Abstract
Background and Aims
The pathophysiology of the inflammatory bowel disease collagenous colitis (CC) is poorly described. Our aim was to use RNA sequencing of mucosal samples from patients with active CC, CC in remission, refractory CC, ulcerative colitis (UC), and control subjects to gain insight into CC pathophysiology, identify genetic signatures linked to CC, and uncover potentially druggable disease pathways.
Methods
We performed whole transcriptome sequencing of CC samples from patients before and during treatment with the corticosteroid drug budesonide, CC steroid-refractory patients, UC patients, and healthy control subjects (n = 9–13). Bulk mucosa and laser-captured microdissected intestinal epithelial cell (IEC) gene expression were analyzed by gene set enrichment and gene set variation analyses to identify significant pathways and cells, respectively, altered in CC. Leading genes and cells were validated using reverse-transcription quantitative polymerase chain reaction or immunohistochemistry.
Results
We identified an activation of the adaptive immune response to bacteria and viruses in active CC that could be mediated by dendritic cells. Moreover, IECs display hyperproliferation and increased antigen presentation in active CC. Further analysis revealed that genes related to the immune response (DUOX2, PLA2G2A, CXCL9), DNA transcription (CTR9), protein processing (JOSD1, URI1), and ion transport (SLC9A3) remained dysregulated even after budesonide-induced remission. Budesonide-refractory CC patients fail to restore normal gene expression, and displayed a transcriptomic profile close to UC.
Conclusions
Our study confirmed the implication of innate and adaptive immune responses in CC, governed by IECs and dendritic cells, respectively, and identified ongoing epithelial damage. Refractory CC could share pathomechanisms with UC.
Keywords: Epithelial Cells, Microscopic Colitis, RNA Sequencing, Ulcerative Colitis
Abbreviations used in this paper: auCC, active/untreated collagenous colitis; aRCC, active/refractory (nonresponding) collagenous colitis; CC, collagenous colitis; DEG, differentially expressed gene; DN, double negative; FDR, false discovery rate; GSEA, gene set enrichment analysis; GSVA, gene set variation analysis; IBD, inflammatory bowel disease; IEC, intestinal epithelial cell; IFN, interferon; IHC, immunohistochemistry; itCC, inactive/treated (responding) collagenous colitis; MMP, matrix metalloproteinase; PBS, phosphate-buffered saline; RNA-seq, RNA-sequencing; RT-qPCR, reverse-transcription quantitative polymerase chain reaction; TIMP, tissue inhibitor of metalloproteinase; UC, ulcerative colitis
Graphical abstract
Summary.
Collagenous colitis is a nondestructive inflammatory bowel disease that involves the innate and adaptive immune responses (ie, intestinal epithelial cell dysfunction and dendritic cell activation). The only effective treatment—the corticosteroid drug budesonide—does not fully restore gene expression.
Collagenous colitis (CC) is a debilitating inflammatory bowel disease (IBD) that causes chronic, nonbloody watery diarrhea, leading to a poor quality of life.1 The macroscopic appearance of the colon is usually normal, and noninvasive biomarkers do not discern between CC and other gut disorders.1 Thus, CC diagnosis relies on histopathological features, including a thickened collagenous band (>10 μm) and increased lymphoplasmacytic infiltrate into the lamina propria.1 The only effective, established treatment is the corticosteroid drug budesonide.2 However, clinical trials report disease relapse in 23%–39% of CC patients during maintenance treatment, and up to 80% after treatment is discontinued.3, 4, 5 In addition, patients can become treatment-refractory.3,6 Therefore, understanding CC pathogenesis is an unmet clinical need, and comprehending the mechanisms of action of budesonide would open the door for new therapeutic opportunities for patients that do not respond to treatment.
Several human leukocyte antigen (HLA) genetic variants have been associated with CC, which indicates activation of the adaptive immune system through antigen presentation events.7 As in the major IBD forms Crohn’s disease and ulcerative colitis (UC), luminal antigens could lead to the activation of an aberrant immune response.8 However, no association has been found between CC and pattern recognition receptor genes to date. Interestingly, nuclear factor κB, a key regulator of inflammatory immune responses involved in cytokine production, is activated in CC, specifically in intestinal epithelial cells (IECs).9 Therefore, these cells might play a role in CC pathogenesis.
Additional pathomechanisms compatible with an intact mucosa include changes in IEC electrophysiology and homeostasis. Transepithelial ion exchange is impaired due to low expression or loss of function of several ion channels in the colonic epithelium, leading to defects in sodium reabsorption.10,11 The decreased osmotic pressure impairs the paracellular reabsorption of water, which is exacerbated by the loss of the water channel aquaporin 8 in the IEC apical membrane; thus, resulting in watery diarrhea.12 Although ion and water channel expression are almost restored during clinical remission, the extent of epithelial dysfunction, immune responses, and dysbiosis contributing to CC pathophysiology, response to treatment, and relapse are still unclear.
It has been suggested that CC shares features with other IBDs, especially UC1,7,13; however, whether the similar clinical presentation of these disorders is caused by common molecular mechanisms is unclear. To address these questions, we investigated the whole transcriptome of colonic mucosa and microdissected IECs from CC patient samples. Our patient cohort, which includes budesonide-treated responding and nonresponding CC patients, has enabled us to propose targets for the development of new treatments for CC patients.
Results
Central Immune Response–Related Genes Are Dysregulated in CC
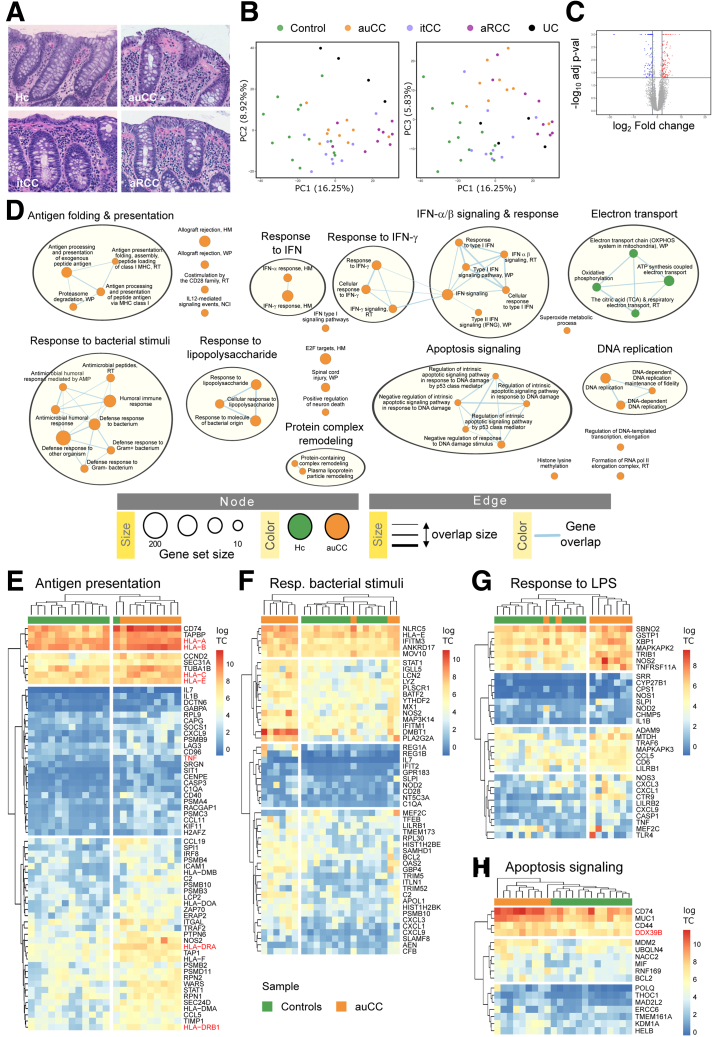
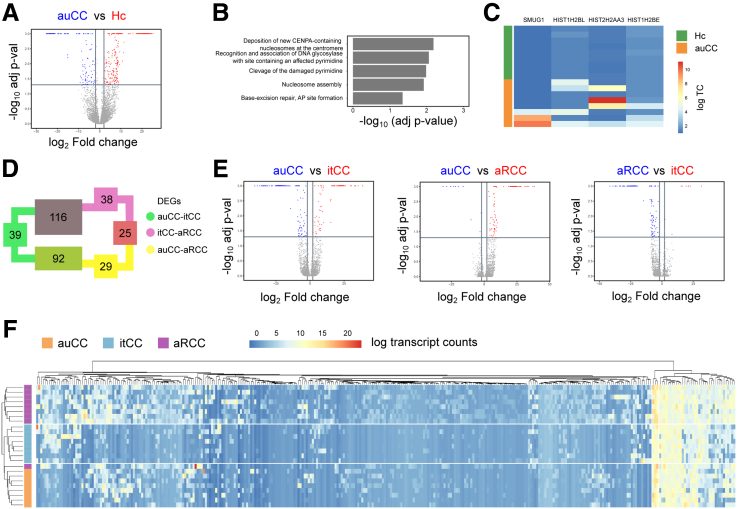
The exact pathophysiology of the chronic diarrheal disorder CC is incompletely understood. We therefore investigated the transcriptome of CC, which features an intact mucosa despite increased lymphoplasmacytic infiltration in the lamina propria (Figure 1A; Table 1). Principal component analyses of bulk biopsy RNA-sequencing (RNA-seq) data separated samples groups into different clusters of gene expression according to the clinical classification (Figure 1B). Active/untreated CC (auCC) mucosa displayed 354 differentially expressed genes (DEGs) compared with healthy control subjects (Figure 1C). Gene set enrichment analysis (GSEA) indicated that auCC-associated genes were related to antigen folding and presentation (HLA, CD74, TAPBP), response to lipopolysaccharide and bacteria (DMBT1, NLRC5, NOS2), apoptosis (CD74/MIF), and DNA replication events (DDX11, HMGA1) (Figure 1D–H and 2; Supplementary Table 1). A substantial number of pathways also contained genes of the response to interferons (IFNs) (Figure 1D and 2; Supplementary Table 1). Of note, human leukocyte antigen (HLA)-I and -II genes that were previously associated with CC7,14 have an increased expression in active CC (Figure 1, Figure 2). Collectively, our results corroborate the genetic association with HLA, and indicate immune response activation and bacterial recognition in CC pathogenesis.
Figure 1.
CC mucosa gene expression indicates activation of central immune response signaling pathways. (A) Representative histology of hematoxylin and eosin–stained paraffin-embedded sections of the human colonic mucosa in a healthy control (Hc) subject, an auCC patient, an itCC patient, and an aRCC patient. (B) Principal component analysis plot of the RNA-seq expression profiles of the different CC subgroups, Hc, and UC samples. (C) Volcano plot showing differentially upregulated (red) and downregulated (blue) genes in auCC compared with control subjects. (D) Enrichment map of gene expression in which orange nodes represent CC and green nodes represent Hc phenotype pathways created with FDR Q value <0.05, and combined coefficient >0.375 with combined constant = 0.5. (E–H) Heatmaps showing normalized log2-transformed fold changes (using the regularized log function in R) of RNA-seq transcript counts from leading genes contributing to the enriched gene pathways in CC colonic mucosa related to (E) antigen folding and presentation, (F) response to bacterial stimuli, (G) response to lipopolysaccharide (LPS), and (H) apoptosis signaling. n = 9–13 samples per group. Hc subjects are shown in green, auCC samples are shown in orange. Genes associated with CC by immunochip are highlighted in red. Heatmap rows and columns are split according to hierarchical clustering. Unless stated otherwise, gene pathways were retrieved from Gene Ontology Biological Process database. HM, Hallmark database; NCI, NCI-Nature curated data; RT, Reactome database; WP, WikiPathways database.
Table 1.
Clinical and Demographic Characteristics of the CC Patient “Exploratory” Cohort and Controls Included in RNA-Seq, Microdissected Intestinal Epithelial Cell RNA-Seq, and Immunohistochemistry Analyses
| Variable | Hc | auCC | itCCa | aRCC | UCb |
|---|---|---|---|---|---|
| Total number of subjects | 13c | 9 | 9 | 9 | 4 |
| On budesonide treatment | No | No | Yes | No | No |
| Steroid responders | — | Yes | Yes | No | — |
| Female, % | 53.85 | 77.78 | 77.78 | 100 | 75 |
| Average age, y | 51 (17–71) | 59 (27–86) | 59 (27–86) | 60 (25–79) | 22 (19–30) |
| Average stools/day | — | 7.56 (6–10) | 1.22 (1–2) | 9.89 (4–20) | — |
| Average watery stools/day | — | 7.56 (6–10) | 0 (N/A)d | 9.89 (4–20) | — |
| Average collagenous band, μm | — | 35.00 (16–52) | 28.33 (5–72)d | 35.56 (10–72) | — |
| Average stool frequency, Mayo score | — | — | — | — | 2.25 (1–3) |
| Average endoscopy, Mayo score | — | — | — | — | 2 (1–3) |
NOTE. Values are n or mean (range), unless otherwise indicated.
auCC, active/untreated collagenous colitis; aRCC, active/steroid-refractory collagenous colitis; CC, collagenous colitis; Hc, healthy control subjects; itCC, inactive/treated collagenous colitis; N/A, not applicable; RNA-seq, RNA sequencing; UC, ulcerative colitis.
Matched samples from itCC patients were collected before and during treatment with budesonide. Note that samples before treatment (active disease) were included in the group of auCC samples, whereas samples during treatment were included as itCC samples. One patient was not included for RNA-seq analysis of microdissected intestinal epithelial cells due to unavailability of paraffin-embedded biopsy sample.
UC disease extension included 1 patient with proctitis, 2 with affection of the descending colon, and 1 with pancolitis. Patients were assessed following the Mayo score. This group was only included for bulk biopsy RNA-seq analysis.
Nine of these patients were included for intestinal epithelial cell microdissection and subsequent RNA-seq analysis.
Note that the average stool frequencies and collagenous band thickness before treatment of itCC patients are nearly the same as the auCC patient group.
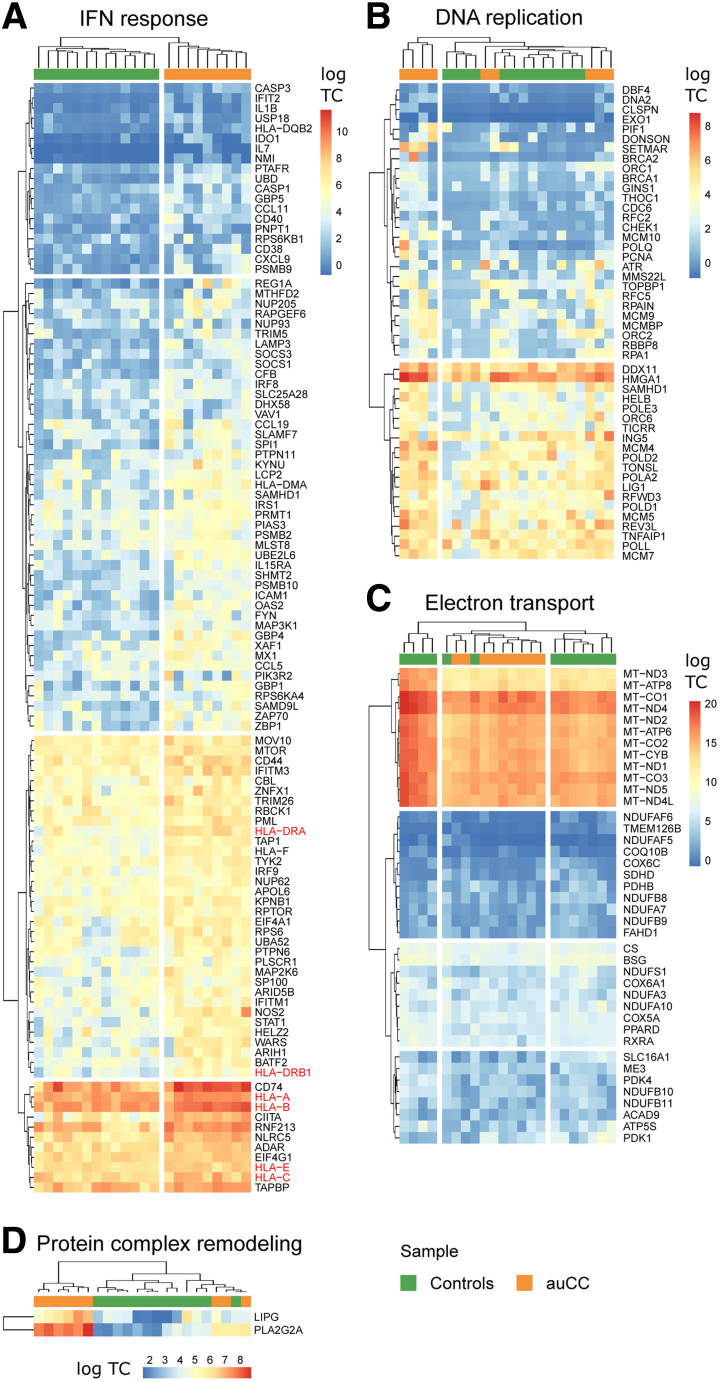
Figure 2.
CC mucosa displays an imbalance in IFN response, DNA replication, and metabolic processes. (A–D) Heatmaps showing normalized log2-transformed fold changes (using the regularized log function in R) of RNA-seq transcript counts (log TC) from leading genes contributing to the enrichment of gene pathways in CC colonic mucosa displayed in Figure 1C related to (A) IFN response, (B) DNA replication, (C) electron transport, and (D) protein complex remodeling. Healthy control (Hc) subjects are shown in green, auCC samples are shown in orange. Genes associated with CC by immunochip5 are highlighted in red. n = 9–13 samples per group.
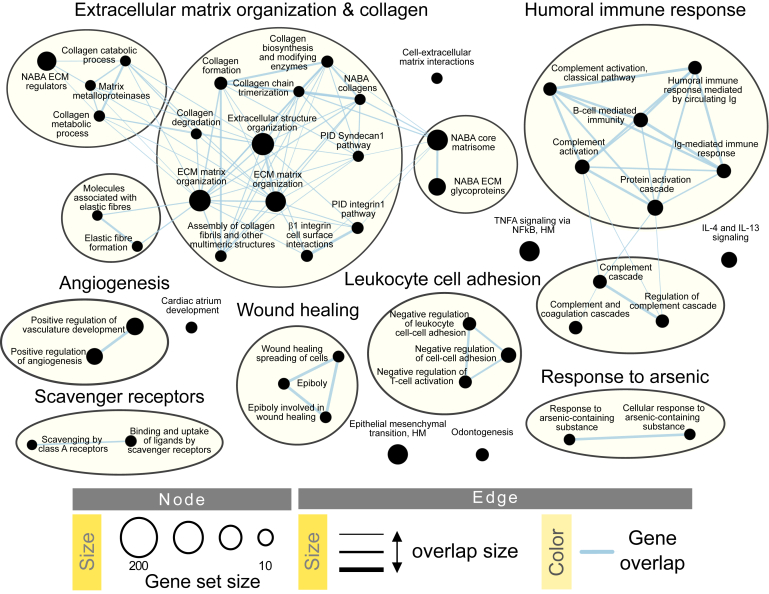
To further explore the idea that CC displays common features with UC, we compared the messenger RNA expression of active CC with UC samples (Table 1, Figure 3). When compared with active CC samples, UC samples differed in the expression of 600 protein-coding genes. In particular, UC mucosal gene expression was enriched for extracellular organization and collagen-related genes, humoral immune response, angiogenesis, wound healing, and leukocyte cell adhesion processes when compared with auCC mucosa (Figure 3). To identify which genes are specifically involved in CC but not in UC pathogenesis, we compared the lists of DEGs between either auCC or UC samples and healthy control subjects, and filtered for those with absolute log2 fold changes above 2. The 161 resulting CC-specific genes were mostly involved in the metabolism of fatty acids and prostaglandins, and in peroxisome proliferator-activated receptor signaling pathways (Table 2; Supplementary Table 2).
Figure 3.
CC differs from UC in gene pathways related to leukocyte adhesion and the humoral immune response, angiogenesis, wound healing, and extracellular matrix (ECM). Enrichment map of gene expression in which nodes represent UC phenotype pathways (normalized enriched scores) created with FDR Q value <0.05, and combined coefficient >0.375 with combined constant = 0.5. Unless otherwise stated, gene pathways were retrieved from Gene Ontology Biological Process database. The analysis did not result in any pathway enriched for auCC phenotype. Based on results from 9 auCC and 4 active UC patients.
Table 2.
Enriched Gene Pathways From CC-Specific Differentially Expressed Genes
| Database | Term | Adjusted P Value | Odds Ratio | Combined Score | Genes |
|---|---|---|---|---|---|
| GO Biological process | cellular protein complex localization (GO:0034629) | .2483 | 20.9082 | 154.6912 | MIOS;NACC2;KLHL21 |
| GO Biological process | protein complex localization (GO:0031503) | .2483 | 19.8068 | 143.7553 | MIOS;NACC2;IFT46 |
| GO Biological process | prostanoid metabolic process (GO:0006692) | .3624 | 24.9421 | 137.5235 | HPGD;ACOX1 |
| GO Biological process | regulation of monooxygenase activity (GO:0032768) | .2483 | 18.8155 | 134.0420 | DDAH2;CALM1;CYGB |
| GO Biological process | alpha-linolenic acid metabolic process (GO:0036109) | .3624 | 22.6735 | 121.3471 | FADS2;ACOX1 |
| GO Biological process | prostaglandin metabolic process (GO:0006693) | .2550 | 17.1033 | 117.5756 | EDN2;HPGD;ACOX1 |
| GO Biological process | unsaturated fatty acid metabolic process (GO:0033559) | .2483 | 12.6108 | 97.8342 | FADS2;SCD;ACOX1;MGLL |
| GO Biological process | unsaturated fatty acid biosynthetic process (GO:0006636) | .3624 | 19.1834 | 97.1681 | HPGD;SCD |
| GO Biological process | peptidyl-threonine dephosphorylation (GO:0035970) | .3624 | 17.8122 | 87.9383 | PPM1A;DUSP10 |
| GO Cellular Components | DNA-directed RNA polymerase II, core complex (GO:0005665) | .3937 | 16.6239 | 80.0785 | URI1;POLR2D |
| KEGG | PPAR signaling pathway | .0046 | 11.2548 | 117.5080 | RXRB;FADS2;GK;ACOX1;SCD;AQP7 |
| KEGG | Biosynthesis of unsaturated fatty acids | .1033 | 15.6764 | 104.1832 | FADS2;SCD;ACOX1 |
| WikiPathways | Estrogen Receptor Pathway WP2881 | .1777 | 22.6735 | 121.3471 | ACOX1;PDK4 |
| WikiPathways | Sulindac Metabolic Pathway WP2542 | .3563 | 30.9922 | 100.0630 | MSRA |
| WikiPathways | ID signaling pathway WP53 | .2153 | 17.8122 | 87.9383 | PAX8;ID3 |
| WikiPathways | PPAR signaling pathway WP3942 | .0308 | 10.2238 | 86.8204 | RXRB;FADS2;ACOX1;SCD;AQP7 |
GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; PPAR, peroxisome proliferator-activated receptor.
Budesonide Modulates CC Transcriptional Program in Responsive Patients
Budesonide is the only effective treatment for CC,1,2,15 but its effects in colitis are not fully understood. We therefore analyzed the transcriptomic profile of active CC patients that volunteered for extra biopsy sampling after achieving clinical remission following budesonide treatment after an average period of 6 weeks (inactive/treated CC [itCC]), and another set of patients who, despite this therapy, never responded to treatment and still suffered from watery diarrhea after 12 weeks (active/refractory CC [aRCC]). Of note, budesonide did not seem to affect the thickness of the collagenous band in most of our patients, independently of the response outcome (Table 1; Figure 1A). RNA sequencing (RNA-seq) analysis of the 9 CC samples obtained after successful treatment (itCC) revealed that 307 protein-coding genes were differentially expressed (DEGs) compared with the matched active disease samples (auCC samples) (Figure 4A and B). In contrast, 92 genes differed between steroid-responsive and nonresponsive patients (itCC vs aRCC) (Figure 4B and C). Based on the expression of all these genes and in comparison with healthy control samples, active disease samples clustered together and displayed very similar expression profiles (Figure 4C). Genes that were associated with active CC forms contributed to DNA regulation and expression, protein synthesis, and trafficking, and to immune responses, as highlighted by GSEAs of itCC samples compared with auCC or aRCC samples (Figure 4D and E; Supplementary Tables 3 and 4). Of note, of the 161 CC-specific genes identified previously, the expression of 149 was normalized after treatment with budesonide, as observed when itCC data were compared with healthy control subjects (Supplementary Table 2).
Figure 4.
Budesonide treatment dampens cell proliferation and the immune response but not in steroid-resistant CC patients. (A) Diagram displaying the numbers of DEGs coding for proteins between CC patient subgroups. The expression of 271 genes changed between auCC and itCC samples (green), while 59 between itCC and aRCC samples (yellow), and 8 between auCC and aRCC (pink). A total of 36 genes changed between different CC subgroups. (B) Volcano plots showing differentially regulated protein-coding genes in auCC compared with itCC (left), and to aRCC (center) samples, and between itCC compared with aRCC samples (right). Red and blue colors indicate the group whose genes are upregulated in. (C) Heatmap showing normalized log2-transformed fold changes (regularized log function in R) of RNA-seq transcript counts from all the differentially expressed protein-coding genes between CC subgroups in comparison to healthy control samples. Heatmap rows and columns are split according to hierarchical clustering. (D, E) Enrichment maps from GSEA performed including all DEGs (E) between auCC and itCC and (E) between itCC and aRCC, ranked based on adjusted P values. GSEA for the comparison of active CC samples (untreated vs refractory) did not reveal any enriched pathway (not shown). n = 9–13 samples per group.
Because the main CC histological feature is a thick collagenous band, we explored the expression of extracellular matrix components, including collagen, matrix metalloproteinases (MMPs), and MMP inhibitors (tissue inhibitors of metalloproteinases [TIMPs]). Of collagens, we not only failed to detect an increase in gene expression but identified a decrease of the COL17A1 gene in active CC forms, which encodes for collagen type XVII α1 chain (Figure 5). In addition, we detected an increase in the expression of TIMPs 1 and 3, mostly in aRCC samples (Figure 5).
Figure 5.
Gene expression of collagen and ECM-related genes in CC mucosa. Normalized log2-transformed fold changes (using the regularized log function in R) of RNA-seq transcript counts. Healthy control subjects (Hc) are shown in green, auCC samples in orange, inactive/treated CC (itCC) samples in blue, and active/refractory CC (aRCC) samples in purple. n = 9–13 samples per group. Statistically significant differences relative to Hc samples are shown as ∗∗P < .01, and ∗∗∗P < .001.
To define the profile of cells present in the mucosa of each CC patient sample group, we computed gene set variation analysis (GSVA) of stroma and immune populations as previously described (Figure 6A; Supplementary Table 5).16 We particularly noticed an apparent decrease of CD34+ GDF10+ stroma cell profile in auCC samples (Figure 6A). In contrast, immune cells such as active dendritic cell and T helper cell profiles were estimated to be increased in active CC, especially in steroid-refractory mucosa (Figure 6A). The dendritic cell infiltration in CC mucosa was confirmed by immunohistochemistry staining for CD1a (Figure 6B). Enteric neuron and innate immune response cells did not change (Figure 6A).
Figure 6.
Immune and stroma cell types in CC mucosa. (A) GSVA computed for all different immune and stroma cell types from RNA-seq data displaying enrichment scores (median with interquartile range). Healthy control subjects (Hc) are shown in green, auCC samples in orange, inactive/treated CC (itCC) samples in blue, and active/refractory CC (aRCC) samples in purple. (B) Representative immunohistochemistry (IHC) images of CD1a staining in paraffin-embedded sections from Hc, auCC, itCC, and aRCC colonic samples. Note the brown staining in active CC forms is stronger than in Hc subjects due to an increased cellular infiltration in the stroma. Fibroblastic stroma cells are subdivided into inflammatory and chemokine-producing cells (IL6 CXCL1), CD34-derived cells, INMT+ cells, proliferative stroma cells (pSC), and other stroma cells (SCx); enteric neurons are subdivided into putative excitatory motor neurons (PEMN), putative inhibitory motor neurons (PIMN), putative interneurons (PIN), putative sensory neurons (PSN), and putative secretomotor/vasodilator neurons (PSVN). n = 9–13 samples per group. Statistically significant differences relative to Hc samples are shown as ∗P < .05, unless other comparison is indicated. activated CD8, activated CD8+ T cells; aDC, activated dendritic cells; iDC, immature dendritic cells; NK, natural killer; regulatory T, regulatory T cells; Tcm cells, central memory T cells; Tem cells, effector memory T cells; Tfh cells, follicular T helper cells; γδT cells, TCRγδ+ T cells.
Of the 11 DEGs identified between auCC and aRCC samples, we opted to validate by reverse-transcription quantitative polymerase chain reaction (RT-qPCR) the 3 genes with higher fold change between these 2 groups using an extended CC patient cohort (Figure 7; Table 3). However, none of them resulted statistically different in auCC when compared with aRCC samples (Figure 7).
Figure 7.
DEGs between auCC and aRCC colonic mucosa. (A, B) Normalized log2-transformed fold changes (using the regularized log function in R) of RNA-seq transcript counts (log TC) from DEGs between auCC and aRCC CC mucosa (A) displayed as a heatmap or (B) as detailed individual plots including samples from Hc subjects and all CC groups (median with interquartile range). Selected genes for further RT-qPCR validation are highlighted in red. (C) Log2 fold changes (–ΔCt values) (median with interquartile range) in gene expression of B3GNT8, DENN4A, and IFIT3 analyzed by quantitative PCR. HPRT1 was used as a housekeeping control. Hc subjects are shown in green, auCC samples in orange, itCC samples in blue, and aRCC samples in purple. n = 9–13 samples per group for RNA-seq analyses; n = 13–20 samples per group for RT-qPCR validation. Statistically significant differences relative to Hc samples are shown as ∗P < .05, ∗∗P < .01, and ∗∗∗P < .001, unless other comparison is indicated; statistically significant differences relative to itCC samples are shown as #P < .05, and ##P < .01.
Table 3.
Clinical and Demographic Characteristics of CC Patient Validation Cohorts and Control Subjects Included in RT-qPCR
| Variable | RT-qPCR With All Groups |
RT-qPCR of itCC Samples: Relapse vs No Relapse |
|||||
|---|---|---|---|---|---|---|---|
| Hc | auCC | itCCa | aRCC | No Relapse (Biopsies and Blood) | Relapse (Biopsies) | Relapse (Blood) | |
| Total number of subjects | 14 | 20 | 14 | 13 | 6 | 8 | 10 |
| Number of subjects included in RNA-seq | 8 | 3 | 6 | 7 | 4 | 2 | 5 |
| On budesonide treatment | No | No | Yes | No | Yes | Yes | Yes |
| Steroid responders | — | Yes | Yes | No | Yes | Yes | Yes |
| Female, % | 42.86 | 75 | 78.57 | 92.31 | 83.33 | 75 | 70 |
| Age, y | 62 (60–71) | 63 (28–86) | 66 (35–86) | 54 (25–75) | 73 (49–86) | 61 (35–76) | 51 (27–73) |
| Stools/day | — | 6.90 (3–12) | 1.43 (1–2) | 8.92 (4–15) | 1.33 (1–2) | 1.50 (1–2) | 1.20 (1–2) |
| Watery stools/day | — | 6.70 (2–12) | 0 (N/A) | 8.38 (4–15) | 0 (N/A) | 0 (N/A) | 0 (N/A) |
| Collagenous band, μm | — | 32.15 (12–52) | 29.57 (2–72) | 33.38 (10–68) | 30.50 (5–72) | 28.88 (2–50) | 22.00 (2–45) |
NOTE. Values are n or mean (range), unless otherwise indicated.
auCC, active/untreated collagenous colitis; aRCC, active/steroid-refractory collagenous colitis; CC, collagenous colitis; Hc, healthy control subjects; itCC, inactive/treated collagenous colitis; N/A, not applicable; RNA-seq, RNA sequencing; RT-qPCR, reverse-transcription quantitative polymerase chain reaction.
Matched samples from itCC patients were collected before and during treatment with budesonide. Note that samples before treatment (active disease) were included in the group of auCC samples, whereas samples during treatment were included as itCC samples.
Taken together, GSEA and GSVA indicate that budesonide affects the immune response in CC by decreasing the protein trafficking and antigen presentation in cells, and possibly decreasing the number of active antigen-presenting cells, but only in steroid-responsive patients. To note, active CC forms (ie, naïve untreated CC and budesonide-refractory CC) do not significantly differ at the transcriptomic level.
Budesonide Fails to Completely Restore the Expression of Dysregulated Immune-Related Genes in Responding CC Patients
Budesonide efficiently maintains clinical remission in 61%–77% of the patients during long-term treatment, but remission continues only in approximately 20% when it is discontinued. To explore possible underlying pathomechanisms, we first aimed to identify CC-associated DEGs that are unaffected by steroid treatment and may thus contribute to the reoccurrence of symptoms. To this end, we analyzed DEGs between healthy control and itCC samples, and selected the genes with higher fold changes between these 2 groups and similar expression pattern in itCC and active CC samples for subsequent validation (Figure 8A). Of the 11 selected genes, qPCR analyses confirmed that several genes related to immune response (DUOX2, PLA2G2A, CXCL9), DNA transcription and protein ubiquitination (CTR9, JOSD1, URI1), and ion transport (SLC9A3) were not restored to normal levels after budesonide treatment (Figure 8B; Table 3).
Figure 8.
DEGs between Hc subjects and itCC colonic mucosa. (A) Normalized log2-transformed fold changes (regularized log function in R) of RNA-seq transcript counts from differentially expressed genes between Hc subjects and itCC mucosa (median with interquartile range). Selected genes for further RT-qPCR validation are highlighted in red. Genes with white background had an absolute log2 fold change value <2 between Hc and itCC samples. (B) Log2 fold changes (–ΔCt values) (median with interquartile range) in gene expression of selected genes analyzed by quantitative RT-qPCR. HPRT1 was used as a housekeeping control. In both panels, Hc are shown in green, auCC samples in orange, itCC samples in blue, and aRCC samples in purple. n = 9–13 samples per group for RNA-seq analyses; n = 13–20 samples per group for RT-qPCR validation. Statistically significant differences relative to Hc samples are shown as ∗P < .05, ∗∗P < .01, and ∗∗∗P < .001, unless other comparison is indicated; statistically significant differences relative to itCC samples are shown as #P < .05, and ##P < .01.
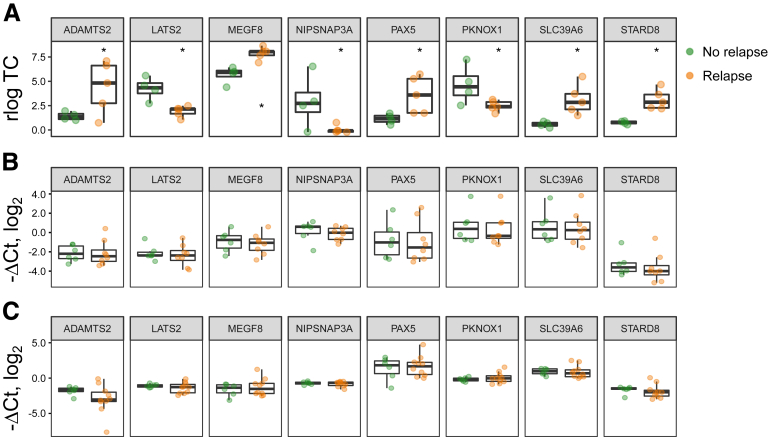
Next, we asked if any DEGs could have predictive value for disease relapse. For this, we separated itCC samples included in our RNA-seq analysis into patients that suffered from a disease relapse or not in the following months and found 8 DEGs (Figure 9A; Table 3). However, we were not able to identify any of them as potential CC relapse biomarkers b RT-qPCR analyses did not show significant changes in gene expression either in biopsy or in peripheral blood samples in the validation cohort (Figure 9B and C; Table 3).
Figure 9.
DEGs in itCC samples from patients that experienced a disease relapse against those who did not. (A) Normalized log2-transformed fold changes (using the regularized log function in R) of RNA-seq transcript counts (log TC) (median with interquartile range) of DEGs between itCC samples from patients that experienced a disease relapse (orange), or did not (green). n = 4–5 samples per group. (B, C) Log2 fold changes (–ΔCt values, median with interquartile range) in gene expression from DEGs identified by RNA-seq in (B) colonic samples or (C) blood of itCC patients that suffered from a disease relapse (orange), or did not (green), as analyzed by RT-qPCR. HPRT1 was used as a housekeeping control. n = 6–10 samples per group for validation analyses. All primers detect all coding transcript variants of the indicated gene. n = 4–5 samples per group. Statistically significant differences are shown as ∗P < .05.
In summary, genes that remain dysregulated despite treatment with budesonide could be targets for new therapies for CC patients, and validation in alternative patient cohorts could be of interest. In contrast, we failed to find a biomarker that predicts CC relapse when patients are under budesonide treatment.
The Intestinal Epithelial Cell Transcriptome Is Altered in CC
Despite the mucosal immune activity in CC, the mucosa is macroscopically intact.1 Synergistic electrolyte and water transport imbalance as well as nuclear factor κB activation implicates involvement of intestinal epithelial cells (IECs) in CC pathogenesis.9, 10, 11, 12 Thus, we microdissected intestinal epithelia from paraffin-embedded tissue sections from samples of our initial patient cohort (Table 1) to explore the role of IECs in CC pathogenesis by RNA-seq. Comparison of active untreated CC samples with healthy control subjects showed only 5 enriched pathways, which belonged to DNA organization in the cell nuclei, with leading genes involved in chromatin remodeling and repair events (Figure 10A–C). The amount of differentially expressed genes between CC subgroups was similar as in bulk biopsies (374 vs 339 DEGs) (Figure 10D and E) and, surprisingly, the matched samples taken before and after budesonide treatment of responsive patients clustered together (Figure 10F).
Figure 10.
CC intestinal epithelial cell gene expression compared with Hc subjects. (A) Volcano plot showing differentially expressed protein-coding genes in auCC (upregulated genes in blue) compared with Hc subjects (upregulated genes in red). (B) GSEA performed including DEGs between auCC and Hc samples, ranked based on adjusted P values. (C) Heatmap showing normalized log2-transformed fold changes (using the regularized log function in R) of RNA-seq transcript counts (log TC) from leading genes contributing to the enriched gene pathways shown in B. (D) Diagram displaying the numbers of DEGs coding for proteins between CC patient subgroups. The expression of 39 genes changed between auCC and itCC samples (green), while 29 did between auCC and aRCC samples (yellow), and 38 did between itCC and aRCC samples (pink). A total of 339 genes changed between different CC subgroups. (E) Volcano plots showing differentially regulated protein-coding genes in auCC compared with itCC samples (left) or to aRCC samples (center), and between itCC compared with aRCC samples (right). Red and blue colors indicate the group were genes are upregulated. (F) Heatmap showing normalized log2-transformed fold changes (regularized log function in R) of RNA-seq transcript counts (log TC) from all the differentially expressed protein-coding genes between CC subgroups in comparison with Hc samples. Heatmap rows and columns are split according to hierarchical clustering. Hc subjects are shown in green, auCC samples in orange, itCC samples in blue, and aRCC samples in purple.
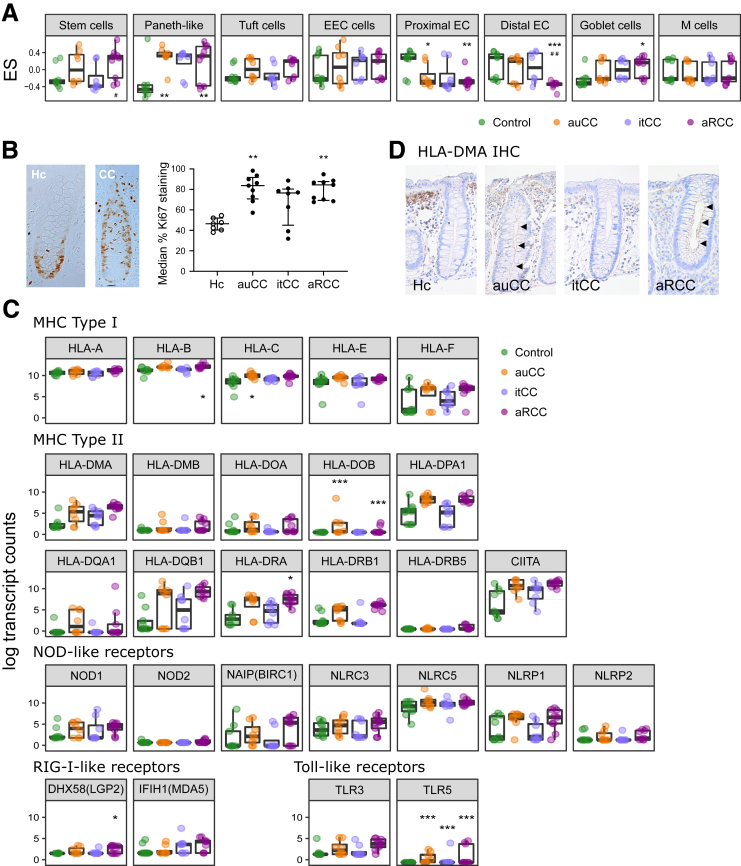
To identify key cellular components, we computed GSVA of 8 IEC populations adapted from the intestinal epithelial atlas produced by Haber et al.17 The enrichment scores of stem cells and Paneth-like cells in active CC forms suggested an increased activity within colonic crypts during the disease (Figure 11A; Supplementary Table 5). However, staining with the proliferation marker Ki67 corroborates an increased proliferation in active CC colonic crypts that is not fully restored by budesonide (Figure 11B). Interestingly, the enterocyte profile is decreased in CC, especially enterocytes from the proximal intestine, but this result should be interpreted carefully due to the extrapolation of these profiles from the small intestine mouse atlas (Figure 11A; Supplementary Table 5).
Figure 11.
CC IECs’ transcriptional profile suggest cell proliferation and recognition of antigens better than IEC from control subjects. (A) GSVA computed for different epithelial cell types from RNA-seq data displaying enrichment scores (ES) (median with interquartile range). Hc subjects are shown in green, auCC samples in orange, itCC samples in blue, and aRCC samples in purple. (B) Representative IHC images of longitudinally sectioned epithelial glands stained for Ki67 proliferation marker (brown) in paraffin-embedded sections of Hc and CC colonic mucosa (left). Analysis of Ki67 relative staining to total crypt length is shown on the right (median with interquartile range, median of 9 crypts/patient). (C) Pattern recognition receptor and HLA gene expression in intestinal epithelial cells from CC mucosa as normalized log2-transformed fold changes (using the regularized log function in R) of RNA-seq transcript counts (median with interquartile range) of pattern recognition receptors including members from the NOD-like, RIG-I-like, and Toll-like families, and HLA genes (divided into genes coding for major histocompatibility complex [MHC] type I and II proteins). Group colors are the same as in A. (D) Representative IHC images of HLA-DMA staining of paraffin-embedded sections from Hc, auCC, itCC, and aRCC colonic samples. Note the brown staining in the apical side of IEC sin crypts from active samples (black arrows). n = 7–13 samples per group. Statistically significant differences relative to Hc samples are shown as ∗P < .05, ∗∗P < .01, and ∗∗∗P < .001; statistically significant differences relative to itCC samples are shown as #P < .05 and ##P < .01. EEC cells, enteroendocrine cells; EC, enterocytes; M cells, microfold cells.
Because some IECs can contribute to immune responses (eg, Paneth-like and goblet cells),18 we explored the expression of pattern recognition receptors and HLA genes in IEC (Figure 11C). Changes in pattern recognition receptors, including NOD-like receptors, accounted for very modest, nonsignificant alterations within all CC subgroups. Moreover, HLA-related genes were prone to increase in active CC samples, which was confirmed by increased HLA-DMA protein levels in the apical side of IECs from active CC samples (Figure 11C and D) Altogether, these data show that CC mucosal transcriptome is altered and that epithelial cells might contribute toward immune responses and disease pathogenesis in CC.
aRCC Shares Similarities With UC
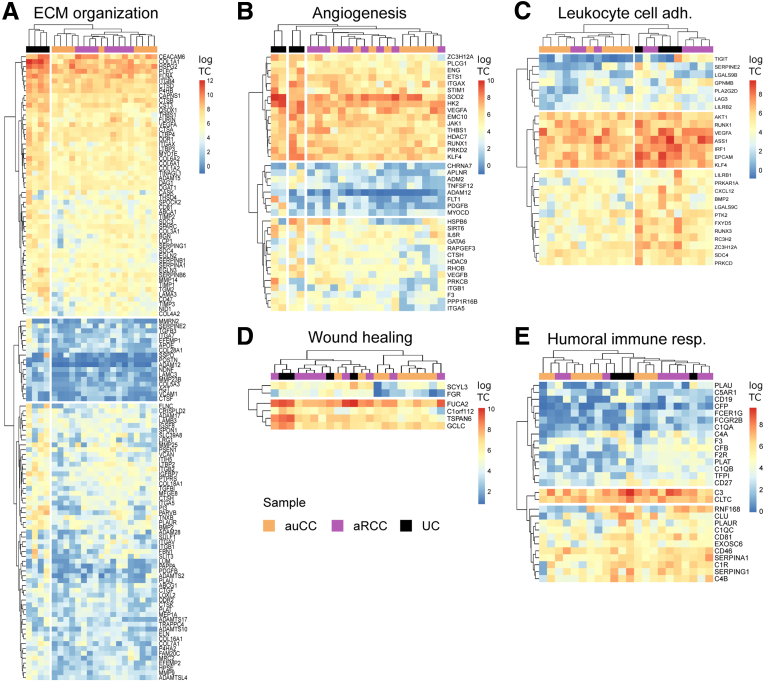
As mentioned previously, CC displays common features with UC (Figure 3), but refractory CC has never been compared with UC. Our GSEA between aRCC and UC samples showed that only the different gene pathway was that regulating vasoconstriction, with ADM and TRPM4 as leading genes (data not shown). When major pathways identified after auCC–UC comparison (from Figure 3) were explored in detail (Figure 12), UC samples segregated from CC samples for genes related to extracellular matrix organization and angiogenesis (Figure 12A and B). Interestingly, leukocyte cell adhesion and wound healing gene pathways clustered UC together with aRCC samples (Figure 12C and D). Therefore, in this scenario, the transcriptional landscape of steroid-refractory CC patients appears similar to UC, which indicates that UC treatments focused on preventing immune cell adhesion or promoting wound healing processes might be useful for treating aRCC.
Figure 12.
Active refractory CC mucosa shares similarities with ulcerative colitis. (A–E) Heatmaps showing regularized log2-transformed fold changes of RNA-seq transcript counts from leading genes contributing to the enriched gene pathways in UC, auCC and aRCC colonic mucosa related to ECM organization and (A) collagen, (B) angiogenesis, (C) leukocyte cell adhesion, (D) wound healing, and (E) and humoral immune response. auCC samples are shown in orange, aRCC samples in purple, and UC samples in black. Heatmap rows and columns are split according to hierarchical clustering. n = 4–9 samples per group.
Discussion
Because the etiology and pathobiology of CC remains poorly understood, here we describe a transcriptional alteration of genes related to antigen presentation, lipopolysaccharide response, and IFN signaling routes, which might point to a role for Gram-negative bacteria and viruses in CC pathogenesis. Dense genotyping of immune-related loci in CC identified HLA-DQ2 as a genetically predisposing factor in CC,1,7 which supports our findings. It has also been suggested that an abnormal translocation of bacteria could trigger the inflammation in CC, and thus attract immune cells into the mucosa.1,19 Microbiota studies so far have identified a decreased concentration of the epithelial-protective Akkermansia muciniphila bacteria (Gram-negative) and the Clostridia-related, butyrate-producing Ruminococcaceae bacteria family (Gram-positive).20,21 Especially a decreased abundance of Ruminococcaceae is, in general, associated with loose stools and is restored after treatment with budesonide, hence the bacterial microbiota might be affected by the lumen content flow.20,22 This profile is shared with other IBD forms,20,22 but whether dysbiosis is causative or consequential to the inflammation remains unknown. Interestingly, the translocation of chemically killed Escherichia coli K12 (Gram-negative) is increased in Ussing chamber mucosal barrier assays using CC biopsy samples,19 and fecal stream diversion has been effective treating nonresponsive CC patients.23,24 On the other side, Epstein-Barr virus has been detected in CC mucosa with even higher DNA detection than the levels reported in UC.25 Because this and other herpes viruses have also been found to be associated with IBD clinical morbidity,26 it would be worth to explore their presence in CC in depth. In addition, we did not find evidence of autoimmune mechanisms, and findings of some autoantibodies have only been reported in small studies and do not correlate with clinical symptoms.27, 28, 29 Altogether, owing to the similarities with classical IBD forms,30 CC can thus be described as an IBD in which mucosal cells overreact against microbiota of diverse nature. Still, we have proposed 161 DEGs that could specifically contribute to CC pathomechanisms, including genes related to fatty acid and prostaglandin metabolism, and peroxisome proliferator-activated receptor signaling. To note, these will require further validation in additional cohorts with larger inflammatory control subsets because the number of UC samples included in this study was limited.
Dendritic cells are professional antigen-presenting cells capable to determine the fate of antigen-specific immune outcomes. In the healthy gut, dendritic cells promote immune tolerance toward nutrients and commensals through regulatory T cell and IgA-producing B cell responses.31 Among the infiltrating cells in CC mucosa, we identified an increase in genes associated with active dendritic cells and an increased infiltration of CD1a+ cells. Similarly, IBD inflamed mucosa displays increased numbers of these cells that overexpress pattern recognition receptors, hence promoting a proinflammatory Th1/Th17 response.31 Usually, dendritic cells rapidly respond to microenvironment changes, such as signals from IECs, and promote a tolerogenic immune response.32,33 However, debilitated IEC tight-junctions, as reported in IBDs, facilitate the passage of invading microorganisms that induce a proinflammatory response.34, 35, 36 In particular, HLA expression in IECs is responsible for Crohn’s disease and UC IEC ability to induce CD4+ T cell proliferation and IFNγ secretion.37 Therefore, CC IECs might exert similar effects through HLA-DMA. In addition, we and others reported an increased IEC proliferation in CC colonic crypts, an increased lysozyme expression, and a possible increase in antigen presentation that, in turn, could disrupt IEC homeostasis and their communication with the underlying stroma cells.34,38,39 Indeed, we also report here an increased expression of metalloprotease inhibitors TIMP1 and TIMP3, which would prevent collagen degradation. Madisch et al40 also associated CC with a genetic variation in the MMP-9 gene and, in our results, we can observe a tendency for MMP-9 messenger RNA upregulation in active CC that is corrected in budesonide-responding CC patients (Figure 5). In addition, we report a potential dysregulation of a subset of CD34+ GDF10+ stroma cells identified in a single-cell RNA-seq mouse atlas that would worth validation. Whether these cells could also be responsible for decreased COL17A1 expression remains unknown and would require a comprehensive analysis of fibroblast—IEC interactions.
Despite identification of active antigen-presenting cells in CC mucosa and increased expression of chemoattractant genes for leukocyte populations (eg, CXCL9), we and others have not found evidence of increase of T cells (ie, CD4+ T helper, CD8+ cytotoxic T cells, or T regulatory cells).41,42 However, our results are limited to the use of gene expression data and the power of GSVA algorithm to estimate cellular population frequencies and due to the extrapolation of mouse gene identifiers to human tissue. Conversely, Kumawat et al38,43 found evidence of increased T cell numbers in CC mucosa, identifying the disease as a mixed Th17/Tc17 and Th1/Tc1 IBD using flow cytometry, which is a more reliable approach. Still, Kumawat et al’s results could not be replicated in the cohort analyzed by Carrasco et al,41 where they only found an increase in CD3+CD4–CD8– double negative (DN) T cells. Despite DN T cell abundance in CC and autoimmune disorders, the function of these cells is not well understood.44 They could be responsible of the increased expression of the pro-inflammatory cytokines found in CC mucosa, but DN T cells have also been attributed with anti-inflammatory properties.41,45,46 Whether the inflammation is effectively restrained by suppressor mechanisms in CC is not clear but will match with the lack of macroscopic mucosal damage1; hence, the exploration of the dendritic cell–IEC and the IEC–T cell interfaces might further clarify CC pathogenesis.
Budesonide is the only established therapy for CC but is an unspecific anti-inflammatory drug with ambiguous effects.1 Besides its anti-inflammatory properties, we observed a generalized decline in the expression of genes related to DNA regulation, protein synthesis and trafficking, and cell cycle regulation when CC patients responded to budesonide, which might be a secondary effect of the tissue restoration to normalcy as budesonide resolves the inflammation. Nonetheless, this inflammation is not completely abrogated because genes involved in innate immunity and cell recruitment remain increased after treatment (eg, DUOX2, PLA2G2A, and CXCL9). Thus, targeting of residual dysregulated genes could support low-dose budesonide therapy to ensure long-term clinical remission of CC patients. Despite our efforts to identify markers in budesonide-responding patients (itCC samples) that could indicate an upcoming disease relapse, we found that RNA-seq DEGs were false positive results after RT-qPCR corroboration, which supports that data validation with different techniques is mandatory when exploiting results for clinical application.
Steroid-refractory patients account for 10%–20% of CC individuals included in clinical trials and represent a clinical challenge.47, 48, 49, 50 In fact, no previous pathological characterization of aRCC has been reported to date. Despite the resemblance of aRCC transcriptome with auCC samples, we observed that leukocyte cell adhesion and wound healing processes are similarly altered in aRCC and UC but differ from auCC samples. This suggest that refractory CC could be a distinct disease entity with potentially unique pathomechanisms. In our previous work, we explored the restoration of water malabsorption in CC colon after budesonide treatment, and also found disparities between water channel aquaporin (AQP) 8 protein levels auCC and aRCC patients.12 Thus, supporting the hypothesis that despite similar to treatment-naïve CC, refractory CC behave differently and that these patients could benefit from alternative treatments, such as the ones available for UC patients. Indeed, immunomodulators and anti-tumor necrosis factor α therapies seem to have positive effects on aRCC patients.1,51,52 In addition, our results suggest that other biological treatments could be worth testing in aRCC. For instance, promising leukocyte/lymphocyte trafficking blocking therapies for UC patients under clinical trial testing include antibodies targeting α4 or β7 integrin subunits on leukocytes, or the mucosal addressin cell adhesion molecule MAdCAM-1 on endothelia, and modulators of the lymphocyte trafficking receptor sphingosine-1-phosphate (S1P).53 Actually, the already approved α4β7 blocking antibody vedolizumab induced clinical remission in almost half of the patients assessed by Rivière et al.54 Thus, novel IBD therapies would be worth to assess in large randomized clinical trials with refractory CC patients.
CC is emerging as a common disorder but, to date, no reliable disease-specific, noninvasive biomarker is available. However, we could propose gene targets that could be addressed for the development of novel therapies (eg, DUOX2, PLA2G2A, CXCL9, CTR9, JOSD1, URI1, and SLC9A3). Particularly, loss of function and excessive activity of enzymes producing reactive oxygen species DUOX2 and NOX1 have been suggested to contribute to gastrointestinal disease progression.55 Thus, NOX/DUOX inhibitors or reactive oxygen species inducers could be of use to develop novel therapies to treat CC. Similarly, other works found increased levels of proinflammatory cytokines and chemokines in CC56,57; hence, targeting the expression of genes related to inflammatory processes (eg, PLA2G2A and CXCL9) could prompt alternative treatment options.
In summary, this study provides a comprehensive landscape of CC pathology. Our results confirm that CC is an immune-mediated IBD in which luminal antigen presentation might occur via dendritic cells. Clinical remission can be achieved after budesonide treatment, but some genes remain dysregulated and may open the door for new treatments (eg, DUOX2, PLA2G2A, CXCL9). Still, budesonide-refractory CC could comprise a transcriptionally distinct disease entity, and owing to its similarities with UC, aRCC patients could benefit from treatments that are under investigation to treat UC.
Materials and Methods
Study Population
Biopsy samples from the descending colon were collected during scheduled colonoscopy in adult patients with CC patients at the Division of Gastroenterology at Linköping University Hospital, Sweden. CC was diagnosed according to the current guidelines,2 primarily clinical history and histopathological features, including a subepithelial collagen band of >10-μm thickness. Active CC was defined as more than 3 bowel movements per day or at least 1 watery bowel movement per day during a 1-week registration period. Clinical remission was defined as less than 3 bowel movements per day and no watery bowel movement within a 1-week period.58 A diagnosis of steroid-refractory CC was reached if patients did not achieve clinical remission after treatment for 12 weeks with 6- to 9-mg/d budesonide.2 Healthy volunteers were recruited from the local colon cancer screening program at Linköping University Hospital (Sweden) or St. Olav’s University Hospital (Norway); these individuals showed normal macro- and microscopic findings upon histopathological assessment, had normal bowel movements, and did not take any medication at the time of colonoscopy. We enrolled treatment-responsive patients with active CC, and some of them agreed to have additional biopsies and blood samples (collected in EDTA tubes; BD Biosciences, San Jose, CA) taken after reaching remission during budesonide treatment (after 6 weeks of treatment on average). We also obtained samples from steroid-refractory CC patients and healthy control subjects following the same bowel preparation procedure and biopsy taking from the descending colon as mentioned above. Active UC samples from patients without medication intake (n = 4) were used for comparison and collected at St. Olav’s University Hospital. These were diagnosed and assessed following the guidelines stated in the Mayo score system.59 Detailed patient characteristics can be found in Tables 1 (exploratory cohort) and 3 (validation cohort). Adjacent biopsy samples from the same mucosal area were stored in AllProtect (Qiagen, Hilden, Germany) or RNAlater (Thermo Fisher Scientific, Waltham, MA) for subsequent RNA extraction, or in phosphate-buffered saline (PBS) for fixation in paraformaldehyde, embedding in paraffin, and analyses using microscopy. Informed written consent was obtained from all subjects, and their data were handled according to current regulations (EU2016/679, corrigendum May 23, 2018). Ethical approval was issued by Linköping’s regional ethical committee to conduct studies in microscopic colitis, including CC (Dnr 2015/31-31), and by St. Olav’s University Hospital with approval from the Central Norway Regional Committee for Medical and Health Research Ethics no 2013/212/REKmidt.
Genome-Wide Messenger RNA-Seq
Biopsies preserved in AllProtect or RNAlater (n = 13 healthy control subjects, n = 9 per CC group, and n = 4 UC) were homogenized in RLT buffer from RNeasy Mini Kit (Qiagen) supplemented with 1% 2-mercaptoethanol using a T10 Ultra Turrax homogenizer (IKA; Thermo Fisher Scientific). Total RNA from homogenized biopsy samples was isolated using RNeasy Mini Kit following the manufacturer’s instructions. RNA from laser capture microdissection material (n = 8–9 per group) was isolated with RNeasy FFPE kit (Qiagen, see the following sections). RNA integrity was assessed using an Agilent RNA 6000 Pico kit on a 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA). The DV200 value, representing the percentage of RNA fragments more than 200 nucleotides long, was used as a measure of RNA quality. The range of DV200 values was 30%–70%.12 RNA sequencing libraries were constructed with SENSE totalRNA with Ribo cop rRNA depletion (Lexogene, Vienna, Austria), and single-read sequenced for 75 cycles to a depth of 25 million base reads on a HiSeq4000 instrument (Illumina, San Diego, CA), according to the manufacturer’s recommendations. FASTQ files were generated using bcl2fastq software v2.18 (Illumina). Data was analyzed using the R Bioconductor software v3.5.1 (R Foundation for Statistical Computing, Vienna, Austria), including SARTools v1.6.6 and DESeq2 v1.22.1 packages.60, 61, 62 Reads were aligned to the Ensembl GRCh38 genome version, release 92. RNA-seq data are available at Gene Expression Omnibus under the accession number GSE159010 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE159010).
RNA-Seq Data Analysis
Differential gene expression from RNA-seq data was determined with linear models using DESeq2 v1.22.1 and significance decided by Benjamini-Hochberg false discovery rate (FDR)–adjusted P values <.05. Principal component analysis was computed after making the data homoscedastic using R. GSEA of CC-specific genes was performed using EnrichR and taking into account the outcome from Gene Ontology, Kyoto Encyclopedia of Genes and Genomes, and WikiPathway databases. GSEA for comparisons of 2 sample groups was performed in GSEA63,64 v4.0.3 (Broad Institute, Cambridge, MA) using updated guidelines.65 DEGs were filtered for genes with <10 counts and ranked according to their differential gene expression log10-adjusted P value and sign of log fold change. Human gene set annotations from Gene Ontology, Reactome, and other databases were downloaded from Bader’s Lab website excluding those inferred from electronic annotations (https://download.baderlab.org/EM_Genesets/, release January 2020). Terms annotating more than 200 or <10 genes were discarded to improve biological interpretation. GSEA results were visualized using EnrichmentMap66 v3.2.0 and a FDR P value of <.05, a Jaccard Overlap Combined index of 0.375, and k constant of 0.5, and annotated using AutoAnnotate v1.3 applications for Cytoscape v3.7.2, and improved in Inkscape v0.92.4. Gene expression was normalized using the regularized logarithmic function in R for subsequent analyses and representation. Markers for stromal (fibroblasts and enteric neurons), immune and epithelial cell subtypes retrieved from publications (Supplementary Table 5)16,17,67,68 were used to compute GSVA using the GSVA69 package in R v3.6.3. Human orthologue annotations of epithelial and fibroblastic cell markers were procured from the Mouse Genome Database, Mouse Genome Informatics (http://www.informatics.jax.org; The Jackson Laboratory, Bar Harbor, ME), April 2020. Paneth-like cells were identified with a combination of Paneth-1 and Paneth-2 biomarkers17 and the inclusion of human Paneth cell–derived defensin genes.70 GSVA was analyzed using linear models using DESeq2 v1.22.1 and significance decided by Benjamin-Hochberg FDR adjusted P values <.05 using R software.
Laser Capture Microdissection
Colonic biopsy samples collected in PBS were fixed in paraformaldehyde and embedded in paraffin (FFPE samples). Matched samples from 8 steroid-responsive CC patients before and during budesonide treatment, 9 steroid-refractory CC patients, and 9 healthy control subjects were used for microdissection. Laser capture microdissection was performed as previously described.12 Briefly, samples were cut into 10-μm sections and mounted on RNase-free MMI Membrane Slides (Molecular Machines and Industry; MMI AG, Eching, Germany); afterward, samples were stained with hematoxylin following standard protocols. The sections were dehydrated with 100% ethanol and xylene, followed by air-drying in a desiccator for at least 30 min. Intestinal epithelial cells (area of 106 μm2, corresponding to approximately 104 cells) were isolated from all samples with a UV-LCM MMI Cellcut device connected to an Olympus IX71 microscope (Olympus, Tokyo, Japan), and collected in MMI isolation caps with diffuser (all from MMI AG), following the manufacturer’s recommendations. Isolated cells were kept in PKD lysis buffer from the RNeasy FFPE kit at –80ºC until RNA was isolated.
Immunohistochemistry
Paraffin-embedded sections (4 μm) were cut in a microtome and deparaffinated with Histolab Clear (Histolab Products, Västra Frölunda, Sweden). Antigen retrieval was performed in 10 mM citric acid pH 6.0 containing 0.05% Tween 20 (Sigma-Aldrich, St. Louis, MO) in a 2100 Retriever (Aptum Biologics, Hampshire, United Kingdom). Samples were incubated with peroxidase 1 and blocked in Background Sniper (both from Biocare Medical, Pacheco, CA). Anti-Ki67 (GTX16667; GeneTex, Irvine, CA), anti-HLA-DMA (HPA012750; Atlas Antibodies, Bromma, Sweden), anti-CD1a (M3571; Dako-Agilent, Santa Clara, CA), rabbit or mouse IgG isotype antibodies (Thermo Fisher Scientific), secondary donkey anti-rabbit IgG biotin conjugated antibody (ab6801-500; Abcam, Cambridge, United Kingdom), and secondary goat anti-mouse IgG biotin conjugated antibody (ab6788; Abcam) used to stain the samples in PBS with 0.1% bovine serum albumin (Sigma-Aldrich). The avidin/biotin-complex kit and DAB peroxidase substrate kit were used to develop the staining (Vector Laboratories, Burlingame, CA). Dehydration was carried out before mounting the slides with EcoMount (Biocare Medical). Images were acquired on an Olympus BX51 microscope.
Reverse-Transcription qPCR
Biopsies preserved in AllProtect were homogenized in RLT buffer from the RNeasy Mini Kit supplemented with 1% 2-mercaptoethanol in a TissueLyser II instrument (all from Qiagen). Total RNA from homogenized biopsy samples was isolated using the RNeasy Mini Kit following the manufacturer’s instructions. Total RNA from frozen blood samples was isolated using TRIzol reagent (Thermo Fisher Scientific) in a 10:1 dilution (TRIzol:blood) following the recommended protocol for RNA isolation and subsequently cleaned with RNAeasy Mini Kit. RNA was quantified using a NanoDrop ND-2000 and reverse-transcribed with a High Capacity cDNA Reverse Transcription Kit (all from Thermo Fisher Scientific). Relative gene expression was quantified by RT-PCR with iTaq Universal SYBR Green Supermix (Bio-Rad, Hercules, CA) following the manufacturer’s instructions and using the primer pairs in Supplementary Table 6. Primers were designed to amplify all transcript coding variants of the selected gene in the Reference Sequence (RefSeq) collection of the National Center for Biotechnology Information (Bethesda, MD), taking the longest transcript sequence as a reference, with primers annealing in different exons for all transcript variants using Primer3Plus v2.4.2 software.71 Quantitative analysis was carried out in a CFX96 Touch Real-Time PCR detection system (Bio-Rad) using the relative quantification –ΔCt method. Hypoxanthine phosphoribosyltransferase (HPRT) 1 was used as a reference gene, and each sample was analyzed in duplicate.
Statistical Analyses
Ki67 median percentage of staining in crypts from human colonic samples were analyzed with the nonparametric Kruskal-Wallis test when different groups were compared among each other. The nonparametric Wilcoxon test was used to compare paired samples from CC patients before and during treatment. Quantitative PCR data (–ΔCt values) were analyzed with the nonparametric Kruskal-Wallis test and Mann-Whitney test when different groups were compared and with the nonparametric Wilcoxon test when paired samples from CC patients before and during treatment were compared, and adjusted according to Benjamin-Hochberg FDR. Statistical analyses were performed and plotted in GraphPad Prism v8.0.1 (GraphPad, San Diego, CA, USA) or R.
All authors had access to the study data and reviewed and approved the final manuscript.
Acknowledgments
Celia Escudero-Hernández is currently affiliated with: Institute of Clinical Molecular Biology, University Hospital Schleswig Holstein, Christian-Albrechts-University Kiel, Kiel, Germany. The authors thank Lena Svensson at Linköping University Hospital and staff of the Gastrointestinal Endoscopy Unit, Department of Gastroenterology and Hepatology, St Olav’s University Hospital for support in collecting human samples. They also thank Arnar Flatberg at the Genomics Core Facility at the Norwegian University of Science and Technology, where the gene expression analysis of RNA sequencing data and differential gene expression analysis was performed. The Genomics Core Facility is funded by the Faculty of Medicine and Health Sciences at Norwegian University of Science and Technology, and the Central Norway Regional Health Authority. The authors thank Claudio Mirabello and Malin Larsson from the drop-in consultations at the National Bioinformatics Infrastructure Sweden in collaboration with Linköping University for helpful discussions about bioinformatics. The authors are grateful to all volunteer patients who agreed to participate in the study.
CRediT Authorship Contributions
Celia Escudero-Hernández, PhD (Conceptualization: Supporting; Data curation: Lead; Formal analysis: Lead; Investigation: Lead; Methodology: Lead; Project administration: Supporting; Software: Equal; Validation: Lead; Visualization: Lead; Writing – original draft: Lead; Writing – review & editing: Lead)
Atle van Beelen Granlund, PhD (Conceptualization: Supporting; Formal analysis: Supporting; Investigation: Supporting; Methodology: Supporting; Writing – review & editing: Supporting)
Torunn Bruland, PhD (Conceptualization: Supporting; Formal analysis: Supporting; Investigation: Supporting; Methodology: Supporting; Writing – review & editing: Supporting)
Arne Kristian Sandvik, MD, PhD (Conceptualization: Supporting; Formal analysis: Supporting; Funding acquisition: Equal; Investigation: Supporting; Resources: Equal; Writing – review & editing: Supporting; Patient enrollment and sampling: Supporting)
Stefan Koch, PhD (Conceptualization: Equal; Formal analysis: Supporting; Investigation: Supporting; Methodology: Supporting; Project administration: Supporting; Resources: Supporting; Software: Supporting; Supervision: Equal; Writing – review & editing: Supporting)
Ann Elisabet Elisabet Østvik, Md, PhD (Conceptualization: Supporting; Project administration: Supporting; Writing – review & editing: Supporting; Patient enrollment and sampling: Supporting)
Andreas Münch, MD, PhD (Conceptualization: Supporting; Funding acquisition: Equal; Investigation: Supporting; Project administration: Supporting; Resources: Equal; Supervision: Supporting; Writing – review & editing: Supporting; Patient enrollment and sampling: Lead)
Footnotes
Conflicts of interest These authors disclose the following: Celia Escudero-Hernández, Stefan Koch, and Andreas Münch have received financial support from Ferring Pharmaceuticals (Switzerland). Andreas Münch has received salary for consultancies from Tillotts Pharma AG, Ferring, Vifor, and Dr Falk Pharma; and speaker honoraria from Tillotts Pharma AG and Vifor. Ann Elisabet Østvik has received support from Tillotts Pharma AG. Ann Elisabet Østvik and Arne Kristian Sandvik have received speaker honoraria from Takeda. The remaining authors disclose no conflicts.
Funding This work was supported by grants from Ferring Pharmaceuticals (Switzerland); ALF (Region Östergötland, Sweden); the Magtarmfonden (Swedish Society of Gastroenterolgy); the Mucosal Infection and Inflammation Centre (Linköping University) postdoctoral fellowship to Celia Escudero-Hernández; the Knut and Alice Wallenberg Foundation (Sweden) grant to Andreas Münch; the Norwegian Research Council grant FRIPRO (#262549) and NTNU Outstanding Academic Fellows Programme to Atle van Beelen Granlund; the Liaison committee between St. Olav’s University Hospital and Faculty of Medicine and Health Sciences at Norwegian University of Science and Technology to Atle van Beelen Granlund, Torunn Bruland, and Ann Elisabet Østvik; the Liaison Committee between the Central Norway Regional Health Authority and the Faculty of Medicine and Health Sciences at Norwegian University of Science and Technology to Torunn Bruland and Ann Elisabet Østvik. These institutions had no role in study design, data collection and analysis, or manuscript preparation.
Supplementary Material
References
- 1.Miehlke S., Verhaegh B., Tontini G.E., Madisch A., Langner C., Munch A. Microscopic colitis: pathophysiology and clinical management. Lancet Gastroenterol Hepatol. 2019;4:305–314. doi: 10.1016/S2468-1253(19)30048-2. [DOI] [PubMed] [Google Scholar]
- 2.Miehlke S., Guagnozzi D., Zabana Y., Tontini G.E., Fiehn A.-M.K., Wildt S., Bohr J., Bonderup O., Bouma G., D’Amato M., Engel P.J.H., Fernandez-Banares F., Macaigne G., Hjortswang H., Hultgren-Hörnquist E., Koulaouzidis A., Kupcinskas J., Landolfi S., Latella G., Lucendo A., Lyutakov I., Madisch A., Magro F., Marlicz W., Mihaly E., Munck L.K., Ostvik A.-E., Patai Á V., Penchev P., Skonieczna-Żydecka K., Verhaegh B., Münch A. European guidelines on microscopic colitis: United European Gastroenterology (UEG) and European Microscopic Colitis Group (EMCG) statements and recommendations. United Eur Gastroenterol J. 2021 Feb 22 doi: 10.1177/2050640620951905. [E-pub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Munch A., Bohr J., Miehlke S., Benoni C., Olesen M., Ost A., Strandberg L., Hellstrom P.M., Hertervig E., Armerding P., Stehlik J., Lindberg G., Bjork J., Lapidus A., Lofberg R., Bonderup O., Avnstrom S., Rossle M., Dilger K., Mueller R., Greinwald R., Tysk C., Strom M., investigators B.U.C. Low-dose budesonide for maintenance of clinical remission in collagenous colitis: a randomised, placebo-controlled, 12-month trial. Gut. 2016;65:47–56. doi: 10.1136/gutjnl-2014-308363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bonderup O.K., Hansen J.B., Teglbjaerg P.S., Christensen L.A., Fallingborg J.F. Long-term budesonide treatment of collagenous colitis: a randomised, double-blind, placebo-controlled trial. Gut. 2009;58:68–72. doi: 10.1136/gut.2008.156513. [DOI] [PubMed] [Google Scholar]
- 5.Stewart M.J., Seow C.H., Storr M.A. Prednisolone and budesonide for short- and long-term treatment of microscopic colitis: systematic review and meta-analysis. Clin Gastroenterol Hepatol. 2011;9:881–890. doi: 10.1016/j.cgh.2011.06.005. [DOI] [PubMed] [Google Scholar]
- 6.Pardi D.S. After budesonide, what next for collagenous colitis? Gut. 2009;58:3–4. doi: 10.1136/gut.2008.163477. [DOI] [PubMed] [Google Scholar]
- 7.Westerlind H., Mellander M.R., Bresso F., Munch A., Bonfiglio F., Assadi G., Rafter J., Hubenthal M., Lieb W., Kallberg H., Brynedal B., Padyukov L., Halfvarson J., Torkvist L., Bjork J., Andreasson A., Agreus L., Almer S., Miehlke S., Madisch A., Ohlsson B., Lofberg R., Hultcrantz R., Franke A., D’Amato M. Dense genotyping of immune-related loci identifies HLA variants associated with increased risk of collagenous colitis. Gut. 2017;66:421–428. doi: 10.1136/gutjnl-2015-309934. [DOI] [PubMed] [Google Scholar]
- 8.Younis N., Zarif R., Mahfouz R. Inflammatory bowel disease: between genetics and microbiota. Mol Biol Rep. 2020;47:3053–3063. doi: 10.1007/s11033-020-05318-5. [DOI] [PubMed] [Google Scholar]
- 9.Andresen L. Activation of nuclear factor B in colonic mucosa from patients with collagenous and ulcerative colitis. Gut. 2005;54:503–509. doi: 10.1136/gut.2003.034165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Field M. Intestinal ion transport and the pathophysiology of diarrhea. J Clin Invest. 2003;111:931–943. doi: 10.1172/JCI18326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Camilleri M., Sellin J.H., Barrett K.E. Pathophysiology, evaluation, and management of chronic watery diarrhea. Gastroenterology. 2017;152:515–532.e2. doi: 10.1053/j.gastro.2016.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Escudero-Hernandez C., Munch A., Koch S. The water channel aquaporin 8 is a critical regulator of intestinal fluid homeostasis in collagenous colitis. J Crohns Colitis. 2020;14:962–973. doi: 10.1093/ecco-jcc/jjaa020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hardin J.A., Wallace L.E., Wong J.F., O’Loughlin E.V., Urbanski S.J., Gall D.G., MacNaughton W.K., Beck P.L. Aquaporin expression is downregulated in a murine model of colitis and in patients with ulcerative colitis, Crohn’s disease and infectious colitis. Cell Tissue Res. 2004;318:313–323. doi: 10.1007/s00441-004-0932-4. [DOI] [PubMed] [Google Scholar]
- 14.Stahl E., Roda G., Dobbyn A., Hu J., Zhang Z., Westerlind H., Bonfiglio F., Raj T., Torres J., Chen A., Petras R., Pardi D.S., Iuga A.C., Levi G.S., Cao W., Jain P., Rieder F., Gordon I.O., Cho J.H., D’Amato M., Harpaz N., Hao K., Colombel J.F., Peter I. Collagenous colitis is associated with HLA signature and shares genetic risks with other immune-mediated diseases. Gastroenterology. 2020;159:549–561.e8. doi: 10.1053/j.gastro.2020.04.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shor J., Churrango G., Hosseini N., Marshall C. Management of microscopic colitis: challenges and solutions. Clin Exp Gastroenterol. 2019;12:111–120. doi: 10.2147/CEG.S165047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tamborero D., Rubio-Perez C., Muinos F., Sabarinathan R., Piulats J.M., Muntasell A., Dienstmann R., Lopez-Bigas N., Gonzalez-Perez A. A pan-cancer landscape of interactions between solid tumors and infiltrating immune cell populations. Clin Cancer Res. 2018;24:3717–3728. doi: 10.1158/1078-0432.CCR-17-3509. [DOI] [PubMed] [Google Scholar]
- 17.Haber A.L., Biton M., Rogel N., Herbst R.H., Shekhar K., Smillie C., Burgin G., Delorey T.M., Howitt M.R., Katz Y., Tirosh I., Beyaz S., Dionne D., Zhang M., Raychowdhury R., Garrett W.S., Rozenblatt-Rosen O., Shi H.N., Yilmaz O., Xavier R.J., Regev A. A single-cell survey of the small intestinal epithelium. Nature. 2017;551:333–339. doi: 10.1038/nature24489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peterson L.W., Artis D. Intestinal epithelial cells: regulators of barrier function and immune homeostasis. Nat Rev Immunol. 2014;14:141–153. doi: 10.1038/nri3608. [DOI] [PubMed] [Google Scholar]
- 19.Münch A., Söderholm J.D., Öst Å., Ström M. Increased transmucosal uptake of E. coli K12 in collagenous colitis persists after budesonide treatment. Am J Gastroenterol. 2009;104:679–685. doi: 10.1038/ajg.2008.95. [DOI] [PubMed] [Google Scholar]
- 20.Carstens A., Dicksved J., Nelson R., Lindqvist M., Andreasson A., Bohr J., Tysk C., Talley N.J., Agreus L., Engstrand L., Halfvarson J. The gut microbiota in collagenous colitis shares characteristics with inflammatory bowel disease-associated dysbiosis. Clin Transl Gastroenterol. 2019;10 doi: 10.14309/ctg.0000000000000065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fischer H., Holst E., Karlsson F., Benoni C., Toth E., Olesen M., Linden M., Sjoberg K. Altered microbiota in microscopic colitis. Gut. 2015;64:1185–1186. doi: 10.1136/gutjnl-2014-308956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rindom Krogsgaard L., Kristian Munck L., Bytzer P., Wildt S. An altered composition of the microbiome in microscopic colitis is driven towards the composition in healthy control subjects by treatment with budesonide. Scand J Gastroenterol. 2019;54:446–452. doi: 10.1080/00365521.2019.1599064. [DOI] [PubMed] [Google Scholar]
- 23.Järnerot G., Tysk C., Bohr J., Eriksson S. Collagenous colitis and fecal stream diversion. Gastroenterology. 1995;109:449–455. doi: 10.1016/0016-5085(95)90332-1. [DOI] [PubMed] [Google Scholar]
- 24.Daferera N., Kumawat A.K., Hultgren-Hornquist E., Ignatova S., Strom M., Munch A. Fecal stream diversion and mucosal cytokine levels in collagenous colitis: A case report. World J Gastroenterol. 2015;21:6065–6071. doi: 10.3748/wjg.v21.i19.6065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rizzo A.G., Orlando A., Gallo E., Bisanti A., Sferrazza S., Montalbano L.M., Macaluso F.S., Cottone M. Is Epstein-Barr virus infection associated with the pathogenesis of microscopic colitis? J Clin Virol. 2017;97:1–3. doi: 10.1016/j.jcv.2017.10.009. [DOI] [PubMed] [Google Scholar]
- 26.Nahar S., Hokama A., Fujita J. Perspectives of cytomegalovirus infection in ulcerative colitis. Polish Arch Intern Med. 1987;11(Suppl 1):16–24. doi: 10.20452/pamw.14835. [DOI] [PubMed] [Google Scholar]
- 27.Bohr J., Tysk C., Yang P., Danielsson D., Jarnerot G. Autoantibodies and immunoglobulins in collagenous colitis. Gut. 1996;39:73–76. doi: 10.1136/gut.39.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roth B., Gustafsson R.J., Ohlsson B. Auto-antibodies and their association with clinical findings in women diagnosed with microscopic colitis. PLoS One. 2013;8 doi: 10.1371/journal.pone.0066088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Glassner K.L., Abraham B.P., Quigley E.M.M. The microbiome and inflammatory bowel disease. J Allergy Clin Immunol. 2020;145:16–27. doi: 10.1016/j.jaci.2019.11.003. [DOI] [PubMed] [Google Scholar]
- 30.Greenson J.K., Giardiello F.M., Lazenby A.J., Peña S.A., Bayless T.M., Yardley J.H. Antireticulin antibodies in collagenous and lymphocytic (microscopic) colitis. Mod Pathol. 1990;3:259–260. [PubMed] [Google Scholar]
- 31.Bernardo D., Chaparro M., Gisbert J.P. Human intestinal dendritic cells in inflammatory bowel diseases. Mol Nutr Food Res. 2018;62 doi: 10.1002/mnfr.201700931. [DOI] [PubMed] [Google Scholar]
- 32.Lee J., Gonzales-Navajas J.M., Raz E. The “polarizing-tolerizing” mechanism of intestinal epithelium: its relevance to colonic homeostasis. Semin Immunopathol. 2008;30:3–9. doi: 10.1007/s00281-007-0099-7. [DOI] [PubMed] [Google Scholar]
- 33.Wells J.M., Rossi O., Meijerink M., van Baarlen P. Epithelial crosstalk at the microbiota-mucosal interface. Proc Natl Acad Sci U S A. 2011;108(Suppl):4607–4614. doi: 10.1073/pnas.1000092107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martini E., Krug S.M., Siegmund B., Neurath M.F., Becker C. Mend your fences: the epithelial barrier and its relationship with mucosal immunity in inflammatory bowel disease. Cell Mol Gastroenterol Hepatol. 2017;4:33–46. doi: 10.1016/j.jcmgh.2017.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Burgel N., Bojarski C., Mankertz J., Zeitz M., Fromm M., Schulzke J.D. Mechanisms of diarrhea in collagenous colitis. Gastroenterology. 2002;123:433–443. doi: 10.1053/gast.2002.34784. [DOI] [PubMed] [Google Scholar]
- 36.Noren E., Mellander M.R., Almer S., Soderman J. Genetic variation and gene expression levels of tight junction genes indicates relationships between PTEN as well as MAGI1 and microscopic colitis. Dig Dis Sci. 2018;63:105–112. doi: 10.1007/s10620-017-4857-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wosen J.E., Mukhopadhyay D., Macaubas C., Mellins E.D. Epithelial MHC class II expression and its role in antigen presentation in the gastrointestinal and respiratory tracts. Front Immunol. 2018;9:2144. doi: 10.3389/fimmu.2018.02144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kumawat A.K., Strid H., Elgbratt K., Tysk C., Bohr J., Hultgren Hornquist E. Microscopic colitis patients have increased proportions of Ki67(+) proliferating and CD45RO(+) active/memory CD8(+) and CD4(+)8(+) mucosal T cells. J Crohns Colitis. 2013;7:694–705. doi: 10.1016/j.crohns.2012.08.014. [DOI] [PubMed] [Google Scholar]
- 39.Rubio C.A. Lysozyme expression in microscopic colitis. J Clin Pathol. 2011;64:510–515. doi: 10.1136/jcp.2010.086850. [DOI] [PubMed] [Google Scholar]
- 40.Madisch A., Hellmig S., Schreiber S., Bethke B., Stolte M., Miehlke S. Allelic variation of the matrix metalloproteinase-9 gene is associated with collagenous colitis. Inflamm Bowel Dis. 2011;17:2295–2298. doi: 10.1002/ibd.21640. [DOI] [PubMed] [Google Scholar]
- 41.Carrasco A., Esteve M., Salas A., Pedrosa E., Rosinach M., Aceituno M., Zabana Y., Fernandez-Banares F. Immunological differences between lymphocytic and collagenous colitis. J Crohns Colitis. 2016;10:1055–1066. doi: 10.1093/ecco-jcc/jjw058. [DOI] [PubMed] [Google Scholar]
- 42.Daferera N., Escudero-Hernández C., Nyström S., Jenmalm M.C., Hjortswang H., Ignatova S., Ström M., Münch A. Collagenous colitis mucosa is characterized by an expansion of nonsuppressive FoxP3+ T helper cells. Inflamm Bowel Dis. 2020 Dec 15 doi: 10.1093/ibd/izaa322. [E-pub ahead of print] [DOI] [PubMed] [Google Scholar]
- 43.Kumawat A.K., Strid H., Tysk C., Bohr J., Hornquist E.H. Microscopic colitis patients demonstrate a mixed Th17/Tc17 and Th1/Tc1 mucosal cytokine profile. Mol Immunol. 2013;55:355–364. doi: 10.1016/j.molimm.2013.03.007. [DOI] [PubMed] [Google Scholar]
- 44.Martina M.N., Noel S., Saxena A., Rabb H., Hamad A.R. Double negative (DN) alphabeta T cells: misperception and overdue recognition. Immunol Cell Biol. 2015;93:305–310. doi: 10.1038/icb.2014.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fischer K., Voelkl S., Heymann J., Przybylski G.K., Mondal K., Laumer M., Kunz-Schughart L., Schmidt C.A., Andreesen R., Mackensen A. Isolation and characterization of human antigen-specific TCR alpha beta+ CD4(-)CD8- double-negative regulatory T cells. Blood. 2005;105:2828–2835. doi: 10.1182/blood-2004-07-2583. [DOI] [PubMed] [Google Scholar]
- 46.D’Acquisto F., Crompton T. CD3+CD4-CD8- (double negative) T cells: saviours or villains of the immune response? Biochem Pharmacol. 2011;82:333–340. doi: 10.1016/j.bcp.2011.05.019. [DOI] [PubMed] [Google Scholar]
- 47.Munch A., Langner C. Microscopic colitis: clinical and pathologic perspectives. Clin Gastroenterol Hepatol. 2015;13:228–236. doi: 10.1016/j.cgh.2013.12.026. [DOI] [PubMed] [Google Scholar]
- 48.Miehlke S., Madisch A., Kupcinskas L., Heptner G., Böhm G., Marks H.J., Neumeyer M., Nathan T., Fernandez Banares F., Greinwald R., Mohrbacher R., Vieth M., Bonderup O. Double-blind, double-dummy, randomised, placebo-controlled, multicenter trial of budesonide and mesalazine in collagenous colitis. Gut. 2012;61:A102. [Google Scholar]
- 49.Münch A., Benoni C., Olesen M., Öst Å., Hjortswang H., Hellström P.M., Strandberg L., Björk J., Hertevig E., Lindberg G., Lapidus A., Löfberg R., Miehlke S., Kirsch C., Armerding P., Bonderup O., Stehlik J., Müller R., Greinwald R., Tysk C., Ström M. Budesonide induces clinical remission and improves quality of life in active collagenous colitis: results from the open-label induction phase of BUD-63/COC (abstr) J Crohns Colitis. 2013;7:S238(P567). [Google Scholar]
- 50.Miehlke S., Madisch A., Kupcinskas L., Petrauskas D., Bohm G., Marks H.J., Neumeyer M., Nathan T., Fernandez-Banares F., Greinwald R., Mohrbacher R., Vieth M., Bonderup O.K., BUC-60/COC Study Group Budesonide is more effective than mesalamine or placebo in short-term treatment of collagenous colitis. Gastroenterology. 2014;146:1222–1230.e1–2. doi: 10.1053/j.gastro.2014.01.019. [DOI] [PubMed] [Google Scholar]
- 51.Park T., Cave D., Marshall C. Microscopic colitis: a review of etiology, treatment and refractory disease. World J Gastroenterol. 2015;21:8804–8810. doi: 10.3748/wjg.v21.i29.8804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Daferera N., Hjortswang H., Ignatova S., Münch A. Single-centre experience with anti-tumour necrosis factor treatment in budesonide-refractory microscopic colitis patients. United Eur Gastroenterol J. 2019;7:1234–1240. doi: 10.1177/2050640619871750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schreiner P., Neurath M.F., Ng S.C., El-Omar E.M., Sharara A.I., Kobayashi T., Hisamatsu T., Hibi T., Rogler G. Mechanism-based treatment strategies for IBD: cytokines, cell adhesion molecules, JAK inhibitors, gut flora, and more. Inflamm Intest Dis. 2019;4:79–96. doi: 10.1159/000500721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rivière P., Münch A., Michetti P., Chande N., de Hertogh G., Schoeters P., Ferrante M., Vermeire S., Van Assche G. Vedolizumab in refractory microscopic colitis: an international case series. J Crohn’s Colitis. 2019;13:337–340. doi: 10.1093/ecco-jcc/jjy169. [DOI] [PubMed] [Google Scholar]
- 55.Dang P.M.-C., Rolas L., El-Benna J. The dual role of reactive oxygen species-generating nicotinamide adenine dinucleotide phosphate oxidases in gastrointestinal inflammation and therapeutic perspectives. Antioxid Redox Signal. 2020;33:354–373. doi: 10.1089/ars.2020.8018. [DOI] [PubMed] [Google Scholar]
- 56.Gunaltay S., Kumawat A.K., Nyhlin N., Bohr J., Tysk C., Hultgren O., Hultgren Hornquist E. Enhanced levels of chemokines and their receptors in the colon of microscopic colitis patients indicate mixed immune cell recruitment. Mediat Inflamm. 2015;2015:132458. doi: 10.1155/2015/132458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu Q., Harpaz N. Expression profiling of inflammatory and immunological genes in collagenous colitis. J Crohn’s Colitis. 2019;13:764–771. doi: 10.1093/ecco-jcc/jjy224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hjortswang H., Tysk C., Bohr J., Benoni C., Kilander A., Larsson L., Vigren L., Ström M. Defining clinical criteria for clinical remission and disease activity in collagenous colitis. Inflamm Bowel Dis. 2009;15:1875–1881. doi: 10.1002/ibd.20977. [DOI] [PubMed] [Google Scholar]
- 59.Magro F., Langner C., Driessen A., Ensari A., Geboes K., Mantzaris G.J., Villanacci V., Becheanu G., Borralho Nunes P., Cathomas G., Fries W., Jouret-Mourin A., Mescoli C., de Petris G., Rubio C.A., Shepherd N.A., Vieth M., Eliakim R., European Society of Pathology; European Crohn’s and Colitis Organization European consensus on the histopathology of inflammatory bowel disease. J Crohns Colitis. 2013;7:827–851. doi: 10.1016/j.crohns.2013.06.001. [DOI] [PubMed] [Google Scholar]
- 60.Anders S., Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Varet H., Brillet-Gueguen L., Coppee J.Y., Dillies M.A. SARTools: a DESeq2- and EdgeR-based R pipeline for comprehensive differential analysis of RNA-seq data. PLoS One. 2016;11 doi: 10.1371/journal.pone.0157022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., Mesirov J.P. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mootha V.K., Lindgren C.M., Eriksson K.F., Subramanian A., Sihag S., Lehar J., Puigserver P., Carlsson E., Ridderstrale M., Laurila E., Houstis N., Daly M.J., Patterson N., Mesirov J.P., Golub T.R., Tamayo P., Spiegelman B., Lander E.S., Hirschhorn J.N., Altshuler D., Groop L.C. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 65.Reimand J., Isserlin R., Voisin V., Kucera M., Tannus-Lopes C., Rostamianfar A., Wadi L., Meyer M., Wong J., Xu C., Merico D., Bader G.D. Pathway enrichment analysis and visualization of omics data using g:Profiler, GSEA, Cytoscape and EnrichmentMap. Nat Protoc. 2019;14:482–517. doi: 10.1038/s41596-018-0103-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Merico D., Isserlin R., Stueker O., Emili A., Bader G.D. Enrichment map: a network-based method for gene set enrichment visualization and interpretation. PLoS One. 2010;5 doi: 10.1371/journal.pone.0013984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pezoldt J., Pasztoi M., Zou M., Wiechers C., Beckstette M., Thierry G.R., Vafadarnejad E., Floess S., Arampatzi P., Buettner M., Schweer J., Fleissner D., Vital M., Pieper D.H., Basic M., Dersch P., Strowig T., Hornef M., Bleich A., Bode U., Pabst O., Bajénoff M., Saliba A.-E., Huehn J. Neonatally imprinted stromal cell subsets induce tolerogenic dendritic cells in mesenteric lymph nodes. Nat Commun. 2018;9:3903. doi: 10.1038/s41467-018-06423-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Drokhlyansky E., Smillie C.S., Van Wittenberghe N., Ericsson M., Griffin G.K., Eraslan G., Dionne D., Cuoco M.S., Goder-Reiser M.N., Sharova T., Kuksenko O., Aguirre A.J., Boland G.M., Graham D., Rozenblatt-Rosen O., Xavier R.J., Regev A. The human and mouse enteric nervous system at single-cell resolution. Cell. 2020;182:1606–1622.e23. doi: 10.1016/j.cell.2020.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hanzelmann S., Castelo R., Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics. 2013;14:7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ehmann D., Wendler J., Koeninger L., Larsen I.S., Klag T., Berger J., Marette A., Schaller M., Stange E.F., Malek N.P., Jensen B.A.H., Wehkamp J. Paneth cell alpha-defensins HD-5 and HD-6 display differential degradation into active antimicrobial fragments. Proc Natl Acad Sci U S A. 2019;116:3746–3751. doi: 10.1073/pnas.1817376116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Untergasser A., Nijveen H., Rao X., Bisseling T., Geurts R., Leunissen J.A. Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res. 2007;35:W71–W74. doi: 10.1093/nar/gkm306. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.