Abstract
Plants of the genus Allium developed a diversity of defense mechanisms against pathogenic fungi of the genus Fusarium, including transcriptional activation of pathogenesis-related (PR) genes. However, the information on the regulation of PR factors in garlic (Allium sativum L.) is limited. In the present study, we identified AsPR genes putatively encoding PR1, PR2, PR4, and PR5 proteins in A. sativum cv. Ershuizao, which may be involved in the defense against Fusarium infection. The promoters of the AsPR1–5 genes contained jasmonic acid-, salicylic acid-, gibberellin-, abscisic acid-, auxin-, ethylene-, and stress-responsive elements associated with the response to plant parasites. The expression of AsPR1c, d, g, k, AsPR2b, AsPR5a, c (in roots), and AsPR4a(c), b, and AsPR2c (in stems and cloves) significantly differed between garlic cultivars resistant and susceptible to Fusarium rot, suggesting that it could define the PR protein-mediated protection against Fusarium infection in garlic. Our results provide insights into the role of PR factors in A. sativum and may be useful for breeding programs to increase the resistance of Allium crops to Fusarium infections.
Keywords: garlic Allium sativum L., pathogenesis-related proteins, biotic stress, Fusarium spp., gene structure, gene expression
1. Introduction
Plants constantly exposed to various pathogens use complex defense mechanisms developed during evolution [1,2,3]. One of the most harmful pathogens is fungi, which developed many sophisticated mechanisms to penetrate and colonize host cells, including hydrolysis of the plant cell wall with pectinases, cellulases, and proteases [4]. The plant cell wall, containing cutin, wax, and lignin, represents the first line of defense against invading pathogens [2]. If it is broken, the plant immune system, which is the second line of defense, is activated through plant receptors that perceive pathogen-associated molecular patterns, including flagellins and components of the fungal cell wall, such as lipopolysaccharides, chitin, and branched β-glucans [5], as well as through the mobilization of resistance-related proteins that recognize pathogen effectors [6]. The result is the induction of the plant hypersensitive response, including the formation of reactive oxygen species (ROS), the induction of salicylic acid (SA) and jasmonic acid (JA) signaling, and the upregulation of pathogenesis-related (PR) genes [3]. The SA pathway, triggered mainly by biotrophic pathogens, is involved in systemic acquired resistance (SAR), which induces the expression of the PR1, PR2, and PR5 genes and prevents the spread of infection to healthy tissues [2], whereas the JA pathway, mainly activated by necrotrophic pathogens, provides local acquired resistance (LAR) through the upregulation of PR3, PR4, and PR12 genes [2].
The PR factors are thermostable, protease-resistant proteins of ~5–43 kDa which are expressed in all plant organs; in the leaves, they constitute about 5–10% of the total protein [7,8]. Currently, 17 PR families differing in structure, mechanisms of action, and specificity to parasites (fungi, bacteria, viruses, insects, or nematodes) are known [9,10]. Among them, PR1–5, PR12, and PR17, such as CAP-domain proteins (pfam00188; PR1), β-1,3-glucanases (PR2), chitinases (PR3), Barwin-domain proteins (PR4), thaumatin-like (PR5), and NtPRp27-like (PR17) proteins, as well as antimicrobial peptides, including defensins (PR12), were shown to be involved in the response to fungal attack [2,7,10,11].
Fusarium pathogens, which are widely spread in the soils of almost all climate zones, vary in nutrition type and cause two of the most destructive fungal diseases, Fusarium basal rot (FBR) and Fusarium wilt (FW), in diverse crop and wild plant species [12,13,14]. Furthermore, they produce multiple mycotoxins that are harmful to humans [15]. Therefore, the mechanisms of protection against Fusarium invasion have been extensively studied both in model plants [16] and in crops such as potato [17,18], tomato [19], and cereals [20,21], and it was found that PR2–5, PR8, and PR11–13 are involved in plant responses to Fusarium infection [2,17,22,23,24].
Fusarium-caused diseases are responsible for significant losses in the yield of garlic (Allium sativum L.)—the second most important bulbous crop in the world, with a production volume of over 30 million tons per year (http://www.fao.org/ (accessed on 5 May 2021)). During garlic cultivation and storage, Fusarium infection may lead to FBR of the bulbs and/or FW of the leaves [14,25]. Both diseases are caused by a specific isolate of F. oxysporum (f. sp. cepae) as well as by other Fusarium species, such as F. acutatum, F. anthophilium, and F. proliferatum [14,26]. Comparative studies of FBR-resistant and susceptible Allium genotypes have disclosed some of the possible biochemical and genetic mechanisms underlying plant susceptibility to FBR [27,28]. In our previous study, we have shown that garlic chitinases belonging to the PR3 family are involved in the immune response against Fusarium infection [28]. However, there is little information regarding the immune defense function of other PR proteins in garlic [29,30].
In the present study, we performed in silico identification and characterization of the genes belonging to the PR1, PR2, PR4, and PR5 families in the A. sativum genome and studied their tissue expression patterns in FBR-resistant and susceptible garlic cultivars infected with F. proliferatum.
2. Results
2.1. Identification of PR1, PR2, PR4, and PR5 Genes in the A. sativum Genome
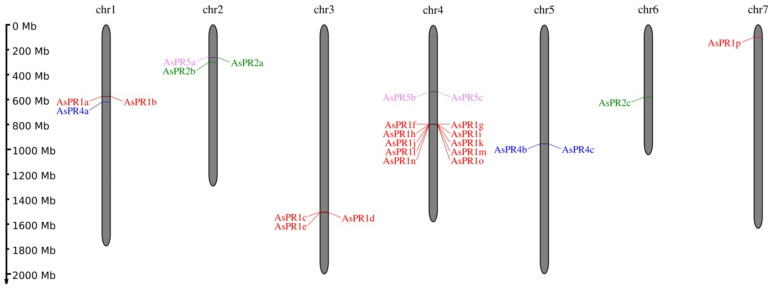
In silico analysis of the A. sativum cv. Ershuizao genome data (PRJNA606385) [31] revealed 16 genes belonging to the PR1 family, and 3 genes in each of the PR2, PR4, and PR5 families (Table 1). The PR1 genes were identified on chromosomes 1 (AsPR1a, b), 3 (AsPR1c-e), 4 (AsPR1f-o, clustered in the 3.34 Mbp region), and 7 (AsPR1p). The PR2 genes were localized on chromosomes 2 (AsPR2a, b) and 6 (AsPR2c), the PR4 genes on chromosomes 1 (AsPR4a) and 5 (AsPR4b, c), and the PR5 genes on chromosomes 2 (AsPR5a) and 4 (AsPR5b, c) (Table 1, Figure 1).
Table 1.
PR1, PR2, PR4, and PR5 genes identified in the genome of A. sativum cv. Ershuizao.
| Gene | Localization | Length (bp) | Number of Exons |
CDS (bp) | Protein (aa) | Transcript ID [31] |
|---|---|---|---|---|---|---|
| PR1 family | ||||||
| AsPR1a | ch1:597808382-597808867 (+) | 486 | 1 | 486 | 161 | Asa1G02133.1 |
| AsPR1b | ch1:597852310-597852792 (+) | 483 | 1 | 483 | 160 | Asa1G02134.1 |
| AsPR1c | ch3:1569741035-1569741535(−) | 501 | 1 | 501 | 166 | Asa3G05742.1 |
| AsPR1d | ch3:1574050049-1574050546 (+) | 498 | 1 | 498 | 165 | Asa3G05767.1 |
| AsPR1e | ch3:1575214795-1575215280 (−) | 486 | 1 | 486 | 161 | Asa3G05770.1 |
| AsPR1f | ch4:828639577-828640059 (−) | 483 | 1 | 483 | 160 | Not detected |
| AsPR1g | ch4:828659513-828659995 (−) | 483 | 1 | 483 | 160 | Not detected |
| AsPR1h | ch4:829079571-829080095 (−) | 525 | 1 | 525 | 174 | Asa4G03112.1 |
| AsPR1i | ch4:829255226-829255750 (−) | 525 | 1 | 525 | 174 | Asa4G03113.1 |
| AsPR1j | ch4:829291631-829292125 (−) | 495 | 1 | 495 | 164 | not detected |
| AsPR1k | ch4:831436248-831436742 (−) | 495 | 1 | 495 | 164 | Asa4G03125.1 |
| AsPR1l | ch4:831555457-831555981 (+) | 525 | 1 | 525 | 174 | Asa4G03126.1 |
| AsPR1m | ch4:831614218-831614742 (+) | 525 | 1 | 525 | 174 | Asa4G03127.1 |
| AsPR1n | ch4:831977068-831977562 (+) | 495 | 1 | 495 | 164 | Asa4G03130.1 |
| AsPR1o | ch4:831982650-831983087 (+) | 438 | 1 | 438 | 145 | Asa4G03131.1 |
| AsPR1p | ch7:103854549-103855043(−) | 495 | 1 | 495 | 164 | Asa7G00352.1 |
| PR2 family | ||||||
| AsPR2a | ch2:272017011-272018179 (−) | 1169 | 2 | 1035 | 344 | Asa2G01057.1 |
| AsPR2b | ch2:311576151-311577239 (−) | 1089 | 2 | 990 | 329 | Asa2G01195.1 |
| AsPR2c | ch6:605851704-605853559 (−) | 1856 | 3 | 945 | 314 | Asa6G06180.1 |
| PR4 family | ||||||
| AsPR4a 1 | ch1:652401564-652402126 (+) | 563 | 2 | 444 | 147 | Asa1G02345.1 |
| AsPR4b | ch5:998122740-998123259 (+) | 520 | 2 | 444 | 147 | Asa5G03281.1 |
| AsPR4c 1 | ch5:999259985-999260547 (+) | 563 | 2 | 444 | 147 | Asa1G02345.1 |
| PR5 family | ||||||
| AsPR5a | ch2:266379091-266379753 (+) | 663 | 1 | 663 | 220 | Asa2G01043.1 |
| AsPR5b | ch4:561214533-561215195 (+) | 663 | 1 | 663 | 220 | Asa4G02099.1 |
| AsPR5c | ch4:561229259-561229921 (+) | 663 | 1 | 663 | 220 | Asa4G02100.1 |
1 These two sequences are 100% identical.
Figure 1.
Chromosomal locations of the AsPR1 (red), AsPR2 (green), AsPR4 (blue), and AsPR5 (purple) genes. The chromosome lengths indicated on the left are based on the A. sativum cv. Ershuizao genome (PRJNA606385) [31]; chr, chromosome.
The sizes of the predicted PR genes varied from 438 bp (AsPR1o) to 1856 bp (AsPR2c). The AsPR1 and AsPR5 genes did not contain introns, whereas the AsPR2 and AsPR4 genes contained 1 (AsPR4a-c, AsPR2a, and AsPR2b) or 2 (AsPR2c) introns (Table 1). The coding sequences (CDSs) of the AsPR genes ranged from 438 bp (AsPR1o) to 1035 bp (AsPR2a), corresponding to proteins from 145 to 344 amino acid residues.
Many of the identified PR genes within each PR family were highly identical: AsPR1a and b (94.0%), AsPR1c and e (91.4%), AsPR1f and g (98.8%), AsPR1h and m (96.0%), AsPR1i and l (99.2%), AsPR4a and c (100%), and AsPR5a and b (98.2%).
2.2. Characterization of Putative PR1, PR2, PR4, and PR5 Proteins in Garlic
The predicted AsPR proteins were characterized for their physicochemical properties and functional annotation in gene ontology terms (Table 2). The molecular weight (MW) ranged from 15.5–15.55 kDa in AsPR4 proteins to 33.53–37.33 kDa in AsPR2 proteins. AsPR1 proteins significantly differed in isoelectric point (pI) (from 4.5 in AsPR1f to 9.25 in AsPR1i), whereas AsPR5 proteins demonstrated close similarity both in pI (4.71–4.74) and MW. The aliphatic index (AI), reflecting the relative number of hydrophobic residues, ranged from 55.00 in AsPR5c to 94.01 in AsPR2a, and almost all AsPR proteins were predicted to be hydrophilic (GRAVY < 0).
Table 2.
Characteristics of PR1, PR2, PR4, and PR5 proteins predicted in A. sativum cv. Ershuizao.
| Protein Symbol | MW (kDa) | pI | AI | GRAVY | Signal Peptide | Catalytic Domain |
Subcellular Localization | Biological Process |
|---|---|---|---|---|---|---|---|---|
| PR1 family | ||||||||
| AsPR1a | 17.34 | 7.55 | 76.40 | 0.038 | 1–23 | CAP (31–149) | Secretory | Defense response (GO:0006952), response to biotic stimulus (GO:0009607) |
| AsPR1b | 17.15 | 6.78 | 77.50 | 0.065 | 1–22 | CAP (30–148) | ||
| AsPR1c | 17.84 | 5.99 | 72.77 | −0.135 | 1–27 | CAP (35–154) | ||
| AsPR1d | 17.60 | 6.14 | 75.64 | −0.155 | 1–26 | CAP (34–153) | ||
| AsPR1e | 17.14 | 6.48 | 68.94 | −0.329 | 1–26 | CAP (34–149) | ||
| AsPR1f | 17.51 | 4.50 | 63.44 | −0.319 | 1–19 | CAP (27–145) | ||
| AsPR1g | 17.45 | 4.63 | 63.44 | −0.300 | ||||
| AsPR1h | 19.38 | 5.33 | 61.15 | −0.533 | 1–23 | CAP (31–149) | ||
| AsPR1i | 19.62 | 9.25 | 71.72 | −0.487 | ||||
| AsPR1j | 18.10 | 7.58 | 61.34 | −0.412 | 1–19 | CAP (27–145) | ||
| AsPR1k | 18.04 | 8.20 | 61.34 | −0.393 | ||||
| AsPR1l | 19.58 | 9.21 | 71.72 | −0.485 | 1–23 | CAP (31–149) | ||
| AsPR1m | 19.35 | 6.41 | 64.54 | −0.501 | ||||
| AsPR1n | 18.07 | 8.20 | 59.57 | −0.407 | 1–19 | CAP (27–145) | ||
| AsPR1o | 16.05 | 7.59 | 68.69 | −0.246 | ||||
| AsPR1p | 18.24 | 6.27 | 85.00 | −0.271 | 1–25 | CAP (34–152) | ||
| PR2 family | ||||||||
| AsPR2a | 34.97 | 7.67 | 94.01 | 0.045 | 1–21 | GH17 (22–329) | Secretory | Carbohydrate metabolic process (GO:0005975) |
| AsPR2b | 37.33 | 5.14 | 92.44 | −0.021 | 1–29 | GH17 (30–343) | Secretory | |
| AsPR2c | 33.53 | 6.41 | 91.02 | −0.008 | n/d | GH17 (5–312) | Nucleus | |
| PR4 family | ||||||||
| AsPR4a | 15.55 | 5.54 | 73.74 | −0.078 | 1–25 | Barwin (31–145) | Secretory | Defense response to fungus (GO:0050832), defense response to bacterium (GO:0042742) |
| AsPR4b | 15.5 | 6.22 | 79.05 | −0.029 | ||||
| AsPR4c | 15.55 | 5.54 | 73.74 | −0.078 | ||||
| PR5 family | ||||||||
| AsPR5a | 23.46 | 4.71 | 58.09 | −0.085 | 1–21 | GH64-TLP-SF (28–220) | Secretory | Defense response (GO:0006952) |
| AsPR5b | 23.50 | 4.71 | 55.86 | −0.126 | ||||
| AsPR5c | 23.59 | 4.74 | 55.00 | −0.164 | ||||
MW, molecular weight; pI, isoelectric point; AI, aliphatic index; GRAVY, grand average hydropathy; n/d, not detected.
In terms of functional activity, the AsPR1, AsPR4, and AsPR5 proteins were associated with plant defense responses, whereas AsPR2 was associated with the carbohydrate metabolic process (Table 2). Except for AsPR2c (localized in the nucleus), all AsPR factors had an N-terminal signal peptide of 19–29 residues and were predicted to be secreted.
2.3. Conserved Domains in AsPR Proteins
All putative AsPR1 proteins contained a functional CAP domain (characteristic for cysteine-rich secretory proteins, antigen 5, and PR1 family members; pfam00188) (Table 2), as well as the conserved motif CxHYTQ[L/V]VWA[N/K]S[V/I]xIGC, which was also specific to the PR1 family [32]. The AsPR2 proteins contained the GH17 domain (glycosyl hydrolase family 17; pfam00332), the AsPR4 proteins contained the Barwin domain (pfam00967), and the AsPR5 proteins, the GH64-TLP-SF domain (glycoside hydrolase family 64 and thaumatin-like proteins superfamily; pfam00314) (Table 2).
2.4. Cis-Acting Elements in the Promoters of the AsPR1, AsPR2, AsPR4, and AsPR5 Genes
In total, 15 hormone-responsive cis-elements were identified: 1–12 in each AsPR gene (except AsPR4a), including elements involved in the response to abscisic acid (ABA) (AsPR1a, d, f, i, k, l, n, AsPR2a–c, AsPR4b, and AsPR5b, c), auxin (AsPR1a, n, AsPR4b, c, and AsPR5b, c), methyl JA (MeJA) (AsPR1b, d, i, l, m, AsPR2a, c AsPR4b, and AsPR5a–c), SA (AsPR1b–d, g–i, l, m, o, p, AsPR2a, c, AsPR4b, and AsPR5a–c), gibberellic acid (GA) (AsPR1b, e, p, AsPR2c, and AsPR5b, c), and ethylene (ET) (AsPR1c–e, j–l, n, and AsPR2b) (Table 3).
Table 3.
Regulatory cis-elements found in silico in the promoters of the PR1, PR2, PR4, and PR5 genes from Allium sativum cv. Ershuizao.
| Motif | Response to 1 | Number of Elements in Promoters | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AsPR1a | AsPR1b | AsPR1c | AsPR1d | AsPR1e | AsPR1f | AsPR1g | AsPR1h | AsPR1i | AsPR1j | AsPR1k | AsPR1l | AsPR1m | AsPR1n | AsPR1o | AsPR1p | AsPR2a | AsPR2b | AsPR2c | AsPR4a | AsPR4b | AsPR4c | AsPR5a | AsPR5b | AsPR5c | ||
| Hormone Response | ||||||||||||||||||||||||||
| ABRE | ABA | 1 | 3 | 1 | 2 | 3 | 2 | 2 | 4 | 1 | 3 | 2 | 2 | 2 | ||||||||||||
| ABRE3a | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |||||||||||||||||
| ABRE4 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |||||||||||||||||
| CARE | 1 | |||||||||||||||||||||||||
| AUXRR-core | Auxin | 1 | 1 | |||||||||||||||||||||||
| AuxRE | 1 | 1 | ||||||||||||||||||||||||
| TGA-element | 1 | 1 | 1 | 1 | ||||||||||||||||||||||
| CGTCA-motif | MeJA | 1 | 1 | 1 | 1 | 1 | 2 | 2 | 2 | 1 | 1 | 1 | ||||||||||||||
| TGACG-motif | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 2 | 1 | 1 | 1 | |||||||||||||||
| AS-1 | SA | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 2 | 1 | 1 | 1 | ||||||||||||||
| TCA-element | 2 | 1 | 2 | |||||||||||||||||||||||
| P-box | 2 | 1 | 1 | 1 | ||||||||||||||||||||||
| TATC-box | GA | 1 | ||||||||||||||||||||||||
| GARE-motif | 1 | 1 | 1 | 1 | 1 | |||||||||||||||||||||
| ERE | ET | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | |||||||||||||||||
| Stress Response | ||||||||||||||||||||||||||
| ARE | Anaerobic conditions |
1 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 2 | 1 | 2 | 2 | |||||||||||||
| DRE1/DRE core | Drought | 1 | ||||||||||||||||||||||||
| MBS | 1 | 1 | 1 | 2 | 2 | 3 | 2 | 2 | 1 | 2 | 3 | 3 | 1 | 1 | 1 | 1 | 1 | |||||||||
| LTR | Cold | 1 | 1 | 1 | 1 | |||||||||||||||||||||
| STRE | Heat, osmotic shock, low pH, starvation | 4 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | ||||||||||||
| F-box | Salt, heavy metals | 1 | ||||||||||||||||||||||||
| TC-rich repeats | Defense | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | ||||||||||||||||
| W-box | Wounding, pathogens | 1 | 2 | |||||||||||||||||||||||
| Wun-motif | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | ||||||||||||||||
| WRE3 | 1 | 1 | 1 | 1 | 1 | |||||||||||||||||||||
| box S | 1 | 1 | ||||||||||||||||||||||||
1 ABA, abscisic acid; MeJA, methyl jasmonate; SA, salicylic acid; GA, gibberellic acid; ET, ethylene.
Furthermore, each AsPR promoter contained 2–7 of the 11 identified cis-elements related to abiotic stresses, such as anaerobic conditions (AsPR1a, b, e, h, o, p, AsPR2a, AsPR4a, b, and AsPR5a–c), drought (all genes except AsPR1a, e–g, p, AsPR2a, AsPR4b, and AsPR5a), cold (AsPR1f, g, and AsPR5b, c), heat, osmotic shock, low pH, and starvation (AsPR1a, c, e, i, k, l, n, o, AsPR2a, c, and AsPR5a–c), salt and heavy metals (AsPR1d), defense (AsPR1g, i, p, AsPR2a–c, AsPR4a, c, and AsPR5a), wounding, and pathogens (all genes except AsPR1a, b, l, n, AsPR2b, AsPR4a, b, and AsPR5b, c) (Table 3).
2.5. Expression Patterns of AsPR Genes
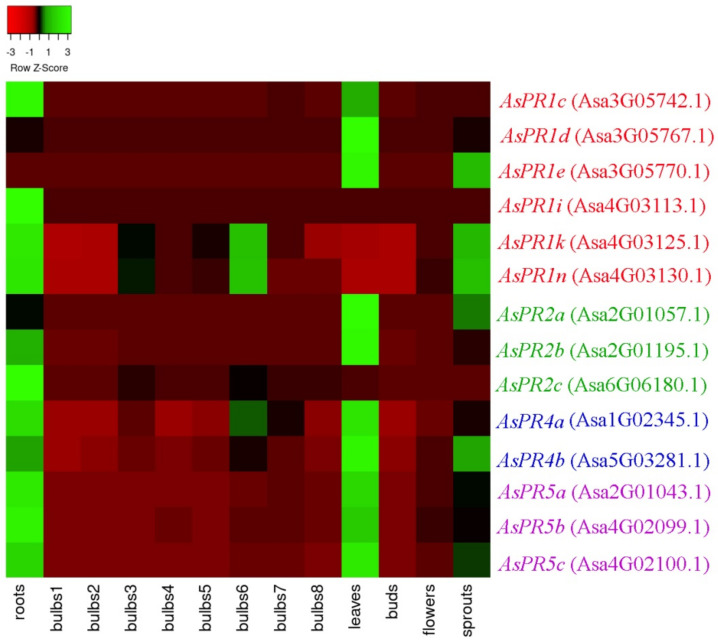
Analysis of the A. sativum cv. Ershuizao transcriptome (PRJNA607255, GSE145455) [31] indicated that all of the identified AsPR genes were transcribed in garlic tissues, including roots, bulbs (8 developmental stages), leaves, buds, flowers, and sprouts. The maximum levels of AsPR1 expression were detected in the roots (AsPR1c, i, k, n), leaves (AsPR1c–e), sprouts (AsPR1i, k, n), and stage 6 bulbs (AsPR1k, n), whereas AsPR1k, n were not expressed in stages 1 and 2 bulbs, leaves, and buds (Figure 2). A strong expression of AsPR2 was observed in the roots (AsPR2a–c), leaves (AsPR2a, b), and sprouts (AsPR2a), of AsPR4, in the roots, leaves, stage 6 bulbs, and sprouts, and of AsPR5, in the roots, leaves, and sprouts (Figure 2). The expressions of AsPR1a, b, h, l, m, o, p, and AsPR4c were extremely low.
Figure 2.
Expression heatmap of PR genes in Allium sativum cv. Ershuizao (GSE145455). The mRNA expression of AsPR1 (red), AsPR2 (green), AsPR4 (blue), and AsPR5 (purple) in the roots, bulbs (1, 2, 3, 4, 5, 6, and 7 correspond to 192-, 197-, 202-, 207-, 212-, 217-, 222-, and 227-day-old bulbs, respectively), leaves, buds, flowers, and sprouts. The color scheme indicates the gene expression gradient from low (red) to high (green).
2.6. AsPR Gene Expression in Response to F. proliferatum Infection
Since the AsPR genes are likely to be involved in the plant defense response (Table 2), their expression was evaluated in two garlic cultivars resistant (cv. Sarmat) and susceptible (cv. Strelets) to FBR. The plants were infected with F. proliferatum and analyzed at two time points (24 and 96 h post-inoculation (hpi)) covering the peak of PR gene expression in response to hemibiotrophic pathogens [33]. The results indicate that FBR symptoms, such as white mycelium on the roots, were observed at 96 hpi only in cv. Strelets.
The expression of the AsPR genes was compared in the roots, stems (basal plates), and cloves of the infected and control bulbs (Figure 3, Figure 4, Figure 5 and Figure 6 and Figure S1).
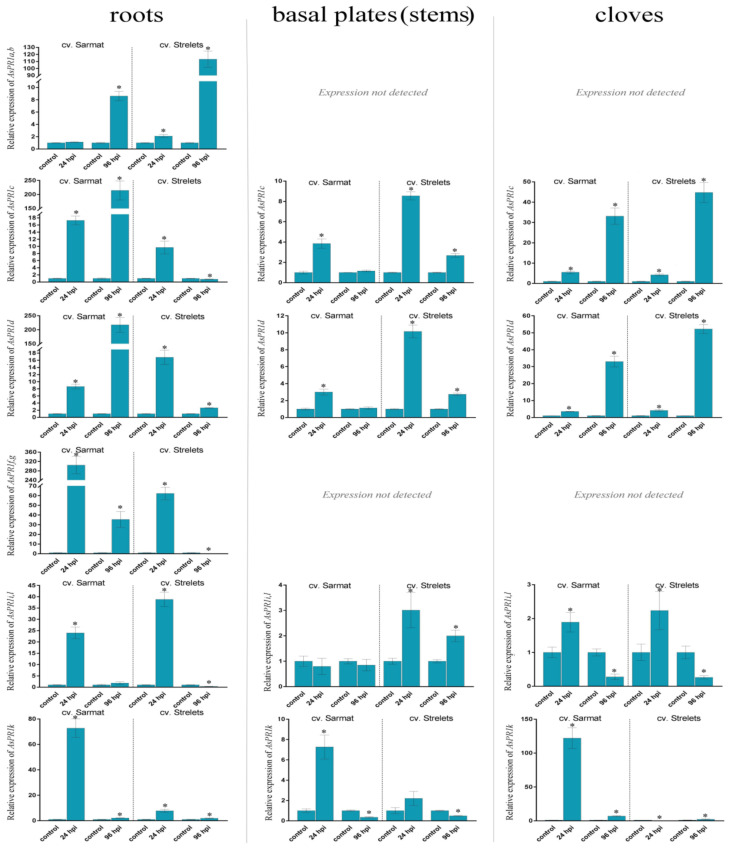
Figure 3.
Transcription of the AsPR1a(b), AsPR1c, AsPR1d, AsPR1f(g), AsPR1i(l), and AsPR1k genes in Allium sativum cv. Sarmat (FBR-resistant) and Strelets (FBR-susceptible) in response to Fusarium proliferatum infection. The plants were incubated with F. proliferatum conidia and analyzed for mRNA levels in the roots, stems, and cloves at 24 and 96 hpi by qRT-PCR. The data were normalized to GAPDH and UBQ mRNA levels and presented as fold change (mean ± SE) of the control taken as 1; * p < 0.01, compared to the uninfected control.
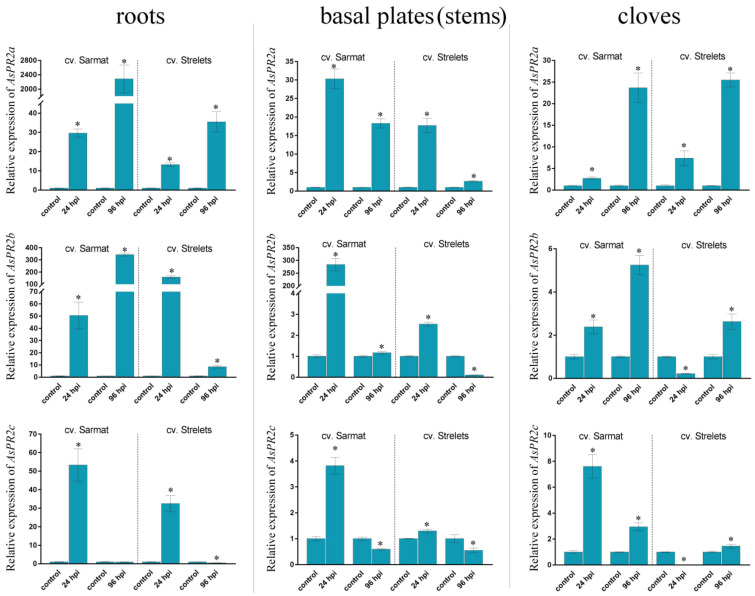
Figure 4.
Expression of the AsPR2a, b, c genes in Allium sativum cv. Sarmat (FBR-resistant) and Strelets (FBR-susceptible) after Fusarium proliferatum infection. The plants were incubated with F. proliferatum conidia and analyzed for mRNA levels in the roots, stems, and cloves at 24 and 96 hpi by qRT-PCR. The data were normalized to GAPDH and UBQ mRNA levels and presented as fold change (mean ± SE) of the control taken as 1; * p < 0.01, compared to the uninfected control.
Figure 5.
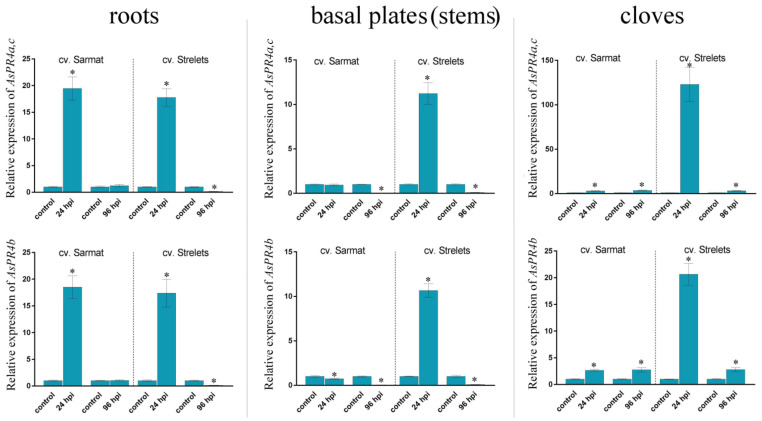
Expression of the AsPR4a(c) and AsPR4b genes in Allium sativum cv. Sarmat (FBR-resistant) and Strelets (FBR-susceptible) infected with Fusarium proliferatum. The plants were incubated with F. proliferatum conidia and analyzed for mRNA levels in the roots, stems, and cloves at 24 and 96 hpi by qRT-PCR. The data were normalized to GAPDH and UBQ mRNA levels and presented as fold change (mean ± SE) of the control taken as 1; * p < 0.01, compared to the uninfected control. Because AsPR2a and AsPR4c are 100% identical, the AsPR4a transcription level also includes that of AsPR4c.
Figure 6.
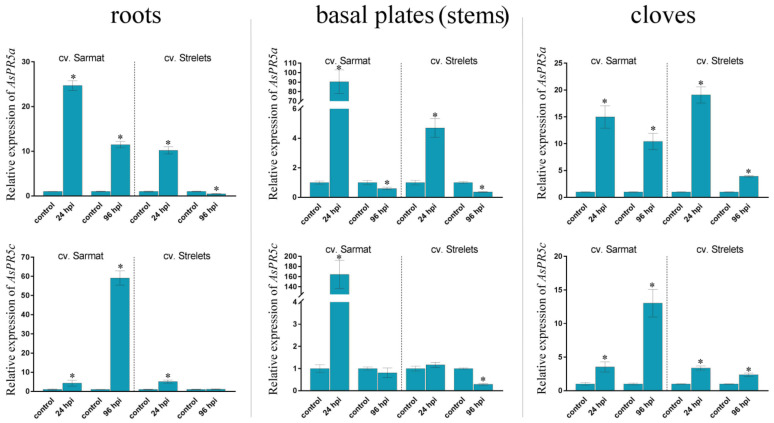
Expression of the AsPR5a, c genes in Allium sativum cv. Sarmat (FBR-resistant) and Strelets (FBR-susceptible) after infection with Fusarium proliferatum. The plants were incubated with F. proliferatum conidia and analyzed for mRNA levels in the roots, stems, and cloves at 24 and 96 hpi by qRT-PCR. The data were normalized to GAPDH and UBQ mRNA levels and presented as fold change (mean ± SE) of the control taken as 1; * p < 0.01 compared to the uninfected control.
2.6.1. AsPR1 Genes
Most of the AsPR1 genes were expressed in the roots, stems, and cloves, except for AsPR1a, b, f, g, which were transcribed only in the roots, and AsPR1p, which was not transcribed in the analyzed organs (Figure 3).
To determine which genes of AsPR1f and AsPR1g were expressed in the tissues of the two garlic cultivars, we performed sequencing of PCR-amplified products, which revealed that only AsPR1g was transcribed.
Overall, the expression of AsPR1 genes was induced by Fusarium infection in both cultivars, albeit to varying degrees. Thus, in cv. Sarmat, AsPR1 genes were upregulated 8–304 times in the roots (except for AsPR1a, b), 3–7 times in the stems (except for AsPR1i), and 2–122 times in the cloves at 24 hpi (Figure 3). At 96 hpi, there was a 200-fold increase in the expression of AsPR1c, d and a 2–35-fold increase in that of the other AsPR1 genes in the roots (except for AsPR1i, whose expression was similar to the control), whereas, in the stems, the AsPR1 expression was significantly decreased compared to 24 hpi, mostly down to the control levels (or even lower in the case of AsPR1k). In the cloves, AsPR1 transcription was increased by 33 (AsPR1c, d), or decreased by 4 (AsPR1k) and 17 (AsPR1i) times (Figure 3).
Compared to FBR-resistant cv. Sarmat, in FBR-susceptible cv. Strelets AsPR1 genes were activated by the infection at 24 hpi to a lesser extent in the roots (except for AsPR1i), to a bigger extent in the stems (except for AsPR1k), and to a similar extent in the cloves (except for AsPR1k). At 24 hpi, AsPR1 transcription was induced by 2–62 times in the roots, 2–10 times in the stems (except for AsPR1k), and 2–4 times in the cloves (except for AsPR1k, the transcription of which decreased 62 times) (Figure 3). At 96 hpi, the expression of AsPR1a, b in the roots was upregulated by 113 times, whereas that of the other AsPR1 genes was induced or downregulated slightly; in the stems, AsPR1 expression was increased by 2.0–2.7 times (except for AsPR1k, showing a 2-fold decrease), and in the cloves, it increased by 2 to 52 times (AsPR1k, AsPR1c, and AsPR1d) or decreased by 4 times (AsPR1i) (Figure 3).
2.6.2. AsPR2 Genes
All three identified AsPR2 genes were transcribed in the roots, stems, and cloves of both cultivars and were mostly upregulated in response to F. proliferatum infection (Figure 4). Specifically, in cv. Sarmat, the expression of the AsPR2a, b, c genes in all organs was significantly increased by infection (except for a decrease in AsPR2c expression, observed at 96 hpi in the roots and stems) (Figure 4). In cv. Strelets, all AsPR2 genes were upregulated at 24 hpi in the roots and stems compared to the control; in the cloves, the expression of AsPR2a was increased, but that of AsPR2b, c decreased. At 96 hpi, the expression of AsPR2a was increased in all organs, whereas that of AsPR2b increased in the roots and cloves but decreased in the stems, and that of AsPR2c decreased in the roots and stems and increased in the cloves (Figure 4).
Comparison of the two cultivars revealed that, overall, the upregulation of AsPR2 genes was much more significant in cv. Sarmat than in cv. Strelets. Thus, at 24 and 96 hpi, AsPR2a transcription in the roots and stems of cv. Sarmat increased by 30 and 2285, and 30 and 18 times, whereas, in those of cv. Strelets, it increased only by 13 and 35, and 18 and 2.6 times, respectively (Figure 4). AsPR2b transcription was increased by 50 and 343 times in the roots of cv. Strelets and by 159 and 8.5 times in those of cv. Strelets at 24 and 96 hpi, respectively. In the stems, AsPR2b expression was increased by 283 and 1.1 times in cv. Sarmat at 24 and 96 hpi, whereas in those of cv. Strelets, it was upregulated by 2.5 times at 24 hpi and downregulated by 10 times at 96 hpi; in the cloves, it was increased by 2.4 and 5.2 times at 24 and 96 hpi in cv. Sarmat, but decreased by 5 times at 24 hpi and increased by 2.6 times at 96 hpi in cv. Strelets (Figure 4).
The expression of AsPR2c gene showed a similar pattern in the roots and stems of the two cultivars. Thus, in the roots, it increased by 53 and 32 times at 24 hpi, and decreased to the control level or below at 96 hpi, whereas in the stems, it increased by 3.8- and 1.3-fold at 24 hpi, and decreased by 1.6- and 1.8-fold at 96 hpi in cv. Sarmat and cv. Strelets, respectively. However, in the cloves of cv. Sarmat, AsPR2c expression increased by 7.6 and 2.9 times, whereas, in those of cv. Strelets, it decreased by 83 times and increased by 1.5 times at 24 and 96 hpi, respectively (Figure 4).
2.6.3. AsPR4 Genes
AsPR4 transcription showed a similar dynamic in response to infection in the roots of both cultivars, where it was significantly upregulated at 24 hpi but dropped to or below the control levels at 96 hpi (Figure 5). In the stems of cv. Sarmat, AsPR4 expression was either similar or significantly lower than in the control, whereas in cv. Strelets, it was markedly upregulated at 24 hpi, but dropped significantly below the control levels at 96 hpi. In the cloves, AsPR4 transcription increased by about 3 times in cv. Sarmat and much more significantly (123-fold for AsPR4a(c) and 20-fold for AsPR4b) in cv. Strelets at 24 hpi, but not at 96 hpi (Figure 5).
These data indicate that, overall, the expression of the AsPR4 genes followed a similar trend in the roots of both cultivars, whereas in stems and cloves, it was strongly activated at 24 hpi only in FBR-susceptible cv. Strelets, showing a stronger response to Fusarium infection, which is probably associated with a faster spread of pathogens compared to the FBR-resistant cv. Sarmat.
2.6.4. AsPR5 Genes
AsPR5a, c genes were expressed in all organs of both cultivars (Figure 6), whereas AsPR5b was not detected.
In the roots, AsPR5a transcription was increased by 25- and 11-fold in cv. Sarmat, and upregulated by 10-fold but downregulated by 2-fold in cv. Strelets at 24 and 96 hpi, respectively, whereas that of AsPR5c was increased by 5-fold in both cultivars at 24 hpi and continued to increase at 96 hpi in cv. Sarmat (by 59-fold) but not in cv. Strelets (Figure 6). In the stems of cv. Sarmat, the levels of AsPR5a and AsPR5c mRNA were significantly increased (by 90- and 164-fold, respectively) at 24 hpi; however, at 96 hpi, they were lower or similar, respectively, to those in the uninfected control. In the stems of cv. Strelets, the expression of AsPR5a was upregulated by 4.7-fold and that of AsPR5c was similar to the control at 24 hpi, and downregulated by 2.7 (AsPR5a) and 3.4 (AsPR5c) times at 96 hpi (Figure 6). In the cloves, the expression of AsPR5a, c was upregulated at both time points in both cultivars. Thus, mRNA levels of AsPR5a and AsPR5c at 24/96 hpi were increased by 15 and 10, and 3.5 and 13 times in cv. Sarmat, and by 19 and 4 and 3.4 and 2.4 times in cv. Strelets, respectively (Figure 6).
These data indicate that overall, the expression of the AsPR5 genes followed a trend similar to that of the other AsPR genes, showing stronger induction by Fusarium infection in resistant cv. Sarmat.
2.7. Cloning and Characterization of CDSs of AsPR Genes Differentially Expressed in FBR-Sensitive and Resistant Cultivars
To examine polymorphisms in AsPR genes between FBR-resistant (Sarmat) and FBR-sensitive (Strelets) cultivars, we amplified, cloned, and sequenced the CDSs of 9 genes (AsPR1c, d, g, k, AsPR2a, b, c, and AsPR5a, c) that showed the strongest transcriptional response to F. proliferatum infection in cv. Sarmat.
Compared to cv. Ershuizao, used as a reference, the AsPR1g of cv. Sarmat and Strelets did not have single nucleotide polymorphisms (SNPs), whereas the other analyzed genes contained 1–10 SNPs (Table 4). There were 13 SNPs shared by cv. Sarmat and Strelets; 5 of them were non-synonymous and led to amino acid substitutions: c. 481G > A to p. V161I in AsPR1c, c. 855G > C to p. L285F in AsPR2c, c. 403G > A to p. G135S and c. 620T > C to p. I207T in AsPR5a, and c. 647A > T to p. D216V in AsPR5c (Table 4). Cultivar-specific SNPs were identified in AsPR1d (c. 200T > G leading to p. I67R) of cv. Sarmat and in AsPR1c (c. 379G > C to p. V127L), AsPR2a (c. 35T > C to p. L12S and c. 559A > C to p. I187L), and AsPR5a (c. 142T > A to p. S48T) in cv. Strelets (Table 4). According to PROVEAN, p. V127L and p. G135S were predicted to be potentially deleterious substitutions, whereas the rest were neutral.
Table 4.
Polymorphisms in the AsPR CDS in cv. Sarmat and Strelets compared to cv. Ershuizao.
| Gene | cv. Sarmat | cv. Strelets |
|---|---|---|
| AsPR1c | c. 481G > A (p. V161I) | c. 379G > C (p. V127L), c. 417T > C, c. 481G > A (p. V161I) |
| AsPR1d | c. 117A > G, c. 200T > G (p. I67R) | |
| AsPR1k | c. 462A > G | |
| AsPR2a | c. 291T > C | c. 35T > C (p. L12S), c. 559A > C p. I187L) |
| AsPR2b | c. 57A > G | |
| AsPR2c | c. 219C > A; c. 855G > C (p. L285F) | |
| AsPR5a | c. 126T > C, c. 159G > C, c. 279A > G, c. 285C > T, c. 403G > A (p. G135S), c. 468G > C, c. 620T > C (p. I207T) |
|
| AsPR5c | c. 516T > C, c. 647A > T (p. D216V) | |
3. Discussion
Fungi dominate among plant parasites in terms of the number of diseases they cause [34]. Infection with the most viable and destructive fungi of the Fusarium genus is accompanied by wilting and the appearance of mycelium and brown necrotic areas (accumulation of dead plant cells); at the molecular level, it is characterized by the production of camalexin and ROS, deposition of callose, and changes in the activity of PR proteins [29,34]. In A. sativum, infection by Fusarium spp. occurs through the roots and basal plates (stems), causing FBR and FW [26,35,36], which are responsible for 60% of the global garlic crop losses at pre- and post-harvest stages [29].
Genes of the PR1, PR2, PR3, PR4, PR5, PR12, and PR13 families encode proteins with antifungal activity, in particular, against Fusarium spp. [2]. In A. sativum, the expression of PR1, PR3, and PR5 genes is considered a positive marker of plant resistance to F. oxysporum f. sp. cepae [29]. Furthermore, PR3 genes AsCHI2, AsCHI3, AsCHI5, and AsCHI7, encoding GH19 class I chitinases, have been shown to be involved in the response to F. proliferatum attack [28].
In this study, we identified and characterized 25 PR genes in A. sativum cv. Ershuizao: 16 PR1, 3 PR2, 3 PR4, and 3 PR5, which encode CAP-domain proteins, β-1,3-glucanases, Barwin-domain proteins, and thaumatin-like proteins, respectively (Table 1). CDSs of 4 AsPR1, 3 AsPR2, and 2 AsPR5 genes were amplified in FBR-resistant cv. Sarmat and FBR-susceptible cv. Strelets; these genes are orthologs of A. sativum PR1 (JN011451.1) and PR5 (KP782043.1), and A. thaliana PR2 (NM_115587.2) and PR4 (NM_111344.6), respectively. The presence of multiple paralogs in each PR family suggests functional redundancy or divergence among PR factors. Paralogs are mostly tandemly clustered on chromosomes (Figure 1), suggesting that AsPR gene families originated as a result of evolutionary tandem duplications.
Our results indicate that the promoters of the AsPR genes contain 15 cis-regulatory elements involved in the response to hormones such as ABA, SA, MeJA, auxin, ET, and GA, as well as 12 elements associated with immune defense and response to elicitors and stresses such as anaerobic conditions, dehydration, low and high temperature, salinization, heavy metals, and wounding (Table 3). This regulatory profile is similar to those of class I chitinase genes of A. sativum (AsPR3) [28] and B. rapa [37]. Stress-responsive elements STRE, W-box, and WUN-motif, known to mediate pathogen- and/or elicitor-inducible transcription of chitinase genes [33,38,39], were found in the promoters of AsPR1, AsPR2, and AsPR5 (STRE), AsPR1f, g (W-box), and AsPR1, AsPR2, and AsPR4 (WUN-motif), confirming that the four identified PR families should be involved in the response to pathogens, including fungi.
Fusarium fungi are hemibiotrophs and pass through a biotrophic phase before switching to necrotrophy [34]. As such, they first elicit systemic (SAR) and then local (LAR) defense responses associated with antagonistic SA and JA signaling, respectively, which activates the transcription of PR1, PR2, and PR5 (SA), and PR3, PR4, and PR11 (JA) [2,40]. Because AsPR genes contain SA-responsive (AsPR1b–d, g–i, l, m, o, p, AsPR2a, c, AsPR4b, and AsPR5a–c) and JA-responsive (AsPR1b, d, i, l, m, AsPR2a, c, AsPR4b, and AsPR5a–c) elements in their promoters (Table 3), they could be involved in both the SAR and the LAR of garlic to Fusarium attack: ASPR1, AsPR2, and AsPR5 genes at the biotrophic phase, and AsPR4 genes at the necrotrophic phase. The presence of ET-responsive elements in AsPR1c–e, j–l, n and AsPR2b, and ABA-responsive elements in AsPR1a, d, f, i, k, l, n, AsPR2a–c, AsPR4b, and AsPR5b, c (Table 3) further confirms the potential roles of these genes in LAR, since ET and ABA synthesis is induced in response to necrotrophic pathogens [40,41]. Moreover, the presence of GA-responsive elements in AsPR1b, e, p, AsPR2c, and AsPR5b, c and auxin-responsive elements in AsPR1a, n, AsPR4b, c, and AsPR5b, c (Table 3) supports the involvement of these genes in LAR because the crosstalk of auxins and GAs with JA mediates the defense against pathogens [42].
Our results indicate that Fusarium infection activates AsPR transcription primarily in the roots (Figure 3, Figure 4, Figure 5 and Figure 6) representing the first barrier to the penetration of soil-dwelling fungi [43], which is consistent with the role of PR proteins in the antifungal defense. However, there was no clear correlation between the timing of gene activation in response to infection (24 or 96 hpi) (Figure 3, Figure 4, Figure 5 and Figure 6) and their putative role in SAR or LAR, especially since the infection signs at 96 hpi were limited to the appearance of mycelium, suggesting that Fusarium infection was still at its biotrophic phase and did not yet proceed to the necrotrophic phase.
At the same time, we found significant variations in gene expression between FBR-resistant and susceptible cultivars (Figure 3, Figure 4, Figure 5 and Figure 6). Compared to the susceptible cv. Strelets, in the resistant cv. Sarmat, the AsPR5 and AsPR2 genes showed stronger activation (the latter especially in the roots and stems), whereas AsPR4 showed weaker activation in the stems and cloves at 24 hpi (Figure 4, Figure 5 and Figure 6). The regulatory mode of the multiple identified AsPR1 genes differed depending on the organ; AsPR1c, d, g, k showed stronger activation in the roots (the latter also in the stems and cloves), whereas AsPR1a, b, i had weaker activation in the roots and AsPR1d, i—in the stems of cv. Sarmat (Figure 3). Given that the encoded PR proteins provide plant protection against fungal infection, these data suggest the mRNA expression profiles of AsPR1c, d, g, k, AsPR2b and AsPR5a, c (in roots), as well as AsPR4a(c), b, and AsPR2c (in stems and cloves), and may serve as a marker of resistance to FBR in garlic cultivars. Thus, sterol-binding PR1 proteins suppress the proliferation of fungal cells because of sterol extraction from their membranes [44], PR2 β-1,3-glucanases, in combination with chitinases, hydrolyze fungal cell walls [45], PR4 Barwin-domain proteins directly inhibit hyphal growth [46], and PR5 thaumatin-like proteins may increase membrane permeability through the destruction of β-1,3-glucans or the inhibition of fungal enzymes, such as xylanases [47]. Compared to other AsPR genes, the AsPR1 family appears to have undergone a particularly high level of expansion (Figure 1). Considering the activation of the expression of at least half of the AsPR1 genes in response to Fusarium infection (Figure 3), the participation of CAP-domain proteins in immune defense may be critical. The presence of a large number of active AsPR1 paralogs suggests their redundancy in the immune response, which may insure the plant against fungal infection even in the event of knockout of individual family members.
The AsPR1e, h, j, l–p, and AsPR5b genes were not functional in cv. Sarmat and Strelets, since their mRNA was not detected irrespectively of F. proliferatum infection; however, AsPR1n and AsPR5b transcripts were observed in the roots and bulbs of cv. Ershuizao (Figure 2), suggesting that these genes are expressed in a cultivar-specific manner.
4. Materials and Methods
4.1. In Silico Identification and Structural Characterization of PR1, PR2, PR4, and PR5 Genes in the Allium Sativum Genome
The search of PR1, PR2, PR4, and PR5 genes was performed in the A. sativum cv. Ershuizao transcriptome (NCBI accession number: PRJNA607255) and whole-genome shotgun contigs (PRJNA606385) [31]. Sequences of garlic AsPR1 (JN011451.1) and AsPR5 (KP782043.1) genes and A. thaliana PR2 (AT3G57270; NM_115587.2) and PR4 (AT3G04720; NM_111344.6) genes were used as references. The selected sequences contained start and stop codons and full-length catalytic domains. Multiple sequence alignment and structural analysis of AsPR genes and the encoded proteins were conducted with MEGA 7.0.26 [48]. To predict exon–intron structures, AsPR genes and CDSs were analyzed with GSDS v2.0 [49]. The predicted proteins were characterized by MW, pI, AI, and GRAVY (ExPASy ProtParam; https://web.expasy.org/protparam/; accessed on 1 March 2021), conserved domains, sites, and motifs (NCBI-CDD, https://www.ncbi.nlm.nih.gov/cdd; accessed on 1 March 2021), biological processes (PANNZER2; http://ekhidna2.biocenter.helsinki.fi/sanspanz/; accessed on 1 March 2021), subcellular localization (BaCello; http://gpcr2.biocomp.unibo.it/; accessed on 1 March 2021), the functional importance of residue substitutions (PROVEAN; [50]), and signal peptide cleavage sites (SignalP 5.0; http://www.cbs.dtu.dk/services/SignalP/; accessed on 1 March 2021). The chromosomal localization map was drawn using MG2C v. 2.1 (http://mg2c.iask.in/mg2c_v2.1/; accessed on 1 March 2021).
4.2. In Silico mRNA Expression Analysis
The expression of AsPR genes in A. sativum cv. Ershuizao tissues (roots, bulbs, stems (basal plates), leaves, buds, flowers, and sprouts) was determined based on RNA-seq data (ID: PRJNA607255), normalized using Fragments Per Kilobase of transcript per Million mapped reads (FPKM) assay [31], and visualized using Heatmapper [51]; only transcripts with an average FPKM value of ≥10 in at least one of the organs were used for heatmap construction.
4.3. Fungi, Plant Material, and F. proliferatum Infection Assay
F. proliferatum was kindly provided by the Group of Experimental Mycology, Winogradsky Institute of Microbiology (Research Center of Biotechnology of the RAS, Moscow, Russia). The strain was previously isolated from cv. Strelets bulbs; the pathogenicity test showed that the first signs of the disease appeared on the surface of the treated cloves after 5 days of infection [52].
Accessions of A. sativum cv. Sarmat and cv. Strelets, resistant and susceptible to FBR, respectively, were kindly provided by the Federal Scientific Vegetable Center (Moscow region, Russia). The choice of cultivars was based on the similarity of morphological characteristics in order to exclude their influence on experimental results [28].
The number of cloves (6) used per biological replicate in the Fusarium infection assay was based on that of cloves in a bulb (5–7 for cv. Strelets and 7–11 for cv. Sarmat). In total, 6 bulbs of each cultivar were used, and 6 cloves from each bulb were processed: 3 for the experiment and 3 for control.
A total of 12 cloves of each cultivar were sterilized in 70% ethanol for 3 min, rinsed with sterile water, placed in Petri dishes with wet filter paper, and incubated at +25 °C in the dark. After 36 h, active root growth was observed and half of the cloves were infected by soaking in F. proliferatum conidial suspension (~106 conidia ml−1) for 5 min, as previously described [35]). The infected cloves (n = 6) were transferred to fresh Petri dishes and incubated at +25 °C in the dark; uninfected cloves (n = 6) were used as a control. The experiment was performed in 3 biological and 2 technical replicates for the infected and uninfected plants per each time point. The roots, stems, and cloves were collected at 24 hpi and 96 hpi, frozen in liquid nitrogen, and stored at −80 °C until analysis. The time points (24 and 96 hpi) were chosen based on the reported peak of PR gene expression, which was observed 1–3 days after inoculation with hemibiotrophic pathogens [53].
4.4. RNA Extraction and Quantitative Real-Time PCR (qRT-PCR) Analysis
Total RNA was extracted from individual roots, stems, and cloves (0.5 g of each tissue) using the RNeasy Plant Mini Kit (QIAGEN, Hilden, Germany), purified from genomic DNA (RNase free DNase set; QIAGEN, Hilden, Germany), qualified by gel electrophoresis, and used for first-strand cDNA synthesis (GoScript Reverse Transcription System; Promega, Madison, USA) with an oligo-dT primer. The RNA and cDNA concentrations were quantified by fluorimetry (Qubit® Fluorometer, Thermo Fisher Scientific, Waltham, MA, USA) and qRT-PCR was performed in a CFX96 Real-Time PCR Detection System (Bio-Rad Laboratories, Hercules, USA) with 3.0 ng of cDNA, SYBR Green RT-PCR mixture (Syntol, Moscow, Russia), and specific primers (Table 5). Gene-specific primers were designed to amplify partial coding sequences; the primers were selected for the most variable parts of the CDS. Given the high similarity (over 98%) of the coding sequences of some paralogous genes (AsPR1a/b; AsPR1f/g; AsPR1i/l; AsPR4a/c; AsPR5a/b), primers that amplify both genes were selected for each pair. The following cycling conditions were used: initial denaturation at 95 °C for 5 min, 40 cycles of denaturation at 95 °C for 15 s, and annealing/extension at 60 °C for 40 s.
Table 5.
The list of primers used in the study.
| Gene | Primer Sequence (5′→3′) | Application |
|---|---|---|
| AsPR1a, b | ATGGAACACGACACTGGCAG GCATACTGACCAGAGTAACTGG |
Gene expression analysis (qRT-PCR) |
| AsPR1c | GGCGGTCCTTATGGTGAAA GCCAGGGTCACATGTGTTA |
|
| AsPR1d | GGCGGTCCTTATGGTGAAA CCAGGGTCACATGTGTTGCT |
|
| AsPR1f, g | CGATCACCACCGCAGTTCA GCGTAGTTCTGTGCGTAATCAG |
|
| AsPR1i | TATGGGGAGAACCTATTCGC AATCTTRACCGACTTAGCCCA |
|
| AsPR1k | GTGTCCGAGAAGCGGTACTAT AGCCGCCAGTGTTGCACC |
|
| AsPR1p | GTCGCAAAATACGCGCAAAGTT GTACTTCACGACATCGGCATC |
|
| AsPR2a | GCTAGAAACCATATCGTTGCCT GCATACCGTAGCATACTCCGA |
|
| AsPR2b | GGTCGCATTTCTCCTAGGCAT GCGTCGCCTGCTGATGGAA |
|
| AsPR2c | GGCCCATTGTCCAGTTCTTG AGGCGCCGTGAATAATGCGTA |
|
| AsPR4a | ATGCCGGCATGTCCCTCG GTCTATGATCCTCACCGTCGTT |
|
| AsPR4b | ATGCCGGCATGTCCCTCG GGTCTATGATCCTCACCGTCAA |
|
| AsPR5a | CATCCGGACACGGCAGCT TCCATGTACTGCTTCAGAGCG |
|
| AsPR5c | GCAAGCAGCTCAACTCAGGA GCCGGTCTGACATCTTCCA |
|
| AsPR1c | ATGGGATCAATCAGTAGTTATA AACGACTGAGTACTCTCAGT |
Gene amplification |
| AsPR1d | ATGGGATCGACCAGTACTTG TAACGTCGTAGTTGTAACGAC |
|
| AsPR1f, g | ATGAAAACGTCATTTCTCTTC ATAAATAGCAGTACACACATAA |
|
| AsPR1k | GCTCAAATTACAATGAAAACGTT CTGTCTGTTTCAGCATGCA |
|
| AsPR2a | TGTGCACCATCGAATTACCTTC CTCTGTCTCCCTTAATAGTAC |
|
| AsPR2b | AAATGCAAGCAAGGAAGCTTG CCCTGGTACATTCATAGTTAAC |
|
| AsPR2c | TTGAAATGGTCATGCATGCCT AAACAGGGCACACATGCAAG |
|
| AsPR5a | ATGTCGACCCAAATTACAGTC ACTCAATCCAAGAAYACAGTTC |
|
| AsPR5c | ATGTCGACCCAAATTACAGTC CTGCAAACTAATTATTCGGTGA |
AsPR gene expression was normalized using two reference garlic genes, GAPDH [54] and UBQ [55], and the qRT-PCR results were statistically analyzed with Graph Pad Prism version 8 (GraphPad Software Inc., San Diego, CA, USA; https://www.graphpad.com/scientific-software/prism/ (accessed on 26 April 2021)). The unpaired t-test was applied to assess differences in gene expression; p < 0.01 was considered to indicate statistical significance. The data were expressed as the mean ± standard error (SE) based on 3 technical replicates of 3 biological replicates for each combination of cDNA and primer pairs.
4.5. Gene Identification
To amplify the CDSs of AsPR genes from garlic cultivars, gene-specific primers were designed based on A. sativum cv. Ershuizao transcriptomic data (NCBI project accession number: PRJNA607255) (Table 5). A manual revision of sequence polymorphisms and an additional evaluation were performed using Primer3 (http://frodo.wi.mit.edu/primer3/; accessed on 15 March 2021). cDNA from the roots of a single plant of each cultivar was used as a template (30 ng) for PCR amplification with the following conditions: initial denaturation at 95 °C for 5 min and 35 cycles of denaturation at 95 °C for 30 s, primer annealing at 55 °C for 30 s, and extension at 72 °C for 2 min followed with a final extension at 72 °C for 5 min. The amplified PCR products of the expected size were purified by using the QIAEX® II Gel Extraction kit (QIAGEN, Hilden, Germany), cloned in the pGEM®-T Easy vector (Promega, Madison, WI, USA), and sequenced (3–5 clones for each accession) on an ABI Prism 3730 DNA Sequencer (Applied Biosystems, Waltham, MA, USA) using the designed primers (Table 5).
4.6. Promoter and 5′-UTR Analysis
The search of specific cis-elements in promoters and 5′-UTRs (1.0 kb regions upstream of the initiation codon) was performed using the PlantCARE database, which provides an evaluation of cis-regulatory elements, enhancers, and repressors [56]; (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/; accessed on 10 March 2021).
5. Conclusions
We identified and characterized 25 genes of the PR1, PR2, PR4, and PR5 families in A. sativum cv. Ershuizao and cloned 4 AsPR1, 3 AsPR2, and 2 AsPR5 homologs from garlic cultivars resistant and susceptible to FBR. The AsPR gene promoters contained hormone- and stress-responsive elements, including those associated with the response to fungal pathogens and their elicitors, indicating that the putative garlic PR proteins AsPR1a, c, e, f, j, k, n, AsPR2b, and AsPR4c may participate in LAR, whereas AsPR1b–d, i, l, p, AsPR2a, c, AsPR4b, and AsPR5a–c participate in both LAR and SAR. The comparison of AsPR transcriptional profiles in FBR-resistant and susceptible garlic cultivars infected with F. proliferatum suggests that the expression profile of AsPR5a, c, AsPR2b, and AsPR1c, d, g, k (in roots), as well as AsPR4a(c), b and AsPR2c (in stems and cloves), could define the difference in the PR-mediated response to Fusarium infection between resistant and susceptible plants. Our results provide useful insights into the functions of PR genes in A. sativum and Allium plants in general, and may be used in breeding programs to increase the resistance of Allium crops to Fusarium infections.
Acknowledgments
We would like to thank Marina Chuenkova for English language editing. This work was performed using the experimental climate control facility in the Institute of Bioengineering (Research Center of Biotechnology, Russian Academy of Sciences).
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/ijms22136688/s1: Figure S1: Expression of AsPR1 (a), AsPR2 (b), AsPR4 (c) and AsPR5 (d) mRNA in Allium sativum cv. Sarmat (FBR-resistant) and Strelets (FBR-susceptible) infected with Fusarium proliferatum. The plants were incubated with F. proliferatum conidia and analyzed for mRNA levels in the roots, stems (basal plate), and cloves at 24 and 96 hpi by qRT-PCR. The data were normalized to GAPDH and UBQ mRNA levels and presented as the mean ± SE (n = 3). * p < 0.01 compared to the same tissue in the other cultivar: blue asterisk – gene expression level in cv. Sarmat is higher than that in cv. Strelets; red asterisk – gene expression level in cv. Strelets is higher than that in cv. Sarmat.
Author Contributions
Performed the experiments: M.A.F. and O.K.A. Analyzed the data: E.Z.K., A.V.S. and M.A.F. Wrote the paper: A.V.S., M.A.F. and E.Z.K. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Russian Foundation for Basic Research (grant no. 20-316-70009) and the Ministry of Science and Higher Education of the Russian Federation.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
PR genes coding sequences of A. sativum cv. Sarmat/cv. Strelets were deposited in NCBI: AsPR1c (MZ216000/MZ216001), AsPR1d (MZ216002/MZ216003), AsPR1g (MZ216004/MZ216005), AsPR1k (MZ216006/MZ216007), AsPR2a (MZ216008/MZ216009), AsPR2b (MZ216010/MZ216011), AsPR2c (MZ216012/MZ216013), AsPR5a (MZ216014/MZ216015), AsPR5c (MZ216016/MZ216017).
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Roux F., Voisin D., Badet T., Balagué C., Barlet X., Huard-Chauveau C., Roby D., Raffaele S. Resistance to phytopathogens e tutti quanti: Placing plant quantitative disease resistance on the map. Mol. Plant Pathol. 2014;15:427–432. doi: 10.1111/mpp.12138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ali S., Ganai B.A., Kamili A.N., Bhat A.A., Mir Z.A., Bhat J.A., Tyagi A., Islam S.T., Mushtaq M., Yadav P., et al. Pathogenesis-related proteins and peptides as promising tools for engineering plants with multiple stress tolerance. Microbiol. Res. 2018;212–213:29–37. doi: 10.1016/j.micres.2018.04.008. [DOI] [PubMed] [Google Scholar]
- 3.Akbudak M.A., Yildiz S., Filiz E. Pathogenesis related protein-1 (PR-1) genes in tomato (Solanum lycopersicum L.): Bioinformatics analyses and expression profiles in response to drought stress. Genomics. 2020;112:4089–4099. doi: 10.1016/j.ygeno.2020.07.004. [DOI] [PubMed] [Google Scholar]
- 4.Dean R., Van Kan J.A., Pretorius Z.A., Hammond-Kosack K.E., Di Pietro A., Spanu P.D., Rudd J.J., Dickman M., Kahmann R., Ellis J., et al. The Top 10 fungal pathogens in molecular plant pathology. Mol. Plant Pathol. 2012;13:414–430. doi: 10.1111/j.1364-3703.2011.00783.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zipfel C., Felix G. Plants and animals: A different taste for microbes? Curr. Opin. Plant Biol. 2005;8:353–360. doi: 10.1016/j.pbi.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 6.Dangl J.L., Jones J.D.G. Plant pathogens and integrated defence responses to infection. Nature. 2001;411:826–833. doi: 10.1038/35081161. [DOI] [PubMed] [Google Scholar]
- 7.Okushima Y., Koizumi N., Kusano T., Sano H. Secreted proteins of tobacco cultured BY2 cells: Identification of a new member of pathogenesis-related proteins. Plant Mol. Biol. 2000;42:479–488. doi: 10.1023/A:1006393326985. [DOI] [PubMed] [Google Scholar]
- 8.Van Loon L.C., Pierpont W.S., Boller T., Conejero V. Recommendations for naming plant pathogenesis-related proteins. Plant Mol. Biol. Rep. 1994;12:245–264. doi: 10.1007/BF02668748. [DOI] [Google Scholar]
- 9.Edreva A. Pathogenesis-related proteins: Research progress in the last 15 years. Gen. Appl. Plant Physiol. 2005;31:105–124. [Google Scholar]
- 10.Sinha M., Singh R.P., Kushwaha G.S., Iqbal N., Singh A., Kaushik S., Kaur P., Sharma S., Singh T.P. Current overview of allergens of plant pathogenesis related protein families. Sci. World J. 2014;2014:543195. doi: 10.1155/2014/543195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guevara-Morato M.A., de Lacoba M.G., García-Luque I., Serra M.T. Characterization of a pathogenesis-related protein 4 (PR-4) induced in Capsicum chinense L3 plants with dual RNase and DNase activities. J. Exp. Bot. 2010;61:3259–3271. doi: 10.1093/jxb/erq148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ma L.J., Geiser D.M., Proctor R.H., Rooney A.P., O’Donnell K., Trail F., Gardiner D.M., Manners J.M., Kazan K. Fusarium pathogenomics. Annu. Rev. Microbiol. 2013;67:399–416. doi: 10.1146/annurev-micro-092412-155650. [DOI] [PubMed] [Google Scholar]
- 13.Summerell B.A. Resolving Fusarium: Current Status of the Genus. Annu. Rev. Phytopathol. 2019;57:323–339. doi: 10.1146/annurev-phyto-082718-100204. [DOI] [PubMed] [Google Scholar]
- 14.Kalman B., Abraham D., Graph S., Perl-Treves R., Meller Harel Y., Degani O. Isolation and Identification of Fusarium spp., the Causal Agents of Onion (Allium cepa) Basal Rot in Northeastern Israel. Biology. 2020;9:69. doi: 10.3390/biology9040069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gagkaeva T., Gavrilova O., Orina A., Lebedin Y., Shanin I., Petukhov P., Eremin S. Analysis of Toxigenic Fusarium Species Associated with Wheat Grain from Three Regions of Russia: Volga, Ural, and West Siberia. Toxins. 2019;11:252. doi: 10.3390/toxins11050252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Masachis S., Segorbe D., Turrà D., Leon-Ruiz M., Fürst U., El Ghalid M., Leonard G., López-Berges M.S., Richards T.A., Felix G., et al. A fungal pathogen secretes plant alkalinizing peptides to increase infection. Nat. Microbiol. 2016;1:16043. doi: 10.1038/nmicrobiol.2016.43. [DOI] [PubMed] [Google Scholar]
- 17.Wrobel-Kwiatkowska M., Lorenc-Kukula K., Starzycki M., Oszmianski J., Kepczynska E., Szopa J. Expression of b-1,3-glucanase in flax causes increased resistance to fungi. Physiol. Mol. Plant Pathol. 2004;65:245–256. doi: 10.1016/j.pmpp.2005.02.008. [DOI] [Google Scholar]
- 18.Samet M., Charfeddine M., Kamoun L., Nouri-Ellouze O., Gargouri-Bouzid R. Effect of compost tea containing phosphogypsum on potato plant growth and protection against Fusarium solani infection. Environ. Sci. Pollut. Res. Int. 2018;25:18921–18937. doi: 10.1007/s11356-018-1960-z. [DOI] [PubMed] [Google Scholar]
- 19.Bharti P., Jyoti P., Kapoor P., Sharma V., Shanmugam V., Yadav S.K. Host-Induced Silencing of Pathogenicity Genes Enhances Resistance to Fusarium oxysporum Wilt in Tomato. Mol. Biotechnol. 2017;59:343–352. doi: 10.1007/s12033-017-0022-y. [DOI] [PubMed] [Google Scholar]
- 20.Li G., Zhou J., Jia H., Gao Z., Fan M., Luo Y., Zhao P., Xue S., Li N., Yuan Y., et al. Mutation of a histidine-rich calcium-binding-protein gene in wheat confers resistance to Fusarium head blight. Nat. Genet. 2019;51:1106–1112. doi: 10.1038/s41588-019-0426-7. [DOI] [PubMed] [Google Scholar]
- 21.Tan R., Collins P.J., Wang J., Wen Z., Boyse J.F., Laurenz R.G., Gu C., Jacobs J.L., Song Q., Chilvers M.I., et al. Different loci associated with root and foliar resistance to sudden death syndrome (Fusarium virguliforme) in soybean. Theor. Appl. Genet. 2019;132:501–513. doi: 10.1007/s00122-018-3237-9. [DOI] [PubMed] [Google Scholar]
- 22.Anand A., Zhou T., Trick H.N., Gill B.S., Bockus W.W., Muthukrishnan S. Greenhouse and field testing of transgenic wheat plants stably expressing genes for thaumatin-likeprotein, chitinase and glucanase against Fusarium graminearum. J. Exp. Bot. 2003;54:1101–1111. doi: 10.1093/jxb/erg110. [DOI] [PubMed] [Google Scholar]
- 23.Mackintosh C.A., Lewis J., Radmer L.E., Shin S., Heinen S.J., Smith L.A., Wyckoff M.N., Dill-Macky R., Evans C.K., Kravchenko S., et al. Overexpression of defense response genes in transgenic wheat enhances resistance to Fusarium head blight. Plant Cell Rep. 2007;26:479–488. doi: 10.1007/s00299-006-0265-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hammad I.A., Abdel-Razik A.B., Soliman E.R., Tawfik E. Transgenic potato (Solanum tuberosum) expressing two antifungal thionin genes confer resistance to Fusarium spp. J. Pharm. Biol. Sci. 2017;12:69–79. doi: 10.9790/3008-1204026979. [DOI] [Google Scholar]
- 25.Cramer C.S. Breeding and genetics of Fusarium basal rot resistance in onion. Euphytica. 2000;115:159–166. doi: 10.1023/A:1004071907642. [DOI] [Google Scholar]
- 26.Gálvez L., Urbaniak M., Waśkiewicz A., Stępień Ł., Palmero D. Fusarium proliferatum—Causal agent of garlic bulb rot in Spain: Genetic variability and mycotoxin production. Food Microbiol. 2017;67:41–48. doi: 10.1016/j.fm.2017.05.006. [DOI] [PubMed] [Google Scholar]
- 27.Galván G.A., Koning-Boucoiran C.F.S., Koopman W.J.M., Burger-Meijer K., González P.H., Waalwijk C., Kik C., Scholten O.E. Genetic variation among Fusarium isolates from onion, and resistance to Fusarium basal rot in related Allium species. Eur. J. Plant Pathol. 2008;121:499–512. doi: 10.1007/s10658-008-9270-9. [DOI] [Google Scholar]
- 28.Filyushin M.A., Anisimova O.K., Kochieva E.Z., Shchennikova A.V. Genome-Wide Identification and Expression of Chitinase Class I Genes in Garlic (Allium sativum L.) Cultivars Resistant and Susceptible to Fusarium proliferatum. Plants. 2021;10:720. doi: 10.3390/plants10040720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chand S.K., Nanda S., Mishra R., Joshi R.K. Multiple garlic (Allium sativum L.) microRNAs regulate the immunity against the basal rot fungus Fusarium oxysporum f. sp. cepae. Plant Sci. 2017;257:9–21. doi: 10.1016/j.plantsci.2017.01.007. [DOI] [PubMed] [Google Scholar]
- 30.Chand S.K., Nanda S., Joshi R.K. Regulation of miR394 in Response to Fusarium oxysporum f. sp. cepae (FOC) Infection in Garlic (Allium sativum L.) Front. Plant Sci. 2016;7:258. doi: 10.3389/fpls.2016.00258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sun X., Zhu S., Li N., Cheng Y., Zhao J., Qiao X., Lu L., Liu S., Wang Y., Liu C., et al. A Chromosome-Level Genome Assembly of Garlic (Allium sativum) Provides Insights into Genome Evolution and Allicin Biosynthesis. Mol. Plant. 2020;13:1328–1339. doi: 10.1016/j.molp.2020.07.019. [DOI] [PubMed] [Google Scholar]
- 32.Van Loon L.C., Van Strien E.A. The families of pathogenesis-related proteins, their activities, and comparative analysis of PR-1 type proteins. Physiol. Mol. Plant Pathol. 1999;55:85–97. doi: 10.1006/pmpp.1999.0213. [DOI] [Google Scholar]
- 33.Bartholomew E.S., Black K., Feng Z., Liu W., Shan N., Zhang X., Wu L., Bailey L., Zhu N., Qi C., et al. Comprehensive Analysis of the Chitinase Gene Family in Cucumber (Cucumis sativus L.): From Gene Identification and Evolution to Expression in Response to Fusarium oxysporum. Int. J. Mol. Sci. 2019;20:5309. doi: 10.3390/ijms20215309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Doehlemann G., Ökmen B., Zhu W., Sharon A. Plant Pathogenic Fungi. Microbiol. Spectr. 2017;5 doi: 10.1128/microbiolspec.FUNK-0023-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leyronas C., Chrétien P.L., Troulet C., Duffaud M., Villeneuve F., Morris C.E., Hunyadi H. First report of Fusarium proliferatum causing garlic clove rot in France. Plant Dis. 2018;102:2658. doi: 10.1094/PDIS-06-18-0962-PDN. [DOI] [Google Scholar]
- 36.Tonti S., Prà M.D., Nipoti P., Prodi A., Alberti I. First Report of Fusarium proliferatum Causing Rot of Stored Garlic Bulbs (Allium sativum L.) in Italy. J. Phytopathol. 2012;160:761–763. doi: 10.1111/jph.12018. [DOI] [Google Scholar]
- 37.Chen J., Piao Y., Liu Y., Li X., Piao Z. Genome-wide identification and expression analysis of chitinase gene family in Brassica rapa reveals its role in clubroot resistance. Plant Sci. 2018;270:257–267. doi: 10.1016/j.plantsci.2018.02.017. [DOI] [PubMed] [Google Scholar]
- 38.Gao Y., Jia S., Wang C., Wang F., Wang F., Zhao K. BjMYB1, a transcription factor implicated in plant defence through activating BjCHI1 chitinase expression by binding to a W-box-like element. J. Exp. Bot. 2016;67:4647–4658. doi: 10.1093/jxb/erw240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gao Y., Zan X., Wu X., Yao L., Chen Y., Jia S., Zhao K. Identification of Fungus-Responsive Cis-Acting Element in the Promoter of Brassica Juncea Chitinase Gene, BjCHI1. Plant Sci. 2014;215–216:190–198. doi: 10.1016/j.plantsci.2013.11.008. [DOI] [PubMed] [Google Scholar]
- 40.Verma V., Ravindran P., Kumar P.P. Plant hormone-mediated regulation of stress responses. BMC Plant Biol. 2016;16:86. doi: 10.1186/s12870-016-0771-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu S., Kracher B., Ziegler J., Birkenbihl R.P., Somssich I.E. Negative regulation of ABA signaling by WRKY33 is critical for Arabidopsis immunity towards Botrytis cinerea 2100. eLife. 2015;4:e07295. doi: 10.7554/eLife.07295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang J., Duan G., Li C., Liu L., Han G., Zhang Y., Wang C. The Crosstalks Between Jasmonic Acid and Other Plant Hormone Signaling Highlight the Involvement of Jasmonic Acid as a Core Component in Plant Response to Biotic and Abiotic Stresses. Front. Plant Sci. 2019;10:1349. doi: 10.3389/fpls.2019.01349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Warman N.M., Aitken E.A.B. The Movement of Fusarium oxysporum f. sp. cubense (Sub-Tropical Race 4) in Susceptible Cultivars of Banana. Front. Plant Sci. 2018;9:1748. doi: 10.3389/fpls.2018.01748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schneiter R., Di Pietro A. The CAP protein superfamily: Function in sterol export and fungal virulence. Biomol. Concepts. 2013;4:519–525. doi: 10.1515/bmc-2013-0021. [DOI] [PubMed] [Google Scholar]
- 45.Balasubramanian V., Vashisht D., Cletus J., Sakthivel N. Plant β-1,3-glucanases: Their biological functions and transgenic expression against phytopathogenic fungi. Biotechnol. Lett. 2012;34:1983–1990. doi: 10.1007/s10529-012-1012-6. [DOI] [PubMed] [Google Scholar]
- 46.Bai S., Dong C., Li B., Dai H. A PR-4 gene identified from Malus domestica is involved in the defense responses against Botryosphaeria dothidea. Plant Physiol. Biochem. 2013;62:23–32. doi: 10.1016/j.plaphy.2012.10.016. [DOI] [PubMed] [Google Scholar]
- 47.De Jesús-Pires C., Ferreira-Neto J.R.C., Pacifico Bezerra-Neto J., Kido E.A., de Oliveira Silva R.L., Pandolfi V., Wanderley-Nogueira A.C., Binneck E., da Costa A.F., Pio-Ribeiro G., et al. Plant Thaumatin-like Proteins: Function, Evolution and Biotechnological Applications. Curr. Protein Pept. Sci. 2020;21:36–51. doi: 10.2174/1389203720666190318164905. [DOI] [PubMed] [Google Scholar]
- 48.Kumar S., Stecher G., Tamura K. MEGA7: Molecular evolutionary genetics analysis version 7.0. Molecular biology and evolution. Mol. Biol. Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hu B., Jin J., Guo A.Y., Zhang H., Luo J., Gao G. GSDS 2.0: An Upgraded Gene Feature Visualization Server. Bioinformatics. 2015;31:1296–1297. doi: 10.1093/bioinformatics/btu817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Choi Y., Chan A.P. PROVEAN web server: A tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics. 2015;31:2745–2747. doi: 10.1093/bioinformatics/btv195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Babicki S., Arndt D., Marcu A., Liang Y., Grant J.R., Maciejewski A., Wishart D.S. Heatmapper: Web-enabled heat mapping for all. Nucl. Acids Res. 2016;44:W147–W153. doi: 10.1093/nar/gkw419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Anisimova O.K., Seredin T.M., Danilova O.A., Filyushin M. First Report of Fusarium proliferatum Causing Garlic clove Rot in Russian Federation. Plant Dis. 2021 doi: 10.1094/PDIS-12-20-2743-PDN. [DOI] [Google Scholar]
- 53.Sugui J.A., Deising H.B. Isolation of infection-specific sequence tags expressed during early stages of maize anthracnose disease development. Mol. Plant Pathol. 2002;3:197–203. doi: 10.1046/j.1364-3703.2002.00113.x. [DOI] [PubMed] [Google Scholar]
- 54.Liu M., Wu Z., Jiang F. Selection and validation of garlic reference genes for quantitative real-time PCR normalization. Plant Cell Tissue Organ Cult. 2015;122:435–444. doi: 10.1007/s11240-015-0780-9. [DOI] [Google Scholar]
- 55.Schwinn K.E., Ngo H., Kenel F., Brummell D.A., Albert N.W., McCallum J.A., Pither-Joyce M., Crowhurst R.N., Eady C., Davies K.M. The onion (Allium cepa L.) R2R3-MYB gene MYB1 regulates anthocyanin biosynthesis. Front. Plant Sci. 2016;7:1865. doi: 10.3389/fpls.2016.01865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lescot M. PlantCARE, a Database of Plant Cis-Acting Regulatory Elements and a Portal to Tools for in Silico Analysis of Promoter Sequences. Nucl. Acids Res. 2002;30:325–327. doi: 10.1093/nar/30.1.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
PR genes coding sequences of A. sativum cv. Sarmat/cv. Strelets were deposited in NCBI: AsPR1c (MZ216000/MZ216001), AsPR1d (MZ216002/MZ216003), AsPR1g (MZ216004/MZ216005), AsPR1k (MZ216006/MZ216007), AsPR2a (MZ216008/MZ216009), AsPR2b (MZ216010/MZ216011), AsPR2c (MZ216012/MZ216013), AsPR5a (MZ216014/MZ216015), AsPR5c (MZ216016/MZ216017).