Abstract
Simple Summary
Colorectal cancer (CRC) is among the most common cancers and the third leading cause of cancer deaths worldwide. Despite the identification of alterations in DNA repair genes and the resulting genomic instability in sub-populations of CRC, therapies that exploit defects in DNA repair pathways or high level of replicative stress have been explored only in breast, ovarian, and other tumor types, but not yet systematically in CRC. Here, we discuss how targeting genes involved in the responses to replication stress and the repair of DNA double-strand breaks (DSBs) could provide new therapeutic opportunities to treat CRCs and have the potential to confer increased sensitivity to current chemotherapy regimens, thus, expanding the spectrum of therapy options, and potentially improving clinical outcomes for CRC patients.
Abstract
Despite the ample improvements of CRC molecular landscape, the therapeutic options still rely on conventional chemotherapy-based regimens for early disease, and few targeted agents are recommended for clinical use in the metastatic setting. Moreover, the impact of cytotoxic, targeted agents, and immunotherapy combinations in the metastatic scenario is not fully satisfactory, especially the outcomes for patients who develop resistance to these treatments need to be improved. Here, we examine the opportunity to consider therapeutic agents targeting DNA repair and DNA replication stress response as strategies to exploit genetic or functional defects in the DNA damage response (DDR) pathways through synthetic lethal mechanisms, still not explored in CRC. These include the multiple actors involved in the repair of DNA double-strand breaks (DSBs) through homologous recombination (HR), classical non-homologous end joining (NHEJ), and microhomology-mediated end-joining (MMEJ), inhibitors of the base excision repair (BER) protein poly (ADP-ribose) polymerase (PARP), as well as inhibitors of the DNA damage kinases ataxia-telangiectasia and Rad3 related (ATR), CHK1, WEE1, and ataxia-telangiectasia mutated (ATM). We also review the biomarkers that guide the use of these agents, and current clinical trials with targeted DDR therapies.
Keywords: colorectal cancer, DNA double strand break repair, replication stress, target therapy
1. Introduction
1.1. Standard of Care of Colorectal Cancer
Despite advances in screening, surveillance, and identification of clinical and molecular features holding significant prognostic value, colorectal cancer (CRC) still ranks the second leading cause of cancer-related deaths worldwide. The 5-year relative survival rate for CRC ranges from 90% for those diagnosed with localized disease to 14% for those with metastatic disease [1]. By 2030, an increase of 60% in the CRC global burden to more than 2.2 million new cases is expected [2], with a worrisome incidence rate increase up to 124% for patients in the age group of 20 to 34 years [3].
Most colorectal tumors occur sporadically and result from a 10 to 15 years sequenced tumorigenic process comprising a persistent accumulation of mutations, also known as adenoma-carcinoma sequence [4,5]. CRC screening through colonoscopy is recommended beginning at age 50 years and then every 10 years in order to detect and remove precursor lesions or, at least, identify the disease in its earliest stages [6]. However, up to 25% of CRC cases are diagnosed when distant metastases are already present [7].
The tumor-node-metastasis (TNM) classification and traditional clinicopathological assessment are commonly used to determine disease prognosis and to drive therapeutic decision-making in CRC. Since survival outcomes between colon and rectum tumors according to the TNM stages are very similar, these diseases therefore share the same staging system [8]. Treatment of stage I CRC (T1-T2, N0, M0) is based on surgical removal and surveillance with colonoscopy, and adjuvant therapy is not recommended. Stages II (T3-T4b, N0, M0) and III (any T, N1–N2b, M0) comprise non-metastatic CRC. For resectable non-metastatic CRC, the preferred surgical procedure is colectomy with removal of the regional lymph nodes [9]. Systemic treatment regimens for stages II and III CRC include doublets or triplets of cytotoxic chemotherapy (5-fluorouracil—5FU or capecitabine, oxaliplatin, and irinotecan). Targeted therapies, including antiangiogenics (bevacizumab, ramucirumab, or ziv-aflibercept) and monoclonal antibodies anti-EGFR (cetuximab, panitumumab) or anti-BRAF (encorafenib) or immune checkpoint inhibitors (anti-PD1/PD-L1 and anti-CTLA4) are the options for metastatic CRC [10].
While for stage III CRC adjuvant systemic therapy is mandatory, stage II CRC must be firstly assessed for potential risk/benefits of therapy and prognosis. Currently, adjuvant chemotherapy for stage II CRC is recommended to patents with less than 12 lymph nodes analyzed after surgery and poor prognostic features (such as poorly differentiated histology, lymphatic/vascular invasion, positive margins, obstruction/perforation, comorbidities) [11]. Currently, the options of adjuvant therapy for high-risk stage II CRC are FOLFOX (5-fluorouracil, leucovorin, and oxaliplatin), CAPEOX (capecitabine and oxaliplatin), capecitabine, or 5-fluorouracil/leucovorin for 3–6 months. The choice between these regimens depends on the patient’s performance status, T stage, the number of high-risk features for recurrence, and patient/physician preferences. In brief, single-agent capecitabine or 5-fluoouracil/leucovorin (5-FU/LV) are indicated for T3 and no high risk-features, while for T3 at high-risk of recurrence or T4 (stage IIB/IIC), FOLFOX, or CAPEOX are the preferred options [12]. Recently, microsatellite instability (MSI) or DNA mismatch repair (MMR) status has been included in risk assessment in stage II disease. For microsatellite-stable (MSS) or MMR-proficient (pMMR) low-risk stage II disease, only observation or adjuvant 5-FU/LV without oxaliplatin is recommended [13,14]. Similarly, stage III CRC is classified into high (T4, N1–2 or any, N2) and low (T1–3, N1) risk of recurrence. In this setting, patients are always referred to adjuvant treatment with CAPEOX (capecitabine, oxaliplatin), FOLFOX or 5-FU/LV if oxaliplatin is not tolerated [15]. This results in a 5-year disease-free survival (DFS) of 63.6% vs. 49.0% in those who did not receive adjuvant chemotherapy [16].
Standardized staging and one-size-fits-all approaches for localized to locally advanced disease fail to identify and deliver the optimal care for a group of patients who will inevitably present a poor prognosis. Inevitably, distinct drug responses and inconsistent survival outcomes are not uncommon within a single stage, which is even more pronounced regarding stages II and III [17]. The existence of a survival paradox between stages IIB/IIC and IIIA (T1–2, N1, M0 or T1, N2, M0) is a widely known phenomenon in CRC patients, and numerous causes had been described to contribute to an inferior survival in IIB/IIC compared with that of stage IIIA [11]. One explanation might be the insufficient use of systemic adjuvant chemotherapy in stage II CRC, since it is part of the standard care of treatment used in stage III CRC [18]. Another inconsistency refers to the distinction between high and low risk in stage II disease, since a significant number of high-risk patients with pathological and or prognostic features of poor outcomes will not develop disease recurrence and vice-versa. Furthermore, there are no data supporting the predictive value of the risk features and response to chemotherapy. Unfortunately, it has been shown that irrespective of the risk of recurrence, 5-year DFS rate in stage II patients undergoing adjuvant treatment was 81.4% (vs. 79.3%) [16], and the improvement of overall survival does not exceed 5% [10,19].
CRC is a heterogeneous disease, but these inconsistent responses to therapy and clinical outcomes may be resolved through incorporation of tumor molecular features to current staging. More recently, the identification of predictive molecular biomarkers has become standard practice for CRC management. Currently, the workup for metastatic disease recommends the investigation of activating mutations in KRAS (exon 2, 3, or 4) or NRAS (exon 2, 3 or 4) and BRAF (V600E), as well MMR status [20]. Within the metastatic disease setting, the presence of KRAS/NRAS mutations provides resistance to cetuximab and panitumumab [21], while BRAFV600E mutation predicts response to anti-EGFR targeted therapies in combination with a BRAF inhibitor [22,23]. In patients with known wild type RAS and BRAF, HER2 testing for amplification or overexpression is recommended. Thus, 3% of all stage IV CRC are eligible for combined anti-HER2 therapies (trastuzumab plus pertuzumab or lapatinib) [24,25]. Finally, the frequency of stage IV MMR-deficient (dMMR) CRC is 3.5% to 5%, and predicts the response to immune checkpoint inhibitors in first and subsequent lines [26,27,28]. Although several clinicopathological and molecular features are known to be determinant on the disease prognosis, the majority of predictive biomarkers, which have reached clinical practice, are limited to metastatic setting.
1.2. Molecular Classification of CRC Tumors
While genetic susceptibility to CRC includes well-defined inherited syndromes—Lynch syndrome or hereditary nonpolyposis CRC (HNPCC), polymerase proofreading–associated polyposis (PPAP) caused by germline missense mutations affecting the proofreading activity of polymerases epsilon (POLE) and delta (POLD1) and NTHL1-and MUTYH-associated polyposis (NAP and MAP, respectively), sporadic CRC is highly heterogeneous on the genetic and gene regulatory levels [29,30,31,32]. Although MSI status and a few numbers of mutations have reached clinical practice due their role on disease prognosis, they are still insufficient to identify all patients with increased risk of recurrence and metastasis. Thus, in order to translate the intra- and inter-tumor heterogeneity, gene expression signatures have been extensively investigated.
Transcriptomic analyses showed the molecular heterogeneity of CRC and established a molecular classification into four consensus molecular subtypes (CMS1–4) [33,34,35]. CMS1 (“immune”) comprises 14% of CRC tumors and is associated with hypermutable characteristics, mutations within the BRAF gene, microsatellite instability (MSI) and strong immune activation. The “canonical” CMS2 (37% of total CRC tumors) is associated with mutated TP53 gene, APC mutations, activated WNT and MYC signaling, chromosomal instability (CIN), and copy number alterations. The “metabolic” CMS3 (13% of tumors), is characterized by metabolic dysregulation and KRAS mutations. The “mesenchymal” CMS4 (23% of tumors) is characterized by upregulation of genes involved in the epithelial to mesenchymal transition (EMT), transforming growth factor-β activation and inflammatory microenvironment. In view of the extensive biological differences between these subtypes, the ability to respond to therapies may also be different for each subtype [36,37]. It is important to note that CMS4 tumors present downregulation of all DNA repair pathways, which is attributed to hypoxia and a stem-like phenotype [38]. CRC CMS4 subtype is often diagnosed in advanced stages. However, it has been also reported that stages II-III patients also present the poorest prognosis among CMS subtypes, mostly due to increased progression rates towards metastatic disease [39]. Although current recommendations for CRC adjuvant treatment include high-risk stage II and all stage III patients, the benefit of chemotherapy in the adjuvant setting for stage II is still a matter of debate [12]. Moreover, while for metastatic disease, CMS4 tumors are resistant to anti-EGFR therapies (irrespective of KRAS mutational status) and to doublet/triplet backbone chemotherapy, the benefit of adjuvant treatment for early and locally advanced CMS4 tumors is not obvious [39,40,41,42]. To date, CMS classification offers richer insights into the molecular heterogeneity of CRC and prognosis, but its role in clinical decision-making is still to be confirmed [36,43].
2. Analyzing the Opportunities to Increase Therapeutic Index for CRC by Using DSB Repair Inhibitors
Cancer chemotherapy and radiotherapy are designed to kill cancer cells mostly by inducing DNA damage and disturbing replication or mitotic machinery. DNA-damaging agents cause various types of DNA lesions, including base modification, intrastrand crosslinks, interstrand crosslinks (ICL), DNA–protein crosslinks, single-strand breaks (SSBs), and double-strand breaks (DSBs). The DNA DSB inducing agents are often exploited in cancer treatment, as DSBs cause greatest genomic instability, leading to cell death in absence of functional repair mechanisms. DSBs are mainly repaired by two pathways: homologous recombination (HR) and non-homologous end-joining (NHEJ) repair [44]. However, DNA repair–deficient cancers often become dependent on backup DNA repair mechanisms, such as microhomology-mediated end-joining (MMEJ) in HR deficient background [45].
Despite the comprehensive advances of the CRC molecular landscape, a limited number of biomarkers have made it to the clinic. Thus, most systemic therapeutic options for CRC still rely on chemotherapy-based regimens for early disease, and few targeted agents are recommended for clinical use in the metastatic setting. Moreover, the impact of cytotoxic, targeted agents and immunotherapy combinations in the metastatic scenario is considered suboptimal rather than life changing. Hence, to improve the outcomes for patients who develop resistance to chemotherapy agents and/or are not eligible for targeted agents or ICB, there is an urgent unmet clinical need in the CRC landscape.
Conventional CRC chemotherapy includes combination regimens with 5-FU, oxaliplatin, and irinotecan. The 5-FU-mediated cytotoxicity relies on thymidylate synthase (TS) inhibition, and misincorporation of fluorouracil and uracil into DNA during replication, requesting multiple DNA repair pathways. Incorporation of fluoronucleotides into RNA leads to impairment of RNA processing and function, contributing to the cytotoxic effect [46]. pMMR CRC cells have increased sensitivity to 5-FU in relation to dMMR CRC cells, because of a futile cycle of repair in attempt to remove the misincorporated bases. Base Excision Repair (BER) pathway, acting by uracil–DNA glycosylase (UDG) and AP endonucleases, creates lesions, such as abasic sites, SSBs and DSBs in response to 5-FU [47]. Moreover, 5-FU was shown to cooperate with DSB-induction in CRC cells by decreasing efficiency of HR interfering with the synthesis of new DNA strands [48]. Oxaliplatin toxicity is characterized by the formation of ICLs and intrastrand cross-links. Moreover, platinum drugs induce DNA-protein cross-links that block DNA replication as well [49,50]. The repair of oxaliplatin-induced ICLs involves the FA pathway, which can use parts of the nucleotide excision repair (NER), HR, and translesion synthesis (TLS) [51]. The topoisomerase I poison Irinotecan induces DNA DSBs during S phase and relies on HR for repair [52].
DNA repair deficiency is a frequent event in tumorigenesis, which results in enhanced mutation rates and genetic instability, thus providing a selective growth advantage, a driving force for tumor evolution. Genetic alterations also modify the cellular response to chemotherapy-induced damage. Defects in DNA repair pathways could provide new therapeutic strategies, still not explored in CRC [53]. Moreover, the use of specific DNA repair inhibitors, targeting essential DNA repair pathways, allows us to provoke failures in DNA Damage Response (DDR) of cancer cells. Recent data have demonstrated that prevalence of somatic DDR defects in CRC ranges between 10% and 30%, and predict worse outcomes and resistance to therapy [53,54]. Of all DDR and DNA repair pathways, genes involved with the repair of DNA double-strand breaks (DSBs) are somatically mutated in up to 20% of CRC [54,55]. Fortunately, evolving evidence obtained from large database analyses of CRC tumors have identified DDR-based signatures, which may underpin for the eligibility of treatment with novel (e.g., inhibitors of ATR, CHK1, WEE1, and ATM), and marketed DNA repair inhibitors (e.g., PARP inhibitors), and immunotherapy [56]. Although MMR deficiency remains the unique relevant DDR-based signature for CRC prognosis and response to therapy, the identification of deficiencies in other DNA repair pathways may unveil mutational signatures and provide insights into new targets.
2.1. HR-Deficient Phenotypes in Colorectal Cancers
Multiple hereditary or somatic cancers are deficient in homologous recombination (HR) repair, which is the main error-free repair pathway involved in DSBs repair in S and G2 phases and reactivation of blocked replication forks [57]. It has been suggested that a subset of CRC patients (6–15%) harbor mutations in HR genes, including ATM, BRCA1/2, MRE11A, FANCC, NBN, PALB2 [54,55,58,59]. Data retrieved from The Cancer Genome Atlas (TCGA) and other databases have shown that genes critical for HR, such as ATM, BRCA1, and BRCA2, are somatically mutated in more than 20% of CRC [55]. Additionally, 8.7% of colorectal tumors harbor non-silent mutations in gene code regions concomitantly in HR and BER or HR and MMR pathways [60]. More recently, it has been described that patients with brain metastases of primary colorectal tumors exhibit mutational signatures of homologous recombination deficiency (HRD) due to mutations on BRCA1, BRCA2, RAD51B, and PAXIP1 [61].
Identification of HR deficiency is classically described in tumors with germline mutations in BRCA1/2, such as breast and ovarian [62]. In these tumors, HR deficiencies grant higher sensitivity to alkylating or platinum-based agents as a result of generation of non-processed and highly toxic DSBs. Data of a study enrolling 108 stage III CRC patients found a germline mutation followed by ATM or BRCA2 somatic mutations in 13.8% and 22.2%, respectively [63]. Moreover, tumors without functional BRCA1/2 are sensitive to poly ADP-ribose polymerase 1 inhibitors (PARPi) [64]. Although PARP inhibitors were found to be selectively active in a subset of tumors harboring BRCA1 or BRCA2 mutations due to the synthetic lethality of PARP inhibition and this particular HRD, it also has a role in BRCA wild type cancers [65,66]. However, HR deficiency phenotype can also occur following genetic and epigenetic inactivation of other HR components in sporadic cancer, which leads to the so-called BRCAness signature [67]. This is particularly noteworthy in the CRC context because it may expand therapeutic options for those who do not benefit from targeted (RAS mutated) or immune therapies (microsatellite stable, MSS). In a recent review article, we discussed the modulation effects of the main DNA repair pathways in the clinical and pathological aspects of CRC and its potential as prognostic and predictive biomarkers [68].
2.2. MMR-Deficient Phenotypes in Colorectal Cancers
The occurrence of CRC due to germline mutations in the MMR genes MLH1, MSH2, MSH6, and/or PMS2 or EpCAM represents 2% to 4% of all CRC cases. About 19% of sporadic colorectal tumors may also present somatic MMR defects following acquired mutations or hypermethylation of the hMLH1 gene promoter [69]. dMMR is clinically equivalent to microsatellite instability-high (MSI-H), whereas pMMR is the same as microsatellite instability-low (MSI-L) or MSS [70]. Impaired MMR function is more frequent in stage II disease (22%) than in stage III disease (12%), whereas in stage IV it occurs in only 3.5% of the cases, suggesting a reduced tendency for distant metastasis [69]. Since discrepancies in the frequency of MMR deficiencies between stages [71], several studies have investigated if this molecular phenotype may be a predictive marker of reduced benefit from adjuvant fluoropyrimidine-based regimens in these settings. Two large retrospective studies have shown that MMR deficiency predicts response to adjuvant 5-FU or capecitabine in these patients [72,73], but more recent studies have failed to prove dMMR as a biomarker of fluoropyrimidines efficacy for stage II disease [74,75]. Therefore, both observation and fluoropyrimidine-based adjuvant chemotherapy are recommended for stage II disease with or without MMR deficiencies and irrespective of recurrence risk. Yet, only for patients at high-risk of recurrence or with dMMR colorectal tumors, adjuvant treatment may include 5-FU/LV or capecitabine in combination with oxaliplatin (FOLFOX, or CAPOX), respectively [36]. In 2021, NCCN recommended the investigation of MMR status for all stages to help with hereditary CRC diagnosis, and in stage II for decision-making if 5-FU-based adjuvant therapy is indicated.
The dMMR sporadic CRC have exclusive features such as delayed onset, frequent BRAFV600E mutations, and better disease prognosis (hazard ratio for overall survival: 0.59; 95% CI: 0.50–0.69) [76]. Additionally, dMMR colorectal tumors are usually infiltrated by intraepithelial and peritumoral lymphocytes in response to a high production of neoantigens generation following the high mutational burden resultant from MSI [55]. Enriched lymphocytic infiltration is associated with boosted anti-tumor immunity. Therefore, it might explain why MSI is observed more often in stages II and III colorectal tumors rather than in metastatic disease, where tumor cells were able to evade immune surveillance [77]. Most of these tumors present loss of MLH1 and PMS2 proteins due to hypermethylation of the hMLH1 gene promoter typically in association with the CpG island methylator phenotype (CIMP) [78]. Genes regulating cell proliferation, survival, cell cycle and DNA repair which harbor microsatellites are disposed to mutations due to dMMR [77]. Several studies have shown that oncogenic RAS activation is associated with replication stress, increased HR-mediated response, increased DNA damage levels, and genomic instability [79,80,81]. Recently, the mutational profile of DDR has been described in the largest transcriptomic study in colorectal tumors [82]. This study included 9321 CRC patients and demonstrated that RAS and BRAF mutational status might have a different impact on the DDR signature in relation to MSI status. From the total, 1290 CRC patients presented mutations in the DDR genes (13.8%) associated with MSI-H/dMMR tumors, with frequency of 76.4% (456/597) vs. 9.5% in MSS/pMMR tumors. The highest mutation frequency between the 29 DDR genes evaluated were detected in ATM (4.5%), BRCA2 (2.7%), PRKDC (1.6%), and CHEK2 (1.2%) genes. The authors have found that RAS-mutant tumors had significantly lower frequency of DDR mutations in relation to the RAS-wild type tumors. However, there was no significant difference in the subgroups (MSS/pMMR or MSI-H/dMMR). Conversely, the BRAF-mutant-type had a significantly higher DDR-mutational profile in comparison to BRAF-wild type tumors (31.1% vs. 12.1%). Likewise, no difference was observed in relation to the MMR proficiency. Amongst the 1529 samples available for CMS analysis the frequency of DDR mutations was highest in the CMS1 subtype (34.8%) as compared to the other subtypes (CMS2—7.1%, CMS3—15.2%, and CMS4—11.8%). In RAS-mutant tumors, differential lower levels of gene expression were observed in CDK12, NBN, BRCA1, MRE11A, and PRKDC, whereas regarding BRAF-mutant tumors, lower gene expression was observed in CDK12, MRE11A, RAD50, ATR, and FANCF [82]. Taken together, these observations suggest the distinctive levels of DDR activity according to the gene expression of HR, NHEJ, DNA damage checkpoint, and Fanconi anemia pathways, as well as MSI status may have a role in the response to therapy in individual patients.
Most patients with a classic Lynch syndrome phenotype carry germline mutations in one of the MMR genes, all of them leading to deficient mismatch repair and consequent MSI in the tumor [83]. The MMR proteins Msh2 and Mlh1 were also used by MMEJ pathway implicated in antibody gene class switch recombination [84]. In sporadic CRC, the presence of MMR deficiency (mostly due to epigenetic inactivation of hMLH1 promoter) is associated with a better prognosis. In this field, our research group has shown that the presence of MSI in sporadic CRC is associated with a reduction in initiation of base excision repair (BER) by the DNA glycosylases OGG1 and MPG, as well a reduction in signaling of DNA single-strand breaks (SSB) repair by PARP1 [85].
The choice of DNA repair pathway determines which type of genomic instability arises from replication stress-associated DNA double-strand breaks [86]. MSI induction in vitro was triggered by replication stress-associated DSBs via DNA repair dependent on both Polθ and PARP1, which is likely mediated by MMEJ in MMR-deficient background [87]. In this scenario, the DSBs are first recognized by HR factors, followed by the HR–MMEJ switch attributed to complex formation. Recent study reported that the MMR complex MSH2-MSH3 at DSB site promotes HR, facilitating long-range resection by EXO1, and inhibits the access of Pol θ [88]. The induction of MSI in MMR-deficient cells is associated with hypermutation phenotype and mutations in cancer-driver genes, resulting in clonal expansion [89]. At the same time that MSI is induced, chromosome instability (CIN) is suppressed in MMR-deficient cells (because of DSB elimination). CIN was proposed to arise when the DSBs are not effectively repaired in S and G2 phases by the HR. Following a defective mitosis, such DNA breaks activate DDR in the G1 phase of the next cell cycle and eventually are ligated to incorrect broken ends by NHEJ, leading to chromosomal alterations [90,91]. Tumors with CIN represent distinct type of genomic instability in CRC (about 60% of all cases), characterized by chromosomal rearrangements and aneuploidy, gene amplifications and deletions, loss of heterozygosity (LOH), and micronuclei [92].
Clinically, mutations in MRE11 occur in the majority of MMR deficient colorectal tumors [93], and are associated with increased susceptibility to CRC [94]. Moreover, it has been suggested that MSI arises from meiotic recombination 11 homolog (MRE11) deficiency due to defective interactions between MLH1 and MRE11 [95]. While upregulation of MRN complex has been associated with a high tumor grade, chemo/radiotherapy resistance, and poor overall and progression-free survival, deficiency in MRE11 predicts a better prognosis [96,97,98,99]. Conversely, for CRC with functional MMR pathway, high MRN expression is associated with earlier tumor grade and good prognosis [100,101]. Recently, it has been reported that MRE11 is an independent favorable prognosis in left-sided, but no right-sided CRC, which may suggest a novel molecular feature according to the disease laterality [102]. Thus, targeting MRE11 to sensitize colorectal tumor cells considering MSI status may be an interesting approach to overcome resistance and expand antineoplastic combinations. Moreover, CRC cells harboring MRE11 deficiencies are more sensitive to PARP inhibitors [103], platinum salts [97], and 5-fluorouracil/leucovorin/ irinotecan combination [96].
3. Cellular Responses to DNA DSB
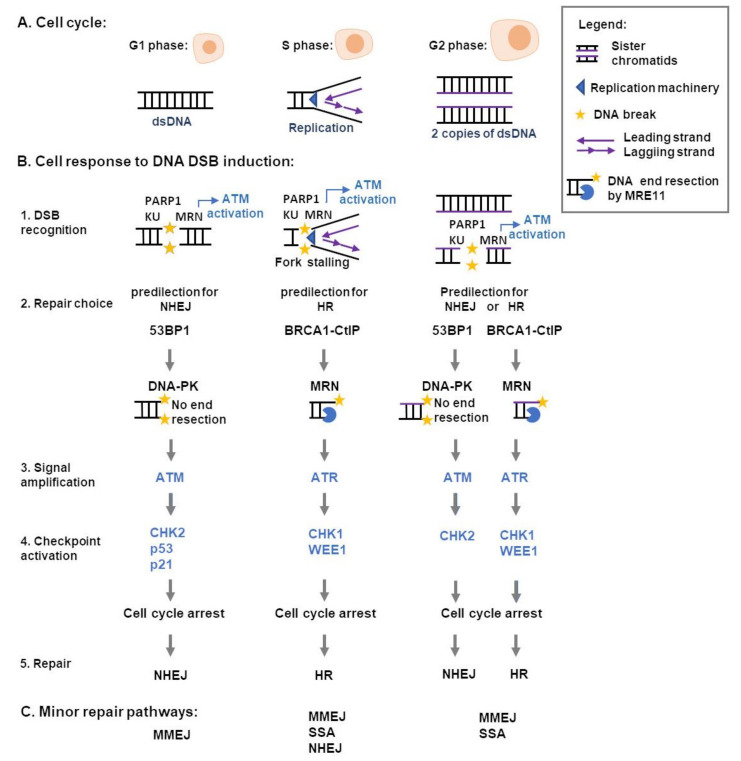
The cell response to DSB formation involves three main steps to enable DNA repair: (1) the recognition of the lesion by sensor proteins (MRE11-RAD50-NBS1 complex, Ku and PARP1); (2) the amplification of the DNA lesion signal by transducer proteins (ATM, ATR, DNA-PKcs) and (3) the cell cycle checkpoint activation by effector kinase proteins (CHK1, CHK2) (Figure 1) [104,105,106].
Figure 1.
Cell cycle-dependent response to DSB formation. Depending on the stage of cell cycle (A), the induction of DSB (B) will lead to distinct cellular ATM or ATR signaling responses and DNA repair pathways. While DNA-PK drives NHEJ repair without resection of DNA ends, the MRN-CtIP removes DNA-PK and HR can function and use sister chromatids as a template for DNA repair synthesis. (C) Upon some context, including HR defect, the MMEJ and SSA repair pathways can act on the resected single-stranded DNA. MMEJ can act as a backup for NHEJ in G1 phase.
When DSB are formed, after gamma-ray ionizing radiation for example, there is an increase in the MRE11-RAD50-NBS1 (MRN) complex formation [107]. The MRN complex binds to the DNA ends of the break site and Nijmegen breakage mutated protein 1 (NBS1) interacts with Ataxia-telangiectasia-mutated (ATM) kinase in such a way that ATM is activated by autophosphorylation of Ser 1981 [108]. The ATM activation, in turn, increases the MRN complex formation [107], and initiates a phosphorylation cascade at key proteins: NBS1-Ser343, CHK2-Thr68, and p53-Ser15 [104]. The DNA repair occurs without induction of CHK2-dependent checkpoint when cells present lesions below a certain threshold level; otherwise, CHK2 is autophosphorylated in Thr387, driving p21 accumulation and cell cycle arrest in G1-phase [109,110,111].
In S and G2 phases MRN transiently activates ATM, then an ATM-to-ATR switch is driven by formation of single-strand DNA coated by RPA after MRN resection of DNA ends. Then the ataxia-telangiectasia and Rad3 related (ATR) activation phosphorylates CHK1 that phosphorylates WEE1 and these events promote cell cycle arrest [112,113,114]. The cell cycle arrest in G2-phase can also be maintained by ATM-CHK2 activation at unresected DSB and 53BP1 keeps the ATM activity to phosphorylate CHK2, thus enhancing ATM-CHK2 signaling (Figure 1B) [115].
Besides MRN, the DNA-PK complex—formed by DNA-PKcs and Ku—also binds to DNA ends during all the cell cycle phases within DSB is formed [105,116]. Ku is a heterodimer (Ku70 and Ku80 subunits) that binds DNA end and recognize DSB simultaneously to the MRN complex [105]. Ku recruits the catalytic subunit of DNA-PK (DNA-PKcs) to form the complex named DNA-PK and drives canonical NHEJ repair. Besides Ku and MRN, PARP1 is recruited to DSB and in S/G2 phase, its enzymatic activity has a role to remove Ku from the DSB site [106]. The pathway choice for DSB repair is cell cycle regulated and depends on DNA end resection (Figure 1B). The NHEJ repair occurs throughout G1, S, and G2 phase, and is preferred for repair of two-ended DSBs with compatible ends, while the HR is preferred for the repair of one-ended DSBs, such as replication-associated lesions, which favoring HR in S phase [117]. It is proposed that Ku plays a role in regulation of arrested replication fork restart, where the initial resection by MRN-Ctp1 is essential for recruitment of HR factors in Schizosaccharomyces pombe. The lack of Ku leads to reduced recruitment of RPA and Rad51 and extensive resection by Exo1 [118]. Moreover, in human cells, the Ku binds to single-ended DSB, and the formation of the DNA-PK complex is necessary to initiate MRN endonuclease activity [105,119]. During S and G2 phases, CDKs, and other factors that promote DNA end resection are activated, and remove Ku from DNA; hence, NHEJ is favored when the resection is limited, as in G1 phase [117]. In the G2 phase, the cell can choose either pathway; however, complex DNA ends or DSBs in transcriptionally active loci preferentially undergo HR repair [117,120]. In addition to HR and NHEJ, which are the main DSB repair pathways, the error-prone repair pathways microhomology end-joining (MMEJ) and single strand annealing (SSA) also can participate in the DSB repair. Both MMEJ and SSA require a prior DSB end resection mediated by CtIP and MRN leading to loss of genetic information. These pathways play a backup role for the HR, while MMEJ also has a role as backup for the NHEJ (Figure 1C) [121,122].
To drive NHEJ repair, 53BP1 is phosphorylated by ATM, protecting the DNA ends from resection [123]. There are other positive regulators of NHEJ repair downstream 53BP1, like RIF1, REV7, and RINN1/2/3, and they inhibit BRCA1 accumulation at the DSB site [124,125]. When the cell drives HR repair, BRCA1 prevents ATM-dependent 53BP1 phosphorylation [123]. The phosphorylation of CtIP by ATM, ATR, or CDK at specific sites—T847 and T859—promotes BRCA1-CtIP interaction, and CtIP stimulates MRN nuclease activity that generates a single-stranded DNA (ssDNA) [119,125]. Moreover, BRCA1-CtIP recruits DNA2 exonuclease and CDK phosphorylates EXO1, which expand the ssDNA in cooperation with DNA2 exonuclease and BLM helicase, removing DNA-PK from DNA (Figure 1B) [126,127,128].
The nuclease activity of MRE11 protein has also a role in the choice of DNA damage repair pathway: the MRE11 endonuclease activity is the initial step for resection in HR repair forming a single-stranded nick, followed by exonuclease activity that generates a ssDNA gap and so prevents NHEJ repair [129]. The inhibition of MRE11 exonuclease activity inhibits HR without increasing NHEJ repair, while inhibition of the endonuclease activity enhances NHEJ by reducing HR [129,130]. Moreover, at collapsed replication forks the MRN complex structurally assists the linking of the end DNA with the sister chromatid through a RAD50 hook that facilitates the homologous recombination repair [131].
4. DNA Replication Fork Arrest, Replication Stress, Checkpoint Activation, and Genome Instability in Cancer
The integrity of the human genome is affected by exogenous insults, such as chemical carcinogens and ionizing radiation, as well as DNA damage that occur during the process of chromosome duplication when the DNA replication forks are slowed down or stalled by varied natural replication barriers, a process referred as replication stress (RS) [132,133]. RS is detected at early stages of carcinogenesis and is considered as a driving power of cancer progression [134,135,136]. RS in cancers frequently results from the oncogene-driven perturbation of the replication initiation program (origin activation and timing) and increased conflicts between replication and transcription. This triggers the generation of under-replicated regions as well as the persistence of stalled and collapsed forks, major sources for chromosomal breakage and chromosome instability [137]. If two converging replication forks stall with no possible compensation by licensed origins in-between, a double fork-stalling event occurs leading to the generation of under-replicated parental DNA and causing chromosome breaks when the cells enter mitosis [137,138]. Collapsed forks lead also to chromosomal breakages and alterations providing a permanent sub-population of cellular variants upon which selection could act, allowing some mutant sub clones to multiply explaining tumor heterogeneity, development, and drug resistance.
While genome instability is associated with poor prognosis, excessive karyotypic instability is deleterious for cancer cell fitness and correlates with improved survival outcome, supporting that a threshold limits extreme risky RS, DSBs, and genome instability in cancers [139]. Consequently, cancer cells need to adapt to the severe replicative defects and the resulting excess DSBs. This is why some of these adaptive mechanisms are currently considered exploited, therapeutically. The first adaptive response to a high level of RS is the ATR-CHK1 checkpoint response, which manages the stability, the repair, and the restart of arrested forks avoiding premature entry into mitosis and allowing the completion of DNA replication [140]. In agreement with the importance of the checkpoint response in cancers for limiting excessive RS, high levels of the checkpoint mediators Chk1, Claspin, and Timeless known to stabilize the stalled replication forks upon RS have been found to correlate with poor patient survival [57,141]. The second relevant adaptive response to RS is the late mitotic DNA synthesis or MiDAS and its critical actor Rad52, a mechanism that differs from semi-conservative DNA replication in S-phase and counteracts lethal chromosome mis-segregation by limiting the persistence of under-replicated DNA in mitosis [138]. As the under-replicated DNA is located ahead of the incoming fork, MiDAS requires replication-coupled repair named break-induced replication (BIR) [142,143]. The third category corresponds to a specialized DNA repair process, an alternative form of end-joining, referred to as microhomology-mediated end-joining (MMEJ; also named polymerase theta-mediated end joining, TMEJ) that substitutes for HR deficiency for the repair of DSBs generated by excessive RS and for which the A-family DNA polymerases Pol θ, encoded by the gene POLQ, holds a key role.
5. Emerging DSB Repair-Targeting Therapies for Colorectal Cancer
Most of the in vitro studies with potential inhibitors that may improve CRC therapy consider biomarkers to predict response. Although some DNA repair inhibitors have not been explored in CRC cell lines, there is scientific basis to be applied (Table 1).
Table 1.
Proteins related to replicative stress and DSB processing: situation of inhibitor studies (in vitro) and relevant biomarkers.
| Inhibitor | Biomarkers | Combined Therapy or Deficiency | Model of Study | Results | Biological Explanation | Reference |
|---|---|---|---|---|---|---|
| Inhibitors in perspective for clinical trial in CRC (inhibitors already tested in clinical trial for another tumor type or condition) | ||||||
| PARPi (ABT-888) | Presence of MSI | MRE11 deficiency | Homozygous MRE11 mutant CRC cell lines (HCT116, LoVo, RKO, SW48) and wild type cell lines (HCT15, SW403, HT29, SW620) | Cell lines with homozygous MRE11 mutation had increased sensitivity to PARPi. | PARPi induces DSB during replication, which is repaired mainly by HR repair. However, the presence of MSI is associated with HR impairment by MRE11 deficiency, which sensitizes cells to PARPi. | [101] |
| ATRi (VX-970) | - | CHK1i (V158411) | CRC cell line (HT29 and U2OS) | Both inhibitors induced loss of viability and the combination of them in low doses increased sensitivity. | Both proteins act on the same cell cycle control axis-repair of DNA DSB forming during S and G2 phases. The inhibition of both reinforces the blocking of the axis, generating replicative stress. | [144] |
| CHK1i (LY2606368) | TP53 mutation and RS markers | - | Primary CRC enriched for cancer stem cells | Sensitive cells displayed signs of replicative stress (P-RPA32, γ-H2AX, and P-ATM) and the effect did not depend on RAS mutation status. | Cells that lack p53 depend on the ATR-CHK1 axis to promote cell cycle arrest when DSB is formed. Therefore, the inhibition of CHK1 kinase in these cells sensitizes them to DSB. Replicative stress induces DSB, therefore cells ongoing RS are sensitive to CHK1i. | [145] |
| WEE1i (AZD1775) | WEE1 overexpression | - | Primary CRC liver metastases endothelial cells | Induction of apoptosis, increase in DNA DSB markers and inhibition of tube formation. | The WEE1 protein is upregulated in CRC liver metastases compared to endothelial cells of the normal adjacent liver and has functional importance by protecting against DNA DSB. The inhibition of WEE1 makes cells vulnerable to DSB. | [146] |
| WEE1i (AZD1775) | TP53 mutation | 5-FU | CRC cell line (HT29) | The WEE1i single treatment had cytotoxic effects by induction of DNA DSB. The co-treatment with 5-FU increased DSB marker (γ-H2AX) and apoptosis compared to the 5-FU single treatment. | The lack of functional p53 prevents G1 arrest, so the cell depends on the G2 checkpoint to repair DNA damage. WEE1 is a kinase that regulates the G2 checkpoint to repair DNA damage, so targeting WEE1 increases DNA damage sensitivity. | [147] |
| WEE1i (AZD1775) | - | PARGi (PDDX-004/PDD00017272) | CRC cell line (HCT116) | The co-treatment increased DNA DSB marker (γ-H2AX) in S-phase dependent manner. | WEE1i induces replication stress and DNA damage while PARGi delays the replication fork restart during replication stress. The combination of both increases DNA damage. | [148] |
| WEE1i (MK1775) | TP53 mutation | Irinotecan | CRC cell line (HT29 and SW480) | The co-treatment increased DNA DSB marker (γ-H2AX) and apoptosis compared to single treatments. | The lack of functional p53 prevents G1 arrest, so the cell depends on the G2 checkpoint to repair DNA damage. WEE1 is a kinase that regulates the G2 checkpoint to repair DNA damage, so targeting WEE1 increases DNA damage sensitivity. | [149] |
| Inhibitors in progress in CRC in vitro studies | ||||||
| PARPi (LT-626) | Presence of MSI | MRE11 deficiency | CRC cell line with a biallelic mutation in MRE11 (HCT116, HCT116/CS, HCT116/C3, RKO, SW48, and LoVo), with monoallelic mutation (DLD1), and wild type (SW837, HT29, and SW480) | Cell lines with a biallelic mutation in MRE11 showed higher sensitivity to PARPi and the knocked-down of MRE11 increased sensitivity to PARPi. | PARPi induces DNA DSB during replication, which is repaired mainly by HR repair. However, the presence of MSI is associated with HR impairment by MRE11 deficiency, which sensitizes cells to PARPi. | [103] |
| RAD51i (RI-1) | KRAS mutation | - | CRC cell line (HCT116, HKe-3) | KRAS-mutant cell (HCT116) was more sensitive to RI-1 than the isogenic wild type HKe-3, which showed a limited response. | KRAS-mutated CRC cells show stalling of the replication fork, which increases HR repair signaling. This occurs because of the hyperactivation of c-MYC. Targeting RAD51, a key protein in HR repair, thus may sensitize KRAS-mutated CRC cells. | [150] |
| RAD51i (B02) | - | - | CRC cell line (SW480) | Induction of apoptosis. | RAD51 expression levels are upregulated in biopsy samples of CRC and may thus support cancer progression. | [48] |
| ATRi (CBP-93872) | TP53 mutation | Oxaliplatin, cisplatin, and 5-FU | CRC cell line (HT29) | ATRi single treatment had no effect, however, the combination with compounds that induce DNA damage as DNA DSB (oxaliplatin and cisplatin) or replication fork arrest (5-FU) induced an increase in apoptosis compared to single therapies. | The lack of functional p53 prevents G1 arrest, so the cell depends on the G2 checkpoint to repair DNA damage. ATR activation regulates the G2 checkpoint to repair DNA damage, so targeting ATR activation may increase DNA damage sensitivity by suppressing the maintenance of the G2 checkpoint. | [151] |
| ATMi (KU55933) | - | PARPi (Olaparib) | CRC cell line (HCT116) | ATMi sensitizes cells to PAPRi and the deletion of p53 increased the co-treatment effect. | PARP1 induces DNA DSB during replication, which activates ATM and so ATR promotes cell cycle arrest and DNA repair. The sensitivity to PARPi increases when the cell lacks ATM levels. | [152] |
| ATMi (AZ31) | - | Irinotecan | CRC cell line (HCT15, HCT116, RKO, LoVo, LS132, and Caco2) and patient-derived xenografts (PDX) | Three (HCT15, HCT116, and RKO) of the six cell lines presented combinational sensitivity to AZ31 and irinotecan compared to single treatments. In CRC PDX models, the co-treatment was effective only in irinotecan-resistant tumors. | Irinotecan induces DNA DSB, which activates ATM to promote cell cycle arrest and DNA repair. The inhibition of ATM may sensitize cells to DNA DSB. | [153] |
| Inhibitors in perspective for in vitro analysis in CRC | ||||||
| MRE11i (mirin) |
RS markers | - | Human myeloma cell lines (MM1S, RPMI-8226, JJN3, U266) and B-cell (LINF903). | Only cells presenting RS markers (RAD51 and γ-H2AX signaling) showed sensitivity to mirin. | Human myeloma cell lines presenting DNA DSB signaling depend on DSB repair pathways for survival. To inhibit DSB repair may sensitize them. | [154] |
| MRE11i (mirin) |
MYCN amplified | - | Neuroblastoma cell line with MYCN-amplification (SHEP, GIMEN, and SK-N-SH) and MYCN single copy (LAN5, IMR32, and KELLY) and LAN5 xenograft in mice. | Only the cell lines with MYCN-amplification showed increased mRNA levels of MRE11 and sensitivity to mirin treatment. Mirin suppressed tumor growth in xenografted mice. | MRE11 is required to restrain replication stress induced by MYCN-amplification. MRE11 inhibition may trigger intolerable levels of RS. | [155] |
| MRE11i (mirin) |
PARP1 upregulation | RAD51i (B02) | CRC-stem cells with upregulation of PARP1, generated by CHK1i (prexasertib) treatment until acquired resistance. | The single treatments with Mirin or B02 were ineffective against CHK1i-resistant cells, but co-treatment killed the cells by induction of mitotic catastrophe and apoptosis. | CHK1i-resistant cells upregulate PARP1 to modulate fork speed and decrease RS levels. MRE11 and RAD51 may cooperate with PARP1 to deal with RS. | [156] |
| RAD52i (F79 aptamer, D-I03) | BRCA1 deficiency | PARPi (talazoparib) |
BRCA1-deficient primary acute myeloid leukemia (AML) xenograft in NSG mice, and BRCA1-deficient solid tumor growth in nude mice. | The combination of PARP inhibitor with RAD52 inhibitors selectively reduced BRCA1-deficient tumor growth. | PARPi induces DNA DSB during replication, which is repaired mainly by HR repair. However, BRCA-deficient cells have HR repair impairment, and single-strand annealing (SSA) is a backup pathway that may support survival. The RAD52 inhibition may sensitize cells to PARPi by the accumulation of DNA DSB in BRCA-deficient tumor cells. | [157] |
| RAD52i (F79 aptamer, 6-OH-Dopa, D-I03) |
BRCA deficiency | PARPi (olaparib, talazoparib) | Several human tumor cell lines | The combination of PARP inhibitors with RAD52 inhibitors selectively killed BRCA-deficient cells. | See description above. | [157] |
| Polθi (Novobiocin) |
BRCA deficiency | PARPi (Olaparib) | Xenograft mice derived from patients with germline BRCA1 mutation and acquired PARPi resistance, and HR-proficient PDX model. | Olaparib single treatment did not reduce tumor growth, while Polθi single treatment reduced tumor growth and the combined therapy was even more efficient. BRCA1 wild type PDX model was resistant to both single and co-treatment. Polθi toxicity depends on the accumulation of RAD51 foci. | PARPi induces DNA DSB during replication, which is repaired mainly by HR repair, but also by MMEJ repair. BRCA-deficient cells have HR repair impairment and respond to PARPi. However, MMEJ has emerged as a backup pathway in PARPi resistant cells. The Polθ inhibition may prevent MMEJ repair and increase PARPi sensitivity in BRCA-deficient tumor cells. | [158] |
MSI: microsatellite instability; CRC: colorectal cancer; DSB: double-strand break; HR: homologous recombination; 5-FU: 5-fluorouracil.
5.1. Homologous Recombination Repair
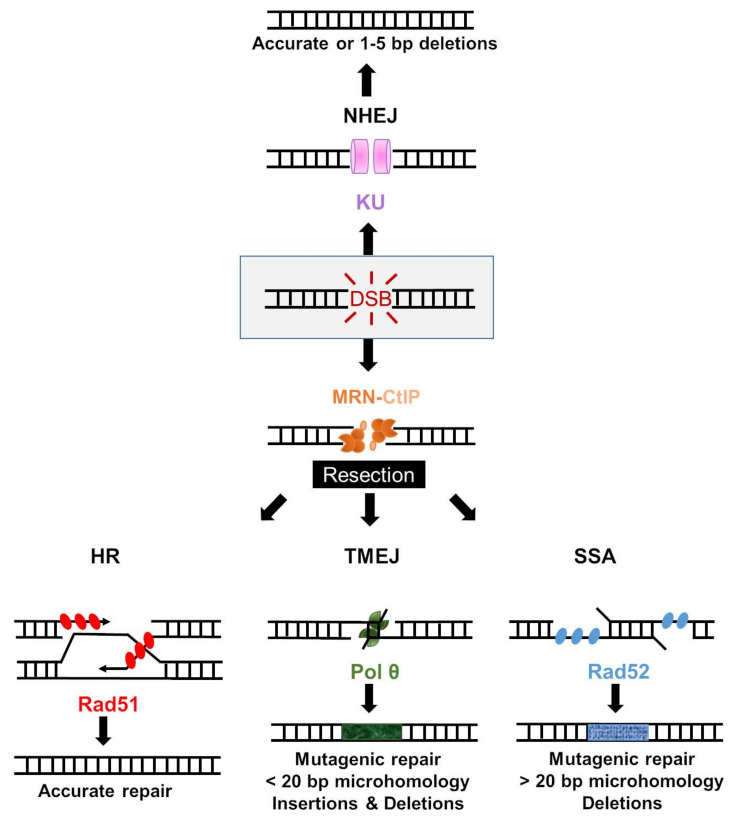
Double-strand breaks formed during the S and G2 phases of cell cycle, where the sister chromatids have been generated, are repaired mainly by the homologous recombination (HR) pathway [159]. In contrast to NHEJ, MMEJ, or SSA, HR is a conservative pathway, which uses the intact sister chromatid as a template and, therefore, restores the DNA sequence without loss of genetic information (Figure 2) [160,161].
Figure 2.
Major double-strand break (DSB) repair pathways in human cells. DSBs in G1 of the cell cycle are primarily repaired by non-homologous end joining (NHEJ) (Top). This pathway is initiated by the Ku heterodimer, which recognizes broken DNA. NHEJ leads to repair with minimal alteration to the original sequence. DSBs in S/G2 phases are subjected to resection leading to stretches of single-stranded DNA. Resected DSBs are substrates for homologous recombination (HR) with a critical role of Rad51 leading to an accurate repair synthesis. When HR is defective (for example when BRCA genes are mutated), an alternative pathway named Pol theta-mediated end-joining (TMEJ) can act as a back-up repair pathway. Polθ promotes the synapsis of the opposing ends, identifies internal microhomologies, which can be annealed, and performs a repair DNA synthesis with poor processivity and frequent aborted synthesis, resulting in a high rate of mutations. Single-strand annealing (SSA) is a HR sub-pathway in mammalian cells with the essential role of RAD52 DNA-binding protein. SSA is an error-prone repair leading to large deletions between the homologous repeats.
HR starts when the MRN complex recognizes the breaks; MRE11 initiates the DNA end resection to produce ssDNA that is then covered by RPA, and the recombinase RAD51 assembles onto ssDNA after RPA displacement [162]. The formation of such RAD51-ssDNA filament is essential for the search of the homologous sequence and its invasion through the formation of a D-loop intermediate to ensure the repair DNA synthesis [163]. It has been reported that expression of the HR proteins MRE11 and RAD51, monitored by immunohistochemistry predicts the response and prognosis of colorectal cancer patients who received oxaliplatin chemotherapy [97]. A significantly better tumor reduction and longer PFS was observed in MRE11- and RAD51-negative cases compared with MRE11 or RAD51 positive cases, suggesting that inhibition of these two main HR actors could improve sensitivity to chemotherapy [97]. In the same way, ATM and BRCA2 somatic mutations are suggested to be biomarkers that predict response to stage III CRC patients that received oxaliplatin-based chemotherapy, and they are associated with recurrence-free survival [63].
5.1.1. Targeting MRE11 in CRC
Genetic instability in MSI tumors, known to result from variation in microsatellite tracts due to a defective MMR process, can be further enhanced by mutations in microsatellite tract repeats in the MRE11 gene [164]. A microsatellite tract of 11(T) located at intron 4 of MRE11 is mutated in approximately 80% of MSI tumors and leads to aberrant splicing and a truncated protein inhibition of MRE11 nuclease activity. Importantly, such mutated MRE11 gene has been shown to sensitize CRC cells to agents causing DNA replication inhibition [165], as well as PARP-1 inhibition [101,103], suggesting that targeting MRE11 in MSS CRC might be an interesting therapeutic strategy. Inhibition of MRE11 by mirin, an inhibitor of its exonuclease activity discovered in 2008 [166], showed promising results in killing myeloma and neuroblastoma cell lines displaying high level of replication stress and endogenous DNA damages, evidenced by high amount of γ-H2AX and RAD51 foci as well high rate of ATR phosphorylation. Enhanced sensitivity of these cells seems specific to HR since they were also sensitive to the RAD51 inhibitor B02, but not the DNA-PK inhibitor NU7441 [154]. Such an effect of mirin was also observed in neuroblastoma cells with high degree of replicative stress. The MRN complex was shown to be important to prevent replicative stress (RS) generated by MYCN during the expansion of cerebellar granule progenitor cells [167]. In some tumor cells, MYCN oncogene is amplified, which increases RS levels [168], and is considered a bad prognosis in neuroblastoma [169]. However, it was observed that MYCN is a transcriptional regulator of MRE11, RAD50, and NBS1 [167], and the MRN complex contributes to the control of DNA damage levels compatible with tumor cell survival, so that high MRE11 expression is related to reduced overall survival in primary human neuroblastoma [155]. The use of MRE11 exonuclease inhibitor mirin increased the RS and DNA damage biomarkers in MYCN-amplified neuroblastoma cells until death in vitro and induced apoptosis in vivo. The inhibitor presented a promising outcome by reducing tumor growth in neuroblastoma-xenografted mice treated for 11 days with encapsulated mirin compared to control [155]. These results suggest that mirin should be explored in other tumors with high RS levels as well. Primary cancer stem cells from colorectal cancer (CRC-SC) isolated from patient samples showed adaptive response to high levels of RS induced by CHK1 inhibition. CRC-SC that were resistant to CHK1i showed PARP-1 upregulation, which decelerated fork progression and decreased RS levels. The combined inhibition of MRE11 and RAD51, by mirin and B02 co-treatment, selectively killed PARP-1-upregulated CRC-SC via mitotic catastrophe [156]. These data suggest that MRE11 and RAD51 cooperate with PARP-1 for RS response. There are other MRE11 inhibitors besides mirin that inhibit endo- (PFM01 and PFM03) or exonuclease (PFM39) activities [129]. However, to date, none is under investigation in clinical trials for CRC patients.
The alternative MMEJ pathway to repair DSBs operates on a common resected HR intermediate and includes the factors involved in the 5′ to 3′ resection (e.g., MRE11, RAD50, NBS1, CtIP, and EXO1). MMEJ requires PARP1 in its early step and starts with the search of short tracts of contiguous microhomology, followed by an annealing step, and then by a processing mechanism of 3′ ssDNA tails and DNA synthesis ensured by the specialized DNA polymerase Pol θ, a major mediator of this pathway [170,171]. MMEJ contributes to 10–20% of DSB repair in mammalian cells, is independent of γ-H2AX signaling, and seems to be poorly requested at the G0 and G1 phases of the cell cycle while it is activated upon S-phase entry [172]. The absence of MRN, the use of mirin, or the expression of the MRE11-H129N nuclease mutant impairs HR and MMEJ to a similar extent [172,173,174].
On the other hand, the use of mirin could also be explored in association with anti-PD-L1 therapy. T lymphocytes can recognize foreign antigens; however, CRC cells with MSI-H or dMMR tumors can upregulate immune checkpoint proteins, such as PD-1 and PD-L1, which permit immune evasion. Currently immunotherapy based on checkpoints inhibition in advanced CRC is limited to MSI-H tumors and other biomarkers being investigated [175]. Both MRN and PD-L1 overexpression are associated with platinum-based chemotherapy resistance [98,176,177]. Recently an association was discovered between PD-L1 and the MRN complex through an immunoprecipitation technique. It was found an interaction of PD-L1 with NBS1, the protein responsible for supporting the MRN complex and for ATM activation. The silencing of NBS1 or PD-L1 sensitized cells resistant to cisplatin, but the silencing of both proteins together showed synergism and induced 80% of cell death; and the synergism was confirmed in vivo [178]. It is known that the ATM/ATR activation induces PD-L1 expression [179], which is one of the reasons that cells with DNA damage repair deficiency respond to PD-L1 blockade [180,181], besides neoantigens formation [182,183]. However, ATM is activated by the MRN complex, so inhibiting the complex could help to inhibit ATM activation, thus explaining the synergism between anti-PD-L1 and NBS1 silencing. The ATM activation does not depend on MRE11 nuclease activity; however, it was shown that mirin inhibits ATM activation by some other way not yet understood [166,184]. Therefore, considering the multiple possibilities of mirin usage, including inhibition of HR repair, MMEJ and ATM activation, this compound could bring beneficial outcomes in CRC or other tumors. It can be employed alone or in combination with other therapies, such as anti-PD-L1 and PARP inhibitors, or in conditions of increased endogenous-DSB formation caused by replicative stress.
5.1.2. RAD51 Inhibition
The inhibition of RAD51 is also utilized as a strategy to impair HR repair in cancer treatments wherein this pathway is important for cell survival. RAD51 participates in HR repair, which is the preferable choice for DSB repair as it is the only one usually error-free pathway [161]. RAD51 also has a role in protection of stalled replication forks by covering the nascent DNA. FANCI-FANCD2 complex directly binds to RAD51 to stabilize the RAD51-ssDNA filament, thus it prevents RAD51 dissociation from DNA and protects DNA end from FEN1, DNA2 and MRE11 nuclease degradation [185,186,187]. RAD51 also enables fork regression by a switch in template strand of stalled replication fork, so that DNA polymerase can bypass the DNA lesion on the leading strand template that is blocking replication fork progression [188]. Thus, the nascent DNA of the opposite branch is used as a template for extension of the leading strand and fork restoration forming a “chicken-foot” structure [189]. The fork regression does not depend on the RAD51 enzymatic function [186], but depends on the proliferating cell nuclear antigen (PCNA) poly-ubiquitination [190,191]. Some compounds focus on inhibiting the RAD51 strand invasion catalysis (via D-loop formation) of HR repair without affecting the ssDNA binding activity of RAD51, which is important for protecting stalled replication forks [192]. The RAD51 mRNA levels analyzed in CRC samples from 48 patients undergoing surgery and without preoperative chemotherapy were upregulated in 2.5-fold compared to non-tumor tissue and associated with T stage [193]. Despite the absence of independent prognostic value of RAD51 mRNA levels in patients without undergoing chemotherapy, in another study, RAD51 expression predicted the response and prognosis of patients who received oxaliplatin chemotherapy; thus, RAD51 may be a candidate for targeted chemotherapies [97,193].
The emergence of RAD51 inhibitors has enabled new therapeutic approaches that target HR repair in vitro, such as in KRAS-mutant CRC cells. Colorectal cancers show a high frequency of activating KRAS mutations [194], and it was found that KRAS-mutated HCT116 cell line (constitutively active KRAS) shows increased stalled replication forks, pRPA32 (S33), and γ-H2AX signaling, besides the increase in RAD51 expression compared with the isogenic wild type cell line (HKe-3); all these signals together suggest a dependency on HR repair [150]. Then Kalimutho et al., (2017) wondered whether RAD51 could be a molecular target to impair HR repair and sensitize CRC cells to death induction. Indeed, the depletion of RAD51 by siRNA in KRAS-mutant HCT116 cell line decreases cell survival without decreasing survival of WT cells, as well as RAD51 inhibition by RI-1 inhibitor [150]. In another study, it was also suggested a CRC dependency on HR repair as it was found a rise in expression levels of HR proteins RAD51 and BRCA2 in CRC biopsy in comparison with normal mucosa biopsies [48]. Interestingly the inhibition of RAD51 in the SW480 CRC cell line by B02 treatment was enough to induce apoptosis. It was not even necessary to induce DSB before B02 treatment to sensitize the cells, indicating a potential therapeutic approach [48]. Besides IR-1 and B02, there are other RAD51 inhibitors yet to be explored in CRC: RI-2, CYT01B, IRB2, and RAD51-IN-2.
5.2. Alternative End-Joining
The alternative end-joining (Alt-EJ) is represented by a variety of repair pathways, genetically different from NHEJ, that repair DSBs by initial end resection generating 3′ single strands. These pathways are intrinsically mutagenic and play an important role in DSBs repair in cancer cells generating deletions and genomic rearrangements [195]. Alt-EJ events start with limited resection by MRN-CtIP, such as HR, and were almost entirely abolished by inhibition of the resection with MRE11i [196]. The Alt-EJ can be classified in relation to the repair proteins that promote the pathway, or by the characteristics of the repair junctions with different mechanisms of formation proposed (summarized in recent review) [197]. It is an independent, tightly regulated pathway that operates, even when the HR and NHEJ are functional. Alt-EJ is active predominantly in S and G2 phase but it also occurs in G1 phase [198,199]. The repair of DNA DSBs by Alt-EJ can be mediated by microhomologies (i.e., MMEJ), typically <20 bp in mammalian cells [195,200]. There is also poorly defined Alt-EJ that can be independent of sequence homology [195]. DNA polymerase theta (Pol θ) is a predominant mediator of the MMEJ pathway (named TMEJ), that generates short insertions and deletions (Figure 2), associated with specific mutational signatures in human cancers [196,201,202].
5.2.1. Polymerase Theta-Mediated End Joining
Microhomology-mediated end-joining (MMEJ), or polymerase theta-mediated end joining (TMEJ), is used at a substantial frequency to repair DSBs in cycling cells even when both NHEJ and HR pathways are available [172]. Unlike HR, MMEJ is intrinsically error-prone and produces deletions and translocations (Figure 2). In mammalian cells, MRE11 nuclease, CtIP, DNA ligase III, and an error-prone multifunctional DNA polymerase theta (Polθ, encoded by the POLQ gene) have critical roles in MMEJ [203]. In the first step of MMEJ in human cells, PARP1 binds to the DSB ends and facilitates the recruitment of the resection factors, CtIP, and MRN complex generating ssDNA and allowing microhomology search. The microhomology annealing induces formation of non-homologous 3′ ends that can be eliminated by Polθ prior to DNA synthesis as recently reported [204], suggesting that Polθ can itself remodel DNA ends to facilitate their repair. The tetrameric Polθ-helicase domain prepares the substrate by microhomology alignment, followed by filling-in synthesis by the Polθ-polymerase domain and ligation by LigIII. Polθ–helicase domain is essential for efficient joining of DNA breaks, acting to dissociate RPA (that prevents spontaneous annealing), and promote the annealing of complementary DNA [205].
5.2.2. Polymerase Theta (Polθ) Inhibition
POLQ was first reported as a powerful biomarker in several solid cancers, where its overexpression correlated significantly to poor patient survival [206]. Later, Polθ has emerged as a promising target for the treatment of various DNA repair–deficient cancers, including those that are deficient in HR [170,207]. Cancers with upregulation of MMEJ pathway display a distinctive mutational signature and present new targets for cancer therapy [208]. The HR factors BRCA1 and BRCA2 play a role in suppressing MMEJ in human cells [209], and BRCA2 protects stalled replication forks from degradation [210]. The depletion of these proteins increased break-induced mutational frequencies and short microhomologies at the break-site. The observed mutational signature may arise from HR failure at a stage after the initiation of resection, and was dependent on the MMEJ factors—CtIP, PARP1, MRE11, and Polθ. Moreover, in this context, the repair may occur in Polθ-dependent MMEJ and possibly by Polθ-independent microhomology-mediated pathways suppressed by RPA [209]. The Polθ-dependent pathway, also called TMEJ, generates unique short flanking DNA microhomologies, which Polθ efficiently locates when present or creates de novo when absent [211].
Interestingly, POLQ expression is increased in cancers, both with and without HR deficiency [212,213]. Moreover, Polθ is coping with replication stress and repairing DSBs upon fork collapse [212,214]. In HR-proficient background, inactivation of POLQ increased cell death in ATR-deficient cells and sensitized breast cancer cells overexpressing POLQ to DNA damaging agents that cause replication stress (camptothecin and etoposide) or fork collapse (ATR inhibitors) [214]. POLQ expression in different cancer types strongly correlated with the expression of factors involved in response to replicative stress (RAD51, FANCD2, and BLM), instead of genes encoding for several core MMEJ proteins, factors involved in DNA end resection or in canonical NHEJ [212].
There is plenty of evidence that deficient DNA repair pathways (HR, MMR, and BER) lead to genomic instability. Several studies have indicated that Polθ may have an important role in limiting excess genomic instability and allowing tumor cell survival when exposed to oncogenic-replicative stress, or when canonical error-free repair pathways are impaired [207,215,216,217]. In fact, the overexpression of POLQ in cases of colorectal, gastric, lung and breast cancers are associated with a worse overall survival [206,218,219].
Genomic sequencing of breast cancer and colorectal adenocarcinomas has identified specific mutations and microhomologies at the rearrangement breakpoints, suggesting involvement of MMEJ [220,221]. Whole-genome sequencing of 2559 samples from 38 tumor types showed that the junction rearrangement could be generated by different formation mechanisms, i.e., NHEJ (no sequence homology at the breakpoint junction), MMEJ (2–7 bp of microhomology) and SSA or others, including microhomology-mediated BIR (10–30 bp of microhomology) [222]. Beyond the NHEJ, synthesis-dependent MMEJ can also form simple deletions and apparent blunt-end junctions [197]. Between those samples, data from 52 colorectal adenocarcinomas showed elevated heterogeneity and high rates of the fragile-site signature [222]. Studies of breakpoint sequences across different tumor types indicated that 27–50% of the genomic rearrangements could be attributed to microhomology-mediated mechanisms [223]. In some tumor types, the breakpoint microhomology has been associated with genomic stability and tumor invasiveness. For example, invasive bladder tumors are characterized by genetic instability and microhomology at rearrangement breakpoints (indication of MMEJ activity), whereas non-invasive tumors do not show microhomology and are genetically stable with error-free DNA repair profile [224].
Moreover, POLQ is largely absent in most normal tissues, representing a promising tumor-specific target for cancer treatment. In this regard, knockdown of POLQ increased the sensitivity of a panel of tumor cell lines from different primary sites to radiation in contrast to the normal tissue cells [225]. POLQ was revealed to be synthetic lethal with DNA repair genes frequently mutated in cancer (such as ATM, ATR, BRCA1/2, FANCD2, Ku70, RAD52, RAD51C, TP53BP1) and extensive efforts for the development of Polθ inhibitors are made by several companies [226]. The antibiotic Novobiocin was shown to inhibit Polθ, thus decreasing viability of HR-deficient tumor cells due to accumulation of toxic RAD51 foci [158]. As HR repair and MMEJ shared a common resection step with participation of MRE11 nuclease [172], Polθ inhibitors could strongly synergize with MRE11 inhibitors. Polθ inhibitors could also synergize with PARPi in HR-deficient cancers and possibly overcome acquired resistance to monotherapy [207,227].
5.3. Single-Strand Annealing
Single Strand Annealing (SSA) is an important DSB repair pathway, which rely on annealing of homologous repeats flanking a DSB, generating loss of genetic information by large deletions (up to several hundred bp) between the repeats (Figure 2) [228]. It was shown that chromosomal translocations at Alu elements, the most numerous family between the repetitive elements that comprise nearly half of the human genome, occur predominantly by SSA [229]. The complementary DNA sequences involved in SSA are more than 20–25 nucleotides that distinguish it from the MMEJ [195]. PARP-1 mediates the recruitment of MRN and CtIP to the DSB for end resection, which is an essential step of SSA. The resultant short single-strand region is rapidly coated by RPA and serves as a binding site for the processive Exo1 or DNA2 exonucleases [195,228]. The resultant long-range resection exposes complementary regions with more than 25 nucleotides. The next steps are annealing by the Rad52 protein that has a robust single annealing activity on single strand complementary sequences, and removal of the non-homologous 3′ DNA tails by ERCC1/XPF complex. After these, the gaps are filled by DNA polymerases and ligated by DNA ligases that are not well defined for SSA.
RAD52 Inhibition
Human RAD52 was recently revealed as a promising candidate for targeted therapy in relation to its important role in replication fork metabolism, promoting cellular viability in cancer cells [230]. RAD52 is a DNA-binding protein involved in the HR sub-pathway single-strand annealing (SSA) in mammalian cells. SSA is an error-prone repair leading to deletions between the homologous repeats, thus increasing genomic instability. Synthetic lethality studies have shown that, in the absence of BRCA2, BRCA1, and PALB2, RAD52 is essential for DSB repair [231,232,233]. The RAD52 N-terminal domain was shown to be essential for maintaining viability of BRCA-deficient cells promoting DSB repair by homology-directed repair (HDR) and SSA [234,235]. The activity of RAD52 responsible for HDR in these cells relies on D-loop formation rather than mediation of RAD51 loading on single-stranded DNA in the presence of RPA. RAD52 plays a role also in replication forks repair and mitotic DNA synthesis (MiDAS) upon replication stress [138,142,230]. Mammalian RAD52 participates in break-induced replication (BIR) repair of collapsed DNA replication forks in cancer cells [236].
RAD52 foci co-localized with FANCD2 into mitotic chromatin following replicative stress (RS), together with RPA and MUS81 [138]. Since RS is the main mechanism of genomic instability in cancer cells, it is extensively studied for identification of targets in cancer therapy. The persistence of stalled forks can lead to fork collapse and chromosome instability (CIN), which is generally associated with poor prognosis, promoting tumor heterogeneity and drug resistance. High RAD52 protein expression in tumor samples from 179 patients who underwent surgery for rectal cancer was associated with worse disease-free survival [233]. None of the cases with RAD52 protein expression was classified as microsatellite instability (MSI)-high (MMR-negative).
Currently there is an increasing clinical interest in the use of RAD52 inhibitors (in addition to inhibitors of PARP, Chk1, and Polθ) in order to exacerbate the RS in human cancers [237]. RAD52 has a limited role in DNA repair of normal cells like Polθ, thus providing a selective therapeutic target. Small-molecule RAD52 inhibitors are discovered by structure-based selection and proposed to be suitable for disruption of RAD52 rings in BRCA-deficient cancers [238]. RAD52 inactivation increased cell death in lung tumors and in BRCA2-deficient cancer cells [138]. Combined disruption of RAD52 and POLQ conferred synthetic reduction in the velocity of replication fork restart and had additive effect on cisplatin toxicity [239]. Moreover, combination of RAD52 and PARP1 inhibitors could improve the response to treatment in HR-deficient cancers [238,240].
5.4. Cell Cycle Checkpoint Inhibition
The cell cycle checkpoint can be impaired by inhibiting the ATM-CHK2-p53 pathway or the ATR-CHK1-WEE1 pathway. Cells that lack functional p53 depend on the G2 checkpoint for DNA repair and survival since the G1 checkpoint is activated by the p53-p21 pathway that is defective in these cells [241]. Inhibition of the G2 checkpoint in p53-deficient cells has shown to increase the sensitivity to some DNA damage inducing agents in vitro [144,151]. Interestingly, there is a high frequency of TP53 mutation in CRC samples. The germline, followed by TP53 somatic mutation (single-nucleotide variants) was found in 70.4% of stage III CRC patients [63]. Moreover, data from 50 CRC patients showed ATR and CHK1 expression significantly increased in the tumor, compared to adjacent mucosa, and ATR expression has significantly increased in the late stages (III and IV), thus ATR and CHK1 appear to be important for tumor progression [242].
5.4.1. ATR Inhibition
In vitro studies indicate that ATR inhibitors can be used in combination with oxaliplatin, cisplatin, or CHK1 inhibitor to inhibit the proliferation of colorectal cancer cells. The inhibition of Nbs1-dependent ATR activation in p53-deficient colorectal cancer cell (HT29) increased sensitivity to oxaliplatin and cisplatin chemotherapeutic agents through a reduction in proliferation, increase in sub-G1 cell population and cleaved-caspase3 after 72 h treatment with the inhibitor CBP-93872 [151]. In this study, the ATR inhibitor suppressed the G2 checkpoint induced by oxaliplatin and cisplatin, as well as suppressed the downstream CHK1 activation [151]. Once these cells are p53-deficient, they depend on G2-checkpoint for survival, which can be suppressed by ATR-CHK1 signaling inhibitors. The ATR inhibition also showed an increase in cytotoxicity when combined with a CHK1 inhibitor in CRC cells in another study. The co-treatment prevented ATR-dependent feedback activation of CHK1 and hence increased the replication stress of cancer cells [144]. The results of this study showed that the CHK1 inhibitor V158411 decreased cell proliferation and induced DNA damage in the CRC cells HT29 and U2OS, assessed by nuclear staining for γ-H2AX, pCHK1 (S317), pCHK2 (T68), and pRPA32 (S4/S8). Moreover, the addition of ATR inhibitor VX-970 with the CHK1 inhibitor increased the DNA damage and the cell growth inhibition in a time and dose dependent manner [144]. The combination of ATR and CHK1 inhibitor reinforce the suppression of checkpoint, which can be lethal for cells undergoing replication stress.
5.4.2. CHK1 Inhibition
The induction of DSB needs to be repaired to cell survival; hence, cell promotes cell cycle arrest. However, cells that lack p53 depend on ATR-CHK1 checkpoint, thus inhibiting this axis is an attractive strategy. In fact, the inhibition of CHK1 by prexasertib (LY2606368) killed primary CRC enriched for cancer stem cells (CRC-SC). However, not all samples showed sensitivity. When they correlated sensitivity with genome sequencing found that TP53 mutations were a biomarker predicting sensitivity, as well as phosphorylation of ATM or RPA32—replicative stress markers—and increased chromosome number. CRC-SC with these markers depend on CHK1 activity, so LY2606368 treatment by inhibiting CHK1 impaired cell cycle checkpoint resulting in lethal replication catastrophe [145]. Interestingly, the inhibition of RAD51 (B02) or MRE11 (Mirin) was able to sensitized resistant CRC-SC to prexasertib by induction of replication stress, while others inhibitors of DDR proteins ATM, ATR, or DNA-PK were ineffective [243]. RAD51 and MRE11 are HR factors, the main pathway activated for resolving DSB during replication, and CRC-SC are tolerable to high replication stress level due to modulation of DNA damage response [156]. These data evidence RAD51 and MRE11 as key regulators of CHK1-resistant CRC-SC survival and support the future development of clinical trials with these treatment regimens.
5.4.3. ATM Inhibition
The ATM-p53 pathway is considered one of the major synthetically lethal partners. ATM is mutated in ~20% of colorectal cancers, which are mostly missense mutations and scattered throughout the coding region [244]. However, functional effects of the ATM mutations are not clear; ataxia telangiectasia (A–T) mutations in ATM are known to induce protein truncation, protein destabilization, and resulting loss of function [245]. In metastatic CRC patients, 15% harbor somatic ATM mutations, which was associated with improved overall survival. Considering CRC patients with TP53 mutations, 69% have a co-mutation in ATM [58]. It was recently reported that 13.8% and 22.2% of stage III CRC patients harbor somatic mutations in ATM or BRCA2, respectively. Moreover, patients carrying ATM, BRCA2, or non-TP53 APC somatic mutations treated with oxaliplatin-based chemotherapy had a better outcome than those without such mutations [63].
The in vitro studies indicate that the ATM inhibition sensitizes CRC cells to chemotherapy, and the p53 status is involved in ATM inhibitor sensitivity [152]. A variety of ATM-deficient cancer cell lines showed sensitivity to PARP inhibitors, including CRC cell lines. Considering this evidence, the use of ATM inhibitor could sensitize cancer cells to the PARP inhibitor Olaparib as well [152,246]; so, the ATM inhibitor KU55933 was tested in the ATM-proficient HCT116 cell line and sensitized it to PARP-inhibitor Olaparib, decreasing the colony formation number after 14 days and the depletion of p53 enhanced the sensitivity [152]. In another study, ATM inhibitor (AZ31) also enhanced the antiproliferative effect in CRC cell lines (HCT15, HCT116, and RKO) and patient derived xenografts with the addition of topoisomerase I inhibitor (SN38), due to a cytostatic effect (increase in G2/M cell cycle arrest without apoptosis induction) [153]. It was found that cell lines sensitive to the co-treatment presented in an intriguing way no p53 protein levels, despite showing increase in p53 phosphorylation (s15) levels after the co-treatment with the ATM inhibitor and irinotecan, while resistant cells presented p53 protein levels and showed little increase in P-p53 (s15) levels after the co-treatment. Interestingly the PDX models that showed sensitivity to combined treatment were irinotecan resistant, while the irinotecan sensitive had little sensitivity to the co-treatment, yet additional study is necessary to identify irinotecan resistance biomarkers [153]. Using samples from metastatic CCR patients the ATM-loss was not associated with improvement in overall survival after irinotecan-based therapy, but after oxaliplatin-based therapy. However, the p53 levels were not evaluated in this study [247]. Notably the use of checkpoint inhibitors depends on the expression of cell damage response biomarkers as well as the damage inducing agents applied.
5.4.4. WEE1 Inhibition
The WEE1 upregulation in colorectal tumor samples has been reported, although whether this upregulation has prognostic value or not remains controversial. Upregulation of WEE1 is suggested as a potential prognostic biomarker for CRC patients [248]. The WEE1 mRNA levels were increased in tumor tissues and the positive staining of WEE1 was found in the most of 102 CRC tissue samples. Both results were correlated with distant metastasis and high TNM stage [248]. In another study, the checkpoint kinase WEE1 was highly expressed in primary CRC obtained from patients of a prospective cohort but it did not reach independent prognostic value [249]. Moreover, an in vitro study has pointed out that WEE1 inhibition selectively kills tumor cells, so WEE1 inhibitors may have a role as targeted therapy for CRC. The inhibitor adavosertib (AZD1775, MK-1755) induced apoptosis in CRC liver metastases endothelial cells (CLMECs) isolated from patients. The WEE1 inhibition had preferential effects on CLMECs than on matched normal liver endothelial cells because CLMECs had WEE1 mRNA and protein levels upregulated in comparison to the normal cells [146]. This particular WEE1 inhibitor also increased 5-FU cytotoxicity in TP53-mutated CRC cell line (HT29) by induction of DSB and apoptosis [147]. Moreover, adavosertib decreased cell survival of CRC cell line (HCT116) in combination to PARG inhibitor (PDDX-004/PDD00017272) in vitro, and in PARG knockout cell clones (HCT116 PARG KO) in xenograft model by induction of DNA damage in S-phase [148]. Adavosertib was also reported to increase apoptosis of the TP53-mutated CRC cell line (HT29 and SW480), and sensitize the cells to irinotecan. These effects may have occurred due to inhibition of the G2 checkpoint so the cell loses the ability to repair the DNA damage induced by irinotecan [149]. The p53-status may be a biomarker and could be considered in clinical trials.
6. Clinical Trials of DDR Inhibitors in CRC Patients
In colorectal tumors with DDR-deficient background, combinations of cytotoxic agents and DSB repair-targeting therapies (e.g., PARP, ATM, ATR, or CHK1inhibitors) may be useful. The studies with checkpoint inhibitors are more advanced than those with DNA repair inhibitors, as some of them have already reached clinical trials (Table 2).
Table 2.
Clinical Trials of DDR inhibitors in CRC patients.
| Inhibitor | NCT Number | Conditions | Primary and Secondary Endpoints | Intervention/Treatment for CRC | Phase(s) | Status | Reference | |
|---|---|---|---|---|---|---|---|---|
| Chk1 Inhibitor | Prexasertib (LY2606368) | NCT02860780 | Advanced/metastatic cancer, including CRC with KRAS and/or BRAF mutations | MTD | Prexasertib + ralimetinib | Phase 1 | Completed | [250] |
| NCT02124148 | Advanced/metastatic cancer, including KRAS wild type CRC, which has failed to oxaliplatin- and irinotecan-based chemotherapy or is intolerant of irinotecan or oxaliplatin | Prexasertib + cetuximab | Phase 1b | Completed | - | |||
| LY2880070 | NCT02632448 | Solid tumors, including CRC | MTD | LY2880070 ± gemcitabine | Phase 1b/2a | Recruiting | [251] | |
| SRA737 | NCT02797964 | Advanced solid tumors (including CRC) and non-Hodgkin’s lymphoma | Subjects with TRAE, MTD, recommended Phase 2 dose, ORR | SRA737 | Phase 1/2 | Completed | [252] | |
| ATM Inhibitor | AZD0156 | NCT02588105 | Advanced solid tumors, including CRC | Subjects with TRAE | AZD0156 + irinotecan/ FOLFIRI |
Phase 1 | Active, not recruiting | [253] |
| ATR Inhibitor | Ceralasertib (AZD6738) | NCT04704661 | Advanced solid tumors, including CRC that have a change (mutation) in the HER2 gene or protein | RP2D, PD profile of tumor tissues between Top1 inhibition and Top1 + ATR dual inhibition | Ceralasertib + trastuzumab deruxtecan | Phase1/1b | Not yet recruiting | - |
| Elimusertib (BAY 1895344) | NCT04535401 | Advanced or metastatic cancers of the stomach and intestines, including CRC, which have previously progressed on irinotecan with and without DDR defects | MTD | Elimusertib + FOLFIRI | Phase 1 | Not yet recruiting | [254] | |
| Berzosertib (M6620, VX-970) | NCT02157792 | Advanced solid tumors, including CRC harboring molecular aberrations, including ATM loss and an ARID1A mutation, achieved complete response, and maintained this response, with a progression-free survival of 29 months at last assessment | Safety (AE, laboratory values, ECG), ORR | M6620 + carboplatin | Phase 1 | Completed | [255,256] | |
| WEE1 inhibitor | Adavosertib (AZD1775, MK-1755) | NCT02906059 | Metastatic CRC with RAS (KRAS or NRAS) or BRAF mutated | DLT and TREA | AZD1775 + irinotecan | Phase Ib | Completed | [257] |
| NCT02465060 | Advanced refractory solid tumors (including CRC), lymphomas, or multiple myeloma | ORR | Adavosertib + targeted therapy according to mutational status | Phase II | Recruiting | [258,259] | ||
| NCT00648648 | Advanced solid tumors, including CRC | DLT, best ORR | Adavosertib + gemcitabine + cisplatin or carboplatin | Phase 1 | Completed | [260] | ||
| ZN-c3 | NCT04158336 | Solid tumors, including CRC | MTD, RP2D, DLR, ORR | ZN-c3 | Phase I/II | Recruiting | [261] | |
CRC, colorectal cancer; MTD: maximum tolerated dose; TRAE: treatment-related adverse event; RP2D: recommended phase 2 dose; ORR: objective response rate; PD: pharmacodynamics; ECG: electrocardiogram; SD: stable disease, DLT: dose limiting toxicities; pCR: pathological clinical response; cCR: clinical complete response; PR: pathological response.
There are currently four candidates in clinical trials acting as ATR-kinase inhibitors—berzosertib, ceralasertib, elimusertib, and M4344. Of these, berzosertib and ceralasertib are the most studied candidates. Although more than 50 clinical trials in phase I/II have been investigating these ATR inhibitors, only one study including CRC patients has published their results so far. The phase I clinical trial of berzosertib in monotherapy or in association with carboplatin included 11 patients with KRAS and BRAF wild type advanced CRC harboring ATM loss and an AT-rich interactive domain 1A (ARID1A) mutation (NCT02157792). Of the three CRC patients treated with monotherapy, one achieved stable disease with an ongoing progression-free survival of 29 months. Baseline tumor analyses of this patients revealed complete absence of ATM protein expression, although there was no evidence of ATM mutation, MMR deficiency (loss of MLH1 and PMS2) and truncating mutations in several DNA repair enzymes, including two heterozygous truncating mutations in ARID1A and heterozygous truncating mutations in CHEK1, RAD50, POLD1 [256].
ARID1A is commonly mutated in CRC (9.4%), which results in loss of protein expression with consequent impairment of SWI/SNF chromatin remodeling complex. In CRC, ARID1A mutation frequency is enriched in tumors with MSI and loss of ARID1A in CRC with late TNM stage, poor pathological classification, and distant metastasis [262,263]. Recently, epigenetic regulator and DNA repair proteins, having synthetic lethality interaction with ARID1A mutation, have been reported, such as inhibition of PARP and ATR. In ARID1A-deficient tumors, decreased accessibility of 53BP1 to DNA lesions leads to reduced NHEJ activity, rendering high sensitivity to PARP inhibitor therapy after exposure to exogenously induced DNA breaks such as ionizing radiation [264]. Moreover, CRC cell lines harboring KRAS mutations are critically dependent on ARID1A function [265]. Interestingly, evidence exists that both NHEJ and HR can be affected by the depletion of the other subunit of the SWI/SNF complex, ARID1B, in ARID1A-deficient cells. A recently study found that an impaired interaction between these two members of SWI/SNF complex and HR enzymes in CRC ARID1A-mutated cell lines contribute to an increased radiosensitivity and reduced RAD51 foci induction, indicating reduced homologous recombination [266]. Taken together, these findings may open up perspectives for new therapeutic combinations aiming to disrupt DSB repair in CRC without an HRD background.
CHK1 inhibitors have a long history in clinical trials, however, their use is associated with elevated toxicity, especially for the low selectivity first-generation CHK1i [267]. From seven CHK1 inhibitors that entered in clinical trials, only three remain active: prexasertib, LY2880070, and SRA737. The phase I/Ib clinical trials of prexasertib for patients with advanced or metastatic solid tumors included CRC patients with KRAS and/or BRAF mutations (NCT02860780) and CRC patients with KRAS wild type CRC who has failed to oxaliplatin- and irinotecan-based chemotherapy (NCT02124148) to evaluate safety of the combinations of prexasertib with ralimetinib or cetuximab, respectively [250]. However, in the study of prexasertib + ralimetinib (a p38 mitogen-activated protein kinase inhibitor), the recommended phase 2 dose (RP2D) of prexasertib were not established due to dose-limiting toxicities [250]. The LY2880070 safety and efficacy of LY2880070 in monotherapy or in combination with gemcitabine (NCT02632448) is under investigation in a phase IB/IIA for solid tumors, including CRC.
The WEE1 inhibitor adavosertib has been under investigation in phases I/II since 2008, and currently, there are 28 clinical trials with this molecule still active. The combination of adavosertib + gemcitabine + cisplatin or carboplatin was assessed in a phase I trial for several solid advanced tumors (NCT00648648). Partial response and stable disease lasting at least 6 weeks as best overall response were observed in 10% and 53% of the patients, respectively. Sixteen patients (8%) presented advanced or metastatic CRC, and one patient had stable disease [260]. A new WEE1 inhibitor, ZN-c3, is under investigation in a phase I/II clinical trial for solid tumors, including CRC (NCT04158336). It has recently been reported that five patients reached stable disease and two had partial response (ovarian cancer and CRC patients). Moreover, a 42% reduction in overall target lesions was observed in CRC patients, which remained about 6 months on study drug before disease progression [261].
Despite CRC high incidence, prevalence and understanding of its molecular etiology, disease staging and tailored therapeutic approaches are still limited. Even for late-stage disease, to which systemic treatment options are biomarker-driven, resistance inevitably occurs [268]. Within this scenario, downregulation of MMR (MLH1 and MLH6) and HR genes (BRCA1/2, RAD51) and upregulation of error-prone DNA polymerases (e.g., Pol iota, Pol kappa) have been implicated as a key mechanism of acquired-resistance to targeted-therapies [269]. A recent study with CRC cell lines and patient-derived xenograft models treated with cetuximab or an anti-BRAF (dabrafenib) has shown that CRC cells can evade targeted therapies by switching off DNA repair pathways, particularly through downregulation of HR. These cells demonstrated compromised DNA repair and persistent elevated DNA damage levels. This observation may reveal another opportunity to elucidate whether anti-VEGF/EGFR-resistant CRC cells may be sensitized by agents, such as PARP inhibitors or the novel agents targeting the ATR-CHK1-WEE1 axis [269].
7. Molecular Selection of CRC Patients for Clinical Trials with DDR Inhibitors
Although DNA repair genes alterations and the resulting genomic instability have been explored in other tumor types, only few DDR inhibitors were employed as monotherapy or combined with chemotherapy in trials enrolling CRC patients, including PARP, WEE1, ATR, and CHK1 inhibitors. Two recent review articles summarizing the ongoing and completed clinical trials concluded that the obtained unsatisfactory results in some trials could be attributed to a lack of molecular selection of patients to be included for any specific treatment [53,270]. Moreover, the lack of characterization of functional repair deficiencies and consensus on the defined set of DDR genes to be tested were appointed as limiting factors. A growing body of evidence suggests that the sensitivity of CRC cells and tumors to therapy induced DNA damage and repair inhibition could differ in relation to the microsatellite instability status and functionality of HR (summarized on Figure 3). Only in one trial with published results—the microsatellite status was used as criteria to stratify 33 patients with disseminated colorectal cancer (20 MSS and 13 MSI-H) enrolled for Olaparib testing, where no statistical difference was found in the median progression-free survival and overall survival between the two groups [271]. However, in this study Olaparib was used as a single-agent and not in combination with DNA damage inducing chemotherapy or radiation therapy. In this respect, in vitro studies have shown a synergistic effect of Olaparib and 5-FU in MSI-H CRC [272], whereas Olaparib monotherapy showed benefits in MSS CRC with non-functional HR, and was proposed as maintenance therapy in patients responsive to oxaliplatin-based regimens [273]. The enrichment of PARPi sensitive MSS cell lines was observed in those with preserved TP53 function and TP53-mediated suppression of RAD51 was appointed as a possible mechanism of action [274]. Further, data from 99 MSS CRC cell lines revealed that functionality of HR repair determined in the RAD51 assay could discriminate for PARPi susceptible CRC tumors, in which no increase in the percentage of RAD51 foci positive nuclei was observed upon radiation [273]. Moreover, this study did not show association between the sensitivity to the PARPi Olaparib and mutational signatures correlated to HR defects or BRCAness, as well as to specific CMS or mutations in KRAS and BRAF. RAD51-low score was found as an indicator of response (to PARP inhibitors and carboplatin) and patient outcome in two studies reported on ESMO Breast Cancer Virtual Congress 2021, and incorporation of RAD51 testing for clinical decision making in triple negative breast cancer was proposed [275,276].
Figure 3.
Possible targets for DDR inhibitors (repair proteins in blue and checkpoint kinases in grey) in CRC chemotherapy with supporting evidence in cancer cell lines and animal models, or candidates “to be tested” (in red), with estimated sensitivity based on results obtained in cells with deficiency of the respective target. HRD—homologous recombination deficiency, IR—ionizing radiation, Topoi—topoisomerase inhibitor, MMS—microsatellite stable, MSI—microsatellite instable, RS—replication stress, dATR-Chk1—deficient ATR-Chk1 signaling, FOLFIRI—folinic acid/fluorouracil/irinotecan chemotherapy regimen.
In order to assess the tumor HR status, a combined HRD score was proposed, calculated as unweighted sum of three independent scores for estimation of genomic instability: telomeric allelic imbalance (TAI), loss of heterozygosity (LOH), and large-scale state transitions (LST), that can be determined on a pretreatment biopsy [277]. Using this DNA-based measure, the authors found that tumors with HR deficiency, with a threshold of HRD score ≥ 42 for breast and ovarian cancers, were responsive to neoadjuvant platinum-based therapy. Recent study extended the utility of HRD analysis for sensitivity to agents inducing RS and DNA double-strand breaks (such as 5-FU, oxaliplatin, irinotecan, topoisomerase inhibitors, between others) to all solid cancers from The Cancer Genome Atlas and Cancer Cell Line Encyclopedia [278]. The authors showed that the HRD cases with history of treatment with DNA-damaging agents (n = 2979) had better overall survival than treated non-HRD cases, while in absence of treatment the HRD cases (n = 6460) had a worse outcome than non-HRD cases. Moreover, the HRD score was higher in tumors with TP53 mutations. The results of this study suggested that personalization of the treatment based on HRD status as a therapeutic biomarker has a potential to improve the precision and efficacy of chemotherapy irrespective of the cancer type [278].
8. Conclusions
In spite of advances in identification of clinical and molecular features with significant prognostic and predictive value in CRC, the therapeutic options still mostly rely on conventional chemotherapy-based regimens rather than in targeted therapies, in particular regarding early stages. Chemotherapy is considered a “one-size-fits-all” approach, which often results in resistance to treatment and suboptimal outcomes. Targeting genes involved in the responses to replication stress and DNA DSBs repair, such as MRE11, RAD51, RAD52, and POLQ, alone or in combination with DNA damaging agents and checkpoint inhibitors, might be a strategy to expand the spectrum of systemic therapeutic options, thus tailoring CRC treatments and improving the disease prognosis. Moreover, molecular selection of alterations in DNA repair genes, and the resulting genomic instability for targeted DDR therapy in sub-populations of CRC patients could be helpful because of tumor heterogeneity. An increasing body of evidence suggests that the sensitivity of CRC cells and tumors to DNA damage, inducing chemotherapy and DNA repair inhibition, could differ in relation to the microsatellite instability status and functionality of HR. Current clinical testing of CRC includes characterization of the microsatellite status, which is considered for selection of chemotherapy regimens and for inclusion in clinical trials, along with mutations in oncogenic driver genes (BRAF, KRAS, etc.), but alterations in DNA DSB repair genes are not investigated. However, recent work has identified additional genomic (i.e., HRD score) and functional assays of DNA repair (i.e., RAD51 assay) that provide new predictive and pharmacodynamic biomarkers for targeted DDR therapies.
Author Contributions
Conceptualization: P.P.T., T.N.G., N.M.L., J.S.H., and J.S.; original draft preparation: P.P.T., T.N.G., N.M.L., S.P., A.-C.B.; review and editing: P.P.T., T.N.G., J.S.H., and J.S. Supervision: J.S.H. and J.S. All authors have read and agreed to the published version of the manuscript.
Funding
P.P.T and J.S were supported by the Brazilian agencies Fundação de Amparo à Pesquisa do Estado do Rio Grande do Sul (FAPERGS) and Conselho Nacional de Desenvolvimento Científico e Tecnológico, (CNPq) –PRONEX-FAPERS/CNPq - Grant no. 16/2551-0000473-0-1.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Siegel R.L., Miller K.D., Goding Sauer A., Fedewa S.A., Butterly L.F., Anderson J.C., Cercek A., Smith R.A., Jemal A. Colorectal cancer statistics, 2020. CA Cancer J. Clin. 2020;70:145–164. doi: 10.3322/caac.21601. [DOI] [PubMed] [Google Scholar]
- 2.Arnold M., Sierra M.S., Laversanne M., Soerjomataram I., Jemal A., Bray F. Global patterns and trends in colorectal cancer incidence and mortality. Gut. 2017;66:683–691. doi: 10.1136/gutjnl-2015-310912. [DOI] [PubMed] [Google Scholar]
- 3.Bailey C.E., Hu C.Y., You Y.N., Bednarski B.K., Rodriguez-Bigas M.A., Skibber J.M., Cantor S.B., Chang G.J. Increasing disparities in the age-related incidences of colon and rectal cancers in the United States, 1975–2010. JAMA Surg. 2015;150:17–22. doi: 10.1001/jamasurg.2014.1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fearon E.R., Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759–767. doi: 10.1016/0092-8674(90)90186-I. [DOI] [PubMed] [Google Scholar]
- 5.Arvelo F., Sojo F., Cotte C. Biology of colorectal cancer. Ecancermedicalscience. 2015;9:520. doi: 10.3332/ecancer.2015.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shaukat A., Kahi C.J., Burke C.A., Rabeneck L., Sauer B.G., Rex D.K. ACG clinical guidelines: Colorectal cancer screening 2021. Am. J. Gastroenterol. 2021;116:458–479. doi: 10.14309/ajg.0000000000001122. [DOI] [PubMed] [Google Scholar]
- 7.Feo L., Polcino M., Nash G.M. Resection of the primary tumor in stage iv colorectal cancer: When is it necessary? Surg. Clin. N. Am. 2017;97:657–669. doi: 10.1016/j.suc.2017.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee Y.C., Lee Y.L., Chuang J.P., Lee J.C. Differences in survival between colon and rectal cancer from SEER data. PLoS ONE. 2013;8:e78709. doi: 10.1371/journal.pone.0078709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.West N.P., Hohenberger W., Weber K., Perrakis A., Finan P.J., Quirke P. Complete mesocolic excision with central vascular ligation produces an oncologically superior specimen compared with standard surgery for carcinoma of the colon. J. Clin. Oncol. 2010;28:272–278. doi: 10.1200/JCO.2009.24.1448. [DOI] [PubMed] [Google Scholar]
- 10.Gill S., Loprinzi C.L., Sargent D.J., Thomé S.D., Alberts S.R., Haller D.G., Benedetti J., Francini G., Shepherd L.E., Seitz J.F., et al. Pooled analysis of fluorouracil-based adjuvant therapy for stage II and III colon cancer: Who benefits and by how much? J. Clin. Oncol. 2004;22:1797–1806. doi: 10.1200/JCO.2004.09.059. [DOI] [PubMed] [Google Scholar]
- 11.Li H., Fu G., Wei W., Huang Y., Wang Z., Liang T., Tian S., Chen H., Zhang W. Re-evaluation of the survival paradox between stage IIB/IIC and stage IIIA colon cancer. Front. Oncol. 2020;10:2468. doi: 10.3389/fonc.2020.595107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Benson A.B., Al-Hawary M.M., Arain M.A., Chen Y.-J., Ciombor K.K., Cohen S., Deming D., Farkas L., Garrido-Laguna I., Grem J.L., et al. NCCN guidelines version 2.2021 colon cancer nccn evidence blocks tm continue nccn guidelines panel disclosures. Cancers. 2021;13:2638. [Google Scholar]
- 13.André T., Boni C., Navarro M., Tabernero J., Hickish T., Topham C., Bonetti A., Clingan P., Bridgewater J., Rivera F., et al. Improved overall survival with oxaliplatin, fluorouracil, and leucovorin as adjuvant treatment in stage II or III colon cancer in the MOSAIC trial. J. Clin. Oncol. 2009;27:3109–3116. doi: 10.1200/JCO.2008.20.6771. [DOI] [PubMed] [Google Scholar]
- 14.André T., De Gramont A., Vernerey D., Chibaudel B., Bonnetain F., Tijeras-Raballand A., Scriva A., Hickish T., Tabernero J., Van Laethem J.L., et al. Adjuvant fluorouracil, leucovorin, and oxaliplatin in stage II to III colon cancer: Updated 10-year survival and outcomes according to BRAF mutation and mismatch repair status of the MOSAIC study. J. Clin. Oncol. 2015;33:4176–4187. doi: 10.1200/JCO.2015.63.4238. [DOI] [PubMed] [Google Scholar]
- 15.Grothey A., Sobrero A.F., Shields A.F., Yoshino T., Paul J., Taieb J., Souglakos J., Shi Q., Kerr R., Labianca R., et al. Duration of adjuvant chemotherapy for stage III colon cancer. N. Engl. J. Med. 2018;378:1177–1188. doi: 10.1056/NEJMoa1713709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Böckelman C., Engelmann B.E., Kaprio T., Hansen T.F., Glimelius B. Risk of recurrence in patients with colon cancer stage II and III: A systematic review and meta-analysis of recent literature. Acta Oncol. 2015;54:5–16. doi: 10.3109/0284186X.2014.975839. [DOI] [PubMed] [Google Scholar]
- 17.Dienstmann R., Mason M.J., Sinicrope F.A., Phipps A.I., Tejpar S., Nesbakken A., Danielsen S.A., Sveen A., Buchanan D.D., Clendenning M., et al. Prediction of overall survival in stage II and III colon cancer beyond TNM system: A retrospective, pooled biomarker study. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2017;28:1023–1031. doi: 10.1093/annonc/mdx052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chu Q.D., Zhou M., Medeiros K.L., Peddi P., Kavanaugh M., Wu X.C. Poor survival in stage IIB/C (T4N0) compared to stage IIIA (T1-2 N1, T1N2a) colon cancer persists even after adjusting for adequate lymph nodes retrieved and receipt of adjuvant chemotherapy. BMC Cancer. 2016;16:460. doi: 10.1186/s12885-016-2446-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Figueredo A., Charette M.L., Maroun J., Brouwers M.C., Zuraw L. Adjuvant therapy for stage II colon cancer: A systematic review from the cancer care Ontario program in evidence-based care’s gastrointestinal cancer disease site group. J. Clin. Oncol. 2004;22:3395–3407. doi: 10.1200/JCO.2004.03.087. [DOI] [PubMed] [Google Scholar]
- 20.Sepulveda A.R., Hamilton S.R., Allegra C.J., Grody W., Cushman-Vokoun A.M., Funkhouser W.K., Kopetz S.E., Lieu C., Lindor N.M., Minsky B.D., et al. Molecular biomarkers for the evaluation of colorectal cancer: Guideline from the american society for clinical pathology, college of american pathologists, association for molecular pathology, and american society of clinical oncology. J. Mol. Diagn. 2017;19:187–225. doi: 10.1016/j.jmoldx.2016.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sorich M.J., Wiese M.D., Rowland A., Kichenadasse G., McKinnon R.A., Karapetis C.S. Extended RAS mutations and anti-EGFR monoclonal antibody survival benefit in metastatic colorectal cancer: A meta-analysis of randomized, controlled trials. Ann. Oncol. 2015;26:13–21. doi: 10.1093/annonc/mdu378. [DOI] [PubMed] [Google Scholar]
- 22.Pietrantonio F., Petrelli F., Coinu A., Di Bartolomeo M., Borgonovo K., Maggi C., Cabiddu M., Iacovelli R., Bossi I., Lonati V., et al. Predictive role of BRAF mutations in patients with advanced colorectal cancer receiving cetuximab and panitumumab: A meta-analysis. Eur. J. Cancer. 2015;51:587–594. doi: 10.1016/j.ejca.2015.01.054. [DOI] [PubMed] [Google Scholar]
- 23.Kopetz S., Grothey A., Yaeger R., Van Cutsem E., Desai J., Yoshino T., Wasan H., Ciardiello F., Loupakis F., Hong Y.S., et al. Encorafenib, binimetinib, and cetuximab in BRAF V600E–Mutated colorectal cancer. N. Engl. J. Med. 2019;381:1632–1643. doi: 10.1056/NEJMoa1908075. [DOI] [PubMed] [Google Scholar]
- 24.Sartore-Bianchi A., Trusolino L., Martino C., Bencardino K., Lonardi S., Bergamo F., Zagonel V., Leone F., Depetris I., Martinelli E., et al. Dual-targeted therapy with trastuzumab and lapatinib in treatment-refractory, KRAS codon 12/13 wild-type, HER2-positive metastatic colorectal cancer (HERACLES): A proof-of-concept, multicentre, open-label, phase 2 trial. Lancet Oncol. 2016;17:738–746. doi: 10.1016/S1470-2045(16)00150-9. [DOI] [PubMed] [Google Scholar]
- 25.Raghav K., Loree J.M., Morris J.S., Overman M.J., Yu R., Meric-Bernstam F., Menter D., Korphaisarn K., Kee B., Muranyi A., et al. Validation of HER2 amplification as a predictive biomarker for anti–epidermal growth factor receptor antibody therapy in metastatic colorectal cancer. JCO Precis. Oncol. 2019;3:1–13. doi: 10.1200/PO.18.00226. [DOI] [PubMed] [Google Scholar]
- 26.Le D.T., Uram J.N., Wang H., Bartlett B.R., Kemberling H., Eyring A.D., Skora A.D., Luber B.S., Azad N.S., Laheru D., et al. PD-1 blockade in tumors with mismatch-repair deficiency. N. Engl. J. Med. 2015;372:2509–2520. doi: 10.1056/NEJMoa1500596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Overman M.J., McDermott R., Leach J.L., Lonardi S., Lenz H.J., Morse M.A., Desai J., Hill A., Axelson M., Moss R.A., et al. Nivolumab in patients with metastatic DNA mismatch repair-deficient or microsatellite instability-high colorectal cancer (CheckMate 142): An open-label, multicentre, phase 2 study. Lancet Oncol. 2017;18:1182–1191. doi: 10.1016/S1470-2045(17)30422-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Overman M.J., Lonardi S., Wong K.Y.M., Lenz H.J., Gelsomino F., Aglietta M., Morse M.A., Van Cutsem E., McDermott R., Hill A., et al. Durable clinical benefit with nivolumab plus ipilimumab in DNA mismatch repair-deficient/microsatellite instability-high metastatic colorectal cancer. J. Clin. Oncol. 2018;36:773–779. doi: 10.1200/JCO.2017.76.9901. [DOI] [PubMed] [Google Scholar]
- 29.Markowitz S.D., Bertagnolli M.M. Molecular basis of colorectal cancer. N. Engl. J. Med. 2009;361:2449–2460. doi: 10.1056/NEJMra0804588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mur P., García-Mulero S., Del Valle J., Magraner-Pardo L., Vidal A., Pineda M., Cinnirella G., Martín-Ramos E., Pons T., López-Doriga A., et al. Role of POLE and POLD1 in familial cancer. Genet. Med. 2020;22:2089–2100. doi: 10.1038/s41436-020-0922-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weren R.D.A., Ligtenberg M.J.L., Kets C.M., De Voer R.M., Verwiel E.T.P., Spruijt L., Van Zelst-Stams W.A.G., Jongmans M.C., Gilissen C., Hehir-Kwa J.Y., et al. A germline homozygous mutation in the base-excision repair gene NTHL1 causes adenomatous polyposis and colorectal cancer. Nat. Genet. 2015;47:668–671. doi: 10.1038/ng.3287. [DOI] [PubMed] [Google Scholar]
- 32.Pilati C., Shinde J., Alexandrov L.B., Assié G., André T., Hélias-Rodzewicz Z., Ducoudray R., Le Corre D., Zucman-Rossi J., Emile J.F., et al. Mutational signature analysis identifies MUTYH deficiency in colorectal cancers and adrenocortical carcinomas. J. Pathol. 2017;242:10–15. doi: 10.1002/path.4880. [DOI] [PubMed] [Google Scholar]
- 33.Guinney J., Dienstmann R., Wang X., De Reyniès A., Schlicker A., Soneson C., Marisa L., Roepman P., Nyamundanda G., Angelino P., et al. The consensus molecular subtypes of colorectal cancer. Nat. Med. 2015;21:1350–1356. doi: 10.1038/nm.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Becht E., De Reyniès A., Giraldo N.A., Pilati C., Buttard B., Lacroix L., Selves J., Sautès-Fridman C., Laurent-Puig P., Fridman W.H. Immune and stromal classification of Colorectal cancer is associated with molecular subtypes and relevant for precision immunotherapy. Clin. Cancer Res. 2016;22:4057–4066. doi: 10.1158/1078-0432.CCR-15-2879. [DOI] [PubMed] [Google Scholar]
- 35.Vodicka P., Vodenkova S., Buchler T., Vodickova L. DNA repair capacity and response to treatment of colon cancer. Pharmacogenomics. 2019;20:1225–1233. doi: 10.2217/pgs-2019-0070. [DOI] [PubMed] [Google Scholar]
- 36.Punt C.J.A., Koopman M., Vermeulen L. From tumour heterogeneity to advances in precision treatment of colorectal cancer. Nat. Rev. Clin. Oncol. 2017;14:235–246. doi: 10.1038/nrclinonc.2016.171. [DOI] [PubMed] [Google Scholar]
- 37.Mouillet-Richard S., Laurent-Puig P. YAP/TAZ signalling in colorectal cancer: Lessons from consensus molecular subtypes. Cancers. 2020;12:3160. doi: 10.3390/cancers12113160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jongen J.M.J., Van der Waals L.M., Trumpi K., Laoukili J., Peters N.A., van Schelven S.J.S., Govaert K.M., Rinkes I.H.M.B., Kranenburg O. Downregulation of DNA repair proteins and increased DNA damage in hypoxic colon cancer cells is a therapeutically exploitable vulnerability. Oncotarget. 2017;8:86296–86311. doi: 10.18632/oncotarget.21145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Coebergh Van Den Braak R.R.J., Ten Hoorn S., Sieuwerts A.M., Tuynman J.B., Smid M., Wilting S.M., Martens J.W.M., Punt C.J.A., Foekens J.A., Medema J.P., et al. Interconnectivity between molecular subtypes and tumor stage in colorectal cancer. BMC Cancer. 2020;20 doi: 10.1186/s12885-020-07316-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lenz H.-J., Ou F.-S., Venook A.P., Hochster H.S., Niedzwiecki D., Goldberg R.M., Mayer R.J., Bertagnolli M.M., Blanke C.D., Zemla T., et al. Impact of consensus molecular subtyping (CMS) on overall survival (OS) and progression free survival (PFS) in patients (pts) with metastatic colorectal cancer (mCRC): Analysis of CALGB/SWOG 80405 (Alliance) J. Clin. Oncol. 2017;35:3511. doi: 10.1200/JCO.2017.35.15_suppl.3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Song N., Pogue-Geile K.L., Gavin P.G., Yothers G., Kim S.R., Johnson N.L., Lipchik C., Allegra C.J., Petrelli N.J., O’Connell M.J., et al. Clinical outcome from oxaliplatin treatment in stage II/III colon cancer according to intrinsic subtypes: Secondary analysis of NSABP C-07/NRG oncology randomized clinical trial. JAMA Oncol. 2016;2:1162–1169. doi: 10.1001/jamaoncol.2016.2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li Y., Yao Q., Zhang L., Mo S., Cai S., Huang D., Peng J. Immunohistochemistry-based consensus molecular subtypes as a prognostic and predictive biomarker for adjuvant chemotherapy in patients with stage II colorectal cancer. Oncologist. 2020;25:e1968–e1979. doi: 10.1002/ONCO.13521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stintzing S., Wirapati P., Lenz H.J., Neureiter D., Fischer von Weikersthal L., Decker T., Kiani A., Kaiser F., Al-Batran S., Heintges T., et al. Consensus molecular subgroups (CMS) of colorectal cancer (CRC) and first-line efficacy of FOLFIRI plus cetuximab or bevacizumab in the FIRE3 (AIO KRK-0306) trial. Ann. Oncol. 2019;30:1796–1803. doi: 10.1093/annonc/mdz387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fong Y.W., Cattoglio C., Tjian R. The intertwined roles of transcription and repair proteins. Mol. Cell. 2013;52:291–302. doi: 10.1016/j.molcel.2013.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Eckelmann B.J., Bacolla A., Wang H., Ye Z., Guerrero E.N., Jiang W., El-Zein R., Hegde M.L., Tomkinson A.E., Tainer J.A., et al. XRCC1 promotes replication restart, nascent fork degradation and mutagenic DNA repair in BRCA2-deficient cells. NAR Cancer. 2020;2 doi: 10.1093/narcan/zcaa013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Negrei C., Hudita A., Ginghina O., Galateanu B., Voicu S.N., Stan M., Costache M., Fenga C., Drakoulis N., Tsatsakis A.M. Colon cancer cells gene expression signature as response to 5-fluorouracil, oxaliplatin, and folinic acid treatment. Front. Pharmacol. 2016;7:172. doi: 10.3389/fphar.2016.00172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wyatt M.D., Wilson D.M. Participation of DNA repair in the response to 5-fluorouracil. Cell. Mol. life Sci. 2009;66:788–799. doi: 10.1007/s00018-008-8557-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Srinivas U.S., Dyczkowski J., Beißbarth T., Gaedcke J., Mansour W.Y., Borgmann K., Dobbelstein M. 5-Fluorouracil sensitizes colorectal tumor cells towards double stranded DNA breaks by interfering with homologous recombination repair. Impact J. 2015;6:12574–12586. doi: 10.18632/oncotarget.3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chválová K., Brabec V., Kašpárková J. Mechanism of the formation of DNA-protein cross-links by antitumor cisplatin. Nucleic Acids Res. 2007;35:1812–1821. doi: 10.1093/nar/gkm032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ming X., Groehler A., Michaelson-Richie E.D., Villalta P.W., Campbell C., Tretyakova N.Y. Mass spectrometry based proteomics study of cisplatin-induced dna-protein cross-linking in human fibrosarcoma (HT1080) cells. Chem. Res. Toxicol. 2017;30:980–995. doi: 10.1021/acs.chemrestox.6b00389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Riddell I.A., Lippard S.J. Metallo-Drugs: Development and Action of Anticancer Agents. Volume 18. De Gruyter; Berlin, Germany: 2018. 7. Medicinal chemistry of gold anticancer metallodrugs; pp. 199–218. [Google Scholar]
- 52.Li F., Jiang T., Li Q., Ling X. Camptothecin (CPT) and its derivatives are known to target topoisomerase I (Top1) as their mechanism of action: Did we miss something in CPT analogue molecular targets for treating human disease such as cancer? Am. J. Cancer Res. 2017;7:2350–2394. [PMC free article] [PubMed] [Google Scholar]
- 53.Reilly N.M., Novara L., Di Nicolantonio F., Bardelli A. Exploiting DNA repair defects in colorectal cancer. Mol. Oncol. 2019;13:681–700. doi: 10.1002/1878-0261.12467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Heeke A.L., Pishvaian M.J., Lynce F., Xiu J., Brody J.R., Chen W.-J., Baker T.M., Marshall J.L., Isaacs C. Prevalence of homologous recombination–related gene mutations across multiple cancer types. JCO Precis. Oncol. 2018;2:1–13. doi: 10.1200/PO.17.00286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Muzny D.M., Bainbridge M.N., Chang K., Dinh H.H., Drummond J.A., Fowler G., Kovar C.L., Lewis L.R., Morgan M.B., Newsham I.F., et al. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lamberti G., Andrini E., Sisi M., Di Federico A., Ricciuti B. Targeting DNA damage response and repair genes to enhance anticancer immunotherapy: Rationale and clinical implication. Future Oncol. 2020;16:1751–1766. doi: 10.2217/fon-2020-0215. [DOI] [PubMed] [Google Scholar]
- 57.Bianco J.N., Bergoglio V., Lin Y.L., Pillaire M.J., Schmitz A.L., Gilhodes J., Lusque A., Mazières J., Lacroix-Triki M., Roumeliotis T.I., et al. Overexpression of claspin and timeless protects cancer cells from replication stress in a checkpoint-independent manner. Nat. Commun. 2019;10 doi: 10.1038/s41467-019-08886-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Randon G., Fucà G., Rossini D., Raimondi A., Pagani F., Perrone F., Tamborini E., Busico A., Peverelli G., Morano F., et al. Prognostic impact of ATM mutations in patients with metastatic colorectal cancer. Sci. Rep. 2019;9 doi: 10.1038/s41598-019-39525-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.AlDubayan S.H., Giannakis M., Moore N.D., Han G.C., Reardon B., Hamada T., Mu X.J., Nishihara R., Qian Z., Liu L., et al. Inherited DNA-repair defects in colorectal cancer. Am. J. Hum. Genet. 2018;102:401–414. doi: 10.1016/j.ajhg.2018.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang Z., Zhao J., Wang G., Zhang F., Zhang Z., Zhang F., Zhang Y., Dong H., Zhao X., Duan J., et al. Comutations in DNA damage response pathways serve as potential biomarkers for immune checkpoint blockade. Cancer Res. 2018;78:6486–6496. doi: 10.1158/0008-5472.CAN-18-1814. [DOI] [PubMed] [Google Scholar]
- 61.Sun J., Wang C., Zhang Y., Xu L., Fang W., Zhu Y., Zheng Y., Chen X., Xie X., Hu X., et al. Genomic signatures reveal DNA damage response deficiency in colorectal cancer brain metastases. Nat. Commun. 2019;10:3190. doi: 10.1038/s41467-019-10987-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chartron E., Theillet C., Guiu S., Jacot W. Targeting homologous repair deficiency in breast and ovarian cancers: Biological pathways, preclinical and clinical data. Crit. Rev. Oncol. Hematol. 2019;133:58–73. doi: 10.1016/j.critrevonc.2018.10.012. [DOI] [PubMed] [Google Scholar]
- 63.Lin P.C., Yeh Y.M., Chan R.H., Lin B.W., Chen P.C., Pan C.C., Shen M.R. Sequential and co-occurring DNA damage response genetic mutations impact survival in stage III colorectal cancer patients receiving adjuvant oxaliplatin-based chemotherapy. BMC Cancer. 2021;21:217. doi: 10.1186/s12885-021-07926-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Faraoni I., Graziani G. Role of BRCA mutations in cancer treatment with poly(ADP-ribose) polymerase (PARP) inhibitors. Cancers. 2018;10:487. doi: 10.3390/cancers10120487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bryant H.E., Schultz N., Thomas H.D., Parker K.M., Flower D., Lopez E., Kyle S., Meuth M., Curtin N.J., Helleday T. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434:913–917. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- 66.Farmer H., McCabe H., Lord C.J., Tutt A.H.J., Johnson D.A., Richardson T.B., Santarosa M., Dillon K.J., Hickson I., Knights C., et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–921. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 67.Lord C.J., Ashworth A. BRCAness revisited. Nat. Rev. Cancer. 2016;16:110–120. doi: 10.1038/nrc.2015.21. [DOI] [PubMed] [Google Scholar]
- 68.Laporte G.A., Leguisamo N.M., Kalil A.N., Saffi J. Clinical importance of DNA repair in sporadic colorectal cancer. Crit. Rev. Oncol. Hematol. 2018;126:168–185. doi: 10.1016/j.critrevonc.2018.03.017. [DOI] [PubMed] [Google Scholar]
- 69.Koopman M., Kortman G.A.M., Mekenkamp L., Ligtenberg M.J.L., Hoogerbrugge N., Antonini N.F., Punt C.J.A., Van Krieken J.H.J.M. Deficient mismatch repair system in patients with sporadic advanced colorectal cancer. Br. J. Cancer. 2009;100:266–273. doi: 10.1038/sj.bjc.6604867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hegde M., Ferber M., Mao R., Samowitz W., Ganguly A. ACMG technical standards and guidelines for genetic testing for inherited colorectal cancer (Lynch syndrome, familial adenomatous polyposis, and MYH-associated polyposis) Genet. Med. 2014;16:101–116. doi: 10.1038/gim.2013.166. [DOI] [PubMed] [Google Scholar]
- 71.Roth A.D., Tejpar S., Delorenzi M., Yan P., Fiocca R., Klingbiel D., Dietrich D., Biesmans B., Bodoky G., Barone C., et al. Prognostic role of KRAS and BRAF in stage II and III resected colon cancer: Results of the translational study on the PETACC-3, EORTC 40993, SAKK 60-00 trial. J. Clin. Oncol. 2010;28:466–474. doi: 10.1200/JCO.2009.23.3452. [DOI] [PubMed] [Google Scholar]
- 72.Ribic C.M., Sargent D.J., Moore M.J., Thibodeau S.N., French A.J., Goldberg R.M., Hamilton S.R., Laurent-Puig P., Gryfe R., Shepherd L.E., et al. Tumor microsatellite-instability status as a predictor of benefit from fluorouracil-based adjuvant chemotherapy for colon cancer. N. Engl. J. Med. 2003;349:247–257. doi: 10.1056/NEJMoa022289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sargent D.J., Marsoni S., Monges G., Thibodeau S.N., Labianca R., Hamilton S.R., French A.J., Kabat B., Foster N.R., Torri V., et al. Defective mismatch repair as a predictive marker for lack of efficacy of fluorouracil-based adjuvant therapy in colon cancer. J. Clin. Oncol. 2010;28:3219–3226. doi: 10.1200/JCO.2009.27.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kim J.E., Hong Y.S., Kim H.J., Kim K.-P., Lee J.L., Park S.J., Lim S.B., Park I.J., Kim C.W., Yoon Y.S., et al. Defective mismatch repair status was not associated with DFS and OS in stage II colon cancer treated with adjuvant chemotherapy. Ann. Surg. Oncol. 2015;22:630–637. doi: 10.1245/s10434-015-4807-6. [DOI] [PubMed] [Google Scholar]
- 75.Hutchins G., Southward K., Handley K., Magill L., Beaumont C., Stahlschmidt J., Richman S., Chambers P., Seymour M., Kerr D., et al. Value of mismatch repair, KRAS, and BRAF mutations in predicting recurrence and benefits from chemotherapy in colorectal cancer. J. Clin. Oncol. 2011;29:1261–1270. doi: 10.1200/JCO.2010.30.1366. [DOI] [PubMed] [Google Scholar]
- 76.Hou J.T., Zhao L.N., Zhang D.J., Lv D.Y., He W.L., Chen B., Li H.B., Li P.R., Chen L.Z., Chen X.L. Prognostic value of mismatch repair genes for patients with colorectal cancer: Meta-analysis. Technol. Cancer Res. Treat. 2018;17 doi: 10.1177/1533033818808507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jin Z., Sinicrope F.A. Prognostic and predictive values of mismatch repair deficiency in non-metastatic colorectal cancer. Cancers. 2021;13:300. doi: 10.3390/cancers13020300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Weisenberger D.J., Siegmund K.D., Campan M., Young J., Long T.I., Faasse M.A., Kang G.H., Widschwendter M., Weener D., Buchanan D., et al. CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat. Genet. 2006;38:787–793. doi: 10.1038/ng1834. [DOI] [PubMed] [Google Scholar]
- 79.Pylayeva-Gupta Y., Grabocka E., Bar-Sagi D. RAS oncogenes: Weaving a tumorigenic web. Nat. Rev. Cancer. 2011;11:761–774. doi: 10.1038/nrc3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hills S.A., Diffley J.F.X. DNA replication and oncogene-induced replicative stress. Curr. Biol. 2014;24:R435–R444. doi: 10.1016/j.cub.2014.04.012. [DOI] [PubMed] [Google Scholar]
- 81.Primo L.M.F., Teixeira L.K. Dna replication stress: Oncogenes in the spotlight. Genet. Mol. Biol. 2020;43:1–14. doi: 10.1590/1678-4685-gmb-2019-0138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Arai H., Elliott A., Xiu J., Wang J., Battaglin F., Kawanishi N., Soni S., Zhang W., Millstein J., Sohal D., et al. The landscape of alterations in DNA damage response pathways in colorectal cancer. Clin. Cancer Res. 2021;27:3234–3242. doi: 10.1158/1078-0432.CCR-20-3635. [DOI] [PubMed] [Google Scholar]
- 83.Boland P.M., Yurgelun M.B., Boland C.R. Recent progress in Lynch syndrome and other familial colorectal cancer syndromes. CA Cancer J. Clin. 2018;68:217–231. doi: 10.3322/caac.21448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sinha S., Villarreal D., Shim E.Y., Lee S.E. Risky business: Microhomology-mediated end joining. Mutat. Res. Fundam. Mol. Mech. Mutagen. 2016;788:17–24. doi: 10.1016/j.mrfmmm.2015.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Leguisamo N.M., Gloria H.C., Kalil A.N., Martins T.V., Azambuja D.B., Meira L.B., Saffi J. Base excision repair imbalance in colorectal cancer has prognostic value and modulates response to chemotherapy. Oncotarget. 2017;8:54199–54214. doi: 10.18632/oncotarget.14909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Fujimori H., Hyodo M., Matsuno Y., Shimizu A., Minakawa Y., Atsumi Y., Nakatsu Y., Tsuzuki T., Murakami Y., Yoshioka K. Ichi mismatch repair dependence of replication stress-associated DSB recognition and repair. Heliyon. 2019;5:e03057. doi: 10.1016/j.heliyon.2019.e03057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Matsuno Y., Atsumi Y., Shimizu A., Katayama K., Fujimori H., Hyodo M., Minakawa Y., Nakatsu Y., Kaneko S., Hamamoto R., et al. Replication stress triggers microsatellite destabilization and hypermutation leading to clonal expansion in vitro. Nat. Commun. 2019;10:3925. doi: 10.1038/s41467-019-11760-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Oh J.-M., Kang Y., Park J., Sung Y., Kim D., Seo Y., Lee E.A., Ra J.S., Amarsanaa E., Park Y.-U., et al. MSH2-MSH3 Promotes DNA end Resection during HR and Blocks TMEJ through Interaction with SMARCAD1 and EXO1. [(accessed on 11 June 2021)]; doi: 10.1093/nar/gkad308. Available online: https://www.biorxiv.org/content/10.1101/2021.04.23.441074v1. [DOI] [PMC free article] [PubMed]
- 89.Yoshioka K.-I., Matsuno Y. Genomic destabilization and its associated mutagenesis increase with senescence-associated phenotype expression. Cancer Sci. 2021;112:515–522. doi: 10.1111/cas.14746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chan Y.W., Fugger K., West S.C. Unresolved recombination intermediates lead to ultra-fine anaphase bridges, chromosome breaks and aberrations. Nat. Cell Biol. 2018;20:92–103. doi: 10.1038/s41556-017-0011-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yoshioka K.I., Matsuno Y., Hyodo M., Fujimori H. Genomic-destabilization-associated mutagenesis and clonal evolution of cells with mutations in tumor-suppressor genes. Cancers. 2019;11:1643. doi: 10.3390/cancers11111643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Thomas J.S., Shi C. Diagnostic Molecular Pathology. Elsevier; Cambridge, MA, USA: 2017. Molecular Testing in Colorectal Cancer; pp. 305–320. [Google Scholar]
- 93.Vilar E., Gruber S.B. Microsatellite instability in colorectal cancerthe stable evidence. Nat. Rev. Clin. Oncol. 2010;7:153–162. doi: 10.1038/nrclinonc.2009.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chubb D., Broderick P., Dobbins S.E., Frampton M., Kinnersley B., Penegar S., Price A., Ma Y.P., Sherborne A.L., Palles C., et al. Rare disruptive mutations and their contribution to the heritable risk of colorectal cancer. Nat. Commun. 2016;7:11883. doi: 10.1038/ncomms11883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Vo A.T., Zhu F., Wu X., Yuan F., Gao Y., Gu L., Li G.M., Lee T.H., Her C. hMRE11 deficiency leads to microsatellite instability and defective DNA mismatch repair. EMBO Rep. 2005;6:438–444. doi: 10.1038/sj.embor.7400392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Pavelitz T., Renfro L., Foster N.R., Caracol A., Welsch P., Lao V.V., Grady W.B., Niedzwiecki D., Saltz L.B., Bertagnolli M.M., et al. MRE11-deficiency associated with improved long-term disease free survival and overall survival in a subset of stage III colon cancer patients in randomized CALGB 89803 trial. PLoS ONE. 2014;9:e108483. doi: 10.1371/journal.pone.0108483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ihara K., Yamaguchi S., Ueno N., Tani Y., Shida Y., Ogata H., Domeki Y., Okamoto K., Nakajima M., Sasaki K., et al. Expression of DNA double-strand break repair proteins predicts the response and prognosis of colorectal cancer patients undergoing oxaliplatin-based chemotherapy. Oncol. Rep. 2016;35:1349–1355. doi: 10.3892/or.2015.4488. [DOI] [PubMed] [Google Scholar]
- 98.Altan B., Yokobori T., Ide M., Bai T., Yanoma T., Kimura A., Kogure N., Suzuki M., Bao P., Mochiki E., et al. High expression of MRE11-RAD50-NBS1 is associated with poor prognosis and chemoresistance in gastric cancer. Int. Inst. Anticancer. Res. 2016;36:5237–5247. doi: 10.21873/anticanres.11094. [DOI] [PubMed] [Google Scholar]
- 99.Ho V., Chung L., Singh A., Lea V., Abubakar A., Lim S.H., Ng W., Lee M., De Souza P., Shin J.-S.S., et al. Overexpression of the MRE11-RAD50-NBS1 (MRN) complex in rectal cancer correlates with poor response to neoadjuvant radiotherapy and prognosis. BMC Cancer. 2018;18:869. doi: 10.1186/s12885-018-4776-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Gao J., Zhang H., Arbman G., Sun X.F. RAD50/MRE11/NBS1 proteins in relation to tumour development and prognosis in patients with microsatellite stable colorectal cancer. Histol. Histopathol. 2008;23:1495–1502. doi: 10.14670/HH-23.1495. [DOI] [PubMed] [Google Scholar]
- 101.Vilar E., Bartnik C.M., Stenzel S.L., Raskin L., Ahn J., Moreno V., Mukherjee B., Iniesta M.D., Morgan M.A., Rennert G., et al. MRE11 deficiency increases sensitivity to poly(ADP-ribose) polymerase inhibition in microsatellite unstable colorectal cancers. Cancer Res. 2011;71:2632–2642. doi: 10.1158/0008-5472.CAN-10-1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Fan C.W., Kopsida M., Liu Y.B., Zhang H., Gao J.F., Arbman G., Cao S.Y.W., Li Y., Zhou Z.G., Sun X.F. Prognostic heterogeneity of MRE11 based on the location of primary colorectal cancer is caused by activation of different immune signals. Front. Oncol. 2020;9:1465. doi: 10.3389/fonc.2019.01465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.McPherson L.A., Shen Y., Ford J.M. Poly (ADP-ribose) polymerase inhibitor LT-626: Sensitivity correlates with MRE11 mutations and synergizes with platinums and irinotecan in colorectal cancer cells. Cancer Lett. 2014;343:217–223. doi: 10.1016/j.canlet.2013.10.034. [DOI] [PubMed] [Google Scholar]
- 104.Kantidze O.L., Velichko A.K., Luzhin A.V., Petrova N.V., Razin S.V. Synthetically lethal interactions of ATM, ATR, and DNA-PKcs. Trends Cancer. 2018;4:755–768. doi: 10.1016/j.trecan.2018.09.007. [DOI] [PubMed] [Google Scholar]
- 105.Chanut P., Britton S., Coates J., Jackson S.P., Calsou P. Coordinated nuclease activities counteract Ku at single-ended DNA double-strand breaks. Nat. Commun. 2016;7:12889. doi: 10.1038/ncomms12889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Yang G., Liu C., Chen S.H., Kassab M.A., Hoff J.D., Walter N.G., Yu X. Super-resolution imaging identifies PARP1 and the Ku complex acting as DNA double-strand break sensors. Nucleic Acids Res. 2018;46:3446–3457. doi: 10.1093/nar/gky088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zhou H., Kawamura K., Yanagihara H., Kobayashi J., Zhang-Akiyama Q.-M. NBS1 is regulated by two kind of mechanisms: ATM-dependent complex formation with MRE11 and RAD50, and cell cycle–dependent degradation of protein. J. Radiat. Res. 2017;58:487–494. doi: 10.1093/jrr/rrx014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Falck J., Coates J., Jackson S.P. Conserved modes of recruitment of ATM, ATR and DNA-PKcs to sites of DNA damage. Nature. 2005;434:605–611. doi: 10.1038/nature03442. [DOI] [PubMed] [Google Scholar]
- 109.Buscemi G., Perego P., Carenini N., Nakanishi M., Chessa L., Chen J., Khanna K.K., Delia D. Activation of ATM and Chk2 kinases in relation to the amount of DNA strand breaks. Oncogene. 2004;23:7691–7700. doi: 10.1038/sj.onc.1207986. [DOI] [PubMed] [Google Scholar]
- 110.Matsuoka S., Rotman G., Ogawa A., Shiloh Y., Tamai K., Elledge S.J. Ataxia telangiectasia-mutated phosphorylates Chk2 in vivo and in vitro. Proc. Natl. Acad. Sci. USA. 2000;97:10389–10394. doi: 10.1073/pnas.190030497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Mak T.W. DNA damage-induced activation of p53 by the checkpoint kinase Chk2. Science. 2000;287:1824–1827. doi: 10.1126/science.287.5459.1824. [DOI] [PubMed] [Google Scholar]
- 112.Shiotani B., Zou L. Single-stranded DNA orchestrates an ATM-to-ATR switch at DNA breaks. Mol. Cell. 2009;33:547–558. doi: 10.1016/j.molcel.2009.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Lee J., Kumagai A., Dunphy W.G. Positive regulation of Wee1 by Chk1 and 14-3-3 proteins. Mol. Biol. Cell. 2001;12:551–563. doi: 10.1091/mbc.12.3.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Rothblum-Oviatt C.J., Ryan C.E., Piwnica-Worms H. 14-3-3 Binding regulates catalytic activity of human Wee1 kinase. Cell Growth Differ. 2001;12:581–589. [PubMed] [Google Scholar]
- 115.Shibata A., Barton O., Noon A.T., Dahm K., Deckbar D., Goodarzi A.A., Löbrich M., Jeggo P.A. Role of ATM and the damage response mediator proteins 53BP1 and MDC1 in the maintenance of G2/M checkpoint arrest. Mol. Cell. Biol. 2010;30:3371–3383. doi: 10.1128/MCB.01644-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Jette N., Lees-Miller S.P. The DNA-dependent protein kinase: A multifunctional protein kinase with roles in DNA double strand break repair and mitosis. Prog. Biophys. Mol. Biol. 2015;117:194–205. doi: 10.1016/j.pbiomolbio.2014.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Katsuki Y., Jeggo P.A., Uchihara Y., Takata M., Shibata A. DNA double-strand break end resection: A critical relay point for determining the pathway of repair and signaling. Genome Instab. Dis. 2020;1:155–171. doi: 10.1007/s42764-020-00017-8. [DOI] [Google Scholar]
- 118.Teixeira-Silva A., Ait Saada A., Hardy J., Iraqui I., Nocente M.C., Fréon K., Lambert S.A.E. The end-joining factor Ku acts in the end-resection of double strand break-free arrested replication forks. Nat. Commun. 2017;8:1982. doi: 10.1038/s41467-017-02144-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Deshpande R.A., Myler L.R., Soniat M.M., Makharashvili N., Lee L., Lees-Miller S.P., Finkelstein I.J., Paull T.T. DNA-dependent protein kinase promotes DNA end processing by MRN and CtIP. Sci. Adv. 2020;6:eaay0922. doi: 10.1126/sciadv.aay0922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Shibata A., Jeggo P.A. Roles for the DNA-PK complex and 53BP1 in protecting ends from resection during DNA double-strand break repair. J. Radiat. Res. 2020;61:718–726. doi: 10.1093/jrr/rraa053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ceccaldi R., Rondinelli B., D’Andrea A.D. Repair pathway choices and consequences at the double-strand break. Trends Cell Biol. 2016;26:52–64. doi: 10.1016/j.tcb.2015.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Iliakis G., Murmann T., Soni A. Alternative end-joining repair pathways are the ultimate backup for abrogated classical non-homologous end-joining and homologous recombination repair: Implications for the formation of chromosome translocations. Mutat. Res.-Genet. Toxicol. Environ. Mutagen. 2015;793:166–175. doi: 10.1016/j.mrgentox.2015.07.001. [DOI] [PubMed] [Google Scholar]
- 123.Feng L., Li N., Li Y., Wang J., Gao M., Wang W., Chen J. Cell cycle-dependent inhibition of 53BP1 signaling by BRCA1. Cell Discov. 2015;1:15019. doi: 10.1038/celldisc.2015.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Gupta R., Somyajit K., Narita T., Maskey E., Stanlie A., Kremer M., Typas D., Lammers M., Mailand N., Nussenzweig A., et al. DNA repair network analysis reveals shieldin as a key regulator of NHEJ and PARP inhibitor sensitivity. Cell. 2018;173:972–988.:e23. doi: 10.1016/j.cell.2018.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Escribano-Díaz C., Orthwein A., Fradet-Turcotte A., Xing M., Young J.T.F., Tkáč J., Cook M.A., Rosebrock A.P., Munro M., Canny M.D., et al. A cell cycle-dependent regulatory circuit composed of 53BP1-RIF1 and BRCA1-CtIP controls DNA repair pathway choice. Mol. Cell. 2013;49:872–883. doi: 10.1016/j.molcel.2013.01.001. [DOI] [PubMed] [Google Scholar]
- 126.Hoa N.N., Kobayashi J., Omura M., Hirakawa M., Yang S.H., Komatsu K., Paull T.T., Takeda S., Sasanuma H. BRCA1 and CtIP are both required to recruit Dna2 at double-strand breaks in homologous recombination. PLoS ONE. 2015;10:e0124495. doi: 10.1371/journal.pone.0124495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Tomimatsu N., Mukherjee B., Harris J.L., Boffo F.L., Hardebeck M.C., Potts P.R., Khanna K.K., Burma S. DNA-damage-induced degradation of EXO1 exonuclease limits DNA end resection to ensure accurate DNA repair. J. Biol. Chem. 2017;292:10779–10790. doi: 10.1074/jbc.M116.772475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Nimonkar A.V., Genschel J., Kinoshita E., Polaczek P., Campbell J.L., Wyman C., Modrich P., Kowalczykowski S.C. BLM-DNA2-RPA-MRN and EXO1-BLM-RPA-MRN constitute two DNA end resection machineries for human DNA break repair. Genes Dev. 2011;25:350–362. doi: 10.1101/gad.2003811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Shibata A., Moiani D., Arvai A.S., Perry J., Harding S.M., Genois M.M., Maity R., van Rossum-Fikkert S., Kertokalio A., Romoli F., et al. DNA double-strand break repair pathway choice is directed by distinct MRE11 nuclease activities. Mol. Cell. 2014;53:7–18. doi: 10.1016/j.molcel.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Moiani D., Ronato D.A., Brosey C.A., Arvai A.S., Syed A., Masson J.-Y., Petricci E., Tainer J.A. Targeting allostery with avatars to design inhibitors assessed by cell activity: Dissecting mre11 endo- and exonuclease activities. Methods Enzymol. 2018;601:205–241. doi: 10.1016/bs.mie.2017.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Zhu M., Zhao H., Limbo O., Russell P. Mre11 complex links sister chromatids to promote repair of a collapsed replication fork. Proc. Natl. Acad. Sci. USA. 2018;115:8793–8798. doi: 10.1073/pnas.1808189115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Macheret M., Halazonetis T.D. DNA replication stress as a hallmark of cancer. Annu. Rev. Pathol. Mech. Dis. 2015;10:425–448. doi: 10.1146/annurev-pathol-012414-040424. [DOI] [PubMed] [Google Scholar]
- 133.Zeman M.K., Cimprich K.A. Causes and consequences of replication stress. Nat. Cell Biol. 2014;16:2–9. doi: 10.1038/ncb2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Negrini S., Gorgoulis V.G., Halazonetis T.D. Genomic instability an evolving hallmark of cancer. Nat. Rev. Mol. Cell Biol. 2010;11:220–228. doi: 10.1038/nrm2858. [DOI] [PubMed] [Google Scholar]
- 135.Bartkova J., Hořejší Z., Koed K., Krämer A., Tort F., Zleger K., Guldberg P., Sehested M., Nesland J.M., Lukas C., et al. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature. 2005;434:864–870. doi: 10.1038/nature03482. [DOI] [PubMed] [Google Scholar]
- 136.Gorgoulis V.G., Vassiliou L.V.F., Karakaidos P., Zacharatos P., Kotsinas A., Liloglou T., Venere M., DiTullio R.A., Kastrinakis N.G., Levy B., et al. Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature. 2005;434:907–913. doi: 10.1038/nature03485. [DOI] [PubMed] [Google Scholar]
- 137.Bertolin A.P., Hoffmann J.S., Gottifredi V. Under-replicated DNA: The byproduct of large genomes? Cancers. 2020;12:2764. doi: 10.3390/cancers12102764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Franchet C., Hoffmann J.S. When RAD52 allows mitosis to accept unscheduled DNA synthesis. Cancers. 2020;12:26. doi: 10.3390/cancers12010026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Sansregret L., Patterson J.O., Dewhurst S., López-García C., Koch A., McGranahan N., Chao W.C.H., Barry D.J., Rowan A., Instrell R., et al. APC/C dysfunction limits excessive cancer chromosomal instability. Cancer Discov. 2017;7:218–233. doi: 10.1158/2159-8290.CD-16-0645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Saldivar J.C., Cortez D., Cimprich K.A. The essential kinase ATR: Ensuring faithful duplication of a challenging genome. Nat. Rev. Mol. Cell Biol. 2017;18:622–636. doi: 10.1038/nrm.2017.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.David L., Fernandez-Vidal A., Bertoli S., Grgurevic S., Lepage B., Deshaies D., Prade N., Carte M., Larrue C., Sarry J.E., et al. CHK1 as a therapeutic target to bypass chemoresistance in AML. Sci. Signal. 2016;9:ra90. doi: 10.1126/scisignal.aac9704. [DOI] [PubMed] [Google Scholar]
- 142.Bhowmick R., Minocherhomji S., Hickson I.D. RAD52 facilitates mitotic DNA synthesis following replication stress. Mol. Cell. 2016;64:1117–1126. doi: 10.1016/j.molcel.2016.10.037. [DOI] [PubMed] [Google Scholar]
- 143.Hickson I.D., Bhowmick R. The “enemies within”: Regions of the genome that are inherently difficult to replicate. F1000Research. 2017;6:666. doi: 10.12688/f1000research.11024.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Massey A.J. Inhibition of ATR-dependent feedback activation of Chk1 sensitises cancer cells to Chk1 inhibitor monotherapy. Cancer Lett. 2016;383:41–52. doi: 10.1016/j.canlet.2016.09.024. [DOI] [PubMed] [Google Scholar]
- 145.Manic G., Signore M., Sistigu A., Russo G., Corradi F., Siteni S., Musella M., Vitale S., De Angelis M.L., Pallocca M., et al. CHK1-targeted therapy to deplete DNA replication-stressed, p53-deficient, hyperdiploid colorectal cancer stem cells. Gut. 2018;67:903–917. doi: 10.1136/gutjnl-2016-312623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Webster P.J., Littlejohns A.T., Gaunt H.J., Young R.S., Rode B., Ritchie J.E., Stead L.F., Harrison S., Droop A., Martin H.L., et al. Upregulated WEE1 protects endothelial cells of colorectal cancer liver metastases. Oncotarget. 2017;8:42288–42299. doi: 10.18632/oncotarget.15039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Webster P.J., Littlejohns A.T., Gaunt H.J., Prasad K.R., Beech D.J., Burke D.A. AZD1775 induces toxicity through double-stranded DNA breaks independently of chemotherapeutic agents in p53-mutated colorectal cancer cells. Cell Cycle. 2017;16:2176–2182. doi: 10.1080/15384101.2017.1301329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Agostini L.C., Jain A., Shupp A., Nevler A., McCarthy G., Bussard K.M., Yeo C.J., Brody J.R. Combined targeting of PARG and Wee1 causes decreased cell survival and DNA damage in an S-phase dependent manner. Mol. Cancer Res. 2021;19:207–214. doi: 10.1158/1541-7786.MCR-20-0708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Yin Y., Shen Q., Tao R., Chang W., Li R., Xie G., Liu W., Zhang P., Tao K. Wee1 inhibition can suppress tumor proliferation and sensitize p53 mutant colonic cancer cells to the anticancer effect of irinotecan. Mol. Med. Rep. 2018;17:3344–3349. doi: 10.3892/mmr.2017.8230. [DOI] [PubMed] [Google Scholar]
- 150.Kalimutho M., Bain A.L., Mukherjee B., Nag P., Nanayakkara D.M., Harten S.K., Harris J.L., Subramanian G.N., Sinha D., Shirasawa S., et al. Enhanced dependency of KRAS-mutant colorectal cancer cells on RAD51-dependent homologous recombination repair identified from genetic interactions in Saccharomyces cerevisiae. Mol. Oncol. 2017;11:470–490. doi: 10.1002/1878-0261.12040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Iwata T., Uchino T., Koyama A., Johmura Y., Koyama K., Saito T., Ishiguro S., Arikawa T., Komatsu S., Miyachi M., et al. The G2 checkpoint inhibitor CBP-93872 increases the sensitivity of colorectal and pancreatic cancer cells to chemotherapy. PLoS ONE. 2017;12:e0178221. doi: 10.1371/journal.pone.0178221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Wang C., Jette N., Moussienko D., Bebb D.G., Lees-Miller S.P. ATM-deficient colorectal cancer cells are sensitive to the PARP inhibitor olaparib. Transl. Oncol. 2017;10:190–196. doi: 10.1016/j.tranon.2017.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Greene J., Nguyen A., Bagby S.M., Jones G.N., Tai W.M., Quackenbush K.S., Schreiber A., Messersmith W.A., Devaraj K.M., Blatchford P., et al. The novel ATM inhibitor (AZ31) enhances antitumor activity in patient derived xenografts that are resistant to irinotecan monotherapy. Oncotarget. 2017;8:110904–110913. doi: 10.18632/oncotarget.22920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Herrero A.B., Gutiérrez N.C. Targeting ongoing DNA damage in multiple myeloma: Effects of DNA damage response inhibitors on plasma cell survival. Front. Oncol. 2017;7:98. doi: 10.3389/fonc.2017.00098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Petroni M., Sardina F., Infante P., Bartolazzi A., Locatelli E., Fabretti F., Di Giulio S., Capalbo C., Cardinali B., Coppa A., et al. MRE11 inhibition highlights a replication stress-dependent vulnerability of MYCN-driven tumors. Cell Death Dis. 2018;9:895. doi: 10.1038/s41419-018-0924-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Manic G., Musella M., Corradi F., Sistigu A., Vitale S., Soliman Abdel Rehim S., Mattiello L., Malacaria E., Galassi C., Signore M., et al. Control of replication stress and mitosis in colorectal cancer stem cells through the interplay of PARP1, MRE11 and RAD51. Cell Death Differ. 2021:1–23. doi: 10.1038/s41418-020-00733-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Sullivan-Reed K., Bolton-Gillespie E., Dasgupta Y., Langer S., Siciliano M., Nieborowska-Skorska M., Hanamshet K., Belyaeva E.A., Bernhardy A.J., Lee J., et al. Simultaneous targeting of PARP1 and RAD52 triggers dual synthetic lethality in BRCA-deficient tumor cells. Cell Rep. 2018;23:3127–3136. doi: 10.1016/j.celrep.2018.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Zhou J., Gelot C., Pantelidou C., Li A., Yücel H., Davis R., Farkkila A., Kochupurakkal B., Syed A., Shapiro G., et al. Polymerase Theta Inhibition Kills Homologous Recombination Deficient Tumors. [(accessed on 22 December 2020)]; Available online: https://www.biorxiv.org/content/10.1101/2020.05.23.111658v1.full.
- 159.Shibata A., Jeggo P. A historical reflection on our understanding of radiation-induced DNA double strand break repair in somatic mammalian cells; interfacing the past with the present. Int. J. Radiat. Biol. 2019;3002:1–12. doi: 10.1080/09553002.2018.1564083. [DOI] [PubMed] [Google Scholar]
- 160.Jeggo P.A., Löbrichf M. How cancer cells hijack DNA double-strand break repair pathways to gain genomic instability. Biochem. J. 2015;471:1–11. doi: 10.1042/BJ20150582. [DOI] [PubMed] [Google Scholar]
- 161.Her J., Bunting S.F. How cells ensure correct repair of DNA double-strand breaks. J. Biol. Chem. 2018;293:10502–10511. doi: 10.1074/jbc.TM118.000371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Ma C.J., Gibb B., Kwon Y., Sung P., Greene E.C. Protein dynamics of human RPA and RAD51 on ssDNA during assembly and disassembly of the RAD51 filament. Nucleic Acids Res. 2016;45:749–761. doi: 10.1093/nar/gkw1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Andriuskevicius T., Kotenko O., Makovets S. Putting together and taking apart: Assembly and disassembly of the Rad51 nucleoprotein filament in DNA repair and genome stability. Cell Stress. 2018;2:96–112. doi: 10.15698/cst2018.05.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Giannini G., Ristori E., Cerignoli F., Rinaldi C., Zani M., Viel A., Ottini L., Crescenzi M., Martinotti S., Bignami M., et al. Human MRE11 is inactivated in mismatch repair-deficient cancers. EMBO Rep. 2002;3:248–254. doi: 10.1093/embo-reports/kvf044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Wen Q., Scorah J., Phear G., Rodgers G., Rodgers S., Meuth M. A mutant allele of MRE11 found in mismatch repair-deficient tumor cells suppresses the cellular response to DNA replication fork stress in a dominant negative manner. Mol. Biol. Cell. 2008;19:1693–1705. doi: 10.1091/mbc.e07-09-0975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Dupré A., Boyer-Chatenet L., Sattler R.M., Modi A.P., Lee J.-H., Nicolette M.L., Kopelovich L., Jasin M., Baer R., Paull T.T., et al. A forward chemical genetic screen reveals an inhibitor of the Mre11–Rad50–Nbs1 complex. Nat. Chem. Biol. 2008;4:119–125. doi: 10.1038/nchembio.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Petroni M., Sardina F., Heil C., Sahún-Roncero M., Colicchia V., Veschi V., Albini S., Fruci D., Ricci B., Soriani A., et al. The MRN complex is transcriptionally regulated by MYCN during neural cell proliferation to control replication stress. Cell Death Differ. 2016;23:197–206. doi: 10.1038/cdd.2015.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Gu L., Chu P., Lingeman R., McDaniel H., Kechichian S., Hickey R.J., Liu Z., Yuan Y.C., Sandoval J.A., Fields G.B., et al. The Mechanism by Which MYCN amplification confers an enhanced sensitivity to a PCNA-derived cell permeable peptide in neuroblastoma cells. EBioMedicine. 2015;2:1923–1931. doi: 10.1016/j.ebiom.2015.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Kushner B.H., Modak S., Kramer K., Laquaglia M.P., Yataghene K., Basu E.M., Roberts S.S., Cheung N.K.V. Striking dichotomy in outcome of MYCN-amplified neuroblastoma in the contemporary era. Cancer. 2014;120:2050–2059. doi: 10.1002/cncr.28687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Mateos-Gomez P.A., Gong F., Nair N., Miller K.M., Lazzerini-Denchi E., Sfeir A. Mammalian polymerase θ promotes alternative NHEJ and suppresses recombination. Nature. 2015;518:254–257. doi: 10.1038/nature14157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Kent T., Chandramouly G., Mcdevitt S.M., Ozdemir A.Y., Pomerantz R.T. Mechanism of microhomology-mediated end-joining promoted by human DNA polymerase θ. Nat. Struct. Mol. Biol. 2015;22:230–237. doi: 10.1038/nsmb.2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Truong L.N., Li Y., Shi L.Z., Hwang P.Y.H., He J., Wang H., Razavian N., Berns M.W., Wu X. Microhomology-mediated end joining and homologous recombination share the initial end resection step to repair DNA double-strand breaks in mammalian cells. Proc. Natl. Acad. Sci. USA. 2013;110:7720–7725. doi: 10.1073/pnas.1213431110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Rahal E.A., Henricksen L.A., Li Y., Williams R.S., Tainer J.A., Dixon K. ATM regulates Mre11-dependent DNA end-degradation and microhomology-mediated end joining. Cell Cycle. 2010;9:2866. doi: 10.4161/cc.9.14.12363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Rass E., Grabarz A., Plo I., Gautier J., Bertrand P., Lopez B.S. Role of Mre11 in chromosomal nonhomologous end joining in mammalian cells. Nat. Struct. Mol. Biol. 2009;16:819–824. doi: 10.1038/nsmb.1641. [DOI] [PubMed] [Google Scholar]
- 175.Golshani G., Zhang Y. Advances in immunotherapy for colorectal cancer: A review. Therap. Adv. Gastroenterol. 2020;13:1–11. doi: 10.1177/1756284820917527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Tran L., Allen C.T., Xiao R., Moore E., Davis R., Park S.J., Spielbauer K., Van Waes C., Schmitt N.C. Cisplatin alters antitumor immunity and synergizes with PD-1/PD-L1 inhibition in head and neck squamous cell carcinoma. Cancer Immunol. Res. 2017;5:1141–1151. doi: 10.1158/2326-6066.CIR-17-0235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Li D., Ye L., Lei Y., Wan J., Chen H. Downregulation of FoxM1 sensitizes nasopharyngeal carcinoma cells to cisplatin via inhibition of MRN-ATM-mediated DNA repair. BMB Rep. 2019;52:208–213. doi: 10.5483/BMBRep.2019.52.3.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Shen B., Huang D., Ramsey A.J., Ig-izevbekhai K., Zhang K., Lajud S.A., O’Malley B.W., Li D. PD-L1 and MRN synergy in platinum-based chemoresistance of head and neck squamous cell carcinoma. Br. J. Cancer. 2019;122:1–8. doi: 10.1038/s41416-019-0697-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Sato H., Niimi A., Yasuhara T., Permata T.B.M., Hagiwara Y., Isono M., Nuryadi E., Sekine R., Oike T., Kakoti S., et al. DNA double-strand break repair pathway regulates PD-L1 expression in cancer cells. Nat. Commun. 2017;8 doi: 10.1038/s41467-017-01883-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Teo M.Y., Seier K., Ostrovnaya I., Regazzi A.M., Kania B.E., Moran M.M., Cipolla C.K., Bluth M.J., Chaim J., Al-Ahmadie H., et al. Alterations in DNA damage response and repair genes as potential marker of clinical benefit from PD-1/PD-L1 blockade in advanced urothelial cancers. J. Clin. Oncol. 2018;36:1685–1694. doi: 10.1200/JCO.2017.75.7740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Shim J.H., Kim H.S., Cha H., Kim S., Kim T.M., Anagnostou V., Choi Y.L., Jung H.A., Sun J.M., Ahn J.S., et al. HLA-corrected tumor mutation burden and homologous recombination deficiency for the prediction of response to PD-(L)1 blockade in advanced non-small-cell lung cancer patients. Ann. Oncol. 2020;31:902–911. doi: 10.1016/j.annonc.2020.04.004. [DOI] [PubMed] [Google Scholar]
- 182.Sato H., Jeggo P.A., Shibata A. Regulation of programmed death-ligand 1 expression in response to DNA damage in cancer cells: Implications for precision medicine. Cancer Sci. 2019;110:3415–3423. doi: 10.1111/cas.14197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Chae Y.K., Anker J.F., Bais P., Namburi S., Giles F.J., Chuang J.H. Mutations in DNA repair genes are associated with increased neo-antigen load and activated T cell infiltration in lung adenocarcinoma. Oncotarget. 2018;9:7949–7960. doi: 10.18632/oncotarget.23742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Lee J.-H., Mand M.R., Deshpande R.A., Kinoshita E., Yang S.-H., Wyman C., Paull T.T. Ataxia telangiectasia-mutated (ATM) kinase activity is regulated by atp-driven conformational changes in the Mre11/Rad50/Nbs1 (MRN) complex. J. Biol. Chem. 2013;288:12840–12851. doi: 10.1074/jbc.M113.460378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Sato K., Shimomuki M., Katsuki Y., Takahashi D., Kobayashi W., Ishiai M., Miyoshi H., Takata M., Kurumizaka H. FANCI-FANCD2 stabilizes the RAD51-DNA complex by binding RAD51 and protects the 5′-DNA end. Nucleic Acids Res. 2016;44:10758–10771. doi: 10.1093/nar/gkw876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Mason J.M., Chan Y.L., Weichselbaum R.W., Bishop D.K. Non-enzymatic roles of human RAD51 at stalled replication forks. Nat. Commun. 2019;10 doi: 10.1038/s41467-019-12297-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Somyajit K., Saxena S., Babu S., Mishra A., Nagaraju G. Mammalian RAD51 paralogs protect nascent DNA at stalled forks and mediate replication restart. Nucleic Acids Res. 2015;43:9835–9855. doi: 10.1093/nar/gkv880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Bugreev D.V., Rossi M.J., Mazin A. V Cooperation of RAD51 and RAD54 in regression of a model replication fork. Nucleic Acids Res. 2011;39:2153–2164. doi: 10.1093/nar/gkq1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Bhat K.P., Cortez D. RPA and RAD51: Fork reversal, fork protection, and genome stability. Nat. Struct. Mol. Biol. 2018;25:446–453. doi: 10.1038/s41594-018-0075-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Slade D. Maneuvers on PCNA rings during DNA replication and repair. Genes. 2018;9:416. doi: 10.3390/genes9080416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Leung W., Baxley R.M., Moldovan G.L., Bielinsky A.K. Mechanisms of DNA damage tolerance: Post-translational regulation of PCNA. Genes. 2019;10:10. doi: 10.3390/genes10010010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Budke B., Tueckmantel W., Miles K., Kozikowski A.P., Connell P.P. Optimization of drug candidates that inhibit the D-loop activity of RAD51 HHS public access. ChemMedChem. 2019;14:1031–1040. doi: 10.1002/cmdc.201900075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Lee J.-H., Bae A.-N., Jung S.-J. Clinicopathological and prognostic characteristics of RAD51 in colorectal cancer. Medicina. 2020;56:48. doi: 10.3390/medicina56020048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Watson R., Liu T.C., Ruzinova M.B. High frequency of KRAS mutation in early onset colorectal adenocarcinoma: Implications for pathogenesis. Hum. Pathol. 2016;56:163–170. doi: 10.1016/j.humpath.2016.06.010. [DOI] [PubMed] [Google Scholar]
- 195.Sallmyr A., Tomkinson A.E. Repair of DNA double-strand breaks by mammalian alternative end-joining pathways. J. Biol. Chem. 2018;293:10536–10549. doi: 10.1074/jbc.TM117.000375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Hussain S.S., Majumdar R., Moore G.M., Narang H., Buechelmaier E.S., Bazil M.J., Ravindran P.T., Leeman J.E., Li Y., Jalan M., et al. Measuring nonhomologous end-joining, homologous recombination and alternative end-joining simultaneously at an endogenous locus in any transfectable human cell. Nucleic Acids Res. 2021 doi: 10.1093/nar/gkab262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Hanscom T., McVey M. Regulation of error-prone DNA double-strand break repair and its impact on genome evolution. Cells. 2020;9:1657. doi: 10.3390/cells9071657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Xiong X., Du Z., Wang Y., Feng Z., Fan P., Yan C., Willers H., Zhang J. 53BP1 promotes microhomology-mediated end-joining in G1-phase cells. Nucleic Acids Res. 2015;43:1659–1670. doi: 10.1093/nar/gku1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Bakr A., Köcher S., Volquardsen J., Petersen C., Borgmann K., Dikomey E., Rothkamm K., Mansour W.Y. Impaired 53BP1/RIF1 DSB mediated end-protection stimulates CtIP-dependent end resection and switches the repair to PARP1-dependent end joining in G1. Oncotarget. 2016;7:57679–57693. doi: 10.18632/oncotarget.11023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Chang H.H.Y., Pannunzio N.R., Adachi N., Lieber M.R. Non-homologous DNA end joining and alternative pathways to double-strand break repair. Nat. Rev. Mol. Cell Biol. 2017;18:495–506. doi: 10.1038/nrm.2017.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Hwang T., Reh S., Dunbayev Y., Zhong Y., Takata Y., Shen J., McBride K.M., Murnane J.P., Bhak J., Lee S., et al. Defining the mutation signatures of DNA polymerase θ in cancer genomes. NAR Cancer. 2020;2:zcaa017. doi: 10.1093/narcan/zcaa017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Maiorano D., Etri J.E., Franchet C., Hoffmann J.S. Translesion synthesis or repair by specialized dna polymerases limits excessive genomic instability upon replication stress. Int. J. Mol. Sci. 2021;22:3924. doi: 10.3390/ijms22083924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Seol J.-H., Shim E.Y., Lee S.E. Microhomology-mediated end joining: Good, bad and ugly. Mutat. Res. Mol. Mech. Mutagen. 2018;809:81–87. doi: 10.1016/j.mrfmmm.2017.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Zahn K.E., Jensen R.B., Wood R.D., Doublié S. Human DNA polymerase θ harbors DNA end-trimming activity critical for DNA repair. Mol. Cell. 2021;81 doi: 10.1016/j.molcel.2021.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 205.Mateos-Gomez P.A., Kent T., Deng S.K., Mcdevitt S., Kashkina E., Hoang T.M., Pomerantz R.T., Sfeir A. The helicase domain of Polθ counteracts RPA to promote alt-NHEJ. Nat. Struct. Mol. Biol. 2017;24:1116–1123. doi: 10.1038/nsmb.3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Lemée F., Bergoglio V., Fernandez-Vidal A., Machado-Silva A., Pillaire M.J., Bieth A., Gentil C., Baker L., Martin A.L., Leduc C., et al. DNA polymerase θ up-regulation is associated with poor survival in breast cancer, perturbs DNA replication, and promotes genetic instability. Proc. Natl. Acad. Sci. USA. 2010;107:13390–13395. doi: 10.1073/pnas.0910759107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Ceccaldi R., Liu J.C., Amunugama R., Hajdu I., Primack B., Petalcorin M.I.R., O’Connor K.W., Konstantinopoulos P.A., Elledge S.J., Boulton S.J., et al. Homologous-recombination-deficient tumours are dependent on Polθ -mediated repair. Nature. 2015;518:258–262. doi: 10.1038/nature14184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Patterson-Fortin J., D’Andrea A.D. Exploiting the microhomology-mediated end-joining pathway in cancer therapy. Cancer Res. 2020;80:4593–4600. doi: 10.1158/0008-5472.CAN-20-1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Ahrabi S., Sarkar S., Pfister S.X., Pirovano G., Higgins G.S., Porter A.C.G., Humphrey T.C. A role for human homologous recombination factors in suppressing microhomology-mediated end joining. Nucleic Acids Res. 2016;44:5743–5757. doi: 10.1093/nar/gkw326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Schlacher K., Christ N., Siaud N., Egashira A., Wu H., Jasin M. Double-strand break repair-independent role for BRCA2 in blocking stalled replication fork degradation by MRE11. Cell. 2011;145:529–542. doi: 10.1016/j.cell.2011.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Carvajal-Garcia J., Cho J.E., Carvajal-Garcia P., Feng W., Wood R.D., Sekelsky J., Gupta G.P., Roberts S.A., Ramsden D.A. Mechanistic basis for microhomology identification and genome scarring by polymerase theta. Proc. Natl. Acad. Sci. USA. 2020;117:8476–8485. doi: 10.1073/pnas.1921791117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Goullet De Rugy T., Bashkurov M., Datti A., Betous R., Guitton-Sert L., Cazaux C., Durocher D., Hoffmann J.S. Excess Polθ functions in response to replicative stress in homologous recombination-proficient cancer cells. Biol. Open. 2016;5:1485–1492. doi: 10.1242/bio.018028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Feng W., Simpson D.A., Carvajal-Garcia J., Price B.A., Kumar R.J., Mose L.E., Wood R.D., Rashid N., Purvis J.E., Parker J.S., et al. Genetic determinants of cellular addiction to DNA polymerase theta. Nat. Commun. 2019;10:4286. doi: 10.1038/s41467-019-12234-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Wang Z., Song Y., Li S., Kurian S., Xiang R., Chiba T., Wu X. DNA polymerase (POLQ) is important for repair of DNA double-strand breaks caused by fork collapse. J. Biol. Chem. 2019;294:3909–3919. doi: 10.1074/jbc.RA118.005188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Yousefzadeh M.J., Wyatt D.W., Takata K.-i., Mu Y., Hensley S.C., Tomida J., Bylund G.O., Doublié S., Johansson E., Ramsden D.A., et al. Mechanism of suppression of chromosomal instability by DNA polymerase POLQ. PLoS Genet. 2014;10:e1004654. doi: 10.1371/journal.pgen.1004654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Newman J.A., Cooper C.D.O., Aitkenhead H., Gileadi O. Structure of the helicase domain of DNA polymerase theta reveals a possible role in the microhomology-mediated end-joining pathway. Structure. 2015;23:2319–2330. doi: 10.1016/j.str.2015.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Wyatt D.W., Feng W., Conlin M.P., Yousefzadeh M.J., Roberts S.A., Mieczkowski P., Wood R.D., Gupta G.P., Ramsden D.A. Essential roles for polymerase θ-mediated end joining in the repair of chromosome breaks. Mol. Cell. 2016;63:662–673. doi: 10.1016/j.molcel.2016.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Kawamura K., Bahar R., Seimiya M., Chiyo M., Wada A., Okada S., Hatano M., Tokuhisa T., Kimura H., Watanabe S., et al. DNA polymerase θ is preferentially expressed in lymphoid tissues and upregulated in human cancers. Int. J. Cancer. 2004;109:9–16. doi: 10.1002/ijc.11666. [DOI] [PubMed] [Google Scholar]
- 219.Pillaire M.J., Selves J., Gordien K., Gouraud P.A., Gentil C., Danjoux M., Do C., Negre V., Bieth A., Guimbaud R., et al. A DNA replication signature of progression and negative outcome in colorectal cancer. Oncogene. 2010;29:876–887. doi: 10.1038/onc.2009.378. [DOI] [PubMed] [Google Scholar]
- 220.Stephens P.J., McBride D.J., Lin M.L., Varela I., Pleasance E.D., Simpson J.T., Stebbings L.A., Leroy C., Edkins S., Mudie L.J., et al. Complex landscapes of somatic rearrangement in human breast cancer genomes. Nature. 2009;462:1005–1010. doi: 10.1038/nature08645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Bass A.J., Lawrence M.S., Brace L.E., Ramos A.H., Drier Y., Cibulskis K., Sougnez C., Voet D., Saksena G., Sivachenko A., et al. Genomic sequencing of colorectal adenocarcinomas identifies a recurrent VTI1A-TCF7L2 fusion. Nat. Genet. 2011;43:964–970. doi: 10.1038/ng.936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Li Y., Roberts N.D., Wala J.A., Shapira O., Schumacher S.E., Kumar K., Khurana E., Waszak S., Korbel J.O., Haber J.E., et al. Patterns of somatic structural variation in human cancer genomes. Nature. 2020;578:112–121. doi: 10.1038/s41586-019-1913-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Ottaviani D., LeCain M., Sheer D. The role of microhomology in genomic structural variation. Trends Genet. 2014;30:85–94. doi: 10.1016/j.tig.2014.01.001. [DOI] [PubMed] [Google Scholar]
- 224.Bentley J., L’Hôte C., Platt F., Hurst C.D., Lowery J., Taylor C., Sak S.C., Harnden P., Knowles M.A., Kiltie A.E. Papillary and muscle invasive bladder tumors with distinct genomic stability profiles have different DNA repair fidelity and KU DNA-binding activities. Genes Chromosom. Cancer. 2009;48:310–321. doi: 10.1002/gcc.20641. [DOI] [PubMed] [Google Scholar]
- 225.Higgins G.S., Prevo R., Lee Y.F., Helleday T., Muschel R.J., Taylor S., Yoshimura M., Hickson I.D., Bernhard E.J., McKenna W.G. A small interfering RNA screen of genes involved in DNA repair identifies tumor-specific radiosensitization by POLQ knockdown. Cancer Res. 2010;70:2984–2993. doi: 10.1158/0008-5472.CAN-09-4040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Schrempf A., Slyskova J., Loizou J.I. Targeting the DNA Repair Enzyme Polymerase θ in Cancer Therapy. Trends Cancer. 2021;7:98–111. doi: 10.1016/j.trecan.2020.09.007. [DOI] [PubMed] [Google Scholar]
- 227.Higgins G.S., Boulton S.J. Beyond PARP—POLu as an anticancer target. Science. 2018;359:1217–1218. doi: 10.1126/science.aar5149. [DOI] [PubMed] [Google Scholar]
- 228.Bhargava R., Onyango D.O., Stark J.M. Regulation of single-strand annealing and its role in genome maintenance. Trends Genet. 2016;32:566–575. doi: 10.1016/j.tig.2016.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Elliott B., Richardson C., Jasin M. Chromosomal translocation mechanisms at intronic Alu elements in mammalian cells. Mol. Cell. 2005;17:885–894. doi: 10.1016/j.molcel.2005.02.028. [DOI] [PubMed] [Google Scholar]
- 230.Malacaria E., Honda M., Franchitto A., Spies M., Pichierri P. Physiological and pathological roles of RAD52 at DNA replication forks. Cancers. 2020:12. doi: 10.3390/cancers12020402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Feng Z., Scott S.P., Bussen W., Sharma G.G., Guo G., Pandita T.K., Powell S.N. Rad52 inactivation is synthetically lethal with BRCA2 deficiency. Proc. Natl. Acad. Sci. USA. 2011;108:686–691. doi: 10.1073/pnas.1010959107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Lok B.H., Carley A.C., Tchang B., Powell S.N. RAD52 inactivation is synthetically lethal with deficiencies in BRCA1 and PALB2 in addition to BRCA2 through RAD51-mediated homologous recombination. Oncogene. 2013;32:3552–3558. doi: 10.1038/onc.2012.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Ho V., Chung L., Singh A., Lea V., Abubakar A., Lim S.H., Chua W., Ng W., Lee M., Roberts T.L., et al. Aberrant expression of RAD52, its prognostic impact in rectal cancer and association with poor survival of patients. Int. J. Mol. Sci. 2020;21:1768. doi: 10.3390/ijms21051768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Hanamshet K., Mazina O.M., Mazin A.V. Reappearance from obscurity: Mammalian Rad52 in homologous recombination. Genes. 2016;7:63. doi: 10.3390/genes7090063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Hanamshet K., Mazin A.V. The function of RAD52 N-terminal domain is essential for viability of BRCA-deficient cells. Nucleic Acids Res. 2020;48:12778–12791. doi: 10.1093/nar/gkaa1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Sotiriou S.K., Kamileri I., Lugli N., Evangelou K., Da-Ré C., Huber F., Padayachy L., Tardy S., Nicati N.L., Barriot S., et al. Mammalian RAD52 functions in break-induced replication repair of collapsed DNA replication forks. Mol. Cell. 2016;64:1127–1134. doi: 10.1016/j.molcel.2016.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Jalan M., Olsen K.S., Powell S.N. Emerging roles of RAD52 in genome maintenance. Cancers. 2019;11:1038. doi: 10.3390/cancers11071038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Nickoloff J.A., Jones D., Lee S.H., Williamson E.A., Hromas R. Drugging the cancers addicted to DNA repair. J. Natl. Cancer Inst. 2017;109:1–13. doi: 10.1093/jnci/djx059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Kelso A.A., Lopezcolorado F.W., Bhargava R., Stark J.M. Distinct roles of RAD52 and POLQ in chromosomal break repair and replication stress response. PLoS Genet. 2019;15:e1008319. doi: 10.1371/journal.pgen.1008319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Toma M., Sullivan-Reed K., Śliwiński T., Skorski T. RAD52 as a potential target for synthetic lethality-based anticancer therapies. Cancers. 2019;11:1561. doi: 10.3390/cancers11101561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Bunz F., Dutriaux A., Lengauer C., Waldman T., Zhou S., Brown J.P., Sedivy J.M., Kinzler K.W., Vogelstein B. Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science. 1998;282:1497–1501. doi: 10.1126/science.282.5393.1497. [DOI] [PubMed] [Google Scholar]
- 242.Aljarbou F., Almousa N., Bazzi M., Aldaihan S., Alanazi M., Alharbi O., Almadi M., Aljebreen A.M., Azzam N.A., Arafa M., et al. The expression of telomere-related proteins and DNA damage response and their association with telomere length in colorectal cancer in Saudi patients. PLoS ONE. 2018;13:e0197154. doi: 10.1371/journal.pone.0197154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Mattiello L., Rehim S.S.A., Musella M., Sistigu A., Guarracino A., Vitale S., Corradi F., Galassi C., Sperati F., Manic G., et al. The targeting of mre11 or rad51 sensitizes colorectal cancer stem cells to chk1 inhibition. Cancers. 2021;13:1957. doi: 10.3390/cancers13081957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Jette N.R., Kumar M., Radhamani S., Arthur G., Goutam S., Yip S., Kolinsky M., Williams G.J., Bose P., Lees-Miller S.P. ATM-deficient cancers provide new opportunities for precision oncology. Cancers. 2020;12:687. doi: 10.3390/cancers12030687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Gilad S., Khosravi R., Shkedy D., Uziel T., Ziv Y., Savitsky K., Rotman G., Smith S., Chessa L., Jorgensen T.J., et al. Predominance of null mutations in ataxia-telangiectasia. Hum. Mol. Genet. 1996;5:433–439. doi: 10.1093/hmg/5.4.433. [DOI] [PubMed] [Google Scholar]
- 246.Mak J.P.Y., Ma H.T., Poon R.Y.C. Synergism between ATM and PARP1 inhibition involves DNA damage and abrogating the G 2 DNA damage checkpoint. Mol. Cancer Ther. 2019;19:123–134. doi: 10.1158/1535-7163.MCT-19-0474. [DOI] [PubMed] [Google Scholar]
- 247.Sundar R., Miranda S., Rodrigues D.N., Chénard-Poirier M., Dolling D., Clarke M., Figueiredo I., Bertan C., Yuan W., Ferreira A., et al. Ataxia telangiectasia mutated protein loss and benefit from oxaliplatin-based chemotherapy in colorectal cancer. Clin. Colorectal Cancer. 2018;17:280–284. doi: 10.1016/j.clcc.2018.05.011. [DOI] [PubMed] [Google Scholar]
- 248.Ge X.C., Wu F., Li W.T., Zhu X.J., Liu J.W., Wang B.L. Upregulation of WEE1 is a potential prognostic biomarker for patients with colorectal cancer. Oncol. Lett. 2017;13:4341–4348. doi: 10.3892/ol.2017.5984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Egeland E.V., Flatmark K., Nesland J.M., Flørenes V.A., Mælandsmo G.M., Boye K. Expression and clinical significance of Wee1 in colorectal cancer. Tumor Biol. 2016;37:12133–12140. doi: 10.1007/s13277-016-5081-3. [DOI] [PubMed] [Google Scholar]
- 250.Bendell J.C., Bischoff H.G., Hwang J., Reinhardt H.C., Zander T., Wang X., Hynes S., Pitou C., Campbell R., Iversen P., et al. A phase 1 dose-escalation study of checkpoint kinase 1 (CHK1) inhibitor prexasertib in combination with p38 mitogen-activated protein kinase (p38 MAPK) inhibitor ralimetinib in patients with advanced or metastatic cancer. Investig. New Drugs. 2020;38:1145–1155. doi: 10.1007/s10637-019-00873-6. [DOI] [PubMed] [Google Scholar]
- 251.Chu Q.S., Jonker D.J., Provencher D.M., Miller W.H., Bouganim N., Shields A.F., Shapiro G., Sawyer M.B., Lheureux S., Samouelian V., et al. A phase Ib study of oral Chk1 inhibitor LY2880070 in combination with gemcitabine in patients with advanced or metastatic cancer. J. Clin. Oncol. 2020;38:3581. doi: 10.1200/JCO.2020.38.15_suppl.3581. [DOI] [Google Scholar]
- 252.Plummer E.R., Kristeleit R.S., Cojocaru E., Haris N.M., Carter L., Jones R.H., Blagden S.P., Evans T.R.J., Arkenau H.-T., Sarker D., et al. A first-in-human phase I/II trial of SRA737 (a Chk1 Inhibitor) in subjects with advanced cancer. J. Clin. Oncol. 2019;37:3094. doi: 10.1200/JCO.2019.37.15_suppl.3094. [DOI] [Google Scholar]
- 253.Abida W., Bang Y.J., Carter L., Azaro A., Krebs M., Im S.-A., Chen Y., Buil-Bruna N., Li Y., Eaton D., et al. Abstract A094: Phase I Modular Study of AZD0156, a First-in-Class Oral Selective Inhibitor of Ataxia Telangiectasia Mutated Protein Kinase (ATM), in Combination with Olaparib (AToM Study, Module 1) [(accessed on 22 April 2021)]; Available online: https://mct.aacrjournals.org/content/17/1_Supplement/A094.
- 254.De Bono J.S., Tan D.S.P., Caldwell R., Terbuch A., Goh B.C., Heong V., Haris N.M., Bashir S., Hong D.S., Meric-Bernstam F., et al. First-in-human trial of the oral ataxia telangiectasia and Rad3-related (ATR) inhibitor BAY 1895344 in patients (pts) with advanced solid tumors. J. Clin. Oncol. 2019;37:3007. doi: 10.1200/JCO.2019.37.15_suppl.3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Terranova N., Jansen M., Falk M., Hendriks B.S. Population pharmacokinetics of ATR inhibitor berzosertib in phase I studies for different cancer types. Cancer Chemother. Pharmacol. 2021;87:185–196. doi: 10.1007/s00280-020-04184-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Yap T.A., O’Carrigan B., Penney M.S., Lim J.S., Brown J.S., De Miguel Luken M.J., Tunariu N., Perez-Lopez R., Rodrigues D.N., Riisnaes R., et al. Phase i trial of first-in-class ATR inhibitor M6620 (VX-970) as monotherapy or in combination with carboplatin in patients with advanced solid tumors. J. Clin. Oncol. 2020;38:3195–3204. doi: 10.1200/JCO.19.02404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Cohen D.J., Grabocka E., Bar-Sagi D., Godin R., Leichman L.P. A Phase Ib Study Combining Irinotecan with AZD1775, a Selective WEE 1 Kinase Inhibitor, in RAS/RAF Mutated Metastatic Colorectal Cancer Patients Who Progressed on First Line Therapy. [(accessed on 22 April 2021)]; Available online: http://ascopubs.org/doi/10.1200/JCO.2017.35.15_suppl.TPS3627.
- 258.Jhaveri K.L., Makker V., Wang X.V., Chen A.P., Flaherty K., Conley B.A., O’Dwyer P.J., Williams P.M., Hamilton S.R., Harris L., et al. Ado-trastuzumab emtansine (T-DM1) in patients (pts) with HER2 amplified (amp) tumors excluding breast and gastric/gastro-esophageal junction (GEJ) adenocarcinomas: Results from the national cancer institute (NCI) molecular analysis for therapy choice (MATCH) trial. J. Clin. Oncol. 2018;36:100. doi: 10.1200/jco.2018.36.15_suppl.100. [DOI] [Google Scholar]
- 259.Cole K.A., Pal S., Kudgus R.A., Ijaz H., Liu X., Minard C.G., Pawel B.R., Maris J.M., Haas-Kogan D.A., Voss S.D., et al. Phase I clinical trial of the WEe1 inhibitor adavosertib (AZD1775) with irinotecan in children with relapsed solid tumors: A COG phase I consortium report (ADVL1312) Clin. Cancer Res. 2020;26:1213–1219. doi: 10.1158/1078-0432.CCR-19-3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Leijen S., Van Geel R.M.J.M., Pavlick A.C., Tibes R., Rosen L., Razak A.R.A., Lam R., Demuth T., Rose S., Lee M.A., et al. Phase I study evaluating WEE1 inhibitor AZD1775 as monotherapy and in combination with gemcitabine, cisplatin, or carboplatin in patients with advanced solid tumors. J. Clin. Oncol. 2016;34:4371–4380. doi: 10.1200/JCO.2016.67.5991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Tolcher A., Mamdani H., Chalasani P., Meric-Bernstam F., Gazdoiu M., Makris L., Pultar P., Voliotis D. Clinical Activity of Single-Agent ZN-c3, an Oral WEE1 Inhibitor, in a Phase 1 Dose-Escalation Trial in Patients with Advanced Solid Tumors. [(accessed on 21 April 2021)]; Available online: https://www.abstractsonline.com/pp8/#!/9325/presentation/5146.
- 262.Tokunaga R., Xiu J., Goldberg R.M., Philip P.A., Seeber A., Battaglin F., Arai H., Lo J.H., Naseem M., Puccini A., et al. The impact of ARID1A mutation on molecular characteristics in colorectal cancer. Eur. J. Cancer. 2020;140:119–129. doi: 10.1016/j.ejca.2020.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Wei X.L., Wang D.S., Xi S.Y., Wu W.J., Chen D.L., Zeng Z.L., Wang R.Y., Huang Y.X., Jin Y., Wang F., et al. Clinicopathologic and prognostic relevance of ARID1A protein loss in colorectal cancer. World J. Gastroenterol. 2014;20:18404–18412. doi: 10.3748/wjg.v20.i48.18404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Park Y., Chui M.H., Rahmanto Y.S., Yu Z.C., Shamanna R.A., Bellani M.A., Gaillard S., Ayhan A., Viswanathan A., Seidman M.M., et al. Loss of ARID1A in tumor cells renders selective vulnerability to combined ionizing radiation and PARP inhibitor therapy. Clin. Cancer Res. 2019;25:5584–5593. doi: 10.1158/1078-0432.CCR-18-4222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Sen M., Wang X., Hamdan F.H., Rapp J., Eggert J., Kosinsky R.L., Wegwitz F., Kutschat A.P., Younesi F.S., Gaedcke J., et al. ARID1A facilitates KRAS signaling-regulated enhancer activity in an AP1-dependent manner in colorectal cancer cells. Clin. Epigenetics. 2019;11 doi: 10.1186/s13148-019-0690-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Niedermaier B., Sak A., Zernickel E., Xu S., Groneberg M., Stuschke M. Targeting ARID1A-mutant colorectal cancer: Depletion of ARID1B increases radiosensitivity and modulates DNA damage response. Sci. Rep. 2019;9:18207. doi: 10.1038/s41598-019-54757-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Gorecki L., Andrs M., Korabecny J. Clinical candidates targeting the atr–chk1–wee1 axis in cancer. Cancers. 2021;13:795. doi: 10.3390/cancers13040795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Parseghian C.M., Napolitano S., Loree J.M., Kopetz S. Mechanisms of innate and acquired resistance to anti-EGFR therapy: A review of current knowledge with a focus on rechallenge therapies. Clin. Cancer Res. 2019;25:6899–6908. doi: 10.1158/1078-0432.CCR-19-0823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Russo M., Crisafulli G., Sogari A., Reilly N.M., Arena S., Lamba S., Bartolini A., Amodio V., Magrì A., Novara L., et al. Adaptive mutability of colorectal cancers in response to targeted therapies. Science. 2019;366:1473–1480. doi: 10.1126/science.aav4474. [DOI] [PubMed] [Google Scholar]
- 270.Mauri G., Arena S., Siena S., Bardelli A., Sartore-Bianchi A. The DNA damage response pathway as a land of therapeutic opportunities for colorectal cancer. Ann. Oncol. 2020;31:1135–1147. doi: 10.1016/j.annonc.2020.05.027. [DOI] [PubMed] [Google Scholar]
- 271.Leichman L., Groshen S., O’Neil B.H., Messersmith W., Berlin J., Chan E., Leichman C.G., Cohen S.J., Cohen D., Lenz H., et al. Phase II study of olaparib (AZD-2281) after standard systemic therapies for disseminated colorectal cancer. Oncologist. 2016;21:172–177. doi: 10.1634/theoncologist.2015-0319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.De Castro e Gloria H., Jesuíno Nogueira L., Bencke Grudzinski P., da Costa Ghignatti P.V., Guecheva T.N., Motta Leguisamo N., Saffi J. Olaparib-mediated enhancement of 5-fluorouracil cytotoxicity in mismatch repair deficient colorectal cancer cells. BMC Cancer. 2021;21:448. doi: 10.1186/s12885-021-08188-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Arena S., Corti G., Durinikova E., Montone M., Reilly N.M., Russo M., Lorenzato A., Arcella P., Lazzari L., Rospo G., et al. A subset of colorectal cancers with cross-sensitivity to olaparib and oxaliplatin. Clin. Cancer Res. 2020;26:1372–1384. doi: 10.1158/1078-0432.CCR-19-2409. [DOI] [PubMed] [Google Scholar]
- 274.Smeby J., Kryeziu K., Berg K.C.G., Eilertsen I.A., Eide P.W., Johannessen B., Guren M.G., Nesbakken A., Bruun J., Lothe R.A., et al. Molecular correlates of sensitivity to PARP inhibition beyond homologous recombination deficiency in pre-clinical models of colorectal cancer point to wild-type TP53 activity. EBioMedicine. 2020;59 doi: 10.1016/j.ebiom.2020.102923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Serra Elizalde V., Llop-Guevara A., Pearson A., Cruz C., Castroviejo-Bermejo M., Chopra N., Tovey H., Toms C., Kriplani D., Gevensleben H., et al. 1O Detection of homologous recombination repair deficiency (HRD) in treatment-naive early triple-negative breast cancer (TNBC) by RAD51 foci and comparison with DNA-based tests. Ann. Oncol. 2021;32:S21–S22. doi: 10.1016/j.annonc.2021.03.015. [DOI] [Google Scholar]
- 276.Llop-Guevara A., Vladimirova V., Schneeweiss A., Villacampa G., Karn T., Zahm D.-M., Herencia-Ropero A., Jank P., van Mackelenbergh M., Fasching P.A., et al. 2O Association of RAD51 with homologous recombination deficiency (HRD) and clinical outcomes in untreated triple-negative breast cancer (TNBC): Analysis of the GeparSixto randomized clinical trial. Ann. Oncol. 2021;32:S22. doi: 10.1016/j.annonc.2021.03.016. [DOI] [PubMed] [Google Scholar]
- 277.Melinda L.T., Kirsten M.T., Julia R., Bryan H., Gordon B.M., Kristin C.J., Zoltan S., William T.B., Eric P.W., Nadine M.T., et al. Homologous recombination deficiency (hrd) score predicts response to platinum-containing neoadjuvant chemotherapy in patients with triple-negative breast cancer. Clin. Cancer Res. 2016;22:3764–3773. doi: 10.1158/1078-0432.CCR-15-2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Shiro Takamatsu A., Brown J., Yamaguchi K., Hamanishi J., Yamanoi K., Takaya H., Kaneyasu T., Mori S., Mandai M., Matsumura N. The utility of homologous recombination deficiency biomarkers across cancer types. medRxiv. 2021 doi: 10.1101/2021.02.18.21251882. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]