Summary
As space travel becomes more accessible, it is important to understand the effects of spaceflight including microgravity, cosmic radiation, and psychological stress. However, the effect on offspring has not been well studied in mammals. Here we investigated the effect of 35 days spaceflight on male germ cells. Male mice that had experienced spaceflight exhibit alterations in binding of transcription factor ATF7, a regulator of heterochromatin formation, on promoter regions in testis, as well as altered small RNA expression in spermatozoa. Offspring of space-traveling males exhibit elevated hepatic expression of genes related to DNA replication. These results indicate that spaceflight has intergenerational effect.
Subject areas: Transcriptomics, Space sciences
Graphical abstract
Highlights
-
•
Rearing mice in outer space alters ATF7-binding profile in testis
-
•
Short-term spaceflight affects epigenetic and small RNA profiles in spermatozoa
-
•
Paternal experience of spaceflight changes transcriptome in offspring tissue
Transcriptomics; Space sciences
Introduction
During spaceflight, astronauts experience various stresses such as hypergravity during launch and microgravity and cosmic radiation in outer space (Demontis et al., 2017). These environmental factors can affect the skeletal, immune, and nervous systems (Williams et al., 2009). In preparation for the coming space age, it is important to evaluate the parental effects of spaceflight on offspring. Studies in fish, amphibians, and birds have found that spaceflight has no effect on the capacity of reproduction (Murata et al., 2015; Aimar et al., 2000; Wentworth and Wentworth, 1996). Effects on the reproductive system of mice have been unclear due to the difficulty of maintaining health in space (Andreev-Andrievskiy et al., 2014; Tavella et al., 2012; Cancedda et al., 2012). In some studies, more than half of experimental mice died due to unexpected stresses in spaceflight, making it difficult to estimate the controlled effects of spaceflight. We recently developed mouse cages specialized for space travel and used them to rear male mice in the International Space Station (ISS)/Kibō for 35 days (Shimbo et al., 2016; Shiba et al., 2017). Male mice were individually housed under either microgravity (MG) or artificial gravity (AG; approximately 1 × g), and all mice returned to Earth alive. Compared with the control group (GC) that remained on Earth, mice that experienced MG had significantly decreased bone volume, whereas the bone volume of mice in the AG group was similar to that of control mice (Shiba et al., 2017). We also observed differences in gene expression profiles in the spleen and retina between GC and MG treatments (Horie et al., 2019; Mao et al., 2018). On the other hand, spermatozoa collected from MG and AG mice had normal morphology and reproduction abilities (Matsumura et al., 2019).
A series of studies in rodents demonstrated that paternal stresses such as nutritional conditions can affect the phenotype of offspring (Youngson and Whitelaw, 2008; Heard and Martienssen, 2014). Paternal consumption of high-fat diet programs β-cell dysfunction and glucose intolerance in female rat offspring (Ng et al., 2010). In mice, offspring of fathers fed a low-protein diet (paternal low-protein diet [pLPD]) exhibit increased expression of cholesterol biosynthesis-related genes and altered cholesterol metabolites in the liver (Carone et al., 2010). Epidemiological studies of human cohorts also suggest that the effects of stress can be passed down to offspring; paternal smoking in childhood correlates with the body mass index of sons, and paternal nutritional surfeit correlates with the diabetes mortality of grandchildren (Kaati et al., 2002; Pembrey et al., 2006). The heritability of non-genetic traits is now recognized as intergenerational or transgenerational inheritance (Perez and Lehner, 2019).
Recently, we found that the transcription factor ATF7 is a key factor for intergenerational inheritance of traits resulting from pLPD in mice (Yoshida et al., 2020). In testicular germ cells (TGCs), ATF7 binds to promoter regions and induces methylation of histone H3 lysine 9 (H3K9me2) via physical interaction with the H3K9me2 methyltransferase G9a, leading to heterochromatin formation. LPD treatment induces release of ATF7 from chromatin via p38-dependent phosphorylation and thereby reduces the amount of H3K9me2. This epigenetic change is maintained by differentiation of TGCs to spermatozoa and also alters the expression of small RNAs such as tRNA-derived small RNA (tsRNA) in spermatozoa.
Results
Spaceflight changes ATF7-binding profile in TGCs
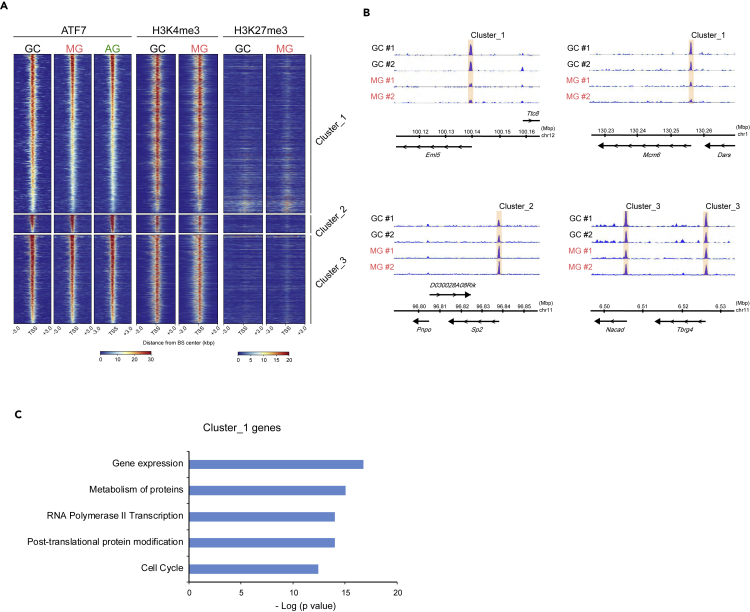
To investigate whether the experience of spaceflight affects ATF7-binding profiles in testis, we performed ATF7 chromatin immunoprecipitation sequencing (ChIP-seq) analysis using TGCs collected from three groups of mice: GC, MG, and AG. We identified 6,102 ATF7-binding sites in the testes of GC mice, and 70.0% of these sites were located in promoter regions (Figure S1A; Table S1). Comparison of ATF7-binding amounts around transcription start sites (TSSs) between GC and MG showed that ATF7-binding TSSs were clustered into three groups, clusters 1–3. ATF7 signals in 59.7% (3,189/5,341 TSSs) of ATF7-binding TSSs in GC mice had reduced ATF7 signals in MG mice (cluster_1); 6.8% (362/5,341 TSSs) had increased ATF7 signals in MG mice (cluster_2); and 33.5% (1,790/5,341 TSSs) had similar ATF7 signals in GC and MG mice (cluster_3) (Figures 1A and 1B). TGCs collected from AG mice also exhibited reduced ATF7 binding around TSSs in cluster_1 (Figure 1A). Accumulative plots and principal-component analysis (PCA) results showed the reproducibility of these tendencies in each individual mouse (Figures S1B and S1C). The expression profiles of spermatogenic marker genes are statistically similar between each group (Kruskal-Wallis one-way analysis of variance) (Figure S1D), indicating that MG/AG stress does not induce drastic change of cell population in testis. Thus, the alteration of spermatogenic cell population did not cause change in ATF7-binding profiles.
Figure 1.
Spaceflight affects ATF7-binding profile in TGCs
(A) Heatmap of signal intensities from ATF7 ChIP-seq data using GC/MG TGCs across ATF7-target genes in GC TGCs. Raw reads were scaled to mapped regions in each sample (reads per million).
(B) Typical ATF7-binding sites for cluster_1 (top), cluster_2 (bottom, left), and cluster_3 (bottom, right). Blue signals indicate the number of raw reads, normalized against total reads in each sample and region. Light orange boxes indicate ATF7-binding sites.
(C) Pathway analysis of cluster_1 genes using Reactome datasets. The top five pathways are shown, ranked by p value (Bonferroni correction for multiple testing).
Most of ATF7 sites were detected in active promoter regions marked with H3K4me3 rather than H3K27me3, and the level of H3K4me3 at these sites was not affected by MG (Figure 1A). The profiles of H3K4me3 and H3K27me3 also did not change in all TSSs including non-ATF7-binding sites (Figure S1E). Motif analysis of ATF7-binding sites showed that the CRE motif (TGACGTCA), which is recognized by ATF7, is significantly enriched in all clusters (Figure S1F). Pathway analysis indicates that ATF7-target genes in cluster_1 include genes related to gene expression, protein metabolism, and the cell cycle (Figure 1C). These data indicate that activation of testicular ATF7, loss of ATF7 on chromatin, was induced by spaceflight (e.g., launching and/or staying in space) rather than the condition of gravity (MG or AG) in space.
Spaceflight affects epigenetic and expression profiles in mature sperm
To answer whether changes in the ATF7-binding profile in TGCs affect epigenetic status in mature sperm, we examined H3K9me2 levels in ATF7 sites (cluster_1) using histone replacement-completed sperm (HRCS) collected from GC and MG mice. H3K9me2 levels at Mcm3 and Mcm6 were decreased in MG mice compared with those in GC, whereas only one of two samples exhibited a similar tendency at the Mcm2, Lig1, and Uhrf1 loci (Figure S2A). A possible explanation for this is that due to the short period of spaceflight, MG mice still have sperm fraction that was not affected.
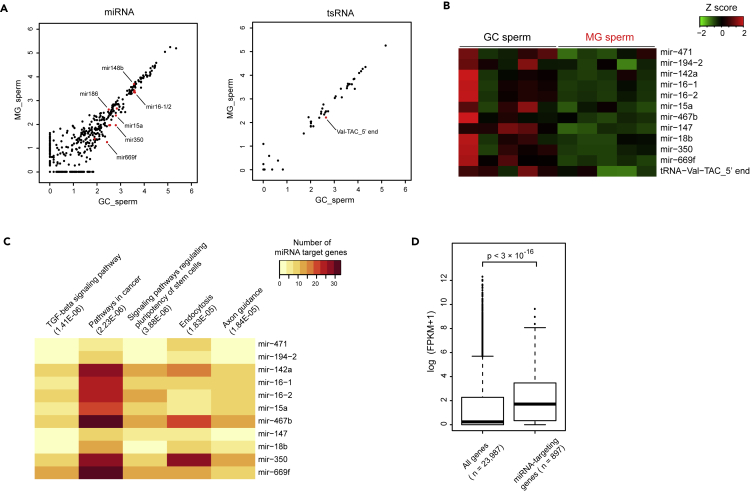
As testicular ATF7 affects the expression profile of small RNAs in spermatozoa in response to LPD (Yoshida et al., 2020), small RNA sequencing (RNA-seq) was performed using HRCS. The results indicate that expression of two micro RNAs (miRNA) increased, whereas expression of 11 miRNAs and one tsRNA decreased (Figures 2A and 2B; Table S2). Although reduction of ATF7 was observed in some tRNA loci (Figure S2B), tRNA expression was not elevated. To estimate the impact of reduced expression of sperm miRNA on offspring phenotype, KEGG pathway analysis for genes targeted by those miRNAs was performed. The result shows that TGF-β signaling and pluripotency were included (Figure 2C; Table S3). Strong candidates of target genes (multiply targeted by at least four miRNAs) tended to be expressed in zygote (Figures 2D and S2C–S2E). In addition to ATF7-induced epigenetic changes in sperm, reduced expression of these small RNAs alternatively can affect the transcriptome in offspring embryo.
Figure 2.
Spaceflight affects small RNA expressions in spermatozoa
(A) Scatterplot of miRNA (left) and tsRNA (right) expression profiles of HRCS in GC and MG mice (n = 5). Each dot indicates log10 (reads per million +1). Red dots indicate DEGs (|FC| > 1.3, FDR <0.05 calculated by Baggerly's test), and the representative DEGs were labeled with “miRNA name” or “name of tRNA origin.”
(B) Heatmap for expression of sperm miRNAs decreased by MG.
(C) Heatmap for number of genes targeted by each miRNA in each pathway. The top five pathways are presented, ranked by p value.
(D) Boxplot of expression level in zygote for all genes or genes targeted by at least 4 of 11 miRNAs. p value was calculated using the Wilcoxon rank-sum test.
Paternal experience of spaceflight changes hepatic expression profiles in offspring
We expected that alteration of epigenetic and small RNA profiles in sperm might change the phenotype of F1 offspring; however, F1 offspring fathered by MG mice do not show obvious defects in growth rates (Matsumura et al., 2019). ATF7 is predominantly activated by environmental stress in spermatocytes during spermatogenesis (Yoshida et al., 2020), and a short-term stay in space (35 days) is sufficient time for differentiation from spermatocyte to mature sperm (Oakberg, 1956).
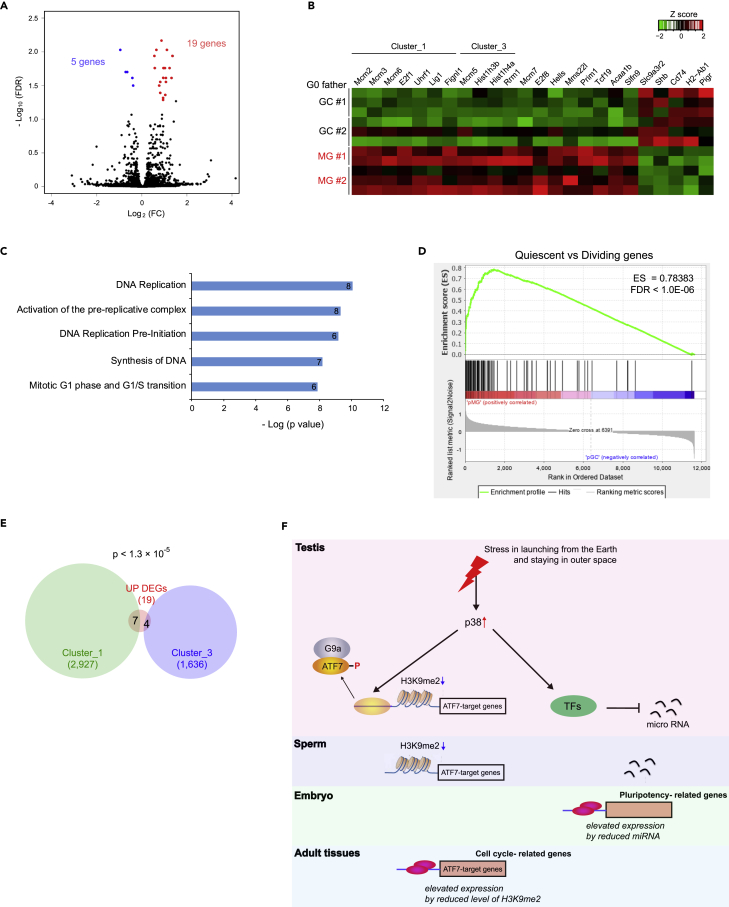
Using sperm collected from two individual GC and MG mice (GC #1/2 and MG #1/2 corresponding to mice in Figure 1B), we produced F1 progeny (pGC and pMG) and compared expression profiles in the liver of F1 males. Nineteen genes with increased expression in pMG offspring and five genes with decreased expression were identified (Figures 3A and 3B; Figure S3A; Table S4). Pathway analysis of genes upregulated by pMG indicates their involvement in DNA replication process (Figure 3C). Results of clustering analysis, cross-comparison, and PCA show distinguishable expressions of DNA replication-related genes between pGC and pMG individually, except for GC#2-2 (Figures S3B–S3D). Gene set enrichment analysis (GSEA) also indicates that proliferation-associated genes are activated in the liver from F1_MG relative to F1_GC (Figure 3D; Table S5), suggesting that cell proliferation could be enhanced in F1_MG mice.
Figure 3.
Spaceflight changes transcriptome in F1 offspring
(A) Volcano plot of RNA-seq results in offspring liver collected from pGC and pMG mice (n = 5–6).
(B) Heatmap for expression (transcripts per million) of DEGs in offspring liver. DEGs overlapping with ATF7-target genes in GC testis are indicated by cluster name.
(C) Pathway analysis of DEGs upregulated by pMG using Reactome datasets. The top five pathways ranked by p value (Bonferroni correction for multiple testing) are presented with the gene number involved in each pathway.
(D) GSEA plot of upregulated genes in dividing cells. GSEA was performed against whole expression profiles in pGC and pMG liver.
(E) Venn diagram between ATF7-target genes in GC testis and DEGs upregulated by pMG.
(F) Expected model of affecting the transcriptome in offspring by paternal stress involved in spaceflight.
Remarkably, most of the genes encoding subunits of the MCM complex were found in upregulated differentially expressed genes (DEGs) (Mcm2, Mcm3, Mcm5, Mcm6, and Mcm7) (Figure S3E). Interestingly, genes upregulated by pMG significantly overlapped with ATF7-target genes belonging to cluster_1/3 (p < 1.3 × 10−5, chi-square test) (Figure 3E). These results suggest that spaceflight affects the transcriptome of the F1 offspring of space-traveling males by a mechanism involving ATF7-dependent epigenetic changes and an altered miRNA expression profile in spermatozoa. Further experiments are required to characterize the phenotypes of offspring in detail.
Discussion
In the present study we show that rearing mice in outer space alters both the ATF7-binding profile in testis and small RNA expression in mature sperm. Offspring male from this sperm exhibit elevated expression of DNA replication-related genes such as Mcm family genes. As the MCM complex plays a pivotal role in the DNA replication process by acting as a DNA helicase, the expression levels of Mcm genes are correlated with resistance to replicative stress (Ibarra et al., 2008). However, cell proliferation is not so active in adult liver. We speculate that excess amount of MCM might function as a fail-safe to induce cell loss by some stress because MCM amount is correlated with proliferation rate, although cell proliferation is not clearly evident. Previously, we and another group reported that pLPD treatment also increases cell cycle-related genes in offspring (Carone et al., 2010; Yoshida et al., 2020). As the intergenerational inheritance of pLPD is regulated by testicular ATF7, molecular mechanism of enhancing proliferation in offspring liver may be sheared between these paternal stresses via ATF7. DEGs upregulated by pLPD are not overlapped with ATF7-binding genes in paternal TGCs (Yoshida et al., 2020). On the other hand, upregulated genes by pMG overlapped significantly with ATF7-binding genes in paternal testis. This difference of the effect on offspring phenotype may be derived from period or strength of paternal stress.
ATF7 is activated by various stresses such as pathogen infection, nutrient stress, and psychological stress via inflammatory cytokines and reactive oxygen species (Yoshida et al., 2015, 2020; Maekawa et al., 2010, 2018). Physiological and psychological stresses during launch and cosmic radiation in the ISS may activate ATF7 in mouse testes via p38 activation, with the likeliest candidate being psychological stress during launching. Recently, it was shown that paternal restraint stress changes the metabolome in Drosophila offspring via dATF2, Drosophila homolog of ATF7. In this case, restraint stress induces the expression of Upd3 (Seong et al., 2020), Drosophila homolog of Il-6. Also, in mouse, similar stress induces Il-6 (Voorhees et al., 2013), which can activate the mitogen-activated protein kinase (Hunter and Jones, 2015). It is reported that freeze-dried mouse spermatozoa stored in ISS for 9 months exhibited increase of DNA damage by cosmic radiation whose dose in ISS is higher than that on ground in earth by roughly 100 times (Wakayama et al., 2017). Thus, the cosmic radiation may also activate p38 slightly. P38 activates ATF7 to decrease H3K9me2 level on ATF7-binding sites and might also interact with other transcription factors to affect miRNA expression, similar to c-MYC with the miR15 family (Bracken et al., 2016; Bueno and Malumbres, 2011). The reduction of ATF7 binding in tRNA loci did not induce tRNA expression in spermatozoa, which suggests that another pathway to produce tsRNA, such as ROS or RNase (Yoshida et al., 2020), was not activated by MG. However, in this trial, these levels were not investigated. Reductions of H3K9me2 and miRNA can be maintained through differentiation of spermatogenesis to spermatozoa (Yoshida et al., 2020), and these changes affect activity of ATF7-target genes (cluster_1, including Mcm genes) in offspring tissue and expression of miRNA-target genes (pluripotency-related genes) in offspring embryo (Figure 3F).
It is also interesting whether epigenetic change induced by spaceflight-related stress can be reversed after one or two cycles of spermatogenesis (∼7–15 weeks) after returning to earth, or not. Current results indicate that this epigenetic change can be regulated by ATF7. Previously we found that once ATF7 is released from chromatin by immunological stress, this release and epigenetic change are maintained for several weeks in macrophage (Yoshida et al., 2015). If ATF7 has similar characteristics in spermatogonia, epigenetic change can be maintained for several cycles of spermatogenesis. If not, TGCs in which epigenetic status is altered by spaceflight can be deposited by newly produced germ cells from spermatogonia.
Limitations of the study
MG/AG group of father males (5 weeks age) were purchased from a vendor different from the one for GC mice. However, rearing conditions (e.g., food composition, humidity, airflow, and noise limit) are similar between the two vendors. Each group of mice were reared with the same condition except for gravity to 12 weeks age, and so it is unlikely that the change of transcriptome in F1 mice is due to a difference of rearing condition during the early 5 weeks age in father mice. At this time, paternal effect of spaceflight was not investigated using Atf7 mutant mice, and so we cannot completely exclude the possibility that another pathway unrelated to ATF7 also alters expressions of some portion of genes in F1 offspring.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-H3K4me3 | MBL | Cat#MABI-0304 (lot:14004); RRID: AB_11123891 |
| Anti-H3K9me2 | MBL | Cat#MABI-0317 (lot:16001) |
| Anti-H3K9me3 | MBL | Cat#MABI-0318 (lot:16001) |
| Anti-H3K27me3 | MBL | Cat#MABI-0323 (lot:15007); RRID: AB_11123929 |
| Anti-ATF7 | B.C. supplied | 2F10 |
| Chemicals, peptides, and recombinant proteins | ||
| Trizol | Thermo Fisher | Cat#15596018 |
| Collagenase | Sigma | Cat#C6138 |
| Trypsin | Wako | Cat#208-17251 |
| DNase I | Takara | Cat#2270A |
| BSA | Sigma | Cat#A7906 |
| Percoll | GE Healthcare | Cat#17-0891-02 |
| protein A-sepharose beads | GE Healthcare | Cat#17-0780-01 |
| anti-mouse IgG-conjugated magnetic beads | Thermo Fisher | Cat#11201D |
| RNase A | Thermo Fisher | Cat#EN0531 |
| proteinase K | Millipore | Cat#1.24568 |
| phenol:chloroform:isoamyl alcohol | Wako | Cat#311-90151 |
| AMPure XP beads | Beckman Coulter | Cat#A63881 |
| Critical commercial assays | ||
| QuantiFast SYBR Green PCR Kit | Qiagen | Cat# 204056 |
| NEBNext Ultra II DNA Library Prep Kit | NEB | Cat#E7645 |
| Illumina NextSeq500 with High Output Kit v2 | Illumina | Cat#FC-404-2005 |
| NEBNext Small RNA Library Prep Kit | NEB | Cat#E7300 |
| Deposited data | ||
| ChIP-seq data | This paper | DDBJ: DRA010655 (https://ddbj.nig.ac.jp/DRASearch/submission?acc=DRA010655) |
| Small RNA-seq data | This paper | DDBJ: DRA010655 (https://ddbj.nig.ac.jp/DRASearch/submission?acc=DRA010655) |
| refGene | UCSC Table Browser | https://genome.ucsc.edu/cgi-bin/hgTables |
| Mouse tRNA sequence | GtRNAdb | http://gtrnadb.ucsc.edu/genomes/eukaryota/Mmusc10/mm10-tRNAs.fa |
| Mouse miRNA sequence | miRBbase | http://www.mirbase.org/ (Release 22) |
| Experimental models: organisms/strains | ||
| Mouse: C57BL6/wild-type | Jackson Laboratory | N/A |
| Oligonucleotides | ||
| Primers for qChIP assay | This paper | Table S6 |
| Software and algorithms | ||
| CLC Genomics Workbench | Qiagen | 10.1.1 |
| BED tool | Quinlan and Hall (2010) | 2.27.0 |
| MACS2 | Feng et al. (2012) | 2.1.1 |
| deepTools2 | Ramírez et al. (2016) | 3.1.2.0.1 |
| PANTHER classification system | Mi et al. (2013) | 14.0 |
| SPOTS1.0 | Shi et al. (2018) | 1.0.5 |
| GSEA | Subramanian et al.(2005) | 4.0.3 |
| mirPath v.3 | Vlachos et al. (2015) | http://snf-515788.vm.okeanos.grnet.gr/ |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to the Lead Contact, Keisuke Yoshida (Keisuke.Yoshida@riken.jp).
Materials availability
This study did not generate new unique reagents.
Data and code availability
All sequencing data from Next Generation Sequencer are deposited in DDBJ (DRA010655).
This study did not generate new computer code.
There is no additional information available for this section.
Experimental model and subject details
Mice
Five-week-age C57BL/6J male mice were purchased from Jackson Laboratory (USA) for experiments in space (MG and AG mice) and from Charles River Laboratories (Japan) for experiments on Earth (GC mice). We note that these groups of mice were reared by the same condition of cage and food after purchase. Mouse experiments were performed as described previously (Shiba et al., 2017). Eight-week-old C57BL/6J female mice were purchased from SLC (Japan) for in vitro fertilization. ICR pseudo-pregnant female mice were purchased from SLC for embryo transfer. The IVF experiment was performed according to standard protocols, and 2-3 male offspring were produced by sperm collected from GC or MG group of individual mice. Experiments were conducted in accordance with the guidelines of the Institutional Animal Care and Use Committee of RIKEN Tsukuba Branch.
Method details
Preparation of fixed TGCs
Testes were collected from each group of mice at 10 weeks age after 35 days of spaceflight. Seminiferous tubules were incubated in RPMI medium containing collagenase (1 mg/ml; Sigma C6138) for 15 minutes at 37°C. Tubules were washed three times with ice-cold RPMI and incubated in RPMI containing trypsin (0.025%; Wako) and DNase I (0.25 U/ml; Takara) for 10 minutes at 37°C. After addition of fetal bovine serum to inactivate trypsin, single cells were extracted from tubules by repeated pipetting on ice. Cells were washed twice with PBS+BSA (PBS containing 5 mg/ml BSA and 2 mM EDTA) and fixed by incubating in 1% formaldehyde in PBS+BSA for 10 minutes at room temperature. The crosslinking reaction was stopped by 0.25 M glycine. After washing cells with PBS+BSA, cell pellets were stored at -80°C. For expression levels of genes of spermatogenic markers (Green et al., 2018), previous RNA-Seq data (Matsumura et al., 2019) in testis collected from GC, MG, or AG mice were re-analyzed. Difference of the expressions were evaluated by Kruskal-Wallis one-way analysis of variance.
X-ChIP in TGCs
X-linked TGCs were washed twice with lysis buffer 1 (50 mM HEPES pH 7.5, 140 mM NaCl, 1 mM EDTA, 10% glycerol, 0.5% NP-40, 0.25% Triton X-100, and complete protease inhibitor cocktail), and then their chromatin was solubilized in elution buffer (50 mM Tris-HCl, pH 8.0, 10 mM EDTA, 1% SDS and protease inhibitors) by sonication. DNA-protein complexes were collected using anti-ATF7 antibody-bound Protein A Sepharose beads, or anti-H3K4me3 (MBL, MABI0304) or anti-H3K27me3 (MBL, MABI0323) antibody-bound anti-mouse IgG-conjugated magnetic beads. Dr. Bruno Chatton in Université de Strasbourg kindly gave us ATF7 monoclonal antibody (2F10). After washing four times with ChIP wash buffer (50 mM HEPES pH 7.0, 0.5 M LiCl, 1 mM EDTA, 0.7% sodium deoxycholate, 1% NP-40) and twice with TE buffer (10 mM Tris-HCl pH 8.0, 1 mM EDTA), the immune complexes were eluted with elution buffer.
Library preparation and sequencing for ChIP-seq analysis
Samples of input and ChIP'd DNA were converted into a sequencing library using NEBNext Ultra II DNA Library Prep Kit (New England Biolabs). Paired-end sequencing (2 × 36 cycles) was performed with an Illumina NextSeq500 system with High Output Kit v2 (Illumina) at Tsukuba i-Laboratory LLP.
Processing read data and peak calling
Paired-end reads were mapped against mouse genome assembly mm9 using the aln function of Burrows-Wheeler Aligner (v 0.7.5) with default settings. Mapped reads were converted to BED files using Samtools (v 0.1.19) and bedtools (v 2.17.0) for downstream analysis. For peak calling, uniquely mapped reads were analyzed by MACS2 at FDR <0.001.
Genome coordinates
Genomic compartments were defined with refGene data (mm9) obtained by UCSC table browser. The features were defined as follows: promoters (1 kb around the annotated TSS), exons, introns, upstream regions (from 5 kb upstream of the TSS), and downstream regions (from 5 kb downstream of the TES).
Preparation of profile plot and heatmap data
The number of raw reads mapped on the 3 kb around the TSS was counted, and read coverage was calculated after normalization to the mapped read number. Matrix data of read coverage against each target promoter region in 50 bp bin size were prepared using deepTools2 (Ramírez et al., 2016). Using this matrix, profile plots were prepared using average values for target regions. The degree of read coverage in each target promoter region is indicated by the heatmap.
Gene functional analysis
Functional analysis of ATF7 target genes or DEGs was performed using PANTHER classification system (Mi et al., 2013). Enrichment terms for Reactome pathway analysis were identified using DEGs from ATF7 target genes from ATF7 ChIP-seq data or expression array data. GSEA was performed using GSEA software (Subramanian et al., 2005; Mootha et al., 2003).
Preparation of HRCS
After thawing frozen sperm, sperm cells were purified by 50% Percoll, followed by treatment with somatic cell lysis buffer (PBS containing 0.1% SDS and 0.5% Triton X-100). HRCS fractions were purified from the total sperm fraction by 82% Percoll after mild sonication (Yoshida et al., 2018).
X-ChIP in HRCS
HRCS cells were fixed with 1% formaldehyde for 10 minutes at room temperature and treated with sperm decondensation buffer (5 mM HEPES pH 8.0, 0.2% NP-40, 10 mM EDTA, 5 mM NaCl, 1.2 M urea, 10 mM DTT, 2× complete protease inhibitor cocktail, 1 mM PMSF) for 5 hours at 42°C. ChIP experiments using decondensed HRCS were performed using the same procedure as for TGCs. Quantitative PCR was performed with QuantiFast SYBR Green PCR Kit (QIAGEN) on a Quant Studio 3 System (Applied Biosystems) using primers listed in Table S6. Technical triplicates were included for all reactions. ChIP efficiency is presented as the percentage of the input sample used for the ChIP lysate.
Small RNA-Seq analysis for mature sperm
HRCS cells (5.0 × 106 cells) were suspended in 50 μl of sperm lysis buffer (PBS containing 0.5% SDS, 10 mM DTT, and 0.2 mg/ml Proteinase K) and incubated for 30 minutes at 37°C. Sperm lysate was thoroughly mixed with 1 ml of Trizol (Invitrogen) by pipetting, and total RNA was purified according to the manufacturer’s protocol. Small RNA library was prepared using the NEBNext Small RNA Library Prep Kit (NEB). Libraries were size-selected with AMPure XP beads (Beckman Coulter) following the library kit instructions. After library quantification and size analysis using the Bioanalyzer DNA High-sensitivity kit (Agilent), 2 × 36 base paired-end sequencing was performed using an Illumina NextSeq 500 system (Illumina) at Tsukuba i-Laboratory.
Small RNA-seq data processing
Adapter trimming was performed to remove the 3′ adapter ‘AGATCGGAAGAGCACACGTCT’ and 5′ adapter ‘GTTCAGAGTTCTACAGTCCGACGATC’ with the default setting of CLC Genomics Workbench (CLC-GW, version 12.0; QIAGEN). We used R2 FASTQ data as the reverse complement using SeqKit tools (https://bioinf.shenwei.me/seqkit/) for the small RNA-seq process. Trimmed reads were aligned and annotated to the genome (UCSC mm10), miRbase_v21, rRNA, tRNA, piRNA, ensembl ncRNA, and rfam 12.3 using the SPORTS1.0 pipeline (https://github.com/junchaoshi/sports1.0) with the default parameters (Shi et al., 2018). We created the raw count tables from the summary count of each small RNA after confirming that the sequence strands were annotated in the correct direction for small RNAs using SPORTS1.0. The raw count tables annotated to each group of tRNA or miRNA were imported into CLC-GW and analyzed by proportion-based statistical analysis (Baggerly’s test) (Baggerly et al., 2003). Genes with |weighted proportions fold-change| > 1.3 and FDR < 0.05 were defined as DEGs. Enrichment of target genes by 5’ or 3’ miRNA portions in each KEGG pathway was analyzed by mirPath v.3 (http://snf-515788.vm.okeanos.grnet.gr/) (Vlachos et al., 2015) with the default setting. To generate scatterplots and heatmaps, read counts of tRNA or miRNA groups with a perfect match to annotation (M.M. = 0) per total reads of tRNA or miRNA, respectively, were calculated. To investigate expression of miRNA-target genes during early embryogenesis, deposited data set (Wu et al., 2016) (GSE66582) was used.
RNA-Seq analysis in liver
RNA was isolated from the livers (median lobe) of individual F1 male mice at 3 weeks age by using Trizol according to the manufacturer’s protocol. Total RNA (500 ng) was converted into RNA-Seq libraries using rRNA depletion (New England Biolabs, E6310) and Next Ultra Directional RNA Library Prep Kit (E7420). Sequencing was performed using a NextSeq500 system (Illumina) to obtain paired-end 2 × 36 base reads. FASTQ files were imported to CLC-GW (v10.1.1), mapped to the mouse reference genome (mm10), and quantified for 49,585 genes. After filtering out annotated genes with low reads (sum of reads in 11 samples < 12), DEGs were distinguished by FDR value < 0.05 computed with the R package “DESeq2”.
Quantification and statistical analysis
In box plots, the center line indicates the median, box limits define the upper and lower quartiles, and whiskers show the 1.5× interquartile range. Outliers are shown as points. For Chi-squared test, expected value was calculated using the total gene number (24,367) obtained from refGene (mm9). For qChIP assay, SD means standard error of qPCR measurement, and n does technical replicates. For RNA-Seq analysis, n means number of individual mice for each group. Investigators were not blinded to experimental conditions, and no randomization or exclusion of data points was used.
Acknowledgments
This work was supported by the Japan Agency for Medical Research and Development (AMED; 18gm0510015h0006; S.I.), the Grant-in-Aid for the Japan Aerospace Exploration Agency (14YPTK-005512; S.T.) and the Grant-in-Aid for Scientific Research (C) from JSPS (18K06189; K.Y.).
Author contributions
K.Y. performed ChIP-seq and qChIP experiments and analyzed the ChIP-seq data. S.-i.F. performed small RNA-seq analysis, and M.M. performed NGS analysis. A.I., M.I., D.S., and M.S. prepared mouse tissue samples. T.K. and S.T. performed the IVF experiment. S.I. conceived and supervised the whole study, and K.Y. and S.I. wrote the manuscript.
Declaration of interests
M.M. declares an association with Tsukuba i-Laboratory as technical consultant.
Published: July 23, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2021.102773.
Supplemental information
References
- Aimar C., Bautz A., Durand D., Membre H., Chardard D., Gualandris-Parisot L., Husson D., Dournon C. Microgravity and hypergravity effects on fertilization of the salamander Pleurodeles waltl (urodele amphibian) Biol. Reprod. 2000;63:551–558. doi: 10.1095/biolreprod63.2.551. [DOI] [PubMed] [Google Scholar]
- Andreev-Andrievskiy A., Popova A., Boyle R., Alberts J., Shenkman B., Vinogradova O., Dolgov O., Anokhin K., Tsvirkun D., Soldatov P. Mice in Bion-M 1 space mission: training and selection. PLoS One. 2014;9:e104830. doi: 10.1371/journal.pone.0104830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baggerly K.A., Deng L., Morris J.S., Aldaz C.M. Differential expression in SAGE: accounting for normal between-library variation. Bioinformatics. 2003;19:1477–1483. doi: 10.1093/bioinformatics/btg173. [DOI] [PubMed] [Google Scholar]
- Bracken C.P., Scott H.S., Goodall G.J. A network-biology perspective of microRNA function and dysfunction in cancer. Nat. Rev. Genet. 2016;17:719–732. doi: 10.1038/nrg.2016.134. [DOI] [PubMed] [Google Scholar]
- Bueno M.J., Malumbres M. MicroRNAs and the cell cycle. Biochim. Biophys. Acta. 2011;1812:592–601. doi: 10.1016/j.bbadis.2011.02.002. [DOI] [PubMed] [Google Scholar]
- Cancedda R., Liu Y., Ruggiu A., Tavella S., Biticchi R., Santucci D., Schwartz S., Ciparelli P., Falcetti G., Tenconi C. The Mice Drawer System (MDS) experiment and the space endurance record-breaking mice. PLoS One. 2012;7:e32243. doi: 10.1371/journal.pone.0032243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carone B.R., Fauquier L., Habib N., Shea J.M., Hart C.E., Li R., Bock C., Li C., Gu H., Zamore P.D. Paternally induced transgenerational environmental reprogramming of metabolic gene expression in mammals. Cell. 2010;143:1084–1096. doi: 10.1016/j.cell.2010.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demontis G.C., Germani M.M., Caiani E.G., Barravecchia I., Passino C., Angeloni D. Human pathophysiological adaptations to the space environment. Front. Physiol. 2017;8:547. doi: 10.3389/fphys.2017.00547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J., Liu T., Qin B., Zhang Y., Liu X.S. Identifying ChIP-seq enrichment using MACS. Nat. Protoc. 2012;7:1728–1740. doi: 10.1038/nprot.2012.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green C.D., Ma Q., Manske G.L., Shami A.N., Zheng X., Marini S., Moritz L., Sultan C., Gurczynski S.J., Moore B.B. A comprehensive roadmap of murine spermatogenesis defined by single-cell RNA-seq. Dev. Cell. 2018;46:651–667.e10. doi: 10.1016/j.devcel.2018.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heard E., Martienssen R.A. Transgenerational epigenetic inheritance: myths and mechanisms. Cell. 2014;157:95–109. doi: 10.1016/j.cell.2014.02.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horie K., Sasanuma H., Kudo T., Fujita S.I., Miyauchi M., Miyao T., Seki T., Akiyama N., Takakura Y., Shimbo M. Down-regulation of GATA1-dependent erythrocyte-related genes in the spleens of mice exposed to a space travel. Sci. Rep. 2019;9:7654. doi: 10.1038/s41598-019-44067-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter C.A., Jones S.A. IL-6 as a keystone cytokine in health and disease. Nat. Immunol. 2015;16:448–457. doi: 10.1038/ni.3153. [DOI] [PubMed] [Google Scholar]
- Ibarra A., Schwob E., Méndez J. Excess MCM proteins protect human cells from replicative stress by licensing backup origins of replication. Proc. Natl. Acad. Sci. U S A. 2008;105:8956–8961. doi: 10.1073/pnas.0803978105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaati G., Bygren L.O., Edvinsson S. Cardiovascular and diabetes mortality determined by nutrition during parents' and grandparents' slow growth period. Eur. J. Hum. Genet. 2002;10:682–688. doi: 10.1038/sj.ejhg.5200859. [DOI] [PubMed] [Google Scholar]
- Maekawa T., Kim S., Nakai D., Makino C., Takagi T., Ogura H., Yamada K., Chatton B., Ishii S. Social isolation stress induces ATF-7 phosphorylation and impairs silencing of the 5-HT 5B receptor gene. EMBO J. 2010;29:196–208. doi: 10.1038/emboj.2009.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maekawa T., Liu B., Nakai D., Yoshida K., Nakamura K.I., Yasukawa M., Koike M., Takubo K., Chatton B., Ishikawa F. ATF7 mediates TNF-α-induced telomere shortening. Nucleic Acids Res. 2018;46:4487–4504. doi: 10.1093/nar/gky155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao X.W., Byrum S., Nishiyama N.C., Pecaut M.J., Sridharan V., Boerma M., Tackett A.J., Shiba D., Shirakawa M., Takahashi S., Delp M.D. Impact of spaceflight and artificial gravity on the mouse retina. Int. J. Mol. Sci. 2018;19:2546. doi: 10.3390/ijms19092546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumura T., Noda T., Muratani M., Okada R., Yamane M., Isotani A., Kudo T., Takahashi S., Ikawa M. Male mice, caged in the International Space Station for 35 days, sire healthy offspring. Sci. Rep. 2019;9:13733. doi: 10.1038/s41598-019-50128-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H., Muruganujan A., Casagrande J.T., Thomas P.D. Large-scale gene function analysis with the PANTHER classification system. Nat. Protoc. 2013;8:1551–1566. doi: 10.1038/nprot.2013.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mootha V.K., Lindgren C.M., Eriksson K.F., Subramanian A., Sihag S., Lehar J., Puigserver P., Carlsson E., Ridderstråle M., Laurila E. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- Murata Y., Yasuda T., Watanabe-Asaka T., Oda S., Mantoku A., Takeyama K., Chatani M., Kudo A., Uchida S., Suzuki H. Histological and transcriptomic analysis of adult Japanese medaka sampled onboard the international space station. PLoS One. 2015;10:e0138799. doi: 10.1371/journal.pone.0138799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng S.F., Lin R.C., Laybutt D.R., Barres R., Owens J.A., Morris M.J. Chronic high-fat diet in fathers programs β-cell dysfunction in female rat offspring. Nature. 2010;467:963–966. doi: 10.1038/nature09491. [DOI] [PubMed] [Google Scholar]
- Oakberg E.F. Duration of spermatogenesis in the mouse and timing of stages of the cycle of the seminiferous epithelium. Am. J. Anat. 1956;99:507–516. doi: 10.1002/aja.1000990307. [DOI] [PubMed] [Google Scholar]
- Pembrey M.E., Bygren L.O., Kaati G., Edvinsson S., Northstone K., Sjöström M., Golding J., ALSPAC Study Team Sex-specific, male-line transgenerational responses in humans. Eur. J. Hum. Genet. 2006;14:159–166. doi: 10.1038/sj.ejhg.5201538. [DOI] [PubMed] [Google Scholar]
- Perez M.F., Lehner B. Intergenerational and transgenerational epigenetic inheritance in animals. Nat. Cell Biol. 2019;21:143–151. doi: 10.1038/s41556-018-0242-9. [DOI] [PubMed] [Google Scholar]
- Quinlan A.R., Hall I.M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez F., Ryan D.P., Grüning B., Bhardwaj V., Kilpert F., Richter A.S., Heyne S., Dündar F., Manke T. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 2016;44:W160–W165. doi: 10.1093/nar/gkw257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seong K.H., Ly N.H., Katou Y., Yokota N., Nakato R., Murakami S., Hirayama A., Fukuda S., Kang S., Soga T. Paternal restraint stress affects offspring metabolism via ATF-2 dependent mechanisms in Drosophila melanogaster germ cells. Commun. Biol. 2020;3:208. doi: 10.1038/s42003-020-0935-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J., Ko E.A., Sanders K.M., Chen Q., Zhou T. SPORTS1.0: a tool for annotating and profiling non-coding RNAs optimized for rRNA- and tRNA-derived small RNAs. Genomics Proteomics Bioinformatics. 2018;16:144–151. doi: 10.1016/j.gpb.2018.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiba D., Mizuno H., Yumoto A., Shimomura M., Kobayashi H., Morita H., Shimbo M., Hamada M., Kudo T., Shinohara M. Development of new experimental platform 'MARS'-Multiple Artificial-gravity Research System-to elucidate the impacts of micro/partial gravity on mice. Sci. Rep. 2017;7:10837. doi: 10.1038/s41598-017-10998-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimbo M., Kudo T., Hamada M., Jeon H., Imamura Y., Asano K., Okada R., Tsunakawa Y., Mizuno S., Yagami K. Ground-based assessment of JAXA mouse habitat cage unit by mouse phenotypic studies. Exp. Anim. 2016;65:175–187. doi: 10.1538/expanim.15-0077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavella S., Ruggiu A., Giuliani A., Brun F., Canciani B., Manescu A., Marozzi K., Cilli M., Costa D., Liu Y. Bone turnover in wild type and pleiotrophin-transgenic mice housed for three months in the International Space Station (ISS) PLoS One. 2012;7:e33179. doi: 10.1371/journal.pone.0033179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlachos I.S., Zagganas K., Paraskevopoulou M.D., Georgakilas G., Karagkouni D., Vergoulis T., Dalamagas T., Hatzigeorgiou A.G. DIANA-miRPath v3.0: deciphering microRNA function with experimental support. Nucleic Acids Res. 2015;43:W460–W466. doi: 10.1093/nar/gkv403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voorhees J.L., Tarr A.J., Wohleb E.S., Godbout J.P., Mo X., Sheridan J.F., Eubank T.D., Marsh C.B. Prolonged restraint stress increases IL-6, reduces IL-10, and causes persistent depressive-like behavior that is reversed by recombinant IL-10. PLoS One. 2013;8:e58488. doi: 10.1371/journal.pone.0058488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakayama S., Kamada Y., Yamanaka K., Kohda T., Suzuki H., Shimazu T., Tada M.N., Osada I., Nagamatsu A., Kamimura S. Healthy offspring from freeze-dried mouse spermatozoa held on the International Space Station for 9 months. Proc. Natl. Acad. Sci. U S A. 2017;114:5988–5993. doi: 10.1073/pnas.1701425114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wentworth B.C., Wentworth A.L. NASA Technical Reports Server; 1996. Fecundity of Quail in Spacelab Microgravity. [Google Scholar]
- Williams D., Kuipers A., Mukai C., Thirsk R. Acclimation during space flight: effects on human physiology. CMAJ. 2009;180:1317–1323. doi: 10.1503/cmaj.090628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Huang B., Chen H., Yin Q., Liu Y., Xiang Y., Zhang B., Liu B., Wang Q., Xia W. The landscape of accessible chromatin in mammalian preimplantation embryos. Nature. 2016;534:652–657. doi: 10.1038/nature18606. [DOI] [PubMed] [Google Scholar]
- Yoshida K., Maekawa T., Ly N.H., Fujita S.I., Muratani M., Ando M., Katou Y., Araki H., Miura F., Shirahige K. ATF7-Dependent epigenetic changes are required for the intergenerational effect of a paternal low-protein diet. Mol. Cell. 2020;78:445–458.e6. doi: 10.1016/j.molcel.2020.02.028. [DOI] [PubMed] [Google Scholar]
- Yoshida K., Maekawa T., Zhu Y., Renard-Guillet C., Chatton B., Inoue K., Uchiyama T., Ishibashi K., Yamada T., Ohno N. The transcription factor ATF7 mediates lipopolysaccharide-induced epigenetic changes in macrophages involved in innate immunological memory. Nat. Immunol. 2015;16:1034–1043. doi: 10.1038/ni.3257. [DOI] [PubMed] [Google Scholar]
- Yoshida K., Muratani M., Araki H., Miura F., Suzuki T., Dohmae N., Katou Y., Shirahige K., Ito T., Ishii S. Mapping of histone-binding sites in histone replacement-completed spermatozoa. Nat. Commun. 2018;9:3885. doi: 10.1038/s41467-018-06243-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youngson N.A., Whitelaw E. Transgenerational epigenetic effects. Annu. Rev. Genomics Hum. Genet. 2008;9:233–257. doi: 10.1146/annurev.genom.9.081307.164445. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequencing data from Next Generation Sequencer are deposited in DDBJ (DRA010655).
This study did not generate new computer code.
There is no additional information available for this section.