Abstract
We demonstrate a novel 3D NNC magic angle spinning (MAS) NMR experiment that generates 15N-15N internuclear contacts in protein systems using an optimized 15N-15N proton assisted recoupling (PAR) mixing period and a 13C dimension for improved resolution. The optimized PAR condition permits the acquisition of high signal-to-noise 3D data that enables backbone chemical shift assignments using a strategy that is complementary to current schemes. The spectra can also provide distance constraints. The utility of the experiment is demonstrated on an M0Ab1–42 fibril sample that yields high-quality data that is readily assigned and interpreted. The 3D NNC experiment therefore provides a powerful platform for solid-state protein studies and is broadly applicable to a variety of systems and experimental conditions.
Graphical Abstract
Protein structure determination using magic angle spinning (MAS) NMR spectroscopy invariably begins with chemical shift assignments, where each distinct resonance is associated with a specific nuclear site. Resonance assignment is typically accomplished using 2D and 3D experiments where magnetization is transferred among 1H/13C/15N nuclei using dipole recoupling experiments designed to reintroduce the coupling attenuated by MAS. While this initial step is of crucial importance, it is often tedious and challenging. Homonuclear dipolar recoupling in 2D and 3D assignment experiments predominantly utilizes 13C-13C magnetization transfer, due to the prevalence of 13C nuclei in biological samples, and to a favorable gyromagnetic ratio of 13C. In principle, it is also possible to use 15N-15N couplings for assignments and structural studies, an approach that is appealing because of the excellent resolution of the 15N dimension. Accordingly, previous approaches employed proton driven spin diffusion (PDSD) techniques to generate 15N-15N correlation spectra of three model peptides1–2, three model proteins3–5, and more recently, oriented membranes6. Additionally, a 15N-15N spectrum obtained using an early version of proton assisted recoupling (PAR) was reported7. Despite the fact that these initial results are very promising, 15N-15N techniques have not evolved to be part of the standard repertoire of MAS protein experiments, primarily for three reasons: (1) the sensitivity in the PDSD and the initial PAR spectra is low and does not permit extension to higher dimensions; (2) cross peaks for long-rang contacts are often missing from the spectra; and (3) the mixing times are 2–5 s and therefore the total experimental time is long. Here we describe a novel 3D NNC experiment (depicted in Figure 1A) that utilizes an optimized 15N-15N PAR protocol that yields excellent 15N sensitivity, and therefore easily extends to a third directly detected 13C dimension to further increase the resolution. Furthermore, the PAR mixing sequence is short (20 ms) and the spectra display an abundance of cross peaks.
Figure 1:
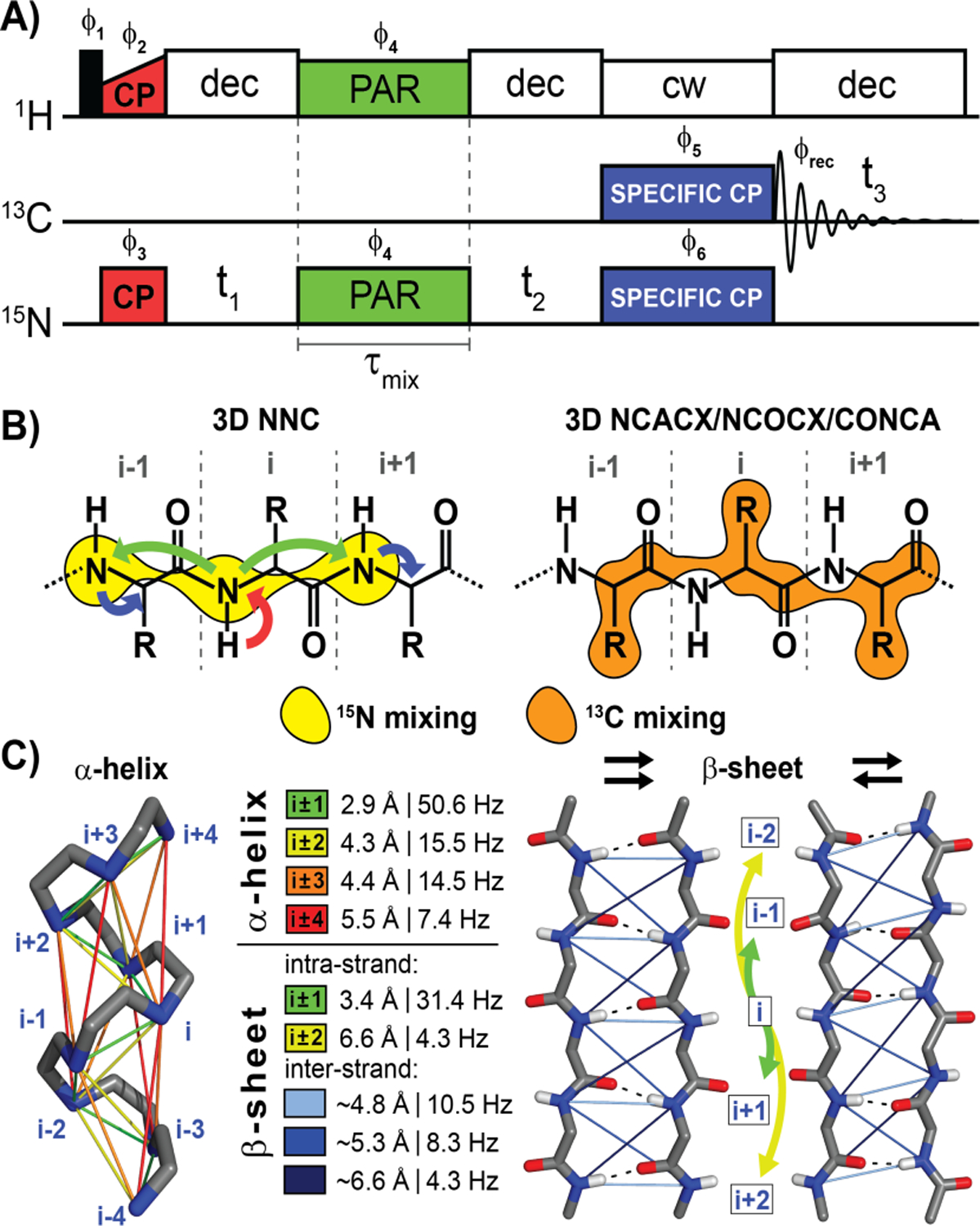
The 3D NNC pulse sequence is shown in A) with phase cycling: ϕ1 = 02, ϕ2 = 0022, ϕ3 = 1, ϕ4 = 2002, ϕ5 = 0123, ϕ6 = 2002 0220, ϕrec = 0123 2301. The CP, PAR, and SPECIFIC-CP blocks are color coded to arrows in (B) illustrating the corresponding magnetization transfer. Additionally, B) contrasts the mode of mixing for the NNC experiment, and commonly used protocols (NCACX, NCOCX, CONCA), illustrating their complementary information due to reliance on separate contacts. C) visualizes the internuclear distances and corresponding dipole coupling values for a range of 15N-15N contacts in α-helices and parallel and anti-parallel β-sheets. The residue interval label (i.e. i ± 1, i ± 2, etc.) is color coded to the lines/arrows illustrating transfer. The 15N-15N inter-strand β-sheet distances are given as approximate averages.
As mentioned above, 3D MAS experiments usually utilize 13C-13C correlations (generated using RFDR8–9, DARR10, PDSD11–12, or PAR13 mixing, amongst others) that are resolved with a 15N dimension. The transfer from 15N to 13C is typically based on SPECIFIC-CP14–16 or other 15N-13C transfer techniques17–19. This combination leads to a suite of experiments used for backbone resonance assignments that includes a combination of NCA/NCACX/NCACB20–25, NCO/NCOCX21–25, and CONCA/CANCO23–25. The 3D NNC, however, allows for the acquisition of non-redundant and complementary information that is otherwise inaccessible and thus facilitates comprehensive assignment strategies when combined with other data sets. This unique sequence shares features with previous 15N-15N correlation experiments in liquids26–27 as well as solid, deuterated samples at high MAS frequencies28. Additionally, direct 15N detection has recently been used for proteins in solution experiments29. However, the NNC is distinguished from the above experiments since it utilizes through-space 15N-15N dipole coupling to mediate coherence transfer and generate homonuclear contacts. However, 15N homonuclear dipolar recoupling remains difficult to implement in non-model samples such as M0Ab1–42 (as opposed to N-f-MLF-OH30 or GB131), the 42 amino acid protein that forms toxic fibril species associated with Alzheimer’s disease32–46. While this system is well suited for solid state NMR analysis because of its monodispersity and small molecular size (enabling high sensitivity), it presents many difficulties for collecting high-quality data as it is significantly more structurally heterogeneous, and less static than any of the usual model systems. It is for this reason that we have chosen M0Ab1–42 to test the efficiency of the NNC protocol.
To demonstrate the importance of PAR mixing we compared the signal-to-noise ratio, the number of observed cross peaks, and maximum distance observed for three available 15N-15N homonuclear mixing schemes: RFDR (tmix = 12.8 ms), PDSD (tmix = 4 s), and PAR (tmix = 20 ms). These results are illustrated in Figure 2. We found that 15N-15N mixing with the first-order recouping sequence RFDR8–9 (Figure 2A), yields no cross-peaks due to the small homonuclear dipolar couplings <50 Hz (Figure 1C). Using 15N-15N PDSD11–12, a second-order recoupling sequence, we observed a limited number of cross peaks (Figure 2B). However, in addition to the minimal quantity of cross-peaks, this experiment requires tmix = 4 s, and all the cross-peaks are assigned to distances <3.5 Å (i±1). In contrast, the 15N-15N PAR experiment, also a second-order recoupling sequence, shows significantly more cross-peaks than the PDSD, and distances of up to 6.8 Å are observed. These are apparent in Figure 2C. Figure 2D compares 1D traces from the RFDR, PDSD, and PAR spectra, clearly showing the superior signal intensity given by PAR, and the presence of additional cross peaks that are attributed to long-range contacts (which are not present in either the RFDR or PDSD spectra). It is also important to note that the total acquisition time for the 2D PAR in Figure 3 was around 21 h, while the PDSD required 39 h, nearly twice as long.
Figure 2:
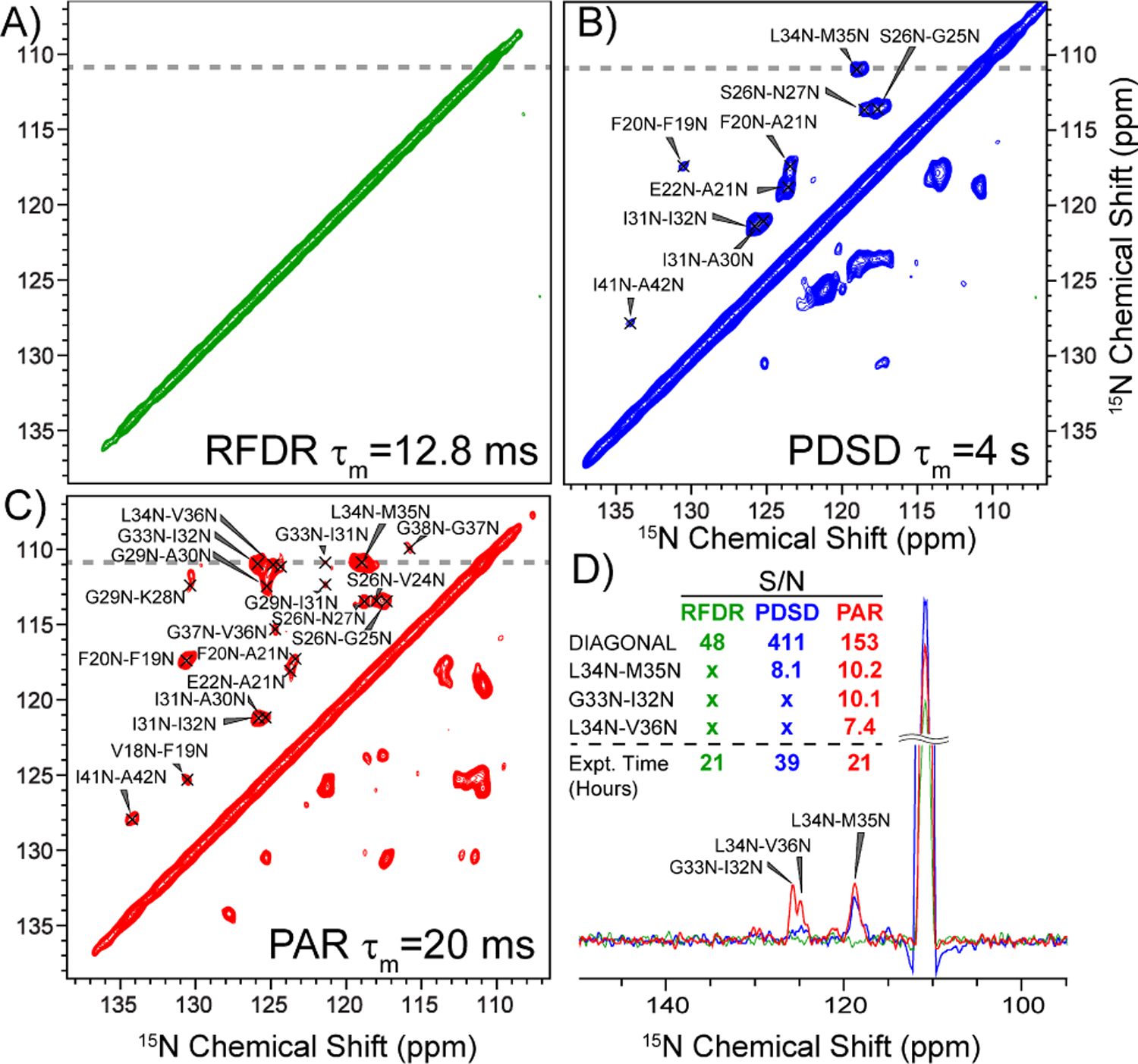
2D 15N-15N homonuclear correlation spectra on M0Aβ1–42. A) gives a comparison of 1D traces extracted from the 2D spectra in B)-D) as indicated by a gray line, with a comparison of measured signal-to-noise (S/N) and total experiment times. Panels B)-D) show a τmix = 12.8 ms 15N-15N RFDR (green), a τmix = 20 ms 15N-15N PAR (red), and a τmix = 4 s 15N-15N PDSD (blue), respectively. Cross-peak assignments are shown.
Figure 3:
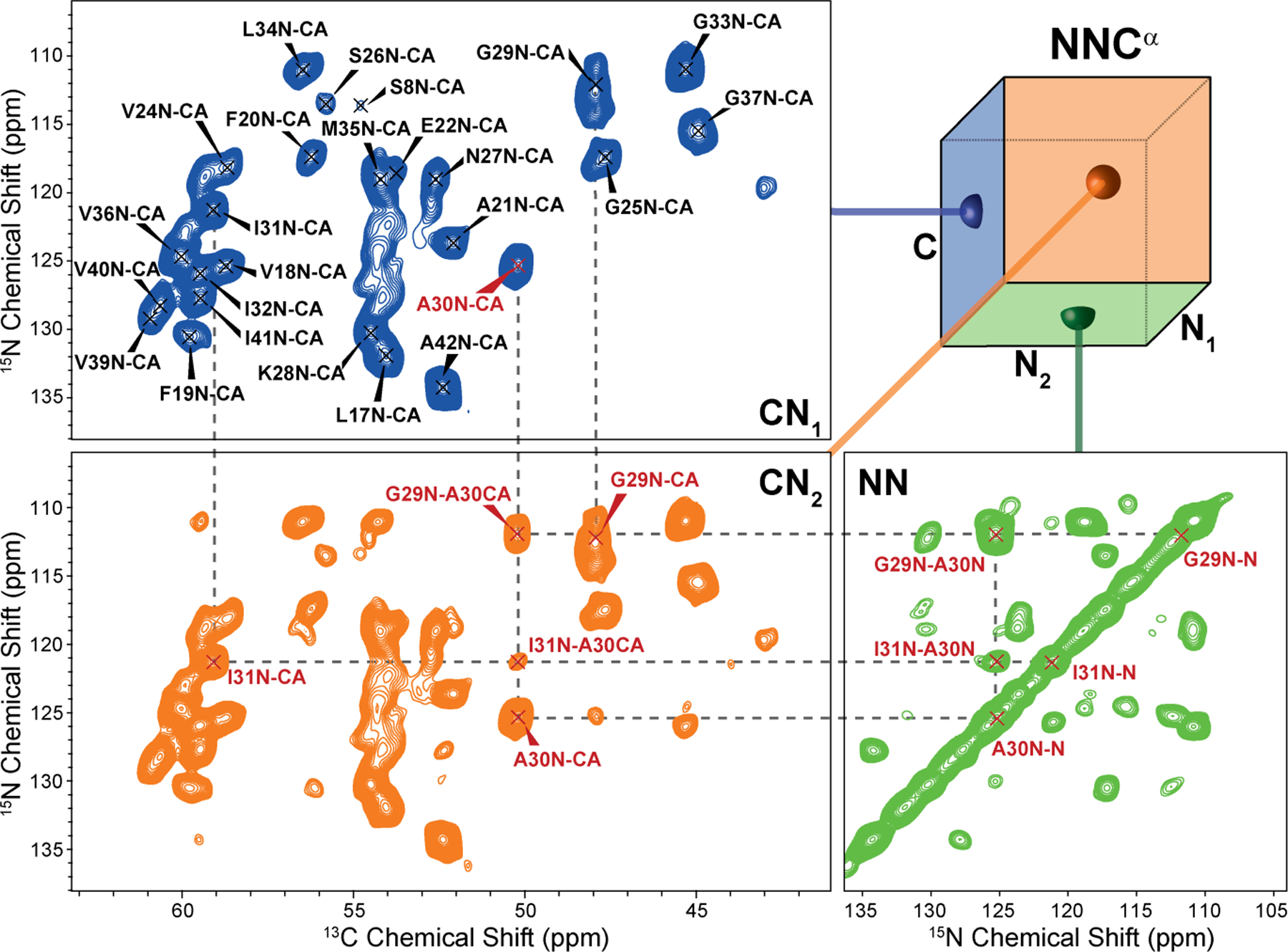
3D NNCα projections of M0Aβ1–42. One CN projection (CN1) shows one peak per residue correlating the nitrogen and the Cα of the same amino acid i. The second CN projection (CN2) is identical to CN1 plus additional peaks due to NN-mixing. For each Cα/N pair (which forms the diagonal peak) there are peaks that correspond to nearby 15N sites, most prominently the i±1 backbone amide 15N nuclei. The third projection (NN) is identical to a 2D 15N-15N PAR spectrum showing the backbone nitrogen of residue i as diagonal and other peaks (mostly backbone nitrogen of residues i±1) as cross-peaks. The 3D spectrum was recorded on a Bruker Avance III 800 MHz spectrometer at ωr/2π = 20 kHz with temperature set to 277 K. A more detailed assignment is shown in Figure S1.
The 2D 15N-15N spectra utilize an optimum PAR matching condition, enabling this innovative approach to backbone resonance assignments on disease-relevant M0Ab1–42. The optimum condition was obtained using a high-throughput protocol to evaluate magnetization transfer across an array of conditions, as described in detail in the Supporting Information. While the PAR mixing period in the NNC experiment generates 15N-15N contacts that are both short-range (between neighboring residues) and long-range (between non-neighboring residues), only the short-range contacts are useful for sequential assignment. Thus, the PAR mixing time can be adjusted to maximize short-range over long-range contacts. Long-range contacts are undesirable for sequential assignment purposes as they may augment spectral congestion and/or obfuscate the assignment process. However, long-range contacts are essential for structural characterization, and can be extracted from NNC spectra collected at longer mixing times. The NNC spectra shown here were collected with a PAR tmix=20 ms, which yields predominantly short-range contacts in M0Ab1–42, with only minor long-range contacts that are easily distinguished by significantly weaker intensities.
The NNC pulse sequence (Figure 1A) consists of three transfer steps: 1H-15N CP, 15N-15N PAR mixing, and 15N-13C SPECIFIC-CP, where the magnetization is selectively transferred to either the Ca or the CO nuclei to generate either an NNCa (see Figures 2 and 3) or an NNCO (see Figures S2 and S3) spectrum. Transfer efficiencies for the 15N-15N PAR mixing alone (i.e. in comparison to 1H-15N CP and 15N-13C SPECIFIC-CP without 15N-15N PAR mixing) are shown in Figure S4 for N-f-MLF-OH and M0Ab1–42. The remaining 15N magnetization after 15N-15N PAR mixing depends on the mixing time and relaxation rates. After 20 ms of PAR mixing, 53% and 20% of the magnetization remain for N-f-MLF-OH and M0Ab1–42, respectively. Besides the 15N-15N PAR mixing, the total efficiency of the 3D NNC experiment depends on the heteronuclear transfer, which is coupled to the selectivity and also the sample and the experimental conditions chosen (vide infra).
A comparison of NNCa and NNCO for M0Ab1–42 shows the NNCa spectrum to have superior resolution due to larger chemical shift dispersion in the Ca spectral region than the CO spectral region. However, the diagonal of the NNCO spectrum contains additional information as it displays correlations between the 15N of residue i and the 13CO of residue i-1, whereas the diagonal of the NNCa shows correlations only between nuclei within the same residue Figure 1B illustrates the internuclear correlations obtained in an NNC experiment vs. commonly used 13C-13C based experiments, and Figure 1C shows internuclear distances (and corresponding dipole coupling values) in standard α-helix as well as parallel and anti-parallel β-sheet structures for the 15N-15N contacts observed with the 3D NNC sequence.
A distinct advantage of the NNC experiment is the increased resolution gained from the inclusion of a 3rd (13C) dimension. The 2D 15N-15N PAR spectrum shown in Figure 3 displays efficient magnetization transfer, but limited resolution. However, both the 3D NNCa as well as the 3D NNCO spectra have immensely improved resolution over their 2D counterparts.
Figure 3 displays 2D projections for each of the three unique faces of the NNC cube. The CN1 projection is virtually identical to an NCA experiment22 or the Ca region of a short-mixing ZF-TEDOR experiment47. Hence, CN1 shows one peak per residue correlating the nitrogen and the Ca of the same amino acid. The CN2 projection is identical to CN1 plus additional peaks due to 15N-15N mixing. For each Ca/N pair (which forms the diagonal peak) there are peaks that correspond to nearby 15N sites, most prominently the i±1 backbone amide 15N nuclei. The third projection (NN) is identical to a 2D 15N-15N PAR spectrum showing the backbone nitrogen of residue i as a diagonal peak, as well as other coherences (mostly backbone nitrogen of residues i±1) as cross-peaks. The correlations between these three projections and the information therein is shown as example for residue A30 of M0Ab1–42 in Figure 2 and in greater detail in Figure S1.
For resonance assignment, each diagonal peak can be arranged in a strip plot (Figure 4) that displays the sequential walk. Each strip shows the diagonal peak (blue) and one or more cross-peaks (red), unless overlapping with other peaks. Typically, the strongest cross-peaks are of i±1 residues. Occasionally, i±2 residues can be present (as discussed earlier), for example between N27 and G29 (6.6 Å, DNN=4.3 Hz), I31 and G33 (6.8 Å, DNN=3.93 Hz), as well as G33 and M35 (5.8 Å, DNN=6.3 Hz).
Figure 4:

Strip plot of the 3D NNCα of M0Aβ1–42. Each strip displays the diagonal peak of residue i (blue) and several cross-peaks (red). The most prominent cross-peaks at 20 ms PAR mixing are from residues i±1, although cross-peaks from residues i±2 can be present (*). The sequential backbone walk is indicated by dashed lines. Colors do not indicate sign and are solely used as visual aids.
In summary, the 3D NNC experiment presented here provides a novel and compelling approach to protein backbone assignments by generating unique data not otherwise accessible. Typically, 13C-13C mixing schemes are used to correlate consecutive residues in proteins while using 15N to resolve spectral crowding. The 3D NNC experiment, however, generates 15N-15N correlations and uses 13C for spectral decongestion. Application of the PAR mixing scheme coupled with a careful optimization of the PAR fields provides efficient 15N homonuclear magnetization transfer, enabling high signal-to-noise data in 3D settings. While this initial demonstration serves as a showcase of the NNC experimental protocol, variations are readily envisaged with different heteronuclear transfer techniques to accomplish the 15N-13C transfer including adiabatic passage48, Lee-Goldburg decoupling49, TEDOR18, PAIN-CP19, and RESPIRATION-CP17, amongst others. These may offer improved selectivity and/or efficiency depending on the sample and the experimental conditions (i.e. spinning frequency and magnetic field). Furthermore, the NNC sequence naturally lends itself to other experiments of higher dimensionality that include additional transfer steps (e.g. a 4D NNCC sequence) and 1H detection. As experimental NMR transitions to higher magnetic fields and faster sample spinning, we expect the NNC protocol to have a continued relevance as PAR has previously shown excellent functionality at higher spinning frequencies (i.e. 65 kHz)50. Finally, while the NNC experiment was developed for sequential assignments, we anticipate that it will provide long-range 15N-15N contacts (using longer mixing times) that are highly valuable for structural characterization. We foresee this approach to be broadly applicable to a large variety of samples including protein crystals and fibrils, nucleic acids, as well as membrane proteins and sedimented proteins.
Supplementary Material
ACKNOWLEDGMENT
The research was supported by grants from the National Institutes of Biomedical Imaging and Bioengineering (EB-001960, EB-002026, and EB-002804) to R.G.G. and by the Swedish Research Council (VR) and a European Research Council (ERC) Advanced Grant to S.L. R.S. is funded by a DFG research fellowship (SI2105/1-1).
Footnotes
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website. The Supporting Information contains a detailed Experimental Section and Supporting Figures S0-S4.
The authors declare no competing financial interests.
REFERENCES
- (1).Reif B; Hohwy M; Jaroniec CP; Rienstra CM; Griffin RG, J. Magn.Res 2000, 145, 132–141. [DOI] [PubMed] [Google Scholar]
- (2).Traaseth NJ; Gopinath T; Veglia G, J. Phys. Chem. B 2010, 114, 13872–13880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).van Rossum B-J; Castellani F; Pauli J; Rehbein K; Hollander J; de Groot HJM; Oschkinat H, J. Biomol NMR 2003, 25, 217–223. [DOI] [PubMed] [Google Scholar]
- (4).Seidel K; Etzkorn M; Heise H; Becker S; Baldus M, ChemBioChem 2005, 6, 1638–1647. [DOI] [PubMed] [Google Scholar]
- (5).Franks WT; Wylie BJ; Stellfox SA; Rienstra CM, J. Am. Chem. Soc 2006, 128, 3154–3155. [DOI] [PubMed] [Google Scholar]
- (6).Mote KR; Gopinath T; Traaseth NJ; Kitchen J; Gor’kov PL; Brey WW; Veglia G, J. Biomol. NMR 2011, 51, 339. [DOI] [PubMed] [Google Scholar]
- (7).Lewandowski JR; Paëpe GD; Eddy MT; Griffin RG, J. Am. Chem. Soc 2009, 131, 5769–5776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Bennett AE; Griffin RG; Ok JH; Vega S, J. Chem. Phys 1992, 96, 8624–8627. [Google Scholar]
- (9).Bennett AE; Rienstra CM; Griffiths JM; Zhen WG; Lansbury PT; Griffin RG, J. Chem. Phys 1998, 108, 9463–9479. [Google Scholar]
- (10).Takegoshi K; Nakamura S; Terao T, Chem. Phys. Lett 2001, 344, 631–637. [Google Scholar]
- (11).Suter D; Ernst RR, Phys. Rev. B 1982, 25, 6038–6041. [Google Scholar]
- (12).Szevernyi N; Sullivan M; Maciel G, J. Magn. Reson 1982, 47, 462–475. [Google Scholar]
- (13).DePaëpe G; Lewandowski JR; Loquet A; Böckmann A; Griffin RG, J. Chem. Phys 2008, 129, 245101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Baldus M; Petkova AT; Herzfeld J; Griffin RG, Mol Phys 1998, 95, 1197–1207. [Google Scholar]
- (15).Petkova AT; Baldus M; Belenky M; Hong M; Griffin RG; Herzfeld J, J. Magn Reson 2003, 160, 1–12. [DOI] [PubMed] [Google Scholar]
- (16).Rienstra CM; Hohwy M; Hong M; Griffin RG, J. Am. Chem. Soc 2000, 122, 10979–10990. [Google Scholar]
- (17).Jain S; Bjerring M; Nielsen NC, J. Phys. Chem, Lett 2012, 3, 703–708. [DOI] [PubMed] [Google Scholar]
- (18).Jaroniec CP; Filip C; Griffin RG, J. Am. Chem. Soc 2002, 124, 10728–10742. [DOI] [PubMed] [Google Scholar]
- (19).Lewandowski JR; De Paëpe G; Griffin RG, J. Am. Chem. Soc 2007, 129, 728–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Igumenova TI; McDermott AE; Zilm KW; Martin RW; Paulson EK; Wand AJ, J. Am. Chem, Soc 2004, 126, 6720–6727. [DOI] [PubMed] [Google Scholar]
- (21).Marulanda D; Tasayco M; McDermott A; Cataldi M; Arriaran V; Polenova T, J. Am. Chem. Soc 2004, 126, 16608–16620. [DOI] [PubMed] [Google Scholar]
- (22).Pauli J; Baldus M; Rossum B. v.; de Groot H; Oschkinat H, ChemBioChem 2001, 2, 272–281. [DOI] [PubMed] [Google Scholar]
- (23).Shi L; Lake E; Ahmed M; Brown L; Ladizhansky V, BBA - Biomembranes 2009, 1788, 2563–2574. [DOI] [PubMed] [Google Scholar]
- (24).Sperling LJ; Nieuwkoop AJ; Lipton AS; Berthold DA; Rienstra CM, J. Biomol. NMR 2010, 46, 149–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Schuetz A; Wasmer C; Habenstein B; Verel R; Greenwald J; Riek R; Böckmann A; Meier BH, ChemBioChem 2010, 11, 1543–1551. [DOI] [PubMed] [Google Scholar]
- (26).Frenkiel T; Bauer C; Carr MD; Birdsall B; Feeney J, J. Magn. Reson 1990, 90, 420–425. [Google Scholar]
- (27).Diercks T; Coles M; Kessler H, J. Biomolecular NMR 1999, 15, 177–180. [DOI] [PubMed] [Google Scholar]
- (28).Andreas LB; Stanek J; Le Marchand T; Bertarello A; Paepe DC-D; Lalli D; Krejčíková M; Doyen C; Öster C; Knott B; Wegner S; Engelke F; Felli IC; Pierattelli R; Dixon NE; Emsley L; Herrmann T; Pintacuda G, J. Biomol. NMR 2015, 62, 253–261. [DOI] [PubMed] [Google Scholar]
- (29).Takeuchi K; Arthanari H; Shimada I; Wagner G, J. Biomol. NMR 2015, 63, 323–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Rienstra CM; Tucker-Kellogg L; Jaroniec CP; Hohwy M; Reif B; McMahon MT; Tidor B; Lozano-Pérez T; Griffin RG, Proc. Nat’l Acad. Sci 2002, 99, 10260–10265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Frericks Schmidt HL; Sperling LJ; Gao YG; Wylie BJ; Boettcher JM; Wilson SR; Rienstra CM, J Phys. Chem. B 2007, 111, 14362–14369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Sachse C; Fändrich M; Grigorieff N, Proc. Nat’l Acad. Sci 2008, 105, 7462–7466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Ahmed M; Davis J; Aucoin D; Sato T; Ahuja S; Aimoto S; Elliott JI; Van Nostrand WE; Smith SO, Nature Struc. Mole. Bio 2010, 17, 561–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Bertini I; Gonnelli L; Luchinat C; Mao J; Nesi A, J. Am. Chem. Soc 2011, 133, 16013–16022. [DOI] [PubMed] [Google Scholar]
- (35).Fändrich M; Schmidt M; Grigorieff N, Trends in Biochem. Sci 2011, 36, 338–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Lopez del Amo J-M; Schneider D; Loquet A; Lange A; Reif B, J. Biomol. NMR 2013, 56, 359–363. [DOI] [PubMed] [Google Scholar]
- (37).Lu J-X; Qiang W; Yau W-M; Schwieters Charles D.; Meredith Stephen C.; Tycko R, Cell 2013, 154, 1257–1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Colvin MT; Silvers R; Frohm B; Su Y; Linse S; Griffin RG, J. Am. Chem. Soc 2015, 137, 7509–7518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Schmidt M; Rohou A; Lasker K; Yadav JK; Schiene-Fischer C; Fändrich M; Grigorieff N, Proc. Nat’l. Acad. Sci 2015, 112, 11858–11863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Schütz AK; Vagt T; Huber M; Ovchinnikova OY; Cadalbert R; Wall J; Güntert P; Böckmann A; Glockshuber R; Meier BH, Angew. Chem. Int. Edit 2015, 54, 331–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Xiao Y; Ma B; McElheny D; Parthasarathy S; Long F; Hoshi M; Nussinov R; Ishii Y, Nat. Struct. Mol. Biol 2015, 22, 499–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Colvin MT; Silvers R; Ni QZ; Can TV; Sergeyev I; Rosay M; Donovan KJ; Michael B; Wall J; Linse S; Griffin RG, J. Am. Chem. Soc 2016, 138, 9663–9674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Elkins MR; Wang T; Nick M; Jo H; Lemmin T; Prusiner SB; DeGrado WF; Stöhr J; Hong M, J. Am. Chem. Soc 2016, 138, 9840–9852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Ravotti F; Wälti MA; Güntert P; Riek R; Böckmann A; Meier BH, Biomol NMR Assign 2016, 10, 269–276. [DOI] [PubMed] [Google Scholar]
- (45).Wälti MA; Ravotti F; Arai H; Glabe CG; Wall JS; Böckmann A; Güntert P; Meier BH; Riek R, Proc. Nat’l Acad. Sci 2016, 113, E4976–E4984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Qiang W; Yau W-M; Lu J-X; Collinge J; Tycko R, Nature 2017, 541, 217–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Jaroniec CP; Filip C; Griffin RG, J. Am. Chem. Soc 2002, 124, 10728–10742. [DOI] [PubMed] [Google Scholar]
- (48).Hediger S; Meier BH; Kurur ND; Bodenhausen G; Ernst RR, Chem Phys Lett 1994, 223, 283–288. [Google Scholar]
- (49).Wu CH; De Angelis AA; Opella SJ, J. Magn. Reson 2014, 246, 1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Lewandowski JR; De Paëpe G; Eddy MT; Struppe J; Maas W; Griffin RG, J. Phys. Chem. B 2009, 113, 9062–9069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Bennett AE; Rienstra CM; Auger M; Lakshmi KV; Griffin RG, J. Chem. Phys 1995, 103, 6951–6958. [Google Scholar]
- (52).Goddard TD; Kneller DG, SPARKY 3.115, University of California, San Francisco. 2008. [Google Scholar]
- (53).Pines A; Gibby MG; Waugh JS, J. Chem. Phys 1973, 59, 569–590. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


