Abstract
Background
Multidrug and extensively drug-resistant tuberculosis (M/XDR-TB) pose major threats to global health. Diagnosis accuracy and delay have been the major drivers for the upsurge of M/XDR-TB. Pyrosequencing (PSQ) is a novel, real-time DNA sequencing for rapid detection of mutations associated with M/XDR-TB. We aimed to systematically synthesize the evidence on the diagnostic accuracy of PSQ for M/XDR-TB.
Methods
We conducted an electronic search of PubMed, Embase, Biosis, Web of Science, and Google Scholar up to March 2020. We used the QUADAS‐2 (Quality Assessment of Diagnostic Accuracy Studies) tool to assess the quality of studies, the BRMA (bivariate random‐effects meta-analysis) model to synthesize diagnostic accuracies, and the Rev-Man 5.4 software to perform the meta-analyses. We analyzed dichotomous data using the risk ratio (RR) with a 95% confidence interval. PROSPERO Registration ID: CRD42020200817.
Results
The analysis included seven studies, with a total sample of 3,165. At 95% confidence interval, the pooled sensitivity and specificity of PSQ were 89.7 (CI: 83.5–93.8) and 97.8 (CI: 94.9–99.1) for Isoniazid, 94.6 (CI: 90.9–96.8) and 98.5 (CI: 96.5–99.3) for Rifampicin, 87.9 (CI: 81.2–92.4) and 98.8 (CI: 97.2–99.5) for Fluoroquinolone, 83.5 (CI: 72.8–90.5) and 99.4 (CI: 98.3–99.8) for Amikacin, 79 (CI: 67–8-87) and 97.9 (CI: 95.5–99) for Capreomycin, and 69.6 (CI: 57–79.8) and 98.2 (CI: 95.9–99.2) for Kanamycin. The overall pooled sensitivity and specificity were 85.8 (CI: 76.7–91.7) and 98.5 (CI: 96.5–99.3), respectively.
Conclusion
According to the pooled data, PSQ is highly sensitive and specific for detecting M/XDR-TB, both from clinical specimens and culture isolates, and within a shorter turnaround time. We suggest a continued synthesis of the evidence on the cost-effectiveness and technical feasibilities of PSQ in low-income countries context, including sub-Saharan Africa.
Keywords: Pyrosequencing (PSQ), Diagnostic, Multi drug-resistant tuberculosis (MDR), Extensively drug-resistant tuberculosis (XDR), Systematic review, Meta-analysis
1. Introduction
Tuberculosis (TB) remained a major threat to global health [1], [2], [3]. According to the World Health Organization’s global TB report 2020 [4], an estimated 10.0 million people fell ill with TB in 2019 and close to half a million people developed rifampicin-resistant TB (RR-TB), of which 78% had multidrug-resistant TB (MDR-TB). The upsurge of multidrug and extensively drug-resistant tuberculosis (M/XDR-TB), as well as the totally drug-resistant TB (TDR-TB)” is hindering the global TB control efforts [5], [6]. The major gap has been the insufficient capability to diagnose M/XDR-TB in a timely and cost-effective manner [7], [8].
Reliable molecular diagnostics, which can detect specific mutations that confer drug resistance, are the most promising technologies for rapid identification of M/XDR-TB. These technologies, most remarkably the Hain MTBDR Plus line probe assay [9], [10] and the Gene Xpert MTB/RIF assay [11], [12], are being used globally to screen for the most prevalent mutations associated with resistance to isoniazid (INH) and/or rifampin (RIF). However, they cannot detect all mutations in specific region and are unable to distinguish silent mutations [13]. Therefore, there has been little progress towards the broad application of these tools for M/XDR-TB.
Pyrosequencing (PSQ) emerges as a real-time assay for rapid sequencing of small segments of genomic DNA to detect M/XDR-TB accurately and reliably [14], [15], [16]. It was reported that it overcomes the limitation of previous M/XDR-TB assays and detects resistance to more drugs in a shorter time [17], [18]. It not only determines the presence or absence of these mutations but also displays the exact sequence data to guide treatment decisions.
However, studies are lacking that synthesized the evidence on the diagnostic accuracy of PSQ for M/XDR-TB. Multiple trials are evaluating novel therapeutic agents, repurposed agents, and novel care cascades to transform the landscape for M/XDR-TB [19], [20], [21], [22]. Looking for new diagnostic tools and following-up of diagnostic accuracy of existing ones are equally important to harness the End-TB pathway. Thus, we aimed to synthesize the evidence on the diagnostic accuracy of PSQ for M/XDR‐TB.
2. Methods
The study followed the Preferred Reporting Items for Systematic Review and Meta-Analysis Protocols (PRISMA-P) 2015 guidelines [23] for reporting results, QUADAS‐2 (Quality Assessment of Diagnostic Accuracy Studies) tool [24] to assess the quality of studies, and the BRMA (bivariate random‐effects meta-analysis) model [25] to synthesize diagnostic accuracies. The protocol has been registered on PROSPERO, ID: CRD42020200817 [26].
2.1. Inclusion criteria
We included studies that met the study’s predefined criteria:
-
•
Types of studies: Multi or single-center, prospective, head-to-head clinical evaluation
-
•
Participants: Patients of any age who had: isoniazid, rifampicin, fluoroquinolones, amikacin, kanamycin and capreomycin resistant TB.
-
•
Index tests: PSQ
-
•
Target conditions: M/XDR-TB
-
•
Reference standard: Phenotypic DST/ BACTEC MGIT960 either on solid or liquid media
2.2. Search strategy
We searched the following databases to find abstracts and their full-text articles: PubMed, Embase, Biosis, Web of Science, and Google Scholar. We searched up to March 2020 using the keywords and search terms “Tuberculosis* AND Pyrosequencing*”, Multidrug-Resistant“, ”Drug Resistance“, ”Drug Resistance, Bacterial“, ”extensively drug resistance“, ”pyrosequencing*“, ”Tuberculosis second-line drug-resistant“, ”PSQ“, ”phenotype DST*“, ”BACTEC MGIT960*“, ”Tuberculosis total drug resistance*“, and ”pyrosequencing, XDR TB“.
2.3. Data collection and analysis
Two review authors independently extracted data and assessed the quality of included studies using QUADAS‐2 tool. We had combined data for PSQ and assessed sensitivity and specificity for anti-M/XDR-TB drugs. We explored the influence on accuracy estimates of individual drugs within a drug class of the same reference standards. We carried out most analyses using a bivariate random‐effects model with culture‐based DST as a reference standard. Where sufficient data were available, we have compiled data applying meta‐analysis. Plotted estimates of sensitivities and specificities in forest plots with 95% Confidence Intervals (Cis). We used Rev-Man 5.4 software to perform the meta-analyses.
2.4. Selection of studies
Two review authors independently examined titles and abstracts of identified citations. Titles and/or abstracts of all citations were screened further for a full-text review using the predefined criteria. For full-text articles, we resolved any discrepancies by consensus.
We extracted results from the selected studies that evaluated the accuracy (sensitivity and specificity) of PSQ for M/XDR-TB in clinical specimens or isolates. We considered the following pre-determined criteria to include studies in our review:
-
•
PSQ was utilized for rapid M/XDR-TB detection
-
•
Detection was performed in clinical specimens or isolates
-
•
The study evaluated accuracy (sensitivity and specificity) of PSQ
-
•
Phenotypic DST on either liquid or solid media used as reference standard
-
•
Sample size should include at least 20, with at least 10 specimens showing M. TB with drug resistance, and at least 10 with drug-susceptible strains.
2.5. Data extraction and management
We customized a data extraction form designed to extract and capture the following information:
-
•
Details of the study: first author; publication year; country where testing was performed; specimen country origin; setting; participant selection procedure; and the number of participants enrolled
-
•
Target conditions: resistance to the following drugs: isoniazid, rifampicin, fluoroquinolones, amikacin, kanamycin and capreomycin.
-
•
Resistances to individual drugs: Isoniazid, rifampin fluoroquinolone, and capreomycin, Kanamycin, and Amikacin.
-
•
Reference standard: type
-
•
Specimen: type; condition (fresh or frozen); definition of a positive test; type of testing
-
•
Outcomes: sensitivity and specificity
2.6. Assessment of methodological quality
Included articles were assessed by the first author, who extracted data using the extraction form. A second reviewer independently extracted data from a subset of the included studies and differences between the two reviewers were reconciled by consensus. Quality of included studies was assessed using the QUADAS criteria.
2.7. Investigations of heterogeneity
Due to the limited number of included studies, it is difficult to assess the source of heterogeneity using meta-regression analysis. We have scrutinized heterogeneity through visual examination of forest plots of sensitivity and specificity. Then, if adequate studies were available, we have explored the possible influence of the following pre-specified categorical covariate: resistance to the following drugs: Isoniazid, Rifampicin, fluoroquinolone, Amikacin, Kanamycin, and capreomycin resistance and based on drug group.
We had planned to explore the effect of HIV status, the condition of the specimen (fresh or frozen), sample volume, and population (patients thought to have MDR‐TB or XDR‐TB) on summary estimates of sensitivity and specificity in a meta‐regression analysis by adding covariate terms to the bivariate model. However, there was inadequate data that specify all the above information for these additional analyses.
2.8. Sensitivity analyses
For our analyses using the culture‐based DST reference standard, we have conducted sensitivity analyses for QUADAS‐2 items to explore methodological quality of the studies, checking whether (Table 3):
-
•
A consecutive or random sample of patients/specimens were enrolled
-
•
A case‐control design was avoided
-
•
Index test results were interpreted without knowledge of the results of the reference standard
-
•
The test was applied in the way recommended by the manufacturer
Table 3.
Sensitivity analysis of included studies.
| Signaling questions | #of studies | Sensitivity | Specificity |
|---|---|---|---|
| Was a consecutive or random sample of patients/specimens enrolled? Yes | 6 | 87% (82.7–90.3) |
98.5% (97.8–98.9) |
| Was a case‐control design avoided? Yes | 7 | 86.6% (82.4–89.9) |
98.5% (97.8–98.9) |
| Were the index test results interpreted without knowledge of the results of the reference standard? Yes | 7 | 86.6% (82.4–89.9) |
98.5% (97.8–98.9) |
| Was the test applied in the way recommended by the manufacturer (index test domain, low concern about applicability)? Yes | 6 | 85.5% (82.1–88.9) |
98.3% (97.5–98.8) |
2.9. Assessment of reporting bias
We did not assess for publication bias of included data as such techniques were not useful for biases within diagnostic test accuracy studies. We summarized data on inter‐reader variability; but not for intra-reader variability as information were not described in the included studies. We had also intended to summarize two patient outcomes, time‐to‐diagnosis, and time‐to‐treatment initiation; however, time‐to‐diagnosis was the only outcome described in the included studies.
3. Result
3.1. Search results
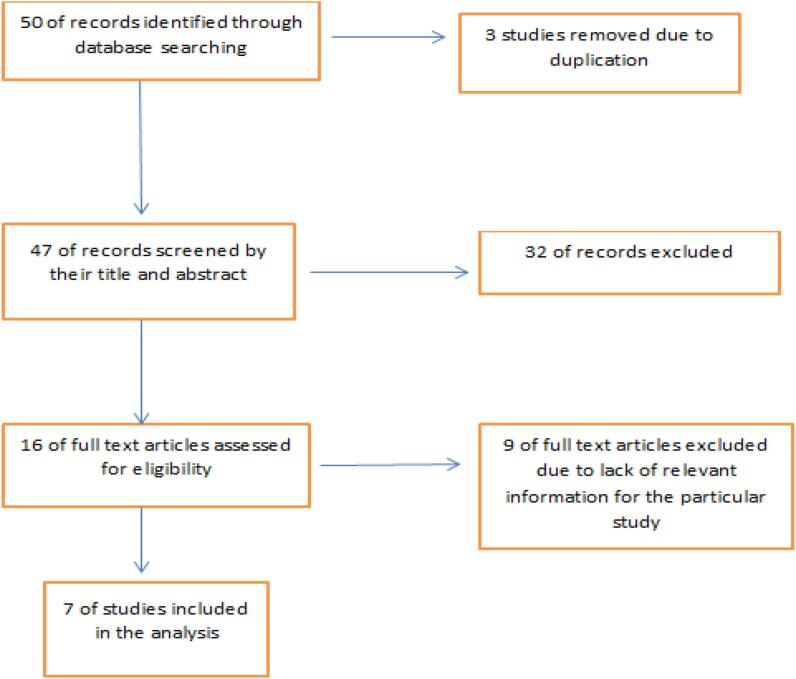
We found 50 studies from the initial search, of which seven [27], [28], [29], [30], [31], [32], [33] met the inclusion criteria of this review (Fig. 1).
Fig. 1.
PRISMA flow diagram of the study.
3.2. Methodological quality
Fig. 2, Fig. 3 show the risk of bias and applicability concerns for each of the 7 included studies. In the patient selection domain, we judged that 1 study (14.3%) had an unclear risk of bias. Since the report on one study did not provide detailed information on the procedure of patient selection, we judged that 6 studies (85.7%) [28], [29], [30], [31], [32], [33] had a low risk of bias in the index test. We have judged that only one study (14.3%) as a low risk of bias in flow and timing of sample collection and study procedure, while the authors of the other studies did not provide information on the timing of sample collection (Fig. 2).
Fig. 2.
Risk of bias and applicability concerns:
Fig. 3.
Risk of bias and applicability concerns summary.
Regarding applicability, six studies [27], [28], [29], [31], [32], [33] had low concern in patient selection and only one study [29] had unclear concerns in the index test domain. In the reference-standard domain, one study (14.3%) [27] had unclear selection and application of reference standard since it was impossible to find out which reference standard had been used (Fig. 3).
3.3. Study characteristics
The seven studies that were included had a total sample of 3165. Of the seven studies, one study reported results using both clinical specimens and clinical isolates, 4 studies (57%) evaluated patients from multi-center of different countries, and the rest (43%) were from a single country. The median sample size (interquartile range) was 593 (57 to 1128). Five (72%) studies reported results of INH, RIF, FQ, AMK, KAN, and CAP. One study [30] did not include the result of AMK, CAP, and KAN, and one study [33] did not include the results of AMK and INH. Table 1 summarized all relevant data extracted from included studies.
Table 1.
Data extracted from all included studies.
| First-line |
Second-line |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sensitivity | Specificity | Sensitivity | Specificity | ||||||||||
| Study | Country | INH | RIF | INH | RIF | FQ | AMK | CAP | KAN | FQ | AMK | CAP | KAN |
| Lin (2013) | USA | 88% | 96% | 100% | 100% | 87% | 100% | 100% | 86% | 100% | 99% | 99% | 100% |
| Ajbani (2014) | India, South Africa, Moldova, Philippines | 94% | 96% | 96% | 100% | 93% | 84% | 88% | 68% | 100% | 100% | 97% | 100% |
| Georghiou (2015) | India | 98% | 98% | 97% | 100% | 96% | 94% | 94% | 93% | 99% | 100% | 99% | 91% |
| Moldova | 94% | 94% | 96% | 100% | 64% | 33% | 40% | 79% | 99% | 99% | 99% | 99% | |
| South Africa | 71% | 77% | 94% | 98% | 90% | 92% | 85% | 92% | 99% | 98% | 98% | 98% | |
| Bravo (2009) | Philippines | 64% | 97% | 100% | 96 | 70% | – | – | – | 100% | – | – | – |
| Catanzaro (2015) | India, Moldova, Philippines, Peru, South Africa, USA | 95% | 94% | 96% | 99% | 94% | 84% | 84% | 50% | 99% | 99% | 99% | 99% |
| Engström (2012) | Sweden | 94% | 95% | 100% | 100% | 87% | 82% | 80% | 84% | 100% | 100% | 97% | 98% |
3.4. Pooled sensitivity and specificity
The overall pooled sensitivity 85.8% (95% CI: 76.7–91.7) and pooled specificity 98.5 (95% CI: 96.5–99.3) (Table 2, Fig. 4, Fig. 5).
Table 2.
Sensitivity and specificity of each drug of included studies.
| Type of drug | Sensitivity | Lower limit | Upper limit | Specificity | Lower limit | |
|---|---|---|---|---|---|---|
| AMK | 83.5 | 72.8 | 90.5 | 99.4 | 98.3 | 99.8 |
| CAP | 79 | 67.8 | 87 | 97.9 | 95.5 | 99 |
| FQ | 87.9 | 81.2 | 92.4 | 98.8 | 97.2 | 99.5 |
| INH | 89.7 | 83.5 | 93.8 | 97.8 | 94.9 | 99.1 |
| KAN | 69.6 | 57 | 79.8 | 98.2 | 95.9 | 99.2 |
| RIF | 94.6 | 90.7 | 96.8 | 98.5 | 96.5 | 99.3 |
Fig. 4.
Sensitivity and specificity of PSQ for each TB-drug included in the studies.
Fig. 5.
Summary ROC Plot of included studies.
3.5. QUADAS‐2 analysis
4. Discussion
Here, we conducted a systematic review and meta-analysis on the value of PSQ for drug susceptibility of M. tuberculosis in direct specimens and isolates of 3265 enrolled patient samples. We found a high overall pooled sensitivity of 85.8% and a pooled specificity of 98.5%.
There is a critical need for rapid and accurate diagnosis of DR-TB. Earlier detection of DR-TB if followed by prompt and appropriate treatment changes could potentially reduce disease progression and transmission. As our pooled analysis indicated, PSQ is highly specific and comparable to diagnostic tools currently in use such as phenotypic DST, LPA, or Xpert. PSQ has an added value as it is more rapid, can be applied directly from the clinical specimen, make available exact data about all known mutations at once, is open and adaptable to new mutations, and allows extensive post-processing analyses [34], [35]. Previous studies reported that PSQ is more affordable for less affluent settings, typically relevant for areas around the world that are highly burdened with drug-susceptible as well as drug-resistant TB, easily distinguish between missense and silent mutation and detect outside the target region compared to phenotypic Compared with DNA sequencing, the concordance rate of PSQ for first-line drugs was 100% [35], [36]. The overall concordance between PSQ and GenoType MTBDR plus in clinical strains was 99.1% [27]. Thus, PSQ for conferring resistance to RIF and other first-line anti-TB drugs is reliable. Compared with other methods, such as PCR-single strand conformational polymorphism assay 20 and LiPA (line probe assay), PSQ is shown to save time and labor [37]. For sequencing, it is possible to run 96 samples in 1–2 h after PCR amplification and can analyze 96 single nucleotide polymorphisms in 10 min [17], [35], [36]. PSQ can therefore be used as a high-throughput and rapid assay for M/XDR-TB detection.
All the included studies reported PSQ as a rapid and accurate method for detecting MTB resistance to INH, RIF, and FQ and SLIs. One study reported low sensitivity of SLIs (KAN and CAP), unlike the other studies. This could be for a reason that PSQ did not detect “eis” mutations. Next-generation PSQ assays have now included “eis” mutations that improved their capacity to confer phenotypic KAN resistance [29].
This review had some limitations. Despite searching several sources, some eligible studies may not have been identified and relatively small numbers of eligible studies were included, hampering the subgroup analysis and meta-regression methods. Furthermore, the study does not address some major first-choice drugs included in the updated WHO recommendations for the treatment of MDRTB, including drug susceptibility testing of bedaquiline, clofazimine, linezolid, delamanid and pretomanid. Another limitation is it does not address some of the variations in techniques used under the umbrella of pyrosequencing; over the last decades, the technique has made several improvements that potentially impact the results.
On contrary, this review has several strengths. It included studies assessing the diagnostic accuracy of PSQ for both first-line and second-line anti-TB drugs, including second-line injectable drugs. The results were based on strict and careful literature searches, study inclusion, and data extraction. The work strictly followed internationally recognized guidelines and procedures.
5. Conclusions
According to the pooled data, PSQ is highly sensitive and specific for detecting M/XDR-TB, both from clinical specimens and culture isolates, and within a shorter turnaround time. We suggest a continued synthesis of the evidence on the cost-effectiveness and technical feasibilities of PSQ in low-income countries context, including sub-Saharan Africa.
Funding
TM received support from the Fogarty International Center and National Institute of Allergy and Infectious Diseases of the U.S. National Institutes of Health under Award Number D43TW009127.
CRediT authorship contribution statement
Emnet Getachew Adere: Conceptualization, Data curation, Formal analysis, Writing - original draft. Tsegaye Adebeta: Data curation, Formal analysis, Writing - original draft. Desye Gebrie: Formal analysis, Writing - original draft. Loveness Charlie: Data curation, Writing - original draft. Bibie Said: Data curation, Writing - original draft. Dawit Getachew Assefa: Data curation, Writing - original draft. Cathrine Lydiah Wanjiru: Data curation, Writing - original draft. Eden Dagnachew Zeleke: Data curation, Writing - original draft. Hanna Amanuel Tesfahunei: Data curation, Writing - original draft. Mekdelawit Abebe: Data curation, Writing - original draft. Michele Joseph: Data curation, Writing - original draft. Tsegahun Manyazewal: Supervision.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgment
The authors are grateful to the Center for Innovative Drug Development and Therapeutic Trials for Africa (CDT-Africa), College of Health Sciences, Addis Ababa University for supporting this review.
References
- 1.Lee A., Xie Y.L., Barry C.E., Chen R.Y. Current and future treatments for tuberculosis. BMJ. 2020;368 doi: 10.1136/bmj.m216. [DOI] [PubMed] [Google Scholar]
- 2.Mohammed H., Oljira L., Roba K.T., Ngadaya E., Ajeme T., Haile T. Burden of tuberculosis and challenges related to screening and diagnosis in Ethiopia. J Clin Tuberc Other Mycobact Dis. 2020;19:100158. doi: 10.1016/j.jctube.2020.100158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Manyazewal T., Woldeamanuel Y., Blumberg H.M., Fekadu A., Marconi V.C. The fight to end tuberculosis must not be forgotten in the COVID-19 outbreak. Nat Med. 2020;26(6):811–812. doi: 10.1038/s41591-020-0917-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.World Health Organization (WHO). Global tuberculosis report 2020. Switzerland, Geneva, WHO; 2020. Available at https://www.who.int/publications/i/item/9789240013131.
- 5.Svadzian A., Sulis G., Gore G., Pai M., Denkinger C.M. Differential yield of universal versus selective drug susceptibility testing of patients with tuberculosis in high-burden countries: a systematic review and meta-analysis. BMJ Glob Health. 2020;5(10):e003438. doi: 10.1136/bmjgh-2020-003438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mussie K.M., Yimer S.A., Manyazewal T., Gradmann C., Torpey K. Exploring local realities: Perceptions and experiences of healthcare workers on the management and control of drug-resistant tuberculosis in Addis Ababa, Ethiopia. PLoS ONE. 2019;14(11):e0224277. doi: 10.1371/journal.pone.0224277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chisompola N.K., Streicher E.M., Muchemwa C.M.K., Warren R.M., Sampson S.L. Molecular epidemiology of drug resistant Mycobacterium tuberculosis in Africa: a systematic review. BMC Infect Dis. 2020;20(1):344. doi: 10.1186/s12879-020-05031-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Murhula Kashongwe I., Mawete F., Anshambi N., Maingowa N., Aloni M., Lukaso L'osenga L. Challenge to treat pre-extensively drug-resistant tuberculosis in a low-income country: a report of 12 cases. J Clin Tuberc Other Mycobact Dis. 2020;21:100192. doi: 10.1016/j.jctube.2020.100192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meaza A., Kebede A., Yaregal Z., Dagne Z., Moga S., Yenew B. Evaluation of genotype MTBDRplus VER 2.0 line probe assay for the detection of MDR-TB in smear positive and negative sputum samples. BMC Infect Dis. 2017;17(1) doi: 10.1186/s12879-017-2389-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.David A., Singh L., Da Silva P., Scott L., Stevens W. The Performance of the Abbott Real Time MTB RIF/INH Compared to the MTBDRplus V2 for the Identification of MDR-TB Among Isolates. Infect Drug Resist. 2020;13:3301–3308. doi: 10.2147/IDR.S247524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bainomugisa A., Gilpin C., Coulter C., Marais B.J. New Xpert MTB/XDR: added value and future in the field. Eur Respir J. 2020;56(5):2003616. doi: 10.1183/13993003.03616-2020. [DOI] [PubMed] [Google Scholar]
- 12.Venter R., Minnies S., Derendinger B., Tshivhula H., de Vos M., Dolby T. Extract from used Xpert MTB/RIF Ultra cartridges is useful for accurate second-line drug-resistant tuberculosis diagnosis with minimal rpoB-amplicon cross-contamination risk. Sci Rep. 2020;10(1) doi: 10.1038/s41598-020-59164-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oommen S., Banaji N. Laboratory diagnosis of tuberculosis: advances in technology and drug susceptibility testing. Indian J Med Microbiol. 2017;35(3):323–331. doi: 10.4103/ijmm.IJMM_16_204. [DOI] [PubMed] [Google Scholar]
- 14.Nambiar R., Shah D., Ajbani K., Kazi M., Sadani M., Shetty A. Evaluation of pyrosequencing for extensive drug resistance-defining anti-tuberculosis drugs for use in public healthcare. Tuberculosis (Edinb) 2018;110:86–90. doi: 10.1016/j.tube.2018.03.006. [DOI] [PubMed] [Google Scholar]
- 15.Molina-Moya B., Lacoma A., García-Sierra N., Blanco S., Haba L., Samper S. PyroTyping, a novel pyrosequencing-based assay for Mycobacterium tuberculosis genotyping. Sci Rep. 2017;7(1) doi: 10.1038/s41598-017-06760-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lacoma A., Molina-Moya B., Prat C., Pimkina E., Diaz J., Dudnyk A. Pyrosequencing for rapid detection of Mycobacterium tuberculosis second-line drugs and ethambutol resistance. Diagn Microbiol Infect Dis. 2015;83(3):263–269. doi: 10.1016/j.diagmicrobio.2015.07.004. [DOI] [PubMed] [Google Scholar]
- 17.Arnold C., Westland L., Mowat G., Underwood A., Magee J., Gharbia S. Single-nucleotide polymorphism-based differentiation and drug resistance detection in Mycobacterium tuberculosis from isolates or directly from sputum. Clin Microbiol Infect. 2005;11(2):122–130. doi: 10.1111/j.1469-0691.2004.01034.x. [DOI] [PubMed] [Google Scholar]
- 18.Marttila H.J., Mäkinen J., Marjamäki M., Soini H. Prospective evaluation of pyrosequencing for the rapid detection of isoniazid and rifampin resistance in clinical Mycobacterium tuberculosis isolates. Eur J Clin Microbiol Infect Dis. 2009;28(1):33–38. doi: 10.1007/s10096-008-0584-5. [DOI] [PubMed] [Google Scholar]
- 19.Lee A., Xie Y.L., Barry C.E., Chen R.Y. Current and future treatments for tuberculosis. BMJ. 2020;2(368) doi: 10.1136/bmj.m216. [DOI] [PubMed] [Google Scholar]
- 20.Subbaraman R., Jhaveri T., Nathavitharana R.R. Closing gaps in the tuberculosis care cascade: an action-oriented research agenda. J Clin Tuberc Other Mycobact Dis. 2020;19:100144. doi: 10.1016/j.jctube.2020.100144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Manyazewal T., Woldeamanuel Y., Holland D.P., Fekadu A., Blumberg H.M., Marconi V.C. Electronic pillbox-enabled self-administered therapy versus standard directly observed therapy for tuberculosis medication adherence and treatment outcomes in Ethiopia (SELFTB): protocol for a multicenter randomized controlled trial. Trials. 2020;21(1):383. doi: 10.1186/s13063-020-04324-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mohammed H., Oljira L., Roba K.T., Yimer G., Fekadu A., Manyazewal T. Containment of COVID-19 in Ethiopia and implications for tuberculosis care and research. Infect Dis Poverty. 2020;9(1):131. doi: 10.1186/s40249-020-00753-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shamseer L., Moher D., Clarke M., Ghersi D., Liberati A., Petticrew M. Preferred reporting items for systematic review and meta-analysis protocols (PRISMA-P) 2015: elaboration and explanation. BMJ. 2015;349(jan02 1):g7647. doi: 10.1136/bmj.g7647. [DOI] [PubMed] [Google Scholar]
- 24.Whiting P.F., Rutjes A.W., Westwood M.E. QUADAS-2: a revised tool for the quality assessment of diagnostic accuracy studies. Ann Intern Med. 2011;155(8):529–536. doi: 10.7326/0003-4819-155-8-201110180-00009. [DOI] [PubMed] [Google Scholar]
- 25.Riley R.D., Abrams K.R., Sutton A.J., Lambert P.C., Thompson J.R. Bivariate random-effects meta-analysis and the estimation of between-study correlation. BMC Med Res Methodol. 2007;7:3. doi: 10.1186/1471-2288-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Getachew E, Tesfai S, Kitui S, Getachew D, Danachew E. Diagnostic accuracy of pyrosequencing versus phenotypic DST for detection of multi and extensively drug-resistant tuberculosis in MDR/XDR Tb Patients. PROSPERO 2020 CRD42020200817 Available from: https://www.crd.york.ac.uk/prospero/display_record.php?ID=CRD42020200817.
- 27.Lin S.-Y.- G., Rodwell T.C., Victor T.C., Rider E.C., Pham L., Catanzaro A. Pyrosequencing for rapid detection of extensively drug-resistant Mycobacterium tuberculosis in clinical isolates and clinical specimens. J Clin Microbiol. 2014;52(2):475–482. doi: 10.1128/JCM.01821-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ajbani K., Lin S.-Y., Rodrigues C., Nguyen D., Arroyo F., Kaping J. Evaluation of pyrosequencing for detecting extensively drug-resistant Mycobacterium tuberculosis among clinical isolates from four high-burden countries. Antimicrob Agents Chemother. 2015;59(1):414–420. doi: 10.1128/AAC.03614-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Catanzaro A., Rodwell T.C., Catanzaro D.G., Garfein R.S., Jackson R.L., Seifert M. Performance comparison of three rapid tests for the diagnosis of drug-resistant tuberculosis. PLoS ONE. 2015;10(8):e0136861. doi: 10.1371/journal.pone.0136861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bravo L.T.C., Tuohy M.J., Ang C., Destura R.V., Mendoza M., Procop G.W. Pyrosequencing for rapid detection of Mycobacterium tuberculosis resistance to rifampin, isoniazid, and fluoroquinolones. J Clin Microbiol. 2009;47(12):3985–3990. doi: 10.1128/JCM.01229-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Engström A., Morcillo N., Imperiale B., Hoffner S.E., Juréen P. Detection of first- and second-line drug resistance in Mycobacterium tuberculosis clinical isolates by pyrosequencing. J Clin Microbiol. 2012;50(6):2026–2033. doi: 10.1128/JCM.06664-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Georghiou S.B., Seifert M., Lin S.-Y., Catanzaro D., Garfein R.S., Jackson R.L. Shedding light on the performance of a pyrosequencing assay for drug-resistant tuberculosis diagnosis. BMC Infect Dis. 2016;16(1) doi: 10.1186/s12879-016-1781-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Govindaswamy A., Sakthi D., Pai R., Jeyaseelan L., Michael J.S. Pyrosequencing: a rapid and effective sequencing method to diagnose drug-resistant tuberculosis. J Med Microbiol. 2018;67(9):1212–1216. doi: 10.1099/jmm.0.000669. [DOI] [PubMed] [Google Scholar]
- 34.Jureen P., Engstrand L., Eriksson S., Alderborn A., Krabbe M., Hoffner S.E. Rapid detection of rifampin resistance in Mycobacterium tuberculosis by Pyrosequencing technology. J Clin Microbiol. 2006;44(6):1925–1929. doi: 10.1128/JCM.02210-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao J.R., Bai Y.J., Zhang Q.H., Wang Y., Luo M., Yan X.J. Pyrosequencing-based approach for rapid detection of rifampin-resistant Mycobacterium tuberculosis. Diagn Microbiol Infect Dis. 2005;51(2):135–137. doi: 10.1016/j.diagmicrobio.2004.10.001. [DOI] [PubMed] [Google Scholar]
- 36.Guo Q., Zheng R.J., Zhu C.T. Pyrosequencing for the rapid detection of rifampicin resistance in Mycobacterium tuberculosis: a meta-analysis. Int J Tuberc Lung Dis. 2013;17(8):1008–1013. doi: 10.5588/ijtld.12.0519. [DOI] [PubMed] [Google Scholar]
- 37.Morgan M., Kalantri S., Flores L., Pai M. A commercial line probe assay for the rapid detection of rifampicin resistance in Mycobacterium tuberculosis: a systematic review and meta-analysis. BMC Infect Dis. 2005;5:62. doi: 10.1186/1471-2334-5-62. [DOI] [PMC free article] [PubMed] [Google Scholar]