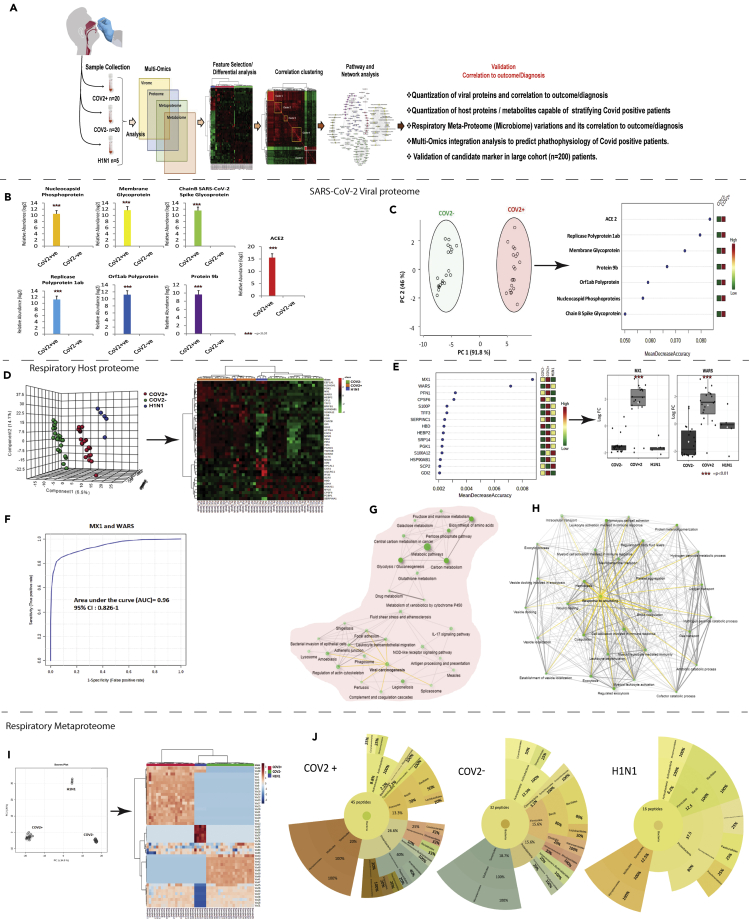
Figure 1.
Quantitative virome profile of the respiratory specimen
(A) Work flow for COVID-19 biomarker identification (discovery phase): Differentially regulated viral/host proteins, metabolites and bacterial peptides were identified which was subjected to random forest and AUROC analysis for identification of candidate for validation. Correlation clustering and pathway analysis provided insight on the pathophysiology of SARS-COV2 infection. Finally the identified candidate indicators were validated in 200 COVID suspect for SARS-CoV-2 infection and outcome prediction.
(B) Log normalized abundance of 6 viral and 1 viral associated proteins identified in the respiratory specimen of COVID-19 positive as compared to COVID-19 negative patients. ∗∗∗ signifies p < 0.05.
(C) Principle component analysis score plot showing segregation of COVID-19 positive patients from COVID-19 negative based on the identified Viral and associated proteins and Random forest analysis showing the mean decrease in accuracy of the viral and associated proteins (Red = upregulated and Green = downregulated) in COVID-19-positive as compared to COVID-19-negative patients.
(D) Partial least square discriminant analysis showing clear segregation of COVID-19-positive patients (Red dots) from COVID-19-negative (Green dots) and influenza A H1N1 pdm 2009 pdm 2009 positive patients (Blue dots) based on host proteomic evaluation and heatmap, hierarchical cluster analysis of top 40 proteins (p < 0.05) capable of segregating COVID-19-positive (orange bar) patients from COVID-19-negative (green bar) or influenza A H1N1 pdm 2009-positive patients (blue bar) (Red = upregulated, Green = downregulated and black = unregulated).
(E) Random forest analysis showing the mean decrease in accuracy of the proteins (Red = upregulated and Green = downregulated and yellow = unchanged) in COVID-19-positive compared to COVID-19-negative or Influenza A H1N1 pdm 2009 pdm 2009-positive patients and relative abundance (Log normalized) for MX1 and WARS showing significant increase in MX1 and WARS levels (p value∗∗ = p < 0.05, ∗∗∗<0.01).
(F) Joint AUROC analysis of MX1 and WARS documenting an AUC = 0.96 CI (0.82–1) p < 0.05 along with prediction class probability score plot showing segregation of CoV2 positive and CoV2 negative.
(G) KEGG pathway analysis of 184 proteins up regulated in COVID-19-positive respiratory specimen as compared to COVID-19-negative or Influenza A H1N1 pdm 2009 pdm 2009-positive cases (FC > 2; p < 0.05, FDR>0.05). Darker nodes are more significantly enriched gene sets. Bigger nodes represent larger gene sets. Thicker edges represent more overlapped genes. Yellow edge show viral linked network.
(H) KEGG pathway analysis of 60 proteins downregulated in COVID-19-positive respiratory specimen as compared to COVID-19-negative or Influenza A H1N1 pdm 2009 pdm 2009-positive cases (FC > 2; p < 0.05, FDR>0.05). Darker nodes are more significantly enriched gene sets. Bigger nodes represent larger gene sets. Thicker edges represent more overlapped genes. Three clusters of edges can be identified: (A) gas transport and oxygen transport cluster (B) wound healing and inflammation cluster and (C) vesicle transport cluster. Yellow edge show wound healing linked network.
(I) Principle component analysis (PCA) showing clear segregation of COVID-19-positive patients based on metaproteins estimations and hierarchical cluster analysis of the metaproteins (p < 0.05) show clear segregation of COVID-19-positive patients (Red bar) from COVID-19-negative (Green bar) or Influenza A H1N1 pdm 2009-positive patients (Blue bar). (Dark brown = upregulated, Blue = downregulated and white = unregulated).
(J) Sunburst plot representative of microbial population difference (phylum: order: family) in COVID-19-positive respiratory specimen as compared to COVID-19 negative or Influenza A H1N1 pdm 2009-positive cases. Percent distribution of identified peptide is provided in each sunburst plot.