Abstract
Lignin is one of the major components of xylem cell walls in tree stems. The lignin in the wood of most flowering plants (dicotyledonous angiosperms) is typically polymerized from three monolignol precursors, coniferyl alcohol, sinapyl alcohol, and p-coumaroyl alcohol, resulting in guaiacyl (G), syringyl (S), and hydroxyphenyl (H) subunits, respectively. In this study, we focus on the transcriptional regulation of a coniferaldehyde 5-hydroxylase (CAld5H2) gene, which encodes a key enzyme for sinapyl alcohol biosynthesis. We carried out a yeast one-hybrid (Y1H) screen to identify candidate upstream transcription factors (TFs) regulating CAld5H2. We obtained 12 upstream TFs as potential regulators of CAld5H2. One of these TF genes, BLH6a, encodes a BEL1-like homeodomain (BLH) protein and negatively regulated the CAld5H2 promoter activity. The direct regulation of CAld5H2 promoter by BLH6a was supported by chromatin immunoprecipitation–quantitative polymerase chain reaction (ChIP–qPCR) and dominant repression of BLH6a in transgenic plants. Luciferase complementation imaging analyses showed extensive protein–protein interactions among these 12 TFs. We propose that BLH6a is a negative regulator of CAld5H2, which acts through combinatorial regulation of multiple TFs for sinapyl alcohol (S monolignol) biosynthesis in poplar.
Keywords: lignin, CAld5H2, BEL1-like homeodomain protein, transcription factor, yeast one hybrid
Introduction
Lignin is a polyphenolic polymer deposited in the secondary cell walls (SCWs) of vascular plants (Sarkanen and Ludwig, 1971). Many different phenylpropanoid subunits may be incorporated into lignin, but the wood of trees within the angiosperms typically contains three predominant lignin subunits, hydroxyphenyl (H), guaiacyl (G), and syringyl (S), derived from p-coumaryl, coniferyl, and sinapyl alcohols, also called monolignols. Lignin composition depends on the relative abundance of monolignols, which are polymerized by radical coupling reactions (Ralph et al., 2004). In angiosperm wood, typically in dicots, lignin is composed primarily of G and S subunits, with minor amounts of H subunits. The proportion of G and S subunits in lignin is different between cell types. Vessel elements are specialized for water transport, and fiber cells provide mechanical support. In the wood of poplar, guaiacyl lignin is predominant in vessel cell walls, whereas S subunits are mainly located in fiber cell walls (Zhou et al., 2011). In gymnosperms, the mature xylem contains one major type of cell (tracheids), which carries out both support and transport. Tracheid lignin has mainly G subunits with only minor amounts of H subunits in their cell walls (Sarkanen and Ludwig, 1971).
In angiosperms, 11 enzyme families comprise a branched grid-like pathway that converts phenylalanine to three major monolignols (Boerjan et al., 2003; Vanholme et al., 2013) (Figure 1). These enzyme families are designated PAL (phenylalanine ammonia-lyase, EC 4.3.1.5), C4H (cinnamate 4-hydroxylase, EC 1.14.13.11), C3H (p-coumaroyl-CoA 3-hydroxylase, EC 1.14.14.1), 4CL (p-coumarate CoA ligase, EC 6.2.1.12), HCT (hydroxycinnamoyltransferase, EC 2.3.1.133), CSE (caffeoyl shikimate esterase, EC 3.1.1-), CCoAOMT (caffeoyl-CoA O-methyltransferase, EC 2.1.1.104), CCR (cinnamoyl-CoA reductase, EC 1.2.1.44), F5H/CAld5H [ferulate/coniferaldehyde 5-hydroxylase (CAld5H2), EC 1.14.13], COMT (caffeic acid 3-O-methyltransferase, EC 2.1.1.6), and CAD (cinnamyl alcohol dehydrogenase, EC 1.1.1.95). In this pathway, coniferaldehyde is converted to coniferyl alcohol by CAD for G monolignol production. Alternatively, coniferaldehyde can be converted to sinapyl alcohol in three steps by three consecutive enzymes, CAld5H/F5H, COMT, and CAD. CAld5H/F5H is a key enzyme in directing S lignin production (Osakabe et al., 1999). Overexpression of F5H driven by a C4H promoter caused an increase (29.5%) in sinapyl alcohol level in hybrid poplar (Stewart et al., 2009).
Figure 1.
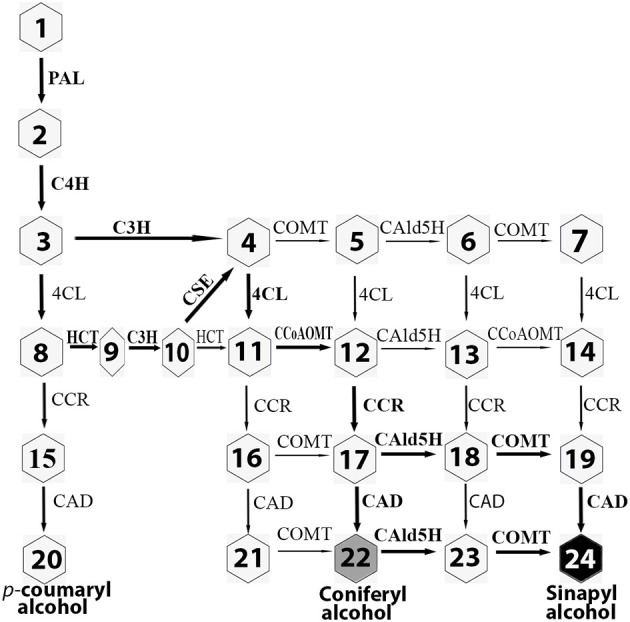
Monolignol biosynthetic pathway in poplar. Bold arrows indicate the main metabolic flux. Eleven enzyme families are PAL, phenylalanine ammonia-lyase; C4H, cinnamate 4-hydroxylase; C3H, p-coumaroyl-CoA 3-hydroxylase; 4CL, p-coumarate CoA ligase; HCT, hydroxycinnamoyltransferase; CSE, caffeoyl shikimate esterase; CCoAOMT, caffeoyl-CoA O-methyltransferase; CCR, cinnamoyl-CoA reductase; CAld5H, coniferaldehyde 5-hydroxylase; COMT, caffeic acid 3-O-methyltransferase; CAD, cinnamyl alcohol dehydrogenase. The metabolites are shown in numbers. 1: phenylalanine; 2: cinnamic acid; 3: 4-hydroxycinnamic acid; 4: caffeic acid; 5: ferulic acid; 6: 5-hydroxyferulic acid; 7: sinapic acid; 8: p-coumaroyl-CoA; 9: p-coumaroyl shikimic acid; 10: caffeoyl shikimic acid; 11: caffeoyl-CoA; 12: feruloyl-CoA; 13: 5-dydroxyferuloyl-CoA; 14: sinapoyl-CoA; 15: p-coumaraldehyde; 16: caffeyl aldehyde; 17: coniferaldehyde; 18: 5-hydroxyconiferaldehyde; 19: sinapaldehyde; 20: p-coumaryl alcohol (H monolignol); 21: caffeyl alcohol; 22: coniferyl alcohol (G monolignol); 23:5-hydroxyconiferyl alcohol; 24: sinapyl alcohol (S monolignol).
Many transcription factors (TFs) in the transcriptional network regulating the SCW synthesis have been characterized (Ko et al., 2014). In the top layer of the gene regulatory network (GRN), NAC [no apical meristem (NAM), arabidopsis transcription activator factor (ATAF1/2), and cup-shaped cotyledon (CUC)] and v-myb avian myeloblastosis viral oncogene homolog (MYB) members, such as secondary wall-associated NAC domain protein1/vascular-related NAC-domain6 (SND1/VND6) (Minoru et al., 2005; Zhong et al., 2006) and MYB46/83 (Zhong et al., 2007; McCarthy et al., 2009; Zhong and Ye, 2012), are master regulators activating SCW synthesis. Most TFs in the GRN are activators and a few are repressors, indicating fine-tuning of the transcriptional control in SCW biosynthesis (Ko et al., 2014). Some TFs in the network directly regulate genes encoding pathway enzymes for cellulose, hemicellulose, and lignin biosynthesis (Ohashiito et al., 2010; Kim et al., 2013). The TFs that directly regulate lignin biosynthesis could act to differentially activate or repress lignin pathway genes. For example, MYB46 directly regulates nine of the 10 Arabidopsis lignin pathway genes characterized (Ko et al., 2014), whereas MYB58 directly activates all 10 of the Arabidopsis lignin pathway genes (Zhou et al., 2009). In Medicago, SND1 directly activates F5H, but the SND1 in Arabidopsis does not (Zhong et al., 2008; Zhao et al., 2010), indicating that the GRN for SCW synthesis may not be the same in different plant species.
Wood provides large quantities of material for the production of pulp, paper, timber, and lignocellulosic biofuels. For pulp, paper, and chemical feedstock production, lignin is a major barrier to such applications and must be removed. The content, composition, and structure of lignin affect the process efficiency of woody biomass (Studer et al., 2011; Li et al., 2014). Lignin composition, calculated as the S/G ratio, is an important factor affecting the pulp yield than the lignin content (Río et al., 2005; Studer et al., 2011). High pulp yield is correlated with a high S/G ratio (Río et al., 2005). Hardwood is generally delignified more readily than softwood due to the abundance of S subunits in the hardwood lignin (Sarkanen and Ludwig, 1971; Chang and Sarkanen, 1973). Wood with a higher proportion of S subunits is preferred for paper/pulping and biofuel production (Wagner et al., 2015).
The favorable properties of lignin with a higher S subunit content have motivated efforts to increase the S lignin content in wood through genetic modification. Overexpression of CAld5H had a 64% increase in the S/G ratio in poplar (Li et al., 2003). Lignin polymers that contain S units have been generated in conifer cells by overexpression of F5H in Pinus radiata tracheary element cultures. Co-transformation of F5H and COMT resulted in a two to three times higher S/G ratio than F5H alone (Wagner et al., 2015).
To learn more about the GRN that determines SCW biosynthesis, we identified and characterized TFs regulating sinapyl alcohol (monolignol) biosynthesis. We conducted a yeast one hybridization (Y1H) screening of a library of TFs from Populus trichocarpa, using the CAld5H2 promoter as bait, and identified 12 candidates as potential targets. Among the 12 TFs, two BEL1-like homeodomain (BLH) 6 proteins, BLH6a and BLH6b, bound CAld5H2 promoter specifically.
We characterized the functions of BLH6a. Transient overexpression and dominant repression of BLH6a regulation inhibited CAld5H2 expression, and extensive protein–protein interactions were detected among the 12 TFs. Our results suggest that a complex GRN with combinatorial and redundant elements may control the CAld5H gene expression for S lignin biosynthesis.
Materials and Methods
Plant Materials
Populus alba × Populus glandulosa and Nicotiana benthamiana plants were grown in a room with long-day conditions (16-h light/8-h dark) at 25 ± 1°C. The P. alba × P. glandulosa sterile plants used for transformation were propagated by microcuttings in bottles and cultured on 0.5 × the Murashige and Skoog (MS) medium.
Yeast One-Hybrid
Total RNAs were extracted from differentiating xylem of 6-month-old P. trichocarpa trees using an RNeasy Plant Mini Kit (QIAGEN Inc., Valencia, CA, USA) and reverse-transcribed to complementary DNAs (cDNAs) by using the PrimeScript™ RT Reagent Kit (TaKaRa, Dalian, China). The cDNAs were used as templates to amplify the coding region of 213 TFs (Supplementary Data Sheet 1) with the Phanta Max Super-Fidelity DNA Polymerase (Vazyme Biotech Co., Ltd., Nanjing, Jiangsu, China). About 202 PCR fragments were digested with corresponding restriction enzymes and cloned into the pGADT7 vectors by T4 DNA ligases. About 11 PCR fragments were cloned into the pGADT7 vector through the recombination method by using the ClonExpressII One Step Cloning Kit (Vazyme Biotech Co., Ltd., Nanjing, Jiangsu, China). The full lengths of coding regions cloned in pGADT7 were sequence confirmed. The coding regions of 14 TFs were synthesized and cloned into pGADT7. Total 227 TF-prey plasmids were constructed.
Genomic DNA was extracted from young leaves using a DNeasy Plant Mini Kit (QIAGEN Inc., Valencia, CA, USA). Gene promoter regions were amplified from genomic DNA and cloned into the pABAi vector by T4 DNA ligases. The resultant DNA-bait plasmid was linearized by BstbI and integrated into the Y1HGold Strain (Clontech Laboratories, Inc., Mountain View, CA, USA). Four transformants were tested on the SD/-ura medium with the addition of Aureobasidin A (AbA) ranging from 100 to 500 μg/ml, and the DNA-bait strain with optimal AbA concentration was selected for Y1H.
The 227 TF-prey plasmids were individually transformed into the DNA-bait strain, and the transformants were selected on the SD/-ura-leu medium. The positive strains were diluted with 0.9% NaCl and screened on the SD/-ura-leu+AbA medium for identifying the TF–DNA interaction.
Effector-Reporter-Based Transactivation/Repression Assays
The 2-kb CAld5H2 promoter was amplified from P. alba × P. glandulosa genomic DNA and cloned into pBG3-LZ004-LUC (luciferase) (Chen et al., 2017) by using the ClonExpressII One Step Cloning Kit (Vazyme Biotech Co., Ltd., Nanjing, Jiangsu, China), generating the reporter construct pBG3-pCAld5H2:LUC. The full-length coding regions of BLH6a, BLH6b, and BLH2 were amplified from P. alba × P. glandulosa xylem cDNA and cloned into the pENTR/D TOPO Vector (Invitrogen Co., Carlsbad, CA, USA). After being verified by sequencing, the genes were Gateway cloned into the pGWB17 Vector by LR Clonase II (Invitrogen), generating the effector constructs pBWB17-35S:BLH6a, pBWB17-35S:BLH6b, and pBWB17-35S:BLH2. The resultant reporter constructs and effector constructs were transformed into Agrobacterium GV3101. Agrobacterium containing an effector construct and a reporter construct were co-injected into the tobacco leaves. After 48 h, the LUC luminescence was detected under a Promega GloMax®-20/20 Luminometer (Promega Corp., Madison, WI, USA). The primers used for the promoter and gene amplification are listed in Supplementary Table 1.
Transcriptional Activation/Inhibition Assays in Yeast
The full-length coding region of BLH6a was amplified from P. alba × P. glandulosa xylem cDNA with the Phanta Max Super-Fidelity DNA Polymerase (Vazyme Biotech Co., Ltd., Nanjing, Jiangsu, China) and cloned into pGBKT7 and pGBKT7-VP16 vector using the ClonExpressII One Step Cloning Kit (Vazyme Biotech Co., Ltd., Nanjing, Jiangsu, China), generating BD-BLH6 and BD-VP16:BLH6a. The constructs were transformed into the yeast strain, yeast two hybridization, Y2H Gold (Clontech Laboratories, Inc., Mountain View, CA, USA). Transformants were grown on the SD/-Trp medium for the selection of positive clones and then transferred to the SD/-Trp-His-Ade medium for the transcriptional activation/inhibition assays with X-α-gal used as an indicator.
Chromatin Immunoprecipitation–Quantitative PCR Assay
The fragment of 3× FLAG was amplified from the pCM1307-N-Flag-HA vector (Zhou et al., 2012), cloned into the pENTR/D TOPO Vector (Invitrogen), and LR ligated into pUC19-35S-RfA-35S-sGFP (Li et al., 2012) to generate pUC19-35S-FLAG-35S-sGFP. BLH6a coding sequences were amplified from P. alba × P. glandulosa xylem cDNA and cloned into pUC19-35S-FLAG-35S-sGFP, generating pUC19-35S-Flag:BLH6a-35S-sGFP. Primers are listed in Supplementary Table 1. Plasmid DNA for the protoplast transfection was prepared using the EndoFree Plasmid Kit (QIAGEN Inc., Valencia, CA, USA). Leaf protoplasts were prepared from mesophyII of leaves in tissue culture bottles following the published protocol (Lee et al., 2017), and xylem protoplasts were prepared as described previously (Lin et al., 2014). Plasmids were transferred into protoplasts, and each chromatin immunoprecipitation (ChIP) assay was performed using 5 × 106 protoplasts with anti-FLAG antibodies as described before (Yan et al., 2019). One-fiftieth of the supernatants before adding antibodies were used as input in the qPCR. qPCR was conducted using the Green Premix Ex Tag II (TaKaRa, Dalian, China) and detected by the Roche LightCycler 480 II (Roche Co., Basel, Switzerland), with 18S as the reference.
RNA in-situ Hybridization
The eighth and nineth internodes of 3-month-old P. alba × P. glandulosa stems were fixed in formalin/acetic acid/alcohol (FAA) and embedded in paraplast. The 8 μm microsections were prepared for the in situ hybridization. PCR fragments of ~250 bp, amplified with gene-specific primers (Supplementary Table 1), were used as templates to synthesize antisense and sense probes with T7 RNA polymerases using the Digoxigenin (DIG) RNA Labeling Kit (Roche). The hybridization and immunological detection were performed as previously described (Liu et al., 2018). The hybridization solution contains 10× in situ salts, deionized formamide, 50% dextran sulfate, 50× Denhardt's solution, and 100 ng/ml tRNA. Signals were detected using anti-DIG antibodies conjugated with alkaline phosphatase, and photographs were taken under an OLYMPUS BX51 Microscope (Olympus Corp., Tokyo, Japan).
Yeast Two Hybridization
The full-length coding regions of BLH6b and BLH2 were amplified from P. alba × P. glandulosa xylem cDNA with the Phanta Max Super-Fidelity DNA Polymerase (Vazyme Biotech Co., Ltd., Nanjing, Jiangsu, China) and cloned into the pGADT7 vector by using the ClonExpressII One Step Cloning Kit (Vazyme Biotech Co., Ltd., Nanjing, Jiangsu, China), generating AD fusion vectors. The full-length coding region of BLH6a was cloned into the pGBKT7 vector, generating the BD-BLH6a vector. The BD-BLH6a and AD fusion vectors were co-transformed into the yeast strain Saccharomyces cerevisiae Y2HGold as described in the Yeast Protocols Handbook (Clontech Laboratories, Inc., Mountain View, CA, USA). Transformants were grown on the SD/-Leu-Trp medium and then screened on the SD/-Ade-His-Leu-Trp medium.
Luciferase Complementation Imaging Assays
The coding region of 12 TFs was amplified from P. alba × P. glandulosa xylem cDNA, and, by using the ClonExpressII One Step Cloning Kit (Vazyme Biotech Co., Ltd., Nanjing, Jiangsu, China), they were cloned into pCAMBIA1300-cLUC and pCAMBIA1300-nLUC, which were pre-linearized by KpnI/SalI (Song et al., 2011), generating TF-nLUC and TF-cLUC. Primers used for the vector construction are shown in Supplementary Table 1. After confirmation by sequencing, the vectors were transformed into Agrobacterium GV3101. Equal concentrations and volumes of Agrobacterium cultures were mixed and co-infiltrated into the N. benthamiana leaves. After incubation for 36 h, the LUC fluorescence was detected under a Berthold NightSHADE LB985 in vivo Plant Imaging System (Berthold Technologies GmbH, Bad Wildbad, Germany).
Vector Construction and Transgenic Production
Two oligos EAR-motif repression domain (SRDX)-F and SRDX-R (Supplementary Table 1) were synthesized. After denature and annealing, the double-strand DNA fragment was ligated into pBI121 at BamHI/SacI, generating pBI121-35S-SRDX. This vector contains a short sequence between 35S promoter and stops codon, encoding a 12 amino acid (LDLDLELRLGFA) plant-specific SRDX (Mitsuda et al., 2011).
The full-length coding region of BLH6a was amplified by 121BLH6a-F/R from P. alba × P. glandulosa xylem cDNA and inserted into pBI121-35S-SRDX using the ClonExpressII One Step Cloning Kit (Vazyme Biotech Co., Ltd., Nanjing, Jiangsu, China), generating pBI121-35S-BLH6a:SRDX. The resultantly plasmid was transformed into Agrobacterium GV3101, and transformation in P. alba × P. glandulosa was conducted by the leaf disk method. The leaves were cut at vein area, immersed in Agrobacterium culture for 15 min. After incubated on the co-cultivation medium (1× MS, pH 5.7–5.9, 5 mg/L 6BA, 0.05 mg/L NAA) in dark for 4 days, the explants were transferred to the selection medium (1× MS, pH 5.7–5.9, 0.5 mg/L 6BA, 0.05 mg/L NAA, 200 mg/L timentin, 50 mg/L kanamycin) under light with the medium substitution every 2 weeks. The generated shoots were cut and transferred to the rooting medium (0.5 × MS, pH 5.7–5.9, 0.2 mg/L NAA, 0.05 mg/L IBA, 200 mg/L timentin, 50 mg/L kanamycin) for the root induction.
Quantitative Reverse Transcription-PCR
Total RNAs were extracted from differentiating xylem of 6-month-old trees using the RNeasy Plant Mini Kit (QIAGEN Inc., Valencia, CA, USA). The total RNAs were reverse transcribed to cDNA using a PrimeScript™ RT Reagent Kit with gDNA Eraser (TaKaRa, Dalian, China). The PCR was conducted using the Green Premix Ex Tag II (TaKaRa, Dalian, China) and detected by the Roche LightCycler 480 II. Actin was used as the reference.
Stem Sectioning and Light Microscopy
The stem fragments, close to the bottom node, were collected from the seventh to 14th internodes, immobilized with glue (LOCTITE 495), and sectioned by an oscillating Leica VT 1000 S Microtome (Leica Microsystems, Wetzlar, Germany). The section thickness was 40 μm. The sections were stained in 0.05% (w/v) toluidine blue (TBO) dye for 1 min and observed under an OLYMPUS BX51 Microscope (Olympus Corp., Tokyo, Japan).
Lignin Content and Composition
The stems below the 20th internode were collected from 9-month-old trees, and bark was removed. Dried wood was ground, lyophilized, and extracted with chloroform/methanol (2:1, v/v). The lignin content was determined by the Klason method as described before (Lu et al., 2013). Lignin compositions were determined by the thioacidolysis method (Lapierre et al., 1995).
Results
Identification of 12 TF Candidates That Bound the P. trichocarpa cAld5H2 Promoter
In P. trichocarpa, two closely related genes CAld5H1 (Potri.005G117500) and CAld5H2 (Potri.007G016400) are key genes in S monolignol biosynthesis with redundant functions (Wang et al., 2012). These enzymes comprise a key branch point in monolignol biosynthesis because they regulate the relative abundance of substrate that becomes coniferyl alcohol as opposed to 5-hydroxyconiferyl alcohol. These enzymes act on coniferyl alcohol directly or they may act on the aldehyde intermediates converting coniferylaldehyde to 5-hydroxyconiferaldehyde. The synthesis of 5-hydroxy intermediates determines the amount of sinapylaldehyde and sinapyl alcohol that is formed and therefore the relative abundance of sinapyl and coniferyl alcohol. The ratio of these alcohols is a major determinant of the ratio of S and G subunits polymerized into lignin (Osakabe et al., 1999; Wang et al., 2014, 2018).
We conducted a Y1H screening to identify TFs that could directly regulate CAld5H genes. Using our previous RNA-seq data (Shi et al., 2017) from xylem, phloem, leaves, and shoot tips from P. trichocarpa, we obtained 227 TFs differentially expressed in xylem compared to three other tissues [fold change >1, and false discovery rate (FDR) <0.05] (Supplementary Data Sheet 1). We generated a TF-prey library by cloning 227 TFs into the yeast expression vector pGADT7. We isolated the 2 kb promoter of CAld5H2 from P. trichocarpa genomic DNA and used it as bait to screen the TF-prey library. The 227 TF-prey plasmids were individually transformed into the bait-strain and selected on the SD/-leu-ura medium containing AbA, we obtained 57 TF containing clones that could grow on the selective medium.
To eliminate the false positives from the 57 TF candidates, we performed co-expression analysis of 57 TFs with CAld5H genes using the AspWood web resource (http://aspwood.popgenie.org). AspWood provides a high-spatial-resolution gene expression profile of 25–28 stem cryosections in P. tremula, from phloem to cambium and then to wood-forming tissues, enabling the co-expression network analysis (Sundell et al., 2017). Of the 57 TF genes that passed the screen, 12 were co-expressed with both CAld5H2 and CAld5H1 transcripts (Supplementary Figure 1, Supplementary Table 2). Their expression levels were low in the cambium (sections 5–9). Starting from section 9 in the wood-forming tissue, their expression levels gradually increased and were maintained at a higher level (Supplementary Figure 2), indicating their roles in xylem cell differentiation and SCW biosynthesis during wood formation. We chose these 12 TFs for further characterization.
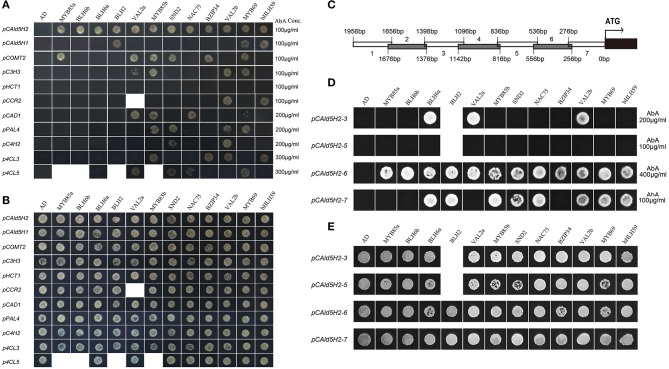
To identify how many TFs among these 12 could interact with the CAld5H2 promoter among the monolignol pathway gene promoters, we conducted Y1H assays to compare these 12 TFs with other monolignol pathway gene promoters isolated from P. trichocarpa, including CAD1, C3H3, COMT2, CCR2, PAL4, HCT1, 4CL3, 4CL5, and C4H2. Among these 12 TFs, only the homologs of BLH6a (Potri.004G159300) and BLH6b (Potri.009G120800) bound CAld5H2 promoter specifically. BLH2 (Potri.005G129500) bound both CAld5H2 and CAld5H1 promoters, but not other promoters (Figures 2A,B). This three TFs belong to the BLH family, indicating that the BLH protein family may be more specific in binding to CAld5H promoters. The other nine TFs recognized promoters of other genes in the monolignol pathway (Figures 2A,B), suggesting that they regulate other steps in monolignol biosynthesis.
Figure 2.
Yeast one hybridization. (A,B) Yeast one-hybrid (Y1H) of 12 transcription factors (TFs) with 11 monolignol pathway genes in Populus trichocarpa. TF-prey plasmids were transformed into bait strains and grown on SD/-ura-leu. We did not obtain transformants for VAL2a transformed into CCR2 promoter strain, and for MYB85a, BLH6b, BLH2, and MYB85b transformed into 4CL5 promoter strain (B). The transformants were selected on SD/-ura-leu+AbA (A). The concentrations of Aureobasidin A (AbA) are shown on right. (C–E) Y1H of 12 TFs with seven fragments of CAld5H2 promoter in P. trichocarpa. (C) The 2 kb P. trichocarpa CAld5H2 promoter was divided into seven fragments. Each fragment is about 300 bp long and two adjacent fragments have a 20–46 bp overlap. (D) Y1H assays of 12 TFs with fragments (3, 5, 6, and 7) of CAld5H2 promoters. The self-activation of fragments 1, 2, and 4 are too higher (1,000 μg/ml) to be removed. The transformants were grown on SD/-ura-leu+AbA for selection. The AbA concentrations were shown on the right. (E) The transformants were grown on SD/-ura-leu.
All 12 Selected TFs Bound the Fragments 6 and 7 of the CAld5H2 Promoter
To identify the specific binding regions of these 12 TFs in the CAld5H2 promoter in P. trichocarpa, we divided the 2 kb promoter into seven fragments (Figure 2C). The Y1H assays were conducted to examine the interactions of 12 TFs with the seven fragments. The self-activation in the bait strains containing fragments 1, 2, and 4 was very strong, and therefore these fragments were removed in the study. The Y1H assays showed that all 12 TFs could bind fragment 6 of CAld5H2 promoter, and eight TFs could bind fragment 7. Fragment 3 of the promoter was recognized by BLH6a, VAL2a, and VAL2b. None of the TFs bound to fragment 5 (Figure 2D).
Fragments 6 and 7 are major binding sites for all 12 TFs. We used PlantPAN 3.0 (http://plantpan.itps.ncku.edu.tw/index.html) to characterize the specificity of cis-acting regulatory DNA elements and to identify possible binding to TF families. Besides the CAAT box and TATA box, multiple cis-acting regulatory elements were found, including WBOXATNPR1, MYBCOREATCYCB1, DOFCOREZM, and SURECOREATSULTR11 (Supplementary Data Sheet 2). The possible TF families that could bind to these motifs include AT-HOOK, Dof, ATA, ALE, WOX, MYB, bZIP, bHLH, NAC, HD-ZIP, and ZF-H (Supplementary Data Sheet 2). Fragments 6 and 7 contain motifs for recognition by the families of 12 TFs, such as the “TGAC” element in motif WBOXATNPR1 for BLH proteins, and the “CANNTG” element in YCCONSENSUSAT motif for bHLH and bZIP TFs (Murre et al., 1989; Ledent and Vervoort, 2001; Kondhare et al., 2019). This analysis supports the binding of 12 TFs to the CAld5H2 promoter.
P. alba × P. glandulosa BLH6a Negatively Regulated CAld5H2 Promoter Activity
Among the three potential CAld5H2-specific upstream regulators, BLH2, also named WBLH2 (Chen et al., 2019), has been implicated as an upstream regulator and was shown to bind the CAld5H1 promoter by ChIP–PCR in P. trichocarpa (Chen et al., 2019). Although BLH6a (also named WBLH3 (Chen et al., 2019) has been implicated as a target of PtrMYB021, the regulatory function of these two BLH6 members on the monolignol pathway genes was not studied before.
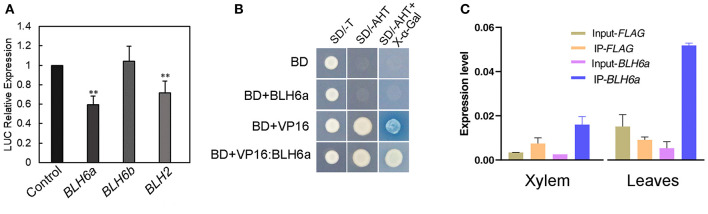
To investigate whether the two BLH6 homologs regulate CAld5H2 in vivo, we conducted effector-reporter-based transactivation/repression assays. The reporter constructs, carrying a LUC luciferase gene under the control of the P. alba × P. glandulosa CAld5H2 promoter and was co-expressed transiently in N. benthamiana leaves with effector construct harboring either BLH6a or BLH6b from the P. alba × P. glandulosa hybrid. BLH6a repressed reporter activity, but BLH6b did not (Figure 3A). BLH2 also inhibited CAld5H2 promoter activity (Figure 3A). We further used the yeast system to detect the activation/repression activity of BLH6a. Yeast with BLH6a overexpression could not grow on the medium lacking histidine (Figure 3B), showing that BLH6a could not activate HIS3 expression. Compared to the VP16 alone, BLH16a:VP16 fusion disturbed the activation of an α-galactosidase gene (Figure 3B), indicating that BLH6a is a transcriptional repressor, not an activator.
Figure 3.
BLH6a is a transcriptional repressor of CAld5H2 in P. alba × P. glandulosa. (A) Effector-reporter-based gene activation/repression assay. LUC gene was driven by the P. alba × P. glandulosa CAld5H2 promoter in the reporter construct. Each TF gene from P. alba × P. glandulosa was driven by the 35S promoter in the effector construct. Effect construct and reporter construct were co-infiltrated in the tobacco leaves by Agrobacterium for the LUC activity determination. The reporter construct alone was the control. (B) Transactivation/repressor activity detections in yeast strain Y2HGold. SD/-T, single dropout medium lacking tryptophan; SD/-AHT, triple dropout medium lacking adenine, histidine, and tryptophan; BD, GAL4-binding domain. (C) Chromatin immunoprecipitation (ChIP) assays in xylem (left) and leaf (right) protoplasts. ChIP was conducted in protoplasts overexpressing FLAG and FLAG:BLH6a, respectively, and followed by qPCR. Input-FLAG and Input-BLH6a were IP using one-fiftieth of the supernatants before adding antibodies in the FLAG and FLAG:BLH6a overexpression protoplasts, respectively. IP-FLAG and IP-BLH6a were IP with FLAG antibodies in FLAG and FLAG:BLH6a overexpression protoplasts, respectively. Error bars represent SD (n = 3). **p < 0.01, determined by Student's t test.
We further used ChIP assays in both xylem and leaf protoplasts to examine the interaction of BLH6a and CAld5H2 promoters. In comparison with a negative control of overexpression of FLAG alone, the CAld5H2 promoter region was significantly enriched in the immunoprecipitation in both xylem and leaf protoplasts overexpressing FLAG:BLH6a fusion (Figure 3C), confirming the direct binding of BLH6a to CAld5H2 promoter.
BLH6a Was Co-expressed With CAld5H Genes in P. alba × P. glandulosa Differentiating Xylem
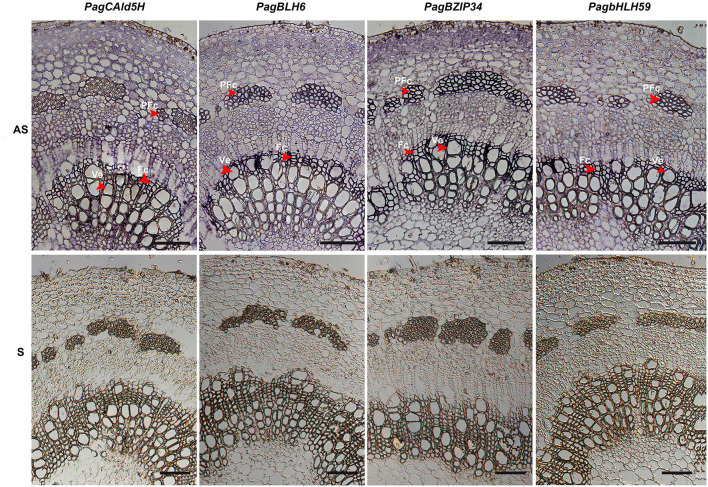
The proposed interaction of BHL6a with the CAld5H2 promoter requires that both genes are expressed in the same cells. We used RNA in situ hybridization (RISH) to examine the co-expression of BLH6a with CAld5H genes in cells of differentiating xylem. BLH6a and BLH6b are a paralogous gene pair, with 90% nucleotide identity; thus, it is difficult to distinguish transcripts of the two genes using an RNA probe. Similarly, RNA probes for CAld5H RISH could not distinguish CAld5H1 and CAld5H2. The RISH detected CAld5H1/2 transcripts in cross-sections of differentiating xylem, including vessels and fiber cells (Figure 4), consistent with the function of CAld5H in monolignol biosynthesis during the SCW biosynthesis. BLH6a/b transcripts were also detected in both vessels and fiber cells, in the early stages of the xylem differentiation (Figure 4). We also selected two TFs (bZIP34 and bHLH59), which did not specifically bind the CAldH2 promoter, for RISH. These two genes showed the same expression pattern as CAld5H1/2 (Figure 4). The RISH showed that bZIP34 and bHLH59 were co-expressed with CAld5H1/2 in all cell types of differentiating xylem, and BLH6a/b were co-expressed with CAld5H1/2 in the early stage of the xylem cell differentiation.
Figure 4.
RNA in situ localization of CAld5H, BLH6, BZIP34, and bHLH59 in the 8–9 internode stems of P. alba × P. glandulosa stems. Bars = 200 μm. Arrows show the signals of the vessels (Ve), fiber cells (Fc), and phloem fiber cell (PFc) in the differentiating xylem where signals of RNA in situ hybridization were intensified. AS and S represent the sections hybridized with antisense and sense probes, respectively.
Protein–Protein Interactions Among 12 TF Candidates
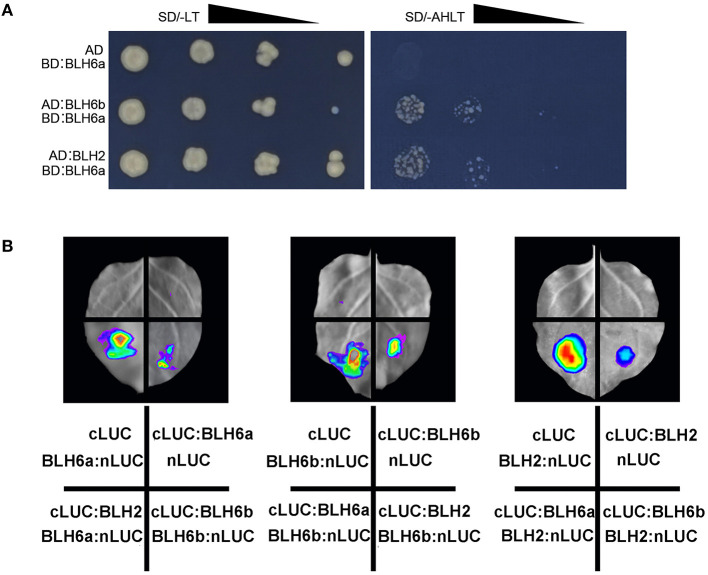
All the identified 12 TFs could bind the 556 bp promoter region (fragments 6–7) of CAld5H2. It would be interesting to know how they cooperatively regulate CAld5H2. We first examined the interactions of BLH6a with other two CAld5H2-specific TFs BLH6b and BLH2 by Y2H, and the assays showed that BLH6a could form a dimer with BLH6b and BLH2 (Figure 5A). We further conducted firefly LCI assays for the three BLH members in the N. benthamiana leaves (Figure 5B). A strong LUC signal was detected when BLH6a-nLUC and cLUC-BLH2 were co-infiltrated, and reciprocal co-infiltration of BLH2-nLUC and cLUC-BLH6a also had strong luminescence. Similarly, when each pair of BLH6a-nLUC/cLUC-BLH6b, BLH6b-nLUC/-cLUC-BLH6a, BLH6b-nLUC/cLUC-BLH2, and BLH2-nLUC/cLUC-BLH6b was co-infiltrated into tobacco leaves, the LUC luminescence was observed. However, no LUC luminescence was observed in negative controls (cLUC/BLH6a-nLUC, cLUC-BLH6a/nLUC, cLUC/BLH6b-nLUC, cLUC-BLH6b/nLUC, cLUC/BLH2-nLUC, and cLUC-BLH2/nLUC). The LCI assays for the interaction among BLH6a, BLH6b, and bHLH2 were repeated four times. All had positive luminescence, showing that these three proteins could interact with each other.
Figure 5.
Protein–protein interactions of BLH6a, BLH6b, and BLH2. (A) Yeast two hybridizations. BLH6a was fused with Gal4-binding domain (BD). BLH6b and BLH2 were fused with activation domain (AD). The resulting AD constructs (AD:BLH6b, AD:BLH2) and BD construct (BD:BLH6a) were co-transfected into a yeast cell, grown on the SD/-Leu-Trp medium (SD/-LT), and selected on the SD/-Ade-His-Leu-Trp medium (SD/-AHLT). (B) Luciferase complementation imaging (LCI) assays for the detection of the interactions between two proteins of BLH6a, BLH6b, and BLH, with one of them being fused to the N-terminal portion of LUC (nLUC), and the other being fused to the C-terminal portion of LUC (cLUC).
We further used the LCI assays to examine the remaining possible interactions between any two TFs among 12 TFs. For each two TFs, we examined their interactions reciprocally (i.e., TF1:nLUC/TF2:cLUC and TF2:nLUC/TF1:cLUC), and each assay was conducted twice. Theoretically, 66 interactions could exist among 12 TFs for the heterodimer formation. Our LCI assay identified 58 interactions (Supplementary Figure 3, Supplementary Table 3). Among 58 interactions, 52 were detected for both combinations (TF1:nLUC/TF2:cLUC and TF2:nLUC/TF1:cLUC), and six interactions were detected for one combination (either TF1:nLUC/TF2:cLUC or TF2:nLUC/TF1:cLUC). Except for MYB85b, all other 11 TFs could form homodimers (Supplementary Figure 3).
Dominant Repression by BLH6a in P. alba × P. glandulosa Inhibited CAld5H Gene Expression and Affected SCW Biosynthesis in Differentiating Xylem
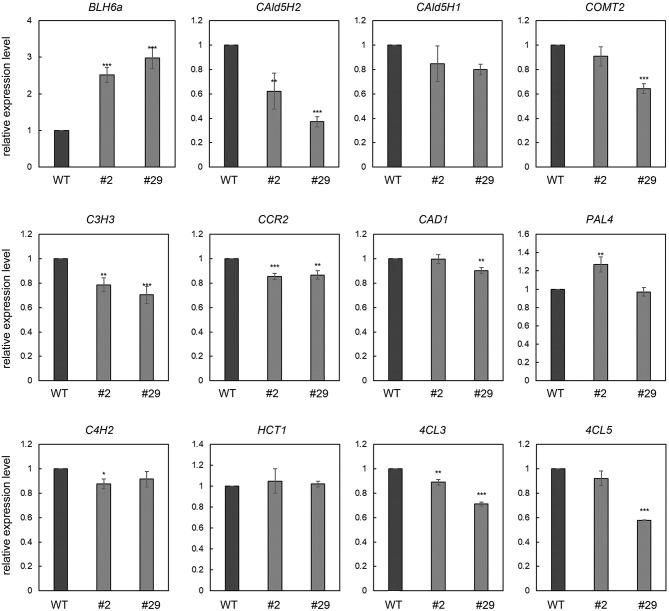
Arabidopsis BLH6 was identified as a transcriptional repressor of REVOLUTA during the SCW formation (Liu et al., 2014). Our results showed that BLH6a is a transcriptional repressor of CAld5H2 in poplar. We identified 12 upstream TFs of CAld5H2, indicating a possibility of redundancy of these TFs. To avoid the redundancy, we exploited the Chimeric REpressor gene Silencing Technology (CRES-T) (Mitsuda et al., 2011) to investigate the regulation of BLH6a on CAld5H2 in vivo. We obtained 36 transgenic P. alba × P. glandulosa lines for overexpression of BLH6a:SRDX, which encoded a chimeric repressor generated by the fusion of the BLH6a protein with a plant-specific SRDX. All transgenic lines grew as normal as wild type (WT). After the quantitative reverse transcription (qRT)-PCR analysis of transgene (BLH6a:SRDX) in the xylem of 36 transgenic lines (Supplementary Figure 4), we selected two lines (#2 and #29) to measure the expression levels of CAld5H2 and CAld5H1, and nine other genes encoding the enzymes of monolignol pathway, including PAL4, CAD1, COMT2, C3H3, C4H2, CCR2, 4CL3, 4CL5, and HCT1. Compared to WT, CAld5H2 transcript levels were significantly reduced in both lines 2 and 29. The reduction in transcript abundance of CAld5H2 in line 29 (62.9%) is greater than line 2 (37.7%), in accordance with the higher transgene expression level in line 29 (Figure 6). The transcript abundance of CAld5H1 in lines 2 and 29 was not changed significantly. Transcript reduction was also observed for COMT2, CAD1, C3H3, CCR2, 4CL3, and 4CL5, but the extent of reduction is much less than that of CAld5H2 (Figure 7). The qRT-PCR results indicate that BLH6a may interact more specifically with the CAld5H2 promoter, consistent with the results of Y1H assays (Figure 2).
Figure 6.
qRT-PCR analysis of monolignol pathway genes in the differentiating xylem of BLH6a:SRDX overexpression transgenic P. alba × P. glandulosa. The transcript abundance in wild type (WT) and transgenic lines 2 and 29 was determined for transgene BLH6a:SRDX and monolignol pathway genes CAld5H2, CAld5H1, COMT2, CAD1, C3H3, CCR2, C4H2, 4CL3, 4CL5, PAL4, and HCT1. Asterisks highlight significant differences by the Student's t-test: *p < 0.5, **p < 0.01; ***p < 0.001.
Figure 7.
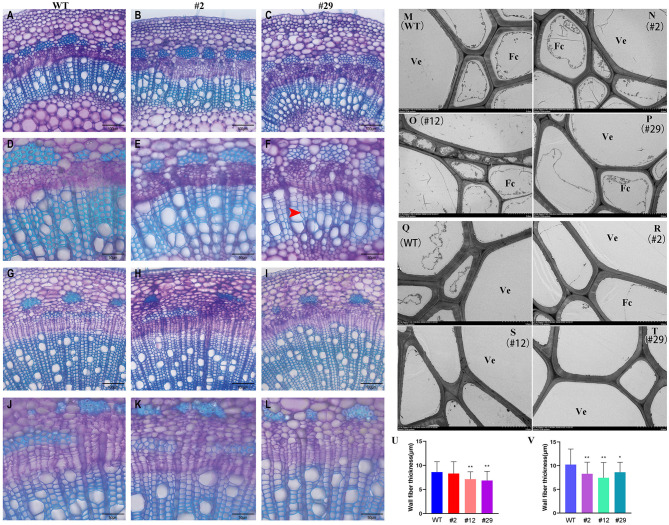
Comparison of xylem structure between P. alba × P. glandulosa BLH6a dominant repression transgenics and WT. (A–L) Light microscopy. (A–C) Cross sections of the seventh internode from WT (A), transgenic lines 2 (B), and 29 (C) at 200 magnification. (D–F) Cross sections of the seventh internode from WT (D), transgenic lines 2 (E), and 29 (F) at 400 magnification. (G–I) Cross sections of the 14th internode from WT (G), transgenic lines 2 (H), and 29 (I) at 200 magnification. (J–L) Cross sections of the 14th internode from WT (J), transgenic lines 2 (K), and 29 (L) at ×400 magnification. Bars = 100 μm in (A–C, G–I) and 50 μm in (D–F, J–L). The red arrow shows the thinner cell wall. (M–T) Transmission electron microscopy. (M–P) Cross-sections of the eighth internode from WT (M), transgenic lines 2 (N), 12 (O), and 29 (P). (Q–T) Cross-sections of the 14th internode from WT (Q), transgenic lines 2 (R), 12 (S), and 29 (T). Bars = 5 μm. Ve, Vessel; Fc, Fiber cell. (U,V) Statistics of fiber cell wall thickness in the eighth internode (U) and the 14th internode (V) of WT, transgenic lines 2, 12, and 29. *p < 0.05, **p < 0.01, determined by the Student's t-test.
To study the effects of dominant repression of BLH6a regulation on the wood formation, we used lines 2 and 29 to examine the cell wall structure at the seventh and the 14th stem internodes using light microscopy. Line 29 had a higher level of BLH6a:SRDX overexpression than line 2. Accordingly, it was observed that the cell wall thickness of vessels and fiber cells in both the seventh and the 14th internodes of line 29 was decreased (Figures 7A–L). We further used transmission electron miscopy to compare the xylem cell wall thickness between transgenic lines (#2, #12, and #29) and WT. Transgenic line 12 had a similar expression level of BLH6a:SRDX as line 29 (Supplementary Figure 4). The measurement of 50 fiber cells showed that the cell wall thickness was significant decreased in both the eighth and the 14th internodes of lines 12 and 29 (Figures 7M–V), indicating that the overexpression of BLH6a:SRDX affected the SCW formation in the differentiating xylem.
To investigate effects on lignin biosynthesis by the BLH6a:SRDX overexpression, we determined lignin content and composition in transgenic lines 2 and 29 (Table 1). We only observed a slight and significant decrease of cellulose content in line 29. Compared to WT, the contents of lignin and the three monomers were not changed significantly, showing that lignin biosynthesis is not affected significantly in the transgenic wood.
Table 1.
Lignin content and compositions in the BLH6a:SRDX overexpression transgenics.
| Line | Cellulose content | Hemicellulose content | Acid-insoluable lignin (%CWR) | Acid-soluable lignin (%CWR) |
H
(μmol/g CWR) |
G
(μmol/g CWR) |
S
(μmol/g CWR) |
|---|---|---|---|---|---|---|---|
| WT | 48.53 ± 0.37 | 16.67 ± 0.34 | 19.88 ± 0.26 | 6.34 ± 0.17 | 2.86 ± 0.11 | 226.86 ± 0.22 | 488.83 ± 10.69 |
| #2 | 47.10 ± 0.29 | 17.18 ± 0.45 | 19.24 ± 0.10 | 6.76 ± 0.01 | 2.60 ± 0.19 | 221.44 ± 4.71 | 547.59 ± 12.91 |
| #29 | 46.21 ± 0.11* | 16.73 ± 0.26 | 20.37 ± 0.69 | 6.45 ± 0.12 | 2.75 ± 0.01 | 241.82 ± 7.17 | 532.77 ± 9.68 |
Values are means ± SE. SEs represent two biological replicates. WT, wild type; #2, BLH6a:SRDX overexpression transgenic poplar line 2; #29, transgenic line 29. The asterisk represents the significant increase of cellulose content in #29 by the Student's t-test (
p < 0.05).
Discussion
Wood is the major resource for timber, paper, and pulping. Because wood is composed of SCW in vascular plants, extensive studies have been carried out to understand the composition, function, and biosynthesis of the SCW, particularly in the model plant Arabidopsis thaliana and several species of poplar. In these model systems, many TFs, including NACs and MYBs, were identified as key regulators during the SCW formation (Zhong et al., 2008; Ye and Zhong, 2015). Hierarchical gene regulatory networks (hGRNs) controlling SCW thickening are under construction using many different bioinformatic and biochemical strategies and techniques. One recent combination of methods used top-down Gaussian Graphical Modeling (GGM) to infer correlated molecular interactions and the direction of the correlated effects (Lin et al., 2013; Chen et al., 2019). Network construction uses relative abundance based on the global transcript sequencing (RNA-seq), chromatin binding to construct a four-layered hGRN directed by PtrSND1-B1, a master regulatory NAC protein affecting wood formation (Lin et al., 2013; Chen et al., 2019). Alternatively, bottom-up GGM was used to build a two-layered hGRN for laccases, which polymerize lignin monomers (Lu et al., 2013). Using enhanced Y1H to identify protein–DNA interactions, networks controlling the vascular cambium development and SCW formation have been constructed (Taylor-Teeples et al., 2015; Xu et al., 2016; Yeh et al., 2019; Smit et al., 2020). Through transcriptional network construction, additional interactions and feedback, and feed-forward regulations, have been described (Ko et al., 2014; Smit et al., 2020). In this study, we constructed a TF-prey library containing 227 xylem-specific TFs and conducted Y1H assays to identify TFs regulating CAld5H2 in hybrid poplar. We identified 12 TF candidates and focused on the characterization of three BLH family proteins. BLH proteins belong to the plant-specific three-amino acid loop extension (TALE) superclass of the homeodomain protein family. In Arabidopsis, BLH6a has been indicated to regulate SCW formation through interacting with KNOTTED ARABIDOPSIS THALIANA7 (KNAT7) (Liu et al., 2014), and BLH2 has been indicated to regulate demethylesterification of homogalacturonan in seed mucilage (Xu et al., 2020). In the Arabidopsis blh6 mutant, expressions of some of CELLULOSE SYNTHASE (CESA) and lignin pathway genes, such as CesA7, CesA8, and F5H, were increased (Liu et al., 2014). Our results confirmed the regulation of BLH6a on CAld5H2 in poplar by transient overexpression, stable dominate repression, and ChIP–qPCR. Our analyses on the activation/repression ability of poplar BLH6a are consistent with the conclusion that Arabidopsis BLH6 is a transcriptional repressor (Liu et al., 2014). Because BLH6a is a transcriptional repressor, overexpression of BLH6a:SRDX would enhance its repression ability, which will cause a similar phenotype of overexpression transgenics. Consistently, decreased cell wall thickness is observed in Arabidopsis overexpressing a BLH6 gene (Liu et al., 2014).
Different methods for the hGRN construction and TF identification have their advantages and limitations. Each method may generate false positives. For network prediction by GGM algorithms, more samples with more variation of the gene expression levels improve the precision of the network. The constructed networks and TFs included in the network by different strategies are not the same. For example, NAC123 directly targets CCoAOMT1 in P. trichocarpa by ChIP–qPCR in xylem protoplasts (Chen et al., 2019). Our Y1H showed that NAC123 (also named SND2 in this study) could bind the promoters of CAld5H2, COMT2, PAL4, C4H2, and 4CL5. In the P. trichocarpa network (Chen et al., 2019), the TF WBLH2 interacts with CAld5H1 and HCT1 promoters, whereas in this study its homolog BLH2 from P. alba × P. glandulosa bound CAld5H2 and CAld5H1 promoters and did not bind the promoters of other monolignol pathway genes. Further experiments, such as ChIP assays in xylem cells (not xylem protoplasts) and electrophoretic mobility shift assays (EMSA) are needed to verify these interactions. The protein–DNA interactions ought to be proven before carrying out experiments to show that they are expressed in the same cells. Based on transcriptome data from a series of xylem cryosections (Sundell et al., 2017), our 12 TF genes are co-expressed with CAld5H2 (Supplementary Figure 1). The RISH has provided evidence of co-expression of BLH6a/b, BZIP34, and bHLH59 with CAld5H1/2. Further experiments, laser capture microdissection, or single-cell RNA sequencing (scRNA-seq) may provide additional information on gene co-expression.
Note that the networks constructed in different species may not be the same. In Arabidopsis, MYB46/83, which is located in the second layer directly regulated by SND1, can directly regulate the bottom layer SCW biosynthetic genes, including three CesA encoding genes (CesA4, 7, and 8), seven monolignol pathway genes (PAL1, 2, 4, 4CL3, CCR, CCoAOMT1, and CAD) and four xylan biosynthetic pathway enzyme encoding genes (FRA8, IRX8, IRX9, and IRX14) (Zhong and Ye, 2012; Kim et al., 2013; Ko et al., 2014). However, in P. trichocarpa, using ChIP assays in xylem protoplasts overexpressing PtrMYB021 (homolog of MYB46) only identifies four bottom layer genes (PtrIRX9, PtrIRX14-L, PtrFRA-1, and PtrPAL2) as the direct targets of PtrMYB021 (Chen et al., 2019). Further ChIP assays on the five TFs (targets of PtrMYB021) in the third layer, including PtrMYB090, PtrMYB161, PtrMYB174, PtrNAC123, PrWBLH1, and PtrWBLH2, identified three CesA gene promoters and 10 monolignol pathway gene promoters (Chen et al., 2019).
The poplar genome has had a historical whole-genome duplication event, generating about 8,000 pairs of duplicated genes (Tuskan et al., 2006). During evolution, both promoter and gene body between the gene pair could have diverged. In Arabidopsis, MYB85 overexpression induces 4CL expression and causes ectopic lignin deposition (Zhong et al., 2008). In our study, the identified 12 TFs contain two poplar MYB85 paralogs (MYB85a and MYB85b). Y1H assays showed that MYB85a only interacted with CAld5H2 and COMT2 promoters, whereas MYB85b interacted with CAdl5H2, COMT2, C3H3, CAD1, PAL4, and 4CL3 promoters, showing the different abilities of these two MYB85 members in activating monolignol biosynthesis. In P. trichocarpa, the two homologs 4CL3 and 4CL5 have different regulatory metabolic activity, indicating that the difference in promotor binding of 4CL3 and 4CL5 may be related to their different metabolic roles (Chen et al., 2013). All 12 TFs could bind the CAld5H2 promoter, but only three TFs could bind the CAld5H1 promoter, indicating promoter divergence.
Many studies have shown that one gene may be regulated by multiple TFs, and multiple TFs binding to the same promoter could regulate target genes by forming protein complexes (Chen et al., 2019). Pairwise interactions among three TFs, PtrMYB090, PtrMYB161, and PtrWBLH1 have been detected in P. trichocarpa, and these three TFs may form a ternary complex regulating CAld5H. Our LCI assays for pairwise interactions among 12 TFs identified extensive interactions between TFs, indicating many TFs may form many dimers. In the BLH6a:SRDX overexpression transgenics, the CAld5H2 gene was downregulated up to 62%, but the CAld5H1 gene was not downregulated significantly. S lignin content was not changed in the BLH6a:SRDX overexpression transgenic wood, which could be due to the redundancy between CAld5H1 and CAld5H2. Thus, it is necessary to perform Y1H to identify the TFs that specifically bind to the CAld5H1 promoter. Although the CRES-T technology could avoid redundancy, using the SRDX system for a transcriptional repressor may not make much difference in some cases, which may be another possible reason for the unchanged S lignin content. When the TF of interest is a transcriptional repressor, transgenic plants expressing a chimeric repressor exhibit an enhanced phenotype (Mitsuda et al., 2011). Further using the CRISPR-CAS9 system to knock out the candidate TF genes regulating CAld5H1/2 in poplar is needed to understand their roles in lignin biosynthesis. Although lignin content and composition did not show alteration in BLH6a:SRDX overexpression transgenic wood compared to WT, we observed that the xylem cell wall thickness was decreased significantly. This phenotype may be caused by the interactions of BLH6a with other TFs. The conserved BELL and SKY domains in BLH proteins comprise a conserved bipartite MID domain, which can interact with the KNAT MEINOX domain to form heterodimers (Bellaoui et al., 2001), and the decrease is likely associated with the repression of REVOLUTA by BLH6 and KNT7 interactions. OVATE FAMILY PROTEIN1 (OFP1) and OFP4 are components of the BLH6-KNAT7 multi-protein complex and may modulate the activity of the BLH6-KNAT7 complex (Liu and Douglas, 2015). Further investigations are needed to study the heterodimer formation among the identified TFs for efficient regulation of CAld5H promoter activity.
Multiple TFs may have the combinatorial coordination to activate or suppress target gene expression. Transcriptional activation and repression strength and sequence specificity of TFs depend on their expression levels. The RISH showed BLH6a transcript abundance in the vessels and fiber cells at the early stages of xylem differentiation. Two other genes BZIP34 and bHLH59, which were expressed throughout the xylem, have the same expression profile with CAld5H1/2. These 12 genes may function at different locations, either activating or suppressing CAld5H genes. Different cells may have different proportions of different protein–protein interactions, thus affecting CAld5H gene expression through different protein complex composition and abundance. Further investigation is needed to determine whether these TFs are activators or repressors and to study their potential for redundant and combinatorial regulation.
Conclusion
We identified 12 upstream candidates of CAld5H2. The regulation of one TF, BLH6a, on CAld5H2 was substantiated by transient effector–reporter assays, dominant repression, and ChIP–qPCR. Further understanding the TFs regulating CAld5H2 and their mechanisms of action will lead to novel strategies for engineering S subunit levels in lignin. Such information will be important for the development of S subunits in gymnosperm wood, which are currently absent in gymnosperms.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Author Contributions
QL designed and supervised the project. QW, XD, HP, YC, XH, HL, and XY performed the experiments. FL, HW, RS, and QL analyzed the data. QW, HW, RS, and QL wrote the manuscript. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Chengcai Chu for the pBG3-LZ004 vector, Dr. Yan Guo for the pCM1307-N-FLAG-HA vector, Dr. Keming Luo for pGBKT7-VP16 vector, Jiaqiang Sun for pGWB17, p1300-35S-nLUC, and p1300-35S-cLUC vectors, and Dr. Wei Li for Populus trichocarpa developing xylem.
Footnotes
Funding. This work was supported by grants from Fundamental Research Funds of the Chinese Academy of Forestry (CAFYBB2017ZY001 and CAFYBB2016ZX001-1), Natural Science Foundation of China (31670667), and the National Key Research and Development Program of China (2016YFD0600103).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.695223/full#supplementary-material
Co-expression analysis of 12 TF genes with CAld5H1 and CAld5H2.
Expression patterns of 12 TFs and two CAld5H genes in stem cryosections of P. tremula.
Protein–protein interactions among 12 TFs by LCI assays.
qRT-PCR of transgene BLH6a:SRDX in the leaves of 36 transgenic lines.
Primers used in this study.
12 TF candidates regulating CAld5H2.
Summary of interactions among 12 TFs.
227 xylem-specific TFs in Populus trichocarpa.
Sequence motif analysis of the fragments 6 and 7 in CAld5H2 promoter.
References
- Bellaoui M., Pidkowich M. S., Samach A., Kushalappa K., Kohalmi S. E., Modrusan Z., et al. (2001). The Arabidopsis BELL1 and KNOX TALE homeodomain proteins interact through a domain conserved between plants and animals. Plant Cell 13, 2455–2470. 10.1105/tpc.010161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boerjan W., Ralph J., Baucher M. (2003). Lignin biosynthesis. Annu. Rev. Plant. Biol. 54, 519–546. 10.1146/annurev.arplant.54.031902.134938 [DOI] [PubMed] [Google Scholar]
- Chang H.-m., Sarkanen K. (1973). Species variation in lignin. TAPPI 56, 132–134. [Google Scholar]
- Chen H., Song J., Williams C. M., Shuford C. M., Liu J., Wang J. P., et al. (2013). Monolignol pathway 4-coumaric acid:CoA ligases in Populus trichocarpa: novel specificity, metabolic regulation, and simulation of CoA ligation fluxes. Plant Physiol. 161, 1501–1516. 10.1104/pp.112.210971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Wang J. P., Liu H., Li H., Lin Y.-C. J., Shi R., et al. (2019). Hierarchical transcription factor and chromatin binding network for wood formation in black cottonwood (Populus trichocarpa). Plant Cell 31, 602–626. 10.1105/tpc.18.00620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Nolan T., Ye H., Zhang M., Tong H., Xin P., et al. (2017). Arabidopsis WRKY46, WRKY54 and WRKY70 transcription factors are involved in brassinosteroid-regulated plant growth and drought response. Plant Cell 29, 1425–1439. 10.1105/tpc.17.00364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W. C., Ko J. H., Kim J. Y., Kim J. M., Bae H. J., Han K. H. (2013). MYB46 directly regulates the gene expression of secondary wall-associated cellulose synthases in Arabidopsis. Plant J. 73, 26–36. 10.1111/j.1365-313x.2012.05124.x [DOI] [PubMed] [Google Scholar]
- Ko J. H., Jeon H. W., Kim W. C., Kim J. Y., Han K. H. (2014). The MYB46/MYB83-mediated transcriptional regulatory programme is a gatekeeper of secondary wall biosynthesis. Ann. Bot. 114:1099. 10.1093/aob/mcu126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondhare K. R., Vetal P. V., Kalsi H. S., Banerjee A. K. (2019). BEL1-like protein (StBEL5) regulates CYCLING DOF FACTOR1 (StCDF1) through tandem TGAC core motifs in potato. J. Plant Physiol. 241:153014. 10.1016/j.jplph.2019.153014 [DOI] [PubMed] [Google Scholar]
- Lapierre C., Pollet B., Rolando C. (1995). New insights into the molecular architecture of hardwood lignins by chemical degradative methods. Res. Chem. Intermediat. 21, 397–412. 10.1007/BF03052266 [DOI] [Google Scholar]
- Ledent V., Vervoort M. (2001). The basic helix-loop-helix protein family: comparative genomics and phylogenetic analysis. Genome Res. 11, 754–770. 10.1101/gr.177001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. H., Jin S., Sun Y. K., Kim W., Ji H. A. (2017). A fast, efficient chromatin immunoprecipitation method for studying protein-DNA binding in Arabidopsis mesophyll protoplasts. Plant Methods 13:42. 10.1186/s13007-017-0192-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Zhou Y., Cheng X., Sun J., Marita J. M., Ralph J., et al. (2003). Combinatorial modification of multiple lignin traits in trees through multigene cotransformation. Proc. Natl. Acad. Sci. U. S. A. 100, 4939–4944. 10.1073/pnas.0831166100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Lin Y.-C., Sun Y.-H., Song J., Chen H., Zhang X.-H., et al. (2012). Splice variant of the SND1 transcription factor is a dominant negative of SND1 members and their regulation in Populus trichocarpa. Proc. Natl. Acad. Sci. U. S. A. 109, 14699–14704. 10.1073/pnas.1212977109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Song J., Peng S., Wang J. P., Qu G. Z., Sederoff R. R., et al. (2014). Plant biotechnology for lignocellulosic biofuel production. Plant Biotechnol. J. 12, 1174–1192. 10.1111/pbi.12273 [DOI] [PubMed] [Google Scholar]
- Lin Y.-C., Li W., Chen H., Li Q., Sun Y.-H., Shi R., et al. (2014). A simple improved-throughput xylem protoplast system for studying wood formation. Nat. Protocols 9, 2194–2205. 10.1038/nprot.2014.147 [DOI] [PubMed] [Google Scholar]
- Lin Y.-C., Li W., Sun Y.-H., Kumari S., Wei H., Li Q., et al. (2013). SND1 transcription factor–directed quantitative functional hierarchical genetic regulatory network in wood formation in Populus trichocarpa. Plant Cell 25, 4324–4341. 10.1105/tpc.113.117697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Xue Z., Tang D., Shen Y., Shi W., Shi W., et al. (2018). Ornithine δ-aminotransferase is critical for floret development and seed setting through mediating nitrogen reutilization in rice. Plant J. 96, 842–854. 10.1111/tpj.14072 [DOI] [PubMed] [Google Scholar]
- Liu Y., Douglas C. J. (2015). A role for OVATE FAMILY PROTEIN1 (OFP1) and OFP4 in a BLH6-KNAT7 multi-protein complex regulating secondary cell wall formation in Arabidopsis thaliana. Plant Signal. Behav. 10:e1033126. 10.1080/15592324.2015.1033126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., You S., Taylor-Teeples M., Li W. L., Schuetz M., Brady S. M., et al. (2014). BEL1-LIKE HOMEODOMAIN6 and KNOTTED ARABIDOPSIS THALIANA7 interact and regulate secondary cell wall formation via repression of REVOLUTA. Plant Cell 26, 4843–4861. 10.1105/tpc.114.128322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S., Li Q., Wei H., Chang M.-J., Tunlaya-Anukit S., Kim H., et al. (2013). Ptr-miR397a is a negative regulator of laccase genes affecting lignin content in Populus trichocarpa. Proc. Natl. Acad. Sci. U. S. A. 110, 10848–10853. 10.1073/pnas.1308936110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy R. L., Zhong R., Ye Z. H. (2009). MYB83 is a direct target of SND1 and acts redundantly with MYB46 in the regulation of secondary cell wall biosynthesis in Arabidopsis. Plant Cell Physiol. 50, 1950–1964. 10.1093/pcp/pcp139 [DOI] [PubMed] [Google Scholar]
- Minoru K., Makiko U., Nobuyuki N., Gorou H., Masatoshi Y., Jun I., et al. (2005). Transcription switches for protoxylem and metaxylem vessel formation. Genes Dev. 19:1855. 10.1101/gad.1331305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsuda N., Matsui K., Ikeda M., Nakata M., Oshima Y., Nagatoshi Y., et al. (2011). CRES-T, an effective gene silencing system utilizing chimeric repressors. Methods Mol. Biol. 754, 87–105. 10.1007/978-1-61779-154-3_5 [DOI] [PubMed] [Google Scholar]
- Murre C., Mccaw P. S., Vaessin H., Caudy M., Lassar A. B. (1989). Interactions between heterologous helix-loop-helix proteins generate complexes that bind specifically to a common DNA sequence. Cell 58, 537–544. 10.1016/0092-8674(89)90434-0 [DOI] [PubMed] [Google Scholar]
- Ohashiito K., Oda Y., Fukuda H. (2010). Arabidopsis VASCULAR-RELATED NAC-DOMAIN6 directly regulates the genes that govern programmed cell death and secondary wall formation during xylem differentiation. Plant Cell 22, 3461–3473. 10.1105/tpc.110.075036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osakabe K., Tsao C. C., Li L., Popko J. L., Umezawa T., Carraway D. T., et al. (1999). Coniferyl aldehyde 5-hydroxylation and methylation direct syringyl lignin biosynthesis in angiosperms. Proc. Natl. Acad. Sci. U. S. A. 96, 8955–8960. 10.1073/pnas.96.16.8955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralph J., Lundquist K., Brunow G., Lu F., Kim H., Schatz P. F., et al. (2004). Lignins: natural polymers from oxidative coupling of 4-hydroxyphenyl- propanoids. Phytochem. Rev. 3, 29–60. 10.1023/B:PHYT.0000047809.65444.a4 [DOI] [Google Scholar]
- Río J. C. D., Gutiérrez A., Hernando M., Landín P., Romero J., Martínez Á. T. (2005). Determining the influence of eucalypt lignin composition in paper pulp yield using Py-GC/MS. J. Anal. Appl. Pyrol. 74, 110–115. 10.1016/j.jaap.2004.10.010 [DOI] [Google Scholar]
- Sarkanen K. V., Ludwig C. H. (1971). Lignins: Occurrence, Formation, Structure and Reactions. New York, NY: Wiley-Interscience. [Google Scholar]
- Shi R., Wang J. P., Lin Y.-C., Li Q., Sun Y.-H., Chen H., et al. (2017). Tissue and cell-type co-expression networks of transcription factors and wood component genes in Populus trichocarpa. Planta 245, 927–938. 10.1007/s00425-016-2640-1 [DOI] [PubMed] [Google Scholar]
- Smit M. E., McGregor S. R., Sun H., Gough C., Bågman A.-M., Soyars C. L., et al. (2020). A PXY-mediated transcriptional network integrates signaling mechanisms to control vascular development in Arabidopsis. Plant Cell 32, 319–335. 10.1105/tpc.19.00562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S., Qi T., Huang H., Ren Q., Wu D., Chang C., et al. (2011). The jasmonate-ZIM domain proteins interact with the R2R3-MYB transcription factors MYB21 and MYB24 to affect jasmonate-regulated stamen development in Arabidopsis. Plant Cell 23, 1000–1013. 10.1105/tpc.111.083089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart J. J., Akiyama T., Chapple C., Ralph J., Mansfield S. D. (2009). The effects on lignin structure of overexpression of ferulate 5-hydroxylase in hybrid poplar1. Plant Physiol. 150, 621–635. 10.1104/pp.109.137059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studer M. H., Demartini J. D., Davis M. F., Sykes R. W., Brian D., Martin K., et al. (2011). Lignin content in natural Populus variants affects sugar release. Proc. Natl. Acad. Sci. U. S. A. 108, 6300–6305. 10.1073/pnas.1009252108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundell D., Street N. R., Kumar M., Mellerowicz E. J., Kucukoglu M., Johnsson C., et al. (2017). AspWood: High-spatial-resolution transcriptome profiles reveal uncharacterized modularity of wood formation in Populus tremula. Plant Cell 29, 1585–1604. 10.1105/tpc.17.00153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor-Teeples M., Lin L., de Lucas M., Turco G., Toal T. W., Gaudinier A., et al. (2015). An Arabidopsis gene regulatory network for secondary cell wall synthesis. Nature 517, 571–575. 10.1038/nature14099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuskan G. A., DiFazio S., Jansson S., Bohlmann J., Grigoriev I., Hellsten U., et al. (2006). The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313, 1596–1604. 10.1126/science.1128691 [DOI] [PubMed] [Google Scholar]
- Vanholme R., Cesarino I., Rataj K., Xiao Y., Sundin L., Goeminne G., et al. (2013). Caffeoyl shikimate esterase (CSE) is an enzyme in the lignin biosynthetic pathway in Arabidopsis. Science 341, 1103–1106. 10.1126/science.1241602 [DOI] [PubMed] [Google Scholar]
- Wagner A., Tobimatsu Y., Phillips L., Flint H., Geddes B., Lu F., et al. (2015). Syringyl lignin production in conifers: Proof of concept in a Pine tracheary element system. Proc. Natl. Acad. Sci. U. S. A. 112:6218. 10.1073/pnas.1411926112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J. P., Matthews M. L., Williams C. M., Rui S., Yang C., Tunlaya-Anukit S., et al. (2018). Improving wood properties for wood utilization through multi-omics integration in lignin biosynthesis. Nat. Comm. 9:1579. 10.1038/s41467-018-03863-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J. P., Naik P. P., Chen H.-C., Shi R., Lin C.-Y., Liu J., et al. (2014). Complete proteomic-based enzyme reaction and inhibition kinetics reveal how monolignol biosynthetic enzyme families affect metabolic flux and lignin in Populus trichocarpa. Plant Cell 26, 894–914. 10.1105/tpc.113.120881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J. P., Shuford C. M., Li Q., Song J., Lin Y. C., Sun Y. H., et al. (2012). Functional redundancy of the two 5-hydroxylases in monolignol biosynthesis of Populus trichocarpa: LC-MS/MS based protein quantification and metabolic flux analysis. Planta 236, 795–808. 10.1007/s00425-012-1663-5 [DOI] [PubMed] [Google Scholar]
- Xu H., Cao D., Feng J., Wu H., Lin J., Wang Y. (2016). Transcriptional regulation of vascular cambium activity during the transition from juvenile to mature stages in Cunninghamia lanceolata. J. Plant Physiol. 200, 7–17. 10.1016/j.jplph.2016.06.003 [DOI] [PubMed] [Google Scholar]
- Xu Y., Wang Y., Wang X., Pei S., Kong Y., Hu R., et al. (2020). Transcription factors BLH2 and BLH4 regulate demethylesterification of homogalacturonan in seed mucilage. Plant Physiol. 183, 96–111. 10.1104/pp.20.00011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan X., Ma L., Pang H., Wang P., Liu L., Cheng Y., et al. (2019). METHIONINE SYNTHASE1 is involved in chromatin silencing by maintaining DNA and histone methylation. Plant Physiol. 181, 249–261. 10.1104/pp.19.00528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Z.-H., Zhong R. (2015). Molecular control of wood formation in trees. J. Exp. Bot. 66, 4119–4131. 10.1093/jxb/erv081 [DOI] [PubMed] [Google Scholar]
- Yeh C.-S., Wang Z., Miao F., Ma H., Kao C.-T., Hsu T.-S., et al. (2019). A novel synthetic-genetic-array–based yeast one-hybrid system for high discovery rate and short processing time. Genome Res. 29, 1343–1351. 10.1101/gr.245951.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q., Wang H., Yin Y., Xu Y., Chen F., Dixon R. A. (2010). Syringyl lignin biosynthesis is directly regulated by a secondary cell wall master switch. Proc. Natl. Acad. Sci. U. S. A. 107, 14496–14501. 10.1073/pnas.1009170107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R., Demura T., Ye Z. H. (2006). SND1, a NAC domain transcription factor, is a key regulator of secondary wall synthesis in fibers of Arabidopsis. Plant Cell 18, 3158–3170. 10.1105/tpc.106.047399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R., Lee C., Zhou J., Mccarthy R. L., Ye Z. H. (2008). A battery of transcription factors involved in the regulation of secondary cell wall biosynthesis in Arabidopsis. Plant Cell 20, 2763–2782. 10.1105/tpc.108.061325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R., Richardson E. A., Ye Z. H. (2007). The MYB46 transcription factor is a direct target of SND1 and regulates secondary wall biosynthesis in Arabidopsis. Plant Cell 19, 2776–2792. 10.1105/tpc.107.053678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R., Ye Z. H. (2012). MYB46 and MYB83 bind to the SMRE sites and directly activate a suite of transcription factors and secondary wall biosynthetic genes. Plant Cell Physiol. 53, 368–380. 10.1093/pcp/pcr185 [DOI] [PubMed] [Google Scholar]
- Zhou C., Li Q., Chiang V. L., Lucia L. A., Griffis D. P. (2011). Chemical and spatial differentiation of syringyl and guaiacyl lignins in poplar wood via time-of-flight secondary ion mass spectrometry. Anal. Chem. 83, 7020–7026. 10.1021/ac200903y [DOI] [PubMed] [Google Scholar]
- Zhou H., Zhao J., Yang Y., Chen C., Liu Y., Jin X., et al. (2012). UBIQUITIN-SPECIFIC PROTEASE16 modulates salt tolerance in Arabidopsis by regulating Na+/H+ antiport activity and serine hydroxymethyltransferase stability. Plant Cell 24, 5106–5122. 10.1105/tpc.112.106393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Lee C., Zhong R., Ye Z. H. (2009). MYB58 and MYB63 are transcriptional activators of the lignin biosynthetic pathway during secondary cell wall formation in Arabidopsis. Plant Cell 21, 248–266. 10.1105/tpc.108.063321 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Co-expression analysis of 12 TF genes with CAld5H1 and CAld5H2.
Expression patterns of 12 TFs and two CAld5H genes in stem cryosections of P. tremula.
Protein–protein interactions among 12 TFs by LCI assays.
qRT-PCR of transgene BLH6a:SRDX in the leaves of 36 transgenic lines.
Primers used in this study.
12 TF candidates regulating CAld5H2.
Summary of interactions among 12 TFs.
227 xylem-specific TFs in Populus trichocarpa.
Sequence motif analysis of the fragments 6 and 7 in CAld5H2 promoter.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.