Abstract
The end TB strategy reinforces the essentiality of readily accessible biomarkers for early tuberculosis diagnosis. Exploration of microRNA (miRNA) and pathway analysis opens an avenue for the discovery of possible therapeutic targets. miRNA is a small, non-coding oligonucleotide characterized by the mechanism of gene regulation, transcription, and immunomodulation. Studies on miRNA define their importance as an immune marker for active disease progression and as an immunomodulator for innate mechanisms, such as apoptosis and autophagy. Monocyte research is highly advancing toward TB pathogenesis and biomarker efficiency because of its innate and adaptive response connectivity. The combination of monocytes/macrophages and their relative miRNA expression furnish newer insight on the unresolved mechanism for Mycobacterium survival, exploitation of host defense, latent infection, and disease resistance. This review deals with miRNA from monocytes, their relative expression in different disease stages of TB, multiple gene regulating mechanisms in shaping immunity against tuberculosis, and their functionality as biomarker and host-mediated therapeutics. Future collaborative efforts involving multidisciplinary approach in various ethnic population with multiple factors (age, gender, mycobacterial strain, disease stage, other chronic lung infections, and inflammatory disease criteria) on these short miRNAs from body fluids and cells could predict the valuable miRNA biosignature network as a potent tool for biomarkers and host-directed therapy.
Keywords: monocyte and macrophage miRNAs, tuberculosis, differential expression, immune regulation, autophagy and biomarkers
Introduction
Tuberculosis being the life-threatening disease caused by Mycobacterium tuberculosis (MTB) is intricate to understand their mycobacterial-mediated host immune subversion. The intracellular nature and delayed cell division of MTB added access to dodge the host microbicidal effect for its survival. The host’s innate defense ability and the pathogen’s strategy in evading the host’s immunity determine the sequel of TB infection (1). MTB establishes infection through multiple modalities, such as i) circumvent phagolysosome fusion and phagocytosis destruction; ii) neutralize the acidic environment (2, 3); iii) blocks the formation of the apoptotic envelope (4); iv) inhibits the plasma membrane repair, leading to the spread of infection through macrophage necrosis (5); v) suppresses activation of immune cells and antigen presentation; vi) limits the proinflammatory response by restricting proinflammatory cytokines; and vii) modulates the disease responsive genes and miRNAs through their targeted pathways. The disease becomes complex as the stages of infection are varied from latency to drug resistance because of the evolution of MTB strains. One third of the population exhibit latent infection, in which MTB remains dormant for a long period and becomes susceptible to the active disease under immune compromised condition. This latency is a menace to mankind as the diagnosis and its effective treatment toward breakdown of the disease in future need unbridled enthusiastic investigations. However, the management of the latent condition can be made possible with public awareness by improving the incidence of TB determinants, such as malnutrition, poverty, smoking, and diabetes, or through the development of new treatment or vaccines (6). The emergence of drug-resistant Mycobacterium due to poor treatment adherence (acquired resistance) and the transmission of drug-resistant strains (primary resistance) is another peril in TB research toward the end TB strategy (7). The multi-drug resistance and its treatment pose multiple challenges as it requires prolonged treatment duration, complex drugs (second-line fluoroquinolones) that may affect adherence along with lower treatment success rate (6). Other co-morbidities, like AIDS and diabetes, intensify TB disease pathogenesis.
Mononuclear cells (monocytes/macrophages) are professional phagocytic defenders against TB infection (8). The disputed behavior of monocytes as a defender against antimycobacterial activity exhibited by CD16neg subset and habitat for MTB promoted by CD16pos subset is well accepted for TB disease (9, 10). The disease-specific perturbation in the mononuclear cell subsets and their immune phenotypes contributed to underlying pathophysiology and as biomarkers for MTB infection. However, the unresolved mechanisms and the pathways affected can be studied through the molecular impression of these subsets from omics platforms in a quest for differentially expressed mRNAs and miRNAs. miRNAs are short, biologically conserved noncoding RNAs that participate in the regulation of inflammatory response, tumorigenesis, and other biological processes. Several studies focused on miRNAs revealed altered miRNA levels during infection and their impact in modulating immune functions within macrophages from TB patients (11–13). Thus, miRNA studies open up new avenues and fascinate the researchers for constructing miRNA-based vaccines, biomarkers, and host-directed therapies. This review is focused on monocyte/macrophage miRNAs, their differential expression, regulatory function, and biomarker utility in tuberculosis disease.
miRNAs
Micro RNAs are discovered as biologically conserved, short noncoding RNAs (14–16) that constitute 18 to 25 nucleotides in length. This groundbreaking innovation by Ambros and Ruvkun prompted the researchers to investigate their functional behavior toward host immune regulation and disease pathogenesis, which resulted in the exponential growth of published studies on miRNA reported by Almeida et al. (17).
miRNAs work as mRNA repressors inhibiting protein synthesis (18), translational activators (19), and molecular decoys for RNA-binding proteins (20), depending on the environment and cell type. The processing, maturation, expression, and action of miRNAs are regulated through multiple mechanisms: a) single-nucleotide polymorphism interfere with the processing and maturation of miRNAs that affect their expression profile (21); b) modulation of epigenetic mechanisms, such as histone acetylation and DNA methylation, influence the transcriptional rate of miRNAs (22); c) impairment in the mRNA-miRNA interactions by the competition of miRNAs with cellular factors and mRNAs with other competitive RNAs (pseudogenes, long non-coding RNAs, and circular RNAs) (23, 24); and d) occurrence of miRNA editing through nucleotide modification by adenosine or cytidine deaminases (21, 25). miRNA research and transcriptomic platform enabled the disease-mediated deregulation of miRNAs and their targeted pathways in multiple diseases, including cancer (26, 27), cardiovascular diseases (28, 29), autoimmune diseases (30, 31), and infectious diseases (32, 33).
Monocyte and Macrophage miRNAs
The disease-oriented modification for any microbial infection is visualized primarily on monocytic cell lineage as being the first-line defenders of innate immunity. Immunological aspect-derived alterations in the subset composition of monocytes/macrophages decipher the role of a pathogen in the peripheral compartment. However, the stimulus for the alteration is better studied through their responsive mRNA and miRNAs. miRNA research for TB is advancing toward a proper understanding of disease mechanism for better prognosis and early prevention. The immune efficiency and other cellular processes of monocyte/macrophages are governed by various miRNAs in both healthy and disease states (34).
Many reports available for the miRNAs mediated monocytic biological functions, such as tissue homeostasis, signaling, cell differentiation, apoptosis, cell motility, cytokine production, inflammatory responses, resolution of inflammation, and other immune responses (35–40). A trio of miRNAs constituting miR-146a, miR-21, and miR-155 are the principal regulators of inflammatory pathways in myeloid cells (41). miR-511 was identified as the putative positive regulator of Toll-like receptor 4 during monocyte differentiation by Tserel et al. (42). miR-214, as suggested by Li et al., targets the phosphatase and tensin homolog in monocyte survival induction during advanced glycation (43). miR-20a, miR-106a, and miR-17 of miR-17/92 and miR-106a/363 clusters are involved in tuning the proinflammatory cytokine production, infiltration of macrophages, and phagocytosis through targeting the expression of signal-regulatory protein alpha (44). Upon Notch activation, miR-148a-3p promotes M1 polarization by hindering M2 activation (45). Myeloid cell differentiation to granulocytes or monocytes is governed by miR-223 with negative control on NLRP3 inflammasome activity (46).
The intense research on miRNA profiling of monocyte subsets delivered their unique profile and regulated functions. Dang et al. deciphered the role of miR-432 in apoptotic potential and miR-19a in cell motility. They also observed that miR-345 was involved in the inflammatory responses by targeting RelA. Besides, upregulated miR-34 in CD16+ monocytes are suggestive of their differentiation ability to dendritic cells by altering the expression of Wingless-Type MMTV Integration Site Family, Member 1 (WNT1), and Jagged 1 (JAG1) (34, 47). Richard et al. focused on the sequencing of miRNAs among monocyte subsets in humans and mice to identify their role in monocyte heterogeneity. From their work, they suggested three miRNAs—miR-21, miR-150, and miR-146a—as immune regulators that mediate resolution of inflammation in the myeloid cells (48). MicroRNA profiling of intermediate monocytes (CD14++ CD16+) yielded a unique miRNA profile, and their connected pathways are involved in gene regulation, TLR, and cytokine-mediated signaling, phagocytosis, antigen processing, and presentation, as well as lipid and triglyceride metabolism (49).
MicroRNA as a Prominent Immune Regulator of Macrophage Mechanisms During TB
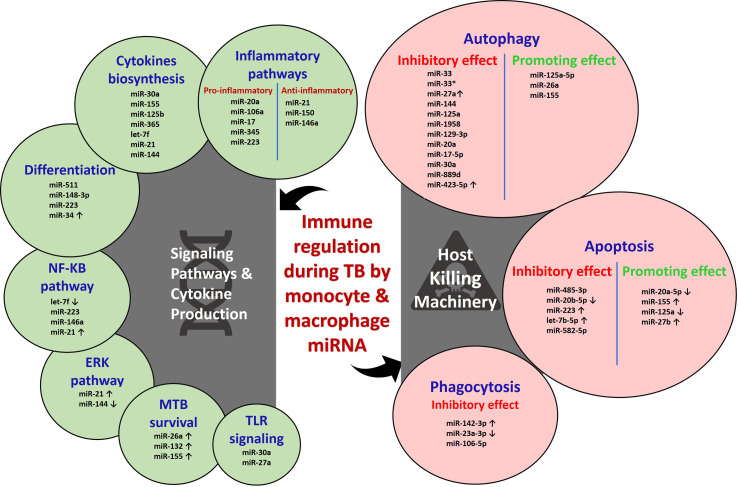
miRNAs regulate about 60% of mammalian genes through its effective binding to 3′ UTR on mRNA and leads to translational repression and mRNA degradation (50, 51). Most of the cellular functions in humans are governed by single or multiple miRNAs. The emergence of miRNA research uncovered the possibility of pathogen (specially their cell wall components) induced alteration of miRNA levels (52). The altered miRNA profile could enhance the disease progression by modulation of the innate and adaptive responses through the hindrance of cell differentiation (53). The distinctive role of miRNA in the maintenance of immune homeostasis and activation of immune defense is largely studied (54). Upon MTB infection, several miRNAs modulate the host mechanism, either favoring the host or the pathogen. In most cases, the underlying causes for host immune evasion by the Mycobacterium are associated with miRNAs. The host signaling pathways, cytokine production, and killing machinery are adversely affected by miRNAs as represented in Figure 1 .
Figure 1.
Host immune regulation by monocyte and macrophage miRNAs during tuberculosis.
miRNAs in Signaling Pathways and Cytokine Production
The prime innate defense recognition starts with the Toll-like receptors (TLRs) upon induction with pathogen-associated molecular patterns (PAMPs). However, this initial priming is affected by multiple miRNAs during MTB infection. TLR/MyD88 activation and cytokine response are inhibited by miR-30a in MTB-infected THP-1 cells (55). TLR3 signaling is attenuated by miR-27a through targeting TICAM1 and c-Abl-BMP signaling (56). Survival of Mycobacterium is favored through the upregulation of miR-26a and miR-132 induced by live and attenuated MTB that negatively controls p300 mRNA in human monocyte-derived macrophages (human MDMs). miR-132 and miR-26a dampen the host responsiveness toward IFN-gamma genes, phagocytosis process, and decreases the HLA-DR and FCgammaR1 levels (57). Inhibition of NF-κB pathway with the hindered downstream secretion of cytokines, chemokines, and NOS is achieved through the increased expression of A20 (TNFAIP3) by downregulated let-7f induction mediated by ESAT-6 in both in vitro and in vivo conditions (58). miR-223 and miR-146a also negatively control the NF-κB pathway in MTB-infected macrophages and suppress the proinflammatory response and the clearance of pathogen (59–62). Infection with BCG induces elevation of miR-21 via NF-κB and ERK pathways that target IL-12p35 mRNA through which it inhibits IL-12 production and T-cell priming function by APCs (63).
The activity of miR-155 is focused on various cell types, such as macrophages, dendritic cells, and T cells. ESAT-6 induces miR-155 in a time- and dose-dependent manner, which downregulates SHIP1, leading to an ultimate increase of the AKT phosphorylation and, thus, exerts pro-survival of MTB on macrophages. Host IL-6 production and Cox-2 activity are limited by upregulated miR-155, as the Cox-2 is essential to prevent necrosis by generating PGE2 and restricting lipoxin A4 (LXA4) (1, 64). The mycobacterial component, such as Lippo Mannan from virulent MTB and M. smeg, induces a differential response in human MDMs. TB-LM induces higher miR-125b expression that targets the TNF mRNA and inhibits TNF biosynthesis through inhibition of TLR-2–mediated miR-155 expression, whereas M. smeg LM induces miR-155 expression and downregulates miR-125b and SHIP1, thereby increasing-PI3K/Akt signaling and TNF production followed by an enhanced proinflammatory response (50). The interpretation of the role of miR-155 in pro-inflammatory responses is quite contradictory as suggested by infection studies with virulent MTB and M. smeg LM (1, 50). This strongly reinforces the synchronized regulatory effect of miR-155 along with a host of miRNAs and, thus, cannot be studied alone (1). An inverse correlation was seen with miR-144 and TPl2 protein levels as the downregulation of miR-144 in MTB-infected human MDMs targets TPL2 mRNA, and their enhancement leads to activation of ERK1/2 phosphorylation and downstream IL-1β, IL-6, and TNF α production (65). Pro-inflammatory cytokine response is suppressed through upregulated miR-32-5p targeting Follistatin-like protein (FSTL1) (66). Downregulation of miR-365 is inversely correlated with IL-6 levels in active TB patients (67).
miRNAs in Host Killing Machinery
The human host has an enormous killing machinery, like phagocytosis, apoptosis, and autophagy, and so on, for the invading pathogen. The intracellular MTB, however, exploits the host defense through various strategies. The recent transcriptomic approach sheds light on miRNA-based modulatory responses by Mycobacterium. The phagocytic function of macrophages is attenuated in the different stages by the Mycobacterium-induced miRNAs. The bacterial encounter and imbibe are affected through N-wasp by miR-142-3p. N-wasp is an actin-binding protein essential for actin dynamics in the phagocytosis process that was negatively regulated by upregulated miR-142-3p in J774A.1 cell line and primary human macrophages during MTB infection (68). Mononuclear cell function and phagocytosis are inhibited in active TB patients, where miR 23a-3p is downregulated. miR-23a-3p targets IRF1/SP1 through TLR4/TNF-α/TGF-β1/IL-10 signaling (69). The principal lysosomal enzyme of phagocytosis process for MTB clearance is cathepsin proteases. miR-106-5p targets the 3′ UTR cathepsin and suppresses the lysosomal activity in MTB-infected macrophages (70).
The downstream killing machinery of phagocytosed pathogen actively occurred through apoptosis of infected macrophages. Macrophages infected with Beijing strain demonstrate its virulence by escaping from host apoptosis and macrophage lysis through miR-485-3p (71). Upon infection with MTB, RAW264.7 macrophages establish attenuated apoptosis through the reduction of miR-20b-5p and elevation of its target Mcl-1 (72). Increased miR-223 expression in macrophages of active TB patients negatively suppresses forkhead box O3 (FOXO3) to inhibit apoptosis (62). The secreted protein MPT64 inhibits apoptosis of RAW264.7 macrophages via NF-κB/miR-21/BCl-2 pathway (73). Inhibition of apoptosis through the downregulation of Fas protein is demonstrated in THP-1 macrophages mediated by upregulated let-7b-5p (74). The decrease in the apoptotic monocytes of active TB patients and decreased apoptosis in THP-1 cells are mediated through the downregulation of FOXO-1 by miR-582-5p (75). Some of the miRNAs positively promote apoptosis for enhanced mycobacterial clearance. For example, reduction of miR-20a-5p is observed in THP-1 macrophages and CD14+ monocytes of active TB patients. Reduced miR-20a-5p inversely increases Bim expression through its target JNK2, which could promote apoptosis (76). Infection of macrophages with M. bovis BCG results in elevated miR-155 expression, which could induce apoptosis through PKA signaling by inhibiting PKI-α (77). Sp110-mediated suppression of miR-125a in RAW264.7 macrophages enhances the expression of Bmf, which could induce apoptosis (78). Upregulated miR-27b enhances p53 signaling, thus favoring apoptosis and bacterial killing by downregulating Bag2 (79).
Autophagy is a highly regulated eukaryotic cellular pathway in which intracellular pathogens are trapped in autophagosomes and degraded in lysosomes. Induction of xenophagy (a selective form of autophagy against microbes) in monocyte-derived macrophages is one of the innate immune mechanisms to intracellular pathogens, such as MTB (80). However, MTB is a successful intracellular pathogen and can escape from host responses by expression of some of the miRNAs and affects autophagy machinery (81). Certain miRNAs control both mycobacterial survival and autophagy pathways by targeting their proteins within macrophages through its altered expression (82, 83). miRNA-33 and miRNA-33* inhibit the fusion of lysosome with bacterial endosome by targeting ATG5, ATG12, LC3B, and LAMP proteins and lipid metabolism by targeting transcription factors FOXO3 and TFEB (84). The occurrence of active TB is suggested because of the suppression of autophagosome-lysosome fusion in macrophages by miR-423-5p through post-transcriptional regulation of VPS33A (85). Active TB patients and MTB-infected mice abundantly express miR-27a, which blocks the Ca2+ signaling through ER-located Ca2+ transporter protein CACN2D. Blockade of Ca2+ signaling inhibits the formation of autophagosome (86). The autophagy protein, DRAM2, promotes PtdInt3K, which initiates the nucleation of auto phagophore formation. In human and murine monocytes or macrophages, MIR144/hsa-miR-144 and miR-125a help in mycobacterial survival by forming a complex with the 3′ UTR of DRAM2 mRNA (87, 88).
TB infection triggered the expression of a new type of miRNA, i.e., miR-1958, which silences the ATG5 in RAW264.7 cells (89). miR-129-3p favors MTB survival by inhibiting ATG4B (90). miR-20a promotes BCG survival by affecting the expression of both ATG7 and ATG16L1 (91). miR-17-5p blocks autophagy by blocking ULK1 in BCG-infected RAW264.7 cells (92). Chen et al. showed that miR-30a inhibits the autophagy pathway and negative correlation between Beclin and miR-30a (93). miR-889d affects the tumor necrosis factor-like weak inducer of apoptosis (TWEAK), which maintains the granuloma formation and promotes the maturation of AMPK (94). miR-125a-5p overexpression was observed in M. avium–infected THP1-derived macrophages and targets STAT-3, which activates the autophagy (95). At the same time, miR-26a targets the KLF4, by which it inhibits MTB survival, and miR-17/PKCδ/STAT3 pathways also attenuate MTB by activating autophagy (96).
According to Wang et al., miR-155 targets Rheb (autophagy blocker) and promotes autophagy (97). PCED1BAS1 is down-regulated in TB patients, which directly binds with miR-155, and subsequently inhibits the activity of miR-155 (98). miR-155 expression helps in the survival of MTB by regulating ATG3 protein in dendritic cells (99). Yang et al. found that the expression of miR-155 was diminished in patients with spinal tuberculosis–induced intervertebral disc destruction and affects its target MMP-11 expression (100).
miRNAs as Biomarkers
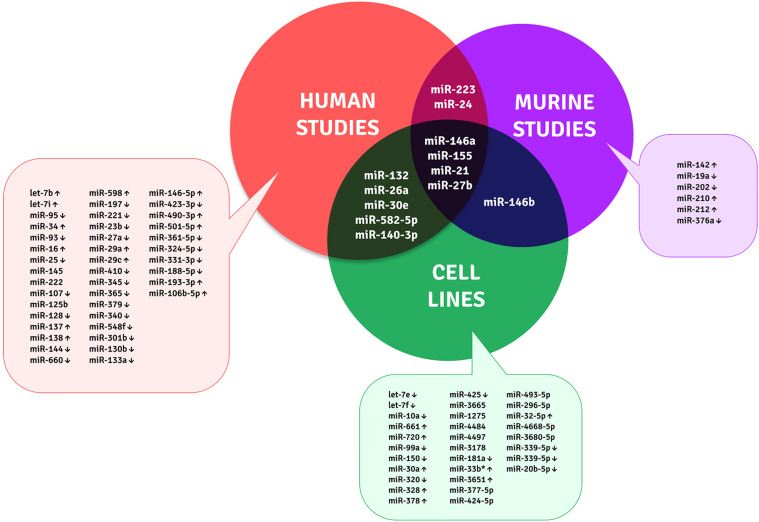
TB biomarker research is ongoing for decades as the disease still causes higher mortality due to multiple factors, such as host immune evasion by MTB, latency condition, drug resistance, and lack of prognostic and protective biomarkers. Many researchers have identified TB-specific–modulated cytokines and genes as biomarkers. However, those are not prominently emerging out since most of them are identified in smaller sample groups that lack sensitivity, differentiation ability, and reproducibility. The potent, robust, minimally invasive, rapid, universally acceptable biomarker is yet to be identified. Immune regulatory miRNAs emerge as a new class of disease-specific diagnostic markers (101, 102). The differential expression of miRNAs in disease phenomenon manifests their biomarker potential. To date, multiple studies are focused on miRNA sequencing from different samples involving PBMCs, serum/plasma, sputum, urine, and exosomes. The candidate biomarkers identified from circulation and PBMCs for discriminating TB from healthy are miR-144* (103), miR155* and miR155 (104); miR-93*, miR-3125, and miR-29a (105); miR-889, miR-576-3p, and miR-361-5p (106); miR-3179, miR-19b*, and miR-147 (11); miR-146a (107); and miR-625-3p (108). A review by Pederson et al. gives a complete biomarker profile on circulating miRNAs (109). However, our focus is on the monocyte/macrophage-based markers since most miRNAs are involved in evading their immune defense. This will help to understand the underlying pathogenesis and for identifying TB-specific biomarkers. The differential expression of miRNAs from MTB infection studies on macrophages and the monocyte-derived macrophages are depicted in Table 1 and Figure 2 .
Table 1.
Monocyte/macrophage-based miRNAs as biomarker candidates for TB.
| Cells | Differentially Expressed miRNAs | Analysis Platform | Reference |
|---|---|---|---|
| Human | |||
| MDMs infected with MTB or BCG | miR-155, miR-146a, miR-145, miR-222, miR-27a, and miR-27b | Taqman low-density array | (110) |
| MDMs from TB patients, LTB, and Healthy individuals | TB vs HC: | Taqman microarray quantitative PCR | (71) |
| Upregulated (hsa-miR-16, hsa-miR-137, hsa-miR-140-3p, hsa- miR-193a-3p, hsa-miR-501-5p, and hsa-miR-598) | |||
| Downregulated (hsa-miR-95) | |||
| LTB vs TB: Upregulated (hsa-miR-101 and hsa-miR-150) | |||
| Unique expression in LTB (miR-146b-3p and hsa-miR-296-5p) | |||
| MDM infected with TB LM | miR-125-b | qPCR | (50) |
| MDM infected with M. smeg LM | miR-155 | ||
| MDM infected with MTB H37Rv | Upregulated (miR-155, miR-21, miR-146a, miR-29a, miR-26a, let-7b, miR-34, miR-132 & miR-138) | Nanostring nCounter miRNA assay | (65) |
| Downregulated (miR-660, miR-144, miR-301b, miR-128, miR-423-3p, miR-410, miR-27a, miR-93, miR-107, miR-345, miR-221, miR-25, miR-23b, miR-361-5p, miR-130b & miR-340) | |||
| MDM infected with MTB | Upregulated (miR-132, miR-146-5p, miR-30e, let-7i, miR-490-3p, miR-29c, miR-26a, miR-21, let-7b & miR-29a) | Nanostring nCounter miRNA assay | (57) |
| Downregulated (miR-25, miR-23b, miR-331-3p, miR-423-3p, miR-548f, miR-340, miR-24, miR-107, miR-93, miR-324-5p, miR-188-5p, miR-130b, miR-410, miR-361-5p, miR-197, mir-27a, miR-128, miR-345, miR-379, miR-133a & miR-221. | |||
| Primary monocytes and MDMs from active TB patients and controls | Upregulated-miR-582-5p | qPCR | (75) |
| Primary macrophages from TB patients vs controls | Upregulated miR-223 | qPCR | (62) |
| Macrophages from TB patients and controls | Downregulated miR-365 | qPCR | (67) |
| MDM infected with MTB | Upregulated miR-106b-5p | qPCR | (70) |
| Mouse | |||
| BMDMs infected with MTB | 6 upregulated (miR-21, miR-21*, miR-146a, miR-146 b, miR210, and miR-155), 1 downregulated (miR-223) | Microarray and qPCR | (111) |
| BMDMs infected with Mtb | 4 upregulated (miR-24, miR-142, miR-155, and miR-212) and 3 downregulated (miR-19a, miR-202, and miR-376a) | Gene expression microarray | (112) |
| BMDMs infected with BCG | miR-21 | Taqman quantitative real-time PCR | (63) |
| BMDMs infected with MTB | Upregulated miR-27b | qPCR | (79) |
| BMDMs infected with MTB | 3 upregulated (miR-155, miR-146a & miR-21) | Taqman low-density arrays | (1) |
| Mouse peritoneal macrophages & BMDMs | Upregulated miR-146a | qPCR | (60) |
| Cell Line | |||
| U937 macrophages | 149 DE (miR-424–5p, miR-493-5p, miR-27 b-3p, miR-296-5p, miR-377–5p, miR-3680–5p) | Microarray | (113) |
| THP-1 cells infected with Beijing/W or non-Beijing/W strains | 13 downregulated (let-7e, let-7f, miR-10a, miR-21, miR-26a, miR-99a, miR-140–3p, miR-150, miR-181a, miR-320, miR-339–5p, miR-425, and miR-582-5p) | Taqman microarray quantitative PCR | (71) |
| THP-1 cells infected with virulent or avirulent Mtb strains | 9 DE (miR-30a, miR-30e, miR-155, miR-1275, miR-3665, miR-3178, miR-4484, miR-4668-5p, and miR- 4497) | Microarray | (114) |
| THP-1 cells infected with MTB HN878 | 12 upregulated (miR-33b*, miR-146a, miR-155, miR-132, miR-146b-5p, miR-720, miR-30e, miR-661, miR-140-3p, miR-3651, miR-328, and miR-378 | (115) | |
| THP-1 cells and U937 cells | Upregulated miR-32-5p | qPCR | (66) |
| THP-1 cells | Upregulated miR-30a | qPCR | (55) |
| RAW264.7 cells and infected with MTB | 3 upregulated (miR-155, miR-146a, and miR-21) | Taqman low-density arrays | (1) |
| RAW264.7 cells infected with MTB | Upregulated miR-27b | qPCR | (79) |
| RAW264.7 cells infected with MTB | Downregulated let-7f | SYBR Green-based miRNA profiling array | (58) |
| RAW264.7 cells | Downregulated miR-20b-5p | Semi quantitative PCR | (72) |
Figure 2.
Unique and shared miRNA biomarker candidates for TB within monocytes and macrophages across human, murine, and cell line studies.
Although many studies are available on the macrophage infection-derived miRNAs, the actual in vivo scenario of a patient is minimal. The limitations of these biomarker candidates are variable between the studies, and each was performed on identifying the miRNA targets for understanding the disease pathology. In the future, the biomarker efficiency of these candidates should be largely examined as multi-centric studies with diverse ethnicities.
miRNAs in Host-Directed Therapy (HDT)
Host-directed therapy is one of the emerging strategies to improve the host immunity and eliminate pathogens in which vitamins, repurposed drugs, cytokines, miRNAs, and, monoclonal antibodies are used as an adjunct with chemotherapy. It helps to control challenges of TB treatments, such as drug resistance, the toxicity of chemotherapy, and immune reconstitute inflammatory syndrome, and so on (116). Induction of autophagy is one of the host-mediated therapy for tuberculosis (117) and is induced by mTOR kinase inhibitors and certain immunomodulators, such as rapamycin and vitamin D3, respectively (118, 119). The PubMed search on miRNAs in HDT for tuberculosis yielded no results. However, many HDT strategies using miRNAs have been proposed by Sabir et al. (96). They suggested direct administration of miRNAs or the use of siRNAs to modulate the host responses. The downregulated anti-mycobacterial miRNAs can be induced by synthetic oligos, and the overexpressed pro-mycobacterial miRNAs can be repressed using anti-miRNA complementary to mature miRNA (120–122). This approach will benefit the host in achieving the proper signaling and their downstream pro-inflammatory responses. Synthetic delivery of miRNAs to macrophages is possible with nanoparticles or liposomes (123, 124). Novel HDT approaches on miRNA-mediated induction of host killing machinery (phagocytosis, apoptosis, and autophagy) could be a beneficial therapy to evade the pathogen strategies and for efficient pathogen clearance.
Future Perspectives
The research of miRNA-mediated regulation of TB is enormous; however, the pro diagnosis and effective therapy for TB are lacking widely. As miRNAs are regulators and modulators of the immune response, the avenue for potential biomarkers and therapeutic possibilities are much promising. Some of the key factors to be considered for future research on miRNA are as follows:
Various circulating miRNAs are available from many studies as biomarkers but research on identifying cell-oriented miRNAs, particularly monocytes and macrophages will help better to understand the evasion of initial defense.
Research on identified miRNAs to investigate their diagnostic efficacy and therapeutic value is highly needed. This will help address whether this differential expression is really specific for TB or overlaps with a disease of similar pathology.
The mycobacterial strain-specific miRNA expression is another concern since there is diversity in TB strains, and the distribution is different in different geographical locations.
Deep single-cell sequencing approach may enable the complete miRNA profile for better understanding their bio-signatures.
Patient samples from all disease stages of TB at diagnosis and during treatment may give the disease-based profile during the entire course of infection for understanding their pathophysiology.
Novel HDT approaches using nanoparticle and siRNAs for direct modulation of these expression signatures to induce the host-mediated defense responses against Mycobacterium will open up a better therapy adjunct with minimal chemotherapy.
More animal studies with miRNA/long non-coding RNA intervention for TB therapeutics should be carried out and explored.
Future collaborative efforts involving multidisciplinary approach in various ethnic population with multiple factors (age, gender, mycobacterial strain, disease stage, other chronic lung infections, and inflammatory disease criteria) on these short miRNAs from body fluids and cells could predict the valuable miRNA biosignature network for biomarker discovery and host-directed therapy.
Author Contributions
PS and KP contributed to the literature collection, writing, drafting, and revision of the manuscript. PS, UR, and RB participated in the conception of the idea, design, drafting, revision, and approval of the manuscript. All authors contributed to the article and approved the submitted version.
Funding
PS has been supported by the DST-INSPIRE fellowship. KP has been supported by the ICMR Fellowship. RB has been supported by the DBT Ramalingaswami Fellowship, Ministry of Science and Technology, Government of India.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- 1. Kumar R, Halder P, Sahu SK, Kumar M, Kumari M, Jana K, et al. Identification of a Novel Role of ESAT-6-Dependent miR-155 Induction During Infection of Macrophages With Mycobacterium Tuberculosis . Cell Microbiol (2012) 14(10):1620–31. 10.1111/j.1462-5822.2012.01827.x [DOI] [PubMed] [Google Scholar]
- 2. Flannagan RS, Cosío G, Grinstein S. Antimicrobial Mechanisms of Phagocytes and Bacterial Evasion Strategies. Nat Rev Microbiol (2009) 7:355–66. 10.1038/nrmicro2128 [DOI] [PubMed] [Google Scholar]
- 3. Meena LS, Rajni. Survival Mechanisms of Pathogenic Mycobacterium Tuberculosis H37Rv. FEBS J (2010) 277:2416–27. 10.1111/j.1742-4658.2010.07666.x [DOI] [PubMed] [Google Scholar]
- 4. Gan H, Lee J, Ren F, Chen M, Kornfeld H, Remold HG. Mycobacterium Tuberculosis Blocks Crosslinking of Annexin-1 and Apoptotic Envelope Formation on Infected Macrophages to Maintain Virulence. Nat Immunol (2008) 9:1189–97. 10.1038/ni.1654PMID:18794848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Divangahi M, Chen M, Gan H, Desjardins D, Hickman TT, Lee DM, et al. Mycobacterium Tuberculosis Evades Macrophage Defenses by Inhibiting Plasma Membrane Repair. Nat Immunol (2009) 10:899– 906. 10.1038/ni.1758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Global Tuberculosis Report . Geneva: World Health Organization; (2020). [Google Scholar]
- 7. Shah N, Wright A, Bai GH. Worldwide Emergence of Extensively Drug-Resistant Tuberculosis (XDR TB): Global Survey of Second-Line Drug Resistance Among Mycobacterium Tuberculosis Isolates. Emerg Infect Dis (2007) 13:380–7. 10.3201/eid1303.061400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sampath P, Moideen K, Ranganathan UD, Bethunaickan R. Monocyte Subsets: Phenotypes and Function in Tuberculosis Infection. Front Immunol (2018) 9:1726. 10.3389/fimmu.2018.01726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Philips JA, Ernst JD. Tuberculosis Pathogenesis and Immunity. Annu Rev Pathol (2012) 7:353–84. 10.1146/annurev-pathol-011811-132458 [DOI] [PubMed] [Google Scholar]
- 10. Balboa L, Barrios-Payan J, González-Domínguez E, Lastrucci C, Lugo-Villarino G, Mata-Espinoza D, et al. Diverging Biological Roles Among Human Monocyte Subsets in the Context of Tuberculosis Infection. Clin Sci (Lond) (2015) 129(4):319–30. 10.1042/CS20150021 [DOI] [PubMed] [Google Scholar]
- 11. Yi Z, Fu Y, Ji R, Li R, Guan Z. Altered microRNA Signatures in Sputum of Patients With Active Pulmonary Tuberculosis. PloS One (2012) 7(8):e43184. 10.1371/journal.pone.0043184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhou L, Guo L, Tang J, Zhang A, Liu X, Xu G. miR- 144 Regulates BCG-and Rapamycin-Induced Autophagy by Targeting Atg4a in RAW264. 7 Cells. Chin J Cell Mol Immunol (2015) 31(2):163–7. [PubMed] [Google Scholar]
- 13. Kleinsteuber K, Heesch K, Schattling S, Kohns M, Sander Julch C, Walzl G, et al. Decreased Expression of miR-21, miR- 26a, miR-29a, and miR-142-3p in CD4 (+) T Cells and Peripheral Blood From Tuberculosis Patients. PloS One (2013) 8(4):e61609. 10.1371/journal.pone.0061609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lee RC, Feinbaum RL, Ambros V. The C. Elegans Heterochronic Gene lin-4 Encodes Small RNAs With Antisense Complementarity to lin-14. Cell (1993) 75:843–54. 10.1016/0092-8674(93)90529-Y [DOI] [PubMed] [Google Scholar]
- 15. Wightman B, Ha I, Ruvkun G. Post Transcriptional Regulation of the Heterochronic Gene lin-14 by lin-4 Mediates Temporal Pattern Formation in C. Elegans. Cell (1993) 75:855–62. 10.1016/0092-8674(93)90530-4 [DOI] [PubMed] [Google Scholar]
- 16. O’Brien J, Hayder H, Zayed Y, Peng C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front Endocrinol (2018) 9:402. 10.3389/fendo.2018.00402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Almeida MI, Reis RM, Calin GA. MicroRNA History: Discovery, Recent Applications, and Next Frontiers. Mutat Res (2011) 717(1-2):1–8. 10.1016/j.mrfmmm.2011.03.009 [DOI] [PubMed] [Google Scholar]
- 18. Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs Predominantly Act to Decrease Target mRNA Levels. Nature (2010) 466(7308):835–40. 10.1038/nature09267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Vasudevan S, Tong Y, Steitz JA. Switching From Repression to Activation: microRNAs can Up-Regulate Translation. Science (2007) 318(5858):1931–4. 10.1126/science.1149460 [DOI] [PubMed] [Google Scholar]
- 20. Eiring AM, Harb JG, Neviani P, Garton C, Oaks JJ, Spizzo R, et al. miR-328 Functions as an RNA Decoy to Modulate hnRNP E2 Regulation of mRNA Translation in Leukemic Blasts. Cell (2010) 140(5):652–65. 10.1016/j.cell.2010.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Correia de Sousa M, Gjorgjieva M, Dolicka D, Sobolewski C, Foti M. Deciphering Mirnas’ Action Through miRNAs Editing. Int J Mol Sci (2019) 20(24):6249. 10.3390/ijms20246249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chuang JC, Jones PA. Epigenetics and microRNAs. Pediatr Res (2007) 61(5 Pt 2):24R–9R. 10.1203/pdr.0b013e3180457684 [DOI] [PubMed] [Google Scholar]
- 23. Sobolewski C, Calo N, Portius D, Foti M. MicroRNAs in Fatty Liver Disease. Semin Liver Dis (2015) Feb 35(1):12–25. 10.1055/s-0034-1397345 [DOI] [PubMed] [Google Scholar]
- 24. Gjorgjieva M, Sobolewski C, Dolicka D, Correia de Sousa M, Foti M. miRNAs and NAFLD: From Pathophysiology to Therapy. Gut (2019) Nov 68(11):2065–79. 10.1136/gutjnl-2018-318146 [DOI] [PubMed] [Google Scholar]
- 25. Blanc V, Davidson NO. APOBEC-1-Mediated RNA Editing. Wiley Interdiscip Rev Syst Biol Med (2010) 2(5):594–602. 10.1002/wsbm.82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Li C, Feng Y, Coukos G, Zhang L. Therapeutic microRNA Strategies in Human Cancer. AAPS J (2009) 11(4):747–57. 10.1208/s12248-009-9145-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, et al. Human microRNA Genes Are Frequently Located at Fragile Sites and Genomic Regions Involved in Cancers. Proc Natl Acad Sci USA (2004) 101(9):2999–3004. 10.1073/pnas.0307323101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. van Rooij E, Sutherland LB, Liu N, Williams AH, McAnally J, Gerard RD, et al. A Signature Pattern of Stress-Responsive microRNAs That Can Evoke Cardiac Hypertrophy and Heart Failure. Proc Natl Acad Sci USA (2006) 103(48):18255–60. 10.1073/pnas.0608791103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Edwards JK, Pasqualini R, Arap W, Calin GA. MicroRNAs and Ultraconserved Genes as Diagnostic Markers and Therapeutic Targets in Cancer and Cardiovascular Diseases. J Cardiovasc Transl Res (2010) 3(3):271–9. 10.1007/s12265-010-9179-5 [DOI] [PubMed] [Google Scholar]
- 30. Stanczyk J, Pedrioli DM, Brentano F, Sanchez-Pernaute O, Kolling C, Gay RE, et al. Altered Expression of MicroRNA in Synovial Fibroblasts and Synovial Tissue in Rheumatoid Arthritis. Arthritis Rheumatol (2008) 58(4):1001–9. 10.1002/art.23386 [DOI] [PubMed] [Google Scholar]
- 31. Dai Y, Huang YS, Tang M, Lv TY, Hu CX, Tan YH, et al. Microarray Analysis of microRNA Expression in Peripheral Blood Cells of Systemic Lupus Erythematosus Patients. Lupus (2007) 16(12):939–46. 10.1177/0961203307084158 [DOI] [PubMed] [Google Scholar]
- 32. Acuña SM, Floeter-Winter LM, Muxel SM. MicroRNAs: Biological Regulators in Pathogen-Host Interactions. Cells (2020) 9(1):113. 10.3390/cells9010113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yang T, Ge B. miRNAs in Immune Responses to Mycobacterium Tuberculosis Infection. Cancer Lett (2018) 431:22–30. 10.1016/j.canlet.2018.05.028 [DOI] [PubMed] [Google Scholar]
- 34. Dang TM, Wong WC, Ong SM, Li P, Lum J, Chen J, et al. MicroRNA Expression Profiling of Human Blood Monocyte Subsets Highlights Functional Differences. Immunology (2015) 145(3):404–16. 10.1111/imm.12456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Self-Fordham JB, Naqvi AR, Uttamani JR, Kulkarni V, Nares S. MicroRNA, Dynamic Regulators of Macrophage Polarization and Plasticity. Front Immunol (2017) 8:1062. 10.3389/fimmu.2017.01062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. O’Connell RM, Rao DS, Baltimore D. microRNA Regulation of Inflammatory Responses. Annu Rev Immunol (2012) 30:295–312. 10.1146/annurev-immunol-020711-075013 [DOI] [PubMed] [Google Scholar]
- 37. Nahid MA, Pauley KM, Satoh M, Chan EKL. miR-146a Is Critical for Endotoxin-Induced Tolerance: Implication in Innate Immunity. J Biol Chem (2009) 284:34590–9. 10.1074/jbc.M109.056317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bazzoni F, Rossato M, Fabbri M, Gaudiosi D, Mirolo M, Mori L, et al. Induction and Regulatory Function of miR-9 in Human Monocytes and Neutrophils Exposed to Proinflammatory Signals. Proc Natl Acad Sci USA (2009) 106:5282–7. 10.1073/pnas.0810909106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bala S, Tilahun Y, Taha O, Alao H, Kodys K, Catalano D, et al. Increased microRNA-155 Expression in the Serum and Peripheral Monocytes in Chronic HCV Infection. J Transl Med (2012) 10:151. 10.1186/1479-5876-10-151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Huang HC, Yu HR, Huang LT, Huang HC, Chen RF, Lin IC, et al. miRNA-125b Regulates TNF-a Production in CD14+ Neonatal Monocytes Via Post-Transcriptional Regulation. J Leukoc Biol (2012) 92:171–82. 10.1189/jlb.1211593 [DOI] [PubMed] [Google Scholar]
- 41. Quinn SR, O’Neill LA. A Trio of microRNAs That Control Toll-Like Receptor Signaling. Int Immunol (2011) 23:421–5. 10.1093/intimm/dxr034 [DOI] [PubMed] [Google Scholar]
- 42. Tserel L, Runnel T, Kisand K, Pihlap M, Bakhoff L, Kolde R, et al. MicroRNA Expression Profiles of Human Blood Monocyte-Derived Dendritic Cells and Macrophages Reveal miR-511 as Putative Positive Regulator of Toll-Like Receptor 4. J Biol Chem (2011) 286:26487–95. 10.1074/jbc.M110.213561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Li L-M, Hou DX, Guo YL, Yang JW, Liu Y, Zhang CY, et al. Role of microRNA-214-Targeting Phosphatase and Tensin Homolog in Advanced Glycation End Product-Induced Apoptosis Delay in Monocytes. J Immunol (2011) 186:2552–60. 10.4049/jimmunol.1001633 [DOI] [PubMed] [Google Scholar]
- 44. Zhu D, Pan C, Li L, Bian Z, Lv Z, Shi L, et al. MicroRNA- 17/20a/106a Modulate Macrophage Inflammatory Responses Through Targeting Signal-Regulatory Protein α. J Allergy Clin Immunol (2013) 132:426–36.e8. 10.1016/j.jaci.2013.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Huang F, Zhao JL, Wang L, Gao CC, Liang SQ, An DJ, et al. miR-148a-3p Mediates Notch Signaling to Promote the Differentiation and M1 Activation of Macrophages. Front Immunol (2017) 8:1327. 10.3389/fimmu.2017.01327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Haneklaus M, Gerlic M, O’Neill LA, Masters SL. miR-223: Infection, Inflammation and Cancer. J Intern Med (2013) 274:215–26. 10.1111/joim.12099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hashimi ST, Fulcher JA, Chang MH, Gov L, Wang S, Lee B, et al. MicroRNA Profiling Identifies miR-34a and miR-21 and Their Target Genes JAG1 and WNT1 in the Coordinate Regulation of Dendritic Cell Differentiation. Blood (2009) 114:404–14. 10.1182/blood-2008-09-179150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Duroux-Richard I, Robin M, Peillex C, Apparailly F. MicroRNAs: Fine Tuners of Monocyte Heterogeneity. Front Immunol (2019) 10:2145. 10.3389/fimmu.2019.02145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zawada AM, Zhang L, Emrich IE, Rogacev KS, Krezdorn N, Rotter B, et al. MicroRNA Profiling of Human Intermediate Monocytes. Immunobiology (2017) 222(3):587–96. 10.1016/j.imbio.2016.11.006 [DOI] [PubMed] [Google Scholar]
- 50. Rajaram MV, Ni B, Morris JD, Brooks MN, Carlson TK, Bakthavachalu B, et al. Mycobacterium Tuberculosis Lipomannan Blocks TNF Biosynthesis by Regulating Macrophage MAPK-Activated Protein Kinase 2 (MK2) and microRNA miR-125b. Proc Natl Acad Sci USA (2011) 108(42):17408–13. 10.1073/pnas.1112660108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-κB-Dependent Induction of microRNA miR-146, an Inhibitor Targeted to Signaling Proteins of Innate Immune Responses. Proc Natl Acad Sci USA (2006) 103:12481–6. 10.1073/pnas.0605298103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Moschos SA, Williams AE, Perry MM, Birrell MA, Belvisi MG, Lindsay MA. Expression Profiling In Vivo Demonstrates Rapid Changes in Lung microRNA Levels Following Lipopolysaccharide-Induced Inflammation But Not in the Anti-Inflammatory Action of Glucocorticoids. BMC Genomics (2007) 240:1–12. 10.1186/1471-2164-8-240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Baltimore D, Boldin MP, O’Connell RM, Rao DS, Taganov KD. MicroRNAs: New Regulators of Immune Cell Development and Function. Nat Immunol (2008) 9:839–45. 10.1038/ni.f.209PMID:18645592 [DOI] [PubMed] [Google Scholar]
- 54. O’Connell RM, Rao DS, Chaudhuri AA, Baltimore D. Physiological and Pathological Roles for Micro- RNAs in the Immune System. Nat Rev Immunol (2010) 10(2):111–22. 10.1038/nri2708 [DOI] [PubMed] [Google Scholar]
- 55. Wu Y, Sun Q, Dai L. Immune Regulation of miR-30 on the Mycobacterium Tu- Berculosis-Induced TLR/MyD88 Signaling Pathway in THP-1 Cells. Exp Ther Med (2017) 14:3299–303. 10.3892/etm.2017.4872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Mahadik K, Prakhar P, Rajmani RS, Singh A, Balaji KN. c-Abl-TWIST1 Epi- Genetically Dysregulate Inflammatory Responses During Mycobacterial Infection by Co-Regulating Bone Morphogenesis Protein and miR27a. Front Immunol (2018) 9:85. 10.3389/fimmu.2018.00085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Ni B, Rajaram MV, Lafuse WP, Landes MB, Schlesinger LS. Mycobacterium Tuberculosis Decreases Human Macrophage IFN-γ Responsiveness Through miR-132 and miR-26a. J Immunol (2014) 193(9):4537–47. 10.4049/jimmunol.1400124 [DOI] [PubMed] [Google Scholar]
- 58. Kumar M, Sahu SK, Kumar R, Subuddhi A, Maji RK, Jana K, et al. MicroRNA Let-7 Modulates the Immune Response to Mycobacterium Tuberculosis Infection Via Control of A20, an Inhibitor of the NF-κb Pathway. Cell Host Microbe (2015) 17(3):345–56. 10.1016/j.chom.2015.01.007 [DOI] [PubMed] [Google Scholar]
- 59. Saba R, Sorensen DL, Booth SA. MicroRNA-146a: A Dominant, Negative Regulator of the Innate Immune Response, Front. Immunol (2014) 5:578. 10.3389/fimmu.2014.00578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Li S, Yue Y, Xu W, Xiong S. MicroRNA-146a Represses Mycobacteria-Induced Inflammatory Response and Facilitates Bacterial Replication Via Targeting IRAK-1 and TRAF-6. PloS One (2013) 8:e81438. 10.1371/journal.pone.0081438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Liu, Wang R, Jiang J, Yang B, Cao Z, Cheng X. miR-223 Is Upregulated in Monocytes From Patients With Tuberculosis and Regulates Function of Monocyte-Derived Macrophages. Mol Immunol (2015) 67:475–81. 10.1016/j.molimm.2015.08.006 [DOI] [PubMed] [Google Scholar]
- 62. Xi X, Zhang C, Han W, Zhao H, Zhang H, Jiao J. MicroRNA-223 Is Upregulated in Active Tuberculosis Patients and Inhibits Apoptosis of Macrophages by Targeting FOXO3, Genet. Test Mol Biomarkers (2015) 19:650–6. 10.1089/gtmb.2015.0090 [DOI] [PubMed] [Google Scholar]
- 63. Wu Z, Lu H, Sheng J, Li L. Inductive microRNA-21 Impairs Anti-Mycobacterial Responses by Targeting IL-12 and Bcl-2. FEBS Lett (2012) 586(16):2459–67. 10.1016/j.febslet.2012.06.004 [DOI] [PubMed] [Google Scholar]
- 64. Chen M, Divangahi M, Gan H, Shin DSJ, Hong S, Lee DM, et al. Lipid Mediators in Innate Immunity Against Tuberculosis: Opposing Roles of PGE2 and LXA4 in the Induction of Macrophage Death. J Exp Med (2008) 205:2791– 2801. 10.1084/jem.20080767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Liu HY. Down-Regulation of miR-144 After Mycobacterium Tuberculosis Infection Promotes Inflammatory Factor Secretion From Macrophages Through the Tpl2/ERK Pathway. Cell Mol Biol (Noisy-le-grand) (2016) 62(2):87–93. [PubMed] [Google Scholar]
- 66. Zhang ZM, Zhang AR, Xu M, Lou J, Qiu WQ. TLR-4/miRNA-32-5p/FSTL1 Signaling Regulates Mycobacterial Survival and Inflammatory Responses in Mycobacterium Tuberculosis-Infected Macrophages. Exp Cell Res (2017) 352:313–21. 10.1016/j.yexcr.2017.02.025 [DOI] [PubMed] [Google Scholar]
- 67. Song Q, Li H, Shao H, Li C, Lu X. MicroRNA-365 in Macrophages Regulates Mycobacterium Tuberculosis-Induced Active Pulmonary Tuberculosis Via Interleukin-6. Int J Clin Exp Med (2015) 8:15458–65. [PMC free article] [PubMed] [Google Scholar]
- 68. Bettencourt P, Marion S, Pires D, Santos LF, Lastrucci C, Carmo N, et al. Actin-Binding Protein Regulation by microRNAs as a Novel Microbial Strategy to Modulate Phagocytosis by Host Cells: The Case of N-Wasp and miR-142-3p. Front Cell Infect Microbiol (2013) 3:19. 10.3389/fcimb.2013.00019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Chen YC, Lee CP, Hsiao CC, Hsu PY, Wang TY, Wu CC, et al. MicroRNA-23a-3p Down-Regulation in Active Pulmonary Tuberculosis Patients With High Bacterial Burden Inhibits Mononuclear Cell Function and Phagocytosis Through TLR4/TNF-α/TGF-β1/Il-10 Signaling Via Targeting IRF1/SP1. Int J Mol Sci (2020) 21(22):8587. 10.3390/ijms21228587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Pires D, Bernard EM, Pombo JP, Carmo N, Fialho C, Gutierrez MG, et al. Mycobacterium Tuberculosis Modulates miR-106b-5p to Control Cathepsin S Expression Resulting in Higher Pathogen Survival and Poor T- Cell Activation. Front Immunol (2017) 8:1819. 10.3389/fimmu.2017.01819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Zheng L, Leung E, Lee N, Lui G, To KF, Chan RC, et al. Differential MicroRNA Expression in Human Macrophages With Mycobacterium Tuberculosis Infection of Beijing/W and Non-Beijing/W Strain Types. PloS One (2015) 10(6):e0126018. 10.1371/journal.pone.0126018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Zhang D, Yi Z, Fu Y. Downregulation of miR-20b-5p Facilitates Mycobacterium Tuberculosis Survival in RAW 264.7 Macrophages Via Attenuating the Cell Apoptosis by Mcl-1 Upregulation. J Cell Biochem (2019) 120(4):5889–96. 10.1002/jcb.27874 [DOI] [PubMed] [Google Scholar]
- 73. Wang Q, Liu S, Tang Y, Liu Q, Yao Y. MPT64 Protein From Mycobacterium Tuberculosis Inhibits Apoptosis of Macrophages Through NF-kB-miRNA21-Bcl-2 Pathway. PloS One (2014) 9(7):e100949. 10.1371/journal.pone.0100949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Tripathi A, Srivastava V, Singh BN. hsa-let-7b-5p Facilitates Mycobacterium Tuberculosis Survival in THP-1 Human Macrophages by Fas Downregulation. FEMS Microbiol Lett (2018) 365(7). 10.1093/femsle/fny040 [DOI] [PubMed] [Google Scholar]
- 75. Liu Y, Jiang J, Wang X, Zhai F, Cheng X. miR-582-5p Is Upregulated in Patients With Active Tuberculosis and Inhibits Apoptosis of Monocytes by Targeting FOXO1. PloS One (2013) 8(10):e78381. 10.1371/journal.pone.0078381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Zhang G, Liu X, Wang W, Cai Y, Li S, Chen Q, et al. Down-Regulation of miR-20a-5p Triggers Cell Apoptosis to Facilitate Mycobacterial Clearance Through Targeting JNK2 in Human Macrophages. Cell Cycle (2016) 15(18):2527–38. 10.1080/15384101.2016.1215386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Ghorpade DS, Leyland R, Kurowska-Stolarska M, Patil SA, Balaji KN. MicroRNA-155 Is Required for Mycobacterium Bovis BCG-Mediated Apoptosis of Macrophages. Mol Cell Biol (2012) 32(12):2239–53. 10.1128/MCB.06597-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Wu Y, Guo Z, Yao K, Miao Y, Liang S, Liu F, et al. The Transcriptional Foundations of Sp110-Mediated Macrophage (RAW264.7) Resistance to Mycobacterium Tuberculosis H37Ra. Sci Rep (2016) 6:22041. 10.1038/srep22041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Liang S, Song Z, Wu Y, Gao Y, Gao M, Liu F, et al. MicroRNA-27b Modulates Inflammatory Response and Apoptosis During Mycobacterium tuberculosis Infection. Infect J Immunol (2018) 200(10):3506–18. 10.4049/jimmunol.1701448 [DOI] [PubMed] [Google Scholar]
- 80. Siqueira MDS, Ribeiro RDM, Travassos LH. Autophagy and Its Interaction With Intracellular Bacterial Pathogens. Front Immunol (2018) 9:935. 10.3389/fimmu.2018.00935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Abdalla AE, Duan X, Deng W, Zeng J, Xie J. MicroRNAs Play Big Roles in Modulating Macrophages Response Toward Mycobacteria Infection. . Infect Genet Evol (2016) 45:378–82. 10.1016/j.meegid.2016.09.023 [DOI] [PubMed] [Google Scholar]
- 82. Kim JK, Kim TS, Basu J, Jo EK. MicroRNA in Innate Immunity and Autophagy During Mycobacterial Infection. Cell Microbiol (2017) 19(1):e12687. 10.1111/cmi.12687 [DOI] [PubMed] [Google Scholar]
- 83. Zhao Y, Wang Z, Zhang W, Zhang L. MicroRNAs Play an Essential Role in Autophagy Regulation in Various Disease Phenotypes. BioFactors (2019) 45(6):844–56. 10.1002/biof.1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Ouimet M, Koster S, Sakowski E, Ramkhelawon B, Van Solingen C, Oldebeken S, et al. Mycobacterium Tuberculosis Induces the miR-33 Locus to Reprogram Autophagy and Host Lipid Metabolism. Nat Immunol (2016) 17(6):677–86. 10.1038/ni.3434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Tu H, Yang S, Jiang T, Wei L, Shi L, Liu C, et al. Elevated Pulmonary Tuberculosis Biomarker miR-423-5p Plays Critical Role in the Occurrence of Active TB by Inhibiting Autophagosome-Lysosome Fusion. Emerg Microbes Infect (2019) 8(1):448–60. 10.1080/22221751.2019.1590129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Liu F, Chen J, Wang P, Li H, Zhou Y, Liu H, et al. MicroRNA-27a Controls the Intracellular Survival of Mycobacterium Tuberculosis by Regulating Calcium-Associated Autophagy. Nat Commun (2018) 9(1):1–14. 10.1038/s41467-018-06836-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Kim JK, Yuk JM, Kim SY, Kim TS, Jin HS, Yang CS, et al. MicroRNA-125a Inhibits Autophagy Activation and Antimicrobial Responses During Mycobacterial Infection. J Immunol (2015) 194(11):5355–65. 10.4049/jimmunol.1402557 [DOI] [PubMed] [Google Scholar]
- 88. Kim JK, Lee HM, Park KS, Shin DM, Kim TS, Kim YS, et al. MIR144* Inhibits Antimicrobial Responses Against Mycobacterium Tuberculosis in Human Monocytes and Macrophages by Targeting the Autophagy Protein DRAM2. Autophagy (2017) 13(2):423–41. 10.1080/15548627.2016.1241922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Ding S, Qu Y, Yang S, Zhao YE, Xu G. Novel miR-1958 Promotes Mycobacterium Tuberculosis Survival in RAW264. 7 Cells by Inhibiting Autophagy Via Atg5. J Microbiol Biotechnol (2019) 29(6):989–98. 10.4014/jmb.1811.11062 [DOI] [PubMed] [Google Scholar]
- 90. Qu Y, Ding S, Ma Z, Jiang D, Xu X, Zhang Y, et al. MiR-129-3p Favors Intracellular BCG Survival in RAW264. 7 Cells by Inhibiting Autophagy Via Atg4b. Cell Immunol (2019) 337:22–32. 10.1016/j.cellimm.2019.01.004 [DOI] [PubMed] [Google Scholar]
- 91. Guo L, Zhao J, Qu Y, Yin R, Gao Q, Ding S, et al. microRNA-20a Inhibits Autophagic Process by Targeting ATG7 and ATG16L1 and Favors Mycobacterial Survival in Macrophage Cells. Front Cell Infect Microbiol (2016) 6:134. 10.3389/fcimb.2016.00134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Duan X, Zhang T, Ding S, Wei J, Su C, Liu H, et al. microRNA-17-5p Modulates Bacille Calmette-Guerin Growth in RAW264. 7 Cells by Targeting ULK1. PloS One (2015) 10(9):e0138011. 10.1371/journal.pone.0138011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Chen Z, Wang T, Liu Z, Zhang G, Wang J, Feng S, et al. Inhibition of Autophagy by MiR-30A Induced by Mycobacteria Tuberculosis as a Possible Mechanism of Immune Escape in Human Macrophages. Jpn J Infect Dis (2015) 68(5):420–4. 10.7883/yoken.JJID.2014.466 [DOI] [PubMed] [Google Scholar]
- 94. Chen DY, Chen YM, Lin CF, Lo CM, Liu HJ, Liao TL. MicroRNA-889 Inhibits Autophagy to Maintain Mycobacterial Survival in Patients With Latent Tuberculosis Infection by Targeting TWEAK. Mbio (2020) 11(1):e03045–19. 10.1128/mBio.03045-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Wang Y, Chen C, Xu XD, Li H, Cheng MH, Liu J, et al. Levels of miR-125a-5p Are Altered in Mycobacterium Avium-Infected Macrophages and Associate With the Triggering of an Autophagic Response. Microbes Infect (2020) 22(1):31–9. 10.1016/j.micinf.2019.07.002 [DOI] [PubMed] [Google Scholar]
- 96. Sabir N, Hussain T, Shah SZA, Peramo A, Zhao D, Zhou X. miRNAs in Tuberculosis: New Avenues for Diagnosis and Host-Directed Therapy. Front Microbiol (2018) 9:602. 10.3389/fmicb.2018.00602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Wang J, Yang K, Zhou L, Wu Y, Zhu M, Lai X, et al. MicroRNA-155 Promotes Autophagy to Eliminate Intracellular Mycobacteria by Targeting Rheb. PloS Pathog (2013) 9(10):e1003697. 10.1371/journal.ppat.1003697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Li M, Cui J, Niu W, Huang J, Feng T, Sun B, et al. Long non-Coding PCED1B-AS1 Regulates Macrophage Apoptosis and Autophagy by Sponging miR-155 in Active Tuberculosis. Biochem Biophys Res Commun (2019) 509(3):803–9. 10.1016/j.bbrc.2019.01.005 [DOI] [PubMed] [Google Scholar]
- 99. Etna MP, Sinigaglia A, Grassi A, Giacomini E, Romagnoli A, Pardini M, et al. Mycobacterium Tuberculosis-Induced miR-155 Subverts Autophagy by Targeting ATG3 in Human Dendritic Cells. PloS Pathog (2018) 14(1):e1006790. 10.1371/journal.ppat.1003697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Yang C, Shi Z, Hu J, Wei R, Yue G, Zhou D. miRNA−155 Expression and Role in Pathogenesis in Spinal Tuberculosis−Induced Intervertebral Disc Destruction. Exp Ther Med (2019) 17(4):3239–46. 10.3892/etm.2019.7313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Rodriguez A, Vigorito E, Clare S, Warren MV, Couttet P, Soond DR, et al. Requirement of bic/microRNA-155 for Normal Immune Function. Science (2007) 316:608–11. 10.1126/science.1139253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Jackson AL, Levin AA. Developing microRNA Therapeutics: Approaching the Unique Complexities. Nucleic Acid Ther (2012) 22:213–25. 10.1089/nat.2012.0356PMID:22913594 [DOI] [PubMed] [Google Scholar]
- 103. Liu Y, Wang X, Jiang J, Cao Z, Yang B, Cheng X. Modulation of T Cell Cytokine Production by miR-144* With Elevated Expression in Patients With Pulmonary Tuberculosis. Mol Immunol (2011) 48:1084–90. 10.1016/j.molimm.2011.02.001 [DOI] [PubMed] [Google Scholar]
- 104. Wu J, Lu C, Diao N, Zhang S, Wang S, Wang F, et al. Analysis of microRNA Expression Profiling Identifies miR-155 and miR-155* as Potential Diagnostic Markers for Active Tuberculosis: A Preliminary Study. Hum Immunol (2012) 73(1):31–7. 10.1016/j.humimm.2011.10.003 [DOI] [PubMed] [Google Scholar]
- 105. Fu Y, Yi Z, Wu X, Li J, Xu F. Circulating microRNAs in Patients With Active Pulmonary Tuberculosis. J Clin Microbiol (2011) 49:4246–51. 10.1128/JCM.05459-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Qi Y, Cui L, Ge Y, Shi Z, Zhao K, Guo X, et al. Altered Serum microRNAs as Biomarkers for the Early Diagnosis of Pulmonary Tuberculosis Infection. BMC Infect Dis (2012) 12:384. 10.1186/1471-2334-12384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Spinelli SV, Diaz A, D’Attilio L, Marchesini MM, Bogue C, Bay ML, et al. Altered microRNA Expression Levels in Mononuclear Cells of Patients With Pulmonary and Pleural Tuberculosis and Their Relation With Components of the Immune Response. Mol Immunol (2013) 53:265–9. 10.1016/j.molimm.2012.08.008 [DOI] [PubMed] [Google Scholar]
- 108. Wang J, Zhu X, Xiong X, Ge P, Liu H, Ren N, et al. Identification of Potential Urine Proteins and microRNA Biomarkers for the Diagnosis of Pulmonary Tuberculosis Patients. Emerg Microbes Infect (2018) 7(1):63. 10.1038/s41426-018-0066-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Pedersen JL, Bokil NJ, Saunders BM. Developing New TB Biomarkers, Are miRNA the Answer? Tuberculosis (Edinb) (2019) 118:101860. 10.1016/j.tube.2019.101860 [DOI] [PubMed] [Google Scholar]
- 110. Furci L, Schena E, Miotto P, Cirillo DM. Alteration of Human Macrophages microRNA Expression Profile Upon Infection With Mycobacterium Tuberculosis. Int J Mycobacteriol (2013) 2(3):128–34. 10.1016/j.ijmyco.2013.04.006 [DOI] [PubMed] [Google Scholar]
- 111. Iwai H, Funatogawa K, Matsumura K, Kato-Miyazawa M, Kirikae F, Kiga K, et al. MicroRNA-155 Knockout Mice Are Susceptible to Mycobacterium Tuberculosis Infection. Tuberculosis (Edinb) (2015) 95(3):246–50. 10.1016/j.tube.2015.03.006 [DOI] [PubMed] [Google Scholar]
- 112. Rothchild AC, Sissons JR, Shafiani S, Plaisier C, Min D, Mai D, et al. MiR-155-Regulated Molecular Network Orchestrates Cell Fate in the Innate and Adaptive Immune Response to Mycobacterium Tuberculosis. Proc Natl Acad Sci USA (2016) 113(41):E6172–81. 10.1073/pnas.1608255113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Meng QL, Liu F, Yang XY, Liu XM, Zhang X, Zhang C, et al. Identification of Latent Tuberculosis Infection-Related microRNAs in Human U937 Macrophages Expressing Mycobacterium Tuberculosis Hsp16.3. BMC Microbiol (2014) 14:37. 10.1186/1471-2180-14-37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Das K, Saikolappan S, Dhandayuthapani S. Differential Expression of miRNAs by Macrophages Infected With Virulent and Avirulent Mycobacterium Tuberculosis. Tuberculosis (2013) 93(Suppl):S47–50. 10.1016/S1472-9792(13)70010-6 [DOI] [PubMed] [Google Scholar]
- 115. Walter ND, Bemis L, Edwards M, Ovrutsky A, Ramamoorthy P, Bai X, et al. Differential Expression of MicroRNA in Mycobacterium Tuberculosis-Infected Human Macrophages. Am J Respir Crit Care Med (2012) 185:A1011. 10.1164/ajrccm-conference.2012.185.1_MeetingAbstracts.A1011 [DOI] [Google Scholar]
- 116. Palucci I, Delogu G. Host Directed Therapies for Tuberculosis: Futures Strategies for an Ancient Disease. Chemotherapy (2018) 63(3):172–80. 10.1159/000490478 [DOI] [PubMed] [Google Scholar]
- 117. Paik S, Kim JK, Chung C, Jo EK. Autophagy: A New Strategy for Host-Directed Therapy of Tuberculosis. Virulence (2019) 10(1):448–59. 10.1080/21505594.2018.1536598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Bento CF, Empadinhas N, Mendes V. Autophagy in the Fight Against Tuberculosis. DNA Cell Biol (2015) 34(4):228–42. 10.1089/dna.2014.2745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Periyasamy KM, Ranganathan UD, Tripathy SP, Bethunaickan R. Vitamin D–A Host Directed Autophagy Mediated Therapy for Tuberculosis. Mol Immunol (2020) 127:238–44. 10.1016/j.molimm.2020.08.007 [DOI] [PubMed] [Google Scholar]
- 120. Meister G, Landthaler M, Dorsett Y, Tuschl T. Sequence-Specific Inhibition of microRNA- and SiRNA-Induced RNA Silencing. RNA (2004) 10:544–50. 10.1261/rna.5235104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Grimm D, Streetz KL, Jopling CL, Storm TA, Pandey K, Davis CR, et al. Fatality in Mice Due to Oversaturation of Cellular microRNA/Short Hairpin RNA Pathways. Nature (2006) 7092:537–41. 10.1038/nature04791 [DOI] [PubMed] [Google Scholar]
- 122. Baumann V, Winkler J. MiRNA-based Therapies: Strategies and Delivery Platforms for Oligonucleotide and Non-Oligonucleotide Agents. Future Med Chem (2014) 17:1967–84. 10.4155/fmc.14.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Duan W, Yang T, Zhou SF, Wang ZL, Zhou ZW, He ZX. Novel Targeting of PEGylated Liposomes for Codelivery of TGF-Beta1 siRNA and Four Antitubercular Drugs to Human Macrophages for the Treatment of Mycobacterial Infection: A Quantitative Proteomic Study. Drug Des Dev Ther (2015) 9:4441–70. 10.2147/DDDT.S79369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Moore LB, Sawyer AJ, Saucier-Sawyer J, Saltzman WM, Kyriakides TR. Nanoparticle Delivery of miR-223 to Attenuate Macrophage Fusion. Biomaterials (2016) 89:127–35. 10.1016/j.biomaterials.2016.02.036 [DOI] [PMC free article] [PubMed] [Google Scholar]