Summary paragraph
Influenza vaccines that confer broad and durable protection against diverse virus strains would have a major impact on global health1. Here we show that computationally designed, two-component nanoparticle immunogens2 induce potently neutralizing and broadly protective antibody responses against a wide variety of influenza viruses. The nanoparticle immunogens display 20 hemagglutinin (HA) trimers in an ordered array, and their assembly in vitro enables precisely controlled co-display of multiple distinct HAs in defined ratios. Nanoparticle immunogens co-displaying the four HAs of licensed quadrivalent influenza vaccines (QIV) elicited antibody responses against vaccine-matched strains that were equivalent or superior to commercial QIV, and simultaneously induced broadly protective antibody responses to heterologous viruses by targeting the subdominant yet conserved HA stem. The combination of potent receptor-blocking and cross-reactive stem-directed antibodies induced by the nanoparticle immunogens make them attractive candidates for a supraseasonal influenza vaccine candidates with potential to replace conventional seasonal vaccines3.
Influenza viruses cause an estimated 290,000–650,000 deaths annually despite the availability of licensed vaccines4, which provide protection against symptomatic infection ranging from about 60% down to less than 10%, varying from year to year5. Current seasonal vaccines primarily elicit antibodies targeting immunodominant hypervariable epitopes in the head domain of the hemagglutinin (HA) glycoprotein, resulting in limited breadth of protection6–8. Epitopes in the HA stem have broader cross-reactive potential, but they are not efficiently targeted by current vaccines. Several antigen design efforts have therefore focused on diminishing responses against the HA head domain to elicit improved stem-focused responses that are otherwise subdominant9–15. Presenting multivalent arrays of HA on self-assembling protein nanoparticles—which are efficiently recognized by the immune system and induce robust humoral responses16–19—has also improved elicitation of stem-directed antibodies. Recently, “mosaic” nanoparticle immunogens, in which multiple related antigens are co-displayed on the same nanoparticle surface, have been suggested as a general approach to improve the breadth of vaccine-elicited antibody responses20–23. However, simple and scalable methods for controllable co-display of multiple oligomeric antigens (such as HA) have not been established. Here we use computationally designed two-component nanoparticles, constructed from multiple copies of two distinct protein building blocks2,24,25, to generate nanoparticle immunogens that controllably co-display diverse influenza HA trimers.
Immunogen design and characterization
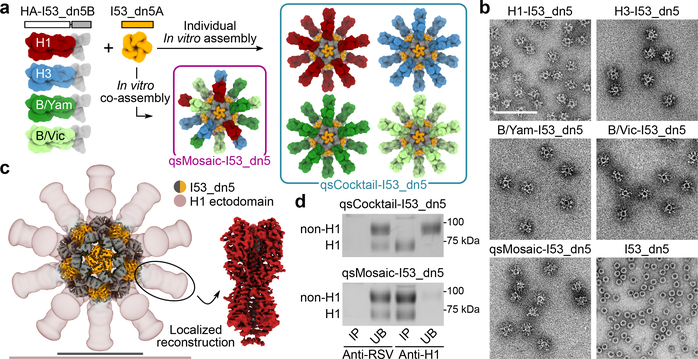
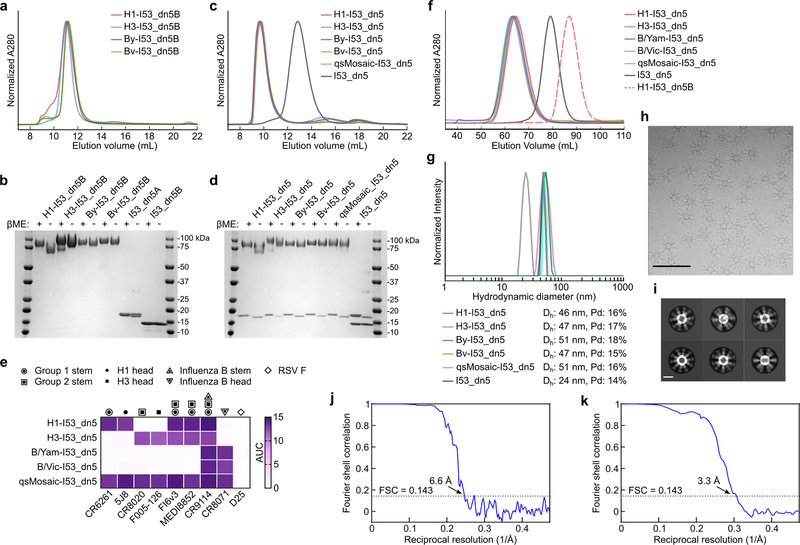
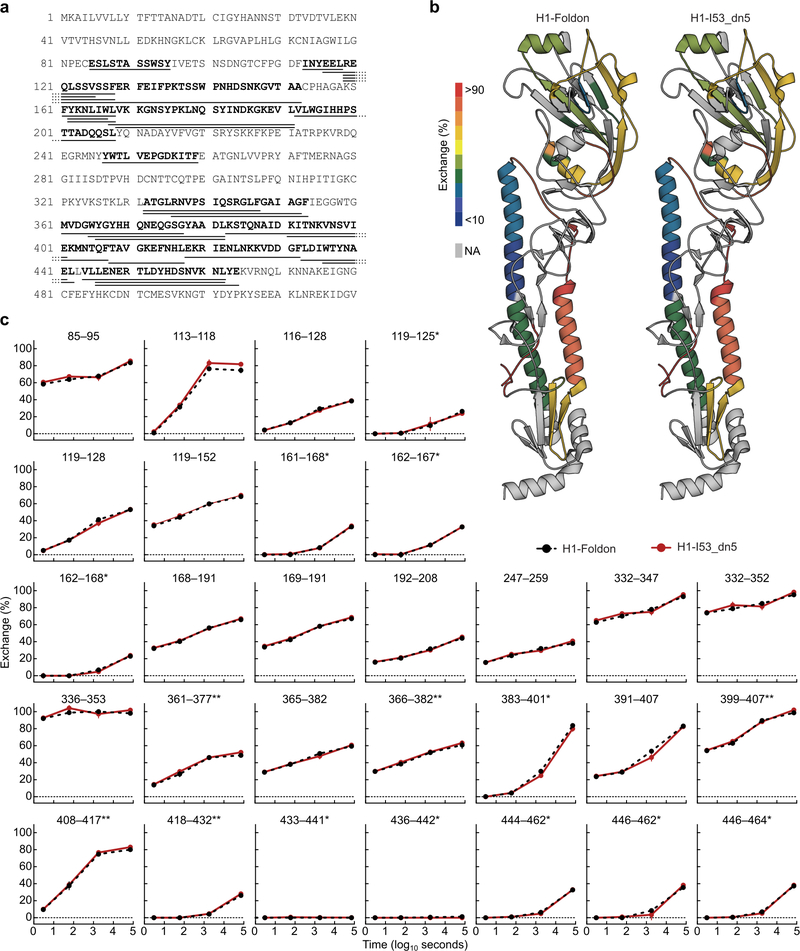
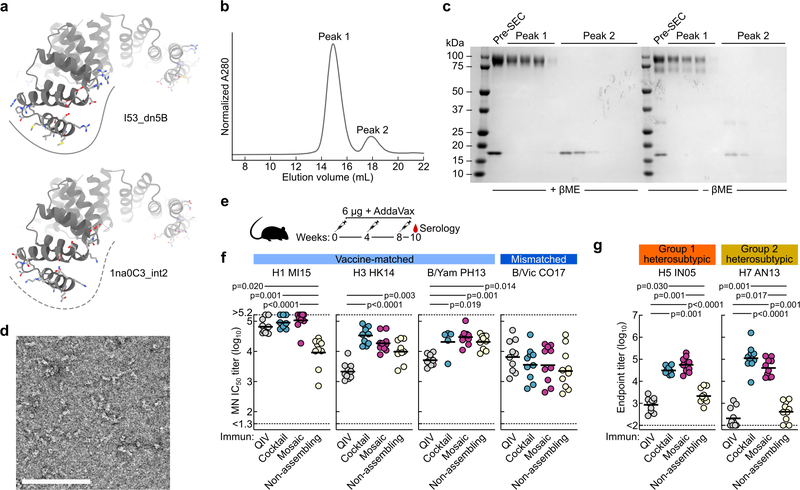
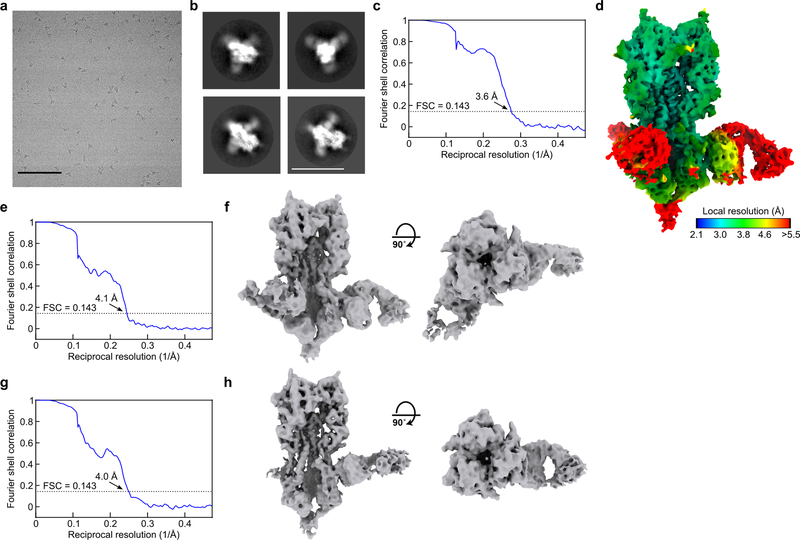
We genetically fused HA ectodomains from the four strains in licensed 2017–2018 seasonal influenza vaccines to the N terminus of I53_dn5B, the trimeric component of the two-component icosahedral nanoparticle I53_dn5, and separately expressed and purified each protein (Fig. 1a, Extended Data Fig. 1a,b, and Supplementary Fig. 1a; ref. 2). All influenza A HA sequences contained the Y98F mutation to facilitate production, which has been shown to minimally impact antigenicity26,27. The purified trimeric HA-I53_dn5B components were mixed in equimolar amounts prior to addition of purified I53_dn5A pentamer to generate a mosaic nanoparticle immunogen co-displaying the four HAs (qsMosaic-I53_dn5; Fig. 1a). In parallel, nanoparticles individually displaying each HA were also produced. Purification by size-exclusion chromatography (SEC) showed that nearly all of the protein eluted in an early peak corresponding to the assembled nanoparticles (Extended Data Fig. 1c,d and Supplementary Fig. 1b), which bound head- and stem-directed mAbs specific to each of the HA trimers displayed (Extended Data Fig. 1e). Comparison of each nanoparticle to I53_dn5 without HA by SEC, dynamic light scattering, and negative stain electron microscopy (NS-EM) confirmed assembly of the intended icosahedral architecture with no evidence of aggregation (Fig. 1b and Extended Data Fig. 1f,g), a result supported by a single-particle cryo-EM reconstruction of the H1-I53_dn5 nanoparticle at 6.6 Å resolution (Fig. 1c and Extended Data Fig. 1h–j). Localized reconstruction of the displayed H1 HA at 3.3 Å resolution (Fig. 1c and Extended Data Fig. 1k) and comparison to an H1 HA-foldon protein by hydrogen-deuterium exchange mass spectrometry (HDX-MS; Extended Data Fig. 2) confirmed full retention of the native structure of the displayed antigen.
Figure 1 |. Design and characterization of HA nanoparticle immunogens.
a, Schematic of in vitro assembly. B/Yam, B/Yamagata/16/1988-like; B/Vic, B/Victoria/2/1987-like. b, Negative stain electron micrographs of purified nanoparticle immunogens and I53_dn5 (scale bar, 200 nm). c, 3D reconstruction of the H1-I53_dn5 nanoparticle and localized reconstruction of H1 HA obtained by single-particle cryo-EM. Scale bars, 23.5 (black) and 50 (pink) nm. d, Immunoprecipitation using RSV F-specific (MPE8) and H1-specific (5J8) mAbs. IP, immunoprecipitated; UB, unbound. All experiments except for cryo-EM were performed at least twice.
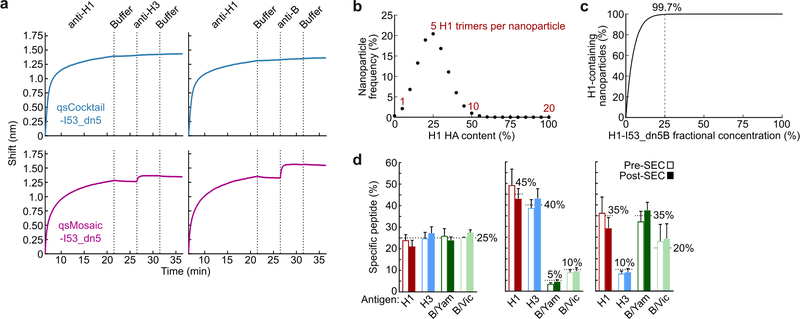
We also prepared a “cocktail” immunogen containing equimolar amounts of the four individual HA-displaying nanoparticles (qsCocktail-I53_dn5; Fig. 1a). While immunoprecipitation of qsCocktail-I53_dn5 nanoparticles with an H1 HA-specific mAb recovered only H1-I53_dn5B, it completely pulled down of all the qsMosaic-I53_dn5 nanoparticles (Fig. 1d and Supplementary Fig. 1c). Likewise, qsMosaic-I53_dn5 nanoparticles immobilized on biolayer interferometry sensors using an H1 HA-specific mAb were subsequently bound by H3- and B HA-specific mAbs, while qsCocktail-I53_dn5 nanoparticles were not (Extended Data Fig. 3a). These results indicated efficient co-assembly of qsMosaic-I53_dn5, matching numerical predictions (Extended Data Fig. 3b,c), and confirmed that subunit exchange did not occur in qsCocktail-I53_dn5. We used quantitative peptide-specific mass spectrometry to confirm that the HA stoichiometry in assembly reactions used to prepare qsMosaic-I53_dn5 nanoparticles of several different compositions was maintained in the SEC-purified nanoparticles (Extended Data Fig. 3d).
Responses against vaccine-matched strains
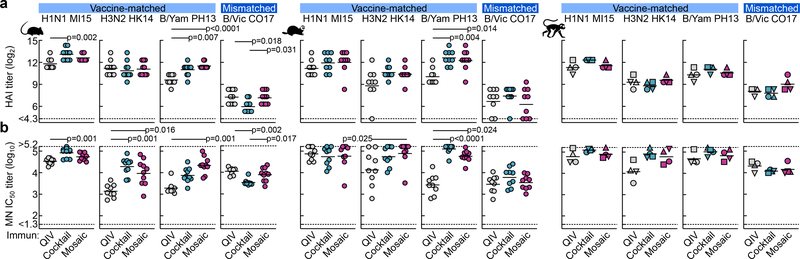
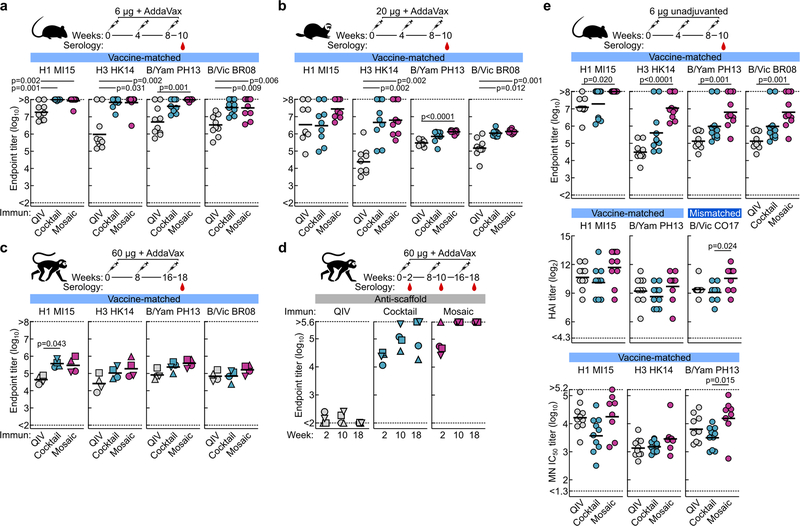
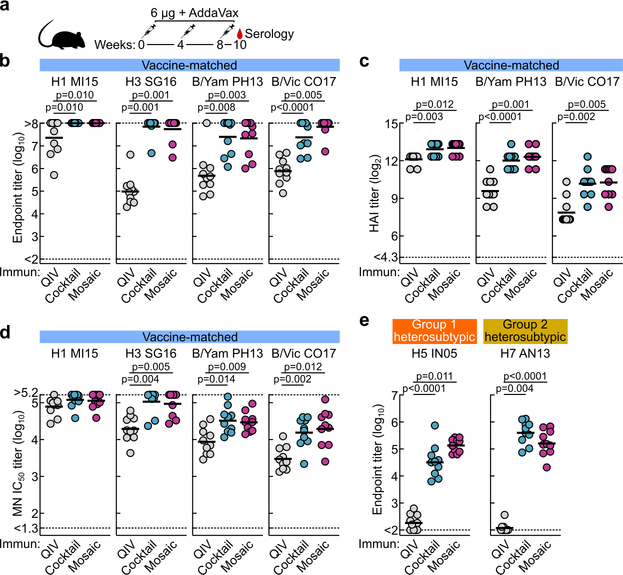
We next compared the immunogenicity of qsCocktail-I53_dn5 and qsMosaic-I53_dn5 to commercial 2017–2018 QIV in mice, ferrets, and nonhuman primates (NHPs), matching the total protein dose of each nanoparticle immunogen to the HA content of QIV. After three immunizations with each immunogen formulated with a squalene oil-in-water adjuvant (AddaVax™), the HA-specific antibody (Extended Data Fig. 4a–c), hemagglutination inhibition (HAI; Fig. 2a), and microneutralization (MN; Fig. 2b) titers induced by both nanoparticle immunogens were equivalent or superior to those induced by QIV. We also observed I53_dn5 nanoparticle scaffold-specific antibodies in NHPs immunized with either nanoparticle immunogen (Extended Data Fig. 4d). Additional immunogenicity studies in mice without adjuvant (Extended Data Fig. 4e) and using updated versions of the three immunogens containing the 2018–2019 vaccine strains (Extended Data Fig. 5a–d) yielded similar results.
Figure 2 |. Vaccine-elicited antibody responses against vaccine-matched viruses in mice, ferrets, and NHPs.
a, Hemagglutination inhibition (HAI) and b, microneutralization (MN) titers in immune sera. Groups of BALB/cJ mice (N = 10), ferrets (N = 9), and rhesus macaques (N = 4) were used in each experiment. Each symbol represents an individual animal and the horizontal bar indicates the geometric mean of the group. Individual NHPs are identified by unique symbols. Statistical analysis was performed using one-sided nonparametric Kruskal–Wallis test with Dunn’s multiple comparisons. All animal experiments except for NHPs were performed at least twice and representative data are shown.
Responses against historical viruses
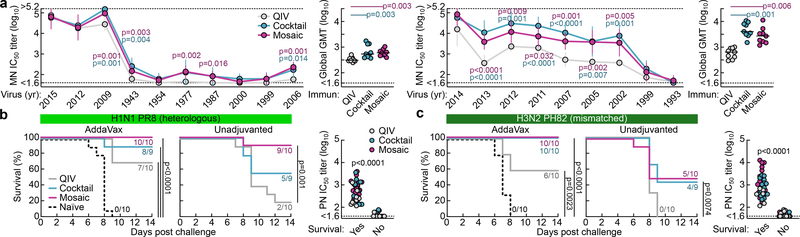
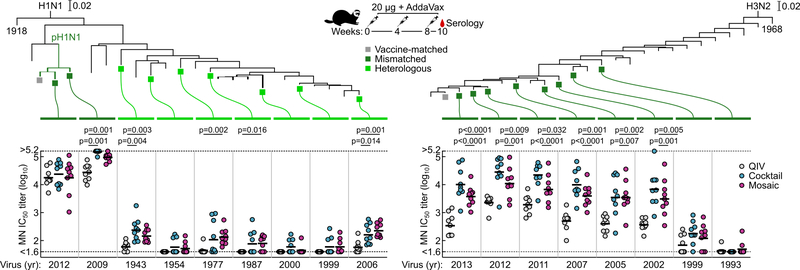
We next tested sera from ferrets immunized with QIV, qsCocktail-I53_dn5, and qsMosaic-I53_dn5 for their ability to neutralize a panel of H1N1 and H3N2 viruses that represent historical antigenic drift and shift28. Both nanoparticle immunogens elicited roughly equivalent or superior neutralizing activity to QIV for all H1N1 strains tested, and ~10-fold higher levels of neutralizing activity against H3N2 viruses dating back to 2002 (Fig. 3a and Extended Data Fig. 6). We then compared the ability of QIV, qsCocktail-I53_dn5, and qsMosaic-I53_dn5 to protect against lethal challenges with heterologous H1N1 (A/Puerto Rico/8/1934) and mismatched H3N2 (A/Philippines/2/1982) viruses in mice (Fig. 3b,c and Supplementary Fig. 3a–c). All mice receiving mock immunizations succumbed to disease and were euthanized by 9 days post-infection. When adjuvanted, both nanoparticle immunogens provided complete or near-complete protection (97% in aggregate), while QIV afforded partial protection against both H1N1 and H3N2 challenges (65%). In the absence of adjuvant, qsMosaic-I53_dn5 provided almost complete (90%) protection from heterologous H1N1 challenge and partial (50%) protection from mismatched H3N2 challenge, while qsCocktail-I53_dn5 provided partial protection in both cases (50% in aggregate). In contrast, unadjuvanted QIV provided negligible protection (10%). Pseudovirus neutralization titers against the challenge strains, measured using sera collected 6 to 8 weeks prior to challenge, showed a clear correlation with protection, with all protected animals having IC50 titers above 100 (Fig. 3b,c). These findings suggest the nanoparticle immunogens might confer multi-season protection without requiring annual vaccine reformulation.
Figure 3 |. Neutralization of and protection against historical H1N1 and H3N2 viruses.
a, Neutralization breadth of ferret immune sera, presented as geometric mean IC50 titers ± geometric s.d. for each group. Statistical analysis was performed using one-sided parametric two-way ANOVA with Tukey’s post-hoc multiple comparisons. Global GMT calculated as the means of geometric mean IC50 titers across 10 H1N1 or 9 H3N2 viruses for each individual animal. Statistical analysis was performed using one-sided nonparametric Kruskal–Wallis test with Dunn’s multiple comparisons. The experiment was performed twice using groups of ferrets (N = 9) and representative data are shown. Heterologous H1N1 (b) and mismatched H3N2 (c) virus challenges in immunized mice. Kaplan–Meier curves were compared using Mantel-Cox log-rank test with Bonferroni correction. Pseudovirus neutralizing (PN) IC50 titers were grouped based on survival outcomes; each symbol represents an individual animal. Statistical analysis was performed using Mann-Whitney test. The challenge experiments were performed once using groups of BALB/cJ mice (N = 10, or N = 9 for qsCocktail-I53_dn5 groups in b and unadjuvanted qsCocktail-I53_dn5 group in c).
Heterosubtypic responses and protection
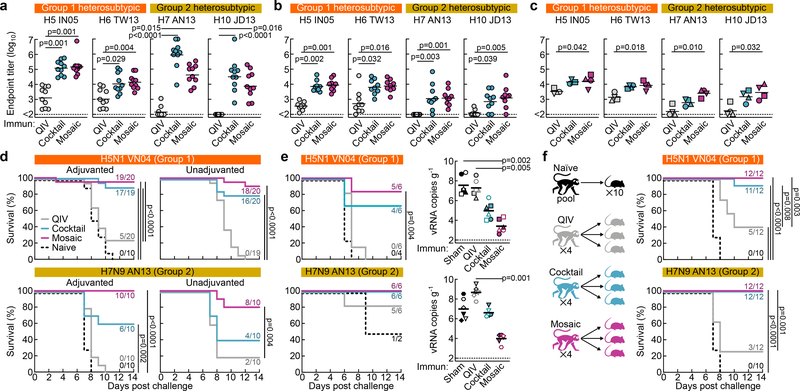
We next compared the ability of QIV, qsCocktail-I53_dn5, and qsMosaic-I53_dn5 to provide immunity against heterosubtypic influenza A viruses. Both nanoparticle immunogens elicited cross-reactive antibody responses to HAs from heterosubtypic group 1 (H5N1 and H6N1) and group 2 (H7N9 and H10N8) viruses, whereas QIV elicited low—in some cases undetectable—levels of such antibodies (Fig. 4a–c and Extended Data Fig. 5e). To assess whether these cross-reactive responses were protective, we first immunized mice with each of the three immunogens with or without AddaVax and challenged them with H5N1 (A/Vietnam/1203/2004) and H7N9 (A/Anhui/1/2013) viruses 8–10 weeks after the last immunization. All animals receiving mock immunizations succumbed to disease, and QIV provided negligible (12%) protection (Fig. 4d and Supplementary Fig. 3d,e). Strikingly, qsCocktail-I53_dn5 conferred partial (73%) and qsMosaic-I53_dn5 nearly complete (92%) protection against these heterosubtypic challenges, even in the absence of adjuvant. The nanoparticle immunogens provided similarly robust protection in ferrets against the same viruses, whereas QIV and mock immunization provided only weak protection (Fig. 4e and Supplementary Fig. 4). RT-qPCR revealed that ferrets receiving qsMosaic-I53_dn5 had significantly lower amounts of H5N1 or H7N9 viral RNA in lung tissues than those immunized with commercial QIV (Fig. 4e, right).
Figure 4 |. Vaccine-elicited heterosubtypic antibody responses and protective immunity.
Cross-reactive antibody responses to heterosubtypic HA antigens in a, BALB/cJ mice (N = 10), b, ferrets (N = 9), and c, rhesus macaques (N = 4). Each symbol represents the endpoint titer (log10) of an individual animal and the horizontal bar indicates the geometric mean of each group. Individual NHPs are identified by unique symbols. All animal immunization experiments except for NHPs were performed at least twice and representative data are shown. Statistical analysis was performed using one-sided nonparametric Kruskal–Wallis test with Dunn’s multiple comparisons. Heterosubtypic influenza virus challenge in immunized mice (d) and ferrets (e). Three ferrets from each group were euthanized 4 days post challenge to measure lung viral RNA (right). Individual ferrets are identified by unique symbols. Right and left caudal lung lobes are indicated as closed and open symbols, respectively. f, Heterosubtypic influenza virus challenge after passive transfer of purified NHP immune Ig in mice. The multiple Kaplan–Meier curves were compared using Mantel-Cox log-rank test with Bonferroni correction. Mouse challenge experiments were performed twice; ferret experiments and passive transfer experiments were performed once.
To determine whether vaccine-elicited serum antibodies alone could confer protection against heterosubtypic challenge, we passively immunized three mice with 10 mg of purified immunoglobulin (Ig) from each immunized NHP 24 h prior to infection with H5N1 or H7N9 virus. All mice that received Ig from qsCocktail-I53_dn5- or qsMosaic-I53_dn5-immunized animals but one, as well as animals that received the stem-directed human broadly neutralizing antibody (bnAb) FI6v3 (ref. 29), showed no weight loss and were protected from disease (Fig. 4f and Supplementary Fig. 5). In contrast, control mice that received Ig purified from influenza-naïve NHPs succumbed to disease and were euthanized, while the mice receiving Ig from QIV-immunized NHPs showed significant weight loss and only partial protection against H5N1 (42%) and H7N9 (25%) challenge.
To better understand the role of nanoparticle display in the antibody responses elicited by qsCocktail-I53_dn5 and qsMosaic-I53_dn5, we immunized mice with a non-assembling version of these immunogens in which the trimeric components lacked the computationally designed interface that drives nanoparticle assembly (Extended Data Fig. 7a–d; ref. 2). While the non-assembling immunogen elicited MN titers against vaccine-matched viruses that were similar to qsCocktail-I53_dn5 and qsMosaic-I53_dn5, the cross-reactive antibody responses elicited against H5N1 and H7N9 HAs were between 10- and 100-fold lower, and were similar to those induced by QIV (Extended Data Fig. 7e–g).
Molecular basis for broad antibody responses
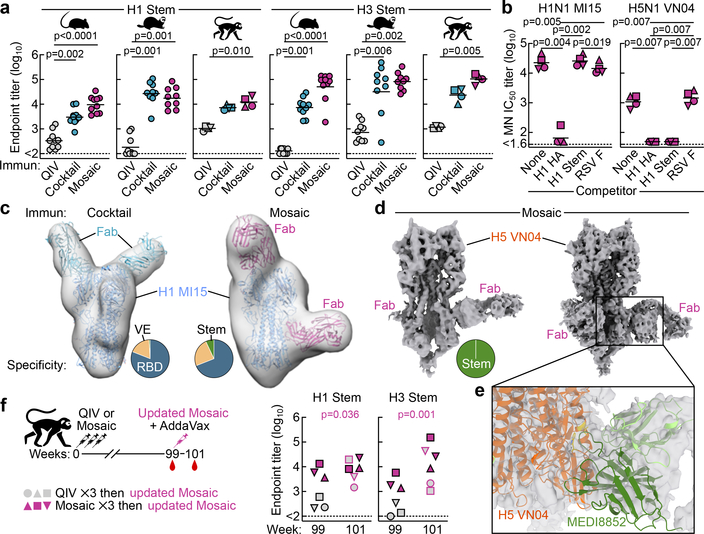
Stem-directed antibodies elicited by both qsCocktail-I53_dn5 and qsMosaic-I53_dn5, measured using stem-only HA proteins9,11, were significantly higher than those induced by QIV in all three animal species (Fig. 5a). MN activity against the vaccine-matched H1N1 virus in sera from NHPs immunized with qsMosaic-I53_dn5 was depleted by vaccine-matched HA ectodomain, but not by an H1 HA stem, suggesting that antibodies targeting epitopes outside of the stem domain account for most of the neutralizing activity as expected (Fig. 5b). In contrast, neutralizing activity against a heterosubtypic H5N1 virus was fully depleted by both vaccine-matched HA and stem-only HA, indicating that stem-directed antibodies are responsible for the observed heterosubtypic neutralization.
Figure 5 |. Molecular basis for nanoparticle-induced protection against heterosubtypic influenza viruses.
a, Serum antibody titers to H1 and H3 stem-only antigens in BALB/cJ mice (N = 10), ferrets (N = 9), and rhesus macaques (N = 4). Each symbol represents the endpoint titer (log10) of an individual animal and the horizontal bar indicates the geometric mean of each group. Individual NHPs are identified by unique symbols. All animal immunization experiments except for NHPs were performed at least twice. b, Serum MN activity against H1N1 and H5N1 viruses in the presence of competitor proteins. Statistical analysis was performed using one-sided nonparametric Kruskal–Wallis test with Dunn’s multiple comparisons. c, Selected NS-EM reconstructions of H1 HA in complex with polyclonal antibody Fab fragments elicited by qsCocktail-I53_dn5 (left) and qsMosaic-I53_dn5 (right). Frequency of complexes observed by EM containing Fab fragments bound to the RBD, VE, and stem are shown as pie charts. d, Two independent cryo-EM reconstructions of H5 HA in complex with polyclonal antibody Fab fragments elicited by qsMosaic-I53_dn5. e, Close-up of one of the dominant antibody classes that resembles MEDI8852 recognition. f, Serum antibody titers to H1 and H3 stem-only antigens in NHPs (N = 6) with pre-existing influenza immunity. Each unique symbol represents the endpoint titer (log10) of an individual animal. Statistical analysis between the two timepoints was performed using paired t test.
Next we directly visualized the nanoparticle-elicited antibodies in individual NHPs in complex with HA using single-particle NS-EM analysis of polyclonal antibodies30. We found that the polyclonal antibodies to vaccine-matched H1 HA target at least three distinct antigenic sites: the receptor-binding domain (RBD), the vestigial esterase (VE) domain, and the stem (Fig. 5c and Extended Data Fig. 8). While antibodies to each antigenic site contained multiple fine specificities and angles of approach, the vast majority of complexes contained antibodies recognizing the RBD. In contrast, single-particle cryo-EM analysis of H5 HA in complex with polyclonal antibody Fab fragments elicited by qsMosaic-I53_dn5 revealed only stem-directed antibodies, unambiguously demonstrating recognition of this conserved supersite (Fig. 5d and Extended Data Fig. 9). The data suggest at least one common class of vaccine-elicited antibodies recognizes the stem in a manner reminiscent of MEDI8852 and 56.a.09, which both belong to the VH6–1+DH3–3 class of multi-donor human bnAbs31,32 (Fig. 5e).
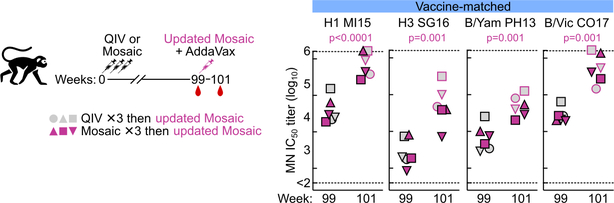
To study how pre-existing influenza immunity could influence antibody responses to the nanoparticle immunogens, NHPs from the QIV and qsMosaic-I53_dn5 groups were boosted 63 weeks later (week 99) with a single dose of an updated 2018–2019 version of qsMosaic-I53_dn5. All animals had high levels of neutralizing antibodies against vaccine-matched strains at week 99 that were strongly boosted upon immunization with updated qsMosaic-I53_dn5 (Extended Data Fig. 10). At week 99, animals pre-immunized with QIV had near-baseline levels of stem-directed antibodies, while animals pre-immunized with qsMosaic-I53_dn5 maintained substantial stem-directed titers (Fig. 5f). Upon boost with an updated qsMosaic-I53_dn5, all animals showed strongly elevated anti-stem antibody titers. These data demonstrate that qsMosaic-I53_dn5 stimulates robust stem-directed antibody responses even in the context of strong pre-existing immunity against the immunodominant HA head.
Discussion
We have developed nanoparticle vaccines that elicit potent vaccine-matched HAI activity as well as protective stem-directed neutralizing antibodies against distantly related—including heterosubtypic—viruses in multiple animal models. These results go beyond previous next-generation influenza vaccine concepts, which have been reported to increase either stem-directed responses9–12,15,33, the potency and breadth of HAI within specific subtypes17,34,35, or both types of responses within specific subtypes36,37. Since both HAI and stem-directed antibodies have been shown as independent immune correlates of protection against influenza infection in humans38, immunogens capable of eliciting both would have advantages over approaches eliciting one or the other and would make attractive candidates for clinical evaluation as supraseasonal vaccines3 that may eventually replace current seasonal vaccines. The broad, antibody-mediated protection conferred by qsCocktail-I53_dn5 and qsMosaic-I53_dn5 suggests that they may be able to provide consistent year-to-year protection against seasonal influenza viruses, even in the event of antigenic mismatches in the hypervariable head domain.
Although defining the immunological or structural basis for the breadth elicited by our nanoparticle vaccines will require further investigation, it is clear that HA presentation on the assembled nanoparticle is vital. Motivated by the data presented here, a variant of qsMosaic-I53_dn5 with updated HAs has been manufactured for a planned Phase I clinical trial. In addition to evaluating safety and reactogenicity, data from this trial should reveal the impact of complex and individualized influenza exposure histories39,40 on the responses elicited by this nanoparticle vaccine.
Methods
Gene synthesis and vector construction
Plasmids for expression of the I53_dn5A pentamer were prepared in pET29b as previously described2. Genes for expression of HA fusions to nanoparticle trimeric components were codon optimized for expression in human cells and cloned into the CMV/R (VRC 8400) mammalian expression vector by Genscript. All HA fusions to the I53_dn5B trimer contained full-length HA ectodomains including native secretion signals, and the H1 and H3 HAs contained an additional mutation (Y98F) to knock out sialic acid binding to facilitate expression and purification27. HA ectodomain sequences preceded a short linker sequence followed by the I53_dn5B trimer sequence with a C-terminal flexible linker, WELQut protease recognition sequence, and a hexa-histidine tag. The amino acid sequences for all proteins used in this study are provided in Supplementary Table 1.
Protein expression and purification
All HA-I53_dn5B trimers, as well as mAbs CR6261 (ref. 41), 5J8 (ref. 42), CR8020 (ref. 43), F005–126 (ref. 44), F045–092 (ref. 45), MEDI8552 (ref. 31), FI6v3 (ref. 29), CR9114 and CR8071 (ref. 46), CT149 (ref. 47), D25 (ref. 48), and MPE8 (ref. 49) were expressed in Expi293F cells (ThermoFisher Scientific) by transient transfection using PEI MAX (Polysciences) or ExpiFectamine™ 293 (ThermoFisher Scientific). mAbs were purified by protein A affinity chromatography using established methods. Recombinant HA ectodomain trimers fused to T4 fibritin foldon were produced and purified as described previously17. The protein-containing supernatants from cells expressing HA-I53_dn5B fusion proteins were further clarified by vacuum filtration (0.22 μm, Millipore Sigma). Prior to immobilized metal affinity chromatography, a background of 50 mM Tris pH 8.0 and 350 mM NaCl was added to the clarified supernatant using concentrated solutions of 1 M Tris pH 8.0 and 5 M NaCl, respectively. For each liter of supernatant, 4 ml of Ni2+ Sepharose Excel resin (GE) was rinsed into PBS using a gravity column and then added to the supernatant, followed by overnight shaking at 4°C. The resin was collected 16–24 h later using a gravity column, then washed twice with 50 mM Tris pH 8.0, 500 mM NaCl, 30 mM imidazole prior to elution of His-tagged protein using 50 mM Tris pH 8.0, 500 mM NaCl, 300 mM imidazole. Eluates were concentrated and applied to a HiLoad 16/600 Superdex 200 pg column or a Superdex 200 Increase 10/300 GL column pre-equilibrated with PBS for preparative size exclusion chromatography. Peaks corresponding to trimeric species were identified based on elution volume and SDS-PAGE (both reducing and non-reducing) of elution fractions. Fractions containing pure HA-I53_dn5B were pooled and the protein quantified using UV/vis spectroscopy. Purified protein was either stored at 4°C until use or flash-frozen in liquid nitrogen and stored at −80°C.
Single colonies of E. coli cells transformed with plasmid encoding the I53_dn5A pentamer were picked and grown in TB medium with 50 μg L−1 kanamycin at 37°C overnight. Subsequently, liquid cultures were diluted 1:40 in TB medium and grown at 37°C until OD600 reached 0.5–0.8. Isopropyl-thio-β-D-galactopyranoside (IPTG) was then added to a concentration of 1 mM and the growth temperature reduced to 18°C to induce protein expression, or cultures were left at 37°C for an additional 5 h before lowering the temperature to 18°C leading to auto-induction by virtue of galactose in the media according to the Studier protocols50. Expression proceeded for 20 h at 18°C, at which point the cell cultures were harvested by centrifugation. Cell pellets were resuspended in 50 mM Tris pH 8.0, 250 mM NaCl, 20 mM imidazole, 1 mM dithiothreitol (DTT), 0.1 mg ml−1 DNase, 0.1 mg ml−1 RNase, and EDTA-free protease inhibitors (Pierce) and lysed by sonication or microfluidization. I53_dn5A protein was purified from the soluble fraction of cell lysates by immobilized metal affinity chromatography using HisTrap HP columns (GE). After application of clarified cell lysate supernatants, the column was washed with 20 column volumes of 50 mM Tris pH 8.0, 250 mM NaCl, 20 mM imidazole, 1 mM DTT. I53_dn5A was eluted using a linear gradient of imidazole up to a final concentration of 500 mM. Protein was concentrated to 1 ml, and 3-[(3-Cholamidopropyl)dimethylammonio]-1-propanesulfonate (CHAPS) was added to 0.75% (w/v) to remove endotoxin. The protein was sterile-filtered at 0.22 μm and purified by preparative SEC using a Superdex 200 Increase 10/300 GL equilibrated in 25 mM Tris pH 8.0, 150 mM NaCl, 5% glycerol. The peak corresponding to the pentamer was identified based on elution volume and SDS-PAGE (both reducing and non-reducing) of eluted fractions. Fractions containing pure I53_dn5A pentamer were pooled and the protein quantified using UV/vis spectroscopy. The samples were confirmed to be low in endotoxin (< 100 EU mg−1) using the Limulus Amebocyte Lysate (LAL) assay (Charles River), then flash-frozen in liquid nitrogen and stored at −80°C within 6 hours of purification to prevent oxidation of cysteines.
In vitro assembly and purification of nanoparticle immunogens
To assemble nanoparticle immunogens bearing multiple copies of single HA antigens (e.g., H1-I53_dn5), individual HA-bearing trimeric components were mixed with pentameric I53_dn5A at a molar ratio of 1:1 (subunit:subunit) at concentrations ranging from 15–40 μM (subunit) by pipetting. Assembly reactions were prepared at room temperature and incubated for 30 min before purification by SEC on a Superose 6 Increase 10/300 GL. The nanoparticle immunogens eluted at the void volume of the column. Fractions were analyzed by SDS-PAGE (both reducing and non-reducing) before pooling and sterile filtering at 0.22 μm.
For H1-I53_dn5 and H3-I53_dn5 nanoparticles, assembly reactions consisted of pentameric components and HA-bearing trimeric components buffered in either PBS or 25 mM Tris pH 8.0, 150 mM NaCl, 5% glycerol. After assembly and incubation, the samples were centrifuged for 10 min at 14,000 rpm at 4°C and the nanoparticle immunogens purified by SEC using a Superose 6 Increase 10/300 GL column pre-equilibrated with 25 mM Tris pH 8.0, 150 mM NaCl, 5% glycerol.
For B/Vic (B/Victoria/2/1987-like)-I53_dn5 and B/Yam (B/Yamagata/16/1988-like)-I53_dn5 nanoparticles, half of the assembly reaction volume consisted of an additional buffer solution with high ionic strength to maintain nanoparticle immunogen solubility. The solutions used were 25 mM Tris pH 8.0, 1.85 M NaCl, 5% glycerol for B/Vic and 25 mM Tris pH 8.0, 3.85 M NaCl, 5% glycerol for B/Yam, which respectively brought NaCl in the assembly reactions to approximately 1 M and 2 M. In these cases the HA-bearing trimeric component was first added to the high-salt buffer prior to addition of the pentameric component. After assembly and incubation, the samples were centrifuged for 10 min at 14,000 rpm at room temperature and the nanoparticle immunogens purified by SEC using a Superose 6 Increase 10/300 GL column pre-equilibrated with either 25 mM Tris pH 8.0, 1 M NaCl, 5% glycerol for B/Vic-I53_dn5 or 25 mM Tris pH 8.0, 2 M NaCl, 5% glycerol for B/Yam-I53_dn5.
For mosaic nanoparticles with equal amounts of each seasonal HA (qsMosaic-I53_dn5), all four HA-bearing trimeric components (in PBS) were first mixed in equimolar amounts. Tris pH 8.0, 1.85 M NaCl, 5% glycerol was added such that the final NaCl in the in vitro assembly reaction would be 1 M. The pentameric component was added and the solution was mixed vigorously by pipetting. After assembly the samples were centrifuged for 10 min at 14,000 rpm at 4°C and the nanoparticle immunogens purified by SEC using a Superose 6 Increase 10/300 GL column pre-equilibrated with 25 mM Tris pH 8.0, 150 mM NaCl, 5% glycerol.
After purification and evaluation of nanoparticle immunogen quality by SDS-PAGE, UV/vis spectroscopy, negative stain EM, DLS, and LAL assay (< 100 EU mg−1), samples were flash-frozen in liquid nitrogen and stored at −80°C.
Dynamic light scattering
Light scattering analysis was conducted using an UNcle (UNchained Labs) at 25°C. For each sample, 10 acquisitions (5 s per acquisition) were obtained using auto-attenuation of the laser. Increased viscosity due to the inclusion of 5% glycerol in the H1-I53_dn5, H3-I53_dn5, B/Yam-I53_dn5, B/Vic-I53_dn5, qsMosaic-I53_dn5 and I53_dn5 nanoparticles was accounted for in the software.
Negative stain and cryo-electron microscopy of immunogens
To image nanoparticles and non-assembling immunogens by negative stain EM, protein samples were diluted to 0.020–0.075 mg ml−1 in 25 mM Tris pH 8.0 with NaCl concentrations ranging from 0.15–2 M. 300 mesh copper grids (Ted Pella) were glow discharged immediately before use. Six μL of sample was applied to the grid for 1 min, then briefly dipped in a droplet of water before blotting away excess liquid with Whatman No. 1 filter paper. Grids were stained with 6 μL of 0.75% (w/v) uranyl formate stain, immediately blotting away excess, then stained again with another 6 μL for 30 s. Grids were imaged on a Morgagni transmission electron microscope with a Gatan camera, and Gatan Digital Micrograph software was used to take images.
To obtain a cryo-EM single particle reconstruction of the H1-I53_dn5 nanoparticle, 3 μl of 0.7 mg ml−1 H1-I53_dn5 was loaded onto a freshly glow-discharged (30 s at 20 mA) Protochips C-flat grid (2.0 μm hole, 200 mesh) by multiple blotting strategy51 prior to plunge freezing using a vitrobot Mark IV (ThermoFisher Scientific) using a blot force of 0 and 7 second blot time at 100% humidity and 25°C. Data was collected using the Leginon software52 on an FEI Titan Krios transmission electron microscope, equipped with a Gatan K2 Summit direct electron detector and Gatan Quantum GIF energy filter, operated in zero-loss mode with a slit width of 20 eV. The dose rate was adjusted to 8 counts pixel−1 s−1, and each movie was acquired in counting mode fractionated in 50 frames of 200 ms. 803 micrographs were collected in a single session with a defocus range between −1.2 μm and −2.5 μm. Movie frame alignment, estimation of the microscope contrast-transfer function parameters and particle picking were carried out using Warp53. 2D and 3D classification (without applying symmetry) were performed using RELION 3.0. A total of 20,827 particles of the best class were selected and subjected to 3D refinement in CryoSPARC applying icosahedral symmetry which led to a reconstruction at 6.6 Å resolution. In Figure 1, high (grey/orange) and low (red) contour 3D representations of the reconstruction are overlaid to enable visualization of the I53_dn5 scaffold and the displayed HA, respectively. The density corresponding to the low contour representation was smoothed using a 16 Å low-pass filter for clarity. We subsequently implemented a localized reconstruction strategy54 (along with extensive 3D classification using Relion) to determine a reconstruction of the H1 MI15 HA at 3.3 Å resolution using Relion (applying C3 symmetry). An atomic model of H1 MI15 HA was built using Coot55, refined using Rosetta56,57, and validated using Molprobity58, Phenix59, Privateer60 and EMRinger61. Reported resolutions are based on the gold-standard Fourier shell correlation (FSC) of 0.143 criterion and Fourier shell correlation curves were corrected for the effects of soft masking by high-resolution noise substitution62. All details for cryo-EM data collection, refinement, and validation statistics are provided in Supplementary Table 2.
Immunoprecipitation
qsCocktail-I53_dn5 and qsMosaic-I53_dn5 samples were mixed with either MPE8 (anti-RSV F) or 5J8 (anti-H1) to final concentrations of 4 μM (each subunit) of immunogen and 0.20 mg ml−1 of mAb. The final buffers contained 2 M NaCl and 0.05% Tween-20 in addition to other buffering agents. The solution was allowed to mix at room temperature for 1 h and then added to recently washed and dried magnetic protein G Dynabeads (Thermo Fisher). The mixture was incubated for 1.5 h and resin was separated from the supernatant magnetically. Beads were washed twice and returned to the same volume used during the binding process, then heated to 95°C for 10 min in the presence of SDS loading dye to detach bound protein. Proteins were then analyzed by SDS-PAGE.
Antigenic characterization
ELISA was used to measure binding of H1-I53_dn5, H3-I53_dn5, B/Yam-I53_dn5, B/Vic-I53_dn5, and qsMosaic-I53_dn5 nanoparticles to mAbs CR6261, 5J8, CR8020, F005–126, MEDI8852, FI6v3, CR9114, CR8071, and D25 (anti-RSV F). 96-well plates were coated with 2 μmol ml−1 nanoparticles (0.1 ml well−1) and incubated at 4°C overnight. Plates were then blocked with PBS containing 5% skim milk at 37°C for 30 min. mAbs were serially diluted in four-fold steps and added to the wells for 1 h. Horseradish peroxidase (HRP)-conjugated anti-human or anti-mouse IgG (Southern Biotech) was added and incubated at 37°C for 30 min. The wells were developed with 3,3′,5′,5-tetramethylbenzidine (TMB) substrate (KPL), and the reactions were stopped by adding 1 M H2SO4 before measuring absorbance at 450 nm with a Spectramax Paradigm plate reader (Molecular Devices).
Mass spectrometry quantification of HA content in qsMosaic-I53_dn5
Label-free quantitation was performed by peptide mass spectrometry to determine the relative abundance of each HA present in the mosaic nanoparticle samples. Each mosaic nanoparticle, either before or after SEC purification, along with a standard mixture of each purified HA-I53_dn5B fusion protein at equimolar concentrations (1:1:1:1), was denatured and reduced using guanidine hydrochloride and DTT. Samples were then alkylated with iodoacetamide, deglycosylated with N-glycanase (New England Biolabs), and digested overnight with LysC protease (ThermoFisher scientific). LC-MS was performed using a Waters Acquity UPLC coupled to a Thermo LTQ-OT using data-dependent acquisition. Peptides were resolved over a Waters CSH C18 1.7 μm, 2.1 × 100 mm column with a linear gradient from 3% to 40% B over 30 minutes (A: 0.1% formic acid; B: acetonitrile with 0.1% formic acid). Peptides were identified from MS/MS data using Protein Prospector using a score cutoff of 15 (http://prospector.ucsf.edu/). Due to the high sequence identity between the HA constructs, only four peptides unique to each specific HA were observed that could be used for label-free quantitation. The integrated peak areas for these peptides relative to the areas from an equimolar mixture of each HA were used to estimate the total abundance of each HA within the mosaic nanoparticle samples (Supplementary Table 3).
Hydrogen-deuterium exchange mass spectrometry
For each timepoint, 30 pM of H1 HA-foldon and H1-I53_dn5 were incubated in deuterated buffer (85% D2O, pH* 7.4) for 3; 60; 1,800; or 72,000 s at room temperature and subsequently mixed with an equal volume of ice-cold quench buffer (4 M urea, 200 mM tris(2-chlorethyl) phosphate (TCEP), 0.2% formic acid) to a final pH* of 2.5. Samples were immediately frozen in liquid nitrogen and stored at 80°C until analysis. Fully deuterated samples were prepared by digesting 30 pmol of undeuterated H1-foldon over a pepsin column, followed by concentration under vacuum, resuspension in deuterated buffer at 65°C for 1 h, then quenching and freezing. Zero timepoint samples were prepared as described previously63. Inline pepsin digestion was performed and analyzed by LC-IMS-MS utilizing a Waters Synapt G2-Si Q-TOF mass spectrometer as previously described63. Deuterium uptake analysis was performed using HD-Examiner (Sierra Analytics) followed by HX-Express v3.13 (refs. 64,65). The percent exchange was normalized to the zero timepoint and fully deuterated reference samples. Internal exchange standards (Pro-Pro-Pro-Ile [PPPI] and Pro-Pro-Pro-Phe [PPPF]) were included in each reaction to ensure that conditions were consistent throughout all of the labeling reactions.
Animal experiments
All animal experiments were reviewed and approved by the Institutional Animal Care and Use Committee of the VRC, NIAID, NIH. All animals were housed and cared for in accordance with local, state, federal, and institutional policies of NIH and American Association for Accreditation of Laboratory Animal Care. The space temperature in the rodent facility is set to 22°C ± 3 degrees. The humidity is maintained between 30% and 70%. The automatic light cycle is a 12 h on/off photo-period.
Immunization and challenge studies
The 2017–2018 and 2018–2019 QIVs used were split virion vaccines manufactured in embryonated chicken eggs (Afluria®, Seqirus). Throughout our studies, we matched the total protein dose of each nanoparticle immunogen to the HA content of QIV. The HA antigens make up approximately 62% of the total peptidic mass of the nanoparticle immunogens. The HA content of commercial QIV was determined by the manufacturer using the standard SRD assay, and the total protein content of the nanoparticle vaccine preparations was measured by UV/vis absorbance. BALB/cJ mice (Jackson Laboratory) were immunized intramuscularly (i.m.) with 6 μg of commercial QIV 2017–2018 (Afluria®, Seqirus), qsCocktail-I53_dn5, or qsMosaic-I53_dn5 in the presence or absence of AddaVax™ (InvivoGen) at weeks 0, 4, and 8. Formulated vaccines were given 50 μl into each hind leg. Serum samples were collected before and after each immunization and used for immunological assays. For challenge studies, mice were infected intranasally at Bioqual with 10×, 25×, 10×, and 10× the experimentally determined 50% lethal dose (LD50) of H1N1, H5N1, H3N2, and H7N9 viruses, respectively (Supplementary Table 4). The animals were monitored twice daily for development of clinical signs and weighed daily for 14 days. Any animals that had lost 20% or more of their initial body weight were euthanized. Finch ferrets (Mustela putorius) were immunized i.m. with 20 μg of commercial QIV 2017–2018, qsCocktail-I53_dn5, or qsMosaic-I53_dn5 with AddaVax three times at weeks 0, 4, and 8. Immunogens were formulated in 500 μL per ferret and injected into limbs. Serum samples were collected periodically before and after immunization and used for immunological assays. Ferrets were infected intranasally at Bioqual with 25× and 10× LD50 of H5N1 and H7N9 viruses, respectively (Supplementary Table 4). Clinical signs of infection, weight, and body temperatures were recorded twice daily for 14 days. Ferrets that showed signs of severe disease (prolonged fever, diarrhea; nasal discharge interfering with eating, drinking or breathing; severe lethargy; or neurological signs) or that had > 20% weight loss were euthanized immediately. Rhesus macaques (Macaca mulatta) were immunized i.m. with 60 μg of commercial QIV 2017–2018, qsCocktail-I53_dn5, or qsMosaic-I53_dn5 with AddaVax three times at weeks 0, 8, and 16. Immunogens were prepared in 1.0 ml volumes per NHP and injected into limbs. Some of the NHPs that were immunized three times with either 2017–2018 commercial QIV (N = 3) or qsMosaic-I53_dn5 (N = 3) were boosted 63 weeks later with a single dose of an updated version of qsMosaic-I53_dn5 containing the 2018–2019 seasonal strains. Serum samples were collected periodically before and after immunization and used for immunological assays.
ELISA
Antigen-specific IgG levels in immune sera were measured by ELISA. The plates were coated with 2 μg ml−1 of recombinant HA-foldon proteins (Supplementary Table 1) and incubated at 4°C overnight. Plates were then blocked with PBS containing 5% skim milk at 37°C for 1 h. mAbs and immune sera were serially diluted in four-fold steps and added to the wells for 1 h. Horseradish peroxidase (HRP)-conjugated anti-human (SouthernBiotech, Catalog 2040–05, used 1/5,000); anti-mouse IgG (SouthernBiotech, Catalog 1080–05, used 1/5000); anti-ferret IgG (Abcam, Catalog Ab112770, used 1/20,000); or anti-monkey IgG (SouthernBiotech, Catalog 4700–05, used 1/5,000) antibody was added and incubated at 37°C for 1 h. The wells were developed with 3,3′,5′,5-tetramethylbenzidine (TMB) substrate (KPL), and the reactions were stopped by adding 1 M H2SO4 before measuring absorbance at 450 nm with a Spectramax Paradigm plate reader (Molecular Devices). Sera from mice immunized with PBS or an irrelevant antigen (DS-Cav1-I53_dn5; ref. 2), ferrets immunized with PBS, and NHPs prior to immunization were used as negative controls, and did not yield signal above background (Supplementary Fig. 2).
Reporter-based microneutralization assay
All reporter viruses were prepared as described previously28. Briefly, all H1N1 and H3N2 viruses were made with a modified PB1 segment expressing the TdKatushka reporter gene (R3ΔPB1) and propagated in MDCK-SIAT-PB1 cells, while H5N1 reporter virus was made with a modified HA segment expressing the reporter (R3ΔHA) and produced in cells stably expressing H5 HA. Replication-restricted reporter influenza viruses encoding influenza B HA and NA coding regions were rescued using plasmids expressing the open reading frames of influenza B HA and NA genes flanked by genome packaging signals of influenza A HA66 and NA segments67, respectively. These viruses have a PB1 segment modified to express the TdKatushka2 reporter gene and encode the internal genes of influenza A (A/WSN/1933, H1N1) virus. Rescued viruses were propagated in MDCK-SIAT1-PB1 in the presence of TPCK-treated trypsin (1 μg ml−1, Sigma) at 34°C. Virus stocks were stored at −80°C. Mouse sera were treated with receptor destroying enzyme (RDE II; Denka Seiken) and heat-inactivated before use in neutralization assays. Immune sera or mAbs were serially diluted and incubated for 1 h at 37°C with pre-titrated viruses (Supplementary Table 4). Serum-virus mixtures were then transferred to 96-well plates (PerkinElmer), and 1.0×104 MDCK-SIAT1-PB1 cells28,68 were added into each well. After overnight incubation at 37°C, the number of fluorescent cells in each well was counted automatically using a Celigo image cytometer (Nexcelom Biosciences). For neutralization competition assays, mouse immune sera were pre-incubated with H1 MI15 HA, H1 CA09 stem HA, or irrelevant RSV F protein at a final concentration of 50 μg ml−1 at room temperature for 1 h prior to use in the reporter-based microneutralization assay described above. IC50 values, defined as the serum dilution or antibody concentration that gives 50% reduction in virus-infected cells, were calculated from neutralization curves using a four-parameter nonlinear regression model and plotted with GraphPad Prism (v8.0).
Pseudovirus neutralization assay
Pseudovirus neutralization assays were carried out using luciferase-encoding lentiviruses pseudotyped with influenza HA and NA, as described previously69,70. The HA and NA sequences used to generate the pseudoviruses were derived from H1N1 PR8 and H3N2 PH82 (Supplementary Table 4). Briefly, mouse sera were treated with receptor destroying enzyme (RDE (II); SEIKEN Accurate Chemical and Scientific) and heat-inactivated before use in assays. Immune sera were serially diluted and incubated with pre-titrated HA-NA pseudotyped viruses for 30 min at room temperature. Serum-pseudovirus mixtures were then transferred to 96-well white/black isoplates (PerkinElmer), and 12,000 293A cells were added into each well of the plate. After overnight incubation at 37°C, wells were supplemented with 100 μl of fresh Dulbecco’s Modified Eagle Medium including 5% fetal bovine serum (Fisher Scientific) and 5,000 units ml−1 penicillin-streptomycin (Gibco), and the plates were incubated in a static 37°C, 5% CO2, humidified incubator for 48 h. Cells were lysed with cell culture lysis buffer (Promega) and luciferase activity in the lysate was measured using Luciferase kit (Promega). Luminescence was measured with a Spectramax L luminometer (Molecular Devices). IC50 values, defined as the serum dilution or antibody concentration that gives 50% reduction in virus-infected cells, were calculated from neutralization curves using a four-parameter nonlinear regression model and plotted with GraphPad Prism (v8.0).
Hemagglutination inhibition assay
HAI titer to vaccine-matched viruses were tested with immune sera. The reporter influenza viruses H1N1 MI15, H3N2 HK14, B/Vic CO17, and B/Yam PH13 (Supplementary Table 4) were propagated in Madin-Darby canine kidney (MDCK) cells. Immune sera were treated with receptor-destroying enzyme (RDE II; Denka Seiken) before use in HAI assays. Immune sera were serially diluted and incubated with viruses (four hemagglutination unit per well) and then incubated with 0.5% turkey or guinea pig (for H3N2 HK14 virus only) red blood cells (Lampire Biological Laboratories) for 30 min at room temperature. The HAI titer of the sample was determined based on the well with the last non-agglutinated appearance, immediately before an agglutination was observed.
Passive transfer
To generate hyper-immune Ig for passive transfer, the immune serum samples from each NHP were diluted 1:50 with PBS, added to protein A columns, and incubated overnight at 4°C. After washing the columns briefly, captured antibodies were eluted with low-pH IgG elution buffer (ThermoFisher Scientific) and the eluates were immediately neutralized by adding 1 M Tris-HCl (pH 8.0) to a final concentration of 100 mM. Purified polyclonal antibodies were dialyzed two times against PBS, concentrated to ~20 mg ml−1 and stored at −80°C until use. BALB/cAnNHsd mice (Envigo) were given intraperitoneally 0.2 mg of FI6v3 (approximately 10 mg kg−1) or 10 mg of purified polyclonal Ig from individual NHPs. Twenty-four hours later, the mice were infected intranasally with 25× or 10× LD50 of H5N1 or H7N9 viruses (Supplementary Table 4) at Bioqual. The animals were monitored twice daily for development of clinical signs of infection and weighed daily for 14 days. Any animals that lost 20% or more of their initial body weight were euthanized.
Preparation of polyclonal immunoglobulin antigen-binding fragments
To generate polyclonal Fab fragments for epitope mapping, the immune serum samples from each NHP were diluted with PBS and applied to protein A columns. After washing the columns, captured antibodies were eluted with 0.1 M glycine pH 3.5, and the eluates were immediately neutralized by adding Tris-HCl (pH 8.0) to a final concentration of 50 mM. Purified IgG was buffer-exchanged into PBS and concentrated to approximately 25 mg ml−1, and 250 μL of 2× digestion buffer (40 mM sodium phosphate pH 6.5, 20 mM EDTA, 40 mM cysteine) was added. 500 μL of resuspended immobilized papain resin (ThermoFisher Scientific) freshly washed in 1× digestion buffer (20 mM sodium phosphate, 10 mM EDTA, 20 mM cysteine, pH 6.5) was further added, and samples were shaken for 5 h at 37°C. The supernatant was separated from resin and mixed with 1 ml of 20 mM Tris pH 8.0. Resin was washed twice with 500 μL of 20 mM Tris pH 8.0 and supernatants from the washes were pooled with the original supernatant to increase sample yield. Pooled supernatants were sterile-filtered at 0.22 μm and applied to protein A columns. Unbound fractions were pooled, concentrated to approximately 10 mg ml−1, and dialyzed twice against 25 mM Tris pH 8.0 to remove excess phosphates and cysteine prior to sample preparation for EM. Final samples were confirmed by SDS-PAGE, flash-frozen, and stored at −80°C.
Electron microscopy polyclonal epitope mapping (EMPEM)
To prepare H1 HA/polyclonal Fab fragment complexes, 150-fold molar excesses of qsCocktail- or qsMosaic-I53_dn5-elicited antibody Fab fragments were incubated with H1 HA-foldon for 1 h at room temperature, and the complexes were purified on a Superdex 200 Increase 10/300 GL column. The purified complexes were adsorbed onto glow-discharged carbon-coated copper mesh grids for 60 s, stained with 2% uranyl formate for 30 s, and allowed to air dry. Grids were imaged using an FEI Tecnai Spirit 120 kV electron microscope equipped with a Gatan Ultrascan 4000 CCD Camera. The pixel size at the specimen level was 1.60 Å. Data collection was performed using Leginon52 with the majority of the data processing carried out in Appion71. 4,112 and 3,237 micrographs were collected for qsCocktail-I53_dn5- and qsMosaic-I53_dn5-elicited Fab/HA complexes respectively. The parameters of the contrast transfer function (CTF) were estimated using CTFFIND4 (ref. 72). All particles were picked in a reference-free manner using DoG Picker73. Reference-free 2D classification was used to select homogeneous subsets of particles using CryoSPARC74. 847,873 and 997,557 particles were subjected for 2D classification of qsCocktail-I53_dn5- and qsMosaic-I53_dn5-elicited Fab/HA complexes, respectively. During the 2D classification, 2D classes were visually inspected and particles from classes not showing clear structural features of Fab/HA complexes were discarded. The remaining particles were subsequently subjected to three rounds of ab initio 3D reconstructions and 3D classifications without any symmetry imposed using CryoSPARC. Only receptor binding domain, vestigial esterase domain, and stem-directed Abs were included in the calculations. Particles from these classes were separately subjected to 3D refinement using CryoSPARC. The head-binding Fabs of the different classes were similar, but most classes showed obvious asymmetric features. All 3D reconstructions were compared to three classes of structurally characterized anti-HA antibodies: (i) receptor binding domain-targeted Abs CH65 (PDB: 5UGY), C05 (PDB: 4FP8), F045–092 (PDB: 4O58), HC63 (PDB: 1KEN), 2G1 (PDB: 4HG4), 8M2 (PDB: 4HFU), 5J8 (PDB: 4M5Z), 1F1 (PDB: 4GXU), and S139/1 (PDB: 4GMS); (ii) vestigial esterase domain-targeted Abs H5M9 (PDB: 4MHJ) and CR8071 (PDB: 4FQJ); and (iii) stem-binding Abs C179 (PDB: 4HLZ), CR6261 (PDB: 3GBN), CR8043 (PDB: 4NM8), CR8020 (PDB: 3SDY), CR9114 (PDB: 4FQI), FI6v3 (PDB: 3ZTJ), MEDI8852 (PDB: 5JW4), and 39.29 (PDB: 4KVN). Estimates of the fraction of particles containing receptor binding domain-, vestigial esterase domain-, and stem-binding Fabs were based on the number of particles clustered in each group. Particles containing Fabs bound to multiple sites were counted against each site. In Figure 5c, the coordinates of an H1 HA crystal structure (PDB 1RUZ) and a Fab fragment (PDB 3GBN) were fitted into the EM densities.
H5 HA/polyclonal Fab fragment complexes were prepared and verified by negative stain electron microscopy as described above and then pooled and concentrated. 3 μl of 0.1 mg ml−1 H5 HA in complex with qsMosaic-I53_dn5-elicited antibody Fab fragments was loaded onto a freshly glow-discharged (30 s at 20 mA) 1.2/1.3 UltraFoil grid (300 mesh) with a thin layer of evaporated continuous carbon prior to plunge freezing using a vitrobot Mark IV (ThermoFisher Scientific) using a blot force of −1 and 2.5 second blot time at 100% humidity and 25°C. Data were acquired using the an FEI Titan Krios transmission electron microscope operated at 300 kV and equipped with a Gatan K2 Summit direct detector and Gatan Quantum GIF energy filter, operated in zero-loss mode with a slit width of 20 eV. Automated data collection was carried out using Leginon at a nominal magnification of 130,000× with a pixel size of 0.525 Å. The dose rate was adjusted to 8 counts/pixel/s, and each movie was acquired in super-resolution mode fractionated in 50 frames of 200 ms. 2,374 micrographs were collected using beam-image shift with a defocus range between −1.0 and −2.5 μm. Movie frame alignment, estimation of the microscope contrast-transfer function parameters, particle picking, and extraction were carried out using Warp. Particle images were extracted with a box size of 800 pixels2 binned to 400 pixels2 yielding a pixel size of 1.05 Å. Two rounds of reference-free 2D classification were performed using CryoSPARC to select well-defined particle images. These selected particles were subjected to 3D refinement in CryoSPARC applying C3 symmetry using a map generated from a crystal structure of H5 HA (PDB 5JW4) low-pass filtered at 30 Å resolution. For beam tilt correction, the micrographs were grouped into beam tilt groups using beam-image shift values from Leginon. Beam tilt refinement was performed in Relion3.0 (ref. 75). CTF refinement was used to refine per-particle defocus values. Particle images were subjected to the Bayesian polishing procedure implemented in Relion3.0 (ref. 76). After determining a refined 3D structure, the particles were then subjected to 3D classification without refining angles and shifts using a soft mask on three Fab regions and with a tau value of 20 using Relion. 3D refinements were carried out using non-uniform refinement along with per-particle defocus refinement in CryoSPARC77. Local resolution estimation, filtering, and sharpening was carried out using CryoSPARC. Reported resolutions are based on the gold-standard Fourier shell correlation (FSC) of 0.143 criterion and Fourier shell correlation curves were corrected for the effects of soft masking by high-resolution noise substitution. In Figure 5e, the coordinates of an H5 HA crystal structure in complex with MEDI8852 Fab (PDB 5JW4) were fitted into the EM densities.
Statistics and Reproducibility
Multi-group comparisons were performed using nonparametric Kruskal-Wallis test with Dunn’s post-hoc analysis in Prism 8 (GraphPad) unless mentioned otherwise. Differences were considered significant when P values were less than 0.05. Statistical test to compare multiple Kaplan-Meier curves was carried out by using Mantel-Cox log-rank test with Bonferroni correction. Statistical methods and P value ranges can be found in the Figures and Figure legends. All figures were compiled in Inkscape (v1.0).
Data availability
All images and data were generated and analyzed by the authors, and will be made available by the corresponding authors (B.S.G., N.P.K., and M.K.) upon reasonable request. Uncropped images of all gels are provided in Supplementary Figure 1. Structural models and density maps have been deposited in the Protein Data Bank and Electron Microscopy Data Bank under accession numbers EMD-22935 (H1-I53_dn5 nanoparticle), EMD-22937 and PDB 7KNA (localized reconstruction of H1 HA), EMD-22940 (H5 HA bound to 3 polyclonal Fabs), EMD-22939 (H5 HA bound to 2 polyclonal Fabs), and EMD-22938 (H5 HA bound to 1 polyclonal Fab).
Extended Data
Extended Data Fig. 1 |. Production and characterization of HA-I53_dn5 components and nanoparticle immunogens.
a, SEC purification of seasonal HAs fused to I53_dn5B trimeric components, using a Superdex 200 Increase 10/300 GL column. b, Reducing and non-reducing SDS-PAGE of SEC-purified trimeric HA-I53_dn5B fusions, pentameric I53_dn5A component, and I53_dn5B trimer lacking fused HA. c, SEC purification of nanoparticle immunogens after in vitro assembly, including I53_dn5 lacking displayed antigen, using a Superose 6 Increase 10/300 GL column. The nanoparticle immunogens elute at the void volume of the column, while I53_dn5 is resolved. Residual, unassembled trimeric and pentameric components elute around 15 mL and 18 mL, respectively. d, Reducing and non-reducing SDS-PAGE of SEC-purified nanoparticle immunogens and I53_dn5. e, Antigenic characterization of purified nanoparticle immunogens by ELISA. Symbols indicate the specificity of each mAb. AUC, area under the curve. f, Analytical SEC of purified nanoparticle immunogens, compared to I53_dn5 nanoparticles lacking displayed antigen and trimeric H1-I53_dn5B, using a Sephacryl S-500 HR 16/60 column. g, Dynamic light scattering (DLS) of SEC-purified nanoparticle immunogens, including I53_dn5. Dh, hydrodynamic diameter; Pd, polydispersity. h, Representative electron micrograph of H1-I53_dn5 embedded in vitreous ice. Scale bar, 100 nm. i, 2D class averages obtained using single-particle cryo-EM. Scale bar, 20 nm. j, Gold-standard Fourier shell correlation curve for the H1-I53_dn5 density map presented in Fig. 1c. k, Gold-standard Fourier shell correlation curve for the localized reconstruction of H1 MI15 presented in Fig. 1c. All experiments except for electron microscopy data collection and processing were performed at least twice.
Extended Data Fig. 2 |. Hydrogen-deuterium exchange mass spectrometry (HDX-MS) of H1-foldon trimer and H1-I53_dn5 nanoparticle.
a, Amino acid sequence of H1 ectodomain expressed as a genetic fusion to both foldon and I53_dn5B. Underlined sequences correspond to peptides analyzed by HDX-MS. b, Hydrogen-deuterium exchange percentages after 20 h for both samples mapped onto the structure of H1 HA (PDB 3LZG). c, Kinetics of hydrogen-deuterium exchange for both samples at multiple timepoints up to 20 h. *, peptides where a negative percent exchange was corrected to zero (< 2% magnitude correction); **, peptides that were missing a replicate at the 30 min timepoint.
Extended Data 3 |. Controllable co-display of multiple antigenic variants on two-component nanoparticle immunogens.
a, Sandwich BLI comparing qsCocktail-I53_dn5 and qsMosaic-I53_dn5. Biotinylated 5J8 immobilized on streptavidin probes was used to capture H1-containing nanoparticles from each sample. The captured particles were then exposed to antibodies specific to H3 (CR8020; left) or influenza B HA (CR8071; right). b, Numerical approximation of the H1 HA content of individual qsMosaic-I53_dn5 nanoparticles assuming an equimolar quadrivalent in vitro assembly reaction (i.e., 25% of the input HA-I53_dn5B trimers bear H1 HA) and random incorporation of each HA-I53_dn5B trimer at each of the 20 trimeric positions into the nanoparticle. A distribution centered on 25% valency (5 H1 HA trimers per nanoparticle) is observed. c, Calculation of the fraction of individual mosaic nanoparticles displaying at least one H1 HA trimer as a function of the fractional concentration of H1-I53-dn5B in the in vitro assembly reaction ([H1]), expressed as: 1 - (1 - [H1])20. At the 25% fractional concentration used to assemble qsMosaic-I53_dn5, 99.7% of the individual nanoparticles are expected to display at least one H1 HA trimer. d, Quantitation of HA antigen content by peptide mass spectrometry in three distinct qsMosaic-I53_dn5 nanoparticles with various antigen ratios before and after preparative SEC. Dashed lines represent the fractional concentration of each HA in the in vitro assembly reactions used to prepare the mosaic nanoparticle immunogens, main bars represent the mean values of four unique peptides from each HA, and error bars represent the standard deviation of measurements across the four unique peptides from each HA. The peptides used to quantify each HA are provided in Supplementary Table 3.
Extended Data Fig. 4 |. Vaccine-elicited antibody responses against vaccine-matched antigens.
HA-specific antibody titers in immunized mice (a), ferrets (b), and NHPs (c). Immunization schemes are shown at the top of each panel. All immunizations were given intramuscularly with AddaVax. Groups of BALB/cJ mice (N = 10), Finch ferrets (N = 9), and rhesus macaques (N = 4) were used in each experiment. ELISA antibody titers are expressed as endpoint dilutions. Each symbol represents an individual animal and the horizontal bar indicates the geometric mean of the group. Individual NHPs are identified by unique symbols. d, Antibody responses against unmodified I53_dn5 nanoparticles lacking displayed HA. Immunization scheme is shown at the top of the panel. Groups of NHPs (N = 4) were immunized three times with either QIV, qsCocktail-I53-dn5, or qsMosaic-I53_dn5 with AddaVax at weeks 0, 8 and 16. Serum samples were collected 2 weeks after each immunization and tested for ELISA binding antibody against unmodified I53_dn5 particles. Antibody titers are expressed as endpoint dilutions. Individual NHPs are identified by unique symbols. The immunization study was performed once. e, Antibody responses against vaccine-matched antigens and viruses elicited by unadjuvanted vaccines in immunized mice. Immunization scheme is shown. All immunizations were given intramuscularly. Groups of BALB/cJ mice (N = 10) were used. HA-specific ELISA binding antibody (top), hemagglutination inhibition (HAI) (middle), and microneutralization titers (bottom) in immune sera are shown. Microneutralization titers are reported as half maximal inhibitory dilution (IC50). Each symbol represents an individual animal and the horizontal bar indicates the geometric mean of the group. Statistical analysis was performed using nonparametric Kruskal–Wallis test with Dunn’s multiple comparisons. All animal experiments except for NHP were performed at least twice and representative data are shown.
Extended Data Fig. 5 |. Antibody responses against vaccine-matched antigens and viruses elicited by 2018–2019 vaccines.
a, Immunization scheme. The commercial QIV, qsCocktail-I53_dn5, and qsMosaic-I53_dn5 vaccines used in this study comprised the WHO-recommended 2018–2019 vaccine strains. Sequences for the HA-I53_dn5B fusion proteins—H1-I53_dn5, SG16-I53_dn5 (updated H3), B/Yam-I53_dn5, and CO17-I53_dn5 (updated B/Vic)—are provided in Supplementary Table 1. All immunizations were given intramuscularly with AddaVax. Groups of BALB/cJ mice (N = 10) were used. b, HA-specific antibody, c, hemagglutination inhibition (HAI), and d, microneutralization titers in immune sera. Microneutralization titers are reported as half maximal inhibitory dilution (IC50). e, Heterosubtypic HA-specific antibody titers in immune sera. Each symbol represents an individual animal and the horizontal bar indicates the geometric mean of the group. Statistical analysis was performed using nonparametric Kruskal–Wallis test with Dunn’s multiple comparisons. The animal experiment was performed once.
Extended Data Fig. 6 |. Neutralization of historical H1N1 and H3N2 viruses.
Immunization scheme for ferret study is shown. Groups of Finch ferrets (N = 9) were used. Phylogenetic trees of HA sequences of human H1N1 (left) and H3N2 (right) viruses are shown (see Supplementary Table 4). Each symbol represents an individual animal and the horizontal bar indicates the geometric mean of the group. Statistical analysis was performed using nonparametric Kruskal–Wallis test with Dunn’s multiple comparisons. The ferret experiment was performed twice and representative data are shown.
Extended Data Fig. 7 |. Antibody responses elicited by a non-assembling immunogen.
a, Model of the I53_dn5B trimer, with the computationally designed interface that drives nanoparticle assembly indicated by the solid line (top), and the 1na0C3_int2 trimer, in which the interface mutations were reverted to their original identities (bottom). The dotted line indicates the inability of this molecule to drive nanoparticle assembly. b, Analytical SEC of the non-assembling immunogen (a mixture of four HA-1na0C3_int2 trimers with pentameric I53_dn5A) using a Superose 6 Increase 10/300 GL column. Only unassembled oligomeric components were observed. c, Reducing and non-reducing SDS-PAGE analysis of the non-assembling immunogen before and after analytical SEC. d, Negative stain EM of the non-assembling immunogen, which confirmed the absence of higher-order structures indicated by analytical SEC. Scale bar, 100 nm. e, Immunization scheme in mice. All immunizations were given intramuscularly with AddaVax. Groups of BALB/cJ mice (N = 10) were used in the experiment. f, Microneutralization titers in immune sera against vaccine-matched or slightly mismatched viruses. Microneutralization titers are reported as half maximal inhibitory dilution (IC50). g, Cross-reactive antibody titers in immune sera. Each symbol represents an individual animal and the horizontal bar indicates the geometric mean of the group. Statistical analysis was performed using nonparametric Kruskal–Wallis test with Dunn’s multiple comparisons. All experiments were performed once.
Extended Data Fig. 8 |. NS-EM analysis of H1 HA complexed with polyclonal antibody Fabs prepared from NHPs immunized with qsCocktail-I53_dn5 or qsMosaic-I53_dn5.
a, NS-EM analysis of Fabs obtained from qsCocktail-I53_dn5-immunized NHPs in complex with recombinant H1 MI15 HA trimers. Two-dimensional classifications were generated using 847,873 particles collected from 4,112 micrographs. The frequencies of complexes containing Fab fragments bound to RBD (81%), VE (18%), or stem (1%) domains are presented as pie charts in Fig. 5c. b, NS-EM analysis of Fabs obtained from qsMosaic-I53_dn5-immunized NHPs in complex with recombinant H1 MI15 HA trimers. Two-dimensional classifications were generated using 997,557 particles collected from 3,237 micrographs. The frequencies of complexes containing Fab fragments bound to RBD (69%), VE (24%), or stem (7%) domains are presented as pie charts in Fig. 5c. Upper part of each panel shows representative reference-free 2D class averages. Scale bars, 20 nm. Lower part of each panel shows seven representative 3D reconstructions of HA–Fab complexes. Single complexes containing Fabs of multiple specificities were counted once against each specificity. The coordinates of an H1 HA crystal structure (PDB 1RUZ) and a Fab fragment (PDB 3GBN) were fitted into the EM densities. Light blue ribbons, H1 HA; cyan or magenta ribbons, Fabs. All experiments were performed once.
Extended Data Fig. 9 |. CryoEM analysis of heterosubtypic H5 HA in complex with polyclonal antibody Fab fragments prepared from NHP immunized with qsMosaic-I53_dn5.
a, Representative cryo-electron micrograph. Scale bar, 100 nm. b, Reference-free 2D class averages. Scale bar, 20 nm. c, Gold-standard Fourier shell correlation (FSC) curve for the asymmetric reconstruction shown in d. d, Asymmetric cryo-EM reconstruction of H5 HA–Fab complexes with Fab fragments bound to all three HA subunits at 3.6 Å resolution. The reconstruction is the same as that shown in the right panel in Fig. 5d, but here is colored by local resolution. e, FSC curve for the asymmetric reconstruction shown in f. f, Two orthogonal orientations of an asymmetric cryo-EM reconstruction of H5 HA–Fab complexes with Fab fragments bound to two HA subunits at 4.1 Å resolution. g, FSC curve for the asymmetric reconstruction shown in h. h, Two orthogonal orientations of an asymmetric cryo-EM reconstruction of H5 HA–Fab complexes with Fab fragments bound to one HA subunit at 4.0 Å resolution. The reconstruction is the same as that shown in the left panel in Fig. 5d. All experiments were performed once.
Extended Data Fig. 10 |. Vaccine-elicited antibody responses against vaccine-matched viruses in NHPs with pre-existing influenza immunity.
Immunization scheme for NHP study shown on the left. NHPs (N = 3) that had been immunized three times with either QIV 2017–2018 or qsMosaic-I53_dn5 2017–2018 were boosted 63 weeks later with a single dose (60 μg) of updated qsMosaic-I53_dn5 2018–2019. All immunizations were given intramuscularly with AddaVax. Microneutralization titers are reported as half maximal inhibitory dilution (IC50). Each symbol represents an individual animal and the horizontal bar indicates the geometric mean of the group. Individual NHPs are identified by unique symbols. Statistical analysis was performed using paired t test.
Supplementary Material
Acknowledgments
The authors thank K. Foulds, A. Noe, S.-F. Kao, V. Ficca, N. Nji, D. Flebbe, and E. McCarthy (VRC) for help with nonhuman primate experiments; A. Taylor, H. Bao, C. Chiedi, M. Dillon, L. Gilman, and G. Sarbador, E. McCarthy, J.-P. Todd, D. Scorpio (VRC) for help with mouse, ferret, and nonhuman primate experiments; H. Andersen, N. Jones, and G. Patel (Bioqual) for help with influenza challenge studies; R. Webby (St. Jude Children’s Research Hospital) for providing influenza reverse genetics plasmids; Y. Tsybovsky and T. Stephens (Frederick National Laboratory for Cancer Research) for initial EM screening; A. Reers and P. Myler (Seattle Children’s Research Institute) for assistance with protein production; and members of the King laboratory and the Influenza Program at the VRC for comments on the manuscript. This study was supported by the intramural research program of the Vaccine Research Center, National Institute of Allergy and Infectious Diseases, National Institutes of Health (M.K. and B.S.G.); a generous gift from the Open Philanthropy Project (D.B. and N.P.K.); a generous gift from the Audacious Project (D.B. and N.P.K.); the Defense Threat Reduction Agency (HDTRA1-18-1-0001; D.B. and N.P.K.); the National Institute of General Medical Sciences (R01GM120553, D.V.); the National Institute of Allergy and Infectious Diseases (DP1AI158186 and HHSN272201700059C, D.V.); a Pew Biomedical Scholars Award (D.V.); an Investigators in the Pathogenesis of Infectious Disease Award from the Burroughs Wellcome Fund (D.V.); and the University of Washington Arnold and Mabel Beckman cryo-EM center. Molecular graphics and analyses performed with UCSF ChimeraX, developed by the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco, with support from National Institutes of Health R01-GM129325 and the Office of Cyber Infrastructure and Computational Biology, National Institute of Allergy and Infectious Diseases.
Footnotes
Competing interests S.B.B., D.E., R.A.G., G.U., B.S.G., N.P.K., and M.K. are listed as inventors on a patent application based on the studies presented in this paper. D.V. is a consultant for Vir Biotechnology Inc. The Veesler laboratory has received an unrelated sponsored research agreement from Vir Biotechnology Inc. N.P.K. is a co-founder, shareholder, and chair of the scientific advisory board of Icosavax, Inc. The King laboratory has received an unrelated sponsored research agreement from Pfizer. D.B. is a co-founder and shareholder of Icosavax, Inc. All other authors declare no competing interests.
References
- 1.Wei C-J et al. Next-generation influenza vaccines: opportunities and challenges. Nat. Rev. Drug Discov. 19, 239–252 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ueda G et al. Tailored design of protein nanoparticle scaffolds for multivalent presentation of viral glycoprotein antigens. Elife 9, e57659 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kanekiyo M & Graham BS Next-Generation Influenza Vaccines. Cold Spring Harb. Perspect. Med. (2020) doi: 10.1101/cshperspect.a038448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Iuliano AD et al. Estimates of global seasonal influenza-associated respiratory mortality: a modelling study. Lancet 391, 1285–1300 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Flannery B et al. Interim Estimates of 2017–18 Seasonal Influenza Vaccine Effectiveness - United States, February 2018. MMWR Morb. Mortal. Wkly. Rep. 67, 180–185 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ellebedy AH et al. Induction of broadly cross-reactive antibody responses to the influenza HA stem region following H5N1 vaccination in humans. Proc. Natl. Acad. Sci. U. S. A. 111, 13133–13138 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Andrews SF et al. Immune history profoundly affects broadly protective B cell responses to influenza. Sci. Transl. Med. 7, 316ra192 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tan H-X et al. Subdominance and poor intrinsic immunogenicity limit humoral immunity targeting influenza HA stem. Journal of Clinical Investigation vol. 129 850–862 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yassine HM et al. Hemagglutinin-stem nanoparticles generate heterosubtypic influenza protection. Nat. Med. 21, 1065–1070 (2015). [DOI] [PubMed] [Google Scholar]
- 10.Impagliazzo A et al. A stable trimeric influenza hemagglutinin stem as a broadly protective immunogen. Science 349, 1301–1306 (2015). [DOI] [PubMed] [Google Scholar]
- 11.Corbett KS et al. Design of Nanoparticulate Group 2 Influenza Virus Hemagglutinin Stem Antigens That Activate Unmutated Ancestor B Cell Receptors of Broadly Neutralizing Antibody Lineages. MBio 10, e02810–18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boyoglu-Barnum S et al. Glycan repositioning of influenza hemagglutinin stem facilitates the elicitation of protective cross-group antibody responses. Nat. Commun. 11, 791 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Steel J et al. Influenza virus vaccine based on the conserved hemagglutinin stalk domain. MBio 1, e00018–10 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bommakanti G et al. Design of an HA2-based Escherichia coli expressed influenza immunogen that protects mice from pathogenic challenge. Proc. Natl. Acad. Sci. U. S. A. 107, 13701–13706 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krammer F, Pica N, Hai R, Margine I & Palese P Chimeric hemagglutinin influenza virus vaccine constructs elicit broadly protective stalk-specific antibodies. J. Virol. 87, 6542–6550 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marcandalli J et al. Induction of Potent Neutralizing Antibody Responses by a Designed Protein Nanoparticle Vaccine for Respiratory Syncytial Virus. Cell 176, 1420–1431.e17 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kanekiyo M et al. Self-assembling influenza nanoparticle vaccines elicit broadly neutralizing H1N1 antibodies. Nature 499, 102–106 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.López-Sagaseta J, Malito E, Rappuoli R & Bottomley MJ Self-assembling protein nanoparticles in the design of vaccines. Comput. Struct. Biotechnol. J. 14, 58–68 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tokatlian T et al. Innate immune recognition of glycans targets HIV nanoparticle immunogens to germinal centers. Science 363, 649–654 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kanekiyo M et al. Mosaic nanoparticle display of diverse influenza virus hemagglutinins elicits broad B cell responses. Nat. Immunol. 20, 362–372 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cohen AA et al. Mosaic nanoparticles elicit cross-reactive immune responses to zoonotic coronaviruses in mice. Science (2021) doi: 10.1126/science.abf6840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Georgiev IS et al. Two-Component Ferritin Nanoparticles for Multimerization of Diverse Trimeric Antigens. ACS Infect Dis 4, 788–796 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cohen AA et al. Construction, characterization, and immunization of nanoparticles that display a diverse array of influenza HA trimers. bioRxiv 2020.01.18.911388 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.King NP et al. Accurate design of co-assembling multi-component protein nanomaterials. Nature 510, 103–108 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bale JB et al. Accurate design of megadalton-scale two-component icosahedral protein complexes. Science 353, 389–394 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martín J et al. Studies of the binding properties of influenza hemagglutinin receptor-site mutants. Virology 241, 101–111 (1998). [DOI] [PubMed] [Google Scholar]
- 27.Whittle JRR et al. Flow cytometry reveals that H5N1 vaccination elicits cross-reactive stem-directed antibodies from multiple Ig heavy-chain lineages. J. Virol. 88, 4047–4057 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Creanga A et al. A comprehensive influenza reporter virus panel for high-throughput deep profiling of neutralizing antibodies. bioRxiv 2020.02.24.963611 (2020) doi: 10.1101/2020.02.24.963611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Corti D et al. A neutralizing antibody selected from plasma cells that binds to group 1 and group 2 influenza A hemagglutinins. Science 333, 850–856 (2011). [DOI] [PubMed] [Google Scholar]
- 30.Bianchi M et al. Electron-Microscopy-Based Epitope Mapping Defines Specificities of Polyclonal Antibodies Elicited during HIV-1 BG505 Envelope Trimer Immunization. Immunity 49, 288–300.e8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kallewaard NL et al. Structure and Function Analysis of an Antibody Recognizing All Influenza A Subtypes. Cell 166, 596–608 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Joyce MG et al. Vaccine-Induced Antibodies that Neutralize Group 1 and Group 2 Influenza A Viruses. Cell 166, 609–623 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wei C-J et al. Induction of broadly neutralizing H1N1 influenza antibodies by vaccination. Science 329, 1060–1064 (2010). [DOI] [PubMed] [Google Scholar]
- 34.Darricarrère N et al. Development of a Pan-H1 Influenza Vaccine. J. Virol. 92, e01349–18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Giles BM & Ross TM A computationally optimized broadly reactive antigen (COBRA) based H5N1 VLP vaccine elicits broadly reactive antibodies in mice and ferrets. Vaccine 29, 3043–3054 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Broecker F et al. A mosaic hemagglutinin-based influenza virus vaccine candidate protects mice from challenge with divergent H3N2 strains. NPJ Vaccines 4, 31 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sun W et al. Development of Influenza B Universal Vaccine Candidates Using the ‘Mosaic’ Hemagglutinin Approach. J. Virol. 93, e00333–19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ng S et al. Novel correlates of protection against pandemic H1N1 influenza A virus infection. Nat. Med. 25, 962–967 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fonville JM et al. Antibody landscapes after influenza virus infection or vaccination. Science 346, 996–1000 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gostic KM, Ambrose M, Worobey M & Lloyd-Smith JO Potent protection against H5N1 and H7N9 influenza via childhood hemagglutinin imprinting. Science 354, 722–726 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Additional References
- 41.Throsby M et al. Heterosubtypic neutralizing monoclonal antibodies cross-protective against H5N1 and H1N1 recovered from human IgM+ memory B cells. PLoS One 3, e3942 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hong M et al. Antibody recognition of the pandemic H1N1 Influenza virus hemagglutinin receptor binding site. J. Virol. 87, 12471–12480 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ekiert DC et al. A highly conserved neutralizing epitope on group 2 influenza A viruses. Science 333, 843–850 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Iba Y et al. Conserved neutralizing epitope at globular head of hemagglutinin in H3N2 influenza viruses. J. Virol. 88, 7130–7144 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee PS et al. Receptor mimicry by antibody F045–092 facilitates universal binding to the H3 subtype of influenza virus. Nat. Commun. 5, 3614 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dreyfus C et al. Highly conserved protective epitopes on influenza B viruses. Science 337, 1343–1348 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu Y et al. A potent broad-spectrum protective human monoclonal antibody crosslinking two haemagglutinin monomers of influenza A virus. Nat. Commun. 6, 7708 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kwakkenbos MJ et al. Generation of stable monoclonal antibody-producing B cell receptor-positive human memory B cells by genetic programming. Nat. Med. 16, 123–128 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Corti D et al. Cross-neutralization of four paramyxoviruses by a human monoclonal antibody. Nature 501, 439–443 (2013). [DOI] [PubMed] [Google Scholar]
- 50.Studier FW & William Studier F Protein production by auto-induction in high-density shaking cultures. Protein Expr. Purif. 41, 207–234 (2005). [DOI] [PubMed] [Google Scholar]
- 51.Snijder J et al. Vitrification after multiple rounds of sample application and blotting improves particle density on cryo-electron microscopy grids. J. Struct. Biol. 198, 38–42 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Suloway C et al. Automated molecular microscopy: the new Leginon system. J. Struct. Biol. 151, 41–60 (2005). [DOI] [PubMed] [Google Scholar]
- 53.Tegunov D & Cramer P Real-time cryo-electron microscopy data preprocessing with Warp. Nat. Methods 16, 1146–1152 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ilca SL et al. Localized reconstruction of subunits from electron cryomicroscopy images of macromolecular complexes. Nat. Commun. 6, 8843 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Emsley P, Lohkamp B, Scott WG & Cowtan K Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Frenz B et al. Automatically Fixing Errors in Glycoprotein Structures with Rosetta. Structure 27, 134–139.e3 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang RY-R et al. Automated structure refinement of macromolecular assemblies from cryo-EM maps using Rosetta. Elife 5, e17219 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen VB et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liebschner D et al. Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Crystallogr D Struct Biol 75, 861–877 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Agirre J et al. Privateer: software for the conformational validation of carbohydrate structures. Nat. Struct. Mol. Biol. 22, 833–834 (2015). [DOI] [PubMed] [Google Scholar]
- 61.Barad BA et al. EMRinger: side chain-directed model and map validation for 3D cryo-electron microscopy. Nat. Methods 12, 943–946 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Scheres SHW & Chen S Prevention of overfitting in cryo-EM structure determination. Nat. Methods 9, 853–854 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Verkerke HP et al. Epitope-Independent Purification of Native-Like Envelope Trimers from Diverse HIV-1 Isolates. J. Virol. 90, 9471–9482 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Guttman M, Weis DD, Engen JR & Lee KK Analysis of overlapped and noisy hydrogen/deuterium exchange mass spectra. J. Am. Soc. Mass Spectrom. 24, 1906–1912 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Weis DD, Engen JR & Kass IJ Semi-automated data processing of hydrogen exchange mass spectra using HX-Express. J. Am. Soc. Mass Spectrom. 17, 1700–1703 (2006). [DOI] [PubMed] [Google Scholar]
- 66.Martínez-Sobrido L et al. Hemagglutinin-pseudotyped green fluorescent protein-expressing influenza viruses for the detection of influenza virus neutralizing antibodies. J. Virol. 84, 2157–2163 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gao Q et al. The influenza A virus PB2, PA, NP, and M segments play a pivotal role during genome packaging. J. Virol. 86, 7043–7051 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bloom JD, Gong LI & Baltimore D Permissive secondary mutations enable the evolution of influenza oseltamivir resistance. Science 328, 1272–1275 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kong W-P et al. Protective immunity to lethal challenge of the 1918 pandemic influenza virus by vaccination. Proc. Natl. Acad. Sci. U. S. A. 103, 15987–15991 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yang Z-Y et al. Immunization by avian H5 influenza hemagglutinin mutants with altered receptor binding specificity. Science 317, 825–828 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lander GC et al. Appion: an integrated, database-driven pipeline to facilitate EM image processing. J. Struct. Biol. 166, 95–102 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rohou A & Grigorieff N CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Voss NR, Yoshioka CK, Radermacher M, Potter CS & Carragher B DoG Picker and TiltPicker: software tools to facilitate particle selection in single particle electron microscopy. J. Struct. Biol. 166, 205–213 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Punjani A, Rubinstein JL, Fleet DJ & Brubaker MA cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017). [DOI] [PubMed] [Google Scholar]
- 75.Zivanov J et al. New tools for automated high-resolution cryo-EM structure determination in RELION-3. Elife 7, e42166 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zivanov J, Nakane T & Scheres SHW A Bayesian approach to beam-induced motion correction in cryo-EM single-particle analysis. IUCrJ 6, 5–17 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Punjani A, Zhang H & Fleet DJ Non-uniform refinement: Adaptive regularization improves single particle cryo-EM reconstruction. Cold Spring Harbor Laboratory 2019.12.15.877092 (2019) doi: 10.1101/2019.12.15.877092. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All images and data were generated and analyzed by the authors, and will be made available by the corresponding authors (B.S.G., N.P.K., and M.K.) upon reasonable request. Uncropped images of all gels are provided in Supplementary Figure 1. Structural models and density maps have been deposited in the Protein Data Bank and Electron Microscopy Data Bank under accession numbers EMD-22935 (H1-I53_dn5 nanoparticle), EMD-22937 and PDB 7KNA (localized reconstruction of H1 HA), EMD-22940 (H5 HA bound to 3 polyclonal Fabs), EMD-22939 (H5 HA bound to 2 polyclonal Fabs), and EMD-22938 (H5 HA bound to 1 polyclonal Fab).