Significance
Symbiosis between single-celled hosts and microalgae is a widespread and ecologically important phenomenon in oceanic plankton. Investigating such relationships can increase understanding of how novel organelles are acquired. Here, we integrate multiple subcellular approaches to show how a unicellular host manipulates the bioenergetic machinery of its algal endosymbionts and maximizes organic carbon production for its own benefit. Symbionts are blocked from dividing, but their photosynthetic machinery and associated mitochondrial network dramatically expand while keeping close physical proximity to each other. Genes involved in photosynthesis and carbon fixation are up-regulated, while those for carbon storage are down-regulated. Such extreme remodeling of microalgae has not been reported in other photosymbioses and provides insight into the putative early steps of plastid acquisition.
Keywords: symbiosis, oceanic plankton, photosynthesis, 3D electron microscopy, single-cell transcriptomics
Abstract
Endosymbioses have shaped the evolutionary trajectory of life and remain ecologically important. Investigating oceanic photosymbioses can illuminate how algal endosymbionts are energetically exploited by their heterotrophic hosts and inform on putative initial steps of plastid acquisition in eukaryotes. By combining three-dimensional subcellular imaging with photophysiology, carbon flux imaging, and transcriptomics, we show that cell division of endosymbionts (Phaeocystis) is blocked within hosts (Acantharia) and that their cellular architecture and bioenergetic machinery are radically altered. Transcriptional evidence indicates that a nutrient-independent mechanism prevents symbiont cell division and decouples nuclear and plastid division. As endosymbiont plastids proliferate, the volume of the photosynthetic machinery volume increases 100-fold in correlation with the expansion of a reticular mitochondrial network in close proximity to plastids. Photosynthetic efficiency tends to increase with cell size, and photon propagation modeling indicates that the networked mitochondrial architecture enhances light capture. This is accompanied by 150-fold higher carbon uptake and up-regulation of genes involved in photosynthesis and carbon fixation, which, in conjunction with a ca.15-fold size increase of pyrenoids demonstrates enhanced primary production in symbiosis. Mass spectrometry imaging revealed major carbon allocation to plastids and transfer to the host cell. As in most photosymbioses, microalgae are contained within a host phagosome (symbiosome), but here, the phagosome invaginates into enlarged microalgal cells, perhaps to optimize metabolic exchange. This observation adds evidence that the algal metamorphosis is irreversible. Hosts, therefore, trigger and benefit from major bioenergetic remodeling of symbiotic microalgae with potential consequences for the oceanic carbon cycle. Unlike other photosymbioses, this interaction represents a so-called cytoklepty, which is a putative initial step toward plastid acquisition.
Living with intracellular microalgae (photosymbiosis) is a globally widespread and ecologically important lifestyle. In the oceanic plankton, various unicellular heterotrophic eukaryotes (e.g., radiolarians, foraminiferans, ciliates) establish symbioses with intracellular microalgae (1, 2). These ubiquitous organisms contribute significantly to primary production (3) and carbon sequestration from surface waters to the deep sea (4–7). Photosymbioses are generally recognized as mutualistic partnerships with hosts providing nutrient-rich and protective microhabitats to algal symbionts, which in return provide energy-rich compounds produced via photosynthesis [e.g., sugars, lipids (8)]. This metabolic crosstalk between a heterotroph and an autotroph provides a competitive advantage in nutrient-poor habitats, such as the open ocean. However, it is difficult to demonstrate whether photosymbioses confer evolutionary advantages on algal symbionts, particularly in marine planktonic symbioses, which cannot be cultured in laboratory conditions and where the costs and benefits for each partner are not clearly defined (9). By contrast, microalgae in their free-living stage can usually be cultured. Therefore, whether photosymbioses are true mutualistic partnerships has long been debated (10).
In the evolutionary history of eukaryotes, photosymbiosis is recognized as a preliminary step that can lead to plastid acquisition (11, 12). Following the establishment of the symbiosis, heterotrophic hosts exploited intracellular prokaryotic (primary endosymbiosis) or plastid-bearing eukaryotic symbionts (secondary or tertiary endosymbiosis). Gradual cellular and genomic reduction of the symbiont then occurred, ultimately leading to plastid acquisition by the host (13–15). However, the underlying cellular mechanisms that allow a host cell to take control and manipulate the bioenergetics of intracellular microalgae remain unknown. Several relevant models have been studied, but reduction of the microalgal cell and horizontal gene transfer to the host has already occurred in each of them [e.g., kleptoplastidy in dinoflagellates (16), Paulinella chromatophora (17)]. Studying contemporary unicellular photosymbioses involving intact microalgae has the potential not only to lead to a better understanding of their ecological success in the ocean but also to provide evolutionary insights into the putative first steps of plastid acquisition and underlying host control.
The cosmopolitan planktonic photosymbiosis between acantharian hosts and intact microalgae (Phaeocystis spp., Haptophyta) is characterized by a morphological transformation of symbionts, wherein algal cell volume increases dramatically, and there is a multiplication of enlarged plastids (18, 19). Free-living Phaeocystis cells can be cultured in laboratory conditions and reach high population densities in many oceanic regions (20, 21), thus confounding any evolutionary advantages symbiosis may confer on this microalga. For the host—uncultivable so far—the ecological success of free-living Phaeocystis is beneficial since it increases opportunities to capture new symbionts throughout the life cycle by horizontal transmission (22). Hence, Acantharia–Phaeocystis photosymbioses are potentially exploitive symbioses with intact symbiont cells and represent an ideal system to investigate this oceanic interaction and bring insight into the early transitional stages of more permanent algal endosymbioses.
To elucidate mechanisms involved in host exploitation of algal cells and the putative initial steps of plastid acquisition, the structural, physiological, and genetic changes of symbiotic microalgae need to be explored at the subcellular and molecular level. Here, the subcellular architecture of Phaeocystis cells outside (i.e., free living in culture) and inside host cells was reconstructed three dimensionally (3D) to quantify structural changes of energy-producing organelles (plastid, mitochondria). In parallel, single-cell transcriptomics compared Phaeocystis gene expression in free-living and symbiotic stages. This combination of quantitative subcellular imaging and transcriptomics showed that the cell cycle of endosymbiotic microalgae is halted, and the bioenergetic machinery is drastically enhanced. We observed a proliferation of plastids with enlarged internal pyrenoids [i.e., compartments containing the CO2-fixing enzyme Rubisco (23)] and correlated extension of reticulated mitochondria in symbionts. These morphological changes were accompanied by the up-regulation of genes involved in photosynthesis and central carbon pathways. Photophysiology, photon propagation modeling, and carbon flux imaging further demonstrated that algal energy production is significantly enhanced, leading to increased production of organic carbon, which is allocated to plastids and transferred to the host cell. This study deciphers mechanisms for how a microalgal cell is morphologically, metabolically, and transcriptionally modified and ultimately exploited by a heterotrophic host cell in the oceanic plankton (i.e., cytoklepty).
Results and Discussion
Phaeocystis Cell Division Is Inhibited in Symbiosis.
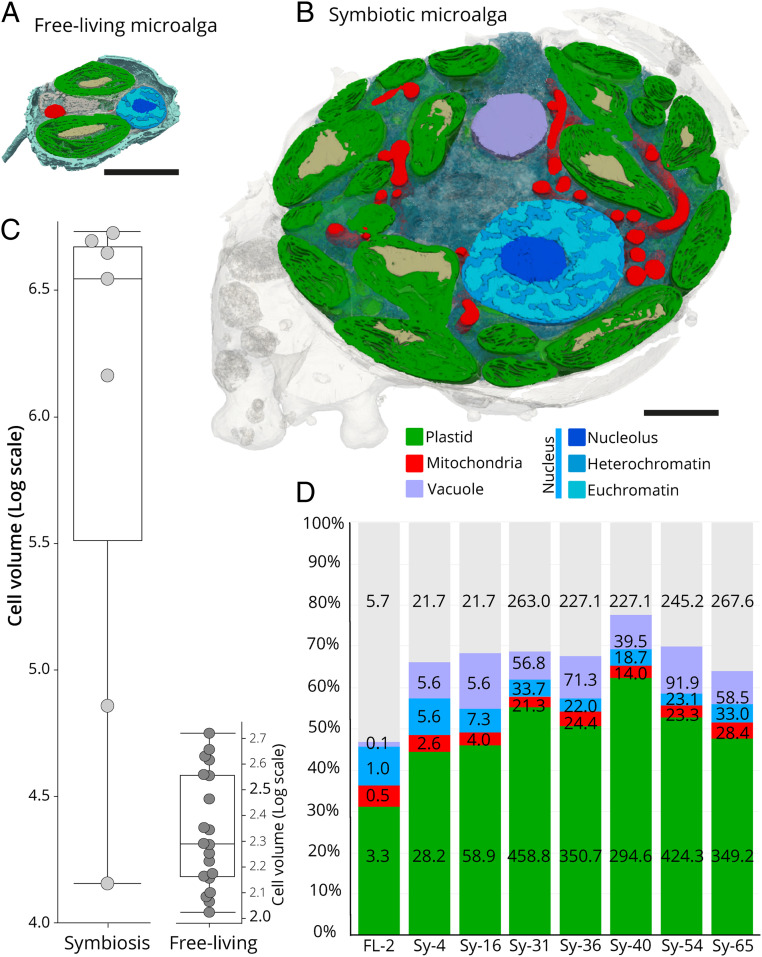
Using 3D electron microscopy (FIB–SEM, Focused Ion Beam–Scanning Electron Microscopy), we reconstructed and performed morphometric analyses of the subcellular architecture of the microalga Phaeocystis in the free-living (n = 20 cells) and endosymbiotic stage within two distinct acantharian hosts (n = 7 cells) collected in the Mediterranean Sea (Fig. 1 and Dataset S1). In addition to the disappearance of flagella and external scales, the total volume of symbiotic Phaeocystis cells increased 6- to 78-fold compared to free-living cells and accommodated more organelles and larger vacuoles (Figs. 1 and 2). Such a dramatic increase in cell size indicates that Phaeocystis cell division might be blocked while living symbiotically (24, 25). Consistent with this hypothesis, single-cell transcriptome analysis on 12 distinct hosts collected in the North Pacific Ocean revealed that key Phaeocystis genes involved in DNA replication and progression through cell-cycle stages (G1, S, G2, and M) were down-regulated in symbiosis (Fig. 2 and SI Appendix, Fig. S1), including genes for DNA polymerase complexes, cyclins, cyclin-dependent kinases, and the anaphase-promoting complex. Additionally, nuclear volume increased in symbiotic cells up to 32-fold (Fig. 2 and Dataset S1), although its relative occupancy decreased from 9.3 ± 1.6% in free-living cells to 4.7 ± 2% in symbiosis (Fig. 1D). Taking advantage of the varying electron densities of compartments within the nucleus, we separately quantified the volumes of the nucleolus (site of ribosome genesis) (26) and eu- and heterochromatin (sites of DNA replication and gene expression) (Fig. 1 A and B) (27). The volume of these three nuclear compartments concomitantly increased in symbiosis (20-fold for the nucleolus, 18-fold for the heterochromatin, 23-fold for euchromatin), and their volume ratios remained relatively stable (SI Appendix, Fig. S1 and Dataset S1). While there is no reorganization of chromatin in symbiosis, the overall increase of its volume suggests that symbionts accumulate DNA. DNA synthesis occurs in symbiosis, but the final steps of mitosis and cytokinesis appear to be prevented. Increased cell and nuclear volume, as well as down-regulation of DNA replication and cell-cycle pathways, compile strong evidence for inhibited cell division in symbiotic Phaeocystis.
Fig. 1.
Morphological transformation of the microalga Phaeocystis cordata in symbiosis unveiled by FIB–SEM. (A and B) Sections of the 3D reconstruction of the free-living (A) and symbiotic (B) Phaeocystis cells as revealed by FIB–SEM, showing multiplication of plastids (green) with immersed pyrenoids (light brown), extension of the mitochondria (red), nuclear compartments (nucleolus in dark blue, heterochromatin in blue, and euchromatin in light blue), and vacuoles (purple). (Scale bar, 2 µm.) See also, SI Appendix, Fig. S1. (C) Box plots showing the increase of the cell volume (log scale, µm3) in symbiotic Phaeocystis (Left) compared to the free-living cells (Right). (D) Relative volume occupancy of different organelles and cellular compartments (plastid, mitochondria, nucleus, and vacuole) as % occupancy in the cell (organelle-volume/cell-volume ratio) in free-living (FL-2 plastids) and seven different symbiotic microalgal cells (Sy) having 4, 16, 31, 36, 40, 54, and 65 plastids. The volumes (µm3) of organelles and cellular compartments are given within respective bar segments and in Dataset S1 (gray bar segments represent the remaining volume of the cell).
Fig. 2.
Increased nuclear volume and inhibited cell cycle in symbiotic Phaeocystis despite lack of evidence for nutrient limitation. (A) 3D reconstruction of algal cells (transparent) and their nucleus (blue) after FIB–SEM imaging, showing increased nuclear volume in symbiotic algal cells with 4, 16, 36, and 65 plastids from Left to Right, compared to the free-living stage (Top left) (see also, SI Appendix, Fig. S1). (Scale bar, 2 µm.) (B) Differential expression of Phaeocystis genes in the KEGG cell-cycle reference pathway (ko04110). Blocks representing protein and enzymes are colored according to differential gene expression results. Solid blue indicates genes that were significantly down-regulated in symbiosis (padj < 0.05, log2FC < −1); transparent blue indicates genes that had a negative fold change in symbiosis but the difference was not statistically significant; transparent red indicates genes that had a positive, but statistically insignificant, fold change in symbiosis; white indicates proteins without annotated genes. (C) Heat map of alkaline phosphatase gene expression (phoA and ehap1-like) as log normalized counts in free-living P. cordata cultures (n = 3) and in individual hosts (n = 12). Alkaline phosphatase expression is a marker for phosphorus limitation in microalgae, and these genes were mostly not expressed in symbiosis. (D) Differential expression of genes in the Nitrogen metabolism KEGG reference pathway (ko00910). Inorganic nitrogen transporters, especially those encoded by Nrt, are marker genes for nitrogen limitation in microalgae. Genes that were significantly down-regulated in symbiosis are colored blue and those that were significantly up-regulated are red. The log2 fold-change values for differentially expressed genes are indicated next to or below the gene name.
Preventing symbiont cell division appears to be one host strategy for managing symbiont populations and limiting symbiont overgrowth (28). In coral symbioses, hosts can regulate symbiont cell division by controlling access to essential nutrients (e.g., nitrogen and phosphorus) (29), which has been previously hypothesized for the Acantharia–Phaeocystis symbiosis. We, therefore, investigated the expression level of marker genes for nutrient deprivation in microalgae: alkaline phosphatase genes for phosphorus limitation (30–32) and nitrate transporter genes for nitrogen limitation (29, 33, 34). Alkaline phosphatase genes (phoA and ehap1-like) that were expressed by free-living cells cultured in nutrient replete conditions were generally not expressed at detectable levels in symbionts, and the nitrate transporter gene (Nrt) was significantly down-regulated in symbiotic cells compared to free-living cells (Fig. 2). Hence, symbiotic Phaeocystis in the host microhabitat does not appear to be limited by these major essential nutrients, despite very low nutrient availability in the waters from which hosts were collected. This is further supported by the increased nucleolar volume observed in symbiotic cells (SI Appendix, Fig. S1), which reflects high ribosomal DNA transcription rates (35) and thus increased ribosome production and protein translation—processes typically reduced under N-limitation (36). Similarly, the translation, translation initiation, and protein folding Gene Ontology (GO) terms, as well as the ribosome Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway, were enriched among the significantly up-regulated genes in symbiosis (SI Appendix, Fig. S2 and Dataset S1). Furthermore, the assimilatory nitrite reductase gene (NirA) was down-regulated in symbiosis. In contrast, genes responsible for ammonia assimilation (GS-GOGAT, CPS1) were up-regulated (Fig. 2D), indicating that ammonium may be readily available to symbionts, as has been shown in several other photosymbioses (37–39). Together, these results suggest that acantharian hosts may rely on a nutrient-independent mechanism to inhibit symbiont cell division and manage intracellular symbiont populations, a strategy that would give hosts finer control over symbionts while ensuring maximal symbiont productivity.
Enhanced Photosynthesis and Carbon Fixation in Symbiosis.
Free-living, flagellated Phaeocystis cells usually have two plastids (Fig. 3A and SI Appendix, Fig. S3). In symbiotic cells, there is a proliferation of enlarged plastids (18), and in this study, we observed 4 to 65 plastids in individual symbiotic Phaeocystis cells. 3D reconstructions allowed us to further determine that plastids occupied 42 to 62% of the total volume of symbiotic cells, compared to 31% in free-living cells, and that both the total volume and surface area of the photosynthetic machinery expanded up to ∼100-fold (Figs. 1 and 3 and Dataset S1). In single-celled algae, plastid division is typically synchronized with cell division and is initiated by the expression of nuclear-encoded plastid division genes during S-phase of the cell cycle (40). Several of these genes (FtsZ, DRP5B, and PDR1) were expressed at similar levels in symbiotic and free-living cells (SI Appendix, Fig. S4), despite cell-cycle genes being significantly down-regulated in symbiosis. Moreover, 3D reconstructions of symbiotic cells revealed several plastids in the process of dividing (SI Appendix, Fig. S4). Continued plastid division without consequent cell division leads to the accumulation of plastids and indicates that plastid division has become decoupled from cell division in symbiotic Phaeocystis. Symbiotic cells may therefore be arrested in S-phase, thus allowing plastids to proliferate and explaining the buildup of chromatin but ultimately keeping symbiont population density constrained.
Fig. 3.
Morphological transformation of the photosynthetic machinery in symbiosis and associated physiological and gene expression activity. (A) Multiplication of plastids in Phaeocystis cells unveiled by FIB–SEM from 2 in free-living stage (Top left) to up to 65 plastids (Sy-65) in symbiosis (see also, SI Appendix, Figs. S3 and S4). (Scale bar, 2 µm.) (B) Single-cell chlorophyll fluorescence imaging showing that the photosynthetic capacity (Fv/Fm parameter) changes as a function of the relative cell size of symbiotic Phaeocystis (symbols). The black line represents the mean Fv/Fm value in free-living cells (± SD, gray lines). (C) Architecture and organization of pyrenoids in plastids of free-living (two types: with and without tubule) and symbiotic Phaeocystis, showing the increased volume of the pyrenoid in symbiosis with multiple thylakoids crossing the pyrenoid (dark green tubules). (Scale bar, 1 µm.) (See also, SI Appendix, Fig. S5.) (D and E) Differential expression results for genes encoding enzymes of the Calvin–Benson cycle (D) and proteins involved in storage molecule biosynthesis (E). The color scale indicates log2 fold change in symbiosis so that positive values (red) represent up-regulation in symbiotic cells and negative values (blue) represent down-regulation in symbiotic cells. Significantly differentially expressed genes (false discovery rate adjusted P value, padj < 0.05) are highlighted with bolded gene names and black boxes in D; all results were significant in E. When several isoforms were expressed for a single gene (Acyl transferase, Ligase 3, and DGAT1), the log2 fold change is shown for each isoform. TAG stands for triacylglycerol. (F) Single isotope probing-NanoSIMS–derived map of carbon relative assimilation [Ka (74)] showing the fraction of carbon (relative to its initial content) assimilated during 5 h of incubation with 13C-labeled bicarbonate in one symbiotic Phaeocystis cell (mainly allocated to plastids) and transferred to the host cytoplasm in specific cellular locations (see also, SI Appendix, Fig. S6). Green arrows indicate the plastids (P) of symbiotic microalgae (surrounded by the dashed circle) showing about 5 atom% of relative carbon assimilation. Mitochondria (M) and the Golgi apparatus (G) also exhibit carbon assimilation. White arrows indicate host areas revealing about 1 to 3 atom% of the assimilated carbon fraction. (Scale bar, 3 µm.)
In plants, plastid proliferation increases photosynthesis more efficiently than plastid enlargement and is recognized as a means of increasing surface exchange, particularly for CO2 diffusion (41). Here, we found that the photosynthesis GO term and KEGG pathway were both enriched among genes up-regulated in symbiotic Phaeocystis cells (SI Appendix, Fig. S2). To further test whether morphological and transcriptional changes in symbiotic cells enhanced photosynthesis, we assessed the relationship between cell size (a proxy for the number of chloroplasts) and photosynthetic efficiency by analyzing photosynthetic parameters in vivo at the single-cell level. We found that photosynthetic capacity increased rapidly with cell size until size had doubled, at which point further increase in photosynthetic capacity was limited (Fig. 3B). This suggests that increased cell size, and therefore expanded photosynthetic machinery, enhances photosynthesis in Phaeocystis. The presence of small and large algal cells within a host indicates that Phaeocystis cells with different photosynthetic capacity coexist (Fv/Fm ranging from 0.4 to 0.78). The smallest symbiotic algae had Fv/Fm values lower than those measured in free-living cells (0.54 ± 0.02). Although it is not possible to conclude here, these algae could represent an early stage during the establishment of symbiosis, in which photosynthesis is transiently repressed, or a late stage when algae start to be degraded.
Expansion of the photosynthetic machinery in symbiotic Phaeocystis should be accompanied by increased carbon fixation and production of organic compounds. Carbon uptake was measured in free-living Phaeocystis cells and symbiotic Phaeocystis within their host incubated with 13C-bicarbonate for 1 h. We found that symbiotic Phaeocystis cells took up ∼150 times more 13C (0.70 ± 0.19 pg of 13C cell−1) than free-living cells (0.0043 ± 0.0004 pg of 13C.cell−1) (Dataset S2). Consistent with this, nearly all nuclear-encoded genes for Calvin–Benson cycle enzymes were up-regulated in symbiosis (Fig. 3D). To further explain enhanced carbon fixation in symbiotic Phaeocystis, we resolved the internal organization of pyrenoids within plastids. Pyrenoids contain the majority of the Rubisco enzyme in plastids and separate it from the surrounding, more-basic stroma to maximize the enzyme’s efficiency and, therefore, the cell’s carbon fixation potential (42). We found that the volume of pyrenoids increased 15-fold from 0.08 ± 0.02 µm3 in free-living cells (n = 27 plastids, 14 cells) to 1.2 ± 0.6 µm3 in symbionts (n = 74 plastids, 7 cells) and that they occupied a larger proportion of plastid volume (13 ± 2.8% in symbiotic cells compared to 5 ± 1.6% in free-living cells, Fig. 3C and SI Appendix, Fig. S5 and Dataset S1). A consequence of enlarged pyrenoids in symbiosis is that their surface area:volume ratio inherently decreases (SI Appendix, Fig. S5), potentially increasing the diffusion barrier for gas and metabolite exchange. Thylakoid tubules deliver CO2 to Rubisco and transport glycerate-3-phosphate product to the stroma, where the remainder of the Calvin–Benson cycles occurs (43, 44). In symbiosis, pyrenoids were always crossed by multiple tubules, whereas pyrenoids in free-living cells either lacked tubules (type 1) or were crossed by one small tubule (type 2) (Fig. 3C). When present, tubules in the pyrenoids of free-living cells were 9.5 times less voluminous than those in symbiotic cells. We hypothesize that multiple tubules in the pyrenoids counterbalance the lower surface area:volume ratio in symbiosis and maintain optimal delivery of CO2. These fine-scale changes in pyrenoid structure, together with up-regulation of Calvin–Benson cycle genes, explain the mechanisms underpinning enhanced carbon fixation in symbiotic Phaeocystis.
Symbionts may themselves take advantage of the additional organic carbon produced from enhanced carbon fixation—either immediately or after synthesizing storage molecules—or they could transfer the additional fixed carbon to hosts. Based on transcriptome analyses, symbiotic cells do not seem to be storing more fixed carbon than free-living cells. Key genes for the biosynthesis of triacylglycerol (TAG)—the preferred storage molecule among microalgae in oligotrophic regions (45)—were significantly down-regulated in symbiosis, including those for acyl-CoA binding proteins and acyltransferases that are involved in the production of TAG precursors (e.g., phosphatidic acid) and diacylglycerol transferase (DGAT), which performs the terminal step in TAG synthesis (Fig. 3E). Likewise, starch synthase was significantly down-regulated in symbiosis (Fig. 3E). Symbiotic cells may store less carbon than free-living cells and instead produce ATP to meet energetic requirements (plastid division, protein synthesis) and relinquish surplus photosynthate to hosts. NanoSIMS (Nanoscale Secondary Ion Mass Spectrometry) analyses incorporating a 5-h incubation with 13C-labeled bicarbonate on four algal cells from two distinct hosts showed that symbiotic Phaeocystis cells mainly allocated carbon to their multiple plastids, mitochondria, Golgi apparatus, and small unidentified vacuoles (Fig. 3F and SI Appendix, Fig. S6). Based on nanoSIMS and 13C uptake data, the estimation of carbon-based growth rates (see Materials and Methods section) shows that the renewal of carbon in the biomass of symbiotic Phaeocystis cells (0.174 ± 0.020%/h) is about 40% higher as compared with free-living cells (0.125 ± 0.006%/h) (Dataset S2). NanoSIMS analyses also demonstrated that photosynthetically fixed carbon is transferred to the host cell (Fig. 3F), thus providing direct evidence that the host benefits from the boosted primary production of its symbiotic microalgae.
Transformation of the Mitochondria into a Reticulated Fine Network Connected to Plastids.
Like plastids, mitochondria underwent an extensive expansion in symbiosis, forming a well-developed fine reticular network with mitochondrial volume and surface area increasing up to 52- and 47-fold, respectively (Fig. 4A and Dataset S1). Such reticular and fused networks are characteristic of actively respiring cells in eukaryotes (46). Yet, in relation to cell volume, the contribution of mitochondria was relatively constant (from 5.3 ± 1.5% in free-living to 3.3 ± 0.6% in symbiosis) (Fig. 1C and Dataset S1). At the suborganelle level, the total volume of mitochondrial cristae (i.e., membrane invaginations, which are respiratory units) increased by up to 38-fold in symbiosis, but volume occupancy was similar in both stages (17.4 ± 3.9% in free-living and 16.3 ± 2.1% in symbiosis) (SI Appendix, Fig. S7). The GO terms for Respiration and Oxidative phosphorylation were enriched among genes down-regulated in symbiosis (SI Appendix, Fig. S2) and the majority of genes in the TCA cycle and respiratory oxidative phosphorylation KEGG pathways were down-regulated in symbiosis (SI Appendix, Fig. S8). The morphology of the mitochondria and down-regulation of respiratory pathways indicates that symbiotic cells may not respire at a higher rate than free-living cells. The mitochondria may, therefore, have a different primary role in symbiotic cells.
Fig. 4.
Expansion of the reticulate mitochondria in symbiosis and its interaction with plastids and role in light capture. (A) Expansion of the mitochondria (red) in symbiotic Phaeocystis cell toward a fine reticular network in contact with plastids. A free-living Phaeocystis cell (Top left) and four symbiotic Phaeocystis cells (Sy) are represented with their 4, 16, 36, and 65 plastids in transparency. Contact surface area (≤ 50 nm, CSA) between the mitochondria and the plastids has been quantified and represented in dark green colors. The mitochondrial surface area in contact with plastids (CSA) increased from 2.6 ± 2% (n = 14) in free living to up to 8.79% in symbiosis (Sy-65). (See also, SI Appendix, Fig. S7.) (Scale bar, 2 µm.) (B) Scatter plot showing the correlation between the volume of the plastid and the volume of the mitochondria in 20 free-living cells (Left Insert) and in 7 symbiotic Phaeocystis cells. (C) Box plot showing the effect of mitochondrial inhibitors (inh: 5 µM Antimycin A and 1 mM Salicylhydroxamic acid-SHAM) on the Fv/Fm parameter of free-living (Left) and symbiotic (Right) Phaeocystis cells. (D) FDTD model calculating the distribution of the light-scattering cross-section (σ) based on 3D architectures of microalgal cells. Light scattering by free-living cells (Left) is forward directed while light scattering by symbiotic cells (Right, 16 plastids) is distributed, yielding an enhanced electric field distribution. Note that due to small light scattering by free-living cells, the scale was adjusted for improved visualization. The outer and inner boxes represent the scattering monitor and the absorption monitor, respectively. Behind the cell, the yellow plane is measuring the transmitted field direction flow (poynting vector). The yellow arrows correspond to the incident plane wave (see also, SI Appendix, Fig. S9).
Other metabolic functions could provide a functional rationale for the morphological modification of mitochondria in symbiotic Phaeocystis. Mitochondrial interaction with plastids is an essential aspect of the bioenergetics of photosynthetic cells (47–49). In symbiotic Phaeocystis, mitochondrial volume tended to increase as plastid volume increased (R2 = 0.74, P = 4.8 E10−11), whereas this relationship was not observed for free-living cells (R2 = 0.07, P = 0.38) (Fig. 4B). We further found that mitochondrial surface area in contact with plastids (≤ 50-nm distance) increased from 2.63 ± 2.1% in free-living (n = 14 cells) to up to 8.8% in the largest symbiotic cell that was imaged (65 plastids, Fig. 4A and Dataset S1), with mitochondria interacting with multiple plastids in symbiotic cells. This indicates that the expansion of the reticulated mitochondria is likely related to plastid proliferation, where mitochondria may serve to maintain minimal diffusion distance between the two organelles and ensure optimal metabolic exchange [e.g., lipids, ammonium, ATP (48, 49)]. Consistent with this possibility, a brief incubation with mitochondrial inhibitors (Antimycin A, Salicylhydroxamic acid) at concentrations blocking respiration (48) decreased the Fv/Fm in symbiosis to 0.55 ± 0.05 (Fig. 4C). We interpret this observation in terms of a reduction of the electron acceptors between the two photosystems, due to increased cellular reducing power upon blocking consumption of reducing equivalents by the respiratory chain, as shown in other microalgae (50). This effect of inhibitors was less pronounced in free-living cells, indicating less interaction between the two organelles in this life stage. Overall, these results show that mitochondria contribute to the enhanced physiological performance of symbiotic Phaeocystis.
Symbiont photosynthesis within multicellular hosts is strongly affected by the light-scattering properties of cells (51, 52). At the cellular level, mitochondria are known to be efficient light scatterers (53). We, therefore, tested whether the well-developed mitochondrial network of symbiotic cells could improve light distribution for photosynthesis. We used Finite-Difference-Time-Domain (FDTD) calculations to model the 3D light propagation in free-living (2 plastids) and symbiotic cells (16 plastids) using the 3D architectures of plastids and mitochondria. We observed that the reticular mitochondrial network homogeneously distributed the incident electric field due to its high scattering cross-section and intricate spatial distribution (Fig. 4D). Contrary to scattering in free-living microalgae, which is small and unidirectional with the incident light, the reticular mitochondrial network of symbiotic microalgae increases the photon pathlength within the algal cell (Fig. 4D and SI Appendix, Fig. S9), consequently enhancing the chance of light absorption (52). This could be advantageous for intracellular microalgae within a host, where light can be rapidly absorbed by the host cytoplasm and can, therefore, become a limiting factor for photosynthesis (e.g., 54). Thus, we show that a possible consequence of the light scattering induced by these large mitochondria could contribute to the enhanced photosynthetic performance of Phaeocystis in symbiosis. This is consistent with earlier measurements, showing that symbiotic cells have improved photosynthetic efficiency under limiting light [the initial slope α of the photosynthesis–irradiance curve (18)] than their free-living counterparts.
Physical Integration of Microalgae into the Host Cell: Symbiosome Invagination into Symbionts.
In photosymbioses, hosts generally phagocytize microalgal cells from the environment and maintain them individually in a vacuole (phagosome), also called a symbiosome, where metabolic exchanges take place. For small symbiotic Phaeocystis cells, the symbiosome surrounds the microalgal cell as observed in other photosymbioses (55) (Fig. 5A). Remarkably, in larger symbiotic cells (>31 plastids), an invagination of the symbiosome vacuole into the microalgal cell was observed in different host cells (Fig. 5B). This invagination can represent a volume of up to 139.4 µm3 (one-fifth of symbiont volume) with a tendency to increase with symbiont size (Fig. 5C). We hypothesize that symbiosome invagination maintains/optimizes metabolic exchanges with very large symbionts that would otherwise have decreased surface area for exchange. Of note, small vesicles were visible in the symbiosome space around symbionts, as well as within the invaginated symbiosome in larger symbionts (Fig. 5 A and B). These vesicles could represent a route for transferring photosynthetic products from symbiont to host or metabolites and signaling molecules from host to symbiont. Indeed, free-living Phaeocystis are often considered “secretory cells” because they excrete vesicles rich in organic carbon (polysaccharides) into the environment (56). Further investigation at higher resolution will be required to better understand the host–symbiont interface in this symbiosis. Several host mitochondria were observed surrounding the symbiotic microalgae and some were closely associated with the symbiosome, as has been observed in other photosymbioses (Fig. 5) (57, 58). Host mitochondria may, therefore, also play a role in the metabolic connection between both partners. Penetration of the symbiosome represents a profound morphological manipulation of the algal cell, not observed in other photosymbioses. This extreme morphological overhaul may be one reason culturing symbionts back to their free-living form has so far been unsuccessful. Thus, the acantharian host hijacks and parasitizes the microalgal cell, ultimately performing “cytoklepty” (or cell farming). Cytoklepty can be defined as an evolutionarily one-sided endosymbiosis, in which the host captures and physiologically exploits endosymbionts, which eventually die and must be replaced by uptake of new endosymbionts from a wild population.
Fig. 5.
Host–symbiont integration and morphometrics of the symbiosome invagination in large symbiotic microalgae. (A) 3D reconstruction following FIB–SEM of a symbiotic microalga with 16 plastids and a cell membrane (light blue) surrounded by mitochondria (orange) and Golgi apparatus (dark purple) of the host. The different organelles and compartments reconstructed in 3D are highlighted in the FIB–SEM electron micrograph frame (Left). A 3D reconstruction (Right) of a large vacuole in the symbiotic microalga (light purple), which is surrounded by a symbiosome (pink) containing small vesicles (dark red). (Scale bar, 2 µm.) (B) A 3D reconstruction of a large symbiotic microalga with 65 plastids and cell membrane (light blue) surrounded by mitochondria (M; orange) and Golgi apparatus (G; dark purple) of the host. The different organelles and compartments reconstructed in 3D are highlighted in the FIB–SEM electron micrograph frame (Left). In large symbionts (>31 plastids), there is an invagination of the symbiosome (S; pink) in the algal cell. Note the presence of small vesicles (Vs; dark red) in the symbiosome and large vacuoles (V; light purple) close to the symbiosome. (Scale bar, 2 µm.) (C) Cell volume of different symbiotic microalgae with 36, 40, and 65 plastids and the associated invaginated symbiosome that increases in volume.
Conclusions
The combination of nanoscale imaging with single-cell transcriptomics in this study illuminates the drastic morphogenetic manipulation of endosymbiotic microalgae involved in a globally distributed and ecologically relevant photosymbiosis. Acantharian hosts prevent Phaeocystis cell division, leading to a complete cellular overhaul that improves symbiont bioenergetic performance, enhances photosynthesis and carbon fixation, and ultimately results in substantial organic carbon production and transfer to hosts. More specifically, there is a structural remodeling of the bioenergetic machinery at multiple scales within the algal cell: plastids proliferate and pyrenoids develop with an immersed thylakoid network, which is accompanied by an extension of reticulated mitochondria that maintains contact with plastids and optimizes light distribution within the algal cell. These modifications are clearly beneficial to single-celled planktonic hosts, which gain an additional energy source and avoid symbiont overgrowth. The observation that host symbiosomes intrude into symbiont cells is unique among marine photosymbioses and provides evidence supporting the idea that the metamorphosis of Phaeocystis is unidirectional, making the relationship an evolutionary dead end for symbionts. From an evolutionary perspective, such extreme manipulation of microalgae may represent a possible step toward a more complete integration of endosymbionts and ultimately plastid acquisition.
Aspects of the symbiont remodeling observed here, such as hypertrophy of the photosynthetic machinery, are also observed in the P. chromatophora endosymbiosis, the most recent known example of primary plastid acquisition (59), and in the Hatena endosymbiosis, a contemporary example of secondary endosymbiosis in progress (60). Expansion of the photosynthetic machinery may, therefore, represent a common stepping-stone toward plastid acquisition. Enlarged nuclei and increased genome ploidy, observed here and in several examples of kleptoplastidy, may represent another commonality in plastid acquisition that serves to support photosynthetic expansion (61, 62). In other planktonic photosymbioses, it remains unknown whether such morphological transformation and energetic exploitation occur, and the diversity of such relationships merits further investigation using subcellular imaging techniques.
The uncoupling between cell-cycle progression and plastid division in symbiotic Phaeocystis suggests that the plastid-to-nucleus signal required to continue cell division is inhibited and deciphering the mechanism involved warrants further attention. Canonical cell-signaling pathways are a promising target for future work, as they have been implicated in mediating other endosymbioses as well as the morphological transformation between flagellate and colonial Phaeocystis cells (63, 64). As this cytoklepty shares important similarities with kleptoplastidy and may represent an early intermediary step between photosymbiosis and more permanent incorporation of the photosynthetic machinery, further elucidation of the mechanisms involved should pave the way to a more complete understanding of how eukaryotes acquire new organelles.
Materials and Methods
Sampling and Preparation for Electron Microscopy.
Symbiotic acantharians harboring intracellular microalgal cells (Phaeocystis cordata) were collected from surface waters as in ref. 18 (Mediterranean Sea, Villefranche-sur-Mer, France). After collection, individual cells were isolated under a microscope with a micropipette, rapidly transferred into filtered natural seawater, and maintained in the same controlled light (100 µmol photons m-2s−1) and temperature (20 °C) conditions as the free-living stage. In parallel, cultures of the haptophyte P. cordata (the symbiont of Acantharia in the Mediterranean Sea) (1) (strain RCC1383 from the Roscoff Culture Collection) were maintained at 20 °C in K5 culture medium and at 100 µmol photons m-2s−1.
Sample preparation protocols were adapted from ref. 18 to optimize the contrast for 3D electron microscopy imaging and facilitate pixel classification. Symbiotic acantharians (with algal endosymbionts) and free-living P. cordata in culture were cryofixed using high-pressure freezing (HPM100, Leica), in which cells were subjected to a pressure of 210 MPa at 196 °C, followed by freeze substitution (FS) (EM ASF2, Leica). Prior to cryofixation, the microalgal cultures were concentrated by gentle centrifugation for 10 min. For FS, a mixture of dried acetone containing 2% osmium tetroxide and 0.5% uranyl acetate were used as contrasting agents. The FS machine was programmed as follows: 60 to 80 h at −90 °C, heating rate of 2 °C h−1 to −60 °C (15 h), 10 to 12 h at −60 °C, heating rate of 2 °C h−1 to −30 °C (15 h), and 10 to 12 h at −30 °C, quickly heated to 0 °C for 1 h to enhance the staining efficiency of osmium tetroxide and uranyl acetate and then back to −30 °C. Cells were then washed in anhydrous acetone for 20 min at 30 °C and gradually embedded in anhydrous Araldite (resin). A graded resin/acetone (volume[vol]/vol) series was used (30, 50, and 70% resin) with each step lasting 2 h at increased temperature: 30% resin/acetone bath from −30 °C to −10 °C, 50% resin/acetone bath from −10 °C to 10 °C, 70% resin/acetone bath from 10 °C to 20 °C. Samples were then placed in 100% resin for 8 to 10 h and in 100% resin with the accelerator BDMA for 8 h at room temperature. Resin polymerization finally occurred at 65 °C for 48 h.
FIB–SEM.
For FIB–SEM, the sample was trimmed with a 90° diamond knife (Diatome) to expose cells at two surfaces (the imaging surface and the surface perpendicular to the FIB) and optimize acquisition (65). For symbiotic P. cordata, trimming was targeted toward the periphery of hosts where microalgae were more abundant. After the sample was trimmed, it was mounted onto the edge of an SEM stub (Agar Scientific) using silver conductive epoxy (CircuitWorks) with the trimmed surfaces facing up and toward the edge of the stub. The sample was gold sputter coated (Quorum Q150RS; 180 s at 30 mA) and placed into the FIB–SEM for acquisition (Crossbeam 540, Carl Zeiss Microscopy GmbH). Once the region of interest (ROI) was located in the sample, Atlas3D software (Fibics Inc. and Carl Zeiss Microscopy GmbH) was used to perform sample preparation and 3D acquisitions. First, a 1-µm platinum protective coat was deposited with a 1.5 nA FIB current. The rough trench was then milled to expose the imaging cross-section with a 15 nA FIB current, followed by a polish at 7 nA. The 3D acquisition milling was conducted with a 1.5 nA FIB current. For SEM imaging, the beam was operated at 1.5 kV/700 pA in analytic mode using an energy-selective backscattered detector (1.1 kV grid collector voltage) at a dwell time of 8 µs with no line averaging. The voxel size used throughout acquisitions was 5 or 10 nm. Datasets were initially aligned by the Fiji plugin “Linear Stack Alignment with SIFT” (66), then fine-tuned by Alignment to Median Smoothed Template (AMST) (67). Raw electron microscopy data are deposited in the Electron Microscopy Public Image Archive (EMPIAR), accession code EMPIAR-10672.
FIB–SEM Image Analysis: Segmentation and Morphometric Analyses.
From the aligned FIB–SEM stack, images were binned in Fiji (https://imagej.net/Fiji), and ROI representing Phaeocystis cells were cropped. Segmentation and 3D reconstruction were performed using the workflow developed in ref. 68, and geometry measurements were provided using algorithms provided at https://gitlab.com/clariaddy/stl_statistics and https://gitlab.com/clariaddy/mindist. Briefly, segmentation of organelles and vacuoles of Phaeocystis (considered as ROI) was carried out with 3D Slicer software (69) (https://www.slicer.org), using a supervised semiautomatic pixel classification mode (3 to 10 slices automatically segmented for each ROI). Each region was “colored” using paint tools and adjusting the threshold range of pixels values of the images. The model maker module from 3D Slicer was then used to generate corresponding 3D models, which were exported in STL format. Three-dimensional reconstructed models were imported into the MeshLab software (70) to clean the model and reduce file size by model decimation. Metrics for volumes, surface area, and the area below the minimum distance between meshes were computed using Numpy-STL (https://pypi.org/project/numpy-stl/) and trimesh (https://trimsh.org/trimesh.html) python packages. Surfaces and volumes were computed using the discrete mesh geometry, the surface being computed directly from mesh triangles, and volume being obtained from the signed volume of individual tetrahedrons, assuming a watertight model. Proximity distance between organelles was calculated based on the closest points between two triangular meshes. The surface area below the proximity distance was quantified based on 1) the minimal distance between each vertex of the plastid mesh to the mitochondria mesh, and 2) matching surface using face data according to a given distance threshold. A distance threshold ≤50 nm was chosen as representative of an interaction between nearby organelles, based on previous morphometric analyses in animal and plant cells (49, 71). Morphometric data are available in Dataset S1.
Single-Cell Chlorophyll Fluorescence Imaging.
Single-cell chlorophyll fluorescence was detected with an imaging system (JBeamBio), mounted on an optical microscope (CKX 53 Olympus). The imaging setup consists of a high sensitivity camera (Orca Flash 4.0 LT) equipped with a near-infrared long-pass filter (RG 695 Schott). ROIs containing host cells were identified using the microscope in transmission mode. The photosynthetic capacity of symbiotic Phaeocystis was quantified as the photochemical quantum yield of Photosystem II (Fv/Fm), calculated as (Fm–Fo)/Fm (72). Cells were excited with blue light pulses (l = 470 ± 12 nm, duration 260 µs) to evaluate minimum fluorescence emission Fo. Short green saturating pulses (l = 520 ± 20 nm, intensity 3,000 µmol photons m−2 s−1, duration 250 ms) were used to reach maximum fluorescence emission (Fm). Fm was evaluated with the same blue pulses used for Fo, fired 10 µs after the saturating light was switched off. We used a 20× (numerical aperture = 0.45) objective to scan the slits. Single algal cells within a given ROI were imaged separately, with a pixel resolution of 1.7 µm2.
Comparative Transcriptomics for Symbiotic and Free-Living Phaeocystis.
Individual acantharians (n = 12) from molecular clade F (all in symbiosis with Phaeocystis) were collected for single-host-cell RNA sequencing from the surface waters of the western subtropical North Pacific (East China Sea), between 9 AM and 5 PM (i.e., during full daylight). RNA-seq was also performed with biological culture replicates (n = 3) of P. cordata CCMP3104 (synonymous to RCC1383) that were filtered and flash frozen in the middle of the photoperiod. Sequencing transcriptomes from cultured, free-living Phaeocystis allowed for the assembly of a de novo reference transcriptome and provided a comparison point for changes in symbiotic gene expression. Raw sequencing reads are available from the NCBI (National Center for Biotechnology Information) Sequencing Read Archive with accession no. PRJNA603434. Detailed descriptions of sampling, culturing, RNA extraction, sequencing, transcriptome assembly and annotation, differential expression, and functional enrichment testing are presented in SI Appendix, Tables S1 and S2 and Fig. S10. All code and data analysis pipelines are available from GitHub (https://maggimars.github.io/PcordataSymbiosisDGE/analysis.html). Genes were considered significantly differentially expressed when the false discovery rate adjusted P value (padj) was less than 0.05 and the log2 fold change was greater than |1|. Likewise, GO and KEGG pathways were considered significantly enriched among up- or down-regulated genes when the padj from the respective enrichment test was less than 0.05.
13C Bulk Enrichment (Elemental Analyzer–Isotope Ratio Mass Spectrometry) and Isotope Analysis.
Symbiotic Acantharia were sampled as described above. Between 54 and 78 individual acantharian cells were pooled together. Two control samples were kept in unspiked filtered seawater (FSW, 0.22 µm) to obtain natural (background) 13C cell content, and two experimental samples were incubated for 1 h in 13C-bicarbonate spiked FSW at 20 °C under constant light (100 µmol photons m−2s−1). To start the incubation, H13CO3 (99% 13C; Cambridge Isotopes Laboratory Inc.) was added to the FSW (0.2 mM, 10% final concentration). After 1 h, acantharian cells were immediately harvested by centrifugation and rinsed three times with FSW and one time with Milli-Q water. All individuals in each sample were then transferred to tin capsules and dried at room temperature for 3 d.
In parallel, cultures of free-living P. cordata RCC1383 maintained at 20 °C in K5 culture medium and at 100 µmol photons m−2s−1 were also incubated for 1 h with 13C-bicarbonate spiked K5 medium (0.2 mM, 10% final concentration). As for symbiotic cells, two samples were used as controls to obtain natural (background) 13C cell content and were kept in unspiked K5 medium. To harvest cells, cultures were centrifuged and rinsed four times with FSW (0.22 µm). Before free-living cells were transferred to tin capsules, an aliquot was taken from each sample to count the number of cells per sample (Dataset S2). Tin capsules were dried for 1 d at 37 °C.
Samples were analyzed for 13C-enrichment using an Elemental Analyzer (Flash 2000, Thermo Scientific) coupled to an Isotopic Ratio Mass Spectrometer (IR–MS Delta V Plus with a Conflo IV Interface, Thermo Scientific). Atom% of samples was derived from isotope ratio data and calculated using the Vienna Pee Dee Belemnite standard (RVPDB = 0.0112372) as follows:
| [1] |
| [2] |
The uptake of 13C (Iisotope) in Acantharia and Phaeocystis cells was obtained from the excess E of the 13C stable isotope in the sample above background (E = atom%Xsample – atom%Xcontrol) and total carbon content in the sample or cell.
| [3] |
To measure 13C uptake of symbiotic Phaeocystis cells in Acantharia, we estimated that one acantharian cell harbors an average of 30 cells [29 ± 27 (1)]. Note that we also added the values calculated for 60 symbiotic Phaeocystis per Acantharia to have conservative estimations of 13C uptake in Acantharia cells (Dataset S2). The uptake of 13C (Iisotope, pg cell−1) for symbiotic Phaeocystis cells was calculated using these estimations. A short incubation (1 h) was chosen to reduce the effect of cell respiration and C-compound exudation on measured 13C uptake values.
NanoSIMS Imaging and Associated Sample Preparation.
Upon collection, symbiotic acantharians were maintained in FSW for 24 h. Cells were then incubated with 13C-labeled bicarbonate (0.4 mM, corresponding to a 16% 13C labeling) for 5 h and cryofixed as for electron microscopy according to an FS protocol from ref. 18. Semithin sections (200 nm) were placed on boron-doped silicon wafers, coated with a 20 nm Au/Pd (80/20) layer, and analyzed with a nanoSIMS 50L (Cameca) at the ProVIS Centre for Chemical Microscopy (Helmholtz Centre for Environmental Research GmbH - UFZ -Leipzig, Germany). The analysis areas involving symbiotic algal cells were defined from SEM observations on consecutive sections. Before analysis, preimplantation with 200 pA of 16 keV Cs+ ion beam was performed for 15 to 20 min in 100 × 100 µm2 area to equilibrate the yield of negative secondary ions. Upon analysis, a 16 keV Cs+ primary ion beam of 1 to 2 pA focused to ∼70 nm was rastered over the sample area between 15 × 15 µm2 and 70 × 70 µm2, with a dwelling time of 2 ms pixel−1 in a 512 × 512 or 1,024 × 1,024 pixel pattern, keeping the physical pixel size well below the beam-spot size in order to avoid an ion-beam–induced surface roughening. Secondary ions extracted from each pixel of the sample surface were analyzed for their mass to charge (m/z) ratio and counted separately with seven electron multiplier detectors. To resolve isobaric interferences, the mass resolving power of the spectrometer was set >8,000 (M/DM) with the entrance slit of 20 × 140 µm (width × height; nominal size), 200 × 200 µm aperture, 40 × 1,800 µm exit slits, and the energy slit cutting off 20% of 12C14N− ions at their high-energy distribution site. Secondary ion species (16O−, 12C2−, 12C13C−, 12C14N−, 13C14N−, 31P−, 32S−) were simultaneously detected in single-ion counting mode. The acquired 40 serial maps of secondary ion count were corrected for lateral drift and accumulated with the Look@NanoSims software (LANS) (73). Isotope ratios were calculated for each ROI defined with LANS either in automatic thresholding mode or by manual drawing over cell compartments recognized in ion-count/ratio maps (normalized by scans and pixel number) and correlative SEM images. The fraction of assimilated carbon relative to its initial content (relative carbon assimilation, Ka) was derived from the changes in carbon-isotope composition as described in ref. 74. We then calculated C-based growth rate considering both bulk IR–MS and nanoSIMS data (Dataset S2). Assuming a constant growth rate of biomass proportional to carbon uptake, its temporal development M(t) may be described as follows:
| [4] |
The KA represents the gain in biomass (relatively to its initial amount) upon incubation. The M0 × KA term represents the biomass gained upon relative assimilation KA.
Division (growth) rate v of each single cell (biomass) can be expressed in KA via following transformation of expression [4]. Dividing the M(t) expression by M0, we obtain the following:
| [5] |
and final expression for the division (growth) rate v in KA:
| [6] |
Photon Propagation Model: FDTD Simulations.
To study the role of light scattering from the 3D mitochondrial network, we performed 3D FDTD calculations using a Maxwell equation solver (Lumerical FDTD Solutions 8.16). FDTD allows for discretizing the real-space time-domain Maxwell equations onto a regular lattice in time and space with equidistant time steps and cubic voxels on the Yee grid. The propagation of the electromagnetic field is modeled by time stepwise forward integration. For plastids, we assumed that the real part of the refractive index was minimally wavelength dependent (between 1.352 to 1.364 for 400 to 700 nm), while the imaginary part of the refractive index (k) was governed by the characteristic absorption profile of chlorophyll a (75). The real refractive index (n) of mitochondria was assumed to be 1.4, which is a moderate estimate for such strongly light-scattering structures (53, 76) with a reduced scattering coefficient about two orders of magnitude greater than the absorption coefficient (µs’ > 100*µa) (53). The background n of the cell was similar to water (n = 1.34). For each cell type (i.e., free-living and symbiotic), we performed calculations in the presence and absence of mitochondria. Additionally, we replaced the mitochondria with a solid sphere of the same volume, to visualize the beneficial light spreading by the intricate mitochondrial network over a solid sphere with the same refractive index properties. For each simulation, excitation was provided by a plane wave that was incident in either the x, y, or z plane. Only one plane is shown as the results were largely independent of the incident plane. The numerical stability of the simulation was ensured by selecting boundary conditions and simulation times (>150 fs) that confirmed that the electric field in the structure decayed prior to the end of the simulation, such that all of the incident excitation was lost from the grid.
Supplementary Material
Acknowledgments
This project received funding from the ATIP-Avenir program, a Défi X-Life grant from the National Center for Scientific Research (France) (CNRS), and the LabEx GRAL (ANR-10-LABX-49-01) and was financed within the University Grenoble Alpes graduate school (Ecoles Universitaires de Recherche; CBH-EUR-GS) (ANR-17-EURE-0003). J.D. was supported by the ATIP-Avenir program. C.U. was supported by a joint University Grenoble Alpes-Eidgenössche Technische Hochschule Zurich PhD grant in the framework of the “Investissements d’avenir” programme (ANR-15-IDEX-02). M.M.B. was supported by a DC1 graduate fellowship awarded by the Japan Society for the Promotion of Science. D.W. was funded by the Gordon and Betty Moore Foundation and L.S. was supported by the Isaac Newton Trust. G.F. and D.F. received funding from the European Research Council: ERC Chloro-mito (Grant No. 833184). This project also received funds from the European Union’s Horizon 2020 research and innovation programme CORBEL under the Grant No. 654248. We thank the institutes that supported the collection of samples: EMBRC (European Marine Biological Resource Centre)-France and the Laboratoire d’Océanographie de Villefranche-sur-Mer (John Dolan and the marine crew). This work used the platforms of the Grenoble Instruct-ERIC centre (ISBG; UMS 3518; CNRS-CEA-UGA-European Molecular Biology Laboratory) within the Grenoble Partnership for Structural Biology, supported by French Infrastructure for Integrated Structural Biology (FRISBI) (ANR-10-INBS-05-02) and GRAL, financed within CBH-EUR-GS (ANR-17-EURE-0003). We thank Guy Schoehn, Christine Moriscot, and the electron microscope facility, which is supported by the Auvergne-Rhône-Alpes Region, the Fondation Recherche Medicale, the fonds FEDER (Fonds européen de développement régional) and the GIS-Infrastructures en Biologie Sante et Agronomie. We are thankful for the use of the analytical facilities of the Centre for Chemical Microscopy (ProVIS) at UFZ Leipzig, which is supported by European Regional Development Funds (Europe funds Saxony) and the Helmholtz Association. Sampling and molecular work for transcriptome analysis was funded by the Marine Biophysics Unit of the Okinawa Institute of Science and Technology (OIST) Graduate University. We thank the captain and crew of the JAMSTEC (Japan Agency for Marine-Earth Science and Technology) R/V Mirai for their assistance and support in sample collection. Hiromi Watanabe, Dhugal Lindsay, and Yuko Hasagawa were instrumental in organizing and facilitating cruise sampling. Lisa Mesrop and Dave Caron collaborated to optimize single-host RNA-seq. Hiroki Goto and the OIST DNA Sequencing Section performed sequencing and provided guidance. We also thank Gaël Guillou, Benoit Lebreton, and the Plateforme de Spectrométrie Isotopique of the La Rochelle University, which is funded by FEDER Poitou-Charentes and the Région Nouvelle Aquitaine (2017 to 2021).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2025252118/-/DCSupplemental.
Data Availability
Code and data analysis pipelines and 3D electron microscopy raw images data have been deposited in GitHub, EMPIAR (transcriptomes: https://maggimars.github.io/PcordataSymbiosisDGE/analysis.html; raw 3D electron microscopy images on EMPIAR: DOI: 10.1101/2020.12.08.416644). All other study data are included in the article and/or supporting information.
References
- 1.Decelle J., et al., An original mode of symbiosis in open ocean plankton. Proc. Natl. Acad. Sci. U.S.A. 109, 18000–18005 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Decelle J., Colin S., Foster R. A., “Photosymbiosis in marine planktonic protists” in Marine Protists: Diversity and Dynamics, Ohtsuka S., Suzaki T., Horiguchi T., Suzuki N., Not F., Eds. (Springer Japan, 2015), pp. 465–500. [Google Scholar]
- 3.Caron D. A., Michaels A. F., Swanberg N. R., Howse F. A., Primary productivity by symbiont-bearing planktonic sarcodines (Acantharia, Radiolaria, Foraminifera) in surface waters near Bermuda. J. Plankton Res. 17, 103–129 (1995). [Google Scholar]
- 4.Michaels A. F., Acantharian abundance and symbiont productivity at the VERTEX seasonal station. J. Plankton Res. 13, 399–418 (1991). [Google Scholar]
- 5.Michaels A. F., Caron D. A., Swanberg N. R., Howse F. A., Michaels C. M., Planktonic sarcodines (Acantharia, Radiolaria, Foraminifera) in surface waters near Bermuda: Abundance, biomass and vertical flux. J. Plankton Res. 17, 131–163 (1995). [Google Scholar]
- 6.Martin P., et al., Sedimentation of acantharian cysts in the Iceland Basin: Strontium as a ballast for deep ocean particle flux, and implications for acantharian reproductive strategies. Limnol. Oceanogr. 55, 604–614 (2010). [Google Scholar]
- 7.Gutierrez-Rodriguez A., et al., High contribution of Rhizaria (Radiolaria) to vertical export in the California current ecosystem revealed by DNA metabarcoding. ISME J. 13, 964–976 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yellowlees D., Rees T. A. V., Leggat W., Metabolic interactions between algal symbionts and invertebrate hosts. Plant Cell Environ. 31, 679–694 (2008). [DOI] [PubMed] [Google Scholar]
- 9.Decelle J., New perspectives on the functioning and evolution of photosymbiosis in plankton: Mutualism or parasitism? Commun. Integr. Biol. 6, e24560 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Douglas A. E., Smith D. C., Are endosymbioses mutualistic? Trends Ecol. Evol. 4, 350–352 (1989). [DOI] [PubMed] [Google Scholar]
- 11.Karkar S., Facchinelli F., Price D. C., Weber A. P. M., Bhattacharya D., Metabolic connectivity as a driver of host and endosymbiont integration. Proc. Natl. Acad. Sci. U.S.A. 112, 10208–10215 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bhattacharya D., Archibald J. M., Weber A. P. M., Reyes-Prieto A., How do endosymbionts become organelles? Understanding early events in plastid evolution. BioEssays 29, 1239–1246 (2007). [DOI] [PubMed] [Google Scholar]
- 13.Keeling P. J., The number, speed, and impact of plastid endosymbioses in eukaryotic evolution. Annu. Rev. Plant Biol. 64, 583–607 (2013). [DOI] [PubMed] [Google Scholar]
- 14.Archibald J. M., Endosymbiosis and eukaryotic cell evolution. Curr. Biol. 25, R911–R921 (2015). [DOI] [PubMed] [Google Scholar]
- 15.Archibald J. M., The puzzle of plastid evolution. Curr. Biol. 19, R81–R88 (2009). [DOI] [PubMed] [Google Scholar]
- 16.Hehenberger E., Gast R. J., Keeling P. J., A kleptoplastidic dinoflagellate and the tipping point between transient and fully integrated plastid endosymbiosis. Proc. Natl. Acad. Sci. U.S.A. 116, 17934–17942 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nowack E. C. M., Melkonian M., Glöckner G., Chromatophore genome sequence of Paulinella sheds light on acquisition of photosynthesis by eukaryotes. Curr. Biol. 18, 410–418 (2008). [DOI] [PubMed] [Google Scholar]
- 18.Decelle J., et al., Algal remodeling in a ubiquitous planktonic photosymbiosis. Curr. Biol. 29, 968–978.e4 (2019). [DOI] [PubMed] [Google Scholar]
- 19.Febvre J., Febvre-Chevalier C., Ultrastructural study of zooxanthellae of three species of Acantharia (Protozoa: Actinopoda), with details of their taxonomic position in the prymnesiales (Prymnesiophyceae, Hibberd, 1976). J. Mar. Biol. Assoc. U. K. 59, 215–226 (1979). [Google Scholar]
- 20.Verity P. G., Smetacek V., Organism life cycles, predation, and the structure of marine pelagic ecosystems. Mar. Ecol. Prog. Ser. 130, 277–293 (1996). [Google Scholar]
- 21.Schoemann V., Becquevort S., Stefels J., Rousseau V., Lancelot C., Phaeocystis blooms in the global ocean and their controlling mechanisms: A review. J. Sea Res. 53, 43–66 (2005). [Google Scholar]
- 22.Mars Brisbin M., Mesrop L. Y., Grossmann M. M., Mitarai S., Intra-host symbiont diversity and extended symbiont maintenance in photosymbiotic Acantharea (Clade F). Front. Microbiol. 9, 1998 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Freeman Rosenzweig E. S., et al., The eukaryotic CO2-concentrating organelle is liquid-like and exhibits dynamic reorganization. Cell 171, 148–162.e19 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jones A. R., et al., Cell-size dependent progression of the cell cycle creates homeostasis and flexibility of plant cell size. Nat. Commun. 8, 15060 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jones A. R., Dewitte W., Cell size and cell cycle progression: The cyclin-dependent kinase link in green algae. J. Exp. Bot. 70, 731–733 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boisvert F.-M., van Koningsbruggen S., Navascués J., Lamond A. I., The multifunctional nucleolus. Nat. Rev. Mol. Cell Biol. 8, 574–585 (2007). [DOI] [PubMed] [Google Scholar]
- 27.Tamaru H., Confining euchromatin/heterochromatin territory: Jumonji crosses the line. Genes Dev. 24, 1465–1478 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Titlyanov E. A., et al., Degradation of zooxanthellae and regulation of their density in hermatypic corals. Mar. Ecol. Prog. Ser. 139, 167–178 (1996). [Google Scholar]
- 29.Xiang T., et al., Symbiont population control by host-symbiont metabolic interaction in Symbiodiniaceae-cnidarian associations. Nat. Commun. 11, 108 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Annis E. R., Cook C. B., Alkaline phosphatase activity in symbiotic dinoflagellates (zooxanthellae) as a biological indicator of environmental phosphate exposure. Mar. Ecol. Prog. Ser. 245, 11–20 (2002). [Google Scholar]
- 31.Alipanah L., et al., Molecular adaptations to phosphorus deprivation and comparison with nitrogen deprivation responses in the diatom Phaeodactylum tricornutum. PLoS One 13, e0193335 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li T., et al., Identification and expression analysis of an atypical alkaline phosphatase in Emiliania huxleyi. Front. Microbiol. 9, 2156 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sanz-Luque E., Chamizo-Ampudia A., Llamas A., Galvan A., Fernandez E., Understanding nitrate assimilation and its regulation in microalgae. Front Plant Sci 6, 899 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alipanah L., Rohloff J., Winge P., Bones A. M., Brembu T., Whole-cell response to nitrogen deprivation in the diatom Phaeodactylum tricornutum. J. Exp. Bot. 66, 6281–6296 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rudra D., Warner J. R., What better measure than ribosome synthesis? Genes Dev. 18, 2431–2436 (2004). [DOI] [PubMed] [Google Scholar]
- 36.Siersma P. W., Chiang K. S., Conservation and degradation of cytoplasmic and chloroplast ribosomes in Chlamydomonas reinhardtii. J. Mol. Biol. 58, 167–185 (1971). [DOI] [PubMed] [Google Scholar]
- 37.LeKieffre C., et al., Ammonium is the preferred source of nitrogen for planktonic foraminifer and their dinoflagellate symbionts. Proc. Biol. Sci. 287, 20200620 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu Z., Mesrop L. Y., Hu S. K., Caron D. A., Transcriptome of Thalassicolla nucleata holobiont reveals details of a radiolarian symbiotic relationship. Front. Mar. Sci. 6, 1–11 (2019). [Google Scholar]
- 39.Maor-Landaw K., van Oppen M. J. H., McFadden G. I., Symbiotic lifestyle triggers drastic changes in the gene expression of the algal endosymbiont Breviolum minutum (Symbiodiniaceae). Ecol. Evol. 10, 451–466 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sumiya N., Mechanism of coordination between cell and chloroplast division in unicellular algae. Plant Morphology 30, 83–89 (2018). [Google Scholar]
- 41.Xiong D., Huang J., Peng S., Li Y., A few enlarged chloroplasts are less efficient in photosynthesis than a large population of small chloroplasts in Arabidopsis thaliana. Sci. Rep. 7, 5782 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moroney J. V., Ynalvez R. A., Proposed carbon dioxide concentrating mechanism in Chlamydomonas reinhardtii. Eukaryot. Cell 6, 1251–1259 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Engel B. D., et al., Native architecture of the Chlamydomonas chloroplast revealed by in situ cryo-electron tomography. eLife 4, e04889 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rochaix J.-D., The pyrenoid: An overlooked organelle comes out of age. Cell 171, 28–29 (2017). [DOI] [PubMed] [Google Scholar]
- 45.Becker K. W., et al., Daily changes in phytoplankton lipidomes reveal mechanisms of energy storage in the open ocean. Nat. Commun. 9, 5179 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Westermann B., Bioenergetic role of mitochondrial fusion and fission. Biochim. Biophys. Acta 1817, 1833–1838 (2012). [DOI] [PubMed] [Google Scholar]
- 47.Spinelli J. B., Haigis M. C., The multifaceted contributions of mitochondria to cellular metabolism. Nat. Cell Biol. 20, 745–754 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bailleul B., et al., Energetic coupling between plastids and mitochondria drives CO2 assimilation in diatoms. Nature 524, 366–369 (2015). [DOI] [PubMed] [Google Scholar]
- 49.Mueller-Schuessele S. J., Michaud M., “Plastid transient and stable interactions with other cell compartments” in Plastids: Methods and Protocols, Maréchal E., Ed. (Springer US, 2018), pp. 87–109. [DOI] [PubMed] [Google Scholar]
- 50.Ghysels B., Godaux D., Matagne R. F., Cardol P., Franck F., Function of the chloroplast hydrogenase in the microalga Chlamydomonas: The role of hydrogenase and state transitions during photosynthetic activation in anaerobiosis. PLoS One 8, e64161 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Holt A. L., Vahidinia S., Gagnon Y. L., Morse D. E., Sweeney A. M., Photosymbiotic giant clams are transformers of solar flux. J. R. Soc. Interface 11, 20140678 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wangpraseurt D., et al., Bionic 3D printed corals. Nat. Commun. 11, 1748 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Beauvoit B., Kitai T., Chance B., Contribution of the mitochondrial compartment to the optical properties of the rat liver: A theoretical and practical approach. Biophys. J. 67, 2501–2510 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wangpraseurt D., Larkum A. W. D., Ralph P. J., Kühl M., Light gradients and optical microniches in coral tissues. Front. Microbiol. 3, 316 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Davy S. K., Allemand D., Weis V. M., Cell biology of cnidarian-dinoflagellate symbiosis. Microbiol. Mol. Biol. Rev. 76, 229–261 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chin W.-C., Orellana M. V., Quesada I., Verdugo P., Secretion in unicellular marine phytoplankton: Demonstration of regulated exocytosis in Phaeocystis globosa. Plant Cell Physiol. 45, 535–542 (2004). [DOI] [PubMed] [Google Scholar]
- 57.Song C., Murata K., Suzaki T., Intracellular symbiosis of algae with possible involvement of mitochondrial dynamics. Sci. Rep. 7, 1221 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kerney R., et al., Intracellular invasion of green algae in a salamander host. Proc. Natl. Acad. Sci. U.S.A. 108, 6497–6502 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nowack E. C. M., Paulinella chromatophora – Rethinking the transition from endosymbiont to organelle. Acta Soc. Bot. Pol. 83, 387–397 (2014). [Google Scholar]
- 60.Okamoto N., Inouye I., A secondary symbiosis in progress? Science 310, 287 (2005). [DOI] [PubMed] [Google Scholar]
- 61.Onuma R., et al., Changes in the transcriptome, ploidy, and optimal light intensity of a cryptomonad upon integration into a kleptoplastic dinoflagellate. ISME J. 14, 2407–2423 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Johnson M. D., The acquisition of phototrophy: Adaptive strategies of hosting endosymbionts and organelles. Photosynth. Res. 107, 117–132 (2011). [DOI] [PubMed] [Google Scholar]
- 63.Zhang S.-F., Zhang K., Cheng H.-M., Lin L., Wang D.-Z., Comparative transcriptomics reveals colony formation mechanism of a harmful algal bloom species Phaeocystis globosa. Sci. Total Environ. 719, 137454 (2020). [DOI] [PubMed] [Google Scholar]
- 64.Mars Brisbin M., Mitarai S., Differential gene expression supports a resource-intensive, defensive role for colony production in the bloom-forming haptophyte, Phaeocystis globosa. J. Eukaryot. Microbiol. 66, 788–801 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Maco B., et al., Semiautomated correlative 3D electron microscopy of in vivo-imaged axons and dendrites. Nat. Protoc. 9, 1354–1366 (2014). [DOI] [PubMed] [Google Scholar]
- 66.Lowe D. G., Distinctive image features from scale-invariant keypoints. Int. J. Comput. Vis. 60, 91–110 (2004). [Google Scholar]
- 67.Hennies J., et al., AMST: Alignment to median smoothed template for focused ion beam scanning electron microscopy image stacks. Sci. Rep. 10, 2004 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Uwizeye C.et al., Morphological bases of phytoplankton energy management and physiological responses unveiled by 3D subcellular imaging. Nat. Commun. 12, 1049 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kikinis R., Pieper S. D., Vosburgh K. G., “3D slicer: A platform for subject-specific image analysis, visualization, and clinical support” in Intraoperative Imaging and Image-Guided Therapy, Jolesz F. A., Ed. (Springer New York, 2014), pp. 277–289. [Google Scholar]
- 70.Cignoni P., et al., “Meshlab: An open-source mesh processing tool” in Eurographics Italian Chapter Conference (Salerno, 2008), pp. 129–136. [Google Scholar]
- 71.Scorrano L., et al., Coming together to define membrane contact sites. Nat. Commun. 10, 1287 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Butler W. L., Energy distribution in the photochemical apparatus of photosynthesis. Annu. Rev. Plant Physiol. 29, 345–378 (1978). [Google Scholar]
- 73.Polerecky L., et al., Look@NanoSIMS'–A tool for the analysis of nanoSIMS data in environmental microbiology. Environ. Microbiol. 14, 1009–1023 (2012). [DOI] [PubMed] [Google Scholar]
- 74.Stryhanyuk H., et al., Calculation of single cell assimilation rates from SIP-NanoSIMS-Derived isotope ratios: A comprehensive approach. Front. Microbiol. 9, 2342 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Qi H., He Z.-Z., Zhao F.-Z., Ruan L.-M., Determination of the spectral complex refractive indices of microalgae cells by light reflectance-transmittance measurement. Int. J. Hydrogen Energy 41, 4941–4956 (2016). [Google Scholar]
- 76.Wilson J. D., Cottrell W. J., Foster T. H., Index-of-refraction-dependent subcellular light scattering observed with organelle-specific dyes. J. Biomed. Opt. 12, 014010 (2007). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Code and data analysis pipelines and 3D electron microscopy raw images data have been deposited in GitHub, EMPIAR (transcriptomes: https://maggimars.github.io/PcordataSymbiosisDGE/analysis.html; raw 3D electron microscopy images on EMPIAR: DOI: 10.1101/2020.12.08.416644). All other study data are included in the article and/or supporting information.