Abstract
The microbiota plays a critical role in regulating organismal health and response to environmental stresses. Intermittent hypoxia and hypercapnia, a condition that represents the main hallmark of obstructive sleep apnea in humans, is known to induce significant alterations in the gut microbiome and metabolism, and promotes the progression of atherosclerosis in mouse models. To further understand the role of the microbiome in the cardiovascular response to intermittent hypoxia and hypercapnia, we developed a new rodent cage system that allows exposure of mice to controlled levels of O2 and CO2 under gnotobiotic conditions. Using this experimental setup, we determined the impact of the microbiome on the transcriptional response to intermittent hypoxia and hypercapnia in the left ventricle of the mouse heart. We identified significant changes in gene expression in both conventionally reared and germ-free mice. Following intermittent hypoxia and hypercapnia exposure, we detected 192 significant changes in conventionally reared mice (96 upregulated and 96 downregulated) and 161 significant changes (70 upregulated and 91 downregulated) in germ-free mice. Only 19 of these differentially expressed transcripts (∼10%) were common to conventionally reared and germ-free mice. Such distinct transcriptional responses imply that the host microbiota plays an important role in regulating the host transcriptional response to intermittent hypoxia and hypercapnia in the mouse heart.
Keywords: gnotobiotics, transcriptome, intermittent hypoxia and hypercapnia, heart, mice
Introduction
Obstructive sleep apnea (OSA) is the most common type of sleep disorder in humans, and OSA is very prevalent in adults and children. It is characterized by repeated episodes of complete or partial obstructions of the upper airways during sleep, which is associated with a reduction in blood oxygen saturation and an increase in blood CO2. It has been demonstrated by experimental and clinical studies that OSA may lead to a number of health consequences including cardiovascular diseases ranging from cardiometabolic disorders and arrhythmogenesis to heart failure (Al Lawati et al., 2009; Baguet et al., 2012; Levy et al., 2013; Drager et al., 2015; Lyons and Bradley, 2015; Hoyos et al., 2017; May et al., 2017).
In addition, evidence from both animal and human studies supports the notion that the gut microbiota plays a critical role in the onset and progression of various cardiovascular diseases (Karbach et al., 2016; Koch et al., 2017; Weis, 2018). Previous studies by others and us have shown that intermittent hypoxia with/without hypercapnia alters the gut microbiome (Moreno-Indias et al., 2015, 2016; Lucking et al., 2018; Tripathi et al., 2018, 2019). In turn, such alterations may induce developmental defects and functional dysregulations. For example, the human oral and intestinal microbiota has been recognized as a significant determinant of cardiovascular disease risk (Ordovas and Mooser, 2006), and suppression of the gut microbiome ameliorates age-dependent arterial dysfunction (Brunt et al., 2019). Recent studies also revealed a potential contribution of gut microbes to certain human cardiometabolic diseases (Vinje et al., 2014) and heart failure (Sandek et al., 2007; Nagatomo and Tang, 2015; Nemet et al., 2020).
Besides impacting cellular signaling, the microbiome has been demonstrated to modify the epigenetic landscape of the host genome, which may influence cellular differentiation and proliferation, leading to new phenotypic traits throughout the age spectrum (Takahashi et al., 2011; Pevsner-Fischer et al., 2017; Qin and Wade, 2018). For example, several studies have demonstrated a direct link between the gut microbiome and host epigenetic states in the context of inflammatory bowel disease, obesity, cancer, and behavioral abnormalities (Kellermayer et al., 2010; Manichanh et al., 2012; Dejea et al., 2014; Paul et al., 2015; Farhana et al., 2018; Sfanos et al., 2018; Chu et al., 2019).
Since gut microbiota plays an important role in regulating host gene expression (Patrignani et al., 2014; Chu et al., 2019; Richards et al., 2019; Weger et al., 2019), in the current study, we tested the hypothesis that the microbiome regulates the transcriptional response to intermittent hypoxia and hypercapnia (IHH) exposure in the heart. Using a newly developed experimental setup to impose IHH under gnotobiotic conditions, we performed 150-bp paired-end RNA-seq and utilized multiple bioinformatic approaches to analyze the transcriptional alterations following IHH treatment in the left ventricle (LV) of conventionally reared (CONV) and germ-free (GF) C57BL/6 mice. In addition, we compared our results with those reported for other organs in male GF mice of similar genetic background and age (Weger et al., 2019) (dataset GSE77221). We found significant and distinct differences in transcriptional responses between CONV and GF mice, demonstrating that, indeed, the microbiome plays an important role in regulating IHH response in the heart.
Materials and Methods
Experimental Animals
GF and CONV C57BL/6 mice that were 8–12 weeks old were used for this experiment. GF status was confirmed weekly by standard microbiological cultures of fecal extracts, using thioglycollate broth and potato dextrose broth for facultative anaerobes including fungi, and cooked meat broth, heart infusion broth, and Columbia blood agar for obligate anaerobes (Moody et al., 2019). All animal protocols were approved by the Animal Care Committee of the University of California, San Diego and followed the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health.
IHH Treatment
All mice were housed in specialized cages for the 2-week treatments (IHH or normoxia; GF or CONV) (Figure 1). These specialized cages were developed by us in the Gnotobiotic Animal Facility at the University of California, San Diego. Each sealed single cage (Sealed Positive Pressure System, Allentown, NJ, United States) was ventilated by a single, HEPA (high-efficiency particulate air)-filtered gas supply pump. The cages were opened and closed for stocking and servicing inside a Class IIa laminar-flow biosafety cabinet. The system allows complete control of gas inflow and outflow, as has been confirmed by measurements of exhaust gases using an O2/CO2 analyzer (Quantek Instruments, Grafton, MA, United States). The IHH condition was maintained in a computer-controlled atmosphere chamber system (OxyCycler, BioSpherix, Redfield, NY, United States) as previously described (Fan et al., 2005; Zhou et al., 2008). IHH exposure was introduced to the mice in short periods of ∼4 min of synchronized reduction in (O2) (from 21 to 8%) and increase in (CO2) (from ∼0.5 to 8%) separated by alternating ∼4-min periods of normoxic room air [(O2) = 21% and (CO2) ≈ 0.5%] with 2 ramp intervals (∼1 min each) for a total of 10 IHH events per hour for 10 h per day during the light cycle. Controls received a flow of room air throughout the entire experimental period. Prior works have shown that the oxygen saturation level (SaO2) ranged from 60 to 80% when mice were subjected to 5–10% hypoxic treatments (Sforza and Roche, 2016; Xue et al., 2017).
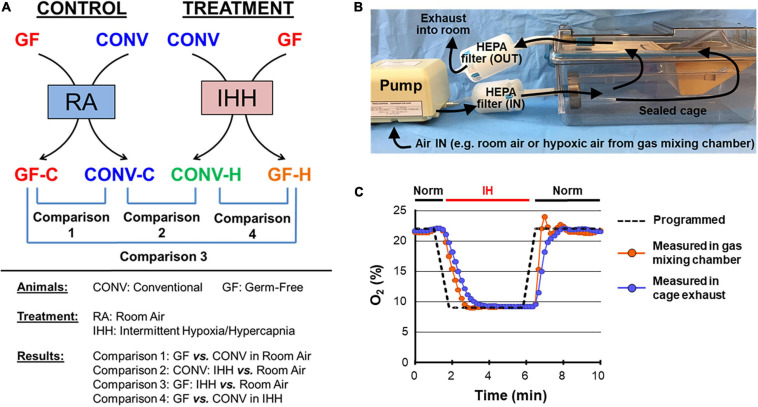
FIGURE 1.
Experiment design and instrumentation. (A) Conventional (CONV) and germ-free (GF) mice were treated with intermittent hypoxia/hypercapnia (IHH) or room air (RA) to determine the impacts of microbiome on heart transcriptome under normoxia and transcriptional response to IHH stress. (B) A tightly sealed cage system was developed to allow control of the cage atmosphere under gnotobiotic conditions by ventilating cages with a single, HEPA-filtered gas. The gnotobiotic chamber for treatment was supplied with IHH gas; room air was supplied to the control chamber. (C) A representative recording of a test run of the experimental setup with intermittent hypoxia (IH). The gas environment inside the cage was controlled by placing the pump inlet into a gas mixing chamber. IH was induced in the cage by obtaining gas from the gas mixing chamber with a programmed cycle of 5 min of 8% O2 followed by 5 min of room air (21% O2) (dashed black line). O2 levels were measured directly in the mixing chamber (red symbols) or in the exhaust gas of the ventilated cage (blue symbols). IH, intermittent hypoxia; Norm, normoxia.
RNA Sequencing and Data Analysis
At the end of the IHH exposure, mice were sacrificed, and the heart was harvested and separated into the right and left ventricle under sterile conditions and then placed into separate tubes containing RNAlater Stabilization Solution (Thermo Fisher Scientific, Ashville, NC, United States). RNA from the LV was extracted using the RNeasy Mini Kit (QIAGEN, Hilden, Germany) and sent to Novogene (Chula Vista, CA, United States) for QC and sequencing (n = 3/group). A 150 nt paired-end sequencing on the Illumina HiSeq 4000 platform (Illumina, San Diego, CA, United States) was performed. Rigorous quality controls of the paired-end reads were assessed using FastQC tools, and the adapter sequences and low-quality bases were trimmed using Cutadapt (Didion et al., 2017). The resulting reads were processed using the Illumina BaseSpace software package using the STAR aligner (Dobin et al., 2013) to align reads against the Mus musculus UCSC mm9 reference genome. Differential expression was calculated using DESeq 2 with default settings (Anders and Huber, 2010), and differentially expressed genes (DEGs) were identified with a threshold of |Fold-Change| > 1.5 and adjusted p-value < 0.05. Bioinformatic data mining of the differentially expressed transcripts was carried out with the tools provided by the GWAS Catalog1 (Buniello et al., 2019), the Database for Annotation, Visualization, and Integrated Discovery (DAVID)2 (Dennis et al., 2003; Huang et al., 2007), the Ingenuity Pathway Analysis (IPA) package (QIAGEN, Redwood City, CA, United States), as well as KEGG3 (Ogata et al., 1999; Kanehisa and Goto, 2000) and the Reactome4 (Fabregat et al., 2017). Multiple testing correction was applied, and the FDR q-values were estimated to correct the p-values for the multiple testing issue.
Data Availability
The paired-end RNA-seq data of n = 12 samples are available at https://www.ncbi.nlm.nih.gov/bioproject/692551 (BioProject ID: PRJNA692551).
Results and Discussion
Experimental Design
The microbiome, particularly in the gut, plays an important role in maintaining systemic homeostasis and contributes to a number of human diseases [for selected reviews, see Lynch and Pedersen (2016), Gilbert et al. (2018), Dominguez-Bello et al. (2019), Harikrishnan (2019), Helmink et al. (2019), Team (2019)]. However, its potential involvement in regulating many physiologic and pathophysiologic responses under stressful conditions remains to be explored. In the current study, we used a prospective case-control study design to determine the impact of the microbiome on cardiac transcriptional responses to IHH in CONV and GF mice (Figure 1A). To maintain GF animals under controlled gas conditions, we developed a custom cage system with minimal dead space and programmable HEPA-filtered gas supply (Figure 1B). Analysis of the exhaust gases of the cage system revealed rapid (<20 s) equilibration of the cage environment to match the input gas mixture (Figure 1C). GF mice could be kept in the system for extended periods (>2 weeks) without compromising their GF state, demonstrating that the newly developed system was well suited to conduct short- and long-term experiments of constant or intermittent hypoxia with or without hypercapnia under gnotobiotic conditions.
GF and CONV mice of the same strain, sex, and age (kept in the same cage system as the GF mice) were either exposed to the IHH condition for 2 weeks to mimic sleep apnea in humans (Kanaan et al., 2006; Xue et al., 2017; Hunyor and Cook, 2018) or kept under normoxia/normocapnia throughout as controls. For a broad characterization of the cardiac gene expression responses, we extracted total RNA from the LV and applied paired-end RNA-seq as a genome-wide discovery approach. Approximately 38–50 million total reads were obtained per sample with over 94% of the reads uniquely mapped to the mouse reference genome (UCSC mm9) (Supplementary Table 1).
Microbiota Impacts Cardiac Gene Expression Under Normoxic Conditions
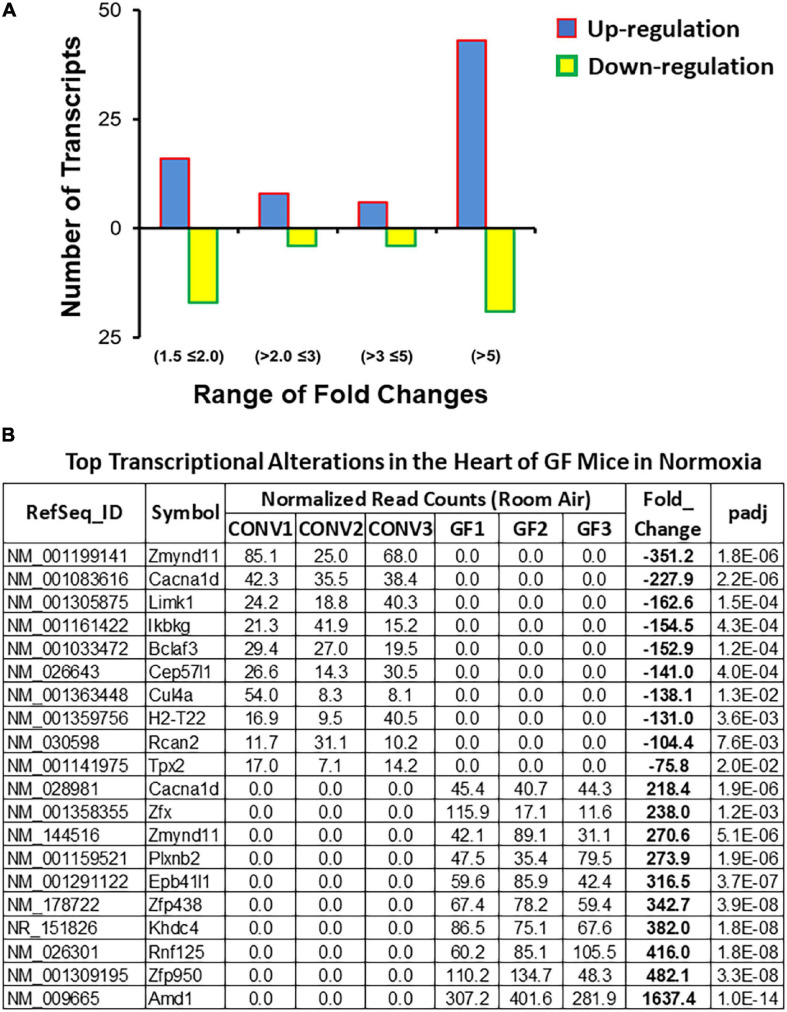
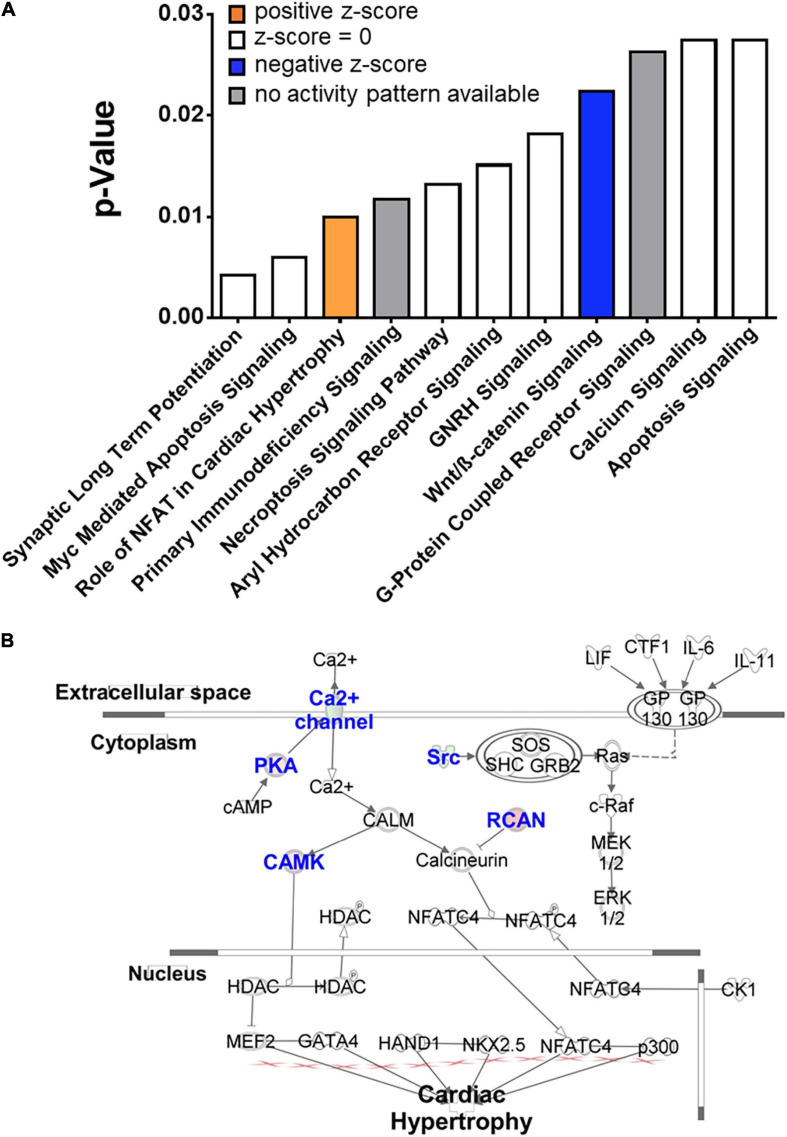
We first determined the impact of the microbiota on cardiac gene expression under normoxic and normocapnic conditions. A total of 117 DEGs were identified between CONV and GF mice. Of these, 73 were upregulated and 44 were downregulated (Fold change > 1.5 and adjusted p-value < 0.05) in the left ventricle of GF mice as compared to CONV controls (Figure 2A and Supplementary Table 2). The most upregulated transcripts included Amd1, Zfp950, Rnf125, and Khdc4, which are involved in polyamine biosynthesis, transcription regulation, ubiquitination, and subsequent proteasomal degradation of target proteins, and pre-mRNA splicing, respectively. In contrast, the most downregulated transcripts included genes encoding a transcription repressor (Zmynd11), a Calcium Voltage-Gated Channel Subunit (Cacna1d), and a serine/threonine kinase (Limk1) (Figure 2B). In addition, several functional implications were suggested by data mining and bioinformatic analyses using the DAVID and IPA tools. Results derived from DAVID functional annotation suggested alterations in transcription regulation, alpha-actinin binding, and sarcolemma organization, as well as pathways that regulate cardiac function and activity, including the GnRH pathway, the MAPK signaling pathway, the Oxytocin signaling pathway, and vascular muscle contraction (Supplementary Table 3A). In addition, Ingenuity analysis showed potential changes in several canonical pathways that are important in cardiac function, such as the mechanisms regulating synaptic long-term potentiation, Myc-mediated apoptosis signaling, and the role of NFAT in cardiac hypertrophy (Figure 3 and Supplementary Table 3B). The different alterations on the “Myc Mediated Apoptosis Signaling” and the “Role of NFAT in Cardiac Hypertrophy” suggested that cell death and proliferation signaling pathways were differently configured in the GF mice (Supplementary Table 3B). Since (1) Myc-mediated apoptosis plays a critical role in response to stress in the myocardium (Hou et al., 2016; Veeraveedu et al., 2017; Guan et al., 2019), (2) NFAT plays a major role in cardiac hypertrophy [for selected reviews see Clerk et al. (2007), Rohini et al. (2010), Nakayama et al. (2013)], and (3) previous studies also shown that gut microbiome regulates the activity of Myc in mouse liver (Kolodziejczyk et al., 2020) and NFAT in colorectal cancer in humans (Peuker et al., 2016), our current findings suggest that alterations of Myc and NFAT signaling in GF mice may modify cardiac activity and stress response.
FIGURE 2.
Summary of transcriptome alterations in the heart of GF mice under normoxic condition. (A) Distribution of fold change of the 117 differentially expressed genes (DEGs) (| Fold change| > 1.5 and Benjamini–Hochberg adjusted p < 0.05) in the left ventricle of GF mice as compared to CONV controls (Supplementary Table 2). (B) List of top 10 significantly up- or downregulated DEGs.
FIGURE 3.
Top significantly altered cellular signaling pathways in the heart of GF mice in normoxia. (A) Transcriptome profiling suggested significant alterations of multiple cellular signaling pathways in the heart of GF mice (right-tailed Fisher’s exact test, p < 0.05, Supplementary Table 3), including (B) activation of the pathway regulating the “role of NFAT in cardiac hypertrophy.” Blue fonts: significantly altered components of the pathway.
Furthermore, we found that 36% of the DEGs (42 out of 117) in GF mice encode nuclear proteins, including transcription factors (e.g., Dach1 and Gtf2ird2), protein kinases (e.g., Clk1 and Limk1), DNA, and histone modification enzymes (e.g., Dnmt3b and Kmt5b). Such a significant enrichment of nuclear proteins suggested differences of transcriptional regulation mechanisms between GF and CONV mice, which may lead to distinct transcriptional responses to stress in the heart.
Distinct Transcriptional Alterations in Different Organs of GF Mice
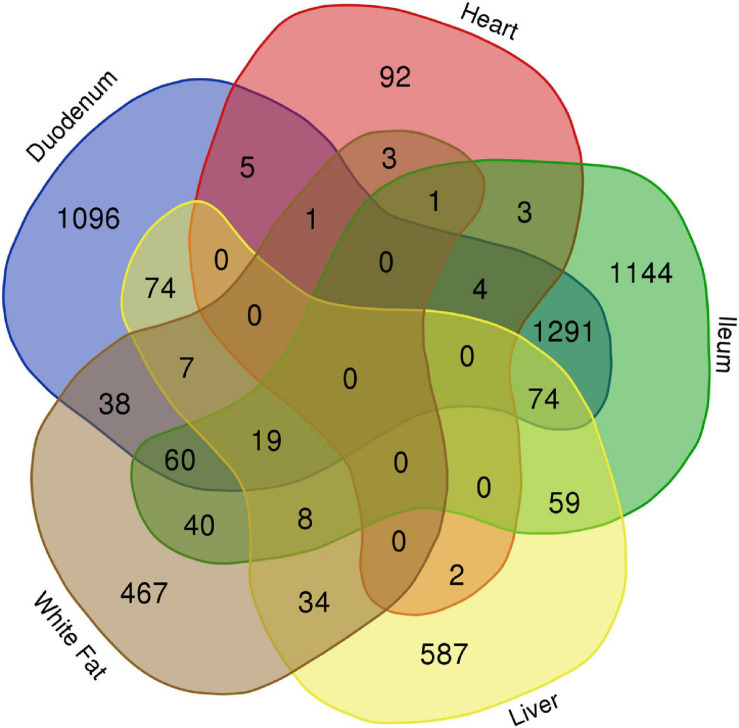
Alterations in gene expression in GF mice have been reported in several organs (Weger et al., 2019; Mills et al., 2020). In order to determine common and distinct transcriptional responses between different organs in GF mice, we compared the reported changes (GSE77221, GEO, NCBI) with those in our study. As shown in Figure 4, each organ displayed a distinct transcriptional profile. Among the total 5,109 significantly altered transcripts (|Fold Change| > 1.5 and FDR < 0.05) in the heart, liver, white adipose tissue, duodenum, and ileum of GF mice, 3,386 (66%) were tissue specific. The largest number of unique changes was observed in the ileum (1,144 genes), followed by duodenum (1,096 genes), liver (587 genes), white fat (467 genes), and heart (92 genes), possibly reflecting the relative vicinity of the respective organs to the abundant gut microbiota. No genes were identified that were altered in all five organs, indicating a lack of common tissue responses to the microbiota. Although the vast majority of cardiac genes with significant expression changes were unique to the heart under GF conditions (79%), the most overlap was seen with the duodenum (5 genes, 4.3% of total DEGs in the heart) and the least with duodenum (or ileum) and white fat (1 gene each, <1% of total DEGs in the heart). Interestingly, the gene cytochrome b-245, beta chain (Cybb) was commonly altered in heart and gut, suggesting that the lack of a microbiome alters the activity of immune cells, especially neutrophils, which may lead to immune dysregulation in these organs.
FIGURE 4.
Distinct transcriptional alterations in different organs of GF mice in normoxia. Little common changes were identified in the differentially expressed genes (DEGs) between the examined organs of the GF mice under normoxic condition, except a significant overlapping between the two gut segments (duodenum and ileum), demonstrating a distinct and organ-specific impact of microbiome on gene expression.
GF Condition Alters Transcriptional Responses to IHH in the Heart
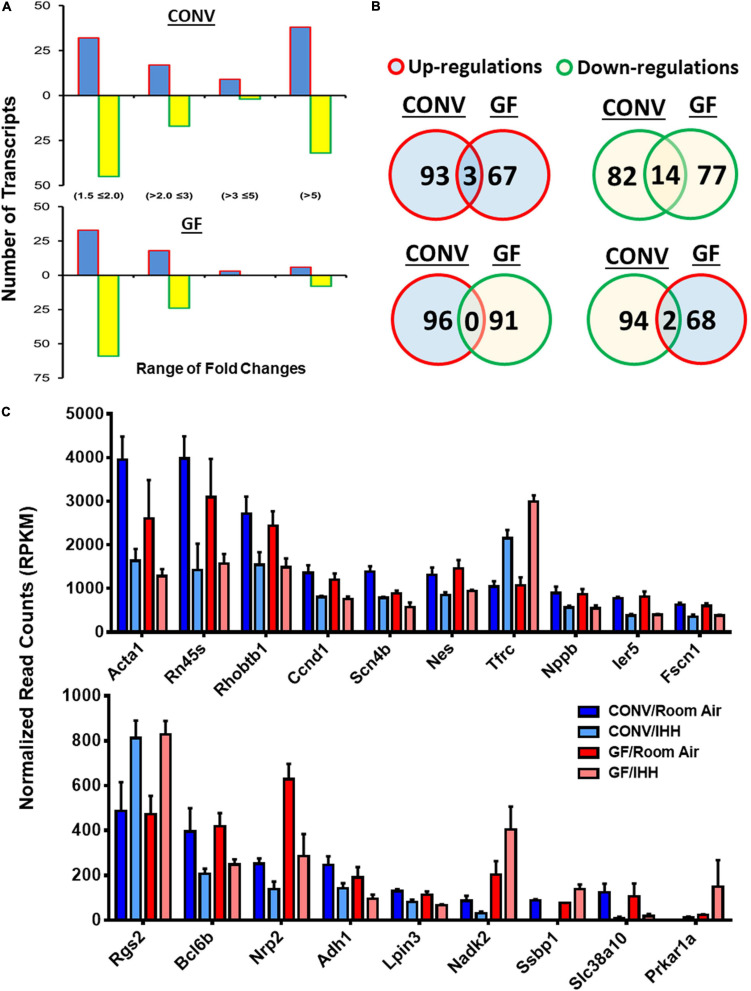
Previous studies have shown that stress can alter the composition of the gut microbiome, and in turn, such changes in gut microbiota may regulate stress response in various organs and tissues, including the central nervous system, the digestive system, and the immune system (Lyte et al., 2020; Park et al., 2020; Rengarajan et al., 2020; Tian et al., 2020; Vuong et al., 2020; Wang et al., 2020; Xu et al., 2020). The current study provides evidence that the microbiome may also affect stress responses in the heart through regulating transcription activities. Indeed, following 2 weeks of IHH exposure, we detected 192 significant changes in CONV mice (96 upregulated and 96 downregulated; | Fold Change| > 1.5 and FDR < 0.05) (Figure 5A and Supplementary Table 4) and 161 significant changes (70 upregulated and 91 downregulated; | Fold Change| > 1.5 and FDR < 0.05) in GF mice (Figure 5B and Supplementary Table 5). As shown in Table 1, the top DEGs in CONV mice include Rnf44, Cyb561a3, Cep250, Asph, Gm14296, and Epb41l1, whereas Kmt5a, Amd1, Tnnt2, Slc44a2, Golga5, and Cpeb3 were among the top changes in GF mice. Interestingly, only 19 transcripts were altered in both CONV and GF mice, representing around 10% of the changes in CONV and GF, respectively, demonstrating that IHH treatment induced distinct and specific transcriptional responses between CONV and GF mice. The majority of these common altered transcripts were downregulated, and the top three downregulated transcripts were Acta1 (Alpha actin 1), Rn45s (45S pre-ribosomal RNA), and Rhobtb1 (Rho-related BTB domain containing 1). On the other hand, Tfrc (transferrin receptor), Rgs2 (regulator of G-protein signaling 2), and Prkar1a (protein kinase, cAMP-dependent regulatory, type I, alpha) were the top three genes that were commonly upregulated in both CONV and GF mice. In contrast, two transcripts, Nadk2 (mitochondrial NAD kinase 2) and Ssbp1 (single-stranded DNA binding protein 1), were downregulated in CONV mice but upregulated in GF mice following IHH exposure (Figure 5C). Such dramatically different transcriptional responses suggested that IHH exposure may lead to different physiological and functional alterations in the heart of CONV and GF mice.
FIGURE 5.
Summary of IHH-induced differentially expressed genes (DEGs) in the hearts of CONV and GF mice. (A) Distribution of DEGs in GF and CONV mice following IHH exposure. Note that a significantly larger proportion of the alterations >5-fold was detected in the CONV mice as compared to that in the GF mice (p < 0.05, chi-square test). Blue bar: up-regulations; yellow bar: down-regulations. (B) IHH exposure-induced common transcriptional alteration between GF and CONV mice, where 3 DEGs were upregulated and 14 DEGs were downregulated in both CONV and GF mice. Two other DEGs were downregulated in CONV but upregulated in GF mice. Red circles with blue fill-in: upregulations; green circles with yellow fill-in: downregulations. Overlaps: number of common changes observed in both CONV and GF mice. (C) Transcriptional abundance of the common DEGs in GF and CONV mice in room air or under IHH condition (| Fold change| > 1.5, Benjamini–Hochberg adjusted p < 0.05).
TABLE 1.
Top transcriptional alterations in CONV and GF mice following IHH exposure.
| RefSeq_ID | Symbol | Normalized read counts | Fold change | Padj | |||||
| Air-1 | Air-2 | Air-3 | IHH-1 | IHH-2 | IHH-3 | ||||
| CONV Mice | |||||||||
| NR_027395 | Rnf44 | 272.4 | 260.7 | 211.7 | 0.0 | 0.0 | 0.0 | −1406.2 | 6.5E-15 |
| NM_201351 | Cyb561a3 | 158.4 | 127.1 | 150.7 | 0.0 | 0.0 | 0.0 | −827.7 | 1.8E-12 |
| NM_008383 | Cep250 | 141.4 | 111.9 | 138.3 | 0.0 | 0.0 | 0.0 | −740.1 | 6.2E-12 |
| NM_001286664 | Ssbp1 | 93.6 | 88.9 | 82.6 | 0.0 | 0.0 | 0.0 | −509.8 | 2.8E-10 |
| NM_001199248 | Kat5 | 123.6 | 107.7 | 17.6 | 0.0 | 0.0 | 0.0 | −471.9 | 8.8E-07 |
| NM_001163009 | Tnik | 62.4 | 73.3 | 97.8 | 0.0 | 0.0 | 0.0 | −443.1 | 2.6E-09 |
| NM_001204277 | Hdac7 | 81.2 | 89.8 | 54.1 | 0.0 | 0.0 | 0.0 | −422.7 | 5.2E-09 |
| NR_130970_1 | Gm14296 | 71.3 | 48.6 | 79.5 | 0.0 | 0.0 | 0.0 | −381.8 | 1.3E-08 |
| NM_008488 | Arhgef1 | 27.4 | 81.0 | 85.5 | 0.0 | 0.0 | 0.0 | −365.4 | 3.4E-07 |
| NM_008642 | Mttp | 61.8 | 46.2 | 74.7 | 0.0 | 0.0 | 0.0 | −349.2 | 3.0E-08 |
| NM_001347201 | Gucd1 | 0.0 | 0.0 | 0.0 | 14.4 | 28.5 | 43.9 | 150.7 | 2.3E-04 |
| NM_001012363 | Slc2a9 | 0.0 | 0.0 | 0.0 | 28.6 | 39.1 | 24.6 | 159.8 | 2.7E-05 |
| NM_001358355 | Zfx | 0.0 | 0.0 | 0.0 | 34.6 | 50.0 | 27.8 | 191.7 | 7.3E-06 |
| NM_178722 | Zfp438 | 0.0 | 0.0 | 0.0 | 38.2 | 42.6 | 55.3 | 234.8 | 6.2E-07 |
| NM_001167886 | Kmt5b | 0.0 | 0.0 | 0.0 | 48.3 | 70.0 | 21.8 | 241.4 | 7.3E-06 |
| NM_001284506 | Plxnb2 | 0.0 | 0.0 | 0.0 | 40.1 | 70.8 | 40.4 | 263.5 | 5.6E-07 |
| NM_001270495 | Tmem254a | 0.0 | 0.0 | 0.0 | 51.6 | 54.4 | 48.3 | 266.2 | 1.3E-07 |
| NM_001291122 | Epb41l1 | 0.0 | 0.0 | 0.0 | 100.8 | 47.7 | 33.0 | 314.3 | 8.4E-07 |
| NR_130970 | Gm14296 | 0.0 | 0.0 | 0.0 | 67.2 | 61.0 | 85.7 | 367.4 | 8.8E-09 |
| NM_001290367 | Asph | 0.0 | 0.0 | 0.0 | 168.1 | 41.5 | 83.9 | 502.3 | 3.9E-08 |
| GF Mice | |||||||||
| NM_001310725 | Kmt5a | 332.6 | 616.9 | 544.7 | 0.0 | 0.0 | 0.0 | −2582.6 | 1.9E-17 |
| NM_009665 | Amd1 | 321.6 | 421.3 | 295.8 | 0.0 | 0.0 | 0.0 | −1800.0 | 3.6E-16 |
| NM_001130180 | Tnnt2 | 0.0 | 69299.9 | 70470.5 | 61.6 | 13.2 | 12.4 | −1602.1 | 2.1E-03 |
| NM_134125 | Trip10 | 92.3 | 146.7 | 489.2 | 0.0 | 0.0 | 0.0 | −1251.8 | 1.0E-07 |
| NM_001102438 | Acbd5 | 107.8 | 132.9 | 183.7 | 0.0 | 0.0 | 0.0 | −734.7 | 1.1E-11 |
| NM_001360080 | Herc2 | 151.3 | 143.0 | 85.8 | 0.0 | 0.0 | 0.0 | −657.8 | 4.6E-11 |
| NM_001329143 | Adamts10 | 94.8 | 120.1 | 117.7 | 0.0 | 0.0 | 0.0 | −571.0 | 4.8E-11 |
| NM_001361696 | Slc38a10 | 170.4 | 58.5 | 88.2 | 10.3 | 29.6 | 12.4 | −6.1 | 9.7E-04 |
| NM_001039087 | Rapgef1 | 767.1 | 518.2 | 501.2 | 156.1 | 160.6 | 283.6 | −3.0 | 2.4E-06 |
| NM_001243837 | C7 | 290.4 | 258.7 | 163.4 | 80.0 | 95.4 | 69.2 | −2.9 | 3.2E-06 |
| NM_011638 | Tfrc | 1034.7 | 887.6 | 1268.0 | 2834.2 | 3037.6 | 3108.3 | 2.8 | 1.7E-28 |
| NM_001304810 | Kcnj3 | 37.4 | 11.6 | 29.9 | 79.4 | 115.0 | 66.8 | 3.3 | 1.7E-02 |
| NM_001277328 | Sox6 | 57.1 | 31.0 | 27.7 | 163.1 | 117.7 | 123.1 | 3.5 | 3.5E-05 |
| NM_001357298 | Tfrc | 11.3 | 65.4 | 25.6 | 136.5 | 133.8 | 147.1 | 4.1 | 1.3E-02 |
| NM_001313974 | Prkar1a | 26.0 | 24.4 | 23.2 | 256.7 | 24.7 | 170.0 | 6.1 | 1.1E-02 |
| NM_001355067 | Ctnnd1 | 0.0 | 0.0 | 0.0 | 145.9 | 152.4 | 18.6 | 600.8 | 1.7E-07 |
| NM_001199308 | Gm14440 | 0.0 | 0.0 | 0.0 | 133.6 | 156.4 | 63.1 | 670.1 | 2.0E-10 |
| NM_198300 | Cpeb3 | 0.0 | 0.0 | 0.0 | 185.3 | 184.8 | 41.5 | 785.4 | 1.5E-09 |
| NM_013747 | Golga5 | 0.0 | 0.0 | 0.0 | 151.5 | 128.8 | 200.1 | 912.2 | 9.2E-13 |
| NM_001199186 | Slc44a2 | 0.0 | 0.0 | 0.0 | 222.6 | 265.2 | 248.5 | 1400.1 | 3.3E-15 |
Distinct Alterations in Cell Signaling in Response to IHH Exposure Between CONV and GF Mice
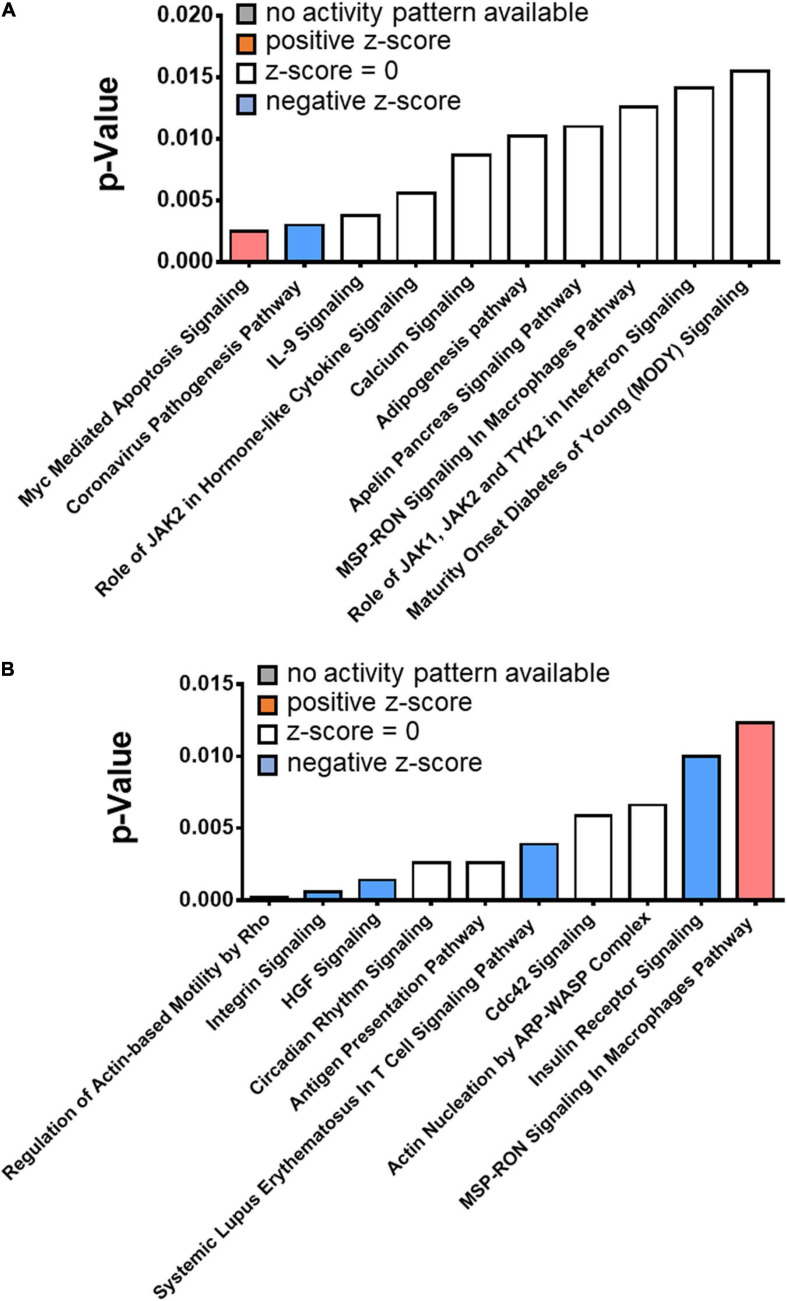
We identified distinct cell signaling mechanisms in the hearts of CONV and GF mice in response to IHH. In the CONV mice, the top significantly altered transcripts encoding a group of nuclear proteins [66/192 (34%), FDR < 0.05] that are involved in “DNA-templated transcription” [GO:0006351, 42/192 (22%), FDR < 0.05] and “regulation of DNA-templated transcription” [GO:0006355, 46/192 (24%), FDR < 0.05] (Supplementary Table 6A). The top altered signaling mechanisms were Myc mediated apoptosis signaling, coronavirus pathogenesis pathway, and IL-9 signaling (Figure 6A and Supplementary Table 6B). In GF mice, on the other hand, the significantly altered genes were involved in the biological processes that regulate cell cycle (GO:0051726, including some cyclins, e.g., Cdkn1a, Ccnd1, and Cables1, FDR < 0.05), biological rhythms (FDR < 0.05), and MHC class II protein complex (GO:0042613, FDR < 0.05) (Supplementary Table 7A). The top representative pathways were “regulation of actin-based motility by Rho,” “integrin signaling,” “HGF signaling,” and “circadian rhythm signaling” (Figure 6B and Supplementary Table 7B).
FIGURE 6.
Top significantly altered pathways in the heart of CONV and GF mice following intermittent hypoxia/hypercapnia exposure. The canonical pathways that were significantly altered by IHH exposure in CONV and GF mice were determined with IPA. (A) The top 10 significantly altered pathways in CONV mice. (B) The top 10 significantly altered pathways in GF mice. The statistical significance was calculated with right-tailed Fisher’s exact test; a p-value < 0.05 was considered significant.
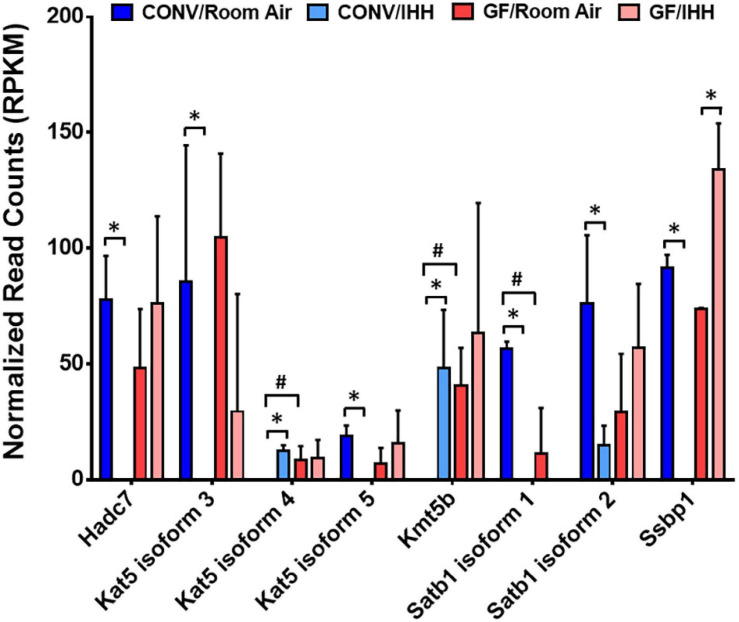
Following IHH exposure, “regulation of DNA-templated transcription” was one of the most significantly altered biological processes represented by 24% of the significantly altered transcripts in CONV mice. These results suggested a fine-tuning of the transcriptome, at least in part, through epigenetic modifications and rebalance of transcription regulators. Indeed, as shown in Figure 7, transcripts encoding several epigenetic modifiers were significantly changed in the CONV mice, but not GF mice, following IHH treatment. In contrast, in the heart of the GF mouse, IHH-induced transcriptional alterations suggested an array of functional/activity adjustments, such as the circadian rhythm and cell cycle genes (e.g., Per2, Clock, Cdkn1a, and Stat3) that regulate the heart rate (Yamamura et al., 2010) or the metabolism, hypertrophy, and apoptosis of cardiac cells (Bonney et al., 2013; Hauck et al., 2016; Harhous et al., 2019).
FIGURE 7.
Intermittent hypoxia/hypercapnia exposure-induced changes of epigenomic modifiers in the heart of CONV mice. The expressional abundance of the differentially expressed genes (DEGs) was presented as mean ± SD for each condition. *: DEGs in CONV mice under IHH (| Fold Change| > 1.5 and Benjamini–Hochberg adjusted p < 0.05). #: DEGs between GF and CONV mice under room air condition (| Fold Change| > 1.5, Benjamini–Hochberg adjusted p < 0.05).
Transcriptomic Indications of Potential Cardiac Disorders in GF and CONV Mice Following IHH Treatment
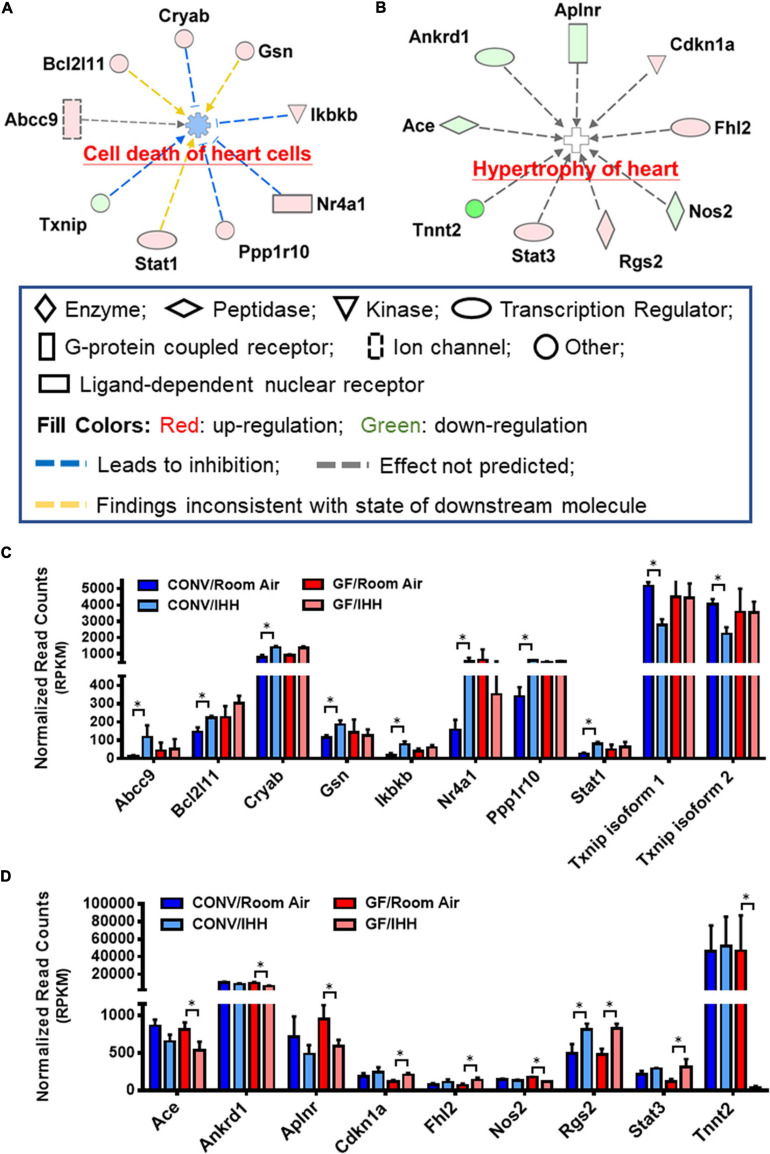
Substantial evidence has been reported that OSA significantly increases the risk of stroke, coronary heart disease, and heart failure in humans (McEvoy et al., 2016; Drager and Lee, 2018; Benjamin et al., 2019). Interestingly, we found that the IHH-induced transcriptional alterations in the GF and CONV mice suggested distinct risks of cardiac disorders, i.e., IHH exposure evoked different sets of well-characterized pathogenetic genes in GF and CONV mice. As shown in Table 2 and Figure 8, in CONV mice, IHH-induced transcriptional alterations mainly affect cardiac necrosis and cell death. In contrast, in the GF mice, the DEGs included several key regulators of cardiac hypertrophy. In the CONV mice, changes of both well-characterized cell death stimulators and inhibitors were detected, which include BCL2-like 1 (Bcl2l1), Crystallin alpha B (Cryab), Gelsolin (Gsn), Inhibitor of nuclear factor kappa B kinase subunit beta (Ikbkb), Nuclear receptor subfamily 4 group A member 1 (Nr4a1), Protein phosphatase 1 regulatory subunit 10 (Ppp1r10), and Signal transducer and activator of transcription 1 (Stat1), as well as the downregulation of Thioredoxin interacting protein (Txnip) [for selected reviews see Wagner and Siddiqui (2009), Li et al. (2012), Ganguly et al. (2014), Wang and Yoshioka (2017), Opferman and Kothari (2018), Herring et al. (2019), Tang et al. (2019)]. In the GF mice, however, most of the disease-related DEGs were cardiac hypertrophy regulators, such as the downregulation of Angiotensin I converting enzyme (Ace), Ankyrin repeat domain 1 (Ankrd1), Apelin receptor (Aplnr), Nitric oxide synthase 2 (Nos2), and Cardiac type Troponin T2 (Tnnt2), as well as the upregulation of Cyclin-dependent kinase inhibitor 1A (Cdkn1a), Four and a half LIM domains 2 (Fhl2), Regulator of G protein signaling 2 (Rgs2), and Signal transducer and activator of transcription 3 (Stat3) [for selected reviews, see Brooks et al. (1998), Drexler (1999), Takeda et al. (2005), Tsang et al. (2010), Haghikia et al. (2011), Liang et al. (2018), Yotti et al. (2019)]. Since several genes in this group are key regulators of hypertension (e.g., Ace, Nos2, and Rgs2) (Li and Forstermann, 2000; Prichard et al., 2001; Tsang et al., 2010), it is reasonable to hypothesize that dysregulation of blood pressure might be an important mechanism regulating IHH-induced hypertrophy in GF mice.
TABLE 2.
Transcriptome indication of potential IHH-induced cardiac disorders in GF and CONV mice.
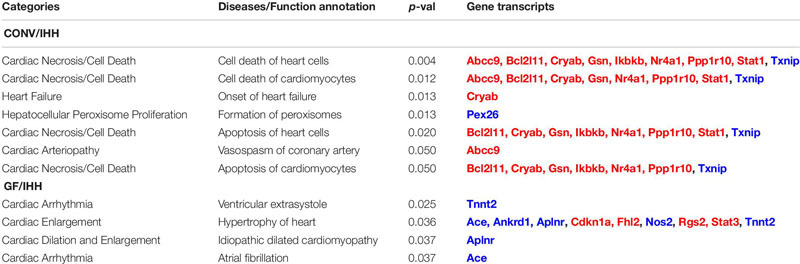 |
Red font: Upregulation. Blue font: Downregulation.
FIGURE 8.
Different set of cardiovascular diseases-related genes was altered in the heart of CONV and GF mice following IHH exposure. Prediction of clinical pathology endpoints driven by IHH-induced differentially expressed genes (DEGs) suggested a significant alteration of cardiac cell death in CONV (A) and cardiac hypertrophy in GF mice (B) under IHH condition (right-tailed Fisher’s exact test, p-value < 0.05). (C,D) Expressional abundance of the related DEGs in CONV and GF mice, respectively (* | Fold Change| > 1.5, Benjamini–Hochberg adjusted p < 0.05).
In summary, our study demonstrated that the microbiome regulates the cardiac transcriptome at baseline and in response to IHH stress. A total of 117 DEGs were identified in the heart of GF mice under normoxic condition. These DEFs encode proteins involved in transcriptional regulation, alpha-actinin binding, and sarcolemma organization. Lack of microbiome also altered cardiac transcriptional responses to IHH stress, since we found 192 IHH-induced DEGs in CONV mice and 161 DEGs in GF mice with little overlap, suggesting that IHH exposure leads to different cardiac responses between CONV and GF mice. Indeed, 24% of the DEGs detected in CONV mice are involved in the “regulation of DNA-templated transcription,” while in GF mice, IHH-induced cardiac DEGs are mostly involved in functional or activity adjustments, such as circadian rhythm and cell cycle. Future functional studies will be needed to define the functional implications of the observed DEGs. For example, IHH-induced DEGs suggest distinct risks of cardiac disorders in GF and CONV mice, since disease-related DEGs mainly affected cardiac necrosis and cell death in CONV mice, while regulators of cardiac hypertrophy were mostly observed in GF mice. Additional studies are also warranted to explore the function of specific microbial species and their metabolites on regulating cardiac gene expression under baseline and stress conditions. Gnotobiotic mouse models are uniquely suited to unravel such microbial contributions to specific physiologic responses.
Conclusion
Even though it has been shown that OSA or IHH exposure may lead to the development of various types of cardiac disorders [for selected reviews, see Baguet et al. (2012), Dumitrascu et al. (2013), Levy et al. (2015), Turnbull (2018)], any contribution of the microbiome to this process is still largely uncharacterized. In the present study, we found that the microbiome plays an important role in the transcriptional responses to IHH stress and identified distinct responsive mechanisms in the heart of GF and CONV mice, suggesting that the microbiome modulates the cardiac function during stressful and disease conditions like OSA. Our study provides insight into the unique and common transcriptional response to IHH that is related to microbial colonization in mice. These transcriptional alterations suggest changes in cellular signaling and interaction networks regulating cardiac function, such as cardiac hypertrophy and cell death. Overall, our study is a step toward a better understanding of the role of microbiota in regulating cardiac health and disease.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics Statement
The animal study was reviewed and approved by the Animal Care Committee of the University of California, San Diego.
Author Contributions
DZ, JX, LE, and GH conceptualized the study, designed the experiments, and wrote the manuscript. DZ, JX, YM, and OP performed the experiments. DZ and JX performed data curation and analysis. LE and GH acquired funding and facilitated access to resources for the study. GH supervised the study. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Iain Hartley and Travis Smith for technical support.
Abbreviations
- OSA
obstructive sleep apnea
- IHH
intermittent hypoxia and hypercapnia
- GF
germ-free
- CONV
conventional
- LV
left ventricle
- HEPA
high-efficiency particulate air
- DAVID
Database for Annotation Visualization and Integrated Discovery
- IPA
Ingenuity Pathway Analysis
- KEGG
Kyoto Encyclopedia of Genes and Genomes.
Funding. This work was partially supported by the NIH grant P30 DK120515 (LE).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2021.680275/full#supplementary-material
References
- Al Lawati N. M., Patel S. R., Ayas N. T. (2009). Epidemiology, risk factors, and consequences of obstructive sleep apnea and short sleep duration. Prog. Cardiovasc. Dis. 51 285–293. 10.1016/j.pcad.2008.08.001 [DOI] [PubMed] [Google Scholar]
- Anders S., Huber W. (2010). Differential expression analysis for sequence count data. Genome Biol. 11:R106. 10.1186/gb-2010-11-10-r106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baguet J. P., Barone-Rochette G., Tamisier R., Levy P., Pepin J. L. (2012). Mechanisms of cardiac dysfunction in obstructive sleep apnea. Nat. Rev. Cardiol. 9 679–688. 10.1038/nrcardio.2012.141 [DOI] [PubMed] [Google Scholar]
- Benjamin E. J., Muntner P., Alonso A., Bittencourt M. S., Callaway C. W., Carson A. P., et al. (2019). Heart Disease and Stroke Statistics-2019 Update: a Report From the American Heart Association. Circulation 139 e56–e528. 10.1161/CIR.0000000000000659 [DOI] [PubMed] [Google Scholar]
- Bonney S., Kominsky D., Brodsky K., Eltzschig H., Walker L., Eckle T. (2013). Cardiac Per2 functions as novel link between fatty acid metabolism and myocardial inflammation during ischemia and reperfusion injury of the heart. PLoS One 8:e71493. 10.1371/journal.pone.0071493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks G., Poolman R. A., Li J. M. (1998). Arresting developments in the cardiac myocyte cell cycle: role of cyclin-dependent kinase inhibitors. Cardiovasc. Res. 39 301–311. 10.1016/s0008-6363(98)00125-4 [DOI] [PubMed] [Google Scholar]
- Brunt V. E., Gioscia-Ryan R. A., Richey J. J., Zigler M. C., Cuevas L. M., Gonzalez A., et al. (2019). Suppression of the gut microbiome ameliorates age-related arterial dysfunction and oxidative stress in mice. J. Physiol. 597 2361–2378. 10.1113/JP277336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buniello A., Macarthur J. A. L., Cerezo M., Harris L. W., Hayhurst J., Malangone C., et al. (2019). The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res. 47 D1005–D1012. 10.1093/nar/gky1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu C., Murdock M. H., Jing D., Won T. H., Chung H., Kressel A. M., et al. (2019). The microbiota regulate neuronal function and fear extinction learning. Nature 574 543–548. 10.1038/s41586-019-1644-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clerk A., Cullingford T. E., Fuller S. J., Giraldo A., Markou T., Pikkarainen S., et al. (2007). Signaling pathways mediating cardiac myocyte gene expression in physiological and stress responses. J. Cell Physiol. 212 311–322. 10.1002/jcp.21094 [DOI] [PubMed] [Google Scholar]
- Dejea C. M., Wick E. C., Hechenbleikner E. M., White J. R., Mark Welch J. L., Rossetti B. J., et al. (2014). Microbiota organization is a distinct feature of proximal colorectal cancers. Proc. Natl. Acad. Sci. U. S. A. 111 18321–18326. 10.1073/pnas.1406199111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis G., Jr., Sherman B. T., Hosack D. A., Yang J., Gao W., Lane H. C., et al. (2003). DAVID: database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 4:P3. [PubMed] [Google Scholar]
- Didion J. P., Martin M., Collins F. S. (2017). Atropos: specific, sensitive, and speedy trimming of sequencing reads. PeerJ 5:e3720. 10.7717/peerj.3720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A., Davis C. A., Schlesinger F., Drenkow J., Zaleski C., Jha S., et al. (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29 15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez-Bello M. G., Godoy-Vitorino F., Knight R., Blaser M. J. (2019). Role of the microbiome in human development. Gut 68 1108–1114. 10.1136/gutjnl-2018-317503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drager L. F., Lee C. H. (2018). Treatment of obstructive sleep apnoea as primary or secondary prevention of cardiovascular disease: where do we stand now? Curr. Opin. Pulm. Med. 24 537–542. 10.1097/MCP.0000000000000523 [DOI] [PubMed] [Google Scholar]
- Drager L. F., Polotsky V. Y., O’donnell C. P., Cravo S. L., Lorenzi-Filho G., Machado B. H. (2015). Translational approaches to understanding metabolic dysfunction and cardiovascular consequences of obstructive sleep apnea. Am. J. Physiol. Heart Circ. Physiol. 309 H1101–H1111. 10.1152/ajpheart.00094.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drexler H. (1999). Nitric oxide synthases in the failing human heart: a doubled-edged sword? Circulation 99 2972–2975. 10.1161/01.cir.99.23.2972 [DOI] [PubMed] [Google Scholar]
- Dumitrascu R., Heitmann J., Seeger W., Weissmann N., Schulz R. (2013). Obstructive sleep apnea, oxidative stress and cardiovascular disease: lessons from animal studies. Oxid. Med. Cell. Longev. 2013:234631. 10.1155/2013/234631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabregat A., Sidiropoulos K., Viteri G., Forner O., Marin-Garcia P., Arnau V., et al. (2017). Reactome pathway analysis: a high-performance in-memory approach. BMC Bioinformatics 18:142. 10.1186/s12859-017-1559-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan C., Iacobas D. A., Zhou D., Chen Q., Lai J. K., Gavrialov O., et al. (2005). Gene expression and phenotypic characterization of mouse heart after chronic constant or intermittent hypoxia. Physiol. Genomics 22 292–307. 10.1152/physiolgenomics.00217.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farhana L., Banerjee H. N., Verma M., Majumdar A. P. N. (2018). Role of Microbiome in Carcinogenesis Process and Epigenetic Regulation of Colorectal Cancer. Methods Mol. Biol. 1856 35–55. 10.1007/978-1-4939-8751-1_3 [DOI] [PubMed] [Google Scholar]
- Ganguly S., Mitra A., Sarkar S. (2014). Role of alpha-crystallin B in regulation of stress induced cardiomyocyte apoptosis. Cardiovasc. Hematol. Agents Med. Chem. 12 60–65. 10.2174/1871525713666150123151731 [DOI] [PubMed] [Google Scholar]
- Gilbert J. A., Blaser M. J., Caporaso J. G., Jansson J. K., Lynch S. V., Knight R. (2018). Current understanding of the human microbiome. Nat. Med. 24 392–400. 10.1038/nm.4517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan F., Yang X., Li J., Dong W., Zhang X., Liu N., et al. (2019). New Molecular Mechanism Underlying Myc-Mediated Cytochrome P450 2E1 Upregulation in Apoptosis and Energy Metabolism in the Myocardium. J. Am. Heart Assoc. 8:e009871. 10.1161/JAHA.118.009871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haghikia A., Stapel B., Hoch M., Hilfiker-Kleiner D. (2011). STAT3 and cardiac remodeling. Heart Fail. Rev. 16 35–47. 10.1007/s10741-010-9170-x [DOI] [PubMed] [Google Scholar]
- Harhous Z., Booz G. W., Ovize M., Bidaux G., Kurdi M. (2019). An Update on the Multifaceted Roles of STAT3 in the Heart. Front. Cardiovasc. Med. 6:150. 10.3389/fcvm.2019.00150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harikrishnan S. (2019). Diet, the Gut Microbiome and Heart Failure. Card. Fail. Rev. 5 119–122. 10.15420/cfr.2018.39.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauck L., Grothe D., Billia F. (2016). p21(CIP1/WAF1)-dependent inhibition of cardiac hypertrophy in response to Angiotensin II involves Akt/Myc and pRb signaling. Peptides 83 38–48. 10.1016/j.peptides.2016.07.003 [DOI] [PubMed] [Google Scholar]
- Helmink B. A., Khan M. A. W., Hermann A., Gopalakrishnan V., Wargo J. A. (2019). The microbiome, cancer, and cancer therapy. Nat. Med. 25 377–388. 10.1038/s41591-019-0377-7 [DOI] [PubMed] [Google Scholar]
- Herring J. A., Elison W. S., Tessem J. S. (2019). Function of Nr4a Orphan Nuclear Receptors in Proliferation, Apoptosis and Fuel Utilization Across Tissues. Cells 8:1373. 10.3390/cells8111373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou N., Ye B., Li X., Margulies K. B., Xu H., Wang X., et al. (2016). Transcription Factor 7-like 2 Mediates Canonical Wnt/beta-Catenin Signaling and c-Myc Upregulation in Heart Failure. Circ. Heart Fail. 9:e003010. 10.1161/CIRCHEARTFAILURE.116.003010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyos C. M., Drager L. F., Patel S. R. (2017). OSA and cardiometabolic risk: what’s the bottom line? Respirology 22 420–429. 10.1111/resp.12984 [DOI] [PubMed] [Google Scholar]
- Huang D. W., Sherman B. T., Tan Q., Collins J. R., Alvord W. G., Roayaei J., et al. (2007). The DAVID Gene Functional Classification Tool: a novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol. 8:R183. 10.1186/gb-2007-8-9-r183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunyor I., Cook K. M. (2018). Models of intermittent hypoxia and obstructive sleep apnea: molecular pathways and their contribution to cancer. Am. J. Physiol. Regul. Integr. Comp. Physiol. 315 R669–R687. 10.1152/ajpregu.00036.2018 [DOI] [PubMed] [Google Scholar]
- Kanaan A., Farahani R., Douglas R. M., Lamanna J. C., Haddad G. G. (2006). Effect of chronic continuous or intermittent hypoxia and reoxygenation on cerebral capillary density and myelination. Am. J. Physiol. Regul. Integr. Comp. Physiol. 290 R1105–R1114. 10.1152/ajpregu.00535.2005 [DOI] [PubMed] [Google Scholar]
- Kanehisa M., Goto S. (2000). KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28 27–30. 10.1093/nar/28.1.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karbach S. H., Schonfelder T., Brandao I., Wilms E., Hormann N., Jackel S., et al. (2016). Gut Microbiota Promote Angiotensin II-Induced Arterial Hypertension and Vascular Dysfunction. J. Am. Heart Assoc. 5:e003698. 10.1161/JAHA.116.003698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellermayer R., Balasa A., Zhang W., Lee S., Mirza S., Chakravarty A., et al. (2010). Epigenetic maturation in colonic mucosa continues beyond infancy in mice. Hum. Mol. Genet. 19 2168–2176. 10.1093/hmg/ddq095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch C. D., Gladwin M. T., Freeman B. A., Lundberg J. O., Weitzberg E., Morris A. (2017). Enterosalivary nitrate metabolism and the microbiome: intersection of microbial metabolism, nitric oxide and diet in cardiac and pulmonary vascular health. Free Radic. Biol. Med. 105 48–67. 10.1016/j.freeradbiomed.2016.12.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolodziejczyk A. A., Federici S., Zmora N., Mohapatra G., Dori-Bachash M., Hornstein S., et al. (2020). Acute liver failure is regulated by MYC- and microbiome-dependent programs. Nat. Med. 26 1899–1911. 10.1038/s41591-020-1102-2 [DOI] [PubMed] [Google Scholar]
- Levy P., Kohler M., Mcnicholas W. T., Barbe F., Mcevoy R. D., Somers V. K., et al. (2015). Obstructive sleep apnoea syndrome. Nat. Rev. Dis. Primers 1:15015. 10.1038/nrdp.2015.15 [DOI] [PubMed] [Google Scholar]
- Levy P., Ryan S., Oldenburg O., Parati G. (2013). Sleep apnoea and the heart. Eur. Respir. Rev. 22 333–352. 10.1183/09059180.00004513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G. H., Arora P. D., Chen Y., Mcculloch C. A., Liu P. (2012). Multifunctional roles of gelsolin in health and diseases. Med. Res. Rev. 32 999–1025. 10.1002/med.20231 [DOI] [PubMed] [Google Scholar]
- Li H., Forstermann U. (2000). Nitric oxide in the pathogenesis of vascular disease. J. Pathol. 190 244–254. [DOI] [PubMed] [Google Scholar]
- Liang Y., Bradford W. H., Zhang J., Sheikh F. (2018). Four and a half LIM domain protein signaling and cardiomyopathy. Biophys. Rev. 10 1073–1085. 10.1007/s12551-018-0434-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucking E. F., O’connor K. M., Strain C. R., Fouhy F., Bastiaanssen T. F. S., Burns D. P., et al. (2018). Chronic intermittent hypoxia disrupts cardiorespiratory homeostasis and gut microbiota composition in adult male guinea-pigs. EBioMedicine 38 191–205. 10.1016/j.ebiom.2018.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch S. V., Pedersen O. (2016). The Human Intestinal Microbiome in Health and Disease. N. Engl. J. Med. 375 2369–2379. 10.1056/NEJMra1600266 [DOI] [PubMed] [Google Scholar]
- Lyons O. D., Bradley T. D. (2015). Heart Failure and Sleep Apnea. Can. J. Cardiol. 31 898–908. 10.1016/j.cjca.2015.04.017 [DOI] [PubMed] [Google Scholar]
- Lyte J. M., Gheorghe C. E., Goodson M. S., Kelley-Loughnane N., Dinan T. G., Cryan J. F., et al. (2020). Gut-brain axis serotonergic responses to acute stress exposure are microbiome-dependent. Neurogastroenterol. Motil. 32:e13881. 10.1111/nmo.13881 [DOI] [PubMed] [Google Scholar]
- Manichanh C., Borruel N., Casellas F., Guarner F. (2012). The gut microbiota in IBD. Nat. Rev. Gastroenterol. Hepatol. 9 599–608. 10.1038/nrgastro.2012.152 [DOI] [PubMed] [Google Scholar]
- May A. M., Van Wagoner D. R., Mehra R. (2017). OSA and Cardiac Arrhythmogenesis: mechanistic Insights. Chest 151 225–241. 10.1016/j.chest.2016.09.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEvoy R. D., Antic N. A., Heeley E., Luo Y., Ou Q., Zhang X., et al. (2016). CPAP for Prevention of Cardiovascular Events in Obstructive Sleep Apnea. N. Engl. J. Med. 375 919–931. 10.1056/NEJMoa1606599 [DOI] [PubMed] [Google Scholar]
- Mills R. H., Wozniak J. M., Vrbanac A., Campeau A., Chassaing B., Gewirtz A., et al. (2020). Organ-level protein networks as a reference for the host effects of the microbiome. Genome Res. 30 276–286. 10.1101/gr.256875.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moody L. V., Miyamoto Y., Ang J., Richter P. J., Eckmann L. (2019). Evaluation of Peroxides and Chlorine Oxides as Disinfectants for Chemical Sterilization of Gnotobiotic Rodent Isolators. J. Am. Assoc. Lab. Anim. Sci. 58 558–568. 10.30802/AALAS-JAALAS-18-000130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Indias I., Torres M., Montserrat J. M., Sanchez-Alcoholado L., Cardona F., Tinahones F. J., et al. (2015). Intermittent hypoxia alters gut microbiota diversity in a mouse model of sleep apnoea. Eur. Respir. J. 45 1055–1065. 10.1183/09031936.00184314 [DOI] [PubMed] [Google Scholar]
- Moreno-Indias I., Torres M., Sanchez-Alcoholado L., Cardona F., Almendros I., Gozal D., et al. (2016). Normoxic Recovery Mimicking Treatment of Sleep Apnea Does Not Reverse Intermittent Hypoxia-Induced Bacterial Dysbiosis and Low-Grade Endotoxemia in Mice. Sleep 39 1891–1897. 10.5665/sleep.6176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagatomo Y., Tang W. H. (2015). Intersections Between Microbiome and Heart Failure: revisiting the Gut Hypothesis. J. Card. Fail. 21 973–980. 10.1016/j.cardfail.2015.09.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama H., Fujio Y., Yamaguchi O. (2013). [Calcium dependent signaling in cardiac hypertrophy and cell death]. Clin. Calcium 23 505–515. CliCa1304505515, [PubMed] [Google Scholar]
- Nemet I., Saha P. P., Gupta N., Zhu W., Romano K. A., Skye S. M., et al. (2020). A Cardiovascular Disease-Linked Gut Microbial Metabolite Acts via Adrenergic Receptors. Cell 180 862–877.e22. 10.1016/j.cell.2020.02.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata H., Goto S., Sato K., Fujibuchi W., Bono H., Kanehisa M. (1999). KEGG: kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 27 29–34. 10.1093/nar/27.1.29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opferman J. T., Kothari A. (2018). Anti-apoptotic BCL-2 family members in development. Cell Death Differ. 25 37–45. 10.1038/cdd.2017.170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ordovas J. M., Mooser V. (2006). Metagenomics: the role of the microbiome in cardiovascular diseases. Curr. Opin. Lipidol. 17 157–161. 10.1097/01.mol.0000217897.75068.ba [DOI] [PubMed] [Google Scholar]
- Park Y. S., Kim S. H., Park J. W., Kho Y., Seok P. R., Shin J. H., et al. (2020). Melatonin in the colon modulates intestinal microbiota in response to stress and sleep deprivation. Intest. Res. 18 325–336. 10.5217/ir.2019.00093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrignani P., Tacconelli S., Bruno A. (2014). Gut microbiota, host gene expression, and aging. J. Clin. Gastroenterol. 48 S28–S31. 10.1097/MCG.0000000000000229 [DOI] [PubMed] [Google Scholar]
- Paul B., Barnes S., Demark-Wahnefried W., Morrow C., Salvador C., Skibola C., et al. (2015). Influences of diet and the gut microbiome on epigenetic modulation in cancer and other diseases. Clin. Epigenetics 7:112. 10.1186/s13148-015-0144-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peuker K., Muff S., Wang J., Kunzel S., Bosse E., Zeissig Y., et al. (2016). Epithelial calcineurin controls microbiota-dependent intestinal tumor development. Nat. Med. 22 506–515. 10.1038/nm.4072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pevsner-Fischer M., Zmora N., Braverman S., Elinav E. (2017). “Epigenetics and the Microbiome,” in Handbook of Nutrition, Diet, and Epigenetics, eds Patel V., Preedy V. (Cham: Springer International Publishing; ), 1–25. 10.1007/978-3-319-31143-2_35-1 [DOI] [Google Scholar]
- Prichard B. N., Cruickshank J. M., Graham B. R. (2001). Beta-adrenergic blocking drugs in the treatment of hypertension. Blood Press. 10 366–386. 10.1080/080370501753400665 [DOI] [PubMed] [Google Scholar]
- Qin Y., Wade P. A. (2018). Crosstalk between the microbiome and epigenome: messages from bugs. J. Biochem. 163 105–112. 10.1093/jb/mvx080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rengarajan S., Knoop K. A., Rengarajan A., Chai J. N., Grajales-Reyes J. G., Samineni V. K., et al. (2020). A Potential Role for Stress-Induced Microbial Alterations in IgA-Associated Irritable Bowel Syndrome with Diarrhea. Cell Rep. Med. 1:100124. 10.1016/j.xcrm.2020.100124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards A. L., Muehlbauer A. L., Alazizi A., Burns M. B., Findley A., Messina F., et al. (2019). Gut Microbiota Has a Widespread and Modifiable Effect on Host Gene Regulation. mSystems 4:e00323. 10.1128/mSystems.00323-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohini A., Agrawal N., Koyani C. N., Singh R. (2010). Molecular targets and regulators of cardiac hypertrophy. Pharmacol. Res. 61 269–280. 10.1016/j.phrs.2009.11.012 [DOI] [PubMed] [Google Scholar]
- Sandek A., Bauditz J., Swidsinski A., Buhner S., Weber-Eibel J., Von Haehling S., et al. (2007). Altered intestinal function in patients with chronic heart failure. J. Am. Coll. Cardiol. 50 1561–1569. 10.1016/j.jacc.2007.07.016 [DOI] [PubMed] [Google Scholar]
- Sfanos K. S., Yegnasubramanian S., Nelson W. G., De Marzo A. M. (2018). The inflammatory microenvironment and microbiome in prostate cancer development. Nat. Rev. Urol. 15 11–24. 10.1038/nrurol.2017.167 [DOI] [PubMed] [Google Scholar]
- Sforza E., Roche F. (2016). Chronic intermittent hypoxia and obstructive sleep apnea: an experimental and clinical approach. Hypoxia 4 99–108. 10.2147/HP.S103091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K., Sugi Y., Nakano K., Tsuda M., Kurihara K., Hosono A., et al. (2011). Epigenetic control of the host gene by commensal bacteria in large intestinal epithelial cells. J. Biol. Chem. 286 35755–35762. 10.1074/jbc.M111.271007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda T., Otsu K., Hori M. (2005). [Calcium channel blocker and cardiac hypertrophy]. Clin. Calcium 15 1645–1649. CliCa051016451649, [PubMed] [Google Scholar]
- Tang D., Kang R., Berghe T. V., Vandenabeele P., Kroemer G. (2019). The molecular machinery of regulated cell death. Cell Res. 29 347–364. 10.1038/s41422-019-0164-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Team N. I. H. H. M. P. A. (2019). A review of 10 years of human microbiome research activities at the US National Institutes of Health, Fiscal Years 2007-2016. Microbiome 7:31. 10.1186/s40168-019-0620-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian P., O’riordan K. J., Lee Y. K., Wang G., Zhao J., Zhang H., et al. (2020). Towards a psychobiotic therapy for depression: bifidobacterium breve CCFM1025 reverses chronic stress-induced depressive symptoms and gut microbial abnormalities in mice. Neurobiol. Stress 12:100216. 10.1016/j.ynstr.2020.100216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi A., Melnik A. V., Xue J., Poulsen O., Meehan M. J., Humphrey G., et al. (2018). Intermittent Hypoxia and Hypercapnia, a Hallmark of Obstructive Sleep Apnea, Alters the Gut Microbiome and Metabolome. mSystems 3 e00020–18. 10.1128/mSystems.00020-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi A., Xu Z. Z., Xue J., Poulsen O., Gonzalez A., Humphrey G., et al. (2019). Intermittent Hypoxia and Hypercapnia Reproducibly Change the Gut Microbiome and Metabolome across Rodent Model Systems. mSystems 4 e00058–19. 10.1128/mSystems.00058-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang S., Woo A. Y., Zhu W., Xiao R. P. (2010). Deregulation of RGS2 in cardiovascular diseases. Front. Biosci. 2:547–557. 10.2741/s84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbull C. D. (2018). Intermittent hypoxia, cardiovascular disease and obstructive sleep apnoea. J. Thorac. Dis. 10 S33–S39. 10.21037/jtd.2017.10.33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veeraveedu P. T., Sanada S., Okuda K., Fu H. Y., Matsuzaki T., Araki R., et al. (2017). Ablation of IL-33 gene exacerbate myocardial remodeling in mice with heart failure induced by mechanical stress. Biochem. Pharmacol. 138 73–80. 10.1016/j.bcp.2017.04.022 [DOI] [PubMed] [Google Scholar]
- Vinje S., Stroes E., Nieuwdorp M., Hazen S. L. (2014). The gut microbiome as novel cardio-metabolic target: the time has come! Eur. Heart J. 35 883–887. 10.1093/eurheartj/eht467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuong H. E., Pronovost G. N., Williams D. W., Coley E. J. L., Siegler E. L., Qiu A., et al. (2020). The maternal microbiome modulates fetal neurodevelopment in mice. Nature 586 281–286. 10.1038/s41586-020-2745-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner M., Siddiqui M. A. (2009). Signaling networks regulating cardiac myocyte survival and death. Curr. Opin. Investig. Drugs 10 928–937. [PubMed] [Google Scholar]
- Wang B. F., Yoshioka J. (2017). The Emerging Role of Thioredoxin-Interacting Protein in Myocardial Ischemia/Reperfusion Injury. J. Cardiovasc. Pharmacol. Ther. 22 219–229. 10.1177/1074248416675731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R. X., Lee J. S., Campbell E. L., Colgan S. P. (2020). Microbiota-derived butyrate dynamically regulates intestinal homeostasis through regulation of actin-associated protein synaptopodin. Proc. Natl. Acad. Sci. U. S. A. 117 11648–11657. 10.1073/pnas.1917597117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weger B. D., Gobet C., Yeung J., Martin E., Jimenez S., Betrisey B., et al. (2019). The Mouse Microbiome Is Required for Sex-Specific Diurnal Rhythms of Gene Expression and Metabolism. Cell Metab. 29 362–382.e8. 10.1016/j.cmet.2018.09.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weis M. (2018). Impact of the gut microbiome in cardiovascular and autoimmune diseases. Clin. Sci. 132 2387–2389. 10.1042/CS20180410 [DOI] [PubMed] [Google Scholar]
- Xu C., Lee S. K., Zhang D., Frenette P. S. (2020). The Gut Microbiome Regulates Psychological-Stress-Induced Inflammation. Immunity 53 417–428.e4. 10.1016/j.immuni.2020.06.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue J., Zhou D., Poulsen O., Imamura T., Hsiao Y. H., Smith T. H., et al. (2017). Intermittent Hypoxia and Hypercapnia Accelerate Atherosclerosis, Partially via Trimethylamine-Oxide. Am. J. Respir. Cell Mol. Biol. 57 581–588. 10.1165/rcmb.2017-0086OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamura Y., Yano I., Kudo T., Shibata S. (2010). Time-dependent inhibitory effect of lipopolysaccharide injection on Per1 and Per2 gene expression in the mouse heart and liver. Chronobiol. Int. 27 213–232. 10.3109/07420521003769111 [DOI] [PubMed] [Google Scholar]
- Yotti R., Seidman C. E., Seidman J. G. (2019). Advances in the Genetic Basis and Pathogenesis of Sarcomere Cardiomyopathies. Annu. Rev. Genomics Hum. Genet. 20 129–153. 10.1146/annurev-genom-083118-015306 [DOI] [PubMed] [Google Scholar]
- Zhou D., Wang J., Zapala M. A., Xue J., Schork N. J., Haddad G. G. (2008). Gene expression in mouse brain following chronic hypoxia: role of sarcospan in glial cell death. Physiol. Genomics 32 370–379. 10.1152/physiolgenomics.00147.2007 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The paired-end RNA-seq data of n = 12 samples are available at https://www.ncbi.nlm.nih.gov/bioproject/692551 (BioProject ID: PRJNA692551).
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.