Significance
Neurofibrillary tangles composed of misfolded and aggregated tau protein are degraded by the autophagy–lysosomal pathway. Microglia play a central role in immune surveillance and neuroinflammation. Here, we reveal that microglial autophagy critically controls microglial metabolic and immune status and also modulates neuronal tau pathology. Autophagy deficiency induces lipid droplet formation and heightened immune response, and these phenotypes can be rescued by activating the lipid efflux system, thus establishing a mechanistic link between lipid accumulation and neuroinflammation. The development of neurofibrillary tangles closely correlates with dementia in Alzheimer’s disease, and our studies provide support that autophagy augmentation may provide therapeutic benefit through targeting both immune cell function and tau pathology.
Keywords: Alzheimer's disease, autophagy, lipid metabolism, microglia, tau
Abstract
The autophagy–lysosomal pathway plays a critical role in intracellular clearance and metabolic homeostasis. While neuronal autophagy is known to participate in the degradation of neurofibrillary tangles composed of hyperphosphorylated and misfolded tau protein in Alzheimer’s disease and other tauopathies, how microglial-specific autophagy regulates microglial intrinsic properties and neuronal tau pathology is not well understood. We report here that Atg7, a key mediator of autophagosome biogenesis, plays an essential role in the regulation of microglial lipid metabolism and neuroinflammation. Microglia-specific deletion of Atg7 leads to the transition of microglia to a proinflammatory status in vivo and to inflammasome activation in vitro. Activation of ApoE and lipid efflux attenuates the lipid droplets accumulation and inhibits cytokine production in microglial cells with Atg7 deficiency. Functionally, we show that the absence of microglial Atg7 enhances intraneuronal tau pathology and its spreading. Our results reveal an essential role for microglial autophagy in regulating lipid homeostasis, neuroinflammation, and tau pathology.
Tauopathies consist of a group of neurodegenerative diseases including frontotemporal dementias (FTD) and Alzheimer’s disease (AD) and are characterized by the intraneuronal accumulation of neurofibrillary tangles (NFTs) composed of misfolded and hyperphosphorylated tau protein (1–3). Overwhelming evidence demonstrate that the NFT pathology closely correlates with neuronal injury and cognitive impairment, supporting the premise that targeting tau and NFT pathology may be therapeutically efficacious.
Although the primary function of tau is to bind and stabilize the microtubules in neuronal axons, hyperphosphorylated and misfolded tau can mislocalize to the cell bodies and dendritic spines and cause synaptic dysfunction under pathological conditions (4, 5). Autophagy (macroautophagy) is a conserved intracellular clearance pathway that functions by engulfing the cellular constituents within the LC3-positive autophagosomes and delivering them to the lysosome for degradation (6, 7). Extensive studies have documented that misfolded and aggregated proteins (8), lipid droplets (9), and mitochondria (10) can be eliminated by the autophagy–lysosomal pathway (ALP) whose deficiency causes premature aging (11), tissue degeneration (12, 13), and proinflammatory responses (14, 15). We and others have shown that activating the ALP leads to a robust effect in attenuating tau and NFT pathology in tauopathy mouse models (16–18).
Besides the intraneuronal pathology, studies using in vitro and in vivo models have established that pathological tau can transfer between neurons and propagate in a prion-like manner (19–24). Interestingly, this form of tau spreading follows a stereotyped pattern through synaptic connections in both animal models and AD brains (25). This pattern of spreading implicates extracellular tau and other cell types in the central nervous system, particularly microglia, which are the resident macrophages with phagocytic ability and crucial mediators of neuroinflammation, in tau pathogenesis. Microglia are known to mediate tau uptake, leading to its degradation (26, 27), or inadvertently promote tau spreading via exosome secretion of nondegraded tau (28). Besides direct protein uptake and clearance, microglial-mediated neuroinflammation have been shown to aggravate tau pathology (29, 30). However, the precise mechanisms regulating these multiple pathways with distinct functional outcomes are not understood, and how microglial autophagy is involved in tauopathy remain elusive.
Here, we examined the role of microglial autophagy by genetically deleting Atg7 which is essential for LC3 lipidation and autophagosome formation, in BV2 cells, in primary cultured microglia, and in microglia of adult mouse brain. We report that microglial Atg7 deficiency results in lipid droplet accumulation and metabolic dysfunction in vitro. This switches microglia to a proinflammatory state under basal conditions and exacerbates neuronal tau spreading and pathology in PS19 tau transgenic mice. The proinflammatory phenotype can be rescued by mobilizing lipid efflux, suggesting that lipid dysregulation induces heightened inflammation.
Results
Atg7-Deficient Microglia Constitute a Proinflammatory State In Vivo.
Autophagy deficiency has been implicated in immunoregulation in the peripheral immune system (14, 31, 32). Since microglia are the resident immune cells in the brain, we wondered whether microglial autophagy may play a similar role. Thus, we crossed an Atg7-floxed allele with Cx3cr1CreER mice to create microglial-specific Atg7 conditional knockout (cKO). Due to the rapid turnover of peripheral monocytes and macrophages, this system has been shown to induce specific gene recombination in the brain without affecting the peripheral immune system after 30 d posttamoxifen administration (33). Immunostaining of brain sections using anti-Iba1 and -p62 antibodies revealed a significant p62 accumulation in Iba1-positive microglia only in Cre-containing and tamoxifen-injected Atg7fl/fl (cKO) mice but not in Cre-negative or vehicle-injected controls (SI Appendix, Fig. S1A), demonstrating that the autophagic blockade is dependent upon tamoxifen-induced Atg7 deletion. To further validate the microglial-specific effect, we sorted microglial and nonmicroglial cells from Atg7 cKO and littermate Cre-negative control mouse brains (34). Western blot analysis showed that the Atg7 protein level was reduced by 70%, accompanied by a twofold increase in p62 protein level in the microglia of Atg7 cKO mouse brains compared to the controls. Neither Atg7 nor p62 showed an appreciable difference in the nonmicroglial population (SI Appendix, Fig. S1 B and C). LC3-II protein level was undetectable in sorted cells, suggesting a rapid LC3-II turnover induced by autophagic flux (SI Appendix, Fig. S1B). These results indicate an efficient microglial-specific Atg7 ablation and autophagic blockade in Atg7 cKO mice.
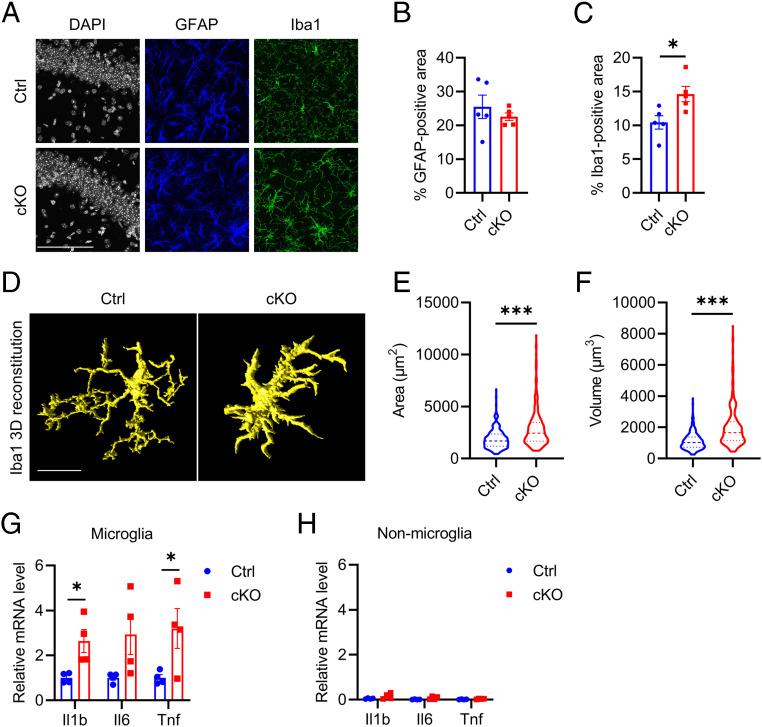
We next examined astrocytes and microglia properties in control and Atg7 cKO mice. While no differences in GFAP immunoreactivity were observed (Fig. 1 A and B), Iba1 immunostaining revealed a significant increase in Iba1-positive area in Atg7 cKO mice compared to the controls (Fig. 1 A and C). Further morphological analysis showed that Atg7-null microglia displayed more amoeboid morphology (Fig. 1D) with increased cell surface area (Fig. 1E) and volume (Fig. 1F), indicative of the activated state. qPCR analysis of sorted microglia revealed significant increases of Il1b and Tnf messenger ribonucleic acid (mRNA) levels, while the upregulation of Il6 mRNA was trending but not statistically significant (Fig. 1G). The expression levels of these cytokines were very low in nonmicroglial population, and no significant alterations were detected between control and cKO samples (Fig. 1H). These results demonstrate that Atg7 plays a physiological role in mediating microglial activation in vivo.
Fig. 1.
Atg7 loss of function induces microgliosis and increases proinflammatory cytokines level in vivo. (A) Representative immunofluorescence images of astroglial marker (GFAP), microglial marker (Iba1) in brains of littermate control, and Atg7 cKO mice 3 mo after tamoxifen injection (5 mo of age). (Scale bar, 100 μm.) (B and C) Quantification of GFAP (B)- and Iba1 (C)-positive area in A. Two-tailed Student’s t test. (n = 4/group). (D) Representative three-dimensional rendering of Iba1 immunostaining in the brains of control and Atg7 cKO mice. (Scale bar, 10 μm.) (E and F) Quantification of microglial cell surface area (E) and cell volume (F). Two-tailed Student’s t test (>200 cells from n = 4/group). (G and H) qPCR measurement of mRNA levels of proinflammatory cytokines (Il1b, Il6, and Tnf) in sorted microglial (G) and nonmicroglial (H) cell population from the brains of control and Atg7 cKO mice 3 wk after tamoxifen injection (10 wk of age). Two-tailed Student’s t test. (n = 4/group). Data are presented as mean ± SEM. *P ≤ 0.05; ***P ≤ 0.001.
Atg7 Deficiency Promotes Proinflammatory Response and Inflammasome Activation In Vitro.
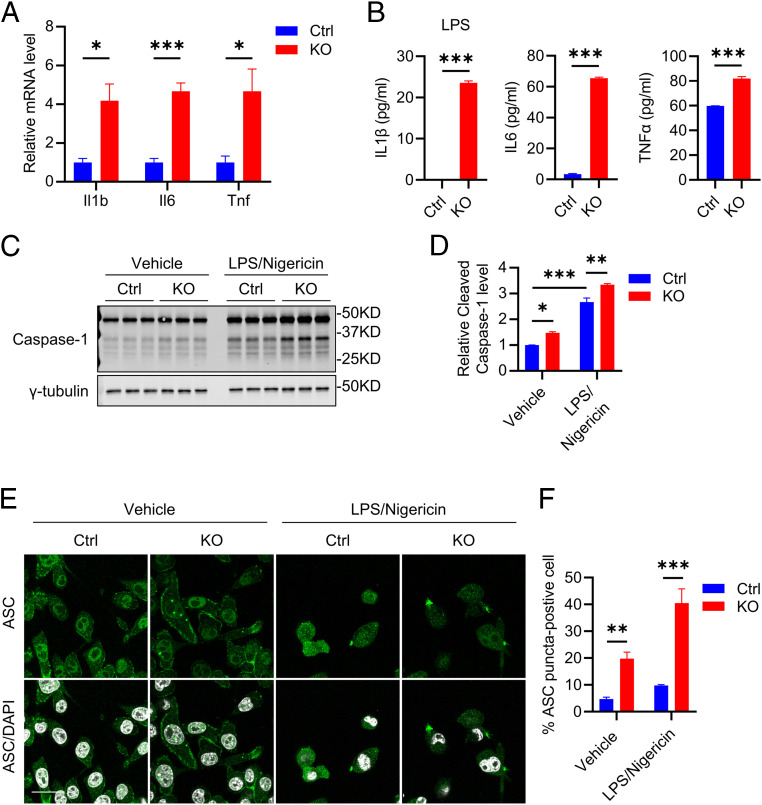
To probe the mechanism of proinflammatory response caused by Atg7 depletion, the CRISPR-Cas9 genome-editing tool was used to knockout the Atg7 gene in BV2 cells, an immortalized murine microglia cell line (SI Appendix, Fig. S2). Western blot analysis showed an almost complete depletion of Atg7 in KO cells, which is associated with a significant accumulation of p62 and absence of LC3-II (SI Appendix, Fig. S2 A and B), consistent with its essential role in LC3 lipidation. As expected, LC3 immunostaining revealed no autophagosome formation (marked as LC3 puncta) in Atg7 KO BV2 cells, while they are readily detectable in control cells (SI Appendix, Fig. S2C). Consistent with our in vivo data, Atg7 deficiency induced robust upregulation of proinflammatory cytokine gene expression (Fig. 2A) and protein secretion under basal (IL6 and TNFα) or lipopolysaccharides (LPS)-induced (IL1β) conditions (Fig. 2B). IL1β was undetectable in both control and Atg7 KO BV2 cells without stimulation.
Fig. 2.
Atg7 KO promotes proinflammatory status in vitro. (A) qPCR measurement of mRNA levels of proinflammatory cytokines (Il1b, Il6, and Tnf) in control and Atg7 KO BV2 cells. Two-tailed Student’s t test (n = 4 of two experiments). (B) ELISA measurement of secreted protein levels of proinflammatory cytokines (IL1β, IL6, and TNFα) in conditioned media of 5 × 104 control and Atg7 KO BV2 cells. The IL1β measurement was performed under 100 ng/mL LPS treatment for 16 h. Two-tailed Student’s t test (n = 3 of two experiments). (C) Western blot image of inflammasome marker Caspase-1 level in control and Atg7 KO BV2 cells treated with vehicle or LPS priming followed by Nigericin treatment. (D) Quantification of cleaved Caspase-1 level in C. Two-way ANOVA was followed by Sidak's multiple comparisons test (n = 3 of two experiments). (E) Representative immunofluorescence images of apoptosis-associated speck-like protein containing a caspase recruitment domain (ACS) in control and Atg7 KO BV2 cells treated with vehicle or LPS priming followed by Nigericin treatment. (Scale bar, 20 μm.) (F) Quantification of ASC speck level in E. Two-way ANOVA was followed by Sidak's multiple comparisons test (n = 10 fields of two experiments). Data are presented as mean ± SEM. *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001.
Since increased IL1β is often associated with inflammasome activation (29, 35), we next tested whether inflammasome is activated in Atg7 KO microglia. Western blot analysis indicated significantly up-regulated cleaved Caspase-1 in Atg7 KO cells compared to the controls (Fig. 2 C and D, Vehicle), with higher ASC puncta by immunostaining (Fig. 2 E and F, Vehicle). Using the LPS priming and Nigericin treatment protocol to activate NLRP3 inflammasome (35), we found a significantly higher cleaved Caspase-1 (Fig. 2 C and D, LPS/Nigericin) and ASC speck occurrence rate (Fig. 2 E and F, LPS/Nigericin) in Atg7 KO cells compared to control cells. These results demonstrate that Atg7 modulates microglial proinflammatory state in vitro.
Atg7 Regulates Cellular Energy and Lipid Homeostasis In Vitro.
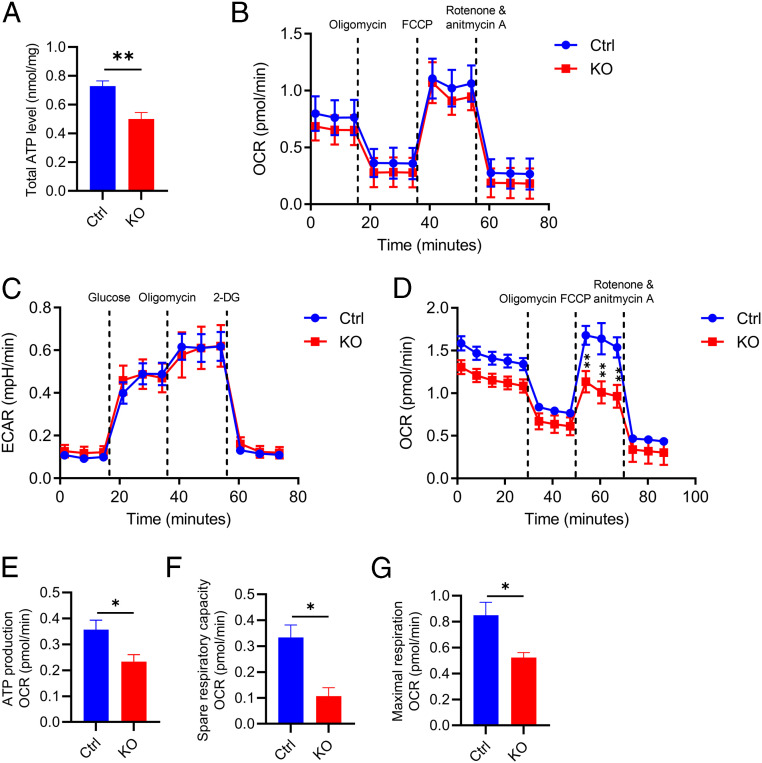
We next assessed whether Atg7 regulates other cellular processes. Measurement of total cellular ATP revealed reduced levels in BV2 cells with Atg7 deficiency (Fig. 3A). To probe the specific pathways contributing to this phenotype, we examined the three major ATP-producing pathways: oxidative phosphorylation (OXPHOS), glycolysis, and the long-chain fatty acid β-oxidation (FAO) by the Seahorse analyzer (36). Neither OXPHOS (Fig. 3B) nor glycolysis (Fig. 3C) were different between the control and Atg7 KO cells. However, when measuring the FAO, which is a major lipid consumption pathway, by feeding the cells with the FAO substrate Palmitate-bovine serum albumin (BSA), the result showed a significantly lower oxygen consumption rate (OCR) in Atg7 KO cells compared to WT controls (Fig. 3D), along with reduced ATP production (Fig. 3E), spare respiratory capacity (Fig. 3F), and maximal respiration (Fig. 3G). This result indicates that reduced FAO-dependent ATP production likely contributed to a lower total ATP level. Treatment with Etomoxir, an irreversible inhibitor of carnitine palmitoyltransferase-1, completely blocked FAO, confirming the reliability of our FAO quantification method (SI Appendix, Fig. S3A).
Fig. 3.
Atg7 regulates lipid metabolic homeostasis in vitro. (A) Total cellular ATP levels in WT and Atg7 KO BV2 cells. Two-tailed Student’s t test (n = 8 of two experiments). (B) OXPHOS measurement of OCR levels in control and Atg7 KO BV2 cells. Two-tailed Student’s t test (n = 3 of two experiments). (C) Glycolysis measurement of ECAR levels in control and Atg7 KO BV2 cells. Two-tailed Student’s t test (n = 3 of two experiments). (D) FAO measurement of OCR levels in control and Atg7 KO BV2 cells. Two-tailed Student’s t test (n = 3 of two experiments). (E–G) The measurement of ATP production (E), spare respiratory capacity (F), and maximal respiration (G) from D. Two-tailed Student’s t test (n = 3 of two experiments). Data are presented as mean ± SEM. *P ≤ 0.05; **P ≤ 0.01.
Since FAO requires mitochondria, we evaluated general mitochondrial properties by staining the control and Atg7 KO BV2 cells with MitoTrackerRed (a mitochondrial membrane potential [ΔΨm]-dependent dye) and MitoSOX (a mitochondria-specific reactive oxygen species [ROS] indicator) respectively. No appreciable differences were detected between the two groups (SI Appendix, Fig. S3 B–E), indicating that the overall mitochondrial function is mostly intact without Atg7. This assessment is consistent with normal OXPHOS in Atg7-null cells. Thus, Atg7 deficiency primarily impinges on the lipid metabolic pathway.
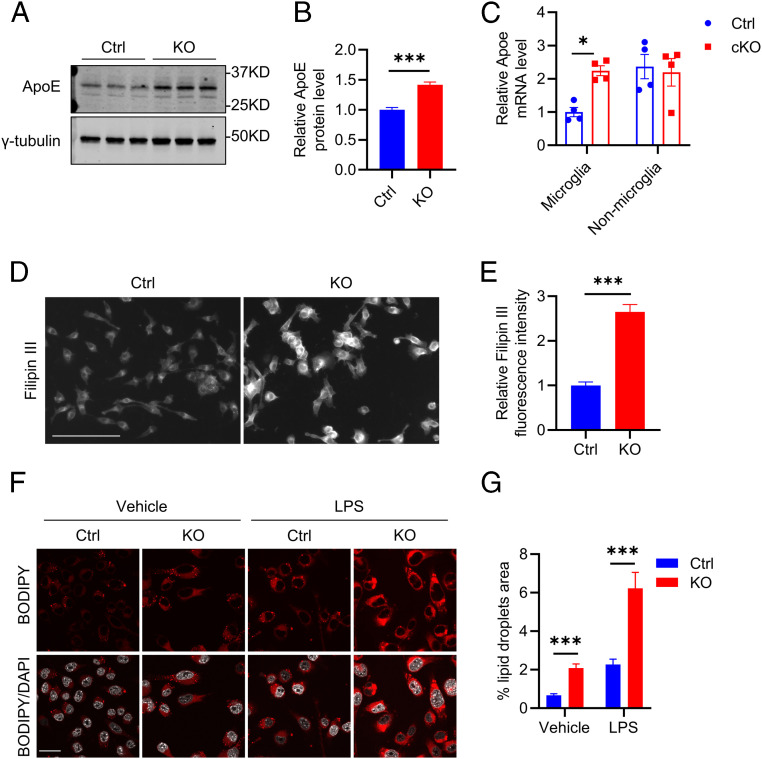
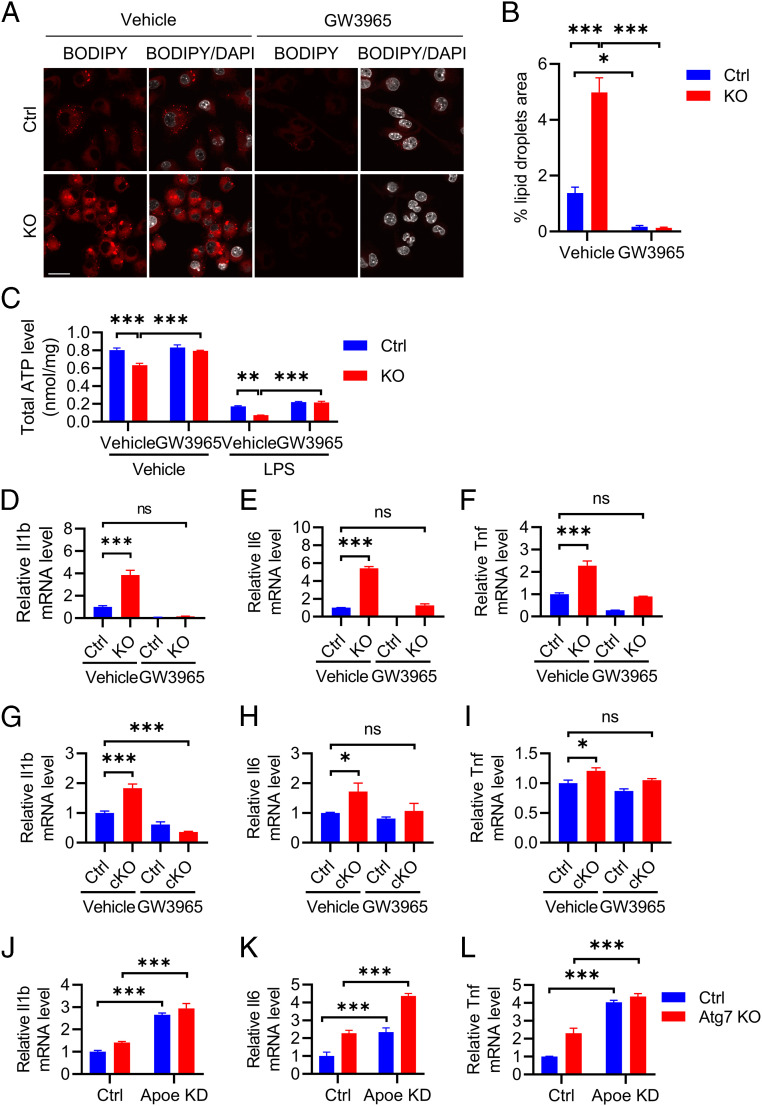
In line with this assessment, we found that apolipoprotein E (ApoE), a major cholesterol and lipid transporter, was elevated in both Atg7 KO BV2 cells detected by Western blotting (Fig. 4 A and B) and in sorted microglia of Atg7 cKO mice measured by qPCR (Fig. 4C). Intriguingly, this was associated with increased intracellular cholesterol as evidenced by higher Filipin III staining (Fig. 4 D and E). This result is corroborated by the higher levels of lipid droplets known to contain neutral lipids and cholesterol revealed by BODIPY C12 labeling in Atg7 KO BV2 cells compared to the controls (Fig. 4 F and G, Vehicle). LPS treatment further increased lipid droplets in both control and Atg7 KO cells (Fig. 4 F and G, LPS). These results suggest that Atg7 regulates lipid homeostasis and that up-regulated ApoE expression in Atg7-null microglia may represent an adaptive mechanism in response to heightened lipid stress.
Fig. 4.
Atg7 KO disrupts lipid homeostasis. (A) Western blot image of ApoE protein level in control and Atg7 KO BV2 cells. (B) Quantification of ApoE level in A. Two-tailed Student’s t test (n = 3 of two experiments). (C) qPCR measurement of mRNA levels of Apoe in the sorted microglial and nonmicroglial cell population from the brains of control and Atg7 cKO mice at 3 wk after tamoxifen injection (10 wk of age). Two-tailed Student’s t test (n = 4/group). (D) Representative fluorescence microscopy images of filipin staining in control and Atg7 KO BV2 cells. (E) Quantification of Filipin fluorescence intensity in D. Two-tailed Student’s t test (n = 8 of two experiments). (F) Representative immunofluorescence images of lipid droplets (BODIPY C12 labeled) in control and Atg7 KO BV2 cells treated with vehicle control or 100 ng/mL LPS for 16 h. (Scale bar, 20 μm.) (G) Quantification of lipid droplets level in F. Two-tailed Student’s t test (n = 50 cells/group of two experiments). Data are presented as mean ± SEM. *P ≤ 0.05; ***P ≤ 0.001.
Mobilizing Lipid Efflux Rescues Proinflammation and Metabolic Dysfunction.
Recent reports have linked age-associated lipid accumulation with proinflammatory response (37–39). Since loss of Atg7 significantly alters fatty acid consumption and cholesterol and lipid droplet accumulation, we sought to test whether stimulating the lipid efflux system can restore the microglial inflammation and metabolic homeostasis. The liver X receptor (LXR) has been shown to control the expression of key lipid transport genes such as Apoe and its binding partners adenosine triphosphate–binding cassette A1 and G1 (39). Therefore, we tested whether the treatment of the LXR agonist GW3965 (40, 41) reduces inflammation and improves metabolism. LPS treatment was used to enhance the lipid droplet formation and to induce cytokine expression. As expected, GW3965 treatment increased Apoe expression at the mRNA level in both control and Atg7 KO BV2 cells (SI Appendix, Fig. S4A). Interestingly, GW3965 not only robustly reduced lipid droplets formation (Fig. 5 A and B) but also normalized the total ATP levels in Atg7 KO BV2 cells (Fig. 5C). Importantly, GW3965 treatment dampened proinflammatory cytokine expressions in BV2 cells (Fig. 5 D–F) and in primary cultured microglia (SI Appendix, Fig. S4B and Fig. 5 G–I) with Atg7 deficiency. Further, to test a functional role of ApoE in this process, we created Apoe knockdown by CRISPR-Cas9 genome editing on control and Atg7 KO BV2 cells (SI Appendix, Fig. S4C). Contrary to the GW3965 treatment, Apoe inactivation resulted in a robust upregulation of proinflammatory cytokine gene expression in both control and Atg7 KO cells (Fig. 5 J–L). The results combined support the notion that promoting lipid efflux rescues proinflammation and metabolic dysfunction caused by Atg7 deficiency, and this process is likely ApoE dependent.
Fig. 5.
Activating lipid efflux rescues proinflammation and metabolic dysregulation. (A) Representative immunofluorescence images of BODIPY C12–labeled lipid droplets in control and Atg7 KO BV2 cells treated with vehicle or 10 µM GW3965 in the presence of 100 ng/mL LPS for 16 h. (Scale bar, 20 μm.) (B) Quantification of lipid droplets level in A. Two-tailed Student’s t test (n = 60 cells/group of two experiments). (C) Total cellular ATP levels in control and Atg7 KO BV2 cells were treated with vehicle control or 10 µM GW3965 with or without 100 ng/mL LPS for 16 h. Two-way ANOVA was followed by Tukey's multiple comparisons test (n = 6 of two experiments). (D–F) qPCR measurement of mRNA levels of proinflammatory cytokines Il1b (D), Il6 (E), and Tnf (F) in WT control and Atg7 KO BV2 cells treated with vehicle or 10 µM GW3965 in the presence of 10 ng/mL LPS for 16 h. One-way ANOVA followed by Dunnett's multiple comparisons test (n = 3 of two experiments). (G–I) qPCR measurement of mRNA levels of proinflammatory cytokines Il1b (G), Il6 (H), and Tnf (I) in WT control and Atg7 cKO primary cultured microglia treated with vehicle or 10 µM GW3965 in the presence of 10 ng/mL LPS for 16 h. One-way ANOVA was followed by Dunnett's multiple comparisons test (n = 3 of two experiments). (J–L) qPCR measurement of mRNA levels of proinflammatory cytokines Il1b (H), Il6 (I), and Tnf (J) in control and Atg7 KO BV2 cells with or without Apoe knockdown (KD). Two-way ANOVA was followed by Sidak's multiple comparisons test. Data are presented as mean ± SEM. ns, not significant, *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001.
Microglial Atg7 Deficiency Augments Tau Pathology and Spreading.
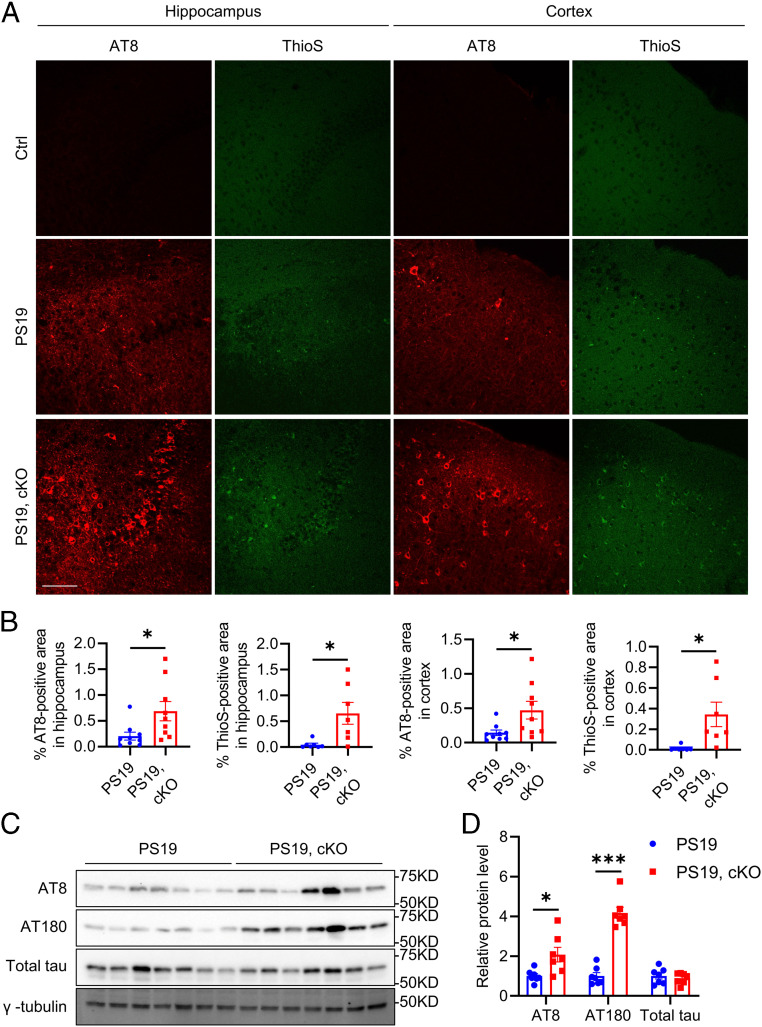
Sustained microglial activation is one of the pathological hallmarks of tauopathy, we sought to test whether this was correlated with altered microglial autophagy. Immunofluorescence staining using the autophagic flux marker p62 revealed minimum p62 immunoreactivity in microglia of control mice but significantly higher fluorescence signal in a group of microglia in the hippocampus of PS19 mice (SI Appendix, Fig. S5 A and B), suggesting a reduced autophagic flux in the microglia of the tauopathy mouse model. Since microglial inflammasome activation and neuroinflammation have been shown to promote tau pathology (29, 30, 42), we thought to determine the functional role of microglial Atg7 on tau pathology in vivo. We generated PS19 tau transgenic mice with microglial Atg7 cKO (PS19) by crossing the PS19 mice with Atg7fl/fl and Cx3cr1CreER mice and treated the mice with tamoxifen at 2 mo of age and performed histological and biochemical characterizations of PS19 and PS19, cKO mice at 12 mo of age. Immunofluorescence staining using the phospho-tau antibody AT8 and staining with the Thioflavin S dye that recognizes the NFTs showed significantly higher fluorescence intensity in both the hippocampus and cortex, areas most affected in the PS19 mouse model and AD patients, of PS19, cKO samples compared to the PS19 mice (Fig. 6 A and B and SI Appendix, Figs. S6 and S7). Consistent with the histological results, immunoblotting revealed significant increases of AT8- and AT180-positive phosphorylated tau in PS19, cKO samples compared to the PS19 controls, while total tau levels remained constant (Fig. 6 C and D). We next assessed the neuronal and synaptic status in PS19 and PS19, cKO mice. Immunofluorescence staining using the neuronal marker NeuN and the presynaptic marker synaptophysin revealed a significant reduction of synaptophysin fluorescence intensity (SI Appendix, Fig. S5 C and E) while no appreciable differences in NeuN immunoreactivity were observed (SI Appendix, Fig. S5 C and D), indicating significant synaptic degeneration but not overt neuronal loss in PS19, cKO samples compared to PS19 mice.
Fig. 6.
Microglial Atg7 cKO exacerbates tau pathology in vivo. (A) Representative immunofluorescence images of the hippocampus and cortex of control, PS19, and PS19, Atg7 cKO mice at 12 mo of age using the AT8 antibody and Thioflavin S dye. (Scale bar, 200 μm.) (B) Quantification of AT8-positive and ThioS-positive area in A. Two-tailed Student’s t test (AT8 staining: n = 9/group; ThioS staining: n = 7/group). (C) Representative Western blot image of phosphorylated tau (AT8, AT180) and total tau protein levels in brains of PS19 and PS19, Atg7 cKO mice at 12 mo of age. (D) Quantification of AT8, AT180, and total tau protein levels in C. Two-tailed Student’s t test (n = 7/group). Data are presented as mean ± SEM. *P ≤ 0.05; ***P ≤ 0.001.
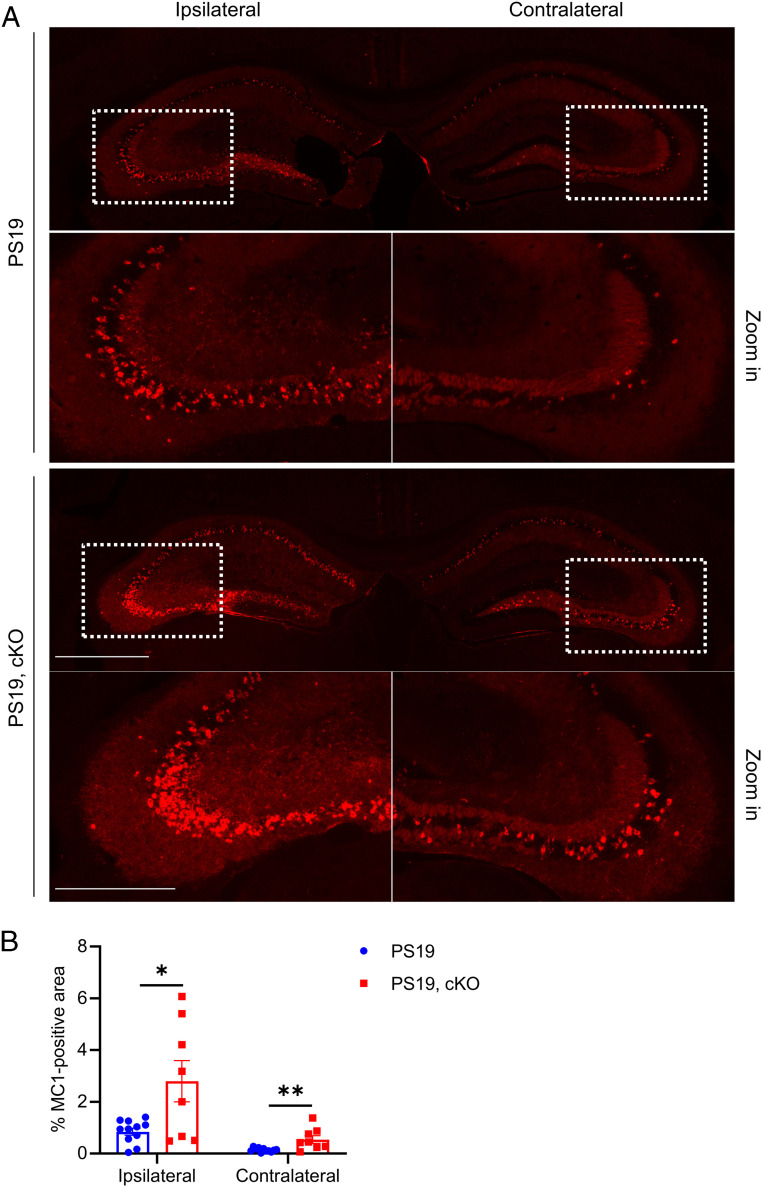
It is well demonstrated that pathological tau undergoes prion-like spreading (23, 24). We next examined the effect of microglial Atg7 cKO in pathological tau spreading. We performed tau spreading experiment by injecting the brain lysate of rTg4510 tau transgenic mice to the left hippocampus of 2- to 3-mo-old PS19 and PS19, cKO mice and analyzed the mice 2 mo later (43). Immunofluorescence staining using the MC1 antibody revealed that the misfolded tau was significantly increased in both the ipsilateral and contralateral hippocampus in PS19, cKO mice compared to PS19 mice (Fig. 7 and SI Appendix, Fig. S8). Thus, microglial-specific depletion of Atg7 exacerbates tau pathology and spreading.
Fig. 7.
Microglial Atg7 cKO augments tau spreading. (A) Representative immunofluorescence images of the ipsilateral and contralateral hippocampus using the misfolded tau MC1 antibody. PS19 and PS19, Atg7 cKO mice were injected with rTg4510 brain lysate at 2 to 3 mo of age. Pathological tau spreading was analyzed 8 wk postinjection. (Scale bar, 1,000 μm; 500 μm in zoom in.) (B) Quantitative analysis of MC1-positive areas in the ipsilateral and contralateral hippocampus of PS19 and PS19, Atg7 cKO mice in A. Two-tailed Student’s t test (n = 11 for PS19; n = 8 for PS19, cKO). Data are presented as mean ± SEM. *P ≤ 0.05; **P ≤ 0.01.
Discussion
The ALP is involved in the degradation of cellular macromolecules and organelles (7). Overwhelming evidence demonstrates an essential activity of neuronal autophagy in the clearance of misfolded and aggregated proteins such as hyperphosphorylated tau and NFTs. However, the role of microglial autophagy is less understood. Here, we show that microglial autophagy regulates lipid metabolism, neuroinflammation, and tau pathology. Blocking autophagy by genetic ablation of Atg7 leads to the buildup of lipid droplets and induces microglial transition to a proinflammatory phenotype. Activating ApoE and lipid efflux effectively rescued the inflammatory phenotypes, providing support that neuroinflammation acts downstream of lipid accumulation.
Both lipid droplets and mitochondria are autophagy substrates and key sources of energy supply. We demonstrate here that Atg7 deficiency is associated with lipid droplet accumulation. Our metabolic measurements revealed a significant reduction of FAO and associated energy production but relatively normal mitochondrial membrane potential and ROS in Atg7 KO cells, suggesting that mitochondrial function is relatively preserved in the absence of autophagy. Since ALP degrades lipid droplets and converts it to free fatty acids to fuel mitochondrial FAO, it is plausible that autophagic deficiency reduces the recycling of free fatty acids, leading to lower FAO and cellular ATP levels. Surprisingly, OXPHOS and glycolysis, the major energy generating pathways, remain unaltered in Atg7-null BV2 cells, suggesting that there is a unique requirement for microglial autophagy in regulating lipid turnover and homeostasis.
Along with lipid droplet formation, we found altered ApoE expression and lipid metabolism in Atg7-null BV2 cells, indicating that ApoE acts as a lipid stress sensor. The fact that increasing ApoE and lipid transport by the LXR agonist GW3965 treatment potently removed lipid droplets demonstrates that ApoE is a key regulator of microglial lipid accumulation. Its upregulation by autophagic blockade may thus function as an adaptive mechanism to counteract intracellular lipid accumulation.
That removing lipid droplets leads to complete rescue of cytokine expression supports the mechanism that deregulated lipid homeostasis is the primary cause for the heightened inflammation by autophagic blockade. The anti-inflammatory activity of GW3965 could be attributed by a general removal of lipid droplets (37) or efflux of specific proinflammatory lipid species (44). Although the precise mechanism remains to be elucidated, this interpretation is consistent with the known anti-inflammatory role of autophagy in peripheral tissues (45). It is also in agreement with Marschallinger et al., who reported a novel class of aging-related lipid droplet–accumulating microglia (LDAM) with increased ROS and proinflammatory cytokines secretion (37). Since the ALP declines with aging, it is tempting to speculate that a subdued autophagic clearance of lipid droplets might contribute to LDAM formation in the aging microglia. However, there are likely substantial differences between LDAM and Atg7 KO microglia. In particular, ApoE was elevated in Atg7-null microglia but unchanged in LDAM. Additional work is needed to understand the nature and functional consequences of the microglial lipid droplets under different pathophysiological conditions.
Tau is localized to the neurons and can also be secreted. Extracellular tau can be taken up by astrocytes and microglia for degradation or transferred to another neuron, a phenomenon termed prion-like spreading. We show that microglial Atg7 deficiency in PS19 mice leads to elevated phospho-tau levels and NFT pathology. This is associated with accelerated spreading induced by tau seeds. Microglial autophagy may impact tau pathogenesis by targeting either intraneuronal or extracellular secreted tau through multiple but not mutually exclusive mechanisms. Specifically, proinflammatory cytokines TNFα, IL1β, and IL6 and inflammasome activation have been shown to directly promote tau phosphorylation in neurons (42, 46, 47). Therefore, microglial Atg7 deficiency could modulate neuronal tau phosphorylation through increased cytokine secretion and inflammasome activation. With regard to extracellular tau, microglial Atg7 may influence its uptake through two distinct mechanisms. First, heightened microglial activation and lipid droplets accumulation have been associated with reduced phagocytosis (37). Accordingly, microglial Atg7 deficiency may impede the extracellular tau uptake and clearance. Second, recent studies demonstrated that Atg7 can target exogenous materials for endocytosis or phagocytosis through LC3-dependent noncanonical autophagy (48–50). Of interest, Aβ has been shown to subject to LC3-associated endocytosis (48). Thus, microglial Atg7 deficiency may impair the endocytosis or phagocytosis of extracellular tau through noncanonical autophagy. One of the limitations of our work is that the mechanism by which cholesterol metabolism alteration contributes to the enhanced tau pathology and spread induced by Atg7 deficiency in microglia remains unknown. In addition, to what degree each of these mechanisms contribute to the overall tau dynamics warrants further investigation. Nevertheless, our results support a beneficial role of microglial autophagy in attenuating tau pathology.
Our finding is in line with a recent publication demonstrating a similar activity of microglial autophagy in addressing α-synuclein pathology (51) but may appear to contradict with Asai et al. documenting that microglia promote tau spreading in an exosome-dependent manner (28). This may be explained by differences in experimental manipulations and outcome measures. In particular, we target a specific microglial degradation pathway whereas Asai et al. employed microglial depletion that ridded all microglial-associated activities. Since there is minimum evidence directly linking autophagy with exosomal secretion, the two experimental systems likely affect tau through distinct mechanisms. Further, since we observed a strong effect of microglial Atg7 deficiency on neuronal tau pathology, changes in the spreading may be a consequence of accelerated pathology. In contrast, Asai et al. implicates microglial exosome-mediated tau spread as a primary mechanism in tau pathogenesis. Nevertheless, additional studies are required to decipher the various microglial pathways in responding and processing the distinct tau species and how they may dictate the functional outcomes in tau pathobiology.
The ALP is known to decline with aging and brain aging is associated with the buildup of LDAM that are proinflammatory (37). Our study demonstrates a beneficial effect of microglial autophagy in tauopathy and establish a causal link between lipid accumulation and trafficking with neuroinflammation. Thus, our study not only offers insights of microglial autophagy in tauopathy but also may have direct implications in aging and age-associated anomalies. In this regard, Cantuti-Castelvetri et al. showed that promoting ApoE-dependent cholesterol efflux by GW3965 leads to improved functional recovery in a demyelination model (39). Thus, it will be interesting to test whether GW3865 offers similar lipid-lowering and anti-inflammatory effect in age-associated LDAM and in neurological diseases with neuroinflammatory underpinnings. Combined with our previous studies showing beneficial effects of tau degradation and exocytosis by neuronal ALP (17, 52) and tau uptake and clearance by astroglial ALP (53), our work overall demonstrates that the ALP acts on multiple cell types to facilitate tau clearance and support autophagy activators as potential therapy for diseases of tauopathy.
Materials and Methods
Mice.
The tau P301S transgenic mouse line (PS19) was purchased from the Jackson Laboratory. Atg7 flox and Cx3cr1CreER mice were kind gifts from Andrea Ballabio’s laboratory (Telethon Institute of Genetics and Medicine) and Wen-biao Gan’s laboratory (New York University School of Medicine), respectively (33, 54). The sample size was determined based on previous studies (17, 43, 52). Both male and female mice were randomly used in the study. Investigators were blinded to the group identities during data collection and analysis. Mice with all genotypes were administered tamoxifen intraperitoneally (180 mg/kg/d for 5 d) at 6 to 8 wk of age (55). All procedures were performed based on the protocols approved by the Institutional Animal Care and Use Committee of Baylor College of Medicine.
Reagents.
MC1 antibody was a kind gift from Peter Davies (Feinstein Institute for Medical Research). Other antibodies and reagents were the following: AT8 (Invitrogen, MN1020), Total tau (DAKO, A0024), γ-tubulin (Sigma, T6557), Atg7 (Sigma, A2856), p62 (Sigma, P0067; Abcam, ab56416), LC3B (Sigma, L7543), GFAP (Millipore, MAB360), Iba1 (Wako, 019-19741), Caspase-1 (AG-20B-0042-C100), ASC (Adipogen, AG-25B-0006-C100), ApoE (Millipore, AB947), NeuN (Millipore, mab377), Synaptophysin (Abcam, ab16659), Il1β enzyme-linked immunosorbent assay (ELISA) kit (R&D systems, DY401), IL6 ELISA kit (R&D systems, DY406), TNFα ELISA kit (R&D systems, DY410), Mito stress kit (Agilent, 103010-100), Glycolysis kit (Agilent, 103017-100), Seahorse XF Palmitate-BSA FAO Substrate (Agilent, 102720-100), Seahorse XFp FluxPak (Agilent, 103022-100), MitoTrackerRed (Invitrogen, M7512), MitoSOX (Invitrogen, M36008), DAPI (Invitrogen, D1306), BODIPY 558/568 C12 (Invitrogen, D3835), GW3965 (Sigma, G6295), HRP-conjugated secondary antibodies (EMD Millipore, AP100P, AP307P), IRDye® secondary antibodies (LI-COR, 926-32211, 926-32212, 926-68070, 926-68073), Alexa Fluor–conjugated secondary antibodies (Thermo Fisher Scientific, A21202, A21422, A21206, A31572), Streptavidin-HRP kit (R&D systems, DY998), lipofectamine 3000 (Thermo Fisher Scientific, L3000015), protease and phosphatase inhibitor mixtures (Roche, 04693116001, 04906845001), ECL (Pierce, 32106, 34096), Nonidet P-40 (Thermo Fisher Scientific, 85124), Pen/Strep (Thermo Fisher Scientific, 15140122), and cell-based cholesterol assay (Abcam, ab133116). The pSpCas9(BB)-2A-GFP (PX458) plasmid was a kind gift from Tong Liang (Baylor College of Medicine). The lentiCRISPR version 2 plasmid was a kind gift from Weiwei Dang (Baylor College of Medicine). The rTg4510 mice brain lysate was described in the previous publication (56).
Cellular Assays.
BV2 cell line maintenance was described as previously (57). To generate the Atg7 KO cell line, two CRISPR guide RNAs (gRNAs) were designed to target mouse Atg7 exon 2 (gRNA1: GAAGTTGAACGAGTACCGCC, gRNA2: TTTAATAGTGCCCTGGACGT). The gRNAs were cloned into the PX458 construct respectively. The transfections were performed according to the manufacturer protocol. Two days after transfection, single GFP-positive cells were sorted using a BD fluorescence-activated cell sorting (FACS) Aria II cell sorter and cloned. The KO efficiency was identified through Atg7 Western blotting. To generate the Apoe knockdown cell line, gRNA: CACAGCCCGCCCTAGCCCTG was cloned into lentiCRISPR version 2 plasmid and cotransfected with VSVG and Gag-Pol plasmids to produce lentivirus. BV2 and BV2 Atg7 KO cells were infected by lentivirus and selected by puromycin treatment (4 μg/mL) for 3 d. The knockdown efficiency was identified through ApoE Western blotting.
Microglia Sorting.
Three weeks after tamoxifen administration, the brains of Atg7 cKO and WT littermate controls mice were collected, and the concurrent brain cell type acquisition assay was performed to sort the microglial and nonmicroglial cells as described previously (34).
Total ATP Assay.
WT and ATG7 KO BV2 cells were seeded in 96-well plates at 5 × 104 cells per well 1 d before assay. After the medium was removed, attached cells were lysed by adding 1× passive lysis buffer (Promega, E1941) in the wells and pipetting for 10 times. The lysates were split equally into two aliquots. One aliquot was transferred to a white opaque 96-well plate for the measurement of ATP concentration using an ATP determination kit (Thermo Fisher Scientific, A22066). The other aliquot was transferred to a clear 96-well plate for the measurement of protein concentration using the bicinchoninic acid assay (BCA) assay (Thermo Fisher Scientific, 23227). The final ATP level was calculated by normalizing the ATP concentration of a well to its corresponding protein concentration.
Metabolic Measurements.
The metabolic measurements follow the protocols of the Mito stress kit or Glycolysis kit. The cells were seeded in Seahorse FluxPaks plates one day before assay. Seahorse FluxPaks sensor cartridges were incubated in calibrating buffer overnight. The drugs’ final concentration was as below: FAO: oligomycin 5 μM, FCCP 2 μM, rotenone/antimycin A 0.5 μM; OXPHOS: oligomycin 1.5 μM, FCCP 0.5 μM, rotenone/antimycin A 0.5 μM; glycolysis: Glucose 10 mM, oligomycin 1.5 μM, 2-DG 50 nM. The oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) were measured by Agilent Seahorse XF HS Mini Analyzer. After analysis, the cells were lysed, and protein concentration was measured as the internal control.
Western Blotting.
The cells or mouse forebrain tissues were homogenized in lysis buffer (Tris-buffered saline [TBS] with 1% Nonidet P-40, 1% sodium deoxycholic acid, 0.1% sodium dodecyl sulfate, and protease and phosphatase inhibitor mixtures). The homogenates were sonicated and centrifuged at 20,000 × g for 10 min to spin down the debris. Supernatants were boiled with the loading buffer for sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to polyvinylidene difluoride (PDVF) or nitrocellulose membrane. After incubation with primary and secondary antibodies, the membranes were developed by the Chemidoc image system (Bio-Rad) or Odyssey image system (LI-COR).
Tau Spreading Assay.
The tau spreading assay was performed as described previously (43). Briefly, PS19 and PS19 Atg7 cKO mice were stereotaxically injected with 2 μL of 10% brain homogenate from 7-mo-old rTg4510 mice at 2 to 3 mo of age into one hippocampal hemisphere (bregma, −2.5 mm, lateral, −2 mm, and depth, −1.8 mm). The mice were euthanized 8 wk later for misfolded tau (MC1) immunofluorescence staining to evaluate tau spreading.
Immunofluorescence Staining.
Mice were perfused with 4% paraformaldehyde (PFA) in TBS followed by overnight fixation. Then, the tissues immersed in 30% sucrose in TBS for dehydration. Brain tissues were cut into 30-μm sections for staining. For BV2 cell lines, cells were fixed in 4% PFA for 20 min at room temperature. Brain sections or cultured cells were incubated with primary antibodies in TBS with 0.4% Triton X-100 and 2% donkey serum overnight, followed by incubating with Alexa Fluor–conjugated secondary antibodies. Brain sections or cells were imaged by confocal microscopy (Leica SPE) or EVOS Cell Imaging System (Thermo Fisher Scientific). To quantify the microglia morphology, Iba1-positive cells were imaged with 63× objective with Z Series by confocal microscopy. Images were analyzed by Surfaces module using the Imaris software (Bitplane). To quantify the percentage area, the images were taken by confocal microscopy or EVOS Cell Imaging System. For brain sections, the hippocampal and cortical regions were identified based on the mouse brain atlas. The images were analyzed by Analyze Particles function after threshold adjustment using ImageJ software. To quantify the cell number in each field, the cell counter plugin of ImageJ was used.
Statistics.
All data are presented as mean ± SEM. For animal experiments, each dot represents one animal. Power analysis was performed using a CI of α = 0.05. Two group comparisons were analyzed using two-tailed Student’s t test, and multiple comparisons were analyzed using one-way or two-way ANOVA followed by post hoc tests as indicated in the figure legends using GraphPad Prism. P values less than or equal to 0.05 were considered statistically significant. *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001.
Supplementary Material
Acknowledgments
We are grateful to the late P. Davies for MC1 antibody and W. Gan (New York University) and A. Ballabio (The Telethon Institute of Genetics and Medicine) for Cx3cr1CreER and Atg7 floxed mice, respectively. We appreciate B. Contreras for expert technical assistance and members of H. Zheng's laboratory for insightful discussions. We thank C. Beeton, J. Sederstrom, and the Baylor College of Medicine Cytometry and Cell Sorting Core supported by Grant NCI-CA125123 for FACS analysis. We thank A. Catic (Baylor College of Medicine), L. Maneix (Baylor College of Medicine), and L. Yan (MD Anderson Cancer Center) for technical assistance with the metabolic measurements. This project was supported by grants from the NIH (R01 NS093652, R01 AG020670, R01 AG057509, RF1 AG054111, and RF1 AG062257 to H.Z.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
See online for related content such as Commentaries.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2023418118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix.
References
- 1.Lee V. M., Goedert M., Trojanowski J. Q., Neurodegenerative tauopathies. Annu. Rev. Neurosci. 24, 1121–1159 (2001). [DOI] [PubMed] [Google Scholar]
- 2.Mandelkow E. M., Mandelkow E., Biochemistry and cell biology of tau protein in neurofibrillary degeneration. Cold Spring Harb. Perspect. Med. 2, a006247 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gendron T. F., Petrucelli L., The role of tau in neurodegeneration. Mol. Neurodegener. 4, 13 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hoover B. R., et al., Tau mislocalization to dendritic spines mediates synaptic dysfunction independently of neurodegeneration. Neuron 68, 1067–1081 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tai H. C., et al., The synaptic accumulation of hyperphosphorylated tau oligomers in Alzheimer disease is associated with dysfunction of the ubiquitin-proteasome system. Am. J. Pathol. 181, 1426–1435 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Klionsky D. J., et al., Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy 12, 1–222 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martini-Stoica H., Xu Y., Ballabio A., Zheng H., The autophagy-lysosomal pathway in neurodegeneration: A TFEB perspective. Trends Neurosci. 39, 221–234 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kaushik S., Cuervo A. M., Proteostasis and aging. Nat. Med. 21, 1406–1415 (2015). [DOI] [PubMed] [Google Scholar]
- 9.Singh R., et al., Autophagy regulates lipid metabolism. Nature 458, 1131–1135 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lazarou M., et al., The ubiquitin kinase PINK1 recruits autophagy receptors to induce mitophagy. Nature 524, 309–314 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rubinsztein D. C., Mariño G., Kroemer G., Autophagy and aging. Cell 146, 682–695 (2011). [DOI] [PubMed] [Google Scholar]
- 12.Komatsu M., et al., Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature 441, 880–884 (2006). [DOI] [PubMed] [Google Scholar]
- 13.Hara T., et al., Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature 441, 885–889 (2006). [DOI] [PubMed] [Google Scholar]
- 14.Martinez J., et al., Noncanonical autophagy inhibits the autoinflammatory, lupus-like response to dying cells. Nature 533, 115–119 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 15.Berglund R., et al., Microglial autophagy-associated phagocytosis is essential for recovery from neuroinflammation. Sci. Immunol. 5, eabb5077 (2020). [DOI] [PubMed] [Google Scholar]
- 16.Xiao Q., et al., Neuronal-targeted TFEB accelerates lysosomal degradation of APP, reducing Aβ generation and amyloid plaque pathogenesis. J. Neurosci. 35, 12137–12151 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Polito V. A., et al., Selective clearance of aberrant tau proteins and rescue of neurotoxicity by transcription factor EB. EMBO Mol. Med. 6, 1142–1160 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lachance V., et al., Autophagy protein NRBF2 has reduced expression in Alzheimer’s brains and modulates memory and amyloid-beta homeostasis in mice. Mol. Neurodegener. 14, 43 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frost B., Jacks R. L., Diamond M. I., Propagation of tau misfolding from the outside to the inside of a cell. J. Biol. Chem. 284, 12845–12852 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guo J. L., Lee V. M., Seeding of normal tau by pathological tau conformers drives pathogenesis of Alzheimer-like tangles. J. Biol. Chem. 286, 15317–15331 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Clavaguera F., et al., Transmission and spreading of tauopathy in transgenic mouse brain. Nat. Cell Biol. 11, 909–913 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.de Calignon A., et al., Propagation of tau pathology in a model of early Alzheimer’s disease. Neuron 73, 685–697 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iba M., et al., Synthetic tau fibrils mediate transmission of neurofibrillary tangles in a transgenic mouse model of Alzheimer’s-like tauopathy. J. Neurosci. 33, 1024–1037 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sanders D. W., et al., Distinct tau prion strains propagate in cells and mice and define different tauopathies. Neuron 82, 1271–1288 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Braak H., Braak E., Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol. 82, 239–259 (1991). [DOI] [PubMed] [Google Scholar]
- 26.Luo W., et al., Microglial internalization and degradation of pathological tau is enhanced by an anti-tau monoclonal antibody. Sci. Rep. 5, 11161 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Audrain M., et al., Integrative approach to sporadic Alzheimer’s disease: Deficiency of TYROBP in a tauopathy mouse model reduces C1q and normalizes clinical phenotype while increasing spread and state of phosphorylation of tau. Mol. Psychiatry 24, 1383–1397 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Asai H., et al., Depletion of microglia and inhibition of exosome synthesis halt tau propagation. Nat. Neurosci. 18, 1584–1593 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ising C., et al., NLRP3 inflammasome activation drives tau pathology. Nature 575, 669–673 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Litvinchuk A., et al., Complement C3aR inactivation attenuates tau pathology and reverses an immune network deregulated in tauopathy models and Alzheimer’s disease. Neuron 100, 1337–1353.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ip W. K. E., Hoshi N., Shouval D. S., Snapper S., Medzhitov R., Anti-inflammatory effect of IL-10 mediated by metabolic reprogramming of macrophages. Science 356, 513–519 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Salem M., Ammitzboell M., Nys K., Seidelin J. B., Nielsen O. H., ATG16L1: A multifunctional susceptibility factor in Crohn disease. Autophagy 11, 585–594 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Parkhurst C. N., et al., Microglia promote learning-dependent synapse formation through brain-derived neurotrophic factor. Cell 155, 1596–1609 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Swartzlander D. B., et al., Concurrent cell type-specific isolation and profiling of mouse brains in inflammation and Alzheimer’s disease. JCI Insight 3, e121109 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Venegas C., et al., Microglia-derived ASC specks cross-seed amyloid-β in Alzheimer’s disease. Nature 552, 355–361 (2017). [DOI] [PubMed] [Google Scholar]
- 36.Russell D. G., Huang L., VanderVen B. C., Immunometabolism at the interface between macrophages and pathogens. Nat. Rev. Immunol. 19, 291–304 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marschallinger J., et al., Lipid-droplet-accumulating microglia represent a dysfunctional and proinflammatory state in the aging brain. Nat. Neurosci. 23, 194–208 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Safaiyan S., et al., Age-related myelin degradation burdens the clearance function of microglia during aging. Nat. Neurosci. 19, 995–998 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cantuti-Castelvetri L., et al., Defective cholesterol clearance limits remyelination in the aged central nervous system. Science 359, 684–688 (2018). [DOI] [PubMed] [Google Scholar]
- 40.Aravindhan K., et al., Assessing the effects of LXR agonists on cellular cholesterol handling: A stable isotope tracer study. J. Lipid Res. 47, 1250–1260 (2006). [DOI] [PubMed] [Google Scholar]
- 41.Groot P. H., et al., Synthetic LXR agonists increase LDL in CETP species. J. Lipid Res. 46, 2182–2191 (2005). [DOI] [PubMed] [Google Scholar]
- 42.Kitazawa M., et al., Blocking IL-1 signaling rescues cognition, attenuates tau pathology, and restores neuronal β-catenin pathway function in an Alzheimer’s disease model. J. Immunol. 187, 6539–6549 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu Y., Zhang S., Zheng H., The cargo receptor SQSTM1 ameliorates neurofibrillary tangle pathology and spreading through selective targeting of pathological MAPT (microtubule associated protein tau). Autophagy 15, 583–598 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Spite M., Serhan C. N., Novel lipid mediators promote resolution of acute inflammation: Impact of aspirin and statins. Circ. Res. 107, 1170–1184 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Qian M., Fang X., Wang X., Autophagy and inflammation. Clin. Transl. Med. 6, 24 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Quintanilla R. A., Orellana D. I., González-Billault C., Maccioni R. B., Interleukin-6 induces Alzheimer-type phosphorylation of tau protein by deregulating the cdk5/p35 pathway. Exp. Cell Res. 295, 245–257 (2004). [DOI] [PubMed] [Google Scholar]
- 47.Bhaskar K., et al., Regulation of tau pathology by the microglial fractalkine receptor. Neuron 68, 19–31 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Heckmann B. L., et al., LC3-associated endocytosis facilitates β-amyloid clearance and mitigates neurodegeneration in murine Alzheimer’s disease. Cell 178, 536–551.e14 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Martinez J., et al., Molecular characterization of LC3-associated phagocytosis reveals distinct roles for Rubicon, NOX2 and autophagy proteins. Nat. Cell Biol. 17, 893–906 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 50.Florey O., Kim S. E., Sandoval C. P., Haynes C. M., Overholtzer M., Autophagy machinery mediates macroendocytic processing and entotic cell death by targeting single membranes. Nat. Cell Biol. 13, 1335–1343 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Choi I., et al., Microglia clear neuron-released α-synuclein via selective autophagy and prevent neurodegeneration. Nat. Commun. 11, 1386 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xu Y., et al., TFEB regulates lysosomal exocytosis of tau and its loss of function exacerbates tau pathology and spreading. Mol. Psychiatry, 10.1038/s41380-020-0738-0 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Martini-Stoica H., et al., TFEB enhances astroglial uptake of extracellular tau species and reduces tau spreading. J. Exp. Med. 215, 2355–2377 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Settembre C., et al., TFEB controls cellular lipid metabolism through a starvation-induced autoregulatory loop. Nat. Cell Biol. 15, 647–658 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gao Z., et al., Neurod1 is essential for the survival and maturation of adult-born neurons. Nat. Neurosci. 12, 1090–1092 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xu Y., Martini-Stoica H., Zheng H., A seeding based cellular assay of tauopathy. Mol. Neurodegener. 11, 32 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wu Y., et al., Microglia and amyloid precursor protein coordinate control of transient Candida cerebritis with memory deficits. Nat. Commun. 10, 58 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.