Abstract
To explore the possible molecular mechanism of reproductive toxicity of Tripterygium wilfordii from the perspective of network pharmacology and bioinformatics.
The compounds of T wilfordii were obtained by querying the relevant Chinese medicine database, the effective compounds were screened and the corresponding targets were obtained, and then compared with the reproductive toxicities related to disease targets obtained from the disease gene database to infer the potential toxic targets of reproductive toxicity of T wilfordii. Then, the key targets of reproductive toxicity of T wilfordii were screened using Search Tool for the Retrieval of Interacting Genes/Protein and Cytoscape. The gene ontology function and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis, as well as module analysis, were performed on the key targets using Database for Annotation, Visualization, and Integrated Discovery and Cytoscape, respectively. Finally, the network between effective compounds-toxic targets was conducted to see how the compounds interacted.
A total of 48 effective compounds and 482 potential toxic targets related to the reproductive toxicity of T wilfordii were screened. The enrichment analysis results showed that the key targets were mainly enriched in biological processes such as response to drug, ionotropic glutamate receptor signaling pathway, and KEGG pathways such as neuroactive ligand-receptor interaction, cAMP signaling pathway. In the protein-protein interaction network of potential toxic targets, there were 78 key targets such as TP53, INS, IL6, AGT, ADCY3, and so on. Enrichment analysis of the top module with 19 genes from module analysis indicated that T wilfordii might cause reproductive toxicity by gene ontology terms and KEGG pathways such as regulation of vasoconstriction, G-protein coupled receptor signaling pathway, inflammatory response, cAMP signaling pathway, and so on. In the network between effective compounds of T wilfordii and key targets, there were 5 compounds with high degree including Tingenone, Wilfordic Acid, Abruslactone A, Nobilin, and Wilforlide B.
The complex molecular mechanism of reproductive toxicity of T wilfordii can be preliminarily elucidated with the help of the network pharmacology method, and the analysis results can provide some reference for the further mechanism research of reproductive toxicity of T wilfordii.
Keywords: molecular mechanism, network pharmacology, reproductive toxicity, Tripterygium wilfordii
1. Introduction
As a traditional Chinese herbal medicine, Tripterygium wilfordii has a long history in the application of traditional Chinese medicine. Modern pharmacological researches[1,2] have shown that the components of T wilfordii are complex, mainly including alkaloids, terpenoids, sugars, and so on and have the functions of regulating immunity, anti-inflammatory, anti-tumor, anti-fertility, anti-angiogenesis, and so on. Due to its outstanding immunomodulatory and anti-inflammatory effects, T wilfordii is widely used in autoimmune diseases. For example, T wilfordii polyglycosides tablets made from extracts of T wilfordii are commonly used in the treatment of rheumatoid arthritis. However, on the one hand, T wilfordii has a good effect; on the other hand, it harms, including hepatotoxicity, nephrotoxicity, reproductive toxicity, and cardiovascular system toxicity,[3] which not only causes harm to patients’ health but also greatly limits the clinical application of T wilfordii. Recent studies[3,4] have suggested that the reproductive toxicity of T wilfordii is related to the atrophy of reproductive organs, function decline, and apoptosis of reproductive cells, whose molecular mechanism is still unclear due to the complex composition.
The purpose of this study is to predict the biological processes and related pathways involved in the reproductive toxicity of T wilfordii by means of network pharmacology, and to explore the molecular mechanism of reproductive toxicity of T wilfordii, so as to provide certain theoretical reference for further molecular experiments and attenuation studies on the reproductive toxicity of T wilfordii.
2. Materials and methods
Since this study is an analysis of data from online databases and the privacy of patients will not be disclosed, so patients’ informed consent and ethical approval are all not required.
2.1. Acquisition of effective compounds and targets of T wilfordii
The chemical compounds contained in T wilfordii and their targets were searched from a Bioinformatics Analysis Tool for Molecular Mechanism of Traditional Chinese Medicine (BATMAN-TCM, http://bionet.ncpsb.org/batman-tcm/),[5] with the score cutoff value of 20 and the adjusted P value of .05. Then, the dataset A of effective compounds and targets of T wilfordii was collected.
2.2. Acquisition of reproductive toxicity targets of T wilfordii
The terms “reproductive toxicity, gonad toxicity, sexual dysfunction, reproductive damage, and gonad damage” were used to retrieve the target genes of reproductive toxicity from the human gene database GeneCards (version5.0, https://www.genecards.org).[6] Reproductive toxicity target dataset B was obtained by excluding the duplicate results. Subsequently, the dataset C of overlapping genes dataset between A and B was gained using a Venn diagram, which was produced by Venny (version2.1, http://bioinfogp.cnb.csic.es/tools/venny/index. html).[7]
2.3. Analysis of GO and KEGG pathway enrichment
For further analysis of the molecular mechanism of reproductive toxicity of T wilfordii, the key targets would be input into Database for Annotation, Visualization and Integrated Discovery (DAVID; version6.8, https://david.ncifcrf.gov/),[8,9] limiting the species as Homo sapiens to conduct the analysis of gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment. It was considered to be statistically significant when P value <.05.
2.4. Construction and module analysis of PPI network
To explore the internal relationship between the potential gene targets of reproductive toxicity of T wilfordii, the target genes in dataset C were uploaded to Search Tool for the Retrieval of Interacting Genes/Proteins database (version11, http://www.string-db.org/),[10] limiting the species as Homo sapiens to obtain the protein interaction relationship of the potential gene targets. Those with interaction scores >0.7 would be selected for further analysis. Then, the results were uploaded to the visualization software Cytoscape (version3.7.1, http://www.cytoscape.org/)[11] to build a protein-protein interaction (PPI) network. Then the analysis of topology characteristics of the PPI network was carried out using Network analyzer plugin in Cytoscape, and key genes were screened and extracted from the network according to the Betweenness Centrality and Closeness Centrality which were larger than the median and the degree which was larger than twice median.
Module analysis on the PPI network of key genes was conducted using the molecular complex detection clustering algorithm in Cytoscape. Module analysis is usually used to identify connected gene groups with common functions and analyze the complex relationship of the nodes in the PPI network. The conditions for the module selection contained: node score cutoff = 4, degree cutoff = 4, maximum depth = 100, and k-core = 2. Additionally, the top module was further analyzed for KEGG pathway enrichment using DAVID, with a cutoff of P < .05.
2.5. Construction and analysis of effective compounds-toxic targets network
The effective compounds and reproductive toxicity targets were uploaded to Cytoscape to build their reaction network. The network between the key genes and the effective compounds was then extracted. The greater the degree value of nodes in the network, the greater their role in the network. A large degree of 1 compound indicates that it can react with many targets; similarly, a large degree of 1 target indicates that it can combine with many compounds.
3. Results
3.1. Effective compounds and targets of T wilfordii
Based on BATMAN database, a total of 48 effective compounds of T wilfordii that met the screening conditions were obtained, and a total of 529 gene targets were predicted.
3.2. Reproductive toxicity targets of T wilfordii
There were totally 13,866 gene targets related to reproductive toxicity retrieved from GeneCards database after detracting and resorting. As shown in Figure 1, there were 482 common gene targets between the active compounds of T wilfordii and reproductive toxicity.
Figure 1.
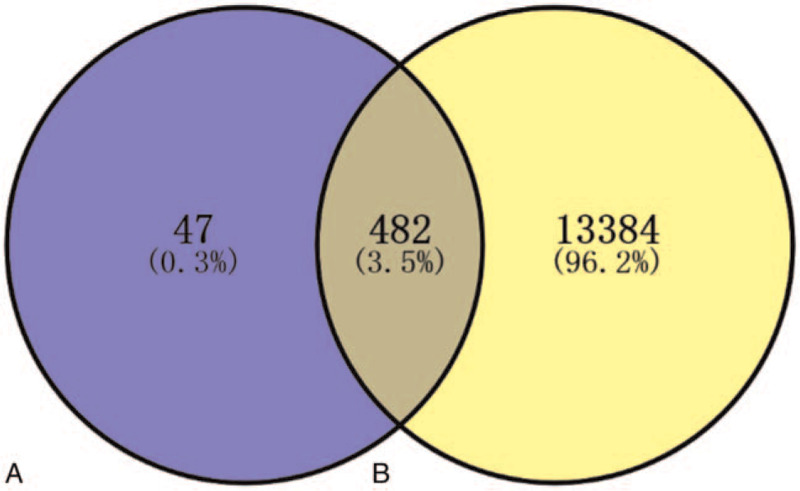
Venn diagram of reproductive toxicity gene targets of Tripterygium wilfordii. (A) Indicates gene targets of T wilfordii. (B) Indicates gene targets related to reproductive toxicity. The common part of A and B indicates gene targets of reproductive toxicity of T wilfordii.
3.3. Enrichment analysis of GO and KEGG
Putting 482 common target genes into the DAVID database, 1154 GO terms and 95 KEGG pathways were enriched (Table 1). The top 5 biological processes in key targets of reproductive toxicity of T wilfordii were a response to drug, ionotropic glutamate receptor signaling pathway, positive regulation of transcription, DNA-templated, positive regulation of gene expression, and positive regulation of transcription from RNA polymerase II promoter. The 5 most enriched KEGG pathways were neuroactive ligand-receptor interaction, nicotine addiction, cAMP signaling pathway, calcium signaling pathway, and amphetamine addiction.
Table 1.
GO analysis in BP and KEGG pathway analysis of reproductive toxicity of Tripterygium wilfordii, including the top 5 terms selected according to the P value.
| Term | Description | Count | P-value |
| Top 5 BPs | |||
| GO:0042493 | response to drug | 57 | 1.25E-29 |
| GO:0035235 | ionotropic glutamate receptor signaling pathway | 18 | 1.14E-21 |
| GO:0045893 | positive regulation of transcription, DNA-templated | 61 | 6.18E-21 |
| GO:0010628 | positive regulation of gene expression | 44 | 6.53E-21 |
| GO:0045944 | positive regulation of transcription from RNA polymerase II promoter | 85 | 3.28E-20 |
| Top 5 KEGG pathways | |||
| hsa04080 | Neuroactive ligand-receptor interaction | 70 | 2.21E-27 |
| hsa05033 | Nicotine addiction | 26 | 6.59E-22 |
| hsa04024 | cAMP signaling pathway | 46 | 2.21E-16 |
| hsa04020 | Calcium signaling pathway | 41 | 2.54E-14 |
| hsa05031 | Amphetamine addiction | 25 | 4.71E-14 |
BPs = biological processes, GO = gene ontology, KEGG = Kyoto Encyclopedia of Genes and Genomes.
3.4. Construction and module analysis of PPI network
Four hundred eighty two potential reproductive toxicity targets of T wilfordii were uploaded to Search Tool for the Retrieval of Interacting Genes/Proteins database to obtain their interaction relationships. The results were opened by Cytoscape and topology analysis of the network was conducted. According to the Betweenness Centrality and Closeness Centrality which were larger than the median and the degree which was larger than twice the median, 78 key genes were screened finally, whose average degree was 26.17, and among which a total of 56 (71.79%) genes had a degree greater than 20. The top 5 of degree among all key genes were TP53, INS, IL6, AGT, and ADCY3 (Table 2).
Table 2.
Top 5 genes with the highest degree of interaction in the PPI network.
| Gene ID | Degree | Closeness Centrality | Betweenness Centrality |
| TP53 | 57 | 0.42209073 | 0.14499223 |
| INS | 55 | 0.42928786 | 0.13712094 |
| IL6 | 51 | 0.42714571 | 0.08306111 |
| AGT | 49 | 0.39194139 | 0.03955276 |
| ADCY3 | 43 | 0.35489221 | 0.03243746 |
ADCY3 = adenylate cyclase 3, AGT = Angiotensinogen, INS = insulin, IL6 = interleukin-6, PPI = protein-protein interaction, TP53 = tumor protein 53.
In molecular complex detection analysis, a total of 5 modules were finally selected (Table 3), including 1 top cluster with 19 nodes and 171 edges (Fig. 2). Enrichment analysis of genes in the top module demonstrated that it may be associated with regulation of vasoconstriction, G-protein coupled receptor signaling pathway, inflammatory response, alpha2-adrenergic receptor activity, epinephrine binding, drug binding, neuroactive ligand-receptor interaction, cGMP-PKG signaling pathway, and cAMP signaling pathway (Table 4).
Table 3.
Five modules from the PPI network satisfied the criteria of MCODE scores >4 and number of nodes >4.
| Cluster | Score | Nodes | Edges | Node IDs |
| 1 | 19 | 19 | 171 | CNR2, CNR1, ADRA2C, PF4, BDKRB2, PTGER3, CX3CR1, ADORA1, ADRA2A, ADCY3, DRD4, CHRM2, C5, ADORA3, ADRA2B, AGT, GPER1, ANXA1, DRD2 |
| 2 | 6.476 | 22 | 68 | GRIN2A, IFNG, TGFB1, GRIA1, CAMK2D, CAMK2A, IL6, ADIPOQ, GRIA2, NCOA1, CALM1, TNF, GRIN1, ESR1, PRKCB, GRIN2B, PRKCD, IL1B, IL10, IL4, BDNF, NGF |
| 3 | 6.429 | 15 | 45 | F2, DRD1, AVPR2, CCL2, INS, FFAR1, ADRB2, GNAS, UTS2R, CRH, PTGS2, CALCA, PTGER1, AVP, KISS1 |
| 4 | 4.333 | 7 | 13 | MED1, AR, SRC, HDAC1, FOS, SIRT1, PPARG |
| 5 | 4 | 6 | 10 | WNT5A, CTNNB1, MTOR, MAPK9, NOTCH1, TP53 |
MCODE = molecular complex detection, PPI = protein-protein interaction.
Figure 2.

Top module from PPI network. PPI = protein-protein interaction.
Table 4.
Enriched GO terms and KEGG pathways of module 1.
| Term | Name | Count | P value | Genes |
| BPs | ||||
| GO:0019229 | Regulation of vasoconstriction | 5 | 4.43E-09 | BDKRB2, ADRA2C, ADRA2B, ADRA2A, AGT |
| GO:0007186 | G-protein coupled receptor signaling pathway | 11 | 5.44E-09 | CX3CR1, CHRM2, C5, GPER1, PTGER3, BDKRB2, ADRA2C, ADRA2B, ADRA2A, AGT, PF4 |
| GO:0006954 | Inflammatory response | 8 | 7.25E-08 | C5, ANXA1, CNR2, GPER1, PTGER3, BDKRB2, ADORA1, PF4 |
| MFs | ||||
| GO:0004938 | Alpha2-adrenergic receptor activity | 3 | 3.22E-06 | ADRA2C, ADRA2B, ADRA2A |
| GO:0051379 | Epinephrine binding | 3 | 1.61E-05 | ADRA2C, ADRA2B, ADRA2A |
| GO:0008144 | Drug binding | 4 | 6.82E-05 | CHRM2, CNR1, DRD2, DRD4 |
| KEGG pathways | ||||
| hsa04080 | Neuroactive ligand-receptor interaction | 12 | 3.71E-12 | CHRM2, CNR2, CNR1, ADORA3, PTGER3, BDKRB2, ADORA1, DRD2, ADRA2C, ADRA2B, ADRA2A, DRD4 |
| hsa04022 | cGMP-PKG signaling pathway | 7 | 1.34E-06 | ADORA3, BDKRB2, ADORA1, ADCY3, ADRA2C, ADRA2B, ADRA2A |
| hsa04024 | cAMP signaling pathway | 5 | 0.001180424 | CHRM2, PTGER3, ADORA1, ADCY3, DRD2 |
BPs = biological processes, KEGG = Kyoto Encyclopedia of Genes and Genomes, MFs = molecular functions.
3.5. Construction and analysis of the network between toxic compounds and reproductive toxicity targets of T wilfordii
The network between effective compounds of T wilfordii and key targets was built by Cytoscape, and then the network between toxic compounds and reproductive toxicity targets of T wilfordii (Figure 3), which contained 111 nodes and 209 edges, involving 33 toxic compounds and 78 reproductive toxicity targets. The top 5 compounds were Tingenone (25 targets), Wilfordic Acid (16 targets), Abruslactone A (16 targets), Nobilin (14 targets), and Wilforlide B (10 targets).
Figure 3.
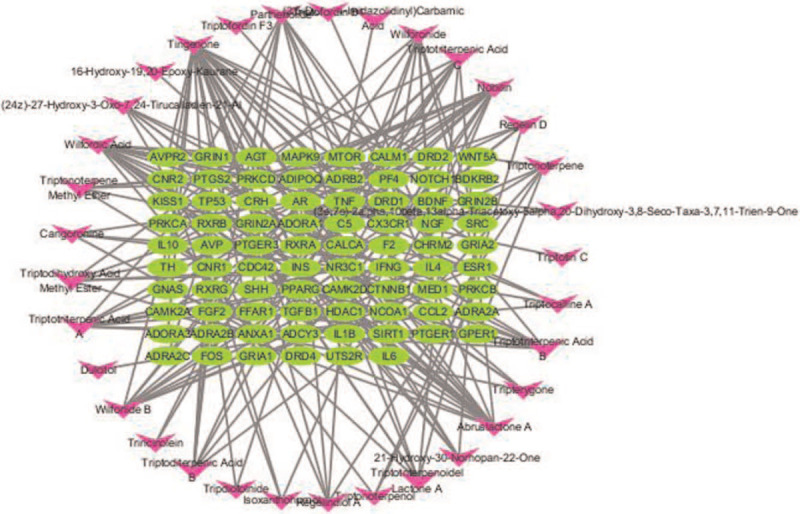
Network of effective compounds and reproductive toxicity targets of Tripterygium wilfordii. The green circles represent targets; the purple arrows represent compounds.
4. Discussion
Not only the decoction pieces of T wilfordii but also the extracts from T wilfordii are widely used in clinical practice. It has been proved that T wilfordii can promote male spermatogenic cell apoptosis, reduce sperm viability, and cause female ovarian function decline.[4] Therefore, it is very important to understand the toxicity mechanism of T wilfordii, and then to develop attenuated strategies for various toxic reactions. This study focused on the reproductive toxicity of T wilfordii, and tried to explore the molecular mechanism based on BATMAN, GeneCards, and other related databases and online tools.
In total, 48 toxic compounds and 482 toxic target genes related to reproductive toxicity of T wilfordii were dugout. Enrichment analysis of these 482 genes preliminary found that the pathogenesis of the reproductive toxicity of T wilfordii might be connected with biological processes such as response to drug, ionotropic glutamate receptor signaling pathway, positive regulation of transcription, DNA-templated, positive regulation of gene expression, positive regulation of transcription from RNA polymerase II promoter, and KEGG pathways such as neuroactive ligand-receptor interaction, nicotine addiction, cAMP signaling pathway, calcium signaling pathway, and amphetamine addiction.
Then, the PPI network of potential toxic targets was further analyzed. Out of them there were 78 key gene targets and Table 2 summarized the top 10 PPI network hub genes including TP53, INS, IL6, AGT, and ADCY3. Most of these genes were associated with reproduction. As a tumor suppressor gene, TP53 can participate in the regulation of the cell cycle, is closely related to cell proliferation and apoptosis, and plays an important role in the occurrence and development of various malignant tumors such as gastric cancer, colon cancer, and ovarian cancer.[12] In animal experiments, knocking out the TP53 gene in mouse ovarian epithelial cells could accelerate cell proliferation and DNA synthesis, and enhance the ability of cell cloning and migration.[13] In addition, clinical studies have found that there is a certain correlation between the TP53 gene and the incidence of male infertility and female infertility.[14,15] INS is an insulin gene, which can regulate the reproductive function of rats through the hypothalamus-pituitary-gonadal axis[16], in humans, insulin resistance changes serum testosterone levels to reduce semen quality and affect male fertility.[17,18] At the same time, improving insulin resistance can improve the reproductive function of polycystic ovary syndrome women.[19] Interleukin-6 (IL-6) gene encodes the cytokine IL-6). Cytokines are a group of molecules that play an important role in the process of cell signal transduction. Studies have shown that cytokines can affect reproductive function by regulating the hypothalamus-pituitary-gonadal axis.[20] Various cytokines including IL-6 and TNF play a certain role in the regulation of ovarian function.[21] AGT gene encodes Angiotensinogen (AGT), a plasma glycoprotein, which is synthesized in the liver, placenta, anterior pituitary, ovary, testis, and other organs. Some studies have shown that AGT deficiency has a certain effect on the fertility of mice.[22] ADCY3 gene encodes adenylate cyclase 3 (ADCY3), which is a membrane integrated protein and is one of the key signal molecules downstream of G protein-coupled receptor. It converts the stimulation of extracellular signals into intracellular signals, regulates the synthesis of cyclic adenosine-3′ and 5′-monophosphate (cAMP), and thus participates in various pathological and physiological processes of the body.[23] However, the current studies on ADCY3 mainly focus on its correlation with diseases such as obesity, fatty liver, and Crohn disease.[24–26]
The module analysis showed that the PPI network of 78 key target genes contained 5 modules. The enrichment results of the top module with 19 genes indicated that T wilfordii might cause reproductive toxicity by GO terms and KEGG pathways such as regulation of vasoconstriction, G-protein coupled receptor signaling pathway, inflammatory response, alpha2-adrenergic receptor activity, epinephrine binding, drug binding, neuroactive ligand-receptor interaction, cGMP-PKG signaling pathway, and cAMP signaling pathway. These may be the underlying mechanisms of reproductive toxicity of T wilfordii.
In addition to the above, the reproductive system damage of T wilfordii is also related to some other genes. An animal experiment showed that tripterygium glycosides had different degrees of damage to the ovaries of female rats at different times, which may be related to its effect on the expression of circadian rhythm genes CLOCK and BMAL1.[27] The reproductive toxicity of T wilfordii was different in different sex rats.[28,29] This suggests that administration time, sex, and other factors may also affect the reproductive toxicity of T wilfordii.
Among top compounds with a high degree in the network between toxic compounds and reproductive toxicity targets of T wilfordii, Tingenone is a pentacyclic triterpene, which can induce peripheral antinociception due to opioidergic activation, NO/cGMP, and ATP-sensitive K(+) channels pathway activation and cannabinoid receptors activation in mice.[30–32] Unfortunately, no studies have been found to explore the direct effect of Tingenone on reproduction. Abruslactone A, also called Wilforlide A, is a triterpenoid from T wilfordii, which has obvious immunosuppressive activity. Abruslactone A can inhibit the activity of adenosine deaminase in HL-60 cells and induce apoptosis.[33,34] The mechanism of Wilforlide A-induced ovarian cell apoptosis may be related to the abnormal expression of ERK/c-fos, and there is a time effect on the expression of apoptosis-related proteins.[35] One study showed that compatibility of T wilfordii and Astragalus membranaceus could downregulate the content of Wilforlide A[36], which suggests that T wilfordii combined with A membranaceus may reduce reproductive toxicity. However, so far, there have been no studies on Wilfordic Acid, Nobilin, or Wilforlide B.
5. Conclusions
The results of this study are consistent with those of published literature, indicating that it is feasible to predict the molecular mechanism from the perspective of bioinformatics by means of network pharmacology. The data from this study may provide greater insight into the molecular mechanisms of reproductive toxicity of T wilfordii. However, because many of the genes identified in this study had not been previously associated with reproductive toxicity of T wilfordii, further studies will be needed to validate the expression of these genes.
Author contributions
Investigation: Qing Ding,Yuanhao Wu.
Methodology: Yuanhao Wu.
Software: Qing Ding.
Validation: Qing Ding.
Visualization: Qing Ding.
Writing – original draft: Qing Ding.
Writing – review & editing: Wei Liu, Yuanhao Wu.
Footnotes
Abbreviations: BATMAN-TCM = Bioinformatics Analysis Tool for Molecular Mechanism of Traditional Chinese Medicine, DAVID = Database for Annotation, Visualization and Integrated Discovery, GO = gene ontology, BPs = biological processes, KEGG = Kyoto Encyclopedia of Genes and Genomes, STRING = Search Tool for the Retrieval of Interacting Genes/Proteins, PPI = protein-protein interaction, MCODE = molecular complex detection, TP53 = tumor protein 53, INS = insulin, IL6 = interleukin-6, AGT = Angiotensinogen, ADCY3 = adenylate cyclase 3.
How to cite this article: Ding Q, Wu Y, Liu W. Molecular mechanism of reproductive toxicity induced by Tripterygium Wilfordii based on network pharmacology. Medicine. 2021;100:27(e26197).
This study was supported by Scientific Research Project of Tianjin Municipal Commission of Education (2019ZD12); High-Level Talent Selection and Training Project of Tianjin Health and Family Planning Industry (02005wuyuanhao); and National Natural Science Foundation of China (81673927).
The authors have no conflicts of interest to disclose.
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
- [1].Hu DJ, Peng ZY, He DC. Research progress on pharmacological action of Tripterygium wilfordii (Chinese). Herald Med 2018;37:586–92. [Google Scholar]
- [2].Zhang Q, Peng GC, Zhu MJ. Research progress on pharmacological action and toxicity of Tripterygium wilfordii (Chinese). Chin J Integr Med Cardio-Cerebrovasc Dis 2016;14:1753–4. [Google Scholar]
- [3].Xu Y, Fan YF, Zhao Y, Lin N. Overview of reproductive toxicity studies on Tripterygium wilfordii in recent 40 years. China J Chin Mater Med 2019;44:3406–14. [DOI] [PubMed] [Google Scholar]
- [4].Xiong W, Chen J. Advances in clinical and animal studies on reproductive toxicity of Tripterygium wilfordii (Chinese). J Mod Clin Med 2014;40:403–5. [Google Scholar]
- [5].Liu Z, Guo F, Wang Y, et al. BATMAN-TCM: a Bioinformatics Analysis Tool for Molecular mechANism of Traditional Chinese Medicine. Sci Rep 2016;6:21146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Stelzer G, Rosen N, Plaschkes I, et al. The GeneCards suite: from gene data mining to disease genome sequence analyses. Curr Protoc Bioinformatics 2016;54:01–30. [DOI] [PubMed] [Google Scholar]
- [7].Oliveros, J.C., Venny. An interactive tool for comparing lists with Venn's diagrams. (2007–2015): https://bioinfogp.cnb.csic.es/tools/venny/index.html. [Google Scholar]
- [8].Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009;4:44–57. [DOI] [PubMed] [Google Scholar]
- [9].Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 2009;37:01–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Szklarczyk D, Gable AL, Lyon D, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res 2019;47(D1):D607–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 2003;13:2498–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].He XW, Xu L. Research progress of cell cycle related genes CDKN2A, TP53, RB1 and BRCA2 in malignant tumors (Chinese). J Mod Oncol 2018;26:153–7. [Google Scholar]
- [13].Zhang T, Fan JW, Li D, et al. Gene knockout of TP53 in primary mouse ovarian epithelial cells and its biological characteristics (Chinese). Lab Anim Sci 2018;35:64–9. [Google Scholar]
- [14].Jin Q. Correlation between PGAM4 gene and TP53 gene and idiopathic male infertility (Chinese). Wannan Medical College 2013. [Google Scholar]
- [15].Tan Y. Correlation of SNPs of TP53 and MDM2 gene with treatment outcomes of infertility and IVF (Chinese). Kunming University of Science and Technology 2013. [Google Scholar]
- [16].Lin JL. Molecular mechanism of spermatogenic dysfunction in immune orchitis and insulin regulation of function of hypothalamus-pituitary-gonadal axis (Chinese). Peking Union Medical College 2018. [Google Scholar]
- [17].Ma J, Han RY, Mei XA, et al. Correlation analysis of insulin resistance index with male reproductive hormone level and semen parameters (Chinese). Natl J Androl 2018;24:695–9. [PubMed] [Google Scholar]
- [18].Ma J, Han RY, Mei XA, et al. Effects of obesity and insulin resistance on semen quality in men (Chinese). Chin J Androl 2018;32:19–24. [Google Scholar]
- [19].Sun XL. Effect of insulin resistance on reproductive function in patients with polycystic ovary syndrome (Chinese). Fudan University 2013. [Google Scholar]
- [20].Ke JW, Zhang T, Duan R. Regulation of hypothalamic-pituitary-gonadal axis (HPG) and hypothalamic-pituitary-adrenal axis (HPAA) by cytokines (Chinese). Jiangxi J Medical Lab Sci 2002;06:379–81. [Google Scholar]
- [21].Li QL, Ni J. Regulation of ovarian function by cytokines (Chinese). Prog Physiol Sci 2000;04:361–3. [Google Scholar]
- [22].Lin Y. Effect of angiotensinogen gene deficiency on fecundity of mice (Chinese). Foreign Med Sci (Obstet Gynecol Fascicle) 2000;27:303. [Google Scholar]
- [23].Yang YM, Yang YQ, Song G, et al. Research progress of adenylate cyclase (Chinese). J Clin Pathol Res 2019;39:390–4. [Google Scholar]
- [24].Pan LY, Cai X, Kong YL, et al. Correlation study between ADCY3 gene polymorphism and fatty liver (Chinese). Acta Nutri Sin 2017;39:337–42. [Google Scholar]
- [25].Sadia S, Amélie B, Filippo T, et al. Loss-of-function mutations in ADCY3 cause monogenic severe obesity. Nat Genet 2018;50: [DOI] [PubMed] [Google Scholar]
- [26].Zhang CB, Zheng H, Chao K, et al. Polymorphism of ADCY3 and NFIL3 genes and their susceptibility to Crohn's disease in Chinese population (Chinese). Chin J Clin Pharmacol 2019;35:743–5. [Google Scholar]
- [27].Liu Z. Experimental study of Tripterygium glycosides CIA rats administered at different times on the function of the reproductive female (Chinese). North China University of Science and Technology 2017. [Google Scholar]
- [28].Fan YY, Xu Y, Su XH, et al. Effect of Tripterygium glycosides tablets on reproductive toxicity in female rats with II type collagen induced arthritis (Chinese). China J Chin Mater Med 2019;44:3486–93. [DOI] [PubMed] [Google Scholar]
- [29].Fan YY, Xu Y, Su XH, et al. Effect of Tripterygium glycosides tablets on reproductive toxicity in male rats with II type collagen induced arthritis (Chinese). China J Chinese Mater Med 2020;45:755–63. [DOI] [PubMed] [Google Scholar]
- [30].Veloso CC, Rodrigues VG, Ferreira RC, et al. Tingenone, a pentacyclic triterpene, induces peripheral antinociception due to opioidergic activation. Planta Med 2014;80:1615–21. [DOI] [PubMed] [Google Scholar]
- [31].de Carvalho VC, Rodrigues VG, Ferreira RC, et al. Tingenone, a pentacyclic triterpene, induces peripheral antinociception due to NO/cGMP and ATP-sensitive K(+) channels pathway activation in mice. Eur J Pharmacol 2015;755:01–5. [DOI] [PubMed] [Google Scholar]
- [32].Veloso CC, Ferreira R, Rodrigues VG, et al. Tingenone, a pentacyclic triterpene, induces peripheral antinociception due to cannabinoid receptors activation in mice. Inflammopharmacology 2018;26:227–33. [DOI] [PubMed] [Google Scholar]
- [33].Ma Z, Liang MX, Zhang Y. Research progress on chemical constituents and pharmacological effects of Tripterygium wilfordii (Chinese). Asia-Pacific Tradit Med 2011;7:157–60. [Google Scholar]
- [34].Wang L, Zhang TL, Fan HH, et al. Various monomers of Tripterygium wilfordii effecting adenosine deaminase activity and inducing HL-60 cell apopotosis (Chinese). Fudan Univ J Med Sci 2007;01:107–10. [Google Scholar]
- [35].Qi AR, Wen SS, Fu B, et al. Effects of different Tripterygium wilfordii on apoptosis of hamster ovary cells (Chinese). Acta Chin Med 2018;33:1510–4. [Google Scholar]
- [36].Xu P, Meng M, Zhang J, Jiang Y, Tang L. Study on wilforlide A before and after compatibility according to “conteract the toxicity of another drug” (Chinese). Chin J Prim Med Pharm 2016;23:166–9. [Google Scholar]


