Abstract
The plant transcription factor WRINKLED1 (WRI1), a member of AP2/EREBP, is involved in the regulation of glycolysis and the expression of genes related to the de novo synthesis of fatty acids in plastids. In this study, the key regulator of seed oil synthesis and accumulation transcription factor gene PoWRI1 was identified and cloned, having a complete open reading frame of 1269 bp and encoding 422 amino acids. Subcellular localization analysis showed that PoWRI1 is located at the nucleus. After the expression vector of PoWRI1 was constructed and transformed into wild-type Arabidopsis thaliana, it was found that the overexpression of PoWRI1 increased the expression level of downstream target genes such as BCCP2, KAS1, and PKP-β1. As a result, the seeds of transgenic plants became larger, the oil content increased significantly, and the unsaturated fatty acid content increased, which provide a scientific theoretical basis for the subsequent use of genetic engineering methods to improve the fatty acid composition and content of plant seeds.
Keywords: Paeonia ostii, WRINKLED1, oil synthesis
1. Introduction
The tree peony, as a famous traditional flower in China, has been cultivated for thousands of years. In addition to its ornamental value, its oil value is also widely recognized. Since being approved as a new resource food by the Ministry of Health of China in 2011 [1], it has been widely promoted by various governments. At present, the tree peony has become the third largest woody oil crop in China after walnut and oil camellia. Paeonia ostii is a perennial woody deciduous shrub belonging to the genus Paeoniae in the Paeoniaceae family, which has the characteristics of large flowering, less tillering, high seed yield and strong ecological adaptability. Currently, P. ostii is not only the most widely cultivated oil peony variety in China, but also is a vital new oil crop integrating ornamental, medicinal, and oil use [2]. The content of unsaturated fatty acids in P. ostii seed oil is rich, among which α-linolenic acid and linoleic acid represent the plant’s advantages over other oil crops because of their higher nutritional value and better economic benefits. To alleviate the contradiction between the production and demand of vegetable oils in China, it is of great significance to improve the oil content and fatty acid composition of woody oil crops by means of biological techniques.
Oil synthesis involves the flow and direction of glycolysis metabolites and the transport coordination of metabolites between different subcellular organelles and cytoplasmic sites. The regulation of plant lipid synthesis is complex, and the research on the lipid metabolism pathway and its structural genes has been relatively clear [3]. In recent years, studies have found that the process of lipid synthesis is strictly regulated at the transcriptional level. At present, some important transcription factors regulating plant lipid metabolism have been found, including five categories: bZIP, B3, NFYB, ASIL, and AP2/EREB transcription factor WRI1 [4]. First, ABI4, a transcription factor of bZIP, can be combined with ABA response elements and can directly combine fatty acids to synthesize the promoter region of catalase DGAT1 in ABA signaling pathway, thus promoting the synthesis of DGAT1 [5]. The GmbZIP123 transcription factor in soybeans can specifically combine the promoter region of SUC1 and SUC5 genes to enhance the transport of endogenous sugar, eventually leading to an increase in oil content [6]. Meanwhile, the overexpression of NsbZIP1 in Nannochloropsis salina could significantly enhance the growth and lipid content of transformants [7]. Second, the B3 transcription factor family plays a vital role in embryo maturation and seed ABA signaling pathway [8]. It has 95, 108, and 81 members in Arabidopsis, soybean, and corn [9], respectively, and can specifically identify DNA sequences containing a conservative RY/Sph structure. The B3 transcription factors mainly include ABSCISIC ACID INSENSITIVE3 (ABI3), FUSCA3 (FUS3), and LEAFY COTYLEDON2 (LEC2) [10]. LEC2 mainly regulates lipid synthesis, and its target genes are ABI3, FUS3, LEC1, and WRI1 [11]. For example, the overexpression of LEC2 in tobacco increased total fatty acid content by 6.8% [12]. Third, among NFYB transcription factors, LEC1 is a subunit encoding CCAAT-Box binding factor HAP3, which is a key regulator of embryonic development. Although the overexpression of ZmLEC1 and ZmWRI1 in maize can increase the corn oil content by 48%, it also affects the normal growth and development of seeds [13]. Fourth, ASIL1 (Arabidopsis 6b-interacting protein 1-like1) protein could indirectly inhibit LEC1, LEC2, FUS2, and ABI3, thus regulating the seed filling process [14].
WRI1 (WRINKLED1) is the master transcription factor found to directly regulate glycolysis and fatty acid synthesis. In Arabidopsis, wri1-1 is a mutant with uneven epidermis and atrophic grain screened from seeds [15]. WRI1 plays a significant role in regulating seed oil content in the complex regulatory network composed of transcription factors [16]. The phosphorylated structure in WRI1 is called PEST-motif. The transcription of PEST-motif can increase the content of triacylglycerol in seeds, and the deletion or mutation of this motif can increase the stability of WRI1 proteins [17]. Moreover, in the early stage of seed development, WRI1 promotes the high expression of genes related to triglyceride synthesis by regulating a variety of genes involved in fatty acid synthesis, glycolysis, and biotin synthesis [18]. Overexpression of the WRI1 gene during seed development could increase the oil content in seeds and other organs [19]. For example, the overexpression of BnWRI1 in Brassica napus could promote the expression of key enzymes in the glycolysis pathway and in fatty acid synthesis, which confirms its ability to enhance the accumulation of oil in plant seeds and leaves [20]. In maize, WRI1 is mainly expressed in the embryo and endosperm of seeds, promoting the increase of fatty acid content in mature maize grains, and does not affect its seeding growth, starch content, or grain yield [13]. Moreover, the expression of the WRI1 gene in the middle and late stages of seed development can promote the accumulation of seed oil and inhibit the accumulation of seed protein, which indicates that WRI1 has a certain role in maintaining the balance of seed oil and protein [21]. P. ostii was used in this study, and the key transcription factor gene PoWRI1 was cloned. After transformed into Arabidopsis, the phenotypic observation, gene expression of the transgenic Arabidopsis seeds, and lipid assay were analyzed in order to study the regulation function of transcription factor PoWRI1 on seed lipid accumulation. Our research lays the foundation for further identification of the PoWRI1 gene and its application in molecular genetic improvement of oil composition.
2. Results
2.1. Isolation and Sequence Analysis of PoWRI1 from Paeonia ostii
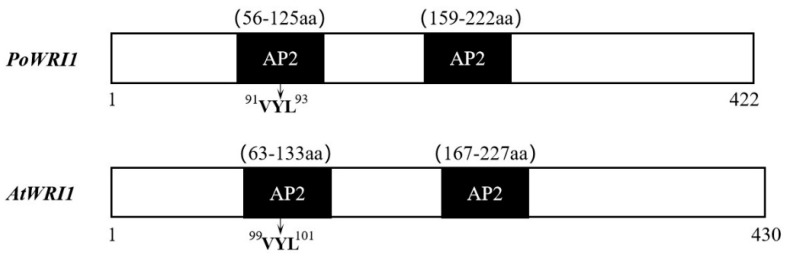
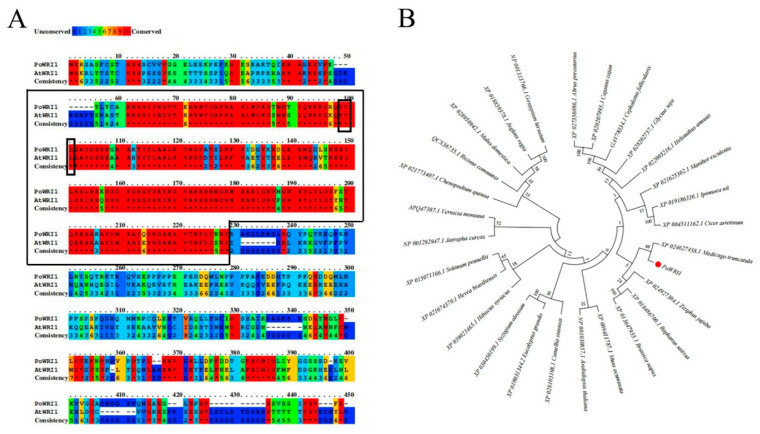
Specific primers were designed according to the full-length cDNA sequence of the WRI1 gene from the transcriptome database (PRJNA317164) [22] (Table S1) and employed to clone the gene named PoWRI1. The total length of the PoWRI1 gene is 1413 bp, which has a complete open reading frame of 1269 bp and encodes 422 amino acids (The accession number is MW930196). After being analyzed by the ProtParam online tool, the isoelectric point of PoWRI1 is 5.68 and the molecular weight of PoWRI1 was 47.0 kDa. It is worth noting that the instability coefficient of PoWRI1 protein was 52.41, so PoWRI1 belongs to the unstable protein. PoWRI1 has two conserved AP2 domains, which are located between amino acids 56 and 125 and between amino acids 159 and 222. The Motif “VYL” encoded by a 9-bp exon is between amino acids 91 and 93, which is similar to the structure of Arabidopsis thaliana (Figure 1). Except the regions of C-terminal and N-terminal, the results of sequences alignment analysis showed that the similarity of the PoWRI1 and WRI1 protein sequence of Arabidopsis thaliana seems high. The regions of the two AP2 conserved domains between PoWRI1 and AtWRI1 protein shared especially high similarity (Figure 2A). In addition, the phylogenetic tree was constructed with the neighbor-joining method by MEGA 7.0 following multiple alignments of protein sequences (Figure 2B). The alignment results showed that the WRI1 protein sequence has a consistency in different plants and PoWRI1 protein has the highest evolutionary similarity with Medicago truncatula (XP_024627458.1).
Figure 1.
The structural characteristics of PoWRI1 (MW930196) and AtWRI1 (NP_001030857.1).
Figure 2.
Sequence analysis of PoWRI1. (A) Alignment of protein sequence of PoWRI1 (MW930196) and AtWRI1 (NP_001030857.1). Most of the similarity between protein sequences of PoWRI1 and AtWRI1 occurs at the AP2 regions of the protein (highlighted by boxes). Conservation of amino acids is denoted by different colors as illustrated by the scale bar. (B) Phylogeny tree of WRI1 homologs including Paeonia ostii (MW930196), Vernicia montana (APQ47387.1), Cephalotus follicularis (GAV78334.1), Arabidopsis thaliana (NP_001030857.1), Jatropha curcas (NP_001292947.1), Gossypium hirsutum (NP_001313766.1), Ricinus communis (QCX36735.1), Cicer arietinum (XP_004511162.1), Musa acuminata (XP_009411787.1), Eucalyptus grandis (XP_010031344.2), Brassica napus (XP_013647955.1), Solanum pennellii (XP_015071166.1), Raphanus sativus (XP_018486700.1), Juglans regia (XP_018859378.1), Ipomoea nil (XP_019186336.1), Cajanus cajan (XP_020207995.1), Manihot esculenta (XP_021625302.1), Hevea brasiliensis (XP_021674370.1), Chenopodium quinoa (XP_021773407.1), Helianthus annuus (XP_022005216.1), Medicago truncatula (XP_024627458.1), Ziziphus jujuba (XP_024927304.1), Abrus precatorius (XP_027356086.1), Camellia sinensis (XP_028103108.1), Glycine soja (XP_028202757.1), Malus domestica (XP_028950942.1), Syzygium oleosum (XP_030450199.1) and Hibiscus syriacus (XP_039023465.1).
2.2. Expression Level Analysis of PoWRI1 in P. ostii
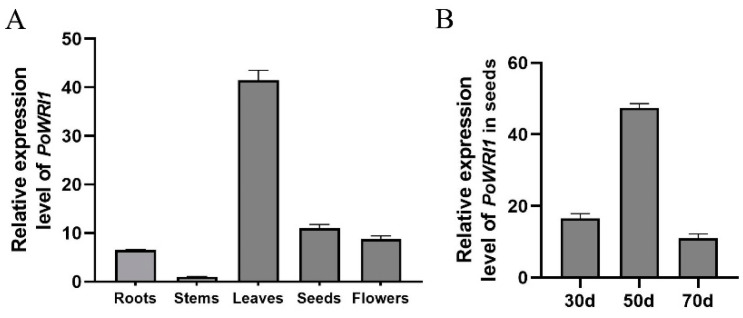
In order to detect the expression of PoWRI1 for different tissues in P. ostii, qRT–PCR was used to analyze the roots, stems, leaves, seeds, and flowers in P. ostii. Ubiquitin (JN699053) was used as the reference gene. As shown in Figure 3A, the expression level of PoWRI1 in different tissues was different, and showed a higher expression level in leaves, followed by seeds, compared with other tissues.
Figure 3.
Expression level analysis of PoWRI1 in P. ostii. (A) Relative expression level of PoWRI1 of different tissues in P. ostii. The reference gene was Ubiquitin (JN699053), the primers were based on the coding sequences of PoWRI1 (MW930196). Total RNA was extracted from roots, stems, leaves, flowers, and seeds (70d) of P. ostii. Values are the means ± SE of three replicates carried out on cDNAs obtained from three independent mRNA extractions. (B) Relative expression level of PoWRI1 in seeds of different stages. The reference gene was Ubiquitin (JN699053), the primers were based on the coding sequences of PoWRI1 (MW930196). Total RNA was extracted from the seeds of 30d, 50d, and 70d. Values are the means ± SE of three replicates carried out on cDNAs obtained from three independent mRNA extractions.
We further investigated the expression level of PoWRI1 in P. ostii seeds at 30d (i.e., days after flowering), 50d, and 70d. qRT–PCR results showed that the expression levels of P. ostii seeds in 50d were higher than the other two stages (Figure 3B). With the development of seeds in P. ostii, the PoWRI1 gene was upregulated and then downregulated.
2.3. Subcellular Localization of the PoWRI1 Protein
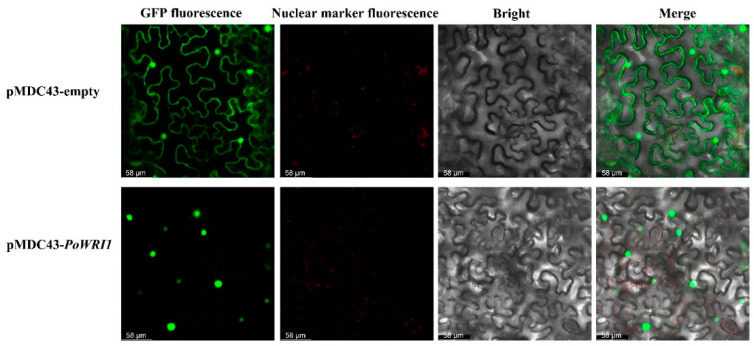
To further determine the subcellular localization of the PoWRI1 protein, the expression vector pMDC43-PoWRI1 was constructed and transformed into N. benthamian by the Fast Agro-mediated Seedling Transformation (FAST) method [23] using an empty vector pMDC43 as a control. As is shown in Figure 4, the GFP green fluorescence sites were widely distributed in the whole plasma membrane system and nucleus in the pMDC43 empty vector. However, the GFP green fluorescence sites of pMDC43-PoWRI1 were located at the nucleus, indicating that PoWRI1 was a nuclear localization protein and consistent with the subcellular localization prediction result.
Figure 4.
Subcellular localization of the PoWRI1 protein. An empty vector pMDC43 and pMDC43-PoWRI1 transiently expressed in N. benthamiana leaves. Bars = 58 μm.
2.4. Genetic Transformation of PoWRI1 Gene in A. thaliana
After pCAMBIA1301-PoWRI1 overexpression, the vector was constructed and transformed into Agrobacterium tumefaciens strain EHA105 by a freeze–thaw method. The inflorescence of wild-type Arabidopsis thaliana was infected. After a series of screening and culture, T3 generation seeds were harvested.
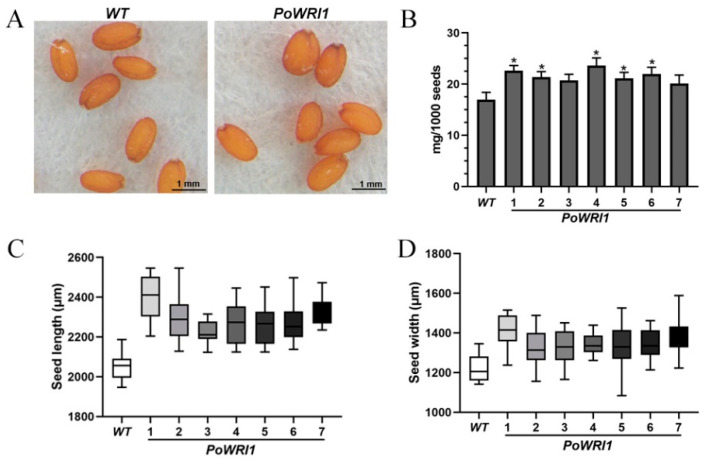
The seeds of T3 generation Arabidopsis thaliana were collected after drying in the oven. The length and width of seven transgenic lines seeds and wild-type Arabidopsis thaliana seeds were measured. As shown in Figure 5A, compared with wild-type ones, the seeds of transgenic Arabidopsis are plumper and larger in shape and darker in color. In addition, the dried Arabidopsis seeds were randomly selected to calculate their average 1000 seed weight and standard deviation. The 1000 seed weight of transgenic Arabidopsis plants significantly higher compared with that of wild-type Arabidopsis (Figure 5B). The average weight of each wild-type seed was 16.97 μg and the average weight of each transgenic seed was 21.64 μg, increased by 27.5% compared with wild-type plants. The length and width of T3 generation seeds of 7 transgenic Arabidopsis plants were significantly larger than those of wild-type ones (Figure 5C,D).
Figure 5.
Phenotypic observation of transgenic Arabidopsis. (A) Dry seeds of WT and PoWRI1 observed under a stereomicroscope. Bars indicate 1 mm. (B) 1000 dry seeds weigh of WT and PoWRI1 (Line 1–7). Error bars are s.e. (n = 3). * indicates significant differences between WT and PoWRI1 (Line 1–7) (p < 0.05). (C) Seed length of WT and PoWRI1 (Line 1–7). Error bars are s.e. (n = 23). (D) Seed width of WT and PoWRI1 (Line 1–7). Error bars are s.e. (n = 23).
2.5. Expression Level Analysis of Transgenic A. thaliana
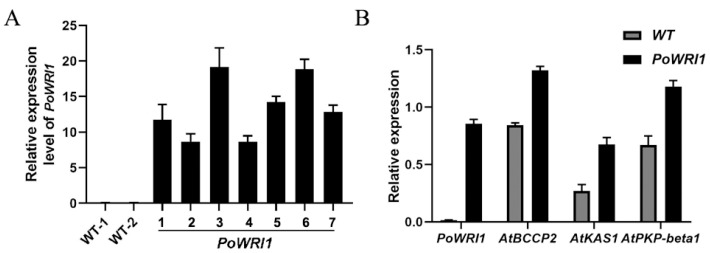
In order to detect the expression of the target gene PoWRI1 in transgenic Arabidopsis, qRT–PCR was used to analyze the transgenic Arabidopsis plants. Taking two wild-type Arabidopsis plants as controls, seven transgenic Arabidopsis plants were selected. As shown in Figure 6A, the expression level of PoWRI1 in different transgenic Arabidopsis plants was different, and showed a higher expression level compared with the control.
Figure 6.
Expression level analysis of transgenic A. thaliana. (A) Relative expression level of PoWRI1. WT-1 and WT-2 is wild-type lines, 1–7 are PoWRI1 transgenic lines. The reference gene was AtActin (AK230311.1), the primers were based on the coding sequences of PoWRI1 (MW930196). Total RNA was extracted from Arabidopsis leaves. Values are the means ± SE of three replicates carried out on cDNAs obtained from three independent mRNA extractions. (B) Relative expression level of WRI1 and downstream target genes in Arabidopsis leaves. The reference gene was AtActin (AK230311.1), the primers were based on the coding sequences of PoWRI1 (MW930196), AtBCCP2 (AT5G15530), AtKAS1 (AT5G46290) and AtPKP-β1 (AT5G52920). Total RNA was extracted from Arabidopsis leaves. Values are the means ± SE of three replicates carried out on cDNAs obtained from three independent mRNA extractions.
To further investigate the function of PoWRI1 in gene expression regulation, specific primers were used to detect the expression level of downstream target genes of WRI1, including biotin carboxyl carrier protein isoform 2 (BCCP2), 3-ketoacyl-acyl carrier protein synthase 1 (KAS1) and plastid pyruvate kinase beta subunit 1 (PKP-β1). qRT–PCR results showed that the expression levels of these genes in transgenic Arabidopsis were higher than those in wild-type Arabidopsis (Figure 6B). The expression of BCCP2 in transgenic Arabidopsis was the highest (56% higher than that in wild-type ones), the expression of KAS1 was 76% higher than that of wild-type ones, and the expression of PKP-β1 was 60% higher than that of wild-type ones. The results showed that the overexpression of PoWRI1 in Arabidopsis increased the expression of downstream genes related to lipid synthesis.
2.6. High Fatty Acid Content in Transgenic Arabidopsis Seeds
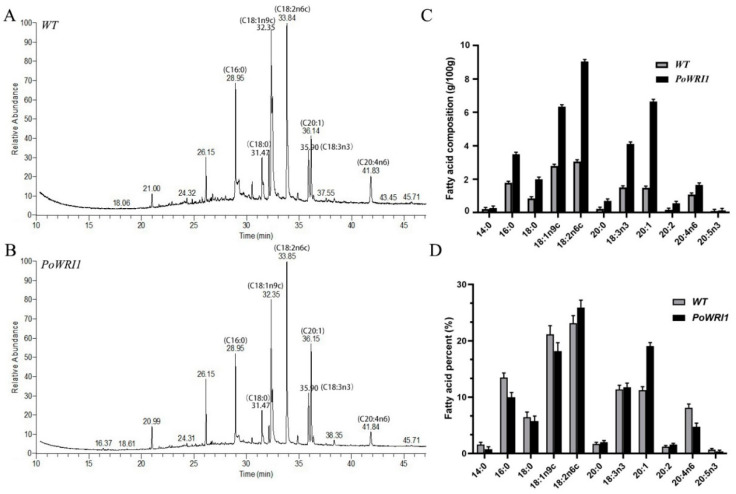
To identify the function of the PoWRI1 gene in fatty acid biosynthesis and oil accumulation, we verified whether the overexpression of the PoWRI1 gene leads to the increase of total fatty acid content in Arabidopsis seeds. Fatty acids in the seeds of transgenic lines (1, 4, 6) and wild-type lines were detected and analyzed by Thermo Trace1310 ISQ GC-MS with nonadecanoic acid (C19:0) as an internal standard. A total of 11 fatty acid were detected in both transgenic and wild-type Arabidopsis (Figure 7A,B). The peak area of the measured results was analyzed by GC-MS and the fatty acid content of Arabidopsis seeds was calculated. As shown in Figure 7C, the content of each fatty acid in transgenic plants increased compared with that in wild type. Notably, the content of long-chain fatty acids and unsaturated fatty acids increased significantly. Among them, oleic acid (C18:1n9c), linoleic acid (C18:2n6c), α-linolenic acid (C18:3n3), and arachidonic acid (C20:0) increased nearly two times compared with wild-type ones. The contents of palmitic acid (C16:0) and stearic acid (C18:0) were nearly doubled compared with wild-type ones. The proportion of each fatty acid component in the seeds of transgenic and wild-type plants was also significantly different (Figure 7D). The proportion of unsaturated fatty acids in transgenic Arabidopsis seeds was higher than that in wild-type ones.
Figure 7.
Analysis of fatty acid content in Arabidopsis seeds. (A,B) GC-MS analysis of fatty acids isolated from the Arabidopsis seeds. (C,D) Fatty acid content and percentages of each component isolated from the Arabidopsis seeds. Error bars are s.e. (n = 3).
3. Discussion
3.1. Structural Characteristics of PoWRI1 and Its Expression Levels in P. ostii
WRI1 was first identified in Arabidopsis [4], and its orthologs have been identified from many plants such as Cocos nucifera [24], Brassica napus [20], Ricinus connunis [25], Glycine max [26], Avena sativa [27], and Zea mays [28]. The full-length cDNA sequence of the PoWRI1 gene of P. ostii was also identified and cloned in our study. It was confirmed that PoWRI1 belongs to the AP2/EREBP transcription factor gene family. In the same way as AtWRI1, PoWRI1 has a PEST motif despite the divergence between its C-terminal sequences (Figure S1), suggesting that this region is conserved and contributes to WRI1 stability [17]. Although the C-terminal regions of AtWRI1 and PoWRI1 are strikingly diverged, bioinformatics analysis showed that PoWRI1 and AtWRI1 shared a 93% sequence similarity in the AP2 domain and a 94% sequence similarity in the second AP2 domain. The differences in these individual amino acids of the AP2 domains may be due to species differences. In addition, studies have shown that there is an important amino acid “VYL”, which is conserved in a number of WRI1 orthologs discovered in many plant species [29]. The mutation of a single amino acid in “VYL” can lead to the impairment of the function of WRI1 protein in Arabidopsis [30]. In this study, the PoWRI1 protein in the first AP2 domain has the same amino acid “VYL” as the WRI1 sequence of Arabidopsis. It can be inferred that the PoWRI1 of P. ostii has the essential role of “VYL” for PoWRI1 function.
During the period of seed development and filling, photosynthetic carbons are partitioned into starch, oil and proteins, and other different storage compounds, which are highly regulated by LEAFY COTYLEDON 1 (LEC1), LEC2, and WRI1 [31]. In P. ostii, the development of seeds can be divided into three stages. The level of fatty acid is relatively low in the first stage. Then there is a period of rapid oil accumulation. Finally, there is a decrease with the seed approaching full maturity [32]. The expression level of PoWRI1 was first upregulated and then downregulated with the development of seeds, which was consistent with the trend of oil accumulation.
3.2. Phenotype of Transgenic Arabidopsis Thaliana and Overexpression of WRI1 Downstream Gene
It is known that the enzymes involved in sugar metabolism and hexose and sucrose accumulation affect the number and size of seed cells during seed development, which in turn affects the quality and size of seeds [33,34]. In addition, as the central regulator of the fatty acid biosynthesis pathway, the WRI1 transcription factor can specifically and positively involved glycolysis and fatty acid biosynthesis [35] through affecting the expression of related genes in glycolysis pathways to regulate key enzymes involved in the process, finally affecting the phenotype of Arabidopsis seeds of glycometabolism. After the preliminary phenotypic observation of transgenic Arabidopsis seeds, it was found that the size and weight of transgenic Arabidopsis seeds were significantly higher than that of wild-type ones, indicating that the PoWRI1 gene has a certain role in promoting the growth and development of plant seeds. Meanwhile, it can be found that the expression levels of target genes of WRI1 downstream including BCCP2, KAS1, and PKP-β1 in transgenic Arabidopsis were higher than those in wild-type Arabidopsis, which were similar to the results that overexpression of RcWRI1 significantly upregulated the expression of pyruvate kinase alpha subunit (PKP-α), acyl carrier protein 1 (ACP1), pyruvate dehydrogenase E1 component alpha subunit (PDH-Elα), BCCP2, KAS1, and PKP-β1 in N. benthamiana leaves [25], regulating the expression of the above essential genes in FA biosynthesis [36,37,38,39]. As the target genes of WRI1, these genes are involved in glycolysis and fatty acid synthesis. The promoter region of WRI1 transcription factor’s downstream gene has a highly conserved sequence, which is called AW-box (CnTnG(n)7CG) or 15 bp element (CAAAAG(T/G)AGG(G/A)APTT). WR11 regulates downstream gene transcription by binding AW (ASML1/WRI1)-box, which plays an important role in regulating the carbon flow from sugar to oil during seed development [35,40].
3.3. Significant Increase of Fatty Acid Content in Transgenic Arabidopsis Thaliana
In fact, WRI1 affects oil accumulation by regulating the metabolic process during seed development, especially the glycolysis pathway [41,42]. Glycolysis provides raw materials for fatty acid synthesis. The overexpression of WRI1 regulates lipid metabolism by up regulating the expression of target genes involved in glycolysis and fatty acid synthesis, thereby promoting the carbon flow from glycolysis to fatty acid synthesis in seeds and improving the content of triacylglycerol in seeds and seedlings [26]. Research has found that all of the WRI1 homologs also induced oil accumulation in N. benthamiana. The increase of unsaturated fatty acids was not obvious [27]. Moreover, the transcription levels of fatty acid synthetase, plastid transporter and key enzymes of plastid glucose metabolism in oil palm with high oil content were enhanced compared with those in date palm. The expression level of the WRI1 transcription factor homologous transcript in oil palm was 57 times higher than that in date palm, while the expression level of enzymes related to triglyceride assembly was similar in both oil palm and date palm. These results further indicated that WRI1 promoted lipid synthesis by regulating plastid pyruvate supply and fatty acid synthesis, but was not directly involved in fatty acid modification and triglyceride synthesis [43]. Our results showed that the content of fatty acids in the seeds of transgenic Arabidopsis was more than twice as high as that of wild-type ones. Therefore, the WRI1 gene significantly promoted the increase of fatty acid content in transgenic Arabidopsis seeds. It’s worth noting that the content of unsaturated fatty acids and long chain fatty acids in transgenic Arabidopsis were also higher than those of wild-type ones. In other words, PoWRI1 has a significant effect on the increase of unsaturated fatty acids, suggesting that PoWRI1 helps the synthesis of an essential enzyme promoting the dehydrogenation of saturated fatty acids to unsaturated fatty acids, such as Fatty acid desaturase (FAD) [44].
Currently, the demand for vegetable oil is increasing. Improving oil content and fatty acid composition of oil crops to meet high demand of vegetable oil market represents a promising strategy, and the heterologous overexpression of WRI1 is an effective measure. For example, transferring the BnWRI1 gene into Arabidopsis will increase the oil content, seed volume, and weight of transgenic Arabidopsis seeds [20], which is similar to the results of this experiment. As a result, our study provides insights into the transcriptional activation of glycolysis and fatty acid biosynthesis pathways in Arabidopsis, and lays a foundation for further elucidating the regulatory network controlling seed oil accumulation.
4. Materials and Methods
4.1. Plant Materials
The seeds (30d, 50d, and 70d), roots, stems, leaves, and flowers of 3-year-old P. ostii from the germplasm repository of the Horticulture and Plant Protection College, Yangzhou University, Jiangsu Province, P.R. China (32°23′31″ N, 119°24′50″ E) were collected and stored in liquid nitrogen. They were stored in −80 °C for RNA extraction.
The transformed plant material is the seed of Arabidopsis thaliana of Columbia Col-0 type and stored at 4 °C (21 days after anthesis).
4.2. Gene Cloning and Sequence Analysis
After the seeds grinding into powder, RNA was separately extracted using the Mini BEST Plant RNA Extraction Kit (TaKaRa, Tokyo, Japan), and all RNA samples were checked using Nanodrop 2000C (Thermo Scientific). According to PrimeScript® RT reagent Kit with gDNA eraser (Perfect Real Time) (TaKaRa, Tokyo, Japan), RNA was reserved into cDNA. The specific primers PoWRI1-F and PoWRI1-R were designed by Primer 5.0 software for the PCR amplification (Table S1). The PCR reaction was as follows: 1 cycle of 94 °C for 5 min; 30 cycles of 94 °C for 30 s, 55 °C for 30 s, 72 °C for 2 min; and one cycle of 72 °C for 10 min. After testing by 1% (w/v) agarose gel electrophoresis, the PoWRI1 PCR products were cloned into the pClone007 Vector and sequenced.
Protparam (http://web.expasy.org/protparam/) (accessed on 30 April 2021) was used to analyze the amino acid composition, relative molecular weight, isoelectric point, and other physical and chemical properties. The conserved domain of PoWRI1 protein was analyzed by CD-Search tool of the National Center for Biotechnology Information site (NCBI, https://www.ncbi.nlm.nih.gov) (accessed on 30 April 2021). The protein sequence of AtWRI1 (NP_001030857.1) was obtained from NCBI and the alignment was analyzed by the PRALINE program (http://www.ibi.vu.nl/programs/pralinewww/) (accessed on 30 April 2021). A Neighbor-Joining phylogenetic tree was generated with MEGA 7.0, and bootstrap values were set as 1000 bootstrap replicates [45].
4.3. Construction of Expression Vector
The specific primers attB-PoWRI1-F and attB-PoWRI1-R were designed (Table S1). The PCR products were then cloned into the Invitrogen GATEWAYTM pDONR/Zeo vector (Thermo Fisher Scientific, Waltham, MA, USA) using the BP reaction and sub-cloned (LR reaction) into the plant GATEWAY™ binary vector: pMDC43 for transient expression in N. benthamiana to determine the subcellular localization of the PoWRI1 protein.
Based on the obtained full-length sequence of PoWRI1, combined with the restriction site of the binary expression vector pCAMBIA1301, the recombinant plant transgenic vector was constructed by T4 DNA ligase Buffer. The Bsa I restriction sites on the polyclonal site of pCAMBIA1301 was used to cleave the vector. At the same time, the coding region of PoWRI1 gene was amplified with primers PoWRI1-Bsa I-F/R (including the corresponding Bsa I restriction sites) (Table S1). The recombinant plasmid pCAMBIA1301-PoWRI1 was constructed by ligating the target fragment to the vector with T4 DNA ligase Buffer.
4.4. Overexpressing PoWRI1 in the Arabidopsis
The expression vector pCAMBIA1301-PoWRI1 plasmids were used for the transformation of competent cells of Agrobacterium tumefaciens strain EHA105. Arabidopsis Col-0 plants were transformed using the floral-dip method [46]. The mature seeds were collected and recorded as T0. The seeds of T1 generation were screened by MS medium containing hygromycin (25 mg/L). When the seedlings grew four rosette leaves, the better growing ones were transplanted into sterilized soil (nutrient soil: perlite: vermiculite = 2:1:1) and cultured at 22 °C for 14 h in light (100–150 μmol m−2 s−1 illumination), 18 °C for 10 h in dark, and the humidity was 70–80%. The seeds were collected and recorded as T2 generation after flowering and fruiting. T2 generation seeds were screened with hygromycin (25 mg/L) until all of them were resistant seedlings, and then were screened and cultured according to the above method until T3 generation seeds were harvested and transgenic plants were identified by PCR. 1–7 transgenic lines were in good growing condition and were selected from the T3 generation seed.
4.5. Phenotypic Observation of Transgenic Arabidopsis
After the flowering and pod ripening of Arabidopsis thaliana, pods were harvested before the coat was split and placed in the oven. After drying at 37 °C for 24 h, the coat of the pod was removed and the seeds of T3 generation Arabidopsis thaliana were collected. The length and width of seeds of different transgenic and wild-type Arabidopsis were measured. Transgenic and wild-type Arabidopsis randomly measured 30 seeds, repeated three times, using SAS/STAT 6.12 statistical analysis software for variance analysis of the results, using GraphPad Prism 8.0.2 software to draw pictures about the length and width of seeds.
1000 dry seeds of transgenic and wild-type Arabidopsis were weighed randomly and repeated for three times. The average seed weight and standard deviation were calculated and plotted by GraphPad Prism 8.0.2 software.
4.6. Quantitative Real-Time PCR Analysis
The total RNA of different tissues and seeds at different stages of P. ostii was extracted, according to the Mini BEST Plant RNA Extraction Kit (TaKaRa, Tokyo, Japan). Then, the RNA was reverse-transcribed into cDNA by the superscript first-strand synthesis system (Prime-Script® RT Reagent Kit With gDNA Eraser, TaKaRa, Tokyo, Japan). Using Ubiquitin (JN699053) as an internal reference, real-time quantitative PCR (qRT-PCR) was introduced to analyze the expression levels of PoWRI1 in different tissues and seeds at different stages of P. ostii with a BIO-RAD CFX Connect Optics Module (Bio-Rad, Des Plaines, IL, USA).
The total RNA of T3 generation transgenic and wild-type Arabidopsis leaves in full bloom was extracted by above mentioned method. Using the Actin (AK230311.1) gene as an internal reference, real-time quantitative PCR (qRT–PCR) was introduced to analyze not only the expression levels of PoWRI1, but also the expression levels of downstream target genes (AtBCCP2, AtKAS1 and AtPKP-β1) of WRI1 in transgenic and wild-type Arabidopsis.
Specific primers were designed using Primer 5.0 (Table S1). The formula 2−ΔΔCt was referred to calculated their values. A final volume of 25 μL (12.5 μL 2 × SYBR Premix Ex Taq, 2 μL cDNA solution, 2 μL mix solution of primers and 8.5 μL ddH2O) was the system to peform qRT-PCR. The reaction conditions were 95 °C for 30 s, 40 cycles at 95 °C for 5 s, 52 °C for 30 s, and 72 °C for 30 s.
4.7. Analysis of Fatty Acids Content in Seeds of Transgenic Arabidopsis
Firstly, the dry wild-type and T3 generation transgenic Arabidopsis seeds, several zeolites, pyrogallic acid, 95% ethanol, and hydrochloric acid solution were added into the flask and mix well. After hydrolysis in water bath and cooling to room temperature, 95% ethanol was added to the hydrolysate. The mixture was then shaken and collected in a constant weight flask. After evaporating and drying in an oven, 2% sodium hydroxide methanol and 14% boron trifluoride methanol were added to the oil extract respectively. Then, n-hexane was added for centrifugation and stratification. The supernatant and n-hexane were dissolved again, and the volume was then fix to 1 mL and mixed well. Finally, after filtration, the membrane was tested. Trace1310 ISQ gas chromatography-mass spectrometry (GC-MS) of Thermo company (Germany) was used to detect fatty acids. The chromatographic column was TG-5MS (30 m × 0.25 mm × 0.25 μm).
5. Conclusions
The PoWRI1 gene, which is located at the nucleus and is associated with the oil accumulation process, was identified and cloned. The overexpression of PoWRI1 in Arabidopsis increased the expression level of downstream target genes such as BCCP2, KAS1, and PKP-β1. Meanwhile, the seeds of transgenic plants became larger, the oil content increased significantly, and the unsaturated fatty acid content increased, which provides a scientific theoretical basis for the subsequent use of genetic engineering methods to improve the fatty acid composition and content of plant seeds.
Acknowledgments
This work was supported by funding from the National Natural Science Foundation of China (31600564), the Modern Agricultural Industrial Technology System in Jiangsu Province (JATS [2020]436), the Natural Science Fund of Jiangsu Province (BK20160460), the program of key members of Yangzhou University outstanding young teachers, the Qing Lan Project of Jiangsu Province and the High-Level Talent Support Program of Yangzhou University. Research on the key technologies of the exploitation and utilization of the characteristic flower resources was based on Leisure Agriculture (CX(20)2030).
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/ijms22136996/s1. Figure S1: PEST motif analysis of AtWRI1 and PoWRI1; Table S1: Primers used for PCR and qRT–PCR.
Author Contributions
Conceptualization, J.S.; data curation, T.C.; formal analysis, J.S.; methodology, J.S. and M.L.; project administration, J.S. and J.T.; resources, M.L.; software, T.C.; validation, D.Z.; visualization, D.Z. and J.T.; writing—original draft, T.C.; writing—review & editing, T.C. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by funding from the National Natural Science Foundation of China (31600564), Modern Agricultural Industrial Technology System in Jiangsu Province (JATS [2020]436), the Natural Science Fund of Jiangsu Province (BK20160460), the program of key members of Yangzhou University outstanding young teachers, Qing Lan Project of Jiangsu Province and High-Level Talent Support Program of Yangzhou University, Research on the key technologies of the exploitation and utilization of the characteristic flower resources based on Leisure Agriculture (CX(20)2030).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data are contained within the article or Supplementary Materials.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Pei Z.S., Hu Y.H., Liu Z., Li B.H. New development and utilization of woody oil crops-oil. For. Ecol. Sci. 2018;34:358–363. [Google Scholar]
- 2.Wang X., Liang H., Guo D., Guo L., Duan X., Jia Q., Hou X. Integrated analysis of transcriptomic and proteomic data from tree peony (P. ostii) seeds reveals key developmental stages and candidate genes related to oil biosynthesis and fatty acid metabolism. Hortic. Res. 2019;6:111. doi: 10.1038/s41438-019-0194-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marchive C., Nikovics K., To A., Lepiniec L., Baud S. Transcriptional regulation of fatty acid production in higher plants: Molecular bases and biotechnological outcomes. Eur. J. Lipid Sci. Techol. 2014;116:1332–1343. doi: 10.1002/ejlt.201400027. [DOI] [Google Scholar]
- 4.Cernac A., Benning C. WRINKLED1 encodes an AP2/EREB domain protein involved in the control of storage compound biosynthesis in Arabidopsis. Plant J. 2004;40:575–585. doi: 10.1111/j.1365-313X.2004.02235.x. [DOI] [PubMed] [Google Scholar]
- 5.Yang Y., Yu X.C., Song L.F., An C.C. ABI4 Activates DGAT1 Expression in Arabidopsis Seedlings during Nitrogen Deficiency. Plant Physiol. 2011;156:873–883. doi: 10.1104/pp.111.175950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Song Q.X., Li Q.T., Liu Y.F., Zhang F.X., Ma B., Zhang W.K., Man W.Q., Du W.G., Wang G.D., Chen S.Y., et al. Soybean GmbZIP123 gene enhances lipid content in the seeds of transgenic Arabidopsis plants. J. Exp. Bot. 2013;64:4329–4341. doi: 10.1093/jxb/ert238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kwon S., Kang N.K., Koh H.G., Shin S.E., Lee B., Jeong B.R., Chang Y.K. Enhancement of biomass and lipid productivity by overexpression of a bZIP transcription factor in Nannochloropsis salina. Biotechnol. Bioeng. 2018;115:331–340. doi: 10.1002/bit.26465. [DOI] [PubMed] [Google Scholar]
- 8.McAtee P., Karim S., Schaffer R., David K. A dynamic interplay between phytohormones is required for fruit development, maturation, and ripening. Front. Plant Sci. 2013;4:79. doi: 10.3389/fpls.2013.00079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Le B.H., Cheng C., Bui A.Q., Wagmaister J.A., Henry K.F., Pelletier J., Kwong L., Belmonte M., Kirkbride R., Horvath S., et al. Global analysis of gene activity during Arabidopsis seed development and identification of seed-specific transcription factors. Proc. Natl. Acad. Sci. USA. 2010;107:8063–8070. doi: 10.1073/pnas.1003530107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Suzuki M., McCarty D.R. Functional symmetry of the B3 network controlling seed development. Curr. Opin. Plant Biol. 2008;11:548–553. doi: 10.1016/j.pbi.2008.06.015. [DOI] [PubMed] [Google Scholar]
- 11.Angeles-Nunez J.G., Tiessen A. Mutation of the transcription factor LEAFY COTYLEDON 2 alters the chemical composition of Arabidopsis seeds, decreasing oil and protein content, while maintaining high levels of starch and sucrose in mature seeds. J. Plant Physiol. 2011;168:1891–1900. doi: 10.1016/j.jplph.2011.05.003. [DOI] [PubMed] [Google Scholar]
- 12.Vanhercke T., El Tahchy A., Liu Q., Zhou X.R., Shrestha P., Divi U.K., Ral J.P., Mansour M.P., Nichols P.D., James C.N., et al. Metabolic engineering of biomass for high energy density: Oilseed-like triacylglycerol yields from plant leaves. Plant. Biotechnol. J. 2014;12:231–239. doi: 10.1111/pbi.12131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shen B., Allen W.B., Zheng P.Z., Li C.J., Glassman K., Ranch J., Nubel D., Tarczynski M.C. Expression of ZmLEC1 and ZmWRI1 Increases Seed Oil Production in Maize. Plant Physiol. 2010;153:980–987. doi: 10.1104/pp.110.157537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gao M.J., Lydiate D.J., Li X., Lui H., Gjetvaj B., Hegedus D.D., Rozwadowski K. Repression of Seed Maturation Genes by a Trihelix Transcriptional Repressor in Arabidopsis Seedlings. Plant Cell. 2009;21:54–71. doi: 10.1105/tpc.108.061309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Focks N., Benning C. wrinkled1: A novel, low-seed-oil mutant of Arabidopsis with a deficiency in the seed-specific regulation of carbohydrate metabolism. Plant Physiol. 1998;118:91–101. doi: 10.1104/pp.118.1.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhai Z.Y., Keereetaweep J., Liu H., Feil R., Lunn J.E., Shanklin J. Expression of a Bacterial Trehalose-6-phosphate Synthase otsA Increases Oil Accumulation in Plant Seeds and Vegetative Tissues. Front. Plant Sci. 2021;12:656962. doi: 10.3389/fpls.2021.656962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ma W., Kong Q., Grix M., Mantyla J.J., Yang Y., Benning C., Ohlrogge J.B. Deletion of a C-terminal intrinsically disordered region of WRINKLED1 affects its stability and enhances oil accumulation in Arabidopsis. Plant J. 2015;83:864–874. doi: 10.1111/tpj.12933. [DOI] [PubMed] [Google Scholar]
- 18.Weselake R.J., Taylor D.C., Rahman M.H., Shah S., Laroche A., McVetty P.B.E., Harwood J.L. Increasing the flow of carbon into seed oil. Biotechnol. Adv. 2009;27:866–878. doi: 10.1016/j.biotechadv.2009.07.001. [DOI] [PubMed] [Google Scholar]
- 19.Li-Beisson Y., Shorrosh B., Beisson F., Andersson M.X., Arondel V., Bates P.D., Baud S., Bird D., Debono A., Durrett T.P., et al. Acyl-lipid metabolism. Arab. Book. 2010;8:e0133. doi: 10.1199/tab.0133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu J., Hua W., Zhan G., Wei F., Wang X., Liu G., Wang H. Increasing seed mass and oil content in transgenic Arabidopsis by the overexpression of wri1-like gene from Brassica napus. Plant Physiol. Biochem. 2010;48:9–15. doi: 10.1016/j.plaphy.2009.09.007. [DOI] [PubMed] [Google Scholar]
- 21.Li Q., Shao J.H., Tang S.H., Shen Q.W., Wang T.H., Chen W.L., Hong Y.Y. Wrinkled1 Accelerates Flowering and Regulates Lipid Homeostasis between Oil Accumulation and Membrane Lipid Anabolism in Brassica napus (vol 6, 1015, 2015) Front. Plant Sci. 2016;6:1015. doi: 10.3389/fpls.2015.01270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xiu Y., Wu G.D., Tang W.S., Peng Z.F., Bu X.P., Chao L.J., Xiong J.N., Zhang H.W., Zhao X.Q., Ding J., et al. Oil biosynthesis and transcriptome profiles in developing endosperm and oil characteristic analyses in Paeonia ostii var. lishizhenii. J. Plant Physiol. 2018;228:121–133. doi: 10.1016/j.jplph.2018.05.011. [DOI] [PubMed] [Google Scholar]
- 23.Weaver J., Goklany S., Rizvi N., Cram E.J., Lee-Parsons C.W. Optimizing the transient Fast Agro-mediated Seedling Transformation (FAST) method in Catharanthus roseus seedlings. Plant Cell Rep. 2014;33:89–97. doi: 10.1007/s00299-013-1514-2. [DOI] [PubMed] [Google Scholar]
- 24.Sun R., Ye R., Gao L., Zhang L., Wang R., Mao T., Zheng Y., Li D., Lin Y. Characterization and Ectopic Expression of CoWRI1, an AP2/EREBP Domain-Containing Transcription Factor from Coconut (Cocos nucifera L.) Endosperm, Changes the Seeds Oil Content in Transgenic Arabidopsis thaliana and Rice (Oryza sativa L.) Front. Plant Sci. 2017;8:63. doi: 10.3389/fpls.2017.00063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ji X.J., Mao X., Hao Q.T., Liu B.L., Xue J.A., Li R.Z. Splice Variants of the Castor WRI1 Gene Upregulate Fatty Acid and Oil Biosynthesis When Expressed in Tobacco Leaves. Int. J. Mol. Sci. 2018;19:146. doi: 10.3390/ijms19010146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen B., Zhang G., Li P., Yang J., Guo L., Benning C., Wang X., Zhao J. Multiple GmWRI1s are redundantly involved in seed filling and nodulation by regulating plastidic glycolysis, lipid biosynthesis and hormone signalling in soybean (Glycine max) Plant Biotechnol. J. 2020;18:155–171. doi: 10.1111/pbi.13183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grimberg A., Carlsson A.S., Marttila S., Bhalerao R., Hofvander P. Transcriptional transitions in Nicotiana benthamiana leaves upon induction of oil synthesis by WRINKLED1 homologs from diverse species and tissues. BMC Plant Biol. 2015;15:192. doi: 10.1186/s12870-015-0579-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pouvreau B., Baud S., Vernoud V., Morin V., Py C., Gendrot G., Pichon J.P., Rouster J., Paul W., Rogowsky P.M. Duplicate maize Wrinkled1 transcription factors activate target genes involved in seed oil biosynthesis. Plant Physiol. 2011;156:674–686. doi: 10.1104/pp.111.173641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ma W., Kong Q., Arondel V., Kilaru A., Bates P.D., Thrower N.A., Benning C., Ohlrogge J.B. Wrinkled1, a ubiquitous regulator in oil accumulating tissues from Arabidopsis embryos to oil palm mesocarp. PLoS ONE. 2013;8:e68887. doi: 10.1371/journal.pone.0068887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krizek B.A. AINTEGUMENTA utilizes a mode of DNA recognition distinct from that used by proteins containing a single AP2 domain. Nucleic Acids Res. 2003;31:1859–1868. doi: 10.1093/nar/gkg292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Manan S., Ahmad M.Z., Zhang G., Chen B., Haq B.U., Yang J., Zhao J. LEAFY COTYLEDON 2 regulates subsets of genes involved in controlling the biosynthesis and catabolism of seed storage substances and seed development. Front. Plant Sci. 2017;8:1604. doi: 10.3389/fpls.2017.01604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li S.S., Wang L.S., Shu Q.Y., Wu J., Chen L.G., Shao S., Yin D.D. Fatty acid composition of developing tree peony (Paeonia section Moutan DC.) seeds and transcriptome analysis during seed development. BMC Genom. 2015;16:208. doi: 10.1186/s12864-015-1429-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weber H., Buchner P., Borisjuk L., Wobus U. Sucrose metabolism during cotyledon development of Vicia faba L. is controlled by the concerted action of both sucrose-phosphate synthase and sucrose synthase: Expression patterns, metabolic regulation and implications for seed development. Plant J. 1996;9:841–850. doi: 10.1046/j.1365-313X.1996.9060841.x. [DOI] [PubMed] [Google Scholar]
- 34.Weber H., Heim U., Golombek S., Borisjuk L., Manteuffel R., Wobus U. Expression of a yeast-derived invertase in developing cotyledons of Vicia narbonensis alters the carbohydrate state and affects storage functions. Plant J. 1998;16:163–172. doi: 10.1046/j.1365-313x.1998.00282.x. [DOI] [PubMed] [Google Scholar]
- 35.Baud S., Wuilleme S., To A., Rochat C., Lepiniec L. Role of WRINKLED1 in the transcriptional regulation of glycolytic and fatty acid biosynthetic genes in Arabidopsis. Plant J. 2009;60:933–947. doi: 10.1111/j.1365-313X.2009.04011.x. [DOI] [PubMed] [Google Scholar]
- 36.Baud S., Feria Bourrellier A.B., Azzopardi M., Berger A., Dechorgnat J., Daniel-Vedele F., Lepiniec L., Miquel M., Rochat C., Hodges M., et al. PII is induced by WRINKLED1 and fine-tunes fatty acid composition in seeds of Arabidopsis thaliana. Plant J. 2010;64:291–303. doi: 10.1111/j.1365-313X.2010.04332.x. [DOI] [PubMed] [Google Scholar]
- 37.Adhikari N.D., Bates P.D., Browse J. WRINKLED1 Rescues Feedback Inhibition of Fatty Acid Synthesis in Hydroxylase-Expressing Seeds. Plant Physiol. 2016;171:179–191. doi: 10.1104/pp.15.01906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.To A., Joubes J., Barthole G., Lecureuil A., Scagnelli A., Jasinski S., Lepiniec L., Baud S. WRINKLED transcription factors orchestrate tissue-specific regulation of fatty acid biosynthesis in Arabidopsis. Plant Cell. 2012;24:5007–5023. doi: 10.1105/tpc.112.106120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li Q.T., Lu X., Song Q.X., Chen H.W., Wei W., Tao J.J., Bian X.H., Shen M., Ma B., Zhang W.K., et al. Selection for a Zinc-Finger Protein Contributes to Seed Oil Increase during Soybean Domestication. Plant Physiol. 2017;173:2208–2224. doi: 10.1104/pp.16.01610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maeo K., Tokuda T., Ayame A., Mitsui N., Kawai T., Tsukagoshi H., Ishiguro S., Nakamura K. An AP2-type transcription factor, WRINKLED1, of Arabidopsis thaliana binds to the AW-box sequence conserved among proximal upstream regions of genes involved in fatty acid synthesis. Plant J. 2009;60:476–487. doi: 10.1111/j.1365-313X.2009.03967.x. [DOI] [PubMed] [Google Scholar]
- 41.Cernac A., Andre C., Hoffmann-Benning S., Benning C. WRI1 is required for seed germination and seedling establishment. Plant Physiol. 2006;141:745–757. doi: 10.1104/pp.106.079574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Baud S., Mendoza M.S., To A., Harscoet E., Lepiniec L., Dubreucq B. WRINKLED1 specifies the regulatory action of LEAFY COTYLEDON2 towards fatty acid metabolism during seed maturation in Arabidopsis. Plant J. 2007;50:825–838. doi: 10.1111/j.1365-313X.2007.03092.x. [DOI] [PubMed] [Google Scholar]
- 43.Bourgis F., Kilaru A., Cao X., Ngando-Ebongue G.F., Drira N., Ohlrogge J.B., Arondel V. Comparative transcriptome and metabolite analysis of oil palm and date palm mesocarp that differ dramatically in carbon partitioning. Proc. Natl. Acad. Sci. USA. 2011;108:18186. doi: 10.1073/pnas.1106502108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sun J., Chen M., Zhu M., Jiang Y., Meng J., Zhao D., Tao J. Cloning, Characterization, and Expression Analysis of Three FAD8 Genes Encoding a Fatty Acid Desaturase from Seeds of Paeonia ostii. Molecules. 2018;23:929. doi: 10.3390/molecules23040929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Saitou N., Nei M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 46.Clough S.J., Bent A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data are contained within the article or Supplementary Materials.