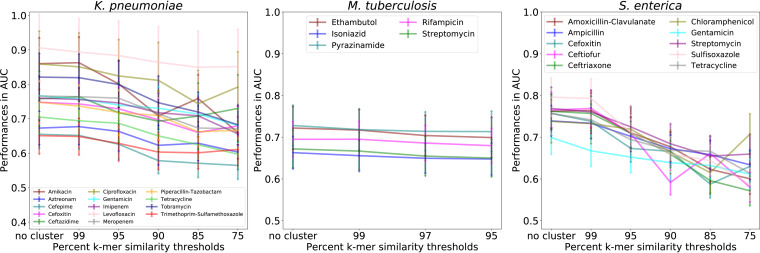
FIG 3.
Results of clustering similar genomes on subalignment model performance. All samples were clustered based on their k-mer similarity in the whole chromosomal alignment at various k-mer identity thresholds. Samples belonging to the same cluster were restricted to either the test or the training sets of each 5-fold cross-validation. Data are the average accuracies with standard deviations of 5-fold cross-validations for 971 Klebsiella pneumoniae (5 kb), 1,066 Salmonella enterica (5 kb), and 441 Mycobacterium tuberculosis (10 kb) subalignments, respectively.