Abstract
Gene editing by use of clustered regularly interspaced short palindromic repeats (CRISPR) has become a powerful tool for crop improvement. However, a common bottleneck in the application of this approach to grain crops, including rice (Oryza sativa), is efficient vector delivery and calli regeneration, which can be hampered by genotype-dependent requirements for plant regeneration. Here, methods for Agrobacterium-mediated and biolistic transformation and regeneration of indica rice were optimized using CRISPR-Cas9 gene-editing of the submergence tolerance regulator SUBMERGENCE 1A-1 gene of the cultivar Ciherang-Sub1. Callus induction and plantlet regeneration methods were optimized for embryogenic calli derived from immature embryos and mature seed-derived calli. Optimized regeneration (95%) and maximal editing efficiency (100%) were obtained from the immature embryo-derived calli. Phenotyping of T1 seeds derived from the edited T0 plants under submergence stress demonstrated inferior phenotype compared to their controls, which phenotypically validates the disruption of SUB1A-1 function. The methods pave the way for rapid CRISPR-Cas9 gene editing of recalcitrant indica rice cultivars.
Keywords: gene editing, CRISPR-Cas9, rice, indica, SUB1A, tissue culture
1. Introduction
The clustered regularly interspaced short palindromic repeats-Cas9 nuclease (CRISPR-Cas9) technology for genome editing has become a powerful tool for gene-targeted crop improvement. This technique has been applied to various crops, including cereals, such as wheat [1,2,3], corn [4,5,6], and rice [7,8,9]. The CRISPR-Cas9 reagents, including the promoter-guide (g)RNA and promoter-Cas9 gene constructs in a single vector [10], are delivered into the host plant where the gRNA-Cas9 complex guides the position of Cas9 protein cleavage in the host genome through non-homologous end-joining (NHEJ) or homologous recombination (HR) [11]. Agrobacterium-mediated transformation via T-DNA insertion is the most common and stable method to deliver transgene vectors into a host plant [12].
In rice (Oryza sativa L.), transformation protocols of japonica cultivars using mature seeds as explants have long been established [13]. Due to poor callus induction and regeneration and low transformation efficiency of seed explants, indica cultivars are more challenging to transform [14,15,16]. The use of immature embryos as explants is a viable alternative, but time and labor are needed to dissect the immature embryos. Hiei and Komari’s protocol [15] requires 74 days, not including growing plants for the immature embryos, to generate the transgenic plants with up to 13% transformation efficiency. Several studies have reported efficient indica rice transformation protocols using mature seeds [17,18,19], but these are largely genotype specific [20,21]. For example, Sahoo’s protocol [19] suggested that the transgenic plants derived from mature seeds of IR64, PB1, CSR10, and Swarna can be obtained in 65 days with up to 46% transformation efficiency and up to 92% regeneration frequency for the untransformed calli and up to 59% for the transformed calli. Meanwhile, Sahoo and Tuteja’s protocol [22] suggested that the transgenic plants derived from mature seeds of IR64 can be obtained in 64 days with 12% transformation efficiency.
Ciherang-Sub1 is an elite indica rice cultivar derived from Ciherang, a popular variety from Indonesia [23]. This submergence tolerant variety has been released in several countries in Southeast and South Asia and used as a new elite parent for further rice improvement [24,25,26,27,28]. Ciherang-Sub1 explants are recalcitrant in shoot regeneration, hampering its improvement via transgene introduction or genome editing. The aim of this study was to optimize for Agrobacterium-mediated transformation and regeneration methods for indica rice from immature embryos as explants. Additionally, we also wanted to compare it with particle bombardment-mediated transformation and regeneration using mature seed-derived calli to avoid having to grow out plants and dissect immature embryos after flowering. By use of Ciherang-Sub1 as a target for gene editing, we demonstrate that disruption of SUB1A-1 alone is sufficient to limit key traits associated with the quiescence strategy of submergence survival [29,30,31,32].
2. Results and Discussion
2.1. Optimization of Callus Induction Medium for Mature Seeds
To optimize callus induction medium (CIM) for mature seeds, several medium components were modified from the previous japonica and indica rice tissue culture protocols [15,19]. First, we compared five different auxin to cytokinin compositions of callus induction formulae (Table A1) as follows: (1) with 3 mg 2,4-Dichlorophenoxyacetic acid (2,4-D) and 0.25 mg 6-Benzylaminopurine (6BA) [19]; (2) 2.5 mg 2,4-D and 0.15 mg 6BA [22]; (3) 3 mg 2,4-D and 0.2 mg 6BA; (4) 3 mg 2,4-D and 0.2 mg 6BA with phytagel increased to 4 g/L; and (5) 3 mg 2,4-D and 0.3 mg 6BA. Calli induced from Formula (1) tended to be darker (Figure S1A). Calli induced from Formulae (1) and (2) were smaller (Figure S1) than Formula (5) (Figure S2). However, calli induction using Formulae (3) and (4) had the smallest calli. Hence, increasing phytagel from 3 g/L to 4 g/L did not show any significant difference between Formulae (3) and (4). This experiment suggests that medium Formula (5) with 3 mg 2,4-D and 0.3 mg 6BA had the best callus induction rate and quality for Ciherang-Sub1.
When seeds were cultured on CIM with an optimal auxin to cytokinin ratio (Formula (5)), nearly all the seeds could be induced for embryogenic calli (Figure S2). After 10–14 days of culturing, roots were removed, and calli were cut into 3 to 4 smaller pieces and sub-cultured for another 14–18 days. These divided-calli could facilitate callus propagation and be beneficial for the transformation process. More than 80% of seeds had propagated new calli after 16 days of sub-culturing in fresh CIM (Figure S2B). The optimal callus for transformation should have a creamy or yellow appearance, with a visible differentiated shape on the surface (Figure 1A). It also should not be too wet, such as a juicy surface or visible water drop on the surface, or too fragile, which could be easily crushed by tweezers.
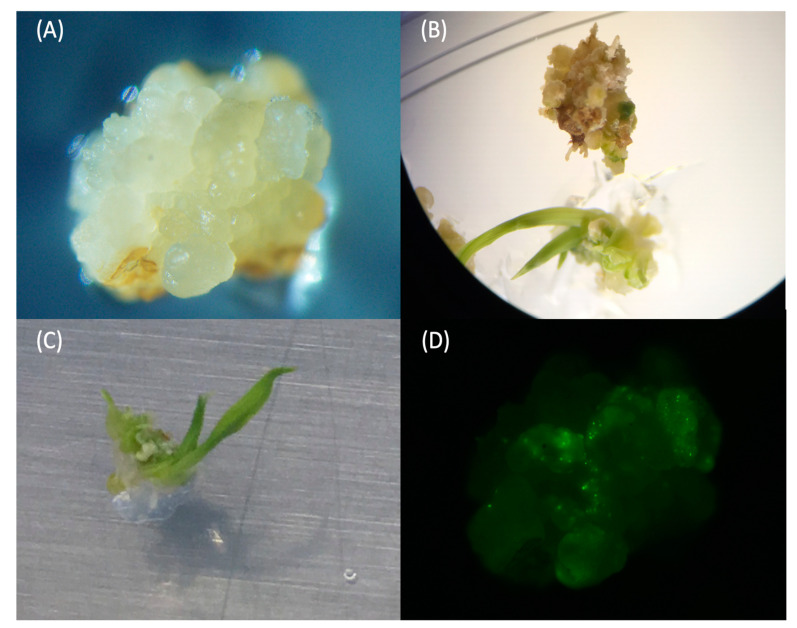
Figure 1.
Calli’s performances under a microscope. (A) The optimal callus for bombardment. (B) Shoot formation of callus on shoot induction medium (SIM). (C) Leaf formation of callus on shoot induction medium (SIM). (D) YFP fluorescence detection in callus five days after biolistic bombardment pPTN-EYFP vector.
Previous studies had revealed that not only different plant hormone ratios but also variation in carbon sources could have significant effects on tissue culture [33]. In our study with Ciherang-Sub1, maltose was the optimal carbon source for the callus induction stage. We observed that calli turned brown and dry when sucrose was used instead of maltose. In this case, sucrose may cause greater osmotic potential than maltose, and it also has a different organogenesis rate [34]. Previous studies also showed that sucrose replacement with maltose could improve callus induction for spring wheat (Triticum aestivum L.) and some indica rice varieties [19,35]. However, for the callus induction in japonica rice cultivars, sucrose is used as the carbon source [15].
In organogenesis, auxin and cytokinin are the major phytohormones, which control the different organogenesis stages. For example, auxin to cytokinin ratio equal to 3:0.2 will induce callus, 0.03:1 can induce shoot regeneration, and 3.0:0.02 can induce roots [36]. Compared to other indica cultivars IR64 and IR64-Sub1, Ciherang-Sub1 has slower callus formation and propagation stages. Therefore, we slightly adjusted the concentration of 2,4-D, a synthetic auxin, and 6BA, a cytokinin source, from two published protocols [19,22]. With a higher 2,4-D supply, the callus became smaller, and more lateral roots differentiated from mature seeds’ main roots. On the other hand, when we slightly increased 6BA concentration from 0.25 mg/L to 0.3 mg/L, the callus formation and propagation became more vigorous. Therefore, we recommend an auxin to cytokinin ratio of 3:0.3 as the optimal ratio for callus induction in Ciherang-Sub1. Our study also suggests that callus with ideal shape developed after 30 days of callus induction is an optimal target for biolistic bombardment. Overall, this particle bombardment protocol using mature seeds as explant requires 80 days from callus induction to the transfer of a transgenic T0 seedling to soil (Figure S3).
2.2. Optimization of Regeneration Medium for Mature Seeds
For the testing of the regeneration medium for shooting, three different cytokinin to auxin ratios were tested (Table A2), including (1) 2 mg kinetin and 0.2 mg 1-Naphthaleneacetic acid (NAA) [19]; (2) 3 mg kinetin and 0.1 mg NAA; and (3) 3 mg kinetin and 0.1 mg NAA with agarose reduced from 8 g/L to 7 g/L (see Method section). The optimal actively dividing calli were selected under a microscope and transferred onto three regeneration media. In the testing phase and the evaluation of regeneration efficiency, all three media had no antibiotics added. The developing and testing processes were conducted using calli derived from mature seeds.
With 2 mg kinetin and 0.2 mg NAA medium (Formula (1)), green tissues could be observed around ten days after transferring onto the regeneration medium; however, around 15% of calli became dry and brown after turning green. In contrast, for 3 mg kinetin and 0.1 mg NAA formula with reduced agarose (Formula (3)), the light green tissue formation could be observed as early as five days after being transferred onto the regeneration medium. After being transferred onto the optimized shoot induction medium (SIM), the yellow or creamy color callus would partially turn light green from the corner. Subsequently, the green became darker, a part expanded, and shoots began to appear. The spikes would elongate and differentiate into leaves (Figure 1A–C). This type of callus was tested for the pPTN-EYFP expression vector beforehand to ensure that we had suitable calli to work with (Figure 1D). In total, we observed 19 out of 20 calli were regenerated after 5–14 days, being transferred onto optimized SIM without antibiotic (Figure S4A, B), indicating that we had a 95% regeneration rate of Ciherang-Sub1. This optimal regeneration medium was also tested for IR64-Sub1 mature seeds, and it worked well, showing its potential to be used for other indica varieties (Figure S4C).
We observed that calli had a similar response to two media (Formulae (2) and (3)) with the same cytokinin to auxin ratio but with different agarose concentrations. Notably, the lower agarose concentration medium allowed calli to turn green 2–3 days earlier and reduce brown calli formation. Hence, the regeneration medium containing 3 mg kinetin and 0.1 mg NAA with 7 g agarose to solidify had the highest percentage of green callus and had faster progress. These results are consistent with previous studies, which revealed that a low auxin:cytokinin ratio generally promotes shoot formation from callus [36,37,38]. We also noticed that reduced agarose concentration from 8 g/L to 7 g/L expedites the regeneration of Ciherang-Sub1.
Many rice tissue culture studies apply fungicide (Benomyl) on the roots or in a hydroponic solution after plants are moved out from the rooting medium [39,40,41]. However, our study used a more convenient protocol where fresh tap water was adequate for rice acclimation. The new roots could be observed 5–7 days after culturing in water (Figure S4D). This is the sign that the plant is ready to be transferred to soil. After moving to the soil, the regenerated plants can be treated as normal plants. No extra cover is needed for maintaining the humidity. Instead, air temperature and aeration are critical points. Air temperature that is too high (>30 °C) or poor aeration will cause plants to grow poorly and eventually die.
2.3. Regeneration Performance of Immature Embryo from Agrobacterium-Mediated Transformations Using the Optimized Shoot Induction Medium
In the first batch of immature embryo transformation, a set of calli were transferred onto the optimized SIM, and another set of calli were transferred onto Hiei and Komari regeneration medium [15]. The results suggested that the optimized SIM works very well for calli induced from the immature embryo (Figure 2A). In contrast, we found that calli on the Hiei and Komari regeneration medium [15] type 4 indica variety regeneration medium tended to generate many tillers from the bottom instead of having normal shoot elongation (Figure 2B). Despite the high genetic background similarity between Ciherang-Sub1 with IR64-Sub1 and IR64, which belong to the type 4 indica variety [15], the regeneration medium for type 4 indica cultivar, IR64, was not suitable for Ciherang-Sub1, presumably due to genotype-dependent specificity in the regeneration protocol.
Figure 2.
Performance of Ciherang-Sub1 calli in regeneration media. (A) Calli in the SIM medium (optimized medium); and (B) calli in regeneration medium for type 4 indica varieties [15].
2.4. Validation of Transgenic Plants Derived from Immature Embryos
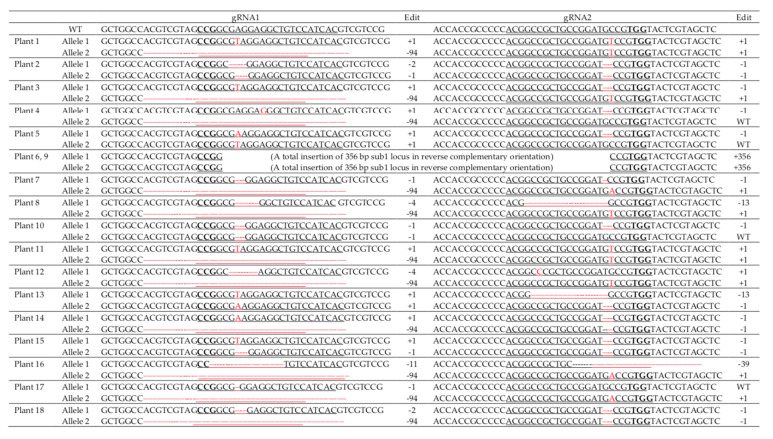
A total of 18 transgenic plants were obtained using immature embryo transformation generated from the optimized regeneration medium. The transgenic plants first were screened by the Cas9 specific primers. Genotyping by PCR confirmed that all 18 hygromycin-resistant transgenics (T0) were positive for Cas9 (Figure S5). Sanger sequencing confirmed edits in the regions of SUB1A targeted by the two gRNAs in all transgenic plants, with biallelic mutations in most cases (Figure 3). T0 plant #1, 3, 4, 7, 8, 11, 12, 14, 16, 17, and 18 had the same 94 bp deletion on the first gRNA region; and plant 8, 13, 16 had deletions of 13 bp and 39 bp on the second gRNA region. Many individual plants had a deletion of the same length in the targeted region. This may indicate an editing hotspot in the target region. Two plants, #6 and 9, had a 355 bp insertion between the two gRNAs, suggesting that larger deletions can be obtained in the region between the two gRNA cutting sites. These results demonstrate 100% editing efficiency using immature embryo calli with Agrobacterium-mediated transformation for the indica cultivar Ciherang-Sub1.
Figure 3.
Sanger sequencing results of transgenic plants derived from immature embryo transformation. Two gRNAs are underlined. PAM sequences are highlighted in bold. gRNA1: 5′-CCGGCGAGGAGGCTGTCCATCAC-3′; gRNA2: 5′-ACGGCCGCTGCCGGATGCCGTGG-3′. gRNA1 and gRNA2 are 350 bp apart. "WT" indicates that the allele was not edited (wild type).
2.5. Optimization of Biolistic Bombardment Procedures
The optimization procedures include testing the optimal calli status, the osmotic medium, setting, and the bombardment system operation. In order to confirm whether the calli are ideal for transformation and validate the biolistic bombardment delivery system, calli were observed and screened under a microscope. The osmotic medium previously reported for bread wheat [42] was tested here for rice calli. To evaluate the transformation, calli were transformed with pPTN-EYFP control vector under 1100 psi helium pressure via Biolistic® PDS-1000/He particle bombardment (BIORAD, Hercules, CA, USA). There are three distances available for the gene gun set up for setting: 3 cm, 6 cm, and 9 cm. We found that 6 cm between calli and the gene gun was the most appropriate distance during the shooting process. The distance of 3 cm was too close, which caused calli to jump out from the medium, whereas the 9 cm distance was too far to allow an efficient transformation. Our results showed that with the 6 cm distance and two shots, the YFP fluorescence could be observed in calli under a fluorescence microscope five days after biolistic bombardment (Figure 1D). This result suggests that the delivery procedure and the callus condition selected are optimal for biolistic bombardment.
2.6. Validation of Transgenic Plants Derived from Mature Seeds
After bombardment and recovery from calli that survived under two selection processes and regenerated from SIM and root induction medium (RIM) with hygromycin added, three plants were obtained. After taking out plants from the RIM, plants were transferred to clean water for acclimation. In the meantime, 2 cm leaf pieces from each plant were collected for DNA extraction and validation. The results showed that all the negative controls had no PCR products amplified. In contrast, the positive control and all the three plants had amplified PCR products with the expected sizes (Figure S6).
2.7. Phenotyping of T1 Transgenic Plants
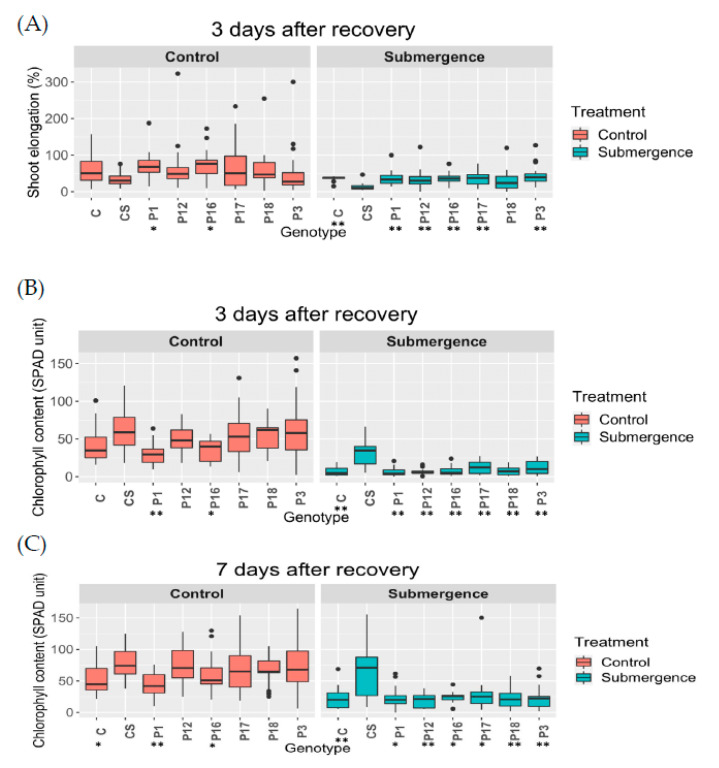
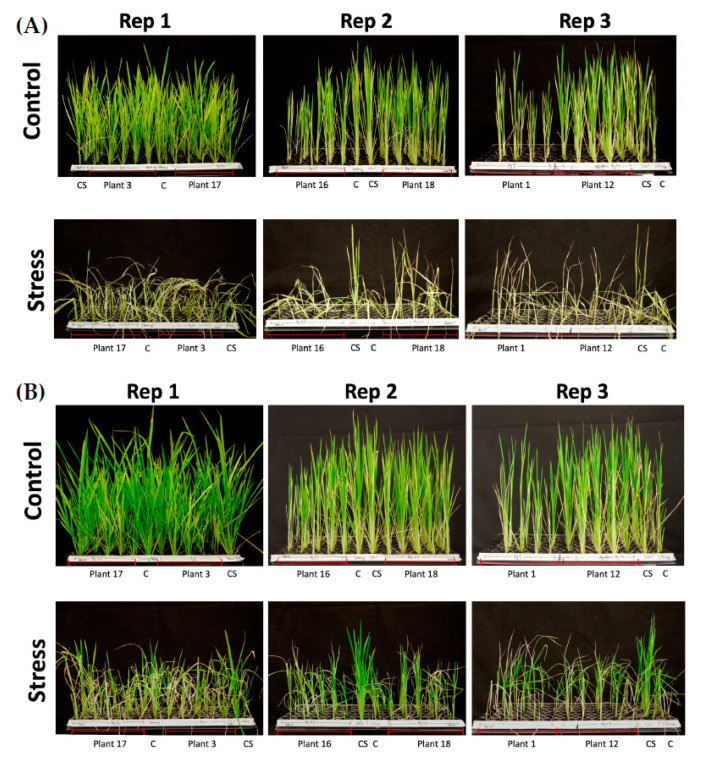
We used T1 seeds derived from the selected six T0-edited plants developed from the immature embryo-derived calli via Agrobacterium-mediated delivery. Two traits were analyzed in the submergence experiment, shoot elongation rate (%) and chlorophyll content. SUB1A-1 encodes an ethylene-responsive factor that provides submergence tolerance. [29]. The presence of SUB1A-1 allows submerged plants to pause shoot elongation during submergence [30]. This phenotype is accompanied with reduced loss of leaf carbohydrates and chlorophyll during submergence. Each tray included 6 Ciherang and 6 Ciherang-Sub1 as negative and positive controls, respectively, along with 30 T1 seeds, each from two selected T0 plants. The same setup was used for both non-submerged (control) and submerged treatments. There were 3 trays/treatment. In general, the edited plants had a similar performance as Ciherang in both shoot elongation rate and chlorophyll content. Additionally, the control (unsubmerged) groups had better growth than submergence groups (Figure 4 and Figure 5). The results of pairwise comparisons at 3 days after recovery suggest that Ciherang-Sub1 had the lower shoot elongation rate than all other genotypes at p < 0.001 except for T1 plants derived from plant #18 (p = 0.09), and the elongation difference also showed between T1 plants derived from plant #18 and those from plant # 3 (p < 0.05) (Figure 4A; Table S1). For chlorophyll content, both control (p < 0.001) and submergence (p < 0.001) groups showed that chlorophyll content increased from 3 days to 7 days after recovery (Figure 4B,C). For the comparison between genotypes, only Ciherang-Sub1 had a higher chlorophyll content than all other genotypes (p < 0.05) (Tables S2 and S3). Overall, the phenotypic data analyses of T1 transgenic plants indicate that the disruption of SUB1A of Ciherang-Sub1 was successful since, in general, the edited plants had Ciherang-like phenotypes.
Figure 4.
Shoot elongation rate and chlorophyll content of control and submerged plants. (A) Shoot elongation rate at three days after desubmergence. Chlorophyll content at three days (B) and seven days (C) after desubmergence. “C” indicates Ciherang; “CS” indicates Ciherang-Sub1; “P1” indicates plant #1; “P3” indicates plant #3; “P12” indicates plant #12; “P16” indicates plant #16; “P17” indicates plant #17; and “P18” indicates plant #18. “*” indicates p-value < 0.05 compared with Ciherang-Sub1; “**” indicates p-value < 0.01 compared with Ciherang-Sub1.
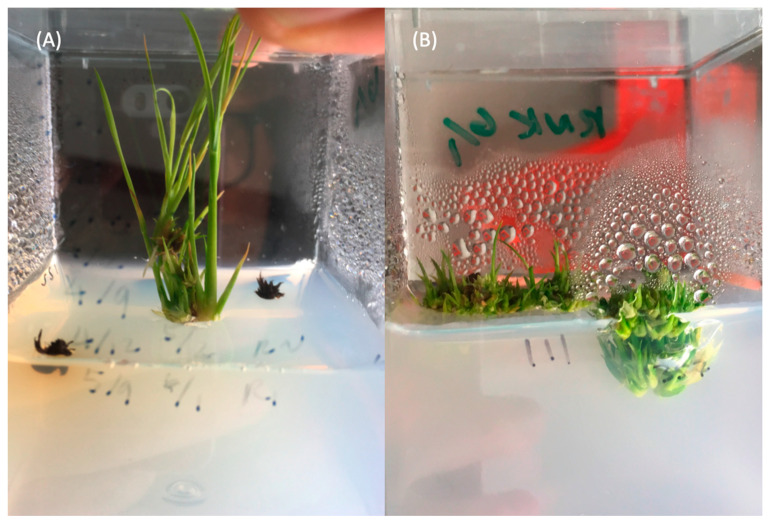
Figure 5.
Performance of submerged Ciherang-Sub1, Ciherang, and T1 seedlings compared to their corresponding controls. (A) Immediately after 14 days of complete submergence, and (B) 7 days post-submergence recovery. C, Ciherang; and CS, Ciherang-Sub1.
3. Materials and Methods
3.1. Plant Materials
The indica rice cultivar Ciherang-Sub1 (IR87424-177-173), derived from a cross of a popular rice variety from Indonesia, Ciherang, and a submergence tolerance donor IR64-Sub1 [31], was used.
3.2. Construction of the CRISPR-Cas9 Vector
The second exon of SUB1A gene was targeted for disruption by editing (Figure S7). Two gRNAs (gRNA1: 5′-CCGGCGAGGAGGCTGTCCATCAC-3′ and gRNA2: 5′-ACGGCCGCTGCCGG ATGCCGTGG-3′) were designed using the design tools CRISPR direct and Cas-OFFinder [43,44]. The gRNAs were inserted into the Golden Gate entry vectors, pYPQ131C (Addgene Plasmid #69284) and pYPQ132C (Addgene Plasmid #69285) [10], that couple an OsU6 promoter to a gRNA, which were then assembled into the Golden Gate recipient vector pYPQ142 (Addgene Plasmid #69294). Next, both pYPQ142 and the Cas9 expression vector (pYPQ150, Addgene Plasmid #69301) were assembled into a binary vector, pMDC32 (Addgene Plasmid #32078), through the LR Clonase II recombination reaction. The final destination vector pMDC32 was transformed into competent E. coli (DH5α) cells by heat shock at 42 °C for 30 s. Transformants were selected on kanamycin and confirmed by DNA sequencing. To prepare plasmid DNA for the bombardment, plasmid DNAs were isolated with the QIAprep Spin Miniprep Kit (QIAGEN, Germantown, MD, USA) and stored in a −20 °C freezer.
3.3. Immature Embryo Harvest, Tissue Culture, and Transformation
Plants were cultivated in a greenhouse (14 h/10 h (day/night) and 29 °C/26 °C (day/night)) in potting soil in College Station, Texas, and embryos were obtained 10–12 days after pollination. Tissue culture and transformation procedures of immature embryo transformation followed the Agrobacterium-mediated transformation procedure for Indica type 4 [15] until the regeneration process. Agrobacterium tumefaciens (EHA105)-delivery transformation followed the Hiei and Komari protocol [15], except that the YEB medium was supplemented with kanamycin (50 mg/L). For co-cultivation, immature embryos, with the scutellum face up, were transferred onto NB-As medium. A drop of 5 uL of Agrobacterium suspension (O.D. = 1.0 at 660 nm) was added onto each immature embryo. These treated embryos were then incubated for 15 min at 25 °C. Selection was performed on a resting medium (CCMC) with 75 mg/L hygromycin.
3.4. Callus Induction from Mature Seeds for Bombardment
The seed coat was removed from healthy mature seeds that were then sterilized with 1.5% (v/v) sodium hypochlorite and a drop of Tween 20 (100%) for 20 min in a mini rotator (Grant-bio, PTR-25; Grant Instruments, Shepreth, Cambridgeshire, UK) at a speed of 3. Seeds were rinsed in sterile water 7–8 times and air-dried on autoclaved filter paper for 5–10 min. For callus induction, 19–20 seeds were placed onto sterile CIM and incubated at 29 °C in the dark for 14 days. For optimization of CIM, we tested several CIM formulae consisting of 4.4 g/L Murashige and Skoog Basal Medium (MS salts) (Sigma-Aldrich, M404), 0.6 g/L proline, 0.3 g/L casein hydrolysate, 3 mg/L, 3g/L phytagel, pH 5.8), either 30 g/L maltose or 30 g/L sucrose, and several different hormone ratios (2,4-D and 6BA). Petri dishes of 90 mm diameter × 15 mm depth with 40 mL medium were used for all subsequent aseptic tissue culture steps. Calli were dissected from roots, cut into small pieces, and sub-cultured on fresh CIM in Petri dishes (90 mm diameter × 20 mm depth) at 29 °C in the dark for 14–20 days before use. A minimum of 4 h or overnight before the bombardment, calli were transferred to a high-osmotic medium (OSM) [42] and incubated at 23 °C in the dark. OSM was prepared using 4.4 g/L MS salts, 5 mg/L 2,4-D, 72.87 g/L mannitol, pH 5.8, and 3.2 g phytagel.
3.5. Plasmid Preparation and Biolistic Bombardment
The biolistic transformation of calli was performed using the PDS-1000/He particle bombardment system with 1.0 µm gold particles at a helium pressure of 1100 psi with a target distance of 6 cm. The bombardment procedures followed the Biolistic® PDS-1000/He Particle Delivery System user manual [45]. Before coating gold particles with DNA, 30 mg of 1.0 µm gold particles were sterilized with 70% (v/v) EtOH and resuspended with 50% (v/v) glycerol. These were stored at −20 °C for up to 2 weeks. The following steps were performed while vortexing continually: A 50 μL aliquot of gold particles was combined with 5 μg of DNA to a total volume of 55 μL using 2.5 M cold CaCl2, after which 20 μL of 0.1 M spermidine was added. The mix was vortexed for 3 min, the gold particles pelleted in a microcentrifuge, and the supernatant discarded. The DNA-coated gold particles were resterilized in 100 mL 100% (v/v) EtOH, pelleted, and resuspended in 48 μL 100% EtOH. These gold particles were vortexed at low speed, and 7 μL was applied onto the dry macrocarriers. Two 1100 psi shots were applied to each plate of tissue. After bombardment, calli were incubated in OSM overnight in the dark at 23 °C. The next day, calli were transferred onto fresh CIM and cultured at 29 °C in the dark for 7–10 days until transfer to selection medium (MSM; CIM containing 50 mg/L hygromycin) and cultured at 27 °C in the dark for 12 days. Brown or black calli were removed, and yellow or cream-colored calli were transferred to fresh MSM for a second selection period of 10–12 days. Sections of small new light-colored calli were transferred onto regeneration medium in Petri dishes (90 mm diameter × 20 mm depth). A third selection was performed sometimes.
3.6. Callus Regeneration for Both Immature Embryos and Mature Seeds
The same shoot regeneration medium was used for calli-induced mature seeds and immature embryos. Shoot induction medium (SIM) was modified from two studies [15,19]. To identify the best growth condition for regeneration medium for Ciherang-Sub1, after selection on hygromycin, calli were transferred onto SIM with 30 mg/L hygromycin (4.4 g/L MS salts, 30 g/L maltose, with two different ratios of kinetin and 1-Naphthaleneacetic acid (NAA), and two different concentration of agarose g/L agarose, pH 5.8) and cultured at 29 °C with a day/night period 14/10 h (light intensity was 405 µmol/m2/s) in Petri dishes (90 mm diameter × 15 mm depth) or Magenta GA-7 vessels (100 mL medium). In the first stage of shooting, calli were transferred onto R1 Petri dishes for 14 days until the calli turned green and shoot differentiation was visible. Calli with green shoots were transferred to fresh R1 in a Magenta vessel for 7–10 days until the plants were 5 cm or taller and three or more tillers.
Plantlets were then transferred to the root induction medium (RIM; containing 2.2 g/L MS salts, 30 g/L sucrose, 3 g/L phytagel, pH 5.8, containing 30 mg/L hygromycin) in Magenta vessels (100 mL) and returned to the growth chamber until the leaves reached the lid (7–10 days). The lid was then removed and replaced by an empty, inverted Magenta GA-7 vessel for another 5–7 days of culture. Plants were removed from the medium, roots were cleaned well with tap water, removed of all the medium residues, and plants were placed into a Magenta GA-7 vessel filled with fresh tap water in a well-ventilated area at 23–25 °C, out of direct sunlight for 7–10 days of acclimation. Once new roots formed, plants were finally transferred to pots containing a 1:1 mixture of Metro Mix 820 Potting soil and Turface Athletics MVP and moved to the greenhouse.
3.7. Validation of Transgenic Plants
DNA was extracted from the young leaves of each regenerated plant using a modified CTAB method [46]. Plants were genotyped using pYPQ150 Cas9 specific primers (Forward: 5′-TCGGTGCTCTTCTTTTCGAT-3′; Reverse: 5′-TTCCGAGATTGGGTGTCTC-3′) and/or hygromycin B phosphotransferase gene (HPT) primers (Forward: 5′-CGCATAA-CAGCGGTCATTGACTGGAGC-3′; Reverse: 5′-GCTGGGGCGTCGGTTTCCACTATCC-G-3′). Plants having positive bands for these markers were further examined by mutations within the target region of SUB1A-1 using gene-specific primers (Forward: 5′-CGGTAGATGCCGAGAAGTGT-3′; Reverse: 5′- GTGGACTATGCGATGTGTGG-3′) and TA cloning-based Sanger sequencing. To confirm the edited region, three colonies from each target were sequenced, totaling six colonies/sample. The expected product size of the HPT marker was 375 bp. DNA of transgenic samples, positive control, and negative controls were diluted to 100–150 ng for the PCR reaction with an annealing temperature of 55 °C.
3.8. Calculation of Regeneration and Transformation Efficiency
The regeneration efficiency was calculated as the number of regenerated calli among the total calli on the regeneration medium. To test the optimal regeneration medium, the regeneration medium was tested using untransformed healthy calli without antibiotics. The editing efficiency was calculated as the number of edited plants among the total number of transgenic plants (i.e., regenerated plants with both selective markers positive).
3.9. Submergence Experiment and Phenotyping of T1 Plants
T1 seeds derived from mutant T0 plants were selected for phenotyping. These were germinated in a 6 × 12 holes plastic tray and grown in a growth chamber with 12/12 h for day/night length and 29 °C/26 °C for day/night, with a light intensity of 600–1000 PAR. On day 14 after germination, seedlings were completely submerged in 100 cm of water in a plastic tank for 16 days. Following de-submergence, trays were returned to the greenhouse bench. Control plants were maintained in the same greenhouse environment. Shoot elongation was measured from the base of the shoot to the tip of the longest leaf. Chlorophyll content was measured in the third leaf in control and treated plants using an Apogee MC-100 Chlorophyll Meter (Apogee Instruments, Inc., Logan, UT, USA)
3.10. Phenotypic Data Analysis
First, the normality of phenotypic data was examined by the Shapiro–Wilk test. The results indicated that both shoot elongation rate and chlorophyll content violated the normal distribution; therefore, a non-parametric Kruskal–Wallis test was used to examine the effects. A Wilcoxon rank-sum test was then used for multiple comparisons.
4. Conclusions
The results of our study show that it is possible to obtain high regeneration and high editing efficiency in Ciherang-Sub1 using CRISPR-Cas9 with immature embryos as explants through Agrobacterium-mediated transformation. In addition, we demonstrated a potential biolistic bombardment transformation protocol using mature indica seeds as explants for CRISPR-Cas9 gene editing. With all the necessary media, including callus induction, osmotic, selection, and regeneration media, the mature seed protocol allowed us to obtain transgenic plants within 80 days. A high regeneration rate of 95% was obtained from indica rice cultivars Ciherang-Sub1 using the optimized SIM, and there is a potential use of this medium for other indica varieties as well. This regeneration medium can be used in both calli derived from mature seeds and immature embryos and can be adapted to both Agrobacterium-mediated and biolistic bombardment transformation procedures. This study will enable further investigation of additional submergence tolerance gene(s) in the background of Ciherang-Sub1 after removing the large effect of the major SUB1A-1 gene. Further, this also provides an opportunity for future efforts to further improve Ciherang-Sub1 and potentially other indica rice varieties via gene-editing and genetic engineering.
Acknowledgments
We thank Marco Molina-Risco and Mayra Faion-Molina for providing the pPTN-EYFP vector.
Abbreviations
| 2:4-D 6BA CIM CRISPR-Cas9 |
2,4-Dichlorophenoxyacetic acid 6-Benzylaminopurine Callus induction medium Clustered regularly interspaced short palindromic repeats-Cas9 |
| gRNA | Guide RNA |
| HR | Homologous recombination |
| MS NAA NEHJ RIM SIM SUB1 |
Murashige and Skoog 1-Naphthaleneacetic acid Non-homologous end-joining Root induction medium Shoot induction medium SUBMERGENCE 1 |
| WT | Wild type |
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/ijms22136989/s1.
Appendix A
Table A1.
Comparison of callus induction media with different cytokinin to auxin ratios.
Table A2.
Comparison of regeneration media with different cytokinin to auxin ratios.
| Components | Formula (1) [19] | Formula (2) | Formula (3) |
|---|---|---|---|
| Kinetin | 2 mg/L | 3 mg/L | 3 mg/L |
| NAA (1-Naphthaleneacetic acid) | 0.2 mg/L | 0.1 mg/L | 0.1 mg/L |
| Agarose | 8 g/L | 8 g/L | 7 g/L |
Author Contributions
Conceptualization, E.M.S., J.B.-S., and Y.L.; methodology, Y.L., S.B., B.K., and E.M.S.; software, Y.L. and S.B.; validation, Y.L., S.B., B.K., J.B.-S., and E.M.S.; formal analysis, Y.L. and S.B.; investigation, Y.L. and S.B.; resources, Y.L., S.B., B.K., J.B.-S., and E.M.S.; data curation, Y.L. and S.B.; writing—original draft preparation, Y.L.; writing—review and editing, J.B.-S. and E.M.S.; visualization, Y.L., J.B.-S., and E.M.S.; supervision, E.M.S.; project administration, Y.L., S.B., and E.M.S.; funding acquisition, E.M.S. and J.B.-S. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Agriculture and Food Research Initiative (2017-67013-26194 to E.M.S. and J.B-S) from the USDA National Institute of Food and Agriculture. Y.L. and S.B.’s Ph.D. studies were supported by the same grant and a Texas A&M graduate research assistantship and teaching assistantship.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Wang Y., Cheng X., Shan Q., Zhang Y., Liu J., Gao C., Qiu J.-L. Simultaneous Editing of Three Homoeoalleles in Hexaploid Bread Wheat Confers Heritable Resistance to Powdery Mildew. [(accessed on 29 June 2017)];Nat. Biotechnol. 2014 32:947. doi: 10.1038/nbt.2969. Available online: https://www.nature.com/articles/nbt.2969#supplementary-information. [DOI] [PubMed] [Google Scholar]
- 2.Shan Q., Wang Y., Li J., Gao C. Genome editing in rice and wheat using the CRISPR/Cas system. Nat. Protoc. 2014;9:2395–2410. doi: 10.1038/nprot.2014.157. [DOI] [PubMed] [Google Scholar]
- 3.Zhang Y., Liang Z., Zong Y., Wang Y., Liu J., Chen K., Qiu J.L., Gao C. Efficient and transgene-free genome editing in wheat through transient expression of CRISPR/Cas9 DNA or RNA. Nat. Commun. 2016;7:12617. doi: 10.1038/ncomms12617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Doll N.M., Gilles L.M., Gérentes M.-F., Richard C., Just J., Fierlej Y., Borrelli V.M., Gendrot G., Ingram G.C., Rogowsky P.M. Single and multiple gene knockouts by CRISPR–Cas9 in maize. Plant Cell Rep. 2019;38:487–501. doi: 10.1007/s00299-019-02378-1. [DOI] [PubMed] [Google Scholar]
- 5.Qi W., Zhu T., Tian Z., Li C., Zhang W., Song R. High-efficiency CRISPR/Cas9 multiplex gene editing using the glycine tRNA-processing system-based strategy in maize. BMC Biotechnol. 2016;16:1–8. doi: 10.1186/s12896-016-0289-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Svitashev S., Schwartz C., Lenderts B., Young J.K., Cigan A.M. Genome editing in maize directed by CRISPR–Cas9 ribonucleoprotein complexes. Nat. Commun. 2016;7:1–7. doi: 10.1038/ncomms13274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li Q., Zhang D., Chen M., Liang W., Wei J., Qi Y., Yuan Z. Development of japonica photo-sensitive genic male sterile rice lines by editing carbon starved anther using CRISPR/Cas9. J. Genet. Genom. 2016 doi: 10.1016/j.jgg.2016.04.011. [DOI] [PubMed] [Google Scholar]
- 8.Miao J., Guo D., Zhang J., Huang Q., Qin G., Zhang X., Wan J., Gu H., Qu L.-J. Targeted mutagenesis in rice using CRISPR-Cas system. Cell Res. 2013;23:1233–1236. doi: 10.1038/cr.2013.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yin X., Biswal A.K., Dionora J., Perdigon K.M., Balahadia C.P., Mazumdar S., Chater C., Lin H.-C., Coe R.A., Kretzschmar T. CRISPR-Cas9 and CRISPR-Cpf1 mediated targeting of a stomatal developmental gene EPFL9 in rice. Plant Cell Rep. 2017;36:745–757. doi: 10.1007/s00299-017-2118-z. [DOI] [PubMed] [Google Scholar]
- 10.Lowder L.G., Zhang D., Baltes N.J., Paul J.W., Tang X., Zheng X., Voytas D.F., Hsieh T.-F., Zhang Y., Qi Y. A CRISPR/Cas9 Toolbox for Multiplexed Plant Genome Editing and Transcriptional Regulation. Plant Physiol. 2015;169:971–985. doi: 10.1104/pp.15.00636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Paul J.W., 3rd, Qi Y. CRISPR/Cas9 for plant genome editing: Accomplishments, problems and prospects. Plant Cell Rep. 2016;35:1417–1427. doi: 10.1007/s00299-016-1985-z. [DOI] [PubMed] [Google Scholar]
- 12.Gelvin S.B. Agrobacterium-Mediated Plant Transformation: The Biology behind the “Gene-Jockeying” Tool. Microbiol. Mol. Biol. Rev. 2003;67:16. doi: 10.1128/MMBR.67.1.16-37.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hiei Y., Ohta S., Komari T., Kumashiro T. Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 1994;6:271–282. doi: 10.1046/j.1365-313X.1994.6020271.x. [DOI] [PubMed] [Google Scholar]
- 14.Aldemita R.R., Hodges T.K. Agrobacterium tumefaciens-mediated transformation of japonica and indica rice varieties. Planta. 1996;199:612–617. doi: 10.1007/BF00195194. [DOI] [Google Scholar]
- 15.Hiei Y., Komari T. Agrobacterium-mediated transformation of rice using immature embryos or calli induced from mature seed. Nat. Protoc. 2008;3:824. doi: 10.1038/nprot.2008.46. [DOI] [PubMed] [Google Scholar]
- 16.Slamet-Loedin I.H., Chadha-Mohanty P., Torrizo L. Agrobacterium-mediated transformation: Rice transformation. In: Henry R., Furtado A., editors. Cereal Genomics. Volume 1099. Humana Press; Totowa, NJ, USA: 2014. pp. 261–271. Methods in Molecular Biology (Methods and Protocols) [DOI] [PubMed] [Google Scholar]
- 17.Kumar K.K., Maruthasalam S., Loganathan M., Sudhakar D., Balasubramanian P. An improvedAgrobacterium-mediated transformation protocol for recalcitrant elite indica rice cultivars. Plant Mol. Biol. Rep. 2005;23:67–73. doi: 10.1007/BF02772648. [DOI] [Google Scholar]
- 18.Lin Y.J., Zhang Q. Optimising the tissue culture conditions for high efficiency transformation of indica rice. Plant Cell Rep. 2005;23:540–547. doi: 10.1007/s00299-004-0843-6. [DOI] [PubMed] [Google Scholar]
- 19.Sahoo K.K., Tripathi A.K., Pareek A., Sopory S.K., Singla-Pareek S.L. An improved protocol for efficient transformation and regeneration of diverse indica rice cultivars. Plant Methods. 2011;7:49. doi: 10.1186/1746-4811-7-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hiei Y., Komari T. Improved protocols for transformation of indica rice mediated by Agrobacterium tumefaciens. Plant Cell Tissue Organ Cult. 2006;85:271–283. doi: 10.1007/s11240-005-9069-8. [DOI] [Google Scholar]
- 21.Zaidi M., Narayanan M., Sardana R., Taga I., Postel S., Johns R., McNulty M., Mottiar Y., Mao J., Loit E. Optimizing tissue culture media for efficient transformation of different indica rice genotypes. Agron. Res. 2006;4:563–575. [Google Scholar]
- 22.Sahoo R.K., Tuteja N. Development of Agrobacterium-mediated transformation technology for mature seed-derived callus tissues of indica rice cultivar IR64. GM Crops Food. 2012;3:123–128. doi: 10.4161/gmcr.20032. [DOI] [PubMed] [Google Scholar]
- 23.Septiningsih E.M., Hidayatun N., Sanchez D.L., Nugraha Y., Carandang J., Pamplona A.M., Collard B.C., Ismail A.M., Mackill D.J. Accelerating the development of new submergence tolerant rice varieties: The case of Ciherang-Sub1 and PSB Rc18-Sub1. Euphytica. 2015;202:259–268. doi: 10.1007/s10681-014-1287-x. [DOI] [Google Scholar]
- 24.Toledo A.M.U., Ignacio J.C.I., Casal C., Jr., Gonzaga Z.J., Mendioro M.S., Septiningsih E.M. Development of Improved Ciherang-Sub1 Having Tolerance to Anaerobic Germination Conditions. Plant Breed. Biotechnol. 2015;32:77–87. doi: 10.9787/PBB.2015.3.2.077. [DOI] [Google Scholar]
- 25.Gonzaga Z.J.C., Carandang J., Singh A., Collard B.C.Y., Thomson M.J., Septiningsih E.M. Mapping QTLs for submergence tolerance in rice using a population fixed for SUB1A tolerant allele. Mol. Breed. 2017;37:47. doi: 10.1007/s11032-017-0637-5. [DOI] [Google Scholar]
- 26.Singh A., Carandang J., Gonzaga Z.J.C., Collard B.C.Y., Ismail A.M., Septiningsih E.M. Identification of QTLs for yield and agronomic traits in rice under stagnant flooding conditions. Rice. 2017;10 doi: 10.1186/s12284-017-0154-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pramudyawardani E.F., Aswidinnoor H., Purwoko B.S., Suwarno W.B., Islam M., Verdeprado H., Collard B.C. Genetic analysis and QTL mapping for agronomic and yield-related traits in Ciherang-Sub1 rice backcross populations. Plant Breed. Biotechnol. 2018;6:177–192. doi: 10.9787/PBB.2018.6.3.177. [DOI] [Google Scholar]
- 28.Septiningsih E.M., Mackill D.J. Genetics and breeding of flooding tolerance in rice. In: Sasaki T., Ashikari M., editors. Rice Genomics, Genetics and Breeding. Springer; Singapore: 2018. pp. 275–295. [DOI] [Google Scholar]
- 29.Xu K., Xu X., Fukao T., Canlas P., Maghirang-Rodriguez R., Heuer S., Ismail A.M., Bailey-Serres J., Ronald P.C., Mackill D.J. Sub1A is an ethylene-response-factor-like gene that confers submergence tolerance to rice. Nature. 2006;442:705–708. doi: 10.1038/nature04920. [DOI] [PubMed] [Google Scholar]
- 30.Fukao T., Xu K., Ronald P.C., Bailey-Serres J. A variable cluster of ethylene response factor–like genes regulates metabolic and developmental acclimation responses to submergence in rice. Plant Cell. 2006;18:2021–2034. doi: 10.1105/tpc.106.043000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Septiningsih E.M., Pamplona A.M., Sanchez D.L., Neeraja C.N., Vergara G.V., Heuer S., Ismail A.M., Mackill D.J. Development of submergence-tolerant rice cultivars: The Sub1 locus and beyond. Ann. Bot. 2009;103:151–160. doi: 10.1093/aob/mcn206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Voesenek L.A., Bailey-Serres J. Flood adaptive traits and processes: An overview. New Phytol. 2015;206:57–73. doi: 10.1111/nph.13209. [DOI] [PubMed] [Google Scholar]
- 33.Mendoza M.G., Kaeppler H.F. Auxin and sugar effects on callus induction and plant regeneration frequencies from mature embryos of wheat (Triticum aestivum L.) Cell. Dev. Biol. Plant. 2002;38:39–45. doi: 10.1079/IVP2001250. [DOI] [Google Scholar]
- 34.de Paiva Neto V.B., Otoni W.C. Carbon sources and their osmotic potential in plant tissue culture: Does it matter? Sci. Hortic. 2003;97:193–202. doi: 10.1016/S0304-4238(02)00231-5. [DOI] [Google Scholar]
- 35.Orshinsky B.R., McGregor L.J., Johnson G.I.E., Hucl P., Kartha K.K. Improved embryoid induction and green shoot regeneration from wheat anthers cultured in medium with maltose. Plant Cell Rep. 1990;9:365–369. doi: 10.1007/BF00232400. [DOI] [PubMed] [Google Scholar]
- 36.Skoog F., Miller C. Chemical regulation of growth and organ formation in plant tissues cultured. Symp. Soc. Exp. Biol. 1957;11:118–131. [PubMed] [Google Scholar]
- 37.Gaspar T., Kevers C., Penel C., Greppin H., Reid D.M., Thorpe T.A. Plant hormones and plant growth regulators in plant tissue culture. In Vitro Cell. Dev. Biol.-Plant. 1996;32:272–289. doi: 10.1007/BF02822700. [DOI] [Google Scholar]
- 38.Skoog F., Tsui C. Chemical Control of Growth and Bud Formation in Tobacco Stem Segments and Callus Cultured In Vitro. Am. J. Bot. 1948;35:782–787. doi: 10.1002/j.1537-2197.1948.tb08148.x. [DOI] [Google Scholar]
- 39.Deb C., Imchen T. An Efficient In vitro Hardening Technique of Tissue Culture Raised Plants. Biotechnology. 2010;9:79–83. doi: 10.3923/biotech.2010.79.83. [DOI] [Google Scholar]
- 40.Hauptmann R.M., Widholm J.M., Paxton J.D. Benomyl: A broad spectrum fungicide for use in plant cell and protoplast culture. Plant Cell Rep. 1985;4:129–132. doi: 10.1007/BF00571298. [DOI] [PubMed] [Google Scholar]
- 41.Thielges B.A., Hoitink H.A.J. Fungicides Aid Rooting of Eastern White Pine Cuttings. For. Sci. 1972;18:54–55. doi: 10.1093/forestscience/18.1.54. [DOI] [Google Scholar]
- 42.Liang Z., Chen K., Zhang Y., Liu J., Yin K., Qiu J.L., Gao C. Genome editing of bread wheat using biolistic delivery of CRISPR/Cas9 in vitro transcripts or ribonucleoproteins. Nat. Protoc. 2018;13:413–430. doi: 10.1038/nprot.2017.145. [DOI] [PubMed] [Google Scholar]
- 43.Bae S., Park J., Kim J.S. Cas-OFFinder: A fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. Bioinformatics. 2014;30:1473–1475. doi: 10.1093/bioinformatics/btu048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bono H., Hino K., Ui-Tei K., Naito Y. CRISPRdirect: Software for Designing CRISPR/Cas Guide RNA with Reduced Off-Target Sites. Bioinformatics. 2014;31:1120–1123. doi: 10.1093/bioinformatics/btu743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Biolistic® PDS-1000/He Particle Delivery System User Manual. [(accessed on 8 June 2021)]; Available online: http://www.bio-rad.com/webroot/web/pdf/lsr/literature/M1652249.pdf.
- 46.Liang Y., Baring M., Wang S., Septiningsih E.M. Mapping QTLs for Leafspot Resistance in Peanut Using SNP-Based Next-Generation Sequencing Markers. Plant Breed. Biotechnol. 2017;5:115–122. doi: 10.9787/PBB.2017.5.2.115. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Not applicable.