Abstract
Background & Aims
Esophageal adenocarcinoma (EAC) develops from its precursor Barrett’s esophagus through intermediate stages of low- and high-grade dysplasia. However, knowledge of genetic drivers and molecular mechanisms implicated in disease progression is limited. Herein, we investigated the effect of Mothers against decapentaplegic homolog 4 (SMAD4) loss on transforming growth factor β (TGF-β) signaling functionality and in vivo tumorigenicity in high-grade dysplastic Barrett’s cells.
Methods
An in vivo xenograft model was used to test tumorigenicity of SMAD4 knockdown or knockout in CP-B high-grade dysplastic Barrett’s cells. RT2 polymerase chain reaction arrays were used to analyze TGF-β signaling functionality, and low-coverage whole-genome sequencing was performed to detect copy number alterations upon SMAD4 loss.
Results
We found that SMAD4 knockout significantly alters the TGF-β pathway target gene expression profile. SMAD4 knockout positively regulates potential oncogenes such as CRYAB, ACTA2, and CDC6, whereas the CDKN2A/B tumor-suppressor locus was regulated negatively. We verified that SMAD4 in combination with CDC6-CDKN2A/B or CRYAB genetic alterations in patient tumors have significant predictive value for poor prognosis. Importantly, we investigated the effect of SMAD4 inactivation in Barrett’s tumorigenesis. We found that genetic knockdown or knockout of SMAD4 was sufficient to promote tumorigenesis in dysplastic Barrett’s esophagus cells in vivo. Progression to invasive EAC was accompanied by distinctive and consistent copy number alterations in SMAD4 knockdown or knockout xenografts.
Conclusions
Altogether, up-regulation of oncogenes, down-regulation of tumor-suppressor genes, and chromosomal instability within the tumors after SMAD4 loss implicates SMAD4 as a protector of genome integrity in EAC development and progression. Foremost, SMAD4 loss promotes tumorigenesis from dysplastic Barrett’s toward EAC.
Keywords: Esophageal Adenocarcinoma (EAC), SMAD4 Loss, Copy Number Alterations (CNA), Barrett’s Tumorigenesis
Abbreviations used in this paper: Cas9, CRISPR associated protein 9; CNA, copy number alteration; ΔΔCT, delta delta cycle threshold; DMEM, Dulbecco's modified Eagle medium; EAC, esophageal adenocarcinoma; FBS, fetal bovine serum; FGA, fraction of the genome altered; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; HGD, high-grade dysplasia; LC-WGS, low-coverage whole-genome sequencing; NSG, NOD-SCID interleukin 2Rγ knockout; PBS, phosphate-buffered saline; PCR, polymerase chain reaction; SDS, sodium dodecyl sulfate; sgRNA, single guide RNA; shRNA, short hairpin RNA; STR, short tandem repeat; TBS, Tris-buffered saline; TGF-β, transforming growth factor β
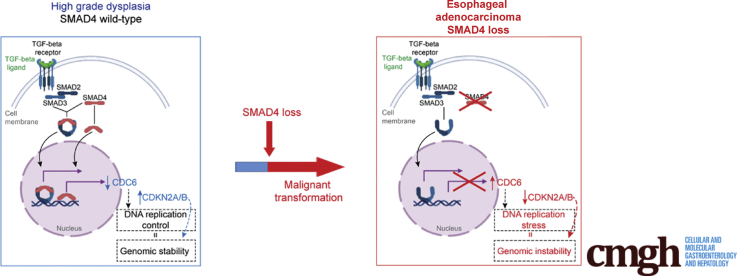
Graphical abstract
Summary.
Loss of the tumor-suppressor gene SMAD4 promotes progression of high-grade Barrett’s esophagus toward esophageal adenocarcinoma. This study provides a novel in vivo model of dysplastic Barrett’s esophagus progression toward invasive esophageal adenocarcinoma upon SMAD4 inactivation.
The incidence of esophageal adenocarcinoma (EAC) has increased rapidly over the past 4 decades.1 Despite the use of multimodality therapy, consisting of chemoradiotherapy combined with surgery, EAC has a high overall mortality rate that has shown only incremental improvements.1, 2, 3 EAC is considered to develop from Barrett’s metaplasia, with studies supporting a linear model of Barrett’s esophagus progression toward EAC through increasing grades of dysplasia.4,5
Intriguingly, we still lack functionally defined molecular drivers of this progression. One of the main barriers to defining molecular drivers is the fact that the majority of recurrent mutations in EAC are present across all stages of disease development. Thus far, the only 2 genes in which mutations have been identified to arise in a stage-specific manner are TP53 and SMAD4.6
TP53 mutations predominantly arise during the progression of nondysplastic Barrett’s esophagus to high-grade dysplasia (HGD).6 As such, TP53 gene mutations already are present in the majority of cases with HGD and patients diagnosed with EAC. In contrast, genetic events leading to SMAD4 loss of function are found almost exclusively in the malignant stage of the disease,6 and SMAD4 mutations or deletions are late branching driver events in the evolution of EAC.7,8 Therefore, TP53 and SMAD4 mutations are likely to have relevance in the progression to EAC. Although significant research efforts are focusing on understanding the functional effects of TP53 mutations and restoring p53 wild-type function,9 herein the focus was on deciphering the effects of SMAD4 loss of function in cancer progression from HGD toward EAC.
Mutations in the gene encoding for SMAD4 are present in approximately 13% of EAC patient samples, whereas SMAD4 protein loss is present in approximately 10% and 44% of primary and metastatic disease, respectively.6,10 SMAD4 represents a common mediator of the transforming growth factor β (TGF-β) pathway, which is known to have tumor-suppressive activity across gastrointestinal tumors, including EAC, whereas once the TGF-β pathway is deregulated, it could have tumor-promoting activity.11 TGF-β pathway–mediated tumor suppression results from transcriptional activation or repression of TGF-β target genes.12 Of importance, SMAD4 loss itself has been associated with increased susceptibility for EAC recurrence and shorter overall survival.10 In addition, Frankel et al13 recently reported SMAD4 mutation or homozygous deletion as a significant negative prognostic indicator in EAC. However, the functional consequences of SMAD4 loss in EAC development and progression has not been elucidated. Notably, recent research has reported common hyperactivation of the TGF-β signaling pathway in 73% of EACs, and it has shown tumor-promoting activity that may be independent of SMAD4.14
To address questions regarding the functional role of SMAD4 loss in EAC, we first investigated the regulation of TGF-β target genes upon SMAD4 loss in the high-grade dysplastic Barrett’s esophageal cell line, CP-B. We report up-regulation of oncogenes and silencing of tumor-suppressor genes after SMAD4 loss in these cells. Based on our initial evidence and previous reports that have shown stage-specific SMAD4 loss of function in EAC,6 we further investigated the functional effect of SMAD4 loss on tumorigenesis. We describe a novel in vivo model of dysplastic Barrett’s esophagus progression toward EAC, suitable for preclinical utility. Finally, we delineate the large-scale genomic events that occur upon SMAD4 loss and progression from dysplastic Barrett’s esophagus to EAC.
Results
SMAD4 Loss Alters Expression of TGF-β Target Genes in Response to Stimulation With TGF-β1
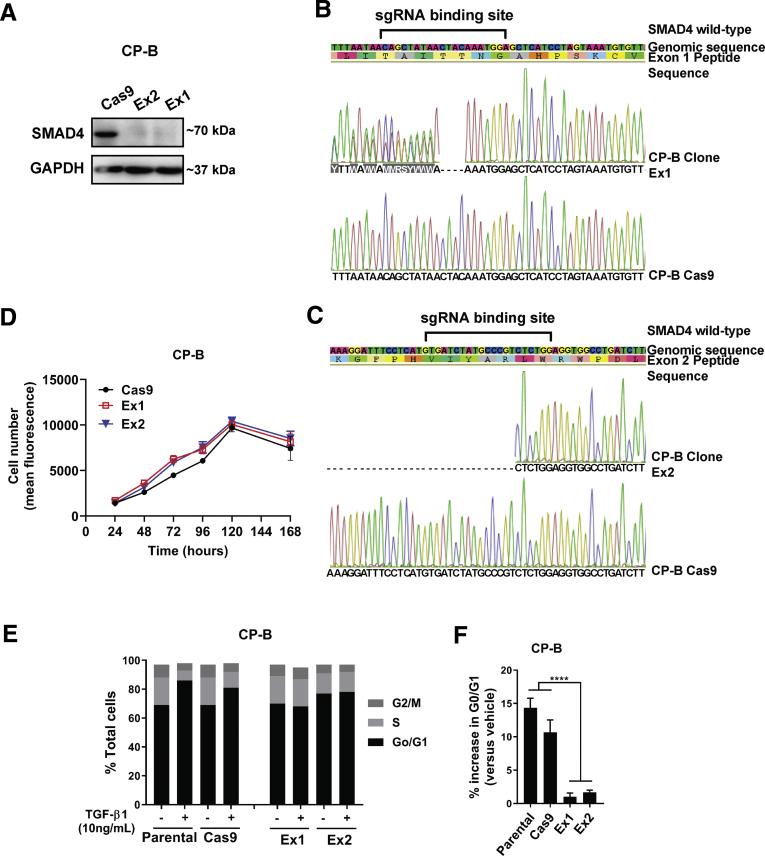
First, a Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) single-guide RNA (sgRNA)/ CRISPR associated protein 9 (Cas9) approach was used to constitutively knockout SMAD4 (Figure 1A) and mimic SMAD4 loss in a high-grade dysplastic Barrett’s esophagus cell line, CP-B. For CRISPR knockouts, single-cell clones were generated from the cell pools (bulk) and SMAD4 protein was absent in a number of clones (Figure 1A). Sanger sequencing confirmed the presence of indel mutations at the sgRNA target sites in exons 1 and 2 of CP-B clones Ex1 and Ex2, respectively (Figure 1B and C). In vitro cell proliferation of SMAD4 knockout cells was not significantly different from complementary SMAD4 wild-type control cells (Figure 1D).
Figure 1.
SMAD4 knockout and its functional role in human esophageal high grade dysplasia CP-B cells. (A) Western blot of SMAD4 protein expression levels in CP-B SMAD4 sgRNA clonal populations Ex1 and Ex2 compared to Cas9 only control cells. Representative results from Sanger sequencing demonstrating indel mutations within (B) SMAD4 exon 1 (Ex1) and (C) exon 2 (Ex2) target sequences in 2 separate cell clones. (D) Relative cell number determined by cell viability assay (Alamar Blue) from 24-168 hours following plating of 5x103 SMAD4 knockout cells (Ex1, Ex2) or Cas9 control cells. All experiments were performed on 3 independent occasions each with 3-6 technical replicates. Data shown represent mean±SEM (N=3). (E) Representative histogram of cell cycle analyses (PI staining) in CP-B SMAD4 wild-type (Parental and Cas9) and SMAD4 knockout (Ex1 and Ex2) cells following treatment with 10 ng/mL TGF-β1 for 24 hours. (F) Percentage increase in cells arrested in G0/G1 following treatment with 10 ng/mL TGF-β1 for 24 hours. N=3 independent experiments, data represent mean ± SEM. Two-tailed Student t-test, ∗∗∗∗P < .0001.
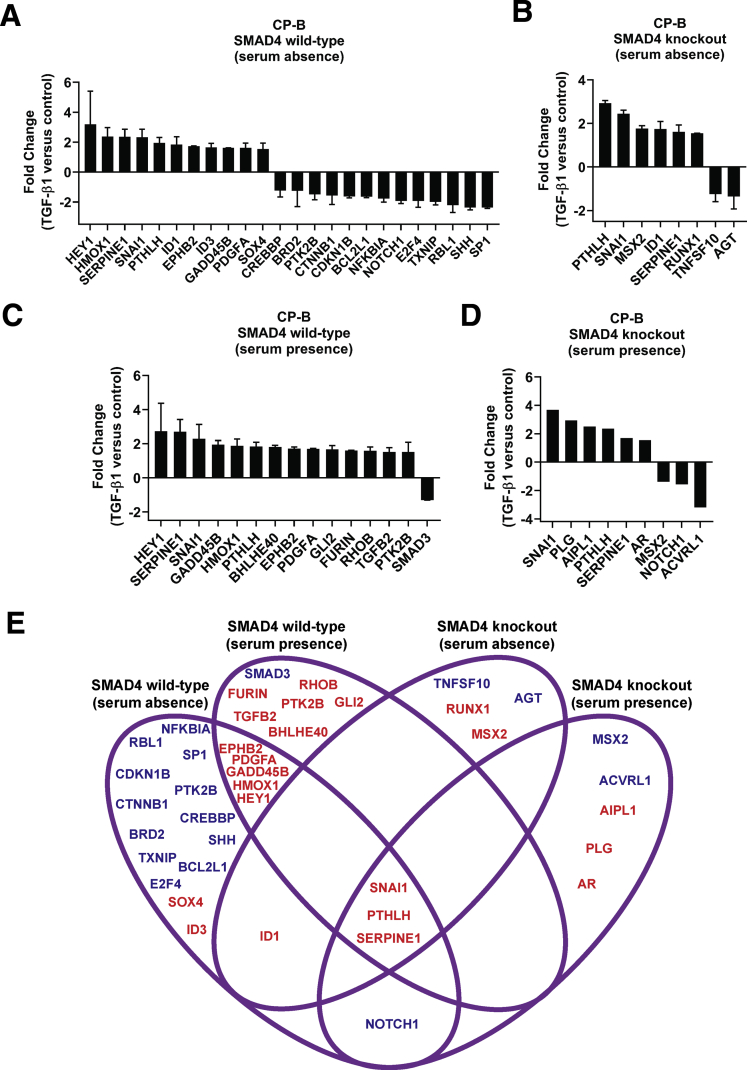
We functionally characterized the response to TGF-β signaling, when its key mediator, SMAD4, is lost in dysplastic Barrett’s cells. TGF-β1 induced G0/G1 cell-cycle arrest in SMAD4 wild-type cells, but not in SMAD4 knockout cells (Figure 1E and F), indicating that deletion of SMAD4 results in deactivation of TGF-β1–mediated cell-cycle arrest. Furthermore, we showed that SMAD4 loss dictates the differential gene expression profile of TGF-β–related target genes upon stimulation with TGF-β1 cytokines compared with basal conditions in the absence (Figure 2A and B) or presence (Figure 2C and D) of serum. A 4-way Venn diagram was created to visualize genes that are regulated in response to TGF-β1 in SMAD4 wild-type cells and/or SMAD4 knockout cells in the absence and/or presence of serum (Figure 2E). For example, TGF-β1–induced up-regulation of GADD45B and down-regulation of SHH in parental cells was lost in SMAD4 knockout cells. In contrast, TGF-β1–induced up-regulation of SNAI1, PTHLH, and SERPINE1 expression, and down-regulation of NOTCH1 expression was maintained in SMAD4 knockout cells, suggesting that TGF-β1–mediated regulation of these genes is SMAD4 independent.
Figure 2.
Altered TGF-β signaling following SMAD4 knockout in CP-B cells. (A-D) Fold change in mRNA expression of TGF-β signaling targets (RT2 Profiler PCR Arrays) in SMAD4 wild-type cells (CP-B Parental and CP-B Cas9) (A, C) and SMAD4 knockout cells (CP-B Ex1 and CP-B Ex2) (B, D) upon treatment with 10 ng/mL TGF-β1 for 16 hours without serum (A, B) or in the presence of serum (C, D) compared to respective vehicle treated cells. Thresholds for fold change ≥1.5 and ≤0.7 were chosen for up-regulated and down-regulated genes, respectively. Reciprocal fold change values are shown (y=0 or fold change 1 (no change) and y=-2 or fold change 0.5). The arithmetic mean of housekeeping genes (ACTB, B2M, GAPDH, HPRT1 and RPLPO) was used to calculate CT values for each gene. N=1 experiment per each cell line (SMAD4 wild-type = mean fold change ± SD of CP-B Parental and Cas9 cells; SMAD4 knockout = mean fold change ± SD of Ex1 and Ex2 clones; or SMAD4 knockout = fold change of Ex2 for graph D). (E) 4 way Venn diagram representing the set of differentially regulated genes following treatment with 10 ng/mL TGF-β1 for 16 hours compared to vehicle treated cells in the presence or absence of serum in SMAD4 wild-type and SMAD4 knockout cells. Gene names in red and blue colour represent up- and down-regulated genes, respectively.
SMAD4 Loss Dictates Up-Regulation of Potential Oncogenes and Down-Regulation of Tumor-Suppressor Genes in High-Grade Barrett’s Esophagus Cells Independent of Exogenous TGF-β1
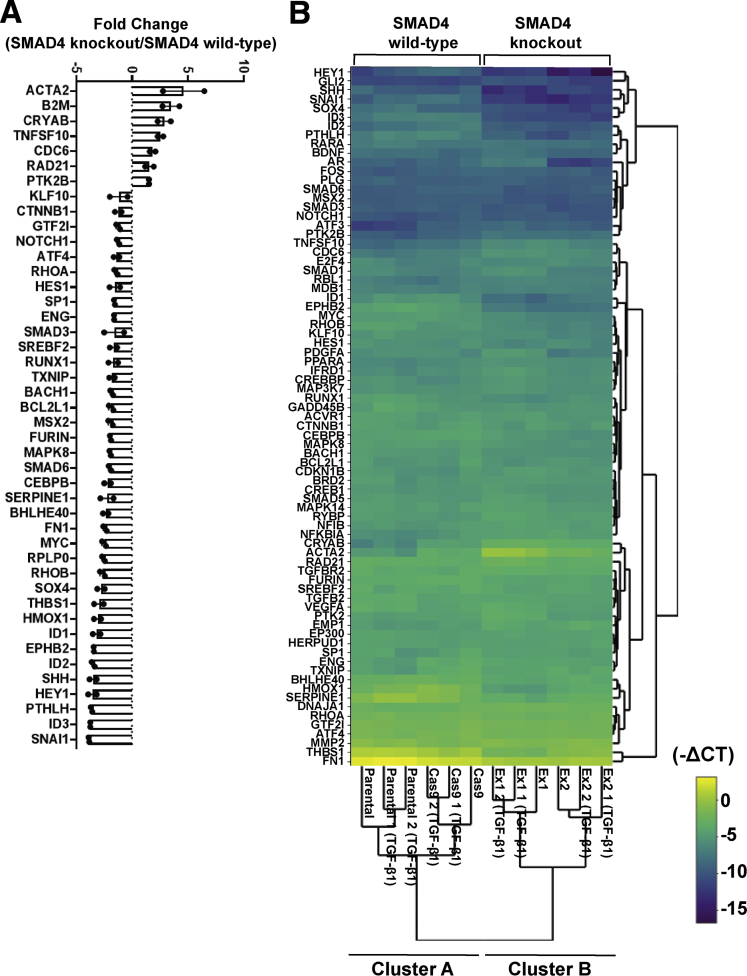
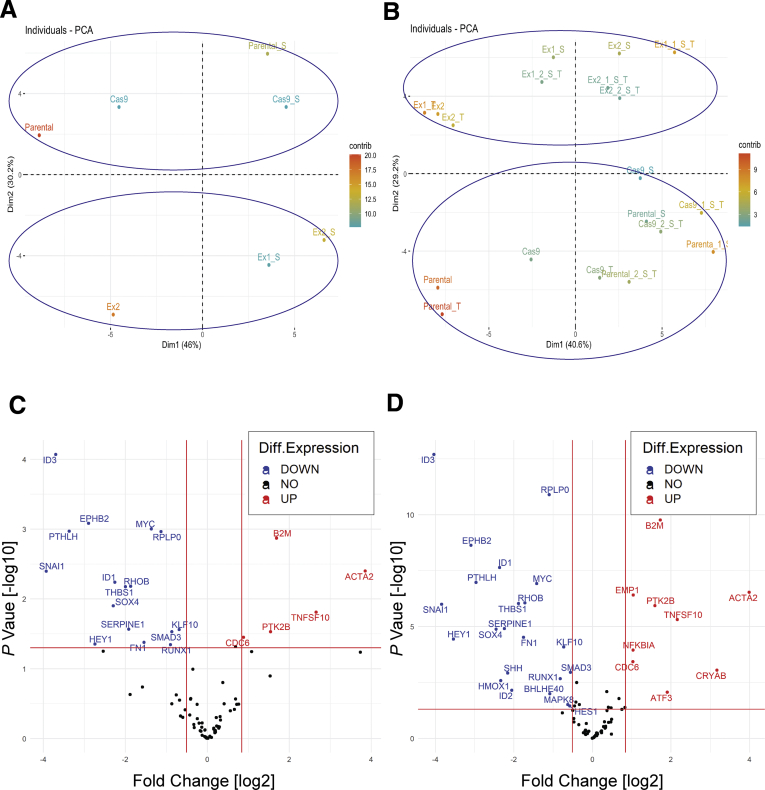
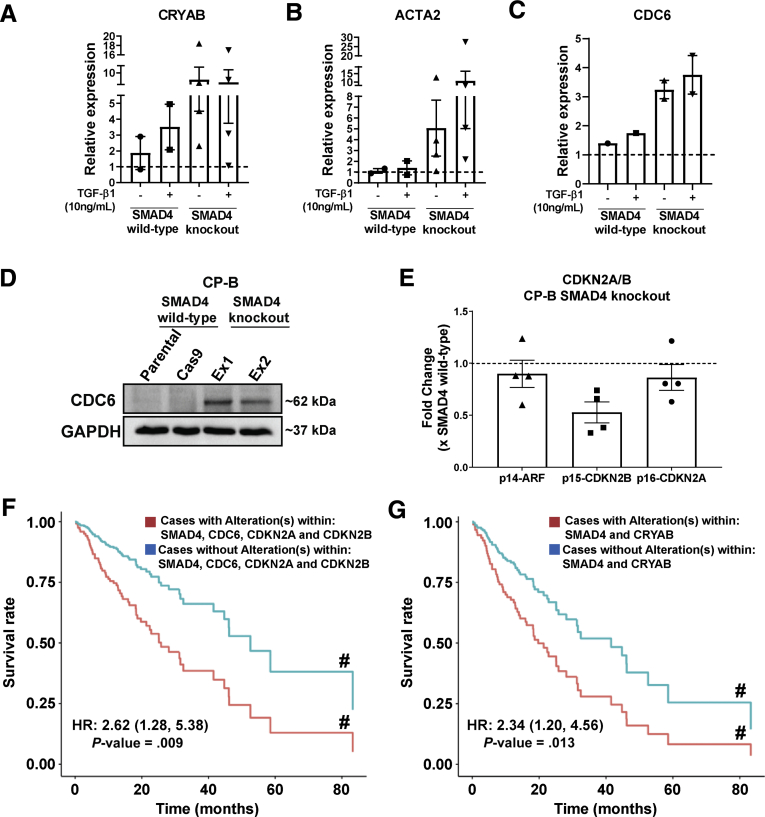
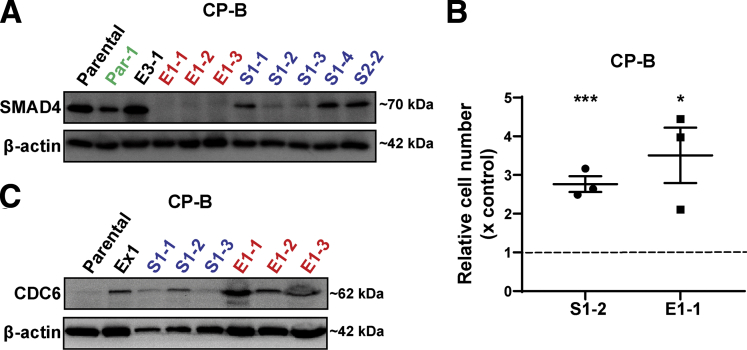
Strikingly, we observed differential gene expression profiles between SMAD4 knockout and SMAD4 wild-type cells under basal conditions, irrespective of TGF-β1 treatment (Figure 3A), as highlighted in clusters A and B (Figure 3B). In addition, principal component analysis showed that the effect of SMAD4 knockout on gene expression was greater than the effect of the presence or absence of serum (Figure 4A) or TGF-β1 treatment (Figure 4B), via clustering SMAD4 wild-type cells from SMAD4 knockout cells (Figure 4A and B). Furthermore, we identified genes responding to SMAD4 loss with statistical significance regardless of the presence or absence of serum (Figure 4C) or treatment with TGF-β1 (Figure 4D). Of note, basal expression of CRYAB, ACTA2, and CDC6 was higher in SMAD4 knockout cells compared with SMAD4 wild-type cells, even without TGF-β1 treatment (Figure 5A–C). Overexpression of these genes in cancer and their potential oncogenic roles have been reported, including in gastrointestinal cancers.15, 16, 17, 18 In addition, Cell division cycle 6 (CDC6) protein expression was almost undetectable in SMAD4 wild-type cells, whereas both SMAD4 knockout cell lines expressed high CDC6 protein levels (Figure 5D). Previous research in mouse embryo fibroblasts showed that up-regulation of CDC6 was associated with down-regulated expression of the CDKN2A/B tumor-suppressor locus genes.19 Consistent with this finding, the expression of CDKN2A/B genes (CDKN2A encodes splice variants p16-CDKN2A and p14-ARF and CDKN2B encodes p15-CDKN2B) was decreased in SMAD4 knockout/CDC6 high-expressing CP-B cells in comparison with SMAD4 wild-type/CDC6 low-expressing CP-B parental cells (Figure 5E).
Figure 3.
Differential expression of TGF-β signaling target genes in SMAD4 knockout compared to SMAD4 wild-type cells. (A) Relative expression levels of TGF-β signaling target genes (RT2 Profiler PCR Array) in SMAD4 knockout cells (CP-B Ex1 and CP-B Ex2) vs SMAD4 wild-type cells (CP-B Cas9 only) under basal conditions (serum deprivation) for 16 hours. Thresholds for fold change ≥1.5 and ≤0.7 are chosen for up-regulated and down-regulated genes, respectively. Reciprocal fold change values are shown (y=0 or fold change 1 (no change) and y=-2 or fold change 0.5). N=1 experiment per each cell line (SMAD4 wild-type = CP-B Cas9 only cells; SMAD4 knockout = Ex1 and Ex2 clones). The comparison was made between CP-B Cas9/Ex1 and CP-B Cas9/Ex2 and mean fold change ± SEM is shown. No error bars indicate SEM less than the symbol size. (B) Hierarchical clustering indicating differentially expressed genes across individual samples, including 2 independent replicates per each of CP-B parental and Cas9 (SMAD4 wild-type), and Ex1 and Ex2 (SMAD4 knockout) cells following treatment with 10ng/mL TGF-β1 in serum deprivation for 16 hours and 1 independent group of untreated cells. Normalized gene expression (-CT) within SMAD4 wild-type and SMAD4 knockout group of cells per each gene was used to generate the clustergram with heatmap. The colour scale indicates -CT values. (A, B) The arithmetic mean of housekeeping genes ACTB, B2M, GAPDH, HPRT1 and RPLPO was used to calculate CT values for each gene.
Figure 4.
Gene expression changes caused by SMAD4 knockout alone, irrespective of the presence or absence of serum or TGF-β1 treatment. Principal component analyses (PCA) of differential gene expression between SMAD4 wild-type and SMAD4 knockout CP-B cells. (A, B) The two-dimensional principal components of gene expression changes caused by SMAD4 knockout. Each dot represents an experimental condition, SMAD4 wild-type (Parental and Cas9) and SMAD4 knockout (Ex1 and Ex2) CP-B cells (A) in the presence [S] or absence of serum and (B) in the presence [S] or absence of serum and treated with TGF-β1 [T]. Experimental conditions with similar expression profile are grouped together. The contribution of a variable (in percentage) to a given principal component is calculated with formula (variable cos2∗100)/(total cos2 of the principal component). (C, D) TGF-β signalling target gene expression in SMAD4 knockout (Ex1 and Ex2) vs. SMAD4 wild-type (Parental and Cas9) cells (C) in serum presence or absence and (D) in serum presence or absence and TGF-β1 treatment. Normalization was performed using the arithmetic mean of ACTB, B2M, GAPDH, HPRT1 and RPLPO genes with each individual sample. The volcano plot displays statistical significance (-log10 P value) on the y axis vs. log2 Fold Change on the x axis. P values were calculated by unpaired Student’s t test of the replicates (Delta CT) values for each gene in the SMAD4 knockout and SMAD4 wild-type groups. Fold-Change was calculated using (2^ (-Delta Delta CT)) method. Red vertical lines represent Fold Change [log2] of gene expression (0.7 and 1.8 for down- and up-regulated genes, respectively), and the horizontal solid line represents a P value [–log10] threshold (P value less than 0.05 is considered as threshold for statistical significance.
Figure 5.
Altered expression of potential oncogenes and tumor suppressor genes in Barrett’s esophagus cells with SMAD4 knockout. (A-C) Relative expression of CRYAB, ACTA2 and CDC6 mRNA in CP-B SMAD4 wild-type and SMAD4 knockout cells assessed by RT-qPCR following treatment with 10ng/mL TGF-β1 for 16 hours in the absence of serum. Data represent the mean ± SEM of pooled results for N=2 independent experiments (CRYAB and ACTA2) or N=1 experiment (CDC6) per each cell type (SMAD4 wild-type = CP-B Cas9 only cells; SMAD4 knockout = mean of Ex1 and Ex2 clones) and are expressed relative to untreated SMAD4 wild-type cells (CP-B Parental). (D) Western blot of CDC6 protein in CP-B SMAD4 wild-type and knockout cells under basal growth conditions. (E) Relative mRNA expression (RT-qPCR) of p14-ARF, p15-CDKN2B and p16-CDKN2A in CP-B SMAD4 knockout cells compared to SMAD4 wild-type cells under basal serum starved conditions. Data represent the mean ± SEM of pooled results for N=2 independent experiments per each cell line (SMAD4 wild-type = CP-B Cas9 only cells; SMAD4 knockout =Ex1 and Ex2 clones). Individual data points represent the comparison between CP-B Cas9/Ex1 and CP-B Cas9/Ex2. Multivariate Cox proportional model showing association of overall survival in cases with (F) a SMAD4 (mutation; deep deletion; mRNA high; mRNA low or multiple alterations), and/or a CDKN2A/B locus (CDKN2A (mutation; deep deletion; mRNA high and multiple alterations) and CDKN2B gene (deep deletion and mRNA high)) and/or a CDC6 (mutation; amplification; mRNA high and multiple alterations) alteration (red group, N=127); (G) a SMAD4 and/or a CRYAB (mutation; deep deletion and mRNA high) alteration (red group, N=43), compared to cases without alterations in any of these genes (blue group, N=55 in panel F, N=139 in panel G) in esophageal cancers (total number of samples, N=182) from the TCGA database. # Survival curves are adjusted for both continuous (age at diagnosis) and categorical (sex, neoplasm histologic grade, primary lymph node presentation assessment, American joint committee on cancer stage code and American joint committee on cancer metastasis stage code) prognostic variables.
Furthermore, we analyzed whether the presence of an alteration (mutation, deep deletion, and/or up-regulated/down-regulated messenger RNA expression) within SMAD4 and/or in the CDKN2A/B locus and/or in the CDC6 gene is prognostic for survival outcomes (Figure 5F) in an esophageal cancer patient cohort (The Cancer Genome Atlas, TCGA), compared with the patients with no alterations in any of these genes (Figure 5F). We also analyzed the survival rate for cases with SMAD4 and/or CRYAB alterations compared with cases without alterations in either of these genes (Figure 5G). Strikingly, multivariate Cox proportional hazard model analyses showed a strong association between the group status and the hazard of death after adjusting for confounding variables (Figure 5G). As such, the hazard of death for the group with alterations in these genes was 2.62 (95% CI, 1.28–5.38; P = .009) (Figure 5F) and 2.34 (95% CI, 1.20–4.56; P = .013) (Figure 5G) times higher than the group without alterations. We also assessed mutual exclusivity and co-occurrence of alterations in SMAD4 and CDKN2A/B genes. Of interest, SMAD4 alterations were mutually exclusive with CDKN2B gene alterations (log2 odds ratio, -1.483; P = .01) encoding p15-CDKN2B in the same patient cohort, suggesting that precancerous cells might rely on either of these events to drive tumorigenesis. Overall, gene deregulation resulting from the absence of wild-type SMAD4 could lead to increased oncogenesis, implicating SMAD4 as a driver of EAC tumorigenesis.
SMAD4 Loss Initiates Tumorigenesis of Dysplastic Barrett’s Esophagus Cells In Vivo
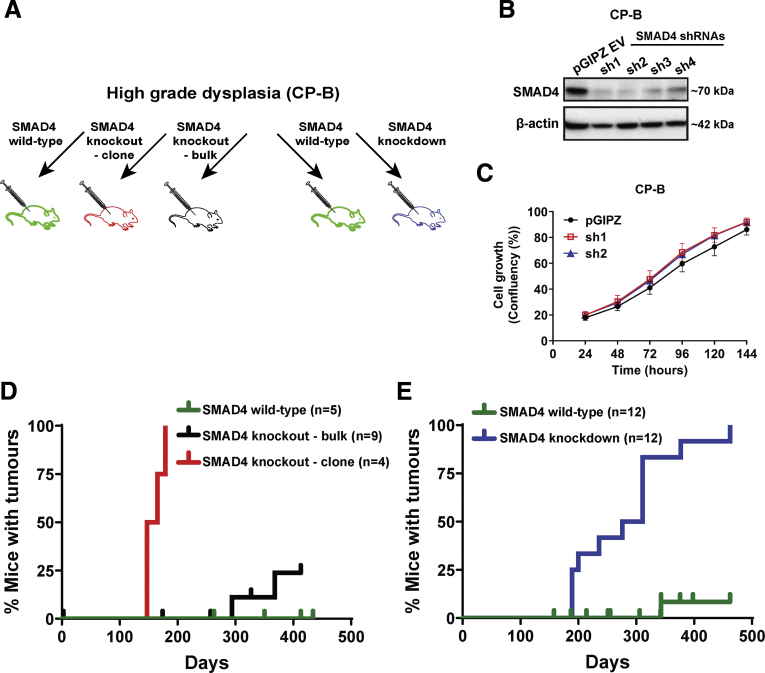
To determine whether SMAD4 loss is a functional driver of Barrett’s esophagus progression, in vivo xenograft experiments using Non-obese diabetic-severe combined immunodeficiency interleukin-2 receptor γ knockout (NSG) mice were performed (Figure 6A). In addition to the CRISPR sgRNA approach to knockout SMAD4, we used SMAD4-targeted short hairpin RNAs (shRNAs) to constitutively knockdown SMAD4. Two of the 4 SMAD4 shRNAs tested, sh1 and sh2, showed high SMAD4 knockdown efficiency and were chosen for future experiments (Figure 6B). These knockdown cells enabled us to explore the dose-dependent effect of SMAD4 expression on tumorigenicity in a bulk cell population. In vitro growth of SMAD4 knockdown cells was not significantly different than complementary empty vector control cells (Figure 6C). Isogenic CP-B SMAD4 knockout, knockdown, or control (parental, pGIPZ vector only, and Cas9) cells were injected subcutaneously into mice. All mice injected with either CP-B SMAD4 knockout or knockdown cells formed palpable tumors. SMAD4 knockout tumors started to be detected around day 150 (Figure 6D), while SMAD4 knockdown cells started to form palpable tumors from day 180 (Figure 6E). In addition, injection of bulk SMAD4 knockout cells resulted in tumor formation in some mice at approximately day 300 (Figure 6D). The decrease in SMAD4 protein in bulk SMAD4 knockout cells was minimal (data not shown), presumably owing to inefficient viral transduction and/or gene editing. This likely explains why bulk SMAD4 knockout cells took longer to form tumors. On the other hand, subcutaneous injections of SMAD4 wild-type control cells did not result in tumor formation, apart from 1 mouse of 17 that formed a tumor at day 340 (Figure 6E). This tumor formed in a mouse injected with CP-B parental cells. The tumor frequency after cell injection in vivo and time to tumor onset are summarized in Table 1.
Figure 6.
Tumor formation following SMAD4 knockout or knockdown in high grade dysplastic Barrett’s cells. (A) Experimental plan of in vivo experiment that includes injection of SMAD4 wild-type control cells (parental, pGIPZ vector only, or Cas9 only expressing CP-B cells), SMAD4 knockout (bulk and clonal CP-B cells), or SMAD4 knockdown (CP-B sh1, CP-B sh2) cells in N=4-12 mice per group. (B) Western blot of SMAD4 protein expression levels in CP-B cells transduced with pGIPZ empty vector or four different SMAD4 shRNA constructs (sh1-4). (C) Cell confluency assay comparing relative cell growth from 24-144 hours following plating of 1x104 cells with pGIPZ empty vector or the two most efficient SMAD4 shRNA constructs (sh1 and sh2). All experiments were performed on 3 independent occasions each with 3-6 technical replicates. Data shown represent mean±SEM (N=3). (D, E) Kaplan-Meier curves showing the percentage of mice with detected tumors following injection with (D) CP-B SMAD4 knockout cells or (E) CP-B SMAD4 knockdown cells compared to SMAD4 wild-type cells. Ticks represent mice that were euthanised due to signs of ill health but did not have a tumor (confirmed at necropsy).
Table 1.
Tumor Frequency In Vivo and Established Cell Lines
| Cells injected | Tumor frequency | Mean time to tumor onset, d (range) | Cell lines established | Cell line names (CP-B) | |
|---|---|---|---|---|---|
| Controls | Parentala | 1/6 | 343 | 1/1 | Par-1 |
| pGIPZ vector onlya | 0/6 | NA | NA | NA | |
| Cas9 onlya | 0/5 | NA | NA | NA | |
| shRNA knockdown | SMAD4 sh1 | 6/6 | 251 (189–311) | 4/6 | S1-1,a S1-2,a S1-3,a S1-4a,b |
| SMAD4 sh2 | 6/6 | 308 (189–462) | 1/6 | S2-2 | |
| CRISPR knockouts | Bulk knockouts | 2/9 | 331 (294–368) | 1/2 | E3-1 |
| Ex1 clonea | 4/4 | 159 (147–179) | 3/4 | E1-1,a,b E1-2,a,b E1-3a |
NOTE. The suffixes S1/S2 and E1/E3 appended to a cell line name indicates that these cells have been developed from sh1- and sh2-mediated SMAD4 knockdown and SMAD4 knockout (E1, clonal; E3, bulk) tumors, respectively. In addition, the suffix “-“ with number 1, 2, 3, or 4 appended to the end of the cell line name indicates the number of the mouse with the tumor. The Par-1 cell line was developed from the only control tumor that arose from an injection of CP-B parental cells.
LC-WGS was performed (see Figures 10 and 11).
Formed spontaneous metastases from xenograft.
Histologic and Functional Characterization of CP-B SMAD4 Knockdown or Knockout Tumors and Derived Cell Lines
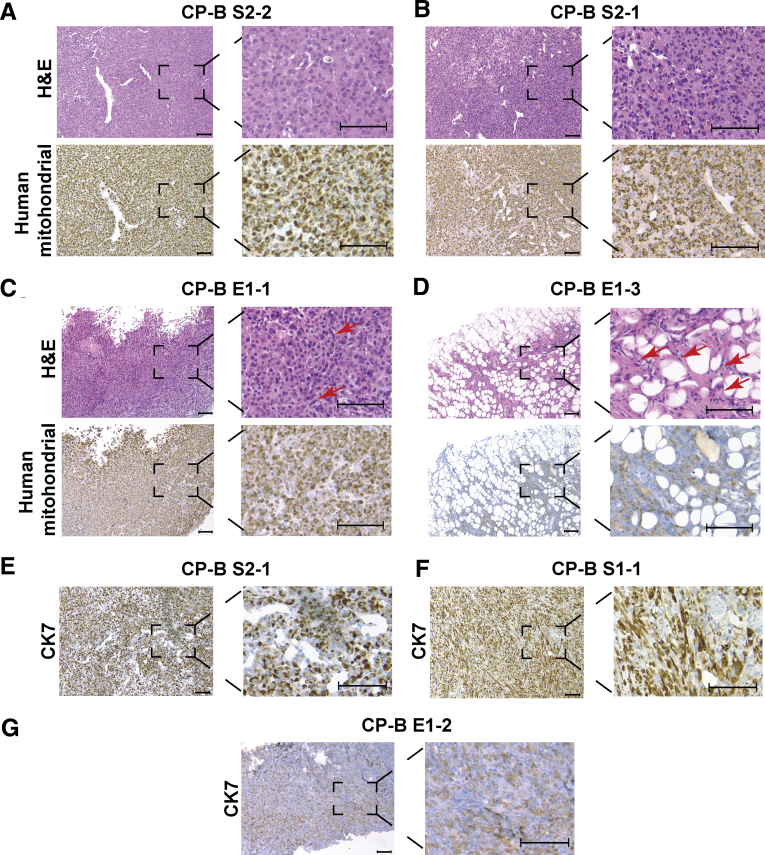
Morphologic features of SMAD4 knockdown tumors (Figure 7A and B, top panels) and SMAD4 knockout tumors (Figure 7C and D, top panels) were largely similar. The tumors were highly cellular, comprising sheets of epithelioid to plasmacytoid cells, in areas resembling a fried egg appearance with round prominent nuclei, dispersed chromatin, moderate amounts of eosinophilic cytoplasm, and a mild degree of pleomorphism. Although the SMAD4 knockout tumors were chiefly composed of epithelioid cells, there was a presence of scattered, intervening spindle cells (Figure 7C and D, top panels), whereas SMAD4 knockdown tumors did not show this morphologic feature (Figure 7A and B, top panels). One SMAD4 knockout tumor (CP-B E1-3) (Figure 7D, top panel), although composed predominantly of spindle cells that were morphologically similar to the other tumors, also had significant interdigitating subcutaneous fat, with both a nodular and septal distribution. Immunohistochemistry using a human-specific mitochondrial antibody showed dot-like positivity, consistent with a mitochondrial staining pattern, in both SMAD4 knockdown (Figure 7A and B, bottom panels) and knockout xenografts (Figure 7C and D, bottom panels), confirming the human origin of the tumors. In addition, SMAD4 knockdown (Figure 7E and F) and knockout (Figure 7G) xenografts showed positivity for cytokeratin 7 (CK7), consistent with an EAC phenotype.
Figure 7.
Histological characterisation of SMAD4 knockdown or knockout xenograft tumors. Representative H&E (upper panels) and human specific mitochondrial staining (lower panels) of (A, B) SMAD4 knockdown tumor xenografts (CP-B S2-2; CP-B S2-1); and (C, D) SMAD4 knockout tumor xenografts (CP-B E1-1; CP-B E1-3). Red arrows point to cells with spindle like morphology. Representative CK7 immunohistochemistry staining across SMAD4 knockdown (E) CP-B S2-1; (F) CP-B S1-1; and SMAD4 knockout (G) CP-B E1-2 tumor xenografts. Scale bars represent 100 µm.
Cell lines were rederived from 10 of the 19 tumors that formed (Table 1). Importantly, cell lines established from SMAD4 knockout clones (E1-1, E1-2, and E1-3) or SMAD4 knockdown (S1-1, S1-2, S1-3, S1-4, and S2-2) tumors still had no or lower SMAD4 protein expression levels compared with CP-B parental cells, respectively. CP-B E3-1 cells derived from a xenograft that arose from bulk knockout cells still expressed SMAD4 (Figure 8A). Interestingly, CP-B Par-1 cells, derived from the only xenograft that arose from the injection of control CP-B SMAD4 wild-type cells, had substantially lower SMAD4 protein levels compared with CP-B parental cells (Figure 8A). We next showed that proliferation of CP-B SMAD4 knockdown (CP-B sh1) and CP-B SMAD4 knockout (CP-B Ex1) cells grown only in in vitro conditions was attenuated significantly compared with cells that were established from corresponding tumor xenografts, CP-B S1-2 and CP-B E1-1 (Figure 8B), respectively. In addition, we showed that besides increased CDC6 expression in SMAD4 knockout cell lines grown only under in vitro conditions (Figure 5D), CDC6 expression also was up-regulated in cell lines rederived from SMAD4 knockdown (S1-1–S1-3) and knockout (E1-1–E1-3) tumor xenografts (Figure 8C). Interestingly, the extent of CDC6 up-regulation seems to be directly proportional to the level of SMAD4 protein down-regulation in SMAD4 knockdown vs knockout tumor xenografts. Overall, it is likely that during the in vivo tumorigenic process, CP-B SMAD4 knockdown and knockout cells acquired additional alterations that led to a functionally altered growth profile compared with the original cells grown only in in vitro conditions.
Figure 8.
Characterisation of SMAD4 knockdown or knockout tumour derived cell lines. (A) Western blot of SMAD4 protein levels across cell lines re-derived from xenografts. (B) Relative cell number determined by a cell viability assay (AlamarBlue) at 144 hours following plating 2x103 of CP-B sh1 SMAD4 knockdown (Control) cells or CP-B Ex1 SMAD4 knockout (Control) cells versus cells re-established from corresponding xenografts CP-B S1-2 and CP-B E1-1, respectively. Cell viability assays were performed on 3 independent occasions each with 6 technical replicates. Data points represent means of technical replicates for each individual experiment, bars represent mean ± SEM for N=3 experiments. ∗P < .05, ∗∗∗P < .001: Two-tailed Student t test. (C) Western blot of CDC6 protein across CP-B SMAD4 wild-type (Parental), SMAD4 knockout grown under in vitro conditions only (Ex1) cells and cell lines re-derived from SMAD4 knockdown (S1-1 – S1-3) and knockout (E1-1 – E1-3) tumor xenografts.
Cell Lines Rederived From CP-B SMAD4 Knockdown and Knockout Xenografts Have Potent Tumorigenic and Metastatic Capabilities
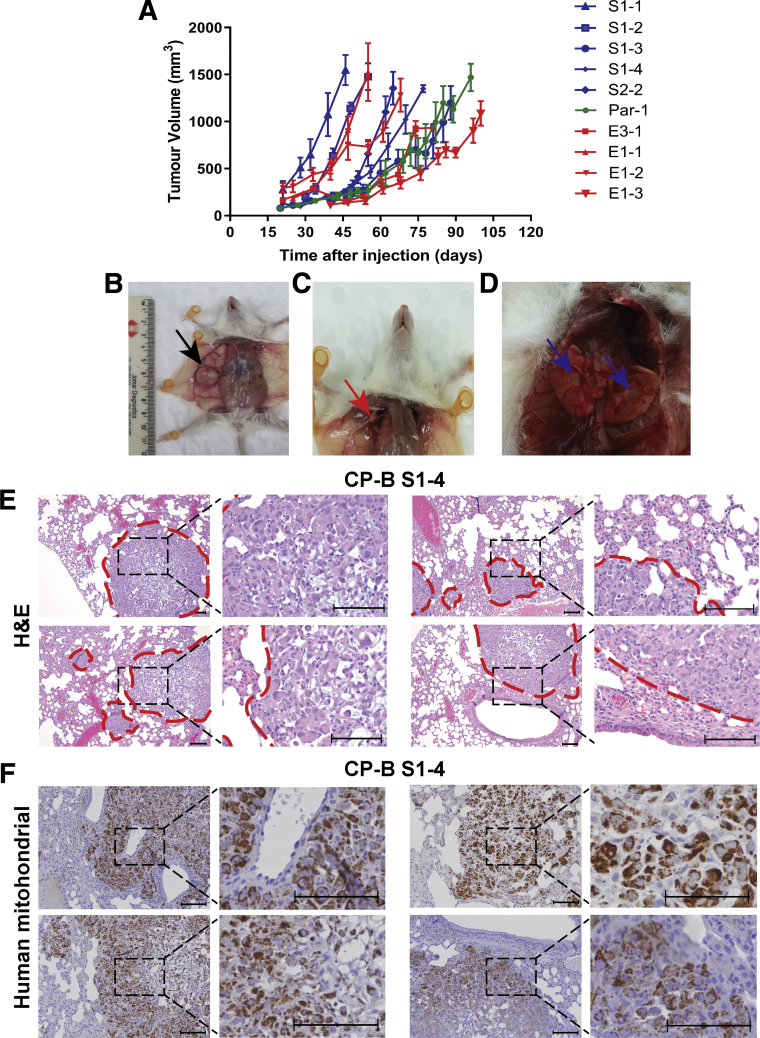
The cell lines derived from tumors were re-injected into mice to confirm their tumorigenicity. Interestingly, not only are these cell lines tumorigenic, but they established palpable tumors and reached ethical size limits in an accelerated timeframe compared with the initial tumors (Figure 9A). The tumor growth rates in vivo were variable between cell lines and there was no consistent relationship between SMAD4 knockdown vs knockout cells and the growth rate. These results suggest that the tumor growth rates are dependent on additional genetic or epigenetic changes that likely arose during tumorigenesis in vivo. In addition, after re-injection of cell lines, it was observed that CP-B E1-1, E1-2, and S1-4 cells acquired spontaneous macrometastatic potential to the lungs and ipsilateral axillary lymph nodes within 8–12 weeks, 2 common metastatic sites in EAC patients (Figure 9B–D and Table 1). The metastatic deposits showed distinctive histologic features characteristic of metastatic sites containing tumor cells (CP-B S1-4) compared with normal lung tissue (Figure 9E), and human-specific mitochondrial antibody showed dot-like positivity (Figure 9F), confirming the human origin of the metastatic cells.
Figure 9.
Potent tumorigenic and metastatic features of CP-B cell lines established from SMAD4 knockdown and knockout xenografts. (A) Tumor growth following injection of cell lines re-derived from xenografts that arose from CP-B SMAD4 knockdown (blue lines), knockout (red lines) or parental (green line) cells. See Table 1 for further details. Data represent mean ± SEM, N=4 mice per group. (B-D) Representative images of necropsy of NSG mouse with primary tumor (B, black arrow), with widespread right axillary lymph node (C, red arrow) and lung (D, blue arrows) macroscopic metastases. (E) H&E counterstaining of metastatic sites within lungs from CP-B S1-4 subcutaneous xenograft. Dashed red lines demarcate the border between the normal lung tissue and metastatic tumor cells. (F) Human specific mitochondrial immunohistochemistry staining of metastatic sites within lungs. Scale bars represent 100 µm.
Distinctive Copy Number Aberrations Characterize Tumorigenesis in CP-B Cells Upon SMAD4 Loss
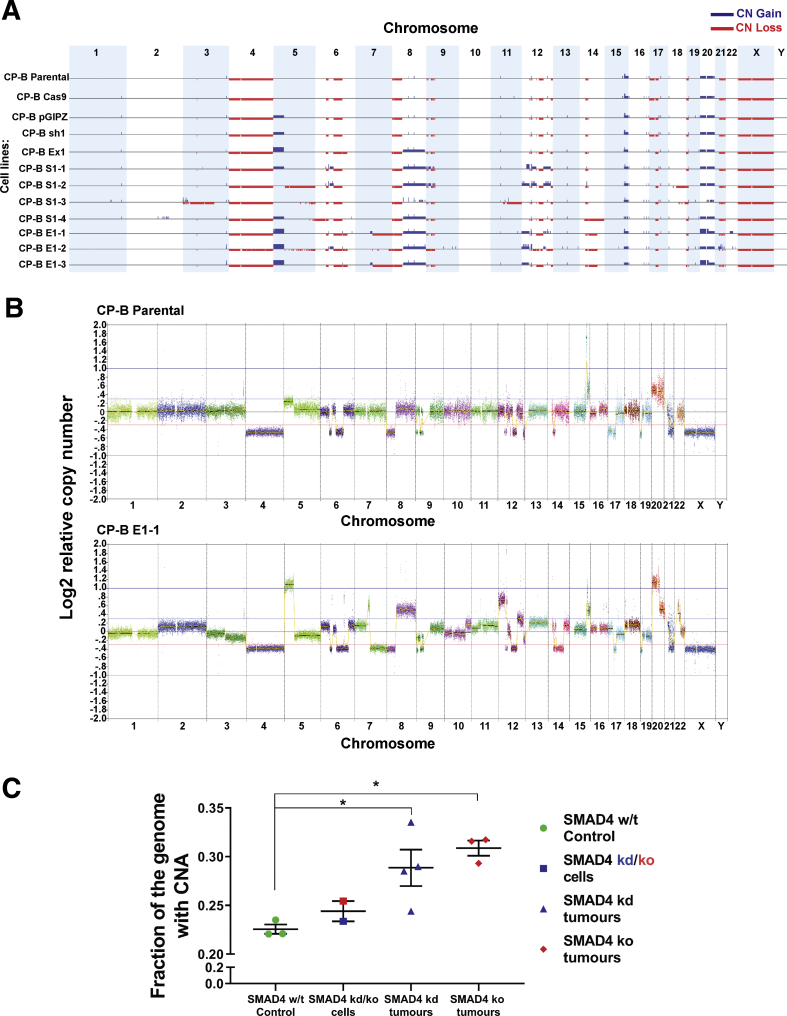
The predominant mechanism of tumorigenesis driven by SMAD4 loss in high-grade dysplasia is not clear. Considering that CP-B SMAD4 knockdown or knockout cells injected in the original experiment took 4–6 months to start growing (Figure 6D and E), whereas cell lines derived from tumors start growing within days after re-injection (Figure 9A), we hypothesized that the original cells needed to acquire additional molecular and genomic alterations before they could begin to form tumors. It has been reported in previous studies that copy number alterations (CNAs) are found more frequently in EACs than in Barrett’s esophagus with dysplasia when comparing paired patient samples.4,20 To acquire a deeper understanding of the genomic perturbations that underlie tumorigenic conversion of the HGD cells in vivo after SMAD4 depletion or loss, we performed low-coverage whole-genome sequencing (LC-WGS) to detect CNA. Initially, we noted the presence of pre-existing CNA within high-grade dysplastic CP-B parental cells, such as loss of whole chromosome 4, 8p, and 21q, and gain of whole chromosome 20 (Figure 10A). More importantly, LC-WGS data showed that the majority of tumorigenic SMAD4 knockdown or knockout CP-B cells showed distinctive and consistent CNA when compared with CP-B SMAD4 wild-type cells (Figure 10A), such as gain of 8q and 12p, partial gain in 6q and 12q, 20p high gain, and loss of 14q21.1-24.3 chromosome regions (Figure 11 and Table 2). These alterations largely happened within the same regions of the genome in each tumor, which might suggest a specific relationship between the observed alterations and loss of SMAD4 expression. Representative copy number profiles of SMAD4 wild-type (CP-B parental) and a tumor cell line with SMAD4 loss (CP-B E1-1) are shown (Figure 10B). In addition, we determined the fraction of the genome altered (FGA) by CNA in sequenced cells. Crucially, tumorigenic SMAD4 knockdown or knockout cells showed significantly higher FGA compared with the CP-B SMAD4 wild-type control cells (Figure 10C).
Figure 10.
Copy number alterations in CP-B cells following SMAD4 knockdown/knockout and tumorigenic transformation in vivo. (A) Genome wide copy number (CN) profile across; SMAD4 wild-type cells (CP-B Parental; CP-B Cas9; CP-B pGIPZ); SMAD4 knockout (CP-B Ex1) and SMAD4 knockdown (CP-B sh1) cells grown in vitro only; and tumorigenic SMAD4 knockdown (CP-B S1-1, S1-2, S1-3, S1-4) and knockout (CP-B E1-1, E1-2, E1-3) cells derived from xenograft tumors. See Table 1 for further details. Summary of the gains (blue) and losses (red) for individual cell lines across all chromosomes. (B) CN profile for 2 representative cell lines: CP-B Parental and CP-B E1-1. Each dot on the graph represents 50 kb normalized read count ratios. Log2 ratio equal to zero corresponds to a copy number of 2. Separate chromosomes from 1 to 22 as well as X and Y are shown and segments from highly repetitive or problematic regions were removed (see methods for details). (C) Fraction of the genome altered by CNA in cells established from SMAD4 knockdown or knockout tumor xenografts and CP-B SMAD4 wild-type controls. CP-B cells compared are SMAD4 wild-type cells, parental, pGIPZ vector only and Cas9 only (SMAD4 w/t Control), original SMAD4 knockdown or knockout (SMAD4 kd/ko cells, CP-B sh1 and CP-B Ex1) cells grown in in vitro conditions only and tumorigenic SMAD4 knockdown (SMAD4 kd tumors, CP-B S1-1 to -4) or knockout (SMAD4 ko tumors, CP-B E1-1 to -3) cells derived from xenograft tumors. Data points represent individual cell lines, bars represent mean ± SEM for each group. ∗P < .05: One way ANOVA with Tukey’s multiple comparison test.
Figure 11.
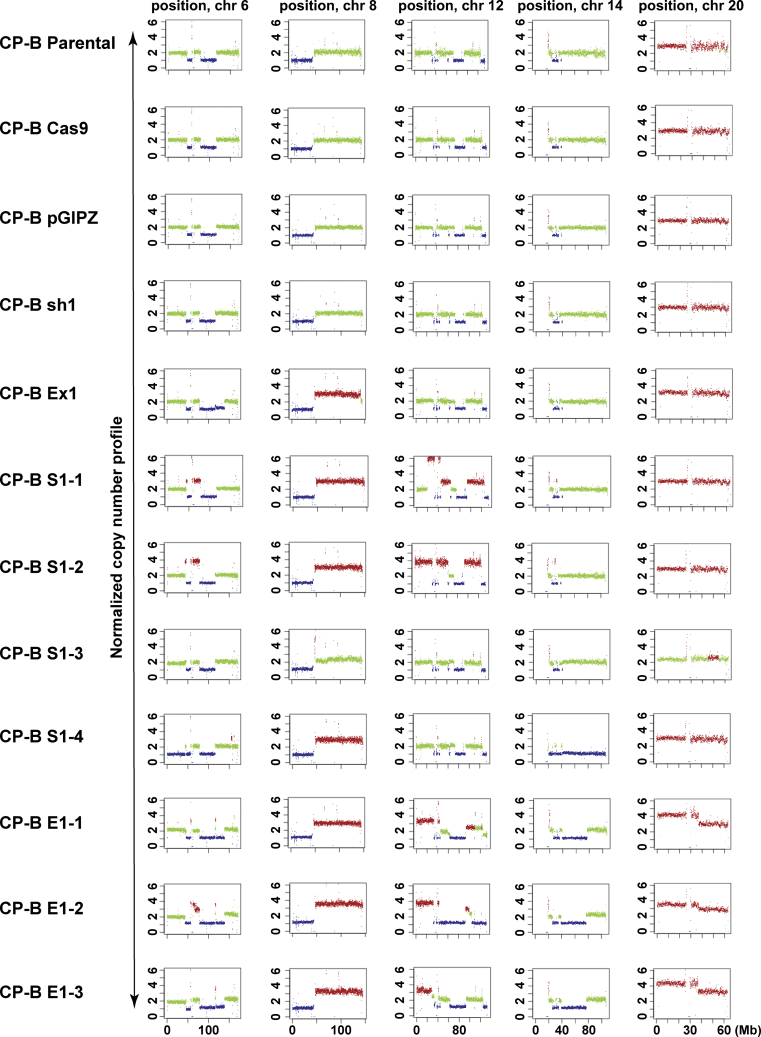
Distinctive copy number alterations across individual chromosomes in CP-B cells following SMAD4 knockdown/knockout and tumorigenic transformation in vivo. Copy number changes for individual chromosomes 6, 8, 12, 14, and 20 in SMAD4 wild-type cells (CP-B Parental; CP-B Cas9; CP-B pGIPZ); SMAD4 knockout (CP-B Ex1) and SMAD4 knockdown (CP-B sh1) cells grown in vitro only; tumorigenic SMAD4 knockdown (CP-B S1-1, S1-2, S1-3, S1-4) and knockout (CP-B E1-1, E1-2, E1-3) cells derived from xenograft tumors (see Table 1 for more details). Plots were constructed via Control-FREEC tool. Red and blue colors represent gains and losses, respectively, whereas green colour represents no changes in copy number. Scale of the x axis is presented in megabase pairs (Mb).
Table 2.
Summary of Frequent CNAs (LC-WGS) Across Cell Lines Rederived From SMAD4 Knockdown and Knockout Xenograft Tumors
| Chromosome | SMAD4 knockdown (in vitro) | SMAD4 knockout (in vitro) | Tumorigenic SMAD4 knockdown | Tumorigenic SMAD4 knockout | |||||
|---|---|---|---|---|---|---|---|---|---|
| Sh1 | Ex1 clone | S1-1 | S1-2 | S1-3 | S1-4a | E1-1a | E1-2a | E1-3 | |
| 6 | 6bq22.31-23.3b | 6p12.1-11.2c | 6p12.1-11.2c | 6q22.31-23.3b, 6q22.1c | 6q22.31-23.3b, 6q22.1c, 6p12.1-11.2c | 6q22.31-23.3b, 6q22.1c | |||
| 7 | 7q11.22-11.23c, 7q11.23-36.3b | 7q11.23-36.3 partialb | 7q11.22-11.23c, 7q11.23-36.3b | ||||||
| 8 | 8qc | 8qc | 8qc | 8q partialc | 8qc | 8qc | 8qc | 8qc | |
| 12 | 12p12.2-11.21 partialc, 12qc | 12p13.33-11.21 partialc, 12qc | 12p13.33-11.21 partialc, 12qc | 12p13.33-11.21 partialc, 12qc | 12p13.33-11.21c | ||||
| 14 | 14q21.1-24.3b | 14q21.1-24.3 | 14q21.1-24.3b | 14q21.1-24.3b | |||||
| 17 | 17pc | 17pc | 17pc | 17pc | 17pc | 17pc | 17pc | 17pc | |
| 20 | 20pc | 20pc | |||||||
Metastatic cell lines.
Copy number loss.
Copy number gain.
Discussion
Although previous genomic studies have identified EAC drivers using bioinformatics approaches,13,21, 22, 23 functional studies of EAC drivers in relevant models of disease development and progression are lacking. In addition, most of the recurrent mutations in EAC already are present at similar frequencies in nondysplastic Barrett’s esophagus, with the exception of mutations in TP53 and SMAD4 tumor-suppressor genes, which arise characteristically in high-grade dysplasia and cancer, respectively.6 Therefore, focusing on deciphering the role of stage-specific mutations and the importance of the mutations not present in Barrett’s esophagus but only in EAC, such as SMAD4 mutations, could provide useful clinical targets for invasive disease.24
This study provides evidence in preclinical models of EAC development that SMAD4 inactivation is sufficient to initiate tumorigenesis in high-grade dysplastic Barrett’s esophagus cells in vivo. Understanding the tumor initiation processes in the setting of SMAD4 loss in precancerous cells is of high importance for discovering mechanisms and clinically discriminatory markers implicated in progression toward EAC. The lack of clinically relevant biomarkers predicting Barrett’s esophagus patients at high risk of progression to EAC makes our findings even more compelling. TP53 mutations are recognized as a risk factor for EAC development in patients with Barrett’s esophagus.25 Therefore, the detection of TP53 in conjunction with SMAD4 mutation or loss could provide clinically relevant biomarkers indicating a high risk of progression to EAC and warranting early intervention.
SMAD4 knockout led to differential expression of TGF-β pathway target genes compared with SMAD4 wild-type parental cells under both basal conditions and TGF-β1 treatment. As such, SMAD4 knockout cells lost the ability to up-regulate expression of genes such as GADD45B, responsible for inducing cell-cycle arrest,26 or down-regulating expression of SHH, aberrant activation of which has been implicated in EAC tumorigenesis.27 However, it should be noted that SMAD4 loss may have a broader impact on gene expression, apart from deregulation of just TGF-β pathway target genes.
Strikingly, the magnitude of differential gene expression changes was more pronounced when comparing SMAD4 wild-type and SMAD4 knockout cells independent of TGF-β1 treatment and time point, or the absence or presence of serum. Of note, up-regulation of potential oncogenes such as ACTA2,15 CDC6,28 CRYAB,16 and TNFSF1029 was observed in SMAD4 knockout cells compared with SMAD4 wild-type cells.
Notwithstanding the essential roles of CDC6 in DNA replication and cell-cycle regulation,30 its deregulation in cancer development and progression has been scarcely studied, including in EAC. We report up-regulation of CDC6 upon SMAD4 loss, and this transcriptional regulation is likely to be through the release of suppressive activity of SMAD4 as a transcription factor. Consistent with this, we showed that the extent of CDC6 expression was dependent on SMAD4 expression levels in CP-B cells. Intriguingly, recently published data showed a novel mechanism of Cdc6 up-regulation in E3 ubiquitin ligase E6AP-/- mouse embryo fibroblasts.19 Of interest, E6-associated protein (E6AP) reduces the E2F1-dependent positive transcription of Cdc6, the potential repressor gene of the Cdkn2a/b (Cdkn2a encodes p16-Cdkn2a and p19-Arf, homolog to p14-ARF in human beings, and Cdkn2b encodes p15-Cdkn2b) tumor-suppressor locus in mouse fibroblasts.19 In our study, expression of CDKN2A/B locus genes was down-regulated significantly in CP-B SMAD4 knockout cells with high CDC6 expression levels compared with parental cells with low CDC6 expression. Given that the CDKN2A/B locus encodes for critical tumor-suppressor genes and frequently is altered across cancers,31 understanding the silencing mechanism of the CDKN2A/B locus via SMAD4 loss/CDC6 up-regulation may aid in deciphering how SMAD4 loss contributes to tumorigenesis. Among other oncogenes, B2M also was up-regulated in SMAD4 knockout cells. Interestingly, B2M has been reported recently as a novel EAC driver gene13 and has been implicated previously in acquired resistance to immunotherapy.32 Altogether, these results led us to investigate the effect of SMAD4 loss on Barrett’s esophagus progression and its potential as an EAC driver.
We found that knockdown or knockout of SMAD4 was sufficient to promote tumorigenesis in high-grade dysplastic CP-B cells. However, the long latency period between introduction of the cells into mice and tumor formation led us to speculate that additional events were required for tumor growth. Overall, SMAD4 knockdown and SMAD4 knockout tumors displayed CNA that frequently are presented across common adenocarcinomas of the gastrointestinal tract, including EACs.20,33,34 Distinctive CNA were present only in cells derived from SMAD4 knockdown/knockout tumor xenografts, but not in CP-B parental cells or SMAD4 knockdown/knockout cells grown only in vitro. Future work is required to more precisely map the genetic mutations and alterations that accompanied tumor formation in this model and to determine potential cooperative drivers with SMAD4 loss.
In addition, we detected spontaneous metastatic potential in some of the re-injected tumorigenic SMAD4 knockdown and SMAD4 knockout cells. Recently, it was reported that SMAD4 loss is present in approximately 44% of patients with locoregional and distant metastasis, suggesting an important role of SMAD4 loss in driving the invasive and metastatic potential of EAC.10 These initial observations will need further functional characterization to understand why SMAD4 loss is advantageous to invasive and metastatic potential in EAC.
Contextual and dual activity of TGF-β signaling as a tumor-promoting or tumor-suppressive pathway has been reported in different cancer types. Nevertheless, TGF-β displays major tumor-suppressive activity across gastrointestinal cancers and the inactivation of TGF-β signaling components has been reported frequently in gastrointestinal tumors.11 However, the mechanistic basis underlying disease development and progression upon disruption of TGF-β signaling components remains unsolved in cancers, including EAC.
Herein, we propose a model of malignant progression from HGD toward EAC upon SMAD4 loss. Altogether, it is likely that SMAD4, as a tumor suppressor, represents an essential gatekeeper of genomic stability within dysplastic Barrett’s cells. In dysplastic Barrett’s cells, wild-type SMAD4 negatively regulates CDC6 and positively regulates CDKN2A/B tumor-suppressor locus expression. Thus, SMAD4 loss may have led to increased genomic instability through a complex network of inactivation of tumor-suppressor genes within the CDKN2A/B locus. Alternatively, CDC6 is involved in the early origin of DNA replication control, which is critical for maintaining cell homeostasis and genomic stability.30 DNA replication stress, including through increased CDC6 activation,35 has been recognized as a significant contributor to genomic instability that may lead to further tumor development.36,37 Thus, our data suggest that up-regulation of the CDC6 oncogene upon SMAD4 loss, leading to genomic instability through increasing replication stress, is implicated in Barrett’s tumorigenesis. To further support this concept, we report increased chromosomal instability, presenting as distinctive CNA, after SMAD4 loss in esophageal HGD cells in vivo. Given the replication stress and increased genomic instability, it may be possible to use vulnerabilities in cells with SMAD4 loss as a therapeutic advantage. As such, targeting additional mechanisms important in maintaining genomic stability could aid in eradicating tumor cells with SMAD4 loss via synthetic lethal interactions.
Implications and Conclusion Highlights of This Study
This study reports that SMAD4 loss promotes progression of high-grade Barrett’s esophagus toward EAC, showing that SMAD4 is an essential tumor-suppressor gene and that its loss leads to the development of EAC. Importantly, we describe a preclinically relevant in vivo model of progression of high-grade Barrett’s esophagus to EAC that is amenable for further functional characterization of tumorigenic, invasive, and metastatic EAC biology. Given that the background genetic and epigenetic alterations in CP-B cells have not been fully characterized and that we used 1 cell model of Barrett’s esophagus progression, some of our findings could be context-dependent. Nevertheless, this work implicates SMAD4 as a protector of genome integrity, opening new possibilities to investigate synthetic lethal interactions and therapeutic targets for EAC. Furthermore, incorporating SMAD4 loss as a clinically relevant biomarker of dysplastic Barrett’s progression toward EAC might be of utmost benefit for early intervention and prevention of this deadly cancer.
Materials and Methods
All authors had access to the study data and reviewed and approved the final manuscript.
CP-B Cell Line Characteristics and Culture
The human telomerase reverse transcriptase (hTERT) immortalized Barrett’s epithelial cell line, CP-B (also identified as CP5273138), was obtained from Professor Peter Rabinovitch (University of Washington, Seattle, WA). The primary cells from which this line was derived were obtained from a region of human Barrett’s esophagus containing HGD, which makes them a suitable cell model system for studying Barrett’s tumorigenesis.38 CP-B cells are reported to be TP53, CDKN2A,38 and KRAS13 mutant, and also are known to have existing genomic copy number alterations.38,39 CP-B parental cells were grown in MCDB-153 medium containing 400 ng/mL hydrocortisone, 20 ng/mL epidermal growth factor, 20 mg/mL adenine, 140 μg/mL bovine pituitary extract, ITS Liquid Media Supplement (100×) for the final concentration of 2.5 μg/mL insulin, 1.4 μg/mL transferrin, 1.3 ng/mL selenium (all from Sigma-Aldrich, Sydney, Australia), 10 nmol/L cholera toxin, 5% v/v fetal bovine serum (FBS), 375 ng/mL fluconazole (Sigma-Aldrich, Sydney, Australia), and 4 mmol/L L-glutamine (Glutamax; Invitrogen, Thermo Fisher Scientific, Carlsbad, CA), adjusted to a pH of 7.2. Cell line authentication was conducted by short tandem repeat (STR) analysis using the PowerPlex 16 genotyping system (Promega, Madison, WI) and cultures were confirmed as mycoplasma free by polymerase chain reaction (PCR) (Cerberus Sciences, Scoresby, Australia).
Cell Viability, Cell Proliferation (Confluency), and Cell-Cycle Assays
AlamarBlue (Life Technologies, Melbourne, Australia) reagent was used to determine cell viability. Cellular proliferation was determined using the IncuCyte Live-Cell Analysis System, which automatically captures and analyzes images of living cells based on algorithms for each cell line designed using proprietary software (Incucyte FLR; Essen BioScience, Ann Arbor, MI). The assays used to assess the cell cycle have been described previously.40 For assessing cell viability, cells were seeded in a 96-well format plate, and allowed to adhere overnight. After treatment or plating only, 20% (v/v) AlamarBlue was added to each well, incubated in humidified incubators for 2 hours at 37°C, and the intensity of fluorescence was measured using a FLUOstar OPTIMA microplate reader 89 (BMG Labtech, Mornington, Australia) at an excitation wavelength of 540 nm and an emission wavelength of 590 nm. The intensity of fluorescence was measured every 24 hours or at the last time point. For cell proliferation experiments, cells were seeded in 96-well plates and allowed to adhere overnight. After treatment or plating only, cells were imaged either every 24 hours or at the last time point. For cell-cycle analyses, 1 × 106 cells were seeded in 10-cm Petri dishes, allowed to adhere and grow for 48 hours, then treated with 10 ng/mL TGF-β1 for 24 hours. After the treatment, all adherent cells were collected, centrifuged, and the supernatant was removed. Cells were washed in cold phosphate-buffered saline (PBS) with 1% FBS, fixed with ice-cold 70% ethanol, then stained for 120 minutes at room temperature in the dark with 25 μg/mL propidium iodide (Molecular Probes, Eugene, OR) and 40 μg/mL RNAse A (Qiagen, Hilden, Germany) dissolved in PBS. At least 1 × 104 single-cell events were detected by flow cytometry (BD FACSCanto II; BD Bioscience, San Jose, Canada) and analyzed using Flowlogic software (Inivai Technologies, Mentone, Australia). The distribution of cells in each cell-cycle phase was presented as a percentage of the total cell population.
Western Blot Analysis
Whole-cell protein extracts were prepared by lysis at 4°C in RIPA buffer (1 mmol/L EDTA, 1% (v/v) nonyl phenoxypolyethoxylethanol (NP40), 0.5% (w/v) sodium deoxycholate, 0.1% (v/v) sodium dodecyl sulfate (SDS), 50 mmol/L sodium fluoride, 1 mmol/L sodium pyrophosphate in PBS containing phosphatase (PhosphoSTOP; Roche, Hawthorn, Australia), and protease (Complete ULTRA; Roche) inhibitors. Protein quantification was performed using a Bio-Rad DC protein assay (Bio-Rad, Hercules, CA). Equivalent amounts of total protein lysates were suspended in 5× SDS sample buffer, and RIPA buffer was added to reach a final concentration of 1× SDS sample buffer (313 mmol/L Tris HCl, pH 6.8, 50% [v/v] glycerol, 10% [v/v] β-mercaptoethanol, 10% [w/v] SDS, and 0.05% [w/v] bromophenol blue), boiled, and resolved by SDS–polyacrylamide gel electrophoresis using 10%–15% (w/v) polyacrylamide gels. After SDS–polyacrylamide gel electrophoresis, proteins were semi–dry transferred to methanol-activated polyvinylidene difluoride membranes. Western blots were blocked for 1 hour in 5% (w/v) skim milk dissolved in Tris-buffered saline (TBS) containing 0.1% (v/v) Tween 20 and then probed overnight at 4°C with the primary antibody, followed by incubation with corresponding peroxidase-conjugated secondary antibody (Agilent Technologies, Santa Clara, CA) for 1 hour at room temperature. The primary and secondary antibodies used for Western blot are listed in Table 3. The horseradish-peroxidase–conjugated secondary antibodies were detected using Western Lightning Enhanced Chemiluminescence (PerkinElmer, Billerica, MA) or ECL Plus Western blot substrate kits (Thermo Scientific, Scoresby, Australia). Membranes were reprobed with anti–β-actin or anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) antibodies to assess protein loading. All blots were washed 3 times in rinsing buffer (0.1% [v/v] Tween 20 in TBS) for 10 minutes between different incubations.
Table 3.
Antibodies for Western Blot and Immunohistochemistry
| Antibody | Origin | Clone | Source | Use |
|---|---|---|---|---|
| Anti-SMAD4 | Rabbit | D3M6U | Cell Signaling Technology | WB |
| Anti-CDC6 | Mouse | sc-9964 | Santa-Cruz | WB |
| Anti-human mitochondrial | Mouse | MAB1273 | Merck Millipore | IHC |
| Anti-CK7 | Mouse | OV-TL 12/30 | Agilent Technologies | IHC |
| Anti–β-actin | Mouse | C4 | MP-Biomedicals | WB |
| Anti-GAPDH | Mouse | 6C5 | Merck Millipore | WB |
| Swine anti-rabbit HRP | Swine | P0217 | Agilent Technologies | WB |
| Goat anti-mouse HRP | Goat | P0447 | Agilent Technologies | WB |
HRP, horseradish peroxidase; IHC, immunohistochemistry; WB, Western blot.
Cell Signaling Technology, MA; Santa-Cruz Biotechnology, Inc, Dallas, TX; Merck Millipore, Bayswater, Australia; MP-Biomedicals, Seven Hills, Australia; and Agilent Technologies, Santa Clara, CA.
shRNA-Mediated Stable SMAD4 Knockdown
Stable SMAD4 knockdown was performed using microRNA-adapted shRNA with the pGIPZ lentiviral vector (GIPZ Lentiviral Vector; Dharmacon, CO) according to the manufacturer's instructions. The following shRNA sequences were used: sh1 (Dharmacon ID: V2LHS_37196) CTGCTAAATTCTATGTTAA; sh2 (Dharmacon ID: V2LHS_37198) GACAATATGTCTATTACGA; sh3 (Dharmacon ID: V3LHS_359404) ACGAGTTGTATCACCTGGA; and sh4 (Dharmacon ID: V3LHS_408444) AGAGAAGTTCTCAAAGTTA.
SMAD4 Knockout Using CRISPR/Cas9 Technology
CRISPR/Cas9-mediated SMAD4 knockout was performed using lentiviral vectors containing a constitutively expressing Cas9 construct and a doxycycline-inducible short guide RNA (sgRNA) construct. The experimental work for genomic knockout of SMAD4 was adapted from previously established protocols.41,42 For production of lentiviral particles, human embryonic kidney 293T (HEK293T) cells were seeded at low confluency in T75 flasks 2 days before transfection, and fresh media was changed 24 hours before transfection. A total of 10 μg vector DNA plasmid together with 10 μg Lenti-X packaging mix (Clontech, Mountain View, CA) and polyethylenimine (4.5 μg/μg DNA; Sigma-Aldrich) in Dulbecco's modified Eagle medium (DMEM) containing tetracycline-free FBS were applied drop-wise onto the HEK293T cells. Culture medium was changed after 24 hours, and viral supernatant was collected at 72 and 96 hours after transfection. Supernatant was filtered through a 0.45-μm filter to make cell-free supernatant and concentrated using an Amicon Ultra-15 Centrifugal Filtration Unit (Merck Millipore, Bayswater, Australia). Before transduction of target cells, 8 μg/mL polybrene (Sigma-Aldrich) was added to each viral aliquot. For genomic knockout of SMAD4, lentiviral vectors containing constitutive-expressing endonuclease Cas9 linked via thosea asigna virus 2A (T2A) peptide to mCherry fluorescent protein or tetracycline-inducible single guide RNA (sgRNA) expression constructs with ubiquitin promoter coupled with tetracycline repressor linked via T2A peptide to green fluorescent protein were transduced concomitantly into CP-B cells, or Cas9-only–expressing construct for control cells. sgRNA expression is dependent on an H1 promoter and Tet-operating site that are regulated negatively by binding tetracycline repressor. The promoter activity is regulated positively by doxycycline induction that relieves the tetracycline repressor from the Tet-operating site. After doxycycline induction (2 μg/mL; Sigma-Aldrich) for 96 hours, expressed sgRNA leads to conformational activation of Cas9 endonuclease and guides Cas9 to the targeted SMAD4 DNA sequence of exon 1 (ATAACAGCTATAACTACAAA), exon 2 (ATGTGATCTATGCCCGTCTC), or exon 3 (GGATTAACACTGCAGAGTAA). This results in DNA cleavage and introduction of double-strand breaks within the DNA region of interest. After successful passages, CP-B cells were sorted by flow cytometry (BD Fusion5; BD Bioscience) to isolate mCherry and green fluorescent protein–positive cells, containing Cas9 and sgRNA, respectively, or Cas9 only. This population subsequently was grown, followed by single-cell sorting and growth of single-cell clones. Single-cell clones were generated from bulk-transduced cells and SMAD4 knockout was confirmed using Western blot and Sanger sequencing.
Sanger Sequencing
Genomic DNA was extracted from cells using the DNA Blood and Tissue Mini kit (Qiagen, Hilden, Germany). The coding regions of exon 1, 2, and 3 of the SMAD4 gene were amplified by PCR and cleaned using ExoSAP-IT (Affymetrix, Santa Clara, CA). Primer sequences were SMAD4 exon 1 (forward: 5’TGTGCCATAGACAAGGTGGA3’; reverse: 5’CTTCCAGAAATTCCCATAATGC3’); exon 2 (forward: 5’TCACTGCAGCCTTGACCTACTG3’; reverse: 5’AAGTCGCGGGCTATCTTCCA3’); and exon 3 (forward: 5’GTGGCTGGTCGGAAAGGATT3’; reverse: 5’TACTGCCTGCCGCTCACAC3’). Cycle sequencing was performed on a 3130 Genetic Analyzer (Thermo Fisher Scientific, Waltham, MA) using the BigDye Terminator v3.1 sequencing kit (Thermo Fisher Scientific).
Animal Experiments
All animal work was conducted according to the regulations of the National Health and Medical Research Council Australian Code of Practice for the Care and Use of Animals for Scientific Purposes and with the approval of the Peter MacCallum Cancer Centre Animal Experimentation Ethics Committee. Female NSG mice were bred in-house or obtained from Australian BioResources (Garvan Institute of Medical Research, New South Wales, Australia).
For in vivo tumorigenesis and tumor growth studies, 5 × 106 cells resuspended in 100 μL of 1:1 PBS and growth factor–reduced Matrigel Matrix (Corning, Corning, NY) were injected subcutaneously into the right flank of 6- to 8-week-old NSG mice. Tumor formation was detected by palpation. Once palpable, tumors were measured with digital calipers and tumor volume was calculated using the following formula: (length × width2)/2. All mice were euthanized at the first signs of ill health (eg, labored breathing, bloated abdomen, or excessive weight loss of >10% of baseline body weight) or when the tumors reached ≥1500 mm3. Necropsy was performed on all animals to harvest the tumors and observe the effects of primary site tumor formation, including macrometastases. The tumors were harvested and used for histology and immunohistochemistry, cryopreservation, and cell line establishment.
Histology and Immunohistochemistry
Immunohistochemistry was performed on formalin-fixed, paraffin-embedded tissues from xenografts after removal of paraffin in xylene and rehydration in graded ethanol. H&E-stained sections of tumors underwent pathologic review. Sections were blocked in high pH Dual Endogenous Enzyme Block (DAKO, Carpinteria, CA), followed by 10% (w/v) bovine serum albumin in TBS containing 0.05% Tween-20 and incubated with primary antibody diluted in 1% (w/v) bovine serum albumin in TBS containing 0.05% Tween-20 for 1 hour at room temperature or overnight at 4ºC. The antibodies used are listed in Table 3. Images of stained sections were captured on a BX-51 microscope (Olympus, Notting Hill, Australia).
Isolation and Establishment of Cell Lines From CP-B SMAD4 Knockdown/Knockout Tumors From In Vivo Model
After harvesting and separation from the surrounding parenchyma, tumors were washed with PBS, finely chopped, and then incubated in 2 mL PBS containing 2 mg/mL collagenase A (Roche, Basel, Switzerland) and 4 mg/mL Dispase (Roche) in a water bath (RATEK Instruments, Boronia, Victoria, Australia) for 2 hours at 37ºC. After addition of 10 mL fresh media, cell suspensions were filtered through 45-μm pore size Membrane Filters (Millipore, Sigma Aldrich) and pelleted by centrifugation (1200 rpm, 5 min). Cell lines were established in DMEM containing 2.5 mmol/L L-glutamine and 4.5 g/L D-glucose (Life Technologies) culture media with 10% (v/v) FBS, and an increased concentration of penicillin (100 U/mL) and streptomycin (100 mg/mL) for at least 2 passages before reducing to standard concentrations of 50 U/mL penicillin and 50 mg/mL streptomycin. Cells were grown in monolayer cultures in humidified incubators at 37°C with 5% CO2. Cells were passaged successfully using 0.25% Trypsin, puromycin selected (SMAD4 shRNA knockdown) or fluorescence-activated cell sorted for mCherry (SMAD4 CRISPR knockout) to remove contaminating host mouse cells. All cell lines were authenticated as being derived from the CP-B parental cell line (>80% compatibility) using the STR Profile Database Matching algorithm from American Type Tissue Collection (Manassas, VA). The STR results are summarized in Tables 4 and 5. For the purpose of the phenotypical and physiological comparison between xenograft-derived cell lines and original cell lines, CP-B parental and CP-B SMAD4 knockdown/knockout isogenic cell lines were adapted to grow in DMEM containing 2.5 mmol/L L-glutamine and 4.5 g/L D-glucose (Life Technologies) and supplemented with 10% v/v FBS, 50 U/mL penicillin, and 50 mg/mL streptomycin (Life Technologies).
Table 4.
STR Analyses of Cell Lines Established From SMAD4 Knockdown Tumors
| Loci | Reference profile | CP-B Par-1 | CP-B S1-1 | CP-B S1-2 | CP-B S1-3 | CP-B S1-4 |
|---|---|---|---|---|---|---|
| CSF1PO | 8, 12 | 8, 12 | 8 | 8 | 8 | 8 |
| D13S317 | 8, 12 | 8, 12 | 8, 12 | 8, 12 | 8, 12 | 8, 12 |
| D16S539 | 10, 13 | 10, 13 | 10, 13 | 10, 13 | 10, 13 | 10, 13 |
| D5S818 | 11, 12 | 11, 12 | 11 | 11 | 11 | 11 |
| D7S820 | 11, 12 | 11, 12 | 11, 12 | 11, 12 | 11, 12 | 11, 12 |
| THO1 | 8, 9 | 8, 9 | 8, 9 | 8, 9 | 8, 9 | 8, 9 |
| TPOX | 8, 9 | 8, 9 | 8, 9 | 8, 9 | 8, 9 | 8, 9 |
| vWA | 18, 20 | 18, 20 | 18, 20 | 18, 20 | 18, 20 | 18, 20 |
| Amelogenin | XY | X | X | X | XY | X |
| Loci matches with reference profile ≥80% | Yes | Yes | Yes | Yes | Yes | Yes |
NOTE. Reference profile: CP-B (ATCC CRL-4028).
Table 5.
STR Analyses of Cell Lines Established From SMAD4 Knockout Tumors
| Loci | Reference profile | CP-B E3-1 | CP-B E1-1 | CP-B E1-2 | CP-B E1-3 |
|---|---|---|---|---|---|
| CSF1PO | 8, 12 | 8, 12 | 8 | 8 | 8 |
| D13S317 | 8, 12 | 8, 12 | 8, 12 | 8, 12 | 8, 12 |
| D16S539 | 10, 13 | 10, 13 | 10, 13 | 10, 13 | 10, 13 |
| D5S818 | 11, 12 | 11, 12 | 11 | 11 | 11 |
| D7S820 | 11, 12 | 11, 12 | 12 | 12 | 12 |
| THO1 | 8, 9 | 8, 9 | 8, 9 | 8, 9 | 8, 9 |
| TPOX | 8, 9 | 8, 9 | 8, 9 | 8, 9 | 8, 9 |
| vWA | 18, 20 | 18, 20 | 18, 20 | 18, 20 | 18, 20 |
| Amelogenin | XY | X | XY | XY | XY |
| Loci matches with reference profile ≥80% | Yes | Yes | Yes | Yes | Yes |
NOTE. Reference profile: CP-B (ATCC CRL-4028).
DNA Purification and Analyses
Total DNA from cultured cells was isolated using the DNeasy Blood and Tissue kit (Qiagen) following the manufacturer’s instructions. DNA concentration was determined by the Invitrogen Qubit 3 Fluorometer (Thermo Fisher Scientific), and purity was estimated by a NanoDrop ND1000 spectrophotometer (Thermo Fisher Scientific).
Library Preparation and LC-WGS Workflow
The preparation of indexed DNA libraries was performed using the NEBNext Ultra II DNA Library Prep Kit (NEB E7645S/L; New England BioLabs, Inc, Ipswich, MA), according to the manufacturer’s instructions. The fragmentation of 100 ng starting DNA material was conducted for 120 seconds in microtubes (Covaris, Inc, Woburn, MA) using the ultrasonication Covaris LE220 system (Covaris). PCR library amplification was performed using the PCR cycling conditions listed in Table 6. After amplification, library DNA concentration was measured using the Invitrogen Qubit 3 Fluorometer (Thermo Fisher Scientific). The sizing analysis of library DNA was performed using a 2200 Tape Station System and High Sensitivity D1000 Screen Tape Assay (2200; Agilent). The prepared libraries were run on a Nextseq 500 Sequencer using a NextSeq 500 High Output Kit (75 paired-end) (Illumina, San Diego, CA). High-output flow cells were used to pool more than 8 samples according to the standard Illumina protocol and sequencing resulted in genome coverage of 1.07–1.70 times per sample. The operation of the Nextseq 500 was conducted by the Molecular Genomics Core Facility, Peter MacCallum Cancer Centre.
Table 6.
PCR Cycling Conditions for DNA Library Amplification
| Cycle step | Temperature | Time | Cycles |
|---|---|---|---|
| Initial denaturation | 98ºC | 30 seconds | 1 |
| Denaturation | 98ºC | 10 seconds | 8a |
| Annealing/extension | 65ºC | 75 seconds | |
| Final extension | 65ºC | 5 seconds | 1 |
| Hold | 4ºC | ∞ |
Optimized for 100 ng input DNA.
Data Analyses for LC-WGS
After removal of sequencing primers by Cutadapt (v1.7.1),43 reads were aligned with bwa mem (v0.7.12-r1039)44 to human genome version 19 (hg19) (GRCh37). We used ControlFREEC (version 6.7)45 to assess the copy number from the LC-WGS data in 50-kb windows across hg19, with default parameters, no matched normal sample, and baseline ploidy set to 2. Blacklisted regions including highly repetitive centromeric regions46,47 were filtered out to yield a genomic profile with scarce noise. Then data were imported into Nexus (v8; BioDiscovery, Inc, Hawthorne, CA) and segmented using SNP-Fast Adaptive States Segmentation Technique (SNP-FASST). CNAs were called if the log2 ratio was ±0.15. The FGA was calculated as described previously.48,49 In brief, the weighted FGA was calculated by the summation of the CNA in base pairs for each chromosome and then dividing by the length of that chromosome. The final FGA for a sample was calculated by taking the average of the percentage of CNAs across all chromosomes.
Quantitative Real-Time PCR
Total cell RNA was isolated using the RNeasy kit according to the manufacturer’s protocol (Qiagen) or the Nucleospin RNA kit (Macherey-Nagel GmbH & Co, KG, Duren, Germany). RNA concentration and purity were measured with a NanoDrop ND1000 spectrophotometer (Thermo Fisher Scientific). Complementary DNA was generated from 1 ug purified RNA using the Transcriptor First-Strand complementary DNA Synthesis Kit (Roche). Gene expression changes were determined by SYBR green real-time quantitative PCR (Lightcycler 480; Roche), analyzed using the comparative delta delta cycle threshold (ΔΔCT) method,50 and normalized against GAPDH. Real-time quantitative PCR primer sequences were as follows: p14-ARF: forward (5'-3'): CCCTCGTGCTGATGCTACTG, and reverse (5'-3'): CATCATGACCTGGTCTTCTAGGAA; p15-CDKN2B: forward (5'-3'): CGGGGACTAGTGGAGAAGGT, and reverse (5'- 3'): CGAAACGGTTGACTCCGTTG; p16-CDKN2A: forward (5'-3'): GGGGGCACCAGAGGCAGT, and reverse (5'-3'): GGTTGTGGCGGGGGCAGTT; p16-CDKN2A: forward (5'-3'): GGGGGCACCAGAGGCAGT, and reverse (5'-3'): GGTTGTGGCGGGGGCAGTT; ACTA2: forward (5'-3'): TCAATGTCCCAGCCATGTAT, and reverse (5'-3'): CAGCACGATGCCAGTTGT; CRYAB: forward (5'-3'): CTTTGACCAGTTCTTCGGAG, and reverse (5'-3'): CCTCAATCACATCTCCCAAC; CDC6: forward (5'-3'): TGGATGTTTGCAGGAGAGCTA, and reverse (5'-3'): GCTCCTTCTTGGCTCAAGGT and GAPDH: forward (5'-3'): GGTGTGAACCATGAGAAG and reverse (5'-3'): CCACAGTTTCCCGGAG.
TGF-β Signaling Targets RT2 Profiler PCR Array
Eighty-four TGF-β signaling target genes were analyzed using the TGF-β Signaling Targets RT2 Profiler PCR Array (PAHS-235Z; Qiagen). RT2 Profiler PCR Arrays were purchased in Format G 384 (4 × 96) suitable for use with the Roche LightCycler 480 (384-well block). Cells were seeded into 6-well plates (9 × 105 cells/well) and grown for 72 hours. After serum starvation for 6 hours, the cells were treated with 10 ng/mL human recombinant TGF-β1 in serum-deprived media and harvested at 4 and 16 hours, after which RNA was extracted as described earlier. Preparation of complementary DNA from 100 ng (for RT2 Profiler PCR Arrays in the presence of serum) and 400 ng (for RT2 Profiler PCR Arrays in the absence of serum) purified RNA was performed using the RT2 First-Strand Kit (Qiagen) suitable for PCR-based gene expression analyses together with RT2 SYBR Green Mastermix (Qiagen), according to the manufacturer's instructions.
Real-time PCR was performed according to the manufacturer's instructions (Qiagen) with the PCR components mix and cycling conditions for the Roche LightCycler 480. The target gene expression levels were quantified relative to the mean arithmetic value obtained for housekeeping genes ACTB, B2M, GAPDH, HPRT1, and RPLPO. The lower limit of detection or CT cut-off value was set to 35. Relative gene expression was calculated using the ΔCT method between the gene of interest and an average of the housekeeping genes, and subsequent ΔΔCT calculations (ΔCT [test group] - ΔCT [control group]). Fold change was obtained using the 2(-ΔΔCT) formula.
Statistics
The Student t test was used to compare 2 groups of interest. For analyses of 3 or more groups, 1-way analysis of variance with the Tukey multiple comparison test was performed. A heat map was generated using the heatmaply R package.51 Hierarchical clustering was performed using the hclust function in R with Manhattan distance and Ward’s (ward.D2) agglomerative method. Principal component analysis and volcano plots were generated in R. Principal component analysis estimation was performed in normalized expression values using the arithmetic mean of housekeeping genes (ACTB, B2M, GAPDH, HPRT1, and RPLPO). The principal components were estimated with the function princomp with default parameters, and visualization was performed using the factoextra package.52
Multivariable Cox proportional hazards models for overall survival were performed to determine whether the alteration status remained significantly associated with survival after adjusting for both continuous (age at diagnosis) and categoric (sex, neoplasm histologic grade, primary lymph node presentation assessment, American Joint Committee on Cancer stage code, and American Joint Committee on Cancer metastasis stage code) prognostic variables. All survival models were fitted using the survival R package.53 Age at diagnosis was included in the Cox model with the use of penalized splines to allow for nonlinear association. The likelihood ratio test was performed to compare the models with and without the nonlinear terms. Proportional hazards assumption was assessed using the Schoenfeld residuals.54 Stratified Cox regression models were considered to alleviate any nonproportional issues with categoric variables.54 The graphical abstract was created with BioRender.com.
Statistical analyses were performed using Prism 7 (GraphPad San Diego, CA) and R statistical software (version 4.0.2; Vienna, Austria).55 Two-sided P values less than .05 were considered statistically significant.
Acknowledgments
CRediT Authorship Contributions
Jovana R Gotovac (Conceptualization: Lead; Data curation: Lead; Formal analysis: Lead; Investigation: Lead; Methodology: Lead; Validation: Lead; Visualization: Lead; Writing – original draft: Lead; Writing – review & editing: Equal)
Tanjina Kader (Formal analysis: Equal)
Julia V Milne (Investigation: Supporting; Methodology: Equal)
Kenji M Fujihara (Conceptualization: Equal; Investigation: Supporting)
Luis Eduardo Lara Gonzales (Formal analysis: Equal)
Kylie L Gorringe (Software: Equal; Writing – review & editing: Supporting)
Sangeetha N Kalimuthu (Methodology: Equal; Writing – review & editing: Equal)
Madawa W Jayawardana (Writing – review & editing: Supporting)
Cuong P Duong (Writing – review & editing: Equal)
Wayne A Phillips (Conceptualization: Supporting; Funding acquisition: Lead; Supervision: Supporting; Writing – review & editing: Supporting)
Nicholas J Clemons (Conceptualization: Equal; Funding acquisition: Lead; Methodology: Supporting; Supervision: Equal; Writing – review & editing: Equal)
Footnotes
Conflicts of interest The authors disclose no conflicts.
Funding This research was supported by National Health and Medical Research Council of Australia Project grant APP1120293 (W.A.P. and N.J.C.) and Ideas grant APP1182525 (N.J.C). Also supported by a Melbourne International Research Scholarship (University of Melbourne) (J.R.G.), and Fellowship MCRF16002 from the Department of Health and Human Services acting through the Victorian Cancer Agency, Victoria, Australia (N.J.C.).
References
- 1.Coleman H.G., Xie S.-H., Lagergren J. The epidemiology of esophageal adenocarcinoma. Gastroenterology. 2018;154:390–405. doi: 10.1053/j.gastro.2017.07.046. [DOI] [PubMed] [Google Scholar]
- 2.Ferlay J., Soerjomataram I., Dikshit R., Eser S., Mathers C., Rebelo M., Parkin D.M., Forman D., Bray F. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 3.Smyth E.C., Lagergren J., Fitzgerald R.C., Lordick F., Shah M.A., Lagergren P., Cunningham D. Oesophageal cancer. Nat Rev Dis Primers. 2017;3:17048. doi: 10.1038/nrdp.2017.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stachler M.D., Taylor-Weiner A., Peng S., McKenna A., Agoston A.T., Odze R.D., Davison J.M., Nason K.S., Loda M., Leshchiner I., Stewart C., Stojanov P., Seepo S., Lawrence M.S., Ferrer-Torres D., Lin J., Chang A.C., Gabriel S.B., Lander E.S., Beer D.G., Getz G., Carter S.L., Bass A.J. Paired exome analysis of Barrett's esophagus and adenocarcinoma. Nat Genet. 2015;47:1047–1055. doi: 10.1038/ng.3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Peters Y., Al-Kaabi A., Shaheen N.J., Chak A., Blum A., Souza R.F., Di Pietro M., Iyer P.G., Pech O., Fitzgerald R.C. Siersema PD. Barrett oesophagus. Nat Rev Dis Primers. 2019;5:1–22. doi: 10.1038/s41572-019-0086-z. [DOI] [PubMed] [Google Scholar]
- 6.Weaver J.M., Ross-Innes C.S., Shannon N., Lynch A.G., Forshew T., Barbera M., Murtaza M., Ong C.-A.J., Lao-Sirieix P., Dunning M.J., Smith L., Smith M.L., Anderson C.L., Carvalho B., O'Donovan M., Underwood T.J., May A.P., Grehan N., Hardwick R., Davies J., Oloumi A., Aparicio S., Caldas C., Eldridge M.D., Edwards P.A.W., Rosenfeld N., Tavaré S., Fitzgerald R.C. Ordering of mutations in preinvasive disease stages of esophageal carcinogenesis. Nat Genet. 2014;46:837–843. doi: 10.1038/ng.3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Murugaesu N., Wilson G.A., Birkbak N.J., Watkins T.B., McGranahan N., Kumar S., Abbassi-Ghadi N., Salm M., Mitter R., Horswell S., Rowan A., Phillimore B., Biggs J., Begum S., Matthews N., Hochhauser D., Hanna G., Swanton C. Tracking the genomic evolution of esophageal adenocarcinoma through neoadjuvant chemotherapy. Cancer Discov. 2015;5:821–831. doi: 10.1158/2159-8290.CD-15-0412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pectasides E., Stachler M.D., Derks S., Liu Y., Maron S., Islam M., Alpert L., Kwak H., Kindler H., Polite B., Sharma M.R., Allen K., O'Day E., Lomnicki S., Maranto M., Kanteti R., Fitzpatrick C., Weber C., Setia N., Xiao S.-Y., Hart J., Nagy R.J., Kim K.-M., Choi M.-G., Min B.-H., Nason K.S., O'Keefe L., Watanabe M., Baba H., Lanman R., Agoston A.T., Oh D.J., Dunford A., Thorner A.R., Ducar M.D., Wollison B.M., Coleman H.A., Ji Y., Posner M.C., Roggin K., Turaga K., Chang P., Hogarth K., Siddiqui U., Gelrud A., Ha G., Freeman S.S., Rhoades J., Reed S., Gydush G., Rotem D., Davison J., Imamura Y., Adalsteinsson V., Lee J., Bass A.J., Catenacci D.V. Genomic heterogeneity as a barrier to precision medicine in gastroesophageal adenocarcinoma. Cancer Discov. 2018;8:37–48. doi: 10.1158/2159-8290.CD-17-0395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu D.S., Read M., Cullinane C., Azar W.J., Fennell C.M., Montgomery K.G., Haupt S., Haupt Y., Wiman K.G., Duong C.P., Clemons N.J., Phillips W.A. APR-246 potently inhibits tumour growth and overcomes chemoresistance in preclinical models of oesophageal adenocarcinoma. Gut. 2015;64:1506–1516. doi: 10.1136/gutjnl-2015-309770. [DOI] [PubMed] [Google Scholar]
- 10.Singhi A.D., Foxwell T.J., Nason K., Cressman K.L., McGrath K.M., Sun W., Bahary N., Zeh H.J., Levy R.M., Luketich J.D., Davidson J.M. Smad4 loss in esophageal adenocarcinoma is associated with an increased propensity for disease recurrence and poor survival. Am J Surg Pathol. 2015;39:487–495. doi: 10.1097/PAS.0000000000000356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gotovac J.R., Fujihara K.M., Phillips W.A., Clemons N.J. TGF-beta signaling and its targeted therapy in gastrointestinal cancers. Discov Med. 2018;26:103–112. [PubMed] [Google Scholar]
- 12.Seoane J., Le H.-V., Shen L., Anderson S.A., Massagué J. Integration of Smad and forkhead pathways in the control of neuroepithelial and glioblastoma cell proliferation. Cell. 2004;117:211–223. doi: 10.1016/s0092-8674(04)00298-3. [DOI] [PubMed] [Google Scholar]
- 13.Frankell A.M., Jammula S., Li X., Contino G., Killcoyne S., Abbas S., Perner J., Bower L., Devonshire G., Ococks E., Grehan N., Mok J., O'Donovan M., MacRae S., Eldridge M.D., Tavaré S., Fitzgerald R.C. The landscape of selection in 551 esophageal adenocarcinomas defines genomic biomarkers for the clinic. Nat Genet. 2019;51:506–516. doi: 10.1038/s41588-018-0331-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blum A.E., Venkitachalam S., Ravillah D., Chelluboyina A.K., Kieber-Emmons A.M., Ravi L., Kresak A., Chandar A.K., Markowitz S.D., Canto M.I., Wang J.S., Shaheen N.J., Guo Y., Willis J.E., Chak A., Varadan V., Guda K. Systems biology analyses show hyperactivation of transforming growth factor-β and JNK signaling pathways in esophageal cancer. Gastroenterology. 2019;156:1761–1774. doi: 10.1053/j.gastro.2019.01.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rees J.R., Onwuegbusi B.A., Save V.E., Alderson D., Fitzgerald R.C. In vivo and in vitro evidence for transforming growth factor-β1-mediated epithelial to mesenchymal transition in esophageal adenocarcinoma. Cancer Res. 2006;66:9583–9590. doi: 10.1158/0008-5472.CAN-06-1842. [DOI] [PubMed] [Google Scholar]
- 16.Chen D., Cao G., Qiao C., Liu G., Zhou H., Liu Q. Alpha B-crystallin promotes the invasion and metastasis of gastric cancer via NF-κB-induced epithelial-mesenchymal transition. J Cell Mol Med. 2018;22:3215–3222. doi: 10.1111/jcmm.13602. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 17.Shi C., Yang X., Bu X., Hou N., Chen P., Liu Q. Alpha B-crystallin promotes the invasion and metastasis of colorectal cancer via epithelial-mesenchymal transition. Biochem Biophys Res Commun. 2017;489:369–374. doi: 10.1016/j.bbrc.2017.05.070. [DOI] [PubMed] [Google Scholar]
- 18.Chen S., Chen X., Xie Ge, He Y., Yan D., Zheng D., Li S., Fu X., Li Y., Pang X., Hu Z., Li H., Tan W., Li J. Cdc6 contributes to cisplatin-resistance by activation of ATR-Chk1 pathway in bladder cancer cells. Oncotarget. 2016;7:40362–40376. doi: 10.18632/oncotarget.9616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gamell C., Gulati T., Levav-Cohen Y., Young R.J., Do H., Pilling P., Takano E., Watkins N., Fox S.B., Russell P., Ginsberg D., Monahan B.J., Wright G., Dobrovic A., Haupt S., Solomon B., Haupt Y. Reduced abundance of the E3 ubiquitin ligase E6AP contributes to decreased expression of the INK4/ARF locus in non–small cell lung cancer. Sci Signal. 2017;10 doi: 10.1126/scisignal.aaf8223. [DOI] [PubMed] [Google Scholar]
- 20.Ross-Innes C.S., Becq J., Warren A., Cheetham R.K., Northen H., O'Donovan M., Malhotra S., di Pietro M., Ivakhno S., He M., Weaver J.M.J., Lynch A.G., Kingsbury Z., Ross M., Humphray S., Bentley D., Fitzgerald R.C. Whole-genome sequencing provides new insights into the clonal architecture of Barrett's esophagus and esophageal adenocarcinoma. Nat Genet. 2015;47:1038. doi: 10.1038/ng.3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin D.-C., Dinh H.Q., Xie J.-J., Mayakonda A., Silva T.C., Jiang Y.-Y., Ding L.-W., He J.-Z., Xu X.-E., Hao J.-J., Wang M.R., Li C., Xu L.Y., Li E.M., Berman B.P., Phillip Koeffler H. Identification of distinct mutational patterns and new driver genes in oesophageal squamous cell carcinomas and adenocarcinomas. Gut. 2018;67:1769–1779. doi: 10.1136/gutjnl-2017-314607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cancer Genome Atlas Research Network Integrated genomic characterization of oesophageal carcinoma. Nature. 2017;541:169–175. doi: 10.1038/nature20805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Secrier M., Li X., De Silva N., Eldridge M.D., Contino G., Bornschein J., MacRae S., Grehan N., O'Donovan M., Miremadi A., Yang T.P., Bower L., Chettouh H., Crawte J., Galeano-Dalmau N., Grabowska A., Saunders J., Underwood T., Waddell N., Barbour A.P., Nutzinger B., Achilleos A., Edwards P.A., Lynch A.G., Tavaré S., Fitzgerald R.C. Mutational signatures in esophageal adenocarcinoma define etiologically distinct subgroups with therapeutic relevance. Nat Genet. 2016;48:1131–1141. doi: 10.1038/ng.3659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Agrawal N., Jiao Y., Bettegowda C., Hutfless S.M., Wang Y., David S., Cheng Y., Twaddell W.S., Latt N.L., Shin E.J., Wang L.D., Wang L., Yang W., Velculescu V.E., Vogelstein B., Papadopoulos N., Kinzler K.W., Meltzer S.J. Comparative genomic analysis of esophageal adenocarcinoma and squamous cell carcinoma. Cancer Discov. 2012;2:899–905. doi: 10.1158/2159-8290.CD-12-0189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reid B.J., Prevo L.J., Galipeau P.C., Sanchez C.A., Longton G., Levine D.S., Blount P.L., Rabinovitch P.S. Predictors of progression in Barrett’s esophagus II: baseline 17p (p53) loss of heterozygosity identifies a patient subset at increased risk for neoplastic progression. Am J Gastroenterol. 2001;96:2839–2848. doi: 10.1111/j.1572-0241.2001.04236.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murai J., Zhang H., Pongor L., Tang S.-W., Jo U., Moribe F., Ma Y., Tomita M., Pommier Y. Chromatin remodeling and immediate early gene activation by SLFN11 in response to replication stress. Cell Rep. 2020;30:4137–4151. e6. doi: 10.1016/j.celrep.2020.02.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang D.H., Clemons N.J., Miyashita T., Dupuy A.J., Zhang W., Szczepny A., Corcoran–Schwartz I.M., Wilburn D.L., Montgomery E.A., Wang J.S., Jenkins N.A., Copeland N.A., Harmon J.W., Phillips W.A., Watkins D.N. Aberrant epithelial–mesenchymal Hedgehog signaling characterizes Barrett's metaplasia. Gastroenterology. 2010;138:1810–1822. e2. doi: 10.1053/j.gastro.2010.01.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu X., Liu Y., Yin L., Peng Y., Peng Y., Gao Y., Yuan B., Zhu Q., Cao T., Xie B., Sun L., Chen Y., Gong Z., Qiu Y., Fan X., Li X. Radiation-promoted CDC6 protein stability contributes to radioresistance by regulating senescence and epithelial to mesenchymal transition. Oncogene. 2019;38:549–563. doi: 10.1038/s41388-018-0460-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hartwig T., Montinaro A., von Karstedt S., Sevko A., Surinova S., Chakravarthy A., Taraborrelli L., Draber P., Lafont E., Vargas F.A., El-Bahrawy M.A., Quezada S.A., Walczak H. The TRAIL-induced cancer secretome promotes a tumor-supportive immune microenvironment via CCR2. Mol Cell. 2017;65:730–742. e5. doi: 10.1016/j.molcel.2017.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kotsantis P., Petermann E., Boulton S.J. Mechanisms of oncogene-induced replication stress: jigsaw falling into place. Cancer Discov. 2018;8:537–555. doi: 10.1158/2159-8290.CD-17-1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim W.Y., Sharpless N.E. The regulation of INK4/ARF in cancer and aging. Cell. 2006;127:265–275. doi: 10.1016/j.cell.2006.10.003. [DOI] [PubMed] [Google Scholar]
- 32.Zaretsky J.M., Garcia-Diaz A., Shin D.S., Escuin-Ordinas H., Hugo W., Hu-Lieskovan S., Torrejon D.Y., Abril-Rodriguez G., Sandoval S., Barthly L., Saco J., Homet Moreno B., Mezzadra R., Chmielowski B., Ruchalski K., Shintaku I.P., Sanchez P.J., Puig-Saus C., Cherry G., Seja E., Kong X., Pang J., Berent-Maoz B., Comin-Anduix B., Graeber T.G., Tumeh P.C., Schumacher T.N.M., Lo R.S., Ribas A. Mutations associated with acquired resistance to PD-1 blockade in melanoma. N Engl J Med. 2016;375:819–829. doi: 10.1056/NEJMoa1604958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Frankel A., Armour N., Nancarrow D., Krause L., Hayward N., Lampe G., Smithers B.M., Barbour A. Genome-wide analysis of esophageal adenocarcinoma yields specific copy number aberrations that correlate with prognosis. Genes Chromosomes Cancer. 2014;53:324–338. doi: 10.1002/gcc.22143. [DOI] [PubMed] [Google Scholar]
- 34.Dulak A.M., Schumacher S.E., Van Lieshout J., Imamura Y., Fox C., Shim B., Ramos A.H., Saksena G., Baca S.C., Baselga J., Tabernero J., Barretina J., Enzinger P.C., Corso G., Roviello F., Lin L., Bandla S., Luketich J.D., Pennathur A., Meyerson M., Ogino S., Shivdasani R.A., Beer D.G., Godfrey T.E., Beroukhim R., Bass A.J. Gastrointestinal adenocarcinomas of the esophagus, stomach, and colon exhibit distinct patterns of genome instability and oncogenesis. Cancer Res. 2012;72:4383–4393. doi: 10.1158/0008-5472.CAN-11-3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Petropoulos M., Tsaniras S.C., Taraviras S., Lygerou Z. Replication licensing aberrations, replication stress, and genomic instability. Trends Biochem Sci. 2019;44:752–764. doi: 10.1016/j.tibs.2019.03.011. [DOI] [PubMed] [Google Scholar]
- 36.Halazonetis T.D., Gorgoulis V.G., Bartek J. An oncogene-induced DNA damage model for cancer development. Science. 2008;319:1352–1355. doi: 10.1126/science.1140735. [DOI] [PubMed] [Google Scholar]
- 37.Di Micco R., Fumagalli M., Cicalese A., Piccinin S., Gasparini P., Luise C., Schurra C., Nuciforo P.G., Bensimon A., Maestro R., Pelicci P.G., di Fagagna d'Adda. Oncogene-induced senescence is a DNA damage response triggered by DNA hyper-replication. Nature. 2006;444:638–642. doi: 10.1038/nature05327. [DOI] [PubMed] [Google Scholar]
- 38.Palanca-Wessels M.C., Barrett M.T., Galipeau P.C., Rohrer K.L., Reid B.J., Rabinovitch P.S. Genetic analysis of long-term Barrett's esophagus epithelial cultures exhibiting cytogenetic and ploidy abnormalities. Gastroenterology. 1998;114:295–304. doi: 10.1016/s0016-5085(98)70480-9. [DOI] [PubMed] [Google Scholar]
- 39.Suchorolski M.T., Paulson T.G., Sanchez C.A., Hockenbery D., Reid B.J. Warburg and Crabtree effects in premalignant Barrett's esophagus cell lines with active mitochondria. PLoS One. 2013;8 doi: 10.1371/journal.pone.0056884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nicoletti I., Migliorati G., Pagliacci M., Grignani F., Riccardi C. A rapid and simple method for measuring thymocyte apoptosis by propidium iodide staining and flow cytometry. J Immunol Methods. 1991;139:271–279. doi: 10.1016/0022-1759(91)90198-o. [DOI] [PubMed] [Google Scholar]
- 41.Ran F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Aubrey B.J., Kelly G.L., Kueh A.J., Brennan M.S., O’Connor L., Milla L., Wilcox S., Tai L., Strasser A., Herold M.J. An inducible lentiviral guide RNA platform enables the identification of tumor-essential genes and tumor-promoting mutations in vivo. Cell Rep. 2015;10:1422–1432. doi: 10.1016/j.celrep.2015.02.002. [DOI] [PubMed] [Google Scholar]
- 43.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011;17:10–12. [Google Scholar]
- 44.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Boeva V., Popova T., Bleakley K., Chiche P., Cappo J., Schleiermacher G., Janoueix-Lerosey I., Delattre O., Barillot E., Control-FREEC a tool for assessing copy number and allelic content using next-generation sequencing data. Bioinformatics. 2012;28:423–425. doi: 10.1093/bioinformatics/btr670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Scheinin I., Sie D., Bengtsson H., van de Wiel M.A., Olshen A.B., van Thuijl H.F., van Essen H.F., Eijk P.P., Rustenburg F., Meijer G.A., Reijneveld J.C., Wesseling P., Pinkel D., Albertson D.G., Ylstra B. DNA copy number analysis of fresh and formalin-fixed specimens by shallow whole-genome sequencing with identification and exclusion of problematic regions in the genome assembly. Genome Res. 2014;24:2022–2032. doi: 10.1101/gr.175141.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kader T., Goode D.L., Wong S.Q., Connaughton J., Rowley S.M., Devereux L., Byrne D., Fox S.B., Arnau G.M., Tothill R.W., Campbell I.G., Gorringe K.L. Copy number analysis by low coverage whole genome sequencing using ultra low-input DNA from formalin-fixed paraffin embedded tumor tissue. Genome Med. 2016;8:1–13. doi: 10.1186/s13073-016-0375-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kader T., Hill P., Zethoven M., Goode D.L., Elder K., Thio N., Doyle M., Semple T., Sufyan W., Byrne D.J., Pang J.B., Murugasu A., Miligy I.M., Green A.R., Rakha E.A., Fox S.B., Mann G.B., Campbell I.G., Gorringe K.L. Atypical ductal hyperplasia is a multipotent precursor of breast carcinoma. J Pathol. 2019;248:326–338. doi: 10.1002/path.5262. [DOI] [PubMed] [Google Scholar]
- 49.Chin S.F., Teschendorff A.E., Marioni J.C., Wang Y., Barbosa-Morais N.L., Thorne N.P., Costa J.L., Pinder S.E., Van de Wiel M.A., Green A.R. High-resolution aCGH and expression profiling identifies a novel genomic subtype of ER negative breast cancer. Genome Biol. 2007;8:R215. doi: 10.1186/gb-2007-8-10-r215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 51.Galili T., O’Callaghan A., Sidi J., Sievert C. heatmaply: an R package for creating interactive cluster heatmaps for online publishing. Bioinformatics. 2017;34:1600–1602. doi: 10.1093/bioinformatics/btx657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kassambara A., Mundt F. Factoextra: extract and visualize the results of multivariate data analyses. R package version 1.0.6. 2017 Available from: https://rpkgs.datanovia.com/factoextra/index.html. [Google Scholar]
- 53.Therneau T. A package for survival analysis in R. R package version 3.2-7, 2020. 2020. https://CRAN.R-project.org/package=survival Available from:
- 54.Therneau T.M., Grambsch P.M. Modeling survival data: extending the Cox model. Springer; New York, NY: 2000. The Cox model; pp. 39–77. [Google Scholar]
- 55.R Core Team R: a language and environment for statistical computing. 2020. Vienna, Austria: R Foundation for Statistical Computing. https://www.R-project.org Available from: