Abstract
Background
A cornerstone of Australia's ability to control COVID-19 has been effective border control with an extensive supervised quarantine programme. However, a rapid recrudescence of COVID-19 was observed in the state of Victoria in June, 2020. We aim to describe the genomic findings that located the source of this second wave and show the role of genomic epidemiology in the successful elimination of COVID-19 for a second time in Australia.
Methods
In this observational, genomic epidemiological study, we did genomic sequencing of all laboratory-confirmed cases of COVID-19 diagnosed in Victoria, Australia between Jan 25, 2020, and Jan 31, 2021. We did phylogenetic analyses, genomic cluster discovery, and integrated results with epidemiological data (detailed information on demographics, risk factors, and exposure) collected via interview by the Victorian Government Department of Health. Genomic transmission networks were used to group multiple genomic clusters when epidemiological and genomic data suggested they arose from a single importation event and diversified within Victoria. To identify transmission of emergent lineages between Victoria and other states or territories in Australia, all publicly available SARS-CoV-2 sequences uploaded before Feb 11, 2021, were obtained from the national sequence sharing programme AusTrakka, and epidemiological data were obtained from the submitting laboratories. We did phylodynamic analyses to estimate the growth rate, doubling time, and number of days from the first local infection to the collection of the first sequenced genome for the dominant local cluster, and compared our growth estimates to previously published estimates from a similar growth phase of lineage B.1.1.7 (also known as the Alpha variant) in the UK.
Findings
Between Jan 25, 2020, and Jan 31, 2021, there were 20 451 laboratory-confirmed cases of COVID-19 in Victoria, Australia, of which 15 431 were submitted for sequencing, and 11 711 met all quality control metrics and were included in our analysis. We identified 595 genomic clusters, with a median of five cases per cluster (IQR 2–11). Overall, samples from 11 503 (98·2%) of 11 711 cases clustered with another sample in Victoria, either within a genomic cluster or transmission network. Genomic analysis revealed that 10 426 cases, including 10 416 (98·4%) of 10 584 locally acquired cases, diagnosed during the second wave (between June and October, 2020) were derived from a single incursion from hotel quarantine, with the outbreak lineage (transmission network G, lineage D.2) rapidly detected in other Australian states and territories. Phylodynamic analyses indicated that the epidemic growth rate of the outbreak lineage in Victoria during the initial growth phase (samples collected between June 4 and July 9, 2020; 47·4 putative transmission events, per branch, per year [1/years; 95% credible interval 26·0–85·0]), was similar to that of other reported variants, such as B.1.1.7 in the UK (mean approximately 71·5 1/years). Strict interventions were implemented, and the outbreak lineage has not been detected in Australia since Oct 29, 2020. Subsequent cases represented independent international or interstate introductions, with limited local spread.
Interpretation
Our study highlights how rapid escalation of clonal outbreaks can occur from a single incursion. However, strict quarantine measures and decisive public health responses to emergent cases are effective, even with high epidemic growth rates. Real-time genomic surveillance can alter the way in which public health agencies view and respond to COVID-19 outbreaks.
Funding
The Victorian Government, the National Health and Medical Research Council Australia, and the Medical Research Future Fund.
Research in context.
Evidence before this study
Since the start of the COVID-19 pandemic, genomic analysis of SARS-CoV-2 has played an important role in tracking the spread of the virus, and in identifying novel and highly transmissible variants. In Australia, the low prevalence of COVID-19 means that samples from all cases of COVID-19 undergo genomic sequencing, which allows unprecedented tracking of the spread of SARS-CoV-2 and enables rapid public health responses. We searched PubMed, medRxiv, and bioRxiv for primary research studies published in English between Jan 1, 2020, and Feb 11, 2021, using combinations of “SARS-CoV-2”, “genomics”, “phylodynamics”, and “public health response”. We identified six articles that used SARS-CoV-2 genomic surveillance in a low-prevalence setting at the time the study was done, including Australia (n=2), New Zealand (n=2), and the Netherlands and China; only two studies describe the comprehensive real-time integration of genomic surveillance data into the public health response in two settings, one in Victoria (Australia) and the other in New Zealand. We did not identify any studies describing the introduction, expansion, and subsequent elimination of a large clonal second wave.
Added value of this study
In this observational, genomic epidemiology study, we present our unique experience of successfully eliminating SARS-CoV-2 nationally for a second time in Australia. Genomics-informed public health responses were important in identification and response to a large second wave of infection in Australia that started in June, 2020. Following initial elimination of the virus in April, 2020, genomic analyses identified Australia's mandatory hotel quarantine system as the source of the second wave. Our analysis directly led to a major structural overhaul of the hotel quarantine programme. Additionally, our phylodynamic analyses indicated an epidemic viral growth rate at the start of the second wave similar to that of contemporaneous emerging variants, such as the B.1.1.7 variant in the UK. However, decisive public health responses, even with high epidemic growth rates, led to elimination of the outbreak strain from Australia by the end of October, 2020. Subsequent cases identified in December, 2020, and January, 2021, represented independent introductions, with limited local spread.
Implications of all the available evidence
Our study highlights the need for ongoing vigilance, even in countries where COVID-19 has effectively been eliminated. Swift, comprehensive public health responses successfully controlled a large second wave of infection in Australia, even when high viral growth rates were observed. The combination of integrated epidemiology and genomics had a substantial effect on the course of the pandemic in Australia, altering the way in which public health and government agencies viewed and responded to the pandemic, and bringing genomic analyses into the mainstream public discourse. Genomic analysis has become a core component of public health responses to COVID-19 in Australia, and, in countries that rely on hotel quarantine systems as a barrier to reintroduction by travellers, genomic sequencing provides an early detection system of possible quarantine breaches.
Introduction
The COVID-19 pandemic, caused by SARS-CoV-2, has resulted in global morbidity, mortality, and socioeconomic disruption on an unprecedented scale. Although many countries have managed to suppress or eliminate a first wave of infections with a range of public health interventions, such as travel restrictions, physical distancing, and mask use, many are encountering severe second, or even, third waves of COVID-19 in 2021.1
A cornerstone of effective control of COVID-19 is the identification of positive cases, primarily through RT-PCR testing of nasopharyngeal swabs. Diagnostic testing is essential to direct the management of cases and their contacts, but it provides no information about the probable source of virus acquisition; hence, control measures rely primarily on epidemiological data. Viral genome sequence analysis (ie, genomics) has emerged as an important tool in understanding the pandemic, both at a global and regional scale.2, 3, 4, 5, 6 When combined with epidemiological data, genomics can provide insights into the source and transmission of SARS-CoV-2 in the community, high-risk workplaces, and health-care facilities.2, 7, 8, 9
Australia has one of the lowest SARS-CoV-2 infection rates globally. A first wave of COVID-19 occurred in March and April, 2020, prompting implementation of nationwide public health interventions, including closing of the Australian international border and restriction of domestic travel. During this initial pandemic phase, SARS-CoV-2 infections were largely attributable to transmission events arising from returning international travellers, evidenced by the co-circulation of multiple genomic lineages of SARS-CoV-2, as described previously.2 Since March 20, 2020, the Australian border has been open by exemption only, with all returning travellers required to complete at least 14 days of quarantine in a hotel or another supervised facility. Following a period of near elimination of COVID-19 in Australia, a second wave occurred between June and October, 2020, concentrated in the state of Victoria, with more than 18 000 cases and 800 deaths.
Here we report the findings of the genomic epidemiological analysis that identified the source of this second wave in Victoria, prompting the rapid institution of strict and extensive public health restrictions,10 and a fundamental restructure of the hotel quarantine programme.
Methods
Setting and data sources
In this observational, genomic epidemiological study, we did genomic sequencing of all laboratory-confirmed cases of COVID-19 in Victoria, Australia between Jan 25, 2020, and Jan 31, 2021. The state of Victoria, which has a population of approximately 6·24 million people, has a consistently high testing rate for SARS-CoV-2.11 All samples testing positive for SARS-CoV-2 by RT-PCR are forwarded to the Doherty Institute Public Health Laboratories (Melbourne, VIC, Australia)12 for real-time, prospective, genomic surveillance of COVID-19, with sequencing usually completed within 7 days. For urgent cases, rapid sequencing and analysis is completed within 24 h. Epidemiological data, including detailed information on demographics, risk factors, and exposure were collected for each case through an interview done by the Victoria Department of Health. Mode of acquisition was categorised as: travel overseas if the individual reported travel in the 14 days before symptom onset; contact with a confirmed case if no overseas travel was reported and case contact had occurred within the same period; or source unknown. A COVID-19 genomics response team was established to integrate and review genomic epidemiological data, as previously described.2 Data were collected in accordance with the Victorian Public Health and Wellbeing Act 2008. Ethical approval was received from the University of Melbourne Human Research Ethics Committee (study number 1954615.3).
Since June, 2020, rapid sharing of SARS-CoV-2 genomic sequence data between Australian states has occurred via AusTrakka, a national sequence sharing programme governed by the Communicable Diseases Genomics Network. All publicly available SARS-CoV-2 sequences available in AusTrakka on Feb 11, 2021, were included, and epidemiological data were obtained from the submitting laboratories.
Procedures
RNA extracted from positive SARS-CoV-2 RT-PCR samples underwent tiled amplicon PCR by use of either ARTIC version 1 or version 3 primers according to published protocols,13 and Illumina sequencing, as previously described.2 Reads were aligned to the reference genome (Wuhan Hu 1; GenBank MN908947.3) and consensus sequences were generated. We applied quality control checks on consensus sequences, which required 95% or higher genome recovery, 42 or fewer single nucleotide polymorphisms from the reference genome, and 30 or fewer ambiguous or missing bases for inclusion in our analyses. A single sequence was selected from each positive patient for phylogenetic analysis.
Data analysis
Genomic clusters were defined as two or more related sequences using a complete linkage hierarchical clustering algorithm of pairwise genetic distances derived from the maximum likelihood phylogenetic tree. Genomic transmission networks were used to group genomic clusters when epidemiological and genomic data suggested that they arose from a single importation event and diversified within Victoria. Detailed methodology for phylogenetic analysis, genomic cluster detection, and epidemiological data collection are provided in appendix 1 (pp 1–2).
Sequencing and clustering analysis was done prospectively, with genomic and epidemiological data examined interactively whenever additional data were available. Findings with potential public health significance were immediately communicated to public health decision makers and presented at relevant incident and outbreak management meetings. Routine reporting of genomic surveillance data was also provided to the same individuals weekly.
Phylodynamic analyses were done by use of BEAST 2.5 software.14 A birth-death tree prior was used to estimate the growth rate with 95% credible intervals, doubling time, and number of days from the first local infection to the collection of the first sequenced genome (ie, the genomic detection lag) of transmission network G, the dominant Victorian wave 2 outbreak strain.15 Our growth rate estimate was compared with recent (January, 2021) estimates from a similar growth phase of lineage B.1.1.7 (also known as the Alpha variant) in the UK.16 To minimise the effect of differences in the proportion of positive samples sequenced between transmission network G and B.1.1.7, each transmission network G sequence was sampled with a probability of 0·1 to match this expected proportion of positive samples in the UK, resulting in a dataset of 121 genomes. The detailed methodology for phylodynamic analyses is provided in appendix 1 (p 3).
Role of the funding source
The funders of the study had no role in study design, data collection, data analysis, data interpretation, or writing of the report. Some study authors were staff at the Victorian Government Department of Health and were involved in data collection and manuscript preparation.
Results
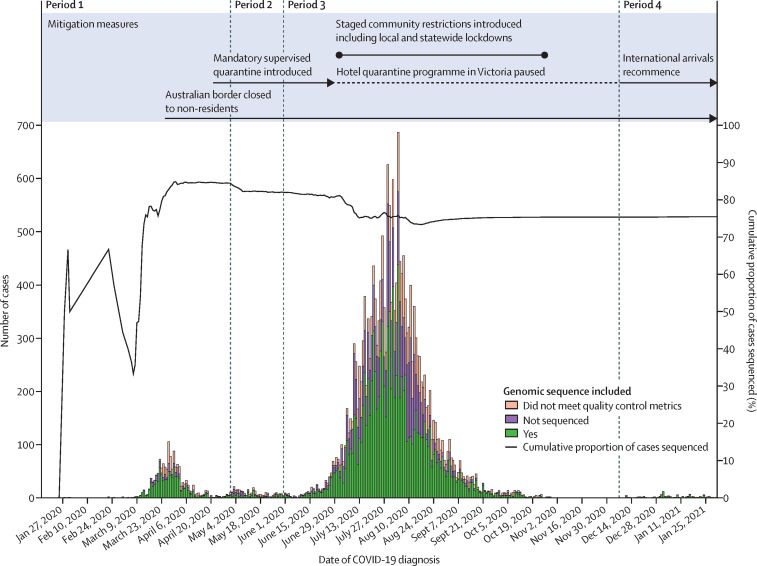
Between Jan 25, 2020, and Jan 31, 2021, 20 451 cases of laboratory-confirmed COVID-19 were diagnosed in Victoria. Cases initially peaked in mid-March, 2020, declined substantially, then surged rapidly between June and July, before declining again (figure 1 ). At the time of our analysis (Feb 9, 2021), samples from 15 431 (75·4%) cases had been submitted for sequencing, of which 11 711 (75·9%) met all quality control metrics and were included in our analysis (figure 1; appendix 2).
Figure 1.
Epidemic curve of COVID-19 in Victoria, Australia between Jan 25, 2020, and Jan 31, 2021
Cases of COVID-19 are plotted by reported date of COVID-19 diagnosis and coloured according to availability of sequence data for inclusion in our analysis. Cases will not have included sequence data if a sample collected from the case was not received at the sequencing laboratory, or the sample was unable to be sequenced due to insufficient volume, or failure of presequencing DNA extraction or library preparation steps, or both.
595 genomic clusters were identified, with a median of five cases per cluster (IQR 2–11; range 2–2193), including 475 genomic clusters within seven local genomic transmission networks. Overall, samples from 11 503 (98·2%) of 11 711 cases clustered with another Victorian sequence, within either a genomic cluster or transmission network.
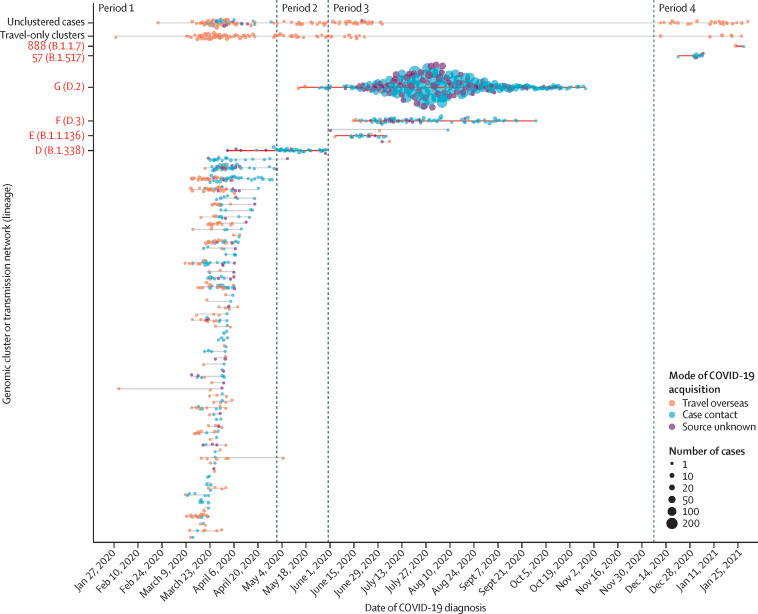
There were four discrete periods of SARS-CoV-2 emergence and transmission in Victoria (figure 2 ). Period 1 (Jan 25 to April 30, 2020) was characterised by a large number (n=114) of small, genetically diverse, travel-associated clusters, consistent with multiple importations by overseas travellers and limited local transmission (table ; appendix 1 p 6).
Figure 2.
Timeline of SARS-CoV-2 genomic clusters in Victoria, Australia between Jan 25, 2020 and Jan 31, 2021
Cases of COVID-19 in Victoria diagnosed between Jan 25, 2020, and Jan 31, 2021, are included. Cases are plotted by diagnosis date and genomic cluster or transmission network. Mode of acquisition was categorised as: travel overseas if the individual reported travel in the 14 days before symptom onset; contact with a confirmed case if no overseas travel was reported and case contact had occurred within the same time period; or source unknown. The size of the circles corresponds to the number of cases diagnosed per day within a genomic cluster or transmission network and with the same mode of acquisition, with larger circle sizes indicating a greater number of cases. Genomic clusters or transmission networks of interest during periods 2, 3, and 4, as discussed in the main text, are labelled on the y-axis (see appendix 1 p 7 for details of all genomic clusters and transmission networks).
Table.
Characteristics of confirmed COVID-19 cases, SARS-CoV-2 sequences, and genomic clusters over the four discrete periods of SARS-CoV-2 emergence and transmission in Victoria, Australia
| Period 1: Jan 25 to April 30, 2020 (n=1365)* | Period 2: May 1 to May 30, 2020 (n=285)* | Period 3: May 31 to Dec 6, 2020 (n=18 694)* | Period 4: Dec 7, 2020, to Jan 31, 2021 (n=107)* | Periods 1–4: Jan 25, 2020, to Jan 31, 2021 (n=20 451) | |
|---|---|---|---|---|---|
| Demographics | |||||
| Age, years | 47 (29–61) | 36 (24–51) | 34 (23–54) | 34 (23–47) | 35 (24–54) |
| Lives in residential care | 7 (0·5%) | 4 (1·4%) | 1946 (10·4%) | 0 | 1957 (9·6%) |
| Health-care worker† | 175 (12·8%) | 20 (7·0%) | 3706 (19·8%) | 3 (2·8%) | 3904 (19·1%) |
| Source of COVID-19 acquisition | |||||
| Case contact | 455 (33·3%) | 155 (54·4%) | 14 988 (80·2%) | 28 (26·2%) | 15 626 (76·4%) |
| Unknown source | 107 (7·8%) | 60 (21·1%) | 3594 (19·2%) | 2 (1·9%) | 3763 (18·4%) |
| Overseas travel | 803 (58·8%) | 70 (24·6%) | 112 (0·6%) | 77 (72·0%) | 1062 (5·2%) |
| Sequence availability | |||||
| Sequence data available | 870 (63·7%) | 120 (42·1%) | 10 646 (56·9%) | 75 (70·1%) | 11 711 (57·3%) |
| Did not pass quality control | 283 (20·7%) | 81 (28·4%) | 3324 (17·8%) | 32 (29·9%) | 3720 (18·2%) |
| Not sequenced | 212 (15·5%) | 84 (29·5%) | 4724 (25·3%) | 0 | 5020 (24·5%) |
| Genomic clustering | |||||
| Genomic clusters or transmission networks circulating‡ | 107/128 (83·6%) | 11/128 (8·6%) | 13/128 (10·2%) | 6/128 (4·7%) | 128/128 (100%) |
| Number of cases per genomic cluster or transmission network§ | 3 (2–6; 1–76) | 3 (2–5; 1–66) | 2 (2–3; 1–10 416) | 4 (2–5; 2–25) | 3 (2–6; 2–10 426) |
| Duration of genomic cluster or transmission network circulation, days§¶ | 7 (2–14) | 3 (1–8) | 5 (2–30) | 5 (1–10) | 7 (3–14) |
| Number of unclustered cases¶‖ | 115/870 (13·2%) | 21/120 (17·5%) | 39/10 646 (0·4%) | 33/75 (44·0%) | 208/11 711 (1·8%) |
| Number of cases in travel-only clusters‖** | 167/870 (19·2%) | 20/120 (16·7%) | 15/10 646 (0·1%) | 12/75 (16·0%) | 214/11 711 (1·8%) |
Data are median (IQR), n (%), median (IQR; range); or n/N (%).
Confirmed cases by time of initial diagnosis.
Includes individuals engaged in both clinical and non-clinical work in a health-care setting.
Includes all clusters with positive cases diagnosed in a particular period; clusters might be counted in more than one period.
Includes clusters only within the relevant time period.
Includes cases that were not clustered at the highest resolution of clustering and that were not part of a larger genomic transmission network.
Presented as a proportion of the number of sequenced positive cases per period.
Genomic clusters in which 100% of cases were suspected to have acquired SARS-CoV-2 from overseas.
Period 2 (May 1 to May 30, 2020) was characterised by near elimination of SARS-CoV-2 in Victoria, as local transmission had stopped in all but one genomic cluster (figure 2). An increase in locally acquired cases in early May, including a large outbreak in a meat processing facility, was attributed to the ongoing spread of this single genomic cluster, which diversified locally to form transmission network D (lineage B.1.338; appendix 1 p 6). An importation event was not identified for transmission network D, and the last case was identified on May 30, 2020.
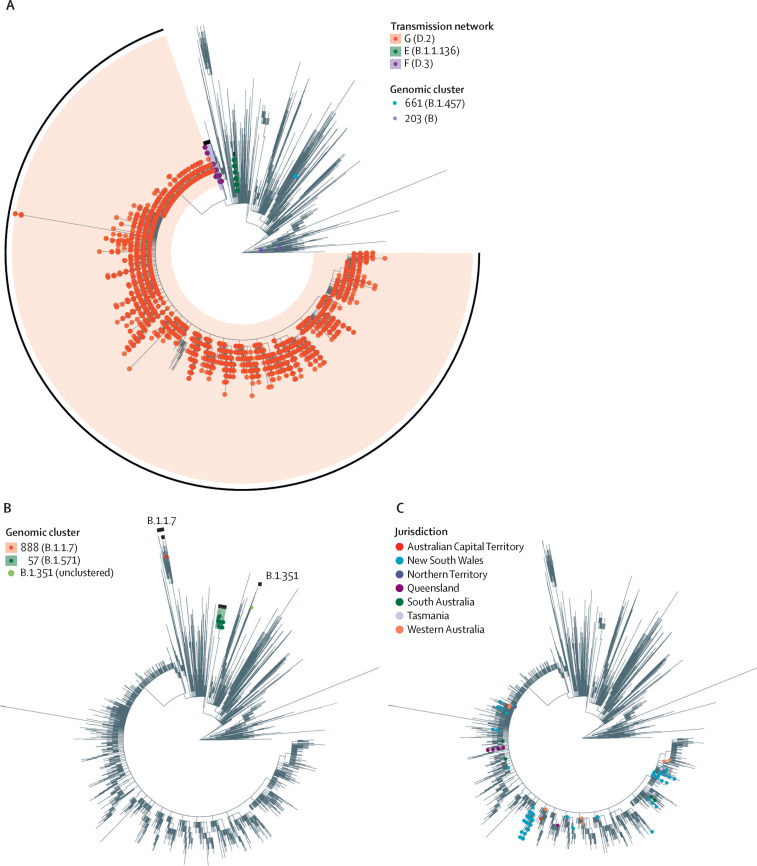
Period 3 (May 31 to Dec 6, 2020) was characterised by a rapidly expanding second wave in Victoria (figure 1). Commencing with a small number of staff at one hotel quarantine site, a surge of cases and outbreaks in the community were identified in mid-June, with no known epidemiological links to each other or to the hotel quarantine programme. Genomic analysis revealed that both staff and community cases were linked to a family of individuals who had returned from Bangladesh, were housed in the hotel staffed by the affected quarantine workers, and were diagnosed with COVID-19 shortly after arrival in Melbourne and 12 days before the first known genomically linked hotel quarantine worker. This single breach from hotel quarantine putatively introduced COVID-19 into the community via hotel quarantine workers infected by returning travellers residing in the hotel, and led to a large transmission network (genomic transmission network G, lineage D.2; Figure 2, Figure 3 ), involving 10 426 cases over 187 days, and accounting for 10 416 (98·4%) of 10 584 locally acquired cases diagnosed during period 3. Two further breaches of hotel quarantine occurred at a separate hotel in early June, resulting in 26 cases (transmission network E, lineage B.1.1.136) and 145 cases (transmission network F, lineage D.3), respectively (Figure 2, Figure 3). Consequently, all international arrivals into Victoria were stopped on June 30, 2020, the Victorian hotel quarantine programme was suspended, and interstate travel within Australia was temporarily restricted. Following the implementation of public health restrictions, no further cases of these transmission networks have been identified since Oct 29, 2020.
Figure 3.
Maximum likelihood phylogenetic trees of Australian SARS-CoV-2 samples
Samples from COVID-19 cases in Victoria diagnosed between Jan 25, 2020, and Jan 31, 2021, and interstate samples from cases uploaded to AusTrakka before Feb 11, 2021, are included. (A) Sequences from Victorian cases diagnosed during period 3 (May 31, 2020, to Dec 6, 2021) identified as within local transmission networks or genomic clusters indicated. Travel-only clusters and unclustered sequences have not been indicated. Further information on genomic clusters 661 and 203 are provided in appendix 1 (p 7). (B) Sequences from Victorian cases diagnosed during period 4 (Dec 7, 2020, to Jan 31, 2021) identified as within local genomic clusters or variants of concern are indicated. Regions of the phylogenetic tree containing B.1.1.7 and B.1.351 variants of concern are also labelled. Travel-only clusters and unclustered sequences have not been indicated. (C) Sequences from interstate samples (excluding Victoria) identified within transmission network G.
Period 4 (Dec 7, 2020, to Jan 31, 2021) was characterised by elimination of local SARS-CoV-2 transmission in Victoria. An extensively modified hotel quarantine programme commenced on Dec 7, 2020, and international travel into Victoria resumed. During period 4, 77 cases were identified in returned travellers through the hotel quarantine programme in Victoria, including 28 cases of the lineage B.1.1.7 variant of concern (figure 3B). Despite this observation, all subsequent transmission within Victoria, introduced either through hotel quarantine (genomic cluster 888, lineage B.1.1.7; figure 2), or interstate spread (genomic cluster 57, lineage B.1.517; figure 2) was successfully controlled with limited local transmission, including one instance of suspected transmission between travellers in a quarantine hotel (genomic cluster 888; figure 2). Pre-departure screening of incoming travellers to Australia was subsequently introduced on Jan 22, 2021.
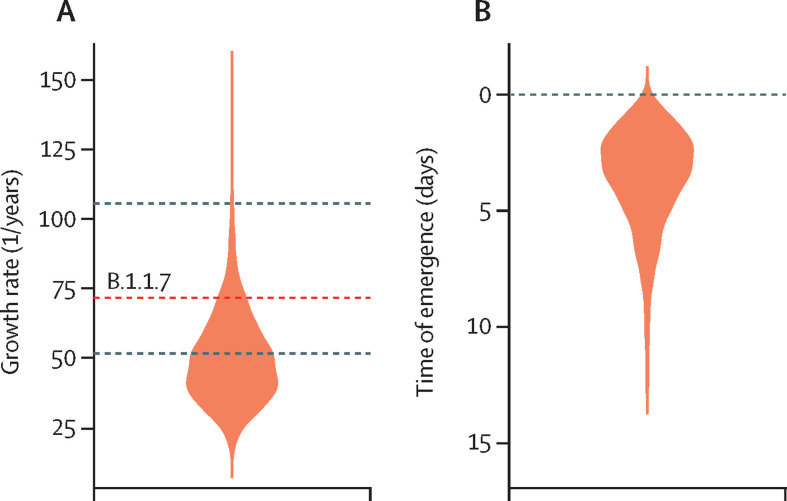
We estimated the growth rate of the early phase (June 4 to July 9, 2020) of the dominant lineage (transmission network G) in Victoria during period 3 at 47·4 putative transmission events, per branch, per year (1/years; 95% credible interval 26·0–85·0), with a corresponding doubling time of 5·3 days (95% credible interval 3·0–9·7). Our growth rate estimate is similar to, albeit somewhat lower than, that of another rapidly emerging outbreak, namely lineage B.1.1.7 in the UK, which has a mean estimated growth rate of 71·5 1/years (figure 4A ).16, 17 We also inferred a detection lag of 3·2 days (95% credible interval 0·6–8·7; figure 4B), which is much shorter than in other countries, such as the UK (14·3 days for the B.1.1.7 variant).15
Figure 4.
Growth rate and genome detection lag of transmission network G
(A) Posterior distribution of the growth rate (as putative transmission events, per branch, per year) for the exponential phase of transmission network G generated with a birth-death model. The sequences were collected from June 4 to July 9, 2020. To compare the growth rate of transmission network G with that of lineage B.1.1.7, a previous estimate of the growth rate of B.1.1.7 is represented by a red dashed line, with the 95% credible interval represented by black dashed lines.16 (B) The genome detection lag, defined as the number of days from the first local infection to the collection of the first sequenced genome, for transmission network G.
To assess the national consequences of the second wave in Victoria, we included 4001 Australian interstate and New Zealand sequences in the phylogenetic analyses (appendix 3). Between July and September, 2020 (in period 3), 1050 cases of COVID-19 were identified in states other than Victoria, of which 750 (71·4%) were thought to be locally acquired.18 In response to the ongoing outbreak, many Australian states closed their internal domestic borders, or imposed heavy restrictions on travel to and from Victoria. Despite these measures, genomic analyses revealed 501 sequences within transmission network G originating in every Australian state and territory, predominantly New South Wales (n=438) and Queensland (n=47; figure 3C). Limited local transmission was detected in three of these jurisdictions. Encouragingly, only seven publicly available sequences of this lineage have been identified outside of Australia, suggesting that the international spread of the outbreak lineage has been minimal.19, 20
Discussion
In this observational, genomic epidemiology study, we describe the rapid re-emergence and subsequent successful control of COVID-19 in Victoria, Australia. Genomics analyses identified the source of the second wave (periods 3 and 4) as hotel quarantine breaches, rather than re-emergence or ongoing cryptic community transmission from cases in the first wave (periods 1 and 2), which would have been assumed in the absence of combined genomic and epidemiological data. This finding substantially changed our understanding of the outbreak in real time and, by showing the effectiveness of previous restrictions in eliminating COVID-19 in Victoria, provided the impetus for the immediate implementation of various long-term and widespread public health interventions, including cessation of international arrivals to Victoria, increasingly widespread community lockdowns, a judicial inquiry into the hotel quarantine breaches, and, eventually, a restructure of the hotel quarantine programme.1, 21, 22
Mandatory supervised quarantine for returning travellers and other international arrivals has been an integral component of Australia's COVID-19 control strategy. Our findings show three independent breaches from hotel quarantine in mid-2020 that subsequently resulted in major modifications to the hotel quarantine programme.21, 22 A judicial inquiry identified the use of inadequately trained, supervised, and monitored private security staff in quarantine hotels as a major contributing factor, and subsequent changes to the programme included restriction of employment for those working in hotel quarantine (to prevent possible transmission from employees working across multiple casual jobs); improved training in infection control practices; amended governance of the programme to ensure transparent accountability; and increased testing of employees working as part of the programme for early detection of possible breaches.21, 23
Our identification of limited ongoing spread from subsequent incursions through the hotel quarantine programme in period 4 suggests that improvements to the programme have been successful, with risk of supervised quarantine escape estimated to be lower than in other similar settings.24 These improvements highlight the importance of stringent infection control practices and priority vaccination of quarantine workers (commenced Feb 22, 2021) to prevent ongoing importation and spread of the virus in the community. These observations also suggest that elimination can be an achievable response strategy in some settings, particularly where strong border controls can be implemented.24, 25 Our findings reinforce the need for constant vigilance, rapid identification, and swift public health responses to any potential transmission once effective elimination has been achieved. The introduction of immediate restrictions, such as so-called short, sharp, circuit breaker-type lockdowns, has been used to successfully control emerging SARS-CoV-2 outbreaks across Australia and New Zealand, where COVID-19 has also previously been eliminated.26, 27, 28
We estimate that one hotel quarantine breach alone was responsible for approximately 98% of cases in Victoria's second wave, with initial growth rates of this lineage similar to those reported for other highly transmissible lineages internationally, most notably the B.1.1.7 lineage in the UK.29, 30 This analysis aimed to compare the outbreak in Victoria with another rapidly expanding lineage; however, the scenarios that produced these two outbreaks differ substantially. The outbreak in Victoria was limited to one dominant lineage, whereas the B.1.1.7 variant was competing simultaneously with several other major lineages in the UK. The B.1.1.7 variant occurred in a population with some previous exposure to SARS-CoV-2, whereas almost all of the population in Victoria were immunologically naive. Sequences in transmission network G did not contain the N501Y mutation in the receptor-binding domain of the spike protein, which is hypothesised to contribute to the increased transmissibility of the B.1.1.7 variant and other reported highly transmissible variants (appendix 1 p 5).17, 29, 31
Although the affected populations and settings, rather than inherent transmissibility of the virus itself, could have contributed to the observed growth rates of the virus in both Victoria and the UK, our genomic, epidemiological, and phylodynamic data suggest that SARS-CoV-2 elimination is possible through rapid and sustained public health measures, even when high growth rates are observed. Additionally, although domestic air travel in July, 2020, was 82% lower than in July, 2019, national spread of this outbreak lineage occurred rapidly before implementation of strict domestic border controls.32
The role of genomic sequencing has changed considerably during the COVID-19 pandemic, particularly in low-prevalence settings such as Australia. Initially, genomic sequencing was used to assign unknown source cases to transmission networks and to clarify cases with ambiguous or multiple epidemiological links.2, 3, 7 With effective elimination now achieved, genomic sequencing analysis is currently used for the rapid identification of potential local transmission, including transmission within quarantine hotels; distinguishing long-term PCR positivity from potential reinfection;33, 34 and identifying highly transmissible variants of concern in returned travellers, who are then required to undergo extended isolation periods on arrival in Australia.17, 23, 29, 31, 35 In effect, rapid genomic epidemiological analysis is used as an enhanced outbreak detection system to inform public health responses, both at national and international levels. In particular, the identification of SARS-CoV-2 variants of concern in the UK, South Africa, Brazil, and India has prompted major changes to international travel policies. For example, the UK restricted travel from several countries in southern Africa and introduced additional testing to prevent the importation and spread of the B.1.351 variant (also known as the Beta variant).36 Similarly, in Australia, travellers identified as having a variant of concern are subject to additional clearance testing before they are allowed to leave isolation to prevent the spread of highly transmissible variants.35
Our study has several strengths and limitations. For maximum utility, rapid turnaround times for genomic sequencing are required, with clear implementation pathways from genomic analyses to public health responses.37 The use of an integrated epidemiological and genomic approach had a substantial effect on the course of the pandemic in Victoria, altering the way in which public health and government agencies viewed and responded to the pandemic and bringing genomic analyses into the mainstream public discourse. Many genomic surveillance studies are hindered by under-sampling and non-representative sequencing. Our expected high case ascertainment, comprehensive sequencing approach, and high proportion of sequenced cases minimises potential biases; however, additional transmission events from alternate sources might not have been identified. In particular, the absence of international detection of the Victorian outbreak lineage responsible for the Victorian second wave could be an underestimation of true international spread due to under-representation of sequence data from some countries and regions. However, this absence of detection might also be anticipated due to restrictions on outgoing travel from Australia. The generalisability of our approach to non-island settings with multiple routes of international entry could be limited, as indicated by the identification of local transmission following an incursion in Victoria from another Australian state after the introduction of hotel quarantine improvements.
In conclusion, our data show that rapid escalation of clonal outbreaks can occur from single importation events, and that swift and comprehensive public health responses to emergent cases are effective, even when high viral growth rates are observed. Genomic analysis has become a core component of public health responses to COVID-19, particularly in settings where effective elimination has been achieved.
For more on AusTrakka see https://www.cdgn.org.au/austrakka
For the ARTIC primers see https://github.com/artic-network/artic-ncov2019/tree/master/primer_schemes/nCoV-2019/V3
For Global Initiative on Sharing All Influenza Data see https://www.gisaid.org/
Data sharing
Consensus sequences, Illumina sequencing reads, and high resolution images of all figures are available at https://github.com/MDU-PHL/COVID19-paper, and on Global Initiative on Sharing All Influenza Data. Sequence ascension numbers can be found in appendices 2 and 3.
Declaration of interests
All authors declare no competing interests.
Acknowledgments
Acknowledgments
This study was funded by the Victorian Government, the National Health and Medical Research Council Australia (APP1149991, APP1196103, and APP1174555), and the Medical Research Future Fund (MRF9200006). We thank the public health, clinical, and microbiology staff across Victoria and Australia who have been involved in the testing, clinical care, and public health responses to COVID-19. We acknowledge The Communicable Diseases Genomics Network, supported by the Australia Commonwealth Office of Health Protection, which has led the national coordination and integration of SARS-CoV-2 genomic data in Australia.
Contributors
BPH, TS, NLS, CRL, MS, KH, SAB, AGdS, TH, PA, MW, and SAJ conceived, designed, and implemented the genomic surveillance programme. BSc, AT, SDo, CA, CL, and NS integrated the surveillance programme into public health responses. All authors were involved in developing the study methods and collecting or analysing epidemiological or microbiological data, or both. TS and KH led the bioinformatics analysis. AFP and SDu did the phylodynamic analysis and assisted with manuscript preparation. CRL, MW, SAJ, AT, SDo, PA, and NLS did the genomic epidemiological analysis. CRL drafted the manuscript, and DAW and BPH edited the manuscript. CRL, NLS, AFP, SDu, KH, PA, MW, AT, SDo, SAJ, DAW, TS, and BPH had full access to and verified all the data in the study. All authors reviewed, approved, and agreed to submit the final version of the manuscript for publication.
Supplementary Materials
References
- 1.Nørgaard SK, Vestergaard LS, Nielsen J, et al. Real-time monitoring shows substantial excess all-cause mortality during second wave of COVID-19 in Europe, October to December 2020. Euro Surveill. 2021;26 doi: 10.2807/1560-7917.ES.2021.26.1.2002023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Seemann T, Lane CR, Sherry NL, et al. Tracking the COVID-19 pandemic in Australia using genomics. Nat Commun. 2020;11:1–9. doi: 10.1038/s41467-020-18314-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Munnink BBO, Nieuwenhuijse DF, Stein M, et al. Rapid SARS-CoV-2 whole-genome sequencing and analysis for informed public health decision-making in the Netherlands. Nat Med. 2020;26:1405–1410. doi: 10.1038/s41591-020-0997-y. [DOI] [PubMed] [Google Scholar]
- 4.Tessema SK, Inzaule SC, Christoffels A, et al. Accelerating genomics-based surveillance for COVID-19 response in Africa. Lancet Microbe. 2020;1:e227–e228. doi: 10.1016/S2666-5247(20)30117-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Geoghegan JL, Ren X, Storey M, et al. Genomic epidemiology reveals transmission patterns and dynamics of SARS-CoV-2 in Aotearoa New Zealand. Nat Commun. 2020;11:1–7. doi: 10.1038/s41467-020-20235-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deng X, Gu W, Federman S, et al. Genomic surveillance reveals multiple introductions of SARS-CoV-2 into Northern California. Science. 2020;369:582–587. doi: 10.1126/science.abb9263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rockett RJ, Arnott A, Lam C, et al. Revealing COVID-19 transmission in Australia by SARS-CoV-2 genome sequencing and agent-based modeling. Nat Med. 2020;26:1398–1404. doi: 10.1038/s41591-020-1000-7. [DOI] [PubMed] [Google Scholar]
- 8.Meredith LW, Hamilton WL, Warne B, et al. Rapid implementation of SARS-CoV-2 sequencing to investigate cases of health-care associated COVID-19: a prospective genomic surveillance study. Lancet Infect Dis. 2020;20:1263–1272. doi: 10.1016/S1473-3099(20)30562-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ladhani SN, Chow JY, Janarthanan R, et al. Increased risk of SARS-CoV-2 infection in staff working across different care homes: enhanced CoVID-19 outbreak investigations in London care homes. J Infect. 2020;81:621–624. doi: 10.1016/j.jinf.2020.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Giles ML, Wallace EM, Alpren C, et al. Suppression of SARS-CoV-2 after a second wave in Victoria, Australia. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa1882. published online Dec 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hasell J, Mathieu E, Beltekian D, et al. A cross-country database of COVID-19 testing. Sci Data. 2020;7:1–7. doi: 10.1038/s41597-020-00688-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Caly L, Druce J, Roberts J, et al. Isolation and rapid sharing of the 2019 novel coronavirus (SARS-CoV-2) from the first patient diagnosed with COVID-19 in Australia. Med J Aust. 2020;212:459–462. doi: 10.5694/mja2.50569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Quick J. nCoV-2019 sequencing protocol v3 (LoCost) V.3. 2020. https://www.protocols.io/view/ncov-2019-sequencing-protocol-v3-locost-bh42j8ye
- 14.Bouckaert R, Vaughan TG, Barido-Sottani J, et al. BEAST 2.5: an advanced software platform for Bayesian evolutionary analysis. PLoS Comput Biol. 2019;15 doi: 10.1371/journal.pcbi.1006650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.du Plessis L, McCrone JT, Zarebski AE, et al. Establishment and lineage dynamics of the SARS-CoV-2 epidemic in the UK. Science. 2021;371:708–712. doi: 10.1126/science.abf2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Volz E, Mishra S, Chand M, et al. Transmission of SARS-CoV-2 lineage B.1.1.7 in England: insights from linking epidemiological and genetic data. medRxiv. 2021 doi: 10.1101/2020.12.30.20249034. published online Jan 4. (preprint). [DOI] [Google Scholar]
- 17.Leung K, Shum MH, Leung GM, Lam TT, Wu JT. Early transmissibility assessment of the N501Y mutant strains of SARS-CoV-2 in the United Kingdom, October to November 2020. Euro Surveill. 2021;26 doi: 10.2807/1560-7917.ES.2020.26.1.2002106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.COVID-19 National Incident Room Surveillance Team COVID-19 Australia: epidemiology report 26. Commun Dis Intell (2018) 2020;44:1–28. [Google Scholar]
- 19.Rambaut A, Holmes EC, O'Toole Á, et al. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat Microbiol. 2020;5:1403–1407. doi: 10.1038/s41564-020-0770-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pango Network Lineage D.2. 2021. https://cov-lineages.org/lineages/lineage_D.2.html
- 21.Coate J. COVID-19 hotel quarantine inquiry: final report and recommendations. Volume I. 2020. https://www.parliament.vic.gov.au/file_uploads/0387_RC_Covid-19_Final_Report_Volume_1_v21_Digital_77QpLQH8.pdf
- 22.State Government of Victoria Victorian Government response to the hotel quarantine inquiry. 2020. https://www.vic.gov.au/hotel-quarantine-inquiry-victorian-government-response
- 23.Victoria State Government Coronavirus update for Victoria — 9 January 2021. 2021. https://www.dhhs.vic.gov.au/coronavirus-update-victoria-9-january-2021
- 24.Grout LM, Katar A, Ouakrim DA, et al. Estimating the failure risk of quarantine systems for preventing COVID-19 outbreaks in Australia and New Zealand. medRxiv. 2021 doi: 10.1101/2021.02.17.21251946. published online April 30. (preprint). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baker MG, Wilson N, Blakely T. Elimination could be the optimal response strategy for covid-19 and other emerging pandemic diseases. BMJ. 2020;371 doi: 10.1136/bmj.m4907. [DOI] [PubMed] [Google Scholar]
- 26.Baker MG, Kvalsvig A, Verrall AJ. New Zealand's COVID-19 elimination strategy. Med J Aust. 2020 doi: 10.5694/mja2.50735. published online Aug 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Flaxman S, Mishra S, Gandy A, et al. Estimating the effects of non-pharmaceutical interventions on COVID-19 in Europe. Nature. 2020;584:257–261. doi: 10.1038/s41586-020-2405-7. [DOI] [PubMed] [Google Scholar]
- 28.Rawson T, Huntingford C, Bonsall M. Temporary “circuit breaker” lockdowns could effectively delay a COVID-19 second wave infection peak to early spring. Front Public Health. 2020;8 doi: 10.3389/fpubh.2020.614945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tegally H, Wilkinson E, Giovanetti M, et al. Emergence and rapid spread of a new severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) lineage with multiple spike mutations in South Africa. medRxiv. 2020 doi: 10.1101/2020.12.21.20248640. published online Dec 22. (preprint). [DOI] [Google Scholar]
- 30.Vöhringer H, Sinnott M, Amato R, et al. Lineage-specific growth of SARS-CoV-2 B.1.1.7 during the English national lockdown. 2020. https://virological.org/t/lineage-specific-growth-of-sars-cov-2-b-1–1-7-during-the-english-national-lockdown/575
- 31.Naveca F, da Costa C, Nascimento V, et al. SARS-CoV-2 reinfection by the new variant of concern (VOC) P.1 in Amazonas, Brazil. 2021. https://virological.org/t/sars-cov-2-reinfection-by-the-new-variant-of-concern-voc-p-1-in-amazonas-brazil/596
- 32.Bureau of Infrastructure and Transport Research Economics. Australian Government Domestic aviation activity, July 2020. 2020. https://www.bitre.gov.au/sites/default/files/documents/domestic-aviation-activity-publication-jul-2020.pdf
- 33.Tillett RL, Sevinsky JR, Hartley PD, et al. Genomic evidence for reinfection with SARS-CoV-2: a case study. Lancet Infect Dis. 2021;21:52–58. doi: 10.1016/S1473-3099(20)30764-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.To KK-W, Hung IF-N, Ip JD, et al. COVID-19 re-infection by a phylogenetically distinct SARS-coronavirus-2 strain confirmed by whole genome sequencing. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa1275. published online Aug 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Australian Government Department of Health Coronavirus Disease 2019 (COVID-19): CDNA national guidelines for public health units. May 11, 2021. https://www1.health.gov.au/internet/main/publishing.nsf/Content/cdna-song-novel-coronavirus.htm
- 36.Department for Transport Travel ban implemented to protect public health following South Africa COVID-19 outbreak. 2020. https://www.gov.uk/government/news/travel-ban-implemented-to-protect-public-health-following-south-africa-covid-19-outbreak
- 37.Bull RA, Adikari TN, Ferguson JM, et al. Analytical validity of nanopore sequencing for rapid SARS-CoV-2 genome analysis. Nat Commun. 2020;11:1–8. doi: 10.1038/s41467-020-20075-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Consensus sequences, Illumina sequencing reads, and high resolution images of all figures are available at https://github.com/MDU-PHL/COVID19-paper, and on Global Initiative on Sharing All Influenza Data. Sequence ascension numbers can be found in appendices 2 and 3.