFLD in an individual based on parent's parameters with a sensitivity of 71.1%, specificity of 64.3%, and accuracy of 67.4% with area under curve = 0.712), emphasizing familial influence on NAFLD. The novelty of this score is that it is based only on parent's parameters, thus risk estimation for sibling and the next generation would be easier and preemptive for regular surveillance. DNA mapping and genetic testing can become a window of opportunity as a noninvasive approach to identify and categorize severity among at-risk individuals.
Limitations
We could not evaluate various other parameters of metabolic syndrome such as fasting insulin levels, homeostatic model assessment of insulin resistance, and serum high-density lipoprotein levels. During history review, we did not analyze detailed dietary routine or calorie intake because of memory bias. Liver biopsy was not carried out as being an invasive procedure for screening parents. The diagnosis of NAFLD was based on USG only, which has low sensitivity for mild steatosis and is subjective also. We could not do magnetic resonance imaging proton density fat fraction and controlled attenuation parameter value assessment due to logistic reasons. We could not use XL probe while doing a fibroscan for individuals with BMI >30 kg/m2 due to its nonavailability. Smaller sample size may have underpowered the study to detect an effective role of genetic inheritance and variable expression of identified gene polymorphism. In the future, a larger study would be needed to refine the concepts behind the genetic role. NAFLD16 score has modest performance in its present form and may need modification.
Challenges
Despite this predictive value, genetic biomarker tests may be neither practical nor cost-effective for large-scale population screening programs. Other problems such as generalizability of results (performance of SNPs) and heterogenicity of allele frequencies in diverse ethnic populations will also hinder the path.
We recommend screening of parents of individuals with NAFLD which will help in the early identification of NAFLD and metabolic risk factors. Evaluation of genetic risk factors should be explored in the future to identify families and stratify them for the risk of NAFLD. NAFLD16 score also estimates the likelihood risk of disease in the next generation, although to prove sensitivity and specificity of this score larger sample size would be needed in the future. Thus, early targeted interventions could prevent future development and progression of NAFLD in such families.
CRediT authorship contribution statement
Shubham Jain: Conceptualization, designing of the manuscript, generation, collection, assembly, analysis, interpretation of data, Writing - original draft, Writing - review & editing. Ravi Thanage: Conceptualization, designing of the manuscript, Writing - original draft. Falguni Panchal: Patatin like phospholipase domain containing 3 (PNPLA3) genetic testing. Pravin M. Rathi: Writing - original draft. Renuka Munshi: Patatin like phospholipase domain containing 3 (PNPLA3) genetic testing. Suhas S. Udgirkar: Conceptualization, designing of the manuscript, Writing - original draft. Qais Q. Contractor: Conceptualization, designing of the manuscript, Writing - original draft, Writing - review & editing. Sanjay J. Chandnani: Writing - original draft. Nair P. Sujit: generation, collection, assembly, analysis. Partha Debnath: generation, collection, assembly, analysis. Anupam Singh: generation, collection, assembly, analysis, interpretation of data.
Conflicts of interest
The authors have none to declare.
Acknowledgments
Funding Authors have received grants for this study from the “Nair Golden Jubilee Research Foundation”.
None.
References
- 1.Younossi Z., Anstee Q.M., Marietti M. Global burden of NAFLD and NASH: trends, predictions, risk factors and prevention. Nat Rev Gastroenterol Hepatol. 2018;15:11–20. doi: 10.1038/nrgastro.2017.109. [DOI] [PubMed] [Google Scholar]
- 2.Williams C.D., Stengel J., Asike M.I. Prevalence of nonalcoholic fatty liver disease and nonalcoholic steatohepatitis among a largely middle-aged population utilizing ultrasound and liver biopsy: a prospective study. Gastroenterology. 2011;140:124–131. doi: 10.1053/j.gastro.2010.09.038. [DOI] [PubMed] [Google Scholar]
- 3.Agrawal S., Duseja A.K. Non-alcoholic fatty liver disease: east versus west. J Clin Exp Hepatol. 2012;2:122–134. doi: 10.1016/S0973-6883(12)60101-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Feldman A., Eder S.K., Felder T.K. Clinical and metabolic characterization of lean caucasian subjects with non-alcoholic fatty liver. Am J Gastroenterol. 2017;112:102–110. doi: 10.1038/ajg.2016.318. [DOI] [PubMed] [Google Scholar]
- 5.Sookoian S., Pirola C.J. Genetics of nonalcoholic fatty liver disease: from pathogenesis to therapeutics. Semin Liver Dis. 2019 May;39:124–140. doi: 10.1055/s-0039-1679920. [DOI] [PubMed] [Google Scholar]
- 6.Kumar A., Kaur Walia G., Gupta V. Genetics of non-alcoholic fatty liver disease in Asian populations. J Genet. 2019;98:29. doi: 10.1007/s12041-019-1071-8. [DOI] [PubMed] [Google Scholar]
- 7.Xu R., Tao A., Zhang S., Deng Y., Chen G. Association between patatin-like phospholipase domain containing 3 gene (PNPLA3) polymorphisms and nonalcoholic fatty liver disease: a HuGE review and meta-analysis. Sci Rep. 2015;5:9284. doi: 10.1038/srep09284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fan J.-G., Kim S.-U., Wong V.W.-S. New trends on obesity and NAFLD in Asia. J Hepatol. 2017;67:862–873. doi: 10.1016/j.jhep.2017.06.003. [DOI] [PubMed] [Google Scholar]
- 9.Sood V., Khanna R., Rawat D., Sharma S., Alam S., Sarin S.K. Study of family clustering and PNPLA3 gene polymorphism in pediatric non alcoholic fatty liver disease. Indian Pediatr. 2018;55:561–567. [PubMed] [Google Scholar]
- 10.Misra A., Chowbey P., Makkar B.M. Consensus statement for diagnosis of obesity, abdominal obesity and the metabolic syndrome for Asian Indians and recommendations for physical activity, medical and surgical management. J Assoc Phys India. 2009;57:163–170. [PubMed] [Google Scholar]
- 11.Das C., Baruah M., Singh D. Imaging of non alcoholic fatty liver disease: a road less travelled. Indian J Endocrinol Metab. 2013;17:990. doi: 10.4103/2230-8210.122606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sterling R.K., Lissen E., Clumeck N. Development of a simple noninvasive index to predict significant fibrosis in patients with HIV/HCV coinfection. Hepatology. 2006;43:1317–1325. doi: 10.1002/hep.21178. [DOI] [PubMed] [Google Scholar]
- 13.Angulo P., Hui J.M., Marchesini G. The NAFLD fibrosis score: a noninvasive system that identifies liver fibrosis in patients with NAFLD. Hepatology. 2007;45:846–854. doi: 10.1002/hep.21496. [DOI] [PubMed] [Google Scholar]
- 14.McPherson S., Stewart S.F., Henderson E., Burt A.D., Day C.P. Simple non-invasive fibrosis scoring systems can reliably exclude advanced fibrosis in patients with non-alcoholic fatty liver disease. Gut. 2010;59:1265–1269. doi: 10.1136/gut.2010.216077. [DOI] [PubMed] [Google Scholar]
- 15.Harrison S.A., Oliver D., Arnold H.L., Gogia S., Neuschwander-Tetri B.A. Development and validation of a simple NAFLD clinical scoring system for identifying patients without advanced disease. Gut. 2008;57:1441–1447. doi: 10.1136/gut.2007.146019. [DOI] [PubMed] [Google Scholar]
- 16.Schwimmer J.B., Celedon M.A., Lavine J.E. Heritability of nonalcoholic fatty liver disease. Gastroenterology. 2009;136:1585–1592. doi: 10.1053/j.gastro.2009.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Struben V.M., Hespenheide E.E., Caldwell S.H. Nonalcoholic steatohepatitis and cryptogenic cirrhosis within kindreds. Am J Med. 2000;108:9–13. doi: 10.1016/s0002-9343(99)00315-0. [DOI] [PubMed] [Google Scholar]
- 18.Siddiqui M.S., Carbone S., Vincent R. Prevalence and severity of nonalcoholic fatty liver disease among caregivers of patients with nonalcoholic fatty liver disease cirrhosis. Clin Gastroenterol Hepatol. 2019;17:2132–2133. doi: 10.1016/j.cgh.2018.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Willner I.R., Waters B., Patil S.R., Reuben A., Morelli J., Riely C.A. Ninety patients with nonalcoholic steatohepatitis: insulin resistance, familial tendency, and severity of disease. Am J Gastroenterol. 2001;96:2957–2961. doi: 10.1111/j.1572-0241.2001.04667.x. [DOI] [PubMed] [Google Scholar]
- 20.Loomba R., Abraham M., Unalp A. Association between diabetes, family history of diabetes, and risk of nonalcoholic steatohepatitis and fibrosis. Hepatology. 2012;56:943–951. doi: 10.1002/hep.25772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bhadoria A.S., Kedarisetty C.K., Bihari C. Impact of family history of metabolic traits on severity of non-alcoholic steatohepatitis related cirrhosis: a cross-sectional study. Liver Int. 2017;37:1397–1404. doi: 10.1111/liv.13396. [DOI] [PubMed] [Google Scholar]
- 22.Dai G., Liu P., Li X., Zhou X., He S. Association between PNPLA3 rs738409 polymorphism and nonalcoholic fatty liver disease (NAFLD) susceptibility and severity. Medicine (Baltim) 2019;98 doi: 10.1097/MD.0000000000014324. e14324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Trépo E., Romeo S., Zucman-Rossi J., Nahon P. PNPLA3 gene in liver diseases. J Hepatol. 2016;65:399–412. doi: 10.1016/j.jhep.2016.03.011. [DOI] [PubMed] [Google Scholar]
- 24.Siddiqui M.S., Vuppalanchi R., Van Natta M.L. Vibration-controlled transient elastography to assess fibrosis and steatosis in patients with nonalcoholic fatty liver disease. Clin Gastroenterol Hepatol. 2019;17:156–163. doi: 10.1016/j.cgh.2018.04.043. e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tovo C.V., Villela-Nogueira C.A., Leite N.C. Transient hepatic elastography has the best performance to evaluate liver fibrosis in non-alcoholic fatty liver disease (NAFLD) Ann Hepatol. 2019;18:445–449. doi: 10.1016/j.aohep.2018.09.003. [DOI] [PubMed] [Google Scholar]
- 26.Vespasiani-Gentilucci U., Gallo P., Dell'Unto C., Volpentesta M., Antonelli-Incalzi R., Picardi A. Promoting genetics in non-alcoholic fatty liver disease: combined risk score through polymorphisms and clinical variables. World J Gastroenterol. 2018;24:4835–4845. doi: 10.3748/wjg.v24.i43.4835. [DOI] [PMC free article] [PubMed] [Google Scholar]