Abstract
Simple Summary
C-X-C chemokine receptor type 4 (CXCR4), a G-protein-coupled receptor, has been demonstrated to stimulate proliferation and invasiveness of many different tumors, including colorectal cancer. Through in vitro evidence, overexpression of CXCR4 has been identified as a negative prognostic factor in colorectal cancer. The identification of prognostic biomarkers can improve the prediction of disease evolution and disease characterization, and guide treatment efforts. This systematic review with a meta-analysis was conducted to pool hazard ratios from prognostic studies on CXCR4, provide an updated estimate of prognostic power of CXCR4, and analyze modalities of evaluating and reporting CXCR4 expression.
Abstract
Background: This study was conducted to provide an updated estimate of the prognostic power of C-X-C chemokine receptor type 4 (CXCR4) in colorectal cancer (CRC), and analyze modalities of evaluating and reporting its expression. Methods: A systematic review with meta-analysis was performed and described according to the Preferred Reporting Items for Systematic Reviews and Meta-Analyses statement. Studies were identified through PubMed and Google Scholar. The pooled hazard ratios (HRs) for overall survival (OS) or progression-free survival (PFS) with 95% confidence interval (CI) were estimated with the random-effect model. Results: Sixteen studies were selected covering a period from 2005 to 2020. An immunohistochemical evaluation of CXCR4 was performed in all studies. Only in three studies assessment of mRNA through RT–PCR was correlated with prognosis; in the remaining studies, the authors identified prognostic categories based on immunohistochemical expression. In pooled analyses, significant associations were found between positive or high or strong expression of CXCR4 and T stage ≥3 (P = 0.0001), and positive or high or strong expression of CXCR4 and left side primary tumor localization (P = 0.0186). The pooled HR for OS was 2.09 (95% CI: 1.30–2.88) in favor of high CXCR4 expression; for PFS, it was 1.42 (95% CI: 1.13–1.71) in favor of high CXCR4 expression. Conclusion: High CXCR4 expression is clearly associated with increased risk of death and progression in CRC. However, strong methodologic heterogeneity in CXCR4 assessment hinders direct translation into clinical practice; thus, a consensus to streamline detection and scoring of CXCR4 expression in CRC is indicated.
Keywords: CXCR4, colorectal cancer, prognosis, overall survival
1. Introduction
C-X-C chemokine receptor type 4 (CXCR4) belongs to the G-protein-coupled receptor superfamily, and it is expressed in a wide variety of cells, predominantly of hematopoietic origin. It binds to C-X-C motif chemokine ligand 1 (CXCL12), also called stromal cell-derived factor-1α (SDF-1α) and mediates a potent chemotactic stimulus [1]. In embryos, it has a major role in processes of neurogenesis, influencing the migration of neurons from neuroprogenitor cells [2]; in adults, one of the most important biological roles of the CXCR4/SDF-1α axis is the regulation of hematopoietic stem cell homing to the bone marrow [3]. However, CXCR4 has been demonstrated to stimulate proliferation and invasiveness of many different tumors including prostate [4], breast [5], lung [6,7,8], melanoma [9,10], glioblastoma [11,12], lymphoma [13,14], and colorectal cancer [15,16]. Through in vitro evidence, overexpression of CXCR4 has been identified as a negative prognostic factor in many different neoplasms [17,18,19,20].
Colorectal cancer (CRC) is the third most common cause of cancer-related deaths worldwide. Survival rates at five years strictly depend on the stage at diagnosis, varying from 90% of American Joint Committeeon Cancer 8th Edition (AJCC) stages I-II to 10% of stage IV [21]. The survival rate of Stage III patients is about 40% and it has been improved in recent years with the administration of adjuvant chemotherapy [22]. The survival of metastatic colorectal cancer (mCRC) patients significantly improved in recent years with the introduction of target-oriented drugs and a better selection of patients based on biologic/molecular characteristics (KRAS/NRAS/BRAF mutations, MSI, HER-2 overexpression, and other molecular markers) [23]. The identification of new prognostic cancer biomarkers is important because it can improve the prediction of disease evolution, enhance disease characterization, and guide treatment efforts. CXCR4 expression is considered a prognostic marker in CRC. However, patients′ risk stratification requires rigorous scientific validation. We previously reported that CXCR4 is able to predict progression-free (PFS) [24] and overall survival (OS) [25] in CRC.
This study was conducted to pool hazard ratios from prognostic studies on CXCR4, provide an updated estimate of prognostic power of CXCR4, and analyze modalities of evaluating and reporting CXCR4 expression.
2. Materials and Methods
2.1. Search Strategy
This meta-analysis was performed and described according to the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) statement [26]. Two-hundred seventy-three studies were identified through PubMed and Google Scholar searching with the following key words algorithm: “colorectal cancer” OR “colorectal tumor” OR “colorectal carcinoma” AND “CXCR4” OR “cxc chemokine receptor type 4” AND “prognosis” OR “disease free survival” OR “progression” OR “survival” (last update on 16 December 2020).
2.2. Study Eligibility
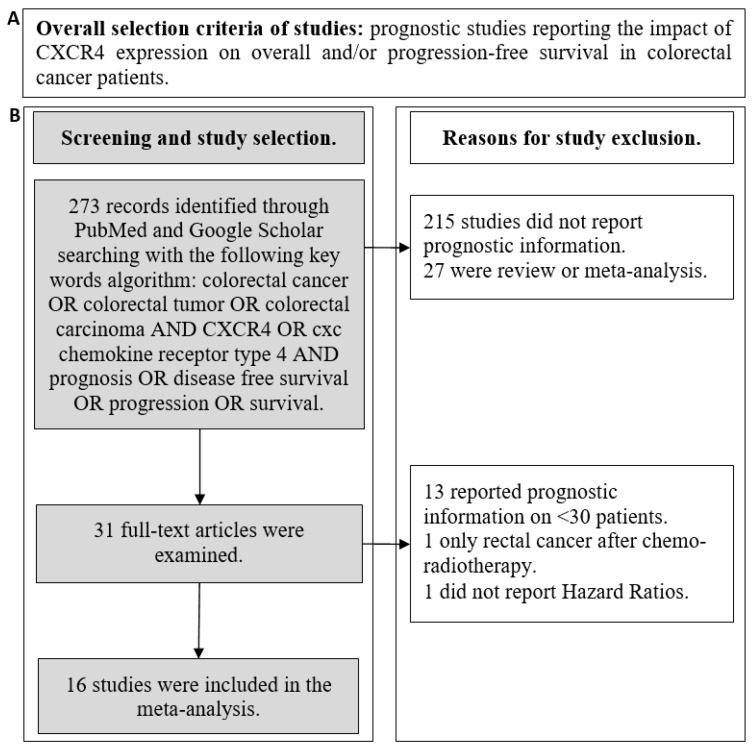
A flowchart summarizing the criteria for studies selection and exclusion is reported in Figure 1. Abstracts of studies in English language that were initially identified were examined to exclude those not reporting prognostic information. Thereafter, all full texts were retrieved and analyzed. Studies were included in the final analysis if they (1) reported prognostic data (association with PFS or OS) about the expression of CXCR4 in CRC patients, (2) reported hazard ratios (HRs) with 95% confidence intervals (CIs), and, (3) had a sample size >30 patients. All the studies presented a score > 6 at the Newcastle–Ottawa Scale for methodology quality assessment [27]. Articles reporting the prognostic role of concomitant biomarker expression (i.e., CXCR4/SDF-1α, CXCR4/CD133, etc.) were excluded.
Figure 1.
Overall selection criteria for study selection (A). Detailed flowchart reporting the criteria for study selection and exclusion (B).
2.3. Data Extraction
The following data were extracted by four investigators for each publication: first author; year of publication; accrual time; number of patients; methods for CXCR4 assessment (including details on IHC scores and eventual fresh tissue evaluation); information about morphologic localization of immunohistochemical CXCR4 expression; eventual presence of ancillary studies; information on study design; association with clinico-pathological variables (age, sex, lymph-nodes involvement, stage, T, side, clinical response, and KRAS mutational status); information on follow-up; HRs of progression and/or death with 95% CIs. Criticisms and/or discordances were discussed between all authors to reach a consensus.
2.4. Hazard Ratio Interpretation
A hazard ratio of 1.0 indicates an identical risk (event probability (EP)) between high and low CXCR4-expressing groups (EP CXCR4 high/EP CXCR4 low). An HR greater than 1.0 indicates that a high CXCR4-expressing group at the numerator has an increased risk of death or progression. When a study reported an HR with low CXCR4 in the numerator (CXCR4 low vs. high), the HR and CI were recalculated (the calculated HRCXC4 high vs. low was 1/HRCXC4 low vs. high) according to Altman et al. [28] in order to harmonize the comparison trajectory (CXCR4 high vs. CXCR4 low).
2.5. Statistical Analysis
The present meta-analysis was performed in order to assess the prognostic impact of CXCR4 expression in terms of OS and PFS in CRC. The secondary end-points were the analysis of the association between CXCR4 and clinico-pathologic variables, and the description of methods and scores used to assess it. Given the significant heterogeneity (see above) among the selected studies, the analysis was performed with the random-effects model. It aims to provide a more conservative estimate of the pooled HR and it is the preferred model when heterogeneity is present. Under the random-effects model, the true effects are assumed to vary between studies, and the summary effect is the weighted average of the effects reported in the different studies [29]. Meta-analysis is depicted in classical forest plots, with point estimates, 95% CIs for each HR, and a final pooled HR.
Heterogeneity was evaluated through I2, that is, the percentage of observed total variation across studies due to real heterogeneity rather than chance. It is calculated as I2 = 100% × (Q − DF)/Q, where Q is Cochran′s heterogeneity statistic and DF is the degrees of freedom. Negative values of I2 are set to be equal to zero so that I2 lies between 0% and 100%. A value of 0% indicates no observed heterogeneity, and larger values show increasing heterogeneity [30]. The risk of publication bias was also evaluated with funnel plot analysis and Egger′s test [31]. The latter is a test for the Y intercept = 0 from a linear regression of normalized effect estimate (estimate divided by its standard error) against precision (reciprocal of the standard error of the estimate). P < 0.005 indicates a significant publication bias.
Associations between CXCR4 expression and clinico-pathological variables were evaluated with the chi-square test.
Analyses were performed with the MedCalc Statistical Software (MedCalc® Statistical Software version 19.6, MedCalc Software Ltd., Ostend, Belgium; https://www.medcalc.org (accessed on 19 December 2020).
3. Results
3.1. Study Characteristics
Sixteen studies were selected covering a period from 2005 to 2020 [24,25,32,33,34,35,36,37,38,39,40,41,42,43,44,45]. The accrual time varied from a minimum of 3 to 14 years. The number of enrolled patients ranged from 31 to 684. Only four studies reported ancillary data including evaluation of CXCR4-related biologic pathways. All studies were retrospective and had a Newcastle–Ottawa Scale score ≥ 6 (Table S1). Most articles described the prognostic power of CXCR4 in stages I-IV or stage IV disease (11/16). The reporting of association with clinico-pathological characteristics was heterogeneous (lymph-nodal status: 13/16; T status: 7/16; side: 5/16); however, very few studies reported data about association with clinical response (1/16) or KRAS status (2/16) (Table S2).
3.2. CXCR4 Expression Methodology
The methodology to assess CXCR4 expression is crucial to identifying prognostic categories and adequately interpreting results. Therefore, we performed a detailed analysis of technical modalities of CXCR4 evaluation (Table 1). Immunohistochemical evaluation of CXCR4 was performed in all studies. Evaluation of fresh tumor tissue was performed only by Kim et al. In eight studies, an mRNA assessment was added (through RT-PCR or FISH). In only three studies assessment of mRNA through RT-PCR was correlated with prognosis; in the remaining studies, the authors identified prognostic categories based on immunohistochemical expression. Ten studies differentiated nuclear versus cytoplasmic CXCR4 expression. Only one study referred to membrane CXCR4 expression. Modalities of building expression scores (number of categories, number of positive cells, and inclusion of staining intensity) were heterogeneous. A detailed description is reported in Table 1.
Table 1.
Description of CXCR4 assessment methodology among selected studies.
| First Author |
Methods | Fresh Tissue Evaluation | Detection Correlated with TTO | Differential Nuclear, Cytoplasmic, Membrane IHC Staining | IHC Distribution Correlated with Prognosis | Scores | No. of Positive Cells for Expression Evaluation | Inclusion of “Staining Intensity” |
|---|---|---|---|---|---|---|---|---|
| Kim J. | IHC, RT-PCR | Yes | RT-PCR | NA | NA | Low vs.High | mRNA median CXCR4 expression | No |
| Ottaiano A. | IHC | No | IHC | Yes | Overall expression | Neg/Low vs. High | ≤50% vs.>50% | No |
| Yoshitake N. | IHC, Western Blot | No | IHC | Yes | Overall expression | Neg vs. Pos | No CXCR4 immunoreactivity vs others | No |
| Speetjens F.M. | RT-PCR | No | RT-PCR | NA | NA | Low vs. High | mRNA median CXCR4 expression | No |
| Speetjens F.M. | IHC | No | IHC | Yes | Nuclear expression | Weak vs. Strong | Examples are included in the work | Yes |
| Ingold B. | IHC | No | IHC | No | Overall expression | 0, 1+, 2+, 3+ | NA | Yes Absent, faint cytoplasmic, moderate cytoplasmic and slight membranous, strong cytoplasmic and strong membranous |
| Wang S.C. | IHC, RT-PCR | No | IHC | Yes | Nuclear expression | 0, 1+, 2+, 3+ | 0; <30%; 30–50%; >50% |
Yes Weak, medium, strong, very strong |
| Yopp A.C. | IHC | No | IHC | Yes | Overall expression | Neg vs. Pos | ≤10% vs.>10% | No |
| Sakai N. | IHC, fluorescence microscopy | No | IHC | Cyto | Cytoplasm | Low vs. High | NA | Yes Relative to the staining intensity of hepatocytes |
| Sakai N. | IHC, fluorescence microscopy | No | IHC | Nucleus | Nuclear | Neg vs. Pos | NA | Yes Relative to the staining intensity of hepatocytes |
| Du C. | IHC | No | IHC | Not Specified | Overall expression | 0, 1+, 2+: Low3+: High | <1%; 1–50% (1+ and 2+); >50% |
Yes Negative, weak, strong |
| Gao Y. | IHC, RT-PCR | No | IHC | No | Overall expression | Sporadic, focal, diffuse | <10%, ≥11<50%, ≥50% | Yes Negative, weak, moderate, strong |
| Stanisavljevic L. (cohort 1) |
IHC, ISH | No | IHC | Yes | Nuclear expression | Low vs. High | 0–20%, >20% | No |
| Stanisavljevic L. (cohort 2) |
IHC | No | IHC | Yes | Nuclear expression | Low vs. High | 0–20%, >20% | No |
| D′Alterio C. | IHC, RT-PCR | No | IHC | Yes | Overall expression | Negative/Low vs. High | 0–50%, >50% | No |
| Wu W. | IHC, RT-PCR | No | RT-PCR | NA | NA | Low vs. High | mRNA median CXCR4 expression | No |
| Weixler B. | IHC | No | IHC | No | Overall expression | Histoscores (a continuous variable) |
(% of positive cells)x (staining intensity) | Yes Negative, 0; weak, 1; moderate, 2; strong, 3 |
| Xu C. | IHC, RT-PCR | No | IHC | No | Membrane | 0, 1, 2, 3, 4 | 0%, 1–25%, 26–50%, 51–75%, >75% | Yes No staining, weak, moderate, strong |
| Ottaiano A. | IHC, RT-PCR | No | IHC | Yes | Overall expression | Neg/Low vs. High | ≥0≤50%, >50% | No |
IHC: Immunohistochemistry; ISH: In situ hybridization; mRNA: messenger ribonucleic acid; NA: not applicable; Neg: negative; Pos: positive; RT-PCR: reverse transcriptase-polymerase chain reaction; TTO: time to outcome.
3.3. Association between CXCR4 Expression and Clinico-Pathological Characteristics of Colorectal Cancer Patients
Exploration of association between a potential biomarker in cancer and patients′ clinico-pathological characteristics is important to generate hypotheses on its biologic role, to improve disease extent prediction, and to prevent biases in subsequent prognostic analyses. Table 2 reports a detailed description of the clinico-pathological characteristics of the patients and tumors according to CXCR4 expression in the selected studies. In a pooled analysis, significant associations were found between positive or high or strong expression of CXCR4 and T stage > 3 (cancer growing outside the muscularis propria) (P = 0.0001), and positive or high or strong expression of CXCR4 and left-side primary tumor localization (P = 0.0186) (Table S3).
Table 2.
Clinico-pathological characteristics according to CXCR4 expression.
| Author | Year | CXCR4 Scores | Age | Sex | T | Side | Lymph nodes | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Young | Old | Male | Female | ≤2 | ≥3 | Left | Right | Involved | Not Involved | |||
| Kim J. | 2005 | Low 44 | 23 | 21 | 18 | 9 | - | - | 8 | 36 | ||
| High 48 | 22 | 26 | 11 | 19 | - | - | 8 | 40 | ||||
| Ottaiano A. | 2006 | Neg 16 | <70:10 | ≥70:6 | 6 | 10 | 5 | 11 | 0 | 15 | ||
| Low 25 | <70:13 | ≥70:12 | 15 | 10 | 13 | 12 | 8 | 18 | ||||
| High 31 | <70:21 | ≥70:10 | 17 | 14 | 11 | 20 | 7 | 24 | ||||
| Yoshitake N. | 2008 | Negative 13 | - | - | 10 | 3 | - | - | - | - | 7 | 6 |
| Positive 47 | - | - | 31 | 16 | - | - | - | - | 38 | 9 | ||
| Speetjens F.M. | 2009 | Low 35 | <68.5:20 | >68.5:15 | 16 | 19 | - | - | 17 | 18 | 12 | 23 |
| High 35 | <68.5:15 | >68.5:20 | 19 | 16 | - | - | 17 | 18 | 11 | 24 | ||
| Speetjens F.M. | 2009 | Strong 43 | <69.7:21 | >69.7:22 | 21 | 22 | - | - | 22 | 21 | 15 | 28 |
| Weak 15 | <69.7:8 | >69.7:7 | 7 | 8 | - | - | 5 | 10 | 4 | 11 | ||
| Ingold B. | 2009 | Negative 267 | ≤65:115 | >65:152 | 145 | 122 | 46 | 206 | - | - | 133 | 119 |
| Positive 135 | ≤65:51 | >65:84 | 69 | 66 | 23 | 108 | - | - | 72 | 59 | ||
| Wang SC. | 2010 | Negative 245 | - | - | 180 | 65 | 138 | 107 | 186 | 59 | 142 | 141 |
| Positive 143 | - | - | 89 | 54 | 47 | 96 | 99 | 44 | 101 | 158 | ||
| Yopp A.C. | 2012 | Negative 28 | ≤60:11 | >60:17 | 13 | 15 | - | - | - | - | - | - |
| Positive 47 | ≤60:21 | >60:26 | 38 | 9 | - | - | - | - | - | - | ||
| Sakai N. | 2012 | Low 56 (cytoplasm) | <60:26 | ≥60:30 | 36 | 20 | - | - | - | - | - | - |
| High 36 (cytoplasm) | <60:10 | ≥60:26 | 22 | 14 | - | - | - | - | - | - | ||
| Du C. | 2014 | Low 89 | <65:39 | ≥65:50 | 51 | 38 | 16 | 73 | 47 | 42 | 6 | 83 |
| High 56 | <65:19 | ≥65:37 | 33 | 23 | 9 | 47 | 29 | 27 | 4 | 52 | ||
| Gao Y. | 2014 | Negative 512 | <55:243 | ≥55:269 | 292 | 220 | - | - | - | - | 201 | 311 |
| Positive 208 | <55:89 | ≥55:119 | 120 | 88 | - | - | - | - | 138 | 70 | ||
| Stanisavljevic L. | 2015 | Low 78 | - | - | 45 | 33 | 3 | 75 | - | - | - | - |
| High 186 | - | - | 93 | 93 | 10 | 176 | - | - | - | - | ||
| Stanisavljevic L. | 2015 | Low 35 | - | - | 20 | 15 | 6 | 29 | - | - | - | - |
| High 190 | - | - | 110 | 80 | 28 | 162 | - | - | - | - | ||
| D′Alterio C. | 2016 | Negative/Low 10 | - | - | - | - | - | - | - | - | - | - |
| High 21 | - | - | - | - | - | - | - | - | - | - | ||
| Wu W. | 2016 | Low 40 | - | - | - | - | - | - | - | - | - | - |
| High 40 | - | - | - | - | - | - | - | - | - | - | ||
| Weixler B. | 2017 | Low 289 | - | - | 145 | 144 | 51 | 230 | 205 | 82 | 142 | 141 |
| High 267 | - | - | 117 | 150 | 60 | 204 | 190 | 77 | 101 | 158 | ||
| Xu C. | 2018 | Low 26 | <60:21 | ≥60:5 | 16 | 10 | 4 | 22 | - | - | 1 | 25 |
| High 22 | <60:20 | ≥60:4 | 18 | 4 | 5 | 17 | - | - | 5 | 17 | ||
| Ottaiano A. | 2020 | Negative/Low 26 | ≤65:12 | >65:14 | 17 | 9 | - | - | 19 | 7 | - | - |
| High 52 | ≤65:20 | >65:32 | 27 | 25 | - | - | 26 | 26 | - | - | ||
3.4. Time to Outcome According to CXCR4 Expression
The primary endpoint of this meta-analysis was to provide a pooled and updated estimate of the prognostic value of CXCR4 expression in CRC. Data regarding timeto outcome (OS and/or PFS) were extracted and are reported in Table 3. Three studies reported both OS and PFS, five reported PFS, and eight reported OS.
Table 3.
Follow-up, time to outcome, and hazard ratios of progression and/or death in selected studies.
| Author | Year | Median Follow-Up (Months) | Median PFS (Months) | HR | CI | P | Median OS (Months) |
HR | CI | P |
|---|---|---|---|---|---|---|---|---|---|---|
| Kim J. | 2005 | 28 | NR | 1.35 | 1.09–1.68 | 0.0065 | High 9; Low 23 |
2.53 | 1.19–5.40 | 0.016 |
| Ottaiano A. | 2006 | 23 | NR | 3.01 | 0.88–5.21 | 0.0991 | NR | |||
| Yoshitake N. | 2008 | NR | 5.08 | 0.65–40.00 | 0.123 | |||||
| Speetjens F.M. | 2009 | NR | NR | 2 | 1.1–3.7 | 0.03 | NR | 1.8 | 1–3.6 | 0.07 |
| Speetjens F.M. | 2009 | NR | NR | 2.6 | 1–6.2 | 0.04 | NR | 3.7 | 1.35–11 | 0.02 |
| Ingold B. | 2009 | 32 | NR | 2.87 | 1.31–6.29 | 0.009 | ||||
| Wang SC. | 2010 | 61 | 5 years DFS rate: High 70%; Low 55% (Nuc CXCR4) |
1.23 | 0.7–2.18 | 0.458 | ||||
| Yopp A.C. | 2012 | 68 | Pos. 15 vs Neg. 73 | 2.2 | 1.2–4.2 | 0.012 | ||||
| Sakai N.* | 2012 | 38 | 3 years OS rate: High 67%; Low 78% |
Cyto: 0.43 | Cyto: 0.18–1.02 | Cyto: 0.056 | ||||
| Sakai N.* | 2012 | 38 | 3 years OS rate: Pos 93%; Neg 67% |
Nuc: 4.05 | Nuc: 1.19–13.8 | Nuc: 0.025 | ||||
| Du C.* | 2014 | 68.5 | 5 years DFS rate: High 76.8%; Low 84.3% |
0.81 | 0.36–1.8 | 0.618 | ||||
| Gao Y. | 2014 | NR | 1.3 | 1.38–1.85 | 0.001 | |||||
| Stanisavljevic L.* | 2015 | Min from 3–5 years | 5 years DFS rate: High 65%; Low 85% |
0.42 | 0.22–0.78 | 0.006 | ||||
| Stanisavljevic L.* | 2015 | Min from 3–5 years | High 82%; Low 89% |
0.89 | 0.31–2.61 | 0.838 | ||||
| D′Alterio C.* | 2016 | 28 | High 14 vs. Neg/Low 46 | 3.405 | 1.70–17.33 | 0.004 | High 28; Neg/Low 46 | 0.079 | 0.062–0.480 | 0.0008 |
| Wu W. | 2016 | Max 60 | NR | 5.38 | 2.42–9.13 | 0.002 | ||||
| Weixler B. | 2017 | NR | 5 years OS rate: High 48%; Low 48% |
0.99 | 0.99–1.0 | 0.322 | ||||
| Xu C.* | 2018 | NR | High 51; Low 54 | 0.188 | 0.03–0.75 | 0.020 | ||||
| Ottaiano A. | 2020 | 53 | High 19; Neg/Low 31 | 3.18 | 2.01–5.02 | 0.0312 |
* HR was transformed in forest plot (see Methods).CI: confidence interval; Cyto: cytoplasmic; DFS: disease-free survival; HR: hazard ratio; Neg: negative; NR: not reported; Nuc: nuclear; OS: overall survival.
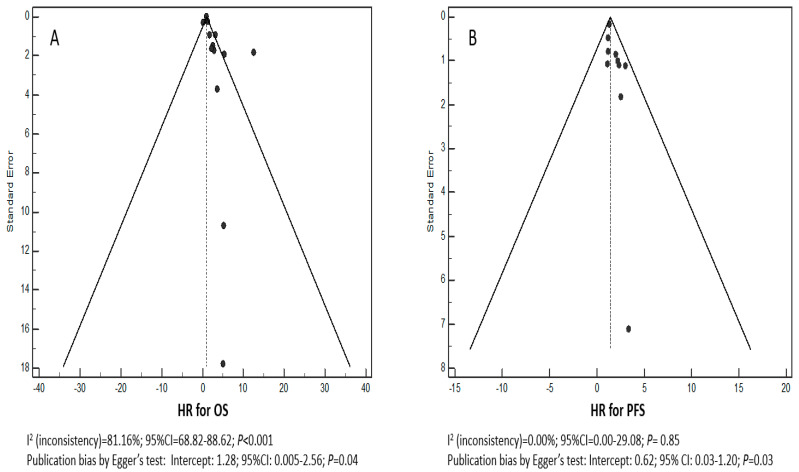
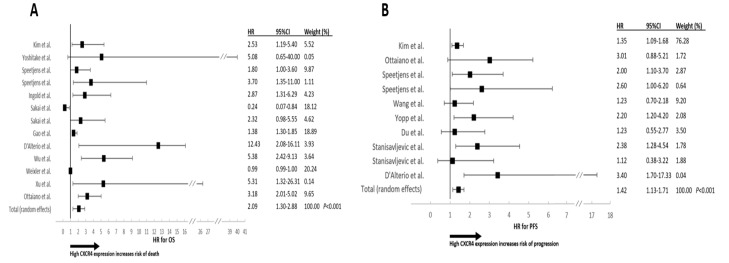
Funnel plots for the HRs of OS and PFS were asymmetric (Figure 2A,B) with a significant I2 test for OS (P < 0.001). Egger′s test was significant for both OS (P = 0.04) and PFS (P = 0.03). The meta-analysis was performed with the random-effects model in order to obtain a more conservative and reliable estimate of the pooled HR, and no attempts were made to conduct a subgroup meta-analysis. A forest plot of treatment effect on OS is shown in Figure 3A. The pooled HR was 2.09 (95% CI: 1.30–2.88) in favor of high CXCR4 expression. The effect on PFS is shown in Figure 3B where the pooled HR is 1.42 (95% CI: 1.13–1.71) in favor of high CXCR4 expression.
Figure 2.
Funnel plots of selected studies for overall survival (OS) (A) and progression-free survival (PFS) (B).
Figure 3.
Forest plots for OS(A) and PFS(B) according to CXCR4 expression. Studies are reported by author′s first name, HRs, 95% CIs and percent of the weighted effects (see Methods). The pooled effect is reported in the last line.
4. Discussion
Identification and validation of predictive and/or prognostic biomarkers in CRC have revolutionized the management of the disease (BRAF, K-, N-RAS, and MSI), and others will also be used in the near future in clinical practice (HER2 and PD-1/PD-L1) [23]. Given relevant biologic reasons, CXCR4 has been explored for many years as a potential prognostic marker in CRC. Our group and others previously reported that CXCR4 expression predicted PFS and OS in CRC [24,25,32,33,34,35,36,37,38,39,40,41,42,43,44,45]. In the present study, we aimed to provide a more accurate and updated estimate of the prognostic power of CXCR4 considering some contradictory results, the maturity of available data, and the intense interest around the role of CXCR4 in modulating the metastatic behavior of CRC.
The search of articles was systematic, and the selection primarily based on a few characteristics indirectly related to the overall quality of the articles (>30 patients enrolled, HRs and CXCR4 expression methods clearly reported, and evaluation of CXCR4 before any treatment).
We found that high CXCR4 expression is clearly associated with increased risk of death and progression (HR OS: 2.09; 95% CI: 1.30–2.88 and HR PFS: 1.42; 95% CI: 1.13–1.71). However, the following major issue deserves to be evidenced and discussed: a strong methodologic heterogeneity in CXCR4 assessment was identified. Table 1 shows a wide heterogeneity in CXCR4 assessment methodology regarding techniques, scores, and categories. Some studies evaluated the membrane or nuclear expression values; however, the evaluation of the total expression value can provide an objective determination of the level of expression of CXCR4. CXCR4 nuclear localization is an atypical compartmentalization of the receptor, likely linked to its still unknown functions, and an IHC determination not sensitive enough to specifically detect the subcellular localization of a molecule, especially in paraffin-embedded tissue can be affected by artifacts and by cell space overlapping. The use of antibodies standardized for IHC studies should be recommended, and standard methods of expression evaluation scores, including the apparent subcellular localization, should be developed with an appropriate consensus between pathologists. Western blotting analysis of protein expression should be avoided because it is a qualitative method that does not distinguish the cell subtype source of the assessed protein and does not assure that the protein has been extracted by cancer cells or other tumor microenvironment cells. qRT-PCR is a quantitative technique, but it requires a strict standardization of the procedure, the selection of the tumor area by a pathologist, and the extraction of RNA from tumor tissues that are often paraffin-embedded, with the consequent decrease inthe RNA sample quality. It is likely that the best procedure would be a world-standardized IHC procedure performed with an automated test. Therefore, this methodological issue in CXCR4 assessment, along with the physiologic heterogeneity in treatments, a downsized sample size (nine studies enrolled <100 patients), and the retrospective nature of all studies are responsible for the large HR confidence intervals in some studies. Finally, the evidence of a publication bias makes these heterogeneities even more relevant. In this regard, we cannot rule out the hypothesis that selection of the studies we applied could have been influenced by this publication bias. Moreover, all selected studies were retrospective. Therefore, their nature is intrinsically biased by mostly unknown and uncontrolled clinical (patients′ location, selection, treatments, etc.) and methodological (techniques, reagents, methods, etc.) biases. Based on these considerations, our group is planning the first prospective evaluation of CXCR4 in both primary and/or metastatic tissues (resected metastases or biopsies) in order to predict the time to relapse after surgery and the response to therapy/subsequent prognosis in CRC. Assessment of CXCR4 will be performed through IHC according to a previously published homogeneous evaluation method (negative, low, high) [24,25]. The hypothesis of the study is based on the following statistical assumptions: (i) HR for high expression of CXCR4 of 2.09 (vs. negative/low), (ii) test power of 80%, (iii) alpha value of the I-type error of 5%, and (iv) median survival of 18 months (in unselected mCRC patients). The survival curves will be depicted with the Kaplan–Meier method, and the statistical significance verified with a two-tailed log-rank test. The final sample will be 200 patients.
5. Conclusions
For the first time, we showed a detailed and critical analysis of technical approaches applied to assess CXCR4 in CRC, finding a wide diversity in the modalities of assessment and the reporting of the receptor expression. This strong methodological heterogeneity hinders direct translation into clinical practice, suggesting that CXCR4 assessment should be revised and harmonized. Based on this, a consensus among experts to harmonize detection and scoring of CXCR4 expression in CRC should be reached; the present work may represent a critical starting point to discussions about methodological issues regarding the assessment of CXCR4 in CRC as a prognostic factor.
Acknowledgments
We are grateful for Alessandra Trocino, Librarian at Istituto Nazionale Tumori di Napoli, IRCCS “G. Pascale”, Italy, for bibliographic assistance. We thank Daniela Capobianco, Istituto Nazionale Tumori di Napoli, IRCCS “G. Pascale”, Italy, for assistance with data management.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/cancers13133284/s1. Table S1: Study characteristics I, Table S2: Study characteristics II, Table S3: Pooled analysis of CXCR4 expression according to clinico-pathological characteristics.
Author Contributions
Conceptualization, A.O. and M.S.; methodology, A.O. and M.S.; software, A.O.; validation, M.C., G.N., and F.P.; formal analysis, A.O. and P.D.P.; investigation, A.O., M.S., P.D.P., and G.N.; resources, G.N.; data curation, A.O.; writing—original draft preparation, A.O. and M.C.; writing—review and editing, S.S., M.S., and G.N.; supervision, G.N. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available on request from the corresponding author.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher′s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Kucia M., Jankowski K., Reca R., Wysoczynski M., Bandura L., Allendorf D.J., Zhang J., Ratajczak J., Ratajczak M.Z. CXCR4-SDF-1 signalling, locomotion, chemotaxis and adhesion. J. Mol. Histol. 2004;35:233–245. doi: 10.1023/B:HIJO.0000032355.66152.b8. [DOI] [PubMed] [Google Scholar]
- 2.Hwang H.M., Ku R.Y., Hashimoto-Torii K. Prenatal environment that affects neuronal migration. Front. Cell. Dev. Biol. 2019;7:138. doi: 10.3389/fcell.2019.00138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kawaguchi N., Zhang T.T., Nakanishi T. Involvement of CXCR4 in Normal and Abnormal Development. Cells. 2019;8:185. doi: 10.3390/cells8020185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Taichman R.S., Cooper C., Keller E.T., Pienta K.J., Taichman N.S., McCauley L.K. Use of the stromal cell-derived factor-1/CXCR4 pathway in prostate cancer metastasis to bone. Cancer Res. 2002;62:1832–1837. [PubMed] [Google Scholar]
- 5.Müller A., Homey B., Soto H., Ge N., Catron D., Buchanan M.E., McClanahan T., Murphy E., Yuan W., Wagner S.N., et al. Involvement of chemokine receptors in breast cancer metastasis. Nature. 2001;41:50–56. doi: 10.1038/35065016. [DOI] [PubMed] [Google Scholar]
- 6.Kijima T., Maulik G., Ma P.C., Tibaldi E.V., Turner R.E., Rollins B., Sattler M., Johnson B.E., Salgia R. Regulation of cellular proliferation, cytoskeletal function, and signal transduction through CXCR4 and c-Kit in small cell lung cancer cells. Cancer Res. 2002;62:6304–6311. [PubMed] [Google Scholar]
- 7.Burger M., Glodek A., Hartmann T., Schmitt-Gräff A., Silberstein L.E., Fujii N., Kipps T.J., Burger. J.A. Functional expression of CXCR4 (CD184) on small-cell lung cancer cells mediates migration, integrin activation, and adhesion to stromal cells. Oncogene. 2003;22:8093–8101. doi: 10.1038/sj.onc.1207097. [DOI] [PubMed] [Google Scholar]
- 8.Hartmann T.N., Burger J.A., Glodek A., Fujii N., Burger M. CXCR4 chemokine receptor and integrin signaling co-operate in mediating adhesion and chemoresistance in small cell lung cancer (SCLC) cells. Oncogene. 2005;24:4462–4471. doi: 10.1038/sj.onc.1208621. [DOI] [PubMed] [Google Scholar]
- 9.Bartolomé R.A., Gálvez B.G., Longo N., Baleux F., Van Muijen G.N., Sánchez-Mateos P., Arroyo A.G., Teixidó J. Stromal cell-derived factor-1alpha promotes melanoma cell invasion across basement membranes involving stimulation of membrane-type 1 matrix metalloproteinase and Rho GTPase activities. Cancer Res. 2004;64:2534–2543. doi: 10.1158/0008-5472.CAN-03-3398. [DOI] [PubMed] [Google Scholar]
- 10.Scala S., Giuliano P., Ascierto P.A., Ieranò C., Franco R., Napolitano M., Ottaiano A., Lombardi M.L., Luongo M., Simeone E., et al. Human melanoma metastases express functional CXCR4. Clin. Cancer Res. 2006;12:2427–2433. doi: 10.1158/1078-0432.CCR-05-1940. [DOI] [PubMed] [Google Scholar]
- 11.Barbero S., Bonavia R., Bajetto A., Porcile C., Pirani P., Ravetti J.L., Zona G.L., Spaziante R., Florio T., Schettini G. Stromal cell-derived factor 1alpha stimulates human glioblastoma cell growth through the activation of both extracellular signal-regulated kinases 1/2 and Akt. Cancer Res. 2003;63:1969–1974. [PubMed] [Google Scholar]
- 12.Rubin J.B., Kung A.L., Klein R.S., Chan J.A., Sun Y., Schmidt K., Kieran M.W., Luster A.D., Segal R.A. A small-molecule antagonist of CXCR4 inhibits intracranial growth of primary brain tumors. Proc. Natl. Acad. Sci. USA. 2003;100:13513–13518. doi: 10.1073/pnas.2235846100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Corcione A., Ottonello L., Tortolina G., Facchetti P., Airoldi I., Guglielmino R., Dadati P., Truini M., Sozzani S., Dallegri F., et al. Stromal cell-derived factor-1 as a chemoattractant for follicular center lymphoma B cells. J. Natl. Cancer Inst. 2000;92:628–635. doi: 10.1093/jnci/92.8.628. [DOI] [PubMed] [Google Scholar]
- 14.Bertolini F., Dell′Agnola C., Mancuso P., Rabascio C., Burlini A., Monestiroli S., Gobbi A., Pruneri G., Martinelli G. CXCR4 neutralization, a novel therapeutic approach for non-Hodgkin′s lymphoma. Cancer Res. 2002;62:3106–3112. [PubMed] [Google Scholar]
- 15.Zeelenberg I.S., Ruuls-Van Stalle L., Roos E. The chemokine receptor CXCR4 is required for outgrowth of colon carcinoma micrometastases. Cancer Res. 2003;63:3833–3839. [PubMed] [Google Scholar]
- 16.Ottaiano A., Di Palma A., Napolitano M., Pisano C., Pignata S., Tatangelo F., Botti G., Acquaviva A.M., Castello G., Ascierto P.A., et al. Inhibitory effects of anti-CXCR4 antibodies on human colon cancer cells. Cancer Immunol. Immunother. 2005;54:781–791. doi: 10.1007/s00262-004-0636-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jiang Q., Sun Y., Liu X. CXCR4 as a prognostic biomarker in gastrointestinal cancer: A meta-analysis. Biomarkers. 2019;24:510–516. doi: 10.1080/1354750X.2019.1637941. [DOI] [PubMed] [Google Scholar]
- 18.Chen Q., Zhong T. The association of CXCR4 expression with clinicopathological significance and potential drug target in prostate cancer: A meta-analysis and literature review. Drug. Des. Devel. Ther. 2015;9:5115–5122. doi: 10.2147/DDDT.S82475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao H., Guo L., Zhao H., Zhao J., Weng H., Zhao B. CXCR4 over-expression and survival in cancer: A system review and meta-analysis. Oncotarget. 2015;6:5022–5040. doi: 10.18632/oncotarget.3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Z., Ni C., Chen W., Wu P., Wang Z., Yin J., Junhua Y., Jian H., Fuming Q. Expression of CXCR4 and breast cancer prognosis: A systematic review and meta-analysis. BMC Cancer. 2014;14:49. doi: 10.1186/1471-2407-14-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ferlay J., Colombet M., Soerjomataram I., Mathers C., Parkin D.M., Piñeros M., Znaor A., Bray F. Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int. J. Cancer. 2019;144:1941–1953. doi: 10.1002/ijc.31937. [DOI] [PubMed] [Google Scholar]
- 22.Iveson T. Adjuvant chemotherapy in colon cancer: State of the art and future perspectives. Cur. Opin. Oncol. 2020;32:370–376. doi: 10.1097/CCO.0000000000000640. [DOI] [PubMed] [Google Scholar]
- 23.Nappi A., Berretta M., Romano C., Tafuto S., Cassata A., Casaretti R., Silvestro L., Divitiis C., Alessandrini L., Fiorica F., et al. Metastatic Colorectal Cancer: Role of Target Therapies and Future Perspectives. Curr. Cancer Drug Targets. 2018;18:421–429. doi: 10.2174/1568009617666170209095143. [DOI] [PubMed] [Google Scholar]
- 24.Ottaiano A., Franco R., Aiello Talamanca A., Liguori G., Tatangelo F., Delrio P., Nasti G., Barletta E., Facchini G., Daniele B., et al. Overexpression of both CXC chemokine receptor 4 and vascular endothelial growth factor proteins predicts early distant relapse in stage II-III colorectal cancer patients. Clin. Cancer Res. 2006;12:2795–2803. doi: 10.1158/1078-0432.CCR-05-2142. [DOI] [PubMed] [Google Scholar]
- 25.Ottaiano A., Scala S., Normanno N., Botti G., Tatangelo F., Di Mauro A., Capozzi M., Facchini S., Tafuto S., Nasti G. Prognostic and Predictive Role of CXC Chemokine Receptor 4 in Metastatic Colorectal Cancer Patients. Appl. Immunohistochem. Mol. Morphol. 2020;28:755–760. doi: 10.1097/PAI.0000000000000828. [DOI] [PubMed] [Google Scholar]
- 26.Liberati A., Altman D.G., Tetzlaff J., Mulrow C., Gotzsche P.C., Ioannidis J.P., Clarke M., Devereaux P.J., Kleijnen J., Moher D. The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate health care interventions: Explanation and elaboration. PLoS Med. 2009;6:e1000100. doi: 10.1371/journal.pmed.1000100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stang A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur. J. Epidemiol. 2010;25:603–605. doi: 10.1007/s10654-010-9491-z. [DOI] [PubMed] [Google Scholar]
- 28.Altman D.G., Machin D., Bryant T.N., Gardner M.J., editors. Statistics with Confidence. 2nd ed. BMJ USA; London, UK: 2000. [Google Scholar]
- 29.DerSimonian R., Laird N. Meta-analysis in clinical trials. Control Clin. Trials. 1986;7:177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 30.Higgins J.P., Thompson S.G., Deeks J.J., Altman D.G. Measuring inconsistency in meta-analyses. BMJ. 2003;327:557–560. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Egger M., Smith G.D., Schneider M., Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim J., Takeuchi H., Lam S.T., Turner R.R., Wang H.J., Kuo C., Foshag L., Bilchik A.J., Hoon D.S. Chemokine receptor CXCR4 expression in colorectal cancer patients increases the risk for recurrence and for poor survival. J. Clin. Oncol. 2005;23:2744–2753. doi: 10.1200/JCO.2005.07.078. [DOI] [PubMed] [Google Scholar]
- 33.Yoshitake N., Fukui H., Yamagishi H., Sekikawa A., Fujii S., Tomita S., Ichikawa K., Imura J., Hiraishi H., Fujimori T. Expression of SDF-1 alpha and nuclear CXCR4 predicts lymph node metastasis in colorectal cancer. Br. J. Cancer. 2008;98:1682–1689. doi: 10.1038/sj.bjc.6604363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Speetjens F.M., Liefers G.J., Korbee C.J., Mesker W.E., Van de Velde C.J., Van Vlierberghe R.L., Morreau H., Tollenaar R.A., Kuppen P.J. Nuclear localization of CXCR4 determines prognosis for colorectal cancer patients. Cancer Microenviron. 2009;2:1–7. doi: 10.1007/s12307-008-0016-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ingold B., Schulz S., Budczies J., Neumann U., Ebert M.P., Weichert W., Röcken C. The role of vascular CXCR4 expression in colorectal carcinoma. Histopathology. 2009;55:576–586. doi: 10.1111/j.1365-2559.2009.03426.x. [DOI] [PubMed] [Google Scholar]
- 36.Wang S.C., Lin J.K., Wang H.S., Yang S.H., Li A.F., Chang S.C. Nuclear expression of CXCR4 is associated with advanced colorectal cancer. Int. J. Colorectal. Dis. 2010;25:1185–1191. doi: 10.1007/s00384-010-0999-1. [DOI] [PubMed] [Google Scholar]
- 37.Yopp A.C., Shia J., Butte J.M., Allen P.J., Fong Y., Jarnagin W.R., De Matteo R.P., D′Angelica M.I. CXCR4 expression predicts patient outcome and recurrence patterns after hepatic resection for colorectal liver metastases. Ann. Surg. Oncol. 2012;19:S339–S346. doi: 10.1245/s10434-011-1774-4. [DOI] [PubMed] [Google Scholar]
- 38.Sakai N., Yoshidome H., Shida T., Kimura F., Shimizu H., Ohtsuka M., Takeuchi D., Sakakibara M., Miyazaki M. CXCR4/CXCL12 expression profile is associated with tumor microenvironment and clinical outcome of liver metastases of colorectal cancer. Clin. Exp. Metastasis. 2012;29:101–110. doi: 10.1007/s10585-011-9433-5. [DOI] [PubMed] [Google Scholar]
- 39.Du C., Yao Y., Xue W., Zhu W.G., Peng Y., Gu J. The expression of chemokine receptors CXCR3 and CXCR4 in predicting postoperative tumour progression in stages I-II colon cancer: A retrospective study. BMJ Open. 2014;4:e005012. doi: 10.1136/bmjopen-2014-005012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gao Y., Li C., Nie M., Lu Y., Lin S., Yuan P., Sun X. CXCR4 as a novel predictive biomarker for metastasis and poor prognosis in colorectal cancer. Tumour. Biol. 2014;35:4171–4175. doi: 10.1007/s13277-013-1545-x. [DOI] [PubMed] [Google Scholar]
- 41.Stanisavljević L., Aßmus J., Storli K.E., Leh S.M., Dahl O., Myklebust M.P. CXCR4, CXCL12 and the relative CXCL12-CXCR4 expression as prognostic factors in colon cancer. Tumour. Biol. 2016;37:7441–7452. doi: 10.1007/s13277-015-4591-8. [DOI] [PubMed] [Google Scholar]
- 42.D′Alterio C., Nasti G., Polimeno M., Ottaiano A., Conson M., Circelli L., Botti G., Scognamiglio G., Santagata S., De Divitiis C., et al. CXCR4-CXCL12-CXCR7, TLR2-TLR4, and PD-1/PD-L1 in colorectal cancer liver metastases from neoadjuvant-treated patients. Oncoimmunology. 2016;5:e1254313. doi: 10.1080/2162402X.2016.1254313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu W., Cao J., Ji Z., Wang J., Jiang T., Ding H. Co-expression of Lgr5 and CXCR4 characterizes cancer stem-like cells of colorectal cancer. Oncotarget. 2016;7:81144–81155. doi: 10.18632/oncotarget.13214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weixler B., Renetseder F., Facile I., Tosti N., Cremonesi E., Tampakis A., Delko T., Eppenberger-Castori S., Tzankov A., Iezzi G., et al. Phosphorylated CXCR4 expression has a positive prognostic impact in colorectal cancer. Cell. Oncol. 2017;40:609–619. doi: 10.1007/s13402-017-0348-2. [DOI] [PubMed] [Google Scholar]
- 45.Xu C., Zheng L., Li D., Chen G., Gu J., Chen J., Yao Q. CXCR4 overexpression is correlated with poor prognosis in colorectal cancer. Life Sci. 2018;208:333–340. doi: 10.1016/j.lfs.2018.04.050. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data presented in this study are available on request from the corresponding author.