Abstract
In the past decade, immunotherapies have been emerging as an effective way to treat cancer. Among several categories of immunotherapies, immune checkpoint inhibitors (ICIs) are the most well-known and widely used options for cancer treatment. Although several studies continue, this treatment option has yet to be developed into a precise application in the clinical setting. Recently, omics as a high-throughput technique for understanding the genome, transcriptome, proteome, and metabolome has revolutionized medical research and led to integrative interpretation to advance our understanding of biological systems. Advanced omics techniques, such as multi-omics, single-cell omics, and typical omics approaches, have been adopted to investigate various cancer immunotherapies. In this review, we highlight metabolomic studies regarding the development of ICIs involved in the discovery of targets or mechanisms of action and assessment of clinical outcomes, including drug response and resistance and propose biomarkers. Furthermore, we also discuss the genomics, proteomics, and advanced omics studies providing insights and comprehensive or novel approaches for ICI development. The overview of ICI studies suggests potential strategies for the development of other cancer immunotherapies using omics techniques in future studies.
Keywords: cancer immunotherapy, immune checkpoint inhibitor, metabolomics, omics, microbiome, immune-related adverse events
1. Introduction
The International Agency for Research on Cancer (IARC), as a part of the World Health Organization (WHO), announced that cancer is the second highest cause of death and is associated with approximately 10 million deaths per year based on the WHO cancer mortality database. To alleviate the mortality of cancer, various therapeutic agents have been developed, ranging from chemotherapy to immunotherapy [1]. Although chemotherapy, as the first generation of cancer therapy, has been the prevalent option for cancer treatment with radiotherapy and surgery in the past decades, the development of tyrosine kinase inhibitors and monoclonal antibodies for cancer, second-generation therapeutic agents, have improved the efficiency of therapies derived by that high-specificity and broad therapeutic window [2]. In recent decades, among several therapeutic approaches, cancer immunotherapy is considered the third generation of cancer therapy, which can overcome the limitations of previous approaches. Cancer immunotherapy, sometimes called immune-oncology, directly or indirectly stimulates the host immune system to control or eliminate cancer [3]. Various strategies have been developed to evoke patients’ own tumor immunity, such as immune checkpoint inhibitors (ICIs), immune cell therapy, and anti-cancer vaccines, but ICIs are the most well-studied category of immunotherapies.
‘Omics’ is a high-throughput technique for the investigation of biological systems, including diverse molecular layers (e.g., genes, proteins, and metabolites), and supports a large number of datasets, such as those of the genome, transcriptome, or metabolome representing biological dynamics [4]. To date, omics-based systems biology has significantly contributed to advances in ontology while considering its numerous applications for cancer research, such as biomarker discovery, therapeutic target suggestions, and prognosis and diagnosis assessment. Likewise, these omics-based platforms have developed immunotherapy known as third-generation cancer therapy. The advantages of omics techniques include (1) supporting reliable high-throughput datasets across several molecular layers, which enable integration for comprehensive biological interpretations, (2) simultaneous achievement of qualification and quantification for many molecules for effective target screening, (3) high availability and applicability using reasonable costs and labor derived from the development of mass spectrometry and sequencing technology. Several studies regarding cancer immunotherapy, especially ICIs, have applied omics and highlighted several outcomes, such as extensive targets for drug discovery and development, determination of mechanisms of action, pre- or post-treatment biomarkers of clinical outcomes, and assessment of immune-related adverse events [5].
In this review, we discuss the recent application of omics techniques in the study of immune checkpoint inhibitors and extend the knowledge of omics-based approaches to advance our understanding of cancer immunotherapy for future studies.
2. Emerging Application of Immunotherapy for Cancer
Since the concept that the immune system can recognize and prevent carcinogenesis at early stages was introduced in 1909 by Paul Erlich, it was incorporated into the ‘cancer immunosurveillance’ theory by Burnet and Thomas in the middle of the 19th century [6]. The controversial theory was based on several studies conducted in various cancer models, providing evidence either confirming or opposing the theory. Following continuous studies, the theory was developed into the concept of ‘cancer immune-editing,’ in that the immune system demonstrates not only host-protection but also tumor-sculpting effects on cancer development [7]. Diverse components of the immune system protect the host against nascent cancer development or improve tumor escape or both by cancer immune-editing [8]. The process is divided into three phases, including elimination, equilibrium, and escape [9]. In the elimination phase, the immune system is able to destroy the tumor through the action of NK, CD4+, and CD8+ cells. However, equilibrium between immune system components and tumor cells leads to the failure of tumor suppression at the second stage. Finally, the tumor acquires immune evasion abilities and becomes detectable in the escape phase.
Recently, the tumor microenvironment (TME), which is the surrounding environment interacting with tumors, has been considered as an emerging field in cancer study and re-establishes drug efficacy and therapeutic strategies in cancer immunotherapy [10]. TME includes various components, such as blood vessels, immune cells, fibroblastic cells, and extracellular matrix [11]. The tumor and TME interact closely, resulting in increased tumor heterogeneity [12]. An improved understanding of the TME demonstrated that immune cell infiltration has a high correlation with anti-cancer immune responses, and it led to the creation of the ‘immune contexture,’ involving organization, composition, and density of immune cell infiltrate [13]. Following the specific categories of the concept, several therapeutic trials were conducted to suggest precision medicine [14]. These findings demonstrated that tumor interaction with immune systems and the TME could serve as a crucial target for cancer immunotherapy.
After the first attempt at using the immune system for cancer therapy in the late 19th century, immune-oncology or immunotherapy has been developed continuously and has become an anticipated field, as evidenced by the Nobel prize for physiology or medicine being awarded T-cell to Drs. Allison and Honjo in 2018 for the discovery of T-cell immune checkpoints, such as CTLA-4 and PD-1 [15,16]. Cancer immunotherapy, regarded as a third-generation cancer therapy, modifies the patient’s own immune system to control or eliminate cancer [17]. To date, the typical types of cancer treatment are chemotherapy, radiation therapy, surgery, and a combination of those options. However, chemotherapy, known as a first-generation anti-cancer treatment, displays many side effects, including fatigue, nausea, vomiting, hair loss, and pain, due to the fact that the treatment cannot distinguish between tumor and host cells [18]. To overcome the limitation of chemotherapy, a second-generation targeted therapy was developed to improve specificity to block cancer growth, progression, and metastasis, followed by an extension of the therapeutic window [19]. Although this strategy increased specificity, universal application was difficult for various types of cancer, and drug resistance occasionally occurs [20]. Meanwhile, cancer immunotherapy activates patients’ own immune systems, which are suppressed by cancer immune-editing, to destroy tumor cells. Thus, it has been used in diverse cancer types without additional gene manipulation in that immune responses are under strict regulation by immune checkpoints, and it increases the quality of treatment in accordance with the extension of survival rate and minimized side effects [21].
Although traditional immune therapy, including tumor vaccines, cytokine therapy, and adaptive cell transfer (ACT), has been used in specific cancers, insufficient effects and severe toxicities of this approach escalated the need for novel cancer therapeutics [22,23]. Following general classification, by which cancer immunotherapies are divided into “inactive (or passive)” and “active” according to their abilities to activate the immune system against tumor cells, tumor-targeting monoclonal antibodies and adoptively transferred T-cells are included in inactive immunotherapy, while anti-cancer vaccines and checkpoint inhibitors are considered as active immunotherapy [24]. Beginning with FDA approval for first-generation cancer immunotherapy, including sipuleucel-T (Provenge®; Dendreon) for prostatic cancer and ipilimumab (Yervoy®; Bristol-Meyers Squibb) for melanoma, immunotherapy has become the fastest-expanding area in cancer therapeutics [25,26]. Further, a blockade of immune checkpoints has been developed through second-generation immunotherapy, including PD-1 and PD-L1 antibodies following ipilimumab (CTLA-4 inhibitor) [27]. As of May 2021, eight immune checkpoint inhibitors have been approved by the FDA (Table 1).
Table 1.
FDA approvals of ICIs from January 2011 to May 2021.
| Drug Name | Active Ingredient | Approval Date | Mechanism of Action | Company | First Approved Indications |
|---|---|---|---|---|---|
| Yervoy | Ipilimumab | 25 March 2011 | CTLA-4-blocker | Bristol-Myers Squibb | Late-Stage Melanoma |
| Keytruda | Pembrolizumab | 4 September 2014 | PD-1 blocker | Merck & Co., Inc. | Advanced or unresectable melanoma |
| Opdivo | Nivolumab | 22 December 2014 | PD-1 blocker | Bristol-Myers Squibb | Unresectable or metastatic melanoma |
| Tecentriq | Atezolizumab | 18 May 2016 | PD-L1 blocker | Genentech Inc. | Urothelial carcinoma, the most common type of bladder cancer |
| Bavencio | Avelumab | 23 March 2017 | PD-L1 blocker | Emd Serono Inc. | Metastatic Merkel cell carcinoma |
| Imfinzi | Durvalumab | 1 May 2017 | PD-L1 blocker | Astrazeneca | Locally advanced or metastatic urothelial carcinoma |
| Libtayo | Cemiplimab-rwlc | 28 September 2017 | PD-1 blocker | Regeneron Pharmaceuticals |
Cutaneous squamous cell carcinoma |
| Jemperli | Dostarlimab-gxly | 22 April 2021 | PD-1 blocker | GlaxoSmithKline | Endometrial cancer |
CTLA-4, cytotoxic T-lymphocyte-associated protein 4; PD-1, Programmed cell death protein 1; PD-L1, Programmed death-ligand 1.
Despite the potential for clinical benefits, low response rates and several resistance mechanisms have not yet been resolved. A critical limitation of immunotherapies is immune-related adverse events (irAEs), which are characterized by host immune activation against healthy cells [28]. The mortality rate due to severe myocarditis, an irAE, was 46% in immunotherapy-treated patients who received ICIs [29]. Moreover, PD-1 and CTLA-4 inhibitors result in the doubling of serious irAEs, although the survival rate of patients increases [30]. Fortunately, several markers for irAEs have been suggested, but the validation of these markers should be performed [31,32]. The insufficient characteristic of second-generation treatments led to the emergence of new generations with various novel therapeutic modalities based on novel strategies, which have been designed to elicit immune responses against tumors [27,33].
Two main strategies are the activation of co-stimulatory receptors and inhibition of immunosuppressive ligands or metabolism [33]. Agonistic monoclonal antibodies targeting tumor necrosis factor receptors, such as OX40, GITR, and CD137 expressed on several immune cells, provoke the extension of CD8+ T-cell survival, increasing tumor-specific T-cell responses, the upregulation of NK cells, and the regulation of regulatory T-cells [34,35,36]. Conversely, several studies have been reported regarding the inhibition of immunosuppressive targets, such as VISTA (v-domain Ig suppressor of T-cell activation), a ligand, and IDO1 (indoleamine 2,3-dioxygenase-1), an enzyme catalyzing the kynurenine pathway as a rate-limiting step for enhancing anti-tumor T-cell responses and inhibition of immune responses by depletion of tryptophan [37,38]. Based on the robust concept of immunotherapy established through past decades, the latest therapeutic trends include improvement of efficacy, management of response and toxicity, and extension of applicable targets [39]. To improve the efficacy using developed immunotherapies, several pharmaceutical and combinational strategies have been applied. Nanotechnology has become an effective option for eliciting immune responses by expanding the therapeutic window and enhancing vaccination or endogenous immune responses [40]. Similarly, PEGlyation, conjugation of polyethylene glycol (PEG) polymer to proteins, has been utilized to increase the half-life of cytokines in vivo [22]. Another method used to enhance efficacy is immunotherapy combined with typical cancer therapy, such as radiotherapy or immunomodulatory drugs [41,42]. Recent studies focus on biomarkers for immune response, including toxicity, as well as the establishment of guidelines for irAE [43,44]. Thus, various studies are continuously performed to discover biomarkers not only for immune responses but also for novel therapeutic targets using omics, which are most widely used in systems biology. Furthermore, many potential drugs are under clinical trials registered with ClinicalTrials.gov (https://clinicaltrials.gov/) (assessed on 21 May 2021) (Table 2).
Table 2.
Potential immune checkpoint inhibitors under clinical trials enrolled in ClinicalTrials.gov. The search conditions were as follows: status, recruiting and not yet recruiting studies; condition of disease, cancer; other terms, PD-1 or PD-L1 or CTLA-4 or checkpoint.
| ClinicalTrials.gov Identifier | Purpose | Study Population | Interventions | Status | Phase | |
|---|---|---|---|---|---|---|
| 1 | NCT02694822 | Evaluation | Advanced solid cancers and Advanced solid cancers refractory to PD-1 | Drug: AGEN1884 | Active, not recruiting | Phase1/2 |
| 2 | NCT03989362 | Combination | Cancer | Drug: Vopratelimab and Ipilimumab | Active, not recruiting | Phase 2 |
| 3 | NCT03515629 | Combination | NSCLC | Drug: REGN2810/Ipilimumab, REGN2810/chemo/Ipilimumab, and Pembrolizumab | Active, not recruiting | Phase 3 |
| 4 | NCT04172454 | Evaluation | Advanced Solid Tumors Melanoma | Drug: AK104 | Not yet recruiting | Phase 1B/2 |
| 5 | NCT03527251 | Combination | NSCLC | Drug: Ipilimumab, SHR-1210 | Unknown | Phase 1 |
| 6 | NCT04326257 | Combination | HNSCC | Drug: Nivolumab/Relatlimab and Nivolumab/Ipilimumab | Recruiting | Phase 2 |
| 7 | NCT04868708 | Combination | Recurrent or Metastatic Cervical Cancer | Biological: AK104 and Bevacizumab, Drug: Paclitaxel and Cisplatin or Carboplatin | Not yet recruiting | Phase 2 |
| 8 | NCT03430063 | Combination | Advanced NSCLS | Drug: SDREGN2810, SDREGN2810/Ipilimumab, and HDREGN2810 | Active, not recruiting | Phase 2 |
| 9 | NCT04140526 | Combination | NSCLC/Advanced Solid Tumor, Metastatic Melanoma, Metastatic Head and Neck Carcinoma, Metastatic RCC/Metastatic CRC, Sarcomas/Metastatic Prostate Cancer, Ovarian Cancer/SCLC, Metastatic Breast Cancer | Drug: ONC-392 and Pembrolizumab | Recruiting | Phase 1 |
| 10 | NCT04544644 | Combination | NSCLC | Drug: AK104/Anlotinib | Not yet recruiting | Phase 2 |
| 11 | NCT02403193 | Evaluation and Combination | NSCLC | Drug: PBF-509_(80~640 mg), PBF-509 (160~640 mg) + PDR001, RP2D with ICIs naïve, Experimental: RP2D with ICIs treated | Active, not recruiting | Phase1/2b |
| 12 | NCT02535078 | Combination | Malignant Melanoma | Drug: IMCgp100, Durvalumab, Tremelimumab | Active, not recruiting | Phase 1B/2 |
| 13 | NCT03388632 | Combination | Metastatic Solid Tumors and Treatment-Refractory Cancers | Drug: rhIL-15, Ipilimumab, and Nivolumab | Recruiting | Phase 1 |
| 14 | NCT03608046 | Combination | Colorectal Neoplasms, Malignant | Drug: Avelumab, Cetuximab Injection, Irinotecan | Recruiting | Phase 2 |
| 15 | NCT03040791 | Expansion | Prostate Cancer | Drug: Nivolumab | Recruiting | Phase 2 |
| 16 | NCT02821754 | Combination | Biliary Tract Neoplasms, Liver Cancer, HCC, Cholangiocarcinoma, and Bile Duct Cancer | Drug: Durvalumab and Tremelimumab, Procedure: TACE, RFA, Cryoablation | Recruiting | Phase 2 |
| 17 | NCT03019003 | Combination | Head and Neck Cancer | Drug: Oral Decitabine and Durvalumab | Recruiting | Phase 1 and Phase 2 |
| 18 | NCT03202758 | Combination | Metastatic CRC | Drug: Durvalumab/Tremelimumab/FOLFOX | Unknown | Phase 1 and Phase 2 |
| 19 | NCT02938793 | Combination | Cancer | Drug: Durvalumab and Tremelimumab | Recruiting | Phase 2 |
| 20 | NCT03925246 | Expansion | High Grade Glioma/ Brain Cancer | Drug: Nivolumab | Active, not recruiting | Phase 2 |
| 21 | NCT03084471 | Combination | Advanced Solid Malignancies | Biological: MEDI4736 and MEDI4736/Tremelimumab | Active, not recruiting | Phase 3 |
| 22 | NCT03608046 | Combination | Colorectal Neoplasms, Malignant | Drug: Avelumab, Cetuximab Injection, and Irinotecan | Recruiting | Phase 2 |
| 23 | NCT03409198 | Combination | Breast Cancer, Hormone Receptor Positive Tumor, and Metastatic Breast Cancer | Drug: Ipilimumab, Nivolumab, Pegylated liposomal doxorubicin, and Cyclophosphamide | Active, not recruiting | Phase 2 |
| 24 | NCT04319224 | Combination | Cancer | Drug: Vopratelimab, Ipilimumab, Nivolumab | Recruiting | Phase 1 and Phase 2 |
| 25 | NCT03526185 | Combination | Metastatic Melanoma | Drug: Tumor Infiltrating Lymphocytes and Nivolumab/Ipilimumab | Active, not recruiting | Early Phase 1 |
| 26 | NCT03911557 | Combination | Tumor, Solid | Drug: Durvalumab/Tremelimumab | Recruiting | Phase 2 |
| 27 | NCT03308396 | Combination | Advanced Kidney Cancer, Kidney Cancer, and Clear Cell RCC | Drug: Guadecitabine, Durvalumab | Active, not recruiting | Phase 1 and Phase 2 |
| 28 | NCT03186326 | Expansion | Metastatic CRC MSI |
Drug: FOLFOX regimen, FOLFIRI Protocol, Avelumab, Panitumumab, Cetuximab, Bevacizumab, and Aflibercept | Active, not recruiting | Phase 2 |
| 29 | NCT03206073 | Combination | CRC, Colorectal Carcinoma, Colorectal Adenocarcinoma, Refractory Cancer, and Colorectal Neoplasms | Drug: Durvalumab and Tremelimumab, Biological: Pexa-Vec | Active, not recruiting | Phase 1 and Phase 2 |
| 30 | NCT03373760 | Combination | Recurrent Squamous Cell Lung Carcinoma and Stage IV Squamous Cell Lung Carcinoma AJCC v7 | Biological: Durvalumab and Tremelimumab, Other: Laboratory Biomarker Analysis | Active, not recruiting | Phase 2 |
| 31 | NCT03959293 | Combination | Gastric Adenocarcinoma and Gastric Cancer | Drug: Durvalumab, Tremelimumab, and FOLFIRI Protocol | Recruiting | Phase 2 |
| 32 | NCT03693612 | Combination | Neoplasms | Drug: Feladilimab, Tremelimumab, Docetaxel, Paclitaxel, and Cetuximab | Active, not recruiting | Phase 1 and Phase 2 |
| 33 | NCT03755739 | Administration | Hepatocarcinoma/Lung Cancer, Melanoma/Renal Cancer, Head and Neck Cancer, and Pancreas Cancer/Ovarian Cancer, CRC/Cervical Cancer/Breast Cancer | Drug: ICIs | Recruiting | Phase 2 and Phase 3 |
| 34 | NCT03033576 | Combination | Advanced Melanoma, Melanoma of Unknown Primary, Mucosal Melanoma, Refractory Melanoma, Stage III Cutaneous Melanoma AJCC v7, Stage IIIA Cutaneous Melanoma AJCC v7, Stage IIIB Cutaneous Melanoma AJCC v7, Stage IIIC Cutaneous Melanoma AJCC v7, Stage IV Cutaneous Melanoma AJCC v6 and v7, Unresectable Cutaneous Melanoma, and Unresectable Melanoma | Biological: Ipilimumab and Nivolumab | Active, not recruiting | Phase 2 |
| 35 | NCT02821754 | Combination | Biliary Tract Neoplasms, Liver Cancer/HCC, Cholangiocarcinoma, and Bile Duct Cancer | Drug: Durvalumab and Tremelimumab, Procedure: TACE, RFA, and Cryoablation | Recruiting | Phase 2 |
Administration, clinical trials for comparison of efficacy following different administration; AGEN1884, anti-CTLA-4 antibody; AK104, humanized IgG1 tetrameric PD-1/CTLA-4 bispecific antibody; chemo, chemotherapy; combination, clinical trials for novel drug combination regimen; CRC, colorectal cancer; evaluation, clinical trials for development of novel drug candidates; expansion, clinical trials for expansion of indication; FOLFOX, combination chemotherapy made up of folinic acid, fluorouracil, and oxaliplatin; HCC, hepatocellular carcinoma; HDREGN2810, high dose cemiplimab; HNSCC, head and neck squamous cell carcinoma; ICI, immune checkpoint inhibitor; IMCgp100, engineered T cell receptor specific for a peptide antigen derived from the protein gp100; MEDI4736, durvalumab; NSCLC, non-small cell lung cancer; ONC-392, a humanized anti-CTLA4 IgG1 monoclonal antibody; PBF-509, Adenosine A2a receptor antagonist; PDR001, anti-PD-1 antibody; pexa-Vec, a thymidine kinase gene-inactivated oncolytic vaccinia virus engineered for the expression of transgenes encoding human granulocyte-macrophage colony-stimulating factor (GM-CSF) and beta-galactosidase; RCC, renal cell carcinoma; REGN2810, cemiplimab; RFA, Radiofrequency ablation; rhIL-15, recombinant interleukin-15; RP2D, PBR-509 + PDR001; SCLC, small cell lung cancer; SDREGN2810, standard dose cemiplimab; SHR-1210, anti-PD-1 antibody; TACE, Transarterial chemoembolization.
3. Development of Omics Workflows for Cancer Immunotherapy Including ICIs
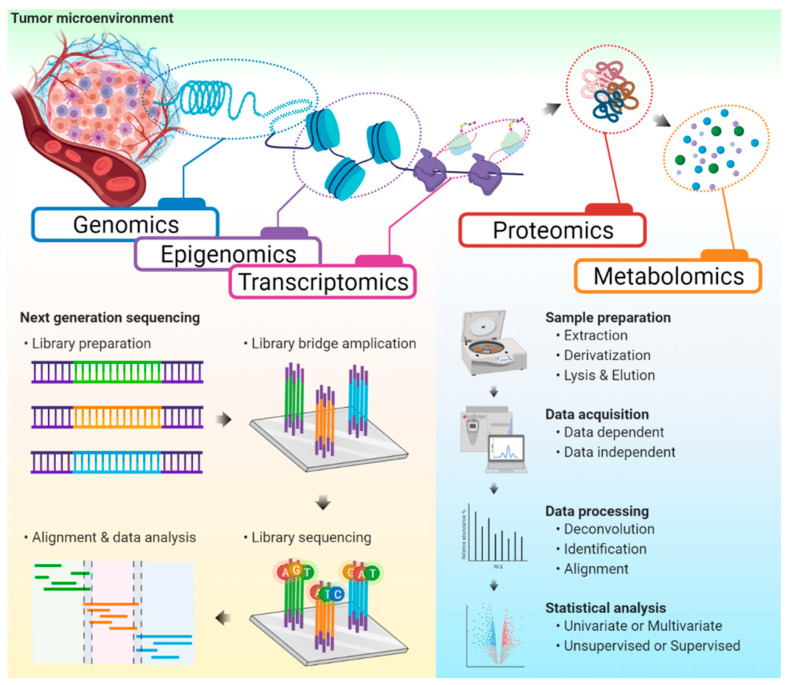
Several omics platforms that provide high-throughput information for biological understanding have been applied to study cancer immunotherapy (Figure 1). In this section, we will deliberate about how these approaches have contributed to the development of ICIs and how they have given insights for future studies involving cancer immunotherapy.
Figure 1.
The workflow of omics platforms. Various omics approaches applied for the study of cancer immunotherapy were demonstrated with general workflows of next-generation sequencing and mass spectrometry-based omics.
3.1. Development of Omics Technologies
Since the human mitochondrial genome was identified about 40 years ago, genetics has been extensively applied to determine the biochemical function of the genome to further study cancer based on human genome sequences [45]. Following the enhancement of demands for high-throughput data, researchers began to realize the importance of the ‘systems biology’ approach for systemic biological interpretation and accelerated the development of several platforms. ‘Omics,’ as a technique to analyze large amounts of data comprising an entire set of analytes, includes various branches, such as genomics, transcriptomics, proteomics, metabolomics (or metabonomics), and lipidomics, and it aims to provide molecular profiles for understanding diverse biological dynamics [46]. In oncology, omics has been frequently used to demonstrate hallmarks, discover biomarker candidates for diagnosis or prognosis, determine target pathways, indicate mechanisms of drug response, and predict or assess toxicity, providing an effective method to develop therapeutic intervention [47,48]. Multiple analytical platforms that support high-throughput data derived from biological samples have been developed over the years. Microarray and next-generation sequencing (NGS), including whole-genome sequencing (WGS) or transcriptome sequencing, which overcame disadvantages associated with the Sanger sequencing method, have been broadly used for DNA and RNA samples. Exome sequencing and epigenome sequencing were additionally developed to improve the quality of target gene analysis. Recently, genotyping using polymerase chain reaction (PCR) or clustered regularly interspaced short palindromic repeats (CRISPR) is also applied for the development of cancer therapies. Moreover, the development of mass spectrometry technology and the method of data acquisition facilitates the broad application of proteomics, metabolomics, and lipidomics. Simultaneously, diverse tools supporting data pre-processing of raw data (e.g., data collection and gap-filling) and post-processing workflow for interpretation (e.g., biomarker analysis, network analysis, and pathway enrichment analysis) have been established [49,50,51,52]. Extensive data from several biochemical analytes are recruited to establish databases or in silico libraries (e.g., TCGA, TCPA, LipidBlast, and FiehnLib) by several societies or analytical teams for future studies [53,54,55,56].
3.2. Advantages of Omics-Based Systems Biology for Oncology
Omics-based systems biological approaches have several advantages in cancer study. First, omics supports reliable high-throughput datasets over several molecular layers (e.g., genome, transcriptome, proteome, and metabolome), which enable the integration for comprehensive biological interpretations, in that it suggests alteration of metabolism, tumor microenvironment (TME), and provides clues regarding tumor mechanisms. For example, diffuse gastric cancers were studied by integration of proteomic and genomic analyses and indicated an association between mRNA-protein abundance and patient survival [57]. Meanwhile, Kang et al. suggested the crucial roles of extracellular cystine in influencing the mechanism of ferroptosis in non-small cell lung cancer through stable isotope labeling-based metabolomics [58]. Second, omics provides the simultaneous achievement of qualitative and quantitative results for many molecules for effective therapeutic target screening, such as biomarkers or pathways associated with pre- or post-treatment clinical outcomes [59]. The clinical outcomes promote precision medicine through positive response or prevention of unintended negative effects, such as including toxicity and drug resistance [60,61]. Third, omics has high availability and applicability using reasonable cost and labor derived from the continuous development of mass spectrometry and sequencing technologies.
3.3. Recent Trends and Advanced Omics Platforms in Cancer Study
Several methodologies have been established for a systems biological approach in oncology to overcome the limitations of typical omics study. Multi-omics, also called pan-omics, is a biological analysis for the simultaneous integrated interpretation of multiple omics data sets [62]. Although comprehensive interpretation based on so-called multi-layer omics, which deduces results according to post-analysis integration, induces a better understanding of biological phenomena than typical single omics, it is complicated and usually conducted by knowledge-based interpretation, including the possibility of bias. Conversely, multi-omics is performed by specialized tools used for combining different omics data sets before further analyses [63]. Therefore, it elucidates potential causative alterations, which may become promising targets for cancer therapy, rather than reactive processes derived from analysis of one omics data set. Despite its powerful support for integrational interpretation, multi-omics need a logical strategy to link each data sets based on the evidence for causation, presenting functional associations between diverse molecular levels and preventing coincidental correlation [64]. Meanwhile, mass spectrometry imaging focuses on visualizing the spatial distribution of molecular targets to overcome the limitations of typical sample preparation methods, which pool all molecules into the same solvent [65]. Therefore, it is especially useful to gain this biochemical information [66]. Regarding mechanistic studies, stable isotope tracing is the most-developed technique to demonstrate the flux of target metabolism [67]. Furthermore, the combination of stable isotope tracing with other techniques, such as metabolomics or MSI, provides novel insights for understanding cancer metabolism and drug development [68,69]. Recently, genome engineering using the CRISPR-Cas9 system, which is the RNA-guided Cas9 nuclease from the microbial clustered regularly interspaced short palindromic repeats, has emerged [70]. Owing to precise genome editing by this technology, genome-wide CRISPR-Cas9 knockout screens were developed to determine the correlation between genotype and phenotype, connected with the development of cancer therapy [71].
3.4. Drug Development for ICIs Based on a Metabolomics Approach
According to the central dogma, genetic information in DNA has passed into protein through RNA, and these proteins regulate the intermediates (e.g., polar metabolites and lipids) associated with metabolic pathways, which can affect phenotypes [72]. In tumorous circumstances, the flow is distorted by several reasons and results in the expression of various cancer hallmarks, demonstrating cellular metabolism that has deviated from normal conditions. Thus, identification or quantification of these intermediates is essential to provide an understanding of metabolic reprogramming to the alteration of phenotypes by cancer. Metabolomics, an omics field, focuses on the study of small molecules, is a high-throughput technique for the parallel assessment of large-scale metabolites. Owing to the remarkable development of instrumental technologies and bioinformatics, the proportion of metabolomics contributing to the systems-level understanding of diseases has increased in the past decades. As well as high accessibility, synergistic effects derived from the inherent importance of the metabolome and flexible applicability based on large-scale datasets lead to frequent use for the identification and validation of metabolic profiles, simultaneous large-scale quantification, and functional analysis of the metabolome [73]. In addition, recent advances regarding metabolome information-based fluxomics and stable isotope tracing enable a greater understanding of disease mechanisms, including cancers [68,74].
Although various methods have been developed and selected by sample matrix, chromatography-coupled mass spectrometry (MS) is the most powerful and broadly used technique for metabolomics. Three important considerations for LC/MS- or GC/MS-based metabolomics are sample preparation, chromatography conditions, and MS compartment. The extraction method and solvent are selected based on the characteristic of interest, and the ultimate goal of optimization is a reproducible technique to extract several metabolites [75,76]. Meanwhile, chromatography conditions determine the separation of numerous metabolites through interaction with stationary and mobile phases and detect metabolites using MS in a time-dependent manner [77,78]. MS coupled with chromatography is an important technique and is most often used in metabolomics. For detection by a mass spectrometer, molecules must be ionized in specific ways, of which ESI and electron impact are frequent methods in LC and GC, respectively.
Recently, metabolomic approaches have been used in several developments related to immunotherapy and highlight changes in downstream molecules (e.g., amino acids, nucleic acids, and lipids) as a result of aberrant upstream signals, which play important roles for metabolic pathways directly related to the expression of crucial phenotypes. Based on this advantage, metabolomics has been applied to investigate novel therapeutic targets for cancer immunotherapy and identify promising metabolic biomarkers for the assessment of post-treatment outcomes or pre-treatment predictors. Further, metabolomics methods are continuously developed and optimized for the study of ICIs [79,80]. Furthermore, this technique has expanded the range of biological interpretation through comprehensive interpretation with other omics approaches (e.g., genomics, proteomics, transcriptomics, and metagenomics) and suggested novel strategies for the development of immunotherapies, such as the association between microbiota metabolites with ICI efficacy and the discovery of immune system-related metabolic pathways (e.g., kynurenine pathway). Herein, we summarized previous studies involving ICIs or focused on the development of ICIs among several studies regarding cancer immunotherapy (Table 3).
Table 3.
Metabolomics-based studies related to ICIs or focusing on the development of ICIs.
| Purpose | Related ICIs | Sample | Methods | Comments | Reference | ||
|---|---|---|---|---|---|---|---|
| Subjects (Number) | Matrix | ||||||
| 1 | Target discovery | - | in vitro | T-cells | LC-MS/MS | PD-1 signaling results in metabolic dysregulation, which suggests considerable metabolic interventions of ICIs’ efficacy. | [81] |
| 2 | Target discovery | - | in vitro | T-cells | LC-MS/MS | Mechanistic association between T-cell senescence and aberrant lipid metabolism was introduced as a novel target for cancer immunotherapy. | [82] |
| 3 | Target discovery | - | in vitro and ex vivo (11 patients with nivolumab and TIL therapy) | T-cells and TILs | LC-MS/MS | Sirt2, associated with reprogramming T-cell metabolism, was identified as a new target of cancer immunotherapy. | [83] |
| 4 | Target discovery | - | in vivo and patients with glioblastoma | tissue | LC-MS/MS and GC-MS | IDO1 inhibition mitigated radiation-induced immunosuppression in glioblastoma. | [84] |
| 5 | Target discovery and Biomarker suggestion | Nivolumab, Pembrolizumab |
ICI-treated patients with NSCLC (23) vs. heathy subjects (20) | plasma | LC-MS/MS | IDO1 inhibitors are a promising treatment for NSCLC considering IDO1 activity seemed to a key role in the primary resistance of ICIs. | [85] |
| 6 | Target discovery | Anti-mouse PC-1, Nivolumab | in vitro, patients with glioblastoma (4), and patients with metastatic melanoma (4) | tissue | LC-MS/MS | ICIs induced the IL4I1, which facilitates tumor progression. | [86] |
| 7 | Target discovery | Anti-PD-1 | in vivo and patients with HCC (196) vs. healthy subjects (176) | urine | LC-MS/MS | PRMT5 inhibition demonstrated a synergistic mechanism enhancing anti-tumor immunity and alleviated the resistance to ICIs. | [87] |
| 8 | Target discovery | Anti-mouse PD-1 | in vivo | tissue | LC-MS/MS | nSMase2 overexpression increased anti-PD-1 efficacy in murine melanoma models. | [88] |
| 9 | Target discovery | - | patients with breast cancer (65) | tissue | MALDI-MSI | The accumulation of PI(18:0/20:3) may affect the PD-1-associated immune checkpoint pathway. | [89] |
| 10 | Target discovery | - | in vivo | plasma | LC-MS/MS and GC-MS |
KEAP1/NRF2 pathway alteration induced reprogramming of pentose phosphate pathway connected with tumorigenesis and tumor regression by immune checkpoint inhibition in NSCLC. | [90] |
| 11 | Target discovery | - | in vitro | Breast cancer cells and PDAC cells | 1H-MRS | Chk-α, COX-2, and TGF-β mediated PD-L1 regulation of metabolism. | [91] |
| 12 | Target discovery | - | patients with breast cancer (58) and patients with HCC (29) | data from previous studies | - | UCD is related to an enhanced response to ICI therapy. | [92] |
| 13 | Target discovery and Biomarker suggestion | Anti-mouse PD-1 and Anti-mouse CTLA-4 | in vitro and patients with PDAC | PDAC cells, serum, and tissue | NMR | IL17 inhibitor enhances ICI sensitivity, and tumor lactate was suggested as a promising early biomarker for efficacy of IL17/PD-1 combination. | [93] |
| 14 | Target discovery and Biomarker suggestion | Nivolumab | nivolumab-treated patients with advanced melanoma (78), nivolumab-treated patients with RCC (485), and everolimus-treated patients with RCC (349) | serum | LC-MS/MS | The combination of a PD-1 inhibitor with IDO/TDO inhibitors was suggested in that worse overall survival associated with simultaneous elevation of resistance and serum kynurenine/tryptophan ratio. | [94] |
| 15 | Biomarker suggestion | Nivolumab and Pembrolizumab | ICI-treated patients with urological cancer (28) | serum | LC-MS/MS | VLCFA-containing lipids are potential predictive biomarkers for ICIs’ response. | [95] |
| 16 | Efficacy evaluation | Nivolumab and Pembrolizumab | ICI-treated patients with NSCLC (19) | plasma | LC-MS/MS | Tryptophan metabolites may become potential predictive biomarkers for the efficacy of the ICIs. | [96] |
| 17 | Biomarker suggestion and Efficacy evaluation | Nivolumab and Pembrolizumab | ICI-treated patients with NSCLC (50) | serum | NMR | The metabolomic fingerprint of serum is a potential biomarker for the response of ICIs. | [97] |
| 18 | Method development | - | patients with melanoma (-) | stool | LC-MS/MS | A comprehensive approach to fecal sample collection and metabolites profiling of gut microbiome were demonstrated. | [98] |
| 19 | Biomarker suggestion | Nivolumab | nivolumab-treated patients with NSCLC (7), NSCLC patients without nivolumabtreatment (4) vs. healthy subjects (8) | stool | GC-MS/SPME and NMR | Microbiota-Linked Biomarkers, including SCFAs, were introduced through network analysis. | [99] |
| 20 | Efficacy evaluation | Nivolumab | nivolumab-treated patients with NSCLC (11) | stool | GC-MS/SPME and 1H-NMR | The identification of microbiota-linked “indicators” is a potential strategy for the prediction of responders, in that gut microbiota metabolic pathways affect the response of ICIs. | [100] |
| 21 | Biomarker suggestion | Nivolumab | nivolumab-treated patients with NSCLC (22) | serum and stool | GC-MS/SPME and NMR | An integrated parameter was proposed to identify good responders for nivolumab treatment. | [101] |
| 22 | Efficacy evaluation | Anti-mouse PD-1, Atezolizumab, Nivolumab, and Pembrolizumab | in vivo and ICI-treated patients with NSCLC (96) vs. healthy subjects (139) | serum and stool | LC-MS/MS | Bifidobacterium bifidum strains make a synergistic effect with ICIs to reduce tumor burden. | [102] |
| 23 | Efficacy evaluation | Nivolumab, and Pembrolizumab | ICI-treated patients of multiple cancers (52) | plasma and stool | LC-MS/MS | Fecal SCFA concentration may affect PD-1 inhibitors’ efficacy. | [103] |
| 24 | Efficacy evaluation | Nivolumab, Pembrolizumab, and Sintilimab | nivolumab-treated patients with NSCLC (4), pembrolizumab-treated patients with NSCLC (42), and sintilimab-treated patients with NSCLC (17) | stool | - | The correlation between intestinal microbiome β-diversity and the response of anti-PD-1 in NSCLC was indicated. | [104] |
Chk-α, choline kinase-α; COX-2, prostaglandin-endoperoxide synthase 2; HCC, hepatocellular carcinoma; ICI, immune checkpoint inhibitor; IDO, indoleamine-2,3-dioxygenase 1; IL4I1, interleukin-4-induced-1; KEAP1, Kelch-like ECH-associated protein 1; KMO, kynurenine monooxygenase; KYNU, kynureninase; LC, liquid chromatography; MALDI, Matrix-Assisted Laser Desorption Ionization; MRS, magnetic resonance spectroscopy; MS/MS, tandem mass spectrometry; MSI, mass spectrometry imaging; NMR, nuclear magnetic resonance; NRF2, nuclear factor erythroid-2-related factor 2; NSCLC, non-small cell lung cancer; PBMC, peripheral blood mononuclear cells; PDAC, pancreatic ductal adenocarcinoma; PRMT5, Protein arginine N-methyltransferase 5; RCC, renal cell carcinoma; SCFA, short-chain fatty acid; Sirt2, NAD+-dependent deacetylase; TDO, tryptophan 2,3-dioxygenase; TGF-β, Transforming growth factor β; TIL, tumor-infiltrating lymphocytes; TN, triple-negative; UCD, urea cycle dysregulation; UV/Vis, UV-Vis spectrophotometer.
3.4.1. Target Discovery
During the development of novel therapeutic approaches for specific diseases, including cancer, the top priority is the identification of suitable and effective targets for therapy. One of the key considerations for effective ICI is the dysregulated metabolism of T-cell-mediated tumor microenvironments, and it provides novel therapeutic targets based on the improved understanding of the interplay between functional states of T-cells and immune metabolism. For example, Palaskas et al. conducted a mass spectrometry-based metabolomics study in vitro to investigate metabolic alterations affected by PD-1 signaling. They demonstrated that PD-1 signaling for non-adherent primary human T-cells prevented de novo nucleoside phosphate synthesis accompanied by decreased mTORC1 signaling, while exogenous purines and pyrimidines failed to rescue the proliferation of PD-L1-treated cells [81]. Liu et al. reported an association between lipid metabolism and T-cell senescence, suggesting that reprogrammed lipid metabolism was triggered by the upregulation of PLA2G4A in cancer cells and regulatory T-cells. Using melanoma and breast cancer in vivo models, they demonstrated enhanced therapeutic efficacy following inhibition of PLA2G4A [82]. Moreover, another study identified the negative correlation between response to tumor-infiltrating lymphocyte (TIL) therapy and upregulation of Sirt2 in human TILs. This study indicated that Sirt2-deficient T-cells increased antitumor activity resulting from upregulated oxidative phosphorylation and glycolysis, following the enhancement of effector functions and proliferation [83]. A remarkable finding regarding immunotherapy indicated that tryptophan metabolites related to the kynurenine pathway and IDO-1 activity were potential targets of novel immunotherapy. Heng et al. performed kynurenine pathway profiling using large-scale clinical samples from patients with several types of breast cancer. They revealed potent immunosuppression by increased anthranilic acid and 2-hydroxylanthranilic acid derived from downregulation of kynurenine monooxygenase and kynureninase in triple-negative and HER2-enriched breast cancer subtypes [105]. Furthermore, many recent studies have reported an association between the immune checkpoint and tryptophan metabolism related to IDO-1/TDO. Some studies have suggested IDO-1 as an indicator or potential target for combination therapy. Kesarwani et al. recommended a combination of IDO-1 inhibitors with radiotherapy for increased therapeutic efficacy by preventing RC-induced immunosuppression [84]. Kocher et al. identified alterations in 67 metabolites in NSCLC patients receiving ICI treatment compared with healthy controls using LC-MS/MS, indicating dysregulation of IDO activity in patients. Based on the results, they suggested tryptophan as a promising biomarker for ICIs [85]. Besides, the discoveries of compensatory or combinational targets for ICIs have been continued. Sadik et al. identified that the activation of aryl hydrocarbon receptor (AHR) reduced anti-tumor immunity, and interleukin-4-induced-1 (IL4I1) was associated with AHR activity more than IDO-1 or TDO2 [86]. Other studies under anti-PD-1 conditions were used to assess the response by drug treatment and investigate novel targets. For example, PRMT5 inhibition demonstrated synergistic mechanisms enhancing anti-tumor immunity and alleviated resistance to ICIs [87]. A lipidomics approach demonstrated that the upregulation of sphingomyelin phosphodiesterase 3 by sphingomyelinase 2 (nSMase2) is a potential strategy to overcome resistance against PD-1 inhibitors according to increased PD-1 inhibitor efficacy following over-expression of wild-type nSMase2 in melanoma [88].
3.4.2. Discovery of Biomarkers and Efficacy Evaluation
Although there is no qualified biomarker related to cancer immunotherapy by U.S. FDA until now, the discovery of biomarkers to evaluate outcomes post-treatment and to predict responses pre-treatment is important for therapy development, as well as finding novel targets. A study using metabolomics for PDAC under anti-PD-1 and anti-CTLA-4 conditions demonstrated increased sensitivity of ICIs by IL17 inhibitor resulting in prevention of cytotoxic CD8 T-cell exclusion from tumors and suggested that tumor lactate may serve as a promising early biomarker for efficacy of IL17/PD-1 combination [93]. Given that the immune mechanism is related to the kynurenine pathway and kynurenine to tryptophan ratio, it contributed to the development of a marker of tumor aggressiveness and metabolic profiling alteration in response to treatment with PD-1 inhibitors (e.g., nivolumab and pembrolizumab). Based on the association between increased serum kynurenine/tryptophan ratio and worse overall survival, the combination of IDO/TDO inhibitors and PD-1 inhibitors has been advocated [94,106]. Additionally, a comprehensive evaluation of metabolites in serum from ICI-treated patients demonstrated that very-long-chain fatty acid (VLCFA) containing lipids predicted efficacy and therapy response [95]. Moreover, biomarkers can be used to evaluate drug efficacy. Karayama et al. analyzed plasma from 19 ICI-treated patients with NSCLC and identified tryptophan metabolites. Based on the interpretation of the metabolite intensities with drug response and survival rate, they suggested tryptophan metabolites as potential predictors of ICI efficacy [96]. Another study also suggested the novel NMR-based metabolomics approach, providing metabolomic serum fingerprints for the predictive assessment of ICI efficacy, and it showed more than 80% accuracy in 50 patients with NSCLC receiving nivolumab and pembrolizumab treatments [97]. Regarding biomarker discovery and efficacy evaluation, metabolomics is most frequently used to study the microbiome under ICI conditions. Stool is a commonly used sample for microbiome studies; thus, several studies have optimized and applied unbiased metabolomic profiling methods for fecal samples [98]. Some studies applied GC-MS/SPME-based metabolomics for the detection of volatile organic compounds (VOCs) and NMR-based metabolomics for non-VOCs to investigate the gut metabolome involved in nivolumab treatment for NSCLC. These studies introduced metabolomic approaches and their network analysis as promising strategies for the management of cancer patients and prediction of good responders using microbiota-linked indicators [99,100]. In addition, another study proposed integrated parameters, including gut metabolites and immunological molecules from serum and stool for identification of nivolumab responders before treatment [101].
3.5. Metabolomics and Metagenomics for Identifying Interactions between the Microbiome and ICIs
Although metabolomics, including lipidomics, play key roles to evaluate the efficacy of therapies and discover predictive biomarkers, it has been focused on studying the response to therapy and highlighting precision medicine in studies of cancer immunotherapy, instead of characteristics of tumor or TME, due to limitations from target molecular layers (e.g., metabolome and lipidome) [107]. The proportion of metabolomic approaches for ICIs is inferior to that of genomics or transcriptomics. However, recent evidence indicated that microbiota as a source of metabolites had been involved in various diseases, including cancers and the immune system related to the tumor microenvironment [108]. Nevertheless, the identification of effects driving alterations to the immune system in tumors and in response to ICIs have not been fully understood yet due to the complexity of bacteria and its metabolites. Fortunately, advanced metabolomics techniques allow for high-throughput data acquisitions to understand this complexity. Given that cancer metabolism, especially the cancer immune system, is affected by microbiota directly or metabolites produced from bacteria indirectly, the identification of the microbiome and metabolome becomes a priority for omics research. Therefore, metabolomics combined with metagenomics are widely used to study the microbiome for cancer immunotherapy. Several previous studies have demonstrated that microbial metabolites and microbiota itself influence the efficacy of immunotherapy [109,110]. Although other omics approaches have contributed to suggest various roles of the microbiome in modulating the response to ICIs, comprehensive interpretation, including metagenomics and metabolomics, have been widely utilized to discover unknown interactions between the microbiome and ICIs [111,112]. Through this approach, specific bacterial strains or mechanisms for enhancing the efficacy of ICIs have been studied. For example, the specific gut microbiome of nivolumab and pembrolizumab responders was identified, while the synergistic effect of anti-PD-1 and B. bifidum strains reportedly reduced cancer growth by modulating the production of IFN- γ by intensifying biosynthesis of immune-stimulating metabolites [102,113]. The development of techniques and information regarding how the microbiome interacts with ICIs have promoted the discovery of microbiota-linked biomarkers for response prediction of ICIs (e.g., indole, aldehydes, and short-chain fatty acids), which could be a promising target for precision medicine [99].
3.6. Omics Approaches for Investigating Upstream Molecular Levels of the Metabolome
In systems biological approaches for cancer immunotherapy, especially for ICIs, proteogenomics and transcriptomics are frequently used because aberrant expression of neoantigens, microsatellite instability (MSI), DNA repair, and TMB are closely associated with abnormal effects of molecules from genome to proteome in the immune system. For example, Anagnostou et al. studied the initial response of ICIs from the comparison of pre-treatment and post-treatment, thus observing an association between genomic alteration and loss of mutation-associated neoantigens in resistant tumors, which demonstrated decreased therapeutic benefits [114]. Additionally, proteomics studies involving the secretome derived from T-cells and B-cells demonstrated specific protein signatures in the exosomes of patients receiving PD-1 inhibitors before treatment and in tumor-associated B-cells, suggesting that these protein signatures can be used as promising predictive markers for PD-1 inhibitors regarding activation of PD-1+ T-cells treated with PD-1 inhibitors [115,116]. Usually, genomics, transcriptomics, and proteomics are simultaneously applied and integrated to obtain synergetic interpretation beyond individual explanations. A previous study generated cell-type immune enrichment scores based on proteogenomic approaches, providing gene and protein expression levels of targets containing PD-1 and evaluating different types of glioblastoma [117]. Further, noninvasive identification methods to assess response at the early stage of ICI treatment were recommended using pre-treatment circulating tumor DNA and peripheral CD8+ T-cell levels to predict the durable clinical benefit of patients based on whole-exome sequencing and RNA-sequencing in non-small cell lung cancer [118]. Meanwhile, several studies reported the key roles of epigenetic markers in oncogenesis and immune-editing [119,120]. Recently, the EPIMMUNE signature was introduced by Duruisseaux et al., which encompassed specific patterns of DNA methylation from nivolumab- or pembrolizumab-treated non-small-cell lung cancer patients and was associated with clinical benefit [121].
Several recent studies using genomics and transcriptomics demonstrated that high MSI and TMB correlate with tumor antigenicity and the response to immunotherapy [122,123]. Evrard et al. mentioned that deficient DNA mismatch repair (dMMR) and MSI display heterogeneity originating from testing methods, and dMMR/MSI screening may be useful with TMB, regarding benefits from immunotherapy in colorectal cancers [124]. Meanwhile, Vanderwalde et al. performed MSI assays using NGS methods to highlight the relationship between MSI, TMB, and PD-L1 using over 11,000 patients across cancer types and suggested MSI as a marker with TMB and PD-L1 expression to determine the use of ICIs [125]. Although previous studies were not able to provide standardized TMB cutoffs among each study and cancer types, TMB is strongly considered as an independent predictive biomarker for the response to ICIs, while genomic techniques for reproducible TMB calculations are in continuous development [122].
4. Multidisciplinary Approaches beyond Fundamental Omics Studies for ICIs
Studies involving ICIs have been performed not only with individual omics but also with various other approaches. In this section, we will demonstrate how other approaches combined with omics have contributed to more comprehensive insights into cancer immunotherapy.
4.1. Multi-Omics and Multi-Layer Omics
Systems biology is an approach to understand the biological system at a diverse level (e.g., genes, proteins, and metabolites) [126]. Systems biology provides a powerful premise and promise based on multiple omics approaches generating high-throughput data sets for biological interpretation. Over the past decades, the development of technologies and reduction of costs facilitate the application of omics. These trends have motivated the use of multi-omics to improve the biological insight of research. Multi-omics or pan-omics is a comprehensive omics approach integrating the data sets from multiple omics (e.g., genomics, transcriptomics, proteomics, and metabolomics) to explain interactions among omics dimensions [127]. Recently, the ‘Australian and New Zealand Metabolomics Conference’ (ANZMET 2018) hosted a peer session on multi-omics, discussed potential limitations of multi-omics, and recommended strategies to overcome these limitations. Furthermore, several strategies to interpret biological meanings, such as top-down and bottom-up data reduction integration, or post-analysis data integration approaches and integrated data analysis approaches, were introduced [63]. Among strategies, post-analysis data integration is widely used and includes the key features on different omics dimensions. In contrast with data integration after analysis, integrated data analysis approaches merge various dimensions of omics data through specialized tools, and several studies have developed the methods and tools using different aspects [63,128].
To obtain valuable insight from massive omics information and complexity of the disease, the optimal integration methods are situationally applied, such as pathway data integration, network analysis, and statistical integration [64]. In oncology, the number of studies using multi-omics has been continuously increased for drug discovery, biomarker signature establishment for prognosis or diagnosis, and assessment of drug response. For example, Lindskrog et al. demonstrated a framework for biomarker discovery based on transcriptomics classification of non-muscle-invasive bladder cancer through a combination of transcriptomics and proteomics [129]. Several data repositories (e.g., TCGA, CPTAC, and METABRIC) about cancer established and have been frequently utilized in multi-omics [62]. Vasaikar et al. also tried to establish the LinkedOmics database containing clinical data derived by multi-omics techniques for 23 cancer types and data from TCGA to support multi-omics platforms for future studies [130]. Additionally, these multi-omics studies have promoted the emergence of precision medicine, considering individual or group variability for disease treatment and prevention [131].
A convenient way to perform comprehensive omics interpretation is the application of established omics databases. Project HOPE (High-tech Omics-based Patient Evaluation) is a clinical research project by the Institutional Review Board of Shizuoka Cancer Center in Japan, including comprehensive whole-exome sequencing and gene expression profiling for 1000 tumor tissues. Using HOPE, Akiyama et al. demonstrated the availability of a multi-omics database for investigating ICI targets, in that this approach provided seven immune response-associated genes and discovered over-expression of PD-L1 in hypermutators [132]. In addition, 13 melanoma data, including two responders from five nivolumab-treated patients in HOPE, were used to suggest upregulation of PD-L1 protein and increase single nucleotide variants after complete remission [133]. While using established omics databases, often the data is integrated with results from additional experiments to expand dataset types. For example, the relationship between VLCFA-containing lipids with ICI response via upregulation of peroxisome signaling in T-cells was indicated based on the integration of transcriptome data in TCGA with a metabolomics dataset from urological cancer patients [95]. In addition, comprehensive interpretation by nuclear magnetic resonance-based metabolomics, proteomic data, and TCGA provided choline kinase-α, cylcooxygenase-2, and transforming growth factor-β as promising options for combinatorial therapies based on ICIs [91].
Established databases can be used by extracting novel results and integrating more than two datasets through statistics or specialized tools for meta-analysis. Integration of TCGA and Gene Expression Omnibus (GEO) datasets involving head and neck cancer concluded that EGFR and PTGS2 were identified as important nodes of the immune phenotype-related network in genetic and epigenetic levels, and EGFR inhibition was recommended as a potential target of combination therapy for ICI non-responders [134]. Similarly, unsupervised clustering of data from 1000 patients with hepatocellular carcinoma from GEO, TCGA-LIHC, and ICGC classified three clusters and revealed potential predictive signatures for response and prognosis assessment of anti-PD-1 and anti-CTLA-4 immunotherapies [135]. Moreover, a previous study established a tool to estimate ICI response predictors using previous large-scale omics data, published ICI trials, non-immunotherapy tumor profiles, and CRISPR screening on the web [136].
Without database utilization, various studies have applied multi-omics approaches to investigate ICIs, including mechanisms of actions, response prediction, and target discovery for combination therapy. Among diverse combinations of omics data, integration of genomics and transcriptomics is widely used. Genomic and transcriptomic features were used as test and validation sets, analyzed separately to represent genomic and transcriptomic characteristics, or integrated before analysis [137,138,139]. Furthermore, transcriptomics data is usually combined with a proteomics approach to demonstrate phenotype differences by transcriptome-affected protein alteration [129]. However, few studies focus on ICIs based on multi-omics, including metabolomics, indicating that the expression of tumor- or immune-related proteins and mechanisms of action are the most effective and important targets for cancer immunotherapy [117].
4.2. Single-Cell Omics
Given that the functions of organs are derived from comprehensive activities of organized individual cells, identifying changes in cells caused by disease is important. Single-cell omics techniques typically analyze specific molecular layers from the identical individual cell and provide cellular heterogeneity, enabling a more profound understanding of key biological mechanisms [140]. Recently, single-cell omics have developed continuously, and some protocols, such as single-cell RNA sequencing, are standardized and widely used [141]. In oncology, the complex interaction between tumor cells and the surrounding microenvironment hampers precise investigation of cellular functions for cancer growth and progression using omics based on bulk analysis. However, single-cell omics can demonstrate functional differences in cellular states within a tumor, which is associated with phenotypic differences driven by particular molecular layers. The usefulness of single-omics techniques, especially single-cell RNA sequencing, for future studies, has been additionally improved by the Human Cell Atlas, established by an international consortium, providing high-throughput data for classification and identification of cells, which can be potentially used for data-driven interpretation of specific diseases [142]. Although current single-cell techniques contain limitations, such as spatial information, scalability, and PCR errors, recent developments (e.g., spatial sequencing and comprehensive joint profiling technology) promise to overcome these limitations [143,144].
In cancer immunology, the TME is considered a complicated mixture of diverse cells (e.g., tumor cells, stromal cells, and immune cells) and non-cellular components (e.g., signaling molecules and extracellular matrix), which play key roles for anti-tumor immunity. To date, many studies have applied single-cell omics, mainly single-cell transcriptomics, to investigate heterogeneity and complicated cross-cellular interactions in the TME [145]. In addition, various trials have been performed to evaluate ICI treatment. Bassez et al. analyzed clinical samples from breast cancer patients who received only anti-PD1 or neoadjuvant chemotherapy before anti-PD1 using single-cell transcriptomics combined with proteome profiling to understand a subset of tumors responding to ICI. This study identified the association of T-cell expansion after anti-PD1 treatment with immunophenotypes and gene sets positively (e.g., expression of PRF1, GZMB, and CXCL13) or negatively (e.g., TCF7+, GZMK+ T-cells, and CX3CR1+, C3+ inhibitory macrophages) [146]. Sade-Feldman et al. applied single-cell RNA sequencing to profile transcriptomes of more than 16,000 individual immune cells derived from 48 patients with melanoma in a discovery set. Through the validation set, including ex vivo and in vivo studies, they demonstrated TCF7+ CD8+ T-cells as a predictive marker for positive clinical outcomes [147]. These studies suggested not only a strategy for discovering predictors and mechanisms of ICI action but also novel targets for the development of cancer immunotherapies.
5. Expanding the Knowledge of Systems Biology Studies for Cancer Immunotherapy
5.1. Evaluation of Immune-Related Adverse Events
Cancer immunotherapies have received attention as a novel generation of cancer treatment. However, unexpected immune-related adverse events (irAE) have been observed following increased use and clinical trials. A network meta-analysis using 36 phase 2 and 3 randomized trials reported that the range of probability is 54% to 76%, and the pooled incidence is 66.4% to 86.8% for all adverse events of five ICIs (atezolizumab, nivolumab, pembrolizumab, ipilimumab, and tremelimumab) [148]. The onset of irAEs has been reported at eight days to more than one year after initial trials of ICI treatment in any organ system [149]. ICIs induce irAEs through various mechanisms, including T-cell activation, cross-reactivity of immune cells and healthy cells, B-cell-mediated autoantibodies, and monoclonal antibody-mediated direct injury [150]. Several guidelines have been recommended for the therapeutic management of irAEs, with steroidal treatment being suggested as a prominent way for irAEs, excluding endocrine irAEs [151,152].
The prevention of irAEs is as important as therapeutic management. To prevent occurrences of irAEs before cancer immunotherapy, several studies suggested diverse biomarkers for response prediction and to determine the mechanisms of irAE as a potential target for prevention. A multi-omics approach demonstrated that lymphocyte cytosolic protein 1 and adenosine diphosphate dependent glucokinase might serve as biomarkers for irAE prediction by evaluating the association between omics data and irAE reporting odds ratios [153]. Meanwhile, Grigoriou et al. focused on transcriptomic reprogramming of regulator T-cells in blood and suggested inflammatory Treg reprogramming as an indicator for irAE development [154]. Another study concentrated on changes observed in B-cell expression after ICI treatment and indicated that early changes in B-cells after treatment may become a marker for risk of irAE in melanoma, based on single-cell RNA sequencing [155].
5.2. Beyond ICIs
In recent decades, the success of ICI-based cancer immunotherapy verified immune system control, resulting in anti-cancer effects and the promotion of further studies identifying novel targets or establishing various methods for immune-oncology treatment. Although ICIs are one of the most well-established methods of immunotherapy, other methods, including immune cell therapy, anti-cancer vaccines, and antibody-drug conjugates, have been developed to overcome the clinical limitation of ICIs [156]. Following this trend, the systems biology approach using omics platforms has been widely used in the development of these methods, as well as those involving ICIs.
The aim of the anti-cancer vaccine is the induction of T-cell responses, usually against specific antigens from tumors [157]. Cancer vaccines combined with ICIs or other immunotherapy may induce their maximized effects [158]. To identify effective neoantigens, Matsushita et al. and Robbins et al. performed whole-exosome sequencing, which identified a mutant marker of sarcoma and a potential correlation between mutated antigens from autologous tumor cells and clinical response in melanoma [159,160]. Unlike the method using anti-cancer vaccines, immune cell therapy using T-cell receptors (TCR), chimeric antigen receptors (CAR), and tumor-infiltrating lymphocytes (TIL) is the treatment of isolated immune cells from patients or genetically engineered immune cells to patients for activating their immune systems. Integrated analysis of proteomic and transcriptomic data sets by a specific algorithm involving acute myeloid leukemia (AML) was conducted by Perna et al. and suggested the concepts of generalizable combinatorial targeting strategies to uncover candidate targets for AML and other cancer studies [161]. High-throughput techniques are also used to enhance therapeutic efficacy and aid the discovery of novel targets. Lu et al. developed novel methods, including single-cell RNA sequencing and co-culture techniques for tumor-infiltrating lymphocytes and autologous antigen-presenting cells to increase the efficacy of adoptive T-cell therapy [162].
6. Concluding Remarks
To date, numerous metabolomics- and other omics-based studies for ICIs have been applied for the development of novel biomarkers, the evaluation or prediction of outcomes, and the identification of mechanisms of action. Furthermore, advanced analytical high-throughput techniques have been developed and optimized for various ICI studies. These studies may provide valuable information for future studies involving various cancer immunotherapy options as well as ICIs.
Acknowledgments
All figures were created with BioRender.
Abbreviations
| ACT | Adaptive cell transfer |
| ANOVA | Analysis of variance |
| APLCNR | Apelin receptor |
| C3 | Complement component 3 |
| CAR | Chimeric antigen receptor |
| CCL | Chemokine antigen receptor |
| CD137 | Tumor necrosis factor receptor superfamily member 9 (TNFRSF9) |
| CD4+ cell | Cluster of differentiation 4-positive cell |
| CD8+ cell | Cluster of differentiation 8-positive cell |
| CRISPR | Clustered regularly interspaced short palindromic repeats |
| CTLA-4 | Cytotoxic T-lymphocyte-associated protein 4 |
| CX3CR1 | CX3C chemokine receptor |
| CXCL13 | Chemokine (C-X-C motif) ligand 13 |
| cyTOF | Mass cytometry |
| dMMR | Deficient DNA mismatch repair |
| EGFR | Epidermal growth factor receptor |
| EI | Electron impact |
| ESI | Electrospray ionization |
| GC | Gas chromatography |
| GCMB | Glial Cells Missing Transcription Factor 2 |
| GEO | Gene Expression Omnibus |
| GITR | Glucocorticoid-induced TNFR family-related gene |
| GZMK | Granzyme K |
| HOPE | High-tech Omics-based Patient Evaluation |
| IARC | International Agency for Research on Cancer |
| ICGC | International Cancer Genome Consortium |
| ICI | Immune checkpoint inhibitor |
| IDO | Indoleamine 2,3-dioxygenase |
| IFN | Interferon |
| IL | Interleukin |
| IL17RA | IL17 receptor A |
| irAE | Immune-related adverse event |
| LC | Liquid chromatography |
| LIHC | Liver hepatocellular carcinoma |
| MS | Mass spectrometry |
| MS/MS | Tandem mass spectrometry |
| MSI | Microsatellite instability |
| NGS | Next-generation sequencing |
| NIST | National Institute of Standards and Technology |
| NK | Natural killer cell |
| nSMase2 | Sphingomyelinase 2 |
| OX40 | Tumor necrosis factor receptor superfamily, member 4 (TNFRSF4) |
| PCA | Principal component analysis |
| PCR | Polymerase chain reaction |
| PD-1 | Programmed cell death protein 1 |
| PD-L1 | Programmed death-ligand 1 |
| PEG | Polyethylene glycol |
| PLA2G4A | Phospholipase A2 group IVA |
| PLS-DA | Partial least squares-discriminant analysis |
| POLE | Polymerase epsilon |
| PRF1 | Perforin 1 |
| PTGS2 | Prostaglandin-endoperoxide synthase 2 |
| PTPN2 | Protein Tyrosine Phosphatase Non-Receptor Type 2 |
| TCF7 | Transcription Factor 7 |
| TCGA | The Cancer Genome Atlas |
| TCPA | The Cancer Proteome Atlas |
| TDO | Tryptophan 2,3-dioxygenase |
| TMB | Tumor mutation burden |
| TME | Tumor microenvironment |
| VEGFA | Vascular endothelial growth factor A |
| VISTA | v-domain Ig suppressor of T-cell activation |
| VLCFA | Very-long-chain fatty acid |
| VOC | Volatile organic compound |
| WGS | Whole-genome sequencing |
| WHO | World Health Organization |
Author Contributions
Conceptualization, S.J.Y. and S.K.B.; writing—original draft preparation, S.J.Y. and S.K.B.; writing—review and editing, S.K.B.; reference collection, C.B.L. and S.U.C., S.J.J.; visualization, S.J.Y. and S.K.B.; supervision, S.K.B. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by a grant of Basic Science Research Program through the National Research Foundation of Korea, funded by the Ministry of Education (2018R1A6A1A03025108), Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health and Welfare (HF20C0002), and Research Fund of the Catholic University of Korea (2019).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Arruebo M., Vilaboa N., Sáez-Gutierrez B., Lambea J., Tres A., Valladares M., González-Fernández Á. Assessment of the Evolution of Cancer Treatment Therapies. Cancers. 2011;3:3279–3330. doi: 10.3390/cancers3033279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zahavi D., Weiner L. Monoclonal Antibodies in Cancer Therapy. Antibodies. 2020;9:34. doi: 10.3390/antib9030034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Waldman A.D., Fritz J.M., Lenardo M.J. A guide to cancer immunotherapy: From T cell basic science to clinical practice. Nat. Rev. Immunol. 2020;20:651–668. doi: 10.1038/s41577-020-0306-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Karczewski K.J., Snyder M.P. Integrative omics for health and disease. Nat. Rev. Genet. 2018;19:299–310. doi: 10.1038/nrg.2018.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guhathakurta D., Sheikh N.A., Meagher T.C., Letarte S., Trager J.B. Applications of systems biology in cancer immunotherapy: From target discovery to biomarkers of clinical outcome. Expert Rev. Clin. Pharmacol. 2013;6:387–401. doi: 10.1586/17512433.2013.811814. [DOI] [PubMed] [Google Scholar]
- 6.Manjili M.H. Revisiting cancer immunoediting by understanding cancer immune complexity. J. Pathol. 2011;224:5–9. doi: 10.1002/path.2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dunn G.P., Bruce A.T., Ikeda H., Old L.J., Schreiber R.D. Cancer immunoediting: From immunosurveillance to tumor escape. Nat. Immunol. 2002;3:991–998. doi: 10.1038/ni1102-991. [DOI] [PubMed] [Google Scholar]
- 8.O’Donnell J.S., Teng M.W.L., Smyth M.J. Cancer immunoediting and resistance to T cell-based immunotherapy. Nat. Rev. Clin. Oncol. 2018;16:151–167. doi: 10.1038/s41571-018-0142-8. [DOI] [PubMed] [Google Scholar]
- 9.Dunn G.P., Old L.J., Schreiber R.D. The Three Es of Cancer Immunoediting. Annu. Rev. Immunol. 2004;22:329–360. doi: 10.1146/annurev.immunol.22.012703.104803. [DOI] [PubMed] [Google Scholar]
- 10.Lim B., Woodward W.A., Wang X., Reuben J.M., Ueno N.T. Inflammatory breast cancer biology: The tumour microenvironment is key. Nat. Rev. Cancer. 2018;18:485–499. doi: 10.1038/s41568-018-0010-y. [DOI] [PubMed] [Google Scholar]
- 11.Quail D.F., Joyce J.A. Microenvironmental regulation of tumor progression and metastasis. Nat. Med. 2013;19:1423–1437. doi: 10.1038/nm.3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Runa F., Hamalian S., Meade K., Shisgal P., Gray P.C., Kelber J.A. Tumor Microenvironment Heterogeneity: Challenges and Opportunities. Curr. Mol. Biol. Rep. 2017;3:218–229. doi: 10.1007/s40610-017-0073-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fridman W.H., Zitvogel L., Sautès–Fridman C., Kroemer G. The immune contexture in cancer prognosis and treatment. Nat. Rev. Clin. Oncol. 2017;14:717–734. doi: 10.1038/nrclinonc.2017.101. [DOI] [PubMed] [Google Scholar]
- 14.Becht E., Giraldo N.A., Dieu-Nosjean M.-C., Sautès-Fridman C., Fridman W.H. Cancer immune contexture and immunotherapy. Curr. Opin. Immunol. 2016;39:7–13. doi: 10.1016/j.coi.2015.11.009. [DOI] [PubMed] [Google Scholar]
- 15.McCarthy E.F. The toxins of William B. Coley and the treatment of bone and soft-tissue sarcomas. Iowa Orthop. J. 2006;26:154–158. [PMC free article] [PubMed] [Google Scholar]
- 16.Smyth M.J., Teng M.W. 2018 Nobel Prize in physiology or medicine. Clin. Transl. Immunol. 2018;7:e1041. doi: 10.1002/cti2.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hiam-Galvez K.J., Allen B.M., Spitzer M.H. Systemic immunity in cancer. Nat. Rev. Cancer. 2021:1–15. doi: 10.1038/s41568-021-00347-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kayl A.E., Meyers C.A. Side-effects of chemotherapy and quality of life in ovarian and breast cancer patients. Curr. Opin. Obstet. Gynecol. 2006;18:24–28. doi: 10.1097/01.gco.0000192996.20040.24. [DOI] [PubMed] [Google Scholar]
- 19.Lee Y.T., Tan Y.J., Oon C.E. Molecular targeted therapy: Treating cancer with specificity. Eur. J. Pharmacol. 2018;834:188–196. doi: 10.1016/j.ejphar.2018.07.034. [DOI] [PubMed] [Google Scholar]
- 20.Curigliano G., Criscitiello C. Successes and Limitations of Targeted Cancer Therapy in Breast Cancer. Successes Limit. Target. Cancer Ther. 2014;41:15–35. doi: 10.1159/000355896. [DOI] [PubMed] [Google Scholar]
- 21.Esfahani K., Roudaia L., Buhlaiga N., Del Rincon S., Papneja N., Miller W. A Review of Cancer Immunotherapy: From the Past, to the Present, to the Future. Curr. Oncol. 2020;27:87–97. doi: 10.3747/co.27.5223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee S., Margolin K. Cytokines in Cancer Immunotherapy. Cancers. 2011;3:3856–3893. doi: 10.3390/cancers3043856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Strizova Z., Bartunkova J., Smrz D. The challenges of adoptive cell transfer in the treatment of human renal cell carcinoma. Cancer Immunol. Immunother. 2019;68:1831–1838. doi: 10.1007/s00262-019-02359-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Galluzzi L., Vacchelli E., Pedro J.M.B.-S., Buqué A., Senovilla L., Baracco E.E., Bloy N., Castoldi F., Abastado J.-P., Agostinis P., et al. Classification of current anticancer immunotherapies. Oncotarget. 2014;5:12472–12508. doi: 10.18632/oncotarget.2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cheever M.A., Higano C.S. PROVENGE (Sipuleucel-T) in Prostate Cancer: The First FDA-Approved Therapeutic Cancer Vaccine. Clin. Cancer Res. 2011;17:3520–3526. doi: 10.1158/1078-0432.CCR-10-3126. [DOI] [PubMed] [Google Scholar]
- 26.Sondak V.K., Smalley K.S.M., Kudchadkar R., Grippon S., Kirkpatrick P. Ipilimumab. Nat. Rev. Drug Discov. 2011;10:411–412. doi: 10.1038/nrd3463. [DOI] [PubMed] [Google Scholar]
- 27.Hoos A. Development of immuno-oncology drugs—From CTLA4 to PD1 to the next generations. Nat. Rev. Drug Discov. 2016;15:235–247. doi: 10.1038/nrd.2015.35. [DOI] [PubMed] [Google Scholar]
- 28.Postow M.A., Sidlow R., Hellmann M.D. Immune-Related Adverse Events Associated with Immune Checkpoint Blockade. N. Engl. J. Med. 2018;378:158–168. doi: 10.1056/NEJMra1703481. [DOI] [PubMed] [Google Scholar]
- 29.Moslehi J.J., Salem J.-E., Sosman J.A., Lebrun-Vignes B., Johnson D.B. Increased reporting of fatal immune checkpoint inhibitor-associated myocarditis. Lancet. 2018;391:933. doi: 10.1016/S0140-6736(18)30533-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hodi F.S., Sileni V.C., Gonzalez R., Grob J.-J., Rutkowski P., Cowey C.L., Lao C.D., Schadendorf D., Wagstaff J., Dummer R., et al. Nivolumab plus ipilimumab or nivolumab alone versus ipilimumab alone in advanced melanoma (CheckMate 067): 4-year outcomes of a multicentre, randomised, phase 3 trial. Lancet Oncol. 2018;19:1480–1492. doi: 10.1016/S1470-2045(18)30700-9. [DOI] [PubMed] [Google Scholar]
- 31.Liudahl S., Coussens L.M. B cells as biomarkers: Predicting immune checkpoint therapy adverse events. J. Clin. Investig. 2018;128:577–579. doi: 10.1172/JCI99036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Diehl A., Yarchoan M., Hopkins A., Jaffee E., Grossman S.A. Relationships between lymphocyte counts and treatment-related toxicities and clinical responses in patients with solid tumors treated with PD-1 checkpoint inhibitors. Oncotarget. 2017;8:114268–114280. doi: 10.18632/oncotarget.23217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dempke W.C.M., Fenchel K., Uciechowski P., Dale S.P. Second- and third-generation drugs for immuno-oncology treatment-The more the better? Eur. J. Cancer. 2017;74:55–72. doi: 10.1016/j.ejca.2017.01.001. [DOI] [PubMed] [Google Scholar]
- 34.Lin W., Bosch-Voskens C., Zhang X., Schindler D.G., Wood A., Burch E., Wei Y., Chen L., Tian G., Tamada K., et al. Fc-dependent expression of CD137 on human NK cells: Insights into “agonistic” effects of anti-CD137 monoclonal antibodies. Blood. 2008;112:699–707. doi: 10.1182/blood-2007-11-122465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Knee D.A., Hewes B., Brogdon J.L. Rationale for anti-GITR cancer immunotherapy. Eur. J. Cancer. 2016;67:1–10. doi: 10.1016/j.ejca.2016.06.028. [DOI] [PubMed] [Google Scholar]
- 36.Ruby C.E., Yates M.A., Hirschhorn-Cymerman D., Chlebeck P., Wolchok J.D., Houghton A.N., Offner H., Weinberg A.D. Cutting Edge: OX40 Agonists Can Drive Regulatory T Cell Expansion if the Cytokine Milieu Is Right. J. Immunol. 2009;183:4853–4857. doi: 10.4049/jimmunol.0901112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lines J.L., Sempere L.F., Broughton T., Wang L., Noelle R. VISTA Is a Novel Broad-Spectrum Negative Checkpoint Regulator for Cancer Immunotherapy. Cancer Immunol. Res. 2014;2:510–517. doi: 10.1158/2326-6066.CIR-14-0072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Uyttenhove C., Pilotte L., Théate I., Stroobant V., Colau D., Parmentier N., Boon T., Eynde B.J.V.D. Evidence for a tumoral immune resistance mechanism based on tryptophan degradation by indoleamine 2,3-dioxygenase. Nat. Med. 2003;9:1269–1274. doi: 10.1038/nm934. [DOI] [PubMed] [Google Scholar]
- 39.Kruger S., Ilmer M., Kobold S., Cadilha B.L., Endres S., Ormanns S., Schuebbe G., Renz B.W., D’Haese J.G., Schloesser H., et al. Advances in cancer immunotherapy 2019—Latest trends. J. Exp. Clin. Cancer Res. 2019;38:268. doi: 10.1186/s13046-019-1266-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Goldberg M.S. Improving cancer immunotherapy through nanotechnology. Nat. Rev. Cancer. 2019;19:587–602. doi: 10.1038/s41568-019-0186-9. [DOI] [PubMed] [Google Scholar]
- 41.Park S., Dong H., Liu X., Harrington S.M., Krco C.J., Grams M.P., Mansfield A.S., Furutani K.M., Olivier K.R., Kwon E.D. PD-1 Restrains Radiotherapy-Induced Abscopal Effect. Cancer Immunol. Res. 2015;3:610–619. doi: 10.1158/2326-6066.CIR-14-0138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.West E.E., Jin H.T., Rasheed A.U., Penaloza-Macmaster P., Ha S.J., Tan W.G., Youngblood B., Freeman G.J., Smith K.A., Ahmed R. PD-L1 blockade synergizes with IL-2 therapy in reinvigorating exhausted T cells. J. Clin. Investig. 2013;123:2604–2615. doi: 10.1172/JCI67008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brahmer J.R., Lacchetti C., Schneider B.J., Atkins M.B., Brassil K.J., Caterino J.M., Chau I., Ernstoff M.S., Gardner J.M., Ginex P., et al. Management of Immune-Related Adverse Events in Patients Treated With Immune Checkpoint Inhibitor Therapy: American Society of Clinical Oncology Clinical Practice Guideline. J. Clin. Oncol. 2018;36:1714–1768. doi: 10.1200/JCO.2017.77.6385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bai R., Lv Z., Xu D., Cui J. Predictive biomarkers for cancer immunotherapy with immune checkpoint inhibitors. Biomark. Res. 2020;8:1–17. doi: 10.1186/s40364-020-00209-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Venter J.C., Adams M.D., Myers E.W., Li P.W., Mural R.J., Sutton G.G., Smith H.O., Yandell M., Evans C.A., Holt R.A., et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 46.Nielsen J. Systems Biology of Metabolism. Annu. Rev. Biochem. 2017;86:245–275. doi: 10.1146/annurev-biochem-061516-044757. [DOI] [PubMed] [Google Scholar]
- 47.Menyhárt O., Győrffy B. Multi-omics approaches in cancer research with applications in tumor subtyping, prognosis, and diagnosis. Comput. Struct. Biotechnol. J. 2021;19:949–960. doi: 10.1016/j.csbj.2021.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xu Y., Dong Q., Li F., Xu Y., Hu C., Wang J., Shang D., Zheng X., Yang H., Zhang C., et al. Identifying subpathway signatures for individualized anticancer drug response by integrating multi-omics data. J. Transl. Med. 2019;17:1–16. doi: 10.1186/s12967-019-2010-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tsugawa H., Cajka T., Kind T., Ma Y., Higgins B., Ikeda K., Kanazawa M., VanderGheynst J., Fiehn O., Arita M. MS-DIAL: Data-independent MS/MS deconvolution for com-prehensive metabolome analysis. Nat. Methods. 2015;12:523–526. doi: 10.1038/nmeth.3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Katajamaa M., Miettinen J., Orešič M. MZmine: Toolbox for processing and visualization of mass spectrometry based molecular profile data. Bioinformatics. 2006;22:634–636. doi: 10.1093/bioinformatics/btk039. [DOI] [PubMed] [Google Scholar]
- 51.Chong J., Wishart D.S., Xia J. Using MetaboAnalyst 4.0 for Comprehensive and Integrative Metabolomics Data Analysis. Curr. Protoc. Bioinform. 2019;68:e86. doi: 10.1002/cpbi.86. [DOI] [PubMed] [Google Scholar]
- 52.Zhou G., Soufan O., Ewald J., Hancock R., Basu N., Xia J. NetworkAnalyst 3.0: A visual analytics platform for comprehensive gene expression profiling and meta-analysis. Nucleic Acids Res. 2019;47:W234–W241. doi: 10.1093/nar/gkz240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li J., Lu Y., Akbani R., Ju Z., Roebuck P.L., Liu W., Yang J.-Y., Broom B.M., Verhaak R.G.W., Kane D.W., et al. TCPA: A resource for cancer functional proteomics data. Nat. Methods. 2013;10:1046–1047. doi: 10.1038/nmeth.2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chang K., Creighton C.J., Davis C., Donehower L., Drummond J., Wheeler D., Ally A., Balasundaram M., Birol I., Butterfield Y.S.N., et al. The Cancer Genome Atlas Pan-Cancer analysis project. Nat. Genet. 2013;45:1113–1120. doi: 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kind T., Liu K.-H., Lee D.Y., DeFelice B., Meissen J.K., Fiehn O. LipidBlast in silico tandem mass spectrometry database for lipid identification. Nat. Methods. 2013;10:755–758. doi: 10.1038/nmeth.2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kind T., Wohlgemuth G., Lee D.Y., Lu Y., Palazoglu M., Shahbaz S., Fiehn O. FiehnLib: Mass Spectral and Retention Index Libraries for Metabolomics Based on Quadrupole and Time-of-Flight Gas Chromatography/Mass Spectrometry. Anal. Chem. 2009;81:10038–10048. doi: 10.1021/ac9019522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mun D.G., Bhin J., Kim S., Kim H., Jung J.H., Jung Y., Jang Y.E., Park J.M., Kim H., Jung Y., et al. Proteogenomic Characterization of Human Early-Onset Gastric Cancer. Cancer Cell. 2019;35:111–124. doi: 10.1016/j.ccell.2018.12.003. [DOI] [PubMed] [Google Scholar]
- 58.Kang Y.P., Mockabee-Macias A., Jiang C., Falzone A., Prieto-Farigua N., Stone E., Harris I.S., DeNicola G.M. Non-canonical Glutamate-Cysteine Ligase Activity Protects against Ferroptosis. Cell Metab. 2021;33:174–189.e7. doi: 10.1016/j.cmet.2020.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Turanli B., Karagoz K., Gulfidan G., Sinha R., Mardinoglu A., Arga K.Y. A Network-Based Cancer Drug Discovery: From Integrated Multi-Omics Approaches to Precision Medicine. Curr. Pharm. Des. 2019;24:3778–3790. doi: 10.2174/1381612824666181106095959. [DOI] [PubMed] [Google Scholar]
- 60.Wilmes A., Bielow C., Ranninger C., Bellwon P., Aschauer L., Limonciel A., Chassaigne H., Kristl T., Aiche S., Huber C.G., et al. Mechanism of cisplatin proximal tubule toxicity revealed by integrating transcriptomics, proteomics, metabolomics and biokinetics. Toxicol. Vitr. 2015;30:117–127. doi: 10.1016/j.tiv.2014.10.006. [DOI] [PubMed] [Google Scholar]
- 61.Shajahan-Haq A.N., Cheema M.S., Clarke R. Application of Metabolomics in Drug Resistant Breast Cancer Research. Metabolites. 2015;5:100–118. doi: 10.3390/metabo5010100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Subramanian I., Verma S., Kumar S., Jere A., Anamika K. Multi-omics Data Integration, Interpretation, and Its Application. Bioinform. Biol. Insights. 2020;14:1177932219899051. doi: 10.1177/1177932219899051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pinu F.R., Beale D.J., Paten A.M., Kouremenos K., Swarup S., Schirra H.J., Wishart D. Systems Biology and Multi-Omics Integration: Viewpoints from the Metabolomics Research Community. Metabolites. 2019;9:76. doi: 10.3390/metabo9040076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hasin Y., Seldin M., Lusis A. Multi-omics approaches to disease. Genome Biol. 2017;18:1–15. doi: 10.1186/s13059-017-1215-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Buchberger A.R., Delaney K., Johnson J., Jillian J. Mass Spectrometry Imaging: A Review of Emerging Advancements and Future Insights. Anal. Chem. 2017;90:240–265. doi: 10.1021/acs.analchem.7b04733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shariatgorji M., Svenningsson P., Andrén P.E. Mass Spectrometry Imaging, an Emerging Technology in Neuropsychopharmacology. Neuropsychopharmacology. 2013;39:34–49. doi: 10.1038/npp.2013.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fernández-García J., Altea-Manzano P., Pranzini E., Fendt S.-M. Stable Isotopes for Tracing Mammalian-Cell Metabolism In Vivo. Trends Biochem. Sci. 2020;45:185–201. doi: 10.1016/j.tibs.2019.12.002. [DOI] [PubMed] [Google Scholar]
- 68.Jang C., Chen L., Rabinowitz J.D. Metabolomics and Isotope Tracing. Cell. 2018;173:822–837. doi: 10.1016/j.cell.2018.03.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lee D.-K., Na E., Park S., Park J.H., Lim J., Kwon S.W. In Vitro Tracking of Intracellular Metabolism-Derived Cancer Volatiles via Isotope Labeling. ACS Central Sci. 2018;4:1037–1044. doi: 10.1021/acscentsci.8b00296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ran F.A., Hsu P.D., Wright J.D., Agarwala V., Scott D.A., Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yin H., Xue W., Anderson D.G. CRISPR–Cas: A tool for cancer research and therapeutics. Nat. Rev. Clin. Oncol. 2019;16:281–295. doi: 10.1038/s41571-019-0166-8. [DOI] [PubMed] [Google Scholar]
- 72.Crick F. Central Dogma of Molecular Biology. Nat. Cell Biol. 1970;227:561–563. doi: 10.1038/227561a0. [DOI] [PubMed] [Google Scholar]
- 73.Johnson C., Ivanisevic J., Siuzdak G. Metabolomics: Beyond biomarkers and towards mechanisms. Nat. Rev. Mol. Cell Biol. 2016;17:451–459. doi: 10.1038/nrm.2016.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Klein S., Heinzle E. Isotope labeling experiments in metabolomics and fluxomics. Wiley Interdiscip. Rev. Syst. Biol. Med. 2012;4:261–272. doi: 10.1002/wsbm.1167. [DOI] [PubMed] [Google Scholar]
- 75.Vale G., Martin S.A., Mitsche M.A., Thompson B., Eckert K.M., McDonald J.G. Three-phase liquid extraction: A simple and fast method for lipidomic workflows. J. Lipid Res. 2019;60:694–706. doi: 10.1194/jlr.D090795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Römisch-Margl W., Prehn C., Bogumil R., Röhring C., Suhre K., Adamski J. Procedure for tissue sample preparation and metabolite extraction for high-throughput targeted metabolomics. Metabolomics. 2011;8:133–142. doi: 10.1007/s11306-011-0293-4. [DOI] [Google Scholar]
- 77.Gika H.G., Theodoridis G.A., Plumb R.S., Wilson I. Current practice of liquid chromatography–mass spectrometry in metabolomics and metabonomics. J. Pharm. Biomed. Anal. 2014;87:12–25. doi: 10.1016/j.jpba.2013.06.032. [DOI] [PubMed] [Google Scholar]
- 78.Karpievitch Y.V., Polpitiya A.D., Anderson G.A., Smith R., Dabney A.R. Liquid chromatography mass spectrometry-based proteomics: Biological and technological aspects. Ann. Appl. Stat. 2010;4:1797–1823. doi: 10.1214/10-AOAS341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang Q., Salzler R., Dore A., Yang J., Ma D., Olson W.C., Liu Y. Multiplex Immuno-Liquid Chromatography-Mass Spectrometry-Parallel Reaction Monitoring (LC-MS-PRM) Quantitation of CD8A, CD4, LAG3, PD1, PD-L1, and PD-L2 in Frozen Human Tissues. J. Proteome Res. 2018;17:3932–3940. doi: 10.1021/acs.jproteome.8b00605. [DOI] [PubMed] [Google Scholar]
- 80.Millet A., Khoudour N., Bros P., Lebert D., Picard G., Machon C., Goldwasser F., Blanchet B., Guitton J. Quantification of nivolumab in human plasma by LC-MS/HRMS and LC-MS/MS, comparison with ELISA. Talanta. 2021;224:121889. doi: 10.1016/j.talanta.2020.121889. [DOI] [PubMed] [Google Scholar]
- 81.Palaskas N.J., Garcia J.D., Shirazi R., Shin D., Puig-Saus C., Braas D., Ribas A., Graeber T.G. Global alteration of T-lymphocyte metabolism by PD-L1 checkpoint involves a block of de novo nucleoside phosphate synthesis. Cell Discov. 2019;5:1–10. doi: 10.1038/s41421-019-0130-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Liu X., Hartman C.L., Li L., Albert C.J., Si F., Gao A., Huang L., Zhao Y., Lin W., Hsueh E.C., et al. Reprogramming lipid metabolism prevents effector T cell senescence and enhances tumor immunotherapy. Sci. Transl. Med. 2021;13:eaaz6314. doi: 10.1126/scitranslmed.aaz6314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hamaidi I., Zhang L., Kim N., Wang M.-H., Iclozan C., Fang B., Liu M., Koomen J.M., Berglund A.E., Yoder S.J., et al. Sirt2 Inhibition Enhances Metabolic Fitness and Effector Functions of Tumor-Reactive T Cells. Cell Metab. 2020;32:420–436.e12. doi: 10.1016/j.cmet.2020.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kesarwani P., Prabhu A., Kant S., Kumar P., Graham S.F., Buelow K.L., Wilson G.D., Miller C.R., Chinnaiyan P. Tryptophan Metabolism Contributes to Radiation-Induced Immune Checkpoint Reactivation in Glioblastoma. Clin. Cancer Res. 2018;24:3632–3643. doi: 10.1158/1078-0432.CCR-18-0041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kocher F., Amann A., Zimmer K., Geisler S., Fuchs D., Pichler R., Wolf D., Kurz K., Seeber A., Pircher A. High indoleamine-2,3-dioxygenase 1 (IDO) activity is linked to primary resistance to immunotherapy in non-small cell lung cancer (NSCLC) Transl. Lung Cancer Res. 2021;10:304–313. doi: 10.21037/tlcr-20-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sadik A., Patterson L.F.S., Öztürk S., Mohapatra S.R., Panitz V., Secker P.F., Pfänder P., Loth S., Salem H., Prentzell M.T., et al. IL4I1 Is a Metabolic Immune Checkpoint that Activates the AHR and Promotes Tumor Progression. Cell. 2020;182:1252–1270.e34. doi: 10.1016/j.cell.2020.07.038. [DOI] [PubMed] [Google Scholar]
- 87.Luo Y., Gao Y., Liu W., Yang Y., Jiang J., Wang Y., Tang W., Yang S., Sun L., Cai J., et al. MYC-Protein Arginine Methyltransferase 5 Axis Defines the Tumorigenesis and Immune Response in Hepatocellular Carcinoma. Hepatology. 2021 doi: 10.1002/hep.31864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Montfort A., Bertrand F., Rochotte J., Gilhodes J., Filleron T., Milhès J., Dufau C., Imbert C., Riond J., Tosolini M., et al. Neutral Sphingomyelinase 2 Heightens Anti-Melanoma Immune Responses and Anti–PD-1 Therapy Efficacy. Cancer Immunol. Res. 2021;9:568–582. doi: 10.1158/2326-6066.CIR-20-0342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kawashima M., Tokiwa M., Nishimura T., Kawata Y., Sugimoto M., Kataoka T.R., Sakurai T., Iwaisako K., Suzuki E., Hagiwara M., et al. High-resolution imaging mass spec-trometry combined with transcriptomic analysis identified a link between fatty acid composition of phosphatidylinositols and the immune checkpoint pathway at the primary tumour site of breast cancer. Br. J. Cancer. 2020;122:245–257. doi: 10.1038/s41416-019-0662-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Best S., DE Souza D., Kersbergen A., Policheni A.N., Dayalan S., Tull D., Rathi V., Gray D., Ritchie M.E., McConville M.J., et al. Synergy between the KEAP1/NRF2 and PI3K Pathways Drives Non-Small-Cell Lung Cancer with an Altered Immune Microenvironment. Cell Metab. 2018;27:935–943.e4. doi: 10.1016/j.cmet.2018.02.006. [DOI] [PubMed] [Google Scholar]
- 91.Pacheco-Torres J., Penet M.-F., Mironchik Y., Krishnamachary B., Bhujwalla Z.M. The PD-L1 metabolic interactome intersects with choline metabolism and inflammation. Cancer Metab. 2021;9:1–16. doi: 10.1186/s40170-021-00245-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lee J.S., Adler L., Karathia H., Carmel N., Rabinovich S., Auslander N., Keshet R., Stettner N., Silberman A., Agemy L., et al. Urea Cycle Dysregulation Generates Clinically Relevant Genomic and Biochemical Signatures. Cell. 2018;174:1559–1570. doi: 10.1016/j.cell.2018.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhang Y., Chandra V., Riquelme Sanchez E., Dutta P., Quesada P.R., Rakoski A., Zoltan M., Arora N., Baydogan S., Horne W., et al. Interleukin-17-induced neutrophil extra-cellular traps mediate resistance to checkpoint blockade in pancreatic cancer. J. Exp. Med. 2020;217:e20190354. doi: 10.1084/jem.20190354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Li H., Bullock K., Gurjao C., Braun D., Shukla S.A., Bossé D., Lalani A.-K.A., Gopal S., Jin C., Horak C., et al. Metabolomic adaptations and correlates of survival to immune checkpoint blockade. Nat. Commun. 2019;10:4346. doi: 10.1038/s41467-019-12361-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Mock A., Zschäbitz S., Kirsten R., Scheffler M., Wolf B., Herold-Mende C., Kramer R., Busch E., Jenzer M., Jäger D., et al. Serum very long-chain fatty acid-containing lipids predict response to immune checkpoint inhibitors in urological cancers. Cancer Immunol. Immunother. 2019;68:2005–2014. doi: 10.1007/s00262-019-02428-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Karayama M., Masuda J., Mori K., Yasui H., Hozumi H., Suzuki Y., Furuhashi K., Fujisawa T., Enomoto N., Nakamura Y., et al. Comprehensive assessment of multiple tryptophan metabolites as potential biomarkers for immune checkpoint inhibitors in patients with non-small cell lung cancer. Clin. Transl. Oncol. 2020;23:418–423. doi: 10.1007/s12094-020-02421-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ghini V., Laera L., Fantechi B., Del Monte F., Benelli M., McCartney A., Leonardo T., Luchinat C., Pozzessere D. Metabolomics to Assess Response to Immune Checkpoint Inhibitors in Patients with Non-Small-Cell Lung Cancer. Cancers. 2020;12:3574. doi: 10.3390/cancers12123574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bui A., Choi Y., Frankel A.E., Koh A.Y. Unbiased Microbiome and Metabolomic Profiling of Fecal Samples from Patients with Melanoma. Methods Mol. Biol. 2021;2265:461–474. doi: 10.1007/978-1-0716-1205-7_33. [DOI] [PubMed] [Google Scholar]
- 99.Vernocchi P., Gili T., Conte F., Del Chierico F., Conta G., Miccheli A., Botticelli A., Paci P., Caldarelli G., Nuti M., et al. Network Analysis of Gut Microbiome and Metabolome to Discover Microbiota-Linked Biomarkers in Patients Affected by Non-Small Cell Lung Cancer. Int. J. Mol. Sci. 2020;21:8730. doi: 10.3390/ijms21228730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Botticelli A., Vernocchi P., Marini F., Quagliariello A., Cerbelli B., Reddel S., Del Chierico F., Di Pietro F., Giusti R., Tomassini A., et al. Gut metabolomics profiling of non-small cell lung cancer (NSCLC) patients under immunotherapy treatment. J. Transl. Med. 2020;18:1–10. doi: 10.1186/s12967-020-02231-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zizzari I.G., Di Filippo A., Scirocchi F., Di Pietro F.R., Rahimi H., Ugolini A., Scagnoli S., Vernocchi P., Del Chierico F., Putignani L., et al. Soluble Immune Checkpoints, Gut Metabolites and Performance Status as Parameters of Response to Nivolumab Treatment in NSCLC Patients. J. Pers. Med. 2020;10:208. doi: 10.3390/jpm10040208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lee S.-H., Cho S.-Y., Yoon Y., Park C., Sohn J., Jeong J.-J., Jeon B.-N., Jang M., An C., Lee S., et al. Bifidobacterium bifidum strains synergize with immune checkpoint inhibitors to reduce tumour burden in mice. Nat. Microbiol. 2021;6:277–288. doi: 10.1038/s41564-020-00831-6. [DOI] [PubMed] [Google Scholar]
- 103.Nomura M., Nagatomo R., Doi K., Shimizu J., Baba K., Saito T., Matsumoto S., Inoue K., Muto M. Association of Short-Chain Fatty Acids in the Gut Microbiome With Clinical Response to Treatment With Nivolumab or Pembrolizumab in Patients With Solid Cancer Tumors. JAMA Netw. Open. 2020;3:e202895. doi: 10.1001/jamanetworkopen.2020.2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Song P., Yang D., Wang H., Cui X., Si X., Zhang X., Zhang L. Relationship between intestinal flora structure and metabolite analysis and immunotherapy efficacy in Chinese NSCLC patients. Thorac. Cancer. 2020;11:1621–1632. doi: 10.1111/1759-7714.13442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Heng B., Bilgin A.A., Lovejoy D.B., Tan V.X., Milioli H.H., Gluch L., Bustamante S., Sabaretnam T., Moscato P., Lim C.K., et al. Differential kynurenine pathway metabolism in highly metastatic aggressive breast cancer subtypes: Beyond IDO1-induced immunosuppression. Breast Cancer Res. 2020;22:1–14. doi: 10.1186/s13058-020-01351-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lucarelli G., Rutigliano M., Ferro M., Giglio A., Intini A., Triggiano F., Palazzo S., Gigante M., Castellano G., Ranieri E., et al. Activation of the kynurenine pathway predicts poor outcome in patients with clear cell renal cell carcinoma. Urol. Oncol. Semin. Orig. Investig. 2017;35:461. doi: 10.1016/j.urolonc.2017.02.011. [DOI] [PubMed] [Google Scholar]
- 107.Grigorieva J., Asmellash S., Net L., Tsypin M., Order H., Roder J. Mass Spectrometry-Based Multivariate Proteomic Tests for Prediction of Outcomes on Immune Checkpoint Blockade Therapy: The Modern Analytical Approach. Int. J. Mol. Sci. 2020;21:838. doi: 10.3390/ijms21030838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Johnson C.H., Spilker M.E., Goetz L., Peterson S.N., Siuzdak G. Metabolite and Microbiome Interplay in Cancer Immunotherapy. Cancer Res. 2016;76:6146–6152. doi: 10.1158/0008-5472.CAN-16-0309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Huo S., Liu L., Liu J., Li Q., Wang J. Modulation of cancer immunotherapy efficacy by gut microbiota. Discov. Med. 2019;27:93–100. [PubMed] [Google Scholar]
- 110.Arpaia N., Campbell C., Fan X., Dikiy S., Van Der Veeken J., DeRoos P., Liu H., Cross J., Pfeffer K., Coffer P.J., et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nat. Cell Biol. 2013;504:451–455. doi: 10.1038/nature12726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Sivan A., Corrales L., Hubert N., Williams J.B., Aquino-Michaels K., Earley Z.M., Benyamin F.W., Lei Y.M., Jabri B., Alegre M.-L., et al. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science. 2015;350:1084–1089. doi: 10.1126/science.aac4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Vétizou M., Pitt J.M., Daillère R., Lepage P., Waldschmitt N., Flament C., Rusakiewicz S., Routy B., Roberti M.P., Duong C., et al. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science. 2015;350:1079–1084. doi: 10.1126/science.aad1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Frankel A.E., Coughlin L.A., Kim J., Froehlich T.W., Xie Y., Frenkel E.P., Koh A. Metagenomic Shotgun Sequencing and Unbiased Metabolomic Profiling Identify Specific Human Gut Microbiota and Metabolites Associated with Immune Checkpoint Therapy Efficacy in Melanoma Patients. Neoplasia. 2017;19:848–855. doi: 10.1016/j.neo.2017.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Anagnostou V., Smith K.N., Forde P.M., Niknafs N., Bhattacharya R., White J., Zhang T., Adleff V., Phallen J., Wali N., et al. Evolution of Neoantigen Landscape during Immune Checkpoint Blockade in Non-Small Cell Lung Cancer. Cancer Discov. 2017;7:264–276. doi: 10.1158/2159-8290.CD-16-0828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Brocco D., Lanuti P., Pieragostino D., Cufaro M., Simeone P., Bologna G., Di Marino P., De Tursi M., Grassadonia A., Irtelli L., et al. Phenotypic and Proteomic Analysis Identifies Hallmarks of Blood Circulating Extracellular Vesicles in NSCLC Responders to Immune Checkpoint Inhibitors. Cancers. 2021;13:585. doi: 10.3390/cancers13040585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Griss J., Bauer W., Wagner C., Simon M., Chen M., Grabmeier-Pfistershammer K., Maurer-Granofszky M., Roka F., Penz T., Bock C., et al. B cells sustain inflammation and predict response to immune checkpoint blockade in human melanoma. Nat. Commun. 2019;10:4186. doi: 10.1038/s41467-019-12160-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wang L.-B., Karpova A., Gritsenko M.A., Kyle J.E., Cao S., Li Y., Rykunov D., Colaprico A., Rothstein J.H., Hong R., et al. Proteogenomic and metabolomic characterization of human glioblastoma. Cancer Cell. 2021;39:509–528.e20. doi: 10.1016/j.ccell.2021.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Nabet B.Y., Esfahani M.S., Moding E.J., Hamilton E.G., Chabon J.J., Rizvi H., Steen C.B., Chaudhuri A.A., Liu C.L., Hui A.B., et al. Noninvasive Early Identification of Therapeutic Benefit from Immune Checkpoint Inhibition. Cell. 2020;183:363–376.e13. doi: 10.1016/j.cell.2020.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Momparler R.L. Cancer epigenetics. Oncogene. 2003;22:6479–6483. doi: 10.1038/sj.onc.1206774. [DOI] [PubMed] [Google Scholar]
- 120.Feng Y., Rudensky A.Y. DNA methylation secures CD4+ and CD8+ T cell lineage borders. Nat. Immunol. 2015;16:681–683. doi: 10.1038/ni.3207. [DOI] [PubMed] [Google Scholar]
- 121.Duruisseaux M., Martínez-Cardús A., Calleja-Cervantes M.E., Moran S., de Moura M.C., Davalos V., Piñeyro D., Sanchez-Cespedes M., Girard N., Brevet M., et al. Epigenetic prediction of response to anti-PD-1 treatment in non-small-cell lung cancer: A multicentre, retrospective analysis. Lancet Respir. Med. 2018;6:771–781. doi: 10.1016/S2213-2600(18)30284-4. [DOI] [PubMed] [Google Scholar]
- 122.Klempner S.J., Fabrizio D., Bane S., Reinhart M., Peoples T., Ali S.M., Sokol E.S., Frampton G., Schrock A.B., Anhorn R., et al. Tumor Mutational Burden as a Predictive Biomarker for Response to Immune Checkpoint Inhibitors: A Review of Current Evidence. Oncologist. 2019;25:e147–e159. doi: 10.1634/theoncologist.2019-0244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Eso Y., Shimizu T., Takeda H., Takai A., Marusawa H. Microsatellite instability and immune checkpoint inhibitors: Toward precision medicine against gastrointestinal and hepato-biliary cancers. J. Gastroenterol. 2020;55:15–26. doi: 10.1007/s00535-019-01620-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Evrard C., Tachon G., Randrian V., Karayan-Tapon L., Tougeron D. Microsatellite Instability: Diagnosis, Heterogeneity, Discordance, and Clinical Impact in Colorectal Cancer. Cancers. 2019;11:1567. doi: 10.3390/cancers11101567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Vanderwalde A., Spetzler D., Xiao N., Gatalica Z., Marshall J. Microsatellite instability status determined by next-generation sequencing and compared with PD-L1 and tumor mutational burden in 11,348 patients. Cancer Med. 2018;7:746–756. doi: 10.1002/cam4.1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Kirschner M.W. The Meaning of Systems Biology. Cell. 2005;121:503–504. doi: 10.1016/j.cell.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 127.Joshi A., Rienks M., Theofilatos K., Mayr M. Systems biology in cardiovascular disease: A multiomics approach. Nat. Rev. Cardiol. 2020;18:313–330. doi: 10.1038/s41569-020-00477-1. [DOI] [PubMed] [Google Scholar]
- 128.Misra B.B., Langefeld C.D., Olivier M., Cox L.A. Integrated Omics: Tools, Advances, and Future Approaches. J. Mol. Endocrinol. 2018 doi: 10.1530/JME-18-0055. [DOI] [PubMed] [Google Scholar]
- 129.Lindskrog S.V., Prip F., Lamy P., Taber A., Groeneveld C.S., Birkenkamp-Demtröder K., Jensen J.B., Strandgaard T., Nordentoft I., Christensen E., et al. An integrated multi-omics analysis identifies prognostic molecular subtypes of non-muscle-invasive bladder cancer. Nat. Commun. 2021;12:1–18. doi: 10.1038/s41467-021-22465-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Vasaikar S., Straub P., Wang J., Zhang B. Abstract 5112: LinkedOmics: Analyzing multi-omics data within and across 32 cancer types. Nucleic Acids Res. 2019 doi: 10.1158/1538-7445.am2019-5112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Olivier M., Asmis R., Hawkins G.A., Howard T.D., Cox L.A. The Need for Multi-Omics Biomarker Signatures in Precision Medicine. Int. J. Mol. Sci. 2019;20:4781. doi: 10.3390/ijms20194781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Akiyama Y., Kondou R., Iizuka A., Ohshima K., Urakami K., Nagashima T., Shimoda Y., Tanabe T., Ohnami S., Ohnami S., et al. Immune response-associated gene analysis of 1,000 cancer patients using whole-exome sequencing and gene expression profiling-Project HOPE. Biomed. Res. 2016;37:233–242. doi: 10.2220/biomedres.37.233. [DOI] [PubMed] [Google Scholar]
- 133.Yoshikawa S., Kiyohara Y., Otsuka M., Kondou R., Nonomura C., Miyata H., Iizuka A., Ohshima K., Urakami K., Nagashima T., et al. Multi-omics Profiling of Patients with Melanoma Treated with Nivolumab in Project HOPE. Anticancer. Res. 2017;37:1321–1328. doi: 10.21873/anticanres.11450. [DOI] [PubMed] [Google Scholar]
- 134.Feng B., Shen Y., Hostench X.P., Bieg M., Plath M., Ishaque N., Eils R., Freier K., Weichert W., Zaoui K., et al. Integrative Analysis of Multi-omics Data Identified EGFR and PTGS2 as Key Nodes in a Gene Regulatory Network Related to Immune Phenotypes in Head and Neck Cancer. Clin. Cancer Res. 2020;26:3616–3628. doi: 10.1158/1078-0432.CCR-19-3997. [DOI] [PubMed] [Google Scholar]
- 135.Liu F., Qin L., Liao Z., Song J., Yuan C., Liu Y., Wang Y., Xu H., Zhang Q., Pei Y., et al. Microenvironment characterization and multi-omics signatures related to prognosis and immunotherapy response of hepatocellular carcinoma. Exp. Hematol. Oncol. 2020;9:1–16. doi: 10.1186/s40164-020-00165-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Fu J., Li K., Zhang W., Wan C., Zhang J., Jiang P., Liu X.S. Large-scale public data reuse to model immunotherapy response and resistance. Genome Med. 2020;12:21–28. doi: 10.1186/s13073-020-0721-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Anagnostou V., Bruhm D.C., Niknafs N., White J.R., Shao X.M., Sidhom J.W., Stein J., Tsai H.-L., Wang H., Belcaid Z., et al. Integrative Tumor and Immune Cell Multi-omic Analyses Predict Response to Immune Checkpoint Blockade in Melanoma. Cell Rep. Med. 2020;1:100139. doi: 10.1016/j.xcrm.2020.100139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Chun H.-J.E., Johann P.D., Milne K., Zapatka M., Buellesbach A., Ishaque N., Iskar M., Erkek S., Wei L., Tessier-Cloutier B., et al. Identification and Analyses of Extra-Cranial and Cranial Rhabdoid Tumor Molecular Subgroups Reveal Tumors with Cytotoxic T Cell Infiltration. Cell Rep. 2019;29:2338–2354. doi: 10.1016/j.celrep.2019.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Xiao Y., Ma D., Zhao S., Suo C., Shi J., Xue M.-Z., Ruan M., Wang H., Zhao J., Li Q., et al. Multi-Omics Profiling Reveals Distinct Microenvironment Characterization and Suggests Immune Escape Mechanisms of Triple-Negative Breast Cancer. Clin. Cancer Res. 2019;25:5002–5014. doi: 10.1158/1078-0432.CCR-18-3524. [DOI] [PubMed] [Google Scholar]
- 140.Chappell L., Russell A.J., Voet T. Protected Single-Cell (Multi)omics Technologies. Annu. Rev. Genom. Hum. Genet. 2018;19:15–41. doi: 10.1146/annurev-genom-091416-035324. [DOI] [PubMed] [Google Scholar]
- 141.Haque A., Engel J., Teichmann S.A., Lönnberg T. A practical guide to single-cell RNA-sequencing for biomedical research and clinical applications. Genome Med. 2017;9:1–12. doi: 10.1186/s13073-017-0467-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Regev A., Teichmann S.A., Lander E.S., Amit I., Benoist C., Birney E., Bodenmiller B., Campbell P., Carninci P., Clatworthy M., et al. The Human Cell Atlas. Elife. 2017;6:e27041. doi: 10.7554/eLife.27041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Rodriques S.G., Stickels R.R., Goeva A., Martin C.A., Murray E., Vanderburg C.R., Welch J., Chen L.M., Chen F., Macosko E.Z. Slide-seq: A scalable technology for measuring genome-wide expression at high spatial resolution. Science. 2019;363:1463–1467. doi: 10.1126/science.aaw1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Cao J., Cusanovich D.A., Ramani V., Aghamirzaie D., Pliner H.A., Hill A.J., Daza R.M., McFaline-Figueroa J.L., Packer J.S., Christiansen L., et al. Joint profiling of chromatin accessibility and gene expression in thousands of single cells. Science. 2018;361:1380–1385. doi: 10.1126/science.aau0730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Guo T., Li W., Cai X. Applications of Single-Cell Omics to Dissect Tumor Microenvironment. Front. Genet. 2020;11 doi: 10.3389/fgene.2020.548719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Bassez A., Vos H., Van Dyck L., Floris G., Arijs I., Desmedt C., Boeckx B., Bempt M.V., Nevelsteen I., Lambein K., et al. A single-cell map of intratumoral changes during anti-PD1 treatment of patients with breast cancer. Nat. Med. 2021;27:820–832. doi: 10.1038/s41591-021-01323-8. [DOI] [PubMed] [Google Scholar]
- 147.Sade-Feldman M., Yizhak K., Bjorgaard S.L., Ray J.P., De Boer C.G., Jenkins R.W., Lieb D.J., Chen J.H., Frederick D.T., Barzily-Rokni M., et al. Defining T Cell States Associated with Response to Checkpoint Immunotherapy in Melanoma. Cell. 2018;175:998–1013.e20. doi: 10.1016/j.cell.2018.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Xu C., Chen Y., Du X.-J., Liu J.-Q., Huang C.-L., Chen L., Zhou G.-Q., Li W.-F., Mao Y.-P., Hsu C., et al. Comparative safety of immune checkpoint inhibitors in cancer: Systematic review and network meta-analysis. BMJ. 2018;363:k4226. doi: 10.1136/bmj.k4226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Yoest J.M. Clinical features, predictive correlates, and pathophysiology of immune-related adverse events in immune checkpoint inhibitor treatments in cancer: A short review. ImmunoTargets Ther. 2017;6:73–82. doi: 10.2147/ITT.S126227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Ramos-Casals M., Brahmer J.R., Callahan M.K., Flores-Chávez A., Keegan N., Khamashta M.A., Lambotte O., Mariette X., Prat A., Suárez-Almazor M.E. Immune-related adverse events of checkpoint inhibitors. Nat. Rev. Dis. Prim. 2020;6:1–21. doi: 10.1038/s41572-020-0160-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Reid P.D., Cifu A.S., Bass A.R. Management of Immunotherapy-Related Toxicities in Patients Treated With Immune Checkpoint Inhibitor Therapy. JAMA. 2021;325:482–483. doi: 10.1001/jama.2020.17308. [DOI] [PubMed] [Google Scholar]
- 152.Thompson J.A., Schneider B.J., Brahmer J., Andrews S., Armand P., Bhatia S., Budde L.E., Costa L., Davies M., Dunnington D., et al. Management of Immunotherapy-Related Toxicities, Version 1.2019. J. Natl. Compr. Canc. Netw. 2019;17:255–289. doi: 10.6004/jnccn.2019.0013. [DOI] [PubMed] [Google Scholar]
- 153.Jing Y., Liu J., Ye Y., Pan L., Deng H., Wang Y., Yang Y., Diao L., Lin S.H., Mills G.B., et al. Multiomics prediction of immune-related adverse events during checkpoint im-munotherapy. Nat. Commun. 2020;11:4946. doi: 10.1038/s41467-020-18742-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Grigoriou M., Banos A., Hatzioannou A., Kloetgen A., Kouzis P., Aggouraki D., Zakopoulou R., Bamias G., Kassi E., Mavroudis D., et al. Regulatory T-cell Transcriptomic Re-programming Characterizes Adverse Events by Checkpoint Inhibitors in Solid Tumors. Cancer Immunol. Res. 2021 doi: 10.1158/2326-6066.CIR-20-0969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Das R., Bar N., Ferreira M., Newman A., Zhang L., Bailur J.K., Bacchiocchi A., Kluger H., Wei W., Halaban R., et al. Early B cell changes predict autoimmunity following combination immune checkpoint blockade. J. Clin. Investig. 2018;128:715–720. doi: 10.1172/JCI96798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Filin I.Y., Solovyeva V.V., Kitaeva K.V., Rutland C.S., Rizvanov A.A. Current Trends in Cancer Immunotherapy. Biomedicines. 2020;8:621. doi: 10.3390/biomedicines8120621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Vermaelen K. Vaccine Strategies to Improve Anti-cancer Cellular Immune Responses. Front. Immunol. 2019;10:8. doi: 10.3389/fimmu.2019.00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Berzofsky J.A., Terabe M., Trepel J.B., Pastan I., Stroncek D.F., Morris J.C., Wood L.V. Cancer vaccine strategies: Translation from mice to human clinical trials. Cancer Immunol. Immunother. 2017;67:1863–1869. doi: 10.1007/s00262-017-2084-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Robbins P.F., Lu Y.-C., El-Gamil M., Li Y.F., Gross C., Gartner J., Lin J.C., Teer J.K., Cliften P., Tycksen E., et al. Mining exomic sequencing data to identify mutated antigens recognized by adoptively transferred tumor-reactive T cells. Nat. Med. 2013;19:747–752. doi: 10.1038/nm.3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Matsushita H., Vesely M., Koboldt D.C., Rickert C.G., Uppaluri R., Magrini V.J., Arthur C.D., White J.M., Chen Y.-S., Shea L.K., et al. Cancer exome analysis reveals a T-cell-dependent mechanism of cancer immunoediting. Nat. Cell Biol. 2012;482:400–404. doi: 10.1038/nature10755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Perna F., Berman S.H., Soni R.K., Mansilla-Soto J., Eyquem J., Hamieh M., Hendrickson R.C., Brennan C.W., Sadelain M. Integrating Proteomics and Transcriptomics for Systematic Combinatorial Chimeric Antigen Receptor Therapy of AML. Cancer Cell. 2017;32:506–519.e5. doi: 10.1016/j.ccell.2017.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Lu Y.C., Zheng Z., Robbins P.F., Tran E., Prickett T.D., Gartner J.J., Li Y.F., Ray S., Franco Z., Bliskovsky V., et al. An Efficient Single-Cell RNA-Seq Approach to Identify Ne-oantigen-Specific T Cell Receptors. Mol. Ther. 2018;26:379–389. doi: 10.1016/j.ymthe.2017.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.