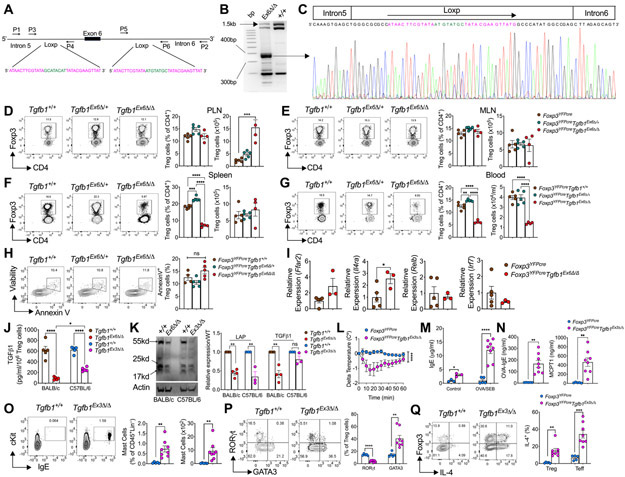
Responding to our recently published report (Turner et al., 2020), Velegraki et al (Velegraki et al., 2021) claimed that the Loxp elements around exon 6 of Tgfb1 allele employed in our studies are predicted not to lead to deletion of the targeted exon but rather to its inversion. They further hypothesized that such an inversion of the targeted sequence would result in the eventual loss of chromosome 7, harboring the targeted Tgfb1 allele, due to generation of an acentric and a dicentric chromosome stemming from asymmetrical chromosomal exchange between the sister chromatids. They further claim that such a phenomenon would lead to the apoptosis during cell cycle and eventually the depletion of regulatory T (Treg) cells, thus phenocopying Foxp3 deficient mice. Accordingly, the phenotype of mice with Treg cell-specific Tgfb1 exon 6 deletion would be the result of Treg cell depletion rather than their Tgfb1 deficiency.
To investigate these claims, we analyzed the targeted genomic Tgfb1 exon 6 region in sorted Treg cells isolated from Foxp3YFPcreTgfb1Exon6Δ/Δ as compared to control Foxp3YFPcre mice, and also sequenced the pre-excision proximal and distal Loxp elements (Figure 1A). We employed PCR primers that bracketed a 1400 base pair (bp) region spanning across the floxed sequence (primers P1 and P2, Figure 1A), whose deletion would be predicted to give rise to a 350 bp sequence. Accordingly, we successfully amplified the predicted post-excision genomic product in the Treg cells of mutant Foxp3YFPcreTgfb1Exon6Δ/Δ but not Foxp3YFPcre mice (Figure 1B, large arrow). Sequencing of this product clearly revealed the transition from an intron 5 sequence immediately 5’ of the original position of the proximal Loxp element, continuing through a retained post-excision composite Loxp sequence, and leading into an intron 6 sequence 3’ to the original distal Loxp sequence (Figure 1C). A correctly oriented DNA fragment corresponding to the sequence targeted for excision by the two Loxp elements was also amplified from the control Treg cells but was much reduced in the mutant Treg cells (Figure 1B, small arrow). No amplified sequences running in the reverse orientation were detected (data not shown). These results refute the claim that the targeted sequence is not deleted in Foxp3YFPCreTgfb1Exon6Δ/Δ Treg cells. They also are in agreement with the results reported in the original report on the generation of the Tgfb1 exon 6 floxed allele, which was successfully deleted by a Lck promoter driven Cre recombinase (Azhar et al., 2009). Extrapolating from our published results of real time PCR quantitation of exon 6 containing Tgfb1 mRNA in Foxp3YFPcreTgfb1Exon6Δ/Δ versus control Foxp3YFPcre Treg cells, we estimate the efficiency of the Cre-mediated Tgfb1 exon 6 excision at about 90%. This estimate is also supported by a virtually identical decrease in secreted TGF-β1 protein production in anti-CD3+anti-CD28 mAb-activated mutant Treg cell cultures as measured by ELISA (Turner et al., 2020).
Figure 1. Differential impact of Tgfb1 exon 6 versus exon 3 deletion in Treg cells.
(A). Schematic presentation of the floxed Tgfb1 exon 6 locus, detailing the proximal and distal Loxp sequences and the positions of the primers used in Sanger sequencing studies. Primers P1 and P2 were used to sequence the entire locus, P3 and P4 for the proximal and P5 and P6 for the distal loxp sites. (B). Gel electrophoresis analysis of P1 and P2 PCR amplicons obtained using genomic DNA of Foxp3YFPCreTgfb1Ex6Δ/Δ and Foxp3YFPCre Treg cells. Long arrow indicates the position of 350bp post Cre excision PCR product, while the short arrow indicates the position of the unexcised full length sequence. (C). Sanger sequencing of the 350bp Cre excision PCR product shown in panel B detailing the position of introns 5 and 6 and the post excision retained Loxp sequence. (D). Representative flow cytometric plots, frequencies and numbers of Treg cells in peripheral lymph nodes (PLN) (D), mesenteric lymph nodes (MLN) (E), Spleen (F), and Blood (G) from Foxp3YFPCre, Foxp3YFPCreTgfb1Ex6Δ/+, and Foxp3YFPCreTgfb1Ex6Δ/Δ mice. (H). Representative flow cytometric plots and frequencies of Annexin V+ Treg cells from the spleens of the respective mouse strains. (I). Real time PCR analysis of relative mRNA expression of Ffar2, Il4ra, Relb, and Irf7 in Foxp3YFPCre and Foxp3YFPCreTgfb1Ex6Δ/Δ Treg cells. (J). ELISA Quantification of TGFβ1 production by anti-CD3+anti-CD28 mAb +IL-2-activated Treg cells isolated from Foxp3YFPCre and Foxp3YFPCreTgfb1Ex6Δ/Δ mice (BALB/c background) and Foxp3YFPCre and Foxp3YFPCreTgfb1Ex3Δ/Δ mice (C57BL/6 background). (K). Immunoblot analysis of TGFβ1 in cell lysate of Treg cells from Foxp3YFPCre and Foxp3YFPCreTgfb1Ex6Δ/Δ (BLAB/c) and Foxp3YFPCre and Foxp3YFPCreTgfb1Ex3Δ/Δ (C57Bl/6) mice. Representative immunoblot, densitometric analysis of latent TGFβ1 and mature TGFβ1 expression in Treg cells. (L). Core body temperature drop following OVA challenge of OVA-SEB sensitized Foxp3YFPCre and Foxp3YFPCreTgfb1Ex3Δ/Δ mice. (M) Serum concentrations of IgE at baseline and post Ova challenge; OVA-specific IgE and MMCP-1 (N-P). Flow cytometric analysis and enumeration of c-Kit+FcεRI+IgE+ mast cells, RORγt+ and GATA3+ Treg cells and IL-4+ Treg and Teff cells (P) in the in the small intestines (SI) of the mouse groups in (L). Results are representative of at least 3 independent experiments. Each symbol represents an independent sample. Numbers in flow plots indicate percentages. Error bars indicate SEM. Statistical tests: Two-way ANOVA (L); One-way ANOVA (D, E, G, H) Student’s t-test (I to K; and M to Q). *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001.
We next addressed the claim that the phenotype of Foxp3YFPcreTgfb1Exon6Δ/Δ mice relates to the depletion of Treg cells. Analysis of Treg cell frequencies and absolute numbers in different lymphoid tissues of Foxp3YFPcreTgfb1Exon6Δ/Δ versus Foxp3YFPcre mice revealed no difference in the frequencies of Treg cells in peripheral and mesenteric lymph nodes (PLN and MLN) in the respective mice, whereas the absolute number of Treg cells were statistically significantly higher in the PLN of Foxp3YFPcreTgfb1Exon6Δ/Δ mice (Figure 1D, E). Whereas the spleens of Foxp3YFPcreTgfb1Exon6Δ/Δ mice exhibited reduced frequencies of Treg cells, their absolute numbers were similar to those found in the spleens of Foxp3YFPcre mice (Figure 1F). Only in the blood were the frequencies and absolute numbers of Treg cells reduced by about 50% in Foxp3YFPcreTgfb1Exon6Δ/Δ mice, most likely reflective of dysregulated homing to lymphoid and inflamed somatic tissues (Figure 1G). Analysis of Foxp3YFPCreTgfb1Exon6Δ/+ mice heterozygous for the floxed allele revealed overall similar frequencies and numbers across the different tissues as those of Foxp3YFPCre mice. Notably, analysis of splenic Treg cell apoptosis by Annexin V staining showed no statistically significant difference between Foxp3YFPcre, Foxp3YFPcreTgfb1Exon6Δ/+ and Foxp3YFPcreTgfb1Exon6Δ/Δ mice (Figure 1H). Furthermore, we analyzed the mRNA expression of several genes along chromosome 7, including Ffar2, Il4ra, Irf7 and Relb. Expression of these genes in the mutant Treg cells was either similar to or increased compared to WT Treg cells, arguing against the loss of chromosome 7 in the targeted Treg cells (Figure 1I). This conclusion is consistent with our previous demonstration in our publication that in heterozygous Foxp3YFPcre/+Tgfb1Exon6fl/fl female mice, the exon 6 deletant mutant Treg cells (Foxp3YFPcreTgfb1Exon6Δ/Δ) had a similar fitness as their exon 6 sufficient (Foxp3+Tgfb1Exon6fl/fl) Treg cell counterparts circulating in the same mice (Turner et al., 2020). The claim that transgenic TGF-β1 expression from the Rosa26 locus failed to rescue the Foxp3YFPcreTgfb1Exon6Δ/Δ excision phenotype cannot be evaluated as evidence of transgenic TGF-β1 expression, analysis of the tissue and cellular phenotypes and the outcome in terms of survival were all not reported, Overall, these results refute the claim the phenotypes of Foxp3YFPcreTgfb1Exon6Δ/+ and Foxp3YFPcreTgfb1Exon6Δ/Δ mice are due to Treg cell depletion, confirming instead that they arise specifically as a result of Treg cell-specific TGF-β1 deficiency
Both Velegraki et al (Velegraki et al., 2021) and Choi et al (Choi et al., 2021) present data that deletion of Tgfb1 exon 2 in Treg cells does not give rise to autoimmune lymphoproliferation. We have previously hypothesized that deletion of proximal exons of Tgfb1 may give rise to hypomorphic phenotypes due to the continued expression of active TGF-β1 from the distal exons 5-7 by means of alternative open reading frames. To validate our hypothesis, we recently evaluated mice with Treg cell specific exon3 deletion (Foxp3YFPcreTgfb1Exon3Δ/Δ) (Jax Lab). Phenotypically, and similar to the exon 2 deletants reported by Velegraki et al (Velegraki et al., 2021) and Choi et al (Choi et al., 2021) the Foxp3YFPcreTgfb1Exon3Δ/Δ mice appeared healthy. Analysis of TGF-β1 production by ELISA revealed that whereas the production of secreted TGF-β1 (both inactive proprotein and mature active TGF-β1) was mostly abrogated in activated Foxp3YFPcreTgfb1Exon6Δ/Δ as compared to similarly treated Foxp3YFPcre Treg cells, substantial residual TGF-β1 production by Foxp3YFPcreTgfb1Exon3Δ/Δ Treg cells could still be detected (Figure 1J). Immunoblot analysis revealed that whereas exon 6 deletion decreased the expression of latency associated peptide (LAP) and mature TGF-β1 (both dimeric and monomeric forms), exon 3 deletion largely spared the latter (Figure 1K).
Notwithstanding their normal appearance, Foxp3YFPcreTgfb1Exon3Δ/Δ mice exhibited allergic dysregulation both at baseline and upon oral sensitization with chicken egg ovalbumin (OVA) as a model food allergen. This phenotype manifested despite these mice being on a C57BL/6 background, normally associated with reduced allergic response. Oral challenge of sensitized Foxp3YFPcreTgfb1Exon3Δ/Δ mice with OVA resulted in intense anaphylaxis as evidenced by the drop in their core body temperature (Figure 1L). Other stigmata of a severe food allergic response in the sensitized Foxp3YFPcreTgfb1Exon3Δ/Δ mice included elevated total and OVA-specific IgE responses, mast cell degranulation as evidenced by elevated serum concentrations of mast cell protease 1(MCPT1) and expansion of mast cell numbers in the small intestine as detected by flow cytometry (Lin−CD45+cKit+FcεR1+IgE+) (Figures 1M-1O). Overall, the allergic dysregulation exhibited by mice with Treg cell-specific homozygous Tgfb1 exon 3 deletion approximated that of mice with Treg cell-specific exon 6 haploinsufficiency or partially penetrant exon 6 deletion using a bacterial artificial chromosome encoded, Foxp3 promoter driven Cre recombinase.
We have previously demonstrated that Tgfb1 exon 6 haploinsufficiency in Treg cells gave rise to allergic dysregulation by a mechanism involving depletion of gut Treg cells expressing the retinoic acid receptor-related orphan receptor γt (RORγt+). Consistent with these findings, analysis of food allergic Foxp3YFPcreTgfb1Exon3Δ/Δ mice revealed decreased frequencies of RORγt+ Treg cells in the gut, with a reciprocal expansion of T helper-2 (Th2) cell-like reprogrammed Treg cells expressing elevated levels of the transcription factor GATA3 and the Th2 cell cytokine IL-4 (Figures 1P and 1Q). Altogether, these results confirm that Treg cell specific Tgfb1 exon 3 deletion gives rise to a hypomorphic phenotype that approximates that observed with Tgfb1 exon 6 haploinsufficiency. The results of Choi et al (Choi et al., 2021) are consistent with an incomplete suppression of TGF-β1 production upon exon 2 deletion, suggesting that these mice may also manifest pronounced allergic dysregulation when tested using the appropriate experimental paradigms. Of note, Tgfb1 exon 6 haploinsufficiency did not give rise to an exaggerated Th1 response, similar to what was revealed in a colitis model employed by Choi et al (data not shown).
In conclusion, the results reported herein reinforce and extended those reported earlier by us to indicate distinct phenotypes associated with Treg cell-specific TGF-β1 deficiency extending from allergic dysregulation to autoimmunity, the nature and magnitude of which is governed by the level of residual TGF-β1 production by Treg cells (Turner et al., 2020). These results warrant further investigations on the molecular regulation of Tgfb1 expression in Treg cells and how it may contribute to disease.
Acknowledgments
This work was supported by NIH NIAID grants 5R01AI126915 and 5R01AI085090 (T.A.C.).
References
- Azhar M, Yin M, Bommireddy R, Duffy JJ, Yang J, Pawlowski SA, Boivin GP, Engle SJ, Sanford LP, Grisham C, et al. (2009). Generation of mice with a conditional allele for transforming growth factor beta 1 gene. Genesis 47, 423–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi G, Kim B-S, Chang J-H, and Chung Y (2021). Defining the role of transforming growth factor b1 in Foxp3+ T regulatory Cells. Immunity 54, 393–394. [DOI] [PubMed] [Google Scholar]
- Turner JA, Stephen-Victor E, Wang S, Rivas MN, Abdel-Gadir A, Harb H, Cui Y, Fanny M, Charbonnier LM, Fong JJH, et al. (2020). Regulatory T Cell-Derived TGF-beta1 Controls Multiple Checkpoints Governing Allergy and Autoimmunity. Immunity 53, 1202–1214 e1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velegraki M, Salem M, Ansaa-Addo EA, Wu BX, and Li Z (2021). Autocrine transforming growth factor b1 in regulatory T cell biology—gone but not missed. Immunity 54, 395–396. [DOI] [PubMed] [Google Scholar]