Abstract
Objective
Immature CD11b + Gr1+ myeloid cells that acquire immunosuppressive capability, also known as myeloid-derived suppressor cells (MDSCs), are a heterogeneous population of cells that regulate immune responses. Our study's objective was to elucidate the role of ovarian cancer microenvironment in regulating the immunosuppressive function of CD11b+Gr1+ myeloid cells.
Methods
All studies were performed using the intraperitoneal ID8 syngeneic epithelial ovarian cancer mouse model. Myeloid cell depletion and immunotherapy were carried out using anti-Gr1 mAb, gemcitabine treatments, and/or anti-PD1 mAb. The treatment effect was assessed by a survival curve, in situ luciferase-guided imaging, and histopathologic evaluation. Adoptive transfer assays were carried out between congenic CD45.2 and CD45.1 mice. Immune surface and intracellular markers were assessed by flow cytometry. ELISA, western blot, and RT-PCR techniques were employed to assess the protein and RNA expression of various markers. Bone marrow-derived myeloid cells were used for ex-vivo studies.
Results
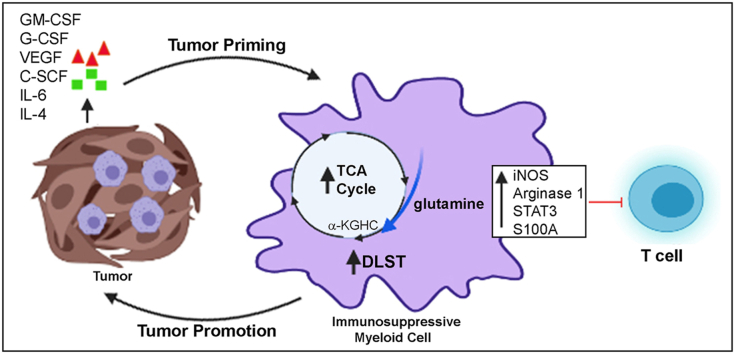
The depletion of Gr1+ immunosuppressive myeloid cells alone and in combination with anti-PD1 immunotherapy inhibited ovarian cancer growth. In addition to the adoptive transfer studies, these findings validate the role of immunosuppressive CD11b+Gr1+ myeloid cells in promoting ovarian cancer. Mechanistic investigations showed that ID8 tumor cells and their microenvironments produced recruitment and regulatory factors for immunosuppressive CD11b+Gr1+ myeloid cells. CD11b+Gr1+ myeloid cells primed by ID8 tumors showed increased immunosuppressive marker expression and acquired an energetic metabolic phenotype promoted primarily by increased oxidative phosphorylation fueled by glutamine. Inhibiting the glutamine metabolic pathway reduced the increased oxidative phosphorylation and decreased immunosuppressive markers’ expression and function. Dihydrolipoamide succinyl transferase (DLST), a subunit of α-KGDC in the TCA cycle, was found to be the most significantly elevated gene in tumor-primed myeloid cells. The inhibition of DLST reduced oxidative phosphorylation, immunosuppressive marker expression and function in myeloid cells.
Conclusion
Our study shows that the ovarian cancer microenvironment can regulate the metabolism and function of immunosuppressive CD11b + Gr1+ myeloid cells and modulate its immune microenvironment. Targeting glutamine metabolism via DLST in immunosuppressive myeloid cells decreased their activity, leading to a reduction in the immunosuppressive tumor microenvironment. Thus, targeting glutamine metabolism has the potential to enhance the success of immunotherapy in ovarian cancer.
Keywords: Ovarian cancer, CD11b+Gr1+ immunosuppressive cells, Glutamine metabolism, DLST, MDSCs, α-KGDC
Graphical abstract
Highlights
-
•
Ovarian cancer microenvironment increases the immunosuppressive ability of myeloid cells.
-
•
Ovarian cancer microenvironment can induce metabolic reprogramming in the immunosuppressive myeloid cells.
-
•
Immunosuppressive myeloid cells upsurge their oxidative metabolism fueled by glutamine in ovarian cancer.
-
•
DLST, a component of alpha-ketoglutarate dehydrogenase complex plays a key role in the immunometabolic regulation of immunosuppressive myeloid cells in ovarian cancer.
1. Introduction
Epithelial ovarian cancer (EOC) remains the most lethal gynecologic cancer in women, yet patient survival has not improved in decades. Immunotherapy is an attractive therapeutic option for ovarian cancer, and evidence suggests that it may be a promising alternative to current therapeutic strategies [1,2]. EOC cells elicit a specific immune response and express many tumor-associated antigens, potentially allowing for susceptibility to immunotherapy [[3], [4], [5]]. High-grade serous ovarian cancer patients with an immunoreactive subtype of tumors exhibit improved survival compared to other subtypes [6,7]. Additionally, in EOC patients, the presence of tumor-infiltrating lymphocytes correlates with longer progression-free and overall survival, while the lack of lymphocytes is associated with poor prognosis [[8], [9], [10], [11]]. Though promising, the results from clinical trials based on checkpoint inhibitors, vaccines, or adoptive T cell therapy are moderate compared to the remarkable developments seen in other cancer types [[12], [13], [14]]. Thus, the efficacy of immunotherapy for EOC has shown only limited improvement in tumor responses or patient survival.
One of the underlying reasons for the limited efficacy of immunotherapy for EOC may be tumor-induced immune suppression [15,16]. Multiple pathways in various cell types inhibit the immune response against tumors; this includes immune suppression by myeloid-derived suppressor cells (MDSCs), which are CD11b+Gr1+ immature myeloid cells that persist under conditions of chronic infection, inflammation, or cancer and play a crucial role in potentiating tumor growth [17]. Their primary function is to suppress various immune responses, especially regarding T cells. In mice, MDSCs are broadly characterized as CD11b+Gr1+ and consist of two subsets called granulocytic/polymorphonuclear MDSCs (PMN-MDSCs) and monocytic MDSCs (M-MDSCs) [18]. PMN-MDSC are similar to neutrophils and are identified as CD11b+Ly6G+Ly6Clo in mice and CD11b + CD14−CD15+ or CD66- in humans. However, M-MDSCs are morphologically similar to monocytes and are recognized as CD11b+Ly6G−Ly6Chi in mice and as CD11b + CD14+HLADR−/loCD15−Lin- in humans [17,18]. Though PMN-MDSCs and M-MDSCs suppress T cells, they do so via distinct mechanisms. PMN-MDSCs produce reactive oxygen species and arginase 1 enzyme, whereas M-MDSCs use nitric oxide synthase 2 and reactive oxygen species to inhibit T-cells [17,18]. Although MDSCs are morphologically and phenotypically similar to neutrophils and monocytes, they have a distinct genomic, biochemical, and functional identity [19,20]. Recently, the similarities between PMN-MDSC and activated neutrophils have blurred the contribution of each cell type in cancer. Tumor-associated neutrophils are pro-inflammatory, promote ovarian cancer metastatic niche, and suppress T-cell function [21,22]. As there are no distinct mouse markers to differentiate between MDSCs and tumor-associated neutrophils, we are using the broad terminology of immunosuppressive CD11b+Gr1+ cells.
Apart from immune suppression, immunosuppressive CD11b+Gr1+ cells can also promote tumor progression by altering the tumor microenvironment; they can produce pro-angiogenic factors, support metastasis, promote chemoresistance, enhance stemness, and, in some cancers, modulate senescence [[23], [24], [25], [26], [27], [28], [29]]. In ovarian cancer, these immunosuppressive myeloid cells have been shown to enhance stemness [26], promote angiogenesis and invasion [23], and influence the outcome of immunotherapy [30,31]. The increased infiltration of immunosuppressive CD11b+Gr1+ cells into tumors was previously noted in cancer patients and is associated with worse outcomes, including in ovarian cancer patients [[8], [9], [10], [11]]. These studies demonstrate that immunosuppressive CD11b+Gr1+ cells play a critical pathological role in altering the immune response against ovarian cancer, ultimately enabling disease progression. Thus, identifying mechanisms to target CD11b+Gr1+ cells may lead to new therapeutic strategies.
Emerging evidence strongly support the role of cellular metabolism, particularly energy metabolism, in regulating the expansion, differentiation, and effector function of various immune cells, including macrophages, T cells, and immunosuppressive CD11b+Gr1+ cells [32,33]. Cells produce ATP, the energy currency mainly generated by glycolysis or oxidative phosphorylation, which are often dysregulated in cancer cells and immune cells. According to the tumor type, the immunosuppressive function and proliferation of immunosuppressive CD11b+Gr1+ cells have been reported to be regulated by various energy pathways. The roles of glycolysis, fatty acid oxidation, and glutaminolysis have been shown in breast cancer, lung and colon cancer, and breast and lung cancer, respectively [[34], [35], [36], [37], [38], [39], [40]].
In this study, we show that immunosuppressive CD11b+Gr1+ myeloid cells play a role in promoting ovarian cancer growth, and their depletion augments the effects of immunotherapy. Mechanistically, we show that the ovarian cancer microenvironment can increase the immunosuppressive ability of these cells by increasing their metabolic fitness through an increase in glutamine metabolism. Collectively, our data provides the framework for developing strategies to target immunosuppressive CD11b+Gr1+ cell metabolism that can be combined with other immunotherapeutic approaches to treat ovarian cancer.
2. Material and methods
2.1. Cell lines and reagents
The mouse ovarian surface-epithelial cancer cell line ID8, transduced with a lentiviral vector expressing firefly luciferase (ID8-luc2), was kindly provided by Dr. John Liao, University of Washington, Seattle, WA [41]. The cell line was maintained in RPMI media (HyClone, Thermo Scientific, Waltham, MA), supplemented with 10% fetal bovine serum (BioAbChem, Ladson, SC). For collecting ID8 conditioned media (ID8 CM), 70% confluent cells were given a media change, which was collected after 48 h. The collected media was filtered, aliquoted, and frozen until use. Metabolic pathway inhibitors BPTES [(1Z, 1′Z)-N′,N''-(5,5'-(thiobis (ethane-2,1-diyl))bis (1,3,4-thiadiazole-5,2-diyl))bis (2-phenylacetimidic acid)], 2-deoxy glucose (2-DG), 6-Diazo-5-oxo-l-norleucine (DON), and etomoxir were purchased from Cayman Chemicals (Ann Arbor, Michigan), and 3 methyl 2 oxalo valeric acid (MOV) was purchased from Sigma Aldrich (St Louis, MO).
2.2. Animal model and experiments
2.2.1. Induction of ovarian cancer
C57/B6 female mice of 6–8 weeks were acquired from Jackson Laboratory (Bar Harbor, ME) and maintained under standard optimized conditions in the animal facility at Henry Ford Hospital (Detroit, MI). Before the start of the study, mice were acclimated to the house for one week. To induce ovarian cancer, mice were injected intraperitoneally (IP) with 107 ID8-luc2 cells in 200 μl of PBS and were observed and weighed every other day. At selected time intervals, mice were imaged to monitor tumor progression. Mice that experienced severe signs of clinical distress (cachexia, anorexia, or increased respiration) or had an accumulation of ascites that interfered with mobility or bodily functions were immediately euthanized [42,43].
2.2.2. Treatments
All treatments began 10 days after tumor injection, and each treatment group consisted of 10 mice. Treatments included (i) anti-mouse Ly6G/Ly6C (anti-Gr-1 mAb) InVivoMab injected IP twice a week at 100 μg/mouse for 3 weeks; (ii) IgG2b isotype control, anti-keyhole limpet hemocyanin InVivoMab injected IP twice a week at 100 μg/kg body weight for 3 weeks; (iii) anti-mouse PD-1 InVivoMab injected intraperitoneally once a week at 100 μg/kg of their body weight for 3 weeks; (iv) gemcitabine injected IP twice a week at 100 mg/kg of body weight for 6 weeks and (v) vehicle PBS injected IP twice a week at 100 μl/mouse for 6 weeks. All InVivoMabs were purchased from BioX Cell (West Lebanon, NH). Gemcitabine was procured from Henry Ford Hospital Pharmacy, Detroit, MI.
2.2.3. Survival Curve
For the survival curve, n = 12 mice per group were injected with ID8 cells to form tumors and allowed to proceed until moribund or their abdominal circumference reached 9–10 cm, according to the IACUC approved endpoint. Survival curves were generated using a Kaplan–Meier analysis.
2.2.4. Bioluminescence imaging
Mice received an intraperitoneal injection of D-luciferin (Perkin Elmer) at 150 mg/kg of body weight (Perkin Elmer, Waltham, MA) 15 min before imaging while under 1–2% inhaled isoflurane anesthesia. The bioluminescence signals of the tumor were recorded using the Xenogen IVIS system 2000 series (Perkin Elmer, Akron, Ohio). Images were acquired over a period of 20 min with a 15s exposure. Living Image software (Perkin Elmer) was used to read and integrate the total bioluminescence signals in each region. Bioluminescence measurements were quantified as total photon flux emission as photons/second in the images acquired at the same exposure time for the various groups.
2.2.5. Adoptive transfer
Three sets of 6- to 8-week-old female C57/BL/6 CD45.1 congenic mice (n = 6/set) and two sets of CD45.2 congenic mice (n = 6/set) were injected intraperitoneally with 107 ID8-luc2 cells suspended in 200 μl of PBS. After 2 weeks, bone marrow (BM) from CD45.2 congenic mice was used to isolate granulocytic myeloid cells (Gr1hiLy6G+; purity ~92%–94%) and non-granulocytic myeloid cells Gr1-Ly6G−; purity ~92%–94%), using an MDSC isolation kit from Miltenyi Biotech (Bergisch Gladbach, Germany). The CD45.1 tumor-bearing mice were separated into three groups; group 1 control did not receive any cells but saline injections, group 2 received two IV injections of 5 × 106 purified granulocytic cells (Gr1+) a week apart, and group 3 received two IV injections of 5 × 106 purified non-granulocytic (Gr1-) cells a week apart [44]. Two weeks after the last transfer, mice were imaged and sacrificed for further analysis.
2.2.6. Ex vivo experiments
For all ex vivo experiments, 6- to 8-week-old female C57/B/6 mice (n = 4) were injected IP with 107 ID8-luc2 cells suspended in 200 μl of PBS. Two weeks later, Gr1+ cells were isolated from BM using BD-IMAG anti-mouse LY6G and LY6C kit (BD Biosciences, San Jose, CA) [45], counted and used for various assays like Seahorse and fluorescence-activated cell sorting.
2.3. Targeted metabolomics
Granulocytic myeloid cells (Gr1+; purity ~92%–94%) were isolated using a BD-IMAG anti-mouse LY6G and LY6C kit (BD Biosciences, San Jose, CA) from the BM of naïve and ID8-luc2 tumor-bearing mice at 2 weeks (n = 4/group). Cells were counted, snap-frozen in liquid nitrogen, and stored at −80 °C until analysis. Metabolites were extracted using 80% methanol, as previously described [46]. Cellular concentrations of metabolites (normalized to cellular protein concentration) were quantitatively determined using an LC-MS/MS-based targeted metabolomics platform that is capable of quantitatively profiling ~ 250 metabolites, as previously described [46]. As described previously [47], data analysis was performed using the MetaboAnalyst 4.0 (www.MetaboAnalyst.ca). Features (metabolites) with >20% missing values (i.e., below the lower limit of quantitation) were removed from the analysis, while the remaining missing values were replaced by 1/5 of the minimum value of a feature. To meet the normality assumption, individual metabolite concentrations were log-transformed and then auto-scaled (mean-centered and divided by the standard deviation of each metabolite). Unsupervised principal component analysis (PCA) was performed to visualize the separation of global metabolic profiles between myeloid cells from naïve and tumor-bearing mice. We used SAM (Significance Analysis of Microarray and Metabolites) to determine the most significant metabolites. A false discovery rate (FDR; i.e., adjusted p-value) of <0.05 was considered statistically significant. Pathway analysis with the significantly changed metabolites was performed to identify the key metabolic pathways involved.
2.3.1. Ethics statement
All protocols were approved by the Henry Ford Hospital Institutional Animal Care and Use Committee before any experiments. All institutional and national guidelines for the care and use of laboratory animals were followed.
2.4. Fluorescence-activated cell sorting analysis
For surface marker staining, lymphocytes from tumors, ascites, blood, BM, or spleens were incubated with fluorochrome-conjugated antibodies to CD3, CD4, CD8, CD1d, CD16, CD11b, Ly6C, Ly6G, and Gr1 at the manufacturer's recommended dilution for 30 min at 4 °C, as before [45]. To analyze intracellular markers like IFNγ, lymphocytes were treated with GolgiPlug (BD Biosciences, San Jose, CA) for 5 h, followed by surface staining with monoclonal antibodies against CD4 or CD8. Cells were washed, fixed, and permeabilized with a cytofix/cytoperm buffer, followed by incubation with an antibody to detect intracellular levels of IFN-γ. Flow cytometric analysis was performed on a BD FACS Calibur (BD Biosciences), and results were analyzed using FACS Diva software (BD Biosciences). Fluorochrome labeled antibodies to anti-mouse CD4 (clone RM4-5), CD8 (clone 53–6.7), CD11b (clone M1/70), Ly6G (clone 1A8), Ly6C (clone HK1.4), CD45 (clone 30-F11), CD45.1 (clone A20) and CD45.2 (clone 104) were purchased from Biolegend (San Diego, CA). Anti-Gr1 (clone RB6-8C5), CD3 (clone 500A2), and IFN-γ (clone XMG1.2) were purchased from BD Biosciences. Antibody details are given in Table S1.
2.5. In vitro T cell suppression assay
CD4 T cells were isolated (more than 92% purity) from the spleen by negative selection using a MojoSort mouse CD4 T cell isolation kit (Biolegend) and labeled with CFSE (carboxyfluorescein diacetate, succinimidyl ester; 5 μM), as described by the manufacturer (Biolegend). GR1+ cells were isolated from the BM of mice harboring ID8 tumors for two weeks using a BD-IMAG anti-mouse LY6G and LY6C kit (>94% purity), in accordance with the manufacturer's instructions (BD Biosciences, San Jose, CA). For the inhibitor studies, isolated MDSCs were pretreated with the respective inhibitor 48h prior to co-culture. Isolated MDSC and CD4 cells were co-cultured at various ratios from 1:1 to 1:10 in 96-well plates coated with anti-CD3 and anti-CD28 antibodies (1 μg/ml). After 72 h, cells were stained for surface markers and intracellular cytokines and analyzed by flow cytometry, as described above [45].
2.6. Ex vivo bone marrow assays
BM cells flushed from the femur and tibia were plated in complete RPMI-1640 (RPMI-1640 includes 10% FBS and 20 mM penicillin/streptomycin) media. For differentiating into myeloid cells, cells were either treated with GM-CSF (20 ng/ml) and IL 6 (10 ng/ml) alone or in the presence of 30% ID8 CM. On day 8, cells were harvested, and GR1+ cells were isolated using an anti-mouse LY6G and LY6C kit according to the manufacturer's instructions (BD Biosciences, San Jose, CA). The purity of the isolated cell populations was determined by flow cytometry, and the frequency of CD11b+Gr1+ cells was >95%. The differentiated BM-MDSCs were treated with indicated inhibitors for 24–48 h. For glutamine deprivation, differentiated BM cells were plated in a low glutamine medium (0.5 mM) or the regular RPMI media containing 2 mM glutamine for 48 h before use for assays.
2.7. Myeloid-derived suppressor PCR array
A myeloid-derived suppressor PCR (Real-Time Primers, MMDSC-I, PA, USA) array was performed after isolating total RNA from the CD11b+Gr1+ myeloid cells (BD-IMAG GR1+) of naïve (no tumor) and ID8 tumor-bearing mice. This library profiles 88 genes involved in immunosuppression. The quantitative expression of the gene was calculated from the cycle threshold (CT) value of each sample and was normalized using the housekeeping genes by using online PCR quantification software provided by the manufacturer.
2.8. Glucose metabolism RT2 profiler PCR array analysis
The total RNA was isolated from the CD11b+Gr1+ myeloid cells using a Qiagen RNeasy® Mini Kit (Qiagen, Germantown, MD) according to the manufacturer's instructions. Single-strand cDNA was synthesized from 1 μg of total RNA by reverse transcription reaction using a Qiagen RT2 First Strand Kit (Qiagen, Germantown, MD) and subjected to the Glucose metabolism array that profiles the expression of 84 genes involved in glucose metabolism (RT2 profiler PCR array-PAMM-006Z, Qiagen). The quantitative expression of the gene was calculated from the cycle CT value of each sample. The samples were analyzed in duplicate and were normalized using the housekeeping genes by using RT2 PCR array data analysis software provided by the manufacturer. The gene list is provided in Supplementary Table 6.
2.9. ELISA
The expression levels of IL-6, IL-1β, IL- 4, and IFN-γ (Biolegend ELISA kits, San Diego, CA) and GM-CSF, G-CSF, VEGF, and S-CSF (R&D Biosystems ELISA kit, Minneapolis, MN) in ascites and cell culture supernatants were evaluated in accordance with the manufacturers’ instructions, as described earlier [43].
2.10. Real-time PCR
The total RNA was extracted from myeloid cells using an RNA assay kit (Qiagen, Germantown, MD) and quantified by Qubit Fluorimeter (Thermo Fisher Scientific, USA). The cDNA was synthesized as above. RT-PCR reactions were performed in a 25 μl mixture (1 μl cDNA, 1 × SYBR green buffer, forward and reverse specified primers) using CFX BioRad Real-time PCR Detection System. Ribosomal protein L27 was used as a housekeeping gene. All the primers were purchased from Integrated DNA Technologies (Coralville, IA). All primers used in the study are detailed in the supplementary material [48]. Primer details are given in Table S2.
2.11. Western blot analysis
The total protein was isolated from myeloid cells by a protein lysis buffer (50 mM Tris–HCl, pH 7.5, 250 mM NaCl, 5 mM EDTA, 50 mM NaF, and 0.5% Nonidet P-40; containing a protease inhibitor cocktail; Sigma, St Louis, MO), as described before [48] and quantified using BCA (Bicinchononic Acid assay kit; Thermo Fisher, Rockford, IL). An equal amount of protein was separated by 10–12% SDS polyacrylamide gel electrophoresis and transferred onto the polyvinylidene difluoride membrane and blocked with 5% skimmed milk. iNOS, Arginase 1, STAT-3, pSTAT3 were purchased from BD Biosciences (San Jose, CA). IL-1β and DLST were purchased from Cell Signaling Technology (Danvers, MA). S100A9 and CSF-R were purchased from Proteintech (Rosemont, IL, USA). The dilutions used for the antibodies are summarized in Table S3.
2.12. Seahorse metabolic analysis
2.12.1. Metabolic phenotype
CD11b+Gr1+ myeloid cells were isolated from naïve and ID8 tumor-bearing mice and were plated at the density of 3 × 105 cells/well in Cell-Tak coated XFe 96 cell plates. The oxidative phosphorylation (OCR) and extracellular acidification (ECAR) rates were measured using an XFe 96 seahorse analyzer (Agilent Seahorse XFe96 Analyzers), as described earlier [48]. For measuring OCR, cells were incubated in an XF base medium supplemented with 2 mmol/L glutamine, 10 mmol/L glucose, and 1 mmol/L pyruvate. Various metabolic inhibitors were injected to observe the effect of the oxidative phosphorylation and extracellular acidification rate. OCR measurements were recorded with port injections of (1) oligomycin (1 μmol), (2) FCCP (0.5 μmol), and a combination of (3) rotenone-antimycin at 1 μmol. ECAR, an indicator of aerobic glycolysis, was measured by incubating cells in an XFe base medium supplemented with 2 mmol/L glutamine. ECAR measurements were recorded after injecting with (1) glucose (10 mM), followed by (2) oligomycin (2 μM), and (3) 2-DG (100 mM).
2.12.2. Mito Fuel Flex test
Mitochondrial fuel dependency and the capacity of CD11b+Gr1+ cells was estimated using a Seahorse XF Mito Fuel Flex test kit (Agilent, USA). Dependency (cells relying on the particular pathway to meet its baseline respiration) and capacity (cells depending on other pathways when the depending pathway is inhibited) were measured for the use of glucose, glutamine, and fatty acid as fuel. BM-derived myeloid cells from naive and tumor-bearing mice were subjected to the fuel test to measure OCR in the presence or absence of carnitine palmityl transferase 1A inhibitor etomoxir (4 μm) to inhibit fatty acid oxidation (FAO) or glutaminase inhibitor BPTES (3 μm) to inhibit glutamine utilization or UK5099 (2 μm) for inhibition of glycolysis. This test determines the cellular dependence of the live cells on the metabolites to fuel their mitochondrial metabolism and capacity for utilizing that substrate when others were blocked. Capacity and dependence were calculated as shown, according to the manufacturer's instructions.
2.13. Histological analysis
Tumor tissue was fixed in 10% formaldehyde for 48 h and paraffin-embedded. Sections were cut and processed for hematoxylin and eosin (H&E) staining and immunohistochemistry to detect Ki-67, CD4, CD8, and cleaved caspase 3. The Ki-67 antibody was purchased from Abcam (Cambridge, MA) and used at a concentration of 1:200. Antibodies against CD4, CD8, and cleaved caspase 3 were obtained from Cell Signaling Technology (Denver, MA), and all were used at a 1:100 dilution. Solutions obtained from Dako Cytomation (Carpinteria, CA) were used for performing immunostaining, using the Dako Autostainer Link 48, as described previously. A minimum of 3–6 slides per group and 6–10 fields per slide were evaluated under a high-power field, and the quantification of stain intensity was performed by blinded pathologists (RA-F and EA) [42].
2.14. Statistical analysis
An unpaired t-test or one-way ANOVA was used where appropriate. A Kaplan Meier analysis was used to determine the survival curve. The significance of survival curves was estimated by using the Gehan Breslow–Wilcox test and the Mantel–Cox tests. All analyses were carried out using Prism 8 (GraphPad Software, La Jolla, CA).
3. Results
3.1. Depletion of CD11b + Gr1+ myeloid cells impedes the progression of ovarian tumors and enhances antitumor immune response
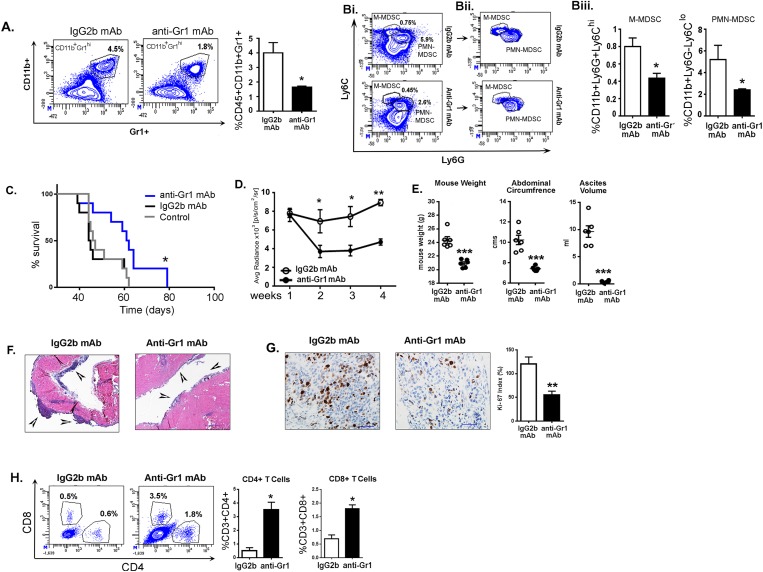
Immunosuppressive CD11b+Gr1+ immature myeloid cells, also identified as MDSCs, have been shown to accumulate in ovarian cancer patients and the ID8 syngeneic model, and the depletion of these cells has previously been demonstrated to suppress the progression of the ID8 tumors [24,49]. Using ID8-luc2 induced tumors, we confirmed that depleting CD11b+Gr1+ myeloid cells by anti-Gr1 mAb treatment depleted ~50% of CD11b+Gr1+ myeloid cells in peripheral blood compared to mice treated with an IgG2b mAb isotype at 6 weeks post-tumor injections (Figure 1A). A significant depletion in both PMN-myeloid cells (CD11b + Ly6GhiLy6Clo) and M-myeloid cells (CD11b+Ly6G−Ly6Chi) was observed (Figure 1B). Survival analysis revealed a significant increase in the median survival of anti-Gr1 mAb treated mice (median survival 61.5 days) compared to the IgG2b mAb isotype (median survival 44.5 days) or control (median survival 46.5 days) groups (Figure 1C). Decreased bioluminescence signals after anti-Gr1 mAb treatment suggested tumor growth was significantly suppressed compared to mice treated with the isotype control mAb (Figure 1D). Furthermore, 6 weeks after tumor inoculation, mice that received the anti-Gr1 mAb had reduced weight, a smaller abdominal circumference, and a decreased ascites volume (Figure 1E). Histological analysis revealed that anti-Gr1 mAb treated mice had smaller tumors than isotype-treated mice, as determined by H&E staining (Figure 1F). Anti-Gr1 mAb treated mice also had a significant reduction in the number of proliferating cells compared to isotype-treated mice, as determined by the Ki-67 index (Figure 1G). CD11b+Gr1+ depletion led to a significant increase in the percentage of CD4+ and CD8+ T cells in the ascites of anti-Gr1mAb-treated mice relative to IgG2b-treated controls (3.5% vs. 0.5% and 1.8% vs. 0.6%, respectively; Figure 1H).
Figure 1.
Depletion of CD11b+Gr1+myeloid cells by anti-Gr1 monoclonal antibody inhibits EOC progression. ID8-luc2 cells were injected intraperitoneally into C57Bl/6 mice. After 10 days of tumor initiation, mice were treated with IgG2b or anti-Gr1 mAb as described in methods. (A) Representative flow cytometry plots and graph from n = 3, of total CD11b+Gr1+ myeloid cells in the peripheral blood of mice treated with IgG2b mAb (left) or anti-Gr1 mAb (right). Numbers represent the frequency of cells in the indicated gate. (Bi) Flow cytometry plots (n = 3) representing LY6C and LY6G population from CD45+CD11b+ cells in mice treated with IgG2b mAb or anti-Gr1 mAb. (Bii) Flow cytometry plots showing only the populations of interest after removing all negative populations for clarity of the gates. (Biii) Bar graphs represent the average percentage of cells in each population. (C) Kaplan–Meier graphs indicating overall survival (n = 12) in ID8-luc2 mice untreated (control) or treated with IgG2b mAb or anti-Gr1 mAb; p = 0.0385 by Gehan-Breslow-Wilcox test and p = 0.0177 by Mantel–Cox test. (D) Average radiance (n = 6) of bioluminescence signal of tumor progression of mice treated with IgG2b mAb or anti-GR1 mAb. The weeks represent post-tumor inoculation time points. (E) Dot plots (n = 6) of mouse weight, abdominal circumference, and ascites volume of ID8-luc2 mice treated with IgG2b mAb or anti-Gr1 mAb. (F) Representative H & E images of tumor tissue in ID8-luc2 mice treated with IgG2b mAb or anti-Gr1 mAb at 4x. (G) Representative images of Ki67 staining at 40× magnification. Tumor sections from all 6 mice/group were examined—representative bar graph for Ki-67 proliferation index. (H) Representative flow plots and bar graphs for CD4+ and CD8+ T cell population in ID8-luc2 mice treated with IgG2b mAb or anti-Gr1 mAb. All analyses were carried out in mice after 5–6 weeks of tumor inoculation. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 anti-Gr1 mAb-treated mice compared to control IgG2b-treated mice as assessed by unpaired T-test.
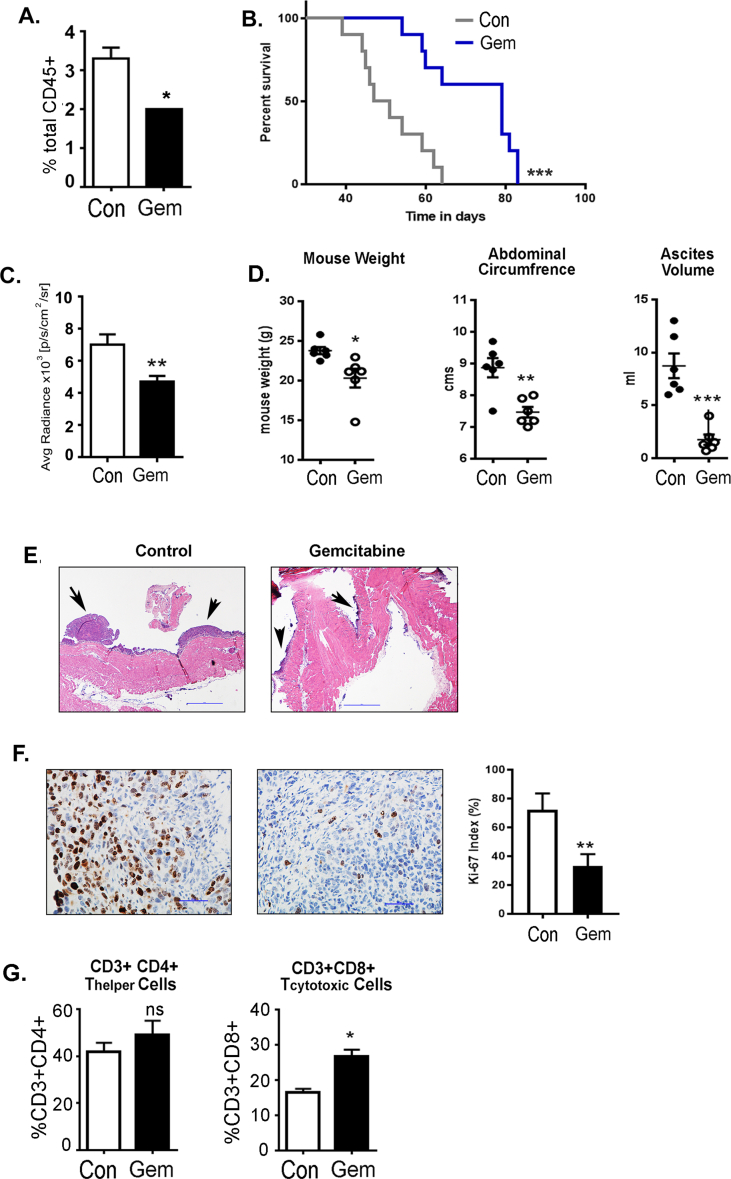
As an alternative treatment approach, we used a pyrimidine analog, gemcitabine, which has been reported to selectively eliminate suppressive myeloid cells without affecting other immune cells [50,51]. Similar to anti-Gr1 mAb treatment, the total number of CD11b+Gr1+ myeloid cells in the peripheral blood was reduced by approximately 50% after 6 weeks post-tumor inoculation in gemcitabine-treated mice (Fig. S1A). Survival analysis revealed a significant increase in the median survival of gemcitabine treated (79 days) mice compared to control mice (49 days; Fig. S1B). Gemcitabine treatment significantly reduced tumor growth, as evident from the quantification of bioluminescence signal after 4 weeks of treatment (Fig. S1C). Gemcitabine treatment also reduced body weight, abdominal circumferences, and ascites volume assessed after 6 weeks post-tumor inoculation (Fig. S1D). The decreased tumor burden in gemcitabine-treated mice was further supported by smaller tumors seen by H&E staining (Fig. S1E) and a lower Ki-67 index (Fig. S1F), indicating gemcitabine-treated mice have a decrease in actively proliferating cancer cells. Gemcitabine treatment also resulted in an increased percentage of total CD4+ and CD8+ T cells in the ascites (Fig. S1G). It is known that gemcitabine treatment induces cancer cell death, and its effect is primarily on the tumor cells and possibly secondary on the suppressive myeloid cells. However, these results further demonstrate the correlation between cancer cell growth and suppressive myeloid cell accumulation. Thus, as observed by us and others, alleviating immunosuppression by depleting immunosuppressive CD11b+Gr1+ cells reduces ID8-Luc2 induced ovarian tumors and restores the antitumor immune response.
3.2. CD11b+Gr1+ depletion enhances the efficacy of PD1 immunotherapy
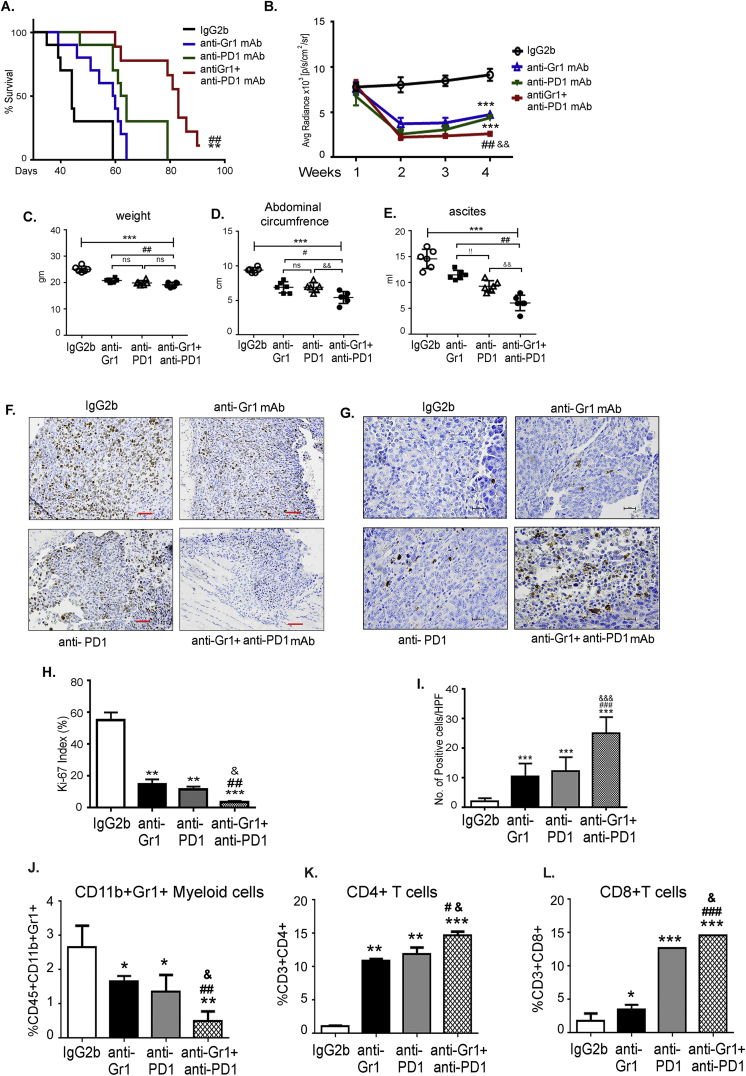
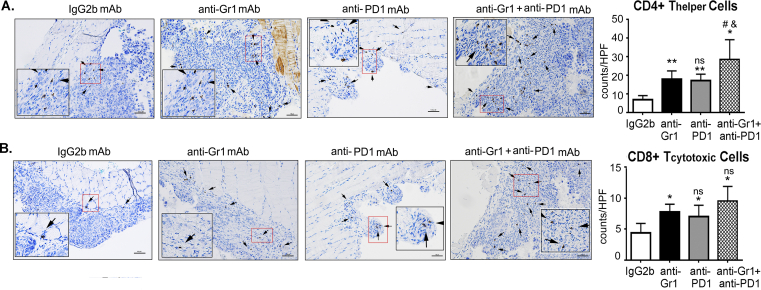
A blockade of PD-1 in mice with ovarian cancer has been shown to reduce the tumor burden in combination treatments [49,52,53]. To evaluate the impact of depleting CD11b+Gr1+ on anti-PD-1 immunotherapy, we performed combination and single treatments of anti-Gr1 mAb and anti-PD1 mAb in mice with ID8-luc2-induced EOC. The median overall survival was significantly increased upon treatment with the anti-Gr1 mAb (59.5 days) and upon the PD1-blockade (63 days) compared to IgG2b mAb treatment (44 days). The combined treatment significantly extended the survival of ID8 tumor-bearing mice (83 days) compared to the treatment alone and control groups (Figure 2A). Furthermore, mice that received the combination treatment had a lower bioluminescence signal each week after the start of the treatment, indicating smaller tumors compared to that in mice in other groups (Figure 2B). The enhanced effects of the combination treatment were also observed with the decrease in mouse weights, abdominal circumference, and ascites volume compared to single or control treatment groups (Figure 2C, D, E). The combination treatment also resulted in a lower Ki-67 index number 6 weeks after tumor inoculation (Figure 2 F, H). Though anti-Gr1 and anti-PD1 mAb treatments alone resulted in significant apoptosis of ID8 tumor cells, a greater increase in apoptotic cells was observed in tumors from mice that received the combination therapy, as evident from the enhanced number of cleaved caspase-3 positive cells in these tumors compared to mice from other groups. (Figure 2 G, I). Thus, targeting CD11b+Gr1+ augments the antitumor effect of the PD1 blockade and results in increased tumor cell death.
Figure 2.
Depletion of CD11b+Gr1+myeloid cells enhances PD1 immunotherapy. ID8-luc2 cells were injected intraperitoneally into C57Bl/6 mice. After 10 days of tumor initiation, mice were treated with IgG2b or anti-Gr1 mAb or anti-PD1 mAb or a combination of both as described in methods. (A) Kaplan–Meier graphs indicating overall survival (n = 12) under various treatments; p = 0.0094 by Gehan-Breslow-Wilcox test and p = 0.0042 by Mantel–Cox test on comparing PD1 alone to combination treatment; p = 0.0011 by Gehan-Breslow-Wilcox test and p = 0.0004 by Mantel–Cox test on comparing Gr1 alone to combination treatment. (B) Average radiance (n = 6) of bioluminescence signal of tumor progression of mice treated with various treatments. Weeks indicate post-tumor injection time points. Dot plots (n = 6) of mouse weight (C), abdominal circumference (D), and ascites volume (E) of ID8-luc2 mice treated with indicated treatments. (F) Representative images of Ki67 staining for assessing tumor cell proliferation. Images are shown at 4× magnification. Tumor sections from all 8 mice/group were examined. Ki-67 proliferation index is plotted as a bar graph (H). (G) Representative images of cleaved caspase 3 staining for assessing tumor cell apoptosis. Images are shown at 20× magnification. Tumor sections from 3 to 4 mice/group were examined. Positive cells were counted from a minimum of 6 fields at HPF from each slide and quantified (I). (J) Bar graph representing the percent of CD11b+Gr1+ myeloid cells in the ascites of mice under various treatments (n = 3). Bar graphs for CD4+ (K) and CD8+ (L) T cell population in ID8-luc2 mice with various treatments. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 compared to IgG2b; #p < 0.05, ##p < 0.01, ###p < 0.001 compared to anti-Gr1 and &p < 0.05, &&&p < 0.001compared to anti-PD1 as assessed by unpaired T-test.
Immune cell profiling of the ascites showed that CD11b+Gr1+ cells were significantly reduced upon treatment with anti-Gr1 mAbs or PD1 mAbs (Figure 2J), with the reduction in the combination group being significantly greater than the other groups (Figure 2J). The decreased frequency of CD11b+Gr1+ was accompanied by an enhanced population of CD4+ and CD8+ T cells in the ascitic environment of the ID8-luc2 tumors of single mAb treated groups that were further increased in mice that received combination therapy (Figure 2K,L). Notably, immunohistochemical analysis of tumor sections showed similar significant differences in the number of CD4+ and CD8+ T cells that had infiltrated the ID8 tumors (Figs. S2A and B). Thus, CD11b+Gr1+ depletion combined with immunotherapy can act as a coordinated approach to improve the immune response against ovarian tumors.
3.3. Adoptive transfer of CD11b+ Gr1+ myeloid cells promotes ovarian cancer progression
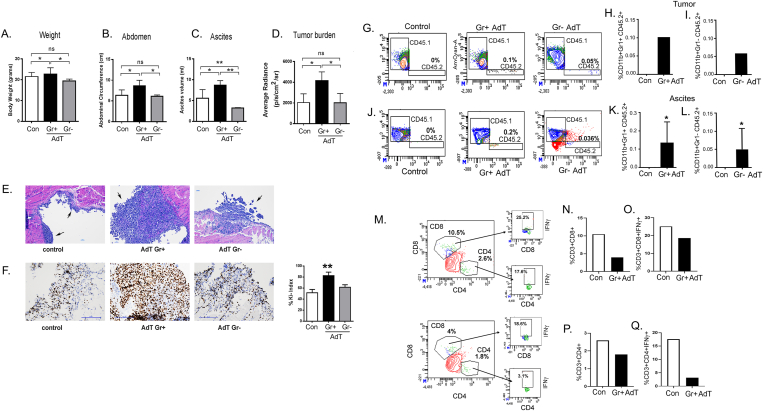
To confirm the role of CD11b+Gr1+ in promoting ovarian cancer, we isolated CD11b+Gr1+ and CD11b+Gr1- cells from the BM of wild-type C57/B6 congenic CD45.2 mice harboring ID8-luc2 cells and adoptively transferred (AdT) them by IV injections into congenic CD45.1 mice that had received ID8-luc2 cells two weeks prior to the transfer of cells. ID8 tumor-bearing recipients of CD11b+Gr1+ AdT had significantly increased bodyweight, abdominal circumference, ascites volume, and tumor burden at 2 weeks post-AdT compared to the control group of mice injected with ID8-luc2 cells but did not receive AdT or mice that received CD11b+Gr1- cells. (Figure 3A–D). Increased tumor burden was also confirmed by larger tumor nodules present in the CD11b+Gr1+ AdT mice, as seen by H&E staining of tumor tissues (Figure 3E) and an increased Ki-67 index, indicating faster proliferating tumor cells (Figure 3F). Flow cytometry analysis detected the donor CD45.2 CD11b+Gr1+ cells in the tumors and ascites of the CD45.1 recipient mice, and they were absent in the control mice that did not receive AdT, as expected. Donor CD45.2 CD11b+Gr1- cells were also detected in the tumors and ascites of the CD45.1 recipient mice, though at lower numbers (Figure 3G,I, J, L). This finding suggests that intravenous AdT CD11b+Gr1+ cells home to the tumor and TME. Concomitant with increased CD11b+Gr1+ cells, AdT mice had a reduction in CD4+ T and CD8+ T cells in tumors (Figure 3M, N, P), signifying immunosuppression. Additionally, CD4+ and CD8+ T cell populations expressed reduced intracellular IFN-γ, indicating decreased effector function (Figure 3O, Q). Thus, the increased number of CD11b+ Gr1+ myeloid cells was shown to enhance ovarian cancer growth and is associated with decreased tumor-infiltrating T cells.
Figure 3.
Adoptive transfer of tumor primed CD11b+Gr1+myeloid cells promotes progression of EOC. CD11b+Gr1+ or CD11b+Gr1- myeloid cells were adoptively transferred (AdT) from ID8-luc2 tumor primed CD45.2 congenic mice into sets of ID8-luc2 tumor-bearing congenic CD45.1 mouse by IV. A control set of mice received sham injections. Bar graphs showing (A) body weight, (B) Abdominal circumference, (C) Ascites volume, and (D) tumor burden measured by the average radiance of bioluminescence signal (n = 6). (E) Representative H &E staining (10x) of tumor tissue from control, CD11b+Gr1+ or CD11b+Gr1- AdT mice 4 week post AdT. (F) Representative Ki67 expression in tumor sections from mice 4 weeks post-AdT and the calculated Ki-67 index. A total of 8 HPF were studied from 3 to 4 tumor sections/group. Representative flow cytometry plots of CD45.2+CD11b+Gr1+ cells in tumors (G, H) and ascites (J, K) from CD45.1 mice with and without tumor. Tumor bar graphs represent the pooled tumor tissue of 3 mice. Ascites bar graph are from n = 3. Representative flow cytometry plots of CD45.2+CD11b+Gr1- cells in tumors (G, I) and ascites (J, L) from CD45.1 mice with and without tumor. Tumor bar graphs represent the pooled tumor tissue of 3 mice. Ascites bar graph are from n = 3. Representative flow plots of CD8+ (M, N)and CD4+ T (M, P) cells infiltrated in tumor and the intracellular IFNγ positive cells from respective CD8 (M, O) and CD4 (M, Q) populations of tumor infiltrates. Tumor bar graphs represent the pooled tumor tissue of 3 mice. ∗p < 0.05, ∗∗p < 0.01 compared to control as assessed by unpaired T-test.
3.4. Ovarian cancer environment enhances the immunosuppressive phenotype of CD11b + Gr1+ myeloid cells
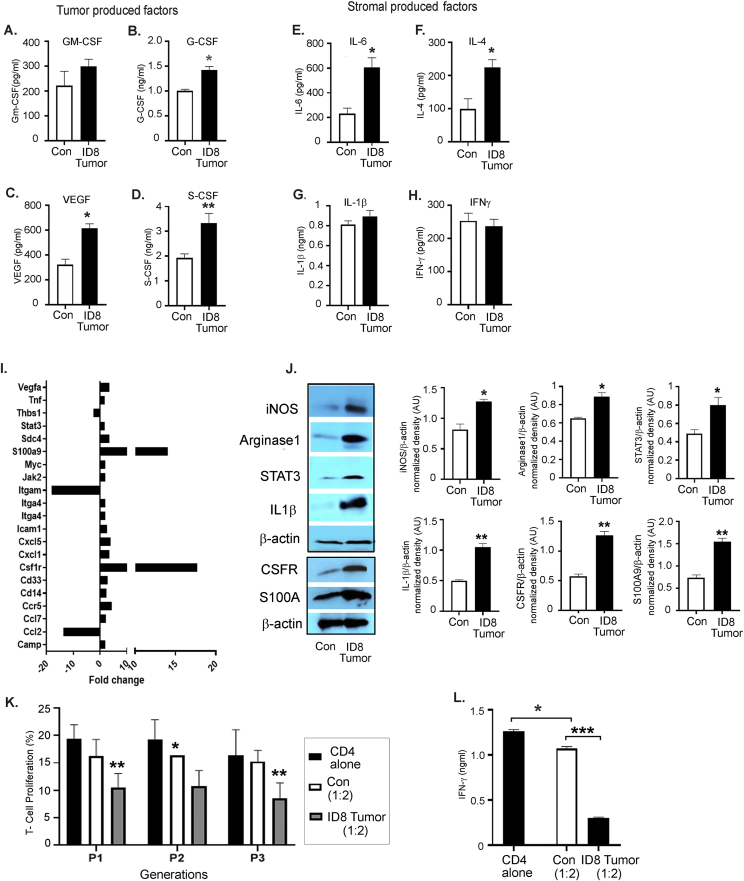
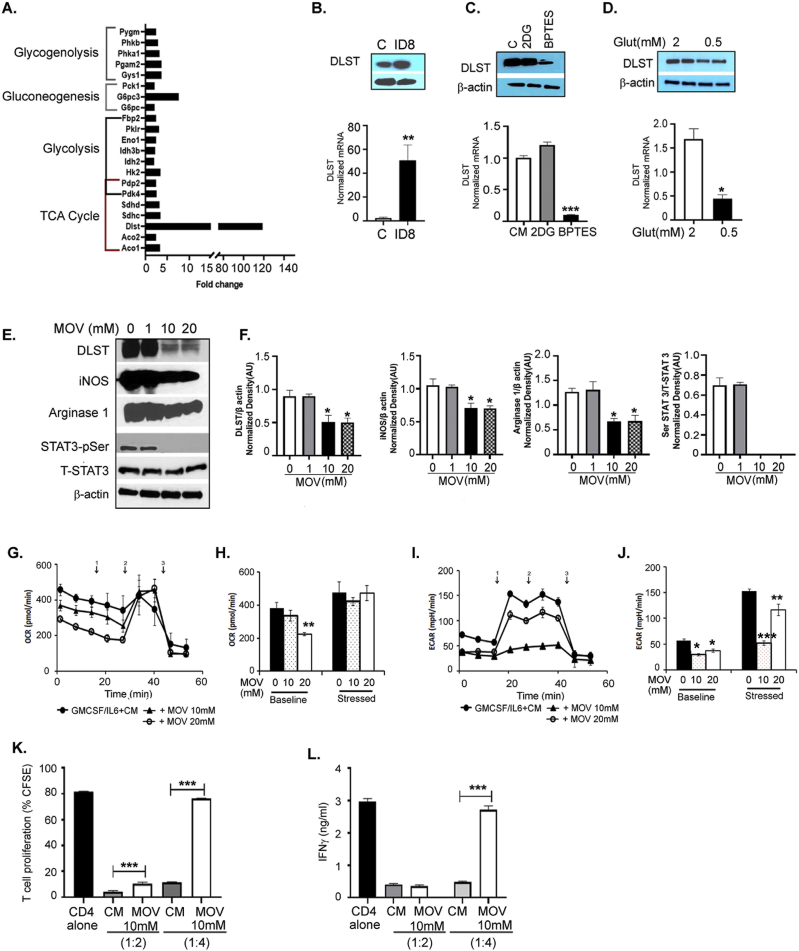
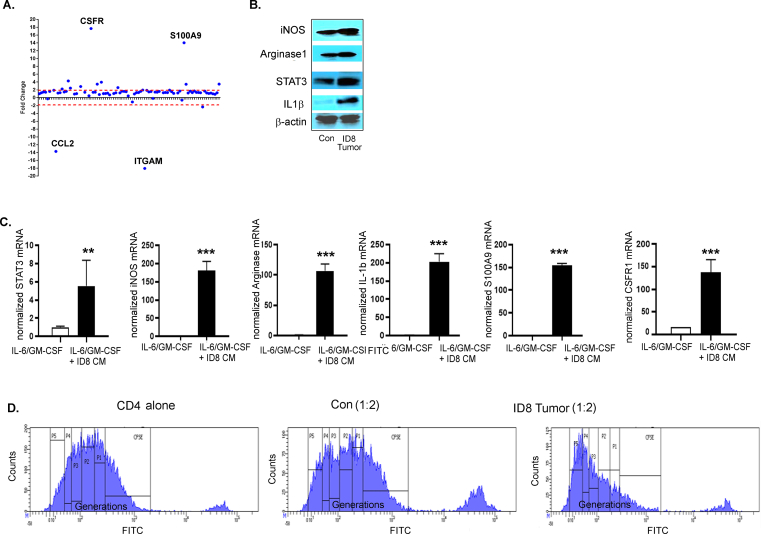
Tumors are known to produce cytokines and chemokines that recruit and promote the differentiation of immature myeloid cells into cells with immunosuppressive capability [54,55]. We observed increased levels of pro-immunosuppressive myeloid cell factors [17,18,54], including GM-CSF (increasing trend), G-CSF, VEGF, and S-CSF, in the ascites collected from mice bearing ID8 -luc2 tumors at 6 weeks (Figure 4A–D). Levels of stromal cell-derived factors, such as IL-6 and IL-4, were significantly increased (Figure 4E,F), while IL-1β and IFN-γ were unchanged (Figure 4G,H). This finding suggests that ovarian cancer cells and the TME produce pro-recruitment and pro-immunosuppressive factors that regulate CD11b+Gr1+ myeloid cells. To confirm that the tumor primed myeloid cells acquire immunosuppressive characteristics, BM-derived CD11b+Gr1+ myeloid cells from naïve mice and mice bearing ID8-Luc2 tumors for 2 weeks were subjected to a Myeloid-Derived Suppressor Real-Time PCR array. We observed a robust increase in the expression of 18 canonical immunosuppression-related markers (Figure 4I, Fig. S3A, Table S4). The most recognized [17,18] upregulated immunosuppressive markers, such as iNOS, arginase1, STAT3, IL-1β, S100A9, and CSFR, were validated by western blots (Figure 4J, Fig. S3B). BM cells were isolated from naïve and differentiated myeloid cells (as described in the methods section) in the absence or presence of ID8 CM to determine whether ex vivo conditions can mimic a similar induction of the immunosuppressive markers, and they were found to increase the expression of the same canonical markers (Fig. S3C). Furthermore, the BM-derived CD11b+Gr1+ myeloid cells from ID8-Luc2 tumor-bearing mice were more effective in suppressing the T cell proliferation, as assessed by a co-culture of CD4 T cells and myeloid cells in a T cell suppression assay, which showed decrease in proliferating T cells and IFN-γ production compared to cells from naïve mice (Figure 4 K, L, Fig. S3D). These data indicate that ovarian cancer cells and TME produce CD11b+Gr1+ recruiting factors and can induce immunosuppressive factors and markers in vivo and ex vivo and are active participants in maintaining a myeloid cell-driven immune-suppressive environment.
Figure 4.
EOC environment enhances the immunosuppressive phenotype of CD11b+Gr1+myeloid cells. Levels of GM-CSF (A), G-CSF (B), VEGF (C), C-SCF (D), IL-6 (E), IL-4 (F), IL- 1β (G), and IFN-γ (H) in the peritoneal wash or ascites from non-tumor (control, con) and ID8-luc2 tumor-bearing mice (ID8 tumor) were measured by ELISA (n = 3) after 6 weeks of tumor growth. (I) Total RNA was isolated from BM-derived CD11b+Gr1+ myeloid cell control and ID8 tumor, and a mouse myeloid-derived suppressor PCR array was performed. Bar graph represents the fold-change of gene expression in tumor primed vs. naïve myeloid cells. (J) Protein expression of iNOS, arginase 1, STAT 3, IL-1β, CSFR, and S100A9 in CD11b+Gr1+ myeloid cells isolated from control and ID8 tumor-bearing mice. Bar graphs represent the normalized density from 3 individual blots. (K) T cell suppression assay performed by co-culturing CD11b+Gr1+ myeloid cells from non-tumor and ID8 tumor-bearing mice with CFSE labeled CD4 cells isolated from naïve mice at 1:2 ratio (n = 3). T cell proliferation was assayed by flow cytometry on day 5 by measuring the CFSE intensity individually in each generation. Data from generation P1–P3 is shown. (L) From the same cells, day 3 supernatant was collected, and IFNγ levels were measured by ELISA. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.01 tumor compared to control as assessed by unpaired T-test. The expanded forms of abbreviations are detailed in the Supplementary Table S4.
3.5. Ovarian cancer microenvironment induces an energetic metabolic state in CD11b+ Gr1+ myeloid cells
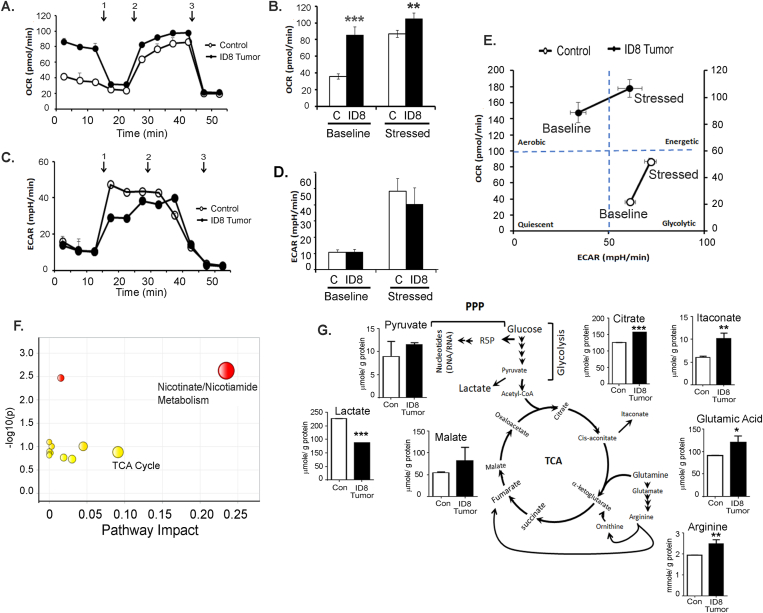
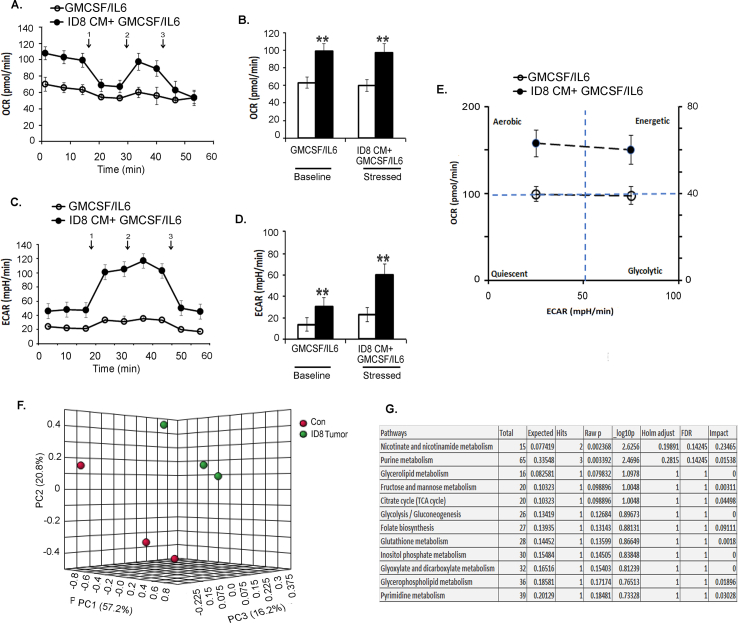
Recent research suggests that the activity of immune cells is defined and modulated by the energy metabolism dictated by the TME [56,57]. To assess whether the ovarian TME could modulate the energy metabolism of the CD11b+Gr1+myeloid cells, BM-derived cells from naïve mice and mice bearing ID8-Luc2 tumors for 2 weeks were analyzed by a Seahorse XFe96 analyzer in real-time (to define their metabolic profile. The tumor-primed CD11b+Gr1+ myeloid cells showed an increased basal and stressed OCR, indicating increased mitochondrial respiration compared to naïve myeloid cells (Figure 5A,B). No significant change was seen in the ECAR at the baseline or stressed conditions, suggesting no glycolysis change under tumor conditions in the myeloid cells (Figure 5C,D). The resulting energy phenotype indicated a basal aerobic energy phenotype in the tumor-primed myeloid cells that further moved to an energetic phenotype when stressed (Figure 5E).
Figure 5.
EOC microenvironment induces an energetic metabolic state in CD11b+Gr1+myeloid cells. CD11b+Gr1+ were isolated from BMs of control (no tumor injection) and mice with ID8-luc2 tumor at 2 weeks and subjected to Seahorse analysis and energy targeted metabolomics. (A) Oxygen consumption rate (OCR) was assessed in real-time using an XFe Seahorse analyzer as described in methods. Port injections of (1) oligomycin, (2) FCCP, and a combination of (3) rotenone-antimycin were given. (B) The bar graph represents basal and stressed OCR (n = 3). (C) Extracellular acidification rate (ECAR) was measured with port injections of (1) glucose, (2) oligomycin, and (3) 2-DG. (D) The bar graph represents basal and stressed ECAR (n = 3). (E) A quadrant plot indicates the metabolic shift of energy phenotype at basal and stressed levels. (F) MetaboAnalyst analysis showing the most significant and impacted pathways in the CD11b+Gr1+ isolated from ID8 tumor mice compared to non-tumor mice. (G) Bar graphs showing the metabolite levels that changed significantly, as quantified by metabolomics in CD11b+Gr1+ cells isolated from control and tumor mice. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.01 tumor compared to control as assessed by unpaired T-test.
Conversely, naïve myeloid cells showed a basal glycolytic phenotype that increased under stressed conditions (Figure 5E). To examine whether similar metabolic changes can be mimicked exvivo, BM cells were analyzed for energy phenotypes when differentiated in the absence or presence of ID8 CM. In the presence of ID8 CM, an increased OCR at the baseline and stressed conditions (Figs. S4A and B) were observed, similar to the in vivo conditions. Interestingly, the ID8 CM also increased ECAR, probably due to the direct and profound exposure to TME (Fig. S4 C, D). Like tumor priming, ID8 CM also conferred a basal aerobic energy phenotype that increased to an energetic phenotype under stressed conditions (Fig. S4E). Thus, both in vivo tumor presence and ex vivo exposure to the ovarian TME can modulate the metabolic phenotype of CD11b+Gr1+ myeloid cells.
To further confirm that mitochondrial respiration is affected, we performed an LC-MS/MS-based targeted metabolomics analysis for the quantitative profiling of ~250 energy pathway metabolites [46,47]. Unsupervised PCA showed that myeloid cells from naïve and tumor-bearing mice could be separated based on their metabolic differences (Fig. S4F). Pathway analysis by MetaboAnalyst 4.0, which uses high-quality KEGG pathways as a knowledge base, revealed 25 pathways with significance (Table S5) and impact value. Most of these pathways were related to mitochondrial respiration pathways. Nicotinate and nicotinamide metabolism and the TCA cycle were identified to be the two pathways with high significance and impact (Figure 5F, Fig. S4G). Nicotinate and nicotinamide are essential for various cellular and metabolic processes, including energy metabolism and the TCA cycle [58]. Charting the individual metabolite levels showed a significant increase in the metabolites of the TCA and associated pathways, including citrate, malate, itaconate, glutamic acid, and arginine. Pyruvate levels showed an increasing trend while lactate levels were decreased, suggesting the incorporation of lactate into pyruvate and its feeding into the TCA cycle (Figure 5G). Thus, ovarian tumor-primed myeloid cells undergo metabolic reprogramming toward applying increased mitochondrial respiration for their immunosuppressive function.
3.6. Enhanced metabolic state and immunosuppressive function of CD11b+ Gr1+ myeloid cells are fueled by the glutamine energy pathway
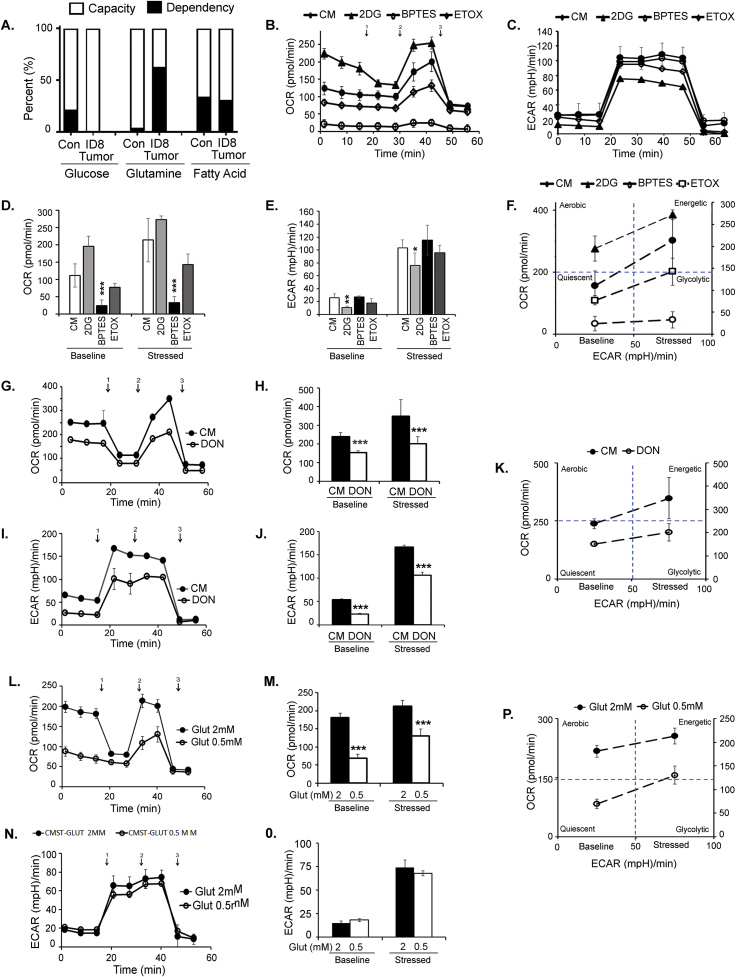
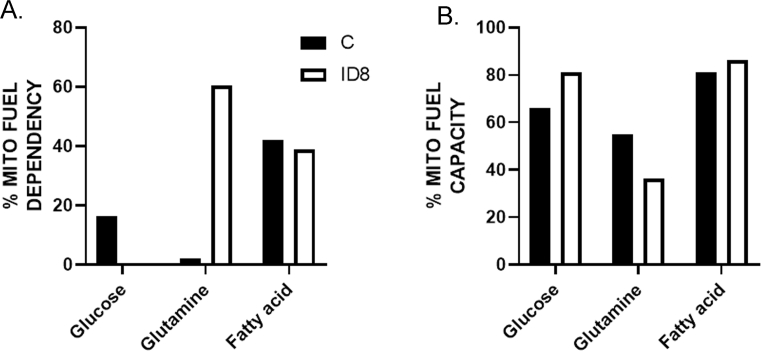
We sought to determine which mitochondrial fuel energy pathway is primarily responsible for the increased mitochondrial respiration and the increased immunosuppressive function of the myeloid cells. BM-derived CD11b+Gr1+ cells from naïve mice and mice bearing ID8-Luc2 tumors for 2 weeks were subjected to the Mito Fuel Flex assay, as described in the methods section. We observed that myeloid cells from tumor-bearing mice were highly dependent on glutamine for meeting their metabolic needs and showed an increased capacity to use glucose in the absence of glutamine (Figures 6A and Fig. 5SA, B). To validate this finding, we measured the effect of the three fuel pathway inhibitors individually on OCR and ECAR profiles in differentiated BM-MDSCs in the absence or presence of ID8 CM. We found that glutaminolysis inhibition by BPTES significantly inhibited OCR (Figure 6 B, D) without affecting ECAR (Figure 6C, E), resulting in a basal quiescent metabolic phenotype that moved to the glycolytic phenotype under stressed conditions (Figure 6 F). Glycolysis inhibition by 2-DG resulted in increased OCR and decreased ECAR, as expected, explaining the basal aerobic metabolic phenotype, which moved to an energetic phenotype under stressed conditions (Figure 6D,E, F). The inhibition of FAO by etomoxir had no significant effect on either pathway. Another glutamine antagonist, DON, also significantly inhibited OCR and ECAR and moved the metabolic phenotype from quiescent under basal conditions to glycolytic under stressed conditions (Figure 6G–K). As further confirmation, we also observed that glutamine deprivation (low glutamine conditions) inhibited mitochondrial respiration (Figure 6 L, M), similar to glutamine pathway inhibitors. However, not much effect on ECAR was observed (Figure 6 N, O). Like the inhibitors, the metabolic phenotypes were quiescent at basal and glycolytic at stressed conditions in the absence of glutamine (Figure 6P). Together, these data strongly suggest that glutamine is an essential mitochondrial respiration fuel for the CD11b+Gr1+ myeloid cells under the influence of ovarian tumors, and its TME and targeting glutamine metabolism in myeloid cells may inhibit their immunosuppressive ability.
Figure 6.
The glutamine energy pathway fuels enhanced metabolic state and immunosuppressive function of CD11b+Gr1+myeloid cells. CD11b+Gr1+ were isolated from BMs of control (no tumor), and mice injected with ID8-luc2 tumor at 2 weeks were subjected to (A) Mito Fuel Flex assay to assess dependency and capacity after blocking the different energy pathways using specific inhibitors UK5009 (glucose inhibitor), BPTES (glutamine inhibitor) and etomoxir (fatty acid oxidation inhibitor). (B) OCR was assessed in real-time using XFe Seahorse analyzer as described in methods, in CD11b+Gr1+ myeloid cells differentiated from BM by exposure of GM-CSF/IL-6 and ID8 conditioned media (CM) in the absence or presence of metabolic inhibitors of 2-DG (glycolysis inhibitor), BPTES (glutaminase inhibitor) and etomoxir (fatty acid oxidation inhibitor). Port injections were given as described before. (D) The bar graph represents basal and stressed OCR (n = 3). (C) Measurement of ECAR profile as described before. (E) The bar graph represents basal and stressed ECAR (n = 3). (F) Energy phenotype shifts under the presence of various metabolic inhibitors. BM-derived CD11b+Gr1+ myeloid cells were treated with glutaminase inhibitor DON, and OCR (G, H) and ECAR (I, J) were assessed similarly (n = 3). (K) Energy phenotype shifts under the presence of glutaminase inhibitor DON. (L–P) Similar metabolic analysis under regular glutamine (2 mm) and low glutamine (0.5 mm). ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001, treatments compared to CM as assessed by unpaired T-test.
3.7. Targeting glutamine metabolism inhibits the immunosuppressive function of CD11b+ Gr1+ myeloid cells
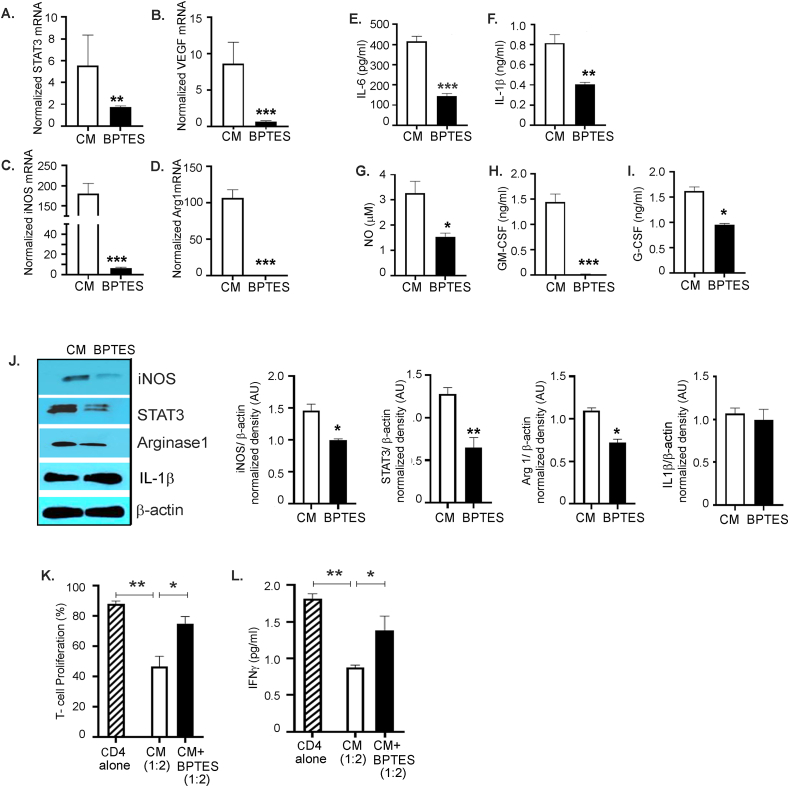
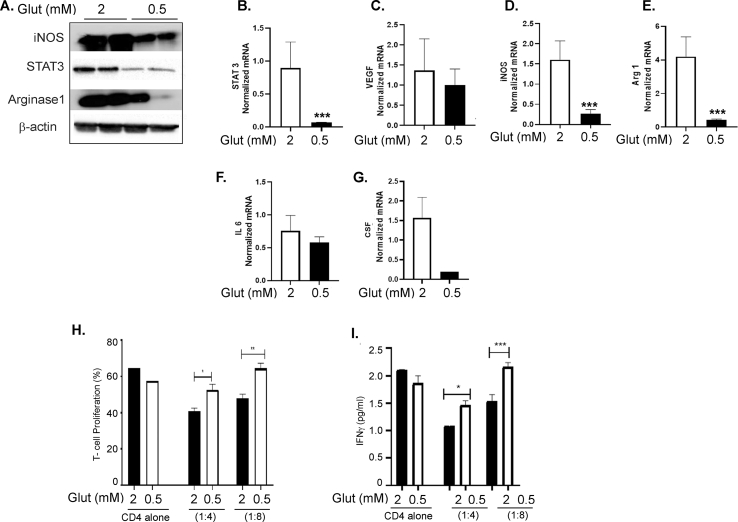
To analyze the contribution of glutamine metabolism, we tested if targeting glutamine metabolism will inhibit the immunosuppressive marker's expression and function. Inhibition of glutaminolysis by BPTES significantly inhibited the expression of immunosuppressive markers including STAT 3, VEGF, iNOS, and arginase 1; (Figure 7A–D), decreased the secretory immunosuppressive markers including IL-6, IL-1β, NO, GM-CSF, and G-CSF; (Figure 7E–I) and inhibited the protein expression of iNOS, STAT 3, and arginase 1, whereas the IL-1β protein level was unaffected (Figure 7J). Furthermore, BM-derived CD11b+Gr1+ myeloid cells treated with BPTES were less efficient in inhibiting T cell proliferation and IFNγ production than ID8 CM exposed cells alone (Figure 7 K, L). As a complementary approach, glutamine deprivation also decreased immunosuppressive marker expressions, such as iNOS, STAT 3, and arginase 1, at the protein level (Fig. S6A) and decreased STAT 3, VEGF, iNOS, arginase 1, IL-6, and CSF at the mRNA level (Figure 6B–G). Myeloid cells pre-exposed to low glutamine were less effective in causing T cell suppression and inhibiting IFN-γ (Fig. S6 H, I) than the cells in regular glutamine media. These data show that inhibiting glutamine utilization by pharmacological inhibitors or glutamine deprivation decreased the immunosuppressive function of ovarian TME-primed CD11b+Gr1+ myeloid cells.
Figure 7.
Targeting glutamine metabolism inhibits the suppressive function of CD11b+Gr1+myeloid cells. CD11b+Gr1+ myeloid cells isolated from the BM and exposed to ID8 conditioned media (CM) were treated with BPTES (10 μmol/L) for 48 h. (A–D) RT-PCR mRNA expression of STAT-3, VEGF, iNOS, and arginase (n = 3). (E–I) Cell culture supernatants from the same cells were collected, and levels of IL-6, IL-1β, GM-CSF, and G-CSF were measured by ELISA. NO levels were (G) measured by Griess reagent (n = 3). (J) Protein expression of iNOS, STAT 3, arginase 1, and IL-1β was measured by immunoblotting. Bar graphs represent the normalized density from 3 independent blots. (K) The immunosuppressive function of myeloid cells was tested by their ability to suppress T-cell proliferation as before and (L) IFNγ production in the absence or presence of BPTES (n = 3). ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001, BPTES treatment compared to CM as assessed by unpaired T-test.
3.8. Increased metabolic activity and function of myeloid cells is due to the increased expression of dihydrolipoamide succinyl transferase, a subunit of the alpha-ketoglutarate dehydrogenase complex
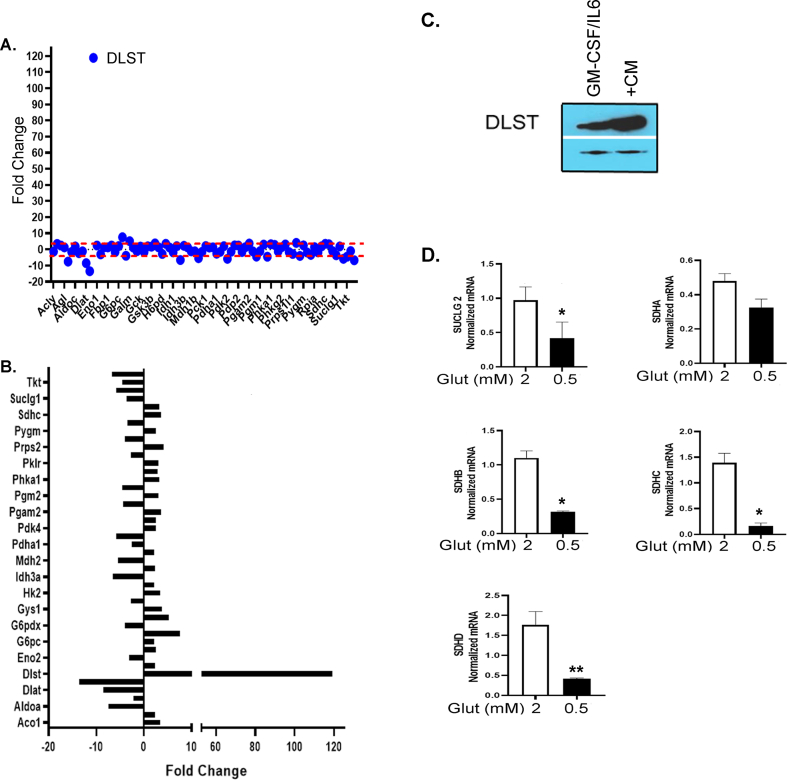
To identify a specific factor playing the pivotal role in the increased glutamine metabolism of CD11b+Gr1+ myeloid cells, we examined the expression of 84 genes involved in 5 different metabolic pathways (glycolysis, TCA cycle, pentose phosphate pathway, gluconeogenesis, and glycogenolysis) using the glucose metabolism PCR array in CD11b+Gr1+ myeloid cells isolated from naïve mice and mice bearing ID8-Luc2 tumors for 2 weeks. Tumor-primed CD11b+Gr1+ myeloid cells showed a significant increase in most genes involved in the TCA cycle and glycolysis (Figure 8 and Fig. S7 A, Table S6), with dihydrolipoamide succinyl transferase (DLST), mitochondrial enzyme and a subunit of the α-KGDC in TCA cycle [59], increasing more than 100-fold (Figure 8A, Fig. S7 A, B). Key TCA cycle upregulated genes were also validated under conditions of glutamine deprivation and were found to be significantly decreased (Fig. S7C), including DLST (Figure 8D). The increase in DLST was validated again at the protein and RNA level in the tumor-primed CD11b+Gr1+ myeloid cells isolated from mouse BM (Figure 8B). In addition, ID8 CM was also able to induce the robust expression of DLST in CD11b+Gr1+ myeloid cells (Fig. S7C), indicating the importance of ovarian TME in modulating the metabolism of myeloid cells. To examine whether the increased DLST expression was due to increased glutamine availability to the myeloid cells, the BM-derived cells were treated with BPTES or subjected to low glutamine in the presence of ID8 CM. Purified CD11b+Gr1+ myeloid cells treated with BPTES showed decreased DLST expression at the protein and mRNA level (Figure 8C). However, 2-DG treatments did not have any effect, indicating the specificity of glutamine availability as the primary determinant of DLST expression.
Figure 8.
Increased metabolic activity and function of myeloid cells are dependent on the increased expression of DLST. CD11b + Gr1+ myeloid cells were isolated from BMs of control (no tumor), and mice injected with ID8-luc2 tumors at 2 weeks were subjected to (A) Glucose metabolism PCR array. The bar graph shows 2-fold and higher upregulated genes in the CD11b+Gr1+ cells from tumor-bearing mice (B) immunoblotting and RT-PCR in CD11b+Gr1+ myeloid cells from control and ID8 mice. (C) Differentiated BM-derived CD11b+Gr1+ myeloid cells exposed to ID8 CM were treated with 2DG or BPTES, and DLST expression was examined by immunoblotting and RT-PCR. (D) Differentiated BM-derived CD11b+Gr1+ myeloid cells exposed to ID8 CM were subjected to low or normal glutamine levels, and DLST expression was examined by immunoblotting and RT-PCR. (E) Differentiated BM-derived CD11b+Gr1+ myeloid cells exposed to ID8 CM were treated with DLST inhibitor, MOV at 1-, 10-, and 20 mM concentrations for 48 h, and immunoblotting was performed to measure DLST, iNOS, arginase 1, pSTAT3, and total STAT-3 expression. (F) Bar graphs represent the normalized density of the western blots from 3 individual blots. (G, H) OCR profile of differentiated BM-derived CD11b+Gr1+ myeloid cells exposed to ID8 CM in the presence or absence of MOV. (I, J) ECAR profile of differentiated BM-derived CD11b+Gr1+ myeloid cells exposed to ID8 CM in the presence or absence of MOV. (K) CD11b+Gr1+ myeloid cells treated with DLST inhibitor were co-cultured with CD4+ T cells in 1:2 and 1:4 ratio to analyze T-cell proliferation (L) IFNγ levels by ELISA in the supernatant on day 3. ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001 treatments compared to control of CM as assessed by unpaired T-test.
Similarly, glutamine deprivation also inhibited DLST expression at protein and RNA levels (Figure 8D). Next, we employed a novel α-KGDC inhibitor, MOV [60], which inhibited DLST expression and the expression of immunosuppressive markers, including iNOS, arginase 1, and pSTAT3 (Figure 8E,F). To test whether inhibition of DLST can alter the metabolic phenotype of myeloid cells, we performed a Seahorse analysis in the absence or presence of the DLST (α-KGDC) inhibitor. MOV treatment reduced the basal OCR but did not affect the stressed OCR levels (Figure 8G,H). MOV's inability to inhibit stressed OCR may be due to the inhibitory mechanism of MOV, which the mitochondria can overcome during the metabolic stress conditions. Basal and stressed ECAR levels decreased in the MOV-treated myeloid cells (Figure 8I,J). MOV-treated myeloid cells also showed a decreased ability to suppress T cell proliferation and increase in IFN-γ production compared to the control ID8 CM group (Figure 8 K, L). Together, these data suggest that targeting glutamine metabolism through DLST (α-2KGDC) inhibition may be a novel approach in inhibiting the immunosuppressive function of CD11b+Gr1+ myeloid cells in the ovarian TME, leading toalleviation of immunosuppression and decreased ovarian tumor growth.
4. Discussion
In this study, we elucidate the critical role of ovarian cancer cells and their microenvironment in promoting the immunosuppressive ability of CD11b+Gr1+ myeloid cells and creating an immunosuppressive environment that promotes its development. As shown by others [16,25,49], we validated that CD11b+Gr1+ myeloid cells encouraged EOC growth, and depleting CD11b+Gr1+ myeloid cells reduced EOC growth, rescued the immune decline, and enhanced immunotherapy (Figure 1, Figure 2, Figure 3), suggesting that targeting CD11b+Gr1+ may be an effective treatment approach alone or in combination. The detrimental role of immunosuppressive CD11b+Gr1+ myeloid cells (also referred to as MDSCs) has been well-established in various other cancer types [61,62].
Since their initial description as vascular leukocytes with an immunosuppressive ability [[63], [64], [65]], there has been a growing understanding of the contributions of the immunosuppressive ability of CD11b+Gr1+ myeloid cells and the complex immunometabolism mechanisms regulating their function. The chemoattractants that recruit the immature myeloid cells are well documented and produced by both the tumor cells and the cells of the microenvironment [17,66]. In EOC, the role of IL-6, 1L-10, prostaglandin E2, stromal-derived factor-1, GM-CSF, G-CSF, IL-4, VEGF, COX2, IFN-γ, TNF-α, and oxidative stress has been shown to aid the recruitment of immunosuppressive CD11b+Gr1+ myeloid cells [55]. In accordance, we found that ovarian TME had increased levels of GM-CSF, G-CSF, VEGF, IL-6, and IL-4 (Figure 4). Not only did the ovarian TME produce recruitment factors, but it also induced various immunosuppressive markers, including canonical markers, such as STAT 3, iNOS, arginase 1, S100A9, CSFR, and IL-1β. In addition, the increased immunosuppressive marker expression translated to increased immunosuppression, as seen by T cell proliferation assays (Figure 4), the primary immunosuppressive function of CD11b+Gr1+ myeloid cells [17,67].
Recently, immunometabolism, whereby immune cell development and function are regulated by their distinct energy metabolism probably cued by the microenvironment, is becoming well-accepted [33,57]. In addition, the dysregulated cancer cell metabolism has been shown to affect the metabolism of immune cells and their function [68]. CD11b+Gr1+ myeloid cells have also been recently shown to rely on energy metabolism pathways for their growth and immunosuppressive function [39]. We also observed that the ovarian TME modulates the energy metabolism of CD11b+Gr1+ myeloid cells. The tumor-primed in vivo isolated myeloid cells and the ex vivo generated ID8 CM stimulated myeloid cells had enhanced oxidative phosphorylation (OXPHOS) compared to myeloid cells that were not exposed to any tumor stimulus at functional and metabolite levels (Figure 5). The Mito Fuel Flex test revealed that the increased OXPHOS is fueled by and is dependent on glutamine (Figure 6), which was corroborated by increased TCA metabolites, including those associated with glutamine metabolism (i.e., glutamic acid and arginine, Figure 5G). We established glutamine as the fuel for increased OXPHOS by showing that only the inhibition of glutaminolysis (and not glycolysis or FAO) can inhibit OXPHOS at both basal and stressed conditions, which was further strengthened by the use of another pharmacological glutaminolysis inhibitor, DON, and glutamine deprivation (Figure 6).
Furthermore, the inhibition of glutamine metabolism resulted in a reduction of immunosuppressive markers and the function of CD11b+Gr1+ myeloid cells (Figure 7). These data strongly suggest that glutamine dependency is central for increasing the metabolic capacity of myeloid cells that translates to increased immunosuppressive ability. Recent studies have reported that targeting glutamine metabolism in many types of cancers, enhanced tumor-specific immunity by changing the function of myeloid cells [35,40,44]. The metabolic regulation by energy pathways in regulating MDSC development and function has been recently investigated. Most of the reports studying the metabolic regulation of CD11b+Gr1+ myeloid cells have investigated how the altered metabolism of cancer cells changes the metabolic activity and function of immunosuppressive CD11b+Gr1+ myeloid cells (referred to as MDSCs in most publications).
Li et al. demonstrated that increased glycolysis in the 4T1 breast cancer model and patient samples regulate the expression of G-CSF and GM-CSF required for MDSC development and immune evasion [35]. Jian et al. showed that MDSCs from the 4T1 breast cancer mouse model exhibit increased levels of glycolysis metabolites essential for MDSC expansion and survival via the prevention of excess reactive oxygen species [36]. Using lung tumor and lymphoma mouse models, mTOR-induced glycolysis was shown to be a driver of M-MDSC expansion and immunosuppressive properties [37,38]. However, the last three studies did not examine functional energy metabolism to assess whether both glycolysis and/or OXPHOS were also increased. Though we observed an increase in the glycolytic enzyme expression in the targeted array studies, the glycolysis inhibitor 2-DG showed a minimal effect on OXPHOS in the immunosuppressive CD11b+Gr1+ myeloid cells in our system (Figure 6). A more recent study demonstrated that M-MDSCs and TAMs present in renal carcinoma TME were imbibing increased glucose compared to any other cells, also translating to increased glycolysis and mitochondrial respiration [69]. We also found that the immunosuppressive CD11b+Gr1+ myeloid cells did exhibit a capacity to use glycolysis in the absence of glutamine (Figure 6), suggesting that glycolysis may also play a role in maintaining the energy metabolism of myeloid cells. Lipid metabolic pathways have also been implicated in regulating the immunosuppressive function of tumor-associated MDSCs. Well-designed studies by the Ochoa group demonstrated that lipid uptake and accumulation fostered by tumor cells resulted in increased OCR via increased FAO, eventually leading to the increased immunosuppressive ability of MDSCs associated with lung, colon, and thymoma tumor models [34,70]. In our current study, the inhibition of FAO by etomoxir did not affect OCR or Mito fuel dependency or capacity (Figure 6), which may suggest a minor role of FAO in ovarian tumor-associated immunosuppressive CD11b+Gr1+ myeloid cells.
Glutamine metabolism feeding into the TCA cycle and regulating the function of immunosuppressive CD11b+Gr1+ myeloid cells has recently been reported in other tumor types. Wu et al. in a comprehensive study using patient-derived and 4T1 breast cancer preclinical model identified glutaminolysis as the critical metabolic pathway regulating the proliferation and activation of immature myeloid cells, regardless of glucose availability [40]. They further showed that restoration of the TCA cycle was due to increased α-ketoglutarate (α-KG) derived from glutamine. Recently, Oh et al. showed that inhibition of glutamine metabolism in 4T1 breast cancer and 3LL lung cancer preclinical models resulted in immunogenic tumor cell death. Inhibition of glutamine metabolism in myeloid cells led to apoptosis of MDSCs and interestingly, converted suppressive MDSCs to a pro-inflammatory anti-tumor phenotype [44].
Thus, these studies, along with ours, show the vital role of glutamine metabolism in regulating the metabolism and function of immunosuppressive CD11b+Gr1+ myeloid cells, ultimately affecting the immune environment of tumors and the antitumor immune response. The glutamine pathway has been established as an upregulated metabolic pathway in many cancers, including ovarian cancer, in which it supports the altered cancer cell metabolism and is crucial for nucleotide and amino acid biosynthesis, glutathione generation for maintaining redox and various other processes [71,72]. Our current study does not distinguish whether the increased glutamine metabolism is due to exogenous glutamine provided by the TME or the upregulation of intrinsic glutamine metabolism of myeloid cells. Our array results and effects of mitochondrial glutaminase inhibitors may suggest that the immunosuppressive CD11b+Gr1+ myeloid cells are able to potentiate their intrinsic glutamine metabolism and fuel the TCA cycle.
To identify a specific target, we performed a metabolism gene expression-based PCR array, which revealed DLST, a component of α−KGDC, increased by more than 100-fold in tumor-primed CD11b+Gr1+ myeloid cells. α-KGDC, an essential enzyme system of the TCA cycle, is a metabolic hub that is a regulator of the signaling metabolite α-KG and regulates cellular redox, and it is interconnected with various metabolic pathways, including glutamine. Glutaminolysis results in the synthesis of α-KG, which may be oxidized by mitochondrial α-KGDC pushing the TCA cycle or reduced and exported to the cytosol and converted to acetyl CoA and aid fatty acid biosynthesis [59]. The importance of α-KGDC in regulating various aspects of cancer cells is well documented; however, the roles of individual components have not been studied [59,73,74]. The α-KGDC comprises of 3 enzyme components (E1-3) that work stepwise to generate succinyl CoA from α-KG. DLST is the E2 enzyme that carries out the reductive succinylation step in the formation of succinyl Co-A [59]. DLST is a non-redundant enzyme, and its deficiency leads to the complete disruption of the TCA cycle [59]. Recently, it has been suggested that DLST might be an essential gene in the etiology of pheochromocytomas and paragangliomas, a type of rare, heritable neuroendocrine tumor that carries mutations in various genes encoding enzymes of the TCA cycle [75]. Anderson et al. showed that DLST inactivation delayed Myc-induced leukemogenesis in a zebrafish model [76]. CPI-613, a DLST/α-KGDC non-specific inhibitor, is being tested in more than 25 clinical trials combined with chemotherapy drugs against various cancer types (ClinicalTrails.gov). In addition, it may be worthwhile to examine whether EOC cells also express increased DLST, as they are also reported to have a glutamine dependency [77].
To the best of our knowledge, this is the first report to demonstrate a connection between immunosuppressive CD11b+Gr1+ myeloid cells and DLST gene expression in EOC. To validate the relationship between the glutamine pathway and DLST expression, we show that glutaminolysis antagonism and glutamine deprivation reduced DLST expression, suggesting that glutamine availability regulates DLST. DLST (α-KGDC) inhibition reduced the immunosuppressive marker expression and OXPHOS and reversed the T cell suppression by CD11b+Gr1+ myeloid cells, suggesting a pivotal role for DLST in regulating the tumor-induced immunosuppressive function of CD11b+Gr1+ myeloid cells.
Overall, our data describe the novel metabolic dependence of the immunosuppressive function of CD11b+Gr1+ myeloid cells mediated by the ovarian TME. Additional work is needed to identify the source of glutamine that fuels the CD11b+Gr1+ myeloid cell metabolism and corroborate the increased DLST expression in patient-derived myeloid cells. This study provides evidence supporting the essential role of glutaminolysis in myeloid cells and identifies DLST as a specific novel target in reducing the immunosuppressive effect of CD11b+Gr1+ myeloid cells and in potentially improving antitumor immunotherapy approaches. Targeting DLST may have the dual benefit of targeting the glutamine pathway in tumor cells and immunosuppressive myeloid cells.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.molmet.2021.101272.
Funding
This work was supported by DOD W81XWH-16-OCRP awarded to RR and Patterson Endowment funds awarded to AM. RR is supported by R01CA249188. MPU is supported by Henry Ford Cancer Institute Post-doctoral Fellowship. SG is supported by National Institutes of Health grants NS112727 and AI144004. The funding sources had no role in study design, data collection, and interpretation, or the decision to submit the work for publication.
Conflict of interest
None declared.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Multimedia component 2
Multimedia component 3
Multimedia component 4
Supplementary figure 1.
Depletion of CD11b+Gr1+myeloid cells by gemcitabine inhibits EOC progression. ID8-luc2 cells were injected intraperitoneally into C57Bl/6 mice. After 10 days of tumor initiation, mice were treated with gemcitabine or PBS as described in methods. (A) Bar graph from n = 3, of total CD11b+Gr1+ myeloid cells in the peripheral blood of mice treated with gemcitabine compared to untreated. (B) Kaplan–Meier graphs indicate overall survival in control and gemcitabine mice (n = 12); p = 0.0005 by Gehan-Breslow-Wilcox test and p = 0.0008 by Mantel–Cox test. (C) The bar graph represents the average radiance of bioluminescence signal at 6 weeks of tumor progression (n = 6). Weeks indicate post-tumor injection time points. (D) Dot plots of mouse weight, abdominal circumference, and ascites volume (n = 6). (E) Representative H & E images of tumor tissue in ID8-luc2 mice treated with gemcitabine compared to untreated control at 10x. (F) Representative images of Ki67 staining for assessing tumor cell proliferation. Images are shown at 40× magnification. Tumor sections from all 8 mice/group were examined and representative bar graph for Ki-67 proliferation index. The number of Ki-67 positive cells were counted per high-power field (HPF; 400x). A total of 8 HPF were studied from 3 to 4 tumor sections/group. (G) The bar graph represents the average frequency of CD4+ and CD8+ T cells in the ascites of mice treated with gemcitabine compared to untreated control (n = 3). ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ns = non-significant, gemcitabine compared to control as assessed by unpaired T-test.
Supplementary figure 2.
Depletion of CD11b+Gr1+myeloid cells and PD1 immunotherapy increase tumor-infiltrating T cells. ID8-luc2 cells were injected intraperitoneally into C57Bl/6 mice. After 10 days of tumor initiation, mice were treated with IgG2b or anti-Gr1 mAbs or anti-PD1 mAb or a combination of both as described in methods. Weeks indicate post-tumor injection time points. Tumor sections were immunostained for CD4 or CD8 expression. Representative images of positive CD4 (A) and CD8 (B) staining at 20x. Inset shows the area magnified to 40×. Quantification was performed by counting the positive cells per HPF from at least 3–4 slides per group with minimum of 6 HPF fields counted per slide ∗p < 0.05, ∗∗p < 0.01 compared to IgG2b; #p < 0.05 compared to anti-Gr1 and &p < 0.05 compared to anti-PD1 as assessed by unpaired T-test.
Supplementary figure 3.
ID8 conditioned media induces immunosuppressive markers in CD11b+Gr1+myeloid cells. (A) Dot Blot representation of all the 88 immunosuppressive gene changes in BM isolated CD11b+Gr1+ myeloid cells from control and ID8 tumor of Figure 4I. (B) Dark exposure of immunoblots in Figure 4J. (C) Total RNA was isolated from BM-MDSC's treated with ID8 CM alone or in the presence of and GM-CSF/ IL 6. Quantitative-PCR mRNA expression of immunosuppressive markers STAT 3, iNOS, arginase 1, IL-1β, S100A9. (D) T cell proliferation was assayed by flow cytometry on day 5 by measuring the CFSE intensity individually in each generation. Representative flow graph is shown.
Supplementary Figure 4.
ID8 conditioned media induces active metabolic phenotype in CD11b+Gr1+myeloid cells. CD11b+Gr1+ myeloid cells differentiated from BM by exposure of GM-CSF/IL-6 and exposed to ID8 conditioned media (CM) were subjected to Seahorse metabolic function analysis. (A) Oxygen consumption rate (OCR) and was assessed in real-time using XFe Seahorse analyzer as described in methods. Port injections of (1) oligomycin, (2) FCCP, and a combination of (3) rotenone-antimycin were given. (B) The bar graph represents basal and stressed OCR (n = 3). (C) Extracellular acidification rate (ECAR) was measured with port injections of (1) glucose, (2) oligomycin, and (3) 2-DG. (D) The bar graph represents basal and stressed ECAR (n = 3). (E) A quadrant plot indicates the metabolic shift of energy phenotype at basal and stressed levels. ∗∗p < 0.01, when compared to GM-CSF/IL6, treated MDSC's as assessed by unpaired T-test. (F) PCA plot showing the separation of CD11b+Gr1+ cells isolated from ID8 tumor mice and of non-tumor mice based on metabolic profile. (G) Table showing the top 12 KEGG metabolic pathways with the highest significance and impact as analyzed by MetaboAnalyst.
Supplementary Figure 5.
Expanded Mito Fuel Flex Assay. Individual values of fuel dependency (A) and capacity (B) are shown in support of Figure 7A. The assay was performed as described in methods and Figure 7.
Supplementary Figure 6.
Glutamine deprivation reduced the immunosuppressive function of CD11b+Gr1+myeloid cells. CD11b+Gr1+ myeloid cells differentiated from BM by exposure of GM-CSF/IL-6 alone and in the presence of ID8 conditioned media (CM) were exposed to regular glutamine (2 mm) and low glutamine (0.5 mm) conditions for 48 h. (A) Representative immunoblots of iNOS, STAT 3, and arginase 1. (B) mRNA expression of (B) STAT-3, (C) VEGF, (D) iNOS, (E) arginase 1, (F) IL-6, and (G) G-CSF as assessed by RT- PCR. (H) Isolated CD11b+Gr1+ myeloid cells were co-cultured with CFSE labeled CD4+ T cells in different ratios at 1:4 and 1:8 for 5 days, and CD4 cell proliferation was analyzed by CFSE intensity using flow cytometry. (I) On day 3, the supernatant was collected from the co-cultures, and IFNγ levels were measured by ELISA. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 compared to regular glutamine control as assessed by unpaired T-test.
Supplementary Figure 7.
Increased DLST expression and effect of glutamine deprivation on upregulated TCA cycle genes. (A) Representative dot plot graph of 5-fold change of all the genes in the glucose metabolism PCR array (B) Bar graph represents the fold change in both downregulated and upregulated genes from glucose metabolism array. (B) Increased DLST expression validated individually by immunoblotting in CD11b+Gr1+ myeloid cells from GMCSF/IL6 and GMCSF/IL6+ ID8 CM and (D) Real-time PCR was performed on key upregulated genes Succinyl Co A ligase (SUCLG2), Succinate dehydrogenases (SDHA-SDHD) (from glucose metabolism array) under conditions of low and regular glutamine conditions (n = 3). ∗p < 0.05 low glutamine compared to regular glutamine as assessed by unpaired T-test.
References
- 1.Bogani G., Lopez S., Mantiero M., Ducceschi M., Bosio S., Ruisi S. Immunotherapy for platinum-resistant ovarian cancer. Gynecologic Oncology. 2020;158(2):484–488. doi: 10.1016/j.ygyno.2020.05.681. [DOI] [PubMed] [Google Scholar]
- 2.Coukos G., Tanyi J., Kandalaft L.E. Opportunities in immunotherapy of ovarian cancer. Annals of Oncology. 2016;27(Suppl 1):i11–i15. doi: 10.1093/annonc/mdw084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu S., Matsuzaki J., Wei L., Tsuji T., Battaglia S., Hu Q. Efficient identification of neoantigen-specific T-cell responses in advanced human ovarian cancer. J Immunother Cancer. 2019;7(1):156. doi: 10.1186/s40425-019-0629-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Odunsi K., Jungbluth A.A., Stockert E., Qian F., Gnjatic S., Tammela J. NY-ESO-1 and LAGE-1 cancer-testis antigens are potential targets for immunotherapy in epithelial ovarian cancer. Cancer Research. 2003;63(18):6076–6083. [PubMed] [Google Scholar]
- 5.Szender J.B., Papanicolau-Sengos A., Eng K.H., Miliotto A.J., Lugade A.A., Gnjatic S. NY-ESO-1 expression predicts an aggressive phenotype of ovarian cancer. Gynecologic Oncology. 2017;145(3):420–425. doi: 10.1016/j.ygyno.2017.03.509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cancer Genome Atlas Research N. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474(7353):609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tothill R.W., Tinker A.V., George J., Brown R., Fox S.B., Lade S. Novel molecular subtypes of serous and endometrioid ovarian cancer linked to clinical outcome. Clinical Cancer Research. 2008;14(16):5198–5208. doi: 10.1158/1078-0432.CCR-08-0196. [DOI] [PubMed] [Google Scholar]
- 8.Adams S.F., Levine D.A., Cadungog M.G., Hammond R., Facciabene A., Olvera N. Intraepithelial T cells and tumor proliferation: impact on the benefit from surgical cytoreduction in advanced serous ovarian cancer. Cancer. 2009;115(13):2891–2902. doi: 10.1002/cncr.24317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Azarianpour S., Corredor G., Bera K., Leo P., Braman N., Fu P. Medical imaging 2020: digital pathology. International Society for Optics and Photonics; 2020. Computer extracted features related to the spatial arrangement of tumor-infiltrating lymphocytes predict overall survival in epithelial ovarian cancer receiving adjuvant chemotherapy. [Google Scholar]
- 10.Sato E., Olson S.H., Ahn J., Bundy B., Nishikawa H., Qian F. Intraepithelial CD8+ tumor-infiltrating lymphocytes and a high CD8+/regulatory T cell ratio are associated with favorable prognosis in ovarian cancer. Proceedings of the National Academy of Sciences of the U S A. 2005;102(51):18538–18543. doi: 10.1073/pnas.0509182102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang L., Conejo-Garcia J.R., Kasaros D., Gimotty P.A., Massobrio M., Regnani G. Intratumoral T cells, recurrence, and survival in epithelial ovarian cancer. New England Journal of Medicine. 2003;348(3):203–213. doi: 10.1056/NEJMoa020177. [DOI] [PubMed] [Google Scholar]
- 12.Brahmer J.R., Tykodi S.S., Chow L.Q.M., Hwu W.-J., Topalian S.L., Hwu P. Safety and activity of anti-PD-L1 antibody in patients with advanced cancer. New England Journal of Medicine. 2012;366(26):2455–2465. doi: 10.1056/NEJMoa1200694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Disis M.L., Patel M.R., Pant S., Infante J.R., Lockhart A.C., Kelly K. Avelumab (MSB0010718C), an anti-PD-L1 antibody, in patients with previously treated, recurrent or refractory ovarian cancer: a phase Ib, open-label expansion trial [abstract] Journal of Clinical Oncology. 2015;33(15 Suppl):5509. [Google Scholar]
- 14.Hamanishi J., Mandai M., Ikeda T., Minami M., Kawaguchi A., Murayama T. Safety and antitumor activity of anti-PD-1 antibody, nivolumab, in patients with platinum-resistant ovarian cancer. Journal of Clinical Oncology. 2015;33(34):4015–4022. doi: 10.1200/JCO.2015.62.3397. [DOI] [PubMed] [Google Scholar]
- 15.Lavoué V., Thédrez A., Levêque J., Foucher F., Henno S., Jauffret V. Immunity of human epithelial ovarian carcinoma: the paradigm of immune suppression in cancer. Journal of Translational Medicine. 2013;11:147. doi: 10.1186/1479-5876-11-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Odunsi K. Immunotherapy in ovarian cancer. Annals of Oncology. 2017;28(Suppl 8) doi: 10.1093/annonc/mdx444. viii1-viii7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gabrilovich D.I. Myeloid-derived suppressor cells. Cancer Immunol Res. 2017;5(1):3–8. doi: 10.1158/2326-6066.CIR-16-0297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bronte V., Brandau S., Chen S.-H., Colombo M.P., Frey A.B., Greten T.F. Recommendations for myeloid-derived suppressor cell nomenclature and characterization standards. Nature Communications. 2016;7:12150. doi: 10.1038/ncomms12150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zilio S., Serafini P. vol. 4. Vaccines; Basel): 2016. Neutrophils and granulocytic MDSC: the janus god of cancer immunotherapy. 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parker K.H., Beury D.W., Ostrand-Rosenberg S. Myeloid-derived suppressor cells: critical cells driving immune suppression in the tumor microenvironment. Advances in Cancer Research. 2015;128:95–139. doi: 10.1016/bs.acr.2015.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jae Lee W., Yi Ko S., Mohamed M.S., Kenny H.A., Lengyel E., Naora H. Neutrophils facilitate ovarian cancer premetastatic niche formation in the omentum. Journal of Experimental Medicine. 2019;216(1):176–194. doi: 10.1084/jem.20181170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Singel K.L., Emmons T.R., Khan A.N.H., Mayor P.C., Shen S., Wong J.T. Mature neutrophils suppress T cell immunity in ovarian cancer microenvironment. JCI Insight. 2019;4(5) doi: 10.1172/jci.insight.122311. PMCID: PMC6483507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Horikawa N., Abiko K., Matsumura N., Hamanishi J., Baba T., Yamaguchi K. Expression of vascular endothelial growth factor in ovarian cancer inhibits tumor immunity through the accumulation of myeloid-derived suppressor cells. Clinical Cancer Research. 2017;23(2):587–599. doi: 10.1158/1078-0432.CCR-16-0387. [DOI] [PubMed] [Google Scholar]
- 24.Horikawa N., Abiko K., Matsumura N., Baba T., Hamanishi J., Yamaguchi K. Anti-VEGF therapy resistance in ovarian cancer is caused by GM-CSF-induced myeloid-derived suppressor cell recruitment. British Journal of Cancer. 2020;122(6):778–788. doi: 10.1038/s41416-019-0725-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Godoy H.E., Khan A.N.H., Vethanayagam R.R., Grimm M.J., Singel K.L., Kolomeyevskaya N. Myeloid-derived suppressor cells modulate immune responses independently of NADPH oxidase in the ovarian tumor microenvironment in mice. PloS One. 2013;8(7) doi: 10.1371/journal.pone.0069631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cui T.X., Kryczek I., Zhao L., Zhao E., Kuick R., Roh M.H. Myeloid-derived suppressor cells enhance stemness of cancer cells by inducing microRNA101 and suppressing the corepressor CtBP2. Immunity. 2013;39(3):611–621. doi: 10.1016/j.immuni.2013.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Di Mitri D., Toso A., Chen J.J., Sarti M., Pinton S., Jost T.R. Tumour-infiltrating Gr-1+ myeloid cells antagonize senescence in cancer. Nature. 2014;515(7525):134–137. doi: 10.1038/nature13638. [DOI] [PubMed] [Google Scholar]
- 28.Hiratsuka S., Watanabe A., Aburatani H., Maru Y. Tumour-mediated upregulation of chemoattractants and recruitment of myeloid cells predetermines lung metastasis. Nature Cell Biology. 2006;8(12):1369–1375. doi: 10.1038/ncb1507. [DOI] [PubMed] [Google Scholar]
- 29.Finke J., Ko J., Rini B., Rayman P., Ireland J., Cohen P. MDSC as a mechanism of tumor escape from sunitinib mediated anti-angiogenic therapy. International Immunopharmacology. 2011;11(7):856–861. doi: 10.1016/j.intimp.2011.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Komura N., Mabuchi S., Shimura K., Yokoi E., Kozasa K., Kuroda H. The role of myeloid-derived suppressor cells in increasing cancer stem-like cells and promoting PD-L1 expression in epithelial ovarian cancer. Cancer Immunology Immunotherapy. 2020;69(12):2477–2499. doi: 10.1007/s00262-020-02628-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Khan A.N.H., Kolomeyevskaya N., Singel K.L., Grimm M.J., Moysich K.B., Daudi S. Targeting myeloid cells in the tumor microenvironment enhances vaccine efficacy in murine epithelial ovarian cancer. Oncotarget. 2015;6(13):11310–11326. doi: 10.18632/oncotarget.3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Domblides C., Lartigue L., Faustin B. Metabolic stress in the immune function of T cells, macrophages and dendritic cells. Cells. 2018;7(7) doi: 10.3390/cells7070068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pearce E.L., Pearce E.J. Metabolic pathways in immune cell activation and quiescence. Immunity. 2013;38(4):633–643. doi: 10.1016/j.immuni.2013.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hossain F., Al-Khami A.A., Wyczechowska D., Hernandez C., Zheng L., Reiss K. Inhibition of fatty acid oxidation modulates immunosuppressive functions of myeloid-derived suppressor cells and enhances cancer therapies. Cancer Immunology Research. 2015;3(11):1236–1247. doi: 10.1158/2326-6066.CIR-15-0036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li W., Tanikawa T., Kryczek I., Xia H., Li G., Wu K. Aerobic glycolysis controls myeloid-derived suppressor cells and tumor immunity via a specific CEBPB isoform in triple-negative breast cancer. Cell Metabolism. 2018;28(1):87–103 e6. doi: 10.1016/j.cmet.2018.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jian S.-L., Chen W.-W., Su Y.-C., Su Y.-W., Chuang T.-H., Hsu S.-C. Glycolysis regulates the expansion of myeloid-derived suppressor cells in tumor-bearing hosts through prevention of ROS-mediated apoptosis. Cell Death & Disease. 2017;8(5):e2779. doi: 10.1038/cddis.2017.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Deng Y., Yang J., Luo F., Qian J., Liu R., Zhang D. mTOR-mediated glycolysis contributes to the enhanced suppressive function of murine tumor-infiltrating monocytic myeloid-derived suppressor cells. Cancer Immunology Immunotherapy. 2018;67(9):1355–1364. doi: 10.1007/s00262-018-2177-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu T., Zhao Y., Wang H., Li Y., Shao L., Wang R. mTOR masters monocytic myeloid-derived suppressor cells in mice with allografts or tumors. Scientific Reports. 2016;6:20250. doi: 10.1038/srep20250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Al-Khami A.A., Rodriguez P.C., Ochoa A.C. Energy metabolic pathways control the fate and function of myeloid immune cells. Journal of Leukocyte Biology. 2017;102(2):369–380. doi: 10.1189/jlb.1VMR1216-535R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu W.C., Sun H.-W., Chen J., OuYang H.-Y., Yu X.-J., Chen H.-T. Immunosuppressive immature myeloid cell generation is controlled by glutamine metabolism in human cancer. Cancer Immunology Research. 2019;7(10):1605–1618. doi: 10.1158/2326-6066.CIR-18-0902. [DOI] [PubMed] [Google Scholar]
- 41.Liao J.B., Ovenell K.J., Curtis E.E.M., Cecil D.L., Koehnlein M.R., Rastetter L.R. Preservation of tumor-host immune interactions with luciferase-tagged imaging in a murine model of ovarian cancer. Journal of Immunotherapy Cancer. 2015;3:16. doi: 10.1186/s40425-015-0060-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Al-Wahab Z., Mert I., Tebbe C., Chhina J., Hijaz M., Morris R.T. Metformin prevents aggressive ovarian cancer growth driven by high-energy diet: similarity with calorie restriction. Oncotarget. 2015;6(13):10908–10923. doi: 10.18632/oncotarget.3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Al-Wahab Z., Tebbe C., Chhina J., Dar S.A., Morris R.T., Ali-Fehmi R. Dietary energy balance modulates ovarian cancer progression and metastasis. Oncotarget. 2014;5(15):6063–6075. doi: 10.18632/oncotarget.2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Oh M.-H., Sun I.-H., Zhao L., Leone R.D., Sun I.-M., Xu W. Targeting glutamine metabolism enhances tumor-specific immunity by modulating suppressive myeloid cells. Journal of Clinical Investigation. 2020;130(7):3865–3884. doi: 10.1172/JCI131859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mangalam A.K., Rattan R., Suhail H., Singh J., Hoda Md N., Deshpande M. AMP-activated protein kinase suppresses autoimmune central nervous system disease by regulating M1-type macrophage-Th17 Axis. The Journal of Immunology. 2016;197(3):747–760. doi: 10.4049/jimmunol.1501549. [DOI] [PubMed] [Google Scholar]
- 46.Bao X., Wu J., Kim .S., LoRusso P., Li J. Pharmacometabolomics reveals irinotecan mechanism of action in cancer patients. The Journal of Clinical Pharmacology. 2019;59(1):20–34. doi: 10.1002/jcph.1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Poisson L.M., Suhail H., Singh J., Datta I., Denic A., Labuzek K. Untargeted plasma metabolomics identifies endogenous metabolite with drug-like properties in chronic animal model of multiple sclerosis. Journal of Biological Chemistry. 2015;290(52):30697–30712. doi: 10.1074/jbc.M115.679068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dar S., Chhina J., Mert I., Chitale D., Buekers T., Kaur H. Bioenergetic adaptations in chemoresistant ovarian cancer cells. Scientific Reports. 2017;7(1):8760. doi: 10.1038/s41598-017-09206-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Duraiswamy J., Freeman G.J., Coukos G. Therapeutic PD-1 pathway blockade augments with other modalities of immunotherapy T-cell function to prevent immune decline in ovarian cancer. Cancer Research. 2013;73(23):6900–6912. doi: 10.1158/0008-5472.CAN-13-1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Le H.K., Graham L., Cha E., Morales J.K., Manjili M.H., Bear H.D. Gemcitabine directly inhibits myeloid derived suppressor cells in BALB/c mice bearing 4T1 mammary carcinoma and augments expansion of T cells from tumor-bearing mice. International Immunopharmacology. 2009;9(7–8):900–909. doi: 10.1016/j.intimp.2009.03.015. [DOI] [PubMed] [Google Scholar]
- 51.Suzuki E., Kapoor V., Jassar A.S., Kaiser L.R., Albelda S.M. Gemcitabine selectively eliminates splenic Gr-1+/CD11b+ myeloid suppressor cells in tumor-bearing animals and enhances antitumor immune activity. Clinical Cancer Research. 2005;11(18):6713–6721. doi: 10.1158/1078-0432.CCR-05-0883. [DOI] [PubMed] [Google Scholar]
- 52.Zeng Y., Li B., Liang Y., Reeves P.M., Qu X., Ran C. Dual blockade of CXCL12-CXCR4 and PD-1-PD-L1 pathways prolongs survival of ovarian tumor-bearing mice by prevention of immunosuppression in the tumor microenvironment. The FASEB Journal. 2019;33(5):6596–6608. doi: 10.1096/fj.201802067RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhu X., Lang J. Programmed death-1 pathway blockade produces a synergistic antitumor effect: combined application in ovarian cancer. J Gynecol Oncol. 2017;28(5):e64. doi: 10.3802/jgo.2017.28.e64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Serafini P., Borrello I., Bronte V. Myeloid suppressor cells in cancer: recruitment, phenotype, properties, and mechanisms of immune suppression. Seminars in Cancer Biology. 2006;16(1):53–65. doi: 10.1016/j.semcancer.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 55.Baert T., Vankerckhoven A., Riva M., Van Hoylandt A., Thirion G., Holger G. Myeloid derived suppressor cells: key drivers of immunosuppression in ovarian cancer. Frontiers in Immunology. 2019;10:1273. doi: 10.3389/fimmu.2019.01273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hu C., Pang B., Lin G., Zhen Y., Yi H. Energy metabolism manipulates the fate and function of tumour myeloid-derived suppressor cells. British Journal of Cancer. 2020;122(1):23–29. doi: 10.1038/s41416-019-0644-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Leone R.D., Powell J.D. Metabolism of immune cells in cancer. Nature Reviews Cancer. 2020;20(9):516–531. doi: 10.1038/s41568-020-0273-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yang Y., Sauve A.A. NAD(+) metabolism: bioenergetics, signaling and manipulation for therapy. Biochimica et Biophysica Acta. 2016;1864(12):1787–1800. doi: 10.1016/j.bbapap.2016.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vatrinet R., Leone G., De Luise M., Girolimetti G., Vidone M., Gasparre G. The alpha-ketoglutarate dehydrogenase complex in cancer metabolic plasticity. Cancer & Metabolism. 2017;5:3. [Google Scholar]
- 60.Young A., Gardiner D., Brosnan M.E., Brosnan J.T., Mailloux R.J. Physiological levels of formate activate mitochondrial superoxide/hydrogen peroxide release from mouse liver mitochondria. FEBS Letters. 2017;591(16):2426–2438. doi: 10.1002/1873-3468.12777. PMID: 28771687. [DOI] [PubMed] [Google Scholar]
- 61.Srivastava M.K., Zhu L., Harris-White M., Kar U.K., Huang M., Johnson M.F. Myeloid suppressor cell depletion augments antitumor activity in lung cancer. PloS One. 2012;7(7) doi: 10.1371/journal.pone.0040677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stromnes I.M., Brockenbrough J.S., Izeradjene K., Carlson M.A., Cuevas C., Simmons R.M. Targeted depletion of an MDSC subset unmasks pancreatic ductal adenocarcinoma to adaptive immunity. Gut. 2014;63(11):1769–1781. doi: 10.1136/gutjnl-2013-306271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bak S.P., Alonso A., Turk M.J., Berwin B. Murine ovarian cancer vascular leukocytes require arginase-1 activity for T cell suppression. Molecular Immunology. 2008;46(2):258–268. doi: 10.1016/j.molimm.2008.08.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Conejo-Garcia J.R., Benencia F., Courreges M.-C., Kang E., Mohamed-Hadley A., Buckanovich R.J. Tumor-infiltrating dendritic cell precursors recruited by a beta-defensin contribute to vasculogenesis under the influence of Vegf-A. Nature Medicine. 2004;10(9):950–958. doi: 10.1038/nm1097. [DOI] [PubMed] [Google Scholar]
- 65.Yang R., Cai Z., Zhang Y., Yutzy W.H., Roby K.F., Roden R.B.S. CD80 in immune suppression by mouse ovarian carcinoma-associated Gr-1+CD11b+ myeloid cells. Cancer Research. 2006;66(13):6807–6815. doi: 10.1158/0008-5472.CAN-05-3755. [DOI] [PubMed] [Google Scholar]
- 66.Galliverti G., Wullschleger S., Tichet M., Murugan D., Zangger N., Horton W. Myeloid cells orchestrate systemic immunosuppression, impairing the efficacy of immunotherapy against HPV(+) cancers. Cancer Immunology Research. 2020;8(1):131–145. doi: 10.1158/2326-6066.CIR-19-0315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Walankiewicz M., Grywalska E., Polak G., Kotarski J., Siwicka-Gieroba D.J., Roliński J. Myeloid-derived suppressor cells in ovarian cancer: friend or foe? Central European Journal of Immunology. 2017;42(4):383–389. doi: 10.5114/ceji.2017.72823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kedia-Mehta N., Finlay D.K. Competition for nutrients and its role in controlling immune responses. Nature Communications. 2019;10(1):2123. doi: 10.1038/s41467-019-10015-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Reinfeld B.I., Madden M.Z., Wolf M.M., Chytil A., Bader J.E., Patterson A.R. Cell-programmed nutrient partitioning in the tumour microenvironment. Nature. 2021;593(7858):282–288. doi: 10.1038/s41586-021-03442-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Al-Khami A.A., Zheng L., Del Valle L., Hossain F., Wyczechowska D., Zabaleta J. Exogenous lipid uptake induces metabolic and functional reprogramming of tumor-associated myeloid-derived suppressor cells. Oncoimmunology. 2017;6 doi: 10.1080/2162402X.2017.1344804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Altman B.J., Stine Z.E., Dang C.V. From Krebs to clinic: glutamine metabolism to cancer therapy. Nature Reviews Cancer. 2016;16(11):749. doi: 10.1038/nrc.2016.114. [DOI] [PubMed] [Google Scholar]
- 72.Yang L., Venneti S., Nagrath D. Glutaminolysis: a hallmark of cancer metabolism. Annual Review of Biomedical Engineering. 2017;19:163–194. doi: 10.1146/annurev-bioeng-071516-044546. [DOI] [PubMed] [Google Scholar]
- 73.Martinez-Outschoorn U.E., Peiris-Pagés M., Pestell R.G., Sotgia F., Lisanti M.P. Cancer metabolism: a therapeutic perspective. Nature Reviews Clinical Oncology. 2017;14(1):11–31. doi: 10.1038/nrclinonc.2016.60. [DOI] [PubMed] [Google Scholar]
- 74.Vander Heiden M.G., DeBerardinis R.J. Understanding the intersections between metabolism and cancer biology. Cell. 2017;168(4):657–669. doi: 10.1016/j.cell.2016.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Remacha L., Pirman D., Mahoney C.E., Coloma J., Calsina B., Currás-Freixes M. Recurrent germline DLST mutations in individuals with multiple pheochromocytomas and paragangliomas. The American Journal of Human Genetics. 2019;104(5):1008–1010. doi: 10.1016/j.ajhg.2019.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Anderson N.M., Li D., Peng H.L., Laroche F.J.F., Mansour M.R., Gjini E. The TCA cycle transferase DLST is important for MYC-mediated leukemogenesis. Leukemia. 2016;30(6):1365–1374. doi: 10.1038/leu.2016.26. PMID: 26876595 PMCID: PMC4889531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Furusawa A., Miyamoto M., Takano M., Tsuda H., Song Y.S., Aoki D. Ovarian cancer therapeutic potential of glutamine depletion based on GS expression. Carcinogenesis. 2018;39(6):758–766. doi: 10.1093/carcin/bgy033. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Multimedia component 2
Multimedia component 3
Multimedia component 4