Abstract
In human neurodegenerative diseases associated with the intracellular aggregation of Tau protein, the ordered cores of Tau filaments adopt distinct folds. Here, we analyze Tau filaments isolated from the brain of individuals affected by Prion-Protein cerebral amyloid angiopathy (PrP-CAA) with a nonsense mutation in the PRNP gene that leads to early termination of translation of PrP (Q160Ter or Q160X), and Gerstmann–Sträussler–Scheinker (GSS) disease, with a missense mutation in the PRNP gene that leads to an amino acid substitution at residue 198 (F198S) of PrP. The clinical and neuropathologic phenotypes associated with these two mutations in PRNP are different; however, the neuropathologic analyses of these two genetic variants have consistently shown the presence of numerous neurofibrillary tangles (NFTs) made of filamentous Tau aggregates in neurons. We report that Tau filaments in PrP-CAA (Q160X) and GSS (F198S) are composed of 3-repeat and 4-repeat Tau isoforms, having a striking similarity to NFTs in Alzheimer disease (AD). In PrP-CAA (Q160X), Tau filaments are made of both paired helical filaments (PHFs) and straight filaments (SFs), while in GSS (F198S), only PHFs were found. Mass spectrometry analyses of Tau filaments extracted from PrP-CAA (Q160X) and GSS (F198S) brains show the presence of post-translational modifications that are comparable to those seen in Tau aggregates from AD. Cryo-EM analysis reveals that the atomic models of the Tau filaments obtained from PrP-CAA (Q160X) and GSS (F198S) are identical to those of the Tau filaments from AD, and are therefore distinct from those of Pick disease, chronic traumatic encephalopathy, and corticobasal degeneration. Our data support the hypothesis that in the presence of extracellular amyloid deposits and regardless of the primary amino acid sequence of the amyloid protein, similar molecular mechanisms are at play in the formation of identical Tau filaments.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00401-021-02336-w.
Keywords: GSS, PrP-CAA, Tau, APrP, Cryo-EM, Neurodegeneration
Introduction
Neurodegenerative diseases with Tau pathology may present with diverse clinical symptoms and have distinct neuropathologies [20]. In fact, in addition to differences in the involvement of anatomic areas and affected cell types, Tau aggregates may also differ in Tau isoform composition and the structure of the Tau filament [3, 10–12, 16, 64]. In the adult brain, alternative mRNA splicing of exons 2, 3, and 10 of the Microtubule-Associated Protein Tau (MAPT) gene yields six Tau isoforms that differ by the absence or presence of one or two acidic N-terminus inserts, and whether they contain three or four repeats (3R, 4R) of a conserved tubulin binding motif at the C-terminus [25]. All six isoforms are found in the insoluble Tau deposits of Alzheimer disease (AD) and chronic traumatic encephalopathy (CTE), whereas only 3R-Tau isoforms are found within those of Pick disease (PiD) and only 4R-Tau isoforms in the deposits of other diseases such as progressive supranuclear palsy (PSP) and corticobasal degeneration (CBD) [20, 24, 26, 27]. Using cryo-electron microscopy (cryo-EM), it has been recently determined that the fold of the core of Tau filaments in sporadic and familial AD differs from those in PiD and CBD [3, 10–12, 16, 64]. Furthermore, although neurofibrillary tangles (NFTs) in AD, CTE, and primary age-related tauopathy (PART) [6, 8, 43] incorporate all six Tau isoforms, the fold of the core of Tau filaments is identical in AD and PART and different from that in CTE [12, 16, 55]. Thus, the Alzheimer Tau fold may be found in the presence (AD) or in the absence (PART) of amyloid β (Aβ); however, the specific role of Aβ in the pathogenesis of Tau aggregation in AD remains undetermined. In addition, whether the conformation and filament fold of Tau varies in NFTs from other neurodegenerative diseases in which the primary amyloid protein deposited is not Aβ [21–23, 59, 60] has yet to be determined.
The Prion-Protein Amyloidoses are dominantly inherited diseases, associated with missense, nonsense, and insertion mutations in the PRNP gene [22]. The co-existence of parenchymal PrP amyloid (APrP) with intraneuronal Tau aggregates has been shown to occur consistently in association with some PRNP mutations [22]; however, in Gerstmann–Sträussler–Scheinker disease (GSS) associated with the P102L mutation, the neuropathologic phenotype has consistently shown present of APrP but not Tau aggregates in neurons [22]. In Prion-Protein Cerebral Amyloid Angiopathies (PrP-CAA), which are associated with nonsense mutations in the PRNP gene [21, 22, 37], the main histologic feature is the co-existence of numerous intraneuronal Tau aggregates with APrP in the vascular compartment. Whether the co-existence of intraneuronal Tau aggregates with intraparenchymal APrP amyloid is dependent of the conformation of the APrP found in the deposits remains to be established [20, 45].
The present work focuses on PrP-CAA with a c.478C > T mutation in PRNP that leads to a truncated PrP protein (p.Q160Ter or Q160X) [17, 37], and GSS with a c.593T > C mutation in PRNP that leads to an amino acid substitution in PrP (p.F198S) [36]. In these two forms of amyloidosis [referred herein as PrP-CAA (Q160X) and GSS (F198S)], APrP coexists with numerous Tau neurofibrillary aggregates made of 3R- and 4R-Tau isoforms, suggesting that different amyloids may share pathogenetic mechanisms leading to Tau aggregation in AD, PrP-CAA, and GSS. Importantly, the anatomical distribution of the respective amyloid protein differs significantly between PrP-CAA, GSS, and AD [21, 22].
Herein, we determined for the first time the biochemical properties and atomic structures of Tau in PrP-CAA (Q160X) and GSS (F198S). This work represents a significant step towards the demonstration of the existence of different conformers of Tau in neurodegenerative diseases with Tau pathology and our understanding of the role of amyloid in the pathogenic mechanism(s) of neurodegeneration.
Materials and methods
Neuropathology
Neuropathology was carried out as previously described [21, 49]. Please see Supplementary Material Methods section for detailed information regarding clinical, genetic, and neuropathologic findings. One half of the brain was fixed in formalin and the other half was frozen at − 80 °C. Hemispheric coronal slabs were selected that included areas of the frontal, insular, temporal, parietal, and occipital lobes. These were submitted for histology and immunohistochemistry. Tissue samples were also obtained from representative brain regions. Brain sections were 8-μm thick and were counterstained with hematoxylin. Thioflavin S (Th-S) was used to demonstrate the presence of amyloid deposits and neurofibrillary tangles. For immunohistochemical studies, primary antibodies (Table S2) were AT8 (Thermo Fisher Scientific MN1020, 1:300), Abeta (NAB 228); anti PrP (Ab 95–108, 1:300); HT7 (1:1000), RD3 (Merck 05–803, 1:3,000), and RD4 (Upstate, 1:100). Double immunohistochemical studies were performed using the Dako En-Vision Doublestain System (Dako) following the manufacturer’s instructions.
Sanger DNA analysis
Genomic DNA was extracted from frozen brain tissue. Polymerase chain reaction (PCR) was performed for the amplification of the PRNP and AβPP genes as described [44, 51]. PCR products were sequenced on a CEQ 2000XL DNA analysis system (Beckman Coulter, Fullerton, CA).
Whole-exome sequencing (WES)
Target enrichment made use of the SureSelectTX human all-exon library (V6, 58 megabase pairs; Agilent) and high-throughput sequencing was carried out using a HiSeq 4,000 (sx75 base-pair paired-end configuration; Illumina). Bioinformatics analyses were performed as described [14].
Tau extraction
Sarkosyl-insoluble Tau was extracted from gray matter of fresh-frozen frontal cortex as previously described [26]. Briefly, 6 g of tissues were homogenized in A68 extraction buffer consisting of 10 mM Tris–HCl, pH 7.4, 0.8 M NaCl, 1 mM EGTA, 5 mM EDTA, and 10% sucrose with protease and phosphatase inhibitors. After a centrifugation at 20,000 × g, the supernatants were brought to 1% sarkosyl and incubated at room temperature (RT) while shaking. The supernatants were spun at 100,000 × g for 1 h at 4 °C, and the sarkosyl-insoluble pellets were resuspended in 10 µl/g tissue 50 mM Tris–HCl, pH 7.4. This resuspended pellet was further purified by 30-fold dilution in A68 extraction buffer, followed by centrifugation at 20,000 × g for 30 min at 4 °C. The pellet, containing large contaminants, was discarded, and the supernatant was centrifuged at 100,000 × g for 1 h at 4 °C. The final pellet was resuspended in 20 mM Tris–HCl, pH 7.4, and 100 mM NaCl, and stored at 4 °C.
Western blotting and dot-blot
For Western blotting, samples were resolved on 10% Tris–glycine gels (Bio-Rad) and blocked in 5% milk in TBS plus 0.1% Tween 20. For dot blots, 2 µl of diluted samples were dotted and absorbed onto 0.22 µm pore size nitrocellulose membranes (Santa Cruz), and then blocked in 5% milk in TBS plus 0.1% Tween 20. Primary antibodies were diluted in TBS plus 0.1% Tween 20 at the following dilutions: AT8 (Thermo Fisher, 1:1000), HT7 (Thermo Fisher, 1:1000), BR133, BR134 and BR135 (1:4000), RD3 and RD4 (Millipore, 1:1000), and 3F4 (Millipore, 1:1000).
Immuno-electron microscopy (EM)
Immunogold labeling was carried out as previously described [26]. Briefly, a 1 µl droplet of 1:50 insoluble tau fraction diluted in PBS was pipetted onto carbon nickel TEM grids (300 mesh, Ted Pella) and blotted dry after 2 min. The grid was blocked in 0.1% gelatin in PBS for 20 min, and then, excess solution blotted off with filter paper. Primary antibody diluted 1:50 in 0.1% gelatin in PBS was added for 1 h at RT. Grids were washed three times in 0.1% gelatin in PBS for 5 min each wash. Grids were then incubated in secondary antibody diluted 1:40 in 0.1% gelatin in PBS for 1 h at RT. Secondary antibodies used were 6 nm anti-mouse immunogold particles or 10 nm anti-rabbit immunogold particles (Electron Microscopy Sciences). Secondary antibody was washed off 3 times in 0.1% gelatin in PBS and blotted dry. Negative staining was performed with NanoVan (Ted Pella) for 5 s at RT. Pronase treatment of fibrils was carried out as described [26]. Briefly, a 5 µl solution of 0.4 mg/ml pronase was dropped onto the grid, and incubated for 5 min at RT. The pronase solution was wicked off with filter paper, and then, 0.1% gelatin solution was added to the grid for blocking. Primary and secondary antibody incubations were carried out as described above. Images were taken on a Tecnai G2 Spirit Twin scope equipped with an AMT CCD Camera.
Tau seeding assay
Tau seeds were prepared from fresh-frozen brain tissue of AD Case 2, GSS (F198S) Case 2, and PrP-CAA (Q160X) case. Tissue was thawed and homogenized in 9 volumes of buffer consisting of 10 mM Tris–HCl, pH7.4, 0.8 M NaCl, and 0.1% sarkosyl, with protease and phosphatase inhibitors. Following a 10 min spin at 10,000×g at 4 °C, the supernatants were brought to 1% sarkosyl and incubated for 1 h at RT while shaking. The supernatants were spun at 300,000×g for 1 h, and the sarkosyl-insoluble pellets were resuspended in 0.1 ml PBS/gram tissue. The resuspended pellets were sonicated with a probe sonicator at 75 W, for a total of 25 × 500 ms pulses. The probe was cleaned with isopropanol and water between samples. This resuspended pellet was then centrifuged at 100,000×g for 30 min at 4 °C. The pellet was resuspended in 20 μl/gram tissue and sonicated again as before. Following a 10 min centrifugation at 10,000×g at 4 °C, the supernatant was removed and stored at –80 °C until used as seeds on cells. HEK 293 T cells stably expressing the aggregation prone repeat domain of Tau with the disease associated P301S mutation, fused at the C-terminus to either CFP or YFP, were obtained from ATCC [33]. Cells were cultured in DMEM supplemented with 10% FBS, 1% pen/strep, and 1% GlutaMax (Invitrogen). Cells were plated at a density of 50,000 cells/well in a 12-well plate, onto coverslips treated with 0.1 mg/ml poly-d-lysine for fixed cell imaging. Cells were incubated overnight at 37 °C and 5% CO2. The following day, 1 µl seed material was combined with Lipofectamine 2000 (Thermo Fisher) and OptiMem medium, and incubated for 20 min at room temp. This transduction complex was then added onto the biosensor cells. Cells were incubated as before for 24–48 h. Cells were then washed with 1X PBS, fixed in 4% paraformaldehyde in PBS, and mounted onto microscope slides for imaging. Images were merged and cropped using ImageJ [53].
Mass spectrometry sample preparation
8 M urea, 50 mM Tris–HCl pH 8.5 (100 µl) was added to 20 µl of insoluble Tau fractions. The sample was reduced with 5 mM Tris(2-carboxyethyl)phosphine hydrochloride (TCEP) and alkylated with 10 mM chloroacetamide (CAM). The sample was diluted with 100 mM Tris–HCl to a final urea concentration of 2 M and digested overnight with 2 µg Trypsin/Lys-C Mix Mass Spectrometry (1:100 protease:substrate ratio, Promega Corporation). Peptides were desalted on a 50 mg Sep-Pak® Vac (Waters Corporation) employing a vacuum manifold (Waters Corporation). After elution from the column in 70% acetonitrile, 0.1% formic acid (FA), peptides were dried by speed vacuum, resuspended in 40 µL of 0.1% FA, and filtered through a 0.2 µm spin filter (Millipore).
Nano-LC–MS/MS analysis
Nano-LC–MS/MS analyses were performed on an EASY-nLC HPLC system (Thermo Fisher Scientific) coupled to an Orbitrap Fusion Lumos mass spectrometer (Thermo Fisher Scientific). One quarter of the sample was loaded onto a reversed phase PepMapTM RSLC C18 column (2 μm, 100 Å, 75 μm × 25 cm) with Easy-Spray tip at 450 nl/min. Peptides were eluted from 3 to 30% B over 85 min, 30 to 80% B over 3 min, 80 to 4% B for 4 min (Mobile phases A: 0.1% FA, water; B: 0.1% FA, 80% Acetonitrile). Mass spectrometer settings include capillary temperature of 300 °C and ion spray voltage was kept at 1.9 kV. Replicate injections were run with the same LC and source methods (CID mode and EtHCD/HCD mode). For the CID method, the mass spectrometer method was operated in positive ion with a 3 s cycle time data-dependent acquisition method with advanced peak determination and Easy-IC (internal calibrant). Precursor scans (m/z 400–1500) were done with an Orbitrap resolution of 120,000, RF lens% 30, maximum inject time 50 ms, 4e5 normalized AGC target, including charges of 2–7 for fragmentation with an intensity threshold of 5e3, and 60 s dynamic exclusion. CID MS2 scans were performed in the ion trap at rapid speed with a 1.6 m/z isolation window, 35% normalized CID collision energy, 2e3 AGC target, and 35 ms maximum IT. The same precursor scan settings were used for the DDA EtHCD/HCD decision tree method, but with a minimum intensity filter of 1e5. The decision tree consisted of either HCD assisted ETD or HCD (30%) activation of charge states, 3, 4, 5, and 6–8 with 2 m/z isolation window, 35 ms max IT, and 1e4 AGC target in rapid ion trap mode. The data were recorded using Thermo Fisher Scientific Xcalibur software (Thermo Fisher Scientific Inc. 2017). Data were analyzed using PEAKS X + Studio [58], including all PTMs and mutations in SPIDER search.
Negative staining for cryo-electron microscopy
Negative stain embedded imaging of Tau fibrils was carried out by applying 3 µl of sample on affinity materials coated lacey carbon grid and incubated for 30 min, then washed 5 times with DI water, followed by addition of 1% uranyl acetate (UA). Then, UA was washed, blotted out and the grid was dried in air for 4–5 min. All negatively stained images were collected on a Tecnai-12 electron microscope.
Graphene oxide coating on Grid
A pyrene solution was used to cover the entire TEM grid surface with a single layer of graphene oxide. This was achieved using a two-step sequence of pyrene solution coating (3 µl of 0.1 mg/ml) that was applied to the dull side of a lacey TEM grid (Ted Pella #01824) followed by 3 µl of 0.1 mg/ml graphene oxide that was applied just before the pyrene solution dried out. The grid was then incubated for 2 min before the dull side of the grid was washed three times with DI water and the shiny side was washed one time with a 3 µl droplet of DI water before drying and storing the hydrophilic grids for pyrene-mediated antibody coating.
Affinity capture and grid freezing
Grids for cryo-EM were prepared using the Affinity Grid technique. A multi-arm Pyrene-PEG-Antibody conjugate was synthesized. Then, a 4 µl solution of Pyrene-PEG-Antibody was applied on a GO-coated grid, incubated for 5 min, and washed three times with Tris buffer. 2.5 µl of Tau sample was applied on the antibody coated side of the grid and incubated for 30 min, and washed 5 times with Tris buffer. The final volume of liquid on the grid was adjusted to ~ 3 µl before blotting for 3 s, followed by plunge freezing into liquid ethane on a cryo-plunger 3 (Gatan) in a biosafety hood.
High-resolution cryo-EM imaging
High-resolution cryo-EM images were collected on an FEI Titan Krios at 300 kV with a Gatan K3- detector in super-resolution mode. The inelastically scattered electrons were removed using a Gatan quantum energy filter with 20 eV slit width. For PrP-CAA (Q160X), we recorded 55 movie frames with an exposure time of 62 ms/frame, with a dose rate of 1.067 electrons per Å2 per frame for a total accumulated dose of 53.35 electrons per Å2 at a pixel size of 1.078 Å. For GSS (F198S), we recorded 50 movie frames with an exposure time of 62 ms/frame, and a dose rate of 1.067 electrons per Å2 per frame for a total accumulated dose of 53.35 electrons per Å2 at a pixel size of 1.078 Å. The datasets for PrP-CAA (Q160X) Tau and GSS (F198S) Tau are composed of 2004 and 1920 micrographs, respectively, with defocus values ranging from − 0.5 to − 2.5 μm. Corrections of super-resolution frames were done for gain reference, binned by a factor of 2, and then, frames were motion-corrected and dose-weighted using MotionCor2 [65]. The contrast transfer function (CTF) of all aligned and non-dose-weighted micrographs were estimated using Gctf [63].
Helical reconstruction
All subsequent image-processing was performed in RELION 3.1 software [31, 52]. Particles were picked manually using RELION helical picker as end-to-end segments. We extracted all the segments with a box size of 768 pixel (827 Å) to cover complete crossover, down scaled to 256 pixels to speed up analysis and an inter-box distance of ~ 10% of the box length. Several rounds of reference-free 2D classifications were carried out to find homogeneous subsets and remove junk particles using a regularization value of T = 1–2. PHFs and SFs in PrP-CAA (Q160X) were visually identified from the reference 2D class averages. For PrP-CAA (Q160X) PHFs and SFs, a new 300-pixel box size without downscaling was used to re-extract with inter-box distance of approximately 15 Å. A box size of 320 pixels was used for GSS (F198S). At this stage, multiple rounds of reference-free 2D classification were carried out to discard suboptimal 2D class averages. The initial 3D reference maps were reconstructed de novo from best 2D class averages of comprising a full helical crossover. The initial round of 3D classification was low-pass-filtered to 60 Å. Several rounds of 3D classification were carried out to remove segments leading to suboptimal 3D class averages with regularization value T = 20, total classes K = 4. The final selected segments were used for final 3D auto-refinement with optimization of the helical twist and rise to yield 3D map showing clearly visible beta-strand separation and side-chain densities. We used a 10% helical z percentage parameter for post-reconstruction application of helical symmetry. The final reconstructions were sharpened using the standard post-processing procedures in RELION. The RELION helix toolbox was used to improve helical symmetry. Optimized twist and rise parameters for PrP-CAA (Q160X) PHFs, PrP-CAA (Q160X) SFs, and GSS (F198S) PHFs are 4.77 Å and – 1.2°, 4.79 Å and – 1.07°, and 4.85 Å and – 1.23°, respectively. Finally, the overall resolution was calculated from Fourier shell correlations at 0.143 between two independently refined half-maps, employing phase randomization for the convolution effects correction of an optimized, soft-edged solvent mask as implemented in the trueFSC.py program of our jspr software [57].
Model building
The previously deposited structure of PHF (PDB 5O3L) was fitted into the sharpened PrP-CAA (Q160X) and GSS (F198S) PHF density maps using Chimera [47]. The same was done for PrP-CAA (Q160X) SF using the deposited structure of SF (PDB 5O3T). The most central chain of each model (e.g., chain E) was adjusted into the electron density of each map by hand in Coot [9]. Each model was then refined using Rosetta according to a previously established procedure [66].
Density map and atomic model analysis
For each unique type of filament, the two half-maps were separately sharpened, aligned, scaled, and matched to the same density level for comparison. If the density of a region was not present in one of the half-maps, it was considered noise and not included for final evaluation. Once noise-free density feature was determined in this process, the full dataset maps for PrP-CAA (Q160X) and GSS (F198S) PHFs were globally sharpened with PHENIX [40], aligned, oriented, scaled, and matched to the density of the corresponding AD PHF map (EMD-3741) for comparison. The PrP-CAA (Q160X) SF density map was also processed and compared against the AD SF (EMD-3743) map in a similar manner. The maps were compared at the full map level, dimer level, and the monomer level.
Data availability
Cryo-EM maps have been deposited in the Electron Microscopy Data Bank (EMDB) under accession numbers EMD-23871 for PrP-CAA (Q160X) PHF, EMD-23890 for PrP-CAA (Q160X) SF, and EMD-23894 for GSS (F198S) PHF case 1. Refined atomic models have been deposited in the Protein Data Bank (PDB) under accession numbers 7MKF for PrP-CAA (Q160X) PHF, 7MKG for PrP-CAA (Q160X) SF, and 7MKH for GSS (F198S) PHF case 1. The mass spectrometry proteomics data generated in this study have been deposited to the ProteomeXchange Consortium [61] via the PRIDE partner repository [46] with the dataset identifier PXD025663. Whole-exome sequencing data have been deposited in the National Institute on Aging Alzheimer’s Disease Data Storage Site (NIAGADS; https://www.niagads.org), under accession number ng00107.
Results
Tau aggregates in Prion-Protein Amyloidoses are composed of hyperphosphorylated Tau containing three- and four-repeat Tau isoforms
One of the hallmarks of the neuropathology of PrP-CAA (Q160X) and GSS (F198S) is the co-existence of APrP and Tau deposits (Fig. 1, Figure S1). Severe Tau neurofibrillary pathology including NFTs and neuropil threads (NTs) in gray structures of the cerebrum and brainstem coexist in the same anatomical areas with APrP deposition. While APrP pathology is severe in the molecular and granule cell layers of the cerebellar cortex, there is no Tau deposition in any of the cerebellar compartments in either of the two diseases. In PrP-CAA (Q160X), APrP angiopathy coexists with severe limbic and neocortical Tau pathology that, as in GSS (F198S), is also characterized by the presence of numerous NFTs and NTs, that are decorated using antibodies recognizing hyperphosphorylated Tau and 3R- and 4R-Tau (Figure S1). APrP fibrillary deposits are decorated by antibodies recognizing PrP and visualized within vessel walls and intimately adherent to them, often forming rosary-like structures. APrP angiopathy involves the walls of small- and medium-sized parenchymal and leptomeningeal blood vessels. In GSS (F198S), NFTs and NTs are decorated using antibodies recognizing hyperphosphorylated Tau, 3R- and 4R-Tau (Figure S1). Tau deposits are seen in neuronal perikarya, in NTs, and in the processes that surround PrP plaques (Fig. 1, Figure S1). PrP plaques may be diffuse or have cores (unicentric or multicentric plaques) that are predominantly in layers 1, 4, 5, and 6 of frontal, insular, temporal, and parietal cortices. It is noteworthy that Tau pathology is equally severe in PrP-CAA (Q160X) and GSS (F198S), even though the APrP pathology is localized in different compartments: i.e., blood vessels walls in PrP-CAA (Q160X) versus neuropile in GSS (F198S).
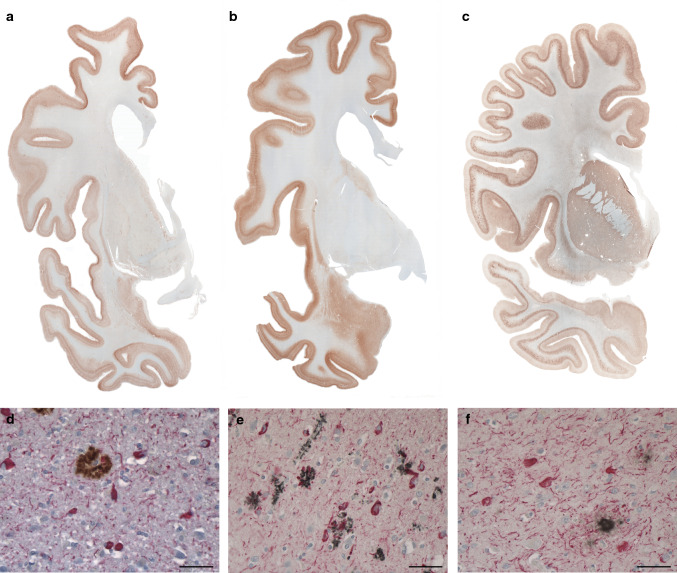
Fig. 1.
Immunohistochemistry in PrP-CAA (Q160X) and GSS (F198S) compared to AD. Hemispheric coronal sections show the distribution of Tau pathology at the level of the cerebral cortex, caudate nucleus and putamen in a case of AD (Case 1, a), cerebral cortex, amygdala, and caudate nucleus in PrP-CAA (Q160X) b, and cerebral cortex, caudate nucleus, putamen, and claustrum in GSS (F198S) (Case 1, c). Double immunohistochemistry of Tau and Aβ in AD (d), Tau and PrP in PrP-CAA (Q160X) (e), and Tau and PrP in GSS (F198S) (f). Nerve cell bodies and NTs are reactive for Tau (red) and are seen in the vicinity of parenchymal Aβ plaques (brown) in AD (d), blood vessels with APrP angiopathy (black) in PrP-CAA (Q160X) (e), or parenchymal APrP plaques (black) in GSS (F198S) (f). a–f, Anti-Tau antibody AT8; d Anti-Aβ antibody NAB 228; e, f Anti-PrP antibody 95–108. Scale bar, 50 µm
Western blot analysis of sarkosyl-insoluble fractions shows the presence of Tau bands with a migration pattern indistinguishable from that seen in AD (Fig. 2, Figure S2a). Western blot analysis also shows that APrP does not co-purify with Tau in our Tau preparations (Fig. 2a). The purified sarkosyl-insoluble fraction enriched for Tau aggregates induces Tau aggregation in a biosensor cell system derived by transducing HEK293T cells with 2 separate lentiviral constructs encoding Tau RD P301S-CFP and Tau RD P301S-YFP [18, 33]. Tau isolated from the frontal cortex of PrP-CAA (Q160X) and GSS (F198S) leads to the formation of aggregates similar to those from Tau isolated from AD brains (Fig. 2b). Aggregation is not seen in samples prepared from frontal cortex (FC) from controls nor from the cerebellum (CB) of PrP-CAA (Q160X) and GSS (F198S) patients, where PrP pathology is severe, but Tau aggregates are not detected.
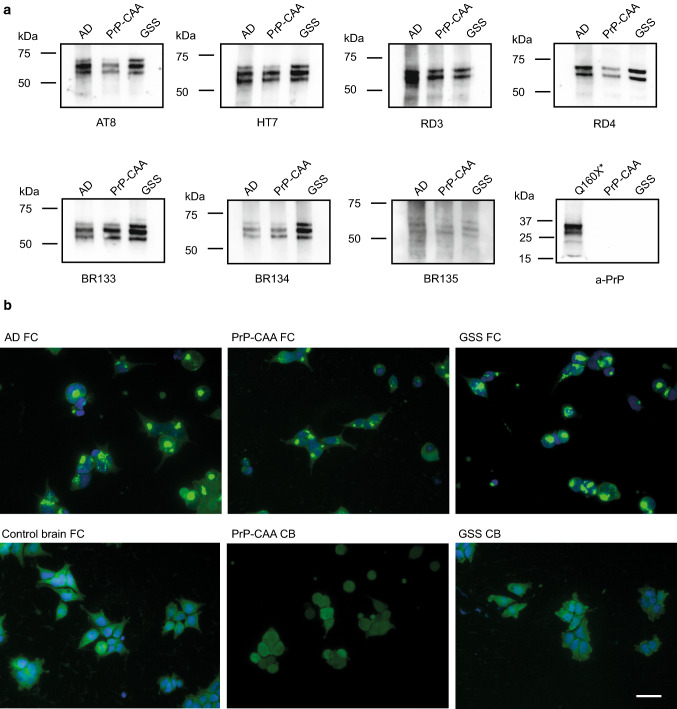
Fig. 2.
PrP-CAA (Q160X) and GSS (F198S) Tau are indistinguishable from AD Tau by Western blot or seeding assay. Western blots of sarkosyl-insoluble Tau fractions from AD (Case 2), PrP-CAA (Q160X), and GSS (F198S) (Case 2) show that Tau aggregates consist of full length, hyperphosphorylated Tau with an identical electrophoretic pattern, consisting of major bands of 60, 64, and 68 kDa. No PrP immunoreactivity is seen in the sarkosyl-insoluble Tau fractions of PrP-CAA (Q160X) and GSS (F198S). Total brain homogenate from PrP-CAA (Q160X*) was used as positive control for PrP (a). Tau biosensor cells incubated with the sarkosyl-insoluble fraction obtained from frontal cortex (FC) of AD (Case 2), PrP-CAA (Q160X) and GSS (F198S) (Case 2) show Tau seeding activity, whereas the insoluble fraction of FC from control brain, and cerebellum (CB) from PrP-CAA (Q160X) and GSS do not seed Tau aggregation in vitro (b). Scale bar: 20 µm
The sarkosyl-insoluble fraction enriched in Tau aggregates from PrP-CAA (Q160X) and GSS (F198S) was incubated in the presence or absence of pronase and evaluated by a dot-blot assay using a variety of antibodies against different epitopes of Tau protein (Figure S2a). We compared the susceptibility of the Tau preparation to pronase digestion in the two diseases to that of Tau fibrils isolated from AD, CTE, CBD, and PiD. Digestion with pronase removes the epitopes recognized by the BR133, HT7 and AT8 antibodies, located in the N-terminal portion of the molecule. It also removes the epitope recognized by the BR134 antibody, located in the C-terminus of Tau. Both N- and C-terminal portions of Tau are also referred to as the “fuzzy coat” [68]. Interestingly, the epitope recognized by the BR135 antibody is partially resistant to pronase digestion, suggesting that this sequence is part of the filament core as in AD, CTE, CBD, and PiD (Figure S2b) [10–12, 16, 64].
Analysis of dispersed preparations of Tau filaments by transmission electron microscopy (TEM) suggests that Tau aggregates from GSS (F198S) may be composed predominantly of paired helical filaments (PHFs) that appear to be identical to those seen in AD (Figure S2c). In PrP-CAA (Q160X), we estimate a ratio of ~ 4:1 PHFs to straight filament (SFs), while SFs could not be detected in the three different GSS (F198S) cases analyzed, suggesting that PHFs may be the predominant (if not the only) type of filaments in GSS (F198S). Using antibodies against different epitopes of Tau protein (Figure S2a) in immuno-EM, we observe that Tau filaments in PrP-CAA (Q160X) and GSS (F198S) are composed of full-length, hyperphosphorylated Tau. Immuno-EM analysis of Tau fibrils after pronase digestion shows that the cores of Tau filaments in PrP-CAA (Q160X) and GSS (F198S) share epitopes located in the R3 and R4 repeat domains. Treatment with pronase removes the fuzzy coat, composed of N- and C-terminal sequences, abolishing the positive labeling observed before pronase treatment. The epitope recognized by antibody BR135, which labels Tau on Western blots of dispersed filaments and in dot-blot analyses, is located in the core of Tau filaments, and is not accessible to the antibody before or after pronase treatment by immuno-EM (Figure S2c). These findings are consistent with the presence of the same Tau sequences in the core of Tau filaments in PrP-CAA (Q160X), GSS (F198S), and AD [11, 16].
Cryo-EM of Tau filaments in PrP-CAA (Q160X) and GSS (F198S)
We determine the structure of Tau filaments at high resolution by cryo-EM imaging and 3D reconstruction. We observe two types of Tau filaments within the sarkosyl-insoluble fraction of PrP-CAA (Q160X), similar to PHFs and SFs of AD (Fig. 3a,c). PHFs were ~ 70% and SFs were ~ 30% of the filament population, both composed of two protofilaments with C-shaped subunits (Figure S3) [16]. Two-dimensional classification readily separated PHF and SF for further processing (Fig. 3d,e). We determine the structure of PHFs and SFs in PrP-CAA (Q160X) using helical reconstruction in RELION3.1 to 3.0 Å resolution (Fig. 3a, c, f) (Table S1). The crossover distance for both types of filaments is approximately between 700 and 850 Å with a width of about 40–300 Å. In GSS (F198S), the protofilament organization of PHFs is identical to that of PrP-CAA protofilaments; however, protofilaments of SF were not visualized. Using helical reconstruction in RELION3.1, we determine a 3.2 Å resolution map of the ordered core of PHFs from GSS (F198S) (Fig. 3b, f), with the same crossover distance and width of PHFs from PrP-CAA (Q160X), and the same monomeric fold as the AD fold, i.e., the C-shaped unit (Figure S3).
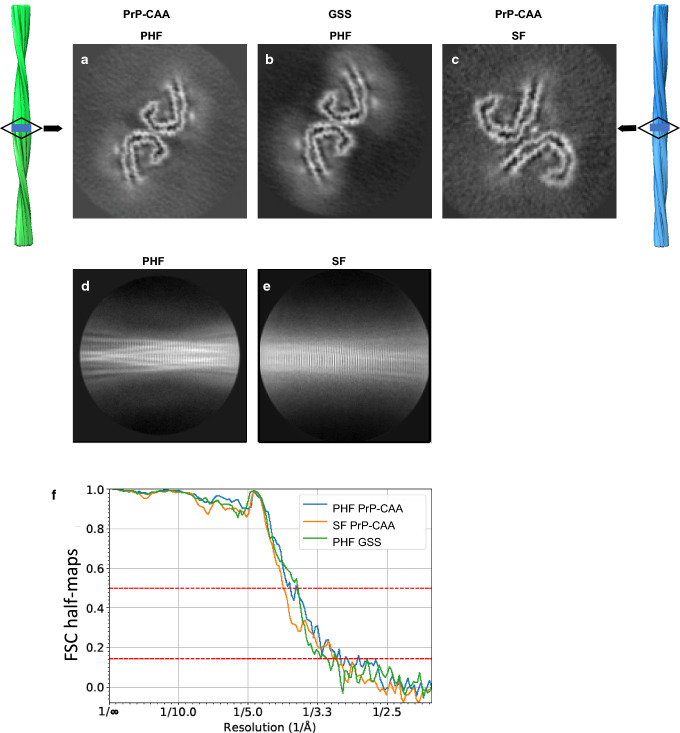
Fig. 3.
Cryo-EM reconstructions of PHF and SF from PrP-CAA (Q160X) and PHF from GSS (F198S). The structures show identical pairs of C-shaped protofilaments and the same inter-protofilament packing between PHFs (a, b) but different packing for SFs (c). The rectangular boxes in the vertical, surface view of the helical reconstructions of PHFs (left, green), and SFs (right, blue) show the location of the cross-sectional densities (a–c). 2D class averages of PHF (d) and SF (e) of PrP-CAA (Q160X). Fourier Shell Correlation (FSC) curves for independently refined half-maps of PrP-CAA (Q160X) PHFs and SFs, and GSS (F198S) PHFs (f)
The cores are composed of eight β-strands (Figure S3, S4a) adopting a C-shaped architecture that is perpendicular to the axial direction of the filament encompassing residues V306 to F378 of Tau in PrP-CAA (Q160X), and K274 to R379 of 3R Tau and S305 to R379 of 4R Tau in GSS (F198S) (Figure S3). The cores include residues located in the R3 and R4 repeat domains, with additional densities extending from the N- and C-terminal regions of the core (Figure S4a). The polar amino acids have side chains mostly facing the solvent (outside the protofilament core) and the non-polar amino acids like valine and isoleucine are facing inwards in the protofilament core forming hydrophobic patches (Fig. 4). PHFs and SFs in PrP-CAA (Q160X) and PHFs in GSS (F198S) have 8 β-strands that pack together to form the fold (Figure S3i-k). The β1 and β2 strands run anti-parallel to β8 (in a cross-β pattern), β3 runs anti-parallel to β7, and β4, β5 and β6 are arranged in a triangular fashion. In between the β-strands, glycine residues forming β-turns and proline residues breaking the β-strands reduce the stress of having 8 β-strands in one protofilament core (Fig. 4). Ordered in-register H-bonding between the residues in the β-strands further reduce this stress. The residues in β1 are mostly hydrophobic, packing with the last few residues of β8 strand (L376 and F378). Contrary to that, the polar residues in the β2 strand are facing inwards to interact with the polar residues of the β8 strand (Figure S3, Fig. 4). A β-turn consisting of G323 and G326 separates the β2 and β3 strands and provides a 90degree turn to the core. The β3 strand is followed by the 332PGGG motif that forms an interface with the other protofilament forming a PHF (Figure S4b) and provides another turn in the core structure. V337 and V339 from the β4 strand interact with I354 of the β6 strand and L357 of β7 strand to form another hydrophobic patch. The β4–β6 strands form a β-helix-like configuration where the three strands form a triangle-like structure (Fig. 4, Figure S3). The inside of the β-helix is filled with hydrophobic amino acid side chains (V339, L344, F346, V350, and I354). The side chains of the polar amino acids are facing outwards and they are mostly alternatively charged. E342–K343 and K347–R349 residues form the two corners of the triangle. G355 provides the turn between β6 and β7 strands. The β7 and β8 strands are interspersed by the 364PGGG motif that provides the 90 degree turn between them. PHFs and SFs differ in how the two C-shaped protofilaments interact with each other (Fig. 4, Figure S4b). In PHFs, the two protofilaments are related by a 21 screw symmetry with only a small shift (~ 2.4 Å) along the filament axis, which results in the two C-shaped subunits appearing to be inverted and symmetrically interacting with each other (Fig. 4). The interface between protofilaments (Figure S4b) is formed by the anti-parallel stacking of residues P332 to Q336, with the G residues 333 to 335 forming H-bondings, and two additional H-bonds between Q336 and K331 from the opposite protofilament. The protofilament surface of PrP-CAA (Q160X) and GSS (F198S) PHFs is similar to the filament surface of PHFs in AD (Figure S4B). The two protofilaments in SF structures are asymmetrically arranged. In SFs, the sidechains of residues P312–K321 of one protofilament are in close proximity with the sidechains of the residues K317–S324 of the other protofilament, while the surface between the two protofilaments is not stabilized by H-bonds like in PHFs.
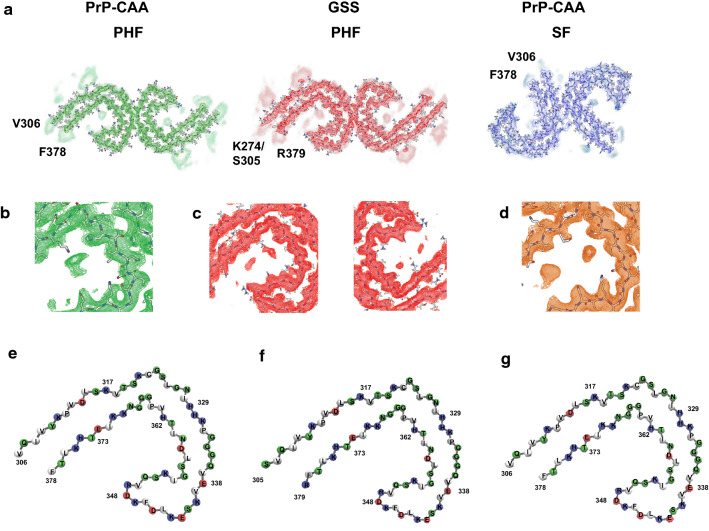
Fig. 4.
Cryo-EM densities and atomic models of PHFs and SFs. Sharpened, high-resolution maps are shown in green (PHF, PrP-CAA (Q160X)), red (PHF, GSS (F198S)), and blue (SF, PrP-CAA (Q160X)). The blurred map regions represent extra densities low-pass-filtered to 5 Å (a). PrP-CAA (Q160X) PHF extra density (b) and GSS (F198S) extra density (c) around the core have different locations and orientations to that of the extra density of AD PHF (d). Schematic view of the PrP-CAA (Q160X) (e) and GSS (F198S) (f) PHF protofilament cores compared to AD (g) PHF (PDB: 5o3l) showing the similarities in the folds. The colors represent the polarity of the amino acids (red: negatively charged, blue: positively charged, white: non-polar)
Additional densities exist around the protofilament core
We observe additional densities around the protofilament cores (Figs. 3, 4). As illustrated in Fig. 4 by the low-pass filtered density map, both N- and C-termini of the cores have disordered densities. In the cores of PHFs in PrP-CAA (Q160X) and GSS (F198S), the sidechains of K317, T319 and K321 interact with an additional density outside the core. Similarly, this strong additional density is seen in the SFs of PrP-CAA (Q160X) at the protofilament interface. Fitzpatrick et al. [5] hypothesized these additional densities to be the 7EFE9 motif at the N-terminus of the Tau protein and proposed that the formation of salt bridges stabilized the interaction between two SF protofilaments. To further investigate the extra densities seen near residues within the core of Tau PHFs and SFs, we perform mass spectrometry (MS) on Tau filaments from AD, PrP-CAA (Q160X) and GSS (F198S). We achieve 100% coverage of the core region of filaments from all 3 diseases. We determine Tau filaments in each disease to be highly post-translationally modified, especially in respect to phosphorylation, acetylation, ubiquitination, and deamidation (Table S3). Specifically, we find peptides in which K317 is ubiquitinated in Tau filaments from all 3 diseases, which may correlate to the large extra density near this residue in both PHFs and SFs, therefore suggesting that ubiquitination of K317 may also contribute to SF stabilization. A density present near residues I371 and T373 in PHFs and SFs in AD is not present in Tau filaments from PrP-CAA (Q160X) (Fig. 4) nor do we find any PTMs on these residues via MS (Table S3). In addition, a density that interacts with the H362 residue side-chain in Tau filaments in AD is seen to interact with the side-chain of K369 in Tau filaments from PrP-CAA (Q160X) and GSS (F198S) (Fig. 4). In fact, our MS results determine that K369 is acetylated in both AD and PrP-CAA (Q160X), whereas no PTMs are observed on H362, suggesting that this extra density may be interacting with K369 as opposed to H362. Interestingly, Tau in GSS (F198S) seems to be less phosphorylated at serine residues (S320, S324 and S356) located in the R3 and R4 region than in Tau filaments from AD and PrP-CAA (Q160X) (Table S3).
Discussion
PrP-CAA (Q160X) and GSS (F198S) are two Prion-Protein Amyloidoses that have distinct clinical and neuropathologic phenotypes. They differ significantly from Creutzfeldt–Jakob disease (CJD), the most common form of prion disease [22]. PrP-CAA (Q160X) and GSS (F198S) are caused by mutations in the PRNP gene that lead to APrP formation and deposition in the vascular compartment in PrP-CAA (Q160X) and in the brain parenchyma in GSS (F198S) [21, 23, 37]. Interestingly, both conditions are neuropathologically characterized by the presence of Tau aggregates. Western blot of Tau aggregates from PrP-CAA (Q160X), GSS (F198S), and AD show identical pattern of migration and immunoreactivity using antibodies against 3R- and 4R-au, strongly suggesting that Tau aggregates in PrP-CAA (Q160X) and GSS (F198S) are composed by 3R- and 4R-au isoforms. By TEM of negative stained preparations, Tau aggregates in PrP-CAA (Q160X) are found to be composed of PHFs and SFs, while in GSS (F198S) Tau aggregates are found to be predominantly, if not entirely, composed of PHFs. Analysis by immuno-EM coupled to pronase treatment suggests that the core of the Tau filaments in PrP-CAA (Q160X) and GSS (F198S) contains the R3 and R4 repeats, as in AD. By cryo-EM, PHFs and SFs are found to be made of two protofilaments with a common cross-β/β-helix C-shaped architecture, as in AD [11, 16]. The core encompasses residues V306 to F378 of Tau in PrP-CAA (Q160X), and K274 to R379 of 3R Tau and S305 to R379 of 4R-Tau in GSS (F198S), with disordered densities sensitive to pronase degradation extending from the N- and C- termini. This is similar to the recently reported high-resolution cryo-EM structures of PHFs and SFs from AD, with R379 and E380 from the sequence after R4 at the C-terminus, and G304 and S305 from R2 in 4R Tau, and G273 and K274 from R1 in 3R-Tau at the N-terminus [11]. Additional densities are identified near the core of Tau filaments from PrP-CAA (Q160X) and GSS (F198S), specifically at the sidechains of K317, T319, and K321. By mass spectrometry analysis, we establish the presence of extensive PTMs in Tau aggregates from PrP-CAA (Q160X) and GSS (F198S). We observe K317 to be ubiquitinated in AD, PrP-CAA (Q160X) and GSS (F198S), suggesting that the extra densities observed by cryo-EM near this residue in PrP-CAA (Q160X) and GSS (F198S) correspond to ubiquitination at this lysine. We also observe PTMs that have been reported to be important in Tau fibrillization and propagation in AD [27, 28], for example, phosphorylation at T231, thought to be central to the initial steps of Tau detachment from microtubules and subsequent aggregation [1, 2], is found in our cases of AD, PrP-CAA (Q160X) and GSS (F198S). We also observe PTMs within the two hexapeptide motifs, 275VQIINK280 and 306VQIVYK310, which are known to be the minimum required motifs for Tau protein aggregation [39]. By MS, we observe that Tau from PrP-CAA (Q160X) is phosphorylated at S199 and S202, while Tau from GSS (F198S) is phosphorylated at S202; however, Tau deposits were immunoreactive for the AT8 phosphoepitope (phosphorylation at residues S202 and T205 but also other epitopes, including S199 and S208) [41] by immunohistochemistry, immunoblotting, and immuno-EM methods. PHF-1 also decorates Tau aggregates in PrP-CAA (Q160X) and GSS (F198S) (not shown), and by MS, we are able to detect the phosphorylated residues S396 and S404, which constitute the epitope recognized by antibody PHF-1 (directed against the doubly phosphorylated epitope S396 and S404) [29, 32]. The MS data also show deamidation of N279, which has been suggested to contribute to the AD folds as opposed to the folds seen in CBD [3, 7]. However, we observe deamidation at this residue only in AD and PrP-CAA (Q160X), but not in GSS (F198S) tryptic peptides, suggesting that this PTM may contribute to PHF over SF formation, and offer explanation as to the lack of SFs in GSS (F198S). In addition, we observe phosphorylation at T175 and S237 in all three diseases. It has recently been reported that singly and doubly ubiquitinated peptides at residues K311 and K317 and phosphorylation at T217 and S262 may differentiate between AD and control groups [67]. We find that all these residues are post-translationally modified in PrP-CAA (Q160X) and GSS (F198S). We observe peptides containing K311 to be either ubiquitinated or acetylated, while K317 is always found ubiquitinated. Although we do not find changes in K280, PTMs in K280 and K311 have been shown to regulate Tau fibrillization in vitro [62]. It has been previously suggested that extra densities near K317 and K321 of AD and CTE Tau may be formed by the 7EFE9 motif of Tau, which may stabilize the protofilament interface of SFs [12, 16]. We identify ubiquitination of K317 in PrP-CAA (Q160X) and GSS (F198S) Tau, suggesting ubiquitin at this site as another possible explanation for the extra density seen via cryo-EM. Additional reports have suggested that ubiquitination of K311 and K317 of the core of Tau filaments would lead to SF formation, rather than PHF [7]; however, our data show that despite ubiquitination at K311 and K317 of tryptic peptides from GSS (F198S), no SFs are found within GSS (F198S) Tau filaments. In addition, we do not find peptides phosphorylated at S320, S324 and S356 in GSS (F198S), which are located in the R3 and R4 region and are phosphorylated in AD and PrP-CAA (Q160X). Our data suggest that these specific PTMs may not be critical for SF formation, and that PTMs may not fully mediate the structural diversity seen between Tau filaments from different diseases. Phosphorylation at T175, S237, and ubiquitination at K281, have been associated with symptomatic AD at Braak VI stage [5, 67]
Amyloid and Tau aggregates coexist in AD and in other diseases in addition to the group of the PrP Amyloidoses [22], two of which are reported here. In fact, in other hereditary cerebral amyloid diseases such as Familial British dementia (FBD) [34, 59] and Familial Danish dementia (FDD) [35, 60], a severe neurofibrillary Tau pathology occurs. Our study shows for the first time that Tau fibrils deposited in the brain of individuals with a brain amyloidosis other than AD are biochemically, antigenically, and structurally identical. Moreover, a recent study shows that Tau fibrils isolated from the brain of individuals with FBD and FDD are also structurally identical to those in AD [56]. The co-existence of Tau aggregates with different types of amyloids suggests a common mechanism through which amyloids, whether Aβ in AD, APrP in Prion diseases, ABri in FBD or ADan in FDD, trigger aggregation of Tau, resulting in Tau filaments with identical structure at their core (Fig. 5). Furthermore, Tau from the brains of patients with AD, GSS (F198S), and PrP-CAA (Q160X) have similar seeding activities in vitro, as has been also seen for brain homogenates from AD and PART [38]. For AD, it has been proposed that Aβ provides a crucial element toward Tau aggregation [4, 30]. This hypothesis has been supported by genetic forms of AD due to mutations in the AβPP, PSEN1, and PSEN2 genes that consistently alter the metabolism of Aβ, with a consequent Tau hyperphosphorylation and formation of Tau aggregates in vitro and in vivo [15, 19, 28]. Altered Tau metabolism in association with APrP has also been observed in in vitro studies [42] and in vivo in mouse models [48, 50]. By determining the structure of the core of Tau filaments from diseases caused by two distinct PRNP mutations, F198S and Q160X, to be identical to the core of Tau filaments from AD, we uncover potential links between amyloid proteins and the resulting Tau aggregation. Structural data are urgently needed for the identification of specific ligands for in vivo imaging of Tau aggregates in a wide range of neurodegenerative diseases.
Fig. 5.
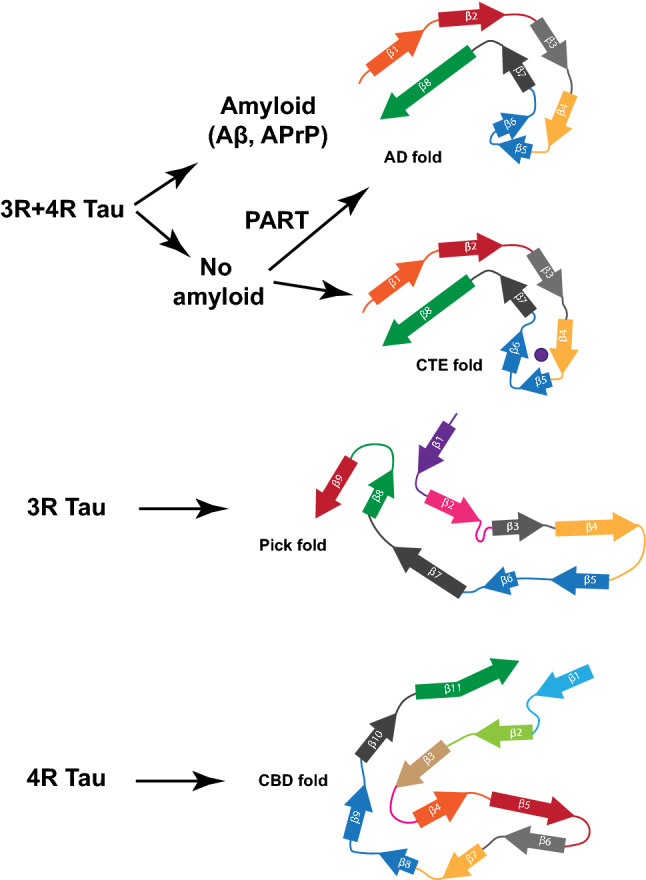
Different folds of Tau that have been identified to-date. Two different Tau folds are associated with 3R and 4R-Tau and make the Tau aggregates in AD, PrP-CAA (Q160X), GSS (F198S), PART, and CTE. A Tau fold associated with 3R-Tau makes the inclusions of Pick disease and a Tau fold associated with 4R-Tau makes the aggregates in CBD. Among the four folds, one known as the Alzheimer Tau fold can occur in the presence or in the absence of an extracellular amyloid deposition. Analysis of additional diseases characterized by 3R-, 4R-, and 3R and 4R-Tau will determine whether this is the complete Tau fold landscape or if additional folds may be found associated with different diseases
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
We thank the families of the patients for donating brain tissues; M. Goedert and S.H.W. Scheres for encouragement, helpful discussions and antibodies; U. Kuederli, M. Jacobsen, F. Epperson, and R.M. Richardson for human brain collection and analysis; F.W. Unverzagt, M.R. Farlow, and M. Geschwind for clinical support; K. Newell for providing control tissue. This work was supported by the US National Institutes of Health (P30-AG010133, U01-NS110437, RF1-AG071177), and the Department of Pathology and Laboratory Medicine, Indiana University School of Medicine. This publication was made possible, in part, by the Stark Neurosciences Research Institute, the Indiana Alzheimer Disease Center, Eli Lilly and Company, and by support from the Indiana Clinical and Translational Sciences Institute funded, in part by Grant Number UL1TR002529 from the National Institutes of Health, National Center for Advancing Translational Sciences, Clinical and Translational Sciences Award. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. We thank the Purdue Cryo-EM Facility (http://cryoem.bio.purdue.edu) for the use of the Titan Krios microscope. Mass spectrometry was provided by the Indiana University School of Medicine Proteomics Core Facility. We acknowledge the Center for Medical Genomics of Indiana University School of Medicine for next-generation DNA sequencing.
Author contributions
GIH, BG, WJ, and RV conceived and coordinated the study; GIH performed protein purifications and biochemical studies; GIH, AF, and RV performed mass spectrometry studies; HJG and RV performed genetic studies; BG performed neuropathologic analyses; MRH performed affinity grid preparation, cryo-EM, and helical reconstruction; MRH, MG, and WJ analyzed cryo-EM data; FSV built the atomic models. GIH, MRH, MG, FSV, AF, HJG, BG, WJ, and RV analyzed the data; GIH, MRH, MG, BG, and RV drafted the images for publication; GIH, MRH, MG, BG, WJ, and RV wrote the manuscript. All authors read and approved the final manuscript.
Declarations
Conflict of interest
The authors declare no competing interests.
Footnotes
These authors jointly supervised this work: Ruben Vidal, Wen Jiang, Bernardino Ghetti.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Grace I. Hallinan and Rejaul Hoq contributed equally to this work.
Contributor Information
Ruben Vidal, Email: rvidal@iupui.edu.
Wen Jiang, Email: jiang12@purdue.edu.
Bernardino Ghetti, Email: bghetti@iupui.edu.
References
- 1.Alonso AD, Di Clerico J, Li B, Corbo CP, Alaniz ME, Grundke-Iqbal I, et al. Phosphorylation of Tau at Thr212, Thr231, and Ser262 combined causes neurodegeneration. J Biol Chem. 2010;285(40):30851–30860. doi: 10.1074/jbc.M110.110957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alquezar C, Arya S, Kao AW. Tau post-translational modifications: dynamic transformers of tau function, degradation, and aggregation. Front Neurol. 2021;11:595532. doi: 10.3389/fneur.2020.595532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arakhamia T, Lee CE, Carlomagno Y, Duong DM, Kundinger SR, Wang K, et al. Posttranslational modifications mediate the structural diversity of tauopathy strains. Cell. 2020;180(4):633–644. doi: 10.1016/j.cell.2020.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beyreuther K, Masters CL. Amyloid precursor protein (APP) and beta A4 amyloid in the etiology of Alzheimer’s disease: precursor-product relationships in the derangement of neuronal function. Brain Pathol. 1991;1(4):241–251. doi: 10.1111/j.1750-3639.1991.tb00667.x. [DOI] [PubMed] [Google Scholar]
- 5.Braak H, Braak E. Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol. 1991;82(4):239–259. doi: 10.1007/BF00308809. [DOI] [PubMed] [Google Scholar]
- 6.Crary JF, Trojanowski JQ, Schneider JA, Abisambra JF, Abner EL, Alafuzoff I, et al. Primary age-related tauopathy (PART): a common pathology associated with human aging. Acta Neuropathol. 2014;128:755–766. doi: 10.1007/s00401-014-1349-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dan A, Takahashi M, Masuda-Suzukake M, Kametani F, Nonaka T, Kondo H, et al. Extensive deamidation at asparagine residue 279 accounts for weak immunoreactivity of tau with RD4 antibody in Alzheimer’s disease brain. Acta Neuropathol Commun. 2013;1(1):54. doi: 10.1186/2051-5960-1-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duyckaerts C, Braak H, Brion JP, Buée L, Del Tredici K, Goedert M, et al. PART is part of Alzheimer disease. Acta Neuropathol. 2015;129(5):749–756. doi: 10.1007/s00401-015-1390-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Cryst. 2010;D66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Falcon B, Zhang W, Murzin AG, Murshudov G, Garringer HJ, Vidal R, et al. Structures of filaments from Pick’s disease reveal a novel tau protein fold. Nature. 2018;561(7721):137–140. doi: 10.1038/s41586-018-0454-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Falcon B, Zhang W, Schweighauser M, Murzin AG, Vidal R, Garringer HJ, et al. Tau filaments from multiple cases of sporadic and inherited Alzheimer’s disease adopt a common fold. Acta Neuropathol. 2018;136(5):699–708. doi: 10.1007/s00401-018-1914-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Falcon B, Zivanov J, Zhang W, Murzin AG, Garringer HJ, Vidal R, et al. Novel tau filament fold in chronic traumatic encephalopathy encloses hydrophobic molecules. Nature. 2019;568(7752):420–423. doi: 10.1038/s41586-019-1026-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Farlow MR, Murrell J, Ghetti B, Unverzagt F, Zeldenrust S, Benson M. Clinical characteristics in a kindred with early-onset Alzheimer's disease and their linkage to a G→T change at position 2149 of the amyloid precursor protein gene. Neurology. 1994;44(1):105–111. doi: 10.1212/WNL.44.1.105. [DOI] [PubMed] [Google Scholar]
- 14.Farlow JL, Robak LA, Hetrick K, Bowling K, Boerwinkle E, Coban-Akademir ZH, et al. Whole-exome sequencing in familial Parkinson disease. JAMA Neurol. 2016;73:68–75. doi: 10.1001/jamaneurol.2015.3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ferrari A, Hoerndli F, Baechi T, Nitsch RM, Götz J. beta-Amyloid induces paired helical filament-like tau filaments in tissue culture. J Biol Chem. 2003;278(41):40162–40168. doi: 10.1074/jbc.M308243200. [DOI] [PubMed] [Google Scholar]
- 16.Fitzpatrick AWP, Falcon B, He S, Murzin AG, Murshudov G, Garringer HJ, et al. Cryo-EM structures of tau filaments from Alzheimer’s disease. Nature. 2017;547(7662):185–190. doi: 10.1038/nature23002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fong JC, Rojas JC, Bang J, Legati A, Rankin KP, Forner S, et al. Genetic prion disease caused by PRNP Q160X mutation presenting with an orbitofrontal syndrome, cyclic diarrhea, and peripheral neuropathy. J Alzheimers Dis. 2017;55(1):249–258. doi: 10.3233/JAD-160300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Furman JL, Holmes BB, Diamond MI. Sensitive detection of proteopathic seeding activity with FRET flow cytometry. J Vis Exp. 2015;106:e53205. doi: 10.3791/53205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Geula C, Wu CK, Saroff D, Lorenzo A, Yuan M, Yankner BA. Aging renders the brain vulnerable to amyloid beta-protein neurotoxicity. Nat Med. 1998;4(7):827–831. doi: 10.1038/nm0798-827. [DOI] [PubMed] [Google Scholar]
- 20.Ghetti B, Oblak AL, Boeve BF, Johnson KA, Dickerson BC, Goedert M. Frontotemporal dementia caused by microtubule-associated protein tau gene (MAPT) mutations: a chameleon for neuropathology and neuroimaging. Neuropathol Appl Neurobiol. 2015;41(1):24–46. doi: 10.1111/nan.12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ghetti B, Piccardo P, Spillantini MG, Ichimiya Y, Porro M, Perini F, et al. Vascular variant of prion protein cerebral amyloidosis with tau positive neurofibrillary tangles: the phenotype of the stop codon 145 mutation in PRNP. Proc Natl Acad Sci USA. 1996;93(2):744–748. doi: 10.1073/pnas.93.2.744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ghetti B, Piccardo P, Zanusso G (2018) Dominantly inherited prion protein cerebral amyloidoses - a modern view of Gerstmann–Sträussler–Scheinker. In: Pocchiari M, Manson J (eds) Handbook of clinical neurology, Elsevier, Amsterdam, Vol 153, pp 243–269 [DOI] [PubMed]
- 23.Ghetti B, Tagliavini F, Masters CL, Beyreuther K, Giaccone G, Verga L, et al (1989) Gerstmann-Sträussler-Scheinker disease. II. Neurofibrillary tangles and plaques with PrP-amyloid coexist in an affected family. Neurology 39(11):1453–1461 [DOI] [PubMed]
- 24.Goedert M, Eisenberg DS, Crowther RA. Propagation of tau aggregates and neurodegeneration. Annu Rev Neurosci. 2017;40:189–210. doi: 10.1146/annurev-neuro-072116-031153. [DOI] [PubMed] [Google Scholar]
- 25.Goedert M, Jakes R. Expression of separate isoforms of human tau protein: correlation with the tau pattern in brain and effects on tubulin polymerization. EMBO J. 1990;9:4225–4230. doi: 10.1002/j.1460-2075.1990.tb07870.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goedert M, Spillantini MG, Cairns NJ, Crowther RA. Tau proteins of Alzheimer paired helical filaments: abnormal phosphorylation of all six brain isoforms. Neuron. 1992;8:159–168. doi: 10.1016/0896-6273(92)90117-V. [DOI] [PubMed] [Google Scholar]
- 27.Goedert M, Spillantini MG, Jakes R, Rutherford D, Crowther RA. Multiple isoforms of human microtubule-associated protein tau: sequences and localization in neurofibrillary tangles of Alzheimer’s disease. Neuron. 1989;3(4):519–526. doi: 10.1016/0896-6273(89)90210-9. [DOI] [PubMed] [Google Scholar]
- 28.Götz J, Streffer JR, David D, Schild A, Hoerndli F, Pennanen L, et al. Transgenic animal models of Alzheimer’s disease and related disorders: histopathology, behavior and therapy. Mol Psychiatry. 2004;9(7):664–683. doi: 10.1038/sj.mp.4001508. [DOI] [PubMed] [Google Scholar]
- 29.Greenberg SG, Davies P, Schein JD, Binder LI. Hydrofluoric acid-treated tau PHF proteins display the same biochemical properties as normal tau. J Biol Chem. 1992;267:564–569. doi: 10.1016/S0021-9258(18)48531-6. [DOI] [PubMed] [Google Scholar]
- 30.Hardy J, Allsop D (1991) Amyloid deposition as the central event in the aetiology of Alzheimer’s disease. Trends Pharmacol Sci. 12(C):383–388 [DOI] [PubMed]
- 31.He S, Scheres SHW. Helical reconstruction in RELION. J Struct Biol. 2017;198(3):163–176. doi: 10.1016/j.jsb.2017.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hoffmann R, Lee VMY, Leight S, Varga I, Otvos L Jr. Unique Alzheimer’s disease paired helical filament specific epitopes involve double phosphorylation at specific sites. Biochemistry. 1997;36:8114–8124. doi: 10.1021/bi970380+. [DOI] [PubMed] [Google Scholar]
- 33.Holmes BB, Furman JL, Mahan TE, Yamasaki TR, Mirbaha H, Eades WC, et al. Proteopathic tau seeding predicts tauopathy in vivo. Proc Natl Acad Sci USA. 2014;111(41):E4376–4385. doi: 10.1073/pnas.1411649111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Holton JL, Ghiso J, Lashley T, Rostagno A, Guerin CJ, Gibb G, et al. Regional distribution of amyloid-Bri deposition and its association with neurofibrillary degeneration in familial British dementia. Am J Pathol. 2001;158(2):515–526. doi: 10.1016/S0002-9440(10)63993-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Holton JL, Lashley T, Ghiso J, Braendgaard H, Vidal R, Guerin CJ, et al. Familial Danish dementia: a novel form of cerebral amyloidosis associated with deposition of both amyloid-Dan and amyloid-beta. J Neuropathol Exp Neurol. 2002;61(3):254–267. doi: 10.1093/jnen/61.3.254. [DOI] [PubMed] [Google Scholar]
- 36.Hsiao K, Dlouhy SR, Farlow MR, Cass C, Costa MDa, Conneally PM,, et al. Mutant prion proteins in Gerstmann-Sträussler-Scheinker disease with neurofibrillary tangles. Nat Genet. 1992;1(1):68–71. doi: 10.1038/ng0492-68. [DOI] [PubMed] [Google Scholar]
- 37.Jayadev S, Nochlin D, Poorkaj P, Steinbart EJ, Mastrianni JA, Montine TJ, et al. Familial prion disease with Alzheimer disease-like tau pathology and clinical phenotype. Ann Neurol. 2011;69(4):712–720. doi: 10.1002/ana.22264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kaufman SK, Del Tredici K, Thomas TL, Braak H, Diamond MI. Tau seeding activity begins in the transentorhinal/entorhinal regions and anticipates phospho-tau pathology in Alzheimer's disease and PART. Acta Neuropathol. 2018;136(1):57–67. doi: 10.1007/s00401-018-1855-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li W, Lee VM. Characterization of two VQIXXK motifs for tau fibrillization in vitro. Biochemistry. 2006;45(51):15692–15701. doi: 10.1021/bi061422+. [DOI] [PubMed] [Google Scholar]
- 40.Liebschner D, Afonine PV, Baker ML, Bunkóczi G, Chen VB, Croll TI, et al. Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Cryst. 2019;D75:861–877. doi: 10.1107/S2059798319011471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Malia TJ, Teplyakov A, Ernst R, Wu SJ, Lacy ER, Liu X, et al. Epitope mapping and structural basis for the recognition of phosphorylated tau by the anti-tau antibody AT8. Proteins. 2016;84(4):427–434. doi: 10.1002/prot.24988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Matamoros-Angles A, Gayosso LM, Richaud-Patin Y, di Domenico A, Vergara C, Hervera A, et al. iPS Cell cultures from a Gerstmann-Sträussler-Scheinker patient with the Y218N PRNP mutation recapitulate tau pathology. Mol Neurobiol. 2018;55(4):3033–3304. doi: 10.1007/s12035-017-0506-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McKee AC, Stern RA, Nowinski CJ, Stein TD, Alvarez VE, Daneshvar DH, et al. The spectrum of disease in chronic traumatic encephalopathy. Brain. 2013;136(Pt 1):43–64. doi: 10.1093/brain/aws307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Murrell J, Farlow M, Ghetti B, Benson MD. A mutation in the amyloid precursor protein associated with hereditary Alzheimer's disease. Science. 1991;254(5028):97–99. doi: 10.1126/science.1925564. [DOI] [PubMed] [Google Scholar]
- 45.Panegyres PK, Toufexis K, Kakulas BA, Cernevakova L, Brown P, Ghetti B, Piccardo P, Dlouhy SR (2001) A new PRNP mutation (G131V) associated with Gerstmann-Sträussler-Scheinker disease. Arch Neurol 58(11):1899–1902 [DOI] [PubMed]
- 46.Perez-Riverol Y, Csordas A, Bai J, Bernal-Llinares M, Hewapathirana S, Kundu DJ, et al (2019) The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 8;47(D1):D442–D450 [DOI] [PMC free article] [PubMed]
- 47.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, et al. UCSF Chimera—A visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 48.Piccardo P, King D, Brown D, Barron RM. Variable tau accumulation in murine models with abnormal prion protein deposits. J Neurol Sci. 2017;383:142–150. doi: 10.1016/j.jns.2017.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Piccardo P, Langeveld JP, Hill AF, Dlouhy SR, Young K, Giaccone G, et al. An antibody raised against a conserved sequence of the prion protein recognizes pathological isoforms in human and animal prion diseases, including Creutzfeldt-Jakob disease and bovine spongiform encephalopathy. Am J Pathol. 1998;152(6):1415–1420. [PMC free article] [PubMed] [Google Scholar]
- 50.Race B, Phillips K, Kraus A, Chesebro B. Phosphorylated human tau associates with mouse prion protein amyloid in scrapie-infected mice but does not increase progression of clinical disease. Prion. 2016;10(4):319–330. doi: 10.1080/19336896.2016.1199313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Risacher SL, Farlow MR, Bateman DR, Epperson F, Tallman EF, Richardson R, et al. Detection of tau in Gerstmann-Sträussler-Scheinker disease (PRNP F198S) by [18F]Flortaucipir PET. Acta Neuropathol Commun. 2018;6(1):114. doi: 10.1186/s40478-018-0608-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Scheres SHW. A Bayesian view on Cryo-EM structure determination. J Mol Biol. 2012;415(2):406–418. doi: 10.1016/j.jmb.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9(7):671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schwalbe M, Kadavath H, Biernat J, Ozenne V, Blackledge M, Mandelkow E, et al. Structural impact of Tau phosphorylation at Threonine 231. Structure. 2015;23(8):1448–1458. doi: 10.1016/j.str.2015.06.002. [DOI] [PubMed] [Google Scholar]
- 55.Shi Y, Murzin AG, Falcon B, Epstein A, Machin J, Tempest P, et al. Cryo-EM structures of tau filaments from Alzheimer's disease with PET ligand APN-1607. Acta Neuropathol. 2021;141(5):697–708. doi: 10.1007/s00401-021-02294-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shi Y, Zhang W, Yang Y, Murzin A, Falcon B, Kotecha A et al (2021) bioRxiv 2021.05.28.446130; doi: 10.1101/2021.05.28.446130
- 57.Sun C, Gonzalez B, Vago FS, Jiang W. High resolution single particle Cryo-EM refinement using JSPR. Prog Biophys Mol Biol. 2021;160:37–42. doi: 10.1016/j.pbiomolbio.2020.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tran NH, Rahman MZ, He L, Xin L, Shan B, Li M (2016) Complete de novo assembly of monoclonal antibody sequences. Sci Rep 6(31730) [DOI] [PMC free article] [PubMed]
- 59.Vidal R, Frangione B, Rostagno A, Mead S, Révész T, Plant G, et al. A stop-codon mutation in the BRI gene associated with familial British dementia. Nature. 1999;399(6738):776–781. doi: 10.1038/21637. [DOI] [PubMed] [Google Scholar]
- 60.Vidal R, Révész T, Rostagno A, Kim E, Holton JL, Bek T, et al. A decamer duplication in the 3' region of the BRI gene originates an amyloid peptide that is associated with dementia in a Danish kindred. Proc Natl Acad Sci USA. 2000;97(9):4920–4925. doi: 10.1073/pnas.080076097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vizcaíno J, Deutsch E, Wang R, et al. ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat Biotechnol. 2014;32:223–226. doi: 10.1038/nbt.2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.von Bergen M, Barghorn S, Li L, Marx A, Biernat J, Mandelkow EM, et al. Mutations of tau protein in frontotemporal dementia promote aggregation of paired helical filaments by enhancing local beta-structure. J Biol Chem. 2001;276(51):48165–48174. doi: 10.1074/jbc.M105196200. [DOI] [PubMed] [Google Scholar]
- 63.Zhang K. Gctf: Real-time CTF determination and correction. J Struct Biol. 2016;193(1):1–12. doi: 10.1016/j.jsb.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang W, Tarutani A, Newell KL, Murzin AG, Matsubara T, Falcon B, et al. Novel tau filament fold in corticobasal degeneration. Nature. 2020;580(7802):283–287. doi: 10.1038/s41586-020-2043-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zheng SQ, Palovcak E, Armache J-P, Verba KA, Cheng Y, Agard DA. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods. 2017;14(4):331–332. doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang RY, Song Y, Barad BA, Cheng Y, Fraser JS, DiMaio F. Automated structure refinement of macromolecular assemblies from cryo-EM maps using Rosetta. Elife. 2016;5:e17219. doi: 10.7554/eLife.17219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wesseling H, Mair W, Kumar M, Schlaffner CN, Tang S, Beerepoot P, et al. Tau PTM Profiles Identify Patient Heterogeneity and Stages of Alzheimer's Disease. Cell. 2020;183(6):1699–1713.e13. doi: 10.1016/j.cell.2020.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wischik CM, Novak M, Edwards PC, Klug A, Tichelaar W, Crowther RA. Structural characterization of the core of the paired helical filament of Alzheimer disease. Proc Natl Acad Sci USA. 1988;85:4884–4888. doi: 10.1073/pnas.85.13.4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Cryo-EM maps have been deposited in the Electron Microscopy Data Bank (EMDB) under accession numbers EMD-23871 for PrP-CAA (Q160X) PHF, EMD-23890 for PrP-CAA (Q160X) SF, and EMD-23894 for GSS (F198S) PHF case 1. Refined atomic models have been deposited in the Protein Data Bank (PDB) under accession numbers 7MKF for PrP-CAA (Q160X) PHF, 7MKG for PrP-CAA (Q160X) SF, and 7MKH for GSS (F198S) PHF case 1. The mass spectrometry proteomics data generated in this study have been deposited to the ProteomeXchange Consortium [61] via the PRIDE partner repository [46] with the dataset identifier PXD025663. Whole-exome sequencing data have been deposited in the National Institute on Aging Alzheimer’s Disease Data Storage Site (NIAGADS; https://www.niagads.org), under accession number ng00107.