Abstract
We read with interest the article by Patel et al. on the identification of potential inhibitors of coronavirus hemagglutinin-esterase. The authors considered hemagglutinin-esterase as a glycoprotein of SARS-CoV-2 and selected hemagglutinin-esterase as a target to identify potential inhibitors using a combination of various computational approaches, and however, SARS-CoV-2 genome lacks hemagglutinin-esterase gene; thus, hemagglutinin-esterase does not exist in SARS-CoV-2 particle.
Graphic abstract
Keywords: Hemagglutinin-esterase, SARS-CoV-2, Genome, Betacoronavirus
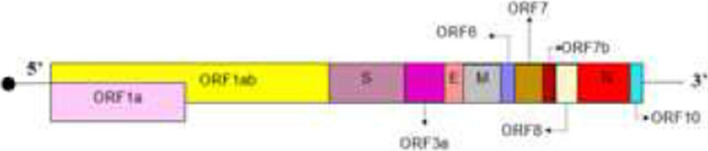
In December 2019, a novel betacoronavirus “severe acute respiratory syndrome coronavirus 2” (SARS-CoV-2) detected in China [1]. SARS-CoV-2 is a member of Coronaviridae family. There are four genera in coronaviruses apha-, beta-, gamma- and deltacoronavirus [2]. Betacoronavirus genera is classified into five subgenera or lineages (Table 1) [3]. SARS-CoV-2 is a positive-sense single-strand RNA virus, and the size of its genome is about 30 kb [4]. The 2/3 of genome of SARS-CoV-2 at 5′ end contains two overlapping open reading frames (ORF1a and ORF1b) which encode two polyprotein precursors including pp1a and pp1b, respectively. Two proteases cleave both pp1a and pp1b to produce non-structural proteins. The remaining 3′ third of the genome encodes four structural proteins including spike (S), envelope (E), matrix (M) and nucleocapsid (N), and also, accessory genes are distributed in among the structural genes[4, 5]. Novel coronavirus (n-CoV) as a betacoronavirus of lineage B does not contain hemagglutinin-esterase (HE) [6–10]. However, betacoronaviruses of lineage A (such as OC43-CoV, HKU1-CoV and Bovine-CoV) harbor HE gene which encodes HE that acts as a receptor-destroying enzyme [11]. Some studies mistakenly reported that SARS-CoV-2 contains HE. Duner et al. stated that some coronaviruses including SARS-COV-2 contain HE [12], and however, they cited a reference which is published in 1998 Aug [13], although SARS-CoV-2 is detected in 2019 Dec and lacks HE. In another article, authors reported that betacoronavirus mostly uses HE to link to sialic acid on the glycoprotein surface [14], and however, betacoronavirus is a genera (Table 1) and among betacoronaviruses, viruses which belong to Embecovirus subgenera can code HE as a glycoprotein surface [15]. Aktas A et al. examined the biological activity of Arbidol in the inhibition of HE and spike glycoproteins of SARS-CoV-2 in silico[16], but according to scientific evidences which are described above, SARS-CoV-2 codes four structural proteins including spike, envelop, membrane and nucleoprotein [17], and therefore, Arbidol has no such interaction with HE in SARS-CoV-2 infection.
Table 1.
Five subgenera and different species of betacoronavirus genus
| Subgenera | ||||
|---|---|---|---|---|
| Embecovirus (Lineage A) | Sarbecovirus (Lineage B) | Merbecovirus (Lineage C) | Nobecovirus (Lineage D) | Hibecovirus |
| Species | ||||
|
Bovine coronavirus Human coronavirus OC43 Human coronavirus HKU1 Mouse hepatitis virus |
SARS-CoV SARS-CoV-2 Bat SARS-like coronavirus WIV1 Bat coronavirus RaTG13 |
MERS-CoV Pipistrellus bat coronavirus HKU5 Hedgehog coronavirus 1 Tylonycteris bat coronavirus HKU4 |
Eidolon bat coronavirus C704 Rousettus bat coronavirus GCCDC1 Rousettus bat coronavirus HKU9 |
Bat Hp-betacoronavirus Zhejiang2013 |
Recently in a published research article, authors presented that genome of SARS-CoV-2 harbors hemagglutinin-esterase gene and encodes HE glycoprotein, authors considered HE as a target for designing drug against SARS-CoV-2 infection [18], however, according to evidence SARS-CoV-2 genome lacks HE gene and cannot encode HE protein.
Funding
No funding.
Declaration
Conflicts of interest
Authors declare no conflicts of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Lai CC, et al. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and coronavirus disease-2019 (COVID-19): the epidemic and the challenges. Int J Antimicrob Agents. 2020;55(3):105924. doi: 10.1016/j.ijantimicag.2020.105924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li H, et al. Coronavirus disease 2019 (COVID-19): current status and future perspectives. Int J Antimicrob Agents. 2020;55(5):105951. doi: 10.1016/j.ijantimicag.2020.105951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wong AC, et al. Global epidemiology of bat coronaviruses. Viruses. 2019;11(2):174. doi: 10.3390/v11020174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rastogi M, et al. SARS coronavirus 2: from genome to infectome. Respir Res. 2020;21(1):1–15. doi: 10.1186/s12931-020-01581-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kelly JA, et al. Structural and functional conservation of the programmed−1 ribosomal frameshift signal of SARS coronavirus 2 (SARS-CoV-2) J Biol Chem. 2020;295(31):10741–10748. doi: 10.1074/jbc.AC120.013449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shi C-S, et al. SARS-Coronavirus Open Reading Frame-8b triggers intracellular stress pathways and activates NLRP3 inflammasomes. Cell Death Discov. 2019;5(1):1–12. doi: 10.1038/s41421-018-0068-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kumar S et al. (2020) Morphology, genome organization, replication, and pathogenesis of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Coronavirus Disease 2019 (COVID-19), p. 23
- 8.Devasena T. Nanotechnology-COVID-19 interface. Singapore: Springer; 2021. The nanotechnology-COVID-19 Interface; pp. 31–58. [Google Scholar]
- 9.Crawford KH, et al. Protocol and reagents for pseudotyping lentiviral particles with SARS-CoV-2 spike protein for neutralization assays. Viruses. 2020;12(5):513. doi: 10.3390/v12050513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zandi M, Behboudi E, Soltani S. Role of glycoprotein hemagglutinin-esterase in COVID-19 pathophysiology? Stem Cell Rev Reports. 2021;28:1–2. doi: 10.1007/s12015-021-10210-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lang Y, et al. Coronavirus hemagglutinin-esterase and spike proteins coevolve for functional balance and optimal virion avidity. Proc Natl Acad Sci. 2020;117(41):25759–25770. doi: 10.1073/pnas.2006299117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Duner P, Salehi A. COVID-19 and possible pharmacological preventive options. J Clin Med Res. 2020;12(12):758. doi: 10.14740/jocmr4383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Haan CA, et al. Coronavirus particle assembly: primary structure requirements of the membrane protein. J Virol. 1998;72(8):6838–6850. doi: 10.1128/JVI.72.8.6838-6850.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frediansyah A, et al. Antivirals for COVID-19: a critical review. Clin Epidemiol Glob Health. 2020;9:90–98. doi: 10.1016/j.cegh.2020.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim C-H. SARS-CoV-2 evolutionary adaptation toward host entry and recognition of receptor O-Acetyl sialylation in virus–host interaction. Int J Mol Sci. 2020;21(12):4549. doi: 10.3390/ijms21124549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aktas A, et al. How do arbidol and its analogs inhibit the SARS-CoV-2? Bratisl Lek Listy. 2020;121(10):705–711. doi: 10.4149/BLL_2020_115. [DOI] [PubMed] [Google Scholar]
- 17.Fani M, et al. The role of miRNAs in COVID-19 disease. Futur Virol. 2021;16(4):301–306. doi: 10.2217/fvl-2020-0389. [DOI] [Google Scholar]
- 18.Patel CN, et al. Identification of potential inhibitors of coronavirus hemagglutinin-esterase using molecular docking, molecular dynamics simulation and binding free energy calculation. Mol Diversity. 2021;25(1):421–433. doi: 10.1007/s11030-020-10135-w. [DOI] [PMC free article] [PubMed] [Google Scholar]