Abstract
Objective
Long non-coding RNAs (lncRNAs) and microRNAs (miRs) are differentially expressed in ovarian cancer (OC) cells and influence OC progression. This study intended to explore the underlying roles of LINC00115 and miR-30a in OC.
Methods
Gene Expression Omnibus database was used to find OC microarray datasets and bioinformatics analysis predicted the potential molecular mechanism of OC. OC stem cells (OCSCs) surface marker was isolated from human OC cell line and identified. CD133+ OCSCs were transfected with LINC00115, miR-30a and SOX9 alone or together to detect sphere-forming ability and apoptosis of OCSCs. Caspase-3 activity and DNA damage in cell supernatant were detected. The levels of CD44, NANOG, POU5F1, LINC00115, CD133, miR-30a and SOX9 were measured. Then sh-LNC00115-treated OCSCs were added with Wnt/β-catenin activator SKL2001 to observe the changes of cell stemness and activity. Finally, animal models were established to evaluate the effect of LINC00115 on OCSC in vivo.
Results
LINC00115 and SOX9 were highly expressed in OC, while miR-30a was lowly expressed. After silencing LINC00115 or overexpressing miR-30a, the sphere-forming rate of CD133+ OCSC and levels of CD133, CD44, NANOG and POU5F1 decreased, while apoptotic rate, Caspase-3 activity and histone-related DNA damage increased. SOX9 reversed these trends. Additionally, LINC00115 could bind to miR-30a and miR-30a could target SOX9. SKL2001 partially reversed cell stemness and activity in sh-LNC00115-treated OCSCs. Finally, silencing LINC00115 could inhibit OCSCs growth in vivo.
Conclusion
LINC00115 promoted stemness and inhibited apoptosis of OCSCs by upregulating SOX9 and in activating the Wnt/β-catenin pathway through competitively binding to miR-30a.
Keywords: Ovarian cancer, LINC00115, Cancer stem cells, CD133+, microRNA-30a, SOX9
Background
Ovarian cancer (OC) is the third most prevalent gynecologic malignancy in the world, but it accounts for the highest mortality rate among these cancers [1]. About 75% of OC patients are diagnosed at an advanced stage due to the asymptomatic features [2]. High-grade serious cancers account for 75% of OC cases and the majority of the mortality [3]. Pelvic inflammatory disease, endometriosis and daily alcohol intake have been proposed as risk factors for OC [4–6]. The 5-year survival rates of OC are less than 50% after diagnosis, because most of the patients present with advanced disease which is largely incurable [7]. The poor outcome for OC patients is due to the high recurrence and chemotherapy-resistance of OC stem cells (OCSCs), and OCSCs have important cancer stemness features, including spherical formation and self-renewal, expressing epithelial-mesenchymal transition markers, enhancing drug resistance, recurrence and tumorigenicity [8, 9]. Herein, it is imperative to explore effective treatments from the aspect of OCSCs.
Long non-coding RNAs (lncRNAs) are important regulators of various biological processes, including cancer stemness and tumorigenesis [10], and their roles in OC cell behaviors are also extensively studied [11]. LINC00115 may be an underlying target for lung cancer by working as a competing endogenous RNA (ceRNA) for regulation of microRNA (miR)-mRNA [12]. However, there is no research on the effect of LINC00115 on OC. Recently, miRs have been reported to be abnormally expressed in several cancers and play a vital part in cancer development, including OC [13]. Notably, existing evidence supports that miRs are pivotal in metastasis, tumor recurrence and drug resistance of CD133+ OCSCs [14]. In the meanwhile, we found LINC00115 could competitively bind to miR-30a. miR-30 family acts as tumor suppressors in several cancers, and their high expression regulated by CD133 promotes migration and invasion in CD133+ pancreatic cancer cells [15]. Besides, miR-30a is evidenced to participate in breast cancer stemness and tumorigenesis [16]. Furthermore, Wang et al. have found that miR-30a-5p expression is substantially decreased in OC cell lines, and miR-30a-5p overexpression inhibits OC cell migration [17]. Based on the above information, we may see some underlying connections of LINC00115 and miR-30a in the treatment of OC. Therefore, in this study, we carried out a series of experiments to identify the underlying mechanisms.
Methods
Ethics statement
This study was approved by the Clinical Ethical Committee of Shengjing Hospital of China Medical University. All animal experiments were complied with the ARRIVE guidelines and carried out in accordance with the National Institutes of Health guide for the care and use of Laboratory animals.
Microarray-based analysis
Data of OC gene/lncRNA microarray datasets (GSE66957/GSE26712/GSE4122) and miR microarray datasets (GSE48485) were downloaded from the Gene Expression Omnibus database (https://www.ncbi.nlm.nih.gov/geo/). R language Affy package (http://www.bioconductor.org/packages/release/bioc/html/affy.html) was used to standardize the microarray dataset expression data, the limma package (http://master.bioconductor.org/packages/release/bioc/html/limma.html) was used for screening differentially expressed genes (DEGs), and the pheatmap package (https://cran.r-project.org/web/packages/pheatmap/index.html) was applied to draw the heat map of DEGs. Firstly, differentially expressed lncRNAs were screened from GSE66957 datasets. Then, the probable regulated miRs of LINC00115 were predicted in the miRcode database (http://www.mircode.org/index.php) and RNAInter database (http://www.rna-society.org/raid/index.html). The predicted results were compared in jvenn (http://jvenn.toulouse.inra.fr/app/example.html), and then compared with the results of GSE48485 to further screen the differentially expressed miRs. TargetScan (http://www.targetscan.org/vert_71/, access date: 11/21/2018) [18], DIANA (http://diana.imis.athena-innovation.gr/DianaTools/index.php?r=microT_CDS/index, access date: 11/21/2018) [19], MicroSearch (http://www.exiqon.com/microrna-target-prediction, access date: 11/23/2018) [20] and miRDB (http://www.mirdb.org/, access date: 11/26/2018) [21]databases were applied to predict the targets of miR-30a. The predicted results were compared with those in GSE26712 and GSE4122 by jvenn.
Isolation, culture and identification of CD133+ OCSCs
OC cells A2780 (Nanjing Cobioer Biotechnology Co., Ltd., Nanjing, Jiangsu, China) were cultured in complete Dulbecco's modified Eagle's medium (DMEM)/F12 (Invitrogen Inc., Carlsbad, CA, USA), and isolated into single cell using trypsin-ethylene diamine tetraacetic acid (EDTA). Then, cells were centrifuged with the supernatant removed, and cells were harvested and washed twice in 2 mL phosphate buffered saline (PBS). After that, cells were added with 1 µL allophycocyanion-labeled mouse anti-human CD133 antibody (APC-CD133) (eBioscience, San Diego, CA, USA), and cultured at 4℃ for 30 min. Finally, cells were washed twice with PBS, and the obtained CD133+ OCSCs were analyzed on a flow cytometer (ACEA Biosciences, San Diego, CA, USA).
The obtained single CD133+ OCSCs were suspended in serum-free medium containing epidermal growth factor (EGF) and basic fibroblast growth factor (bFGF) (PeproTech, Rocky Hill, NJ, USA). Next, the suspended cells were inoculated into the low-adhesive 6-well plates (Corning Inc, Corning, New York, NY, USA) at 1 × 104 cells/well. Ten days later, CD133+ OCSCs were observed in serum-free mammary epithelial basal medium (MEBM) (Lonza Group Ltd., Basel, Switzerland) [9], and collected for subsequent experiments.
Cell transfection and grouping
The cultured CD133+ OCSCs were assigned into control shRNA (Ctrl), sh-LINC00115-1 (sh1), sh-LINC00115-2 (sh2), mimic-negative control (NC), miR-30a mimic, mimic-NC + vector-NC, miR-30a mimic + vector-NC, mimic-NC + SOX9-vector, miR-30a mimic + SOX9-vector, sh1 + inhibitor-NC, sh1 + miR-30a inhibitor, sh1 + vector-NC, sh1 + SOX9-vector, sh1 + PBS and sh1 + SKL2001 (a specific activator of the Wnt/β-catenin pathway) groups. miR-30a mimic, miR-30a inhibitor and their controls were purchased from Invitrogen Inc., SOX9-vector (the correct sequence of SOX9 was inserted into pCEP4 plasmid to construct pCEP4/SOX9 eukaryotic expression vector) and its NC (pCEP4 empty plasmid, named vector-NC), sh1, sh2 (two shRNA expression vectors, sh1 and sh2, were constructed using pRNAT-U6.1/neo plasmid) (Table 1) and their controls (pRNAT-U6.1/neo empty plasmid, named Ctrl) were purchased from Guangzhou RiboBio Co., Ltd, (Guangzhou, Guangdong, China).
Table 1.
Sequences of 2 shRNAs of LINC00115
| shRNA sequence | |
|---|---|
| Sh1 | GAAGAAUGGUACAAAUCCAAG |
| Sh2 | CUUAAAGGAACCAAUGAGUCC |
CD133+ OCSCs were seeded into 24-well plastics culture plates containing 10% fetal bovine serum in DMEM at 24 h before transfection, with about 1 × 105 cells per well. The plasmids (4 µg) and liposome Lipofectamine® 3000 (L3000001, ThermoFisher Scientific, Waltham, MA, USA) were diluted with 50 µL Opti-MEM before transfection. CD133+ OCSCs were transfected with Lipofectamine® 3000 when the cell confluence reached 85%-90%. Cells were transfected with 80 nM plasmids, and the culture medium was refreshed after 6-h transfection. After 48-h culture, cells were collected for subsequent experiments. SKL2001 was purchased from Selleck Chemicals (Houston, TX, USA, 5 mM, Cat: S832001).
Sphere-forming assay
CD133+ OCSC single cell suspension with density of 5.0 × 103 cells/mL was inoculated into poly(2-hydroxyethyl methacrylate) (P3932-25G, Sigma-Aldrich, Merck KGaA, Darmstadt, Germany)-coated Petri dishes. Serum-free MEBM containing 2 mL-glutamine, 100 U/mL penicillin and 100 mg/mL streptomycin, 5 mg/mL insulin, 0.5 mg/mL hydrocortisone, 1 U/mL heparin, 2% B27, 20 ng/mL EGF, and 20 ng/mL bFGF (Invitrogen) was added to the Petri dishes. After 10 days of culture, the sphere-forming rate = the number of spheroidized cells/the number of accessing cells [22].
Reverse transcription quantitative polymerase chain reaction (RT-qPCR)
The total RNA of OC A2780 cells and CD133+ OCSCs was extracted using TRIzol (Invitrogen), and 5 µg RNA was reversely transcribed into DNA according to the instructions of the cDNA kit (ThermoFisher Scientific). TaqMan microRNA assay (Ambion, Austin, Texas, USA) was used to detect miR-30a expression with U6 as a reference. PrimeScript RT-PCR kits (Roche, Basel, Switzerland) were used to detect the expression of LINC00115, CD133 and other genes with glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as the loading control. All primers were synthesized by Takara Holdings Inc., (Kyoto, Japan) (Table 2). The 2−ΔΔCt indicated the ratio of target gene expression between the experiment and control groups. ΔΔCT = ΔCt experimental group—ΔCt control group, and ΔCt = Ct target gene—Ct control gene. The experiment was repeated three times [23].
Table 2.
Primer sequence of RT-qPCR
| Gene | Sequence 5′–3′ | |
|---|---|---|
| CD133 | F: AGTCGGAAACTGGCAGATAGC | R: AGTCGGAAACTGGCAGATAGC |
| LINC00115 | F: TGGCTTGTCTTCCATCGTCC | R: GCACGAGGGTTGTTACAGGA |
| CD44 | F: CGGACACCATGGACAAGTTT | R: CCGTCCGAGAGATGCTGTAG |
| GAPDH | F: CAAGATCATCACCAATGCCT | R: CCCATCACGCCACAGTTTCC |
| NANOG | F: TTTGTGGGCCTGAAGAAAACT | R: AGGGCTGTCCTGAATAAGCAG |
| POU5F1 | F: CTGGGTTGATCCTCGGACCT | R: CCATCGGAGTTGCTCTCCA |
| miR-30a | F: GCGACTGTAAACATCCTCGAC | R: CAGCTGCAAACATCCGACTG |
| SOX9 | F: AGGAAGTCGGTGAAGAACGG | R: AGGAAGTCGGTGAAGAACGG |
| SOX9-3’UTR-Primer | F: CGGACTAGTACAGACCTTAATCTTAATTA | R: CCCAAGCTTTTTTTAATGCAATGTATATT |
| SOX9-mut-Primer | F: GACTAGTTATTAACAGTTTTAGAAGTC | R: CAAGCTTAATGCAATGTATATTTATTG |
| Luc-LINC00115wt-Primer | F: CGGACTAGTATCAGCAGAGATATGGAGGA | R: CCCAAGCTTGACAATCCAGGTCATCCTTC |
| Luc-LINC00115mut-Primer | F: GACTAGTGTAGCAGAGAGGGCAGCAGA | R: CAAGCTTTTGATACTTATACTTAGTTG |
| U6 | F: CTCGCTTCGGCAGCACATATACT | R: ACGCTTCACGAATTTGCGTGTC |
RT-qPCR, reverse transcription quantitative polymerase chain reaction; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; miR-30a, microRNA-30a
Western blot analysis
Cells were collected and washed with pre-cooled PBS for 2–3 times, and then cells were lysed by protein extraction lysate to extract total protein. The protein concentration was measured using the bicinchoninic acid kit (ThermoFisher Scientific). Next, proteins were run on 12% sodium dodecyl sulfate–polyacrylamide gel electrophoresis, then moved into polyvinylidene fluoride membranes, and sealed with skim milk for 1 h. Subsequently, the membranes were cultivated with rabbit anti-human antibodies SOX9 (1 µg/mL, ab185966), CD44 (1:5000, ab51037), CD133 (2 µg/mL, ab19898), NANOG (1:2000, ab109250), POU5F1 (1:500, ab107643), Wnt-3 (1:2000, ab32249), Wnt-6 (1:500, ab50030), p-β-catenin (1:500, ab75777, Abcam) and β-catenin (1:1000, ab16051) at 4℃ overnight. Then, the membranes were washed with tris-buffered saline-tween (TBST) three times (each for 10 min), and then added with goat anti-rabbit secondary antibody (1:2000, ab6721) for 1 h. After 3 TBST washings, the membranes were rinsed in enhanced chemiluminescence reagent for X-ray, development, fixing and result analysis. Chemidoc XRS + system (Bio-Rad, USA) was used to collect images, and the optical density value of each band was measured using ImageJ software. The optical density value = 1 g (average gray value of the brightest area of electrophoretic film/average gray value of the target strip to be tested). GAPDH (1:10,000, ab8245) or β-tubulin (1:1000, ab52623, Abcam) was selected as a reference. The relative level of the protein was calculated as the gray value ratio of the target protein to the reference protein. All antibodies were from Abcam Inc., (Cambridge, MA, USA).
Flow cytometry
Cell apoptosis was measured using annexin V-fluorescein isothiocyanate (FITC)/propidium iodide (PI) double staining. Cells were incubated at 37℃ for 48 h and washed with precooled PBS, and then re-suspended in 1 × binding buffer. According to the provided instructions, apoptotic cells were stained with annexin V-FITC/PI apoptotic detection kit (Becton Dickinson BD Biosciences, San Jose, CA, USA). NovoCyte flow cytometer and NovoExpress 1.0.2 software (ACEA Biosciences) were used to detect apoptotic cells [24].
Determination of Caspase-3 activity
The transfected cells (2 × 106) were collected and lysed in 50 µL lysis buffer on the ice for 10 min, and then centrifuged at 14,000 rpm at 4℃ for 5 min. The supernatant (50 µL) was collected, and cultured with 2 × reaction/Dithiothreitol (DTT) Mix containing N-acetyl Asp-Glu-Val-Asp (DVED)-p-nitroanilde (pNA) (Caspase 3 chromogenic substrate) (50 µM) at 37℃ for 3 h. Finally, the absorbance at a wavelength of 405 nm was detected using the ApoAlert Caspase Colorimetric Kit (Clontech Laboratories, Mountain View, CA, USA) [24].
Analysis of DNA damage
Enzyme-linked immunosorbent assay kit (Roche Diagnostics, Mannheim, Germany) was applied to quantitatively cleave nucleosome DNA. Cell supernatant (20 µL) was detected at a wavelength of 405 nm using a microplate reader (Tecan, Salzburg, Austria). After subtracting background values, optical density values of DNA fragments representing nucleosomes in Hsp90 inhibitor treated samples were compared with those in control cells and expressed as the percentage of control cells [24].
Dual-luciferase reporter gene assay
Artificially synthesized SOX9 3’ untranslated region (3'UTR) gene fragments were introduced into pMIR-reporter (Beijing Huayueyang Biotechnology Inc., Beijing, China) using endonuclease sites SpeI and Hind III. Complementary sequence mutation sites of seed sequences were designed on SOX9 wild type (WT), and digested by restriction endonuclease. T4 DNA ligase was used to insert the target fragment into the pMIR-reporter plasmid. The correctly sequenced luciferase reporter plasmids SOX9 3'-UTR WT and SOX9 3'-UTR mutant type (MUT) were co-transfected into HEK-293 T cells (CRL-1415, Shanghai Xinyu Biotechnology Co., Ltd., Shanghai, China) with miR-30a, respectively. After 48 h of transfection, cells were collected and lysed. Luciferase activity was detected using the Luciferase Detection Kit (RG005, Beyotime Biotechnology Co., Ltd, Shanghai, China) and a Glomax 20/20 luminometer (Promega Corporation, Madison, Wisconsin, USA). The relationship between lncRNA LINC00115 and miR-30a was detected with the same method.
RNA pull-down assay
OC cells were transfected with 50 nM biotin-labeled Bio-miR-30a-WT and Bio-miR-30a-MUT and corresponding Bio-NC. After 48 h, cells were harvested and washed with PBS. Cells were incubated in specific lysis buffer (Ambion) for 10 min and then centrifuged at 14,000g to obtain the supernatant. Protein lysate was incubated with RNase-free bovine serum albumin and M-280 streptavidin beads (S3762, Sigma-Aldrich) pre-coated with yeast tRNA (TRNABAK-RO, Sigma-Aldrich). The beads were incubated at 4 ℃ for 3 h, washed twice with precooled lysis buffer, three times with low-salt buffer and one time with high-salt buffer. The binding RNA was purified by Trizol and then expression of miR-30a and LINC00115 was detected by RT-qPCR.
RNA immunoprecipitation (RIP) assay
Cells were lysed in lysis buffer containing RNasin (Takara) and protease inhibitor mixture (B14001a, Roche Diagnostics GmbH, Indianapolis, IN, USA) (25 mM Tris–HCl pH 7.4, 150 mM NaCl, 0.5% NP-40, 2 mM EDTA, 1 mM NaF and 0.5 mM dithiothreotol). The supernatant was centrifuged at 1200 g for 30 min and then supplemented with anti-Ago2 magnetic beads (130–061-101, Shanghai Univ Biotechnology Co., Ltd., Shanghai, China), while the control group was added with anti-immunoglobulin G magnetic beads. The beads were incubated at 4℃ for 4 h and washed three times with washing buffer (50 mM Tris–HCl, 300 mM NaCl pH 7.4, 1 mM MgCl2, and 0.1% NP-40). Finally, RNA was extracted from magnetic beads by TRIzol method. LINC00115 and miR-30a were detected using RT-qPCR.
Animal experiments
Fifteen female BALB/c nude mice at specific pathogen-free grade, aged 4–5 weeks, were purchased from Hunan SJA Laboratory Animal Co., Ltd., (Changsha, Hunan, China, SCXK (Hunan) 2016–0002). After 3 days of adaptive feeding, the mice were randomly assigned into three groups, 5 mice each, namely Ctrl group (mice were injected with OCSCs in the Ctrl group), sh1 group (mice were injected with OCSCs in the sh1 group) and sh2 group (mice were injected with OCSCs in the sh2 group). Cells in logarithmic growth phase were collected, detached by trypsin, counted, and centrifuged to discard the supernatant. Next, single cell suspension was prepared using PBS, and cell concentration was adjusted to 1 × 105 cells/mL. Each nude mouse was injected with 100 µL cell suspension at the axillary mammary gland. After the injection, the spirit, diet, drinking, defecation, motility and growth of subcutaneous tumors were observed regularly. At the same time, the length diameter (a) and the short diameter (b) of the subcutaneous tumors were estimated using a vernier caliper, and tumor volume (a × b2/2) was calculated, the growth curve of the tumors was drawn. The nude mice were euthanized after 44 days of cell injection. Tumor tissue was dissected and weighed. Subsequently, immunohistochemical staining was carried out according to the previous literature [25]. The primary antibody anti-Ki67 (ab15580, Abcam) and the secondary anti-rabbit antibody (1:1,000; ab6721; Abcam) labeled with horseradish peroxidase were used in this experiment.
Statistical analysis
SPSS 22.0 statistical software (IBM Corp. Armonk, NY, USA) was used to process the data. Measurement data were described as mean ± standard deviation. The unpaired t test was applied to analyze comparisons between two groups, and one-way analysis of variance (ANOVA) to analyze comparisons among multi-groups. Tukey’s multiple comparison test as post hoc were applied. A probability value of p < 0.05 indicated the difference was significant.
Results
Identification of CD133+ OCSC
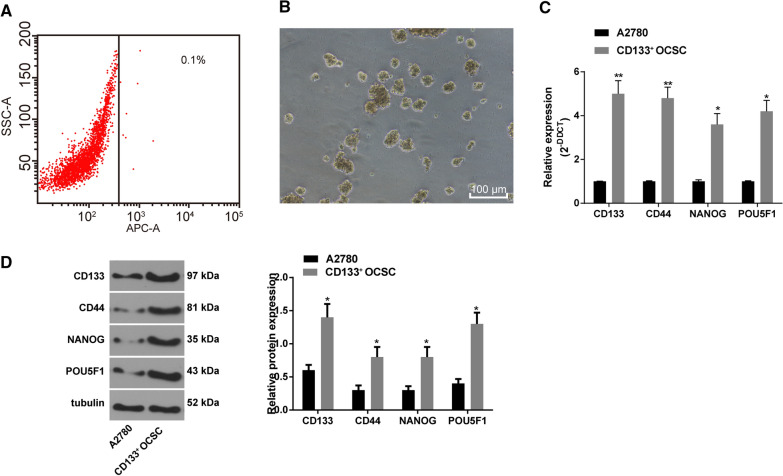
OC A2780 cells were labeled with APC-CD133 and then sorted with flow cytometry. The sorting results revealed CD133+ OCSCs only accounted for 0.1% of A2780 cells (Fig. 1A). The separated CD133+ OCSCs were suspended in serum-free medium and observed under an optical microscope. After 10 days of suspension culture, the cells aggregated and grew into spheres, and the spheres became larger gradually, showing a convergent three-dimensional structure (Fig. 1B). The expression of CD133, CD44, NANOG and POU5F1 in A2780 cells and CD133+ OCSCs was detected by RT-qPCR and Western blot analysis. It demonstrated that their levels in CD133+ OCSCs were evidently higher than those in A2780 cells (Fig. 1C-D) (all p < 0.05). These results indicated that CD133+ OCSCs were separated successfully.
Fig. 1.
Successful separation of CD133+ OCSCs. A A2780 cells were stained with APC-labeled CD133 antibody (x-axis), and CD133+ OCSCs were sorted using flow cytometry; B The morphological characteristics of CD133+ OCSCs cultured in serum-free medium for 5–6 days were observed under the microscope (100 ×); C, D Levels of CD133, CD44, NANOG and POU5F1 in A2780 cells and CD133+ OCSCs were detected by RT-qPCR and Western blot analysis. Data were analyzed using the unpaired t test. Compared with the A2780 cells, *p < 0.05, **p < 0.01. The experiment was performed in triplicate
LINC00115 promotes the maintenance of OCSC stemness and inhibits its apoptosis
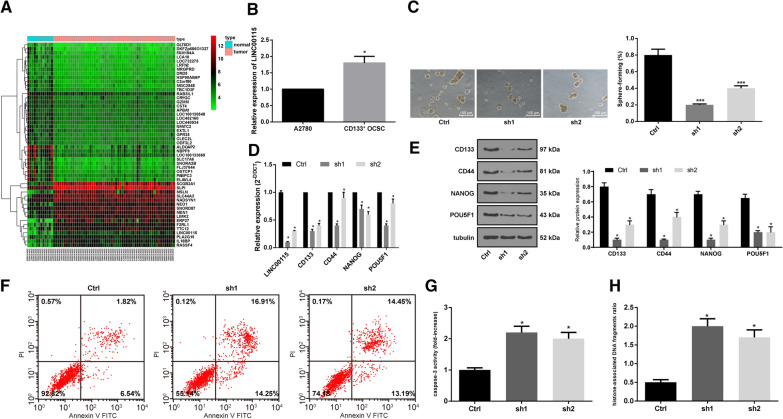
R language was used to analyze the differential expression of microarray datasets. Fifty DEGs/lncRNAs were mapped on GSE66957. LINC00115 expression was higher in OC tissues than in normal tissues (Fig. 2A). Meanwhile, LINC00115 expression in A2780 cells and CD133+ OCSCs was detected using RT-qPCR. LINC00115 expression in CD133+ OCSCs was notably higher than that in A2780 cells (Fig. 2B) (p < 0.05). Next, we used two different shRNAs to silence LINC00115 expression and analyzed its effect on CD133+ OCSCs. RT-qPCR showed that both shRNAs played an interference role (Fig. 2D). The results elicited that after silencing LINC00115, the sphere-forming rate of CD133+ OCSCs was decreased greatly (Fig. 2C) (p < 0.05). Then, the expression patterns of LINC00115, CD133, CD44, NANOG and POU5F1 were detected using RT-qPCR and Western blot analysis. It showed that silencing LINC00115 could significantly reduce the expression of CD133, CD44, NANOG and POU5F1 (Fig. 2D-E) (all p < 0.05). Flow cytometry indicated that the apoptotic rate of cells with silencing LINC00115 was increased markedly (Fig. 2F). In addition, Caspase-3 activity and histone-related DNA damage were increased significantly after silencing LINC00115 (Fig. 2G-H) (both p < 0.05). These results suggested that LINC00115 could promote the maintenance of cell viability and inhibit apoptosis of OCSCs.
Fig. 2.
LINC00115 promotes the maintenance of OCSC stemness and inhibits its apoptosis. Two sh-LINC00115 vectors (sh1 and sh2) and their negative control (Ctrl) were used to transfect CD133+ OCSCs cells. A The expression heat map of 50 DEGs/lncRNAs on GSE66957, in which Y-axis represents DEGs/lncRNAs. The upper right histogram is the color order, and each rectangle corresponds to a sample expression value; B Relative expression of LINC00115 in A2780 cells and CD133+ OCSCs detected by RT-qPCR, compared with the A2780 cells, *p < 0.05; C The sphere-forming rate of CD133+ OCSCs was observed under a microscope, VS the Ctrl group, ***p < 0.001; D, E. Relative expression of LINC00115, CD133, CD44, NANOG and POU5F1 in CD133+ OCSCs was detected using RT-qPCR and Western blot analysis, vs the Ctrl group, *p < 0.05; F Cell apoptosis was measured using flow cytometry; G. Relative Caspase-3 activity in cells of each group, VS the Ctrl group, *p < 0.05; H. Percentage of DNA damage in cell supernatant of each group, VS the Ctrl group, *p < 0.05. Data in panel B were analyzed using the unpaired t test, and the data in panels C-H were analyzed using one-way ANOVA and Tukey’s multiple comparison test. The experiment was performed in triplicate and data were expressed as means ± standard deviation
LINC00115 sponges miR-30a
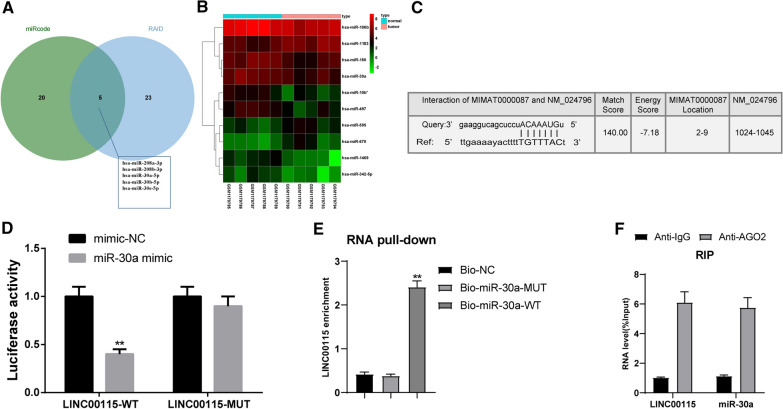
The possible targeting miRs of LINC00115 were predicted in the miRcode and RNAInter databases [26, 27]. The Venn diagram (Fig. 3A) exhibited that there were 5 intersecting miRs: hsa-miR-208a-3p, hsa-miR-208b-3p, hsa-miR-30a-5p, hsa-miR-30b-5p, hsa-miR-30b-5p and hsa-miR-30e-5p, which may be the targeting miRs of LINC00115. Additionally, 10 differentially expressed miRs were mapped in differential analysis of GSE48485 (Fig. 3B). Among them, hsa-miR-30a showed weak expression in OC. miR-30a-5p has been found to show weak expression in cisplatin-resistant OC cells [28]. According to RNAInter prediction analysis, LINC00115 had binding sites to miR-30a (Fig. 3C). The results of dual-luciferase reporter gene assay, RNA pull-down assay and RIP assay all verified that LNC00115 could bind to miR-30a (Fig. 3D-F) (all p < 0.05).
Fig. 3.
LINC00115 could bind to miR-30a. A Comparing the predicted results of LINC00115-targeted miRs from the miRcode and RNAInter databases, there were five intersecting miRs; B The expression heat map of 10 differentially expressed miRs on GSE48485, in which Y-axis represents miRs. The upper right histogram is the color order, and each rectangle corresponds to a sample expression value; C The binding sites between LINC00115 and miR-30a were analyzed by RNAInter database; D-F. LNC00115 could bind to miR-30a verified by dual luciferase reporter gene assay (D, vs the mimic-NC group, **p < 0.01), RNA pull-down assay (E, vs the Bio-miR-30a-mut group, **p < 0.01) and RIP assay (F, vs the IgG group, ***p < 0.001). Data in panel D were analyzed using the unpaired t test, and data in panels E–F were analyzed using one-way ANOVA and Tukey’s multiple comparison test. Repetitions = 3. The experiment was performed in triplicate and data were expressed as means ± standard deviation
miR-30a targets SOX9
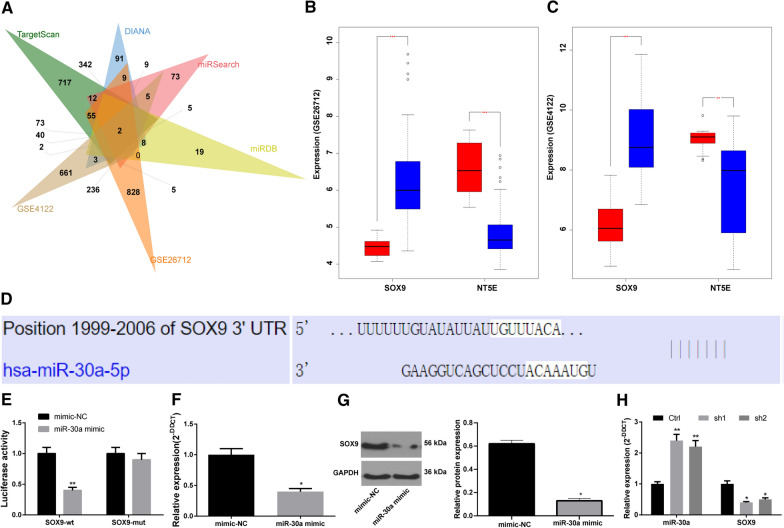
The TargetScan, DIANA, miRSearch and miRDB databases were used to predict the target genes of miR-30a, and the prediction results were compared with the DEGs on GSE26712 and GSE4122 microarray datasets. Two intersecting genes SOX9 and ecto-5'-nucleotidase (NT5E) were found (Fig. 4A). The differential expression of the two genes may be regulated by miR-30a. SOX9 is highly expressed in OC, while NT5E is poorly expressed (Fig. 4BC). TargetScan (http://www.targetscan.org/vert_72/) showed that miR-30a could target SOX9 (Fig. 4D). The results of dual luciferase reporter gene assay suggested that versus the mimic-NC group, the luciferase activity of co-transfection group of miR-30a mimic and SOX9-MUT did not change significantly (p > 0.05), but the luciferase activity of co-transfection group of miR-30a mimic and SOX9-WT was decreased notably (Fig. 4E) (p < 0.05). As RT-qPCR and Western blot analysis showed, relative to the mimic-NC group, SOX9 expression in the miR-30a mimic group was decreased noticeably (Fig. 4F, G) (p < 0.05). In addition, RT-qPCR results revealed that miR-30a expression in CD133+ OCSCs was significantly increased after silencing LINC00115, while SOX9 expression was decreased (Fig. 4H) (both p < 0.05). Briefly, miR-30a could target SOX9, and LINC00115 could act as a sponge to adsorb miR-30a and promote SOX9 expression.
Fig. 4.
miR-30a could target SOX9. A TargetScan, DIANA, miRSearch and miRDB databases predicted the target genes of miR-30a, and the predicted results were compared with the DEGs on GSE26712 and GSE4122; B, C Differential expression of SOX9 and NT5E on GSE26712 and GSE4122; D TargetScan software predicted the potential binding sites of miR-30a to SOX9; E Luciferase activity detected the targeting relationship between miR-30a and SOX9; vs the mimic-NC cells, **p < 0.01; F-G: Relative SOX9 expression in CD133+ OCSCs detected by RT-qPCR and Western blot analysis, vs mimic-NC cells, *p < 0.05; H. Relative expression of SOX9 and miR-30a in CD133+ OCSCs after silencing LINC00115 detected by RT-qPCR. Compared with the Ctrl group, *p < 0.05, **p < 0.01. Data in panels E–G were analyzed using the unpaired t test, while data in panel H were analyzed using one-way ANOVA and Tukey’s multiple comparison test. The experiment was performed in triplicate and data were expressed as means ± standard deviation
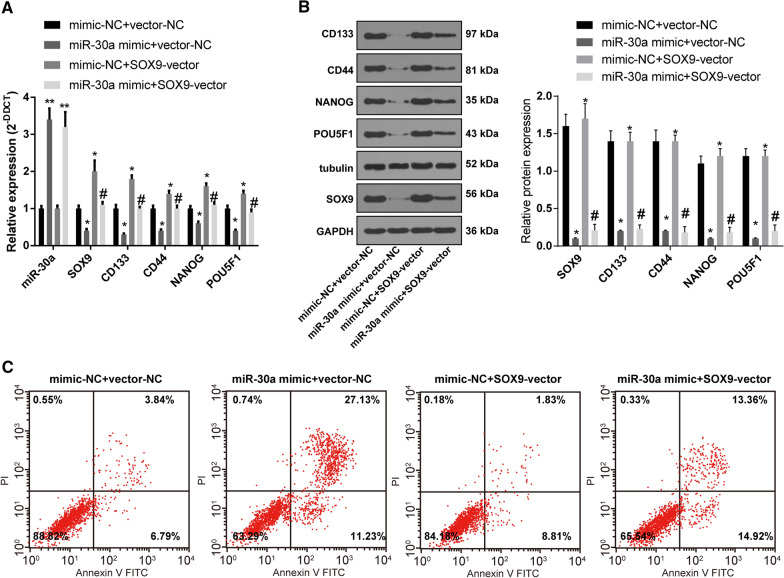
miR-30a inhibits OCSC stemness and promotes apoptosis by targeting SOX9
Based on the above results, miR-30a and SOX9 were both overexpressed in CD133+ OCSC respectively. The results of RT-qPCR, Western blot analysis and flow cytometry showed that compared with the mimic-NC + vector-NC group, miR-30a expression and apoptotic rate were increased, and levels of SOX9, CD133, CD44, NANOG and POU5F1 were decreased significantly in OCSCs treated with miR-30a mimic + vector-NC, while opposite trends were exhibited in the cells treated with mimic-NC + SOX9-vector. The levels of SOX9, CD133, CD44, NANOG and POU5F1 were evidently elevated while apoptotic rate was declined in the miR-30a mimic + SOX9-vector group relative to those in the miR-30a mimic + vector-NC group (Fig. 5A-C) (all p < 0.05). In conclusion, miR-30a can target SOX9 to inhibit maintenance and promote apoptosis of OCSCs.
Fig. 5.
miR-30a can target SOX9 to inhibit maintenance and promote apoptosis of OCSCs. A, B Levels of miR-30a SOX9, CD133, CD44, NANOG and POU5F1 in OCSCs were detected by RT-qPCR and Western blot analysis; C. Cell apoptosis was measured by flow cytometry. VS the mimic-NC + vector-NC group, *p < 0.05, **p < 0.01; vs the miR-30a mimic + vector-NC group, # p < 0.05. Data in panels A and B were analyzed using one-way ANOVA and Tukey’s multiple comparison test. The experiment was performed in triplicate and data were expressed as means ± standard deviation
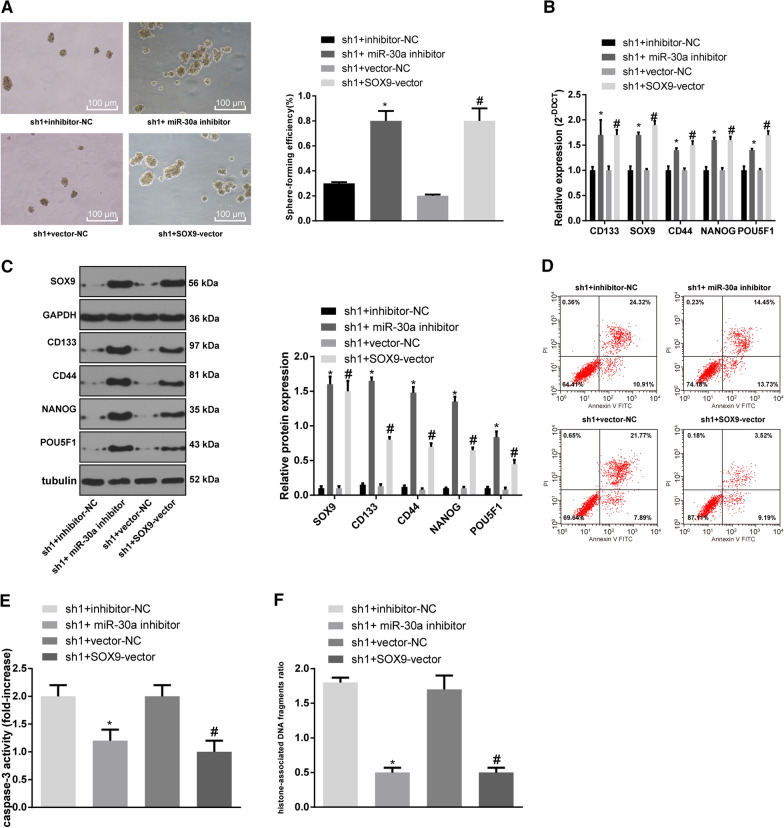
LINC00115 participates in the maintenance of OCSC stemness through the miR-30a/SOX9 axis
The sh1 with higher interference efficiency was selected to co-transfect with miR-30a inhibitor or SOX9 vector for subsequent functional rescue experiments. Sphere-forming assay was then performed to evaluate OCSCs’ capability of sphere formation. Compared with the OCSCs treated with sh1 + inhibitor-NC, the sphere-forming rate of OCSCs treated with sh1 + miR-30a inhibitor was increased significantly, and that of sh1 + SOX9-vector treatment cells was increased relative to the sh1 + vector-NC treatment (Fig. 6A) (both p < 0.05). The results of RT-qPCR, Western blot analysis and flow cytometry elicited that compared with the sh-LINC00115 + inhibitor-NC group, the levels of stemness-related genes (CD133, SOX9, CD44, NANOG, POU5F1) in the sh1 + miR-30a inhibitor group were significantly elevated, while apoptotic rate was reduced; relative to the sh1 + vector-NC group, the levels of stemness-related genes in the sh1 + SOX9-vector group were enhanced while apoptotic rate was decreased significantly (Fig. 6B–D) (all p < 0.05). In addition, compared with the sh1 + inhibitor-NC group, Caspase-3 activity and histone-related DNA damage in the sh1 + miR-30a inhibitor group were significantly reduced. Relative to the sh1 + vector-NC group, Caspase-3 activity and histone-related DNA damage in the sh1 + SOX9-vector group were noticeably decreased (Fig. 6EF) (all p < 0.05). In brief, LINC00115 participated in the maintenance and apoptosis of OCSCs through modulating the miR-30a/SOX9 axis.
Fig. 6.
LINC00115 participates in the maintenance and apoptosis of OCSCs through the miR-30a/SOX9 axis. A Microscopic observation of spheres (100 ×) and sphere-forming rate of OCSCs measured by sphere-forming assay; B, C Relative levels of CD133, SOX9, CD44, NANOG, and POU5F1 were detected by RT-qPCR and Western blot analysis; D Cell apoptosis was measured by flow cytometry; E Relative Caspase-3 activity; F Relative percentage of DNA damage in cell supernatant of each group. VS the sh1 + inhibitor-NC group, * p < 0.05; vs the sh1 + vector-NC group, # p < 0.05. Data were analyzed using one-way ANOVA and Tukey’s multiple comparison test. The experiment was performed in triplicate and data were expressed as means ± standard deviation
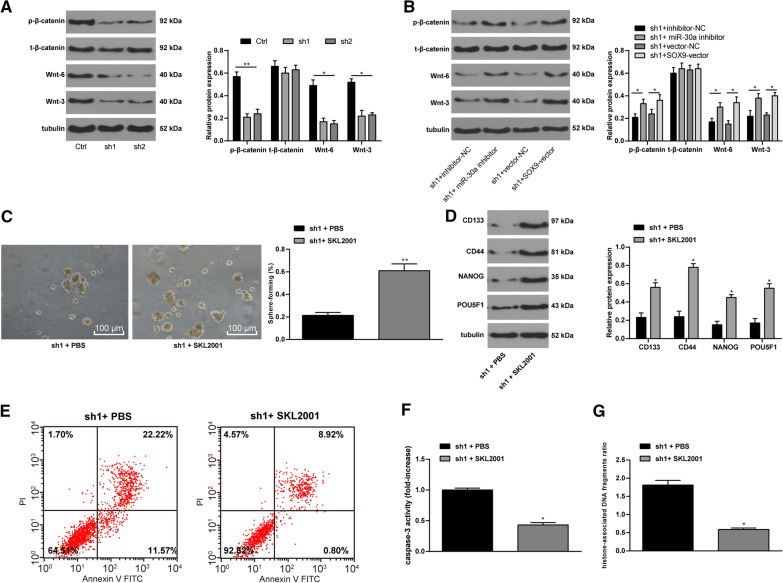
LINC00115 upregulates the Wnt/β-catenin pathway by regulating the miR-30a/SOX9 axis
Wnt/β-catenin pathway is involved in OC development and the maintenance of OCSC stemness [29]. Then, we detected the expression of Wnt/β-catenin pathway in OCSCs in the Ctrl, sh1, and sh2 groups. It revealed that silencing LINC00115 resulted in the down-regulation of Wnt/β-catenin in OCSCs (Fig. 7A). Subsequently, the stable sh1-transfected OCSCs was delivered with miR-30a inhibitor or SOX9 vector. Expectedly, further silencing of miR-30a or overexpression of SOX9 partly averted the levels of sh1-alerted Wnt/β-catenin (Fig. 7B) (all p < 0.05). Therefore, to further verify the role of Wnt/β-catenin pathway in the maintenance of OCSC stemness, we added Wnt/β-catenin specific activator SKL2001 to sh1-treated OCSCs [30–32]. It turned out that the activity and stemness of OCSCs were reversed partially, and Caspase-3 activity and DNA damage were decreased (Fig. 7C-G) (all p < 0.05).
Fig. 7.
Wnt/β-catenin pathway participates in the maintenance of OCSCs. OCSCs stably transfected with sh1 were delivered miR-30a inhibitor or SOX9-vector, or 5 mM Wnt/β-catenin specific activator SKL2001. A, B. Western blot analysis detected the expression of Wnt/β-catenin pathway; C. Microscopic observation of spheres (100 ×) and sphere-forming rate of OCSCs measured by sphere-forming assay; D Relative levels of CD133, SOX9, CD44, NANOG, and POU5F1 in CD133+ OCSCs were measured using Western blot analysis; E Cell apoptosis was measured by flow cytometry; F Relative Caspase-3 activity; G. Relative percentage of DNA damage in cell supernatant of each group. VS the sh1 + PBS group, * p < 0.05. Data in panels A and B were analyzed using one-way ANOVA and Tukey’s multiple comparison test, while data in panels C and F were analyzed using the unpaired t test. The experiment was performed in triplicate and data were expressed as means ± standard deviation
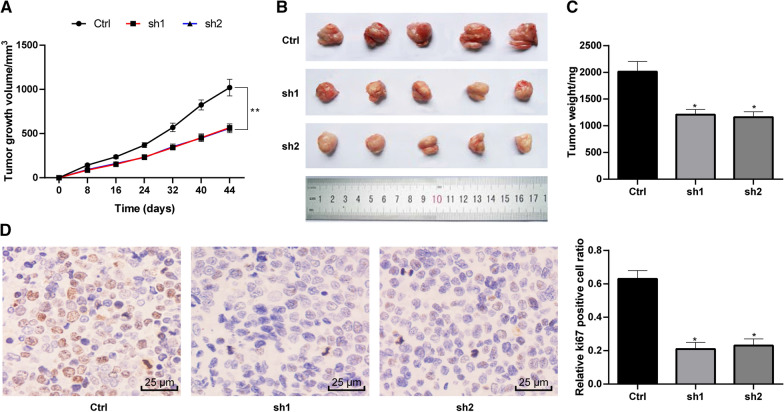
LINC00115 promotes OCSC growth in vivo
In animal experiments, we found that intervention with LINC00115 could slow down the growth rate of OCSCs (Fig. 8A-C) in nude mice. Immunohistochemical staining of Ki67 showed that LINC00115 also reduced Ki67 positive cells (Fig. 8D) (p < 0.05).
Fig. 8.
LINC00115 promotes OCSC growth in vivo. Nude mice were randomly assigned into three groups, 6 mice in each group, namely Ctrl group (injected with OCSCs), sh1 group (injected with sh1-treated OCSCs) and sh2 group (injected with sh2-treated OCSCs). A–C In vivo OCSC growth (including tumor size and weight) in nude mice transplanted with tumor cells; D Immunohistochemical staining of Ki67. Data were analyzed using one-way ANOVA and Tukey’s multiple comparison test. VS the Ctrl group, *p < 0.05; **p < 0.01
Discussion
In spite of improvements in treatment of OC in the past several decades, its prognosis is still undesirable, which is largely due to chemotherapeutic resistance because of the OCSC properties [33]. Therefore, identifying the potential mechanisms for inducing and maintaining OCSC properties will improve the efficacy and prognosis of OC patients undergoing chemotherapy. In this study, we probed the interactions among LINC00115, miR-30a and SOX9 in OCSCs. Collectively, we came to a conclusion that LINC00115 could sponge miR-30a to upregulate SOX9 and the Wnt/β-catenin pathway, thus promoting OCSC stemness and preventing apoptosis (Fig. 9).
Fig. 9.
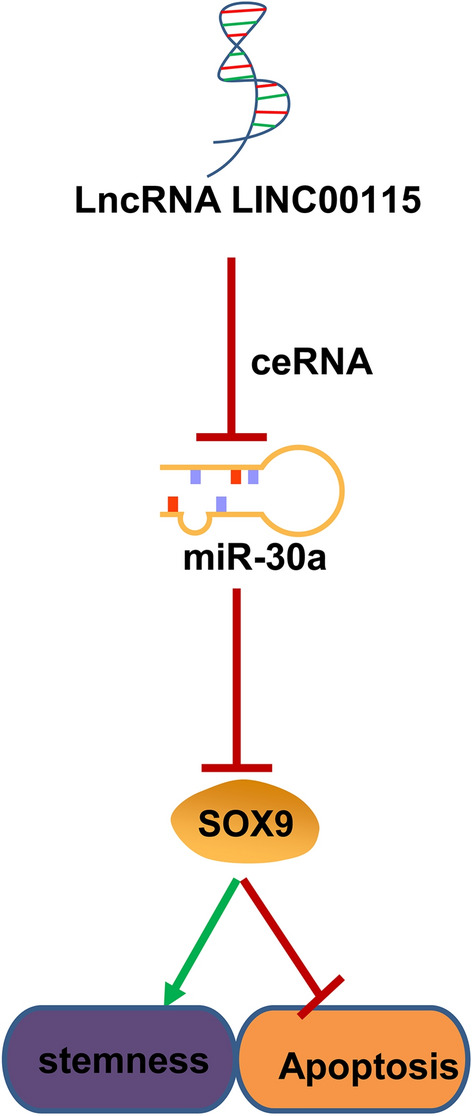
LINC00115 sponges miR-30a to regulate SOX9 expression and participates in the maintenance of ovarian cancer stem cell stemness and anti-apoptosis
Firstly, we found the expression of CD133, CD44, NANOG and POU5F1 in A2780 cells and CD133+ OCSCs was significantly enhanced. CD133+ and spheroid formation are reported to the main features of OCSCs [34]. Cioffi et al. demonstrated that CD133+ OC cells have stronger drug resistance, tumor metastasis, and OCSC sphere formation [35]. Michela Lupia et al. recorded that the recognized cancer-initiating cell markers, CD44, NANOG, POU5F1 and SOX2, were at higher levels in OC cells, which may drive tumor onset and recurrence, thus contributing to the high death/incidence ratio of OC [22]. In addition, LINC00115 was highly expressed in CD133+ OCSCs, and silencing LINC00115 could significantly reduce the expression of CD133, CD44, NANOG and POU5F1, increase apoptotic rate, Caspase-3 activity and histone-related DNA damage. The stemness-associated genes such as NANOG, POU5F1/OCT4, and SOX2, encode transcription factors that drive self-renewal and multipotency and are associated with OC progression and chemoresistance and poor prognosis [36, 37]. These results suggested that LINC00115 could promote the maintenance of stem cell viability and inhibit apoptosis of OCSCs.
Additionally, through the dual-luciferase reporter gene assay, RNA pull-down assay and RIP assay, we found LINC00115 could bind to miR-30a, and miR-30a could target SOX9. Evidence indicated that overexpression of miR-30 inhibited the self-renewal capability of breast tumor-initiating cells and induced apoptosis [38]. Another study has revealed that miR-30a showed weak expression in OC, and miR-30a-5p showed weak expression in cisplatin-resistant OC cells [28]. Similarly, higher expression of miR-30 family was associated with elevated overall survival and progression-free survival [39], and provided diagnostic, prognostic and therapeutic approaches for different types of OC patients [40]. Similarly, elevated level of SOX2 enhanced tumor sphere potential, the levels of stemness-related genes, and tumor-initiating capacity, but inhibited OCSC apoptosis [36]. SOX9, as a stem cell regulator, is necessary for stem cell maintenance, and might encourage new methods for treating stem cell-caused diseases and cancers [41]. Abnormal SOX9 expression has been demonstrated to promote OC cell survival and drug resistance in hypoxic condition and to affect OC aggressiveness [42]. Michelle K. Y. Siu et al. found that downregulation of SOX9 was related to linited migration, invasion, and sphere-forming abilities, and diminished stemness of OC cells [43]. The interaction between miR-30a and SOX9 was also found in osteoarthritis, in which miR-30a promoted extracellular matrix degradation by targeting SOX9 [44]. Furthermore, SOX9, CD133, CD44, NANOG and POU5F1 levels were decreased significantly in OCSCs treated with miR-30a mimic or LINC00115 inhibitor. LINC00115 silencing sponged miR-30a to downregulate SOX9, thus inhibiting stemness and promoting apoptosis of OCSCs. Similar to our study, it is reported that LINC00115 might also act as a ceRNA to sponge miR-7 and regulate fibroblast growth factor 2 in lung adenocarcinoma [12].
Moreover, Wnt/β-catenin pathway promoted stemness and repressed apoptosis of OCSCs. It is well-recognized that the Wnt/β-catenin pathway is pivotal to induce and maintain OCSC features, and induce stemness and chemoresistance [33]. Haibo Pan et al. demonstrated that suppression of the Wnt/β-catenin pathway was beneficial to reduce tumor sphere formation and stemness and eliminate OCSCs [12]. Interestingly, SOX9 seems to be a target of Wnt, and low levels of SOX9 regulate stem cell proliferation in a Wnt-dependent manner [45]. A recent study has demonstrated that overexpression of miR-30-5p reduced sphere formation rate, cell viability, cell stemness, inhibits chemoresistance in colorectal cancer cells through the Wnt pathway [46].
To summarize, this is an innovative study investigating the ceRNA network of LINC00115/miR-30a/SOX9 in OCSCs. We identified that silencing LINC00115 downregulated SOX9 and inactivated the Wnt/β-catenin pathway through competitively binding to miR-30a, thus maintaining OCSC stemness and promoting the apoptosis. Our research may provide a new perspective to evaluate the function of LINC00115 in OCSCs through an alteration in miR expression profiles, and thus offering cancer prevention and therapeutic strategies. Further investigations should be performed to validate our findings and figure out the clinical values.
Acknowledgements
Not applicable
Abbreviations
- ANOVA
Analysis of variance
- APC
Antigen presenting cells
- bFGF
Basic fibroblast growth factor
- ceRNA
Competing endogenous RNA
- EDTA
Ethylene diamine tetraacetic acid
- EGF
Epidermal growth factor
- FITC
Fluorescein isothiocyanate
- GAPDH
Glyceraldehyde-3-phosphate dehydrogenase
- lncRNA
Long non-coding RNA
- MEBM
Mammary epithelial basal medium; miRs: microRNAs
- MUT
Mutant type
- NC
Negative control
- NT5E
Ecto-5'-nucleotidase
- OC
Ovarian
Authors' contributions
LJ contributed to the study concepts, study design; RH contributed to the literature research, clinical studies, data acquisition, manuscript preparation; LJ and RH contributed to the definition of intellectual content, experimental studies, data analysis, statistical analysis, manuscript editing, manuscript review. Both authors read and approved the final manuscript.
Funding
This work was partially supported by Scientific Research Project of Liaoning Provincial Education Department (JCZR2020011) and 345 Talent Project of Shengjing Hospital.
Availability of data and materials
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Declarations
Ethics approval and consent to participate
This study was approved by the Clinical Ethical Committee of Shengjing Hospital of China Medical University. All animal experiments were complied with the ARRIVE guidelines and carried out in accordance with the National Institutes of Health guide for the care and use of Laboratory animals.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
This article has been retracted. Please see the retraction notice for more detail: https://doi.org/10.1186/s12935-024-03561-5
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
11/8/2024
This article has been retracted. Please see the Retraction Notice for more detail: 10.1186/s12935-024-03561-5
References
- 1.Kuroki L, Guntupalli SR. Treatment of epithelial ovarian cancer. BMJ. 2020;371:m3773. [DOI] [PubMed] [Google Scholar]
- 2.Lheureux S, Gourley C, Vergote I, Oza AM. Epithelial ovarian cancer. Lancet. 2019;393(10177):1240–53. [DOI] [PubMed] [Google Scholar]
- 3.Menon U, Karpinskyj C, Gentry-Maharaj A. Ovarian Cancer Prevention and Screening. Obstet Gynecol. 2018;131(5):909–27. [DOI] [PubMed] [Google Scholar]
- 4.Rasmussen CB, Jensen A, Albieri V, Andersen KK, Kjaer SK. Is pelvic inflammatory disease a risk factor for ovarian cancer? Cancer Epidemiol Biomarkers Prev. 2017;26(1):104–9. [DOI] [PubMed] [Google Scholar]
- 5.Taniguchi F. New knowledge and insights about the malignant transformation of endometriosis. J Obstet Gynaecol Res. 2017;43(7):1093–100. [DOI] [PubMed] [Google Scholar]
- 6.Ugai T, Kelemen LE, Mizuno M, Ong JS, Webb PM, Chenevix-Trench G, Australian Ovarian Cancer Study G, Wicklund KG, Doherty JA, Rossing MA et al: Ovarian cancer risk, ALDH2 polymorphism and alcohol drinking: Asian data from the Ovarian Cancer Association Consortium. Cancer Sci 2018, 109(2):435–445. [DOI] [PMC free article] [PubMed]
- 7.Bull CJ, Yarmolinsky J, Wade KH. Commentary: Mendelian randomization analysis identifies circulating vitamin D as a causal risk factor for ovarian cancer. Int J Epidemiol. 2016;45(5):1631–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tung SL, Huang WC, Hsu FC, Yang ZP, Jang TH, Chang JW, Chuang CM, Lai CR, Wang LH. miRNA-34c-5p inhibits amphiregulin-induced ovarian cancer stemness and drug resistance via downregulation of the AREG-EGFR-ERK pathway. Oncogenesis. 2017;6(5):e326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang C, Li B, Yu J, Yang F, Cai K, Chen Z. Ultrasound microbubbles mediated miR-let-7b delivery into CD133(+) ovarian cancer stem cells. Biosci Rep. 2018;38:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lu MY, Liao YW, Chen PY, Hsieh PL, Fang CY, Wu CY, Yen ML, Peng BY, Wang DP, Cheng HC, et al. Targeting LncRNA HOTAIR suppresses cancer stemness and metastasis in oral carcinomas stem cells through modulation of EMT. Oncotarget. 2017;8(58):98542–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mitra R, Chen X, Greenawalt EJ, Maulik U, Jiang W, Zhao Z, Eischen CM. Decoding critical long non-coding RNA in ovarian cancer epithelial-to-mesenchymal transition. Nat Commun. 2017;8(1):1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li DS, Ainiwaer JL, Sheyhiding I, Zhang Z, Zhang LW. Identification of key long non-coding RNAs as competing endogenous RNAs for miRNA-mRNA in lung adenocarcinoma. Eur Rev Med Pharmacol Sci. 2016;20(11):2285–95. [PubMed] [Google Scholar]
- 13.Zhou B, Xu H, Xia M, Sun C, Li N, Guo E, Guo L, Shan W, Lu H, Wu Y, et al. Overexpressed miR-9 promotes tumor metastasis via targeting E-cadherin in serous ovarian cancer. Front Med. 2017;11(2):214–22. [DOI] [PubMed] [Google Scholar]
- 14.Wu Q, Guo R, Lin M, Zhou B, Wang Y. MicroRNA-200a inhibits CD133/1+ ovarian cancer stem cells migration and invasion by targeting E-cadherin repressor ZEB2. Gynecol Oncol. 2011;122(1):149–54. [DOI] [PubMed] [Google Scholar]
- 15.Tsukasa K, Ding Q, Miyazaki Y, Matsubara S, Natsugoe S, Takao S. miR-30 family promotes migratory and invasive abilities in CD133(+) pancreatic cancer stem-like cells. Hum Cell. 2016;29(3):130–7. [DOI] [PubMed] [Google Scholar]
- 16.Zhang Z, Sun L, Zhang Y, Lu G, Li Y, Wei Z. Long non-coding RNA FEZF1-AS1 promotes breast cancer stemness and tumorigenesis via targeting miR-30a/Nanog axis. J Cell Physiol. 2018;233(11):8630–8. [DOI] [PubMed] [Google Scholar]
- 17.Wang L, Zhao S, Yu M. Mechanism of Low Expression of miR-30a-5p on Epithelial-Mesenchymal Transition and Metastasis in Ovarian Cancer. DNA Cell Biol. 2019;38(4):341–51. [DOI] [PubMed] [Google Scholar]
- 18.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120(1):15–20. [DOI] [PubMed] [Google Scholar]
- 19.LaFace DM, Peck AB. Reciprocal allogeneic bone marrow transplantation between NOD mice and diabetes-nonsusceptible mice associated with transfer and prevention of autoimmune diabetes. Diabetes. 1989;38(7):894–901. [DOI] [PubMed] [Google Scholar]
- 20.Qiu L, Wang J, Chen M, Chen F, Tu W. Exosomal microRNA146a derived from mesenchymal stem cells increases the sensitivity of ovarian cancer cells to docetaxel and taxane via a LAMC2mediated PI3K/Akt axis. Int J Mol Med. 2020;46(2):609–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wong N, Wang X. miRDB: an online resource for microRNA target prediction and functional annotations. Nucleic Acids Res. 2015;43(Database issue):146–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lupia M, Angiolini F, Bertalot G, Freddi S, Sachsenmeier KF, Chisci E, Kutryb-Zajac B, Confalonieri S, Smolenski RT, Giovannoni R, et al. CD73 Regulates stemness and epithelial-mesenchymal transition in ovarian cancer-initiating cells. Stem Cell Rep. 2018;10(4):1412–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ghuwalewala S, Ghatak D, Das P, Dey S, Sarkar S, Alam N, Panda CK, Roychoudhury S. CD44(high)CD24(low) molecular signature determines the Cancer Stem Cell and EMT phenotype in Oral Squamous Cell Carcinoma. Stem Cell Res. 2016;16(2):405–17. [DOI] [PubMed] [Google Scholar]
- 24.Lee HG, Shin SJ, Chung HW, Kwon SH, Cha SD, Lee JE, Cho CH. Salinomycin reduces stemness and induces apoptosis on human ovarian cancer stem cell. J Gynecol Oncol. 2017;28(2):e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Campbell PS, Mavingire N, Khan S, Rowland LK, Wooten JV, Opoku-Agyeman A, Guevara A, Soto U, Cavalli F, Loaiza-Perez AI, et al. AhR ligand aminoflavone suppresses alpha6-integrin-Src-Akt signaling to attenuate tamoxifen resistance in breast cancer cells. J Cell Physiol. 2018;234(1):108–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jeggari A, Marks DS, Larsson E. miRcode: a map of putative microRNA target sites in the long non-coding transcriptome. Bioinformatics. 2012;28(15):2062–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin Y, Liu T, Cui T, Wang Z, Zhang Y, Tan P, Huang Y, Yu J, Wang D. RNAInter in 2020: RNA interactome repository with increased coverage and annotation. Nucleic Acids Res. 2020;48(D1):D189–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Han X, Zhen S, Ye Z, Lu J, Wang L, Li P, Li J, Zheng X, Li H, Chen W, et al. A feedback loop between miR-30a/c-5p and DNMT1 mediates cisplatin resistance in ovarian cancer cells. Cell Physiol Biochem. 2017;41(3):973–86. [DOI] [PubMed] [Google Scholar]
- 29.Marson A, Foreman R, Chevalier B, Bilodeau S, Kahn M, Young RA, Jaenisch R. Wnt signaling promotes reprogramming of somatic cells to pluripotency. Cell Stem Cell. 2008;3(2):132–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li B, Cao Y, Meng G, Qian L, Xu T, Yan C, Luo O, Wang S, Wei J, Ding Y, et al. Targeting glutaminase 1 attenuates stemness properties in hepatocellular carcinoma by increasing reactive oxygen species and suppressing Wnt/beta-catenin pathway. EBioMedicine. 2019;39:239–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qiao W, Wang H, Zhang X, Luo K. Proline-rich protein 11 silencing inhibits hepatocellular carcinoma growth and epithelial-mesenchymal transition through beta-catenin signaling. Gene. 2019;681:7–14. [DOI] [PubMed] [Google Scholar]
- 32.Su Z, Wang C, Chang D, Zhu X, Sai C, Pei J. Limonin attenuates the stemness of breast cancer cells via suppressing MIR216A methylation. Biomed Pharmacother. 2019;112:108699. [DOI] [PubMed] [Google Scholar]
- 33.Ruan X, Liu A, Zhong M, Wei J, Zhang W, Rong Y, Liu W, Li M, Qing X, Chen G, et al. Silencing LGR6 attenuates stemness and chemoresistance via inhibiting Wnt/beta-catenin signaling in ovarian cancer. Mol Ther Oncolytics. 2019;14:94–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nam EJ, Lee M, Yim GW, Kim JH, Kim S, Kim SW, Kim YT. MicroRNA profiling of a CD133(+) spheroid-forming subpopulation of the OVCAR3 human ovarian cancer cell line. BMC Med Genomics. 2012;5:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cioffi M, D’Alterio C, Camerlingo R, Tirino V, Consales C, Riccio A, Ierano C, Cecere SC, Losito NS, Greggi S, et al. Identification of a distinct population of CD133(+)CXCR4(+) cancer stem cells in ovarian cancer. Sci Rep. 2015;5:10357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bareiss PM, Paczulla A, Wang H, Schairer R, Wiehr S, Kohlhofer U, Rothfuss OC, Fischer A, Perner S, Staebler A, et al. SOX2 expression associates with stem cell state in human ovarian carcinoma. Cancer Res. 2013;73(17):5544–55. [DOI] [PubMed] [Google Scholar]
- 37.Peng S, Maihle NJ, Huang Y. Pluripotency factors Lin28 and Oct4 identify a sub-population of stem cell-like cells in ovarian cancer. Oncogene. 2010;29(14):2153–9. [DOI] [PubMed] [Google Scholar]
- 38.Yu F, Deng H, Yao H, Liu Q, Su F, Song E. Mir-30 reduction maintains self-renewal and inhibits apoptosis in breast tumor-initiating cells. Oncogene. 2010;29(29):4194–204. [DOI] [PubMed] [Google Scholar]
- 39.Shi M, Mu Y, Zhang H, Liu M, Wan J, Qin X, Li C. MicroRNA-200 and microRNA-30 family as prognostic molecular signatures in ovarian cancer: A meta-analysis. Medicine (Baltimore). 2018;97(32):e11505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhao H, Ding Y, Tie B, Sun ZF, Jiang JY, Zhao J, Lin X, Cui S. miRNA expression pattern associated with prognosis in elderly patients with advanced OPSC and OCC. Int J Oncol. 2013;43(3):839–49. [DOI] [PubMed] [Google Scholar]
- 41.Jo A, Denduluri S, Zhang B, Wang Z, Yin L, Yan Z, Kang R, Shi LL, Mok J, Lee MJ, et al. The versatile functions of Sox9 in development, stem cells, and human diseases. Genes Dis. 2014;1(2):149–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Raspaglio G, Petrillo M, Martinelli E, Li Puma DD, Mariani M, De Donato M, Filippetti F, Mozzetti S, Prislei S, Zannoni GF, et al. Sox9 and Hif-2alpha regulate TUBB3 gene expression and affect ovarian cancer aggressiveness. Gene. 2014;542(2):173–81. [DOI] [PubMed] [Google Scholar]
- 43.Siu MKY, Jiang YX, Wang JJ, Leung THY, Han CY, Tsang BK, Cheung ANY, Ngan HYS, Chan KKL. Hexokinase 2 regulates ovarian cancer cell migration, invasion and stemness via FAK/ERK1/2/MMP9/NANOG/SOX9 Signaling Cascades. Cancers (Basel). 2019;11:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chang T, Xie J, Li H, Li D, Liu P, Hu Y. MicroRNA-30a promotes extracellular matrix degradation in articular cartilage via downregulation of Sox9. Cell Prolif. 2016;49(2):207–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Formeister EJ, Sionas AL, Lorance DK, Barkley CL, Lee GH, Magness ST. Distinct SOX9 levels differentially mark stem/progenitor populations and enteroendocrine cells of the small intestine epithelium. Am J Physiol Gastrointest Liver Physiol. 2009;296(5):G1108-1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jiang S, Miao D, Wang M, Lv J, Wang Y, Tong J. MiR-30-5p suppresses cell chemoresistance and stemness in colorectal cancer through USP22/Wnt/beta-catenin signaling axis. J Cell Mol Med. 2019;23(1):630–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.