Abstract
Drought stress impacts seedling establishment, survival and whole-plant productivity. Molecular responses to drought stress have been most extensively studied in herbaceous species, mostly considering only aboveground tissues. Coniferous tree species dominate boreal forests, which are predicted to be exposed to more frequent and acute drought as a result of ongoing climate change. The associated impact at all stages of the forest tree life cycle is expected to have large-scale ecological and economic impacts. However, the molecular response to drought has not been comprehensively profiled for coniferous species. We assayed the physiological and transcriptional response of Picea abies (L.) H. Karst seedling needles and roots after exposure to mild and severe drought. Shoots and needles showed an extensive reversible plasticity for physiological measures indicative of drought-response mechanisms, including changes in stomatal conductance (gs), shoot water potential and abscisic acid (ABA). In both tissues, the most commonly observed expression profiles in response to drought were highly correlated with the ABA levels. Still, root and needle transcriptional responses contrasted, with extensive root-specific down-regulation of growth. Comparison between previously characterized Arabidopsis thaliana L. drought-response genes and P. abies revealed both conservation and divergence of transcriptional response to drought. In P. abies, transcription factors belonging to the ABA responsive element(ABRE) binding/ABRE binding factors ABA-dependent pathway had a more limited role. These results highlight the importance of profiling both above- and belowground tissues, and provide a comprehensive framework to advance the understanding of the drought response of P. abies. The results demonstrate that a short-term, severe drought induces severe physiological responses coupled to extensive transcriptome modulation and highlight the susceptibility of Norway spruce seedlings to such drought events.
Keywords: ABA, drought stress, gene expression, Norway spruce, Picea abies, RNA-Seq, transcriptome
Introduction
Boreal forests are dominated by coniferous trees (Shorohova et al. 2011) and represent approximately one-third of terrestrial forests (FAO 2010). They are sinks of atmospheric CO2 and carbon emissions from anthropogenic sources, and are important in balancing the global carbon cycle (Kirschbaum and Fischlin 1996, Pan et al. 2011). Growth conditions in the boreal zone are limiting, with a cold climate and long periods of sub-zero temperatures leading to short growing seasons (Troeng and Linder 1982, Burton et al. 2010, Kneeshaw et al. 2011). Recently, climate change has caused a mean annual temperature increase of 1.5 °C in high-latitude boreal forests, with temperatures predicted to rise by an additional 5 °C by 2100 (IPCC 2013). Observations made in the boreal forest biomes over Fennoscandia, North America and Russia identified accelerated growth in response to higher temperatures and longer growing seasons (Lapenis et al. 2005, Zhang et al. 2008, Kauppi et al. 2014). However, such positive growth responses are dependent on adequate water availability, and the same studies also reported severe increases in summer drought and evapotranspiration in boreal regions (Zhang et al. 2008, Allen et al. 2010, Beck et al. 2011, Peng et al. 2011). Drought stress is a major cause of tree mortality and a threat to boreal forest trees, including Norway spruce (Picea abies (L.) H. Karst) (Kellomäki et al. 2008, Ge et al. 2011).
Major insights to biochemical and molecular mechanisms important for plant drought response have been gained from studies of herbaceous plants such as Arabidopsis (Arabidopsis thaliana L.), rice (Oryza sativa L.) and maize (Zea maize L.). To avoid or tolerate water deficit, maintenance of cellular turgor is of high priority and plants therefore accumulate metabolites such as sugars, amino acids and amines (Tabaeizadeh 1998). In addition, cellular components such as proteins and membranes require stabilization, which involves late embryogenesis-abundant (LEA) proteins, heat shock proteins (HSPs) and dehydrins, together with the above metabolites, which also function as antioxidants (Harfouche et al. 2014). The phytohormone abscisic acid (ABA) is an important signal leading to the expression of various stress-responsive genes and transcriptional networks (Yamaguchi-Shinozaki and Shinozaki 2006). Transcription factors (TFs) from the ABA responsive element(ABRE)-binding proteins (AREB)/ABRE binding factor (ABF), MYB/MYC, NAC, WRKY and NF-Y families have been shown to regulate ABA-responsive gene expression under drought stress (Singh and Laxmi 2015). In addition, an ABA-independent pathway for signal transduction under drought stress exists, involving the dehydration-responsive element-binding protein (DREB)/C-repeat binding factor (CBF) TFs, a subfamily of APETALA 2 (AP2)/ethylene-responsive element-binding factor (ERF) transcriptional activators (Nakashima et al. 2009).
Physiological changes, such as stomatal closure, are the primary cause of drought-induced decreases in productivity. Stomatal closure is stimulated by increasing levels of endogenous ABA during drought stress (Iuchi et al. 2001), which consequently results in reduced availability of CO2 for photosynthesis. Plants differ in their sensitivity to dehydration, with substantial intra- and inter-specific variation in response to drought. Herbaceous and woody ang iosperm species are thought to be mainly anisohydric in that they display a risk-taking behavior, maintaining stomatal conductance (gs) under drought stress in order to maintain productivity (Sade et al. 2012). In contrast, isohydric species, including most gymnosperms of the pinophyte, such as Norway spruce, initiate stomatal closure early during drought and maintain high levels of ABA in the foliage, restricting fast recovery (Brodribb and McAdam 2013, Brodribb et al. 2014). Maintenance of an efficient hydraulic system to ensure transport of water from the soil and to reduce water loss from leaves is especially important in trees, which are long-lived, for optimal plant hydration under seasonally and annually fluctuating environmental conditions (Raven 1977, Edwards et al. 1998). Differences in the regulation of water status can determine survival of trees in response to drought, with improper response mechanisms resulting in mortality either from carbon starvation or hydraulic failure in isohydric and anisohydric species, respectively. It is important to understand how species will respond within this continuum of response mechanisms in light of predicted patterns of intensity, duration and frequency of future drought events (McDowell et al. 2008), which will particularly reduce productivity in ecosystems where the isohydric Pinophyta dominate (Roman et al. 2015).
Roots have an essential role in determining water uptake from the environment and are the first to sense soil water deficit. Norway spruce is often found to have a shallow root system (Kalliokoski 2011), with the majority of fine roots located in the upper soil layers (Børja et al. 2008). As such, Norway spruce, and especially seedlings, are highly sensitive to summers with lower precipitation and high evapotranspiration, which will become more prevalent as a result of climate change. The role of ABA production in roots and transport to the foliage to actively reduce guard cell turgor is generally accepted as the primary mechanism mediating stomatal closure during drought stress in herbaceous plants (Schroeder et al. 2001, Roelfsema and Hedrich 2005). However, in trees, the time required for transport of ABA to the leaves via the vascular tissue exceeds the observed time needed to close stomata (Zimmermann and Brown 1971, Schulze and Hall 1982), and evidence suggests that foliar-derived ABA is sufficient to explain stomatal responses in conifers (Mitchell et al. 2017). The importance of root-derived ABA is therefore questionable (Wilkinson and Davies 2002), and the relative importance and role of the ABA pools in different plant tissues remains unresolved. The molecular basis of isohydrism has been less studied, but in gymnosperm species, it includes a high sensitivity to ABA (Brodribb and McAdam 2013, Brodribb et al. 2014).
A number of studies have examined the physiological and transcriptional drought responses of various coniferous species (Lorenz et al. 2011, Moran et al. 2017, Du et al. 2018, Fox et al. 2018, de María et al. 2020, Klápště et al. 2020). These have established that ABA has a conserved role in controlling drought-response mechanisms (Brodribb and McAdam 2013, Brodribb et al. 2014, Pashkovskiy et al. 2019) and that there is genetically controlled variation in drought response within and between species (Moran et al. 2017, Trujillo-Moya et al. 2018, de María et al. 2020, Klápště et al. 2020). A common feature of these previous transcriptomics studies was the use of sequence similarity to assign functional descriptions on the basis of the highest sequence similarity homolog. These functional descriptions were nearly exclusively assigned on the basis of work performed using angiosperm species, in particular Arabidopsis. Contemporary studies of drought response in extant species must consider the extensive divergence time of the gymnosperm and angiosperm lineages. Subsequent evolution in both lineages after the split may have resulted in the divergence of both gene regulation and function. As extremely few genes have been functionally characterized in gymnosperm species, caution is needed when using such a sequence similarity-based transfer of functional information.
This study used RNA-sequencing (RNA-Seq) to assay the transcriptional response of Norway spruce seedlings experiencing a mild and severe water stress and samples after re-irrigation to determine the extent of expression conservation to typified angiosperm drought-response mechanisms. There was extensive reversible plasticity for physiological responses, including gs, shoot water potential and ABA levels. Needles and roots showed markedly contrasting transcriptome remodulation in response to drought, including differences in plasticity upon re-watering. Additionally, comparison of the transcriptional response with a set of well-characterized Arabidopsis drought-response genes identified regulators with both divergence and conservation at the expression level.
Materials and methods
Data and materials availability
Sequencing data have been deposited at the European Nucleotide Archive (ENA, https://www.ebi.ac.uk/ena) under the accession ID PRJEB26933. Associated scripts are available from the git repository https://github.com/loalon/2020-spruce-drought-stress and the data can be interactively explored at http://terra.upsc.se/2020-spruce-drought-stress/.
Experimental design
Three-year-old Norway spruce seedlings of the seed provenance Lilla Istad (56° 30′ N) were grown in a growth room at 18 h light (120 μmol m−2 s−1 at plant height) and 20 °C in 3-l pots filled with peat. For a controlled drought treatment, the field capacity (FC) of the soil was determined as the difference in the weight of wet soil and dry weight after oven-drying at 70 °C. Seedlings of the control treatment were kept well watered and monitored gravimetrically to maintain the soil moisture at 80% FC. For a mild drought treatment, water was withheld from the seedlings until soil moisture reduced to 30% FC, 5 days after the initiation of the experiment. Seedlings were kept in this stage of mild drought stress for 7 days by adding the daily evapotranspirational water loss. This was a non-lethal drought stress and above a point where leaf death occurred. A severe drought stress was then imposed by completely withholding water until symptoms of severe dysfunction, as measured by photosynthetic assimilation rates, were observed. At this moment, the severe drought stress was extended for 3 days, bringing plants closer to catastrophic hydraulic failure, before re-irrigation was started. After 4 days of re-watering, soil moisture had returned to the initial well-watered conditions at 80% FC. Needles and fine and small lateral roots (<4 mm) of three to four seedlings were sampled at control (Day 0), mild (2, 4, 5 and 13 days) and severe (18 and 21 days) drought and after re-irrigation (25 days) for RNA-Seq analysis. A set of plants that were not re-irrigated exhibited needle death after one additional Day of the severe drought stress (indicated by an X in Figure 1a), highlighting the severity of the stress and that the seedlings were close to catastrophic hydraulic failure when sampled.
Figure 1.
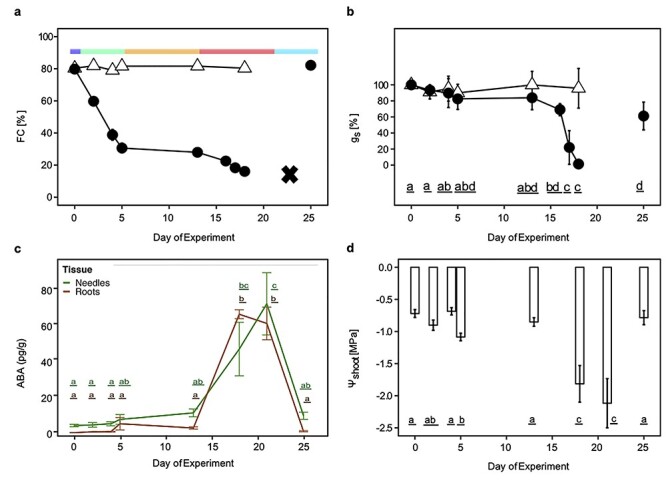
Physiological response of Norway spruce seedlings to drought. Norway spruce seedlings were subjected to increasing levels of water deficit (filled circles) or kept in well-watered control conditions (clear triangles). (a) Colored bars above the plot represent the following treatment conditions: soil moisture dropped from 80% (dark blue portion of the colored bar) to 30% FC after withholding water for 5 days and was regarded as mild water deficit (green portion of the colored bar). After maintaining this condition for 7 days (orange portion of the colored bar), a more severe drought stress was induced (red portion of the colored bar), again by withholding water. After 21 days, water-stressed plants were re-irrigated (cyan portion of the colored bar) and the soil moisture levels recovered to the level of well-watered plants. Colors indicating these experimental conditions are used in subsequent figures. All plants were visually identical, with no visible effects of drought for the sampled time points. An additional set of plants (n = 5) were not re-irrigated after the severe drought stress. All needles on these plants died after 23 days, indicated by a black cross. (b) Needle stomatal conductance (gs) in control (clear triangles) and drought-stressed (filled circles) seedlings. (c) Abscisic acid levels of needles (green) and roots (brown) of drought-stressed seedlings. (d) Midday water potential (ψshoot) measured on shoots of three to five independent plants per time point in drought stress. The result of a one-way ANOVA performed on the average values of three measurements per plant is indicated. For all panels, data are means and error bars represent the ±95% CIs. Letter represent statistically significant differences (P < 0.05; Tukey’s HSD) between time points of the drought stress.
Gas exchange and water potential measurements
Gas exchange measurements were performed with a portable infrared gas analyser (model LI-6400XT; Licor, Lincoln, NE, USA) at Days 0, 2, 4, 5, 13, 16, 17, 18 and 25 of the drought treatment. Leaf photosynthesis at light saturation (Amax) and gs were measured between 10:00 and 15:00h on three control and three drought-treated plants not used for RNA-Seq in a randomized sequence. Photosynthetic photon flux density inside the cuvette was maintained at 1500 μmol m−2 s−1 and CO2 concentration was maintained at 400 μmol mol−1. Furthermore, leaf-to-air vapor pressure deficit was between 1 and 1.5 kPa, leaf temperature was close to 22 °C and humidity in the cuvette was maintained above 60%. Photosynthetic leaf area of the shoots used for gas exchange measurements was determined using ImageJ (Schneider et al. 2012, Rueden et al. 2017). Water potential (Ψshoot) was measured at midday on three lateral shoots from each of three to five plants on Days 0, 2, 4, 5, 13, 18, 21 and 25 of the treatment. The cut shoots were sealed in a plastic bag and stored in an insulated container until measurement with a Scholander-type pressure chamber (SKPM 1400, Skye instruments Ltd, Powys, UK).
Visualization and statistical analysis was performed in R 3.4.3 (R Development Core Team 2018). Values for Amax and gs were set relative to Day 0. Differences in Amax or gs over time in the treated plants were tested with one-way repeated measure analysis of variance (ANOVA) using the package nlme (Pinheiro et al. 2018). Post hoc tests were performed using the multcomp package (Hothorn et al. 2008) with Tukey’s honestly significant difference (HSD) test. Differences in water potential were assessed with a one-way ANOVA and with time as the factor. A per-time comparison t-test was then applied to test significant differences between measurements of control plants and water-stressed plants. Significant differences were assigned at P < 0.05.
Abscisic acid measurements
For sample extraction, ~50 mg of each ground root and needle sample used for RNA extraction was placed into 1.5 ml Eppendorf tubes together with 1 ml of ice-cold extraction medium (methanol/water/acetic acid, 80:19:1), including 2 ng of [2H6]-ABA (Olchemim Ltd, Olomouc, Czech Republic) as an internal standard. The samples were extracted in a mixer mill (Retsch GmbH and Co. KG, Haan, Germany) at 30 Hz for 3 min, including a 3-mm tungsten carbide bead in the tube. The bead was removed, and after centrifugation (10 min, 14,000 rpm, 4 °C), the supernatants were transferred into clean test tubes. The extraction solvent was evaporated using a SpeedVac concentrator for 1.5 h (Savant Instrument, Farming dale, NY, USA) until approximately 100 μl of solvent was left. After evaporation, 5 μl of 1 M HCl was added prior to solid-phase extraction (SPE) analysis.
For SPE, purification of the samples was performed using a Biotage Pressure +48 manifold (Biotage, Uppsala, Sweden). All samples were purified using a reverse-phased C8 column (Isolute®C8 (EC), 100 mg/3 ml Biotage). The SPE sorbent was activated and equilibrated using 1 ml of 10% methanol. The samples were washed with 1 ml of 10% methanol, and thereafter, ABA was eluted with 2 ml of 80% methanol containing 1% acetic acid. After SPE, the solvent was evaporated using a SpeedVac concentrator and stored at −20 °C until further analysis.
Prior to the Ultra High Performance Liquid Chromatography tandem Mass Spectrometry (UHPLC–MS/MS) analysis, the samples were resuspended in 50 μl of 40% methanol and transferred to LC–MS vials. A six-point calibration curve was prepared from 0.025 to 4 ng/μl, including 2 ng/μl of [2H6]-ABA in all calibration levels.
Abscisic acid was analysed using a 1290 Infinity system from Agilent Technologies (Waldbronn, Germany) coupled to an Agilent Technologies 6495 triple quadrupolequadruple tandem mass spectrometer (Agilent, Santa Clara, CA, USA) equipped with a jet stream electrospray ion source operated in the negative mode. Twenty microliter of each sample was injected onto an Acquity UPLC® HSST3 1.8 μm, 2.1 × 50 mm column (Waters, Milford, MA, USA) held at 40 °C in a column oven. The gradient eluents used were 0.1% formic acid (A) and acetonitrile with 0.1% formic acid (B), with a flow rate of 500 μl min−1. The initial conditions consisted of 0.1% B, hold between 0 and 2 min, linear increments between 0 and 2 min to 10% B, 2 and 7 min to 99% B and hold between 9 and 9.30 min at 99% B. Thereafter, the column was re-equilibrated back to 0.1% B.
The mass spectrometry was performed under the conditions of 150 °C gas temperature, gas flow at 12 l min−1, nebulizer 35 psi, sheath gas temperature 400 °C, sheath gas flow 12 l min−1, capillary voltage at 3500 V and collision energy at 11 V. The instrument was running in multiple reactions monitoring mode (MRM) with a dwell time of 200 ms for each MRM transition. The following MRM transitions were recorded: ABA 263 → 219, 263 → 201, 263 → 153; [2H6]-ABA 269 → 225, 269 → 207 and 269 → 159.
RNA extraction and sequencing
The same plants used for water potential and ABA measurements were sampled for the analysis of the drought transcriptome. At Days 0, 2, 4, 5, 13, 18, 21 and 25 of the treatment, needles and roots of three to four seedlings were collected on dry ice and stored at −80 °C until further processing. They were ground manually in liquid nitrogen, and total RNA was prepared after Chang et al. (1993) with modifications as detailed in Street et al. (2006). The RNA was further purified using the RNeasy mini kit (QIAGEN, Hilden, Germany), following the manufacturer’s instructions. A DNase digestion with the RNase-free DNase set (QIAGEN) was performed. RNA was eluted in 40 μl of RNase-free water for 2 min at room temperature. Integrity of total RNA was assessed with the Agilent RNA 6000 Nano kit (Agilent Technologies, Waldbronn, Germany) on a Bioanalyzer 2100 (Agilent Technologies) and purity was measured with a NanoDrop 2000 spectrophotometer (Nanodrop Technologies, Wilmington, DE, USA). High-quality total RNA with a RNA Integrity Number (RIN) ≥7.5, Optical Density(OD) 260/280 ratio of ≥2.0 and concentrations ≥50 ng μl−1 was sequenced by SciLifeLab (Stockholm, Sweden) for paired-end (2 × 125 bp) sequencing on a HiSeq 2000 platform using standard Illumina protocols. The sequencing library preparation included an enrichment for poly-adenylated mRNAs, and all samples each yielded >12.4 million read pairs.
Sequencing data analysis
Raw reads were pre-processed following Delhomme et al. (2014). Briefly, initial quality of raw data was assessed using FastQC (v0.11.2; https://www.bioinformatics.babraham.ac.uk/projects/fastqc/), and ribosomal RNA was removed with SortMeRNA (v2.0) (Kopylova et al. 2012). Sequencing adapters and low-quality regions were cut using Trimmomatic (v0.36), with the setting ILLUMINACLIP:2:30:10 (Bolger et al. 2014). Reads were then aligned to the Norway spruce reference genome (P. abies v1.0; Nystedt et al. 2013) using STAR (v2.4.0f1) set to −outSAMmapqUnique 254 −quantMode TranscriptomeSAM −outFilterMultimapN-max 100 −chimSegmentMin 1 (Dobin et al. 2013), and a count table was generated with HTSeq (v0.6.1) and settings −m intersection-nonempty −s yes −t exon −i Parent (Anders et al. 2015).
Count tables of needle and root data were filtered separately for genes with more than one sequencing read in at least two biological replicates, leaving 43,639 needle and 47,880 root genes in the tissue-specific drought stress transcriptomes. Expression data were normalized using variance stabilization transformation (vst) in DESeq2 (v1.16.1) (Love et al. 2014).
Principal component analysis (PCA) was used to provide a visual overview of the treatment effect on the transcriptome. On the basis of visual inspection, one of the needle samples at Day 5 was identified as an outlier and was removed. On the basis of the PCA plot, samples were grouped into mild (2, 4 and 5 days after start of the experiment), severe (Days 18 and 21 of the treatment) stress and rehydrated samples (Day 25) for calling differentially expressed (DE) genes (DEGs), and pairwise comparison between the groups and well-watered samples at Day 0 for each tissue separately was performed using DESeq2. Results were filtered for a Log2 fold change >2 and an adjusted P-value <0.01.
Clusters of DEGs were identified using ComplexHeatmap (Gu et al. 2016) in R, after calculating the mean expression for each sampling point using vst values. Default method for clustering was ‘hclust’, and ‘euclidean’ distance and vst values were scaled by row means. Thirteen clusters separated up- and down-regulated genes by tissue and time point. Gene Ontology (GO) annotations were obtained from the Conifer Genome Integrative Explorer (ConGenIE; http://congenie.org) (Sundell et al. 2015). GOSlim annotations were built for Norway spruce by the Map2Slim function of Owltools (https://github.com/owlcollab/owltools/wiki/Map2Slim) using the plant slim subset provided by the (GO Consortium (Ashburner et al. 2000, Beike et al. 2015). Enrichment analysis was performed using GeneMerge (Castillo-Davis and Hartl 2003). The background geneset comprised genes with vst >4 (transcriptome size needles 42,821, roots 47,880 genes), and enrichment assigned if P < 0.05.
A single co-expression network was inferred for the needle and root data using the per gene vst-normalized expression values. Genes with low variance were removed by calculating the median absolute deviation (MAD) and by excluding all genes with MAD = 0. Eleven network inference methods were computed for each dataset using the Seidr v0.12 toolkit (Schiffthaler et al. 2018)—ARACNE, CLR, Elastic Net, GENIE3, Narromi, PCor, Pearson, PLSNET, Spearman, SVM and Tigress. In case of non-symmetrical scoring by the method, the higher score for every symmetric edge pair was kept. The networks were aggregated using the inverse rank product method (Zhong et al. 2014) and the edges were filtered according to the noise-corrected backbone (Coscia and Neffke 2017) at a sigma of 2.32 (which roughly corresponds to a P-value of 1%). Network clusters were identified using InfoMap 0.19.26 (Rosvall and Bergstrom 2008) with markov-time of 1.4. Gene Ontology and MAPMAN (Thimm et al. 2004) annotations were obtained using gopher, an in-house tool available at DOI 10.5281/zenodo.3731544.
Sequence homology BLASTp searches of differentially expressed P. abies genes were performed against A. thaliana, other gymnosperms (Picea glauca, Picea sitchensis, Pinus pinaster, Pinus sylvestris, Pinus taeda, Pseudotsuga menziesii, Gnetum montanum, Taxus baccata, Cycas micholitzii and Gingko biloba) and further angiosperm species (Populus trichocarpa, O. sativa ssp. japonica and Amborella trichopoda) in addition to Physcomitrella patens. Orthologs were detected using Gymno PLAZA 1.0 using OrthoMCL and Best-Hits-and-Inparalogs (BHIF) predictions (Proost et al. 2015). Previously recognized genes functioning in drought response in A. thaliana from the DroughtDB (Alter et al. 2015) and from drought-related GO terms: ‘response to water deprivation’ (GO:0009414), ‘cellular response to water deprivation’ (GO:0042631) and ‘regulation of response to water deprivation’ (GO:2000070), and all child categories of these were used for a collective search of conserved function within the differentially expressed transcripts in samples of needles and roots of Norway spruce.
Transcription factors and their corresponding families were obtained from PlantTFDB 3.0 (Jin et al. 2014). Hypergeometric tests were performed using the ‘phyper’ function in the base R package stats and enrichment for TF families in the background of all TF in the transcriptomes and of singletons (if no sequence homology of any kind existed) in the set of DEG in comparison with the transcriptome sizes calculated, and significance assigned if P < 0.05.
Motif analysis was performed on 1 kb promoter regions classified as being upstream sequences of a start codon. Genes with shorter upstream sequences or without stop codons were excluded of the analysis. In total, 37,621 upstream regions were analysed. Counts of the different analysed motifs were obtained with the stand-alone version of PatMatch (Yan et al. 2005). Gene expression profiles of genes with promoter regions containing a motif over-represented involved in the ABA-dependent pathway were plotted using the ComplexHeatmap R package. A motif was considered over-represented if it was present in an upstream region more times than the respective confidence interval (CI) upper bound (Chawade et al. 2007). The motifs analysed represented consensus target sequences for AP2/ERF, MYB, MYC and bZIP TFs (YAACR/CANNTG, respectively) and ABRE cis elements that are part of the ABA-dependent pathway (Shinozaki and Yamaguchi-Shinozaki 2007). Thus, when the ABRE motif was present twofold or more-fold in a promoter and the less promiscuous (i.e., fewer global occurrences) CANNTG/YAACR motifs were one or more times present, they were considered as over-represented.
Results
The physiological drought response of P. abies seedlings
Treatment severity increased during the first 5 days, with a 10% decrease in FC each Day until 30% (Figure 1a), after which FC was maintained at 30% for 7 days. Thereafter, the seedlings were exposed to a more severe drought stress by withholding water until catastrophic dysfunction of stomatal conductance, and Amax was detected in the aboveground plant tissues (Figure 1b). The gs decreased non-significantly within the first 5 days, remained stable at 30% FC and significantly decreased in response to severe drought stress (P < 0.01) when the soil water content was 20% FC (Figure 1b). Abscisic acid levels were low in both needles and roots throughout the mild drought stress (Days 2, 4, 5 and 13), increased dramatically during the severe drought stress (Days 18 and 21) and returned to control levels after re-irrigation (Figure 1c). A small, transient but non-significant increase in root ABA was observed at Day 5, with levels dropping to control values after water was added to maintain FC at 30%. During the same period, ABA levels in needles showed a small, gradual but non-significant increase. Shoot water potential changed significantly after 5 days of mild water stress (P < 0.05) (Figure 1d), but it temporarily recovered during the phase when FC was held at 30%. After initiation of the severe drought stress, shoot water potential showed a further significant decrease (P < 0.01). These results demonstrated that Norway spruce is an isohydric conifer species, reducing gs over a very narrow range of water potential (between −1.1 and −1.8 MPa) in order to maintain midday shoot water potential at a reduced but relatively constant level (−2.1 MPa). Both gs and Amax (Figure S1 available as Supplementary data at Tree Physiology Online) were maximally limited after 18 days, 5 days after initiation of the severe drought stress. Seedlings in the severe drought treatment could be recovered by re-watering after Day 21 (Figure 1d). However, continuation of the severe drought for an additional 2 days resulted in defoliation and death of the seedlings (not shown). Four Days after re-watering (at Day 21), the shoot water potential and ABA levels returned to pre-stress levels; gs recovered to ~60% and Amax to ~80% of the levels prior to initiation of the drought treatment. The gs and Amax of control seedlings were unchanged throughout the period of the experiment (Figure 1b and c, and Figure S1 available as Supplementary data at Tree Physiology Online).
Needles and roots have distinct transcriptional responses to drought
Needle and root samples revealed only limited changes in relative transcript abundance in response to the mild drought treatment, while the severe drought induced a more extensive transcriptional response, particularly in roots (Figure 2a and Figures S2 and S3 available as Supplementary data at Tree Physiology Online). Re-irrigation reversed the transcriptional changes in needles, but the transcriptome of roots remained distinct after recovery (Figure 2a and Figure S2 available as Supplementary data at Tree Physiology Online). Mild drought induced differential expression of 98 genes in needles, of which 91 were up- and 7 were down-regulated (Figure 2b and c). Similarly, 98 genes were DE in roots, with 90 up-and eight down-regulated genes (Figure 2b and c). Three of the genes DE during the mild drought stress were commonly up-regulated between the tissues (Figure 2c). In contrast, 1304 genes were DE in response to severe drought stress in needles, with 871 of these up- and 433 down-regulated, while in roots, there were 5835 DEGs, of which 1926 were up- and 3909 down-regulated. Extensive down-regulation was therefore a notable component of the transcriptional response mechanism of roots during severe drought stress, and active remodulation of the transcriptome was far more prominent in roots than in needles; 440 and 129 of the genes DE during the severe drought stress were commonly up- or down-regulated between the tissues, respectively (Figure 2c). After re-irrigation, 23 genes were DE in needles, 14 up- and 9 down-regulated, whereas in roots, 853 genes were DE, 603 up- and 250 down-regulated (Figure 2b and c). Two of the genes DE during re-irrigation were commonly down-regulated in both tissues, and one gene was commonly up-regulated (Figure 2c).
Figure 2.
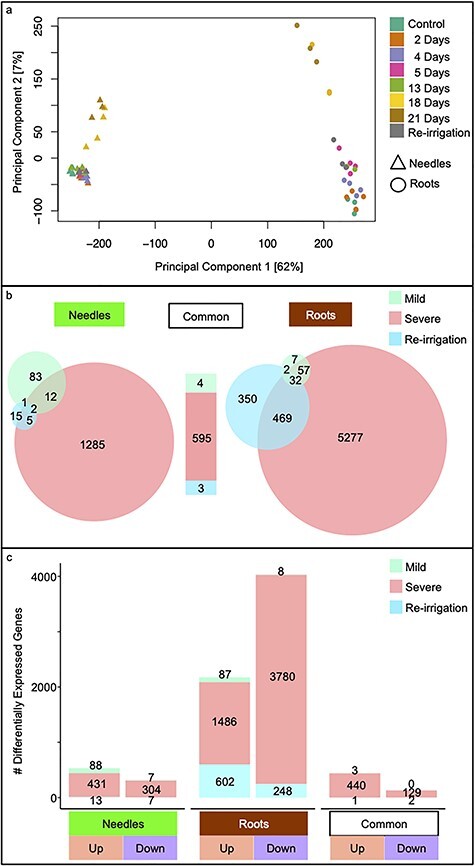
Effect of drought on the transcriptomes of needles and roots in Norway spruce. Principal component analysis plot of transcriptomic data of drought-stressed needle and root (a) samples. Expression data of control, mild and severe drought-stressed seedlings and after re-irrigation were normalized using variance stabilizing transformation before ordination analysis. Three to four plants were sampled at each time point and were used for RNA-Seq. The first two components of the PCA are shown with samples colored by soil water percentage FC. (b) Venn diagrams of DEGs in needles (left) and roots (right) after comparing the mild and severe drought stages and after re-irrigation separately against well-watered expression levels. Independently, the number of DEGs and the number of these in common between the tissues are displayed separately for each stage of the treatment. Conditions are colored as in Figure 1c). Genes either up- or down-regulated in needles and roots and with the same expression in the tissues (common) are displayed in a bar graph and colored by treatment according to Figure 1.
The transcriptional response of roots involves extensive down-regulation of growth-related processes
To explore the biological processes involved in the transcriptome response to drought, 13 clusters representing co-expressed sets of genes were identified and functional annotations were assigned to these (Figures 3a and 4a, and Tables S1 and S2, available as Supplementary data at Tree Physiology Online, for gene clusters primarily up- or down-regulated in response to drought, respectively). Gene Ontology terms are assigned to ~34% of the DEGs, and GOSlim terms were used to provide a global overview of the biological processes induced in response to drought in needles and roots (Figures 3b and 4b). The GOSlim term ‘response to stress’ (GO:0006950) was significantly enriched (P < 0.05) in Cluster 3 (Figure 3b), which contained genes commonly up-regulated in both needles and roots in response to severe drought. Cluster 10 (Figure 4a) comprised a large set of genes specifically down-regulated in roots during severe drought, with this down-regulation reversed upon re-irrigation, indicating recovery from water stress. This cluster was significantly enriched (P < 0.05) for GOSlim terms associated with growth including ‘anatomical structure development’ (GO:0048856), ‘transport’ (GO:0006810), ‘reproduction’ (GO:0000003), ‘carbohydrate metabolic process’ (GO:0005975), ‘cell wall organisation or biogenesis’ (GO:0071554), ‘DNA metabolic process’ (GO:0006259), ‘chromosome organization’ (GO:0051276), ‘cell differentiation’ (GO:0030154), ‘cell cycle’ (GO:0007049), ‘growth’ (GO:0040007), ‘anatomical structure formation involved in morphogenesis’ (GO:0048646), ‘cell morphogenesis’ (GO:0000902), ‘developmental maturation’ (GO:0021700), ‘cell division’ (GO:0051301), ‘cell proliferation’ (GO:0008283) and ‘pigmentation’ (GO:0043473). As such, active down-regulation of growth appeared to be an important component of the drought-response mechanisms in the roots of Norway spruce seedlings. Finally, ‘response to stress’ (GO:0006950) was significantly enriched (P < 0.05) in Cluster 11 (Figure 4b), combining DE genes down-regulated in roots both during severe drought and after re-irrigation.
Figure 3.
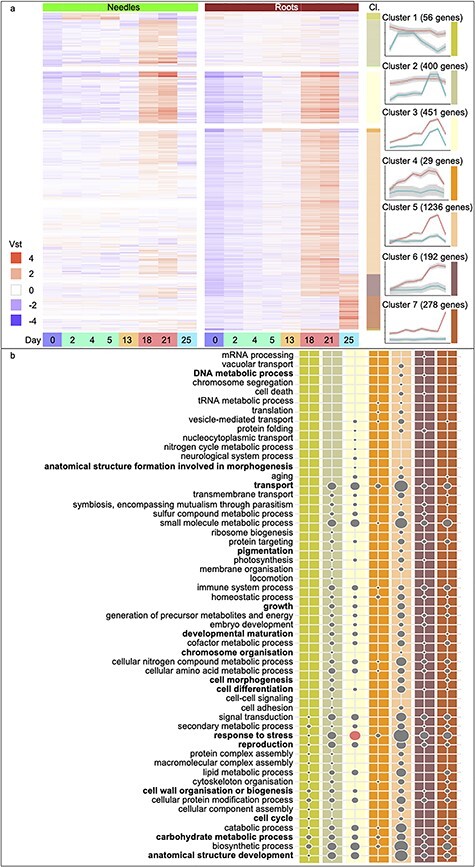
Gene Ontology Slim enrichment analysis of biological processes up-regulated in response to drought stress. Heatmaps of DEGs with expression over the eight sampling time points (Days 0, 2, 4, 5, 13, 18, 21 and 25 and colored as in Figure 1), separated by tissue, needles (green) and roots (brown). (a) Up-regulated genes of needle- (Clusters 1 and 2) and root-specific (Clusters 4–7) clusters or in common between the tissues (Cluster 3). Displayed are the Variance Stabilizing Transformation (vst) values scaled by row means. The seven highly populated clusters (Cl.) are detailed on the side, and the average trend of gene expression and a 95% CI are indicated for both tissues. Colored bars to the right of the heatmap correspond to the clusters represented in the line graphs as indicated by a colored bar to the right of each cluster line graph. (b) The lower panel of the figure lists all GOSlim biological processes categories represented in the clusters shown in (a), with columns colored by cluster. Bold text indicates categories that are significantly enriched. Circle sizes are relative to the category containing the largest number of genes; ‘response to stress’ (76 up-regulated genes). Circles in red represent significantly enriched categories (false discovery rate-adjusted P-value <0.05).
Figure 4.
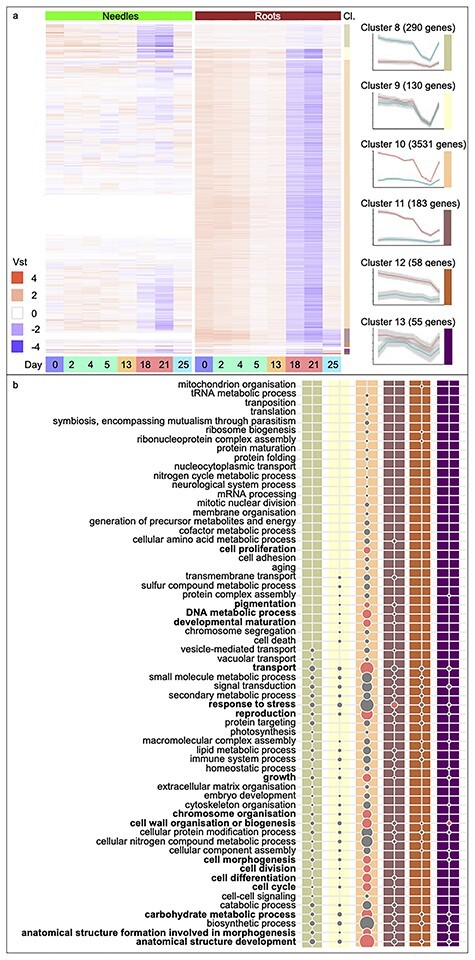
Gene Ontology Slim enrichment analysis of biological processes down-regulated in response to drought stress. Heatmaps of DEGs with expression over the eight sampling time points (Days 0, 2, 4, 5, 13, 18, 21 and 25 and colored as in Figure 1), separated by tissue, needles (green) and roots (brown). (a) Down-regulated genes of needle- (Cluster 8) and root-specific (Clusters 10--12) Clusters or common between the tissues (Clusters 9 and 13). Displayed are the vst values scaled by row means. The six highly populated clusters (Cl.) are detailed on the side, and the average trend of gene expression and a 95% CI are indicated for both tissues. (b) The lower panel of the figure lists all GOSlim biological processes categories represented in the clusters shown in (a), with columns colored by cluster. Bold text indicates categories that were significantly enriched. Circle sizes are relative to the category containing the largest number of genes; ‘anatomical structure development’ (220 down-regulated genes). Circles in red represent significantly enriched categories (false discovery rate-adjusted P-value <0.05).
In roots, re-irrigation resulted in up-regulation (P > 0.05) of metabolism (e.g., ‘small molecule metabolic process’ (GO:0044281), ‘carbohydrate metabolic process’ (GO:0005975) and ‘biosynthetic process’ (GO:0009058) (Figure 3b, Cluster 7), indicating an ongoing recovery from the severe stress. However, ‘response to stress’ (GO:0006950) remained induced in roots (P > 0.05) after re-irrigation (Figure 3b, Cluster 6). Re-irrigation also induced additional genes associated with a stress response to hypoxia in roots (Figure 3a, Cluster 7) (e.g., ADH1 and LOX2, see Table S3 available as Supplementary data at Tree Physiology Online for details), suggesting that a high level of responsiveness was maintained in roots even after release of drought stress.
Functional analysis of the Norway spruce transcriptome in comparison to Arabidopsis
Arabidopsis is the most comprehensively characterized species for a range of abiotic stresses, including drought. To ascertain how relevant biological annotation of genes in this model system is for informing studies in Norway spruce, sequence homology analysis was performed. Arabidopsis orthologs were identified for 13% (184 of 1400) and 14% (888 of 6160) of DEGs in needles and roots, respectively, and homologs were identified for 57 and 55%, respectively (Table 1 and Table S3 available as Supplementary data at Tree Physiology Online, see methods for details of ortholog/homolog classification). An additional 18 and 19%, respectively, were homologous to gymnosperm or other angiosperm species available at the PLAZA resource (Proost et al. 2015), and 12% (166 in needles and 762 in roots) of the drought-responsive genes had no homology match and were termed Norway spruce-specific singletons. There was no significant enrichment for singletons (P > 0.05; Fisher’s exact test).
Table 1.
Percentage of drought-responsive transcripts in Norway spruce with sequence conservation in plants.
| Homology (DE) | NeedleDE (%) | RootDE (%) | Common |
|---|---|---|---|
| (1400) | (6160) | (634) | |
| Ortholog (Ath) | 13.1 (184) | 14.4 (888) | (n = 82) |
| Homolog (Ath) | 57.1 (800) | 54.7 (3369) | (n = 369) |
| Angiosperm | 0.0 (0) | 0.1 (4) | (n = 0) |
| Gymnosperm | 15.7 (220) | 15.7 (970) | (n = 100) |
| Others | 2.1 (30) | 2.7 (167) | (n = 13) |
| Singletons (Pab) | 11.9 (166) | 12.4 (762) | (n = 70) |
Differentially expressed genes in response to drought of needle and root samples were searched for orthologs and homologs in A. thaliana (Ath). Non-matching genes were searched to be specific to other angiosperms, other gymnosperms or both or defined as P. abies (Pab)-specific singletons.
To determine whether singletons were fully integrated within the drought-response network, had particular network statistic characteristics or if there were specific transcriptional modules enriched for singletons, which could represent Norway spruce or conifer specific components of a stress response, a gene co-expression network was inferred, and network modules (NMs) were identified (Table S1 available as Supplementary data at Tree Physiology Online; NM expression profiles and functional enrichment results are available in Supplementary Note S1, available as Supplementary data at Tree Physiology Online) and associated network connectivity statistics were calculated. Examination of the location of the conserved and singleton genes within the co-expression network (Figure S4 available as Supplementary data at Tree Physiology Online) revealed no enrichment of either category within specific NMs. Genes from both categories (conserved/singleton) were integrated across the network and across the range of connectivity measures. As an example, in NM 1:2, the top 10 genes ranked by PageRank, as a measure of network importance/centrality, included two genes with no identified Arabidopsis homolog.
To examine whether extensively characterized Arabidopsis drought-response genes displayed similar transcriptional response to drought in Norway spruce, a manually curated set of 373 genes was further considered (Table S3 available as Supplementary data at Tree Physiology Online). Orthologs or homologs were identified for 335 (89.9%) of these, represented by 979 Norway spruce gene models (Table 2 and Figure S5 available as Supplementary data at Tree Physiology Online), of which >87% were expressed in the current samples. A subset of these were DE in needles (10.5%; 39 genes) or roots (31.4%; 117 genes), and only this subset was therefore considered to have evidence of conserved function (indicated by transcriptional response) in Norway spruce seedlings. These DEGs were assigned into the clusters detailed in Figures 3a and 4a based on their expression profiles (Table S3 available as Supplementary data at Tree Physiology Online). Among this set of genes, conservation existed for genes functioning in the induction of solute accumulation, such as for sugars and amino acids (e.g., GolS1, RS5/SIP1, PCK1, MGL, Figure 3a, Cluster 3; and GolS2, BAM1/BMY7, AVP1/AVP3, HKL3 specifically in roots, Figure 3a; Cluster 5), reactive oxygen species (ROS) scavenging (e.g., GSTU19/GST8, Figure 3a, Cluster 3 and GPX3, Figure 3a, Cluster 5), dehydrins (e.g., LTI29/ERD10/LTI45, Figure 3a, Cluster 3 and LEA7, LEA4-1 specifically in roots; Figure 3a, Cluster 5) and ABA biosynthesis and signaling (e.g., NCED3/SIS7/STO1, NCED5, PP2CA/AHG3, CIPK6/SNRK3.14/SIP3, AFP2, Figure 3a, Cluster 3). Additionally, genes that restrict proline catabolism (e.g., ERD5/PDH1, PDH2, Figure 4a, Cluster 10), ROS production (e.g., RBOH F, RBOH D, Figure 4a, Cluster 9; and GSTF10/ERD13, GSTF6/GST1/ERD11/GSTF3, GST30/ERD9/GSTU17, Figure 4a, Cluster 10) and auxin and cytokinin signaling (e.g., UGT74E2, Figure 4a, Cluster 8; and UGT76C2, WOL/CRE1/AHK4, Figure 4a, Cluster 10) displayed conserved transcriptional responses.
Table 2.
Drought-responsive genes of A. thaliana identified as P. abies homologs and orthologs.
| Plant species | Genome | Needles | Roots | Common |
|---|---|---|---|---|
| Transcriptome/DE | Transcriptome/DE | |||
| Arabidopsis (373 genes) | 335 (89.8%) | 325/39 (87.1/10.5%) | 328/117 (87.9/31.4%) | 322/33 (86.3/8.9%) |
| Norway spruce | 979 | 782/43 | 826/164 | 758/24 |
An overview of the gene numbers in the needle and root transcriptome and in the set of DEGs, respectively, with sequence homology to drought genes in A. thaliana is given. Three hundred and seventy-three Arabidopsis genes with well-characterized roles in drought response were considered. Orthologs or homologs of these genes (indicated in the genome column) were identified in Norway spruce. Identified orthologs/homologs were then classified based on whether they showed conservation of differential expression in response to drought in the needles and roots of Norway spruce seedlings or were commonly DE in both tissues.
Transcription factors regulating drought stress in Norway spruce
Among the DEGs in needles and roots were 74 and 240 TFs, respectively (Figure 5). These TFs included members of 20 and 33 TF families, respectively, from the 54 TF families annotated in the Norway spruce genome (Figure S6 and Table S4 available as Supplementary data at Tree Physiology Online). Among all TF families, there was significant enrichment (P < 0.05; Fisher’s exact test) of ERF, C2H2, MYB-related and NAC TF families in needles and ERF, MYB and SAP in roots (Figure 5 and Figure S6 available as Supplementary data at Tree Physiology Online). Four TFs in needles and 26 in roots had a known drought function in Arabidopsis (Tables S3 and S4 available as Supplementary data at Tree Physiology Online), three of which were DE in both tissues. Notably, the bZIP AREB/ABF (ABRE binding/ABFs) TFs of the ABA-dependent pathway, which are key regulators of angiosperm drought-response, were not DE. Two genes with sequence similarity to Arabidopsis ABFs were expressed (MA_18107g0010, MA_61649g0010), but both displayed small, non-significant increases in expression in response to severe drought in both needles and roots. The canonical ABF binding motif was present in the available promoter region of several genes; however, these were not consistently DE in response to drought (Figure S7 available as Supplementary data at Tree Physiology Online).
Figure 5.

Heatmap diagram of TFs differentially expressed in Norway spruce in response to drought. Displayed are the differentially expressed TFs over the eight sampling time points (Days 0, 2, 4, 5, 13, 18, 21 and 25 and colored as in Figure 1) and are separated by tissue, needles (green) and roots (brown). (a) Up-regulated TFs were clustered (Cl.) by expression and sorted by the same color coding as in Figure 3a and b). Down-regulated TFs were clustered by expression and sorted by the same color coding as in Figure 4a. Displayed are the vst values scaled by row means. In addition, information on sequence homology (Homol.) is visualized on the right side, with genes orthologous to Arabidopsis colored dark green, homologous to Arabidopsis in light blue, homologous to other gymnosperms in light green and Norway spruce-specific singletons in dark blue. Furthermore, TF families significantly enriched: ERFs, MYB-related, C2H2s and NACs in needle samples, and ERFs, MYBs and SAPs in roots are color-coded.
Components of ABA-independent signaling via DREB2B and ERF53 were DE in response to drought in roots (see below). In needles, TF ZF2 was up-regulated in Cluster 2 (Figure 3a). In Cluster 3 and NM 1:2, both ABA-dependent NAC TFs (NAC3/NAC055, RD26) and ABA-independent ERFs (DREB19, ERF7) were commonly up-regulated in both tissues. In roots, the earliest transcriptional response was up-regulation of the ABA-dependent MYB2 in Cluster 4 after 5 days, the expression of which remained high under severe water stress (Figure 3a). Severe drought in roots additionally activated NAC019, TFs within the ERF family (DREB2B, ERF53, RAP2.1, RAP2.4 and RAP2.6 L), a WRKY TF (WRKY33) and MYBs (MYB96, MYBR1) (Figure 3a, Cluster 5). ARR-B TFs (RR10, RR1) and ZFHD1/ZFHD11/HB29 were also up-regulated after severe drought in roots. Down-regulation of TFs in roots after severe drought occurred for ERF family members (CBF1/DREB1B, CBF3/DREB1A, CBF4/DREB1D, HRD, DDF1, RAP2.1 and SHN1/WIN1, Figure 4a, Cluster 10). Also, MYB TFs (MYB61, MYBR1), a BHLH TF (BHLH100) and a HD-ZIP TF (HB6) were down-regulated. After re-irrigation, increased expression of TFs, such as of MYB15, RAP2.6 or RAP2.6 L and NAC3/NAC055 and RD26, was observed in roots (Figure 3a, Clusters 6 and 7). Drought-responsive TFs included several Norway spruce-specific singletons (Table S4 available as Supplementary data at Tree Physiology Online).
It should be noted that exact ortholog inference is currently challenging for Norway spruce due to the fragmented nature of the genome assembly (Nystedt et al. 2013). As such, caution is needed in interpreting inference of functional ortholog conservation. This likely underlies many of the cases where multiple copies of Norway spruce genes had the highest sequence similarity to a single Arabidopsis gene. For some such cases, there were differences in which gene copy was DE between the tissues. While these potentially represent two genuine copies that have undergone sub-functionalization, such observations require detailed follow-up studies or reconsidering upon the availability of an improved genome assembly or functional validation.
Discussion
Drought stress typically induces short-term physiological and molecular responses that enable plants to survive periods of limited or fluctuating water availability. In roots, these responses are initiated by the limitation of soil water uptake, with root-level responses to this initial limitation being an important component in determining the whole-plant drought tolerance and survival. Despite the importance of root responses, most previous studies have focused on the response of aboveground tissues. In this study, drought-induced transcriptional changes of both roots and needles of Norway spruce seedlings were assayed by subjecting seedlings to a progressively increasing soil water deficit, followed by a recovery phase after re-irrigation. The induced transcriptional responses were placed within a framework of physiological responses observed in needles (Figure 6). To explore the extent of conservation in the transcriptional drought response and to determine the appropriateness of utilizing functional annotations derived from other species, the transcriptional response of Norway spruce was compared with previous results in Arabidopsis as the most comprehensively studied model system for drought and other abiotic stress responses and the primary source of gene functional annotation.
Figure 6.
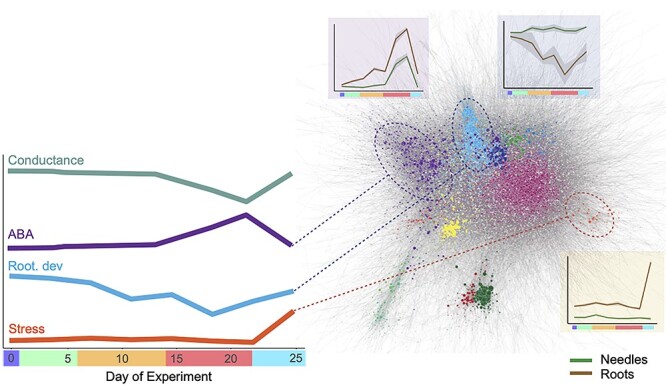
Overview representation of physiological and gene expression responses to drought in Norway spruce seedlings. The left panel indicates representative profiles indicating changes in physiological indicators of drought stress. Profiles are representative and based on the data presented in Figure 1 (for stomatal conductance and ABA) or gene expression values (for root development and stress). On the right, the co-expression network is indicated with nodes (genes) colored to indicate their assigned network cluster. For three clusters, associated with ABA, root development or stress response (indicated by cluster gene ontology biological process enrichment results), expression profile plots are shown in which green profiles represent the expression in needles and brown represent the expression in roots.
Needle gs and Amax were at their observed minimums at Day 18, and midday shoot water potential was as low as −2.1 MPa at Day 21, before which there was little observed physiological response to water stress in the needle samples (Figure 1d). In a similar study of red spruce seedlings (Picea rubens Sarg.) photosynthesis continued until water potential decreased to −3.0 MPa and was rapidly recovered to 70% of well-watered plants after re-irrigation (Seiler and Cazell 1990). In the current experiment, re-irrigation after 21 days resulted in 80% recovery of the photosynthetic rates (Figure 1c). Nevertheless, the treatment was regarded as severe, with death occurring in a set of seedlings that experienced an additional 2 days of severe drought (indicated by an X symbol in Figure 1a). In agreement with this observation, it was reported earlier that Norway spruce seedlings die after crossing a critical point of dehydration (Ditmarova et al. 2010) and that drought results in the mortality of Norway spruce seedlings (Ivanov et al. 2019). These findings highlight the severe negative effects that could result from the changing patterns of soil water availability as a result of continued climate change.
The limited physiological response of needles to severe drought was reflected in the transcriptome, with substantially fewer DEGs in needles than in roots (Figure 2b). This was despite very similar increases in ABA levels in both tissues, suggesting either that the observed increase in ABA was coincidental rather than controlling the induced transcriptional responses or that the two tissues have differential sensitivity to ABA. The known importance of ABA in regulating drought response together with the observed correlation between ABA levels and the predominant expression profile induced by severe drought in both tissues points toward different transcriptional responses being induced by the common ABA signal in the two tissues. In contrast to the needle response, changes in the transcriptome of roots were observed starting at 30% FC, with the response being particularly prominent in response to severe drought (Day 18; Figure S2b available as Supplementary data at Tree Physiology Online). Correspondingly, a greater number of biological processes were significantly altered in roots, particularly categories associated with growth and development. Nevertheless, there was a common enrichment in both tissues of up-regulated genes for ‘response to stress’, indicating a core stress response program around which a root-specific extensive down-regulation of genes involved in growth was induced. This is in line with the finding that drought response in conifers is highly dependent on established roots, with the development of new roots being restricted due to carbon costs (Mackay et al. 2020) and with previous observations of decreased root growth of Norway spruce seedlings under drought stress (Zlobin et al. 2018). In needles, metabolism remained largely unchanged, with no enrichment for terms associated with the primary physiological mechanisms of photosynthesis. Pashkovskiy et al. (2019) reported ABA levels in Norway spruce and Scots pine roots and needles in response to drought, finding significantly higher baseline ABA levels and more substantial induction in Norway spruce needles than roots. The current results contrast in that similar ABA levels and extents of increase were recorded in both tissues (Figure 1c). Further experiments will be needed to determine whether the differences observed in the two studies arise from the substantially different experimental setups used to induce drought stress (water deprivation in the current study and polyethylene glycol within a hydroponic system for Pashkovskiy et al.) or result from genotypic variation.
Comparison of transcriptional regulation between species is important to determine the evolution of stress responses and to ascertain the applicability of transferring biological knowledge (i.e., functional annotation descriptions) and selection of targets across species. While a number of studies have examined transcriptome responses to drought in conifer species (Lorenz et al. 2011, Du et al. 2018, Fox et al. 2018, de María et al. 2020), finding variation between species and among genotypes (de María et al. 2020), it is also of interest to know how extensively functional knowledge generated using angiosperm species translates to coniferous species. Here, indications of both conservation and divergence in transcriptional responses were observed between Arabidopsis and Norway spruce. The conserved component of the Norway spruce response included up-regulation of solute accumulation, ROS scavenging, ABA biosynthesis and signaling, with many genes involved in these processes highly ranked within NMs. In contrast, proline catabolism, ROS production and auxin and cytokinin signaling responses were limited. However, a large proportion of the homologs or orthologs of the characterized Arabidopsis drought-responsive genes considered were not DE in response to drought, suggesting that there are extensive differences as to which genes regulate the induced, common components of the conserved stress response transcriptional program between the two species. For this subset of genes in particular, functional descriptions derived in Arabidopsis potentially do not reflect function in Norway spruce, highlighting the need for species- or lineage-specific functional characterization. Determining whether these diverged transcriptional responses reflect the differences between lineages or is a specific component of the contrasting isohydric versus anisohydric drought-response strategies also requires further studies. In addition to this set of sequence-conserved genes, there were several Norway spruce-specific genes actively regulated in response to drought as has been reported by previous studies of drought stress in a range of conifer species (Lorenz et al. 2011, Du et al. 2018, Fox et al. 2018, de María et al. 2020). These genes lack functional annotation based on homology and were fully integrated within the co-expression network, making them interesting targets for future characterization studies.
Differences and parallels between species were identified in the transcriptional response of genes within the ABA signaling pathway. Signaling by plant hormones plays an important role in the early response to drought stress, and production of ABA is a well-recognized component of the typical response to water deficit (Schachtman and Goodger 2008) as well as in conifers more specifically (Brodribb and McAdam 2013, Brodribb et al. 2014, Pashkovskiy et al. 2019). In the current study, there were numerous ABA-associated genes present in Cluster 3/NM 1:2, suggesting an expected role for ABA in inducing coordinated gene expression changes. The serine/threonine kinase SnRK2/OST1, involved in ABA signaling, serves as a useful example of the potential complications presented when transferring ortholog-inferred annotations across divergent genomes. In Arabidopsis, OST1 functions in the control of stomatal closure, while in the current experiment, the Norway spruce ortholog (MA_10430272g0020) exhibited high and constant expression in needles with no observed change in expression in response to drought. In contrast, expression in roots was highly responsive and correlated to changes in ABA, with a transient, small increase at Day 5 and a dramatic increase during the severe drought. There were also numerous examples, as exemplified by RD26 and RD22, where multiple Norway spruce genes have maximal sequence similarity to a single Arabidopsis ortholog and with those multiple Norway spruce genes having divergent expression between tissues and in response to drought. In contrast to Arabidopsis, members of the bZIP TFs of the ABA-dependent pathway, the AREB/ ABFs (Singh and Laxmi 2015), were not DE. While expression of homologs to AREB1/ABF2, AREB2/ABF4 and ABF3 increased, these changes were small and not significant, and many of the potential target genes of the ABFs were not DE (Figure S6 available as Supplementary data at Tree Physiology Online). As homologs of the ABA-responsive bZIPs are present but were not DE in response to drought, it will be interesting for future work to ascertain whether those bZIPs and their targets have evolved an alternative function in Norway spruce. In contrast, TFs of the ABA-independent DREB1/CBF and DREB2 regulons were significantly DE in roots. DREB1/CBF type genes have mainly been reported to be responsive to low temperatures in Arabidopsis (Nakashima et al. 2009) but were also up-regulated during water deficit in maize shoots and, to an even greater extent, in roots (Liu et al. 2013), with similar expression changes reported in poplar and loblolly pine roots (Cohen et al. 2010, Lorenz et al. 2011). In the current study, expression of DREB1/CBF genes was strongly down-regulated in roots and only DREB2B, which has a well-defined function in drought stress (Nakashima et al. 2009), was up-regulated in roots. Similarly, expression of ERF53, which has been proposed to function in a similar manner to DREB2A via regulation of drought stress-responsive genes (Cheng et al. 2012), increased under drought stress. Altogether, these findings indicate that there are potential differences in the regulators controlling transcriptional drought response in Norway spruce seedlings compared with the typical response of Arabidopsis.
While many genes differed in their response to drought between Arabidopsis, several genes with known functional roles in drought tolerance were DE and showed a transcriptional response in needles and roots, suggesting conserved function and making these robust targets for either breeding and selection or genome engineering. The observed drought response involved expression-conserved up-regulation of solute synthesis, such as of malate (PCK1) (Penfield et al. 2012), raffinose (RS5/SIP1, GolS1) (Taji et al. 2002, Nishizawa et al. 2008, Egert et al. 2013, Rasheed et al. 2016) or amino acids (MGL) (Joshi and Jander 2009), and protection of cellular components via expression of dehydrins (LTI29/ERD10/LTI45) (Kasuga et al. 2004) and ROS scavengers (GSTU19/GST8 and GST30/ERD9/GSTU17) (Chen et al. 2012, Xu et al. 2016) in both tissues. Furthermore, other regulatory genes had a conserved function in Norway spruce. For example, NAC and MYB TFs, such as ANAC072/RD26, which were here up-regulated in needles and roots, have been shown to improve drought stress tolerance in Arabidopsis when overexpressed (Nakashima et al. 2012). The ANAC019 and ANAC072/RD26 genes, here with root-specific expression, have been classified as stress-responsive NAC (SNAC) TFs (Tran et al. 2004). Expression changes of many MYB TFs have been reported during drought stress responses in plants (Baldoni et al. 2015), including in conifer species (Lorenz et al. 2011, Du et al. 2018, de María et al. 2020), although their functional role has not been elucidated. MYB2 is known to mediate response to abiotic stress and regulate gene expression of ABA-inducible genes (Abe et al. 2003) and to be expressed in both the shoots and roots of Arabidopsis and rice (Yang et al. 2012, Baldoni et al. 2015). This gene was DE early on in roots in the current study, suggesting functional conservation, indicating early activation of ABA-induced response mechanisms despite the non-significant increase in ABA levels. Severe drought stress induced MYB96 in roots, which may function to arrest lateral root growth in a similar manner to the described role in Arabidopsis (Seo et al. 2009). This response is suggested to enable maximal resource allocation to primary root growth to facilitate rapid recovery after cessation of water shortage (Comas et al. 2013). During recovery, MYB15 was here expressed in roots and, in Arabidopsis, this gene has been reported as a negative regulator of DREB1/CBF genes during cold stress (Agarwal et al. 2006). Potentially, this gene may therefore have a similar function in Norway spruce prior to and during recovery.
The observed drought-response mechanism highlights the potential for Norway spruce seedlings to experience severe negative effects when exposed to relatively short periods of soil drying. Given the combination of a typically shallow root system and the expected increase in both the frequency and severity of such drought events, there could be substantial ecological and economic implications. This sensitivity to drought is especially relevant to the context of Norway spruce forestry and planting after clear-cut. As a late successional species, Norway spruce is increasingly likely to have maladapted abiotic stress mechanisms when grown in a clear-cut context where conditions are considerably different from those where the Norway spruce seedlings would typically establish. This study provides a comprehensive characterization of the transcriptional drought-response mechanisms in Norway spruce roots and needles, highlighting the substantial difference in the transcriptional response to drought between the two tissues. The extensive down-regulation of genes related to the growth and metabolism in roots would result in a large impact on the seedling growth, establishment and biomass accumulation. There are also associated potential implications that we do not explore in the current study, such as the effects of alternations to microbial community interactions as a result of altered root growth. In contrast to roots, little transcriptional response was observed in needles, reflecting the limited physiological changes in water potential and stomatal conductance shown until severe drought was experienced. Combined with reduced root growth, these effects could have substantial ecological impacts and economic implications, including the effects on carbon sequestration.
Authors’ contribution
J.C.H., V.H. and N.R.S. planned and designed the study. J.C.H. performed the experiments and analysed the data together with A.V., V.H. and N.R.S. A.R.S. performed network analyses. S.M. performed ABA profiling. J.C.H. wrote the initial draft of the manuscript. J.C.H., A.V., V.H. and N.R.S. edited the manuscript into its final form. All authors read and approved the final manuscript.
Supplementary Material
Acknowledgments
The authors thank the UPSC bioinformatics facility (https://bioinfomatics.upsc.se) for technical support, the greenhouse staff at the Umeå Plant Science Centre for horticultural services and Catherine Campbell for support with the gas exchange measurements.
Contributor Information
Julia C Haas, Umeå Plant Science Centre, Department of Plant Physiology, Umeå University, SE-901 87 Umeå, Sweden.
Alexander Vergara, Umeå Plant Science Centre, Department of Forest Genetics and Plant Physiology, Swedish University of Agricultural Sciences (SLU), SE-901 83 Umeå, Sweden.
Alonso R Serrano, Umeå Plant Science Centre, Department of Forest Genetics and Plant Physiology, Swedish University of Agricultural Sciences (SLU), SE-901 83 Umeå, Sweden.
Sanatkumar Mishra, Umeå Plant Science Centre, Department of Forest Genetics and Plant Physiology, Swedish University of Agricultural Sciences (SLU), SE-901 83 Umeå, Sweden.
Vaughan Hurry, Umeå Plant Science Centre, Department of Forest Genetics and Plant Physiology, Swedish University of Agricultural Sciences (SLU), SE-901 83 Umeå, Sweden.
Nathaniel R Street, Umeå Plant Science Centre, Department of Plant Physiology, Umeå University, SE-901 87 Umeå, Sweden.
Funding
This work was supported by funding from the HolmenSkog AB and Berzelii Centre for Forest Biotechnology to V.H. and from the Swedish University of Agricultural Science’s Trees and Crops for the Future (TC4F) program to V.H. and N.R.S.
Conflict of interest
None declared.
References
- Abe H, Urao T, Ito T, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2003) Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 15:63–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal M, Hao YJ, Kapoor A, Dong CH, Fujii H, Zheng XW, Zhu JK (2006) A R2R3 type MYB transcription factor is involved in the cold regulation of CBF genes and in acquired freezing tolerance. J Biol Chem 281:37636–37645. [DOI] [PubMed] [Google Scholar]
- Allen CD, Macalady AK, Chenchouni H et al. (2010) A global overview of drought and heat-induced tree mortality reveals emerging climate change risks for forests. For Ecol Manage 259:660–684. [Google Scholar]
- Alter S, Bader KC, Spannagl M, Wang Y, Bauer E, Schon CC, Mayer KF (2015) DroughtDB: an expert-curated compilation of plant drought stress genes and their homologs in nine species. Database (Oxford) 2015:bav046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Pyl PT, Huber W (2015) HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics 31:166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA et al. (2000) Gene Ontology: tool for the unification of biology. Nat Genet 25:25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldoni E, Genga A, Cominelli E (2015) Plant MYB transcription factors: their role in drought response mechanisms. Int J Mol Sci 16:15811–15851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck PSA, Juday GP, Alix C, Barber VA, Winslow SE, Sousa EE, Heiser P, Herriges JD, Goetz SJ (2011) Changes in forest productivity across Alaska consistent with biome shift. Ecol Lett 14:373–379. [DOI] [PubMed] [Google Scholar]
- Beike AK, Lang D, Zimmer AD, Wust F, Trautmann D, Wiedemann G, Beyer P, Decker EL, Reski R (2015) Insights from the cold transcriptome of Physcomitrella patens: global specialization pattern of conserved transcriptional regulators and identification of orphan genes involved in cold acclimation. New Phytol 205:869–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Børja I, de Wit HA, Steffenrem A, Majdi H (2008) Stand age and fine root biomass, distribution and morphology in a Norway spruce chronosequence in southeast Norway. Tree Physiol 28:773–784. [DOI] [PubMed] [Google Scholar]
- Brodribb TJ, McAdam SAM (2013) Abscisic acid mediates a divergence in the drought response of two conifers. Plant Physiol 162:1370–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodribb TJ, McAdam SAM, Jordan GJ, Martins SCV (2014) Conifer species adapt to low-rainfall climates by following one of two divergent pathways. Proc Natl Acad Sci USA 111:14489–14493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton PJ, Bergeron Y, Bogdanski BEC et al. (2010) Sustainability of boreal forests and forestry in a changing environment. In: Mery G, Katila P, Galloway G, Alfaro RI, Kanninen M, Lobovikov M, Varjo J (eds) Forests and society - responding to global drivers of change. International Union of Forest Research Organizations (IUFRO), Vienna, Austria, IUFRO World Series, pp 249–282. [Google Scholar]
- Castillo-Davis CI, Hartl DL (2003) GeneMerge--post-genomic analysis, data mining, and hypothesis testing. Bioinformatics 19:891–892. [DOI] [PubMed] [Google Scholar]
- Chang S, Puryear J, Cairney J (1993) A simple and efficient method for isolating RNA from pine trees. Plant Mol Biol Rep 11:113–116. [Google Scholar]
- Chawade A, Bräutigam M, Lindlöf A, Olsson O, Olsson B (2007) Putative cold acclimation pathways in Arabidopsis thaliana identified by a combined analysis of mRNA co-expression patterns, promoter motifs and transcription factors. BMC Genomics 8:304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JH, Jiang HW, Hsieh EJ, Chen HY, Chien CT, Hsieh HL, Lin TP (2012) Drought and salt stress tolerance of an Arabidopsis glutathione S-transferase U17 knockout mutant are attributed to the combined effect of glutathione and abscisic acid. Plant Physiol 158:340–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng M-C, Hsieh E-J, Chen J-H, Chen H-Y, Lin T-P (2012). Arabidopsis RGLG2, Functioning as a RING E3 Ligase, Interacts with AtERF53 and Negatively Regulates the Plant Drought Stress Response. Plant Physiology 158 (1) 363-375; DOI: 10.1104/pp.111.189738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen D, Bogeat-Triboulot MB, Tisserant E et al. (2010) Comparative transcriptomics of drought responses in Populus: a meta-analysis of genome-wide expression profiling in mature leaves and root apices across two genotypes. BMC Genom 11:630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comas LH, Becker SR, Cruz VV, Byrne PF, Dierig DA (2013) Root traits contributing to plant productivity under drought. Front Plant Sci 4:442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coscia M, Neffke F (2017) Network backboning with noisy data. 425–436.
- Delhomme N, Mähler N, Schiffthaler B, Sundell D, Mannapperuma C, Hvidsten TR, Street NR (2014) Guidelines for RNA-Seq data analysis. In: EpyGeneSys Protocol (prot 67). https://http://www.epigenesys.eu/en/protocols/bio-informatics/1283-guidelines-for-rna-seq-data-analysis, 15 September 2018, date last accessed).
- Ditmarova L, Kurjak D, Palmroth S, Kmet J, Strelcova K (2010) Physiological responses of Norway spruce (Picea abies) seedlings to drought stress. Tree Physiol 30:205–213. [DOI] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR (2013) STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du M, Ding G, Cai Q (2018) The transcriptomic responses of Pinus massoniana to drought stress. Forests 9:326. [Google Scholar]
- Edwards D, Kerp H, Hass H (1998) Stomata in early land plants: an anatomical and ecophysiological approach. J Exp Bot 49:255–278. [Google Scholar]
- Egert A, Keller F, Peters S (2013) Abiotic stress-induced accumulation of raffinose in Arabidopsis leaves is mediated by a single raffinose synthase (RS5, At5g40390). BMC Plant Biol 13:218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FAO (2010) Global Forest Resources Assessment 2010. FAO, Food and Agriculture Organization of the United Nations, Rome. [Google Scholar]
- Fox H, Doron-Faigenboim A, Kelly G, Bourstein R, Attia Z, Zhou J, Moshe Y, Moshelion M, David-Schwartz R (2018) Transcriptome analysis of Pinus halepensis under drought stress and during recovery. Tree Physiol 38:423–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge ZM, Kellomäki S, Peltola H, Zhou X, Wang KY, Väisänen H (2011) Impacts of changing climate on the productivity of Norway spruce dominant stands with a mixture of Scots pine and birch in relation to water availability in southern and northern Finland. Tree Physiol 31:323–338. [DOI] [PubMed] [Google Scholar]
- Gu Z, Eils R, Schlesner M (2016) Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 32:2847–2849. [DOI] [PubMed] [Google Scholar]
- Harfouche A, Meilan R, Altman A (2014) Molecular and physiological responses to abiotic stress in forest trees and their relevance to tree improvement. Tree Physiol 34:1181–1198. [DOI] [PubMed] [Google Scholar]
- Hothorn T, Bretz F, Westfall P (2008) Simultaneous inference in general parametric models. Biom J 50:346–363. [DOI] [PubMed] [Google Scholar]
- IPCC (2013) Climate change 2013: the physical science basis. In: Stocker TF, Qin D, Plattner G-K et al. (eds) Contribution of working group I to the fifth assessment report of the intergovernmental panel on climate change. Cambridge University Press, Cambridge, UK and New York, NY, USA, p 1535. [Google Scholar]
- Iuchi S, Kobayashi M, Taji T et al. (2001) Regulation of drought tolerance by gene manipulation of 9-cis-epoxycarotenoid dioxygenase, a key enzyme in abscisic acid biosynthesis in Arabidopsis. Plant J 27:325–333. [DOI] [PubMed] [Google Scholar]
- Ivanov YV, Kartashov AV, Zlobin IE, Sarvin B, Stavrianidi AN, Kuznetsov VV (2019) Water deficit-dependent changes in non-structural carbohydrate profiles, growth and mortality of pine and spruce seedlings in hydroculture. Environ Exp Bot 157:151–160. [Google Scholar]
- Jin J, Zhang H, Kong L, Gao G, Luo J (2014) PlantTFDB 3.0: a portal for the functional and evolutionary study of plant transcription factors. Nucleic Acids Res 42:D1182–D1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi V, Jander G (2009) Arabidopsis methionine gamma-lyase is regulated according to isoleucine biosynthesis needs but plays a subordinate role to threonine deaminase. Plant Physiol 151:367–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalliokoski T (2011) Root system traits of Norway spruce, Scots pine, and silver birch in mixed boreal forests: an analysis of root architecture, morphology, and anatomy Dissertationes Forestales 121. PhD thesis,. Faculty of Agriculture and Forestry, University of Helsinki, Helsinki, p 67. [Google Scholar]
- Kasuga M, Miura S, Shinozaki K, Yamaguchi-Shinozaki K (2004) A combination of the Arabidopsis DREB1A gene and stress-inducible rd29A promoter improved drought- and low-temperature stress tolerance in tobacco by gene transfer. Plant Cell Physiol 45:346–350. [DOI] [PubMed] [Google Scholar]
- Kauppi PE, Posch M, Pirinen P (2014) Large impacts of climatic warming on growth of boreal forests since 1960. PloS One 9:e111340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellomäki S, Peltola H, Nuutinen T, Korhonen KT, Strandman H (2008) Sensitivity of managed boreal forests in Finland to climate change, with implications for adaptive management. Philos Trans R Soc Lond B Biol Sci 363:2341–2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirschbaum M, Fischlin A (1996) Climate change impacts on forests. In: Watson R, Zinyowera MC, Moss RH (eds) Climate change 1995: impacts, adaptations and mitigation of climate change. Scientific-technical analysis. Contribution of working group II to the second assessment report of the intergovernmental panel of climate change. Cambridge University Press, Cambridge, UK and New York, NY, USA, pp 95–129. [Google Scholar]
- Klápště J, Lecoy J, del Rosario García-Gil M (2020) Drought stress adaptation in Norway spruce and related genomics work. In: Porth IM, De, Torre AR (eds) The spruce genome. Springer International Publishing, Cham, pp 129–153. [Google Scholar]
- Kneeshaw D, Bergeron Y, Kuuluvainen T (2011) Forest ecosystem structure and disturbance dynamics across the circumboreal forest. In: Millington M, Blumler M, Schickhoff U (eds) The SAGE handbook of biogeography. SAGE Publications Ltd., London, pp 261–278. [Google Scholar]
- Kopylova E, Noe L, Touzet H (2012) SortMeRNA: fast and accurate filtering of ribosomal RNAs in metatranscriptomic data. Bioinformatics 28:3211–3217. [DOI] [PubMed] [Google Scholar]
- Lapenis A, Shvidenko A, Shepaschenko D, Nilsson S, Aiyyer A (2005) Acclimation of Russian forests to recent changes in climate. Glob Chang Biol 11:2090–2102. [DOI] [PubMed] [Google Scholar]
- Liu SX, Wang XL, Wang HW et al. (2013) Genome-wide analysis of ZmDREB genes and their association with natural variation in drought tolerance at seedling stage of Zea mays L. PLoS Genet 9:e1003790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz WW, Alba R, Yu YS, Bordeaux JM, Simões M, Dean JFD (2011) Microarray analysis and scale-free gene networks identify candidate regulators in drought-stressed roots of loblolly pine (P. taeda L.). BMC Genomics 12:264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay DS, Savoy PR, Grossiord C, Tai X, Pleban JR, Wang DR, McDowell NG, Adams HD, Sperry JS (2020) Conifers depend on established roots during drought: results from a coupled model of carbon allocation and hydraulics. New Phytol 225:679–692. [DOI] [PubMed] [Google Scholar]
- de María N, Guevara MÁ, Perdiguero P et al. (2020) Molecular study of drought response in the Mediterranean conifer Pinus pinaster Ait.: differential transcriptomic profiling reveals constitutive water deficit-independent drought tolerance mechanisms. Ecol Evol 10:9788–9807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDowell N, Pockman WT, Allen CD et al. (2008) Mechanisms of plant survival and mortality during drought: why do some plants survive while others succumb to drought? New Phytol 178:719–739. [DOI] [PubMed] [Google Scholar]
- Mitchell PJ, McAdam SA, Pinkard EA, Brodribb TJ (2017) Significant contribution from foliage-derived ABA in regulating gas exchange in Pinus radiata. Tree Physiol 37:236–245. [DOI] [PubMed] [Google Scholar]
- Moran E, Lauder J, Musser C, Stathos A, Shu M (2017) The genetics of drought tolerance in conifers. New Phytol 216:1034–1048. [DOI] [PubMed] [Google Scholar]
- Nakashima K, Ito Y, Yamaguchi-Shinozaki K (2009) Transcriptional regulatory networks in response to abiotic stresses in Arabidopsis and grasses. Plant Physiol 149:88–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima K, Takasaki H, Mizoi J, Shinozaki K, Yamaguchi-Shinozaki K (2012) NAC transcription factors in plant abiotic stress responses. Biochim Biophys Acta Gene Regul Mech 1819:97–103. [DOI] [PubMed] [Google Scholar]
- Nishizawa A, Yabuta Y, Shigeoka S (2008) Galactinol and raffinose constitute a novel function to protect plants from oxidative damage. Plant Physiol 147:1251–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nystedt B, Street NR, Wetterbom A et al. (2013) The Norway spruce genome sequence and conifer genome evolution. Nature 497:579–584. [DOI] [PubMed] [Google Scholar]
- Pan YD, Birdsey RA, Fang JY et al. (2011) A large and persistent carbon sink in the world’s forests. Science 333:988–993. [DOI] [PubMed] [Google Scholar]
- Pashkovskiy PP, Vankova R, Zlobin IE, Dobrev P, Ivanov YV, Kartashov AV, Kuznetsov VV (2019) Comparative analysis of abscisic acid levels and expression of abscisic acid-related genes in Scots pine and Norway spruce seedlings under water deficit. Plant Physiol Biochem 140:105–112. [DOI] [PubMed] [Google Scholar]
- Penfield S, Clements S, Bailey KJ, Gilday AD, Leegood RC, Gray JE, Graham IA (2012) Expression and manipulation of PHOSPHOENOLPYRUVATE CARBOXYKINASE 1 identifies a role for malate metabolism in stomatal closure. Plant J 69:679–688. [DOI] [PubMed] [Google Scholar]
- Peng CH, Ma ZH, Lei XD et al. (2011) A drought-induced pervasive increase in tree mortality across Canada’s boreal forests. Nat Clim Change 1:467–471. [Google Scholar]
- Pinheiro J, Bates D, DebRoy S, Sarkar D; R Core (2018) nlme: linear and nonlinear mixed effects models. R package version 3.1-137.
- Proost S, Van Bel M, Vaneechoutte D, Van de Peer Y, Inze D, Mueller-Roeber B, Vandepoele K (2015) PLAZA 3.0: an access point for plant comparative genomics. Nucleic Acids Res 43:D974–D981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team (2018) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- Rasheed S, Bashir K, Matsui A, Tanaka M, Seki M (2016) Transcriptomic analysis of soil-grown Arabidopsis thaliana roots and shoots in response to a drought stress. Front Plant Sci 7:180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raven JA (1977) The evolution of vascular land plants to supracellular transport processes. In: Woolhouse HW (ed) Advances in botanical research. Elsevier, Academic Press, London, pp 153–219. [Google Scholar]
- Roelfsema MRG, Hedrich R (2005) In the light of stomatal opening: new insights into ‘the Watergate’. New Phytol 167:665–691. [DOI] [PubMed] [Google Scholar]
- Roman DT, Novick KA, Brzostek ER, Dragoni D, Rahman F, Phillips RP (2015) The role of isohydric and anisohydric species in determining ecosystem-scale response to severe drought. Oecologia 179:641–654. [DOI] [PubMed] [Google Scholar]
- Rosvall M, Bergstrom CT (2008) Maps of random walks on complex networks reveal community structure. Proc Natl Acad Sci USA 105:1118–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rueden CT, Schindelin J, Hiner MC, DeZonia BE, Walter AE, Arena ET, Eliceiri KW (2017) ImageJ2: ImageJ for the next generation of scientific image data. BMC Bioinformatics 18:529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sade N, Gebremedhin A, Moshelion M (2012) Risk-taking plants: anisohydric behavior as a stress-resistance trait. Plant Signal Behav 7:767–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schachtman DP, Goodger JQD (2008) Chemical root to shoot signaling under drought. Trends Plant Sci 13:281–287. [DOI] [PubMed] [Google Scholar]
- Schiffthaler B, Serrano A, Delhomme N, Street NR (2018) Seidr: a toolkit for calculation of crowd networks. bioRxiv:250696 . [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW (2012) NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9:671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder JI, Allen GJ, Hugouvieux V, Kwak JM, Waner D (2001) Guard cell signal transduction. Annu Rev Plant Physiol Plant Mol Biol 52:627–658. [DOI] [PubMed] [Google Scholar]
- Schulze E-D, Hall AE (1982) Stomatal responses, water loss and CO2 assimilation rates of plants in contrasting environments. In: Pirson A, Zimmermann MH (eds) Encyclopedia of plant physiology, new series. Springer-Verlag, Berlin, pp 181–230. [Google Scholar]
- Seiler JR, Cazell BH (1990) Influence of water-stress on the physiology and growth of red spruce seedlings. Tree Physiol 6:69–77. [DOI] [PubMed] [Google Scholar]
- Seo PJ, Xiang FN, Qiao M, Park JY, Lee YN, Kim SG, Lee YH, Park WJ, Park CM (2009) The MYB96 transcription factor mediates abscisic acid signaling during drought stress response in Arabidopsis. Plant Physiol 151:275–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinozaki K, Yamaguchi-Shinozaki K (2007) Gene networks involved in drought stress response and tolerance. J Exp Bot 58:221–227. [DOI] [PubMed] [Google Scholar]
- Shorohova E, Kneeshaw D, Kuuluvainen T, Gauthier S (2011) Variability and dynamics of old-growth forests in the circumboreal zone: implications for conservation, restoration and management. Silva Fenn 45:785–806. [Google Scholar]
- Singh D, Laxmi A (2015) Transcriptional regulation of drought response: a tortuous network of transcriptional factors. Front Plant Sci 6:895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Street NR, Skogström O, Sjödin A, Tucker J, Rodríguez-Acosta M, Nilsson P, Jansson S, Taylor G (2006) The genetics and genomics of the drought response in Populus. Plant J 48:321–341. [DOI] [PubMed] [Google Scholar]
- Sundell D, Mannapperuma C, Netotea S et al. (2015) The plant genome integrative explorer resource: PlantGenIE.Org. New Phytol 208:1149–1156. [DOI] [PubMed] [Google Scholar]
- Tabaeizadeh Z (1998) Drought-induced responses in plant cells. In: Jeon KW (ed) International Review of Cytology Volume 182, Pages 193-247 10.1016/S0074-7696(08)62170-1. [DOI] [PubMed]
- Taji T, Ohsumi C, Iuchi S, Seki M, Kasuga M, Kobayashi M, Yamaguchi-Shinozaki K, Shinozaki K (2002) Important roles of drought- and cold-inducible genes for galactinol synthase in stress tolerance in Arabidopsis thaliana. Plant J 29:417–426. [DOI] [PubMed] [Google Scholar]
- Thimm O, Bläsing O, Gibon Y et al. (2004) MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J 37:914–939. [DOI] [PubMed] [Google Scholar]
- Tran LSP, Nakashima K, Sakuma Y et al. (2004) Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell 16:2481–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troeng E, Linder S (1982) Gas-exchange in a 20-year-old stand of Scots pine. 1. Net photosynthesis of current and one-year-old shoots within and between seasons. Physiol Plant 54:7–14. [Google Scholar]
- Trujillo-Moya C, George JP, Fluch S et al. (2018) Drought sensitivity of Norway spruce at the species’ warmest fringe: quantitative and molecular analysis reveals high genetic variation among and within provenances. G3 (Bethesda) 8:1225–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson S, Davies WJ (2002) ABA-based chemical signalling: the co-ordination of responses to stress in plants. Plant Cell Environ 25:195–210. [DOI] [PubMed] [Google Scholar]
- Xu J, Tian YS, Xing XJ, Peng RH, Zhu B, Gao JJ, Yao QH (2016) Over-expression of AtGSTU19 provides tolerance to salt, drought and methyl viologen stresses in Arabidopsis. Physiol Plant 156:164–175. [DOI] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K (2006) Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu Rev Plant Biol 57:781–803. [DOI] [PubMed] [Google Scholar]
- Yan T, Yoo D, Berardini TZ, Mueller LA, Weems DC, Weng S, Cherry JM, Rhee SY (2005) PatMatch: a program for finding patterns in peptide and nucleotide sequences. Nucleic Acids Res 33:W262–W266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang A, Dai XY, Zhang WH (2012) A R2R3-type MYB gene, OsMYB2, is involved in salt, cold, and dehydration tolerance in rice. J Exp Bot 63:2541–2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Kimball JS, Hogg EH, Zhao M, Oechel WC, Cassano JJ, Running SW (2008) Satellite-based model detection of recent climate-driven changes in northern high-latitude vegetation productivity. J Geophys Res Biogeosci 113:G03033. [Google Scholar]
- Zhong R, Allen JD, Xiao G, Xie Y (2014) Ensemble-based network aggregation improves the accuracy of gene network reconstruction. PLoS One. 9:e106319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann MH, Brown CL (1971) Trees - structure and function. Springer-Verlag, New York, NY. [Google Scholar]
- Zlobin IE, Ivanov YV, Kartashov AV, Kuznetsov VV (2018) Impact of drought stress induced by polyethylene glycol on growth, water relations and cell viability of Norway spruce seedlings. Environ Sci Pollut Res Int 25:8951–8962. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


