Abstract
Gastric cancer (GC) is a common cancerous tumor, and is the third leading cause of cancer mortality worldwide. Although comprehensive therapies of GC have been widely used in clinical set ups, advanced gastric cancer carries is characterized by poor prognosis, probably due to lack of effective prognostic biomarkers. Mammalian histone deacetylase family, histone deacetylases (HDACs), play significant roles in initiation and progression of tumors. Aberrant expression of HDACs is reported in many cancer types including gastric cancer, and may serve as candidate biomarkers or therapeutic targets for GC patients.
Gene Expression Profiling Interactive Analysis was used to explore mRNA levels of HDACs in GC. Kaplan–Meier plotter was used to determine the prognostic value of HDACs mRNA expression in GC. Genomic profiles including mutations of HDACs were retrieved from cBioPortal webserver. A protein–protein interaction network was constructed using STRING database. GeneMANIA was used to retrieve additional genes or proteins related to HDACs. R software was used for functional enrichment analyses.
Analysis of mRNA levels of HDAC1/2/4/8/9 showed that they were upregulated in GC tissues, whereas HDAC6/10 was downregulated in GC tissues. Aberrant expression of HDAC1/3/4/5/6/7/8/10/11 was all correlated with prognosis in GC. In addition, expression levels of HDACs were correlated with different Lauren classifications, and clinical stages, lymph node status, treatment, and human epidermal growth factor receptor 2 status in GC.
The findings of this study showed that HDAC members are potential biomarkers for diagnosis or prognosis of gastric cancer. However, further studies should be conducted to validate these findings.
Keywords: bioinformatic analysis, gastric cancer, HDAC, prognostic value
1. Introduction
Gastric cancer (GC) originates from epithelium mucosa of the stomach, and is the fifth most prevalent diagnosed cancer and the third leading cause of cancer-related mortality worldwide.[1] The 5-year survival of gastric cancer ranges approximately from 57% to 18% in different countries, owing to its high malignancy, invasion, and recurrence.[2,3] Currently, clinicians rely mainly on American Joint Committee on Cancer staging system for prognostic prediction of gastric cancer.[4] Despite advances in comprehensive therapies of GC including surgery, chemotherapy, radiation, and target therapy, the prognostic outcome remains poor. Therefore, there is need to explore novel diagnostic and prognostic indicators, to improve treatment of GC patients.
Mammalian histone deacetylase (HDAC) family comprises 18 members, which are subdivided into 4 classes based on their sequence homology and cofactor specificity. These classes include Class I histone deacetylases (HDACs) (HDAC1/2/3/8, homologous with yeast Rpd3), Class II HDACs (HDAC4/5/6/7/9/10, homologous with yeast Hda1), Class III HDACs or Sirtuins (SIRT1–7, homologous with yeast Sir2), and Class IV HDACs (HDAC11, homologous with both class I and class II).[5–8] Class I, II, and IV are referred to as “classical” HDACs, and their activities are Zn+-dependent, whereas Class III HDACs activity is NAD+-dependent (this family will not be subject of discussion in this article).[9,10] Previous studies report that HDACs are involved in proliferation, apoptosis, invasion, and migration of various cancer cell types, including gastric cancer cells, thus modulating carcinogenesis and cancer progression.[11–13] In addition, aberrant expression of HDACs is reported in gastric cancer, and may serve as candidate biomarkers and therapeutic targets for GC patients.[12,14–18] The potential role of HDACs in gastric cancer has been receiving increasing attention.
Bioinformatic analyses are widely used in genomics as a result of advances in microarray technology and establishment of online databases. In this study, bioinformatic analyses were performed using online databases to explore expression profiles and prognostic values of HDACs in GC.
2. Materials and methods
2.1. Gene expression analysis
mRNA levels of HDACs in GC were analyzed using Gene Expression Profiling Interactive Analysis (GEPIA) (http://gepia.cancer-pku.cn). GEPIA contains RNA sequence expression data of 9736 tumors and 8587 normal tissue samples.[19] Expression levels of HDACs between tumor and normal tissues were compared using Student's t test. Expression of HDACs in different pathological stages of GC was compared using F test. Statistical significance was defined as P < .01 and fold change > 2.
2.2. Prognosis analysis
Kaplan–Meier Plotter (http://www.kmplot.com) was used to analyze the prognostic value of HDACs mRNA expression. Kaplan–Meier Plotter contains gene expression data and survival information of 1440 clinical GC patients. To explore the overall survival (OS) of GC patients, samples were divided into low and high expression groups based on median mRNA levels, with a hazard ratio (HR) with 95% confidence intervals and log-rank P value. Kaplan–Meier survival analysis was then performed on the 2 groups. Log-rank P value < .05 was used to show statistical significance. Univariate cox analysis was conducted with adjustments to different Lauren classification, clinical stages, lymph node status, treatment, and human epidermal growth factor receptor 2 (HER2) status of GC.
2.3. Analysis of gene alteration and associated network construction
To explore gene alterations of HDACs in GC patients, genomic profiles including mutations were obtained from cBioPortal webserver for Cancer Genomics (http://www.cbioportal.org). Protein–protein interaction network analysis was performed on HDAC members using STRING database (https://string-db.org/), to explore potential interactions between HDACs. GeneMANIA tool (http://www.genemania.org) was used to retrieve additional genes or proteins related to HDACs.
2.4. Functional enrichment analysis
Functional enrichment analysis of HDACs were performed using gene ontology (GO) terms and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway were conducted and visualized in R software using “org.Hs.eg.db,” “clusterProfiler,” “pathview,” “Goplot,” and “ggplot2” packages. Level of significance was set at P-value < .05.
2.5. Ethical statement
All data were retrieved from open-access databases, not directly from patients or animals. Therefore, ethical approval was not necessary.
3. Results
3.1. Gene expression of HDACs in patients with GC
mRNA expression levels of HDACs in GC tissues were compared with those in normal tissues using GEPIA tool, which contains 408 GC samples and 211 normal gastric samples. Analysis showed differential expression of HDACs in GC tissues compared with normal tissues (Fig. 1). mRNA levels of HDAC1, HDAC2, HDAC4, HDAC8, and HDAC9 were upregulated in GC tissues compared with normal tissues (Fig. 2). On the other hand, analysis of mRNA expression showed that HDAC6 and HDAC10 were downregulated in GC tissue compared with normal tissues. Notably, box plots showed significance increase in mRNA expression levels of HDAC2 and HDAC4 mRNA levels. Analysis showed no differences for mRNA expression levels of the HDAC3/5/7/11 between GC and normal tissues. Moreover, expression of HDACs in different pathological stages of GC was explored. Expression levels of HDAC9 and HDAC11 varied significantly in different pathological stages, whereas expression of other HDAC members in various stages was not significantly different across pathological stages (Fig. 3).
Figure 1.

The relative level of HDACs in STAD (GEPIA). GEPIA = Gene Expression Profiling Interactive Analysis; HDACs = histone deacetylases.
Figure 2.
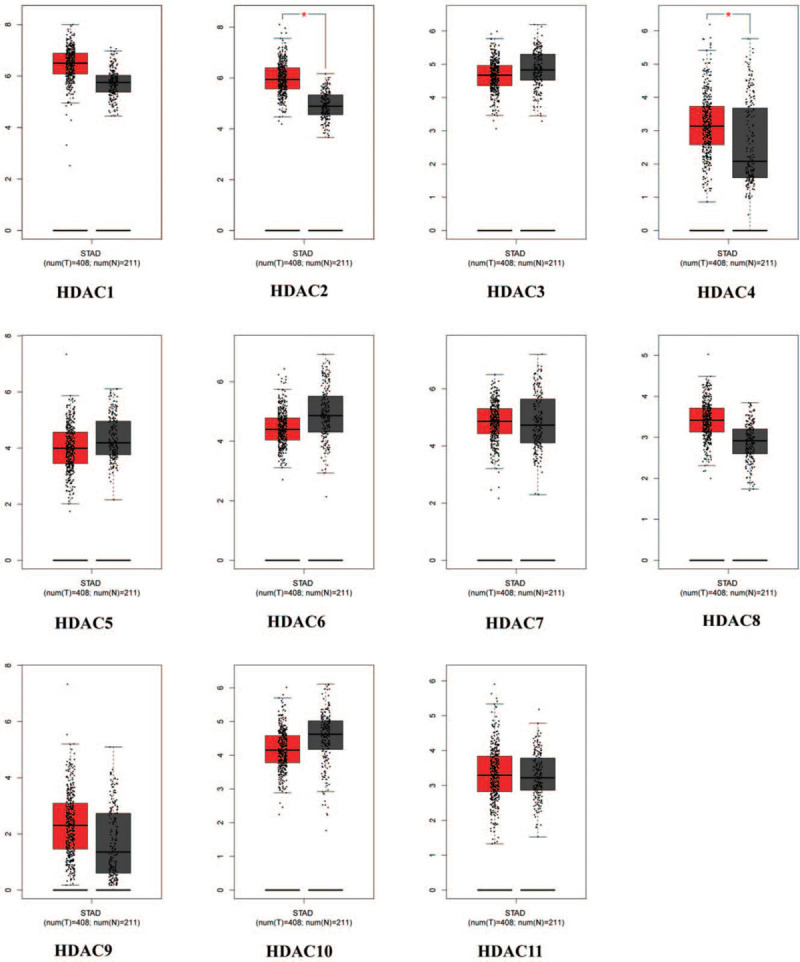
The mRNA expression of HDACs in GC patients (GEPIA). GC = gastric cancer; GEPIA = Gene Expression Profiling Interactive Analysis; HDACs = histone deacetylases.
Figure 3.
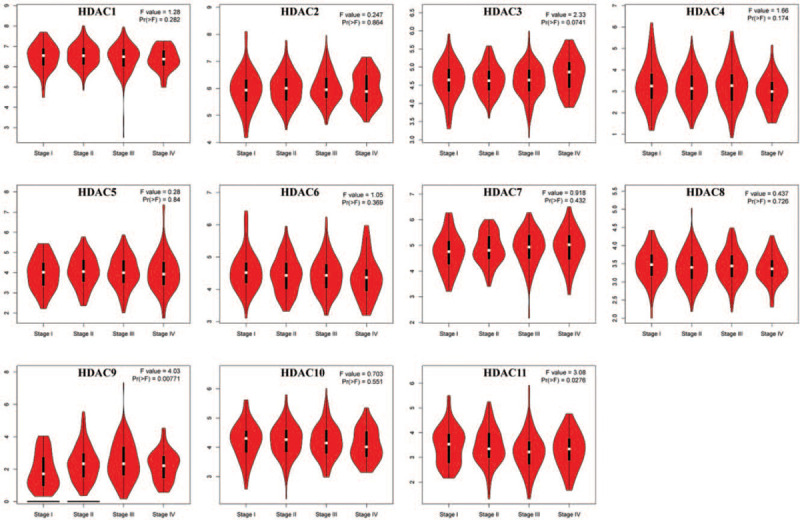
The expression of HDACs in different tumor stage of GC patients (GEPIA). GC = gastric cancer; GEPIA = Gene Expression Profiling Interactive Analysis; HDACs = histone deacetylases.
3.2. Prognostic value of HDACs in gastric cancer
Correlation analysis of the expression of HDACs and prognosis of GC patients was performed using Kaplan–Meier plotter. The findings showed that low mRNA expression of HDAC3 was correlated with favorable OS in all gastric cancer cases. Similarly, low expression levels of HDAC4/5/6/7/8/10/11 were correlated with favorable OS for GC patients (Fig. 4). GC patients with high HDAC1 mRNA levels were predicted to have a good prognosis, whereas mRNA expression levels of HDAC2 and HDAC9 were not statistically correlated with OS of GC patients (Fig. 4).
Figure 4.
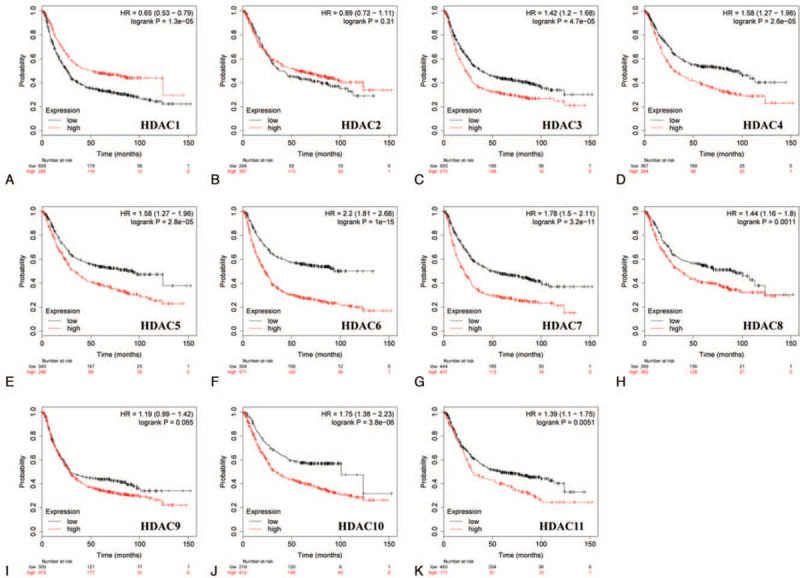
Correlation of HDAC mRNA expression with OS in all GC patients (Kaplan–Meier plotter). GC = gastric cancer; OS = overal survival.
Furthermore, prognostic significance of HDACs in different Lauren classification of GC, including intestinal, diffuse, and mixed type was determined. GC patients with decreased mRNA expression of HDAC3, HDAC5, HDAC6, HDAC7, HDAC8, HDAC9, and HDAC10 showed longer OS, whereas patients with decreased HDAC1 and HDAC4 mRNA expression showed shorter OS in intestinal gastric cancer (Fig. 5). Low HDAC4/5/6/7/8/9/10/11 mRNA expression levels or increased HDAC1 mRNA levels were correlated with good prognosis in GC patients of diffuse type (Fig. 6).
Figure 5.
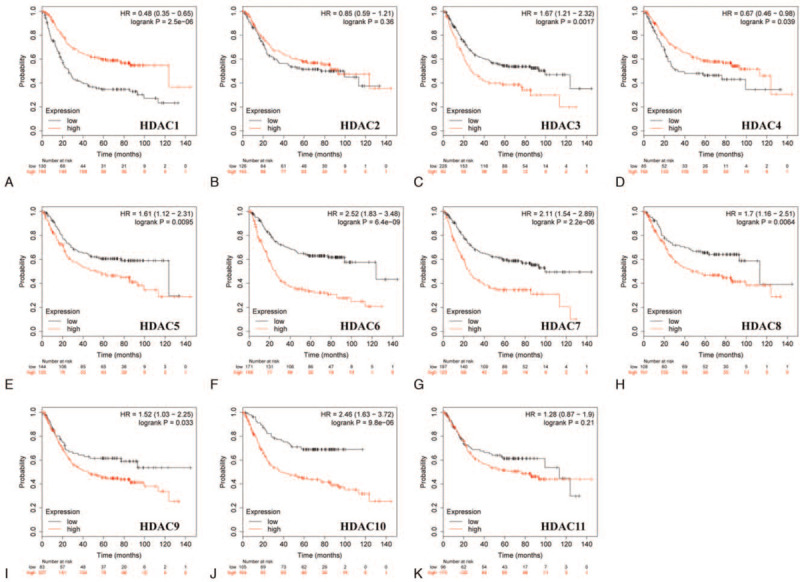
Correlation of HDAC mRNA expression with OS in intestinal gastric cancer patients (Kaplan–Meier plotter). OS = overal survival.
Figure 6.
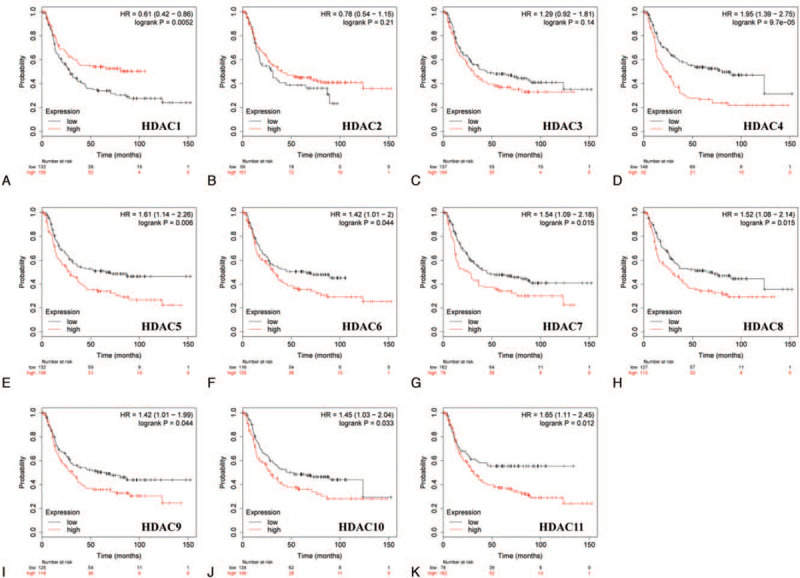
Correlation of HDAC mRNA expression with OS in diffuse gastric cancer patients (Kaplan–Meier plotter). OS = overal survival.
3.3. Prognostic value of HDACs in GC patients with different clinicopathological characteristics
Clinical stages, lymph node status, treatment, and HER2 status of patients with GC were compared to explore prognostic values of HDACs in patients with different clinicopathological characteristics. Low HDAC3/6 mRNA levels were correlated with favorable OS in stage 1 gastric cancer patients, whereas low expression of HDAC3/5/6/7/8/9 was correlated with good prognosis in stage 2 patients (Table 1). In stage 3 GC, low mRNA expression levels of HDAC3/5/6/7/8/10/11 were correlated with longer OS, whereas low HDAC1 mRNA expression levels were correlated with shorter OS. In addition, high expression of HDAC1/3 or low expression of HDAC4/5/7/9/10 was correlated with a good prognosis of stage 4 GC patients.
Table 1.
Correlation of HDAC mRNA expression with clinical stages of GC patients.
| HDACs | Clinical stages | Cases | HR (95%CI) | P value |
| HDAC1 | 1 | 69 | 0.37 (0.12–1.15) | .074 |
| 2 | 145 | 0.72 (0.37–1.38) | .32 | |
| 3 | 319 | 0.5 (0.35–0.69) | .000032∗ | |
| 4 | 152 | 0.64 (0.43–0.94) | .023∗ | |
| HDAC2 | 1 | 69 | 0.35 (0.12–1.08) | .057 |
| 2 | 145 | 1.79 (0.96–3.33) | .062 | |
| 3 | 319 | 1.25 (0.86–1.83) | .24 | |
| 4 | 152 | 0.81 (0.54–1.2) | .3 | |
| HDAC3 | 1 | 69 | 2.89 (1.08–7.73) | .027∗ |
| 2 | 145 | 3.29 (1.29–8.35) | .0081∗ | |
| 3 | 319 | 1.54 (1.16–2.05) | .0029∗ | |
| 4 | 152 | 0.65 (0.43–0.99) | .044∗ | |
| HDAC4 | 1 | 69 | 1.95 (0.59–6.38) | .26 |
| 2 | 145 | 1.6 (0.85–2.99) | .14 | |
| 3 | 319 | 0.69 (0.46–1.04) | .075 | |
| 4 | 152 | 1.66 (1.11–2.47) | .013∗ | |
| HDAC5 | 1 | 69 | 0.45 (0.13–1.57) | .2 |
| 2 | 145 | 2.12 (1.14–3.94) | .015∗ | |
| 3 | 319 | 2.08 (1.39–3.12) | .00026∗ | |
| 4 | 152 | 1.62 (1.09–2.41) | .016∗ | |
| HDAC6 | 1 | 69 | 4.75 (1.72–13.08) | .00089∗ |
| 2 | 145 | 2.08 (1.13–3.82) | .016∗ | |
| 3 | 319 | 2.29 (1.57–3.34) | .0000098∗ | |
| 4 | 152 | 1.37 (0.93–2.01) | .11 | |
| HDAC7 | 1 | 69 | 0.56 (0.2–1.56) | .26 |
| 2 | 145 | 2.06 (1.12–3.77) | .018∗ | |
| 3 | 319 | 1.68 (1.26–2.23) | .00034∗ | |
| 4 | 152 | 1.58 (1.07–2.32) | .019∗ | |
| HDAC8 | 1 | 69 | 1.61 (0.54–4.83) | .39 |
| 2 | 145 | 2.09 (1.12–3.92) | .018∗ | |
| 3 | 319 | 1.71 (1.16–2.51) | .006∗ | |
| 4 | 152 | 1.36 (0.91–2.03) | .14 | |
| HDAC9 | 1 | 69 | 0.54 (0.17–1.73) | .3 |
| 2 | 145 | 2.06 (1.02–4.18) | .041∗ | |
| 3 | 319 | 0.79 (0.59–1.05) | .1 | |
| 4 | 152 | 1.54 (1.05–2.26) | .026∗ | |
| HDAC10 | 1 | 69 | 2 (0.59–6.74) | .26 |
| 2 | 145 | 1.73 (0.84–3.54) | .13 | |
| 3 | 319 | 1.82 (1.25–2.65) | .0014∗ | |
| 4 | 152 | 1.59 (1.05–2.42) | .029∗ | |
| HDAC11 | 1 | 69 | 4.11 (0.53–31.66) | .14 |
| 2 | 145 | 1.88 (0.92–3.86) | .079 | |
| 3 | 319 | 1.83 (1.23–2.71) | .0023∗ | |
| 4 | 152 | 0.73 (0.47–1.13) | .15 |
GC = gastric cancer; HDACs = histone deacetylases; HR = hazard ratio.
P value < .05.
Analysis of lymph node negative GC patients showed that high HDAC1 mRNA levels, and low HDAC6/9 mRNA levels were significantly correlated with a good prognosis. A positive lymph node status was correlated with increased overall survival in patients with high expression level of HDAC1 or low expression levels of HDAC4/5/6/7/8/10/11, as shown in Table 2.
Table 2.
Correlation of HDAC mRNA expression with lymph node status of GC patients.
| HDACs | Lymph node status | Cases | HR (95%CI) | P value |
| HDAC1 | Negative | 76 | 0.4 (0.17–0.92) | .025∗ |
| Positive | 437 | 0.54 (0.41–0.7) | .0000028∗ | |
| HDAC2 | Negative | 76 | 2.44 (0.72–8.28) | .14 |
| Positive | 437 | 0.84 (0.64–1.1) | .12 | |
| HDAC3 | Negative | 76 | 1.49 (0.66–3.4) | .34 |
| Positive | 437 | 1.2 (0.92–1.56) | .18 | |
| HDAC4 | Negative | 76 | 1.9 (0.82–4.38) | .13 |
| Positive | 437 | 1.79 (1.36–2.36) | .000026∗ | |
| HDAC5 | Negative | 76 | 0.54 (0.22–1.32) | .17 |
| Positive | 437 | 1.74 (1.34–2.27) | .000026∗ | |
| HDAC6 | Negative | 76 | 2.93 (1.26–6.85) | .0094∗ |
| Positive | 437 | 1.83 (1.41–2.38) | .0000051∗ | |
| HDAC7 | Negative | 76 | 1.77 (0.69–4.49) | .23 |
| Positive | 437 | 1.55 (1.18–2.04) | .0014∗ | |
| HDAC8 | Negative | 76 | 1.84 (0.8–4.21) | .14 |
| Positive | 437 | 1.58 (1.2–2.06) | .00084∗ | |
| HDAC9 | Negative | 76 | 3.56 (1.39–9.12) | .0049∗ |
| Positive | 437 | 1.28 (0.95–1.71) | .1 | |
| HDAC10 | Negative | 76 | 1.85 (0.62–5.54) | .26 |
| Positive | 437 | 1.91 (1.45–2.52) | .0000028∗ | |
| HDAC11 | Negative | 76 | 2.08 (0.91–4.73) | .075 |
| Positive | 437 | 1.4 (1.05–1.86) | .02∗ |
GC = gastric cancer; HDACs = histone deacetylases; HR = hazard ratio.
P value < .05.
Prognostic values of HDACs in GC patients with 2 different treatments, including surgery alone and 5 FU based adjuvant were analyzed (Table 3). Analysis of the surgery-alone group, patients with decreased mRNA expression levels of HDAC4/5/7/8/9 or increased mRNA expression level of HDAC1 showed better OS. High HDAC2/9 mRNA or low HDAC1/3/6/11 mRNA levels were correlated with longer OS for GC patients treated with 5 FU based adjuvant.
Table 3.
Correlation of HDAC mRNA expression with treatment of GC patients.
| HDACs | Treatment | Cases | HR (95%CI) | P value |
| HDAC1 | Surgery alone | 393 | 0.69 (0.51–0.93) | .013∗ |
| 5 FU based adjuvant | 157 | 2.03 (1.36–3.05) | .00048∗ | |
| HDAC2 | Surgery alone | 393 | 0.88 (0.66–1.18) | .39 |
| 5 FU based adjuvant | 157 | 0.39 (0.15–1.01) | .046∗ | |
| HDAC3 | Surgery alone | 393 | 0.83 (0.62–1.12) | .22 |
| 5 FU based adjuvant | 157 | 1.65 (1.16–2.35) | .0052∗ | |
| HDAC4 | Surgery alone | 393 | 1.59 (1.19–2.12) | .0016∗ |
| 5 FU based adjuvant | 157 | 1.9 (0.74–4.86) | .17 | |
| HDAC5 | Surgery alone | 393 | 1.54 (1.15–2.05) | .0032∗ |
| 5 FU based adjuvant | 157 | 1.61 (0.63–4.1) | .31 | |
| HDAC6 | Surgery alone | 393 | 1.36 (0.98–1.87) | .062 |
| 5 FU based adjuvant | 157 | 1.63 (1.13–2.36) | .009∗ | |
| HDAC7 | Surgery alone | 393 | 1.5 (1.09–2.04) | .011∗ |
| 5 FU based adjuvant | 157 | 1.39 (0.98–1.97) | .062 | |
| HDAC8 | Surgery alone | 393 | 1.43 (1.07–1.92) | .016∗ |
| 5 FU based adjuvant | 157 | 2.08 (0.78–5.5) | .13 | |
| HDAC9 | Surgery alone | 393 | 1.51 (1.1–2.08) | .011∗ |
| 5 FU based adjuvant | 157 | 0.64 (0.45–0.91) | .012∗ | |
| HDAC10 | Surgery alone | 393 | 1.36 (0.97–1.9) | .074 |
| 5 FU based adjuvant | 157 | 3.43 (0.78–15) | .082 | |
| HDAC11 | Surgery alone | 393 | 1.36 (0.99–1.85) | .054 |
| 5 FU based adjuvant | 157 | 3.05 (1.06–8.79) | .031∗ |
5 FU = 5 fluorouracil; GC = gastric cancer; HDACs = histone deacetylases; HR = hazard ratio.
P value < .05.
Analysis based on HER2 status showed that expression levels of all HDACs, except for HDAC2, were correlated with overall survival in HER2-negative GC patients (Table 4). Low expression of HDAC3/4/5/6/7/8/9/10/11 or high HDAC1 expression was correlated with good prognosis. In HER2-positive GC patients, low mRNA levels of HDAC3/6/7/8/10 were correlated with favorable OS.
Table 4.
Correlation of HDAC mRNA expression with HER2 status of GC patients.
| HDACs | HER2 status | Cases | HR (95%CI) | P value |
| HDAC1 | Negative | 641 | 0.56 (0.43–0.74) | .00002∗ |
| Positive | 424 | 0.82 (0.62–1.09) | .17 | |
| HDAC2 | Negative | 641 | 0.82 (0.63–1.07) | .14 |
| Positive | 424 | 1.36 (0.87–2.14) | .18 | |
| HDAC3 | Negative | 641 | 1.37 (1.09–1.72) | .0064∗ |
| Positive | 424 | 1.43 (1.08–1.89) | .011∗ | |
| HDAC4 | Negative | 641 | 1.86 (1.42–2.43) | .0000036∗ |
| Positive | 424 | 0.8 (0.53–1.21) | .29 | |
| HDAC5 | Negative | 641 | 1.63 (1.25–2.12) | .00027∗ |
| Positive | 424 | 1.43 (0.98–2.09) | .064 | |
| HDAC6 | Negative | 641 | 2.1 (1.64–2.67) | .0000000012∗ |
| Positive | 424 | 1.88 (1.36–2.6) | .00011∗ | |
| HDAC7 | Negative | 641 | 1.69 (1.34–2.13) | .0000061∗ |
| Positive | 424 | 1.83 (1.38–2.42) | .000017∗ | |
| HDAC8 | Negative | 641 | 1.38 (1.05–1.8) | .019∗ |
| Positive | 424 | 1.74 (1.18–2.56) | .0049∗ | |
| HDAC9 | Negative | 641 | 1.33 (1.04–1.7) | .023∗ |
| Positive | 424 | 1.2 (0.89–1.62) | .23 | |
| HDAC10 | Negative | 641 | 1.66 (1.25–2.2) | .00034∗ |
| Positive | 424 | 1.79 (1.16–2.77) | .0076∗ | |
| HDAC11 | Negative | 641 | 1.38 (1.03–1.84) | .03∗ |
| Positive | 424 | 1.42 (0.98–2.06) | .063 |
CI = confidence intervals; GC = gastric cancer; HDACs = histone deacetylases; HER2 = human epidermal growth factor receptor 2; HR = hazard ratio.
P value < .05.
3.4. Genetic alterations and functional prediction of HDACs in GC
Genetic alterations of HDACs in GC patients were analyzed using cBioPortal tool. A total of 147 samples out of 708 (21%; data not shown) with stomach adenocarcinoma showed altered expression levels of at least 1 HDAC. Percentages of alterations in HDACs among 5 GC datasets ranged from 1.3% to 6% for individual genes (HDAC1, 2.1%; HDAC2, 3%; HDAC3, 1.3%; HDAC4, 6%; HDAC5, 4%; HDAC6, 2.8%; HDAC7, 2.2%; HDAC8, 1.6%; HDAC9, 4%; HDAC10, 2.4%; HDAC11, 1.9%; Fig. 7A). Analysis showed no significant association for OS and disease free survival between cases with or without HDAC alteration in gastric cancer (P = .258 and P = .510, respectively; Fig. 7B and C). To explore the potential interactions between HDAC members, a protein–protein interaction network was constructed using STRING database. A total of 11 nodes and 55 edges of 55 were observed in the protein–protein interaction network network (Fig. 7D). In addition, a network for HDACs with the structure or function of neighboring genes constructed using GeneMANIA showed that other 20 genes including DAXX, MEF2A, BRAP, USP39, USP3, USP20, USP51, USP45, USP22, USP33, USP16, USP49, USP44, BCOR, ANKRA2, RBBP4, USP5, USP13, RFXANK, and NRIP1 were closely associated with HDACs (Fig. 7E).
Figure 7.
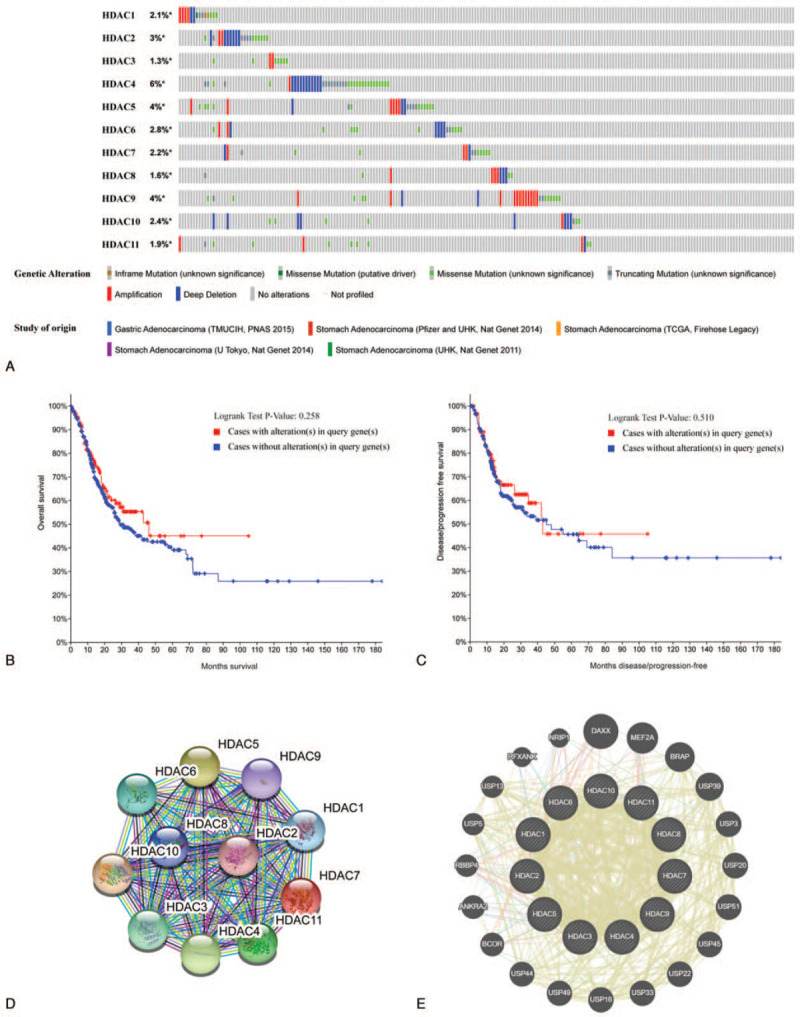
Alterations and network of HDACs (cBioPortal, String, and GeneMANIA). HDACs = histone deacetylases.
To further explore the biological functions of HDACs, functional enrichment analysis including GO terms (BP: biological process; CC: cellular component; MF: molecular function) and KEGG pathway were conducted using “org.Hs.eg.db,” “clusterProfiler,” “pathview,” “Goplot,” “ggplot2” packages. Significantly enriched GO terms and KEGG pathways of HDACs, based on adjusted P-values are presented in Figure 8. GO analysis of the top 12 GO terms showed that in molecular function category, these genes were mainly enriched in deacetylase activity, on both nicotinamide adenine dinucleotide-dependent and non-dependent, for histone and non-histone proteins, with H3-K14 having the highest activity (Supplementary Table 1). In biological process group, HDACs were mainly associated with histone H3 deacetylation, histone deacetylation, and protein deacetylation. For cellular component term, histone deacetylase complex was the highest enriched cell component. The top 12 enriched KEGG pathway of HDACs showed that HDACs were significantly enriched in 12 pathways, including alcohol metabolism, viral carcinogenesis, thyroid hormone signaling pathway, microRNAs in cancer, notch signaling pathway, longevity regulating pathway-multiple species, amphetamine addiction, chronic myeloid leukemia, cell cycle, apelin signaling pathway, transcriptional misregulation in cancer, and Epstein-Barr virus infection (Supplementary Table 2).
Figure 8.
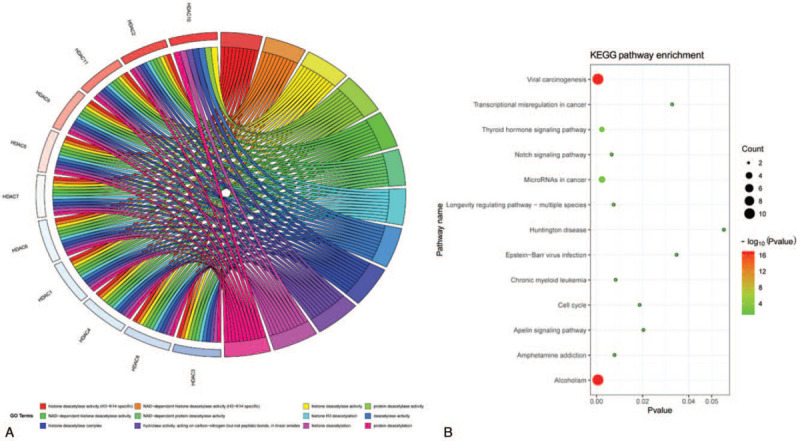
The enrichment analysis of HDACs (R software). HDACs = histone deacetylases.
4. Discussion
HDACs are involved in deacetylation of histones and several non-histone proteins including transcription factors and other abundant cellular proteins. They maintain equilibrium between histone acetylation and deacetylation, thus regulating proliferation and cell cycle progression, apoptosis and metastasis.[20–22] Recent studies reported a crucial role of HDACs in carcinogenesis and cancer progression, implying that HDACs are potential targets for cancer therapy. However, only few HDAC family members have been explored in GC. The present study is the first to explore expression levels, prognostic values, genetic alterations, and biological functions of HDACs using bioinformatics analysis.
Aberrant overexpression of HDAC1 is reported in various cancers, and studies report that it is a potential therapeutic target in several tumors.[23–27] Recent studies reported that HDAC1 expression is associated with gastric cancer, where it plays a significant role in distant metastasis and poor patient prognosis.[28,29] A study by Sun et al reported that HDAC1 knockdown repressed the proliferative potential of GC cells and promoted apoptosis induction, implying that HDAC1 is a promising target for gastric cancer therapy.[30] Lin et al reported inhibitory effects of HDAC1-downregulation on metastatic ability in GC cells by targeting the miRNA-34a/CD44 pathway-axis.[27] The findings of the present study showed that HDAC1 was upregulated in GC tissues compared with normal tissues. However, in the current study, high HDAC1 expression was correlated with a good prognosis in GC patients including intestinal and diffuse type, mainly in clinical stage 3 or 4 GC patients, which was not consistent with the previous reports. Therefore, further studies should be conducted to explore the prognostic value of HDAC1 in gastric cancer.
HDAC2, a member of the class I HDACs, is highly upregulated in several cancers.[31] A study by Jung et al reported that inhibition of HDAC2 enhanced expression of p21 (WAF1/Cip1) and repressed expression of cyclin D1, cyclin E2, and CDK2 in cell cycle regulation.[32] Wagner et al reported that high expression of HDAC2 inhibits pro-apoptotic functions of p53, thus inhibiting apoptosis and promoting tumorigenesis.[31] Furthermore, abnormal HDAC2 expression promotes aggressiveness of GC.[33] Moreover, in oral squamous cell carcinoma, HDAC2 increases cell migration and invasion through stabilization of hypoxic induction factor-1α at protein modification level.[34] The findings of the current study showed that expression of HDAC2 was significantly upregulated in GC tissues; however, the expression level was not correlated with overall survival in GC patients. Further studies should be conducted to estimate the prognostic role of HDAC2 in GC.
Previous studies report a negative correlation between expression of HDAC3 and overall survival in some cancer types such as gastric, pancreatic cancer, and glioma.[35–37] In addition HDAC3 is implicated in promoting gastric carcinogenesis through the miR-454-mediated pathway by targeting CHD5.[35] Consistent with findings from previous studies, the findings of the current study showed that low expression of HDAC3 was correlated with favorable OS of clinical stage 1 to 4 GC patients. Analysis showed a significant role of HDAC3 as a candidate biomarker for predicting prognosis of GC.
HDAC4, a key member of class IIa family of HDACs, is highly expressed in a variety of tissues and is associated with cancer progression.[38] Mottet et al performed a study to explore the mechanism of action and reported that HDAC4 is implicated in suppression of p21 (WAF1/Cip1) and has Sp1/Sp3-binding sites in glioblastoma.[39] Jin et al reported that miR-520b inhibits growth of lung cancer cells by targeting HDAC4.[40] Furthermore, HDAC4 promotes cell growth and metastasis by inhibiting expression of p21, p27, E-cadherin, and β catenin, and enhancing expression of CDK2 and vimentin in gastric, glioma, and esophageal carcinoma.[18,41,42] Consistently, the findings of this study showed that HDAC4 was significantly overexpressed in GC tissues, and high expression of HDAC4 was correlated with poor OS in GC patients mainly in clinical stage 4 patients and lymph node positive patients. Therefore, the findings of this study imply that HDAC4 can be used as a diagnostic or prognostic indicator in GC.
Studies on HDAC5 and HDAC7 in GC are limited. Liu et al carried out a study on non-small cell lung cancer, and reported an important role of miR-589-5p/HDAC5 pathway-axis in growth and metastasis of tumor cells, implying that HDAC5 is a potential oncogene.[43] HDAC7 modulates cell cycle progression by regulation of c-Myc expression.[44] Furthermore, Yu et al reported that HDAC7 expression was negatively correlated with overall survival in patients with GC.[45] The findings of the current study showed that elevated HDAC5/7 expression was correlated with poor prognosis in GC including intestinal and diffuse type, mainly those classified as clinical stage 2/3/4 and lymph node positive patients. However, no significant differences were observed for expression levels of HDAC5/7 between GC and normal tissues.
Aberrant expression of HDAC6 has been reported in several diseases, such as neurodegenerative diseases, cardiovascular disease, and cancer.[46,47] Previous studies reported that HDAC6 is implicated in cancer initiation and progression by modulating cell proliferation, angiogenesis, motility, invasion, and metatasis of tumor cells.[48] Li et al explored the mechanism of action of HDAC6 and reported that targeting of non-histone proteins such as α-tubulin, cortactin, and heat shock protein 90 by HDAC6 contributed to tumorigenesis and cancer progression. Moreover, HDAC6 affected immune system by regulating program death receptor-1 and program death receptor ligand-1 receptor.[49] Aberrant expression of HDAC6 has been reported in GC samples, and studies report that it is an oncogene.[50,51] In the present study, HDAC6 was downregulated in GC tissues, and patients with low HDAC6 expression level showed longer OS including intestinal and diffuse type, clinical stage 1/2/3, lymph node positive/negative patients. These findings imply that HDAC6 is a potential prognostic biomarker or therapeutic target in gastric cancer.
Numerous studies report aberrant overexpression of HDAC8 in a variety of cancers, including gastric, liver, breast, and oral squamous cell carcinoma.[52–56] For example, HDAC8 was reported as an oncogene that promotes malignant progression of breast cancer. Menbari et al reported that HDAC8 may exert its activity by protecting Notch1 from Fbwx7-facilitated protein degradation, resulting in activation of breast cancer stem cells.[52,53] A study on HCC reported that knockdown of HDAC8 inhibited tumor growth and induced apoptosis by upregulating expression of p53 and acetylation of p53 at Lys382.[54] In addition, a study by Song reported that HDAC8 plays a role in promoting tumorigenesis in GC.[55] Consistent with the findings from these studies, the findings from the current study exhibited that HDAC8 was predominantly overexpressed in GC tissues, and it was significantly correlated with poor prognosis in patients with GC mainly in clinical stage 2/3 and lymph node positive patients. Therefore, HDAC8 is a potential target for anti-GC therapeutics.
HDAC9 plays a critical role in progression of tumor. Xiong et al reported a negative correlation between expression of HDAC9 and OS in GC patients[12]; downregulation of miRNA-383-5p was correlated with poor patient survival and metastasis in GC by targeting HDAC9.[57] In addition, targeting of HDAC9 by miR-377 promoted cell migration and enhanced the proliferative potential of oral squamous cell carcinoma.[58] Consistent with findings from previous studies, analysis in the current study showed that HDAC9 was highly expressed in GC tissues, and expression level of HDAC9 varied significantly in different pathological stages. Furthermore, high HDAC9 expression level was correlated with poor prognosis in intestinal and diffuse type GC patients, as well as clinical stage 2/4 and lymph node negative patients.
Only a few studies have explored the roles of HDAC10 or HDAC11 in tumorigenesis and cancer progression. Jin et al reported that HDAC10 was downregulated in GC tissues, which was correlated with poor prognosis in GC patients.[59] This was consistent with findings of the present study that expression of HDAC10 was downregulated in GC tissues; however, low expression of HDAC10 was correlated with good OS, which was not consistent with previous reports. Low HDAC11 expression level was correlated with favorable OS in GC patients mainly for the diffuse type. However, differential HDAC11 expression was not observed between GC and normal tissues. Further studies should be conducted to explore the roles of HDAC10/HDAC11 in gastric cancer.
As promising tumor suppressors or oncogenes, genetic alterations of HDACs may be correlated with carcinogenesis, progression, and prognosis of GC. Relatively low consistent levels of alterations were observed in each HDAC in GC, which had no impact on OS or disease free survival. Previous studies report that HDAC alterations may not affect GC prognosis. To further explore the biological functions of HDACs, network analysis was performed for HDAC family members. The genes were mainly enriched in deacetylase activity, tumor-related pathways, and growth-related pathways such as microRNAs in cancer, notch signaling pathway, longevity regulating pathway-multiple species, cell cycle, transcriptional misregulation in cancer. The findings of this study show the potential role of HDACs as therapeutic targets in gastric cancer.
In view of the important roles of HDACs in a variety of biological processes in carcinogenesis and cancer progression, a number of clinical trials of HDAC inhibitors (HDACis) have been carried out. To date, 4 HDACis including vorinostat (SAHA), romidepsin (FK228), panobinostat ((LBH589), and belinostat (PXD101) have been approved by the US Food and Drug Administration in the treatment of several hematological malignancies and lymphomas,[60–63] while numerous clinical trials are ongoing for advanced or refractory tumors. For instance, San-Miguel et al reported a phase III trial of the pan-HDACi panobinostat in combination with bortezomib and dexamethasone and schedules in 768 patients with relapsed multiple myeloma; the median OS of panobinostat group was 40.3 months while that of placebo group was 35.8 months.[64] Similar encouraging results have been reported on a phase I to II clinical trial of the cyclic peptide HDACi romidepsin combined with dexamethasone and bortezomib.[65] Similarly, a phase III trial found that, compared with the placebo-controlled group, the subtype-selective HDACi tucidinostat (also named chidamide) plus exemestane regimen could improve progression-free survival for postmenopausal patients with advanced, hormone receptor-positive breast cancer, which may serve as a new treatment option.[66] The results of current trial revealed that HDAC inhibitors could be a promising avenue for cancer treatment; however, studies on HDACis in gastric cancer therapy are limited, which requires further research.
5. Conclusion
In summary, expression of HDAC2/4 was significantly upregulated in GC, and aberrant expression of HDAC1/3/4/5/6/7/8/10/11 was associated with prognosis in GC. Moreover, expression levels of the HDACs were correlated with different Lauren classifications, and clinical stages, lymph node status, treatment, and HER2 status. The findings of this study show the role of HDACs as potential diagnostic or prognostic biomarkers and therapeutic targets in GC. However, further research is required to validate these findings.
Author contributions
Conceptualization: Lei Pan.
Data curation: Luting Chen, Yuchang Fei, Yurong Zhao.
Formal analysis: Luting Chen, Yuchang Fei, Yurong Zhao.
Methodology: Luting Chen, Yuchang Fei.
Visualization: Luting Chen, Yuchang Fei, Yurong Zhao.
Writing – original draft: Luting Chen, Quan Chen.
Writing – review & editing: Peifeng Chen, Lei Pan.
Supplementary Material
Footnotes
Abbreviations: GC = gastric cancer, GEPIA = Gene Expression Profiling Interactive Analysis, GO = gene ontology, HDACis = HDAC inhibitors, HDACs = histone deacetylases, HER2 = human epidermal growth factor receptor 2, KEGG = Kyoto Encyclopedia of Genes and Genomes, OS = overall survival.
How to cite this article: Chen L, Fei Y, Zhao Y, Chen Q, Chen P, Pan L. Expression and prognostic analyses of HDACs in human gastric cancer based on bioinformatic analysis. Medicine. 2021;100:27(e26554).
This work is supported by the Zhejiang Provincial Natural Science Foundation of China (grant no. LY20H270009) and 2019 School-level Young and Middle-aged Scientific Research Innovation Fundation of Zhejiang Chinese Medical University (KC201929).
The authors have no conflicts of interest to disclose.
The datasets generated during and/or analyzed during the current study are publicly available.
Supplemental digital content is available for this article.
References
- [1].Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394–424. [DOI] [PubMed] [Google Scholar]
- [2].Allemani C, Weir HK, Carreira H, et al. Global surveillance of cancer survival 1995–2009: analysis of individual data for 25,676,887 patients from 279 population-based registries in 67 countries (CONCORD-2). Lancet 2015;385:977–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Park JM, Lee HJ, Yoo JH, Ko WJ, Cho YJ, Hahm KB. Overview of gastrointestinal cancer prevention in Asia. Best Pract Res Clin Gastroenterol 2015;29:855–67. [DOI] [PubMed] [Google Scholar]
- [4].Yamamoto M, Rashid OM, Wong J. Surgical management of gastric cancer: the East vs. West perspective. J Gastrointest Oncol 2015;6:79–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Gregoretti IV, Lee YM, Goodson HV. Molecular evolution of the histone deacetylase family: functional implications of phylogenetic analysis. J Mol Biol 2004;338:17–31. [DOI] [PubMed] [Google Scholar]
- [6].Witt O, Deubzer HE, Milde T, Oehme I. HDAC family: what are the cancer relevant targets? Cancer Lett 2009;277:08–21. [DOI] [PubMed] [Google Scholar]
- [7].Haberland M, Montgomery RL, Olson EN. The many roles of histone deacetylases in development and physiology: implications for disease and therapy. Nat Rev Genet 2009;10:32–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Peng L, Seto E. Deacetylation of nonhistone proteins by HDACs and the implications in cancer. Handb Exp Pharmacol 2011;206:39–56. [DOI] [PubMed] [Google Scholar]
- [9].Yang XJ, Seto E. The Rpd3/Hda1 family of lysine deacetylases: from bacteria and yeast to mice and men. Nat Rev Mol Cell Biol 2008;9:206–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Ropero S, Esteller M. The role of histone deacetylases (HDACs) in human cancer. Mol Oncol 2007;1:19–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Shen Q, Tang W, Sun J, Feng L, Jin H, Wang X. Regulation of CRADD-caspase 2 cascade by histone deacetylase 1 in gastric cancer. Am J Transl Res 2014;6:538–47. [PMC free article] [PubMed] [Google Scholar]
- [12].Xiong K, Zhang H, Du Y, Tian J, Ding S. Identification of HDAC9 as a viable therapeutic target for the treatment of gastric cancer. Exp Mol Med 2019;51:01–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Mrakovcic M, Bohner L, Hanisch M, Hanisch M. Epigenetic targeting of autophagy via HDAC inhibition in tumor cells: role of p53. Int J Mol Sci 2018;19:3952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Zhang X, Yashiro M, Ren J, Hirakawa K. Histone deacetylase inhibitor, trichostatin A, increases the chemosensitivity of anticancer drugs in gastric cancer cell lines. Oncol Rep 2006;16:563–8. [PubMed] [Google Scholar]
- [15].Deng R, Zhang P, Liu W, et al. HDAC is indispensable for IFN-gamma-induced B7-H1 expression in gastric cancer. Clin Epigenetics 2018;10:153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Shin H, Lee YS, Lee YC. Sodium butyrate-induced DAPK-mediated apoptosis in human gastric cancer cells. Oncol Rep 2012;27:1111–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Choi JH, Kwon HJ, Yoon BI, et al. Expression profile of histone deacetylase 1 in gastric cancer tissues. Jpn J Cancer Res 2001;92:1300–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Kang ZH, Wang CY, Zhang WL, et al. Histone deacetylase HDAC4 promotes gastric cancer SGC-7901 cells progression via p21 repression. PLoS One 2014;9:e98894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res 2017;45:W98–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Glozak MA, Seto E. Histone deacetylases and cancer. Oncogene 2007;26:5420–32. [DOI] [PubMed] [Google Scholar]
- [21].Telles E, Seto E. Modulation of cell cycle regulators by HDACs. Front Biosci (Schol Ed) 2012;4:831–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Hagelkruys A, Sawicka A, Rennmayr M, Seiser C. The biology of HDAC in cancer: the nuclear and epigenetic components. Handb Exp Pharmacol 2011;206:13–37. [DOI] [PubMed] [Google Scholar]
- [23].Xiong W, Yang S, Zhang W, Chen Y, Wang F. MiR-761 inhibits colorectal cancer cell proliferation and invasion through targeting HDAC1. Pharmazie 2019;74:111–4. [DOI] [PubMed] [Google Scholar]
- [24].Gao DJ, Xu M, Zhang YQ, et al. Upregulated histone deacetylase 1 expression in pancreatic ductal adenocarcinoma and specific siRNA inhibits the growth of cancer cells. Pancreas 2010;39:994–1001. [DOI] [PubMed] [Google Scholar]
- [25].Zhang L, Bu L, Hu J, et al. HDAC1 knockdown inhibits invasion and induces apoptosis in non-small cell lung cancer cells. Biol Chem 2018;399:603–10. [DOI] [PubMed] [Google Scholar]
- [26].Sudo T, Mimori K, Nishida N, et al. Histone deacetylase 1 expression in gastric cancer. Oncol Rep 2011;26:777–82. [DOI] [PubMed] [Google Scholar]
- [27].Lin L, Jiang H, Huang M, et al. Depletion of histone deacetylase 1 inhibits metastatic abilities of gastric cancer cells by regulating the miR-34a/CD44 pathway. Oncol Rep 2015;34:663–72. [DOI] [PubMed] [Google Scholar]
- [28].Jiang Z, Yang H, Zhang X, Wang Z, Rong R, Wang X. Histone deacetylase-1 as a prognostic factor and mediator of gastric cancer progression by enhancing glycolysis. Hum Pathol 2019;85:194–201. [DOI] [PubMed] [Google Scholar]
- [29].Cao LL, Yue Z, Liu L, et al. The expression of histone deacetylase HDAC1 correlates with the progression and prognosis of gastrointestinal malignancy. Oncotarget 2017;8:39241–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Sun MY, Zhang H, Tao J, Ni ZH, Wu QX, Tang QF. Expression and biological function of rhotekin in gastric cancer through regulating p53 pathway. Cancer Manag Res 2019;11:1069–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Wagner T, Brand P, Heinzel T, Kramer OH. Histone deacetylase 2 controls p53 and is a critical factor in tumorigenesis. Biochim Biophys Acta 2014;1846:524–38. [DOI] [PubMed] [Google Scholar]
- [32].Jung KH, Noh JH, Kim JK, et al. HDAC2 overexpression confers oncogenic potential to human lung cancer cells by deregulating expression of apoptosis and cell cycle proteins. J Cell Biochem 2012;113:2167–77. [DOI] [PubMed] [Google Scholar]
- [33].Song J, Noh JH, Lee JH, et al. Increased expression of histone deacetylase 2 is found in human gastric cancer. APMIS 2005;113:264–8. [DOI] [PubMed] [Google Scholar]
- [34].Chang CC, Lin BR, Chen ST, Hsieh TH, Li YJ, Kuo MY. HDAC2 promotes cell migration/invasion abilities through HIF-1alpha stabilization in human oral squamous cell carcinoma. J Oral Pathol Med 2011;40:567–75. [DOI] [PubMed] [Google Scholar]
- [35].Xu G, Zhu H, Zhang M, Xu J. Histone deacetylase 3 is associated with gastric cancer cell growth via the miR-454-mediated targeting of CHD5. Int J Mol Med 2018;41:155–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Jiao F, Hu H, Han T, et al. Aberrant expression of nuclear HDAC3 and cytoplasmic CDH1 predict a poor prognosis for patients with pancreatic cancer. Oncotarget 2016;7:16505–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Zhong S, Fan Y, Wu B, et al. HDAC3 expression correlates with the prognosis and grade of patients with glioma: a diversification analysis based on transcriptome and clinical evidence. World Neurosurg 2018;119:e145–58. [DOI] [PubMed] [Google Scholar]
- [38].Wang Z, Qin G, Zhao TC. HDAC4: mechanism of regulation and biological functions. Epigenomics 2014;6:139–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Mottet D, Pirotte S, Lamour V, et al. HDAC4 represses p21 (WAF1/Cip1) expression in human cancer cells through a Sp1-dependent, p53-independent mechanism. Oncogene 2009;28:243–56. [DOI] [PubMed] [Google Scholar]
- [40].Jin K, Zhao W, Xie X, Pan Y, Wang K, Zhang H. MiR-520b restrains cell growth by targeting HDAC4 in lung cancer. Thorac Cancer 2018;9:1249–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Cai JY, Xu TT, Wang Y, et al. Histone deacetylase HDAC4 promotes the proliferation and invasion of glioma cells. Int J Oncol 2018;53:2758–68. [DOI] [PubMed] [Google Scholar]
- [42].Zeng LS, Yang XZ, Wen YF, et al. Overexpressed HDAC4 is associated with poor survival and promotes tumor progression in esophageal carcinoma. Aging (Albany NY) 2016;8:1236–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Liu C, Lv D, Li M, et al. Hypermethylation of miRNA-589 promoter leads to upregulation of HDAC5 which promotes malignancy in non-small cell lung cancer. Int J Oncol 2017;50:2079–90. [DOI] [PubMed] [Google Scholar]
- [44].Zhu C, Chen Q, Xie Z, et al. The role of histone deacetylase 7 (HDAC7) in cancer cell proliferation: regulation on c-Myc. J Mol Med (Berl) 2011;89:279–89. [DOI] [PubMed] [Google Scholar]
- [45].Yu Y, Cao F, Yu X, et al. The expression of HDAC7 in cancerous gastric tissues is positively associated with distant metastasis and poor patient prognosis. Clin Transl Oncol 2017;19:1045–54. [DOI] [PubMed] [Google Scholar]
- [46].Batchu SN, Brijmohan AS, Advani A. The therapeutic hope for HDAC6 inhibitors in malignancy and chronic disease. Clin Sci (Lond) 2016;130:987–1003. [DOI] [PubMed] [Google Scholar]
- [47].McLendon PM, Ferguson BS, Osinska H, et al. Tubulin hyperacetylation is adaptive in cardiac proteotoxicity by promoting autophagy. Proc Natl Acad Sci U S A 2014;111:E5178–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Lernoux M, Schnekenburger M, Dicato M, Diederich M. Anti-cancer effects of naturally derived compounds targeting histone deacetylase 6-related pathways. Pharmacol Res 2018;129:337–56. [DOI] [PubMed] [Google Scholar]
- [49].Li T, Zhang C, Hassan S, et al. Histone deacetylase 6 in cancer. J Hematol Oncol 2018;11:111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Park SJ, Kim JK, Bae HJ, et al. HDAC6 sustains growth stimulation by prolonging the activation of EGF receptor through the inhibition of rabaptin-5-mediated early endosome fusion in gastric cancer. Cancer Lett 2014;354:97–106. [DOI] [PubMed] [Google Scholar]
- [51].He Q, Li G, Wang X, et al. A decrease of histone deacetylase 6 expression caused by Helicobacter Pylori infection is associated with oncogenic transformation in gastric cancer. Cell Physiol Biochem 2017;42:1326–35. [DOI] [PubMed] [Google Scholar]
- [52].Menbari MN, Rahimi K, Ahmadi A, et al. MiR-216b-5p inhibits cell proliferation in human breast cancer by down-regulating HDAC8 expression. Life Sci 2019;237:116945. [DOI] [PubMed] [Google Scholar]
- [53].Chao MW, Chu PC, Chuang HC, et al. Non-epigenetic function of HDAC8 in regulating breast cancer stem cells by maintaining Notch1 protein stability. Oncotarget 2016;7:1796–807. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [54].Wu J, Du C, Lv Z, et al. The up-regulation of histone deacetylase 8 promotes proliferation and inhibits apoptosis in hepatocellular carcinoma. Dig Dis Sci 2013;58:3545–53. [DOI] [PubMed] [Google Scholar]
- [55].Song S, Wang Y, Xu P, et al. The inhibition of histone deacetylase 8 suppresses proliferation and inhibits apoptosis in gastric adenocarcinoma. Int J Oncol 2015;47:1819–28. [DOI] [PubMed] [Google Scholar]
- [56].Ahn MY, Yoon JH. Histone deacetylase 8 as a novel therapeutic target in oral squamous cell carcinoma. Oncol Rep 2017;37:540–6. [DOI] [PubMed] [Google Scholar]
- [57].Xu G, Li N, Zhang Y, Zhang J, Xu R, Wu Y. MicroRNA-383-5p inhibits the progression of gastric carcinoma via targeting HDAC9 expression. Braz J Med Biol Res 2019;52:e8341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Rastogi B, Kumar A, Raut SK, et al. Downregulation of miR-377 promotes oral squamous cell carcinoma growth and migration by targeting HDAC9. Cancer Invest 2017;35:152–62. [DOI] [PubMed] [Google Scholar]
- [59].Jin Z, Jiang W, Jiao F, et al. Decreased expression of histone deacetylase 10 predicts poor prognosis of gastric cancer patients. Int J Clin Exp Pathol 2014;7:5872–9. [PMC free article] [PubMed] [Google Scholar]
- [60].Verza FA, Das U, Fachin AL, Dimmock JR, Marins M. Roles of histone deacetylases and inhibitors in anticancer therapy. Cancers (Basel) 2020;12:1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Li G, Tian Y, Zhu WG. The roles of histone deacetylases and their inhibitors in cancer therapy. Front Cell Dev Biol 2020;8:576946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Cosenza M, Pozzi S. The therapeutic strategy of HDAC6 inhibitors in lymphoproliferative disease. Int J Mol Sci 2018;19:2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Suraweera A, O’Byrne KJ, Richard DJ. Combination therapy with histone deacetylase inhibitors (HDACi) for the treatment of cancer: achieving the full therapeutic potential of HDACi. Front Oncol 2018;8:92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].San-Miguel JF, Hungria VT, Yoon SS, et al. Overall survival of patients with relapsed multiple myeloma treated with panobinostat or placebo plus bortezomib and dexamethasone (the PANORAMA 1 trial): a randomised, placebo-controlled, phase 3 trial. Lancet Haematol 2016;3:e506–15. [DOI] [PubMed] [Google Scholar]
- [65].Harrison SJ, Quach H, Yuen K, et al. High response rates with the combination of bortezomib, dexamethasone and the pan-histone deacetylase inhibitor romidepsin in patients with relapsed or refractory multiple myeloma in a phase I/II clinical trial. Blood 2008;112:1267. [Google Scholar]
- [66].Jiang Z, Li W, Hu X, et al. Tucidinostat plus exemestane for postmenopausal patients with advanced, hormone receptor-positive breast cancer (ACE): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol 2019;20:806–15. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


