Significance
Meropenem is a broad-spectrum carbapenem antibiotic widely used in the clinic. Its inhibition of l,d-transpeptidase cell-wall biosynthetic enzymes is increasingly recognized as a critical aspect of its broad-spectrum activity, particularly against mycobacterial species including Mycobacterium tuberculosis. We have demonstrated unexpected, reversible, and nonhydrolytic off-loading reactions of meropenem with one of its key targets, LdtMt2, representative of the l,d-transpeptidase superfamily. These findings belie the tacit assumption of irreversible inhibition with an exclusively hydrolytic off mechanism for carbapenems against their classical target, the penicillin-binding proteins. While modestly effective against tuberculosis, existing carbapenems have not been optimized for potency against l,d-transpeptidases. The knowledge gained here of two nonhydrolytic off-loading mechanisms provides important insights to help guide the design and synthesis of improved carbapenems.
Keywords: meropenem; l,d-transpeptidase; beta-lactam; beta-lactone; drug mode of action
Abstract
The carbapenem family of β-lactam antibiotics displays a remarkably broad spectrum of bactericidal activity, exemplified by meropenem’s phase II clinical trial success in patients with pulmonary tuberculosis, a devastating disease for which β-lactam drugs historically have been notoriously ineffective. The discovery and validation of l,d-transpeptidases (Ldts) as critical drug targets of bacterial cell-wall biosynthesis, which are only potently inhibited by the carbapenem and penem structural classes, gave an enzymological basis for the effectiveness of the first antitubercular β-lactams. Decades of study have delineated mechanisms of β-lactam inhibition of their canonical targets, the penicillin-binding proteins; however, open questions remain regarding the mechanisms of Ldt inhibition that underlie programs in drug design, particularly the optimization of kinetic behavior and potency. We have investigated critical features of mycobacterial Ldt inhibition and demonstrate here that the covalent inhibitor meropenem undergoes both reversible reaction and nonhydrolytic off-loading reactions from the cysteine transpeptidase LdtMt2 through a high-energy thioester adduct. Next-generation carbapenem optimization strategies should minimize adduct loss from unproductive mechanisms of Ldt adducts that reduce effective drug concentration.
The β-lactam family of antibiotics act by inhibiting biosynthesis of the peptidoglycan (PG) portion of the bacterial cell wall canonically by irreversible, covalent inhibition of penicillin-binding proteins (PBPs) (1). The PBPs known as d,d-transpeptidases (Ddts) generate 4 → 3 cross-links between amino acids in PG stems, while those known as d,d-carboxypeptidases hydrolyze the PG peptide stem C-terminal d-alanyl-d-alanyl amide bonds (2). The PBPs were long thought to be the sole target of β-lactams. However, in 1974, mycobacterial PG was shown to additionally contain 3 → 3 cross-links (3), and Streptococcus faecalis membrane preparations were demonstrated to catalyze l,d-type 3 → 3 cross-linking reactions in a penicillin-insensitive manner (4). The first characterized l,d-transpeptidase (Ldt) enzyme, Ldtfm, was identified in Enterococcus faecium in 2005 (5), and the dominant cross-link type in mycobacteria was demonstrated to be of type 3 → 3 soon after (6). Discovery of the primary l,d-transpeptidase in Mycobacterium tuberculosis, LdtMt2, and its validation as a drug target followed (7). Ldts are widespread in bacteria; both pathogenic and harmless mycobacterial species contain multiple paralogues (8). The Ldt and PBP families display convergent evolution of PG transpeptidase activity: the PBPs use a serine nucleophile to cross-link cell walls, and the alternately folded Ldts catalyze cross-linking reactions with an active-site cysteine. Within the β-lactam class of drugs, only the related carbapenem and penem classes potently inhibit l,d-transpeptidases (9–12). Carbapenems have been in clinical use since the approval of imipenem in 1985 (13) and were identified as antimycobacterial compounds in 2009 (14), a unique property among β-lactams. A phase II clinical trial has demonstrated that meropenem, in combination with amoxicillin and clavulanic acid, is effective at reducing mycobacterial burden in adults with pulmonary tuberculosis over 14 d (15), and a clinical trial is underway for use of meropenem in adults with rifampin-resistant tuberculosis (https://clinicaltrials.gov, no. NCT03174184). However, key mechanistic questions about the mode of action of carbapenems antimycobacterial activity remain open.
Based on mechanistic studies with PBPs, β-lactams were initially assumed to undergo an irreversible reaction, in which the only pathway to regenerate active enzyme after drug acylation is hydrolysis and thus destruction of the covalent warhead of the antibiotic (16, 17). Kinetic models of carbapenem Ldt inactivation have been built upon the assumption of irreversibility (10, 18). However, universal irreversibility of β-lactam acylation of enzymes has been challenged by the discovery of the reversible action against Ldts of the cephalosporin nitrocefin. Ldt–carbapenem reversibility requires re-formation of a β-lactam ring in the active site of an enzyme, an energetically challenging reaction but precedented in the light of nitrocefin-reversibility and biosynthesis of certain β-lactam containing natural products (19).
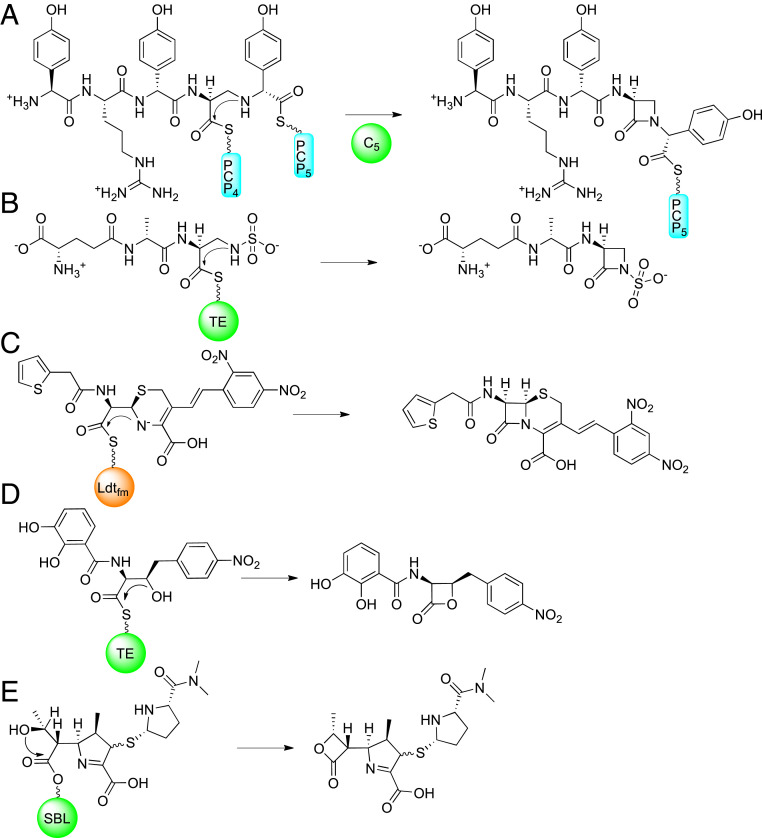
Four distinct enzyme catalyzed mechanisms of β-lactam ring closure are known from natural product biosynthesis. Penicillins and cephalosporins form their bicycle within one nonheme iron-dependent enzyme, powered by the exergonic reduction of diatomic oxygen to water (20). The three remaining mechanisms are nonoxidative, transacylation reactions (Fig. 1). Carbapenems, clavams, and tabtoxinine are synthesized from an asparagine synthetase homolog, in which ATP is consumed to produce an AMP-adenylate, primed for intramolecular attack of an amine to generate the four-membered ring (21–23). The β-lactam in carbapenems is synthesized through a carbapenam intermediate prior to desaturation of the five-membered ring and, to our knowledge, there is little precedence, synthetically or biosynthetically, for direct ring closure of a desaturated pyrroline to form a strained carbapenem directly. A monocyclic β-lactam family, the nocardicins, exploit a high-energy pantetheine thioester bond, seen in most nonribosomal peptide synthetase (NRPS) intermediates, to close their ring in a condensation domain (24). The other family of monocyclic β-lactams, monobactams, also form their eponymous rings while bound to an NRPS but on an atypical type 1 thioesterase (T1 TE) bearing a catalytic cysteine. The cysteine of the monobactam T1 TE catalyzes product release from the NRPS system by β-lactam formation instead of the hydrolysis or unstrained macrocyclization reactions characteristically observed for serine T1 TEs (25). Interestingly, replacement of the catalytic cysteine of the monobactam sulfazecin T1 TE with serine reverts the TE into a hydrolase, emphasizing the need for a high-energy intermediate, such as a thioester, to enable β-lactam formation (26).
Fig. 1.
Key four-membered ring formations. In monocyclic β-lactams, the azetidinone ring is formed in A by a condensation domain while attached as thioesters to peptidyl carrier proteins in the case of nocardicin G (the nocardicins), or (B) a thioesterase as shown in sulfazecin (the monobactams). (C) Nitrocefin re-forms its β-lactam in the active site of Ldtfm using its conjugated, anionic amine to substitute the enzyme thioester. (D) The obafluorin β-lactone ring is formed on a thioester. The meropenem-derived lactone (E) is formed by attack of the C6 hydroxyethyl group into the ester of class D, serine β-lactamases (SBLs). The condensation domain responsible for monocyclic β-lactam formation is depicted as a green circle, labeled C5, as it is in the “module” responsible for incorporating the fifth amino acid in nocardicin G biosynthesis. The two peptidyl carrier proteins that deliver nocardicin biosynthetic intermediate to the condensation domain within module 5 are represented as cyan rectangles labeled PCP4 and PCP5. The thioesterases used to form the sulfazecin β-lactam and obafluorin β-lactone is a green circle labeled “TE.” The l,d-transpeptidase from E. faecium, Ldtfm, is an orange circle. The C3′ position of nitrocefin is labeled with a black arrow. The class D β-lactamase is depicted as a green circle labeled “SBL.”
The Ldt from E. faecium (Ldtfm) reacts reversibly with nitrocefin, which, unlike most cephalosporins, does not have a C3′ leaving group to drive the reaction forward irreversibly (27). Instead, the C3′ sidechain contains a conjugated chromogen that remains deprotonated, giving a colorimetric signal concurrent with β-lactam scission. This resonance-stabilized anion can attack back into the Ldt thioester, re-forming nitrocefin’s original β-lactam ring, and releasing itself from Ldtfm, the sole characterized reversible reaction of a β-lactam with a transpeptidase (28). In contrast, Ldtfm was proposed to be irreversibly inhibited by carbapenems due to apparent rapid protonation of the pyrroline nitrogen in the ring-open enzyme–adduct form (28–30). However, the pKa of the nitrogen of a pyrolline-2-carboxylic acid is 6.2 (31) and, while this study was done on a small molecule in solution rather than an enzyme adduct, it is reasonable that a portion of the pyrroline nitrogens in the ensemble of Ldt–carbapenem adducts remain unprotonated for nucleophilic β-lactam ring closure in our reaction conditions at pH 8.0. Analogous to biosynthesis of the monocyclic β-lactams, the Ldt cysteine nucleophile forms a thioester upon drug acylation, rather than a more stable PBP oxyester. Given the resemblance among the Ldt–carbapenem thioester adduct, the biosynthetic thioester intermediates, and the Ldtfm–nitrocefin adduct, we reasoned that, although strained, carbapenem drugs may have a reversible mechanism of action with Ldt enzymes through re-formation of the β-lactam ring. Chemically, this proposal would follow from the principle of microscopic reversibility.
The β-lactone family of natural products is chemically related to the β-lactam family, and the known biosyntheses are analogous. They may proceed through AMP-adenylates on β-lactone synthetases (32), as in carbapenems and clavams, or in obafluorin biosynthesis, a T1 TE thioester on an NRPS (33), thus mirroring monobactam biosynthesis. The class D β-lactamases, a family of serine-based β-lactamases, are notable for both their ability to degrade carbapenem drugs and the unusual carbapenem β-lactone formation, which, along with hydrolysis, defines their mechanism of antibiotic degradation (34). Meropenem’s nucleophilic hydroxyethyl combined with a 1-β-methyl capable of directing a conformation favoring 4-exo-trig cyclization sets the stage for kinetically favorable β-lactone ring formation while meropenem is bonded to an enzyme, and, in the case of Ldt–meropenem adducts, the Ldt thioester lowers the energy barrier for β-lactone formation further.
Results
We wished to demonstrate the instability of Ldt–carbapenem adducts by direct, mass spectrometry (MS)-based observation. Hydrolysis, re-formation of a β-lactam ring, and/or formation of a β-lactone will all result in transient apo-enzyme, even with excess carbapenem in solution. Replacement of one inhibitor-derived adduct with a second inhibitor can thus occur if the relative kinetics favor the second molecule (Fig. 2A). The second-order LdtMt2 inactivation rates by faropenem and meropenem have previously been measured at pH 6.0 in phosphate buffer demonstrating faropenem to be the more rapid inactivator of LdtMt2 (18). For consistency, we measured LdtMt2 inactivation rates by faropenem and meropenem. We determined the rates to be 0.04 and 0.02 μM−1 ⋅ m−1, respectively, in our standard reaction conditions of HEPES buffer at pH 8.0 confirming the faster LdtMt2 inactivation rate by faropenem in assay conditions used throughout this work (SI Appendix, Fig. S1).
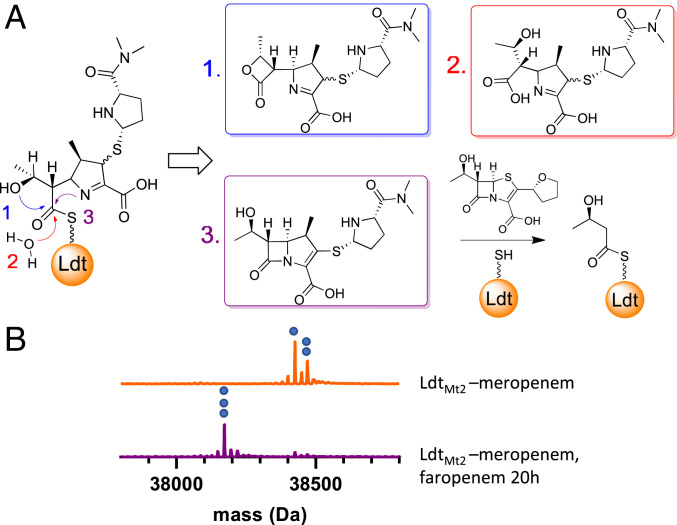
Fig. 2.
Intact protein UPLC–MS data demonstrate Ldt–meropenem adduct lability. (A) Replacement of meropenem on LdtMt2 by faropenem can proceed through three mechanisms: 1) formation of a β-lactone product, 2) hydrolysis of the protein-drug thioester, and/or 3) reclosure of the meropenem β-lactam ring. (B) LdtMt2-meropenem (decarboxylated, 38,425 Da, indicated with one blue circle; intact, 38,469 Da, indicated with two blue circles) was replaced by faropenem (38,172 Da, indicated with three blue circles) after prolonged incubation.
To test the stability of the LdtMt2–meropenem adduct, LdtMt2 was first inactivated with 25 equivalents of meropenem, followed by addition of 25 equivalents of faropenem, to the fully formed LdtMt2–meropenem adduct. Intact protein ultra-performance liquid chromatography–MS (UPLC–MS) analysis of LdtMt2 was performed after 20 h and revealed replacement of the meropenem adduct with a +86 Da adduct (Fig. 2B and SI Appendix, Table S1), which is known to be derived from fragmentation of faropenem when it covalently labels various cysteine enzymes, including LdtMt2, as thioesters (35, 36). We observed consistent decarboxylation of enzyme–meropenem adducts during electrospray ionization. This favorable gas-phase reaction is commonly observed in carbapenems, particularly while in the ring-open form (37). Protein crystallography data of Ldt–carbapenems, including meropenem, clearly reveal the presence of the carboxylate form on the enzyme (38), and we found the ratio of the decarboxylated meropenem adduct to the intact meropenem adduct to be approximately constant over time (SI Appendix, Fig. S13), supporting our hypothesis that decarboxylation occurs during electrospray ionization, rather than in solution. Therefore, we added the intensity of decarboxylated and intact meropenem adducts together for both LdtMt2 and DacB2 adducts during quantification of percent-bound adducts.
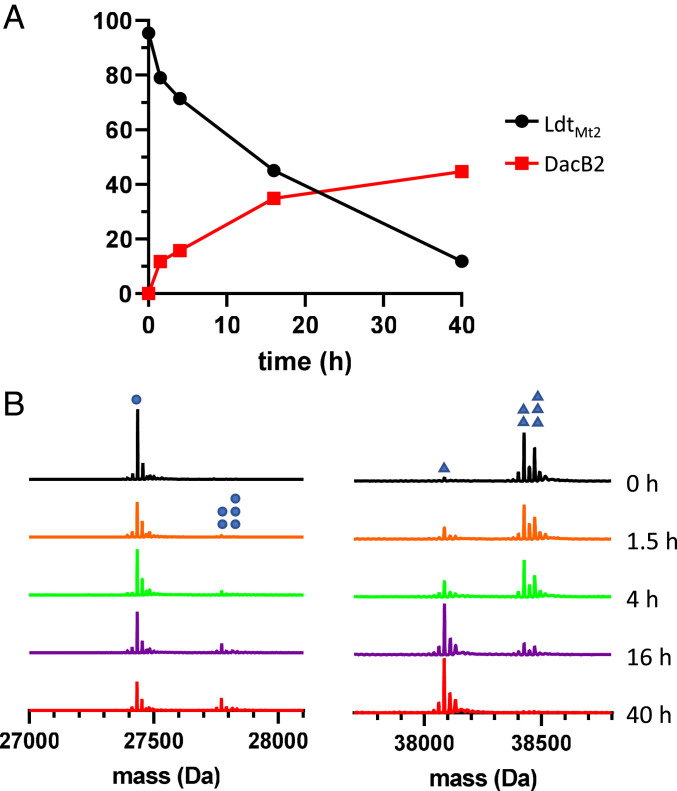
To determine if the observed transient apo-enzyme could be generated by the reverse reaction of meropenem’s acylation of LdtMt2, that is, reclosure of the β-lactam ring in the active site, we performed an acyl-transfer experiment from LdtMt2 to a serine-based mycobacterial PBP, the d,d-carboxypeptidase DacB2 (39). Titration of DacB2 with meropenem demonstrated nearly stoichiometric covalent labeling in the micromolar concentration range (SI Appendix, Fig. S9 and Table S3), signifying that DacB2 would provide sensitive detection of any meropenem released into solution. First, the LdtMt2–meropenem adduct was formed by the addition of two equivalents of meropenem, followed by buffer exchange to remove free drug, and then adding equimolar DacB2 to the LdtMt2–meropenem adduct. Over time, LdtMt2–meropenem reverted to apo-LdtMt2, with concurrent formation of a DacB2-meropenem adduct. Catalytically inactive LdtMt2C354A and meropenem in buffer alone were also subjected to buffer exchange and added to DacB2 to control for inefficient washing of drug or noncovalent association of meropenem to the enzyme. Approximately 40% of the meropenem initially bonded to LdtMt2 transferred to DacB2 over 40 h (Fig. 3 and SI Appendix, Fig. S8 and Table S2), while residual meropenem in both negative controls bound to ∼10% of DacB2 and did not increase over time (SI Appendix, Fig. S9 and Table S3).
Fig. 3.
Adduct transfer from LdtMt2 to DacB2. (A) The percent bound of LdtMt2 and DacB2 are plotted versus time since admixture of LdtMt2 and DacB2. Each point is a single measurement and curve is composed of straight lines, connecting contiguous points. (B) Spectra of DacB2 (Left) collected at time points after adding LdtMt2–meropenem show apo-DacB2 (27,435 Da, single blue dot) gradually decrease as the two DacB2–meropenem species (27,774 Da, decarboxylated, two blue dots; 27,818 Da, intact, three blue dots) increase. Spectra of LdtMt2 (Right) show apo-LdtMt2 (single blue triangle, 38,086 Da) appear as LdtMt2–meropenem (38,425 Da, decarboxylated, two triangles; 38,469 Da, intact, three triangles) is consumed.
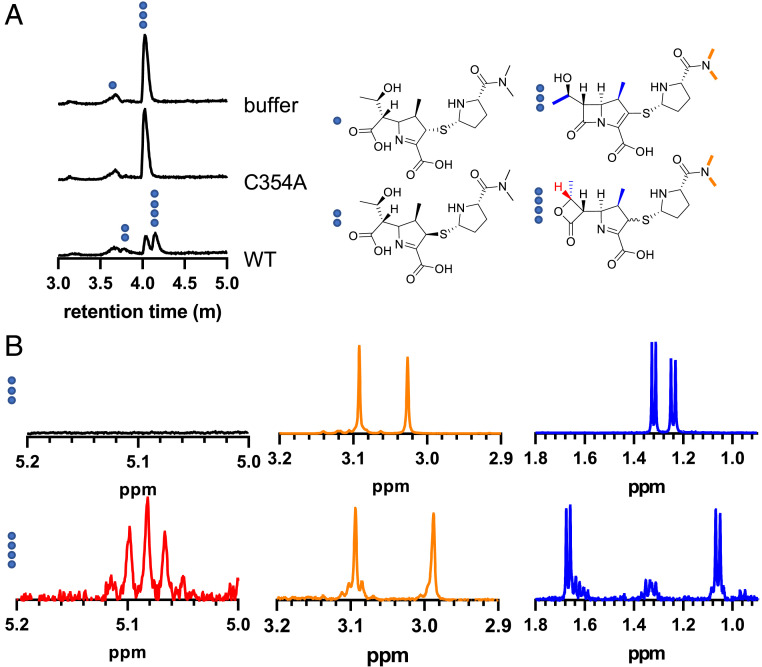
The loss of mass balance in the DacB2 trapping experiment implied partitioning of reversible and irreversible degradation processes. To identify the degradation products of LdtMt2–meropenem, LdtMt2 was incubated with two equivalents of meropenem for 25 h, and the small molecules in solution were compared to those formed both when meropenem was incubated with a catalytically incompetent mutant of LdtMt2, LdtMt2C354A, and meropenem in 25 mM HEPES pH 8.0 buffer alone. The only identified and assigned product from meropenem without native LdtMt2 in solution was a single hydrolysis product, assigned as the more stable, (2S) diastereomer (Fig. 4A and SI Appendix, Fig. S4). A second meropenem hydrolysis product appeared when wild-type LdtMt2 was present (Fig. 4A and SI Appendix, Fig. S5), attributed to the kinetically favorable (2R) diastereomer of hydrolyzed meropenem, which has been characterized in β-lactamase hydrolysis of carbapenems (40). Alongside hydrolysis products, a distinct compound with the same parent mass as meropenem, but at a later UPLC retention time, appeared as meropenem was consumed (Fig. 4A and SI Appendix, Fig. S3).
Fig. 4.
Incubation of LdtMt2 with meropenem results in a β-lactone product. (A) UPLC–MS total ion chromatograms demonstrate appearance of a lactone product with a later retention time than meropenem, as well as an additional hydrolysis peak, assigned as the less stable (2R) diastereomer of hydrolyzed meropenem. Compounds found under each peak are labeled with blue dots. (B) 1H-NMR analysis of meropenem, Top traces, and a meropenem lactone product, Bottom traces. The β-lactone C8 hydrogen, shown in red, gives an apparent pentet with a coupling constant of J = 6.3 Hz. The sidechain amide methyl groups, in orange, appear in the region of δ2.9 to 3.1 for both meropenem and meropenem lactone. The methyl doublets are drawn in blue. The hydroxyethyl methyl group peak is seen at ∼1.32 ppm in meropenem and is downfield at ∼1.68 ppm in the meropenem lactone product, while the 1-β-methyl group moves upfield from ∼1.24 ppm in meropenem to ∼1.06 ppm in the meropenem lactone product.
Meropenem undergoes in-source fragmentation during electrospray ionization MS, and the masses of these fragments can be highly diagnostic. Therefore, analysis of various electrospray ionization-derived fragments was performed to illuminate differences between the two observed compounds with the same parent mass. In the earlier eluting isomer known to be intact meropenem, along with the parent mass of 384.159 m/z, we observed a mass of 298.122 m/z, corresponding to a retro [2+2] cyclization, releasing a ketene and an imine (41), which is diagnostic of the presence of a β-lactam ring, along with a single decarboxylation, typical of carbapenems (SI Appendix, Fig. S2). The later-eluting isomer of meropenem did not contain the β-lactam–specific retro [2+2] fragment in its mass spectrum, but instead, a fragment corresponding to two decarboxylations was observed (SI Appendix, Fig. S3). The second decarboxylation was attributed to the well-known retro [2+2] loss of CO2 from the β-lactone functional group (42–44), a facile reaction that hampered gas chromatography–MS identification of biological β-lactone intermediates in cis-olefin biosynthesis (32).
A large-scale enzymic reaction was performed to obtain enough β-lactone for collection of 1H-NMR data. Two equivalents of meropenem were added to LdtMt2, and the reaction was halted by removal of enzyme after 60 h, when meropenem was no longer detectable by UPLC–MS (SI Appendix, Fig. S7). 1H-NMR analysis of the partially purified reaction mixture showed a doublet ∼0.35 ppm downfield of the meropenem C6 sidechain methyl hydrogens and an apparent pentet at 5.09 ppm for the adjacent C8 hydrogen (Fig. 4B and SI Appendix, Fig. S6), fully in accord with previously characterized carbapenem-derived β-lactones (34). As carbapenem β-lactones can acylate select β-lactamases (34), and a meropenem β-lactone enzyme adduct would be indistinguishable from a meropenem-derived adduct, we added the meropenem β-lactone to DacB2 and LdtMt2 to detect whether the observed acyl transfer from LdtMt2 to DacB2 could be caused by release of the β-lactone product. We observed no productive adduct formation between the meropenem-derived lactone and either LdtMt2 or DacB2 (SI Appendix, Fig. S10 and Table S4). As a portion of the (2R) diastereomer of the meropenem lactone product is transiently formed by class D β-lactamases before isomerizing into the more stable (2S) diastereomer, we wished to rule out loss of an active diastereomer that could transfer to DacB2 but had finished isomerizing during our purification step. Therefore, we combined LdtMt2 and meropenem in a 1:1 ratio until a substantial amount of lactone was formed, no meropenem remained, but LdtMt2–meropenem adduct was still present, releasing fresh lactone into solution (SI Appendix, Fig. S11A and Table S5). Rapid removal of enzyme by centrifugal filtration and immediate addition of DacB2 demonstrated no DacB2 adduct formation (SI Appendix, Fig. S11B and Table S5). We thus attribute transfer of meropenem from LdtMt2 to DacB2 to reversible re-formation of meropenem’s β-lactam ring.
The quantification of the relative amount of hydrolysis and lactone products of meropenem involves challenges in MS-based measurements of two potential C-2 stereoisomers of both hydrolyzed meropenem and the meropenem lactone product. We approximated the relative amount of each compound by incubating LdtMt2 with a substoichiometric amount of meropenem and, separately, meropenem in buffer alone. We then compared the amount of meropenem and hydrolyzed meropenem in buffer (SI Appendix, Fig. S12A) to the amount of products in the presence of excess LdtMt2 by UPLC–MS (SI Appendix, Fig. S12B and Table S6). LdtMt2 was subjected to amino acid analysis (45) to improve the accuracy of the calculated extinction coefficient of LdtMt2 and thus the enzyme concentration for this quantitative experiment. We found that at both 48 and 72 h, the ratio of lactone to hydrolyzed products was ∼1.7 (SI Appendix, Fig. S12B). As the experiment progressed, the ratio of the (2R) diastereomer of hydrolyzed meropenem to the (2S) diastereomer gradually shifted from majority (2R) to majority (2S), consistent with previous observations with carbapenemases that while the (2R) diastereomer of hydrolyzed meropenem is formed faster, the (2S) product is thermodynamically more stable (40).
The rate of meropenems loss from all combined processes, namely, lactone formation, hydrolysis, and re-formation of the meropenem β-lactam ring were measured by faropenem competition. LdtMt2–meropenem was formed and washed of free drug by two sequential buffer exchange steps. A large excess faropenem was added to immediately trap apo-LdtMt2 as it forms, similarly to PBP–meropenem off-rate experiments performed by replacing radio-labeled meropenem with cold meropenem (46). We found that loss of the meropenem adduct by all processes had a half-life of 6.3 h (SI Appendix, Fig. S14 and Table S7), compared to a previously obtained degradative half-life, excluding reversibility, of 16.5 h (18). The combined process lifetime is in general agreement with the slightly slower replacement with faropenem under equimolar (carba)penem conditions (Fig. 2), which resulted in 20% remaining LdtMt2–meropenem adduct after 20 h.
Discussion
β-lactam inhibitors of transpeptidases are known to be irreversible for the serine-based PBPs as a consequence of their strained azetidinone four-membered rings. However, β-lactam formation within an enzyme active site is relatively widespread and necessary for biosynthesis of these natural products. Thus, it is not unexpected for β-lactams to react reversibly if the energy of the acyl-enzyme adduct is relatively high. Ldt adducts are high-energy cysteine thioesters, as opposed to serine oxyesters, and thus parallel monocyclic β-lactam formation. High-energy thioesters also enable β-lactone formation, and in the case of meropenem, both nucleophilic nitrogen and oxygen atoms are available for intramolecular azetidinone and oxetanone ring formations, respectively. Precedence has established that even a lower energy β-lactamase oxyester can catalyze attack of the C8-OH to form a β-lactone. LdtMt2 appears to catalyze both the reverse reaction, that is, β-lactam re-formation, and degradation of meropenem through β-lactone formation. LdtMt2 thus aligns with two precedented high-energy thioester biosynthetic processes that lead to strained four-membered rings.
The catalytic promiscuity of Ldt enzymes has already been exploited in dramatic technological advances in visualization of bacterial cell walls by incorporating fluorescent d-amino acids by nucleophilic attack into endogenous peptidoglycan substrates (47). Our additional insights into four-membered ring formation again showcase the ability of Ldts to catalyze transacylation reactions with a wide range of nucleophiles. Meropenem inhibition of Ldt drug targets is a critical component of its antitubercular activity and thus clinical success. Design of novel inhibitors requires detailed information on mechanism. While reversible, covalent mechanisms have many virtues, as seen in the β-lactamase inhibitor avibactam, the partial reversibility observed here will still partition to degradation to inactive meropenem β-lactones and hydolyzed forms. Lack of sustained inhibition could be of particular relevance in slow-growing mycobacteria such as M. tuberculosis, which displays a doubling time of ∼20 h, supported by the recent discovery of the slow, reversible covalent mechanism of d-cycloserine inhibition of Mtb d-alanine racemase, and the proposal that this slow reversible activity explains the tempered antibiotic effectiveness of d-cycloserine against fast-growing bacteria relative to slow growers (48). The 1-β-methyl–containing carbapenems increased tendency to form β-lactone compounds on class D β-lactamases has been studied computationally and experimentally (34). Our present work demonstrates the importance of continuing to explore the determining chemical features present in structural variants of carbapenems that may minimize or eliminate unproductive enzyme-mediated β-lactone formation in favor of sustained or purely reversible inhibition.
Materials and Methods
Carbapenems and reagents were purchased from Sigma-Aldrich. Plasmids pET28a+ containing Tobacco Etch Virus protease-cleavable, N-terminal hexahistidine-tag bearing constructs of LdtMt2C354A(ΔN55), LdtMt2 (ΔN55), and DacB2(ΔN27) were transformed into Escherichia coli BL21(DE3) (Novagen) cells by electroporation and overexpressed as described previously (49). All experiments were performed at 22 °C. Enzyme concentration was determined using the Beer–Lambert law by ultraviolet-visible spectroscopy at A280 in 7 M guanidinium chloride and calculated extinction coefficients for each construct (50), except where indicated.
UPLC–High Resolution MS.
UPLC–high resolution MS (UPLC–HRMS) experiments were analyzed on a Waters Acquity H-Class UPLC system equipped with a multiwavelength UV-Vis diode array detector in conjunction with a Waters Acquity BEH-300 μL UPLC column packed with either a C4 stationary phase (2.1 × 50 mm; 1.7 μm) to analyze intact proteins or a C18 stationary phase (2.1 × 50 mm; 1.7 μm) to analyze small molecules, in tandem with HRMS analysis by a Waters Xevo-G2 quadrupole-time of flight electrospray ionization MS.
UPLC–MS Intact Protein Analysis.
Enzyme samples were separated at 60 °C to enhance peak resolution with a flow rate of 0.3 mL/min and the following mobile phase: 0 to 1 min 90% water, 10% ACN, 0.1% formic acid (FA); 1 to 7.5 min gradient up to 20% water, 80% ACN, 0.1% FA; 7.5 to 8.4 min 20% water, 80% ACN, 0.1% FA; 8.4 to 8.5 min linear gradient up to 90% water, 10% ACN, 0.1% FA; 8.5 to 10 min 90% water + 10% ACN, 0.1% FA. The first minute of eluate was discarded to remove salts and buffer online. Samples were analyzed in positive mode. The MaxEnt1 algorithm within MassLynx was used to deconvolute each m/z spectrum, and the resulting averaged mass spectrum was normalized to the sum of the intensities. Mass spectra were separately deconvoluted, peaks identified, integrated, and the resulting integrated intensities and parts per million error recorded with BiopharmaLynx. Adduct percentages were calculated by summing the integrated intensities of decarboxylated and intact enzyme adducts of meropenem and dividing by the sum of integrated intensities of the meropenem-bound, apo, and, in trapping experiments, faropenem-bound forms.
UPLC–MS Small-Molecule Method A.
Small-molecule analysis MS was performed in positive mode with a gradient mobile phase consisting of 0.1% FA, flow rate 0.3 mL/min: 0 to 1 min 100% water, 1 to 7.5 min gradient from 0 to 80% ACN, 7.5 to 8.4 min isocratic 80% ACN, 8.4 to 10 min 100% water. The first minute of eluate was discarded.
UPLC–MS Small-Molecule Method B.
Small-molecule analysis MS was performed in positive mode with a gradient mobile phase consisting of 0.1% FA, flow rate 0.3 mL/min: 0 to 1 min 100% water, 1 to 7.5 min gradient from 0 to 80% ACN, 7.5 to 8.4 min isocratic 80% ACN, 8.4 to 10 min 100% water.
1H-NMR of Meropenem and Partially Purified β-lactone.
NMR spectra were acquired on a 400 MHz Bruker Avance I equipped with a 5 mM double resonance broadband observe probe with Z-gradient. Spectra were acquired in D2O at 298 K.
Inactivation Kinetics of LdtMt2.
Inactivation kinetics of LdtMt2 were followed by UV-Vis spectroscopy, similar to previously described (18), but notably, carried out on a Cary 50 UV-Vis Spectrophotometer as opposed to a stopped-flow apparatus. An enzyme solution in 25 mM HEPES pH 8.0 was used to blank the spectrophotomer. A solution of 25 μM LdtMt2 was prepared with meropenem or faropenem at 50, 75, 100, 125, 150, and 175 μM at 22 °C. Samples were immediately mixed and monitored at 297 nM for meropenem or 307 nM for faropenem. ΔAbs = EoΔε(1 − e−kt) was fit to the data. Without a stopped-flow apparatus, a variable amount of enzyme was inactivated before data were recorded; thus, the initial enzyme amount, Eo, was fit alongside the rate. The change in extinction coefficients, Δε, used were 10,000 M−1 ⋅ cm−1 for meropenem and 5,000 M−1 ⋅ cm−1 for faropenem.
Carbapenem Replacement Assays.
Meropenem (50 μM) was incubated with LdtMt2 (2 μM) for 5 h at 22 °C in 25 mM HEPES pH 8.0, at which time complete covalent labeling of enzyme was verified by UPLC–MS intact protein analysis. Faropenem was subsequently added to a final concentration of 50 μM, and intact protein analysis was conducted 20 h after addition of faropenem.
Acyl Transfer.
A solution of 40 μM meropenem in 25 mM HEPES pH 8.0 was incubated with either 20 μM wild-type LdtMt2, 20 μM LdtMt2 C354A, or buffer alone for 90 min at 22 °C, and adduct formation with wild-type LdtMt2 was confirmed by UPLC–MS intact protein analysis. Free meropenem was removed from all three conditions by subjecting each solution to two sequential buffer exchanges with Thermo Scientific Zeba 7 kDa spin desalting columns, which had been pre-equilibrated with 25 mM HEPES pH 8.0 buffer. The concentration of wild-type LdtMt2-meropenem after both buffer exchanges was determined to be 12.3 μM; thus, DacB2 in 25 mM HEPES pH 8.0 was diluted to 12.3 μM and solutions were added 1:1 for a solution of 6.15 μM of both LdtMt2 and DacB2. Solutions were subjected to UPLC–MS intact protein analysis at various time points. Spectra were deconvoluted with MaxEnt1 in MassLynx to generate spectra plots, and apo versus bound forms percentages were quantified in BiopharmaLynx.
LdtMt2-derived Reaction Products.
To a 400 μL solution of 25 mM HEPES containing 40 μM LdtMt2, 40 μM LdtMt2 C354A catalytically incompetent mutant, or buffer alone was added 400 μL 80 μM meropenem for final solutions of 40 μM meropenem and 20 μM enzyme. Reactions were incubated for 25 h at 22 °C before removing enzyme through a 10 kDa Amicon Ultra 0.5 mL centrifugal filter and diluting the enzyme-free flow through 1:2 with a solution of 20 μM internal standard doripenem in ddH2O for a final concentration of 10 μM doripenem. Filtered reactions were subjected to UPLC–MS analysis A and a solution of 10 kDa Amicon Ultra 0.5 mL centrifugal filtered 25 mM HEPES, with addition of 10 μM doripenem was background subtracted from total ion chromatograms of reaction products.
Preparation of β-lactone Product.
A 10 mL solution containing 25 mM HEPES pH 8.0, 100 μM LdtMt2, and 200 μM meropenem were incubated at 22 °C, and samples were periodically removed, subjected to filtration through a 10 kDa Amicon Ultra 0.5 mL centrifugal filter, and the filtrate was analyzed by small-molecule UPLC–MS method A until meropenem was no longer detectable. Enzyme was removed with a 10 kD Amicon Ultra 4-mL centrifugal filter, the sample was flash frozen on liquid N2 and lyophilized. To remove HEPES buffer and salts, the residue was taken up in ddH2O and run over a Varian Bond Elut C18 solid-phase extraction column, pre-equilibrated with ddH2O, washed with 5 mL water, and eluted with 1:1 acetonitrile:water. The resultant partially purified meropenem lactone product was diluted 1:50 in ddH2O and, alongside a 20 μM solution of meropenem in ddH2O for comparison, subjected to small-molecule analysis B. Acetonitrile was removed in vacuo, the sample frozen in liquid nitrogen, and lyophilized before dissolution in D2O for 1H-NMR analysis.
Activity of β-lactone Product.
Both meropenem and the isolated β-lactone compound were separately incubated at ∼50 μM with 2 μM LdtMt2 or 2 μM DacB2 at 22 °C. Intact protein UPLC–MS analysis was performed at 2, 5, and 24 h to determine potential adduct formation of the lactone product with LdtMt2 or DacB2. To control for DacB2 activity of a nascent (2R) lactone that would have isomerized to the more stable (2S) lactone diastereomer during partial purification, we added 100 μM meropenem to 100 μM LdtMt2. Samples were taken at 24 and 42 h and diluted 1:100, and percent LdtMt2 bound to meropenem was determined by intact protein UPLC–MS analysis. At the same time points, protein was removed by filtration through a 10 kDa Amicon Ultra 0.5-mL centrifugal filter, and the filtrate was analyzed by small-molecule analysis A to determine amount of lactone and absence of meropenem. After 42 h, sufficient lactone was judged to have been produced, and LdtMt2 was removed by filtration through a 10 kDa Amicon Ultra 0.5-mL centrifugal filter, and DacB2 was immediately added to the filtrate. DacB2 was analyzed by intact protein UPLC–MS to determine any adduct formation with the (2R) lactone product at 2 and 5 h.
Quantitative Analysis of LdtMt2–Meropenem Products.
For this experiment, LdtMt2 concentration was calculated with an extinction coefficient of 84,000 M−1 ⋅ cm−1 obtained by amino acid analysis performed by the University of California, Davis Molecular Structure Facility. A total of 60 μM meropenem was incubated with and without 80 μM LdtMt2 at 22 °C, and samples were removed at 1, 24, 48, and 72 h, filtered through a 10 kDa Amicon Ultra 0.5-mL centrifugal filter, and diluted 1:2 with a 20 μM solution of doripenem in ddH2O for a final concentration of 10 μM doripenem internal standard before being subjected to small-molecule analysis A. A solution of 25 mM HEPES pH 8.0, filtered through a 10 kDa Amicon Ultra 0.5 mL centrifugal filter and diluted 1:2 with a 20 μM solution of doripenem in ddH2O, was also analyzed by small-molecule analysis A before each time point and used for background subtraction of each experimental spectrum to allow integration of key species without interference from the internal standard, contaminants from the centrifugal filter, and general background from the UPLC–MS system. Solutions that included LdtMt2 had samples removed, were diluted 1:80 into 25 mM HEPES pH 8.0, and were subjected to UPLC–MS intact protein analysis at each time point. Quantification of all products was performed as described in SI Appendix, Materials and Methods. Additionally, we quantified the percentage of LdtMt2–meropenem adducts, which were decarboxylated from this experiment at each time point.
Off-Rate Determination of LdtMt2–Meropenem.
A solution of 20 μM LdtMt2 and 40 μM meropenem was incubated for 1 h at 22 °C, and adduct formation was measured by UPLC–MS intact protein MS. LdtMt2–meropenem was subjected to two sequential rounds of buffer exchange in Thermo Scientific Zeba 7 kDa desalting columns, which had been pre-equilibrated with 25 mM HEPES pH 8.0 buffer, and faropenem was added to a concentration of 5 mM. Samples were removed, diluted 1:10 into 25 mM HEPES pH 8.0, and percent bound to meropenem was measured by UPLC–MS intact protein analysis by dividing the combined intensity of decarboxylated and intact meropenem adducts by the total LdtMt2 intensity.
Supplementary Material
Acknowledgments
We gratefully thank Professor G. Lamichhane (School of Medicine, Johns Hopkins University) for plasmids containing LdtMt2, LdtMt2C354A, and DacB2 and Professor L. A. Brammer Basta (Chemistry Department, United States Naval Academy) for the critical reading of the manuscript. This work was supported by the NIH through Grant Nos. R01AI137329 (C.A.T.) and T32GM008403 (T.A.Z.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. M.A. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2008610118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix.
References
- 1.Page M. I., The mechanisms of reactions of β-lactam antibiotics. Acc. Chem. Res. 17, 144–151 (1984). [Google Scholar]
- 2.Sauvage E., Kerff F., Terrak M., Ayala J. A., Charlier P., The penicillin-binding proteins: Structure and role in peptidoglycan biosynthesis. FEMS Microbiol. Rev. 32, 234–258 (2008). [DOI] [PubMed] [Google Scholar]
- 3.Wietzerbin J., et al., Occurrence of D-alanyl-(D)-meso-diaminopimelic acid and meso-diaminopimelyl-meso-diaminopimelic acid interpeptide linkages in the peptidoglycan of Mycobacteria. Biochemistry 13, 3471–3476 (1974). [DOI] [PubMed] [Google Scholar]
- 4.Coyette J., Perkins H. R., Polacheck I., Shockman G. D., Ghuysen J. M., Membrane-bound DD-carboxypeptidase and LD-transpeptidase of Streptococcus faecalis ATCC 9790. Eur. J. Biochem. 44, 459–468 (1974). [DOI] [PubMed] [Google Scholar]
- 5.Mainardi J. L., et al., A novel peptidoglycan cross-linking enzyme for a β-lactam-resistant transpeptidation pathway. J. Biol. Chem. 280, 38146–38152 (2005). [DOI] [PubMed] [Google Scholar]
- 6.Lavollay M., et al., The peptidoglycan of stationary-phase Mycobacterium tuberculosis predominantly contains cross-links generated by L,D-transpeptidation. J. Bacteriol. 190, 4360–4366 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gupta R., et al., The Mycobacterium tuberculosis protein LdtMt2 is a nonclassical transpeptidase required for virulence and resistance to amoxicillin. Nat. Med. 16, 466–469 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zandi T. A., Marshburn R. L., Stateler P. K., Brammer Basta L. A., Phylogenetic and biochemical analyses of mycobacterial L,D-transpeptidases reveal a distinct enzyme class that is preferentially acylated by meropenem. ACS Infect. Dis. 5, 2047–2054 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Triboulet S., et al., Kinetic features of L,D-transpeptidase inactivation critical for β-lactam antibacterial activity. PLoS One 8, e67831 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dubée V., et al., Inactivation of Mycobacterium tuberculosis l,d-transpeptidase LdtMt1 by carbapenems and cephalosporins. Antimicrob. Agents Chemother. 56, 4189–4195 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cordillot M., et al., In vitro cross-linking of Mycobacterium tuberculosis peptidoglycan by L,D-transpeptidases and inactivation of these enzymes by carbapenems. Antimicrob. Agents Chemother. 57, 5940–5945 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Triboulet S., et al., Inactivation kinetics of a new target of β-lactam antibiotics. J. Biol. Chem. 286, 22777–22784 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lyon J. A., Imipenem/cilastatin: The first carbapenem antibiotic. Drug Intell. Clin. Pharm. 19, 895–899 (1985). [PubMed] [Google Scholar]
- 14.Hugonnet J. E., Tremblay L. W., Boshoff H. I., Barry C. E., Blanchard J. S., Meropenem-clavulanate is effective against extensively drug-resistant Mycobacterium tuberculosis. Science 323, 1215–1218 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Diacon A. H., et al., β-Lactams against tuberculosis—New trick for an old dog? N. Engl. J. Med. 375, 393–394 (2016). [DOI] [PubMed] [Google Scholar]
- 16.Rowley D., Cooper P. D., Roberts P. W., Smith E. L., The site of action of penicillin. 1. Uptake of penicillin on bacteria. Biochem. J. 46, 157–161 (1950). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frère J.-M., Leyh-Bouille M., Ghuysen J. M., Perkins H. R., Interaction between β-lactam antibiotics and exocellular DD-carboxypeptidase-transpeptidase of Streptomyces R61. Eur. J. Biochem. 50, 203–214 (1974). [DOI] [PubMed] [Google Scholar]
- 18.Dhar N., et al., Rapid cytolysis of Mycobacterium tuberculosis by faropenem, an orally bioavailable β-lactam antibiotic. Antimicrob. Agents Chemother. 59, 1308–1319 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Townsend C. A., Convergent biosynthetic pathways to β-lactam antibiotics. Curr. Opin. Chem. Biol. 35, 97–108 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Byford M. F., Baldwin J. E., Shiau C. Y., Schofield C. J., The mechanism of ACV synthetase. Chem. Rev. 97, 2631–2650 (1997). [DOI] [PubMed] [Google Scholar]
- 21.Bachmann B. O., Li R., Townsend C. A., β-Lactam synthetase: A new biosynthetic enzyme. Proc. Natl. Acad. Sci. U.S.A. 95, 9082–9086 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gerratana B., Stapon A., Townsend C. A., Inhibition and alternate substrate studies on the mechanism of carbapenam synthetase from Erwinia carotovora. Biochemistry 42, 7836–7847 (2003). [DOI] [PubMed] [Google Scholar]
- 23.Kinscherf T. G., Willis D. K., The biosynthetic gene cluster for the β-lactam antibiotic tabtoxin in Pseudomonas syringae. J. Antibiot. (Tokyo) 58, 817–821 (2005). [DOI] [PubMed] [Google Scholar]
- 24.Gaudelli N. M., Long D. H., Townsend C. A., β-Lactam formation by a non-ribosomal peptide synthetase during antibiotic biosynthesis. Nature 520, 383–387 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Horsman M. E., Hari T. P. A., Boddy C. N., Polyketide synthase and non-ribosomal peptide synthetase thioesterase selectivity: Logic gate or a victim of fate? Nat. Prod. Rep. 33, 183–202 (2016). [DOI] [PubMed] [Google Scholar]
- 26.Oliver R. A., Li R., Townsend C. A., Monobactam formation in sulfazecin by a nonribosomal peptide synthetase thioesterase. Nat. Chem. Biol. 14, 5–7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Page M. I., Proctor P., Mechanism of β-lactam ring opening in cephalosporins. J. Am. Chem. Soc. 106, 3820–3825 (1984). [Google Scholar]
- 28.Edoo Z., Arthur M., Hugonnet J. E., Reversible inactivation of a peptidoglycan transpeptidase by a β-lactam antibiotic mediated by β-lactam-ring recyclization in the enzyme active site. Sci. Rep. 7, 9136 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bhattacharjee N., et al., Negative impact of carbapenem methylation on the reactivity of β-lactams for cysteine acylation as revealed by quantum calculations and kinetic analyses. Antimicrob. Agents Chemother. 63, e02039-e18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Triboulet S., et al., Tryptophan fluorescence quenching in β-lactam-interacting proteins is modulated by the structure of intermediates and final products of the acylation reaction. ACS Infect. Dis. 5, 1169–1176 (2019). [DOI] [PubMed] [Google Scholar]
- 31.Lewis M. L., et al., The effect of pH on the solution structure of Δ1-pyrroline-2-carboxylic acid as revealed by NMR and electrospray mass spectroscopy. Bioorg. Med. Chem. Lett. 3, 1193–1196 (1993). [Google Scholar]
- 32.Christenson J. K., et al., β-Lactone synthetase found in the olefin biosynthesis pathway. Biochemistry 56, 348–351 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schaffer J. E., Reck M. R., Prasad N. K., Wencewicz T. A., β-Lactone formation during product release from a nonribosomal peptide synthetase. Nat. Chem. Biol. 13, 737–744 (2017). [DOI] [PubMed] [Google Scholar]
- 34.Lohans C. T., et al., A new mechanism for β-lactamases: Class D enzymes degrade 1β-methyl carbapenems through lactone formation. Angew. Chem. Int. Ed. Engl. 57, 1282–1285 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kumar P., et al., Non-classical transpeptidases yield insight into new antibacterials. Nat. Chem. Biol. 13, 54–61 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lohans C. T., et al., Non-hydrolytic β-lactam antibiotic fragmentation by L,D-transpeptidases and serine β-lactamase cysteine variants. Angew. Chem. Int. Ed. Engl. 58, 1990–1994 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.de Souza Barbosa F., et al., Stability in clinical use and stress testing of meropenem antibiotic by direct infusion ESI-Q-TOF: Quantitative method and identification of degradation products. J. Pharm. Biomed. Anal. 179, 112973 (2020). [DOI] [PubMed] [Google Scholar]
- 38.Li W.-J., et al., Crystal structure of L,D-transpeptidase LdtMt2 in complex with meropenem reveals the mechanism of carbapenem against Mycobacterium tuberculosis. Cell Res. 23, 728–731 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kumar P., et al., Meropenem inhibits D,D-carboxypeptidase activity in Mycobacterium tuberculosis. Mol. Microbiol. 86, 367–381 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lohans C. T., et al., Mechanistic insights into β-lactamase-catalysed carbapenem degradation through product characterisation. Sci. Rep. 9, 13608 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mitscher L. A., Showalter H. D. H., Shirahata K., Foltz R. L., Chemical-ionization mass spectrometry of β-lactam antibiotics. J. Antibiot. (Tokyo) 28, 668–675 (1975). [DOI] [PubMed] [Google Scholar]
- 42.Einhorn A., Ueber derivate der orthonitrozimmtsäure. Ber. Dtsch. Chem. Ges. 16, 2208–2216 (1883). [Google Scholar]
- 43.Liang H. T., Bartlett P. D., Kinetics and mechanism of the reactions of β-isovalerolactone in water. J. Am. Chem. Soc. 80, 3585–3590 (1958). [Google Scholar]
- 44.Noyce D. S., Banitt E. H., The stereochemistry of the decarboxylation of β-lactones to form olefins. J. Org. Chem. 31, 4043–4047 (1966). [Google Scholar]
- 45.Ozols J., “Amino acid analysis” in Guide to Protein Purification, Deutscher M. P., Ed. (Academic Press, 1990), pp. 587–601. [Google Scholar]
- 46.Sumita Y., Fukasawa M., Okuda T., Affinities of SM-7338 for penicillin-binding proteins and its release from these proteins in Staphylococcus aureus. Antimicrob. Agents Chemother. 34, 484–486 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kuru E., et al., In situ probing of newly synthesized peptidoglycan in live bacteria with fluorescent D-amino acids. Angew. Chem. Int. Ed. Engl. 51, 12519–12523 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.de Chiara C., et al., D-Cycloserine destruction by alanine racemase and the limit of irreversible inhibition. Nat. Chem. Biol. 16, 686–694 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brammer Basta L. A., et al., Loss of a functionally and structurally distinct L,D-transpeptidase, LdtMt5, compromises cell wall integrity in Mycobacterium tuberculosis. J. Biol. Chem. 290, 25670–25685 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Edelhoch H., Spectroscopic determination of tryptophan and tyrosine in proteins. Biochemistry 6, 1948–1954 (1967). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.