Abstract
Background:
Canine babesiosis is one of the mainly worldwide-distributed tick-borne haemoprotozoan parasitic diseases in dogs.
Methods:
A total of 43 blood samples were randomly collected from naturally infected dogs in seven villages from different geographical areas of Meshkin Shahr, Ardabil Province, Iran. The presence of Babesia species detected with standard methods including parasitological and gene sequencing techniques targeting the 18S rRNA gene.
Results:
Our results revealed that four dogs 9.3% (4/43) including one female and three male dogs were infected with Babesia. All four Babesia-infected dogs were confirmed B. canis by the molecular-based method. Sequence alignments comparison of the B. canis genotypes A and B, it was revealed that all B. canis isolates belonged to genotype B.
Conclusion:
This study provides essential data for subsequently define the critical importance of the molecular studies in management and prevention of the canine babesiosis in Iran.
Keywords: Babesia canis, Babesiosis, Dogs, Genotyping, RNA, Iran
Introduction
Canine babesiosis is a tick-borne parasitic disease with worldwide importance and caused by intra-erythrocytic Babesia species. The identification of each Babesia species routinely is based on the host specificity and the morphological characteristics of piroplasmids (1). The differences in geographical distribution, vector specificity, antigenic properties, genetic characteristics and severity of the clinical manifestations sub divided the former species into the three subspecies, namely B. canis canis (3–5μm) is transmitted by Dermacentor reticulatus in Europe, B. canis vogeli transmitted by Rhipicephalus sanguineus sensu lato in tropical and sub-tropical regions, and B. canis rossi transmitted by Haemaphysalis leachi in South Africa (1, 2).
Babesia gibsoni (1.5–2.5μm) is present in Asia, North America, Africa, Australia and Europe (3-6). The geographical distributions of both species of D. marginatus and D. reticulatus in Europe range from Portugal to Ukraine (continue to the east of Kazakhstan), Turkey and probably to the northern parts of Iran (7–10). The first report of Dermacentor ticks in Iran was documented in 1971 by Mazlum (11). This study performed among the 30 provinces and the results defined that, Dermacentor ticks were found only in six provinces (Semnan, Khorasan, Kurdistan, Ardabil, East Azerbaijan, and Zanjan) with the highest rate of distribution in Ardabil in which ticks was found to be restricted to four species; D. niveus, D. marginatus, D. raskemensis and D. daghestanicus (12–17). In a study 200 adult Dermacentor ticks (139 D. niveus and 61 D. marginatus) were collected from a sheep babesiosis infected in the Ardabil region of Iran, B. ovis was detected in ticks by semi-nested PCR. Based on the results obtained D. niveus and D. marginatus, which are distributed in Ardabil region of Iran, might play a crucial role in the transmission of Babesia infection to domestic animals (12). To date, there are no reports of D. reticulatus in Iran. Dermacentor ticks usually occurred in the mountainous area with cold climatic conditions and high altitude (15). A cross sectional study in southeastern Iran, among tick-infested dogs, three dogs were infected with B. gibsoni. In this study, ticks were identified and belonged to R. sanguineus sensu lato and no Babesia DNA was detected. This study first record of B. gibsoni in dogs in Iran (18). It seems that B. canis and B. gibsoni were the major species infecting dogs and causing various clinical symptoms in Iran (19–21). Based on the morphological features of the infection, it has documented that tick species including R. bursa, R. sanguineus, R. turanicus and D. marginatus have distributed in different regions of Iran. R. bursa and R. sanguineus may play the major roles as the vector of the parasite respectively in the case of animal babesiosis in Iran (13). The babesiosis infection was detected with molecular and serological methods in dogs and other wild canine (22–27). The molecular-based techniques enable differentiation of morphologically undistinguishable Babesia species (28). The molecular diagnostic methods (for example PCR) are cost-effective with high sensitivity and specificity and the most reliable techniques for Babesia DNA detection in blood (29–31). Easy application and accessibility of databases and growing amount of annotated genomic sequences in databanks caused an improvement in the phylogenetic studies on B. canis (31). Limited information on the canine babesiosis has been documented, while a high number of suspicious clinical cases reported in dogs from Iran. This study provides essential and valuable data to insight into the prevalence and distribution of canine babesiosis in Iran. Thereby, regarding to raise of knowledge on this parasite, the detection and characterization of the Babesia species and subspecies from canine babesiosis in Meshkin shahr has a great importance through application of PCR and sequencing of 18S rRNA gene sequences.
Materials and Methods
Study area, blood and spleen samples
From July 2017 to February 2018, a total of 43 blood samples were randomly collected from shepherd dogs (Canis familiaris) (32 males and 11 females, 9 months to 7 years old) in Meshkin Shahr, Ardabil Province, Northwest of Iran. Blood samples were collected into 0.001M EDTA-containing tubes, and transported in iceboxes to the laboratory of protozoology, faculty of medicine, Iran University of medical sciences. Blood samples aliquoted, smears were prepared from EDTA-sampled, blood air-dried, and stained with Giemsa. Genomic DNA was extracted from each blood samples using a DNA extraction kit (Qiagen DNA Blood Mini-Kit, Germany). All samples were identified and followed the detection process using PCR.
Molecular analysis and characterization of the isolated Babesia species
DNA was extracted from whole blood samples using the DNA extraction kit from blood (Qiagen, Hilden, Germany) through following manufacturer instructions as previously described (22, 31). To detect Babesia species, the gene fragment (∼550bp) from 18S rRNA was amplified and sequenced using the primers BAB GF2 (5′-GYYTTGTAATTGGAATGA TGG-3′) and BAB GR2 (5′- CCAAAGAC TTTGA TTTCTCTC-3′). All stages were performed using the previously described PCR protocol (23). Generally, reactions were performed in a total of 25μl, including 2.5μl of 10X PCR buffer, 2.0μl of dNTP (2.5mM each), 1.25U of Taq DNA polymerase (SinaColon Co. Iran) 1.0μl of template DNA, 1.0μl of each primer (10pmol), and 16.25μl of double distilled water (Sina Colon Co. Iran). The PCR reaction was 95 °C (3min), [95 °C (30s), 55 °C (30s), 72 °C (90s)]× 35 cycles, 72 °C (5min). In the second round of PCR 418bp of DNA fragments were generated using another pair of primers, PIRO-nest F (5′-GGATAACCGTGST AATTSTAGGGC-3′) and PIRO-nest R (5′-GTGTGTACAAAGGG CAGGGACG-3′) (4). The amplified PCR products were maintained at −20 °C until analyzed. The products were run on electrophoresis in a 1.5% agarose gel containing 0.2μg of safe stain/ ml in Trisacetate-EDTA buffer at 120V for 30 min and consequently transilluminated under UV light.
Ethical approve
This study was approved by admission with the ethics procedures and guidelines of the respective national the animal ethics use committees of research issued by the council of the Iran University of Medical Sciences (IR. IUMS.REC. 27899.).
Sequences analysis
Sequences subjected to online BLAST algorithm and were compared with previously registered sequences in the GenBank database. To confirm the classification of the parasite, large fragments of the 18S rRNA gene were amplified from each sample that was positive for Babesia. The18S rRNA genes sequences were analyzed by standard technique using a sequencer and BioEdit software (Perkin-Elmer, USA) (32). Analysis of DNA sequences and phylogenetic relationships for B. canis isolates and the group of isolates from dogs were aligned using Clustal W software (33). A phylogenetic tree was created using alignments performed with neighbor joining (NJ) phylogenetic tree using Kimura-2-Parameter algorithm with bootstrap as the tree construction method (34). Furthermore, phylogenetic analysis of gene sequences were performed with maximum likelihood method with MEGA 7.0 software. The representative sequence was annotated in the GenBank database with accession number MN173220, MN173221, MN173222 and MN 173223 (Table 1). To assess B. canis genotypes (4, 35), the obtained sequences were compared with the members from genotype A (AY 703072) and genotype B (AY649326).
Table 1.
Principle information on the animals sampled and the Babesia species isolated
| Samples | Pathogen | Clinical symptoms | Accession number (s) | Blood analysis* | Diagnostic investigation | Location and Coordinates | |||
|---|---|---|---|---|---|---|---|---|---|
| Breed | ID of animal | Age | Sex | ||||||
| Mongrel | Mesh 1 | 1 year | Female | B. canis | Fever, splenomegaly | MN173220 | RBC: 4.3 HGB: 99 HCT: 27.6% Direct Bilirubin: 1.08 |
PCR: positive Blood smear: positive Tissue: positive |
Ag bolagh 38°20′57″N 47°39′57″E |
| Kurd Mastiff (Pshdar) | Mesh 2 | 3 years | Male | B. canis | Fever, vomitin, lethargy | MN173221 | RBC: 4.8 HGB: 90 HCT: 26.1% Direct Bilirubin: 1.01 |
PCR: positive Blood smear: positive Tissue: positive |
Parikhan 38°24′51″N 47°38′38″E |
| Anato-lian Karabas | Mesh 3 | 4 years | Male | B. canis | Fever, icter, splenomegaly | MN173222 | RBC: 4.3 HGB: 91 HCT: 29.1% Direct Bilirubin: 1.15 |
PCR: positive Blood smear: Not tested Tissue: positive |
Qurt tappeh 38°25′43″N 47°37′29″E |
| Mongrel | Mesh 4 | 14 months | Male | B. canis | Cough, splenomegaly | MN173223 | RBC: 4.1 HGB: 97 HCT: 26.6% Direct Bilirubin: 1.12 |
PCR: positive Blood smear: positive Tissue: positive |
Qara darvish 38°56′43″N 47°28′48″E |
Normal ranges: Red blood cell (RBC) count, 4.6–10×109/L; Hemoglobin concentration (HGB), 93–153g/L; Haematocrit (HCT), 28–49%; Direct bilirubin, 0.15±0.01
Results
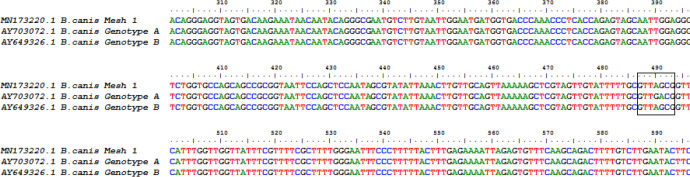
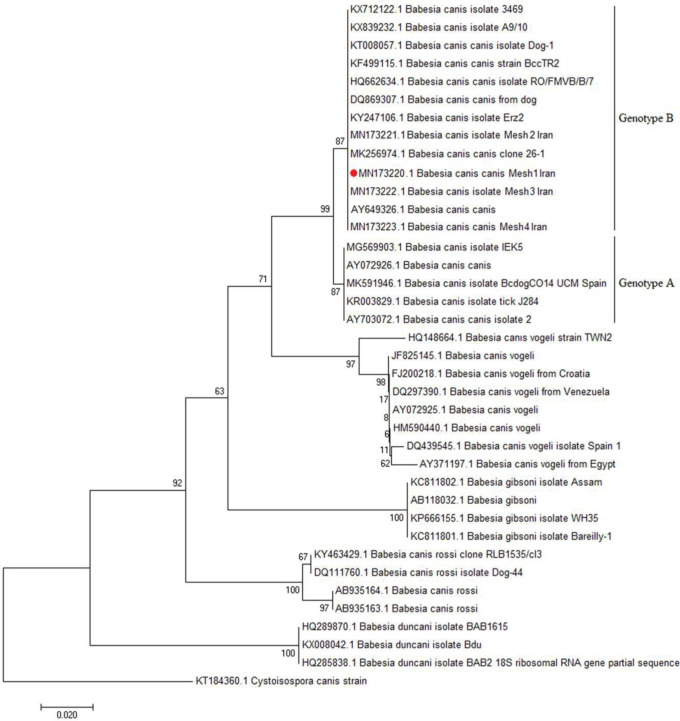
In the direct microscopic diagnostic investigation of blood smear and molecular study of blood samples revealed that four dogs 9.3% (4/43) including one female and three male dogs were infected with B. canis (Fig 1). In the clinical examinations, all four dogs had major symptoms of babesiosis and most of the infected dogs had fever and splenomegaly. In addition, blood parameters including hemoglobin concentration, haematocrit, RBC count, and direct bilirubin had increased (Table 1). Out of the total samples subjected to PCR, four dogs was Babesia-positive including one female and three males. DNA was purified from all blood samples of the collected dogs and used as the PCR template, which a 550bp band was observed in the analysis. The results of sequence analysis were the same as with the other previously annotated sequences. The nucleotide sequences from canine samples were identical to each other and had shown a 99.6–100% identity with B. canis derived from dogs in Gen-Bank reference sequences originated from different countries such as Turkey (KY247106 and KF499115), China (MK256974), Slovakia (DQ869307), Estonia (KT008057), Romania (HQ662634), and Croatia (AY072926). The comparison of B. canis nucleotide sequences obtained in this study with genotypes A and B revealed that all our isolates were classified as genotype B and the main difference was observed in positions 490 and 491. The difference of the two genotypes in the row of adenine and guanine nucleotides are in genotype B as AG and GA in genotype A (Fig. 2). The results of phylogenetic analysis revealed that the 18S rRNA gene sequences obtained in this study matched with B. canis and alignments showed that all B. canis isolates belonged in the category of genotype B (Fig. 3). In present study, the common ancestor of genotype A and B was obtained with confidence level 99%. Genetic confidence intervals can help to better understand genealogical relationships to DNA matches.
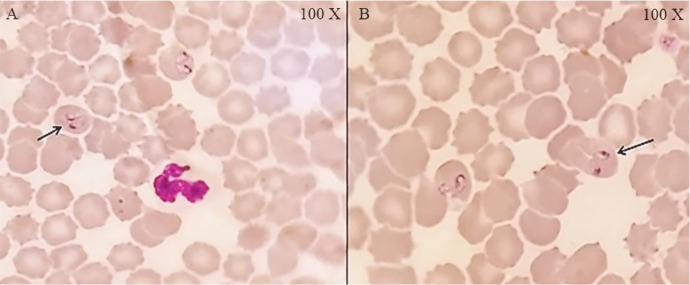
Fig. 1.
Direct microscopic detection of Babesia canis in the blood of naturally infected dogs. Field-Giemsa stained thin smears showing various forms of B. canis in erythrocytes. A: Closed angle pyri form bodies of B. canis and B: Wide angles B. canis near the margin of the infected RBCs
Fig. 2.
Multiple sequence alignment of the partial 18S rRNA gene and the sequences of genotype A and B. In this position nucleotide changes can be seen
Fig. 3.
Neighbor-joining analysis of canine Babesia sequences obtained from samples submitted. A 550bp fragment of the 18S rRNA was aligned with representative sequences derived from GenBank. Bootstrap values (1000 replications) are shown in the phylogenetic tree. Comparison of the B. canis sequences obtained in this study with genotypes A and B. Samples sequenced in the present study are marked with red cycle (MN173220- MN173223). The tree was inferred using the neighbor joining method of MEGA7, bootstrap values are shown at each branch point
Discussion
In this study due to the most availability of 18S rDNA sequences from B. canis in the Gen-Bank, the 18S rDNA was used to search for the intraspecific variability and the most available abundant B. canis sequence. Out of the 43 samples subjected to PCR, 9.3% (4/43) were found to be positive for Babesia infection. This is the first study of a molecular detection and identification of B. canis infection in dogs from Iran and our results revealed that B. canis was prevalent in Meshkin Shahr, Iran. On the basis of 18S rRNA gene sequence analysis, genetic heterogeneity of B. canis has been reported in Poland, Croatia, Estonia, Lithuania, Hungary and china (4, 22, 35). Two genotypes of B. canis, includes A and B, have been documented so far, and have shown to have variable virulences (4, 36). The results provided principle information toward a better understanding of the epidemiology of canine babesiosis in Iran and prepared the situation for implementation of an effective control planning on babesiosis. A variable interspecies pathogenicity of the B. canis genotypes stated by previous studies (1, 4, 25, 28). The clinical manifestations of B. canis infection are mild to acute, and the severity of disease has a significant relationship with the species of Babesia causing infection (4). There are few reports and studies on Babesia spp. in Iran, while a widespread distribution of the parasite vector and suitable weather condition were observed in some areas of Iran (18, 37). Therefore, there is the probability canine babesiosis and establishment of an infection chain in some geographic areas of Iran. Niak et al. (1973) studied the blood parasites of 155 dogs and one fox (Vulpes vulpes) in the north of Iran, B. canis was just found in one splenectomized dog and B. gibsoni was found from fox (21). Jalali et al. (2013) applied a PCR method in the study and documented that the prevalence of canine babesiosis was 0.36% (20). In another study, Akhtardanesh et al. (2016) detected 60 tick-infested anemic dogs, among which three dogs (5%) were positive through using a genus-specific PCR and all infected with B. gibsoni. None of the collected ticks was positive at the Babesia specific PCR (18). In the collected blood samples of dogs from seven regions of Shiraz in south of Iran, only one positive sample was infected with B. canis (19). The results provided useful data on the distribution of B. canis genotypes in dogs from Iran, and showed the necessity to use a molecular-based analysis for an accurate diagnosis of canine babesiosis. The PCR-based analysis demonstrated that the molecular techniques can a highly sensitive easy to use and cost-effective tools for the simultaneous detection and differentiation of B. canis genotypes. However, since a limited number of target gene sequences are currently available for molecular detection of this parasite, any consideration on the population genetics of Babesia in the study areas would be highly scrupulously (38). Genetic intraspecific variability is a vital mechanism for piroplasm parasite survival in hosts (39). It is proven that B. canis transmitted by D. reticulatus, and the distribution area of the parasite is directly related to the presence of this tick species. Although B. canis has been observed in dogs in Iran (19–21, 37), But so far we have no reports of D. reticulatus ticks in Iran. These results demonstrate that probably B. canis and D. reticulatus have infested a dog’s population, at least in the northern part of Iran. Dermacentor reticulatus, not occurring in the Mediterranean climatic zone, is a tick of some cool regions generally in wooded areas. This ticks has a wide spread geographical overlap with D. marginatus. Preferred habitats are forests and swamps zones where it can survive for long periods (8). The main clinic pathological sings in Babesia infections were a moderate to acute disease haemoglobinuria and a mild to very severe normochromic normocytic haemolytic anemia but, the symptoms of the disease are classified based on clinical sings and severity of the infection (40).
In the present study, the main clinical signs in Babesia infected dogs were fever, splenomegaly, vomiting, cough. Haemoglobinuria and haemolytic anemia was not seen, which may be due to host immune system status, age and stages of infection. Based on clinical signs and mortality rates, genotype B is more virulent than geno-type A (4). Considering that, all of the positive cases of Babesiosis in this study were of geno-type B and all of them had typical clinical symptoms of the disease, the results of this study are consistent with those of other studies (4, 36, 40).
The babesiosis infection is detected with molecular and serological methods in domestic dogs and other wild canine in the world. The basic method of diagnosis is the observing intracellular parasite, however, this method has limitations such as false positive, co infections and non-identification of the species (41). Serological analysis is a very useful diagnostic method, but has some limitations such as cross-reactivity between different Babesia species and it cannot differentiate between acute infection and prior exposure with the parasites (40, 41). The molecular-based techniques enable differentiation of morphologically undistinguishable Babesia species and the most reliable techniques for Babesial DNA detection in blood and tissue (36 , 39, 42). Ardabil Province and especially Meshkin Shahr region have cold and mountainous climate with forest and swamp conditions. The presence probability of this tick, because of the proof of B. canis not far-fetched. It has proven that global warming and climate change will lead to a further spread of the vectors and transmitted pathogens (42). Indeed, the climate change is a global challenge, which may explain not only the increase of density and scattering of tick vectors, but also the pattern distribution their hosts, changes in periods of activity, and variations in geographical distribution (42). The studies suggest a possible role of Dermacentor spp. as vectors of tick-borne pathogens that affect human and animal health (12, 19). Fast diagnostic technique is necessary for the accurate determination of Babesia in canine that could be carried and possibly transmitted by Dermacentor or other related spp. More studies are needed to increase the knowledge in the epizootiology, ecology and epidemiology of canine babesiosis in Ardabil area. Epidemio-molecular studies are necessary to provide important data for the development of new vaccines and effective therapies against canine babesiosis (43). Through a novel diagnostic strategy, our study could characterize B. canis infection in dogs in Meshkin Shahr, Iran. Due to the increasing numbers of piroplasm species, infected dogs may state a drastic health position threat to dog’s population and prevalence of animal infectious disease in Iran.
Conclusion
Our study identified the presence of B. canis in dogs in Meshkin Shahr, Iran, but further studies are needed on the prevalence of Babesia spp. in large sample dog populations from extended areas in Iran to understand better about the epidemiology of canine babesiosis and to promote an effective control program to determine the tick species diversity in dogs in different areas of Iran. The finding of this study provides essential data for subsequently define the critical importance of the molecular studies in management and prevention of the canine babesiosis in Iran.
Acknowledgements
The authors are pleased to thank Prof Mehdi Mohebali and Dr Zabihollah Zareei (Department of Medical Parasitology and Mycology, Tehran University of Medical Sciences, Iran) and Prof Lameh Akhlaghi (Department of Medical Parasitology and Mycology, Iran University of Medical Sciences, Iran), for their technical advises and valuable assistances. The authors declare that they have no competing interests.
References
- 1.Solano-Gallego L, Sainz A, Roura X, Estrada-Pena A, Miro G. (2016) A review of canine babesiosis: the European perspective. Parasit Vectors. 9(1): 336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Uilenberg G. (2006) Babesia a historical overview. Vet Parasitol. 138(1–2): 3–10. [DOI] [PubMed] [Google Scholar]
- 3.Rene-Martellet M, Moro CV, Chene J, Bourdoiseau G, Chabanne L, Mavingui P. (2015) Update on epidemiology of canine babesiosis in southern France. BMC Vet Res. 11: 223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang J, Liu J, Yang J, Liu Z, Wang X, Li Y, Luo J, Guan G, Yin H. (2019) Molecular detection and genetic diversity of Babesia canis canis in pet dogs in Henan Province, China. Parasitol Int. 71: 37–40. [DOI] [PubMed] [Google Scholar]
- 5.Zygner W, Gorski P, Wedrychowicz H. (2009) New localities of Dermacentor reticulatus tick (vector of Babesia canis canis) in central and eastern Poland. Pol J Vet Sci. 12(4): 549–555. [PubMed] [Google Scholar]
- 6.Vichova B, Miterpakova M, Iglodyova A. (2014) Molecular detection of co-infections with Anaplasma phagocytophilum and/or Babesia canis canis in Dirofilaria-positive dogs from Slovakia. Vet Parasitol. 203(1–2): 167–172. [DOI] [PubMed] [Google Scholar]
- 7.Rubel F, Brugger K, Pfeffer M, Chitimia-Dobler L, Didyk YM, Leverenz S, Hans D, Olaf K. (2016) Geographical distribution of Dermacentor marginatus and Dermacentor reticulatus in Europe. Ticks Tick Borne Dis. 7(1): 224–233. [DOI] [PubMed] [Google Scholar]
- 8.Rar VA, Maksimova TG, Zakharenko LP, Bolykhina SA, Dobrotvorsky AK, Morozova OV. (2005) Babesia DNA detection in canine blood and Dermacentor reticulatus ticks in southwestern Siberia, Russia. Vector Borne Zoonotic Dis. 5(3): 285–287. [DOI] [PubMed] [Google Scholar]
- 9.Orkun O, Karaer Z. (2017) Molecular characterization of Babesia species in wild animals and their ticks in Turkey. Infect Genet Evol. 55: 8–13. [DOI] [PubMed] [Google Scholar]
- 10.Aktas M, Ozubek S, Altay K, Ipek ND, Balkaya I, Utuk AE, Erdem Utuk A, Kırbas A, Şimsek S, Dumanlı N. (2015) Molecular detection of tick-borne rickettsial and protozoan pathogens in domestic dogs from Turkey. Parasit Vectors. 8: 157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mazloum Z. (1971) Ticks of domestic animals in Iran: Geographical distribution, host relation and seasonal activity. Iran J Vet Med. 27: 1–32. [Google Scholar]
- 12.Gholamreza S, Somaieh M, Roya S, Alireza B, Ghazale A, Yasin B. (2017) First detection of Babesia ovis in Dermacentor spp in Ardabil area, northwest of Iran. J Vector Borne Dis. 54(3): 277–81. [DOI] [PubMed] [Google Scholar]
- 13.Esmaeilnejad B TM, Asri-Rezaei S, Dalir-Naghadeh B, Mardani K, Golabi M, Arjmand J, Kazemnia A, Jalilzadeh G. (2015) Determination of prevalence and risk factors of infection with Babesia ovis in small ruminants from West Azerbaijan Province, Iran by polymerase chain reaction. J Arthropod Borne Dis. 9(2): 246–252. [PMC free article] [PubMed] [Google Scholar]
- 14.Nabian S, Rahbari S, Shayan P, Hadadzadeh HR. (2008) Identification of tick species of dermacentor in some localities of Iran. J Vet Res. 63(3): 123–126. [Google Scholar]
- 15.Rahbari S, Nabian S, Shayan P. (2007) Primary report on distribution of tick fauna in Iran. Parasitol Res. 101 Suppl 2: S175–7. [DOI] [PubMed] [Google Scholar]
- 16.Razmi GR, Glinsharifodini M, Sarvi S. (2007) Prevalence of ixodid ticks on cattle in Mazandaran Province, Iran. Korean J Parasitol. 45(4): 307–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nabian S, Shayan P, Haddadzadeh HR. (2007) Current status of tick fauna in north of Iran. Iran J Parasitol. 2(1): 12–17. [Google Scholar]
- 18.Akhtardanesh B, Saberi M, Nurollahifard SR, Aghazamani M. (2016) Molecular detection of Babesia spp. in tick-infested dogs in southeastern Iran. J Dis Glob Health. 8(2): 72–77. [Google Scholar]
- 19.Bigdeli M, Rafie SM, Namavari MM, Jamshidi S. (2012) Report of Theileria annulata and Babesia canis infections in dogs. Comp Clin Path. 21(3): 375–377. [Google Scholar]
- 20.Jalali R, Mosallanejad MH, Avizeh B, Alborzi R, Hamidi Nejat AR, Taghipour H. (2013) Babesia infection in urban and rural dogs in Ahvaz District, Southwest of Iran. Arch Razi Inst. 68(1): 37–42. [Google Scholar]
- 21.Niak A, Anwar M, Khatibi S. (1973) Canine babesiosis in Iran. Trop Anim Health Prod. 5(3): 200–201. [DOI] [PubMed] [Google Scholar]
- 22.Augustine S, Sabu L, Lakshmanan B. (2017) Molecular identification of Babesia spp. in naturally infected dogs of Kerala, South India. J Parasit Dis. 41(2): 459–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bajer A, Rodo A, Bednarska M, Mierzejewska E, Welc-Faleciak R. (2013) Babesia canis and tick-borne encephalitis virus (TBEV) co-infection in a sled dog. Ann Agric Environ Med. 20(3): 426–430. [PubMed] [Google Scholar]
- 24.Beck A, Huber D, Polkinghorne A, Kurilj AG, Benko V, Mrljak V. (2017) The prevalence and impact of Babesia canis and Theileria sp. in free-ranging grey wolf (Canis lupus) populations in Croatia. Parasit Vectors. 10(1): 168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ionita M, Mitrea IL, Pfister K, Hamel D, Buzatu CM, Silaghi C. (2012) Canine babesiosis in Romania due to Babesia canis and Babesia vogeli: a molecular approach. Parasitol Res. 110(5): 1659–1664. [DOI] [PubMed] [Google Scholar]
- 26.Juwaid S, Sukara R, Penezic A, Mihaljica D, Veinovic G, Kavallieratos NG, Ćirović D, Tomanović S. (2019) First evidence of tick-borne protozoan pathogens, Babesia sp. and Hepatozoon canis, in red foxes (vulpes vulpes) in Serbia. Acta Vet Hung. 67(1): 70–80. [DOI] [PubMed] [Google Scholar]
- 27.Sukara R, Chochlakis D, Cirovic D, Penezic A, Mihaljica D, Cakic S, Valčić M, Tselentis Y, Psaroulaki A, Tomanović S. (2018) Golden jackals (Canis aureus) as hosts for ticks and tick-borne pathogens in Serbia. Ticks Tick Borne Dis. 9(5): 1090–1097. [DOI] [PubMed] [Google Scholar]
- 28.Solano-Gallego L, Baneth G. (2011) Babesiosis in dogs and cats-expanding parasitological and clinical spectra. Vet parasitol. 181(1): 48–60. [DOI] [PubMed] [Google Scholar]
- 29.Baneth G, Kenny MJ, Tasker S, Anug Y, Shkap V, Levy A, Shaw SE. (2004) Infection with a proposed new subspecies of Babesia canis, Babesia canis subsp. presentii, in domestic cats. J Clin Micro-biol. 42(1): 99–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brkljacic M, Matijatko V, Kis I, Kucer N, Forsek J, Rafaj RB, Grden D, Torti M, Mayer I, Mrljak V. (2010) Molecular evidence of natural infection with Babesia canis canis in Croatia. Acta Vet Hung. 58(1): 39–46. [DOI] [PubMed] [Google Scholar]
- 31.Lyp P, Adaszek L, Furmaga B, Winiarczyk S. (2015) Identification of new 18S rRNA strains of Babesia canis isolated from dogs with subclinical babesiosis. Pol J Vet Sci. 18(3): 573–577. [DOI] [PubMed] [Google Scholar]
- 32.Sanchez-Villeda H, Schroeder S, Flint-Garcia S, Guill KE, Yamasaki M, McMullen MD. (2008) DNA Align Editor: DNA alignment editor tool. BMC Bioinformatics. 9: 154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thompson JD, Higgins DG, Gibson TJ. (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22(22): 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nishimaki T, Sato K. (2019) An Extension of the Kimura Two-Parameter Model to the Natural Evolutionary Process. J Mol Evol. 87(1): 60–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Adaszek L, Winiarczyk S. (2008) Molecular characterization of Babesia canis canis isolates from naturally infected dogs in Poland. Vet Parasitol. 152(3–4): 235–241. [DOI] [PubMed] [Google Scholar]
- 36.Hrazdilova K, Mysliwy I, Hildebrand J, Bunkowska-Gawlik K, Janaczyk B, Perec-Matysiak A, Modrýa D. (2019) Paralogs vs. genotypes, variability of Babesia canis assessed by 18S rDNA and two mitochon drial markers. Vet Parasitol. 266: 103–110. [DOI] [PubMed] [Google Scholar]
- 37.Ashrafi H, Haddadzadeh H, Shirani D, Khazraiinia P, Mostofi S. (2001) Histopathologic, hematologic and clinical study on canine babesiosis. Iran J Vet Med. 56: 93–96. [Google Scholar]
- 38.Annoscia G, Latrofa MS, Cantacessi C, Olivieri E, Manfredi MT, Dantas-Torres F, Otranto D. (2017) A new PCR assay for the detection and differentiation of Babesia canis and Babesia vogeli. Ticks Tick Borne Dis. 8(6): 862–865. [DOI] [PubMed] [Google Scholar]
- 39.Milanović Z, Vekić J, Radonjić V, Ilić Božović A, Zeljković A, Janac J, Spasojević-Kalimanovska V, Buch J, Chandrashekar R, Bojić-Trbojević Ž, Hajduković L, Christopher MM, Kovačević Filipović M. (2019) Association of acute Babesia canis infection and serum lipid, lipoprotein, and apoprotein concentrations in dogs. J Vet Intern Med. 33(4): 1686–1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Davitkov D, Vucicevic M, Stevanovic J, Krstic V, Tomanovic S, Glavinic U, Stanimirovic Z. (2015) Clinical babesiosis and molecular identification of Babe-sia canis and Babesia gibsoni infections in dogs from Serbia. Acta Vet Hung. 63 (2): 199–208. [DOI] [PubMed] [Google Scholar]
- 41.Kovacevic Filipovic MM, Beletic AD, Ilic Bozovic AV, Milanovic Z, Tyrrell P, Buch J, Breitschwerdt EB, Birkenheuer AJ, Chandrashekar R. (2018) Molecular and serological prevalence of Anaplasma phagocytophilum, A. platys, Ehrlichia canis, E. chaffeenses, E. ewingii, Borrelia burgdorferi, Babesia canis, B. gibsoni and B. vogeli among clinically healthy outdoor dogs in Serbia. Vet Parasitol Reg Stud Reports. 14: 117–122. [DOI] [PubMed] [Google Scholar]
- 42.Beugnet F, Chalvet-Monfray K. (2013) Impact of climate change in the epidemiology of vector-borne diseases in domestic carnivores. Comp Immunol Microbiol Infect Dis. 36(6): 559–566. [DOI] [PubMed] [Google Scholar]
- 43.Baneth G. (2018) Antiprotozoal treatment of canine babesiosis. Vet Parasitol. 254: 58–63. [DOI] [PubMed] [Google Scholar]