Abstract
COVID-19 pandemic exerts a health care emergency around the world. The illness severity is heterogeneous. It is mostly unknown why some individuals who are positive for SARS-CoV-2 antibodies stay asymptomatic while others show moderate to severe disease symptoms. Reliable biomarkers for early detection of the disease are urgently needed to attenuate the virus’s spread and help make early treatment decisions. Bioactive sphingolipids play a crucial role in the regulation of viral infections and pro-inflammatory responses involved in the severity of COVID-19. However, any roles of sphingolipids in COVID-19 development or detection remain unknown. In this study, lipidomics measurement of serum sphingolipids demonstrated that reduced sphingosine levels are highly associated with the development of symptomatic COVID-19 in the majority (99.24%) SARS-CoV-2-infected patients compared to asymptomatic counterparts. The majority of asymptomatic individuals (73%) exhibited increased acid ceramidase (AC) in their serum, measured by Western blotting, consistent with elevated sphingosine levels compared to SARS-CoV-2 antibody negative controls. AC protein was also reduced in almost all of the symptomatic patients’ serum, linked to reduced sphingosine levels, measured in longitudinal acute or convalescent COVID-19 samples. Thus, reduced sphingosine levels provide a sensitive and selective serologic biomarker for the early identification of asymptomatic versus symptomatic COVID-19 patients.
Subject terms: Biochemistry, Lipidomics, Biomarkers, Predictive markers
Introduction
The Coronavirus disease 2019, COVID-19, pandemic is an urgent national health emergency due to the outbreak of the novel coronavirus (SARS-CoV-2) infection that continues spreading at dangerous levels with a high level of morbidity in the United States and around the world. Recent studies suggest that mortality due to COVID-19 is mainly attributed to the hyperinflammatory response leading to cytokine storm and acute respiratory distress syndrome (ARDS) in infected patients1–3. It is known that in severe cases of COVID-19, patients showed increased serum cytokine levels of IL-2, TNFα, IL-1β, IFNγ, MCP-1, MIP1α, IL-10, and IL-6. ARDS was reported to be developed more often in elderly COVID-19 patients due to a cytokine storm4–8.
The severity of COVID-19 disease and the infection course is heterogeneous and appears to be more severe in the elderly and individuals with underlying comorbidities, including cancer9,10, pregnant women11, hypertension12–14, and obesity15. However, modifiers of the immune response and their mechanisms remain unknown. We also do not know why some patients develop severe COVID-19, while others stay asymptomatic. Thus, while the infected patient’s overall immunity cannot explain this broad spectrum in disease presentation, differences in immune responses and metabolic alterations might be involved in the disease’s progression to severe stages. Although successful vaccines against SARS-CoV-2 are now in place for disease prevention, early diagnosis, and isolation of mild or asymptomatic COVID-19 patients using selective biomarkers is essential to control the spread of SARS-CoV-2. However, the clinical characteristics of these asymptomatic individuals remain elusive.
The differences in specific immune responses required to eliminate the virus and prevent disease progression to severe stages might be involved. For example, it has been shown that after the SARS-CoV-2 infection, CD4+T lymphocytes are rapidly activated to become pathogenic T helper (Th) 1 cells and generate GM-CSF, which then induces inflammatory CD14+CD16+ monocytes with high expression of IL-6 and accelerate the inflammation16,17. These inflammatory monocytes then enter the pulmonary circulation in huge numbers and play a damaging immune role, causing lung functional disability and quick mortality. The cytokine release syndrome (CRS) was reported to affect patients with severe conditions8. While effector T cells exhibit increased aerobic glycolysis18, memory T cells utilize oxidative phosphorylation (OXPHOS)19. Furthermore, molecules such as AMPK20, HIF1α21, and Foxo122,23 dictate the balance between effector and memory T cells. The dependence of memory T cells on fatty acid oxidation and lysosomal lipolysis24 has been shown. In addition to mitochondrial biogenesis, the mitochondria’s quality, as observed by the cristae organization, also influences T cell fitness and its ability to control tumors and viral infections25. Recently, Th17 cells gained increased attention because their “stem cell-like” characteristics enable them to persist longer in the host18,26. Increased effector T cell function controls tumors and exhibits superior anti-viral activity to control SARS-CoV-2/COVID-19 is mainly associated with increased glutaminolysis and increased dependence on mitochondrial metabolism27. Thus, determining metabolic modifiers, including lipid metabolism, will likely help define the mechanisms of immune responses elicited by SARS-CoV-2 viral infection.
Sphingolipid metabolism is known to regulate inflammation and immune response. Previous studies demonstrated that sphingolipid metabolism, through the conversion of sphingosine to sphingosine 1-phosphate (S1P), induces immune response and inflammation by creating an S1P gradient, resulting in the egress of lymphocytes from lymphoid organs to the bloodstream28,29. S1P analogs or antagonists that inhibit inflammatory signaling in Mycobacterium tuberculosis (H37Ra) infected macrophages prevented pro-inflammatory response and resulted in the clearance of bacterial load30. Additionally, S1P signaling was identified as a potential target to play essential roles in pulmonary disorder in response to H1N1 influenza virus infection in mice with decreased IFNγ, TNFα, CCL2, CCL5, CXCL10, IL-1rα, and IL-6 secretion31,32. Mechanistically, while S1P induces PI3K/AKT and NFκB activation, sphingosine plays opposing roles and inhibits pro-inflammatory cytokine release via mostly dephosphorylation of AKT and p65 subunit of NFκB33. Recently, FTY720 (Fingolimod, Novartis), a sphingosine analog and immune modulator drug was introduced in the clinical trial to determine its role as an immunomodulator in COVID-19 patients34,35. Interestingly, a recent study demonstrated that sphingosine inhibited Spike protein’s interaction with ACE receptors, suggesting its potential role in preventing SARS-CoV-2 viral infection36. Sphingosine can be generated by the metabolism of ceramide with the function of ceramidases, including acid ceramidase (AC, encoded by ASAH1 gene) known to be secreted to the serum in its active form37,38. However, the clinical significance of sphingolipid alterations concerning the COVID-19 disease outcomes has not been reported previously.
Thus, in this study, we investigate the potential clinical utility of serologic sphingolipid levels measured using mass spectrometry-based lipidomics. Our goal is early detection of individuals who are positive for an antibody that recognizes SARS-CoV-2-specific Spike protein to exhibit asymptomatic versus symptomatic disease outcomes based on their serologic sphingosine (lipid) and active form of acid ceramidase (AC protein) levels.
Results
Reduced serum sphingosine is associated with symptomatic COVID-19 patients compared to asymptomatic donors who are SARS-CoV-2 antibody-positive
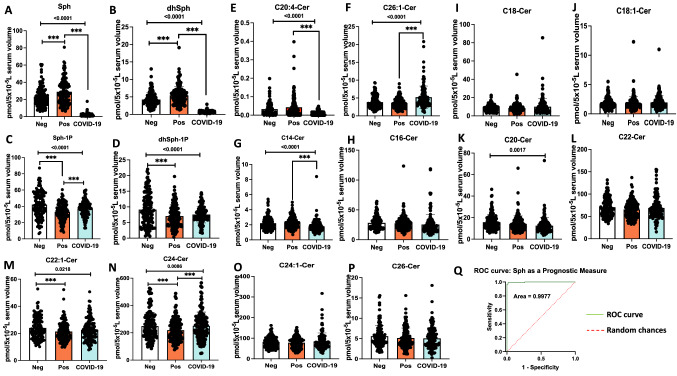
To assess whether there are any alterations in sphingolipid metabolism in asymptomatic individuals who are serologically positive versus negative for the SARS-CoV-2 antibody, we measured ceramides, sphingosine, and S1P levels in their serum samples using quantitative high performance liquid chromatography-tandem mass spectrometry (HPLC–MS/MS)-based lipidomics approach. The data showed that there is a slight but significant increase in the levels of sphingosine (p < 0.05) in individuals who are antibody positive (n = 134) compared to negative (n = 130), with sphingosine levels 28.96 versus 23.25 pmol/5 × 10−5 L serum, respectively (Fig. 1A). We then determined whether serum sphingosine levels also altered in individuals who were serologically positive for the SARS-CoV-2 antibody and clinically symptomatic (n = 131) for COVID-19 versus asymptomatic donors. The data showed that COVID-19 patients’ serum sphingosine levels were around 15-fold decreased compared to that of asymptomatic donors from 28.96 to 1.88 pmol/5 × 10−5 L serum, respectively (Fig. 1A). These results were also consistent with an around sevenfold decrease in dihydro(dh)-sphingosine levels in serum samples obtained from COVID-19 patients compared to asymptomatic donors with 0.74 and 5.4 pmol/5 × 10−5 L serum, respectively (Fig. 1B). Decreased serum sphingosine in COVID-19 patients compared to asymptomatic donors was not associated with S1P levels in serum (Fig. 1C,D), which were comparable, suggesting that metabolism of S1P might not play a role in regulating sphingosine levels in COVID-19 patients or asymptomatic donors.
Figure 1.
Decreased serum sphingosine level is linked with COVID-19 symptoms. Lipidomics measurements based on High-Performance Liquid Chromatography-tandem Mass Spectrometry (LC–MS/MS) for (A) Sphingosine (Sph), (B) dihydro-sphingosine (dhSph), (C) sphingosine 1-phosphate (Sph-1p), and (D) dihydro-sphingosine 1-phosphate (dhSph-1p). LC–MS/MS ceramide (Cer) measurements for (E) C20:4-Cer, and (F) C26:1 Cer. Moreover, LC–MS/MS ceramide (Cer) measurements for (G) C14-Cer, (H) C16-Cer, (I) C18-Cer, (J) C18:1-Cer, (K) C20-Cer, (L) C22-Cer, (M) C22:1-Cer, (N) C24-Cer, (O) C24:1-Cer, and (P) C26-Cer. (Q) Graphical ROC (receiving-operating characteristics) curve for Sph as a prognostic measure, produced by plotting Sensitivity(true positive rate) against 1-Specificity (false positive rate). Area under the ROC curve is 0.9977 or 99.77%. The light green line (ROC curve) shows combinations of sensitivity and specificity for threshold values, while the red dotted line (Random chances) indicates a reference cut-off point for a useful model. Neg negative antibody test (n = 130), Pos asymptomatic positive antibody test (n = 134), COVID-19 symptomatic patients (n = 131). Data are means of ± SEM, and P < 0.05 is considered significant. Comparisons significant at the 0.05 level are indicated by ***.
Sphingosine can be generated in response to ceramide's hydrolysis by ceramidases, including acid ceramidase (AC, encoded by ASAH1 gene), which is known to be secreted in its active form to regulate blood levels of sphingosine in various models. Thus, we also measured ceramides with different fatty acyl chain lengths, including short, long, and very-long-chain fatty acyl chains (C14/16-, C18/C20-, and C22/C26-ceramides, respectively). Interestingly, while there were no considerable changes in short-chain ceramides, the levels of C20:4-ceramide were decreased, and C26:1-ceramide were increased (Fig. 1E–P) in COVID-19 patients compared to asymptomatic donors. These data suggest that the metabolic balance between ceramide and sphingosine might play a role in altered serum sphingosine levels in COVID-19 patients or asymptomatic donors. Additionally, our analysis of the ROC (receiving-operating characteristics) curve for reduced sphingosine levels in COVID-19 patients gave rise to 99.77% area under the curve (Fig. 1Q). From the ROC analysis, a sphingosine threshold (or cut-off) value of 8.2 pmol/5 × 10−5 L resulted in 98.47% (95% CI 94.60–99.73%) sensitivity and 98.51% (95% CI 94.72–99.73%) specificity, suggesting that serum sphingosine level provides a selective and sensitive biomarker to identify symptomatic patients versus asymptomatic donors who are positive for SARS-CoV-2 antibody.
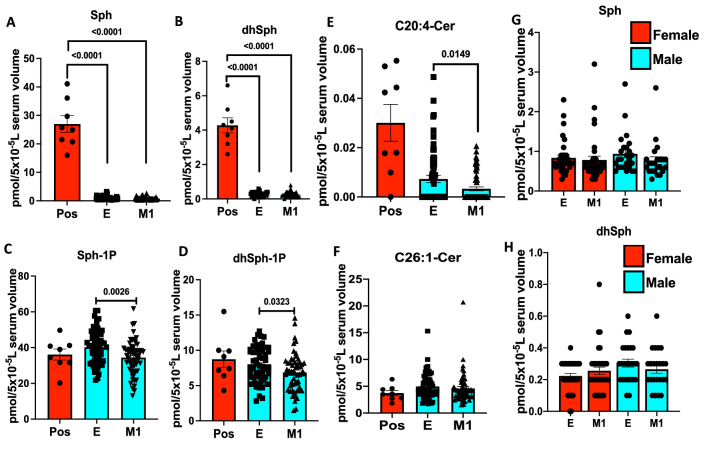
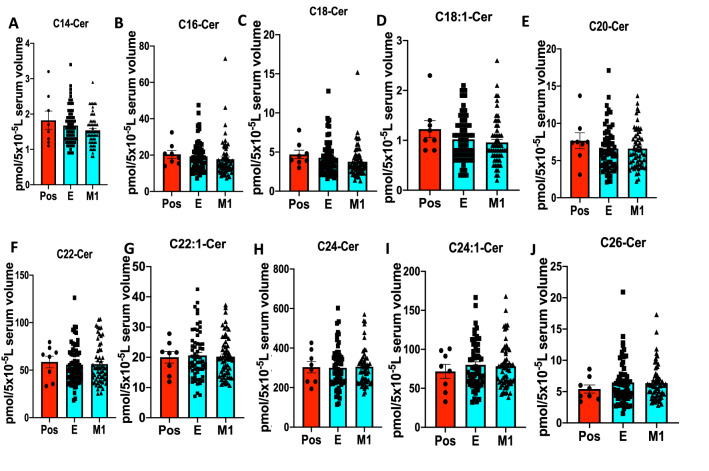
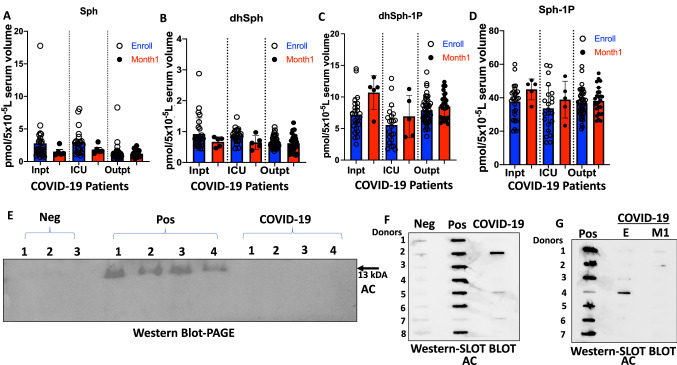
To further examine whether decreased serum sphingosine and dh-sphingosine levels in COVID-19 patients is linked to disease severity, we obtained longitudinal serum samples from symptomatic patients enrolled at the time of diagnosis (E) or after 1 month of recovery (M1) (Figs. 2A–H and 3A–J). The data showed that serum levels of sphingosine and dh-sphingosine of COVID-19 patients at the time of diagnosis or after 1 month of recovery were decreased at a comparable rate regardless of the gender, about a 15-fold reduction to that of asymptomatic donors (Fig. 2A–H). Also, reduced sphingosine and dh-sphingosine levels were detected at similar levels in COVID-19 patients who were treated as an inpatient, outpatient, or intensive care unit (ICU) who had < 5 pmol or < 1 pmol/5 × 10−5 L serum levels of sphingosine or dh-sphingosine, respectively (Fig. 4A–D). These data suggest that while reduced serologic sphingosine and dh-sphingosine levels are associated with symptomatic COVID-19, they do not appear to monitor the disease's severity.
Figure 2.
Reduced serum sphingosine level in COVID-19 patients, is not associated with the diseases’ severity. LC–MS/MS measurements for (A) Sphingosine (Sph), (B) dihydro-sphingosine (dhSph), (C) sphingosine 1-phosphate (Sph-1p), and (D) dihydro-sphingosine 1-phosphate (dhSph-1p). LC–MS/MS ceramide (Cer) measurements for (E) C20:4-Cer, and (F) C26:1 Cer. (G,H) Sph (G) and dhSph (H) levels of symptomatic COVID-19 patients in male and female patients. Pos asymptomatic positive antibody test (n = 8), E enrolled at time of diagnosis (n = 60), M1 one month of recovery (n = 59). Data are means of ± SEM, and P < 0.05 is considered significant.
Figure 3.
Serum ceramide levels with different fatty acyl chain lengths in COVID-19 longitudinal samples compared to asymptomatic positives. LC–MS/MS ceramide (Cer) measurements for (A) C14-Cer, (B) C16-Cer, (C) C18-Cer, (D) C18:1-Cer, (E) C20-Cer, (F) C22-Cer, (G) C22:1-Cer, (H) C24-Cer, (I) C24:1-Cer, and (J) C26-Cer. Pos Asymptomatic Positive antibody test (n = 8), E enrolled at time of diagnosis (n = 60), M1 one month of recovery (n = 59). Data are means of ± SEM, and P < 0.05 is considered significant.
Figure 4.
Elevated acid ceramidase level in asymptomatic serum is positively correlated with sphingosine levels. LC–MS/MS measurements for (A) sphingosine (Sph), (B) dihydro-sphingosine (dhSph), (C) dihydro-sphingosine 1-phosphate (dhSph-1p), and (D) sphingosine 1-phosphate (Sph-1p). (E) Representative western blot image detecting AC (mature alpha-subunit) in equal volumes (24 µl) of serum for Neg (n = 18), Pos (n = 22), and COVID-19 (n = 22). Representative slot blot images detecting AC in equal volumes (15 µl) of serum for (F) Neg (n = 23), Pos (n = 114), and COVID-19 (n = 23), and (G) Pos (n = 59), E (n = 59), and M1 (n = 59). Inpt inpatient (E, n = 27; M1, n = 5), ICU Intensive Care Unit (E, n = 22; M1, n = 5); Outpt outpatient (E, n = 46; M1, n = 26). Pos asymptomatic positive antibody test, E enrolled at time of diagnosis, M1 one month of recovery, Neg negative antibody test, COVID-19 symptomatic patients. Data are means of ± SEM, and P < 0.05 is considered significant.
Increased sphingosine in asymptomatic donors is associated with elevated AC in the serum
To determine if serologic sphingosine/ceramide metabolism in COVID-19 patients and/or asymptomatic individuals regulated by AC, we measured secreted and active AC protein39 in the serum by Western blotting using PAGE (Fig. 4E) or slot blot apparatus using a commercially available anti-AC antibody (Fig. 4F,G). The data showed that out of 114 asymptomatic donors, 84 were positive for serum AC (73.7%), whereas only 2 out of 23 COVID-19 patients were positive for AC protein in their serum samples (8.6%), which were similar to the AC positive donors who were negative for the SARS-CoV-2 antibody (6 out of 23, 26%) (Fig. 4F). In addition, among longitudinal COVID-19 patient serum samples, we did not detect any upregulation of AC in E or outpatient samples after 1-month recovery or severity of COVID-19 disease, compared to asymptomatic donors (Fig. 4G). The original blots shown in Fig. 4E–G are included in Supplemental Fig. S1A–C. Thus, these data suggest that serologic AC protein levels are highly increased in most asymptomatic donors. However, most COVID-19 patients, regardless of disease severity, are negative for serum AC, which is also consistent with their decreased sphingosine and dh-sphingosine levels compared to asymptomatic donors.
Discussion
COVID-19 pandemic continues to exert worldwide a health care emergency, and how some individuals stay asymptomatic despite SARS-CoV-2 infection remains largely unknown2,3. In this study, we compared the metabolic alterations concerning sphingolipids in SARS-CoV-2 antibody-positive donors who were asymptomatic versus symptomatic for COVID-19. Our data revealed that while most asymptomatic donors have a slight increase in their serum sphingosine, consistent with the presence of AC protein in their serum, symptomatic COVID-19 patients exhibited a robust decrease in their serum sphingosine levels, almost 15-fold reduction, compared to asymptomatic donors’s levels. In addition to the ROC analysis, our data demonstrated that most individuals (99.24%) whose serum sphingosine levels are between 0–8 pmol/5 × 10−5 L serum have developed COVID-19 symptoms compared to individuals whose serum sphingosine levels are between 8.4–23.25 pmol/5 × 10−5 L serum, supporting that serum sphingosine levels could be used as a sensitive and selective biomarker to predict positive COVD-19 symptomatic and asymptomatic patients. Although it is known that sphingosine and S1P levels are associated with increase in inflammation state locally, and that SARS-CoV-2 infections also induces inflammation, however, we didn’t observe a positive correlation between serum sphingosine, S1P, and SARS-CoV-2 infection.
Previous studies reported that the SARS-CoV-2 viral loads are usually similar in asymptomatic and symptomatic individuals, while asymptomatic individuals have higher immune cell counts, including T lymphocytes, B cells, or natural killer cells40,41. Although COVID-19 patients have decreased CD4+ and CD8+ T cells, increased IL-6 and IL-10 levels, associated with cytokine storm, are linked to disease severity42,43. However, the mechanisms behind why some SARS-CoV-2-infected individuals stay asymptomatic while others develop severe COVID-19 remain mostly unknown. It is also known that there is cross-reactivity among memory T cells against SARS-CoV-2 and common cold coronavirus epitopes, which may play a role in asymptomatic response in some individuals9,44. However, it will be essential to identify novel serological biomarkers for early identification of SARS-CoV-2-infected patients who would likely stay asymptomatic or develop COVID-19 symptoms.
It is also known that pro-inflammation and cytokine storm is associated with the severity of COVID-19 patients compared to asymptomatic individuals45,46. In addition to inflammatory cytokines, lipid metabolism alterations are also known to play a role in inflammation in response to bacterial and viral infections47,48. However, serologic alterations of biochemical metabolites, including sphingolipids, in SARS-CoV-2-infected symptomatic versus asymptomatic individuals have not been reported previously. To this end, our study provides a novel and selective serologic biomarker, reduction of sphingosine, which we believe would be clinically significant for the early detection of symptomatic versus asymptomatic individuals who are positive for SARS-CoV-2 antibody by providing early treatment decisions in the fight to stop the spread of the virus globally49,50.
One of the biochemical biomarkers, lactate dehydrogenase (LDH), was highly elevated in symptomatic patients with an increased mortality rate51,52. Interestingly, reduced sphingosine levels were not associated with disease severity in COVID-19 patients. These data also suggest that increased or sustained serum sphingosine levels, linked to higher serum AC protein, might prevent COVID-19 disease, while reduced sphingosine could result in enhanced inflammation and symptomatic response in some individuals. This is consistent with the effects of exogenous exposure of sphingosine on inhibiting the association between Spike protein and ACE receptors in vitro36, supporting the possible preventive roles of sphingosine in SARS-CoV-2 viral infection. There are reports which also showed that sphingosine plays a role in inhibiting pulmonary Pseudomonas and Staphylococcus aureus infections in cystic fibrosis53–55.
There are some limitations that need to be considered for the data presented in this work. For example, this study was performed retrospectively. It will be important to conduct similar studies in a prospective study with a larger cohort of donors to identify serologic sphingosine and AC measurements' efficiency in predicting future COVID-19 development in SARS-CoV-2-infected individuals. Also, since we don’t have symptoms onset information for the symptomatic COVID-19 patients, we don’t know exactly the delay between the onset of the symptoms and lipidomics measurements, which could influence sphingosine and acid ceramidase levels. Whether sphingosine and sphingolipid signaling play any roles and their mechanism of action in preventing viral infection or pro-inflammation/cytokine storm in individuals who are SARS-CoV-2 antibody-positive remains unknown and needs to be determined. Lastly, measurement of serum sphingolipid levels, particularly sphingosine, using mass spectrometry is relatively more expensive than simple ELISA-based assays. To this end, based on recent evidence that reduced serum S1P levels are highly associated with the severity of COVID-19 in patients56, it might be helpful to include S1P measurements also. These data are also consistent with recent studies which reported altered lipid metabolism associated with COVID-19 pathogenesis. For example, increased lipid accumulation was reported in SARS-CoV-2 infected cells and in the lungs of COVID-19 patients, suggesting a role for lipids in SARS-CoV-2 pathogenesis57. Also, low apo B/apo A1 ratio and low density lipoprotein-C (LDL-C) are observed to be predictive of renal deterioration in COVID-19 patients58, supporting the roles of alterations of lipid metabolism in the pathogenesis of COVID-19.
Overall, recent advances in lipidomics provide cost- and time-effective biomarker development and detection at a sizeable high-throughput scale to selectively identify symptomatic versus asymptomatic individuals who are positive for the SARS-CoV-2 antibodies. In conclusion, we report here a new and reliable serologic biomarker, sphingosine, whose reduced levels beyond 8.3 pmol/5 × 10−5 L serum is highly (99.24%) associated with symptomatic COVID-19 development. In contrast, SARS-CoV-2 antibody-positive individuals whose sphingosine levels are between 8.4 and > 23.25 pmol/5 × 10−5 serum will (99.2–99.9%) likely be asymptomatic. These results have vital biological implications to early detect symptomatic and asymptomatic individuals that could be important to make early treatment and/or social distancing decisions on a larger scale.
Materials and methods
Patients and serum samples
COVID-19 symptomatic patient samples were received from the MUSC COVID-19 Biorepository between May and July 2020. The samples were then heat inactivated at 56 °C in a water bath incubator for 1 h before further processing. The negatives and asymptomatic positives donor samples were received from the MUSC Center for Cellular Therapy (CCT)—that collects donor sample stocks from The Blood Connection (Charleston, SC). The Blood Connection is a blood center that collects donor samples from three US Southeast regions—South Carolina, North Carolina, and Georgia. Samples were collected from 46 counties in South Carolina, 56 counties in North Carolina, and 15 counties in Georgia. The negative control serum samples also known as true negatives, were collected from healthy donors by The Blood Connection at least more than a year before the COVID-19 pandemic started. See Tables 1, 2 and 3 for patient’s characteristics.
Table 1.
Serum samples from healthy negative individuals, asymptomatic positives, and symptomatic COVID-19 patients.
| SARS-CoV-2 negative individuals (Neg) | SARS-CoV-2 positive asymptomatic patients (Pos) | Symptomatic patients (COVID-19) | |
|---|---|---|---|
| Total number of serum samples | 130 | 134 | 131 |
| Sex (male:female:Unk) | 50:80:0 | 56:70:8 | 44:48:39 |
| Age range, years | 17–85 | 16–76 | 20–94 |
| Mean age, years (SD) | 46.4 (15.6) | 45.8 (16.1) | 50.8 (17.6) |
| Race (W:B:A:Unk) | 112:2:1:14 | 90:9:4:18 | 55:28:0:5 |
| Hispanic or Latino | 1 | 13 | 6 |
White (W), Caucasian/European; Black (B), African American/African; Asian (A); Unk (unknown); SD standard deviation.
Table 2.
One-month longitudinal serum samples from symptomatic COVID-19 patients.
| Male | Female | |||
|---|---|---|---|---|
| Enroll (E) | Month 1 (M1) | Enroll (E) | Month 1 (M1) | |
| Total number of serum samples | 25 | 23 | 35 | 36 |
| White | 19 | 17 | 26 | 27 |
| Black | 4 | 4 | 8 | 8 |
| Asian | 0 | 0 | 1 | 1 |
| Unknown | 2 | 2 | 0 | 0 |
White, Caucasian/European; Black, African American/African.
Table 3.
Symptomatic COVID-19 serum samples from outpatients, inpatients, and patients in intensive care units.
| Outpatient (Outpt) | Inpatient (Inpt) | Intensive Care Unit (ICU) | |
|---|---|---|---|
| Total number of serum samples | 46 | 27 | 23 |
| Sex (male:female:Unk) | 15:25:6 | 14:9:4 | 12:11:0 |
| Mean age, years (SD) | 43.3 (13.6) | 51.9 (17.7) | 59.4 (15.0) |
| Race(W:B:A:Unk) | 34:4:0:2 | 13:7:0:0 | 9:14:0:0 |
| Hispanic or Latino | 2 | 2 | 0 |
| Died | 0 | 0 | 3 |
| Mean time from symptoms to admission (day) | n/a | 11.4 | 5.9 |
| Ventilatory support | 0 | 0 | 15 |
| ARDS severity (range) | n/a | n/a | 24–146 |
ARDS severity defined as PaO2/FiO2 (only reported in 5 subjects). White (W), Caucasian/European; Black (B), African American/African; Asian (A); Unk (unknown); SD standard deviation, n/a not available, ARDS acute respiratory distress syndrome, PaO2 oxygen in arterial blood, FiO2 oxygen in the inspired air.
Inclusion and exclusion criteria
The negative and asymptomatic positive groups were determined based on IgG to SARS-CoV-2 spike protein Optical Density 490 (OD 490) values obtained from a microplate reader. OD 490 value less than 0.45 was defined as negatives, while OD 490 value of 0.45 or higher was defined as asymptomatic positives. Detail description of the SARS-CoV-2 spike protein assays used to classify negatives and asymptomatic positives for the same study cohort is reported in our recent publication59. However, in this study we only included values of OD 490 ≥ 1 as asymptomatic positive, while the asymptomatic positive OD 490 values between 0.45 and 1 were excluded since they were categorized as low positives. Both negative and asymptomatic positive individuals didn’t exhibit any COVID-19 related symptoms at the time of serum collection, while the COVID-19 individuals had common symptoms, including fever, cough, asthenia, shortness of breath, new loss of taste or smell, and/or sore throat with a positive PCR test at time of sample collection (Table 1). The 1-month longitudinal study included only positive symptomatic COVID-19 patients at time of their enrollment (Table 2). Finally, the outpatients, inpatients, and patients in intensive care units were all symptomatic and tested positive for COVID-19 virus (Table 3). The outpatients visited MUSC hospital for treatment but were not admitted, while inpatients were admitted to the university hospital for treatments. Patients in intensive care units were in critical conditions needing constant monitoring and medical supports like the ventilatory support.
Ethics statement
All patient information’s were de-identified, and studies were performed under the approved protocols by the MUSC’s Institutional Review Board for Human Research (IRB, #: Pro00100487) and Institutional Biosafety Committee (IBC, #: IBC-2020-01064). Overall, all the studies reported here were performed based on the protocols approved by the institutional and/or licensing committee approving according to relevant guidelines and regulations. Studies involving specimens obtained from human participants were performed by approved protocols by the IRB at MUSC in accordance with relevant guidelines/regulations, and informed consent was obtained from all participants and/or their legal guardians by the COVID-19 Biorepository at MUSC.
Mass spectrometry-based lipidomics
Separations for sphingolipids were performed by HPLC–MS/MS analyses at the MUSC Lipidomics Shared Resource as described60. The equipment consisted of a Thermo Scientific Vanquish uHPLC system coupled to a Thermo Scientific Quantum Access Max triple quadrupole mass spectrometer equipped with an ESI probe operating in the multiple reactions monitoring positive ion mode (MRM). Chromatographic separations were obtained under a gradient elution on a C8 column using a mobile phase with ammonium formate, formic acid in the water, and methanol, as previously described. Before analysis, samples undergo an ethyl acetate/isopropanol liquid–liquid extraction. Quantitative analyses of sphingolipids are based on eight-point calibration curves generated for each target analyte. The synthetic standards and a set of internal standards are spiked into an artificial matrix; they are then subjected to identical extraction procedure as the biological samples. These extracted standards are then analyzed with the samples by the HPLC–MS/MS. Peaks for the target analytes and internal standards are recorded and processed using the instrument’s software. Plotting the analyte/internal standard peak area ratios against analyte concentrations generates the sphingolipid specific calibration curves. Any sphingolipid for which no standards are available is quantitated using the calibration curve of its closest counterpart.
Western blotting
AC protein, secreted to the serum in its active 13 kDa form (mature alpha-subunit), was detected using commercially available anti-AC antibody (BD Biosciences, #612302) by Western blotting after PAGE or slot-blot apparatus in serum samples obtained from SARS-CoV-2 antibody-negative or positive donors, who were asymptomatic or symptomatic.
Statistical analysis
Data analysis represents Standard Error of the Mean (SEM) using Graph Pad Prism 9.0.0, and p < 0.05 were considered statistically significant for all comparisons. Normality and variance homogeneity assumptions were assessed. All continuous outcomes among 3 independent groups are compared using ANOVA tests. If a significant result is discovered, all possible pairwise comparisons are performed adjusting for multiple comparisons using Turkey’s method. For measures taken from the same subject at different time points, a paired t test is used to compare the mean difference at baseline and 1 month later. Unpaired t test is used to compare the mean difference of unrelated subjects. The ROC curve analysis was done using Graph Pad Prism 9.0.0, and the Wilson/Brown method was used to calculate the 95% confidence interval.
Supplementary Information
Acknowledgements
We thank Drs. Aiping Bai, Min Cai and Zdzislaw Szulc and Mrs. Octavia Mitchell at the Lipidomics Core Facility (MUSC) for their helpful discussions and assistance.
Author contributions
B.O. and S.M. initiated the project and analyzed the data. A.H.J., M.F.K. and J.S.P. performed the lipidomics and AC detection experiments; C.J.D and P.C. carried out the SARS-CoV-2 antibody detection by ELISA; P.A.F and S.N.N. provided specimens used in the project. H.L. performed statistical analysis. The manuscript was written by A.H.J. and B.O. All authors reviewed and approved the manuscript.
Funding
This work was supported in part by the Medical University of South Carolina’s Lipidomics Shared Resource through the funding of laboratory space for the Analytical Unit located in 505 Children’s Research Institute (CRI): Hollings Cancer Center (P30 CA138313), the Lipidomics Shared Resource in the South Carolina Lipidomics and Pathobiology COBRE (P30 GM103339); MUSC Department of Biochemistry (P20 RR017677), the National Center for Research Resources and Office of the Director of the National Institutes of Health (C06 RR018823)”, Hollings Cancer Center Support Grant (P30 CA138313), and individual research grants (CA214641, DE016572, and P01 CA203628 to BO) from the National Institutes of Health.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-93857-7.
References
- 1.Allegra A, Pioggia G, Tonacci A, Musolino C, Gangemi S. Cancer and SARS-CoV-2 infection: Diagnostic and therapeutic challenges. Cancers. 2020;12:1581. doi: 10.3390/cancers12061581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amanat F, Krammer F. SARS-CoV-2 vaccines: Status report. Immunity. 2020;52:583–589. doi: 10.1016/j.immuni.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cao X. COVID-19: Immunopathology and its implications for therapy. Nat. Rev. Immunol. 2020;20:269–270. doi: 10.1038/s41577-020-0308-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen G, et al. Clinical and immunological features of severe and moderate coronavirus disease 2019. J. Clin. Investig. 2020;130:2620–2629. doi: 10.1172/JCI137244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Diao B, et al. Reduction and functional exhaustion of T cells in patients with coronavirus disease 2019 (COVID-19) Front. Immunol. 2020;11:827. doi: 10.3389/fimmu.2020.00827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang J, Jiang M, Chen X, Montaner LJ. Cytokine storm and leukocyte changes in mild versus severe SARS-CoV-2 infection: Review of 3939 COVID-19 patients in China and emerging pathogenesis and therapy concepts. J. Leukoc. Biol. 2020;108:17–41. doi: 10.1002/JLB.3COVR0520-272R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ye Q, Wang B, Mao J. The pathogenesis and treatment of the ‘Cytokine Storm’ in COVID-19. J. Infect. 2020;80:607–613. doi: 10.1016/j.jinf.2020.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Song P, Li W, Xie J, Hou Y, You C. Cytokine storm induced by SARS-CoV-2. Clin. Chim. Acta. 2020;509:280–287. doi: 10.1016/j.cca.2020.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grifoni A, et al. Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell. 2020;181:1489–1501.e1415. doi: 10.1016/j.cell.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beigel JH, et al. Remdesivir for the treatment of Covid-19—Preliminary report. N. Engl. J. Med. 2020;383:1813–1826. doi: 10.1056/NEJMoa2007764. [DOI] [PubMed] [Google Scholar]
- 11.Hapshy V, et al. COVID-19 and pregnancy: Risk, symptoms, diagnosis, and treatment. Compr. Clin. Med. 2021 doi: 10.1007/s42399-021-00915-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sahu KK, George L, Jones N, Mangla A. COVID-19 in patients with sickle cell disease: A single center experience from Ohio, United States. J. Med. Virol. 2021;93:2591–2594. doi: 10.1002/jmv.26816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sheppard JP, et al. Association between blood pressure control and coronavirus disease 2019 outcomes in 45 418 symptomatic patients with hypertension: An observational cohort study. Hypertension. 2021;77:846–855. doi: 10.1161/HYPERTENSIONAHA.120.16472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nishiga M, Wang DW, Han Y, Lewis DB, Wu JC. COVID-19 and cardiovascular disease: From basic mechanisms to clinical perspectives. Nat. Rev. Cardiol. 2020;17:543–558. doi: 10.1038/s41569-020-0413-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuehn BM. More severe obesity leads to more severe COVID-19 in study. JAMA. 2021;325:1603–1603. doi: 10.1001/jama.2021.4853. [DOI] [PubMed] [Google Scholar]
- 16.Karki R, et al. Synergism of TNF-alpha and IFN-gamma triggers inflammatory cell death, tissue damage, and mortality in SARS-CoV-2 infection and cytokine shock syndromes. Cell. 2021;184:149–168 e117. doi: 10.1016/j.cell.2020.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mangalmurti N, Hunter CA. Cytokine storms: Understanding COVID-19. Immunity. 2020;53:19–25. doi: 10.1016/j.immuni.2020.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Muranski P, et al. Th17 cells are long lived and retain a stem cell-like molecular signature. Immunity. 2011;35:972–985. doi: 10.1016/j.immuni.2011.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van der Windt GJ, Pearce EL. Metabolic switching and fuel choice during T-cell differentiation and memory development. Immunol. Rev. 2012;249:27–42. doi: 10.1111/j.1600-065X.2012.01150.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rolf J, et al. AMPK α1: A glucose sensor that controls CD 8 T-cell memory. Eur. J. Immunol. 2013;43:889–896. doi: 10.1002/eji.201243008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Doedens AL, et al. Hypoxia-inducible factors enhance the effector responses of CD8+ T cells to persistent antigen. Nat. Immunol. 2013;14:1173. doi: 10.1038/ni.2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rao RR, Li Q, Gubbels Bupp MR, Shrikant PA. Transcription factor Foxo1 represses T-bet-mediated effector functions and promotes memory CD8(+) T cell differentiation. Immunity. 2012;36:374–387. doi: 10.1016/j.immuni.2012.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hess Michelini R, Doedens AL, Goldrath AW, Hedrick SM. Differentiation of CD8 memory T cells depends on Foxo1. J. Exp. Med. 2013;210:1189–1200. doi: 10.1084/jem.20130392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chang C-H, Pearce EL. Emerging concepts of T cell metabolism as a target of immunotherapy. Nat. Immunol. 2016;17:364–368. doi: 10.1038/ni.3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Buck MD, et al. Mitochondrial dynamics controls T cell fate through metabolic programming. Cell. 2016;166:63–76. doi: 10.1016/j.cell.2016.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kryczek I, et al. Human TH17 cells are long-lived effector memory cells. Sci. Transl. Med. 2011;3:104ra100. doi: 10.1126/scitranslmed.3002949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chatterjee S, et al. CD38-NAD(+)axis regulates immunotherapeutic anti-tumor T cell response. Cell Metab. 2018;27:85–100 e108. doi: 10.1016/j.cmet.2017.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ogretmen B. Sphingolipid metabolism in cancer signalling and therapy. Nat. Rev. Cancer. 2018;18:33–50. doi: 10.1038/nrc.2017.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cartier A, Hla T. Sphingosine 1-phosphate: Lipid signaling in pathology and therapy. Science. 2019 doi: 10.1126/science.aar5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weigert A, Olesch C, Brune B. Sphingosine-1-phosphate and macrophage biology-how the sphinx tames the big eater. Front. Immunol. 2019;10:1706. doi: 10.3389/fimmu.2019.01706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Walsh KB, et al. Suppression of cytokine storm with a sphingosine analog provides protection against pathogenic influenza virus. Proc. Natl. Acad. Sci. U.S.A. 2011;108:12018–12023. doi: 10.1073/pnas.1107024108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao J, et al. Combination of sphingosine-1-phosphate receptor 1 (S1PR1) agonist and antiviral drug: A potential therapy against pathogenic influenza virus. Sci. Rep. 2019;9:5272. doi: 10.1038/s41598-019-41760-7. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 33.Maceyka M, Spiegel S. Sphingolipid metabolites in inflammatory disease. Nature. 2014;510:58–67. doi: 10.1038/nature13475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Foerch C, Friedauer L, Bauer B, Wolf T, Adam EH. Severe COVID-19 infection in a patient with multiple sclerosis treated with fingolimod. Mult. Scler. Relat. Disord. 2020;42:102180. doi: 10.1016/j.msard.2020.102180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bollo L, et al. Seroconversion and indolent course of COVID-19 in patients with multiple sclerosis treated with fingolimod and teriflunomide. J. Neurol. Sci. 2020;416:117011. doi: 10.1016/j.jns.2020.117011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Edwards MJ, et al. Sphingosine prevents binding of SARS-CoV-2 spike to its cellular receptor ACE2. J. Biol. Chem. 2020;295:15174–15182. doi: 10.1074/jbc.RA120.015249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cheng JC, et al. Radiation-induced acid ceramidase confers prostate cancer resistance and tumor relapse. J. Clin. Investig. 2013;123:4344–4358. doi: 10.1172/JCI64791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Eliyahu E, et al. Acid ceramidase improves the quality of oocytes and embryos and the outcome of in vitro fertilization. FASEB J. 2010;24:1229–1238. doi: 10.1096/fj.09-145508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ramsubir S, et al. In vivo delivery of human acid ceramidase via cord blood transplantation and direct injection of lentivirus as novel treatment approaches for Farber disease. Mol. Genet. Metab. 2008;95:133–141. doi: 10.1016/j.ymgme.2008.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.De Biasi S, et al. Marked T cell activation, senescence, exhaustion and skewing towards TH17 in patients with COVID-19 pneumonia. Nat. Commun. 2020;11:3434. doi: 10.1038/s41467-020-17292-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weiskopf D, et al. Phenotype and kinetics of SARS-CoV-2-specific T cells in COVID-19 patients with acute respiratory distress syndrome. Sci. Immunol. 2020 doi: 10.1126/sciimmunol.abd2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guirao JJ, Cabrera CM, Jimenez N, Rincon L, Urra JM. High serum IL-6 values increase the risk of mortality and the severity of pneumonia in patients diagnosed with COVID-19. Mol. Immunol. 2020;128:64–68. doi: 10.1016/j.molimm.2020.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Copaescu A, Smibert O, Gibson A, Phillips EJ, Trubiano JA. The role of IL-6 and other mediators in the cytokine storm associated with SARS-CoV-2 infection. J. Allergy Clin. Immunol. 2020;146:518–534 e511. doi: 10.1016/j.jaci.2020.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nelde A, et al. SARS-CoV-2-derived peptides define heterologous and COVID-19-induced T cell recognition. Nat. Immunol. 2021;22:74–85. doi: 10.1038/s41590-020-00808-x. [DOI] [PubMed] [Google Scholar]
- 45.Nasi A, et al. Reactive oxygen species as an initiator of toxic innate immune responses in retort to SARS-CoV-2 in an ageing population, consider N-acetylcysteine as early therapeutic intervention. Toxicol. Rep. 2020;7:768–771. doi: 10.1016/j.toxrep.2020.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Delgado-Roche L, Mesta F. Oxidative stress as key player in severe acute respiratory syndrome coronavirus (SARS-CoV) infection. Arch. Med. Res. 2020;51:384–387. doi: 10.1016/j.arcmed.2020.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tanner JE, Alfieri C. The fatty acid lipid metabolism nexus in COVID-19. Viruses. 2021;13:90. doi: 10.3390/v13010090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rioux V, Record M. Special Section Involvement of lipids in the occurrence of COVID-19. Biochimie. 2020;179:228. doi: 10.1016/j.biochi.2020.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Van Der Made CI, et al. Presence of genetic variants among young men with severe COVID-19. JAMA. 2020;324:663–673. doi: 10.1001/jama.2020.13719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fontanet A, et al. SARS-CoV-2 variants and ending the COVID-19 pandemic. Lancet. 2021;397:952–954. doi: 10.1016/S0140-6736(21)00370-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cure E, Cumhur Cure M. COVID-19 may affect the endocrine pancreas by activating Na+/H+ exchanger 2 and increasing lactate levels. J. Endocrinol. Investig. 2020;43:1167–1168. doi: 10.1007/s40618-020-01307-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu M-Y, et al. Clinical evaluation of potential usefulness of serum lactate dehydrogenase (LDH) in 2019 novel coronavirus (COVID-19) pneumonia. Respir. Res. 2020;21:1–6. doi: 10.1186/s12931-019-1261-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rice TC, et al. Sphingosine rescues aged mice from pulmonary pseudomonas infection. J. Surg. Res. 2017;219:354–359. doi: 10.1016/j.jss.2017.06.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Carstens H, et al. Clinical development of sphingosine as anti-bacterial drug: Inhalation of sphingosine in mini pigs has no adverse side effects. Cell. Physiol. Biochem. 2019;53:1015–1028. doi: 10.33594/000000194. [DOI] [PubMed] [Google Scholar]
- 55.Tabazavareh ST, et al. Lack of sphingosine causes susceptibility to pulmonary Staphylococcusaureus infections in cystic fibrosis. Cell. Physiol. Biochem. 2016;38:2094–2102. doi: 10.1159/000445567. [DOI] [PubMed] [Google Scholar]
- 56.Marfia G, et al. Decreased serum level of sphingosine-1-phosphate: A novel predictor of clinical severity in COVID-19. EMBO Mol. Med. 2021;13:e13424. doi: 10.15252/emmm.202013424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nardacci R, et al. Evidences for lipid involvement in SARS-CoV-2 cytopathogenesis. Cell Death Dis. 2021;12:1–12. doi: 10.1038/s41419-021-03527-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Henry BM, et al. Alterations in the lipid profile associate with a dysregulated inflammatory, prothrombotic, anti-fibrinolytic state and development of severe acute kidney injury in coronavirus disease 2019 (COVID-19): A study from Cincinnati, USA. Diabetes Metab. Syndr. 2021;15:863–868. doi: 10.1016/j.dsx.2021.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dwyer CJ, et al. Comparative analysis of antibodies to SARS-CoV-2 between asymptomatic and convalescent patients. Iscience. 2021 doi: 10.1016/j.isci.2021.102489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bielawski J, et al. Sphingolipid analysis by high performance liquid chromatography-tandem mass spectrometry (HPLC-MS/MS) Adv. Exp. Med. Biol. 2010;688:46–59. doi: 10.1007/978-1-4419-6741-1_3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.