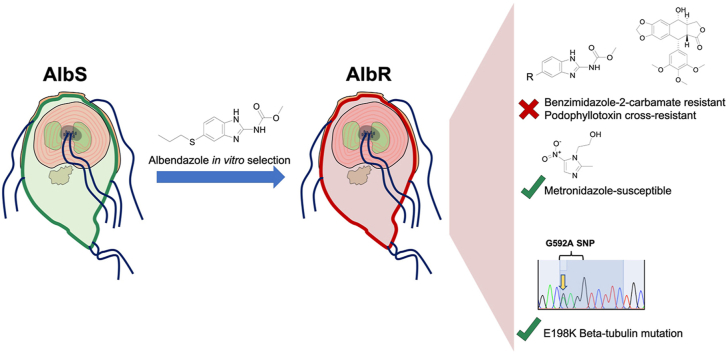
Abstract
Benzimidazole-2-carbamate (BZ) compounds, including Albendazole (Alb), are one of just two drug classes approved to treat the gastrointestinal protist Giardia duodenalis. Benzimidazoles bind to the tubulin dimer interface overlapping the colchicine binding site (CBS) of β-tubulin, thereby inhibiting microtubule polymerisation and disrupting microtubule networks. These BZ compounds are widely used as anthelmintic, anti-fungal and anti-giardial drugs. However, in helminths and fungi, BZ-resistance is widespread and caused by specific point mutations primarily occurring at F167, E198 and F200 in β-tubulin isoform 1. BZ-resistance in Giardia is reported clinically and readily generated in vitro, with significant implications for Giardia control. In Giardia, BZ mode of action (MOA) and resistance mechanisms are presumed but not proven, and no mutations in β-tubulin have been reported in association with Alb resistance (AlbR). Herein, we undertook detailed in vitro drug-susceptibility screens of 13 BZ compounds and 7 Alb structural analogues in isogenic G. duodenalis isolates selected for AlbR and podophyllotoxin, another β-tubulin inhibitor, as well as explored cross-resistance to structurally unrelated, metronidazole (Mtz). AlbR lines exhibited co-resistance to many structural variants in the BZ-pharmacophore, and cross-resistance to podophyllotoxin. AlbR lines were not cross-resistant to Mtz, but MtzR lines had enhanced survival in Alb. Lastly, Alb analogues with longer thioether substituents had decreased potency against our AlbR lines. In silico modelling indicated the Alb-β-tubulin interaction in Giardia partially overlaps the CBS and corresponds to residues associated with BZ-resistance in helminths and fungi (F167, E198, F200). Sequencing of Giardia β-tubulin identified a single nucleotide polymorphism resulting in a mutation from glutamic acid to lysine at amino acid 198 (E198K). To our knowledge, this is the first β-tubulin mutation reported for protistan BZ-resistance. This study provides insight into BZ mode of action and resistance in Giardia, and presents a potential avenue for a genetic test for clinically resistance isolates.
Keywords: Giardia duodenalis, Albendazole, Benzimidazoles, Drug-resistance, Tubulin
Abbreviations: Alb, albendazole; AlbR, albendazole resistance; BZ, benzimidazole-2-carbamate derivative; CBS, colchicine binding site; Mtz, metronidazole; MtzR, metronidazole resistance; MT, microtubule
Graphical abstract
Highlights
-
•
In vitro albendazole-resistant Giardia were broadly resistant to benzimidazole-2-carbamates.
-
•
Cross-resistance to structurally unrelated microtubule inhibitors was observed.
-
•
The predicted Giardia benzimidazole binding overlaps the colchicine binding site.
-
•
The E198K β-tubulin mutation was identified in the albendazole-resistant line.
-
•
Metronidazole-resistance may enhance acquisition of albendazole-resistance.
1. Introduction
Giardia duodenalis (syn. G. lamblia, G. intestinalis) is a gastrointestinal protist causing ~180 million symptomatic infections (Lane and Lloyd, 2002) and a major cause of diarrheal diseases linked to significant mortality and malnutrition in child under 5 years of age (Kotloff et al., 2013). Giardia infection may cause chronic, post-infectious gastrointestinal diseases (PI-GID), most notably irritable bowel syndrome and chronic fatigue syndrome (Hanevik et al., 2014; Hunskar et al., 2016; Litleskare et al., 2018). Treatment of giardiasis is limited to nitroheterocyclics (e.g. metronidazole, Mtz) and benzimidazole-2-carbamate derivatives (BZs) (e.g. albendazole (Alb) or mebendazole) (Meloni et al., 1990; Gardner and Hill, 2001; Solaymani-Mohammadi et al., 2010). Resistance to both classes has been demonstrated to develop in vitro (Ansell et al., 2015) and these observations have increasingly been reflected in clinical failures for both Mtz (Nabarro et al., 2015; Lalle and Hanevik, 2018) and Alb (Farbey et al., 1995; Lemee et al., 2000). Considering the limited treatment options (Ansell et al., 2015; Carter et al., 2017; Lalle and Hanevik, 2018), drug-resistance poses a significant risk to reducing disease burdens associated with G. duodenalis infection.
In nematodes, BZs are predicted to selectively bind near the colchicine binding site (CBS) of β-tubulin (Russell et al., 1992; Keeling and Doolittle, 1996; Chatterji et al., 2011; Wang et al., 2016). The anti-fungal carbendazim, the parent BZ carbamate, binds β-tubulin at a site overlapping the CBS in Botrytis cinerea and related fungi (Lacey et al., 1987; Yarden and Katan, 1993; Leroux et al., 1999; Cai et al., 2015; Liu et al., 2019). The colchicine binding site (CBS) occurs at the interface between α and β-tubulin, and is shared by many natural and synthetic compound classes including colchicine, podophyllotoxin, and BZs, and is also distinct from the vinblastine binding site, itself shared by maytansines and phomopsin (Lacey, 1988; Wang et al., 2016; McLoughlin and O'Boyle, 2020). Based on their site of action, CBS inhibitors prevent the soluble tubulin dimers adding to the growing insoluble microtubule to cause destabilisation of the mitotic cytoskeleton and failure of cell division, so-called metaphase arrest (McLoughlin and O'Boyle, 2020). BZ-resistance in helminths is clearly associated with three primary mutations in β-tubulin, including F167Y, E198A, F200Y (Lacey and Prichard, 1986; Lubega and Prichard, 1991; Elard and Humbert, 1999; Ghisi et al., 2007; Rufener et al., 2009; Beech et al., 2011), whereas several mutational variants of E198 (E198A, E198K, E198V) confer anti-fungal BZ-resistance (Yarden and Katan, 1993; Banno et al., 2008; Cai et al., 2015; Liu et al., 2019).
Tubulin is also implicated as the conserved target of BZ compounds in Giardia. BZ compounds bind to Giardia's β-tubulin dense cytoskeletal structures (Chavez et al., 1992), and cause dramatic and deleterious effects on its microtubule-based cytoskeleton (Morgan et al., 1993; Upcroft et al., 1996; Mariante et al., 2005). MacDonald et al. (2004) also demonstrated BZ compounds bound with high affinity to recombinant Giardia β-tubulin and arrested microtubule (MT) polymerisation. Together, these data support a conserved BZ-β-tubulin binding model and mode of action blocking MT polymerisation, which would correspond to anthelmintic mechanisms (Lacey, 1988; Aguayo-Ortiz et al., 2013a, 2013b). Despite this, mechanisms of Alb resistance have not been robustly demonstrated in Giardia, and it remains unclear if BZ resistance mechanisms in this parasite are consistent with those described for helminths or fungi. Notably, despite having conserved orthology with the specific β-tubulin residues correlated with BZ-anthelmintic resistance (Katiyar et al., 1994), no BZ-resistance association mutations have been reported for Giardia to date (Upcroft et al., 1996; Arguello-Garcia et al., 2009). Furthermore, several transcriptome and proteome studies have highlighted tubulin-independent responses in Alb resistant (AlbR) Giardia within in vitro culture (Arguello-Garcia et al., 2009; Paz-Maldonado et al., 2013; Argüello-García et al., 2015), while others have hypothesised tubulin-independent BZ-targets among cytoskeletal Giardia-specific gene families (e.g. NEKs, Protein 21.1, giardins) (Manning et al., 2011; Hagen et al., 2020; Hennessey et al., 2020).
Overall, our limited understanding of protistan BZ-resistance is a concerning impediment for deployment of effective clinical surveillance. Herein, we have robustly explored BZ resistance in Giardia from the view of clarifying unresolved hypotheses regarding β-tubulin as the target of BZs and mechanism of resistance. We have performed the most extensive BZ compound screen in Giardia to date, including dissecting relationships between AlbR and MtzR cross-resistance, and used this to guide computational modelling linking the molecular mode of action between β-tubulin, BZs, and BZ resistance. We have performed both capillary and amplicon sequencing of β-tubulin, and note we are only the third study to do so in Giardia (Upcroft et al., 1996; Arguello-Garcia et al., 2009). By taking this targeted approach, this study consolidates our understanding of the mode of action of BZs in Giardia with nematodes and fungi, and lays the platform for overcoming the most significant risk to their continued use, drug-resistance.
2. Materials and methods
2.1. Trophozoite in vitro culture and induction of Albendazole resistance
Giardia duodenalis trophozoites from the WB-1B isolate (Capon et al., 1989) were generously provided by Professor Jacqueline Upcroft and Professor Peter Upcroft. Alb-sensitive trophozoites were maintained in flat-sided 10 mL tubes (Nunclon delta) filled with TYI-S33 medium modified for Giardia (Davids and Gillin, 2011), containing 0.5 g/L bovine bile (Sigma Aldrich) and bovine serum (10%), capped and sub-cultured twice weekly. AlbR induction in WB-1B were attempted via pulse and continuous sublethal exposures, with pulse-based inductions ceased after multiple unsuccessful attempts.
AlbR was promoted through pulse selection (Lindquist, 1996), with trophozoite cultures exposed to Alb at the approximated IC90 of 0.16 μM over 24 h (‘pulse’), following by replacing cultures with fresh media containing lower, sublethal levels of Alb (0.1 μM). Pulse cultures were allowed to recover and grow, and then subjected to higher pulse concentrations and increased Alb during subculturing with successive pulse rounds. Pulse-based inductions were ceased after multiple unsuccessful attempts. AlbR induction via continuous sublethal exposure was performed over a period of approximately 400 days. An Alb stock (Sigma Aldrich; 1 mM stock dissolved in dimethyl sulfoxide (DMSO)) was added to the WB-1B line at a starting concentration of half the compound IC50 in the susceptible line at 0.1 μM (Upcroft et al., 1996), with continuous exposure maintained and drug concentration incrementally increased between 0.1 and 0.5 μM, allowing for parasite survival and growth to stabilise at each concentration. After induction to 0.5 μM was completed, cultures were maintained at 0.5 μM to ensure stable growth over several months. For the AlbS line, the WB-1B line was maintained alongside the AblR line in complete media with equivalent DMSO vehicle.
2.2. Drug-sensitivity testing in AlbR and AlbS isogenic lines
All synthetic BZ compounds including the commercially available drugs nocodazole, flubendazole, mebendazole, albendazole, parbendazole, fenbendazole, ciclobendazole, oxibendazole, ricobendazole, oxfendazole and thiabendazole were sourced from the in-house compound library at Microbial Screening Technologies. The BZs were dissolved in DMSO to a stock concentration of 1 mM while Mtz (Sigma Aldrich) was dissolved as a 100 mM stock dissolved in DMSO.
Drug-screening in isogenic lines was performed as previously described (Ansell et al., 2017). Briefly, growth and cell viability were assessed using CellTiter-Glo (Promega) reagent to measure ATP-based luminescence (corresponding to live cells/well). 10-point dilutions of BZ compounds were performed between 10 and 0.02 μM, and 100–0.2 μM for Mtz in 96-well clear-bottom plate (Corning #3610) with a well volume of 50 μL, and 2000 trophozoites/well used for AlbS and 4000/well used for AlbR lines. Screening was performed in triplicate plates. The baseline for 100% inhibition (‘kill’ well) was subtracted and data transformed to a proportion of the value for control (DMSO-only, ‘uninhibited’) wells. Prism software (Graphpad) was used to plot dose-response curves and calculate IC50 values for AlbS and AlbR, via the “log(inhibitor) vs. normalized response—variable slope (four parameters)” module, with the top constrained to 100 and the bottom constraint to < 0.
2.3. Molecular modelling of Albendazole with Giardia β-tubulin
2.3.1. Template and binding-site identification and homology modelling
The protein sequence of Giardia β-tubulin (GL50803_101291) was retrieved from the Predictein web-server (http://www.predictein.org/) (Ansell et al., 2019). The Giardia β-tubulin protein structure was submitted to the COACH meta-server (https://zhanglab.ccmb.med.umich.edu/COACH/) to predict the protein-ligand binding sites and templates (Yang et al., 2013b, a). Templates were selected based on ligand-binding sites which overlapped with CBS, as well as proximal residues reported as mutated in BZ anthelmintic resistance (i.e. F167, E198, F200) (Aguayo-Ortiz et al., 2013a). The Protein DataBank (PDB) structures 1SA0_B, 3N2G_D and 5C8Y_D were identified as potential templates, and were used to perform template based homology modelling to predict Giardia β-tubulin monomer using SWISS-MODEL (Waterhouse et al., 2018).
2.3.2. Molecular docking between Albendazole compounds and β-tubulin
The structure of Alb molecule was downloaded from DRUGBANK (https://go.drugbank.com), whereas the structures of the Alb analogues for the 5-hexylthio- and 5-octylthio- analogues were prepared using Chemdraw v19.1. The albendazole molecule was subjected to molecular docking with the homology model of the Giardia β-tubulin monomer by using AutoDock Vina (Trott and Olson, 2010). Autodock Tools v.1.5.6 (Morris et al., 2009) was used to add polar hydrogen atoms to the Giardia β-tubulin model and to detect root and choose torsions in the Alb molecule to allow it to rotate freely. Finally, a grid-box was defined in Autodock Tools, surrounding to residues F167, E198 and F200 in the receptor molecule (Giardia β-tubulin) for site specific docking in AutoDock Vina.
2.3.3. Molecular docking between Albendazole compounds and β-tubulin with the E198K mutation
Mutagenesis in Giardia β-tubulin monomer was performed by using PyMOL v2.3.0. Giardia β-tubulin monomer with E198K mutation was then further docked with Alb, and the 5-hexylthio- and 5-octylthio- analogues as described above.
2.4. Sequencing of β-tubulin in WB AlbR and AlbS isogenic lines
2.4.1. Capillary sequencing
gDNA was isolated from both AlbS and AlbR cell pellets using DNeasy PowerBiofilm kit (Qiagen) and quantified by Qubit dsDNA HS kit. The β-tubulin (GL50803_136020) amplicon was generated using primers from Arguello-Garcia et al. (2009) which spanned nucleotides 125-682. PCR reactions were standardized in 21 containing 10 GoTaq Green enzyme mix, 1 gDNA (~100 ng), 1 105′ overhang primer, 1 103′ overhang primer and 8 nuclease-free water. PCR cycling conditions were performed as: a) 95 °C 2 min, b) [95 °C 15 s, 60 °C 30 s, 72 °C 30 s] × 18 cycles, c) 72 °C 7 min and d) 4 °C hold. PCR amplicons were then purified using 1.0 × Ampure Beads (Beckman Coulter) and amplicon size distribution was evaluated using 1% agarose gel electrophoresis. Conventional sanger sequencing was then performed by Australian Genome Research Facility (AGRF, Melbourne) and sequences were analysed using 4peaks version 1.8.
2.4.2. Amplicon sequencing
CRISPR overhang sequencing libraries (Aubrey et al., 2015) was prepared including primary amplification of Cas9-induced InDels, and secondary index incorporation. As the larger size of the amplicon (~500bp) and the exceeded the sequencing range of the Illumina MiSeq platform is 200–300 bp, three primer sets of overhang-primers were used to cover the entire amplicon, including pairs which covered 210 bp at 5′ end, 3′ end, and middle part of β-tubulin amplicon respectively (Table 1). For primary amplification, PCR reactions were standardized in 20 containing 10 GoTaq Green enzyme mix, 1 gDNA (~100 ng), 0.5 105′ overhang primer, 0.5 103′ overhang primer and 8 nuclease-free water. PCR cycling conditions were performed as above for capillary sequencing. PCR amplicons were purified using 1.0 × Ampure Beads (Beckman Coulter) and amplicon size and distribution evaluated using 1% agarose gel electrophoresis.
Table 1.
Primer sets used for amplicon sequencing of Giardia β-tubulin (GL50803_136020). Overhang sequencing is underlined.
| Name | Direction | Primer sequence | Amplicon length (bp) | Sequence length (bp) |
|---|---|---|---|---|
| F-OH | 5′ 3′ | GTGACCTATGAACTCAGGAGTCTCCAGATCGAGAGGATCAACGT | 5’-[1-557]-3′ | 5’-[1-200]-3′ |
| R-OH | CTGAGACTTGCACATCGCAGCGGTGGTTGAGGTCTCCGTAGGT | |||
| F2A | 5′ 3′ | GTGACCTATGAACTCAGGAGTCCGAAGGGCCACTACACGGAG | 5’-[200-557]-3′ | 5’-[201-400]-3′ |
| R-OH | CTGAGACTTGCACATCGCAGCGGTGGTTGAGGTCTCCGTAGGT | |||
| flipF-OH | 3′ 5′ | GTGACCTATGAACTCAGGAGTCGGTGGTTGAGGTCTCCGTAGGT | 5’-[1-557]-3′ | 5’-[400-557]-3′ |
| flipR-OH | CTGAGACTTGCACATCGCAGCTCCAGATCGAGAGGATCAACGT |
Secondary amplification was performed to incorporate index primer required for sequencing derived from the Illumina Nextera design as sample barcodes. The 12 forward index primers and 8 reverse index primers were designed as: SRT1_OH1 5′- CAAGCAGAAGACGGCATACGAGATCCGGTCTCGGCATTCCTGCTGAACCGCTCTTC.
2.5. CGATCTNNNNNNNNGTGACCTATGAACTCAGGAGTC-2′; SRT2-OH2: 5′ - AATGATACGGCGACCACCGAGATCT
ACACTCTTTCCCTACACGACGCTCTTCCGATCTNNNNNNNNCTGAGACTTGCACATCGCAGC-3′. where sequence NNNNNNNN indicates where individual indexes are placed in the oligo design. PCR reaction were standardized in 21 containing 10 GoTaq Green enzyme mix, 10 PRC1 product, 0.5 10forward index primer and 0.5 10reverse index primer. PCR cycling conditions were performed as: a) 95 °C 3 min, b) [95 °C 15 s, 60 °C 30 s, 72 °C 30 s] × 24 cycles, c) 72 °C 7 min and d) 4 °C hold. PCR amplicons were then purified using 1.0 × Ampure Beads (Beckman Coulter) and amplicon size distribution was ascertained using 1% agarose gel electrophoresis. Amplicon sequencing of the final pooled library was performed at the Genomics Hub located within the Walter and Eliza Hall Institute of Medical Research (Melbourne, Australia).
Variant calling was performed on three technical in vitro culture replicates of AlbR and AlbS samples to determine the frequency of wild type and resistant allele associated with the β-tubulin structure of Giardia. The raw reads were filtered based on a quality score of 30, and then were mapped to nucleotide sequence of β-tubulin protein of Giardia by using BWA v.0.7 (Li and Durbin, 2009) with default parameters. Next, we used Genome Analysis Toolkit's (GATK v3.7.0) (McKenna et al., 2010) to identify SNPs using GATK best practices pipeline (Van der Auwera et al., 2013).
2.6. Drug-sensitivity testing in MtzR and MtzS isogenic lines
Isogenic MtzR lines from Ansell et al. (2017); Emery et al. (2018) were maintained as previously described in 30 μM Mtz and sub cultured as above (Section 2.1). Mtz-sensitive lines included WB-1B isolate (WB-MtzS), BRIS/83/HEPU/106 (106-MtzS) and BRIS/87/HEPU/713 (713-MtzS) along with their Mtz-resistant progeny lines WB-M3 (WB-MtzR), BRIS/83/HEPU/106-2ID10 (106-MtzR) and BRIS/83/HEPU/713-M3 (713-MtzR) (Boreham et al., 1984, 1988; Capon et al., 1989; Townson et al., 1992). Drug-sensitivity testing was performed as above for 10-point dilutions for Alb (2–0.0039 μM), and for Mtz (100–0.2 μM), with data transformation, normalisation and analysis performed as described above. (Methods 2.4).
3. Results
3.1. Induction of a new AlbR line from the WB genetic background
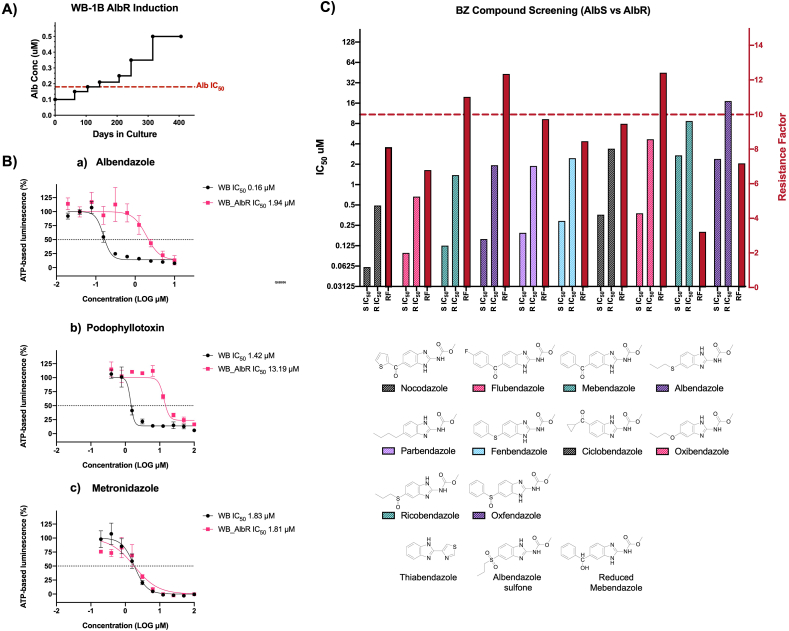
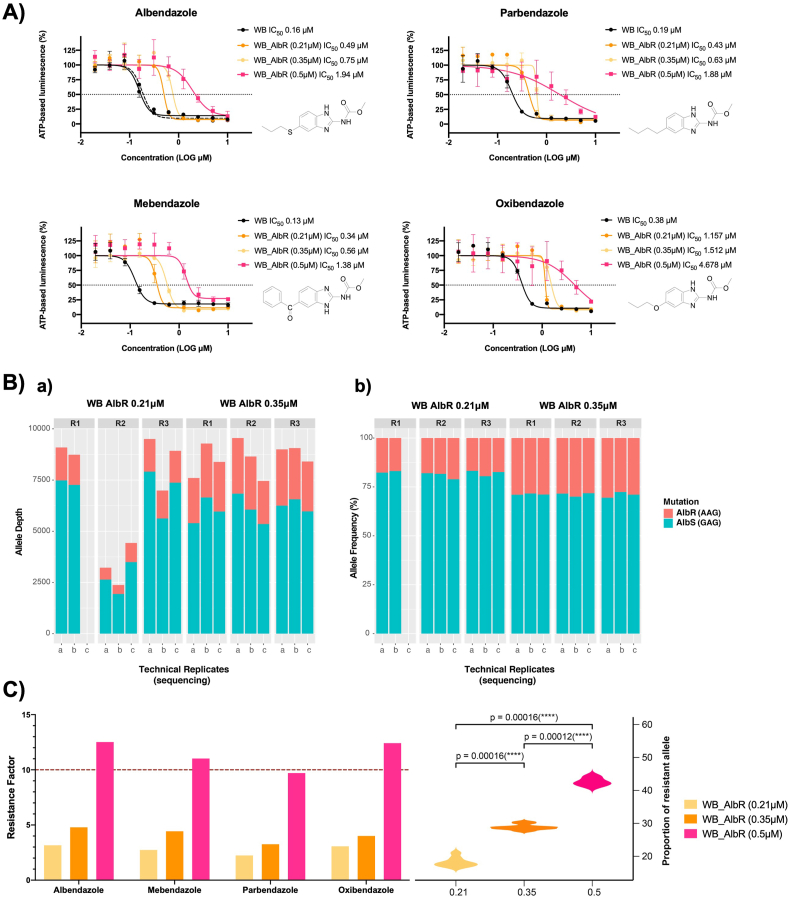
We selected isogenic albendazole-resistant (AlbR) from a susceptible (AlbS) line of the WB-1B isolate (Capon et al., 1989) via continuous sublethal exposure to 0.5 μM. Trophozoite cultures were subjected to continuous, exposures from 0.1 μM, with concentrations increased after periods of adaptation and tolerance based on parameters of parasite confluence, adherence and growth rate. Induction occurred over ~400 days (Fig. 1A), with increasing selection to 0.5 μM Alb between 0 and 315 days, after which the isolate was maintained stably over ~100 days. Continuous selection resulted in an increase in Alb IC50, from 0.16 μM in AlbS to 1.94 μM in AlbR lines, representing a resistance factor (RF) of 12.1 (Fig. 1B). This is consistent with previous observations where G. duodenalis Alb IC50 was higher than its tolerance levels under continuous exposure (Upcroft et al., 1996; Arguello-Garcia et al., 2009). Prior attempts at the induction of AlbR via continuous sublethal exposure has been met with variable success for this parasite. Upcroft et al. (1996) were unable to generate AlbR isolates from WB-1B isolate through this approach. Arguello-Garcia et al. (2009) were able to adapt a WB clone to 0.18 μM Alb through continuous sublethal exposure, however this took over 86 weeks, compared to 15 weeks in our AlbR line. In contrast, our attempts at AlbR induction using pulse selection for the same WB-1B lines were unsuccessful, despite previously described successes (Lindquist, 1996), with no WB-1B cultures surviving beyond a maximum of 6 rounds of pulse exposure.
Fig. 1.
Generation of an AlbR line and BZ compound screens: A) Time in culture and induction of AlbR in a WB-1B line starting at 0.1 μM and finishing at 0.5 μM over 400 days. B) IC50 dose response curves for albendazole, podophyllotoxin and metronidazole in AlbS and AlbR isogenic lines, demonstrating a substantial shift in IC50 in the AlbR for albendazole and podophyllotoxin but not for metronidazole. C) Bar chart of IC50 values for AlbS and AlbR isogenic lines (left axis) as well as the calculated RF for the AlbR (right axis) for BZ compounds. Compounds are listed from left to right in order of potency. The full dose response curves of all BZ compounds are in Supplementary Fig. S1. Thiabendazole, albendazole sulfone and reduced mebendazole were not sufficiently active in both lines at 10 μM to calculate an IC50, and therefore has not been shown.
3.2. AlbR line is broadly resistant to benzimidazole-2-carbamates
In total we investigated 13 BZ compounds (Supplementary Fig. S1), of which 12 were benzimidazole-2-carbamates with differing 5-position substituents via a single spacer (e.g. oxygen, suphur, sulphoxide, carbonyl). All BZs had a sub-micromolar IC50 in the WB-1B line (AlbS), except for those that are active only as in vivo BZ metabolites, consistent with observations from nematode testing (Dayan, 2003). The single non-carbamate BZ, thiabendazole, was not inhibitory at the highest doses tested (10 μM), consistent with previous reported bioassays (Edlind et al., 1990).
The relative sensitivity of G. duodenalis to active BZ compounds were nocodazole < flubendazole < mebendazole < albendazole < parbendazole < fenbendazole < ciclobendazole < oxibendazole < ricobendazole < oxfendazole over a 0.02–10 μM concentration range. The relevant order of potency was consistent between the AlbS and AlbR lines. Sulfoxide metabolites, including ricobendazole (albendazole sulphoxide; AlbS IC50 2.71 μM) and oxfendazole (febendazole sulphoxide; AlbS IC50 = 2.39 μM), were least potent, which is consist with previous reports (Morgan et al., 1993). Similarly, the oxidised sulfone metabolites were nearly inactive at 10 μM, including albendazole sulfone, and the in vivo metabolite of mebendazole formed by the reduction of the 5-benzoyl moiety to the 5-benzylhydrol (reduced mebendazole). Overall, our observations were consistent with binding affinity experiments using recombinant Giardia β-tubulin (MacDonald et al., 2004), which classified BZ-compounds as high-affinity (Alb, fenbendazole, mebendazole), medium- (albendazole-sulphoxide, oxibendazole, parbendazole), and low-affinity (thiabendazole). These sensitivities are also similar to gastrointestinal nematodes, where high potency BZ compounds have carbonyl (C=O; nocodozole, flubendazole, mebendazole, ciclobendazole) or sulfur (Alb, fenbendazole) bridges to 5-position aromatic or alkyl or cycloalkyl moieties that undergo metabolic conversion leading to more polar analogues which are less potent (Lacey, 1988; Agarwal et al., 1993).
For all BZ compounds, we noted the AlbS line never reached complete inhibition at the highest BZ screening concentrations (Supplementary Fig. 1), which are consistent with reversible drug-target binding which have static, not cidal, effects (Edlind et al., 1990; Upcroft et al., 1996). Nonetheless, increased IC50 and RFs were observed across all BZ compounds tested in the AlbR line (Fig. 1C), with highest differences in potency observed for the most active inhibitors, thus albendazole (RF = 12.3), mebendazole (RF = 11.0) and oxibendazole (RF = 12.4). This demonstrated Alb selection conferred resistance across the BZ class, regardless of the structural variation in the 5-position substituent of twelve BZs tested (Fig. 1C). The AlbR line was also cross-resistant to podophyllotoxin, a structurally independent CBS inhibitor, with an IC50 shift from 1.42 μM to 13.19 μM, a RF = 9.3 (Fig. 1B). The shared cross-resistance of two chemicals classes of CBS-binding MT inhibitors (Cortese et al., 1977; Schonbrunn et al., 1999; Wang et al., 2016), provided compelling evidence for a β-tubulin dependent resistance mechanism in the AlbR line that overlapped and excluded other CBS inhibitors.
3.3. Exploring AlbR derived from MtzR backgrounds
Our AlbR line was equally sensitive to Mtz as its AlbS parent (Fig. 1B), which indicated no cross-resistance to nitroheterocyclics. In contrast, when Upcroft et al. (1996) attempted AlbR induction in five G. duodenalis isolates, only the MtzR isolate WB-M3 acquired AlbR and tolerated high concentrations of 2 μM Alb. Of the remaining Giardia isolates in Upcroft et al. (1996), two failed to display a stable or highly resistant phenotype at > 0.2 μM Alb, while another two isolates did not survive repeated Alb exposures. Interestingly, WB-1B was one of these failed isolates, which was the parent isolate of WB-M3 (Townson et al., 1992) as well as our AlbR line selected herein.
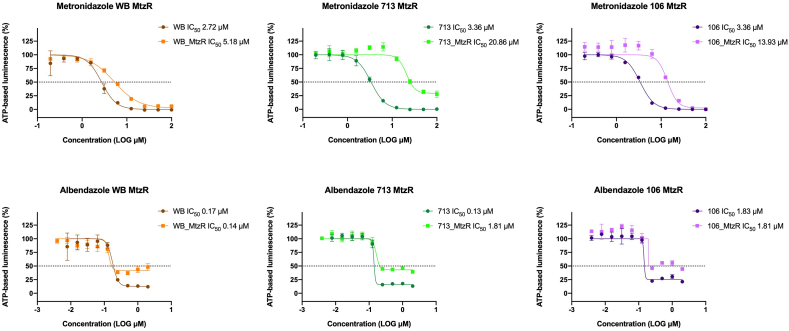
Based on this, we hypothesised cross-resistance phenotypes were one-directional: that MtzR could confer AlbR, but AlbR could not confer MtzR. To explore this we investigated Alb drug sensitivity in three isogenic MtzR lines derived from BRIS/83/HEPU/106, BRIS/87/HEPU/713 and WB-1B (Fig. 2), noting the latter's MtzR line is WB-M3 (Townson et al., 1992). Our independently derived Mtz IC50 values and RF for MtzR lines were consistent with previous reports (Ansell et al., 2017). However, Alb dose-response curves in MtzR lines demonstrated increased survival of parasites at drug concentrations of 2–0.25 μM. In particular, levels of ATP-based luminescence in the AlbR line (i.e. metabolically active trophozoites) were approximately half of uninhibited controls (Fig. 2), although no shift in the dose-response curves was observed, neither were differences observed in the Alb IC50 between MtzS and MtzR lines. This suggested MtzR isolates had increased tolerance or enhanced survivability for Alb, including in the WB-M3 line which had previously achieved high levels of AlbR in Upcroft et al. (1996). This may also explain why Upcroft et al. (1996) did not report β-tubulin mutations in the WB-M3-Alb line, as its AlbR mechanisms may have been derived from pre-existing mechanisms in its Mtz-resistant background. Based on similarity of the dose response curves but the differing viability at high concentration, we propose the survival of Giardia in MtzR lines exposed to Alb is a tolerance that is unrelated to Alb-selected resistance. Therefore, we hypothesised two broad AlbR phenotypes which accounted for observed directionality of cross-resistance: AlbR derived from MtzR (i.e., Mtz-dependent) and AlbR derived through BZ compounds (i.e., Alb-dependent and Mtz-independent). We proposed the latter of these was derived from a tubulin dependent mechanism with structural specificity consistent with known classes of CBS binding compounds.
Fig. 2.
Albendazole susceptibility in MtzR lines: IC50 dose response curves for albendazole and metronidazole in MtzR isogenic lines (Ansell et al., 2017) derived from WB-1B, BRIS/83/HEPU/106 and BRIS/87/HEPU/713. These demonstrate the expected shift in dose response for Mtz in all 3 isolates (above), and an enhanced survival in MtzR lines in Alb between substantial shift 2–0.25 μM, but no change in IC50 or shift in the dose response, indicative of enhanced survival at higher Alb concentration.
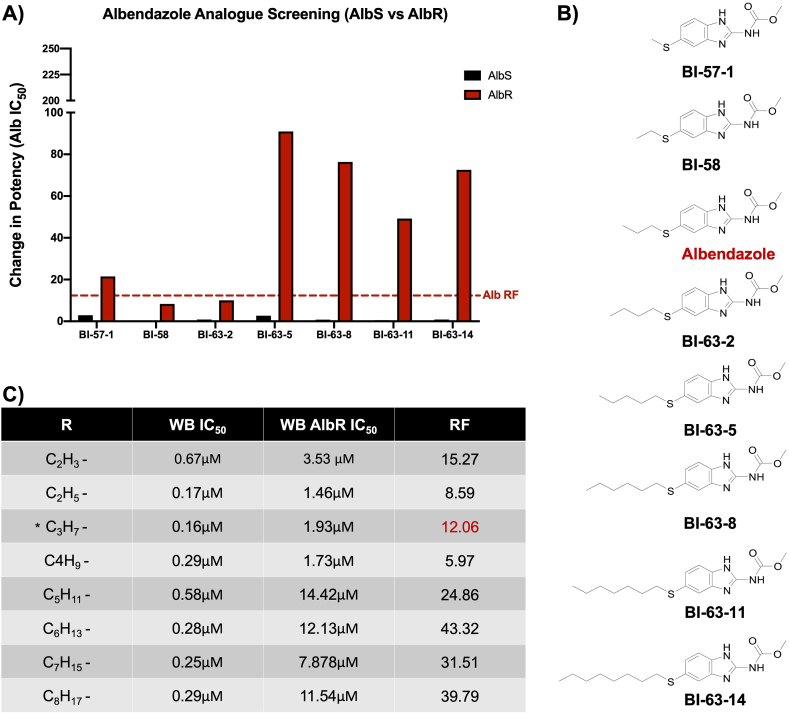
3.4. Albendazole structural modifications affect AlbR inhibitory activity and resistance factor
The insensitivity of the AlbR line to all BZ compounds, as well as another β-tubulin inhibitor, podophyllotoxin, proposed to share the same binding site, implicated a change in binding site conformation. We explored this in more detail using a range of Alb analogues to assess structural modifications of the Alb parent molecule for altered efficacy specific to the AlbR line (Supplementary Fig. 2). We calculated changed potency for analogues in AlbS and AlbR lines based on the Alb IC50 (0.16 μM) (Fig. 3A). This analysis included thioether substituents from 5-methylthio- (CH3S-) to 5-octylthio- (C8H17S-) both shorter and longer than Alb, which is the propylthio- (C3H7S-) (Fig. 3B). Overall, changes in potency were minimal in the AlbS line, where decreased potency ranged from only 0.05–2.9 μM (1.24 ± 1.02 μM, mean ± SD). However, in the AlbR line, increasing the size to C5H11S- to C8H17S- dramatically reduced potency, with RF exceeding those observed for Alb by at least two-fold (Fig. 3C). Decreased potency in these longer thioethers in the AlbR line, but not the AlbS, highlighted that the resistance mechanisms were dependent on the length or shape of the alkyl chains, which further credited a change in the AlbR binding site.
Fig. 3.
Screening of BZ analogues and Albendazole thioesters: A) Change in potency for BZ analogues based on the Alb IC50 (0.16 μM). Compounds include albendazole and 7 additional albendazole thioethers. B) Structures of albendazole carbamate thioethers (methylthio-to octylthio-) C) Table of IC50 in WB AlbS and AlbR lines and their respective resistance factors. albendazole (*) corresponds to the R group -SC3H7 in the thiolether series.
3.5. Molecular modelling of Alb and β-tubulin drug interactions overlaps with the CBS
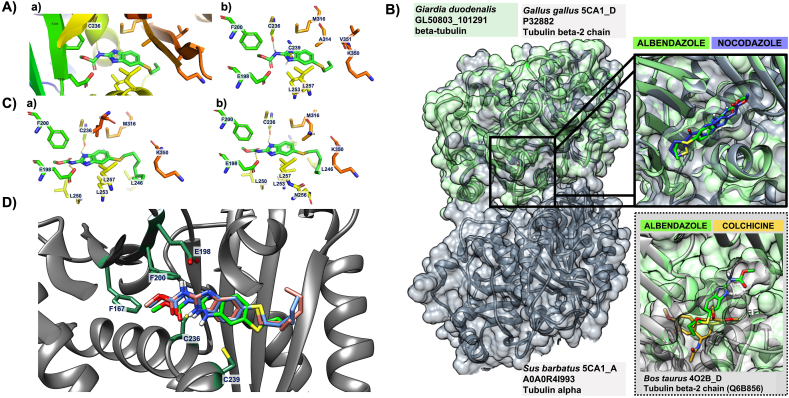
Our results from detailed drug-susceptibility screens implicated conserved β-tubulin mechanisms of BZs (Fig. 1), and a change in the Alb binding site in our AlbR line (Fig. 3). We contextualised these results through computational modelling which explored the BZ ligand binding site in Giardia β-tubulin, and how this site overlapped with the CBS together with mutations demonstrated in anthelmintic and anti-fungal resistance. Using the Giardia β-tubulin (GL50803_101291) protein sequence we identified β-tubulin templates through the COACH meta-server which (1) had ligand binding sites of interest (i.e. CBS), and (2) could be used to generate homology models of Giardia β-tubulin through SWISS-MODEL (Supplementary Table 1). Of the three templates the highest GMQE score (Global Model Quality Estimation) for accuracy of the tertiary structure was the β-tubulin template 5C8Y_D, a crystal structure of tubulin complexed with another CBS inhibitor, Plinabulin (Wang et al., 2016).
Based on the 5C8Y_D template, COACH predicted the binding site residues in the model of Giardia β-tubulin. These included Y50, Q134, C165, F167, E198, F200, C236, T237, C239, L240, L246, L250, L253, L257, A314, A315, M316, K350, V351, S352, I368. Many of these residues were highly conserved across Giardia, helminths and fungal species; all of which are susceptible to BZ compounds or related classes (Supplementary Fig. 3). We noted in our model, and similar to a previous Giardia homology model (Aguayo-Ortiz et al., 2013b), multiple cysteines in the predicted binding site (which can act as hydrogen bond donors). More specifically, three cysteines (C165, C236, C239) occur in the predicted Giardia β-tubulin binding site, of which only C239 is conserved in other BZ-susceptible organisms (Supplementary Fig. 3). All residues in the predicted binding site in the 3D structure of Giardia β-tubulin occurred at the tubulin heterodimer interface and overlapped the conserved CBS (Wang et al., 2016). They also overlapped the of single nucleotide polymorphisms (SNPs) in anthelmintic and anti-fungal BZ resistance (F167, E198, F200) (Lubega and Prichard, 1991; Elard and Humbert, 1999; Rufener et al., 2009; Beech et al., 2011), as well as residues predicted in both molecular models of BZ-binding in helminths (Robinson et al., 2004; Aguayo-Ortiz et al., 2013a) and BZ binding in fungal species (Cai et al., 2015; Vela-Corcía et al., 2018).
We docked Alb to the Giardia β-tubulin model using AutoDock Vina. The binding mode of the Alb was selected based on lowest energy conformation and preferable hydrogen-bond formation, noting that there is no experimental binding affinity that can be used for direct comparison with the calculated binding energy. The best docked pose of Alb had a binding affinity −5.6 kcal/mol and formed a hydrogen bond with the Giardia-specific C236 residue (Fig. 4A; Supplementary Fig. 3). This bond occurred between the carbamate and C236 in Giardia, whereas in helminths species the similar bonds between C239 occurred with the BZ 5-substituents (Aguayo-Ortiz et al., 2013a, 2013b), resulting in differences in BZ position and orientation between these BZ-susceptible organisms. We compared our model of Alb docked to Giardia β-tubulin with an experimentally derived crystal structure of αβ-tubulin heterodimers complexed with nocodazole (PDB ID: 4O2B_C/D) (Wang et al., 2016), as well as colchicine (PDB ID: 4O2B_C/D) (Prota et al., 2014) (Fig. 4B), which highlighted that docked Alb occurred at the αβ-tubulin interface, where it would disrupt protofilament formation and favour depolymerisation. Furthermore, Alb in our Giardia models occurred in similar orientation and position as nocodazole derived from crystal structures, and partially overlapped with the binding position of colchicine. These predictions are consistent with crystal structures (Wang et al., 2016), and molecular models (Aguayo-Ortiz et al., 2013a), which highlight the BZ binding site as separate, but in close proximity to, the CBS.
Fig. 4.
Molecular modelling of Alb with Giardia β-tubulin A) Illustrations of the binding mode of albendazole with Giardia β-tubulin. Surface view to represent binding pocket for Alb (left) and with the ribbon structure demonstrating polar contact (middle), with C236 and its hydrogen bond interaction indicated with an arror. The predicted binding modes of Alb inside the proposed binding site with the amino acid residues involved in hydrogen bond formation and non-bonded interaction are shown on the right. B) Overlapped binding sites of the experimentally derived crystal structure of T2R-TTL-nocodazole complex (PDB ID: 5CA1_C/D) shown in grey and our model of G. duodenalis β-tubulin shown in green. A close-up inserted image in the top right compared the orientation of nocodazole (blue) from the crystal structure (5CA1_D) and the docked Alb (green) structure from our molecular modelling of Giardia, and demonstrates that nocodozole and Alb share the same binding position and direction. In contrast, the inserted structure on the bottom left of the experimentally derived crystal structure of the tubulin-colchicine complex (PDB ID: 4O2B_C/D) in grey shows that the colchicine (yellow) molecule partially overlaps with docked Alb (green) in our structure from Giardia. C) Ribbon structures demonstrating polar contact between Giardia β-tubulin and heptylthioether (BI-63-8; left) and octyl-thiol (BI-63-14; right) analogues associated with decreased potency in the AlbR line. Orientation of the molecular is the same as in Alb, but feature an additional hydrogen bond with E198 for both molecules as indicated by the arrow.
Besides C326, no additional hydrogen bonds were observed in our docked Alb models. Although all residues linked to anthelmintic and anti-fungal BZ resistance were in close proximity to Alb in our models, we only observed an additional non-bonded interaction with F200. Despite a different docked pose of Alb (binding energy −5.3 kcal/mol) predicted non-bonded interactions with F167, E198 and F200, none formed hydrogen bond interactions (Supplementary Fig. 4). However, when we modelled binding modes of two Alb thioether analogues (Fig. 3), a single hydrogen bond was formed between thioethers and E198 with the carbamate (Fig. 4C). These docked hexyl- and octyl-thioethers, both which had decreased potency in the AlbR line (Fig. 4C), had similar binding affinities to Alb, calculated at −5.1 kcal/mol and −5.2 kcal/mol, respectively. The position and orientation of these analogues were the same as Alb (Fig. 4D), although the hydrogen bond with C236 shifted to the benzimidazole (Fig. 4A). Overall, these models highlighted residues associated with SNPs in BZ resistance were located within the binding site, and that some Alb analogues interacted with E198. Bonded interactions with E198 and BZ compounds have been reported in experimental and modelled structures (Robinson et al., 2004; Aguayo-Ortiz et al., 2013a; Wang et al., 2016), and E198 mutations are common in both anthelmintic resistance (Rufener et al., 2009) and anti-fungal resistance (Cai et al., 2015; Liu et al., 2019).
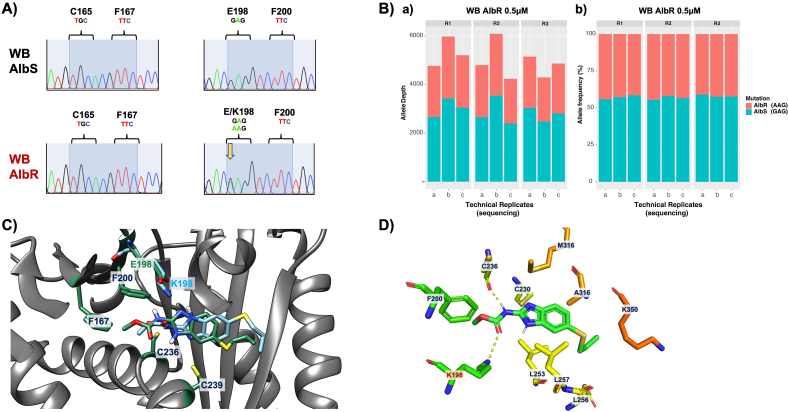
3.6. Identification of E198K mutation in β-tubulin sequences from the AlbR line
Our bioassay and molecular models support the hypothesis that the AlbR line had undergone a mutation in its β-tubulin sequence. To investigate this, we performed capillary and amplicon sequencing of a 600 bp region of Giardia β-tubulin as based on Arguello-Garcia et al. (2009). Firstly, chromatograms from capillary sequencing of Giardia β-tubulin were inspected, including nucleotides corresponding to amino acids 50, 134, 165, 167, 198 and 200, which are residues reported in BZ resistance in other species. From this we identified a clear heterozygous SNP at position 440 of G/A in each AlbR replicate, whereas only G occurred in DMSO replicates. This altered the sequence from GAG to AAG, and resulted in a E198K mutation (Fig. 5A). No other SNPs were observed (Supplementary Fig. 5). We performed variant calling analyses of amplicon sequences to confirm these SNPs, and detected the G592A SNP which encoded the E198K mutation in the nucleotide sequence of all three AlbR samples, but not the AlbS samples (Fig. 5B).
Fig. 5.
Identification and modelling of an E198K mutation in tha AlbR line A) Representative sequence chromatograms from capillary sequencing of Giardia β-tubulin from AlbS and AlbR lines highlighting nucleotides corresponding to amino acids 165, 167, 198 and 200. A double peak at nucleotide position 440 G/A is highlighted with a yellow arrow in the AlbR isolate is indicative of a single nucleotide polymorphism which would correspond to a E198K mutation. Chromatograms from the three technical replicates in each AlbR and AlbS line can be seen in Supplementary Fig. S5. B) Results of amplicon sequencing analysing the G592A SNP in the AlbS and AlbR lines in replicates from in vitro culture (R1, R2, R3), as well as sequencing replicates (a, b, c). The figure shows both the number of reads aligned at this position for the G592A SNP (a), as well as the relative frequency of the G592A SNP (b). C) Orientation of Alb in the binding pocket where E198 is present (green) compared to K198 (blue), the latter which binds slightly shallower and in a twisted orientation. Side chains of other key residues are also shown, including cysteines which can be hydrogen bond donors, and amino acids implicated in anthelmintic resistance. D) Ribbon structures demonstrating polar contact between Giardia β-tubulin and albendazole with the E198K mutation. An additional hydrogen bond with K198 is observed, as is the hydrogen bond with C236, which was observed with Alb in models with E198.
To our knowledge, this is the first report of a β-tubulin mutation in protistan BZ-resistance. To understand the molecular impacts of mutation at E198K for BZ-binding, we performed additional modelling for Alb and Giardia β-tubulin with the E198K mutation (Fig. 5C). We noted the longer side chain of lysine and pronounced increased basicity compared to shorter and acidic glutamate would have impacted both the size and ionisation balance within the binding pocket, and we observed Alb docked in a shallower position with a different orientation in our K198 model. Both E198 and K198 models of docked Alb favoured the hydrogen bond between C236 and the Alb carbamoyl moiety (Figs. 3A and 5C), although we observed a new hydrogen bond between K198 and the carbamate in the K198 docked albendazole (Fig. 5D). The presence of two hydrogen bonds resulted in a slightly higher binding affinity −6.0 kcal/mol as compared to our E198 models (−5.6 kcal/mol), where only a single hydrogen bond was observed with C236 and docked Alb.
Given our first models of Alb binding Giardia β-tubulin did not favour two E198 hydrogen bonds with BZ compounds, similar binding affinities between E198 and K198 models were expected. In contrast, molecular models which reported two E198 hydrogen bonds with BZ compounds also reported lower binding affinities in models with E198 mutations. In particular, the E198A mutation in helminths resulted in loss of two E198 hydrogen bonds in the A198 mutant (Aguayo-Ortiz et al., 2013a, 2013b), whereas fungi with E198K mutations reported reduction to only one, less favourable, hydrogen bond with the K198 mutant (Cai et al., 2015). Although our hydrogen bond interactions were dissimilar to these previous models, the E198K mutation altered the binding orientation, position and affinity of Alb in our models, indicative of changes to interactions between key residues which bind and orientate the BZ within the β-tubulin site (Aguayo-Ortiz et al., 2013a, 2013b) (Fig. 6).
fig. 6.
BZ-resistance phenotypes and genotypes across AlbR induction. A) IC50 dose response curves for albendazole, mebendazole, parbendazole and oxibendendazole in AlbS and AlbR isogenic lines grown at 0.21 μM, 0.35 μM and 0.5 μM drug, demonstrating a shift in IC50 in all the AlbR lines relative to Alb concentration. B) Results of amplicon sequencing analysing the G592A SNP in the 0.21 μM and the 0.35 μM AlbR line in replicates from in vitro culture (R1, R2, R3), as well as sequencing replicates (a, b, c). The figure shows both the number of reads aligned at this position for the G592A SNP (a), as well as the relative allele frequency of the G592A SNP (b). C) Correlation between BZ-resistance phenotypes and the G592A SNP in the 0.21 μM, 0.35 μM and 0.5 μM lines. The left axis for the bar chart demonstrates the calculated RF for the AlbR lines, whereas the right axis for the violin plot demonstrates the allele frequency from in vitro culture as well as sequencing replicates, with statistical significance established via Wilcoxon signed-rank test.
4. Discussion
BZs have been widely successful as antiparasitic and antifungal drugs in medical, veterinary and agrochemical fields for several decades. These compounds are well-tolerated, highly effective, often in single doses, and extremely low-cost. For Giardia, BZs are one of the few alternatives to frontline nitroheterocyclics, including Mtz or tinidazole. As Mtz-resistance increases in frequency (Nabarro et al., 2015; Lalle and Hanevik, 2018) the reports of clinical Alb resistance (Farbey et al., 1995; Lemee et al., 2000) are a pressing concern. Yet, our understanding of the underlying mechanisms of BZ-resistance in Giardia are poorly understood. Most importantly, while the evidence favours a conserved BZ-tubulin mechanism of action in Giardia (Chavez et al., 1992; MacDonald et al., 2004), the link between BZ-resistance and tubulin in Giardia has received little attention compared to our understanding of β-tubulin mutations in BZ-resistant strains of nematodes and fungi.
Upon review, the lack of data exploring BZ resistance in Giardia has been driven by challenges associated with its induction. Multiple studies report unsuccessful attempts to culture trophozoites after in vitro Alb exposure (i.e. pulse exposure) (Meloni et al., 1990; Chavez et al., 1992) and the failure to induce AlbR in vitro (Morgan et al., 1993; Upcroft et al., 1996). While, among the successful attempts at in vitro selection of AlbR Giardia (Lindquist, 1996; Upcroft et al., 1996; Jimenez-Cardoso et al., 2004; Arguello-Garcia et al., 2009) there are no consistent predictors of success, such as selection method (pulse vs. sublethal), along with large variability in the adaptation times and maximum doses tolerated. This variability in AlbR induction could indicate potential variation in resistance mechanisms among lab lines, which is common in MtzR (Ansell et al., 2015). However in contrast to Mtz, all in vitro stable and highly BZ-resistant lines are from the Giardia WB line or its clones (Lindquist, 1996; Upcroft et al., 1996; Arguello-Garcia et al., 2009), whereas in vitro Mtz-resistance has been readily generated across many genome, lab and clinical isolates (Ansell et al., 2015).
Overall, our data were consistent with β-tubulin dependent mechanism of BZ-resistance in Giardia, and our bioassay, computational modelling and sequencing data collectively implicated a change in the BZ site of action. This data included overlapping resistances across BZ 5-position structural variants, as well as cross-resistance to structurally independent CBS inhibitors. This highlighted these molecules could not be accommodated or correctly orientated in a shared binding site. Computational modelling predicted the BZ binding site as overlapping the CBS at the αβ-tubulin interface, in agreement with by our studies of BZ structural activity relationships (SAR), and consistent with our identification of a known BZ-resistance allele in this predicted BZ-binding site. However, we also observed several species-specific differences in the Giardia CBS that made it difficult to resolve the exact orientation and position of BZ-binding in Giardia, and further affected our modelling of the impacts of the identified mutation. Notably, multiple cysteines which can interact with BZs occur in the predicted Giardia binding site, including C165 and C236, which do not occur in other BZ-susceptible species. In these other species, C239 is predicted to interact with the carbonyl, sulfur or sulfoxide groups of BZ 5-position substituents (Aguayo-Ortiz et al., 2013a; Wang et al., 2016), whereas our models consistently favoured C236 interactions. A previous homology model of Giardia β-tubulin with BZ compounds predicted these interactions instead with C165, resulting in a different orientation of the BZ molecule (Aguayo-Ortiz et al., 2013b). The main impact between C236 hydrogen bonding as compared to C165 in Giardia was the formation of two additional hydrogen bonds between E198 in models favouring C165. We further note two hydrogen bonds for E198 have been observed in models of helminths with BZ compounds (Aguayo-Ortiz et al., 2013a, 2013b), models of fungi with carbendazim (Cai et al., 2015), and in experimentally derived crystal structures of BZ-tubulin complexes (Wang et al., 2016). The absence of these bonds in our models affected our downstream modelling of the identified E198K mutation, as this mutation should reduce hydrogen bonding capacity to a single, less favourable bond (Cai et al., 2015). Therefore, resolving the Giardia-specific model of BZ-binding relative to unique amino acids in its CBS is required to complete our understanding of the molecular basis BZ-resistance based on β-tubulin mechanisms.
Our identification of an E198K mutation is the first report of a β-tubulin mutation in Giardia BZ-resistance. Importantly, this mutation in Giardia is not a primary site of mutation associated with antihelmintic resistance (i.e., F167Y, E198A, F200Y) (Lubega and Prichard, 1991; Kwa et al., 1994; Beech et al., 2011), but a common mutational variant conferring anti-fungal resistance to the parent BZ, carbendazim (Yarden and Katan, 1993; Banno et al., 2008; Cai et al., 2015; Liu et al., 2019). In fungi, E198K mutations are specifically associated with higher levels of carbendazim resistance compared to the other E198 mutations (E198A, E198V), and cross-resistance to other β-tubulin inhibitors (Cai et al., 2015). Our AlbR line also displayed high resistance factors to all BZ compounds tested, and cross-resistance to podophyllotoxin. Therefore, not only are our results the first evidence of conserved BZ-tubulin mechanisms in Giardia BZ-resistance, they also suggest mutations may not coincide with those observed in anthelmintic resistance. This could reflect the differences in the BZ binding pocket of Giardia β-tubulin compared to helminths, noted herein and previous studies (Aguayo-Ortiz et al., 2013b). Unfortunately, investigations of β-tubulin mutations are few in the context of anti-giardial resistance (Upcroft et al., 1996; Argüello-García et al., 2015), and therefore more sequencing data is required to confirm if E198K is a favoured mutation, or if additional mutated alleles occur at E198 or elsewhere, and in what proportions these occur.
Based on observations of enhanced survivability of MtzR isolates in Alb, we hypothesised MtzR backgrounds may enhance acquisition of Alb resistance. Intriguingly, cross-resistance was directional, and AlbR could not confer MtzR. While the molecular basis of this cross-resistance requires exploration, it is possible that complex MtzR phenotypes could provide metabolic routes reducing the toxicity of BZ compounds, given MtzR parasites have significant differential activity and expression of glycolytic and antioxidant enzymes (Ansell et al., 2015, 2017; Emery et al., 2018). Notably, Argüello-García et al. (2015) have previously demonstrated reductions in Alb sulfoxide and sulfone metabolites in AlbR lines which have elevated antioxidant responses, but lack β-tubulin mutations. Theoretically, similar altered Alb metabolism could occur in MtzR lines and enhance emergence of ‘persister’ cells (Barrett et al., 2019), producing Alb-refractory populations that may underpin chronic infections and treatment failures. This hypothesis is concerning given clinical MtzR and AlbR correlations have been reported which could correspond to our observed in vitro Mtz-dependent AlbR. In particular, low cure rates (>20%) have been demonstrated for Alb in patients which failed nitroheterocyclic treatment (Meltzer et al., 2014), and co-occurrence of AlbR and MtzR in clinical isolates (Lemee et al., 2000). While β-tubulin markers for AlbR would help dissect these cross-resistances, we note that consistent and robust MtzR markers for Giardia are also required, and such markers have proven indefinable despite multiple dedicated explorations.
Prior to this study, no SNPs had been identified in the β-tubulin gene in BZ-resistant Giardia. We propose this has been partly due to under-sampling, and highlighted extenuating factors regarding sequences from MtzR backgrounds. However, given the complex microtubule system in this parasite, there certainly may be alternative targets for BZ drugs and alternative mechanisms of resistance. Indeed, Argüello-García et al. (2015) have highlighted metabolic AlbR phenotypes operating in absence of β-tubulin mutations, and we further hypothesise the possibility of non-genetic mechanisms which preceded the acquisition of the identified β-tubulin mutations during our Alb-selection as possible ‘persister’ phenotypes (Barrett et al., 2019). Nonetheless, our data were wholly consistent with β-tubulin as both the BZ-target and BZ-resistance mechanism in Giardia, and note BZ-resistant Giardia share common mutations with anti-fungal, but not anthelmintic, BZ-resistance. Currently, several loop-mediated isothermal amplification (LAMP) assays are able to rapidly and sensitively detect mutated E198 alleles in fungi in the field (Fan et al., 2019, Liu et al., 2019), highlighting potential that similar approaches could be optimised for clinical surveillance of Giardia BZ-resistance. Based on this, we suggest efforts be consolidated to elucidate the frequency and diversity of mutational alleles in Giardia tubulin towards validating markers for clinical diagnosis and surveillance of drug-resistant strains.
Declaration of competing interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: All authors listed have contributed to the research, have no conflict of interest or competing interests to declare, and have approved the manuscript for submission to IJPDDR. The funders of this study have had no role in its design, data collection and interpretation, and are listed in the manuscript’s acknowledgements.
Acknowledgements
SJE, QS, ST, LB and AJ are supported by the Victorian State Government Operational Infrastructure Support and Australian Government National Health and Medical Research Council Independent Research Institute Infrastructure Support Scheme. AJ is supported by an NHMRC Career Development Fellowship (APP1126395).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ijpddr.2021.05.003.
Supplementary Information
All supplementary data, including Supplementary Table 1 and Supplementary Figs. 1–5, have been compiled and can be found in the Supplementary Information.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Agarwal S.K., Sharma S., Bhaduri A.P., Katiyar J.C., Chatterjee R.K. Segregation of activity profile in benzimidazoles: effect of spacers at 5(6)-position of methyl benzimidazole-2-carbamates [1] Z. Naturforsch. C Biosci. 1993;48:829. [Google Scholar]
- Aguayo-Ortiz R., Mendez-Lucio O., Medina-Franco J.L., Castillo R., Yepez-Mulia L., Hernandez-Luis F., Hernandez-Campos A. Towards the identification of the binding site of benzimidazoles to beta-tubulin of Trichinella spiralis: insights from computational and experimental data. J. Mol. Graph. Model. 2013;41:12–19. doi: 10.1016/j.jmgm.2013.01.007. [DOI] [PubMed] [Google Scholar]
- Aguayo-Ortiz R., Méndez-Lucio O., Romo-Mancillas A., Castillo R., Yépez-Mulia L., Medina-Franco J.L., Hernández-Campos A. Molecular basis for benzimidazole resistance from a novel β-tubulin binding site model. J. Mol. Graph. Model. 2013;45:26–37. doi: 10.1016/j.jmgm.2013.07.008. [DOI] [PubMed] [Google Scholar]
- Ansell B.R., Baker L., Emery S.J., McConville M.J., Svard S.G., Gasser R.B., Jex A.R. Transcriptomics indicates active and passive metronidazole resistance mechanisms in three seminal giardia lines. Front. Microbiol. 2017;8:398. doi: 10.3389/fmicb.2017.00398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansell B.R., McConville M.J., Ma'ayeh S.Y., Dagley M.J., Gasser R.B., Svard S.G., Jex A.R. Drug resistance in Giardia duodenalis. Biotechnol. Adv. 2015;33:888–901. doi: 10.1016/j.biotechadv.2015.04.009. [DOI] [PubMed] [Google Scholar]
- Ansell B.R.E., Pope B.J., Georgeson P., Emery-Corbin S.J., Jex A.R. Annotation of the Giardia proteome through structure-based homology and machine learning. GigaScience. 2019;8 doi: 10.1093/gigascience/giy150. giy150-giy150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argüello-García R., Cruz-Soto M., González-Trejo R., Paz-Maldonado L.M.T., Bazán-Tejeda M.L., Mendoza-Hernández G., Ortega-Pierres G. An antioxidant response is involved in resistance of Giardia duodenalis to albendazole. Front. Microbiol. 2015;6 doi: 10.3389/fmicb.2015.00286. 286-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arguello-Garcia R., Cruz-Soto M., Romero-Montoya L., Ortega-Pierres G. In vitro resistance to 5-nitroimidazoles and benzimidazoles in Giardia duodenalis: variability and variation in gene expression. Infect. Genet. Evol. 2009;9:1057–1064. doi: 10.1016/j.meegid.2009.05.015. [DOI] [PubMed] [Google Scholar]
- Aubrey B.J., Kelly G.L., Kueh A.J., Brennan M.S., O'Connor L., Milla L., Wilcox S., Tai L., Strasser A., Herold M.J. An inducible lentiviral guide RNA platform enables the identification of tumor-essential genes and tumor-promoting mutations in vivo. Cell Rep. 2015;10:1422–1432. doi: 10.1016/j.celrep.2015.02.002. [DOI] [PubMed] [Google Scholar]
- Banno S., Fukumori F., Ichiishi A., Okada K., Uekusa H., Kimura M., Fujimura M. Genotyping of benzimidazole-resistant and dicarboximide-resistant mutations in Botrytis cinerea using real-time polymerase chain reaction assays. Phytopathology. 2008;98:397–404. doi: 10.1094/PHYTO-98-4-0397. [DOI] [PubMed] [Google Scholar]
- Barrett M.P., Kyle D.E., Sibley L.D., Radke J.B., Tarleton R.L. Protozoan persister-like cells and drug treatment failure. Nat. Rev. Microbiol. 2019;17:607–620. doi: 10.1038/s41579-019-0238-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beech R.N., Skuce P., Bartley D.J., Martin R.J., Prichard R.K., Gilleard J.S. Anthelmintic resistance: markers for resistance, or susceptibility? Parasitology. 2011;138:160–174. doi: 10.1017/S0031182010001198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boreham P.F., Phillips R.E., Shepherd R.W. The sensitivity of Giardia intestinalis to drugs in vitro. J. Antimicrob. Chemother. 1984;14:449–461. doi: 10.1093/jac/14.5.449. [DOI] [PubMed] [Google Scholar]
- Boreham P.F., Phillips R.E., Shepherd R.W. Altered uptake of metronidazole in vitro by stocks of Giardia intestinalis with different drug sensitivities. Trans. R. Soc. Trop. Med. Hyg. 1988;82:104–106. doi: 10.1016/0035-9203(88)90278-7. [DOI] [PubMed] [Google Scholar]
- Cai M., Lin D., Chen L., Bi Y., Xiao L., Liu X.L. M233I mutation in the beta-tubulin of Botrytis cinerea confers resistance to zoxamide. Sci. Rep. 2015;5 doi: 10.1038/srep16881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capon A.G., Upcroft J.A., Boreham P.F., Cottis L.E., Bundesen P.G. Similarities of Giardia antigens derived from human and animal sources. Int. J. Parasitol. 1989;19:91–98. doi: 10.1016/0020-7519(89)90026-x. [DOI] [PubMed] [Google Scholar]
- Carter E.R., Nabarro L.E., Hedley L., Chiodini P.L. Nitroimidazole-refractory giardiasis; a growing problem requiring rational solutions. Clin. Microbiol. Infect. 2017;24(1):37–42. doi: 10.1016/j.cmi.2017.05.028. [DOI] [PubMed] [Google Scholar]
- Chatterji B.P., Jindal B., Srivastava S., Panda D. Microtubules as antifungal and antiparasitic drug targets. Expert Opin. Ther. Pat. 2011;21:167–186. doi: 10.1517/13543776.2011.545349. [DOI] [PubMed] [Google Scholar]
- Chavez B., Cedillo-Rivera R., Martinez-Palomo A. Giardia lamblia: ultrastructural study of the in vitro effect of benzimidazoles. J. Protozool. 1992;39:510–515. doi: 10.1111/j.1550-7408.1992.tb04841.x. [DOI] [PubMed] [Google Scholar]
- Cortese F., Bhattacharyya B., Wolff J. Podophyllotoxin as a probe for the colchicine binding site of tubulin. J. Biol. Chem. 1977;252:1134–1140. [PubMed] [Google Scholar]
- Davids B., Gillin F. In: Methods for Giardia Culture, Cryopreservation, Encystation, and Excystation in Vitro. Luján H., Svärd S., editors. Springer Vienna; 2011. pp. 381–394. (Giardia). [Google Scholar]
- Dayan A.D. Albendazole, mebendazole and praziquantel. Review of non-clinical toxicity and pharmacokinetics. Acta Trop. 2003;86:141–159. doi: 10.1016/s0001-706x(03)00031-7. [DOI] [PubMed] [Google Scholar]
- Edlind T.D., Hang T.L., Chakraborty P.R. Activity of the anthelmintic benzimidazoles against Giardia lamblia in vitro. J. Infect. Dis. 1990;162:1408–1411. doi: 10.1093/infdis/162.6.1408. [DOI] [PubMed] [Google Scholar]
- Elard L., Humbert J.F. Importance of the mutation of amino acid 200 of the isotype 1 β-tubulin gene in the benzimidazole resistance of the small-ruminant parasite Teladorsagia circumcincta. Parasitol. Res. 1999;85:452–456. doi: 10.1007/s004360050577. [DOI] [PubMed] [Google Scholar]
- Emery S.J., Baker L., Ansell B.R.E., Mirzaei M., Haynes P.A., McConville M.J., Svard S.G., Jex A.R. Differential protein expression and post-translational modifications in metronidazole-resistant Giardia duodenalis. GigaScience. 2018;7 doi: 10.1093/gigascience/giy024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Fei, Hahn Matthias, Li Guo-Qing, Lin Yang, Luo Chao-Xi. Rapid detection of benzimidazole resistance in Botrytis cinerea by loop-mediated isothermal amplification. Phytopathology Research. 2019;1(1):10. doi: 10.1186/s42483-019-0016-8. [DOI] [Google Scholar]
- Farbey M.D., Reynoldson J.A., Thompson R.C. In vitro drug susceptibility of 29 isolates of Giardia duodenalis from humans as assessed by an adhesion assay. Int. J. Parasitol. 1995;25:593–599. doi: 10.1016/0020-7519(94)00174-m. [DOI] [PubMed] [Google Scholar]
- Gardner T.B., Hill D.R. Treatment of giardiasis. Clin. Microbiol. Rev. 2001;14:114–128. doi: 10.1128/CMR.14.1.114-128.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghisi M., Kaminsky R., Maser P. Phenotyping and genotyping of Haemonchus contortus isolates reveals a new putative candidate mutation for benzimidazole resistance in nematodes. Vet. Parasitol. 2007;144:313–320. doi: 10.1016/j.vetpar.2006.10.003. [DOI] [PubMed] [Google Scholar]
- Hagen K.D., McInally S.G., Hilton N.D., Dawson S.C. Microtubule organelles in giardia. Adv. Parasitol. 2020;107:25–96. doi: 10.1016/bs.apar.2019.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanevik K., Wensaas K.A., Rortveit G., Eide G.E., Morch K., Langeland N. Irritable bowel syndrome and chronic fatigue 6 years after giardia infection: a controlled prospective cohort study. Clin. Infect. Dis. 2014;59:1394–1400. doi: 10.1093/cid/ciu629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennessey K.M., Alas G.C.M., Rogiers I., Li R., Merritt E.A., Paredez A.R. Nek8445, a protein kinase required for microtubule regulation and cytokinesis in Giardia lamblia. Mol. Biol. Cell. 2020;31:1611–1622. doi: 10.1091/mbc.E19-07-0406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunskar G.S., Bjorvatn B., Wensaas K.A., Hanevik K., Eide G.E., Langeland N., Rortveit G. Excessive daytime sleepiness, sleep need and insomnia 3 years after Giardia infection: a cohort study. Sleep Health. 2016;2:154–158. doi: 10.1016/j.sleh.2016.03.005. [DOI] [PubMed] [Google Scholar]
- Jimenez-Cardoso E., Flores-Luna A., Perez-Urizar J. In vitro activity of two phenyl-carbamate derivatives, singly and in combination with albendazole against albendazole-resistant Giardia intestinalis. Acta Trop. 2004;92:237–244. doi: 10.1016/j.actatropica.2004.08.003. [DOI] [PubMed] [Google Scholar]
- Katiyar S.K., Gordon V.R., McLaughlin G.L., Edlind T.D. Antiprotozoal activities of benzimidazoles and correlations with beta-tubulin sequence. Antimicrob. Agents Chemother. 1994;38:2086–2090. doi: 10.1128/aac.38.9.2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeling P.J., Doolittle W.F. Alpha-tubulin from early-diverging eukaryotic lineages and the evolution of the tubulin family. Mol. Biol. Evol. 1996;13:1297–1305. doi: 10.1093/oxfordjournals.molbev.a025576. [DOI] [PubMed] [Google Scholar]
- Kotloff K.L., Nataro J.P., Blackwelder W.C., Nasrin D., Farag T.H., Panchalingam S., Wu Y., Sow S.O., Sur D., Breiman R.F. Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): a prospective, case-control study. Lancet. 2013;382:209–222. doi: 10.1016/S0140-6736(13)60844-2. [DOI] [PubMed] [Google Scholar]
- Kwa M.S.G., Veenstra J.G., Roos M.H. Benzimidazole resistance in Haemonchus contortus is correlated with a conserved mutation at amino acid 200 in β-tubulin isotype 1. Mol. Biochem. Parasitol. 1994;63:299–303. doi: 10.1016/0166-6851(94)90066-3. [DOI] [PubMed] [Google Scholar]
- Lacey E. The role of the cytoskeletal protein, tubulin, in the mode of action and mechanism of drug resistance to benzimidazoles. Int. J. Parasitol. 1988;18:885–936. doi: 10.1016/0020-7519(88)90175-0. [DOI] [PubMed] [Google Scholar]
- Lacey E., Edgar J.A., Culvenor C.C. Interaction of phomopsin A and related compounds with purified sheep brain tubulin. Biochem. Pharmacol. 1987;36:2133–2138. doi: 10.1016/0006-2952(87)90141-9. [DOI] [PubMed] [Google Scholar]
- Lacey E., Prichard R.K. Interactions of benzimidazoles (BZ) with tubulin from BZ-sensitive and BZ-resistant isolates of Haemonchus contortus. Mol. Biochem. Parasitol. 1986;19:171–181. doi: 10.1016/0166-6851(86)90122-2. [DOI] [PubMed] [Google Scholar]
- Lalle M., Hanevik K. Treatment-refractory giardiasis: challenges and solutions. Infect. Drug Resist. 2018;11:1921–1933. doi: 10.2147/IDR.S141468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane S., Lloyd D. Current trends in research into the waterborne parasite Giardia. Crit. Rev. Microbiol. 2002;28:123–147. doi: 10.1080/1040-840291046713. [DOI] [PubMed] [Google Scholar]
- Lemee V., Zaharia I., Nevez G., Rabodonirina M., Brasseur P., Ballet J.J., Favennec L. Metronidazole and albendazole susceptibility of 11 clinical isolates of Giardia duodenalis from France. J. Antimicrob. Chemother. 2000;46:819–821. doi: 10.1093/jac/46.5.819. [DOI] [PubMed] [Google Scholar]
- Leroux P., Chapeland F., Desbrosses D., Gredt M. Patterns of cross-resistance to fungicides in Botryotinia fuckeliana (Botrytis cinerea) isolates from French vineyards. Crop Protect. 1999;18:687–697. [Google Scholar]
- Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindquist H.D. Induction of albendazole resistance in Giardia lamblia. Microb. Drug Resist. 1996;2:433–434. doi: 10.1089/mdr.1996.2.433. [DOI] [PubMed] [Google Scholar]
- Litleskare S., Rortveit G., Eide G.E., Hanevik K., Langeland N., Wensaas K.A. Prevalence of irritable bowel syndrome and chronic fatigue 10 Years after giardia infection. Clin. Gastroenterol. Hepatol. 2018;16:1064–1072. doi: 10.1016/j.cgh.2018.01.022. e1064. [DOI] [PubMed] [Google Scholar]
- Liu Y.H., Yuan S.K., Hu X.R., Zhang C.Q. Shift of sensitivity in Botrytis cinerea to benzimidazole fungicides in strawberry greenhouse ascribing to the rising-lowering of E198A subpopulation and its visual, on-site monitoring by loop-mediated isothermal amplification. Sci. Rep. 2019;9 doi: 10.1038/s41598-019-48264-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubega G.W., Prichard R.K. Specific interaction of benzimidazole anthelmintics with tubulin from developing stages of thiabendazole-susceptible and -resistant Haemonchus contortus. Biochem. Pharmacol. 1991;41:93–101. doi: 10.1016/0006-2952(91)90015-w. [DOI] [PubMed] [Google Scholar]
- MacDonald L.M., Armson A., Thompson A.R., Reynoldson J.A. Characterisation of benzimidazole binding with recombinant tubulin from Giardia duodenalis, Encephalitozoon intestinalis, and Cryptosporidium parvum. Mol. Biochem. Parasitol. 2004;138:89–96. doi: 10.1016/j.molbiopara.2004.08.001. [DOI] [PubMed] [Google Scholar]
- Manning G., Reiner D.S., Lauwaet T., Dacre M., Smith A., Zhai Y., Svard S., Gillin F.D. The minimal kinome of Giardia lamblia illuminates early kinase evolution and unique parasite biology. Genome Biol. 2011;12:R66. doi: 10.1186/gb-2011-12-7-r66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariante R.M., Vancini R.G., Melo A.L., Benchimol M. Giardia lamblia: evaluation of the in vitro effects of nocodazole and colchicine on trophozoites. Exp. Parasitol. 2005;110:62–72. doi: 10.1016/j.exppara.2005.01.007. [DOI] [PubMed] [Google Scholar]
- McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., DePristo M.A. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLoughlin E.C., O'Boyle N.M. Colchicine-binding site inhibitors from chemistry to clinic: a review. Pharmaceuticals. 2020;13 doi: 10.3390/ph13010008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meloni B.P., Thompson R.C., Reynoldson J.A., Seville P. Albendazole: a more effective antigiardial agent in vitro than metronidazole or tinidazole. Trans. R. Soc. Trop. Med. Hyg. 1990;84:375–379. doi: 10.1016/0035-9203(90)90324-8. [DOI] [PubMed] [Google Scholar]
- Meltzer E., Lachish T., Schwartz E. Treatment of giardiasis after nonresponse to nitroimidazole. Emerg. Infect. Dis. 2014;20:1742–1744. doi: 10.3201/eid2010.140073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan U.M., Reynoldson J.A., Thompson R.C. Activities of several benzimidazoles and tubulin inhibitors against Giardia spp. in vitro. Antimicrob. Agents Chemother. 1993;37:328–331. doi: 10.1128/aac.37.2.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris G.M., Huey R., Lindstrom W., Sanner M.F., Belew R.K., Goodsell D.S., Olson A.J. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J. Comput. Chem. 2009;30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabarro L.E., Lever R.A., Armstrong M., Chiodini P.L. Increased incidence of nitroimidazole-refractory giardiasis at the hospital for tropical diseases, london: 2008-2013. Clin. Microbiol. Infect. 2015;21:791–796. doi: 10.1016/j.cmi.2015.04.019. [DOI] [PubMed] [Google Scholar]
- Paz-Maldonado M.T., Arguello-Garcia R., Cruz-Soto M., Mendoza-Hernandez G., Ortega-Pierres G. Proteomic and transcriptional analyses of genes differentially expressed in Giardia duodenalis clones resistant to albendazole. Infect. Genet. Evol. 2013;15:10–17. doi: 10.1016/j.meegid.2012.08.021. [DOI] [PubMed] [Google Scholar]
- Prota A.E., Danel F., Bachmann F., Bargsten K., Buey R.M., Pohlmann J., Reinelt S., Lane H., Steinmetz M.O. The novel microtubule-destabilizing drug BAL27862 binds to the colchicine site of tubulin with distinct effects on microtubule organization. J. Mol. Biol. 2014;426:1848–1860. doi: 10.1016/j.jmb.2014.02.005. [DOI] [PubMed] [Google Scholar]
- Robinson M.W., McFerran N., Trudgett A., Hoey L., Fairweather I. A possible model of benzimidazole binding to beta-tubulin disclosed by invoking an inter-domain movement. J. Mol. Graph. Model. 2004;23:275–284. doi: 10.1016/j.jmgm.2004.08.001. [DOI] [PubMed] [Google Scholar]
- Rufener L., Kaminsky R., Mäser P. In vitro selection of Haemonchus contortus for benzimidazole resistance reveals a mutation at amino acid 198 of β-tubulin. Mol. Biochem. Parasitol. 2009;168:120–122. doi: 10.1016/j.molbiopara.2009.07.002. [DOI] [PubMed] [Google Scholar]
- Russell G.J., Gill J.H., Lacey E. Binding of [3H]benzimidazole carbamates to mammalian brain tubulin and the mechanism of selective toxicity of the benzimidazole anthelmintics. Biochem. Pharmacol. 1992;43:1095–1100. doi: 10.1016/0006-2952(92)90617-r. [DOI] [PubMed] [Google Scholar]
- Schonbrunn E., Phlippen W., Trinczek B., Sack S., Eschenburg S., Mandelkow E.M., Mandelkow E. Crystallization of a macromolecular ring assembly of tubulin liganded with the anti-mitotic drug podophyllotoxin. J. Struct. Biol. 1999;128:211–215. doi: 10.1006/jsbi.1999.4183. [DOI] [PubMed] [Google Scholar]
- Solaymani-Mohammadi S., Genkinger J.M., Loffredo C.A., Singer S.M. A meta-analysis of the effectiveness of albendazole compared with metronidazole as treatments for infections with Giardia duodenalis. PLoS Neglected Trop. Dis. 2010;4 doi: 10.1371/journal.pntd.0000682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townson S.M., Laqua H., Upcroft P., Boreham P.F., Upcroft J.A. Induction of metronidazole and furazolidone resistance in Giardia. Trans. R. Soc. Trop. Med. Hyg. 1992;86:521–522. doi: 10.1016/0035-9203(92)90095-t. [DOI] [PubMed] [Google Scholar]
- Trott O., Olson A.J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upcroft J., Mitchell R., Chen N., Upcroft P. Albendazole resistance in Giardia is correlated with cytoskeletal changes but not with a mutation at amino acid 200 in beta-tubulin. Microb. Drug Resist. 1996;2:303–308. doi: 10.1089/mdr.1996.2.303. [DOI] [PubMed] [Google Scholar]
- Van der Auwera G.A., Carneiro M.O., Hartl C., Poplin R., Del Angel G., Levy-Moonshine A., Jordan T., Shakir K., Roazen D., Thibault J., Banks E., Garimella K.V., Altshuler D., Gabriel S., DePristo M.A. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinformatics. 2013;43 doi: 10.1002/0471250953.bi1110s43. 11 10 11-11 10 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vela-Corcía D., Romero D., de Vicente A., Pérez-García A. Analysis of β-tubulin-carbendazim interaction reveals that binding site for MBC fungicides does not include residues involved in fungicide resistance. Sci. Rep. 2018;8:7161. doi: 10.1038/s41598-018-25336-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Zhang H., Gigant B., Yu Y., Wu Y., Chen X., Lai Q., Yang Z., Chen Q., Yang J. Structures of a diverse set of colchicine binding site inhibitors in complex with tubulin provide a rationale for drug discovery. FEBS J. 2016;283:102–111. doi: 10.1111/febs.13555. [DOI] [PubMed] [Google Scholar]
- Waterhouse A., Bertoni M., Bienert S., Studer G., Tauriello G., Gumienny R., Heer F.T., de Beer T.A.P., Rempfer C., Bordoli L., Lepore R., Schwede T. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46:W296–W303. doi: 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Roy A., Zhang Y. BioLiP: a semi-manually curated database for biologically relevant ligand-protein interactions. Nucleic Acids Res. 2013;41:D1096–D1103. doi: 10.1093/nar/gks966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Roy A., Zhang Y. Protein-ligand binding site recognition using complementary binding-specific substructure comparison and sequence profile alignment. Bioinformatics. 2013;29:2588–2595. doi: 10.1093/bioinformatics/btt447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarden O., Katan T. Mutations leading to substitutions at amino acids 198 and 200 of beta-tubulin that correlate with benomyl-resistance phenotypes of field strains of Botrytis cinerea. Phytopathology. 1993;83:1478–1483. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.