Significance
Susceptibility (S) genes are plant genes that facilitate pathogen infection. Inactivation of S genes has been considered a promising strategy to obtain broad-spectrum and durable resistance in crops. We characterized two orthologs of the Arabidopsis S gene DMR6 in tomato: SlDMR6-1 and SlDMR6-2. We show that SlDMR6-1, but not SlDMR6-2, is associated with plant immunity. Remarkably, Sldmr6-1 mutants display enhanced resistance to bacterial, oomycete, and fungal pathogens. This phenotype correlates with increased levels of the defense hormone salicylic acid (SA) and enhanced transcriptional activation of plant immune responses. We also demonstrate that SlDMR6-1 and SlDMR6-2 convert SA into its inactive form, 2,5-DHBA, indicating that they play a role in SA homeostasis.
Keywords: DMR6, disease resistance, CRISPR/Cas9 technology, salicylic acid, crop engineering
Abstract
Plant diseases are among the major causes of crop yield losses around the world. To confer disease resistance, conventional breeding relies on the deployment of single resistance (R) genes. However, this strategy has been easily overcome by constantly evolving pathogens. Disabling susceptibility (S) genes is a promising alternative to R genes in breeding programs, as it usually offers durable and broad-spectrum disease resistance. In Arabidopsis, the S gene DMR6 (AtDMR6) encodes an enzyme identified as a susceptibility factor to bacterial and oomycete pathogens. Here, we present a model-to-crop translational work in which we characterize two AtDMR6 orthologs in tomato, SlDMR6-1 and SlDMR6-2. We show that SlDMR6-1, but not SlDMR6-2, is up-regulated by pathogen infection. In agreement, Sldmr6-1 mutants display enhanced resistance against different classes of pathogens, such as bacteria, oomycete, and fungi. Notably, disease resistance correlates with increased salicylic acid (SA) levels and transcriptional activation of immune responses. Furthermore, we demonstrate that SlDMR6-1 and SlDMR6-2 display SA-5 hydroxylase activity, thus contributing to the elucidation of the enzymatic function of DMR6. We then propose that SlDMR6 duplication in tomato resulted in subsequent subfunctionalization, in which SlDMR6-2 specialized in balancing SA levels in flowers/fruits, while SlDMR6-1 conserved the ability to fine-tune SA levels during pathogen infection of the plant vegetative tissues. Overall, this work not only corroborates a mechanism underlying SA homeostasis in plants, but also presents a promising strategy for engineering broad-spectrum and durable disease resistance in crops.
Crop pathogens reduce the yield and quality of agricultural production and pose a threat to global food security (1). Plant diseases are often difficult to control and require an integrated approach that includes the use of appropriate cultural practices, agrochemical use, and resistant varieties (2). The development of resistant varieties typically relies on the deployment of dominant resistance (R) genes, whose products mediate the recognition and protection against specific pathogen strains. However, resistance mediated by a single R gene frequently lacks durability, because pathogens can easily lose or mutate their R gene’s cognate molecule (effector) (3). Lasting disease resistance has been a central goal for crop improvement. A promising strategy to overcome a single R gene breakdown is the simultaneous introduction of multiple R genes, a process called gene stacking. By combining multiple R genes in one genotype, resistance cannot be easily overcome as a pathogen is unlikely able to alter or lose all corresponding effector genes concomitantly (4, 5).
Lasting disease resistance can also be achieved by the inactivation of susceptibility (S) genes, which are plant genes that can facilitate pathogen infection and proliferation (6). A classic example of S genes is the MILDEW RESISTANCE LOCUS O (MLO), whose inactivation provides broad, durable resistance against the powdery mildew pathogen Blumeria graminis f. sp. hordei. Notably, mlo-based resistance has been effectively used in European barley agriculture for about four decades (7). In comparison to R gene stacking, disruption of a single S gene is a faster and simpler process to achieve broad-spectrum and durable disease resistance in crops. However, since S genes have functions other than being a compatibility factor for the pathogen, the pleiotropic effects caused by their inactivation need to be evaluated one by one for the effectiveness of their application in the field (8).
S genes are usually conserved among plant species (9). Once an S gene is identified and characterized in one plant, it can guide the identification of its homologs in other species. The S gene DOWNY MILDEW RESISTANCE 6 (DMR6) has been identified in a loss-of-susceptibility ethyl methanesulfonate mutant screen in Arabidopsis thaliana (10). DMR6 belongs to the superfamily of 2-oxoglutarate Fe(II)-dependent dioxygenases (2-ODDs), and it is up-regulated during pathogen infection (11, 12). Inactivation of Arabidopsis DMR6 (AtDMR6) results in increased salicylic acid (SA; 2-hydroxybenzoic acid) levels and confers resistance to different classes of pathogens, including bacteria and oomycetes (12).
SA is synthesized via two distinct pathways in plants: the phenylalanine ammonia-lyase (PAL) pathway and the isochorismate synthase (ICS) pathway (13). Both pathways require chorismate, the end product of the shikimate pathway, as a precursor for SA biosynthesis. In Arabidopsis, most of the SA produced in response to pathogen infection comes from the ICS pathway, but for some plant species the PAL pathway seems to have more significant participation. Once synthesized, SA can undergo a series of chemical modifications, such as glycosylation and hydroxylation, that are important for the proper maintenance of SA homeostasis (14, 15). Although the pathways involved in SA biosynthesis and catabolism in plants have been known for a while, some of the enzymes catalyzing these pathways have only recently been identified.
Despite the early report of DMR6 as a disease susceptibility factor in Arabidopsis (10), its biochemical mechanism remained unknown for years. Recently, contrasting evidence has been presented for DMR6 enzymatic activity and the cause of increased SA levels in the dmr6 mutants (16, 17). Falcone Ferreyra et al. (16) described Arabidopsis DMR6 as a flavone synthase (FNS) that catalyzes the conversion of naringenin to apigenin. This metabolic pathway would compete for chorismate, the precursor for both SA pathways, thus explaining the increased SA levels in the mutant. Zhang et al. (17), however, reported that DMR6 catalyzes the hydroxylation of SA at carbon-5, producing the metabolite 2,5-dihydroxybenzoic acid (2,5-DHBA). This compound is the intermediate of SA’s storage form 2,5-DHBA sugar conjugates, and inactivation of DMR6 would compromise SA catabolism, explaining the accumulation of this phytohormone in the dmr6 mutants.
Despite all the advances in the study of the plant immune system and the understanding of SA metabolism, it is still challenging to convert this accumulated knowledge from model plants into suitable applications for crop improvement. This may be a result of the variable biological complexity of different plant species as well as the impact of environmental factors on disease and immunity (18). Here, we present a model-to-crop translational work, in which we identify two DMR6 orthologs in tomato and show a comprehensive characterization of the effects of their CRISPR inactivation under laboratory and field conditions. Based on biochemical assays, we show that both tomato DMR6 proteins are SA-5 hydroxylases (S5H) that catalyze the formation of 2,5-DHBA, demonstrating that these proteins are indeed important players in the SA catabolic pathway. These results not only contribute to the understanding of SA homeostasis in plant–pathogen interactions, but also constitute a promising strategy for engineering broad-spectrum and long-lasting disease resistance in crops.
Results
Identification of DMR6 Orthologs and Generation of Stable Mutants in Tomato.
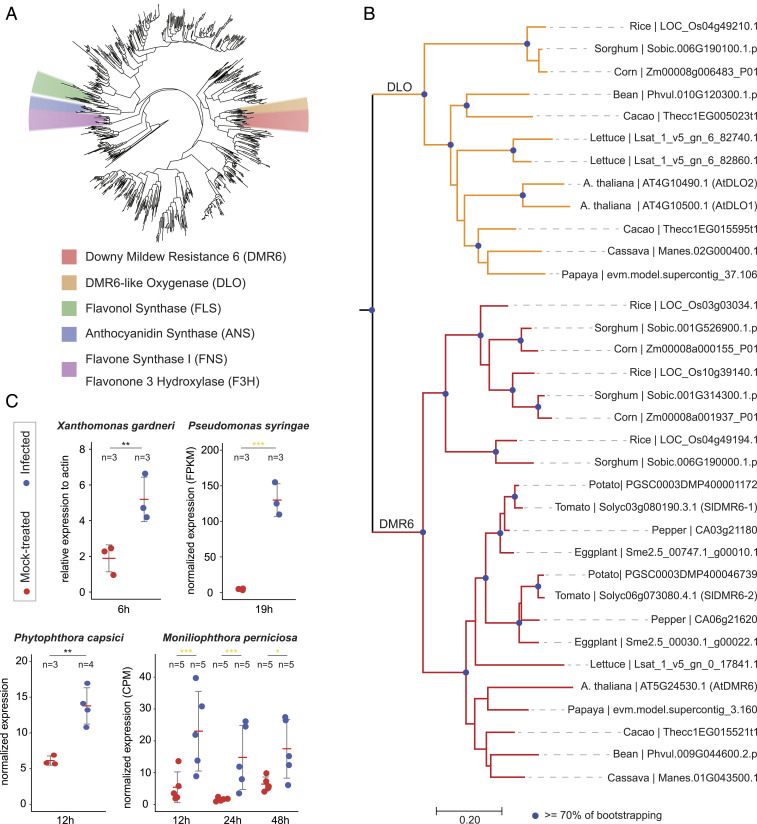
DMR6 belongs to the superfamily of 2-ODDs (Fig. 1A), and is broadly present in flowering plants, ranging from monocots to dicots (Fig. 1B) (12). In addition to DMR6, many plant species have a close DMR6 paralog named DLO1 (DMR6-like oxygenase 1) (Fig. 1 A and B) (12). As with AtDMR6 in A. thaliana, inactivation of AtDLO1 (Arabidopsis DLO1) results in enhanced disease resistance (12). To identify AtDMR6 and AtDLO1 orthologous genes in tomato, we performed a phylogenetic analysis of the 2-ODD superfamily using 13 different plant species (Fig. 1 A and B). In tomato, the 2-ODD superfamily comprises more than 140 members and includes two AtDMR6 orthologous genes: SlDMR6-1 (Solyc03g080190.3) and SlDMR6-2 (Solyc06g073080.4). Conversely, no DLO1 has been identified either in tomato or in any other Solanaceae species analyzed in this work (Fig. 1B).
Fig. 1.
Identification and expression analyses of AtDMR6 orthologs in tomato. (A) Phylogenetic tree of the 2-ODD in plants. The 2-ODD homologs collected from 13 different plant species were used to infer the tree. The clades that contain functionally known 2-ODD members, including DMR6, DLO, FLS, ANS, FNS, and F3H, are annotated in different colors. (B) Phylogenetic tree of the DMR6 and DLO clades. The DMR6 and DLO clades were zoomed in from the phylogenetic tree given in A. The blue dots on the nodes indicate bootstrap values ≥ 70. (C) Up-regulation of SlDMR6-1 gene in response to different pathogens: X. gardneri (P = 0.0012), P. syringae (FDR = 2.69E-95), P. capsici (P = 0.00419), and M. perniciosa (FDR12h = 6E-4, FDR24h = 3.27E-7, FDR48h = 0.045). SlDMR6-2 was not expressed or did not appear among the DEGs under the tested conditions. FDRs and P values are represented by yellow and gray asterisks, respectively. ns, not significant, P/FDR ≥ 0.05; *P/FDR < 0.05; **P/FDR < 0.01; ***P/FDR <0.001. Gene-expression values for P. syringae, P. capsici, and M. perniciosa were obtained from public transcriptome data (43–45) and were differentially normalized. X. gardneri gene expression was obtained by qPCR (SI Appendix, Materials and Methods).
AtDMR6 and AtDLO1 are induced after infection with different classes of pathogens, including bacteria, oomycetes, and fungi, which is consistent with their involvement in plant immunity (12). In tomato, while SlDMR6-2 expression was not detectable even upon pathogen infection, SlDMR6-1 expression is activated in response to several pathogens, such as the bacteria Xanthomonas gardneri and Pseudomonas syringae pv. tomato, the oomycete Phytophthora capsici, and the fungus Moniliophthora perniciosa (Fig. 1C). These results indicate that SlDMR6-1, but not SlDMR6-2, might play a role in plant immunity. Based on this evidence, we employed the CRISPR/Cas9 system (19) to induce mutations in the SlDMR6-1 gene and engineer disease resistance in tomato. Two single-guide RNAs (sgRNAs) targeting exon 2 (gRNA 1) and exon 3 (gRNA 2) of SlDMR6-1 were designed and successfully tested by transient gene-expression assays in Nicotiana benthamiana (SI Appendix, Fig. S1A and Table S1). After confirming their activity in planta (SI Appendix, Fig. S1A), both gRNAs were used to produce stable tomato transformants. In total, 5 of 61 T0 plants (8.2%) were homozygous mutants with frameshift deletion alleles that truncate the protein and disrupt the DMR6 active site. Two independently generated homozygous mutants were named Sldmr6-1.1 (from gRNA 1 targeting exon 2) and Sldmr6-1.2 (from gRNA 2 targeting exon 3) (SI Appendix, Fig. S1B).
Inactivation of SlDMR6-1 Results in Enhanced Disease Resistance with No Evident Growth Penalty.
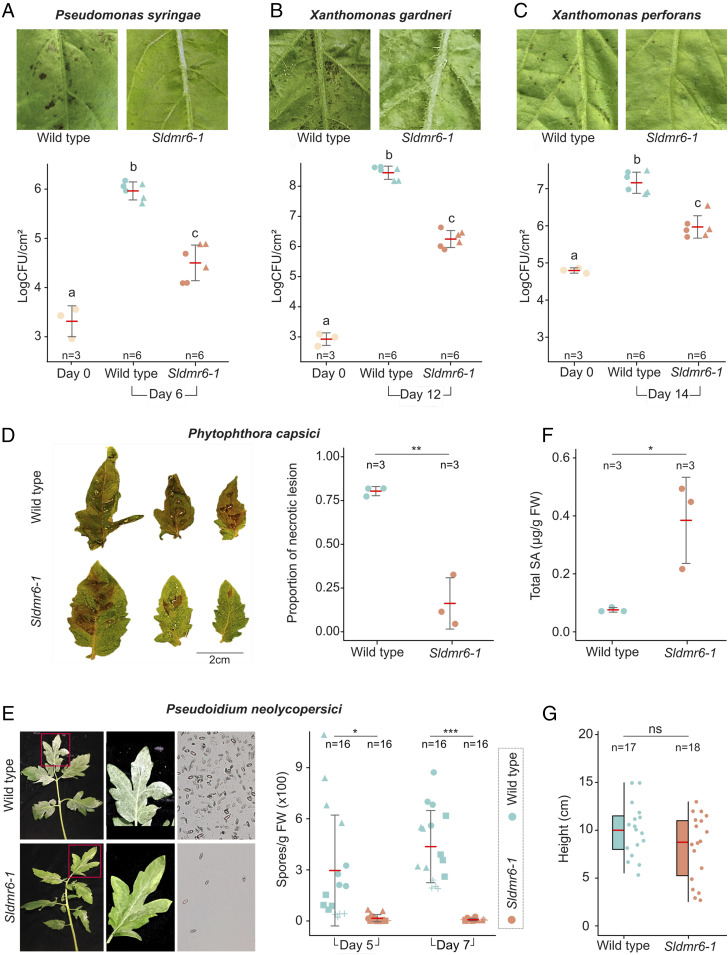
Disease resistance assays with the bacterial pathogens P. syringae pv. tomato, X. gardneri, and Xanthomonas perforans showed impaired growth of all three pathogens and reduced disease severity in the Sldmr6-1 mutants in comparison to wild-type plants (Fig. 2 A–C). Similar results were observed in infection assays with the oomycete P. capsici, in which disease symptoms (i.e., necrotic lesions) developed earlier and were more severe in wild-type plants than in mutant lines (Fig. 2D). We also tested the tomato powdery mildew pathogen Pseudoidium neolycopersici. Similar to the assay with the other pathogens, wild-type plants displayed more intense symptoms with a considerably higher density of white powdery spores on the top of the leaf surfaces (Fig. 2E). Overall, these results show that the Sldmr6-1 mutants display enhanced resistance against three evolutionarily distinct classes of pathogens: bacteria, oomycetes, and fungi. Notably, the enhanced pathogen resistance observed in the Sldmr6-1 mutants correlated with increased levels of SA (SI Appendix, Materials and Methods), suggesting that disease resistance may be associated with the activation of SA-mediated immune responses (Fig. 2F).
Fig. 2.
SlDMR6-1 inactivation in tomato leads to broad-spectrum disease resistance. Disease symptoms and resistance assays with the bacterial pathogens (A) P. syringae pv. tomato DC3000, (B) X. gardneri 153, and (C) X. perforans 4b, (D) the oomycete P. capsici LT1534 isolate (P = 0.00172), and (E) the fungus P. neolycopersici MF-1 isolate (PDay5 = 0.0037, PDay7 = 1.5E-06). Fungal spores in E were imaged using an epifluorescence microscope with 5× lens. All these pathogens show reduced growth or cause less disease symptoms in the Sldmr6-1 mutants. The letters indicate significant differences between the conditions as determined using a one-way ANOVA followed by a Tukey honest significant difference (HSD) test (P < 0.05). Symbols (circles, triangles, and squares) represent leaves from different plants. (F) Total SA content is shown for Fla. 8000 wild-type plants and Sldmr6-1 mutants (P value = 0.023) in response to X. gardneri infection. (G) Quantification of the heights of wild-type and Sldmr6-1 lines (P = 0.121) grown under laboratory conditions. Asterisks represent significant P values from t tests: ns, not significant, P ≥ 0.05; *P < 0.05; **P < 0.01; ***P < 0.001.
To assess the potential of using Sldmr6-1 mutants in plant breeding, we also evaluated the effect of SlDMR6-1 impairment on plant growth. We measured the height of Sldmr6-1 mutants and wild-type plants growing under growth chamber conditions. No clear phenotypic differences were observed under the tested conditions, except that some Sldmr6-1 plants seemed to display a minor decrease in size. However, these differences were not statistically significant (Fig. 2G and SI Appendix, Fig. S2).
SlDMR6-1 Up-Regulation Is Mostly Localized in the Pathogen Inoculation Site.
To analyze the tissue specificity of SlDMR6-1 expression in response to pathogen infection, we generated transgenic tomato lines containing a construct with the GUS reporter gene driven by the SlDMR6-1 promoter (proSlDMR6-1:GUS). We evaluated GUS expression 8 h after infiltrating tomato leaves with mock solution and X. gardneri suspension cells. Although a significant infiltration effect was observed on the proSlDMR6-1:GUS expression (possibly due to tissue damage), we were able to quantify a slightly higher GUS staining in the site inoculated with the pathogen. Therefore, we confirmed the up-regulation of SlDMR6-1 by pathogen infection and showed that its expression mostly occurs in the pathogen inoculation site (SI Appendix, Fig. S3).
Sldmr6-1 Mutants Display Enhanced Activation of Defense Genes upon Pathogen Infection.
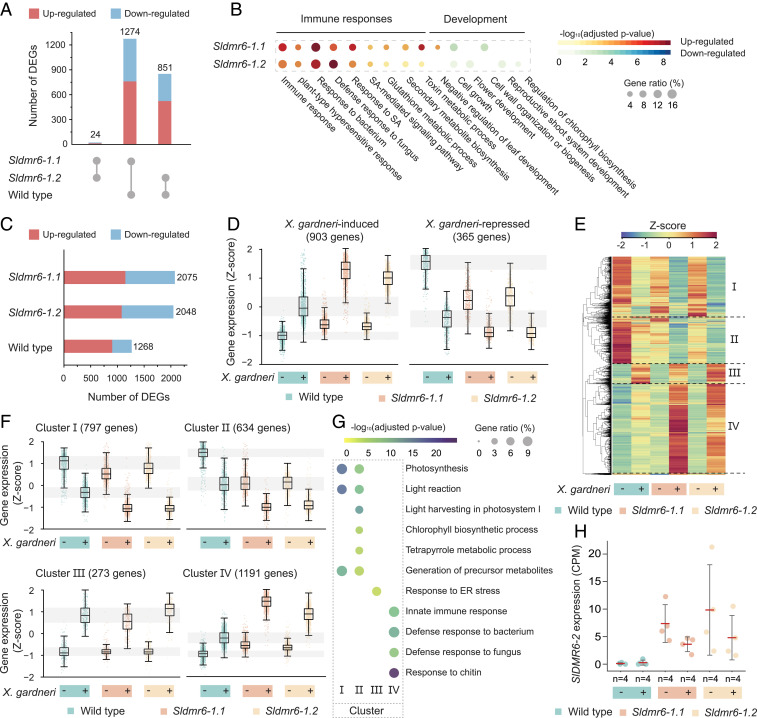
Having evidence that SlDMR6-1 plays a role in plant immunity, we performed a comprehensive gene-expression analysis to understand the molecular changes that occur in the Sldmr6-1 mutants in response to pathogen infection. For transcriptome sequencing, we collected leaf punches of three different genotypes (wild-type, Sldmr6-1.1, and Sldmr6-1.2) that were exposed to two different treatments (mock-inoculated and X. gardneri–inoculated leaves) for 6 h. We used four biological replicates for each genotype/treatment. Only a few differentially expressed genes (DEGs) were observed in the pairwise comparison of the mutants Sldmr6-1.1 and Sldmr6-1.2 in both mock and X. gardneri treatments (false-discovery rate [FDR] < 0.05 and |log2 [fold-change]| ≥ 1), confirming that these two independent mutants produced essentially identical outcomes (SI Appendix, Table S2).
To investigate the precise effect of SlDMR6-1 mutation, we first compared the transcriptomes of wild-type and both Sldmr6-1 mutants in the absence of pathogen infection (mock-inoculated leaves) (Fig. 3 A and B). In comparison to the wild-type, each Sldmr6-1 mutant showed approximately 1,000 DEGs (Fig. 3A, SI Appendix, Table S2, and Dataset S1), indicating that these mutants undergo significant changes as a consequence of SlDMR6-1 inactivation. Among these changes, we verified a constitutive activation of plant immune responses and suppression of key developmental processes (Fig. 3B and Datasets S1 and S2). Gene ontology (GO) enrichment analysis revealed that up-regulated DEGs were enriched in biological processes related to plant immunity, such as SA response and innate immune response, thus indicating that plant defenses are activated even in the absence of a pathogen. This conclusion was supported by the up-regulation of genes encoding immune receptors (receptor-like kinases or NB-LRRs), members of the WRKY family of transcription factors (WRKY 46, 80, and 81) and antimicrobial proteins of the pathogenesis-related (PR) superfamily (Datasets S1 and S2). The SlDMR6-1 mutation also affected the expression of genes associated with plant development and photosynthetic capacity. Some down-regulated biological processes included flower morphogenesis, developmental growth involved in morphogenesis, regulation of chlorophyll biosynthetic process, regulation of developmental vegetative growth, chlorophyll metabolic process, regulation of meristem growth, cell growth, cell morphogenesis, and cell wall organization or biogenesis (Fig. 3B and Dataset S2). Notably, despite the suppression of some metabolic processes associated with development, these plants did not show any compromised growth (Fig. 2G and SI Appendix, Fig. S2), indicating that the level of suppression of these genes was not sufficient to affect plant development under the tested laboratory conditions.
Fig. 3.
Large-scale reprogramming of the tomato transcriptome as a result of SlDMR6-1 inactivation. (A) Comparison of the number of DEGs among the wild-type, Sldmr6-1.1, and Sldmr6-1.2 lines. (B) Selected GO terms that are differentially represented in the Sldmr6-1 mutants. For a complete list of the GO enrichment analysis, please refer to Dataset S2. (C) Number of DEGs in the wild-type and Sldmr6-1 mutant lines in response to X. gardneri infection. (D) Genes up-regulated by pathogen infection are more expressed in the Sldmr6-1 lines than in wild-type plants. Similarly, down-regulated genes are less expressed in the mutants. (E) Hierarchical clustering showing groups of genes that respond to pathogen infection in the absence and presence of X. gardneri. The DEGs were classified into four clusters (I to IV). (F) Details of each hierarchical cluster. Clusters I and II consist of genes that are down-regulated by the pathogen infection in all three genotypes. In particular, genes in cluster II are repressed in the Sldmr6-1 mutants even before infection. Similarly, clusters III and IV show genes that are up-regulated by pathogen. Particularly, genes in cluster IV are slightly activated in the Sldmr6-1 mutants before infection and more expressed in response to pathogen infection. (G) Selected GO terms that are differentially represented in the four different clusters from E and F. For a complete list of the GO enrichment analysis, please refer to Dataset S5. (H) Comparison of SlDMR6-2 expression in the wild-type, Sldmr6-1.1, and Sldmr6-1.2 lines (FDR < 0.05). Units are shown as counts per millions (CPMs) normalized using the trimmed mean of M-values (TMM) in edgeR.
Next, we analyzed the effect of pathogen infection on the transcriptional response of the wild-type, Sldmr6-1.1, and Sldmr6-1.2 lines (SI Appendix, Table S2 and Dataset S3). A comparison between mock-inoculated and X. gardneri–inoculated leaves revealed 1,268, 2,075, and 2,048 DEGs in the wild-type, Sldmr6-1.1, and Sldmr6-1.2 lines, respectively (Fig. 3C, SI Appendix, Table S2, and Dataset S3). These results show that pathogen infection produced approximately twice more DEGs in the mutant lines than in wild-type plants, indicating that the plant response to pathogen attack is quantitatively intensified by the SlDMR6-1 mutation (Fig. 3C, SI Appendix, Table S2, and Dataset S3). In addition, comparing the responses to X. gardneri infection across the genotypes, we verified that many X. gardneri–induced genes were more intensely expressed in the Sldmr6-1 lines than in wild-type plants, whereas several X. gardneri–repressed genes were even less expressed in the mutants (Fig. 3D, SI Appendix, Fig. S4A, and Datasets S3 and S4). Moreover, GO terms associated with plant immunity were enriched in the set of DEGs that are up-regulated in the infected Sldmr6-1 mutants relative to infected wild-type plants (SI Appendix, Fig. S4B and Dataset S4). Together, these results indicate that the mutant plants show a qualitatively and quantitatively intensified response to pathogen attack. This amplified transcriptional response occurring mostly upon pathogen infection explains the increased disease-resistance phenotype of Sldmr6-1 mutants and agrees with the milder detrimental effect on growth and development verified for these mutants under laboratory conditions (Fig. 2G and SI Appendix, Fig. S2).
A hierarchical clustering analysis using genes that respond to pathogen infection revealed four clusters of genes (I to IV) with similar expression patterns (Fig. 3E and Dataset S5). While clusters I and II represent genes that are down-regulated by pathogen infection in all three genotypes (wild-type and both Sldmr6-1 mutants), clusters III and IV show genes that are up-regulated by infection (Fig. 3 E and F). In particular, cluster II contains genes that are repressed in the Sldmr6-1 mutants even before infection. In contrast, cluster IV consists of genes that are slightly activated in the mutants before infection and display a more intense expression in the infected leaves (Fig. 3 E and F). GO analysis of each cluster revealed that the enriched biological processes of clusters I and II are related to plant development, especially with the plant photosynthetic capacity (Fig. 3G and Dataset S5). In contrast, differentially represented GO terms in clusters III and IV are mostly comprised of processes associated with plant immune responses (Fig. 3G and Dataset S5).
SlDMR6-2 Does Not Have a Role in Immunity of the Vegetative Tissues.
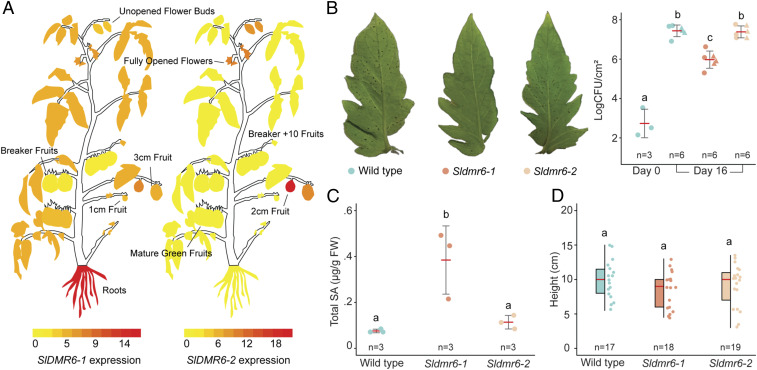
Analysis of public transcriptome data showed that SlDMR6-2 expression is mainly restricted to the plant reproductive organs, such as flowers and immature fruits (2- to 3-cm fruits) (Fig. 4A). In agreement, RT-PCR results confirmed that SlDMR6-2 expression is detected in tomato flowers, but not in leaf tissues (SI Appendix, Fig. S5). In contrast, SlDMR6-1 expression is observed in most plant structures—including leaves, roots, flowers, and fruits—suggesting that these genes might have different regulatory elements (Fig. 4A). A comparison between the promoter regions of SlDMR6-1 and SlDMR6-2 revealed that they do not share any homology or conserved DNA motifs representing putative transcription binding sites (SI Appendix, Fig. S6). Despite the need for experimental validation, these data suggest that SlDMR6-2, unlike SlDMR6-1, may not be involved in plant-defense responses of the vegetative tissues.
Fig. 4.
SlDMR6-2 does not have immunity-related functions. (A) Expression pattern of SlDMR6-1 and SlDMR6-2 in different tissues/organs of wild-type tomato plants. The figure was generated using the ePlant Tomato Tool (bar.utoronto.ca/eplant_tomato/). (B) Disease symptoms and resistance assay with the bacteria X. gardneri (strain 153) showing that SlDMR6-2 inactivation does not interfere with disease resistance. (C) Total SA content is shown for Fla. 8000 wild-type plants, Sldmr6-1 and Sldmr6-2 mutants, in response to X. gardneri infection. (D) Quantification of the heights of wild-type, Sldmr6-1, and Sldmr6-2 lines grown under laboratory conditions. The letters indicate significant differences between the conditions as determined using a one-way ANOVA followed by a Tukey HSD test (P < 0.05).
Although SlDMR6-2 expression is not detected in leaves of wild-type plants (Fig. 4A and SI Appendix, Fig. S5), RNA-sequencing (RNA-seq) data showed that its expression is activated in leaves of the Sldmr6-1 mutants (Fig. 3H). Therefore, we used the CRISPR/Cas9 system to inactivate the SlDMR6-2 gene in the wild-type and in the Sldmr6-1 backgrounds. Two gRNAs were designed to independently generate mutations in exon 1 of the SlDMR6-2 gene. The efficiency of both gRNAs was successfully confirmed using transient expression assays in N. benthamiana (SI Appendix, Fig. S7A). Three homozygous plants containing mutations in the SlDMR6-2 gene were successfully regenerated when the wild-type background was transformed, and homozygous mutants with frameshift deletion alleles that truncate the protein and disrupt the DMR6 active site were obtained in the first generation (T0 plants) (SI Appendix, Fig. S7B). In contrast, when the Sldmr6-1 background was transformed, only two sterile plants with heterozygous SlDMR6-2 mutations were obtained, which impaired our subsequent analyses of the double-mutant Sldmr6-1 Sldmr6-2.
Disease resistance assays with the bacterial pathogen X. gardneri showed that Sldmr6-2 lines do not show increased pathogen resistance (Fig. 4B). Moreover, in contrast to the Sldmr6-1 lines, the Sldmr6-2 mutants do not accumulate more SA than wild-type plants upon pathogen infection (Fig. 4C). Also, comparing the Sldmr6-2 mutants with wild-type plants, we did not observe differences in plant height or other phenotypic characteristics under laboratory conditions (i.e., growth chamber and greenhouse) (Fig. 4D).
SlDMR6-1 and SlDMR6-2 Have S5H Activity.
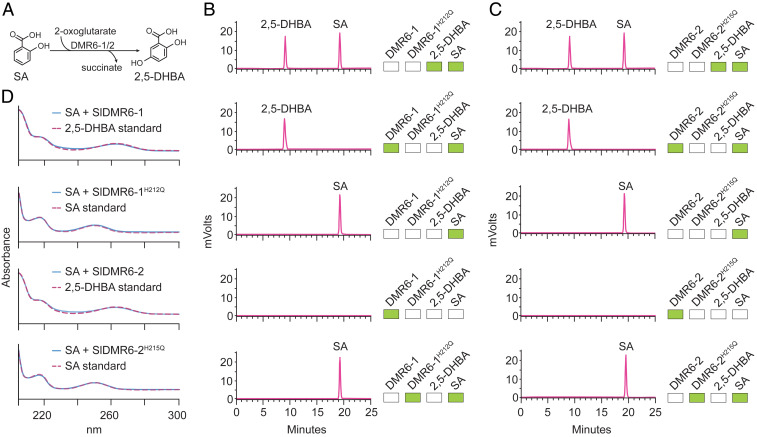
To investigate the enzymatic properties of SlDMR6-1 and SlDMR6-2, the full open reading frame of each gene was cloned in the pET28a vector. These proteins were independently expressed in Escherichia coli as N-terminal fusion proteins with a His-6 tag. Purified proteins were used in enzyme activity assays and the products were analyzed by HPLC. As controls, we also purified the A. thaliana enzymes AtDLO1 and AtDMR6, as well as the catalytic mutants SlDMR6-1 (H212Q) and SlDMR6-2 (H215Q). These mutants contain a substitution of an important histidine residue, which is part of the HDH triad that binds to the iron (FeII) ion presumably required for DMR6 catalysis (12).
Previous studies have proposed distinct substrates for AtDMR6. Falcone Ferreyra (16) described AtDMR6 as an FNS that converts naringenin to apigenin, while Zhang et al. (17) reported that AtDMR6 has S5H activity, catalyzing the hydroxylation of SA to 2,5-DHBA (16, 17). Our results using the recombinant protein AtDMR6 agree with the data presented by Zhang et al. (17). Under the tested conditions, SA is converted to 2,5-DHBA in the presence of AtDMR6, whereas, when naringenin is used as a substrate, no other compound is formed (SI Appendix, Fig. S8 A and B). In addition, as reported by Zhang et al. (20), SA is converted to 2,3-DHBA and 2,5-DHBA in the presence of the recombinant protein AtDLO1 (SI Appendix, Fig. S8A). For the tomato enzymes, we also tested both substrates that have been proposed for AtDMR6: naringenin and SA. Under the tested conditions, both SlDMR6-1 and SlDMR6-2 converted SA into its hydroxylated form 2,5-DHBA (Fig. 5), but they failed to metabolize naringenin (SI Appendix, Fig. S8C). As expected, SlDMR6-1 (H212Q) and SlDMR6-2 (H215Q) did not show any catalytic activity (Fig. 5 B and C). These results demonstrate that purified DMR6 proteins from tomato display S5H activity, but no FNS activity.
Fig. 5.
SlDMR6-1 and SlDMR6-2 catalyze the conversion of SA to 2,5-DHBA. (A) The reaction catalyzed by SlDMR6-1 and SlDMR6-2 showing the hydroxylation of SA at carbon 5 with the subsequent formation of 2,5-DHBA. (B) HPLC profile of the standards 2,5-DHBA and SA (first panel). SA is converted to 2,5-DHBA by the recombinant SlDMR6-1 protein (second panel), but not in the control reaction with no enzyme (third panel). Enzyme preparation contained no contaminants (fourth panel). Mutation of the active site of SlDMR6-1 (SlDMR6-1 H212Q) prevents the conversion of SA to 2,5-DHBA (fifth panel). (C) HPLC profile of the standards 2,5-DHBA and SA (first panel). SA is converted to 2,5-DHBA by the recombinant SlDMR6-2 protein (second panel), but not in the control reaction with no enzyme (third panel). Enzyme preparation contained no contaminants (fourth panel). Mutation of the active site of SlDMR6-2 (SlDMR6-2 H215Q) prevents the conversion of SA to 2,5-DHBA (fifth panel). The green boxes in B and C indicate the presence of that compound in the reaction mixture. (D) Comparison of the absorbance spectra of SA/2,5-DHBA standards to the products of the enzyme assays. The absorbance spectra of the enzymatic product 2,5-DHBA from B and C are identical to that of the 2,5-DHBA standards. All reactions were repeated at least three times and representative data are shown.
To gather structural insights on the substrate specificity of these enzymes, the three-dimensional structures of SlDMR6-1 and SlDMR6-2 (81% sequence identity to each other) were modeled using the crystallographic structure of A. thaliana anthocyanidin synthase (AtANS) as a template (PDB ID code 1GP6; 35 to 30% sequence identity with target proteins) (21) (SI Appendix, Fig. S9 A–C). Molecular docking of SA or naringenin in the conserved active site of the predicted SlDMR6-1 and SlDMR6-2 structures are in accordance with the enzymatic activity identified for these proteins (SI Appendix, Figs. S9 and S10). For SA, we found two possible orientations that fit into the substrate-binding site and place the C-5 carbon near Fe(II), which is compatible with the S5H activity detected for these enzymes (SI Appendix, Fig. S9 D and E). In contrast, the docking results failed to reproduce the naringenin positioning observed in the crystal structure of AtANS (21), thus indicating that the substrate-binding sites of SlDMR6-1/2 enzymes do not fit this substrate properly (SI Appendix, Fig. S10). Structural comparisons between AtANS and SlDMR6-1/2 indicate several amino acid replacements in the substrate-binding site, including the F144R substitution that seems to impose a steric barrier for naringenin positioning in the SlMDR6 enzymes (SI Appendix, Fig. S10). This arginine residue (R128 in SlDMR6-1) is conserved in homologous proteins with sequence identity as low as 50% to SlDMR6-1, indicating that it might play a crucial role in substrate selectivity. Thus, according to these structural analyses, the small substrate-binding site of SlDMR6-1/2, narrowed by a conserved arginine residue, seems to allow the binding of SA but impairs the accommodation of larger molecules, such as naringenin (SI Appendix, Figs. S9 and S10).
SlDMR6-1 Mutations Lead to Pathogen Resistance in the Field.
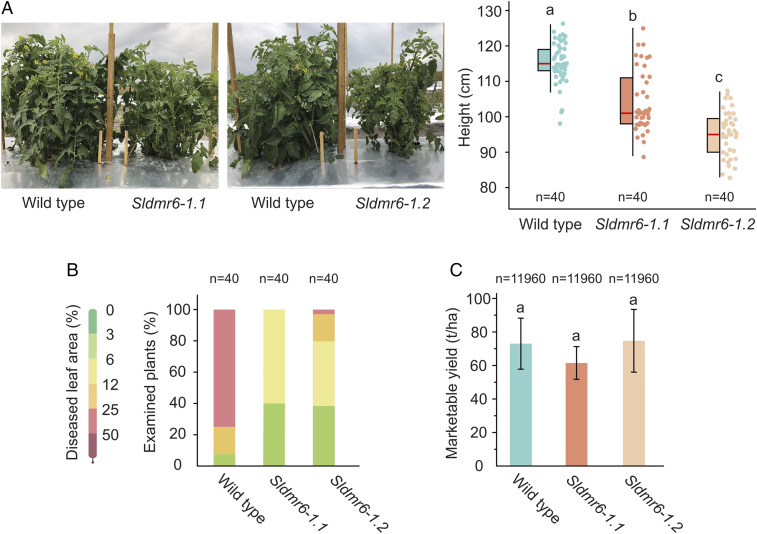
The performance and morphological characteristics of the Sldmr6-1 mutants were evaluated under commercial-type growth conditions in a spring field trial in Florida. Field conditions were typical for the season, with warm weather and very few rainfall events until late in the season (22). Disease severity data were collected after the onset of summer rains, when disease pressure was the highest (SI Appendix, Materials and Methods). Using the Horsfall–Barratt rating scale (23), we estimated the severity of bacterial spot disease in wild-type and Sldmr6-1 lines. From a total of 40 individuals of each genotype, 30 wild-type plants showed 25 to 50% diseased leaf area (index 6 of the Horsfall–Barratt rating scale), while no Sldmr6-1.1 and a single Sldmr6-1.2 plant showed this same disease index. Moreover, most Sldmr6-1 mutants (40 of 40 Sldmr6-1.1 and 31 of 40 Sldmr6-1.2 plants) displayed only 3 to 12% diseased leaf area, which corresponds to indices 3 and 4 of the Horsfall–Barratt rating scale (Fig. 6B). Therefore, the trial demonstrated that Sldmr6-1 mutants display enhanced resistance to bacterial spot disease caused by X. perforans under field conditions (Fig. 6B).
Fig. 6.
Sldmr6-1 mutants are resistant to Xanthomonas infection in the field. (A) Growth phenotype and quantification of the heights of wild-type, Sldmr6-1.1, and Sldmr6-1.2 lines. (B) Disease resistance assays with the bacteria X. perforans race T4 (106 CFU per milliliter of each of strains GEV904, GEV917, GEV1001, GEV1063). (C) Comparison of the total marketable yield of wild-type plants and Sldmr6-1.1 and Sldmr6-1.2 mutant lines. Total marketable yield includes the medium, large, and extra-large fruit categories, which are defined according to the US Department of Agriculture specifications (49). No significant differences in total marketable yield were observed in this field trial (P > 0.05). Error bars represent SD. The letters indicate significant differences between the conditions as determined using a one-way ANOVA followed by a Tukey HSD test (P < 0.05).
Tomato cultivation areas in Florida are nearly always affected by naturally existing Xanthomonas. As a consequence, experiments involving healthy uninfected plants are not feasible. Therefore, the results presented here are based on infected plants of the three genotypes: wild-type, Sldmr6-1.1, and Sldmr6-1.2 lines. Relative to wild-type, Sldmr6-1 mutants displayed a moderately stunted phenotype, affecting height (Fig. 6A), internode length and leaf size (SI Appendix, Fig. S11), as well as fruit size (SI Appendix, Fig. S12). Despite these characteristics, no significant differences were observed between these mutants and wild-type plants regarding the total marketable yield, which is based on the weight of medium, large, and extra-large fruit categories (P > 0.05) (Fig. 6C). Although important results on the performance of Sldmr6-1 mutants in the field have been presented, future experiments are still necessary to evaluate the effect of SlDMR6-1 mutation on healthy uninfected plants under field conditions.
Discussion
The S gene DMR6 facilitates infection by oomycete and bacteria pathogens (10–12). It was first discovered in A. thaliana, but it is widely conserved among flowering plants, including economically important crops, such as tomato, cacao, and cassava (Fig. 1B) (12). Arabidopsis DMR6 (AtDMR6) as well as its close paralog DLO1 (AtDLO1) have been shown to be partially redundant, and both genes encode negative regulators of immunity that act by suppressing plant defenses to pathogens (12). Two AtDMR6 orthologs were identified in tomato, which were named SlDMR6-1 and SlDMR6-2. Conversely, no AtDLO1 orthologous gene was found in either tomato or other members of the Solanaceae family (Fig. 1B). Like several genes encoding negative regulators of immunity (24, 25), AtDMR6 and AtDLO1 are up-regulated during pathogen infection (12). In tomato, SlDMR6-1 is activated by different classes of pathogens, including X. gardneri, P. syringae, P. capsici, and M. perniciosa (Fig. 1C). In contrast, SlDMR6-2 expression is not altered by infection (Fig. 1C), indicating that only SlDMR6-1 might play a role in immunity.
A remarkable trait of the Sldmr6-1 mutants is the broad-spectrum disease-resistance phenotype. This is a highly desirable trait for crop plants, because it confers resistance to more than one pathogen species or to most races or strains of the same pathogen (26). Notably, the Sldmr6-1 mutants showed enhanced disease resistance against different classes of tomato pathogens, such as bacteria (P. syringae, X. gardneri, and. X. perforans), oomycete (P. capsici), and fungi (P. neolycopersici) (Fig. 2 A–E). Furthermore, pathogen resistance correlated with higher accumulation of SA in the Sldmr6-1 mutants upon infection (Fig. 2F), suggesting that disease resistance is associated with SA-mediated activation of plant immune responses. SA is a major defense signal molecule against biotrophic and hemibiotrophic pathogens (27). Although being resistant to several pathogens, plants with constitutive activation of immune responses typically display a dwarf phenotype (28, 29). This growth penalty makes this approach undesirable for resistance breeding in crops (29). Sldmr6-1 mutants, however, did not display any detectable reduction on growth under the tested laboratory conditions (Fig. 2G and SI Appendix, Fig. S2). In agreement with our findings, Kieu et al. (30) have also reported that inactivation of the Solanum tuberosum DMR6-1 gene (StDMR6-1) results in potato plants with increased resistance to Phytophthora infestans, but with no obvious detrimental effect on plant growth.
RNA-seq of wild-type and Sldmr6-1 lines provided a comprehensive view of the large-scale reprogramming of the plant transcriptome associated with SlDMR6-1 inactivation (Fig. 3). Interestingly, even in the absence of pathogen infection, the Sldmr6-1 mutants displayed significant transcriptional changes in comparison to wild-type plants, with around 1,000 DEGs associated with SlDMR6-1 mutation (Fig. 3A, SI Appendix, Table S2, and Dataset S1). Among these changes, the most remarkable alterations were a weak activation of plant immune responses (including genes encoding immune receptors, members of the WRKY family, and PR superfamily) and a slight down-regulation of genes associated with plant development and photosynthetic capacity (Fig. 3B and Datasets S1 and S2). Therefore, despite the great number of DEGs, the small amplitude of gene induction/repression in the Sldmr6-1 mutants (Fig. 3D and Dataset S1) agrees with their morphologically indistinguishable phenotype from wild-type plants under laboratory conditions (Fig. 2G and SI Appendix, Fig. S2). On the other hand, when challenged by a pathogen, the Sldmr6-1 lines showed a remarkable transcriptional reprogramming with twice more DEGs than wild-type plants (Fig. 3C and Dataset S3). Moreover, we verified that the pathogen effect on gene expression is considerably intensified in the Sldmr6-1 mutants, which show higher/lower expression of genes that respond to the pathogen than wild-type tomatoes (Fig. 3 D–G, SI Appendix, Fig. S4, and Datasets S3–S5). Based on these data, we concluded that Sldmr6-1 mutants display a weak preactivation of plant defenses in the absence of a pathogen. However, with pathogen infection, a quantitatively and qualitatively amplified transcriptional response occurs in the Sldmr6-1 mutants in comparison to the wild-type (Fig. 3 D–G, SI Appendix, Fig. S4, and Datasets S3–S5). This amplified immune response explains the increased disease resistance phenotype of the Sldmr6-1 lines. Interestingly, mutants displaying constitutively active immunity (e.g., constitutive expressor of PR-genes or cpr mutants) usually show strongly compromised growth and lesion-mimic phenotypes (31). However, the fact that this intense immune response of Sldmr6-1 is mainly triggered by pathogen infection is in agreement with the morphologically indistinguishable phenotype of these mutants in relation to wild-type lines under laboratory conditions (Fig. 2G and SI Appendix, Fig. S2).
Among the alterations of the plant transcriptome, we also detected the activation of SlDMR6-2 expression in leaves of the Sldmr6-1 mutants, either in the absence or presence of pathogen infection (Fig. 3H). We then decided to employ the CRISPR/Cas9 system to inactivate the SlDMR6-2 gene in the wild-type and in the Sldmr6-1 backgrounds. Although our attempts to generate Sldmr6-1 Sldmr6-2 mutants were not successful, Sldmr6-2 single mutants were successfully generated (SI Appendix, Fig. S7). Recently, Kieu et al. (30) reported that knockout mutants of the S. tuberosum DMR6-2 gene (StDMR6-2) do not show increased resistance to pathogen but exhibit significantly lower plant height and fresh weight. Similarly, the Sldmr6-2 lines did not show increased pathogen resistance against X. gardneri (Fig. 4B). However, unlike Stdmr6-2 mutants, Sldmr6-2 mutants did not exhibit any obvious detrimental effect on growth, or accumulation of higher SA levels as a response to infection (Fig. 4 C and D). These results are consistent with the expression pattern of SlDMR6-2, which is restricted to the plant reproductive organs (i.e., flowers and immature fruits), whereas SlDMR6-1 expression is observed in most plant tissues (Fig. 4A and SI Appendix, Fig. S5). Interestingly, SlDMR6-2 has similar characteristics to AtDLO2, another DMR6 paralog in Arabidopsis. AtDLO2 is not up-regulated in response to pathogen infection, and is not expressed in any tissue, except for the plant reproductive structures, such as siliques and developing seeds (12). Confirming the distinct expression pattern of the SlDMR6-1 and SlDMR6-2 genes, no homology was found between their promoter regions (SI Appendix, Fig. S6). This distinct expression pattern suggests that SlDMR6 duplication in tomato resulted in subsequent subfunctionalization, where SlDMR6-2 might have specialized in balancing the SA levels during flowering/fruit development, while SlDMR6-1 conserved the ability to fine-tune the SA levels during pathogen infection.
Despite the evident involvement of DMR6 in plant immunity, the literature shows controversial results for its precise enzymatic activity (16, 17). Zhang et al. (17) characterized AtDMR6 as a S5H that catalyzes the conversion of SA into its hydroxylated form 2,5-DHBA. Conversely, Falcone Ferreyra et al. (16) proposed that AtDMR6 has FNS activity, catalyzing the formation of the flavone apigenin from naringenin. FNS activity would interfere with immunity by competing with the SA biosynthetic pathway for precursors. In most flavone-producing plants, flavone biosynthesis is catalyzed by a cytochrome P450-dependent monooxygenase named FNS II. However, in a few Apiaceae species, it is catalyzed by a 2-ODD enzyme named FNS I, which appears to have evolved by gene duplication from another 2-ODD, the flavanone 3β-hydroxylase (F3H or FHT) (32). Although DMR6 is also a 2-ODD enzyme, it belongs to a different clade than F3H and FNS I, and it is more closely related to DLO1 (Fig. 1 A and B), which is a 2-ODD involved in SA catabolism during leaf senescence (20). DLO1 has been characterized as an SA 3-hydroxylase (S3H) that hydroxylates SA to form 2,3-DHBA, a precursor of 2,3-DHBA sugar conjugates, which can be stored in the plant vacuoles (20). In fact, DLO1 also forms 2,5-DHBA in vitro, but only 2,3-DHBA in vivo (SI Appendix, Fig. S8A) (20). To gather insights about the substrate specificity of DMR6 enzymes, we used the tomato SlDMR6 enzymes. As experimental controls, we used the purified Arabidopsis DMR6 and DLO1 enzymes. Under our assay conditions, we confirmed that DLO1 converts SA to 2,3-DHBA and 2,5-DHBA, and Arabidopsis DMR6 hydroxylates SA to form 2,5-DHBA (SI Appendix, Fig. S8A). Arabidopsis DMR6 did not show any activity when naringenin was used as a substrate (SI Appendix, Fig. S8B). Remarkably, we demonstrated that SlDMR6-1 and SlDMR6-2 also convert SA to its inactive form 2,5-DHBA, acting as an S5H (Fig. 5 B and C). Therefore, the increased SA levels are caused by the reduction of 2,5-DHBA production, confirming the results from Zhang et al. (17) in Arabidopsis.
Our work also shows that the Sldmr6-1 lines have the potential to be used as a strategy to mitigate the negative impact of diseases in the field. Field conditions consist of a complex environment with multiple abiotic and biotic factors that can affect crop performance (33). We tested the resistance of the Sldmr6-1 lines to the bacterial pathogen X. perforans under field conditions in Florida. In agreement with the results of our growth-chamber experiments (Fig. 2C), the mutant lines displayed increased pathogen resistance (Fig. 6B). In addition, although the Sldmr6-1 lines showed a somewhat stunted growth as well as a reduction in fruit size (i.e., lower yield of extra-large fruits) in this field trial (Fig. 6A and SI Appendix, Figs. S11 and S12), the total yield of marketable tomatoes was not affected (Fig. 6C). Notably, these results resemble the findings on the barley Mlo gene. Like the Sldmr6-1 lines, mlo mutants show durable broad-spectrum resistance against most isolates of the powdery mildew pathogen B. graminis f. sp. hordei (34). However, under particular field conditions, mlo mutants can exhibit pleiotropic phenotypes, such as the formation of spontaneous leaf lesions, which can impact crop yield and performance (35, 36). These effects may be attenuated by the use of different host genotypes and can be overcompensated by the benefit of decreased pathogen growth in the mutants (7).
The field trial with the Sldmr6-1 mutants was performed under conditions that favor bacterial infection (warm and humid tropical climate) (37). Yet, the disease severity in this trial was not high. In addition, we were not able to evaluate the effect of SlDMR6-1 mutation on healthy uninfected plants in the field due to experimental limitations (see Results, SlDMR6-1 Mutations Lead to Pathogen Resistance in the Field). Therefore, although this study has shed light on the potential efficacy of SlDMR6-1 inactivation against bacterial spot on tomato, further trials under high disease pressures, as well as trials including healthy uninfected plants, are still necessary to better address the practical benefits of this strategy in the field. Strategies for disease management based on the activation of the plant immune system usually require a careful evaluation given that host responses can be affected by many factors, such as environmental conditions and host genotype (18, 38–40). As an example, studies on the agrochemical acibenzolar-S-methyl, a functional analog of SA used for crop protection, require extensive studies and dosage adjustments to confer resistance without significantly reducing growth and yield (41). Similarly, subsequent work on DMR6 will also include the identification of mutations in the promoter regulatory elements (42) that down-regulate SlDMR6-1 with the aim of maintaining the increased pathogen resistance, while minimizing the detrimental effects on plant growth and development in the field. Therefore, future research might include an integrated approach, in which biotic, abiotic factors, and gene dosage will be considered to provide a more comprehensive assessment of the impact of SlDMR6-1 inactivation in tomato production under field conditions and evaluate its potential as a strategy for tomato breeding. Finally, given that DMR6 is up-regulated by pathogen infection in other evolutionary distant plant species, such as cacao and cassava (SI Appendix, Fig. S13), our results might be extended to other crops, making DMR6 orthologs promising targets to engineering broad-spectrum disease resistance in additional crop species.
Materials and Methods
Details of the methods used in this work, including phylogenetic and promoter analyses, plant transformation and Cas9-mediated inactivation of SlDMR6-1 and SlDMR6-2 genes, pathogen and plant growth conditions, gene expression and RNA-seq analysis, pathogen assays, measurement of tomato growth, and histochemical GUS staining are described in SI Appendix, Materials and Methods. Information about the LC-MS/MS analysis, recombinant protein expression and purification, enzyme activity assays, HPLC quantification, homology modeling and docking analysis, and field trial assays are also provided in SI Appendix, Materials and Methods.
Supplementary Material
Acknowledgments
We thank the QB3 MacroLab facility at University of California, Berkeley, which performed recombinant protein expression and purification; the University of North Carolina Department of Chemistry Mass Spectrometry Core Laboratory, which performed the mass spectrometry measurements; the University of Nebraska’s Plant Transformation Core Research Facility, which generated the Sldmr6-1 and Sldmr6-2 mutants; the Barker DNA Sequencing Facility and the Vincent J. Coates Genomics Sequencing Laboratory at University of California, Berkeley, which performed the Sanger and the next-generation sequencing, respectively; the greenhouse staff, in particular, Christina Wistrom, for the support with plant cultivation; undergraduate students Quinton Brail and Catherine Quinto for the great help with plant genotyping and tomato harvesting; and Paulo José Pereira Lima Teixeira for critical comments on the manuscript. Research reported here was supported by the Two Blades Foundation and the Innovative Genomics Institute Director’s fund. D.P.T.T. received The Pew Latin American Fellows Program in the Biomedical Sciences Fellowship (Contract ID 00027358).
Footnotes
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2026152118/-/DCSupplemental.
Data Availability
RNA-seq data (raw sequence for transcriptomic experiments) reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database (accession no. GSE157154). All other study data are included in the article and supporting information. Previously published data were used for this work (43–48).
References
- 1.Savary S., et al., The global burden of pathogens and pests on major food crops. Nat. Ecol. Evol. 3, 430–439 (2019). [DOI] [PubMed] [Google Scholar]
- 2.van Esse H. P., Reuber T. L., van der Does D., Genetic modification to improve disease resistance in crops. New Phytol. 225, 70–86 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dangl J. L., Horvath D. M., Staskawicz B. J., Pivoting the plant immune system from dissection to deployment. Science 341, 746–751 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fuchs M., Pyramiding resistance-conferring gene sequences in crops. Curr. Opin. Virol. 26, 36–42 (2017). [DOI] [PubMed] [Google Scholar]
- 5.Mundt C. C., Pyramiding for resistance durability: Theory and practice. Phytopathology 108, 792–802 (2018). [DOI] [PubMed] [Google Scholar]
- 6.Zaidi S. S.-A., Mukhtar M. S., Mansoor S., Genome editing: Targeting susceptibility genes for plant disease resistance. Trends Biotechnol. 36, 898–906 (2018). [DOI] [PubMed] [Google Scholar]
- 7.Acevedo-Garcia J., Kusch S., Panstruga R., Magical mystery tour: MLO proteins in plant immunity and beyond. New Phytol. 204, 273–281 (2014). [DOI] [PubMed] [Google Scholar]
- 8.van Schie C. C. N., Takken F. L. W., Susceptibility genes 101: How to be a good host. Annu. Rev. Phytopathol. 52, 551–581 (2014). [DOI] [PubMed] [Google Scholar]
- 9.Dong O. X., Ronald P. C., Genetic engineering for disease resistance in plants: Recent progress and future perspectives. Plant Physiol. 180, 26–38 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Van Damme M., et al., Identification of Arabidopsis loci required for susceptibility to the downy mildew pathogen Hyaloperonospora parasitica. Mol. Plant Microbe Interact. 18, 583–592 (2005). [DOI] [PubMed] [Google Scholar]
- 11.van Damme M., Huibers R. P., Elberse J., Van den Ackerveken G., Arabidopsis DMR6 encodes a putative 2OG-Fe(II) oxygenase that is defense-associated but required for susceptibility to downy mildew. Plant J. 54, 785–793 (2008). [DOI] [PubMed] [Google Scholar]
- 12.Zeilmaker T., et al., DOWNY MILDEW RESISTANT 6 and DMR6-LIKE OXYGENASE 1 are partially redundant but distinct suppressors of immunity in Arabidopsis. Plant J. 81, 210–222 (2015). [DOI] [PubMed] [Google Scholar]
- 13.Dempsey D. A., Vlot A. C., Wildermuth M. C., Klessig D. F., Salicylic Acid Biosynthesis and Metabolism (The American Society of Plant Biologists, 2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dean J. V., Shah R. P., Mohammed L. A., Formation and vacuolar localization of salicylic acid glucose conjugates in soybean cell suspension cultures. Physiol. Plant. 118, 328–336 (2003). [DOI] [PubMed] [Google Scholar]
- 15.Zhang Z., et al., Dual regulation role of GH3.5 in salicylic acid and auxin signaling during Arabidopsis-Pseudomonas syringae interaction. Plant Physiol. 145, 450–464 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Falcone Ferreyra M. L., et al., The identification of maize and Arabidopsis type I flavone synthases links flavones with hormones and biotic interactions. Plant Physiol. 169, 1090–1107 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Y., et al., S5H/DMR6 encodes a salicylic acid 5-hydroxylase that fine-tunes salicylic acid homeostasis. Plant Physiol. 175, 1082–1093 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huot B., et al., Dual impact of elevated temperature on plant defence and bacterial virulence in Arabidopsis. Nat. Commun. 8, 1808 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jinek M., et al., A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang K., Halitschke R., Yin C., Liu C.-J., Gan S.-S., Salicylic acid 3-hydroxylase regulates Arabidopsis leaf longevity by mediating salicylic acid catabolism. Proc. Natl. Acad. Sci. U.S.A. 110, 14807–14812 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wilmouth R. C., et al., Structure and mechanism of anthocyanidin synthase from Arabidopsis thaliana. Structure 10, 93–103 (2002). [DOI] [PubMed] [Google Scholar]
- 22.Horvath D. M., et al., Transgenic resistance confers effective field level control of bacterial spot disease in tomato. PLoS One 7, e42036 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Horsfall J. G., Barratt R. W., An improved grading system for measuring plant diseases (abstract). Phytopathology 35, 655 (1945). [Google Scholar]
- 24.Ge X., et al., AtNUDT7, a negative regulator of basal immunity in Arabidopsis, modulates two distinct defense response pathways and is involved in maintaining redox homeostasis. Plant Physiol. 145, 204–215 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xing D.-H., et al., Stress- and pathogen-induced Arabidopsis WRKY48 is a transcriptional activator that represses plant basal defense. Mol. Plant 1, 459–470 (2008). [DOI] [PubMed] [Google Scholar]
- 26.Li W., Deng Y., Ning Y., He Z., Wang G.-L., Exploiting broad-spectrum disease resistance in crops: From molecular dissection to breeding. Annu. Rev. Plant Biol. 71, 575–603 (2020). [DOI] [PubMed] [Google Scholar]
- 27.Boatwright J. L., Pajerowska-Mukhtar K., Salicylic acid: An old hormone up to new tricks. Mol. Plant Pathol. 14, 623–634 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huot B., Yao J., Montgomery B. L., He S. Y., Growth-defense tradeoffs in plants: A balancing act to optimize fitness. Mol. Plant 7, 1267–1287 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van Butselaar T., Van den Ackerveken G., Salicylic acid steers the growth-immunity tradeoff. Trends Plant Sci. 25, 566–576 (2020). [DOI] [PubMed] [Google Scholar]
- 30.Kieu N. P., Lenman M., Wang E. S., Petersen B. L., Andreasson E., Mutations introduced in susceptibility genes through CRISPR/Cas9 genome editing confer increased late blight resistance in potatoes. Sci. Rep. 11, 4487 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bruggeman Q., Raynaud C., Benhamed M., Delarue M., To die or not to die? Lessons from lesion mimic mutants. Front. Plant Sci. 6, 24 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gebhardt Y., et al., Molecular evolution of flavonoid dioxygenases in the family Apiaceae. Phytochemistry 66, 1273–1284 (2005). [DOI] [PubMed] [Google Scholar]
- 33.Atkinson N. J., Urwin P. E., The interaction of plant biotic and abiotic stresses: From genes to the field. J. Exp. Bot. 63, 3523–3543 (2012). [DOI] [PubMed] [Google Scholar]
- 34.Jørgensen I. H., Discovery, characterization and exploitation of Mlo powdery mildew resistance in barley. Euphytica 63, 141–152 (1992). [Google Scholar]
- 35.Kusch S., Panstruga R., mlo-based resistance: An apparently universal “weapon” to defeat powdery mildew disease. Mol. Plant Microbe Interact. 30, 179–189 (2017). [DOI] [PubMed] [Google Scholar]
- 36.Makepeace J. C., et al., Associations between fungal and abiotic leaf spotting and the presence of mlo alleles in barley. Plant Pathol. 56, 934–942 (2007). [Google Scholar]
- 37.Potnis N., et al., Bacterial spot of tomato and pepper: Diverse Xanthomonas species with a wide variety of virulence factors posing a worldwide challenge. Mol. Plant Pathol. 16, 907–920 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang C.-H., et al., Effect of application frequency and reduced rates of acibenzolar-S-methyl on the field efficacy of induced resistance against bacterial spot on tomato. Plant Dis. 96, 221–227 (2012). [DOI] [PubMed] [Google Scholar]
- 39.Egel D. S., Kleczewski N. M., Mumtaz F., Foster R., Acibenzolar-S-methyl is associated with yield reduction when used for managing bacterial wilt (Erwinia tracheiphila) in cantaloupe. Crop Prot. 109, 136–141 (2018). [Google Scholar]
- 40.Romero A. M., Kousik C. S., Ritchie D. F., Resistance to bacterial spot in bell pepper induced by acibenzolar-S-methyl. Plant Dis. 85, 189–194 (2001). [DOI] [PubMed] [Google Scholar]
- 41.Louws F. J., et al., Field control of bacterial spot and bacterial speck of tomato using a plant activator. Plant Dis. 85, 481–488 (2001). [DOI] [PubMed] [Google Scholar]
- 42.Rodríguez-Leal D., Lemmon Z. H., Man J., Bartlett M. E., Lippman Z. B., Engineering quantitative trait variation for crop improvement by genome editing. Cell 171, 470–480.e8 (2017). [DOI] [PubMed] [Google Scholar]
- 43.Yang Y.-X., et al., RNA-seq analysis reveals the role of red light in resistance against Pseudomonas syringae pv. tomato DC3000 in tomato plants. BMC Genomics 16, 120 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jupe J., et al., Phytophthora capsici-tomato interaction features dramatic shifts in gene expression associated with a hemi-biotrophic lifestyle. Genome Biol. 14, R63 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Costa J. L., et al., Moniliophthora perniciosa, the causal agent of witches’ broom disease of cacao, interferes with cytokinin metabolism during infection of Micro-Tom tomato and promotes symptom development. New Phytologist 231, 365–381 (2021). [DOI] [PubMed] [Google Scholar]
- 46.Teixeira P. J. P. L., et al., High-resolution transcript profiling of the atypical biotrophic interaction between Theobroma cacao and the fungal pathogen Moniliophthora perniciosa. Plant Cell 26, 4245–4269 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fister A. S., et al., Theobroma cacao L. pathogenesis-related gene tandem array members show diverse expression dynamics in response to pathogen colonization. BMC Genomics 17, 363 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cohn M., et al., Xanthomonas axonopodis virulence is promoted by a transcription activator-like effector–mediated induction of a SWEET sugar transporter in cassava. MPMI 27, 1186–1198 (2014). [DOI] [PubMed] [Google Scholar]
- 49.USDA , United States Standards for Grades of Fresh Tomatoes (US Department of Agriculture, 1991). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq data (raw sequence for transcriptomic experiments) reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database (accession no. GSE157154). All other study data are included in the article and supporting information. Previously published data were used for this work (43–48).