ABSTRACT
The DOE Joint Genome Institute (JGI) Metagenome Workflow performs metagenome data processing, including assembly; structural, functional, and taxonomic annotation; and binning of metagenomic data sets that are subsequently included into the Integrated Microbial Genomes and Microbiomes (IMG/M) (I.-M. A. Chen, K. Chu, K. Palaniappan, A. Ratner, et al., Nucleic Acids Res, 49:D751–D763, 2021, https://doi.org/10.1093/nar/gkaa939) comparative analysis system and provided for download via the JGI data portal (https://genome.jgi.doe.gov/portal/). This workflow scales to run on thousands of metagenome samples per year, which can vary by the complexity of microbial communities and sequencing depth. Here, we describe the different tools, databases, and parameters used at different steps of the workflow to help with the interpretation of metagenome data available in IMG and to enable researchers to apply this workflow to their own data. We use 20 publicly available sediment metagenomes to illustrate the computing requirements for the different steps and highlight the typical results of data processing. The workflow modules for read filtering and metagenome assembly are available as a workflow description language (WDL) file (https://code.jgi.doe.gov/BFoster/jgi_meta_wdl). The workflow modules for annotation and binning are provided as a service to the user community at https://img.jgi.doe.gov/submit and require filling out the project and associated metadata descriptions in the Genomes OnLine Database (GOLD) (S. Mukherjee, D. Stamatis, J. Bertsch, G. Ovchinnikova, et al., Nucleic Acids Res, 49:D723–D733, 2021, https://doi.org/10.1093/nar/gkaa983).
IMPORTANCE The DOE JGI Metagenome Workflow is designed for processing metagenomic data sets starting from Illumina fastq files. It performs data preprocessing, error correction, assembly, structural and functional annotation, and binning. The results of processing are provided in several standard formats, such as fasta and gff, and can be used for subsequent integration into the Integrated Microbial Genomes and Microbiomes (IMG/M) system where they can be compared to a comprehensive set of publicly available metagenomes. As of 30 July 2020, 7,155 JGI metagenomes have been processed by the DOE JGI Metagenome Workflow. Here, we present a metagenome workflow developed at the JGI that generates rich data in standard formats and has been optimized for downstream analyses ranging from assessment of the functional and taxonomic composition of microbial communities to genome-resolved metagenomics and the identification and characterization of novel taxa. This workflow is currently being used to analyze thousands of metagenomic data sets in a consistent and standardized manner.
KEYWORDS: metagenomics, assembly, annotation, binning, SOP, IMG, JGI
INTRODUCTION
Metagenomics, the study of the genetic content of natural microbial communities, provides a wealth of information about the structure, dynamics, perturbation, and resilience of ecosystems. Many tools are available for processing and analyzing metagenomic data sets, including metaSPAdes (1) and MEGAHIT (2) for assembly, Prokka (3) and MG-RAST (4) for annotation, and Kraken 2 (5) for taxonomic identification, as are integrated workflows such as SqueezeMeta (6) and MGnify (7). Some tools process data sets without any a priori information, while tools like MetaPhlAn2 (8) for taxonomic profiling and HUMAnN2 (9) for functional profiling use advanced reference-based techniques. The Joint Genome Institute (JGI) Metagenome Workflow is an integrated workflow focused on the analysis of assembled data and includes read filtering, read error correction and assembly, structural and functional annotation of assembled contigs, and contig-based binning.
RESULTS
The DOE JGI Metagenome Workflow aims to provide consistently processed metagenome data in standard formats suitable for a wide variety of analyses and interpretations across many studies and environmental samples. The workflow performs multiple quality checks and artifact removal and provides a variety of summary statistics to assist users with the assessment of data quality and consistency. We illustrate the workflow using microbiomes from the Loxahatchee Nature Preserve in the Florida Everglades (10) as an example. In this follow-up study, sediment samples were collected and DNA was isolated by the students of Boca Raton Community High School, Boca Raton, FL, from 4 different sites in the Loxahatchee Nature Preserve with 5 replicates at each site, as previously described. DNA isolated from these samples was sequenced at the JGI using the Illumina NovaSeq platform and standard library and sequencing protocols (Kapa HyperPrep library preparation kit) (see Materials and Methods). Raw 2 × 150 reads were then processed by the DOE JGI Metagenome Workflow. The metadata for these samples can be found in the Genomes OnLine Database (GOLD) (11) under GOLD study identifier Gs0136122. Raw reads, as well as intermediate results and final assembly and annotation data, can be found in the JGI data portal (https://genome.jgi.doe.gov) using JGI sequencing project identifiers linked to the GOLD study and Integrated Microbial Genome (IMG) (12) taxon identifiers provided in Table 1.
TABLE 1.
Sequencing and assembly statistics for 20 samples (4 sites, with 5 replicates each) from the Loxahatchee Nature Preserve
| Sample name | IMG taxon ID | Latitude/longitude | No. of filtered reads (million) | No. of contigs (million) | Contig size (Mb) | Contig L50 | % of reads mapped to assembly (%) |
|---|---|---|---|---|---|---|---|
| Lox_West_1 | 3300038551 | 26.469/−80.443 | 432.41 | 6.37 | 4,281.10 | 783 | 61.64 |
| Lox_West_2 | 3300038408 | 26.469/−80.443 | 335.90 | 5.01 | 3,329.57 | 763 | 58.92 |
| Lox_West_3 | 3300038552 | 26.469/−80.443 | 478.21 | 7.22 | 4,968.38 | 814 | 65.04 |
| Lox_West_4 | 3300038469 | 26.469/−80.443 | 447.07 | 6.49 | 4,393.53 | 792 | 62.92 |
| Lox_West_5 | 3300038470 | 26.469/−80.443 | 347.74 | 4.89 | 3,172.10 | 734 | 53.95 |
| Lox_North_1 | 3300038409 | 26.677/−80.375 | 265.39 | 3.12 | 2,017.05 | 736 | 52.06 |
| Lox_North_2 | 3300038421 | 26.677/−80.375 | 294.36 | 3.60 | 2,255.03 | 697 | 52.26 |
| Lox_North_3 | 3300038558 | 26.677/−80.375 | 355.61 | 4.86 | 2,909.37 | 646 | 44.28 |
| Lox_North_4 | 3300038550 | 26.677/−80.375 | 296.91 | 3.86 | 2,361.02 | 666 | 43.15 |
| Lox_North_5 | 3300038422 | 26.677/−80.375 | 240.01 | 3.14 | 1,896.85 | 654 | 41.56 |
| Lox_South_1 | 3300038401 | 26.358/−80.298 | 241.50 | 2.87 | 1,328.12 | 445 | 23.17 |
| Lox_South_2 | 3300038549 | 26.358/−80.298 | 335.62 | 4.83 | 2,379.73 | 481 | 31.57 |
| Lox_South_3 | 3300038402 | 26.358/−80.298 | 240.39 | 2.93 | 1,406.77 | 469 | 25.33 |
| Lox_South_4 | 3300038403 | 26.358/−80.298 | 244.71 | 3.00 | 1,514.26 | 496 | 27.91 |
| Lox_South_5 | 3300038663 | 26.358/−80.298 | 253.01 | 3.31 | 1,771.86 | 538 | 33.78 |
| Lox_East_1 | 3300038454 | 26.502/−80.223 | 299.62 | 3.99 | 2,746.17 | 819 | 54.72 |
| Lox_East_2 | 3300038455 | 26.502/−80.223 | 322.84 | 4.18 | 2,834.88 | 795 | 52.17 |
| Lox_East_3 | 3300038431 | 26.502/−80.223 | 292.44 | 3.65 | 2,385.22 | 740 | 46.35 |
| Lox_East_4 | 3300038410 | 26.502/−80.223 | 247.69 | 3.49 | 2,320.95 | 761 | 52.70 |
| Lox_East_5 | 3300038468 | 26.502/−80.223 | 266.29 | 3.75 | 2,317.21 | 670 | 46.14 |
Read prefiltering and assembly results.
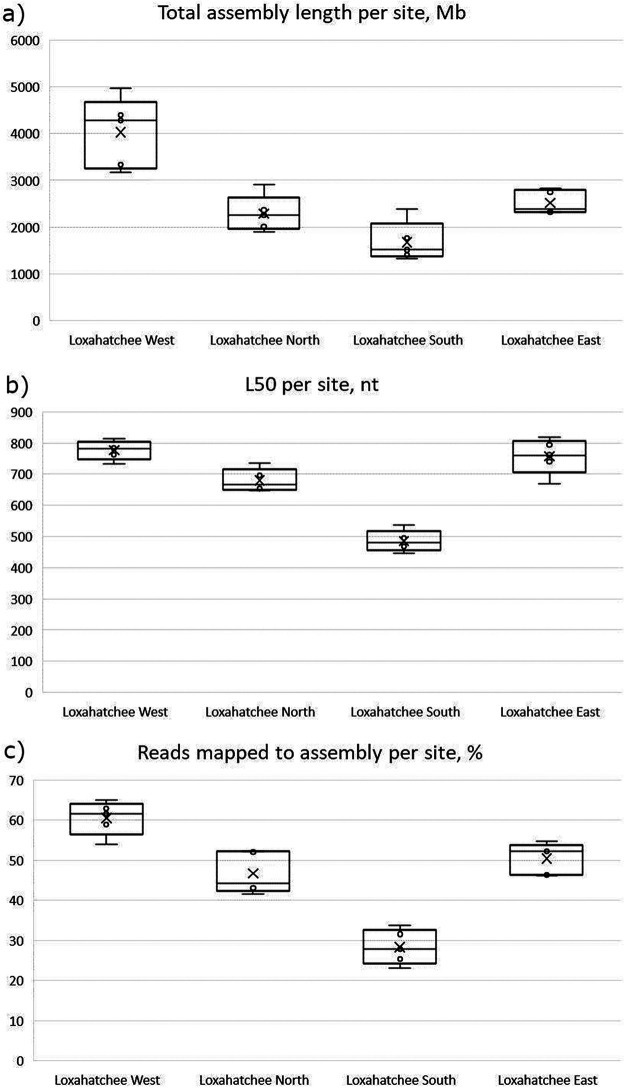
The target amount of raw sequence data was 45 Gb per sample (300 million reads). The numbers of high-quality (HQ) raw reads per sample after quality trimming, filtering, and artifact and contamination removal are shown in Table 1. While the replicates from Loxahatchee West were sequenced somewhat more deeply than other samples, there is no significant difference in the amount of sequence generated for the other three sites. The prefiltering and assembly modules of the workflow automatically generate several conventional measures of assembly quality that are provided in README files and can be accessed via the JGI data portal. A subset of these measures, which helps with assessing the consistency of the samples and identifying outliers and artifacts, is shown in Table 1. Despite the fact that the samples from Loxahatchee F1 North, South, and East received very similar amounts of sequence, assembly statistics indicate that the replicates collected at the South site differ from the rest, as shown in Fig. 1a. Box-and-whisker plots for the L50 metric (the shortest length of contigs for which the sum of lengths makes up half of the data set size) (Fig. 1b) and the percentage of reads mapped to the assembly (Fig. 1c) demonstrate that assemblies of South site replicates are significantly more fragmented, as indicated by the much lower L50, and have fewer reads mapped to them. This may be due to the fact that the sediment at the South site has a large amount of sand, which hindered the isolation of sufficient quantities of high-quality DNA (Jonathan B. Benskin, personal communication), thereby resulting in a suboptimal library and poor assembly. Variation of library quality due to the quality and quantity of the source DNA may not be immediately obvious with a functional and/or taxonomic analysis of unassembled reads but is prominently brought to the researcher’s attention by the DOE JGI Metagenome Workflow. It highlighted the differences between the South site and other sites due to the inconsistent performance of a sampling protocol, which may confound statistical analysis and obfuscate the true differences in functional and taxonomic composition.
FIG 1.
Plots of sequencing and assembly statistics for 4 sites in the Loxahatchee Nature Preserve. (a) Total assembly length per site, in megabases. (b) L50 (the smallest length of contigs whose sum of lengths makes up half of the data set size) per site, in nucleotides. (c) Reads mapped to the assembly as a percentage of the total number of reads generated per sample, per site.
Annotation results.
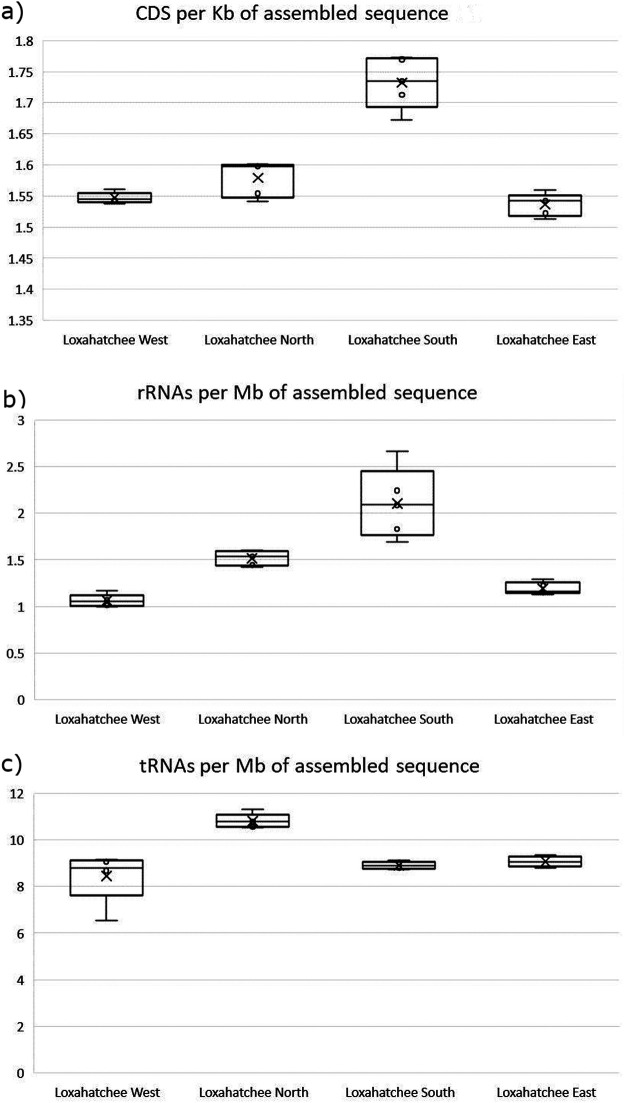
The DOE JGI Metagenome Workflow performs feature prediction (also known as structural annotation) on the assembled sequences and functional annotation of the coding sequences (CDSs). Similar to the filtering and assembly modules, the annotation module generates summary statistics helpful for the identification of artifacts and outlier samples. These statistics are provided in README files via the JGI data portal and can be also found in the IMG database on the metagenome details page of each data set. A subset of the annotation measures for Loxahatchee samples is provided in Table 2. The results of functional annotation of CDSs appear to be highly consistent across the four sites, with 65.75% ± 1.2% of all CDSs assigned to Clusters of Orthologous Genes (COGs) (13), 14.25% ± 0.44% assigned to TIGRFAMs (14), 62.65% ± 0.81% assigned to Pfams (15), and 40.2% ± 1.85% assigned to KEGG Orthology (KO) terms (16). However, the results of feature prediction summarized in Fig. 2 paint a different picture. Again, the South site is different from the other three sites, having more predicted CDSs per kilobase of assembled sequence (Fig. 2a) and a much higher number of predicted rRNAs per megabase of assembled sequence (Fig. 2b). Remarkably, there is no significant difference in tRNA counts (Fig. 2c). These observations are consistent with the lower contiguity of South site assemblies, as reflected in their lower L50 (Fig. 1b), which in turn results in the fragmentation of longer protein-coding genes as well as long 16S/18S and 23S/28S rRNA genes. On the other hand, tRNAs, which are on average <100 nucleotides (nt) long, are largely unaffected by the fragmentation of assembled sequences. Importantly, protein-coding genes, which span a large interval of sequence lengths, will be affected unevenly, with the copy number of longer proteins appearing to be higher in more fragmented assemblies, while shorter proteins will show no differences. These factors have to be taken into account when comparing the functional compositions of different samples and attempting to correlate them with various environmental factors. The feature prediction and functional annotation module of the DOE JGI Metagenome Workflow provides other indicators of the quality and consistency of metagenomic data: the counts of eukaryotic 18S and 28S rRNAs suggest the presence and abundance of eukaryotic genomes in the sample, which could derive from the eukaryotic members of the microbial community and/or host DNA in host-associated microbiomes. On the other hand, the relatively low percentage of CDSs assigned to COGs and Pfams may indicate the presence of a large viral fraction in the community since viral proteins are poorly represented in these protein and domain classification systems. All of these characteristics of the assembled metagenome need to be taken into account in comparative analyses as they may affect the results of the taxonomic and functional annotation of the communities.
TABLE 2.
Annotation statistics for 20 samples (4 sites, with 5 replicates each) from the Loxahatchee Nature Preserve
| Sample name | IMG taxon ID | Contig size (Mb) | No. of CRISPR elements | Predicted count |
% of CDSs assigned to database (% of total) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CDSs (million) | 16S rRNA | 18S rRNA | 23S rRNA | 28S rRNA | 5S rRNA | tRNAs | COGs | TIGRFAM | Pfam | KEGG | ||||
| Lox_West_1 | 3300038551 | 2,859.7 | 391 | 4.413 | 943 | 2 | 1,559 | 8 | 384 | 18,675 | 67 | 15 | 63 | 39 |
| Lox_West_2 | 3300038408 | 2,204.4 | 250 | 3.412 | 742 | 8 | 1,209 | 14 | 377 | 19,124 | 65 | 14 | 63 | 39 |
| Lox_West_3 | 3300038552 | 3,396.0 | 458 | 5.245 | 1,084 | 8 | 1,735 | 12 | 560 | 29,892 | 64 | 14 | 62 | 38 |
| Lox_West_4 | 3300038469 | 2,949.2 | 420 | 4.534 | 957 | 5 | 1,612 | 9 | 529 | 27,020 | 65 | 14 | 62 | 39 |
| Lox_West_5 | 3300038470 | 2,061.2 | 242 | 3.218 | 722 | 6 | 1,292 | 11 | 384 | 18,675 | 66 | 15 | 63 | 40 |
| Lox_North_1 | 3300038409 | 1,293.3 | 339 | 1.994 | 574 | 15 | 973 | 22 | 289 | 13,655 | 65 | 14 | 62 | 39 |
| Lox_North_2 | 3300038421 | 1,408.6 | 372 | 2.189 | 644 | 16 | 1,029 | 20 | 292 | 14,843 | 65 | 14 | 62 | 39 |
| Lox_North_3 | 3300038558 | 1,761.8 | 255 | 2.818 | 877 | 11 | 1,432 | 9 | 382 | 19,094 | 65 | 14 | 62 | 41 |
| Lox_North_4 | 3300038550 | 1,460.0 | 171 | 2.333 | 736 | 9 | 1,209 | 13 | 345 | 16,512 | 65 | 14 | 62 | 40 |
| Lox_North_5 | 3300038422 | 1,161.4 | 145 | 1.860 | 589 | 9 | 978 | 12 | 268 | 12,534 | 66 | 14 | 62 | 40 |
| Lox_South_1 | 3300038401 | 571.3 | 58 | 1.011 | 454 | 21 | 863 | 33 | 150 | 4,992 | 67 | 14 | 63 | 44 |
| Lox_South_2 | 3300038549 | 1,139.7 | 137 | 1.977 | 622 | 15 | 1,187 | 27 | 237 | 10,120 | 67 | 14 | 63 | 42 |
| Lox_South_3 | 3300038402 | 653.7 | 83 | 1.159 | 421 | 18 | 854 | 33 | 140 | 5,752 | 68 | 14 | 64 | 44 |
| Lox_South_4 | 3300038403 | 750.8 | 105 | 1.286 | 465 | 14 | 895 | 20 | 174 | 6,767 | 67 | 14 | 63 | 43 |
| Lox_South_5 | 3300038663 | 950.1 | 87 | 1.589 | 493 | 5 | 911 | 7 | 190 | 8,662 | 68 | 14 | 64 | 42 |
| Lox_East_1 | 3300038454 | 1,852.5 | 219 | 2.803 | 691 | 11 | 1,041 | 15 | 334 | 16,789 | 65 | 15 | 63 | 39 |
| Lox_East_2 | 3300038455 | 1,891.4 | 259 | 2.879 | 678 | 10 | 1,158 | 20 | 322 | 17,682 | 65 | 15 | 63 | 39 |
| Lox_East_3 | 3300038431 | 1,551.8 | 156 | 2.396 | 615 | 8 | 1,020 | 12 | 249 | 13,642 | 66 | 15 | 64 | 40 |
| Lox_East_4 | 3300038410 | 1,529.8 | 208 | 2.359 | 557 | 8 | 942 | 12 | 246 | 14,059 | 65 | 14 | 62 | 39 |
| Lox_East_5 | 3300038468 | 1,431.5 | 196 | 2.232 | 581 | 13 | 966 | 18 | 271 | 12,773 | 64 | 14 | 61 | 38 |
FIG 2.
Plots summarizing the results of structural annotation for 20 samples (4 sites, with 5 replicates each) from the Loxahatchee Nature Preserve. (a) Number of predicted CDSs per kilobase of assembled sequence. (b) Number of predicted rRNA genes per megabase of assembled sequence. (c) Number of predicted tRNA genes per megabase of assembled sequence.
Binning results.
The DOE JGI Metagenome Workflow includes automated binning of assembled sequences as well as an initial characterization of bins in terms of completeness, contamination, and quality. The bins are assigned to high-quality (HQ) and medium-quality (MQ) categories based on Minimum Information about a Metagenome-Assembled Genome (MIMAG) standards (17). Bins that do not meet the standards for HQ or MQ are discarded. For HQ and MQ bins, additional data processing is performed: bins are assigned a predicted lineage based on the NCBI (18) and GTDB-tk (19) taxonomies. The results of genome binning for the Loxahatchee samples are summarized in Table 3. The vast majority of the bins generated for these data sets are MQ and represent a minor portion of the total assembly typical of high-complexity metagenomes from soil and sediment samples. Binning results for each data set can be accessed via the JGI data portal and in the IMG database, where a number of tools for searching, analysis, and comparison of metagenome bins are available.
TABLE 3.
Binning statistics for 20 samples (4 sites, with 5 replicates each) from the Loxahatchee Nature Preserve
| Sample name | IMG taxon ID | High-quality bins |
Medium-quality bins |
||||
|---|---|---|---|---|---|---|---|
| No. of bins | Size (Mb) | No. of contigs | No. of bins | Size (Mb) | No. of contigs | ||
| Lox_West_1 | 3300038551 | 0 | 0 | 0 | 9 | 18.97 | 3,041 |
| Lox_West_2 | 3300038408 | 0 | 0 | 0 | 12 | 24.90 | 3,854 |
| Lox_West_3 | 3300038552 | 0 | 0 | 0 | 11 | 27.55 | 3,251 |
| Lox_West_4 | 3300038469 | 0 | 0 | 0 | 10 | 19.56 | 2,542 |
| Lox_West_5 | 3300038470 | 0 | 0 | 0 | 6 | 16.68 | 2,542 |
| Lox_North_1 | 3300038409 | 0 | 0 | 0 | 4 | 12.25 | 2,100 |
| Lox_North_2 | 3300038421 | 0 | 0 | 0 | 4 | 15.51 | 2,241 |
| Lox_North_3 | 3300038558 | 1 | 1.25 | 35 | 12 | 22.80 | 3,749 |
| Lox_North_4 | 3300038550 | 1 | 1.29 | 46 | 6 | 7.24 | 1,180 |
| Lox_North_5 | 3300038422 | 1 | 1.26 | 39 | 6 | 10.36 | 1,751 |
| Lox_South_1 | 3300038401 | 0 | 0 | 0 | 1 | 3.14 | 498 |
| Lox_South_2 | 3300038549 | 1 | 7.34 | 152 | 3 | 4.06 | 711 |
| Lox_South_3 | 3300038402 | 0 | 0 | 0 | 0 | 0 | 0 |
| Lox_South_4 | 3300038403 | 0 | 0 | 0 | 1 | 0.83 | 103 |
| Lox_South_5 | 3300038663 | 0 | 0 | 0 | 2 | 3.50 | 528 |
| Lox_East_1 | 3300038454 | 2 | 4.16 | 365 | 6 | 18.80 | 2,485 |
| Lox_East_2 | 3300038455 | 0 | 0 | 0 | 4 | 8.41 | 1,150 |
| Lox_East_3 | 3300038431 | 0 | 0 | 0 | 7 | 16.21 | 2,177 |
| Lox_East_4 | 3300038410 | 0 | 0 | 0 | 8 | 22.64 | 3,269 |
| Lox_East_5 | 3300038468 | 0 | 0 | 0 | 10 | 21.20 | 2,753 |
Run times.
We illustrate the typical computational requirements of the DOE JGI Metagenome Workflow on 20 samples from Loxahatchee Nature Preserve in Table 4. Filtering was done using Intel Xeon Gold 6140 processors using 32 virtual centralized processing unit (vCPU) and 324 GB of random access memory (RAM). For error correction, assembly, and mapping, a mix of configurations was used. Some data sets were run on Intel Xeon Platinum 8000 series processors with different amounts of memory depending on the stage (16 vCPU and 128 GB of random access memory (RAM). RAM for error correction, 64 vCPU and 512 GB of RAM for assembly, and 32 vCPU and 256 GB of RAM for mapping). For others, Intel Xeon Gold 6140 processors were used, with 72 vCPU, 1.5 TB of RAM, and 5 TB of local disk. The run time assembly in Table 4 represents CPU hours for filtering, error correction, assembly, and mapping. For annotation, assembled metagenomic sequences were split into 10-MB shards. The splitting is performed by a wrapper script for the optimal utilization of the JGI compute infrastructure and is not required to run the workflow. These 10-MB shards were then processed in parallel, with each shard running on its own 2.3-GHz Haswell processor node with 128 GB of RAM. Binning was run on 2.3-GHz Haswell processor nodes with 128 GB of RAM.
TABLE 4.
CPU hours for different modules in the JGI Metagenome Workflow on 20 samples from Loxahatchee Nature Preserve
| Sample name | IMG taxon ID | CPU h |
|||
|---|---|---|---|---|---|
| Assembly | Feature prediction | Functional annotation | Binning | ||
| Lox_West_1 | 3300038551 | 3,576.16 | 12,423.68 | 8,980.48 | 264.9 |
| Lox_West_2 | 3300038408 | 2,751.16 | 12,572.8 | 6,836.48 | 110.3 |
| Lox_West_3 | 3300038552 | 4,155.04 | 13,522.56 | 10,065.92 | 367.6 |
| Lox_West_4 | 3300038469 | 3,699.6 | 12,163.84 | 9,695.36 | 225.9 |
| Lox_West_5 | 3300038470 | 2,713.03 | 8,332.16 | 7,274.88 | 90.0 |
| Lox_North_1 | 3300038409 | 1,801.75 | 5,659.52 | 3,489.28 | 23.9 |
| Lox_North_2 | 3300038421 | 2,064.19 | 6,092.85 | 3,990.40 | 23.5 |
| Lox_North_3 | 3300038558 | 2,455.81 | 7,430.4 | 6,223.36 | 14.9 |
| Lox_North_4 | 3300038550 | 1,944.75 | 6,147.2 | 4,270.08 | 11.0 |
| Lox_North_5 | 3300038422 | 1,692.39 | 5,338.8 | 3,429.76 | 9.3 |
| Lox_South_1 | 3300038401 | 1,540.82 | 62.72 | 29.30 | 2.1 |
| Lox_South_2 | 3300038549 | 1,534.45 | 88.55 | 62.23 | 7.1 |
| Lox_South_3 | 3300038402 | 1,556.06 | 78.19 | 33.38 | 1.9 |
| Lox_South_4 | 3300038403 | 1,621.84 | 61.65 | 36.12 | 5.7 |
| Lox_South_5 | 3300038663 | 1,771.97 | 72.76 | 53.28 | 7.4 |
| Lox_East_1 | 3300038454 | 2,086.37 | 114.67 | 99.84 | 59.3 |
| Lox_East_2 | 3300038455 | 2,298.94 | 117.02 | 89.79 | 62.5 |
| Lox_East_3 | 3300038431 | 2,153.02 | 102.98 | 100.34 | 31.7 |
| Lox_East_4 | 3300038410 | 1,877.78 | 99.47 | 84.15 | 35.4 |
| Lox_East_5 | 3300038468 | 1,795.02 | 101.5 | 66.69 | 25.3 |
DISCUSSION
The DOE JGI Metagenome Workflow provides automatic assembly, annotation, and binning of metagenome data sets. It is largely based on publicly available software and databases supplemented with custom scripts and wrappers to control the workflow and enable the seamless integration of the input and output of different programs. Filtering, read correction, assembly, and mapping use a median of 2,004 CPU hours for current metagenomes such as the Loxahatchee sediment metagenomes and can be performed on standard high-performance computing nodes such as the Intel Xeon Platinum 8000 series processor with 256 GB of memory. On average, the annotation module of the workflow (feature prediction, functional annotation, and product name assignment) can process 1 million bp in 9 CPU hours on a 2.3-GHz Haswell processor (Intel Xeon Processor E5-2698 v3) node with 128 GB of RAM. On the same Haswell node, the entire binning workflow, from initial bin prediction to scaffold-level cleanup, bin-level phylogenetic prediction, and estimation of contamination and completion, can process 100,000 scaffolds in an average of 13 CPU hours. The workflow modules for read filtering and metagenome assembly are available as a workflow description language (WDL) file (https://code.jgi.doe.gov/BFoster/jgi_meta_wdl). The annotation and binning modules of the workflow are publicly available via the IMG system’s submission site (https://img.jgi.doe.gov/submit), which accepts assembled metagenome sequences in fasta format and requires the submission of the sample and project metadata as a condition of annotation and binning services. We plan to continue to improve the workflow by updating reference database versions, extending the existing software, and adding new tools that allow the identification and characterization of more features in the metagenome data sets, as well as improving the performance by making changes geared toward exploiting the specific infrastructure that the workflow is utilizing. Describing features in IMG that allow comparative analysis of data sets is described separately in the IMG/M version 6.0 publication (12).
MATERIALS AND METHODS
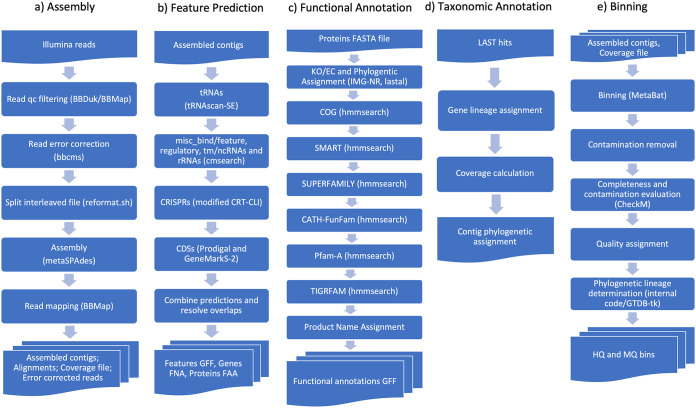
Figure 3 shows workflow diagrams for computational tasks.
FIG 3.
Workflow diagrams of the components of the DOE JGI Metagenome Workflow. (a) Assembly, which produces assembled contigs and an alignment of reads to assembled contigs. qc, quality control. (b) Feature prediction, which produces features in general feature format (GFF), genes in FASTA nucleic acid (FNA) format, and proteins in FASTA amino acid (FAA) format. (c) Functional annotation, which produces product name assignments. (d) Taxonomic annotation, which produces contig-level phylogenetic assignment. (e) Binning, which produces high- and medium-quality genome bins.
Data input.
Standard metagenomes at JGI currently use 100 ng of genomic DNA, sheared to 300 bp using the Covaris LE220 instrument and size selected with SPRI using TotalPure NGS beads (Omega Bio-tek). The fragments are treated with end repair, A-tailing, and ligation of Illumina-compatible adapters (IDT, Inc.) using the Kapa-HyperPrep kit (Kapa Biosystems) to create an unamplified Illumina library, which is then sequenced 2 × 150 bp on the Illumina NovaSeq 6000 platform using S4 flow cells. The workflow can be used on paired-end Illumina data sets; kmer sizes for assembly should be adjusted if reads are shorter than 150 bp.
Sequence data preprocessing.
Data are processed using Real-Time Analysis (RTA) version 3.4.4 (https://support.illumina.com/downloads.html). BBDuk version 38.79 from the BBTools package (https://jgi.doe.gov/data-and-tools/bbtools/) is used to remove contamination, trim reads that contain an adapter sequence, and quality trim reads where quality drops to zero. Furthermore, it is used to remove reads that contain 4 or more “N” bases, have an average quality score across the read of <3, or have a minimum length of ≤51 bp or 33% of the full read length. Homopolymer stretches of 5 G’s or more at the ends of reads are removed. Reads that can be mapped with BBMap from BBTools to masked human, cat, dog, and mouse references at 93% identity are separated into a “chaff” file and not used in the assembly. In an abundance of caution, reads aligned to common microbial contaminants described in the literature, such as Ralstonia pickettii and Acinetobacter calcoaceticus (20–23), are also separated into a chaff file. Masked references can be found at https://portal.nersc.gov/dna/microbial/assembly/bushnell/fusedERPBBmasked2.fa.gz. For convenience, chaff files are provided on JGI’s data portal.
Assembly.
Filtered reads are error corrected using bbcms version 38.44 from BBTools with a minimum count of 2 and a high-count fraction of 0.6. bbcms uses a count-min sketch to store kmer counts, making it a scalable solution for error correction of metagenomic data sets. For computational efficiency, interleaved fastq files are split into two separate files. These split-error-corrected files are assembled with metaSPAdes version 3.13.0 using the “metagenome” flag, running the assembly module only (i.e., without error correction) with kmer sizes of 33, 55, 77, 99, and 127. Contigs that are smaller than 200 bp are discarded. Filtered reads are mapped back to contigs larger than 200 bp using BBMap 38.44 with “interleaved” as true, “ambiguous” as random, and the “covstats” option specifying a contig coverage file for subsequent analysis of the abundances of various populations and genes. The coverage file contains information on the average fold coverage, length, GC content, percentage of bases covered, number of reads by strand, read GC, median fold, and standard deviation of coverage. No further analysis is performed on unassembled reads.
Feature prediction.
The assembled contigs are passed on to the annotation module of the workflow, which first predicts noncoding RNA (ncRNA) genes (tRNAs, rRNAs, and other RNAs), followed by the identification of clustered regularly interspaced short palindromic repeats (CRISPR) and protein-coding genes (CDSs), as shown in Fig. 3b. Prediction of tRNAs is performed using tRNAscan-SE 2.0.6 (24) in “bacterial” and “archaeal” search modes. This allows the workflow to select the best annotation mode and ensure higher annotation accuracy for metagenomic contigs of different taxonomic origins since many archaeal tRNAs cannot be predicted in “bacterial” or “general” modes. For each contig, the numbers of tRNAs with a known isotype returned by each mode are compared. The results from the mode with the higher number of tRNAs with a known isotype are reported, and if both modes have returned the same number, the results from the bacterial mode are included in the final annotation. rRNA genes (5S, 16S, and 23S) as well as other ncRNA genes, including transfer-messenger RNA (tmRNA) and antisense RNAs, etc., and RNA regulatory features, such as various binding sites and motifs (“misc_bind,” “misc_feature,” and “regulatory”), are identified by comparing the contigs via cmsearch from the INFERNAL 1.1.3 package (25) against the Rfam 13.0 database (26) using the trusted cutoffs parameter (–cut_tc). If any reported hits are overlapping even by 1 bp and they belong to the same Rfam class, the lower scoring of the two is discarded. CRISPR elements are identified using a version of CRT-CLI 1.2 modified in-house as described previously (27). For the search parameter, the minimum and maximum repeat lengths are set to 20 and 50 bp, respectively, whereas the minimum and maximum spacer lengths are set to 20 and 60 bp, respectively. The search window size is set to 7 bp, and an element needs to have at least three repeats to be reported. Protein-coding genes are predicted via a combination of Prodigal 2.6.3 (28) and GeneMarkS-2 1.07 (29). Prodigal is executed in “meta” mode and with the “-m” argument so that genes will not be built across runs of N’s. GeneMark is run with “–Meta mgm_11.mod” and “–incomplete_at_gaps 30.” CDSs shorter than 75 bp (25 amino acids) are discarded. The last step of the feature prediction combines the results from all tools and attempts to resolve overlaps between features of different types. Two features are considered to overlap if they share more than 10 bp or more than 90 bp in the case of two CDSs. The regulatory RNA features (misc_bind, misc_feature, and regulatory) are allowed to overlap any other feature type. In the case of an overlap between other types of features, the lower-ranked feature is removed. The feature ranking order is rRNA > tRNA > ncRNA, tmRNA > CRISPR > GeneMarkS-2 > Prodigal. Before deleting a CDS that overlaps another feature over its 5′ end, an attempt is first made to find an alternative start site for the protein-coding gene that removes the overlap. Functional annotations of RNA features are based on their descriptions provided by the tool or database used to predict them: tRNA isotype (amino acid and codon) as well as potential pseudogene annotations are provided by tRNAscan-SE, while product names for rRNAs, ncRNAs, and regulatory RNA features are derived from the corresponding Rfam models. Functional annotation and product name assignment for protein sequences of the nonoverlapping CDSs are performed by the functional annotation module.
Functional annotation.
Functional annotation for metagenomes consists of associating protein-coding genes with KO terms, Enzyme Commission (EC) numbers, COG assignments, SMART domains, SUPERFAMILY assignments, CATH-FunFam annotations, Pfams, and TIGRFAM annotations, as shown in Fig. 3c. Genes are associated with KO terms and EC numbers based on the results of a sequence similarity search of metagenome proteins against a reference database of isolate proteomes using lastal 1066 from the LAST package (30), with default parameters. The reference database of isolate proteomes (IMG-NR) is composed of all nonredundant protein sequences encoded by public, high-quality genomes in the current version of the IMG database. For each metagenome protein, the top five LAST hits are considered. At least two of the top five hits need to have a KO assignment, and all hits that have a KO assignment need to list the same combination of KO terms. If both conditions are met, the same combination of KO terms is assigned to the query gene if the alignment length for any of the hits with a KO assignment covers at least 70% of the shorter one of query and subject. Proteins are associated with COGs by comparing protein sequences to the COG hidden Markov models (HMMs) created from the updated 2014 models using HMMER 3.1b2 (31) and a thread-optimized version of hmmsearch (32), with a per-domain E value cutoff (–domE) of 0.01. Since an alignment of a protein to the model may be fragmented, i.e., there may be multiple aligned segments of the two, these are concatenated, and their cumulative alignment length is calculated. If the cumulative alignment length is less than 70% of the shorter of the two (the protein or the model), such a hit is discarded. In addition, if a protein has hits to different COG models and their alignments overlap significantly (by more than 10% of the length of the shorter model), the hit to the model with the lower full-sequence bit score is discarded; for significantly overlapping hits with the same bit score, the hit with the higher E value is removed. The same thread-optimized version of hmmsearch as well as parameters, filtering, and overlap resolution rules are used to assign protein sequences to the 01_06_2016 version of the SMART database (33), the 1.75 version of the SUPERFAMILY database (34), and the frozen set of the 4.2.0 version of the CATH-FunFam database (35). Proteins are associated with Pfam-A by comparing protein sequences to version 30 of the Pfam database using the thread-optimized version of hmmsearch from HMMER 3.1b2. Model-specific trusted cutoffs are used with the –cut_tc option in hmmsearch, and for overlapping hits that belong to the same Pfam clan, the lower-scoring one is removed. Proteins are associated with TIGRFAMs using version 15.0 of the TIGRFAM database and hmmsearch with a per-domain E value cutoff (–domE) of 0.01. All hits that do not cover at least 70% of the shorter protein or model are discarded. Furthermore, if two hits overlap for more than 10% of the length of the shorter model, the hit to the lower-scoring model (by bit score) is discarded. Protein product names are assigned based on the name of their associated protein families in the order of priority KO term > TIGRFAM > COG > Pfam. If multiple TIGRFAMs with different isology types are associated with a protein, only one TIGRFAM is assigned in the order equivalog > hypoth_equivalog > paralog > exception > equivalog_domain > hypoth_equivalog_domain > paralog_domain > subfamily > superfamily > subfamily_domain > domain > signature > repeat. Proteins without any of the above-mentioned assignments are annotated as a “hypothetical protein.” Proteins associated with multiple protein families of the same type (KO term, TIGRFAM, COG, or Pfam) are annotated with a product name consisting of concatenation of individual protein family names joined with “/.” Multiple repetitions of the same protein family are collapsed into a single instance. The contig coverage information is used to calculate so-called “estimated gene copies,” whereby the number of genes in a certain group, such as a COG or Pfam protein family, is multiplied by the average coverage of the contigs from which these genes were predicted. This step is important for accurate estimation of the abundance of protein families and takes into account the different abundances of populations found in the assembled metagenome sequences.
Taxonomic annotation.
For the taxonomic annotation of metagenomes, the best LAST (30) hits of CDSs, computed as described above for KO term assignment, are used. The taxonomy of the best hit is assigned to each metagenome protein. The taxonomy of metagenome contigs (“scaffold lineage”) is predicted based on the majority rule, whereby the lineage at the lowest taxonomic rank to which at least 50% of CDSs encoded by the metagenomic contig have hits is assigned. Similar to protein family annotations, contig coverage information is used to estimate the abundance of various lineages in the community by multiplying contig counts by their average coverage.
Binning.
The assembled contigs and coverage file generated per metagenome are used as the input to the MetaBAT v2.12.1 (36) program to generate genome bins based on the consistency of coverage and tetranucleotide frequency. The genome bins then undergo contamination removal, wherein the per-scaffold phylum information generated by the annotation module (“scaffold lineage”) is used to remove scaffolds per bin that are not assigned to the predominant phylum. The postprocessed bins are fed to the CheckM v1.0.12 (37) program to determine genome completion and contamination estimates. These estimates along with the per-scaffold rRNA and tRNA information generated by the annotation module are used to assign an HQ or MQ value to each bin, per MIMAG standards. The HQ and MQ bins are then subjected to phylogenetic lineage determination by two methods. First, an internal IMG program computes the phylogenetic lineage per genome bin using the per-scaffold lineage generated by the annotation module. Next, the GTDB-tk v0.2.2 program computes per-bin lineage by placing them into domain-specific, concatenated protein reference trees. The high- and medium-quality bins, along with the corresponding data processing metadata, are loaded into the IMG database for user access and download.
Preformatted tables.
To assist with preparing publications, 9 tables are generated. Information on what is contained in each table is described in Table 5.
TABLE 5.
Preformatted tables
| Table no. | Table information |
|---|---|
| 1 | Study information |
| 2 | Sample information |
| 3 | Library information |
| 4 | Sequence process |
| 5 | Assembly statistics |
| 6 | Annotation parameters |
| 7 | Functional diversity |
| 8 | Metagenome properties |
| 9 | Taxonomic composition |
Availability of data.
The metadata for these samples can be found in GOLD (https://gold.jgi.doe.gov/) under GOLD study identifier Gs0136122. Raw reads, as well as intermediate results and final assembly and annotation data, can be found in the JGI data portal (https://genome.jgi.doe.gov) by following links from the GOLD study or by using IMG taxon identifiers provided in Table 1. A WDL for filtering and genome assembly (v1.0) is available at https://code.jgi.doe.gov/BFoster/jgi_meta_wdl. IMG for annotation (v5.0.19) and binning (v1.0) is available at https://img.jgi.doe.gov/. For information and tutorials on using GOLD and submitting metadata, see the IMG-GOLDWebinar:DataSubmissionandManagement or the GOLDhelppage. For information on IMG, seeIMGhelp page or the IMGWebinarSeries.
ACKNOWLEDGMENTS
The work conducted by the U.S. Department of Energy Joint Genome Institute, a DOE Office of Science User Facility, is supported by the Office of Science of the U.S. Department of Energy under contract no. DE-AC02-05CH11231.
We thank the Advanced International Certificate of Education (AICE) biology class at Florida’s Boca Raton Community High School and Jonathan B. Benskin for partnering with the JGI on the Everglades metagenome studies.
Contributor Information
Alicia Clum, Email: aclum@lbl.gov.
Nicola Segata, University of Trento.
REFERENCES
- 1.Nurk S, Meleshko D, Korobeynikov A, Pevzner PA. 2017. metaSPAdes: a new versatile metagenomic assembler. Genome Res 27:824–834. doi: 10.1101/gr.213959.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li D, Liu CM, Luo R, Sadakane K, Lam TW. 2015. MEGAHIT: an ultrafast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 31:1674–1676. doi: 10.1093/bioinformatics/btv033. [DOI] [PubMed] [Google Scholar]
- 3.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 4.Keegan KP, Glass EM, Meyer F. 2016. MG-RAST, a metagenomics service for analysis of microbial community structure and function. Methods Mol Biol 1399:207–233. doi: 10.1007/978-1-4939-3369-3_13. [DOI] [PubMed] [Google Scholar]
- 5.Wood DE, Lu J, Langmead B. 2019. Improved metagenomic analysis with Kraken 2. Genome Biol 20:257. doi: 10.1186/s13059-019-1891-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tamames J, Puente-Sánchez F. 2019. SqueezeMeta, a highly portable, fully automatic metagenomic analysis pipeline. Front Microbiol 9:3349. doi: 10.3389/fmicb.2018.03349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mitchell AL, Almeida A, Beracochea M, Boland M, Burgin J, Cochrane G, Crusoe MR, Kale V, Potter SC, Richardson LJ, Sakharova E, Scheremetjew M, Korobeynikov A, Shlemov A, Kunyavskaya O, Lapidus A, Finn RD. 2020. MGnify: the microbiome analysis resource in 2020. Nucleic Acids Res 48:D570–D578. doi: 10.1093/nar/gkz1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Segata N, Waldron L, Ballarini A, Narasimhan V, Jousson O, Huttenhower C. 2012. Metagenomic microbial community profiling using unique clade-specific marker genes. Nat Methods 9:811–814. doi: 10.1038/nmeth.2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Franzosa EA, McIver LJ, Rahnavard G, Thompson LR, Schirmer M, Weingart G, Lipson KS, Knight R, Caporaso JG, Segata N, Huttenhower C. 2018. Species-level functional profiling of metagenomes and metatranscriptomes. Nat Methods 15:962–968. doi: 10.1038/s41592-018-0176-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Abraham BS, Caglayan D, Carrillo NV, Chapman MC, Hagan CT, Hansen ST, Jeanty RO, Klimczak AA, Klingler MJ, Kutcher TP, Levy SH, Millard-Bruzos AA, Moore TB, Prentice DJ, Prescott ME, Roehm R, Rose JA, Yin M, Hyodo A, Lail K, Daum C, Clum A, Copeland A, Seshadri R, del Rio TG, Eloe-Fadrosh EA, Benskin JB. 2020. Shotgun metagenomic analysis of microbial communities from the Loxahatchee nature preserve in the Florida Everglades. Environ Microbiome 15:2. doi: 10.1186/s40793-019-0352-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mukherjee S, Stamatis D, Bertsch J, Ovchinnikova G, Sundaramurthi J, Lee J, Kandimalla M, Chen IMA, Kyrpides NC, Reddy TBK. 2021. Genomes OnLine Database (GOLD) v.8: overview and updates. Nucleic Acids Res 49:D723–D733. doi: 10.1093/nar/gkaa983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen IMA, Chu K, Palaniappan K, Ratner A, Huang J, Huntemann M, Hajek P, Ritter S, Varghese N, Seshadri R, Roux S, Woyke T, Eloe-Fadrosh EA, Ivanova NN, Kyrpides N. 2021. IMG/M data management and analysis system v.6.0: new tools and advanced capabilities. Nucleic Acids Res 49:D751–D763. doi: 10.1093/nar/gkaa939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Galperin MY, Makarova KS, Wolf YI, Koonin EV. 2015. Expanded microbial genome coverage and improved protein family annotation in the COG database. Nucleic Acids Res 43:D261–D269. doi: 10.1093/nar/gku1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haft DH, Selengut JD, Richter RA, Harkins D, Basu MK, Beck E. 2013. TIGRFAMs and genome properties in 2013. Nucleic Acids Res 41:D387–D395. doi: 10.1093/nar/gks1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Finn RD, Coggill P, Eberhardt RY, Eddy SR, Mistry J, Mitchell AL, Potter SC, Punta M, Qureshi M, Sangrador-Vegas A, Salazar GA, Tate J, Bateman A. 2016. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res 44:D279–D285. doi: 10.1093/nar/gkv1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K. 2017. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res 44:D353–D361. doi: 10.1093/nar/gkw1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bowers RM, Kyrpides NC, Stepanauskas R, Harmon-Smith M, Doud D, Reddy T, Schulz F, Jarett J, Rivers AR, Eloe-Fadrosh EA, Tringe SG, Ivanova NN, Copeland A, Clum A, Becraft ED, Malmstrom RR, Birren B, Podar M, Bork P, Weinstock GM, Garrity GM, Dodsworth JA, Yooseph S, Sutton G, Glöckner FO, Gilbert JA, Nelson WC, Hallam SJ, Jungbluth SP, Ettema TJG, Tighe S, Konstantinidis KT, Liu W-T, Baker BJ, Rattei T, Eisen JA, Hedlund B, McMahon KD, Fierer N, Knight R, Finn R, Cochrane G, Karsch-Mizrachi I, Tyson GW, Rinke C, Genome Standards Consortium, Lapidus A, Meyer F, Yilmaz P, Parks DH, et al. 2017. Minimum information about a single amplified genome (MISAG) and a metagenome-assembled genome (MIMAG) of bacteria and archaea. Nat Biotechnol 35:725–731. doi: 10.1038/nbt.3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Federhen S. 2012. The NCBI Taxonomy database. Nucleic Acids Res 40:D136–D143. doi: 10.1093/nar/gkr1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chaumeil PA, Mussig AJ, Hugenholtz P, Parks DH. 2019. GTDB-Tk: a toolkit to classify genomes with the Genome Taxonomy Database. Bioinformatics 36:1925–1927. doi: 10.1093/bioinformatics/btz848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Salter SJ, Cox MJ, Turek EM, Calus ST, Cookson WO, Moffatt MF, Turner P, Parkhill J, Loman NJ, Walker AW. 2014. Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol 12:87. doi: 10.1186/s12915-014-0087-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tanner MA, Goebel BM, Dojka MA, Pace NR. 1998. Specific ribosomal DNA sequences from diverse environmental settings correlate with experimental contaminants. Appl Environ Microbiol 64:3110–3113. doi: 10.1128/AEM.64.8.3110-3113.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Onstott TC, Moser DP, Pfiffner SM, Fredrickson JK, Brockman FJ, Phelps TJ, White DC, Peacock A, Balkwill D, Hoover R, Krumholz LR, Borscik M, Kieft TL, Wilson R. 2003. Indigenous and contaminant microbes in ultradeep mines. Environ Microbiol 5:1168–1191. doi: 10.1046/j.1462-2920.2003.00512.x. [DOI] [PubMed] [Google Scholar]
- 23.Kulakov LA, McAlister MB, Ogden KL, Larkin MJ, O’Hanlon JF. 2002. Analysis of bacteria contaminating ultrapure water in industrial systems. Appl Environ Microbiol 68:1548–1555. doi: 10.1128/AEM.68.4.1548-1555.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chan PP, Lowe TM. 2019. tRNAscan-SE: searching for tRNA genes in genomic sequences. Methods Mol Biol 1962:1–14. doi: 10.1007/978-1-4939-9173-0_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nawrocki EP, Eddy SR. 2013. Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics 29:2933–2935. doi: 10.1093/bioinformatics/btt509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kalvari I, Argasinska J, Quinones-Olvera N, Nawrocki EP, Rivas E, Eddy SR, Bateman A, Finn RD, Petrov AI. 2018. Rfam 13.0: shifting to a genome-centric resource for non-coding RNA families. Nucleic Acids Res 46:D335–D342. doi: 10.1093/nar/gkx1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bland C, Ramsey TL, Sabree F, Lowe M, Brown K, Kyrpides NC, Hugenholtz P. 2007. CRISPR recognition tool (CRT): a tool for automatic detection of clustered regularly interspaced palindromic repeats. BMC Bioinformatics 8:209. doi: 10.1186/1471-2105-8-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hyatt D, Chen GL, Locascio PF, Land ML, Larimer FW, Hauser LJ. 2010. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lomsadze A, Gemayel K, Tang S, Borodovsky M. 2018. Modeling leaderless transcription and atypical genes results in more accurate gene prediction in prokaryotes. Genome Res 46:1079–1089. doi: 10.1101/gr.230615.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kielbasa SM, Wan R, Sato K, Horton P, Frith MC. 2011. Adaptive seeds tame genomic sequence comparison. Genome Res 21:487–493. doi: 10.1101/gr.113985.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mistry J, Finn RD, Eddy SR, Bateman A, Punta M. 2013. Challenges in homology search: HMMER3 and convergent evolution of coiled-coil regions. Nucleic Acids Res 41:e121. doi: 10.1093/nar/gkt263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Arndt W. 2016. Modifying HMMER3 to run efficiently on the Cori supercomputer using OpenMP tasking, p 239–246. In Proceedings of the 2018 IEEE International Parallel and Distributed Processing Symposium Workshops (IPDPSW). IEEE, Piscataway, NJ. [Google Scholar]
- 33.Letunic I, Bork P. 2018. 20 years of the SMART protein domain annotation resource. Nucleic Acids Res 46:D493–D496. doi: 10.1093/nar/gkx922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gough J, Karplus K, Hughey R, Chothia C. 2001. Assignment of homology to genome sequences using a library of hidden Markov models that represent all proteins of known structure. J Mol Biol 313:903–919. doi: 10.1006/jmbi.2001.5080. [DOI] [PubMed] [Google Scholar]
- 35.Sillitoe I, Dawson N, Lewis TE, Das S, Lees JG, Ashford P, Tolulope A, Scholes HM, Senatorov I, Bujan A, Ceballos Rodriguez-Conde F, Dowling B, Thornton J, Orengo CA. 2019. CATH: expanding the horizons of structure-based functional annotations for genome sequences. Nucleic Acids Res 47:D280–D284. doi: 10.1093/nar/gky1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kang DD, Froula J, Egan R, Wang Z. 2015. MetaBAT, an efficient tool for accurately reconstructing single genomes from complex microbial communities. PeerJ 3:e1165. doi: 10.7717/peerj.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. 2015. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res 25:1043–1055. doi: 10.1101/gr.186072.114. [DOI] [PMC free article] [PubMed] [Google Scholar]