Abstract
Background
Red blood cell invasion by the Plasmodium vivax merozoite requires interaction between the Duffy antigen receptor for chemokines (DARC) and the P. vivax Duffy-binding protein II (PvDBPII). Given that the disruption of this interaction prevents P. vivax blood-stage infection, a PvDBP-based vaccine development has been well recognized. However, the polymorphic nature of PvDBPII prevents a strain transcending immune response and complicates attempts to design a vaccine.
Methods
Twenty-three P. vivax clinical isolates collected from three areas of Ethiopia were sequenced at the pvdbpII locus. A total of 392 global pvdbpII sequences from seven P. vivax endemic countries were also retrieved from the NCBI archive for comparative analysis of genetic diversity, departure from neutrality, linkage disequilibrium, genetic differentiation, PvDBP polymorphisms, recombination and population structure of the parasite population. To establish a haplotype relationship a network was constructed using the median joining algorithm.
Results
A total of 110 variable sites were found, of which 44 were parsimony informative. For Ethiopian isolates there were 12 variable sites of which 10 were parsimony informative. These parsimony informative variants resulted in 10 nonsynonymous mutations. The overall haplotype diversity for global isolates was 0.9596; however, the haplotype diversity was 0.874 for Ethiopia. Fst values for genetic revealed Ethiopian isolates were closest to Indian isolates as well as to Sri Lankan and Sudanese isolates but further away from Mexican, Papua New Guinean and South Korean isolates. There was a total of 136 haplotypes from the 415 global isolates included for this study. Haplotype prevalence ranged from 36.76% to 0.7%, from this 74.2% were represented by single parasite isolates. None of the Ethiopian isolates grouped with the Sal I reference haplotype. From the total observed nonsynonymous mutations 13 mapped to experimentally verified epitope sequences. Including 10 non-synonymous mutations from Ethiopia. However, all the polymorphic regions in Ethiopian isolates were located away from DARC, responsible for junction formation.
Conclusion
The results of this study are concurrent with the multivalent vaccine approach to design an effective treatment. However, the presence of novel haplotypes in Ethiopian isolates that were not shared by other global sequences warrant further investigation.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12936-021-03843-7.
Keywords: Plasmodium vivax, PvDBPII, DARC, Polymorphism, Mutations
Background
Of the Plasmodium species infecting human, Plasmodium vivax is responsible for the most geographically widespread form of malaria. Once considered benign, P. vivax is now known to cause recurrent infections that can sometimes lead to severe life-threatening pathologies [1, 2]. Though chemotherapies are available, there is as yet a vaccine due to challenges with polymorphisms and the presence of multiple strains in endemic regions that restrict strain-transcending global protection [3–6]. The pathological outcomes of P. vivax infection depend on the parasite's ability to identify, invade human erythrocytes and develop clinical levels of parasitaemia. The molecular mechanisms of invasion of immature red blood cells by P. vivax requires a Duffy Binding Protein II (PvDBPII) ligand, which interacts with its human receptor on the surface of erythrocytes, the Duffy Antigen Receptor for Chemokines (DARC). As PvDBP-DARC interaction is critical, individuals lacking the Duffy antigen on their erythrocyte surface (Duffy negatives) are resistant or have reduced susceptibility to P. vivax infection [7, 8]. Moreover, acquired antibodies against PvDBPII can inhibit PvDBP-DARC interaction and P. vivax invasion, partly qualifying PvDBPII as a leading asexual blood stage vaccine candidate against P. vivax. However, PvDBPII like other Duffy-binding orthologues, is particularly polymorphic especially at its amino terminal cysteine-rich region II (PvDBPII). This hampers the rational design of potent DBP-based vaccines and necessitates the identification of globally conserved epitopes across parasite populations for evaluation in possible multivalent vaccine designs. Therefore, knowledge of the nature and levels of genetic polymorphisms in PvDBPII among global P. vivax isolates is important toward vaccine development.
Analysis of PvDBPII protein sequences identified seven haplotypes represented in 60% of global P. vivax populations [9]. African isolates were poorly represented in this data partly due to relative dominance of Plasmodium falciparum cases in Africa and mostly Duffy-negative human populations that are resistant to P. vivax infection [10]. Though the Duffy blood group antigen (Fya or Fyb) is not expressed on red blood cells (RBCs) of the majority of individuals of African origin [11], low levels of P. vivax malaria has now been reported across much of Africa using sensitive molecular diagnosis methods [7, 12–15]. This has raised questions on the expansion of the infection across more African population as P. falciparum is targeted and eliminated. This risk is particularly high in populations where transmission of P. falciparum and P. vivax is sympatric and at significant levels, such as Ethiopia and population across the Sahara. In Ethiopia, P. vivax accounts for almost 30% of all malaria cases indicating the great public health significance of this neglected parasite [16]. Future P. vivax vaccines based on PvDBPII must, therefore, assess these populations for global efficacy.
In Ethiopia, evidence of pvdbpII gene amplification (three and eight copies) in two Duffy-negative individuals has been reported [17]. Worryingly, a study has also shown that parasites with increased pvdbpII copy number are able to infect erythrocytes even in the presence of naturally acquired antibodies that would have otherwise blocked binding of PvDBP to DARC [18]. Upon this is antigenic variation, i.e., the ability to produce variable antigens, which enables the parasite to evade host immunity [6]. Thus, a multivalent PvDBPII based vaccine will be most favorable and its design will require knowledge of most strains of P. vivax circulating globally. This study describes genetic diversity of pvdbpII in isolates from Ethiopia, which is the fourth highest contributor to global vivax infections.
Methods
Blood samples, DNA extraction and P. vivax diagnosis
A total of 30 P. vivax samples were collected in a cross-sectional study conducted from Nov. 2018 to Dec. 2018 in East Shoa Zone, Ethiopia. The blood samples were collected from clinical malaria patients attending health facilities. Thick and thin blood films prepared for species identification. Blood samples were spotted onto filter papers for parasite DNA extraction. Vivax malaria-infected patients were treated by chloroquine. Genomic DNA was extracted from DBS using QIAamp DNA Mini kit (QIAGEN, Hilden, Germany) according to manufacturers’ protocol. DNA extracts were then used to confirm P. vivax infections by qPCR using the GENESIG Plasmodium vivax standard kit (PrimerDesign Ltd, Chandler’s Ford, UK) according to manufacturers’ protocol. Cycling conditions on the CFX96TM Real-Time PCR detection system (Bio-Rad Laboratories Ltd, California, USA) include enzyme activation at 95 °C for 2 m, followed by 50 cycles of denaturation at 95 °C for 10 s and annealing/data acquisition on the FAM channel at 60 °C for 60 s.
Amplification and sequencing of the PvDBPII
The PvDBP region was amplified using a modified version of previously published PCR conditions [18] and comparing amplicons from two different forward primers (PvDBP_N1_fwd 5′- GATAAAACTGGGGAGGAAAAAGAT-3′ and PvDBP_N2_fwd 5′- CCTCGAATGGTGGCAATCCT-3′) in combination with a single reverse primer (PvDBP_N1_rev 5′- CTTATCGGATTTGAATTGGTGGC-3′) to give amplicons of approximately 1243 bp and 1062 bp respectively. Final amplification conditions contained 0.02U/µl of Q5® High-Fidelity DNA Polymerase enzyme in 1X Q5® High-Fidelity DNA Polymerase Buffer (New England Biolabs, Hitchin, UK), 0.2 mM dNTPs, 0.1 ng/ml of Bovine serum albumin (BSA), 0.5µM of the forward and reverse primers and 5µl of template DNA. Cycling conditions were 98 °C of enzyme activation for 30 secs, followed by 35 cycles of denaturation at 98 °C for 10 s, annealing at 62 °C (for amplicon 1–1243 bp) or 67 °C (for amplicon 2–1062 bp) for 30 s, elongation at 72 °C for 60 s and a final extension at 72 °C for 120 s. Amplicons were purified using Sephadex G-50 (Merck, Darmstadt, Germany) and purified amplicons were then used as templates for cycle sequencing reaction, using the Applied Biosystems™ BigDye™ Terminator v3.1 Cycle Sequencing Kit, according to manufacturers’ protocols. Sequences were detected on the SeqStudio genetic analyzer (Applied Biosystems, California, USA). PvdbpII sequences were successfully obtained for 23 Ethiopian sequences. For each isolate, trace files for pvdbpII were curated and assembled by Seqman pro in Lasergene v 15.3 (DNASTAR) to generate consensus sequences. The latter were aligned using ClustalW alignment algorithm in MEGA X. For comparative analysis, a total of 392 global pvdbpII sequences from 7 P. vivax endemic countries were retrieved from the NCBI archive. These included 100 isolates from India (FJ491142.1-FJ491241.1), 35 from Mexico (KP759780-KP759814), 12 from Myanmar (JN25576.1-JN255587.1), 89 from PNG (AF469515-AF469602), 13 from South Korea (JN989472.1-JN989484.1), 100 from Sri-lanka (GU143914-GU143949), and 43 from Sudan (MG805616-MG805657). The reference sequence from Sal-1 (DQ156512) was included in the final analysis.
Genetic diversity of pvdbpII sequences
DNA sequence polymorphisms and diversity statistics were analysed using DnaSP v 6.12. The following genetic diversity parameters were assessed: number of segregating sites (S), nucleotide diversity (π), haplotype diversity (Hd), average number of pair wise nucleotide differences (K) within the population, and total number of mutations (). To test if population diversity at pvdbpII deviated from neutrality, Tajimas D and Fu and Lis F indices were calculated for the overall protein region sequenced as well as by scanning across the regions using the sliding window approach (window size of 90 bp and a step size of 3 bp). Departures from neutrality were further assessed using the rates of synonymous (dS) and non-synonymous (dN) mutations, estimated and compared by the Z-test (P < 0.05) in MEGAx using the Nei and Gojobori method with jukes cantor correction and 1000 bootstrap replications.
Linkage disequilibrium (LD), effective population size, probability of recombination between adjacent nucleotides per generation and minimum number of recombination events (Rm) were measured using DnaSP. Genetic differences between populations were determined using Wier and Cockerham’s Fst. To visualize the global haplotype network between the isolates, the median joining algorithm as implemented on NETWORK v10. STRUCTURE v 2.3.4 was used to resolve the genetic structure of P. vivax haplotypes. For STRUCTURE, data were run for values K2-K9 using admixture ancestry model, each run had a burn in period of 50,000 iterations and 100,000 Markov chain Monte carlo replications. The most probable K value was then calculated according to Evannos method by using the STRUCTURE harvester website. CLUMPAK was used to graphically display results produced by the STRUCTURE software. To investigate the effects of host immune pressure on the diversity of B and T cell epitopes, the genetic diversity in in-silico predicted and known B and T cell epitopes and MHC-binding regions in PvDBPII were examined. These epitopes were mapped against the tertiary structure of PvDBPII extracted from the Protein data bank (6R2S).and 3RRC which shows crystal structure of region ii from P. vivax Duffy binding protein (Dimerization of P. vivax DBP is induced upon receptor binding and drives recognition of DARC).
Results
High level of diversity in PvDBPII
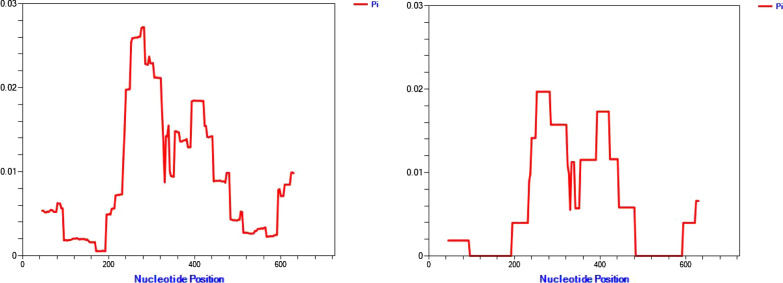
Out of the 30 samples that tested positive for P. vivax, pvdbpII was successfully amplified and sequenced from 23 P. vivax single infections as verified using qPCR. The 23 sequences ranged from 958 to 1190 bp. Further analysis was constrained to a 675 bp region that aligned across all isolates from Ethiopia and the global source from NCBI (total of 415 sequences). From these, 110 variable sites were detected, of which 44 were parsimony informative. For Ethiopian isolates only, there were 12 variable sites of which 10 were parsimony informative (contains at least two types of nucleotides, or at least two of them occur with a minimum frequency of two). These parsimony informative variants resulted in 10 nonsynonymous mutations. Overall global isolates in this study showed 66 singleton mutations, i.e., observed in only one isolate. The overall haplotype diversity for global isolates was 0.9596, while for Ethiopia it was 0.874. The observed nucleotide diversity for Ethiopian isolates was 0.00608 (Table 1) with highest peaks of nucleotide diversity with the pvdbpII coming between the middle intersection of the alignment (196–366) base pairs. Sliding window analysis reveals that nucleotide diversity (π) ranged from 0.014 to 0.019 in this region.
Table 1.
Nucleotide diversity parameters for pvdbpII gene in eight P. vivax endemic countries
| Countries | No. of isolates | S | H | Hd | K | π |
|---|---|---|---|---|---|---|
| Ethiopia | 23 | 12 | 9 | 0.87352 | 4.08696 | 0.00608 |
| India | 100 | 36 | 35 | 0.92141 | 5.90222 | 0.00885 |
| Mexico | 35 | 10 | 7 | 0.55294 | 2.68908 | 0.00400 |
| Myanmar | 12 | 27 | 12 | 1.000 | 8.25758 | 0.001229 |
| PNG | 89 | 61 | 42 | 0.91267 | 6.29826 | 0.00937 |
| S. Korea | 13 | 14 | 12 | 0.98718 | 3.69231 | 0.00549 |
| Sri Lanka | 100 | 27 | 39 | 0.92242 | 6.54020 | 0.00973 |
| Sudan | 42 | 17 | 15 | 0.89547 | 4.01742 | 0.00598 |
S Segregating sites, Hd Haplotype diversity, K Average number of pair- wise nucleotide differences, π Nucleotide diversity
Balancing selection signatures map around PvDBPII immune epitopes
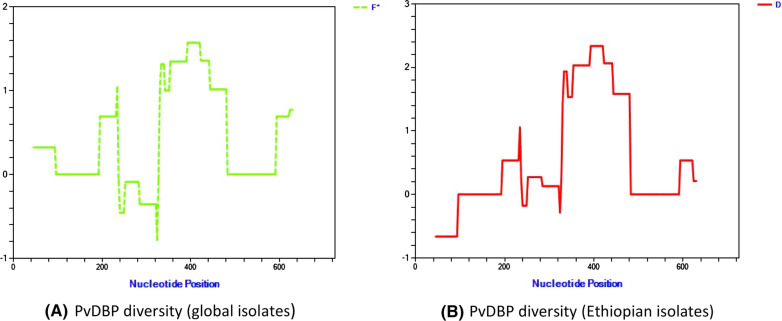
Overall tests for neutrality by Fu and Lis’ F and Tajimas’ D were positive for pvdbpII from all populations, with values of X and Y, respectively for Ethiopian isolates. Sequence diversity was heterogeneous across the region with highest Pi values around nucleotides 200 to 500 (Figs. 1 and 2). Both Fu and Lis D and F tests detected significant positive values in Ethiopian isolates in the region that spans from 265–465 bp (1.5704 P < 0.10). Significant positive values were also observed for Tajimas D value in the region 289–384 (1.9349 P < 0.10) for global isolates and 310–486 (2.0651 P < 0.05) for Ethiopian isolates.
Fig. 1.
Sliding window analysis (window size 90, step size 3) for Nucleotide diversity (π) in Global (left) and Ethiopian (right) using pvdbpII sequence
Fig. 2.
Sliding window analysis (Window size 90, step size 3) results for Departure from Neutrality using Fu and Lis F (left) and Tajimas’ D (right) for Ethiopian pvdbpII’ sequences
The regions with higher signatures of balancing selection were dominated by non-synonymous mutations. The global set of isolates exhibited 30 nonsynonymous mutations, of which 13 mapped to epitope sequences (Table 2). The observed 10 non-synonymous mutations in Ethiopia equally mapped against regions of balancing selection and mapped/corresponded to experimentally verified epitopes.
Table 2.
Polymorphic regions (highlighted) that map to known epitopes sequences
| Epitope name | Epitope | Sequence |
|---|---|---|
| 5 | T/B | VNNTDTNFHRDITFR |
| 45 | B | SIFGTGEKAQQHRKQ |
| 48 | B | EKAQQHRWWNESK |
| 66 | T | ICKINVAVNIEPQIY |
| Ia | MHC class I | GNFIWICKI |
| Ic | MHC class I | VLSNKFKSVKNAEK |
| IIa | MHC class II | YSVKKRLKGN |
| IIb | MHC class II | FIWICKINY |
The source of genetic variation was tested for recombination using the minimum recombination events as implemented in DnaSP. In global P. vivax populations the minimum numbers of recombination events between adjacent polymorphic sites (Rm) was high in Indian, Myanmar and Sri Lankan isolates all of which were > 5 (Table 3). In compliment LD index R2 declined across analysed region in Ethiopian isolates an indicator of intragenic recombination as a possible means for increased diversity (Fig. 3).
Table 3.
Estimates of recombination events in pvdbpII gene in eight global P. vivax endemic countries
| Country (No. of isolates) | Ra | Rb | Rm |
|---|---|---|---|
| Ethiopia (23) | 0.0261 | 17.5 | 2 |
| India (100) | 0.0240 | 16.1 | 10 |
| Mexico (35) | 0.000 | 0.001 | 1 |
| Myanmar (12) | 0.2623 | 176 | 7 |
| PNG (89) | 0.0146 | 9.8 | 3 |
| South Korea (13) | 0.0741 | 49.7 | 1 |
| Sri Lanka (100) | 0.0206 | 13.8 | 9 |
| Sudan (42) | 0.0310 | 20.8 | 3 |
Ra recombinant parameter between adjacent sites, Rb recombination parameter for whole gene, Rm minimum number of recombination events
Fig. 3.
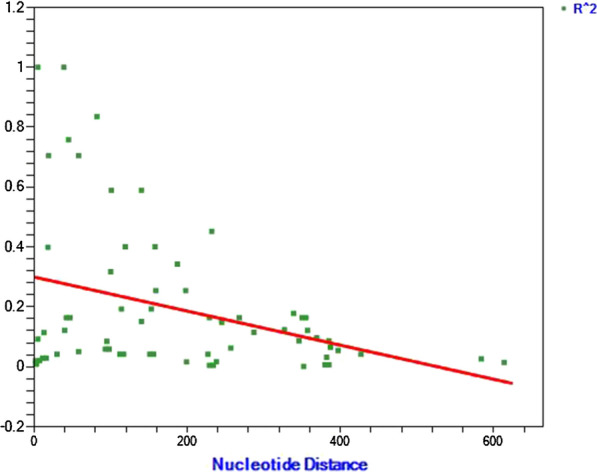
Linkage disequilibrium plot of R2 for pvdbpII gene from Ethiopian isolates
Differentiation at PvDBPII between populations
Pairwise population Fst values at PvDBPs between the eight P. vivax endemic populations ranged from 0.0193 between Sri Lanka and India to 0.3647 between South Korea and Mexico (Table 4). Ethiopian isolates were closer to Indian and Sri Lankan compared to Sudanese isolates despite geographic proximity between Sudan and Ethiopia. They were most diverged from Mexican, Papua New Guinean and South Korean isolates.
Table 4.
Genetic differentiation (Fst) values of the pvdbpII gene among eight P. vivax endemic geographical populations
| Ethiopia | India | Mexico | Myanmar | PNG | South K | Sri lanka | Sudan |
|---|---|---|---|---|---|---|---|
| India | 0.0422 | ||||||
| Mexico | 0.1940 | 0.2500 | |||||
| Myanmar | 0.0846 | 0.0499 | 0.1458 | ||||
| PNG | 0.1275 | 0.1180 | 0.2766 | 0.1314 | |||
| South K | 0.1301 | 0.1226 | 0.3647 | 0.1441 | 0.2454 | ||
| Sri_lanka | 0.0529 | 0.0193 | 0.2653 | 0.0826 | 0.1302 | 0.1158 | |
| Sudan | 0.0945 | 0.1328 | 0.0673 | 0.0660 | 0.2239 | 0.1927 | 0.1495 |
Nine distinct haplotypes of pvdbpII were detected in Ethiopian isolates (Fig. 4). Of the 9 haplotypes, 6 had the highest frequency with 6 isolates followed by haplotypes 2 with 5, haplotypes 5 and 7 had 3 isolates each whereas there were 2 isolates for haplotype 1, while the rest had one isolate each (Fig. 5). There was a total of 136 haplotypes from the 415 global isolates analysed. Haplotype prevalence ranged from 36.76% to 0.7%, with 74.2% represented by a single parasite isolate. None of the Ethiopian isolates grouped with the Sal I reference haplotype; instead two formed their own cluster (1.47%), five clustered together in the dominant haplotype type represented in 50 isolates (36.76%) from India, Sri Lanka and Sudan. Another five Ethiopian isolates grouped in a cluster of 39 isolates from Mexico and Sudan (28.67%), three grouped with 18 isolates from Sri Lanka and Sudan (13.24%), further three grouped with 17 Indian and Sudanese isolates (12.4%), whereas four isolates formed their own unique single haplotype cluster each.
Fig. 4.
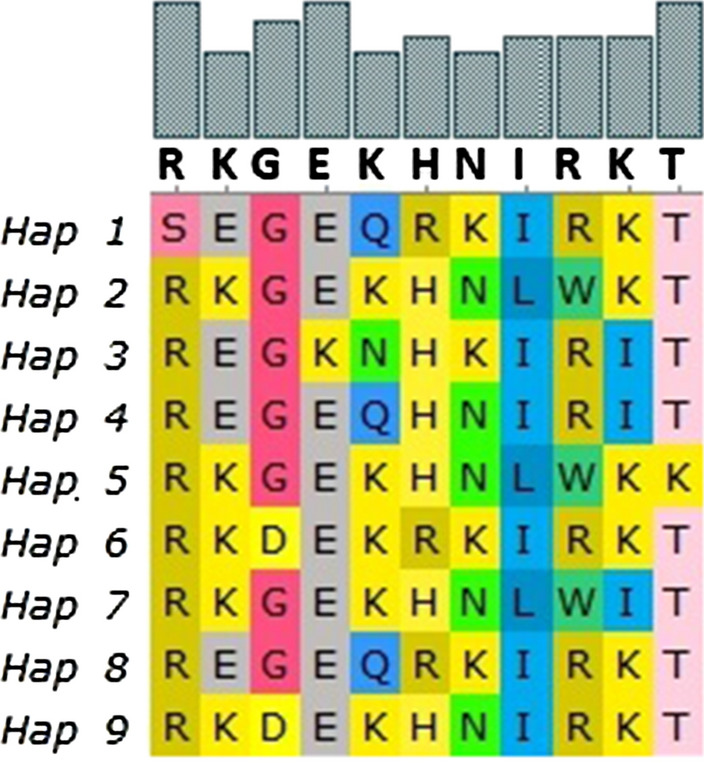
PvdbpII haplotypes observed among Ethiopian isolates.There were nine haplotypes observed from the 23 Ethiopian isolates (Fig. 4)
Fig. 5.
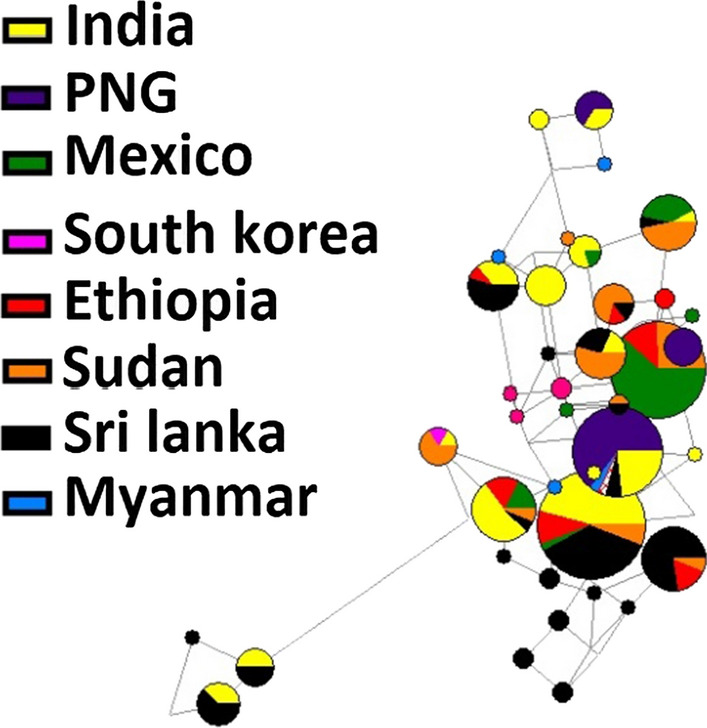
Network analysis of pvdbpII haplotypes using the median joining algorithm implemented in Network software. The Pies represent the haplotypes with lines indicating the connections, size of each pie corresponds to frequency, and colors indicate the geographic populations
All the polymorphic regions in Ethiopian isolates were located away from the Duffy antigen receptor for chemokines responsible for junction formation (Additional file 1). In sharp contrast the polymorphic region from other isolates showed that a mutation is present in this critical region.
Discussion
The interaction between the Duffy antigen for chemokine receptors in humans and the Duffy binding protein in P. vivax is critical for the subsequent invasion of reticulocytes. This pathway is currently the only known way the parasite colonizes its human host. However, the loss of this receptors in erythrocytes has been shown to reduce susceptibility or complete resistance to P. vivax infection [7, 8]. As a result studies have targeted the Duffy binding protein as a potential vaccine candidate, which would be capable of blocking entrance to its host [4]. In support of this hypothesis, several studies have shown that the polymorphic sites of the Duffy binding protein are localized and that most of the sequence remains conserved [9]. This is largely down to the structural biology of the protein and the nature of its junction formation with the Duffy antigen for chemokine receptors [19]. In order to maintain this junction, the protein needs to maintain its structural rigidity as an alteration could lead to a loss of forming this critical network with its receptor [20]. Therefore, much of the region responsible for attachment is highly conserved. Instead, the protein shows polymorphism away from these regions, mostly on B and T cell epitopes or around them. The protein uses these polymorphic sites to exercise antigenic diversity, providing the protein the capability of immune evasion. Therefore, to use this protein as a vaccine, a multivalent vaccine approach is critical, i.e., the ability to cover the diversity of circulating strains globally. However much of these studies have focused on non-African strains, instead targeting only on Asian, South American and Australian strains. This is counterproductive as Africa and particularly Ethiopia is the one of the highest contributors to morbidity rates in the world [16]. Much remains unclear in terms of understanding the nature of the Duffy binding protein in Ethiopia, particularly circulating strains, nature of haplotypes and genetic diversity. Here for the first time, the haplotype diversity of pvdbpII and natural selection of Ethiopian PvDBPII and the haplotype prevalence based on predominant, non-synonymous single nucleotide polymorphisms (SNPs) are reported.
There were 10 nonsynonymous mutations in Ethiopian isolates, all of which mapped exclusively to epitopes regions. This was concurrent with observation with other global isolates; however, other isolates exhibited nonsynonymous mutation in other regions as well, albeit near the epitopes. This agrees with the hypothesis that immune pressure from the host is responsible for the diversifying selection occurring on this region. However, the extent of immune selection in Ethiopian isolates seems to be much more pronounced as evidenced from the departure of neutrality test. Indeed, significant positive values would indicate diversifying selection, whereas significantly negative values would indicate purifying selection. Analysis of Ethiopian isolates reveals significant positive values for both Tajimas D and Fu and Lis f tests. Furthermore, both tests showed that selection was operating in the same location as the epitopes, confirming the immune selection hypothesis. This trend was also similar for global sequences although significantly positive values were not found for Ethiopian isolates. In compliment, genetic diversity (π) was highest in two regions particularly that are found in the central domain of the protein. For Ethiopian isolates this ranged from 0.014 to 0.019, a mark of localized diversity. This was also in complement to the departure of neutrality test which found significantly negative values in regions away from this peak diversity. It has been indicated that higher levels of polymorphism may also reflect the artifact of differences in sampling, PCR and/or sequencing, which may have caused South Korean isolates to have higher genetic diversity as compared to countries endemic with high vivax malaria such as Colombia and PNG [21, 22]. However, nucleotide diversity was moderate in Ethiopian isolates in relation to other countries, and only specific regions appeared to have significantly higher nucleotide diversity as such it is not expected to be a result of sampling and/or sequencing artifacts.
Meiotic recombination has previously been indicated as a potential source of genetic variation in pvdbpII. Here recombination was evaluated using DnaSP minimum recombination event parameter and as an indirect indicator of Linkage disequilibrium. In the event of meiotic recombination, the frequency of linkage disequilibrium declines significantly with increasing distance. Indeed, linkage disequilibrium showed significant decline in the analysed region suggesting a role for recombination. Similarly, DnaSP found regions that were recombining, although to a lesser extent in Ethiopian and South Korean isolates. It has been suggested that this could be a result of a declining P. vivax transmission or an indication of malaria intervention (could be related to vivax seasonality or transmission strength) [22]. However other global population showed higher recombination values indicating the contribution of Recombination towards genetic diversity. This is in agreement with several other studies, which examined the effects of recombination in P. vivax endemic countries. As such it is also likely that the test used to detect recombination for Ethiopian isolates may not have sufficient power to detect low recombination events. Low transmission can explain why recombination is low while balancing selection maintains the already high genetic diversity observed in Ethiopia, indeed samples were taken from low transmission setting and hence might explain why genetic diversity is high while recombination is low.
Fst index was used to assess the degree of population differentiation and gene flow in the eight P. vivax endemic countries. Ethiopian isolates showed lower degree of differentiation with Indian isolates, perhaps unsurprisingly given the expanding P. vivax population and/or extensive gene flow supported by intense human migration between the two countries. On the other hand, little differentiation was also seen in Ethiopian and Sri Lankan isolates, Ethiopian and Sudanese isolates. It is also likely that the strong selection pressure resulted in low Fst values, as fixation of certain alleles may also affect the result [5]. Moreover, relatively high degree of differentiation was exhibited between Ethiopian and Mexican, Ethiopian and Papua New Guinean, Ethiopian and South Korean isolates. This could be due to the geographic barrier between the countries inhibiting gene flow. Indeed, Fst values exceeded 0.25 which would refer to highly differentiated populations, in isolates from India PNG and South Korea vs Mexican isolates showing a reduced gene flow. Similarly, it is quite possible that the strong selection pressure exerted in this gene may have caused a fixation of alleles in certain geographic regions while remaining absent in others. It should be noted that Fst values are affected by factors such as population structure, where the test assumes a stable population structure which may not be the case for P. vivax infections. However other studies have indeed found population clustering in relation to geographic origin and distance, as confirmed using AMOVA and STRUCTURE analysis, similarly the same held true for Ethiopian isolates using STRUCTURE analysis.
High numbers of haplotypes were found in the Ethiopian datasets, interestingly, this was like a recent global pvdbpII haplotype analysis study which previously identified seven haplotypes that can cover 60% parasite population in 8 malaria endemic countries [9]. However, the previous study used isolates from non-African countries and recommended an inclusion of representative haplotypes. Accordingly, few studies have explored genetic diversity of pvdbpII in African countries, such as Sudan. The study in Sudanese isolates found that only one unique haplotype was not included in global haplotype diversity [23]. The finding of this study show four unique haplotypes in the Ethiopian isolates that were not shared with other global isolates, perhaps indicating that there are haplotypes that have not been considered for vaccine trials. Indeed, the frequency of this haplotypes in this study in was not significant as three of the four only had one representative. Even so the low number of samples in this study compared to the number of haplotypes observed, will warrant further study. As such further studies on the frequency of circulating haplotypes in Ethiopia will give a clear indication as to which are the dominant strains that need be included for global vaccine trials.
Haplotype diversity was indeed high in most of the population included in this study, although it is much more pronounced in Sri Lankan and Indian isolates. In the Sri Lankan isolates haplotype diversity reach 1, showing the extent of variability in the population. In Ethiopia, however, haplotype diversity was intermediate in relation to the other included population sets. Indeed, again a small number of samples included could have limited the power of the analysis, but even so the analysis found nine types of haplotypes from 23 Ethiopian isolates. Five of these haplotypes were part of the major haplotype cluster responsible for 36.7% of the global population included for this study. A total of 136 haplotypes were results of singleton mutations, hence the results support the previously mentioned study which identified seven haplotypes that can cover 60% parasite population in 8 malaria endemic countries [9].
Subsequently this region was mapped to a recently experimentally validated tertiary structure of PvDBPII [20]. This structure was determined using a reliable X-ray diffraction method, as opposed to a previous homology modelled structure. This was advantageous as homology modelling is not as reliable as x ray diffraction. This may have affected a 2010 study which found a high genetic diversity and residues under strong positive selection near the DARC receptor domain [24]. This would result in the blockage of junction formation, virtually inhibiting red cell invasion. Apart from this, the study stipulated that antibodies involved in invasion inhibitory activity also block PvDBP-DARC interaction. However recent studies on pvdbpII copy number amplification have shown that they are able to infect even in the presence of naturally acquired antibodies [18]. This would at least indicate the presence of other mechanism for immune evasion such as antigenic variation. This is also supported by subsequent worldwide studies, which have shown many mutations to be predominantly placed on epitopes which agreed with the results of this study [5, 9]. This result is further supported by departure of neutrality tests and now a reliable tertiary structure of PvDBPII which placed non-synonymous mutations away from the DARC recognition site.
Conclusion
This study provides the first description on genetic diversity; the prevalence and nature of pvdbpII sequence polymorphisms in Ethiopian field P. vivax isolates. The results of this study support the multivalent vaccine approach to design an effective treatment. However, the presence of novel haplotypes that were not shared by other global sequences warrant further investigation. As such additional studies need to be made to assess pvdbpII sequence polymorphism by collecting large number of clinical isolates from various heterogenous regions of Ethiopia. This will be an important step in designing and developing an efficacious PvDBPII vaccine capable of covering global strains.
Supplementary Information
Additional file 1. 3-D structure of P. vivax Duffy binding protein bound to human antibody and its polymorphic sites.
Acknowledgements
This work was supported through the DELTAS Africa Initiative [DELGEME grant 107740/Z/15/Z]. The DELTAS Africa Initiative is an independent funding scheme of the African Academy of Sciences (AAS)’s Alliance for Accelerating Excellence in Science in Africa (AESA) and supported by the New Partnership for Africa’s Development Planning and Coordinating Agency (NEPAD Agency) with funding from the Wellcome Trust [DELGEME grant 107740/Z/15/Z] and the UK government. The views expressed in this publication are those of the author(s) and not necessarily those of AAS, NEPAD Agency, Wellcome Trust or the UK government. The funder has no role in the design of the study and collection, analysis, and interpretation of data and in writing this manuscript.
Abbreviations
- DARC
Duffy antigen receptor for chemokines
- Hd
Haplotype diversity
- K
Average number of pair wise nucleotide differences within the population
- LD
Linkage disequilibrium
Total number of mutations
- π
Nucleotide diversity
- SNP
Single nucleotide polymorphism
- S
Segregating sites
- dS
Rates of synonymous
- dN
Rates of non-synonymous mutations
- PCR
Polymerase chain reaction
- PvDBPII
Plasmodium vivax Duffy binding protein II
- RBCs
Red blood cells
Authors’ contributions
LG and AAN did the study design, AM analysed the data and did the initial draft, ECO and LG did the laboratory work and ECO and AAN revised the manuscript. All authors read and approved the final manuscript.
Availability of data and materials
Not applicable.
Declarations
Ethics approval and consent to participate
Ethical clearance was obtained from the institutional review board of Aklilu Lemma Institute of Pathobiology, Addis Ababa University. Information about the objective of the study, and its possible risks and benefits were explained to the participants or their guardians using the local language. Figure prink blood samples were taken after obtaining written informed consent and/or assent from parents or guardian’s. Confidentiality was maintained throughout the study.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Kevin BJ. Evidence and implications of mortality associated with acute Plasmodium vivax malaria. Clin Microbiol Rev. 2013;26:36–57. doi: 10.1128/CMR.00074-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Howes RE, Mioramalala SA, Ramiranirina B, Franchard T, Rakotorahalahy AJ, Bisanzio D, et al. Contemporary epidemiological overview of malaria in Madagascar: operational utility of reported routine case data for malaria control planning. Malar J. 2016;15:502. doi: 10.1186/s12936-016-1556-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen E, Salinas ND, Huang Y, Ntumngia F, Plasencia MD, Gross ML, et al. Broadly neutralizing epitopes in the Plasmodium vivax vaccine candidate duffy binding protein. Proc Natl Acad Sci USA. 2016;113:6277–6282. doi: 10.1073/pnas.1600488113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ntumngia FB, Schloegel J, Barnes SJ, McHenry AM, Singh S, King CL, et al. Conserved and variant epitopes of Plasmodium vivax Duffy binding protein as targets of inhibitory monoclonal antibodies. Infect Immun. 2012;80:1203–1208. doi: 10.1128/IAI.05924-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cole-Tobian J, King CL. Diversity and natural selection in Plasmodium vivax Duffy binding protein gene. Mol Biochem Parasitol. 2003;127:121–132. doi: 10.1016/S0166-6851(02)00327-4. [DOI] [PubMed] [Google Scholar]
- 6.VanBuskirk KM, Cole-Tobian JL, Baisor M, Sevova ES, Bockarie M, King CL, et al. Antigenic drift in the ligand domain of Plasmodium vivax duffy binding protein confers resistance to inhibitory antibodies. J Infect Dis. 2004;190:1556–1562. doi: 10.1086/424852. [DOI] [PubMed] [Google Scholar]
- 7.Miller LH, Mason SJ, Clyde DF, McGinniss MH. The resistance factor to Plasmodium vivax in blacks. N Engl J Med. 1976;295:302–304. doi: 10.1056/NEJM197608052950602. [DOI] [PubMed] [Google Scholar]
- 8.Twohig KA, Pfeffer DA, Baird JK, Price RN, Zimmerman PA, Hay SI, et al. Growing evidence of Plasmodium vivax across malaria-endemic Africa. PLoS Negl Trop Dis. 2019;13:e0007140. doi: 10.1371/journal.pntd.0007140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Sousa TN, Carvalho LH, de Brito CFA. Worldwide genetic variability of the Duffy binding protein: insights into Plasmodium vivax vaccine development. PLoS ONE. 2011;6:e22944. doi: 10.1371/journal.pone.0022944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Howes RE, Patil AP, Piel FB, Nyangiri OA, Kabaria CW, Gething PW, et al. The global distribution of the Duffy blood group. Nat Commun. 2011;2:266. doi: 10.1038/ncomms1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mason SJ, Miller LH, Shiroishi T, Dvorak JA, Mcginniss MH. The Duffy Blood Group determinants: their role in the susceptibility of human and animal erythrocytes to Plasmodium knowlesi malaria. Br J Haematol. 1977;36:327–335. doi: 10.1111/j.1365-2141.1977.tb00656.x. [DOI] [PubMed] [Google Scholar]
- 12.Cavasini CE, de Mattos LC, Couto AAD, Bonini-Domingos CR, Valencia SH, Neiras WCS, et al. Plasmodium vivax infection among Duffy antigen-negative individuals from the Brazilian Amazon region: an exception? Trans R Soc Trop Med Hyg. 2007;101:1042–1044. doi: 10.1016/j.trstmh.2007.04.011. [DOI] [PubMed] [Google Scholar]
- 13.Culleton RL, Mita T, Ndounga M, Unger H, Cravo PVL, Paganotti GM, et al. Failure to detect Plasmodium vivax in West and Central Africa by PCR species typing. Malar J. 2008;7:174. doi: 10.1186/1475-2875-7-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carvalho TAA, Queiroz MG, Cardoso GL, Diniz IG, Silva ANLM, Pinto AYN, et al. Plasmodium vivax infection in Anajás, State of Pará: no differential resistance profile among Duffy-negative and Duffy-positive individuals. Malar J. 2012;11:430. doi: 10.1186/1475-2875-11-430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zimmerman PA. Plasmodium vivax infection in Duffy-negative people in Africa. Am J Trop Med Hyg. 2017;97:636–638. doi: 10.4269/ajtmh.17-0461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.WHO. World malaria report 2018. Geneva: World Health Organization. 2018. http://apps.who.int/iris/bitstream/handle/10665/275867/9789241565653-eng.pdf?ua=1.
- 17.Lo E, Hostetler JB, Yewhalaw D, Pearson RD, Hamid MMA, Gunalan K, et al. Frequent expansion of Plasmodium vivax Duffy binding protein in Ethiopia and its epidemiological significance. PLoS Negl Trop Dis. 2019;13:e0007222. doi: 10.1371/journal.pntd.0007222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Popovici J, Roesch C, Carias LL, Khim N, Kim S, Vantaux A, et al. Amplification of Duffy-binding protein-encoding gene allows Plasmodium vivax to evade host anti-DBP humoral immunity. Nat Commun. 2020;11:953. doi: 10.1038/s41467-020-14574-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Duffy PE. Structure solves the problem with malaria merozoite vaccines. Trends Parasitol. 2019;35:855–857. doi: 10.1016/j.pt.2019.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rawlinson TA, Barber NM, Mohring F, Cho JS, Kosaisavee V, Gérard SF, et al. Structural basis for inhibition of Plasmodium vivax invasion by a broadly neutralizing vaccine-induced human antibody. Nat Microbiol. 2019;4:1497–1507. doi: 10.1038/s41564-019-0462-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ju HL, Kang JM, Moon SUY, Bahk YY, Cho PY, Sohn WM, et al. Genetic diversity and natural selection of Duffy binding protein of Plasmodium vivax Korean isolates. Acta Trop. 2013;125:67–74. doi: 10.1016/j.actatropica.2012.09.016. [DOI] [PubMed] [Google Scholar]
- 22.Hu Y, Wang L, Mbenda HGN, Soe MT, Yu C, Feng H, et al. Genetic diversity, natural selection and haplotype grouping of Plasmodium vivax Duffy-binding protein genes from eastern and western Myanmar borders. Parasit Vectors. 2019;12:546. doi: 10.1186/s13071-019-3803-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoque MR, Elfaki MMA, Ahmed MA, Lee SK, Muh F, Albsheer MMA, et al. Diversity pattern of duffy-binding protein sequence among duffy- negatives and duffy-positives in Sudan. Malar J. 2018;17:297. doi: 10.1186/s12936-018-2425-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sousa TN, Tarazona-Santos EM, Wilson DJ, Madureira AP, Falcão PRK, Fontes CJF, et al. Genetic variability and natural selection at the ligand domain of the Duffy binding protein in Brazilian Plasmodium vivax populations. Malar J. 2010;9:334. doi: 10.1186/1475-2875-9-334. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. 3-D structure of P. vivax Duffy binding protein bound to human antibody and its polymorphic sites.
Data Availability Statement
Not applicable.