Summary
During morphogenesis, cellular sheets undergo dynamic folding to build functional forms. Here, we develop an image-based quantitative morphology field (QMorF) protocol that quantifies the morphological features of cellular structures and associated distributions. Using feather shafts with different rigidities as examples, QMorF performs coarse-graining statistical measurements of the fitted cellular objects over a micro-image stack, revealing underlying mechanical coupling and developmental clues. These images give intuitive representations of mechanical forces and should be useful for analyzing tissue images showing clear cellular features.
For complete details on the use and execution of this protocol, please refer to Chang et al. (2019).
Graphical abstract

Highlights
-
•
Protocol describes how to quantify the morphological features of a cellular structure
-
•
Cellular morphology is characterized by the ellipse fitting of cellular cross sections
-
•
Coarse-graining averaging of quantified morphology improves the measuring statistics
-
•
QMorF heat map reveals the mechanical coupling and developmental clues in tissues
During morphogenesis, cellular sheets undergo dynamic folding to build functional forms. Here, we develop an image-based quantitative morphology field (QMorF) protocol that quantifies the morphological features of cellular structures and associated distributions. Using feather shafts with different rigidities as examples, QMorF performs coarse-graining statistical measurements of the fitted cellular objects over a micro-image stack, revealing underlying mechanical coupling and developmental clues. These images give intuitive representations of mechanical forces and should be useful for analyzing tissue images showing clear cellular features.
Before you begin
Rationale of quantitative morphology field
Quantitative morphology field (QMorF) is an image-based analysis that quantifies the morphological features of a cellular structure as distributions, which are displayed as a heat map (Chang et al. 2019). The morphology is quantified by fitting two-dimensional (2D) cross-sections of cells to ellipses and characterizing these fitting ellipses (see “Fitting elliptical object”). Coarse graining is used to average the quantified cross-sectional morphology over randomly packed three-dimensional (3D) tissue structures, and the spatial distribution of the averaged measurement is output in an intuitive heat map to reveal the QMorF of the tissue (see “Calculation and analysis of the quantitative morphology field”). Horizontal coarse graining is achieved by sampling fitting objects within a window in the pre-processed cross-sectional image. Longitudinal coarse graining is based on a stack of aligned and pre-processed images from serial sectioning (see “Acquiring the cross-sectional micro-image stack” and “Pre-processing the tissue section images”). QMorF results in an innovative spatial presentation of a quantified and analyzed cellular morphology based on robust statistics and geometric fitting. The current protocol starts from a simple 2D physical section (see “Sectioning the rachis tissue sample”) and can be easily performed in a typical biological laboratory. It provides the quantitative morphology characterization of cellular structures for various tissue-level research (Chen et al. 2021, Chang et al. 2019).
Sectioning the rachis tissue sample
Timing: 12–24 h
The cellular structure of the medulla in a feather shaft is a good model for demonstrating our image-based quantitative morphology field (QMorF) analysis. First, we require a stack of sectional micro-images of the feather shaft tissue. As a sample segment of the feather shaft, the rachis is prepared by a process derived from typical paraffin wax embedding and sectioning of the biological tissue, forming multiple fixed tissue sections. At the end of this stage, the sections from a localized region of the rachis are fixed and sealed on microscope slides for serial imaging under an optical microscope.
-
1.
Preparation of sample segments
At each localized region of interest (ROI) to be analyzed, the rachis is cut into a sample segment (typically < 4 mm in length) that fits within the tissue-embedding mold.
-
2.
Sample rachis segment dehydration
To ensure complete infiltration of the tissue-supporting medium, the sample rachis segment is treated in ethanol with a stepwise increase in concentration.
Note: The usual treatment is 70% ethanol for 1 h, 90% ethanol for another h, then 100% ethanol for more than 12 h.
-
3.Paraffin wax infiltration
-
a.Before embedding, the sample rachis segments are infiltrated with a tissue-supporting medium that supports the porous medulla in section.
-
b.The dehydrated tissue sample is first immersed in pure xylene for 1 h, followed by immersion in fresh xylene for another hour.
-
c.The xylene-immersed tissue sample is then immersed in paraffin wax for at least 12 h to allow paraffin wax infiltration.
-
a.
Note: The usual supporting medium is paraffin wax. Leica-Paraplast PLUS paraffin wax was used in our experiment.
-
4.
Paraffin wax embedding
The infiltrated sample segment is placed in the paraffin wax-filled embedment mold with the desired sectioning surface facing downward.
Note: Multiple sample segments can be embedded within the same mold. We also intentionally misaligned multiple segments in the mold to avoid localized dulling of the blade during sectioning.
-
5.
Multiple sectioning of the sample segment
Starting from the desired position, multiple intact sections (of thickness 0.5–10 μm) are sliced from a 100 μm-long localized region of the sample rachis segment.
Note: The slicing is performed by a rotary microtome (PATHO CUTTER R 35) and placed on the microscope slide glass (New Silane III). A continuous section of the rachis tissue is ideal but not essential. More than five good quality sections per 100 μm are acceptable for the later image stacking and QMorF analysis. Provided that no overlap occurs, multiple sections can be placed on the same glass slide.
-
6.
Deparaffinization of the sections
The paraffin section slides are dipped into a xylene-filled container for at most one second to avoid drifting and flattening of the sections on the microscope slides.
-
7.
Sealing of the slides
The sectioned rachis tissue samples on the microscope slides are sealed with cover glass using regular slide seal adhesive.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Biological samples | ||
| Chicken feather rachis tissue sample | NCHU iEGG Center | http://iegg.nchu.edu.tw |
| Feather rachis tissue sectioning | Taiwan Mouse Clinic | http://tmc.sinica.edu.tw |
| Deposited data | ||
| Raw and analyzed data | This paper | Mendeley Data: https://doi.org/10.17632/g35h3y3396.1 |
| Experimental models: Organisms/strains | ||
| Leghorn chicken | NCHU iEGG Center | http://iegg.nchu.edu.tw |
| Software and algorithms | ||
| MATLAB | MathWorks | Versions R2014a |
| Photoshop | Adobe | Versions 13.0 20120315.r.428 |
| Other | ||
| Olympus BX51 Fluorescence Microscope | Olympus Co. | n/a |
| Canon EOS 50D Digital Camera | Canon Inc. | n/a |
| Olympus PLN 10×/0.25 Objective | Olympus Co. | N1215800 |
| Olympus LMPLFLN 10×/0.25 Objective | Olympus Co. | LMPLFLN10X |
| Olympus LMPLFLN 20×/0.40 Objective | Olympus Co. | LMPLFLN20X |
| Leica-Paraplast PLUS paraffin wax | Leica Biosystems | Cat# 39602004 |
| PATHO CUTTER R 35 microtome blade | Erma Inc. | Cat# 08-636-0 |
| New Silane III microscope slide glass | Muto Pure Chemical Co. | Cat# 511618 |
| Continuous images, analysis, and resources related to feather rachis structures. | This paper | n/a |
Alternatives: The above table lists the equipment used in our experiment. This equipment can be replaced with any microscope that can clearly acquire undistorted cellular structures.
Materials and equipment
Image processing toolbox
| MATLAB toolbox name | Purpose | Step | Description |
|---|---|---|---|
| imgaussfilt() | Applies a Gaussian filter to the image to reduce possible noise in later image processing steps. | Pre-processing the tissue section images | https://www.mathworks.com/help/images/ref/imgaussfilt.html |
| imcomplement() | Complements the image gray level to facilitate later image processing. | Pre-processing the tissue section images | https://www.mathworks.com/help/images/ref/imcomplement.html |
| adapthisteq() | Uses the contrast- limited adaptive histogram equalization (CLAHE) method to improve the clarity of the mesh network of the cellular border signal for later mesh and pore image segmentation. | Pre-processing the tissue section images | https://www.mathworks.com/help/images/ref/adapthisteq.html |
| im2bw() | Binarizes the image to reduce the amount of image information. | Pre-processing the tissue section images | https://www.mathworks.com/help/images/ref/im2bw.html |
| imopen() | Isolates the image noises attached to the mesh network. | Pre-processing the tissue section images | https://www.mathworks.com/help/images/ref/imopen.html |
| bwareaopen() | Eliminates image noise below a threshold pixel size. | Pre-processing the tissue section images | https://www.mathworks.com/help/images/ref/bwareaopen.html |
| imclose() | Closes the locally broken mesh network. | Pre-processing the tissue section images | https://www.mathworks.com/help/images/ref/imclose.html |
| bwboundaries() | Identifies and records the edge pixels of the cellular pore patch. | Fitting elliptical object | https://www.mathworks.com/help/images/ref/bwboundaries.html |
| fit_ellipse() | Fits the cellular pore into an elliptical object based on randomly sampled edge pixels. | Fitting elliptical object | https://www.mathworks.com/matlabcentral/fileexchange/3215-fit_ellipse |
Step-by-step method details
Acquiring the cross-sectional micro-image stack
Timing: ∼ 4 h (for 20 tissue sections)
A successful QMorF analysis requires a series of 2-dimensional (2D) micro-images of rachis tissue sections from a localized region of the rachis (usually < 100 μm, depending on the required resolution). A set of good quality micro-images, showing clear cellular morphology in each section, is requisite for the entire QMorF protocol. A qualified raw image, shown by Figure 5 in the expected outcomes and Figure 9 in the problem 1 of the troubleshooting section, not only saves effort in the later image pre-processing steps but also ensures an accurate analysis. Here we introduce the standard acquisition procedure for obtaining a stack of qualified micro-images of multiple intact tissue sections fixed and sealed on the slide from a 100 μm-long localized region of a rachis (Figures 1A and 1B).
-
1.Marking the reference rachis medulla contour for pre-alignment.
-
a.The least significant distortion/deformation tissue section is first chosen as the reference medulla contour.
-
b.Record the outline of this reference medulla contour by simply marking the contour on the display screen as the reference for the later whole image-stack acquisition.
-
a.
Note: Over the entire micro-image stack, the medulla contours of the tissue sections must be well aligned to correctly reconstruct the three-dimensional (3D) relationships among the internal cellular structures in the rachis medulla. In practice, maintaining the relative position and orientation of each tissue section on a sealed slide is not trivial. A manual alignment is required before taking the micro-image of each section to obtain a well-aligned image stack.
-
2.Acquiring a micro-image stack of the pre-aligned section slides.
-
a.Using the recorded outline of the reference medulla contour in step 1 as the reference, manually pre-align each rachis tissue section sealed on the microscope slide by adjusting the position and orientation of each section according to the marked reference medulla contour in the field of view for acquiring the micro image of each section under an Olympus BX51 microscope.
-
b.Bright-field images of the rachis tissue structure are taken under a 10× or 20× objective lens with a Canon EOS 50D digital camera under a transmissive light source.
-
c.The acquired images are saved as 8-bit monochrome digital images in BMP or uncompressed TIFF format to simplify the later image pre-processing step.
-
a.
Note: To amplify the object–background contrast, we adjust the brightness of the image by tuning the microscope illumination and the camera exposure time. After adjustment, the grayscale indices of the background and object signals are approximately 90–180 and 50–80, respectively. A typical image and its histogram are shown in the upper panel of Figure 2.
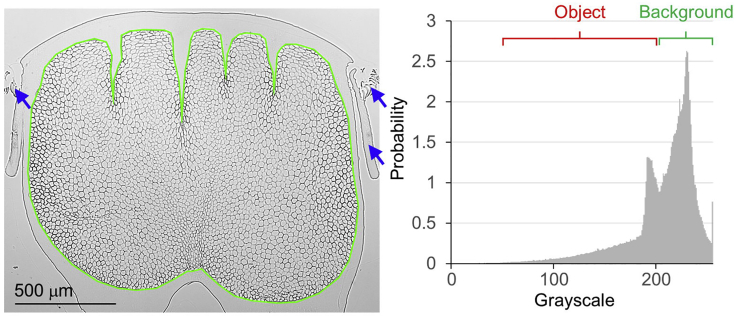
Figure 5.
Typical cross-sectional micro-image of the rachis tissue
The image is obtained by “Acquiring the cross-sectional micro-image stack” after a standard brightness and contrast enhancement for better visualization (Left panel), and the histogram (right panel). Scale bar, 500 μm.
Figure 9.
Examples of disqualified micro-images
(A) Qualified micro-image showing a clear and complete cross-section of the rachis tissue suitable for QMorF analysis. If the tissue section is folded (B and C) or distorted (D), part of the tissue image is not distinguishable, and the micro-image is disqualified. Scale bar, 500 μm.
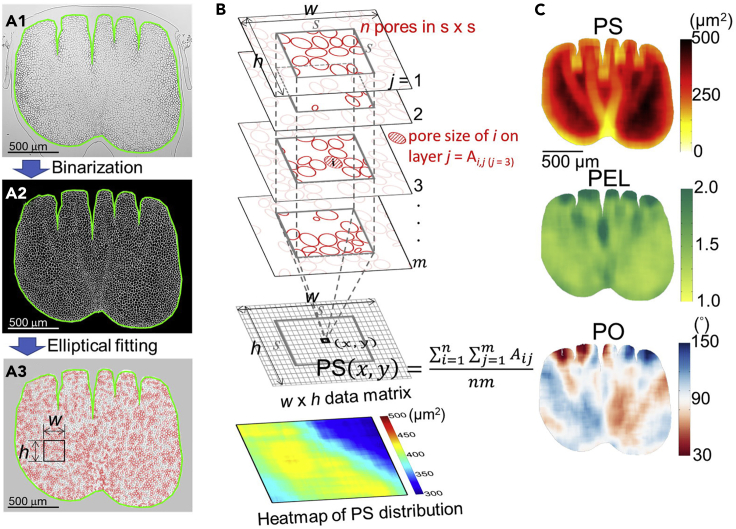
Figure 1.
Acquisition of the cross-sectional micro-image stack and the image pre-processing in the QMorF analysis
(A) A stack of multiple cross-sectional tissue sample images obtained along a finite length of the rachis.
(B) Each sectional image shows the mesh network of the sectioned porous medulla surrounded by the dense cortex.
(C) Pre-process the tissue section image for cellular patch identification in a later process (Scale bar, 100 μm). The left panels in (C) are demonstrative results of the first image (j= 1) of sectional rachis tissue after each step. The center panels are zoom-ins of the yellow framed regions in the low-magnification views at left. The right panel is a flowchart of the whole process, showing the purpose of each step.
Figure 2.
Demonstration of the object/background contrast change after each image pre-processing step
(A) and (B) show the result of four processed regional images and the corresponding histograms of the rachis medulla from Figure 1C after each step of the image pre-process protocol. The variation of the distributions of object and background signals after each step is displayed. Arrows indicate the reference grayscale levels for the parameter determination of the corresponding step. Scale bar, 50 μm.
Note: The cross-sectional tissue images could be intentionally taken slightly out of focus to clarify the mesh network of the medulla cross-section (see illustration in the next major step), and hence improve the 2D cellular pore patch identification.
Note: Besides the manual alignment method, other image alignment algorithms could also be used to align images in the stack. SIFT (Scale-Invariant Feature Transform) is one of the examples.
CRITICAL: Disqualified cross-sectional micro-images should be discarded in this step for accurate QMorF analysis. Please see problem 1 of the troubleshooting section for the examples of disqualified micro-images and the potential solution.
Pre-processing the tissue section images
Timing: 1–4 h (depending on image size and image quantity)
The micro-images of the sectioned rachis tissue are pre-processed through a series of complementation, contrast enhancement, and segmentation steps. At the end of this major step, 2D cellular pore patches distinguishable for elliptical fitting are obtained. Here we illustrate the pre-processing procedures sequentially applied to a stack of micro-images in our standard QMorF analysis of rachis tissue (Figure 1C). The result of this stage is a stack of binary images of the medulla section, showing black cellular pore patches isolated by a white mesh network of the cellular boarders.
-
3.
Grayscale image complementation: To facilitate the gray level operation in the later image processing, which identifies the contours of cellular pore patches, the micro-image of the cross-section rachis tissue is first processed using the grayscale image complement (“imcomplement” toolbox in MATLAB).
Note: The keratinized components of the rachis are optically opaque. In a bright-field micro-image of a rachis cross-section, the 2D cross-section of the 3D porous medulla appears as a dark mesh of keratinized cellular membrane boarder, and the 3D vacuoles composing the medulla appear as transparent bright 2D cellular pore patches.
Note: After this step, the mesh network and pore areas are converted into high and low gray levels, respectively. A higher gray level of the mesh signal provides wider dynamic ranges for the following operations.
-
4.
Image enhancement: To improve the contrast of the complemented image for later segmentation between the cellular pore patch and mesh network of cellular boarder, apply the contrast limited adaptive histogram equalization (CLAHE) enhancement method (“adapthisteq” toolbox in MATLAB) to the complement images.
Note: This method adjusts the contrast value of each tissue section. Unlike ordinary histogram equalization, the CLAHE enhancement method prevents the over-enhancement of localized noise signals, and brings the mesh signal to a similar level over the entire image.
-
5.
Image binarization: The complemented grayscale images are converted into black and white images (“im2bw” toolbox in MATLAB) to exclude the unnecessary morphological information.
Note: Binary images are suitable for most of the MATLAB toolboxes applied in later processes and simplify the pore-mesh segmentation. The mesh network and cellular pore patches in each image are then clearly distinguished for later morphological-based processing.
-
6.Medulla region identification and noise reduction.
-
a.To improve the accuracy of elliptical fitting in the next major step, the cortex portion of the rachis and the signals from the debris objects are manually removed from the binarized image using conventional image processing software (e.g., Photoshop or ImageJ). Only image signals contributed by the medulla portion are preserved for later stages.
-
b.Small noise patches on the image are automatically eliminated if their size is below a specified threshold (∼200 μm2) (“bwareaopen” toolbox in MATLAB).
-
a.
-
7.
Morphological closing: To clarify the pixel patch of each pore structure (i.e., the cellular patch) for accurate elliptical fitting in the next stage, a morphological closing process (“imclose” toolbox in MATLAB) is applied to the medulla in the binary image of the last step.
Note: Step 7 morphologically closes the locally broken mesh network and isolates the cellular patch of each pore in the binary image. These isolated cellular pore patches can be morphologically characterized by geometric fitting into an elliptical object. The parameters of these patches are extracted from the fitting objects in the following major step.
Note: The demonstrative graphs in Figure 1C show the anticipated result of each step. The histogram of each step shown in Figure 2 illustrates the enhanced signal quality through the variation of the gray-level distribution.
CRITICAL: To avoid elliptical-fitting failure in next major step, the pre-processed image must present an interconnected mesh network for the cellular border and isolated cellular patches. Please see problem 2 of the troubleshooting section for the example of poor pore-mesh segmentation and potential solutions.
Fitting elliptical object
Timing: 1–12 h (depending on image size and image quantity)
To characterize the morphological features of the cellular patches isolated by the interconnecting mesh network, the contour of each cellular patch is fitted into an ellipse. The location, orientation, and geometric parameters of the fitted ellipses are easily obtained. These fitted elliptical objects fairly represent the morphology of the original cellular patches over the cross-section of the medulla, and allow a quantitative description of this morphology (see Figure 3).
-
8.Elliptical fitting of the cellular patch.
-
a.Applying the edge-finding method (“bwboundaries” toolbox in MATLAB) to the cellular patch, the edge pixels of each 2D cellular pore patch are identified.
-
b.Input 30 randomly sampled edge pixels of a cellular patch into the open source MATLAB module “fit_ellipse,” the “fit_ellipse” module outputs an ellipse representing the original cellular patch.
-
a.
-
9.
Record the ellipse parameter: During the elliptical fitting of a cellular patch in step 8, the position, lengths of the major and minor axes (2a and 2b, respectively), and tilting angle Θ of the ellipse are recorded (Figure 3B). These recorded parameters preliminarily quantify the morphology of the corresponding cellular patch.
-
10.
Obtain the elliptical-fitting object image: Applying the fitting to every distinguishable cellular pore patch over the entire pre-processed micro-image stack, we obtain the plurality of elliptical-fitting objects and their recorded parameters.
Note: After this step, the parameters that quantify the morphologies of the original cellular pores in each medulla cross-section are obtained. In non-technical terms, the original grayscale micro-image stack is converted into a new stack of “images” composed of “elliptical-fitting objects.” This new “image” stack is actually a 3D matrix containing the morphological features of the cellular pore patches in a stack of rachis cross-sections distributed over the 100 μm-long rachis segment.
Note: Theoretically, the elliptical fitting requires at least three randomly sampled edge pixels. Obviously, sampling more edge pixels will improve the accuracy of the fitting, but will increase the computational time. To balance the fitting accuracy and computational resources, we recommend the sampling of 30 edge pixels from each cellular patch. The optimization of the number of sampled edge pixels and the fitting accuracy are detailed in Figure 3D.
CRITICAL: Most of the elliptical objects should be accurately fitted on their represented cellular patches. The small portions of the incorrect fitted objects can be manually deleted. Please see problem 3 of the troubleshooting section for the example of incorrect fitted objects and the potential solution.
Figure 3.
Quantification of the cellular morphology through fitting the cellular pore patch to an ellipse
(A) Cross-sectional micro-image of rachis tissue from a chicken flight feather (Scale bar, 500 μm). The green loop highlights the cortex–medulla interface.
(B) Zoomed-in image of the rectangular region enclosed by the black frame in (A). One of the cellular patches in the medulla cross-section is fitted to an elliptical object, from which the parameters for quantifying the cellular patch morphology can be obtained.
(C) Scheme of elliptical fitting on a group of N pixels randomly sampled from the edge of the cellular pore patch. The blue crosses in the left panel show the 10 randomly sampled edge elements and the red ellipse is the corresponding fitting result. The red ellipses in the three right panels are the fitting results for different numbers of sampling pixels (N = 5, 10, and 30).
(D) Area distributions of the fitted ellipse with different N after 5000 random sampling trials along the contour of a chosen cellular pore patch. Increasing N increases the computational power but also ensures rapid convergence toward a narrow distribution. It was found that N= 30 achieves a good balance between the fitting accuracy and computational time.
Calculation and analysis of the quantitative morphology field
Timing: 1–4 h (depending on image size)
The quantitative morphology of the cellular structure in a localized region of the medulla tissue is achieved by coarse-grain averaging the quantitative characters of the elliptical-fitting objects within a grain (Figure 4). In this stage, the parameters and/or calculated results are statistically averaged over the correctly fitted elliptical objects within a specific coarse-graining volume in the 3D medulla centered at (x, y). The quantitative morphology is thus measured at (x, y). The spatial variation of the quantitative morphology measurements over a specific morphological feature are presented as a two-dimensional distribution graph called the quantitative morphology field (QMorF). The QMorF reveals the collective cellular deformation under the spatially varying mechanical stress imposed on the local cells during morphogenesis (Chang et al. 2019).
-
11.Align and overlap the fitting object image stack: To sample the elliptical-fitting objects in a 3D coarse-graining volume in the image stack, the elliptical-fitting object images in the stack are aligned first.
-
a.Overlap the plural elliptical-fitting object images within the 100 μm-long rachis segment from which the raw image stack was acquired. The overlap is performed by best manual alignment of the medulla contour of each frame.
-
b.When the tissue section has been slightly distorted and the medulla contours cannot be perfectly aligned, use a small ROI or fine morphological features as reference coordinates for the alignment.
-
a.
Note: This alignment precisely reconstructs the spatial arrangement of the cellular pore patches, and therefore their corresponding elliptical-fitting objects, in three dimensions.
-
12.Coarse-graining calculation.
-
a.The quantified morphological feature at a specific position (x, y) is calculated from the parameters of the elliptical-fitting objects within a pre-defined coarse-graining volume centered at (x, y).
-
i.The regional averaged ellipse size PS(x, y) is obtained by calculating <πab> .
-
ii.The regional averaged ellipse elongation rate PEL(x, y) is obtained by calculating <a/b>.
-
iii.The regional averaged ellipse orientation angle PO(x, y) is obtained by calculating <Θ>.Note: Here <> denotes the mean of the measurements within the coarse-graining volume (Chang et al. 2019). The size of the coarse-graining volume is defined as an s × s region on the cross-sectional plain multiplied by the vertical dimension contributed by the m images composing the image stack.
-
i.
-
b.After conducting the above calculation over the rachis cross-section, a matrix recording the local quantified morphological features of the cellular patches (averaged size PS, elongation rate PEL, or orientation angle PO) at each pixel position is obtained.
-
a.
-
13.
QMorF result output: To visualize the computationally calculated data and reveal the spatial features of the cellular morphology, the matrix obtained in step 12 is output in 2D heat map format.
Note: This output heat map is particularly useful for side-by-side comparison between the original grayscale cross-sectional image and the associated fine morphological variations. The range of the scale color bar depends on the data type and visual purpose. In the QMorF analysis of a typical chicken flight feather rachis, the ranges PS, PEL, and PO are 0–500 μm2, 1.0–2.5, and 30°–150°, respectively.
CRITICAL: If the output QMorF heat map is blurry and cannot reveal the desired morphological features, please see problem 4 of the troubleshooting section for example heat maps and the potential solution.
Figure 4.
Major steps in the QMorF analysis and their results
Panels (A1) to (A3) show how the cross-sectional micro-image of a flight feather rachis tissue (A1) is pre-processed (A2) to obtain an elliptical object fitting image (A3). Panel (B) demonstrates the QMorF analysis on the w × h ROI within the black frame in (A3). After overlapping the entire stack of elliptical-fitting objects, we calculate the desired QMorF by averaging the quantified morphological features of the elliptical-fitting objects within a coarse-grained volume. For example, the pore size, i.e., the area of a cellular patch in the binary image, is computed by averaging the pore areas Aij over the virtual volume defined as an s × s region on the cross-sectional plain multiplied by the vertical dimension contributed by the m images composing the image stack, where Aij is the pore area of the ith elliptical-fitting object in the jth section, and s is the horizontal dimension of the grain size. PS(x, y) represents the coarse-grained averaged pore size at position (x, y). The calculation is repeated over the entire ROI and the measured PS(x, y) matrix is presented as a heatmap. The regional PSs obtained by QMorF are displayed in the bottom panel of (B). Extracting the coarse-grained averaged pore sizes (PS ≡ <πab>), pore elongation rates (PEL≡ <a/b>), and pore orientation angles (PO≡ <Θ>) over the whole set of rachis tissue sections, we obtain different distributions of the morphological features from the chicken flight feather rachis tissue. The results are shown in (C). The heat maps in panel (C) are adopted and modified from (Chang et al. 2019). Scale bar, 500 μm
Expected outcomes
A typical cross-sectional micro-image of the rachis tissue obtained by “Acquiring the cross-sectional micro-image stack” is shown in the left panel of Figure 5. The green loop is the medulla contour, i.e., the interface between the outer cortex and inner porous medulla. This reference micro-image shows a clear dark mesh network of the keratinized cellular membrane boarder and the dense outer cortex in a 5 μm-thick chicken flight feather rachis section taken under a bright-field microscope. The blue arrows indicate the debris of the feather bar and barbules after the sectioning. The histogram in the right panel shows the signal-background difference. The red and green brackets in the histogram indicate the grayscale level and background of the object, respectively.
A typical binary image generated by “Pre-processing the tissue section images” is shown in Figure 6. The reference pre-processed binary image clarifies a white mesh network of the keratinized cellular membrane border from a 5 μm-thick chicken flight feather rachis section. The outer cortex region and debris have been manually removed. Most of the black mesh pores are isolated by the white mesh, providing distinct cellular pore patches for the following stage, “Elliptical object fitting.”
Figure 6.
Typical binary image generated by “Pre-processing the tissue section images”
Scale bar, 500 μm.
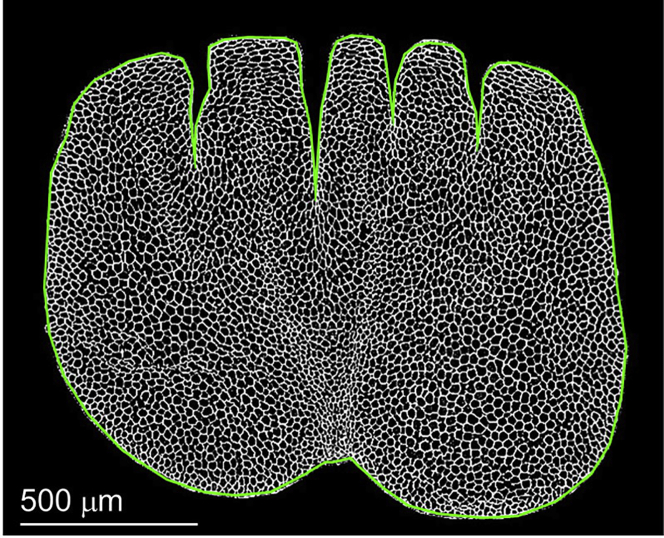
A typical distribution of elliptical-fitting objects overlain on the original gray level micro-image of the rachis tissue cross-section is shown in Figure 7. Open red ellipses show where the underlying cellular patches are properly fitted. Because the local mesh network of the cellular border is imperfectly connected, the “Fitting elliptical object” fails in some cellular patches. Those mis-fitting objects, which are usually characterized by extreme sizes, are automatically removed. Although the fitting objects do not cover the whole medulla region, they are expected to be randomly distributed in the cross-sectional image. Therefore, when the fitting object images are stacked, the object distribution should be pervasive. The elliptical-fitting object image converted from an original micro-image with considerable object density is ready for the final stage, “Calculation and Analysis of the Quantitative Morphology Field.”
Figure 7.
Distribution of elliptical-fitting objects overlain on the original gray-level micro-image of the rachis tissue cross-section
Scale bar, 500 μm.
The outcome heat map of the QMorF analysis in Figure 8 shows the spatial matrix of the quantified cellular pore size distribution obtained through coarse-grained averaging. For details, see “Calculation and Analysis of the Quantitative Morphology Field.” The cells are heterogeneously arranged in the rachis tissue and a periodic cell band appears (Chang et al. 2019). The QMorF result of the pore size was obtained from an original image stack of 10 micro-images. Each micro-image was obtained from a 5 μm-thick rachis tissue section along a 60 μm-long rachis segment in the middle of the chicken flight feather.
Figure 8.
Typical heat map generated by QMorF
The heat map shows the spatial matrix of the quantified cellular pore size distribution. The figure is adopted and modified from (Chang et al. 2019). Scale bar, 500 μm.
Limitations
-
•
Insufficient statistics due to an insufficient number of micro-images: A successful QMorF analysis relies on the sufficient number of elliptical-fitting objects from the cellular patches in the rachis tissue. The statistics are obtained from a considerable number of ensembles to eliminate sampling bias. Therefore, the sample tissue within a finite volume of interest must be sampled multiple times to acquire sufficiently many elliptical-fitting objects and ensure an accurate QMorF result. We suggest that at least five sections along a 100 μm-long rachis segment will achieve a smooth and accurate result.
-
•
Insufficient contrast of the images: Due to limitations of image processing, the grayscale intensity of the micro-image must be distributed over a reasonable range, leaving a margin for the image enhancing and object segmentation processes. Applying the image pre-process module to images of marginal quality, such as overexposure or underexposed images, may degrade the cellular patch identification.
-
•
Insufficient image resolution: As the cellular morphology is quantified by elliptically fitting the edge pixels of the cellular patch, the micro-image resolution must be sufficiently fine to ensure a viable number of randomly selected edge elements. Small cellular patches with a large contour-to-area ratio in a micro-image tend to be erroneously fitted. To ensure an accurate and efficient elliptical fitting, we recommend an averaged cellular patch size exceeding 250 pixels in the micro-image. At least 30 edge pixels should be sampled from each patch.
Troubleshooting
Problem 1: Micro-image of distorted tissue section in “sectioning the rachis tissue sample” (step 2)
Figure 9 shows the examples of disqualified micro-images with folded and distorted tissue sections. These cases frequently happen during the serial sectioning of the stiff rachis tissue.
Potential solution
If the number of high-quality sections is sufficient (i.e., more than five cross-sectional images show clear and complete tissue along the 100 μm rachis length), the sections with severe deformation or distortion can be simply discarded. If the number of complete cross-sectional images is insufficient due to technical difficulties, the user can manually remove the problematic portions from the original images during the “Pre-processing the tissue section images” stage, and proceed with the analysis. Note that in this case, the accuracy of the final QMorF distribution will be non-uniform over the cross-section because after removing the distorted portions, the cellular patches in the tissue micro-image will be unevenly sampled.
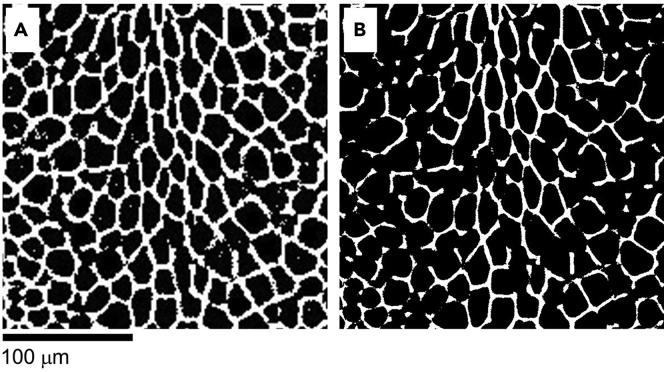
Problem 2: Poor pore-mesh segmentation in “pre-processing the tissue section images” (step 7)
Because of the variation in the sample characteristics, preparation, degradation, and instrumentation, pre-processing a micro-image successfully requires effort. The right panel in Figure 10 shows a failed pre-processed image with a broken mesh network for the cellular border and interconnected cellular patches. The poor pore-mesh segmentation is a major cause of elliptical-fitting failure.
Figure 10.
Typical pre-processed images showing cellular patches
A pre-processed image suitable for elliptical fitting (A), and a failed pre-processed image with a broken mesh network for the cellular border and interconnected cellular patches (B). Scale bar, 100 μm.
Potential solution
-
•
Lowering the threshold of the binarization process will preserve more of the mesh network information in the binary image. Note that the noise could also increase and may interfere with the later elliptical fitting.
-
•
Increasing the size of the structuring element in the MATLAB toolbox for morphological closing will improve the connectivity of the mesh network. Note that aggressive morphological closing can significantly alter the geometry of the cellular patch, cause elliptical fitting on distorted patches, and reduce the accuracy of the morphology quantification.
-
•
Applying a Gaussian filter (“imgaussfilt“ toolbox in MATLAB) on the original image will homogenize the mesh signals of the original image and reduce the noise, but this solution is unlikely to benefit images with poor contrast.
Note: The above potential solutions can be applied non-exclusively depending on the quality of the original image.
Problem 3: Inaccurate elliptical fitting in “fitting elliptical object” (step 8)
Inaccurate elliptical fittings (as shown in the right panel of Figure 11) may occur even when the pore-mesh segmentation is successful.
Figure 11.
Demonstrative elliptical-fitting results
Correct (left) and inaccurate (right) elliptical-fitting result. Scale bar, 100 μm.
Potential solution
This problem can be resolved by increasing the number of randomly sampled edge pixels in the elliptical object fitting. In images with low resolution, the number of pixel coordinates cannot be increased, and the image should be recaptured at higher resolution or under a higher magnification.
Problem 4: Detail-losing heatmap in “calculation and analysis of the quantitative morphology field” (step 13)
The output QMorF heatmap is blurry (as shown in the right panel in Figure 12). The anticipated details of the cellular morphology are not well resolved.
Figure 12.
Spatial feature of the morphology revealed by QMorF
Typical heat map showing the prominent features of the medulla morphology distribution (A), and a detail-losing heat map showing blurry morphological features (B). Panel (A) is adopted and modified from (Chang et al., 2019). Scale bar, 500 μm.
Potential solution
This problem can be resolved by adjusting the coarse-graining size s in the QMorF analysis. Reducing the coarse-graining region usually preserves more morphological details but also increases the fluctuating signals. On the contrary, enlarging the coarse-graining region will reduce the noise but blur the fine morphological features.
Resource availability
Lead contact
Further information and requests for resources should be directed to and will be fulfilled by the lead contact, Wen-Tau Juan (wtjuancmu@gmail.com).
Materials availability
This study did not generate new unique reagents.
Data and code availability
The original image and the outcome of each stage generated during this study have been deposited to Mendeley Data: https://doi.org/10.17632/g35h3y3396.1.The MATLAB codes supporting the current study have not been deposited in a public repository because they are still in the patenting process, but are available from the corresponding author on request.
Acknowledgments
The authors thank Professor Chih-Feng Chen and Pin-Chi Tang for providing the chicken feather samples and Professor Kai-Jung Chi for her inspiring discussion. This work was supported by the Minister of Science and Technology, Taiwan (MOST 109-2112-M-039 -002 -MY3 and MOST 109-2311-B-039 -002 -MY3), and China Medical University in Taiwan (CMU109-MF-17); the Integrative Stem Cell Center, China Medical University and University Hospital, Taiwan; the Integrative Evolutionary Galliform Genomics Center (iEGG) and the Avian Genetic Resource and Animal Biotechnology Center supported by Feature Areas Research Center Program within the framework of the Higher Education Sprout Project by the MOE in Taiwan (MOE-107-S-0023-G&H&I), National Chung Hsing University, Taiwan. C.-M.C. is supported by US National Institute of Arthritis and Musculoskeletal and Skin Diseases (AR60306) and a research contract between CMU and USC (USC 5351285884).
Author contributions
H.W., Y.-K.C., and W.-T.J. conceived and designed the protocol. H.W., Y.-K.C., J.-C.T., C.-M.C., and W.-T.J. developed the method. H.W. and Y.-K.C. prepared feather samples and analyzed data. H.W. and W.-T.J. co-wrote the manuscript.
Declaration of interests
Applications for patents on our main methodology, referred to as QMorF in the manuscript, are pending in Taiwan and in the United States of America. C.-M.C. is a non-paid science advisor of Integrative Stem Cell Center, CMU/hospital.
References
- Chang W.L., Wu H., Chiu Y.K., Wang S., Jiang T.X., Luo Z.L., Lin Y.C., Li A., Hsu J.T., Huang H.L. The making of a flight feather: Bio-architectural principles and adaptation. Cell. 2019;179:1409–1423.e17. doi: 10.1016/j.cell.2019.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.K., Juan W.T., Liang Y.C., Wu P., Chuong C.M. Making region-specific integumentary organs in birds: evolution and modifications. Curr. Opin. Genet. Dev. 2021;69:103–111. doi: 10.1016/j.gde.2021.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The original image and the outcome of each stage generated during this study have been deposited to Mendeley Data: https://doi.org/10.17632/g35h3y3396.1.The MATLAB codes supporting the current study have not been deposited in a public repository because they are still in the patenting process, but are available from the corresponding author on request.