Significance
Cytosolic peptide:N-glycanase (NGLY1) is a widely conserved enzyme involved in de–N-glycosylation of N-glycosylated proteins. Mutations in the human NGLY1 gene cause global developmental delay and multisystemic symptoms, but the molecular mechanism underlying pathogenesis remains poorly understood. FBXO6/FBS2, a subunit of the SCF (SKP1–CUL1–F-box protein) ubiquitin ligase complex, recognizes N-glycans of cytosolic glycoproteins in the endoplasmic reticulum–associated degradation (ERAD) pathway. This paper reports that high levels of ERAD glycoprotein substrates abnormally ubiquitinated by SCFFBS2 in the absence of NGLY1 impair the proteasome, contributing to the pathogenesis of NGLY1 deficiency. Importantly, knockout of Fbxo6/Fbs2 rescued the lethality of NGLY1 deficiency in mice, suggesting a strategy for developing therapeutics for this intractable disease.
Keywords: ERAD, FBXO6/FBS2, NGLY1, proteasome, ubiquitination
Abstract
Mutations in the human peptide:N-glycanase gene (NGLY1), which encodes a cytosolic de–N-glycosylating enzyme, cause a congenital autosomal recessive disorder. In rodents, the loss of Ngly1 results in severe developmental delay or lethality, but the underlying mechanism remains unknown. In this study, we found that deletion of Fbxo6 (also known as Fbs2), which encodes a ubiquitin ligase subunit that recognizes glycoproteins, rescued the lethality-related defects in Ngly1-KO mice. In NGLY1-KO cells, FBS2 overexpression resulted in the substantial inhibition of proteasome activity, causing cytotoxicity. Nuclear factor, erythroid 2–like 1 (NFE2L1, also known as NRF1), an endoplasmic reticulum–associated transcriptional factor involved in expression of proteasome subunits, was also abnormally ubiquitinated by SCFFBS2 in NGLY1-KO cells, resulting in its retention in the cytosol. However, the cytotoxicity caused by FBS2 was restored by the overexpression of “glycan-less” NRF1 mutants, regardless of their transcriptional activity, or by the deletion of NRF1 in NGLY1-KO cells. We conclude that the proteasome dysfunction caused by the accumulation of N-glycoproteins, primarily NRF1, ubiquitinated by SCFFBS2 accounts for the pathogenesis resulting from NGLY1 deficiency.
The ubiquitin–proteasome system (UPS) is the major pathway for selective protein degradation in eukaryotes (1). The UPS maintains proteostasis, thus avoiding the generation of cytotoxic aggregates caused by accumulation of misfolded proteins (2, 3). Reduction in proteasome activity causes neurodegenerative disorders such as Parkinson’s disease and Alzheimer’s disease (4). Hence, it is necessary to understand the mechanisms underlying the decline in proteasome function during aging and disease.
Nuclear Factor, Erythroid 2–like 1 (NFE2L1, also known as NRF1) is a transcription factor that regulates proteasome activity through a so-called “bounce-back” mechanism (5, 6). NRF1 is an N-glycosylated, single-pass type II membrane protein whose DNA-binding domain usually faces the lumen of the endoplasmic reticulum (ER) (7). In general, misfolded or unassembled proteins in the ER are retrotranslocated into the cytosol, followed by proteasomal degradation, a process referred to as ER-associated degradation (ERAD) (8, 9). Remarkably, at steady state, NRF1, like its Caenorhabditis elegans functional ortholog SKN-1A, is constitutively targeted to the ERAD pathway, and the amount of the NRF1 is maintained at low basal levels in the cell (5, 6). During retrotranslocation, NRF1 is ubiquitinated by HRD1 and deglycosylated by a peptide:N-glycanase (NGLY1) while it undergoes proteasomal degradation. Under conditions in which proteasomal activity is compromised, however, retrotranslocated NRF1 is stabilized and cleaved by an aspartyl protease, DNA-damage inducible 1 homolog 2 (DDI2), thereby generating activated NRF1. The activated NRF1 is translocated into the nucleus, where it transcriptionally activates proteasome subunits (10, 11). The deglycosylation activity of NGLY1 is essential for NRF1 activation (i.e., processing, nuclear translocation, and transcriptional activity) (11–13).
NGLY1 is an evolutionarily conserved enzyme that is responsible for removing Asn-linked oligosaccharides (N-glycans) from glycoproteins in the cytosol (14). Mutations in NGLY1 result in NGLY1 deficiency (OMIM # 615273: NGLY-1-CDDG), a rare autosomal recessive congenital disorder with multiorgan symptoms (15, 16). Symptoms include global developmental delay, movement disorders, hypotonia, hypolacrimia/alacrimia, scoliosis, and peripheral neuropathy (16, 17). Nearly all pathogenic NGLY1 mutations examined to date are associated with decreases in both NGLY1 protein levels and enzymatic activity (18). In mice, Ngly1 knockout (KO) in the C57BL/6 background is embryonically lethal, and Ngly1-KO Sprague-Dawley rats exhibit severe developmental delay and impaired motor functions, consistent with the symptoms observed in human patients (19, 20). Although additional deletion of the gene encoding endo-β-N-acetylglucosaminidase (Engase), another deglycosylating enzyme in the cytosol, partially rescues the embryonic lethality of Ngly1-KO mice (19), these animals still develop progressive defects such as trembling or bent spine, indicating that Engase deletion has a limited ability to suppress phenotypes caused by Ngly1 deletion. The molecular mechanism underlying the defects caused by the loss of NGLY1 activity remains unknown, and no therapeutic treatment for NGLY1 deficiency is currently available.
We previously reported that three F-box proteins that recognize sugar chains (FBS; FBS1, FBS2, and FBS3, also known as FBXO2, FBXO6, and FBXO27, respectively) are present in the cytosol (21–23). F-box proteins are receptors for substrates of the SCF (SKP1–CUL1–F-box protein–RBX1) ubiquitin ligase complex (24), and SCFFBS complexes ubiquitinate N-glycoproteins. FBS1 and FBS2 function in the ERAD pathway, recognizing the innermost Man3GlcNAc2 structure in high mannose-type glycans as a hallmark of misfolded proteins (25–27). In addition, FBS3 binds to membranes via N-myristoylation and ubiquitinates glycoproteins that are exposed to the cytosol upon lysosomal damage, thereby inducing lysophagy (23).
In this study, we found that Fbs2;Ngly1 double-KO (dKO) mice are born and exhibit no overt abnormalities, suggesting that FBS2 activity is the primary cause of the lethality and progressive defects in Ngly1-KO mice. We also observed that FBS2 overexpression induces cell death in NGLY1-KO cells through a mechanism involving proteasomal dysfunction. Our findings reveal the molecular mechanisms underlying NGLY1 deficiency and suggest that FBS2 inhibitors could serve as potential therapeutic agents for patients with NGLY1 deficiency.
Results
Fbs2;Ngly1 dKO Mice Are Viable and Exhibit Normal Motor Function.
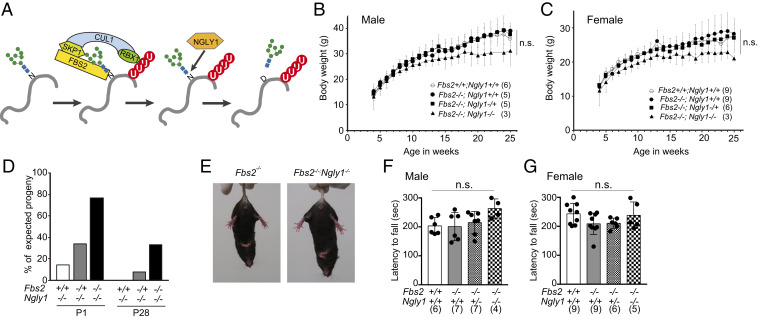
FBS2 and NGLY1 act on cytosolic glycoproteins in the ERAD pathway; consequently, FBS2 cannot recognize ERAD substrates if they are deglycosylated by NGLY1 (Fig. 1A). To elucidate the genetic interactions between FBS2 and NGLY1, we generated Fbs2;Ngly1 dKO mice. The KO construct for Fbs2 is shown in SI Appendix, Fig. S1 A and B; the construct for Ngly1 was reported previously (19). Fbs2−/− mice developed and bred normally (Fig. 1 B and C), whereas Ngly1−/− mice with the C57BL/6 background were embryonic/perinatal lethal (SI Appendix, Table S1) (19). Fbs2 expression is detected in various organs of adult mice (22) and was relatively high in heart and liver at later stages of embryonic development (SI Appendix, Fig. S1C). We first generated double-heterozygous mice (Fbs2−/+;Ngly1−/+), which were then crossed to generate dKO (Fbs2−/−;Ngly1−/−) mice. Surprisingly, the Fbs2−/−;Ngly1−/− mice were viable, indicating that the deletion of Fbs2 rescued the embryonic lethality caused by the defect in Ngly1 (Fig. 1D and SI Appendix, Table S1). The survival ratio at P0 was much higher (1.9-fold) than in Engase;Ngly1 dKO mice (19), suggesting that Fbs2−/− exerts a stronger effect in suppressing the embryonic defect of Ngly1−/− mice. Consistent with this observation, Fbs2−/+;Ngly1−/− mice were also born, albeit at a reduced ratio, whereas no viable Engase−/+;Ngly1−/− mice were obtained (19). This result clearly indicates that even a 50% reduction in the Fbs2 dose can alleviate the lethality of Ngly1−/− mice.
Fig. 1.
Fbs2;Ngly1 dKO mice are viable and have normal motor function. (A) The role of FBS2 and NGLY1 in the ERAD pathway. SCFFBS2 recognizes the innermost positions of high mannose-type glycans of misfolded glycoproteins and ubiquitinates the glycoproteins. NGLY1 removes N-glycans from glycoproteins in which the N-glycosylated Asn (N) are converted to Asp (D) prior to degradation by the proteasome. (B and C) Body weights of Fbs2;Ngly1 dKO male (B) and female (C) mice. Mice were weighed at weekly intervals after weaning. Values represent means ± SD. The number of mice weighed is noted in parentheses. Statistical significance among four groups was determined using one-way ANOVA. n.s., not significant. (D) Suppression of embryonic lethality of Ngly1-KO mice by Fbs2 deletion. The ratios of observed to expected frequencies of progeny obtained by crossing of Fbs2−/+;Ngly1−/+ mice are shown. Refer to SI Appendix, Table S1 for actual count. (E) Fbs2;Ngly1 dKO mice, like wild-type mice, exhibited no sign of abnormal hindlimb clasping when suspended by the tail. Left shows Fbs2 KO male (control, 25 wk); right shows Fbs2;Ngly1 dKO male (25 wk). Compare with the example of the mouse exhibiting typical hindlimb clasping in SI Appendix, Fig. S1C. (F and G) Rotarod testing of motor coordination of male (F) and female (G) mice at 5 wk of age. The time until the mouse dropped from the accelerating rod (4.5 to 45 rpm in 4 min) is shown. Values represent means ± SD. The number of mice examined is shown in parentheses. Statistical significance among four groups was determined using one-way ANOVA. n.s., not significant.
Although Engase−/−;Ngly1−/− mice exhibited motor function defects such as hindlimb clasping (SI Appendix, Fig. S1D), Fbs2−/−;Ngly1−/− mice exhibited no motor function defects (Fig. 1 E–G), even at 22 mo after birth (Movies S1 and S2). Unlike Ngly1−/− mice with a C57BL/6 background, Ngly1−/− rats survived to adulthood (20). However, these animals still develop pathological abnormalities in the central and peripheral nervous systems (20), indicating that mammalian NGLY1 plays critical roles in the nervous system after the embryonic stage. Notably in this regard, no obvious pathological abnormality was detected in the brains of dKO mice (SI Appendix, Fig. S1E). These findings indicate that the lethality and progressive defects caused by the Ngly1 deletion in mice are due to Fbs2 activity.
SCFFBS2 Ubiquitinates NRF1 in the Cytosol in NGLY1-KO Cells.
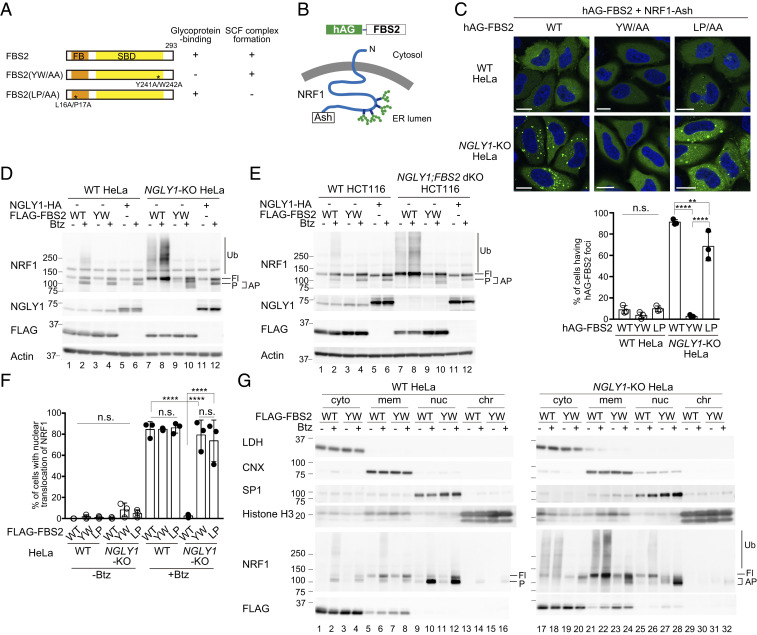
To obtain further insight into the effect of FBS2 on the pathology of NGLY1-KO cells, we generated NGLY1-KO HeLa cells using CRISPR/Cas-9 technology. We first investigated whether FBS2 could interact with glycoprotein ERAD substrates more efficiently in the absence of NGLY1. NRF1, an ER-associated transcription factor, is a well-characterized endogenous substrate of NGLY1 and is constitutively degraded through the ERAD pathway (11, 13). Therefore, we examined the interaction between FBS2 and NRF1 by Fluoppi assays (28). Fluoppi is a technique for visualizing protein–protein interactions in mammalian cells as fluorescent foci; in this method, the interacting proteins of interest are fused with either tetrameric fluorescent protein (humanized Azami Green; hAG) or an oligomeric assembly helper tag (Ash-tag). We constructed expression plasmids encoding C-terminally Ash-tagged NRF1 and hAG-tagged FBS2 derivatives. FBS2 YW/AA is a lectin activity mutant with a defect that prevents it from recognizing glycoproteins, whereas the LP/AA mutant fails to form the SCF ubiquitin ligase complex (Fig. 2A and SI Appendix, Fig. S2A). Topologically, the C-terminal Ash-tag on NRF1 is in the ER lumen and is therefore separated from cytosolic FBS2 by the ER membrane (Fig. 2B). Accordingly, only a few wild-type HeLa cells expressing hAG-FBS2 and NRF1-Ash formed Fluoppi-driven foci (Fig. 2C). However, in NGLY1-KO HeLa cells, hAG-FBS2 with glycoprotein-binding activity effectively formed foci with NRF1-Ash, indicating that FBS2 recognizes NRF1 through N-glycans in the cytosol.
Fig. 2.
Ubiquitination of NRF1 by SCFFBS2 suppresses its activation in response to a proteasome inhibitor in NGLY1-KO cells. (A) Schematic representation of FBS2 and its mutants used in this study. Glycoprotein-binding and SCF complex formation abilities (SI Appendix, Fig. S2A) are also summarized. FB: F-box domain, SBD: sugar-binding domain. (B) Topology of NRF1-Ash and hAG-FBS2, used in the Fluoppi assay. (C) Fluoppi assay demonstrating an interaction between FBS2 and NRF1 in NGLY1-KO HeLa cells. (Upper) Wild-type (WT) or NGLY1-KO HeLa cells were transiently transfected in combination with hAG-FBS2 or its mutants and NRF1-Ash. Foci of hAG-FBS2 or its mutants were observed by confocal microscopy. (Scale bars, 20 μm.) (Lower) Quantification of cells with hAG-FBS2 foci. Percentages of cells with more than three foci are shown. Error bars show means ± SD of three biological replicates. Over 120 cells were counted in each of three replicate dishes. (D and E) Ubiquitination of NRF1 by SCFFBS2 in NGLY1-KO cells (D: HeLa cells; E: HCT116 cells). Cells stably expressing FLAG-FBS2, FLAG-FBS2 YW/AA mutant (equivalent to the no-expression control shown in SI Appendix, Fig. S2C), or NGLY1-HA were treated with or without 20 nM bortezomib (Btz) for 5 h before harvesting. Cell lysates (15 μg each) were analyzed by immunoblotting. Fl, unprocessed full-length NRF1; P, processed NRF1; AP, abnormally processed NRF1; and vertical lines (Ub), ubiquitinated NRF1. (F) Nuclear translocation of NRF1-HA in response to bortezomib (Btz) treatment in WT and NGLY1-KO cells overexpressing FLAG-FBS2 or mutants. Quantification of cells with nuclear translocation of NRF1 is based on the image in SI Appendix, Fig. S2D, with three biological replicates. Positive nuclear translocation of NRF1 was confirmed by detection of prominent unstained nucleoli in nuclei immunostained with anti-HA antibody. Over 110 cells were counted in each of three replicate dishes. (G) Subcellular distribution of endogenous NRF1 in WT and NGLY1-KO cells overexpressing FLAG-FBS2 or its lectin mutant (YW). Fl, unprocessed full-length NRF1; P, processed NRF1; AP, abnormally processed NRF1; vertical line (Ub), ubiquitinated NRF1; cyto, cytosol fraction; mem, membrane fraction; nuc, nuclear soluble fraction; and chr, chromatin-bound fraction. LDH, lactate dehydrogenase (cytosol marker); CNX, calnexin (ER marker); SP1, soluble nuclear protein marker; and Histone H3 (chromatin fraction marker). Statistical significance was determined using one-way ANOVA with Tukey’s posttest (E and F). ****P < 0.0001, **P < 0.002, n.s., not significant.
When the proteasome is inhibited, retrotranslocated NRF1 processed by DDI2 is translocated into the nucleus where it induces the transcription of proteasome subunits (5, 6, 10, 11). Hence, we next examined the effect of FBS2 on the processing and ubiquitination of NRF1 in the presence of a proteasome inhibitor. In wild-type HeLa and HCT116 cells, processed NRF1 was detected in the presence of the proteasome inhibitor Btz, as previously reported (6), and FBS2 overexpression had little effect on its processing and ubiquitination (Fig. 2 D and E). Importantly, we failed to generate NGLY1-KO HCT116 cells; this result is consistent with the results of genome-scale CRISPR guide RNA (gRNA) library screens showing that NGLY1 is a fitness-related gene in HCT116 cells (29). Although endogenous FBS2 expression was not detected in HeLa cells (30), it was detected in HCT116 cells (SI Appendix, Fig. S2B), and we were able to successfully establish NGLY1;FBS2 dKO in this cell line by first deleting FBS2, suggesting that the FBS2 expression is harmful in NGLY1-KO HCT116 cells. Although the processing of NRF1 in the presence of Btz was observed in both NGLY1-KO HeLa and HCT116 cells, with or without overexpression of the FBS2 YW/AA mutant, the processing pattern for NRF1 was slightly different (Fig. 2 D and E, compare lanes 4 and 10 NRF1 panels; SI Appendix, Fig. S2C; see the extent of FBS2 and NGLY1 overexpression in SI Appendix, Fig. S2D). The overexpression of NGLY1 in NGLY1-KO cells restored the normal processing of NRF1, similar to that observed in Btz-treated wild-type cells (Fig. 2 D and E, lane 12), confirming that the abnormal processing of NRF1 was due to a lack of NGLY1. Notably, in NGLY1-KO cells, FBS2 not only induced NRF1 ubiquitination but also inhibited processing of NRF1, even in the presence of Btz (Fig. 2 D and E, lane 8), suggesting that NRF1 activation by DDI2 and translocation to the nucleus is suppressed by FBS2.
Ubiquitination of NRF1 by SCFFBS2 Inhibits Its Nuclear Transport in NGLY1-KO Cells.
We next analyzed the subcellular localization of NRF1 by immunofluorescence microscopy using wild-type and NGLY1-KO HeLa cells stably expressing NRF1-HA (Fig. 2F and SI Appendix, Fig. S2E). The level of NRF1 was low in wild-type cells at a steady state, but NRF1 accumulated in the nucleus in the presence of a proteasome inhibitor, as previously reported (13). In NGLY1-KO cells overexpressing inactive FBS2 mutants (YW/AA or LP/AA) treated with Btz, the NRF1 that escaped from proteasomal degradation was detected in both the nucleus and the cytoplasm (cytosol and membrane fraction) (SI Appendix, Fig. S2E). On the other hand, FBS2 overexpression induced K48-linked ubiquitination of NRF1 and strongly inhibited the nuclear localization of NRF1 in NGLY1-KO cells (Fig. 2F and SI Appendix, Fig. S2F), suggesting that NRF1 ubiquitination by SCFFBS2 inhibits its nuclear translocation.
We next examined the subcellular distribution of endogenous NRF1 (Fig. 2G). In wild-type cells without Btz treatment, full-length NRF1 was detected only in the membrane fraction, which contains the ER membrane (Fig. 2G, lanes 5 and 7). In the presence of Btz, processed NRF1 predominantly accumulated in the nuclear fraction but was detected in both the cytoplasm and the chromatin-bound fraction (Fig. 2G lanes 2, 4, 6, 8, 10, 12, 14, and 16). In NGLY1-KO cells overexpressing the YW/AA mutant treated with Btz, the abnormally processed NRF1 accumulated in the nuclear fraction (Fig. 2G, lane 28). The level of processed NRF1 in the nuclear fraction in the wild-type and NGLY1-KO cells was essentially the same (SI Appendix, Fig. S2G, lanes 12 and 16), suggesting that NGLY1 activity has no significant effect on the nuclear transport of NRF1. Importantly, FBS2 overexpression induced the accumulation of ubiquitinated NRF1 in the cytoplasm regardless of whether cells were treated with a proteasome inhibitor (Fig. 2G, lanes 17, 18, 21, and 22); however, processed NRF1 was present only at very low levels in the nucleus (Fig. 2G, lanes 25 and 26). Because most of the slow-migrating NRF1 appeared to be membrane-associated in FBS2-overexpressing NGLY1-KO cells, it is unlikely that NRF1 is processed by DDI2. These results indicate that the ubiquitination by FBS2 critically impairs the processing and nuclear translocation of NRF1.
FBS2 Overexpression Impairs Proteasome Activity and Causes Cytotoxicity in NGLY1-KO Cells.
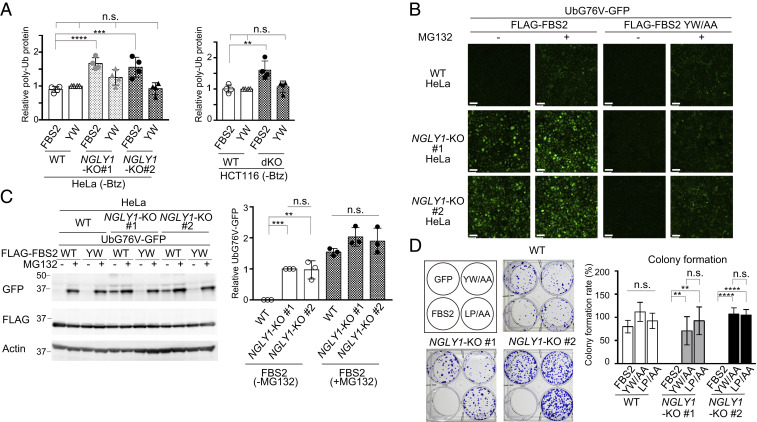
We noticed that in addition to ubiquitinated NRF1, overexpression of FBS2 increased the levels of ubiquitinated proteins in cell lysates from both NGLY1-KO HeLa and HCT116 cells, even without Btz treatment (Figs. 2 D and E, lanes 7 and 8 and 3A; SI Appendix, Fig. S3 A and B). This result led us to hypothesize that FBS2 may impair proteasome function in NGLY1-KO cells, thereby inducing cytotoxicity. To validate this hypothesis, we examined the proteasome activity in NGLY1-KO HeLa and HCT116 cells using UbG76V-GFP, a proteasome reporter that undergoes rapid proteasome-mediated degradation (31). In wild-type cells, green fluorescence was observed only in the presence of the proteasome inhibitor MG132 (Fig. 3B and SI Appendix, Fig. S3C). Remarkably, however, FBS2 overexpression induced a strong GFP signal in NGLY1-KO cells even in the absence of MG132. The accumulation of UbG76V-GFP by FBS2 in NGLY1-KO cells was confirmed by immunoblotting (Fig. 3C and SI Appendix, Fig. S3D). A subset of well-known, short-lived, nonglycosylated protein substrates of the proteasome, c-Jun, p53, and p27, also accumulated in NGLY1-KO cells upon FBS2 overexpression (SI Appendix, Fig. S3 E and F). Collectively, these results indicate that the accumulation of glycoproteins ubiquitinated by SCFFBS2 results in inhibition of proteasome activity specifically in NGLY1-KO cells.
Fig. 3.
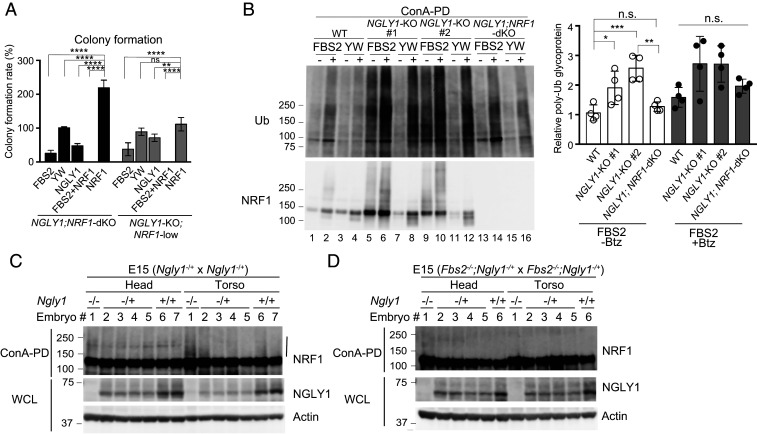
FBS2 overexpression induces proteasomal dysfunction in NGLY1-KO cells. (A) Accumulation of ubiquitinated proteins in NGLY1-KO HeLa (#1, exon 6 mutant; #2, exon 11 mutant) and HCT116 (FBS2;NGLY1-dKO) cells by FBS2 overexpression. Cells were treated with or without 20 nM bortezomib (Btz) for 5 h prior to harvest. The relative levels of ubiquitinated proteins in cell lysates in SI Appendix, Fig. S3 A and B were quantified. The intensity of the lane with wild-type (WT) cells overexpressing the YW/AA mutant was defined as 1. Error bars show means ± SD of four biological replicates. (B) Low-magnification fluorescence micrographs of WT and NGLY1-KO HeLa cells coexpressing UbG76V-GFP and FLAG-FBS2 (WT or YW/AA mutant) treated with or without 50 μM MG132. (Scale bars, 100 µm.) (C) Detection of UbG76V-GFP accumulation by immunoblotting. Accumulation of UbG76V-GFP in FBS2-overexpressing NGLY1-KO cells. (Left) Cell lysates in (B) were analyzed with anti-GFP antibody. (Right) Relative levels of UbG76V-GFP in FBS2-overexpressing WT and NGLY1-KO cells were quantified; the intensity of the band of FBS2-overexpressing NGLY1-KO #1 cells without MG132 treatment was defined as 1. Error bars show means ± SD of three biological replicates. (D) Colony formation assays in WT and NGLY1-KO HeLa cells. (Left) Colony formation assay in WT and NGLY1-KO HeLa cells (#1, exon 6 mutant; #2, exon 11 mutant) overexpressing the indicated proteins. Recombinant retrovirus for the expression of the indicated proteins, produced by pMXs-puro, was used to infect WT and NGLY1-KO HeLa cells. After infection for 36 h, 500 cells were plated onto 6-well dishes in the presence of 1 μg/mL puromycin. After culture for 11 to 14 d, the colonies were stained with crystal violet and counted. (Right) Colony formation rates were calculated relative to the number of colonies formed by GFP-overexpressing cells. Cells were counted in each of four replicates. Statistical significance was determined by one-way ANOVA with Dunnett’s posttest (A) or one-way ANOVA with Tukey’s posttest (C and D). ****P < 0.0001; ***P < 0.0002; **P < 0.002; n.s., not significant.
It is well known that the disruption of proteasome activity induces an apoptotic cascade that leads to growth arrest and eventually to cell death (32, 33). To examine the effect of FBS2 on cell viability in NGLY1-KO cells, we performed colony formation assays (34). For these experiments, we infected wild-type and NGLY1-KO HeLa cells with a retrovirus generated using a pMXs-puro vector encoding wild-type or mutant FBS2. Equal numbers of infected cells were seeded and cultured in the presence of puromycin. In wild-type cells, wild-type or mutant FBS2 had little effect on colony formation (Fig. 3D). On the other hand, in NGLY1-KO cells, colony-forming efficiency was unaffected by both the YW/AA and LP/AA mutants, whereas FBS2-overexpressing cells formed no colonies. These results clearly indicate that prolonged proteasome inhibition caused by the accumulation of glycoproteins ubiquitinated by SCFFBS2 leads to cell death in NGLY1-KO cells.
“Glycan-Less” NRF1 Mutants Restore Cell Growth of FBS2-Overexpressing NGLY1-KO Cells.
NGLY1 not only removes N-glycans from glycoproteins but also converts the glycosylated asparagine (N) into aspartic acid (D); this amino acid “editing” of SKN-1A, the functional homolog of NRF1 in C. elegans, is critical for the activation of this transcription factor (12). Hence, we constructed a “sequence-edited” NRF1 8N/D mutant in which all eight of the putative N-glycosylation sites in the Asn/Ser/Thr-rich (NST) region were replaced with Asp (35) (SI Appendix, Fig. S4A). The NRF1 8N/D mutant itself was normally processed by DDI2 and migrated to nuclei in NGLY1-KO cells treated with Btz. Unexpectedly, however, FBS2, which was previously thought to act only on N-glycoproteins, partially suppressed its processing and nuclear transport (SI Appendix, Fig. S4B, lanes 6, 7, 12, and 13; SI Appendix, Fig. S4C). We identified another putative N-glycosylation site (N574) outside the NST region and mutated it; the NRF1 9N/D mutant was processed normally and redistributed in NGLY1-KO cells after treatment with Btz, regardless of FBS2 overexpression (SI Appendix, Fig. S4 A and B, lanes 8, 9, 14, and 15; SI Appendix, Fig. S4D), implying that 9N/D but not 8N/D represents the “sequence-edited” form of human NRF1 by NGLY1.
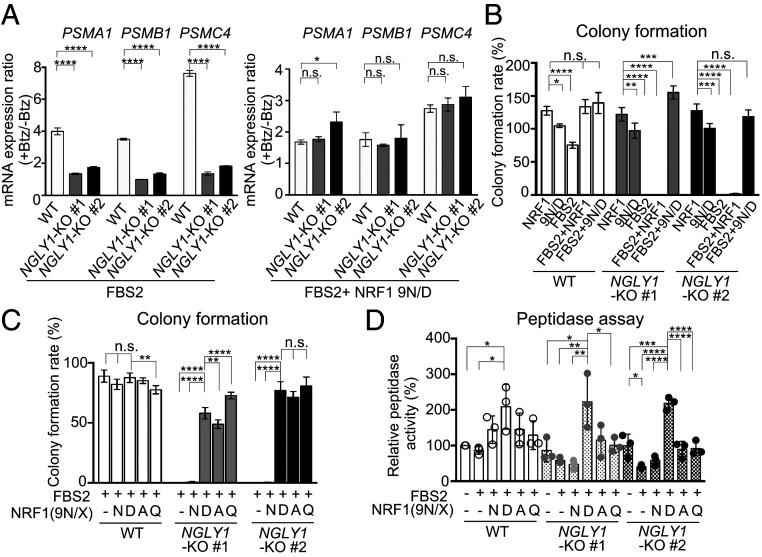
We then investigated whether the sequence-edited NRF1 9N/D enhanced the “bounce-back” response after proteasome inhibition. In wild-type cells, Btz treatment increased the levels of messenger RNAs (mRNAs) encoding the proteasome subunits PSMA1, PSMB1, and PSMC4 (Fig. 4A, Left). As previously reported, this response was weaker in NGLY1-KO cells (13), but exogenous 9N/D increased the expression of proteasome subunits following Btz treatment (SI Appendix, Fig. S5A). The response in FBS2-overexpressing NGLY1-KO cells was restored to the wild-type level by 9N/D coexpression (Fig. 4A, Right and SI Appendix, Fig. S5B). We then investigated whether 9N/D could rescue FBS2-induced cell death in NGLY1-KO cells. The results clearly indicated that coexpression of 9N/D but not wild-type NRF1 restored colony formation (Fig. 4B and SI Appendix, Fig. S5C), suggesting that proteasomes that were newly synthesized in response to NRF1 9N/D could rescue lethality by FBS2 in NGLY1-KO cells.
Fig. 4.
“Glycan-less” NRF1 mutants restore cell growth of FBS2-overexpressing NGLY1-KO cells. (A) Restoration of the bounce-back response of NGLY1-KO HeLa cells by sequence-edited NRF1 9N/D in the presence of proteasome inhibitor. Wild-type (WT) and NGLY1-KO HeLa cells overexpressing FBS2 (Left) or FBS2 and NRF1 9N/D (Right) were treated with Btz for 16 h. mRNA levels of the proteasome genes PSMA1, PSMB1, and PSMC4 in WT and NGLY1-KO HeLa cells were then quantified by qPCR. mRNA levels of target genes were normalized against the corresponding levels of GAPDH mRNA. Error bars show means ± SD of three biological replicates (SI Appendix, Fig. S5B). The ratio of relative mRNA expression of cells between Btz-treated and untreated are shown. (B and C) Colony formation assays in WT and NGLY1-KO HeLa cells overexpressing the indicated proteins. Colony formation rates were calculated relative to the number of colonies in GFP-overexpressing cells. Cells were counted in each of four replicates. (D) Chymotryptic peptidase activity of the proteasome in lysates of WT and NGLY1-KO HeLa cells or cells overexpressing FBS2 with or without NRF1 or NRF1 mutants. Error bars show means ± SD of three biological replicates. Statistical significance was determined by one-way ANOVA with Dunnett’s posttest (A and B) or one-way ANOVA with Tukey’s posttest (C and D). ****P < 0.0001; ***P < 0.0002; **P < 0.002; *P < 0.03, n.s., not significant.
We next investigated whether N-to-D conversion of NRF1 is required for alleviation of the cytotoxicity of FBS2 in NGLY1-KO cells. To this end, we constructed additional “glycan-less” NRF1 mutants, 9N/A and 9N/Q, in which all of the N-glycosylation sites were replaced with alanine and glutamine, respectively. Surprisingly, coexpression of these inactive NRF1 mutants, as well as the edited 9N/D mutant, restored colony formation in FBS2-overexpressing NGLY1-KO cells (Fig. 4C and SI Appendix, Fig. S5D). Unlike 9N/D, 9N/A and 9N/Q failed to promote the expression of proteasome subunits in NGLY1-KO cells in the presence of Btz (SI Appendix, Fig. S5E). Furthermore, 9N/D overexpression significantly increased the chymotryptic peptidase activity of the proteasomes both in wild-type and NGLY1-KO cells coexpressing FBS2, but neither 9N/A nor 9N/Q affected peptidase activity in NGLY1-KO cells (Fig. 4D). These results suggest that the recovery from the detrimental effects of FBS2 on NGLY1-KO cells does not necessarily depend on the expression of newly synthesized proteasomes induced by 9N/D. Rather, ubiquitination of N-glycosylated NRF1 by SCFFBS2 underlies the cytotoxicity in NGLY1-KO cells.
The SCFFBS2-Mediated Ubiquitination of NRF1 Is Primarily Responsible for the Induction of Cytotoxicity in NGLY1-KO Cells.
Because NRF1 is constitutively targeted to the ERAD pathway at steady state, NRF1 itself may burden proteasomes with an increased load as a substrate when it is highly ubiquitinated by SCFFBS2. Hence, we investigated whether the deletion of NRF1 could ameliorate the cytotoxic effect of FBS2 in NGLY1-KO cells. To this end, we established NGLY1;NRF1-dKO HeLa cells. The proliferation of NGLY1;NRF1-dKO was more efficient than NRF1-KO cells, although the colony formation of dKO cells was also dependent on the exogenous NRF1 (SI Appendix, Fig. S6A). On the other hand, the growth of NGLY1-KO;NRF1-low (generated during the process of establishing NGLY1;NRF1-dKO cells; SI Appendix, Materials and Methods for details), in which a low level of NRF1 expression was observed, was normal and not dependent on the exogenous NRF1 (Fig. 5A and SI Appendix, Fig. S6 A and B), suggesting that NRF1 expression is critical for efficient proliferation of HeLa cells, especially in the presence of NGLY1. Surprisingly, colony formation by both NGLY1-KO;NRF1-low and NGLY1;NRF1-dKO cells was obvious even when they overexpressed FBS2 (Fig. 5A and SI Appendix, Fig. S6A). By contrast, FBS2 was detrimental to both types of cells when NRF1 was restored, indicating that the accumulation of ubiquitinated NRF1 by SCFFBS2 is primarily responsible for the cytotoxicity in NGLY1-KO cells. Although the recovery of proliferation by coexpression of glycan-less NRF1 in FBS2-overexpressing NGLY1-KO cells was complete (Fig. 4C), the recovery effect by the deletion of NRF1 was partial. These results suggest that glycan-less NRF1, even 9N/A or 9N/Q mutants, may activate NRF1 targets other than proteasome subunits, such as genes related to mitophagy or oxidative stress response (36), to promote survival of FBS2-overexpressing NGLY1-KO cells.
Fig. 5.
NRF1 is primarily responsible for the induction of cytotoxicity in NGLY1-KO cells. (A) Colony formation assays in NGLY1;NRF1-dKO and NGLY1-KO;NRF1-low HeLa cells overexpressing the indicated proteins. Colony formation rates were calculated relative to the number of colonies in GFP-overexpressing cells. Cells were counted in each of four replicates. (B) Accumulation of ubiquitinated glycoproteins in wild-type (WT), NGLY1-KO, and NGLY1;NRF1-dKO HeLa cells overexpressing FBS2. (Left) Ubiquitination of glycoproteins concentrated with concanavalin A (ConA)–conjugated agarose were analyzed by immunoblotting with anti-ubiquitin (Ub) antibody. (Right) Ubiquitinated glycoprotein levels (average intensities between 100 and 250 kDa) were normalized against the level of glycoprotein ubiquitination in FBS2 YW/AA–overexpressing WT cells. Error bars show means ± SD from four biological replicates. (C and D) Ubiquitination of NRF1 in Ngly1-KO (C) or Fbs2;Ngly1 dKO (D) day 15 embryos. ConA-bound glycoproteins from whole-cell lysates (WCL) were subjected to immunoblotting with anti-NRF1 antibody. Vertical line: ubiquitinated NRF1. Statistical significance was determined by one-way ANOVA with Dunnett’s posttest (A) or one-way ANOVA with Tukey’s posttest (B). ****P < 0.0001; ***P < 0.0002; **P < 0.002; *P < 0.03, n.s., not significant.
We next compared the ubiquitination level of concanavalin A–enriched glycoproteins among wild-type, NGLY1-KO, and NGLY1;NRF1-dKO cells (Fig. 5B). In FBS2 YW/AA mutant–overexpressing control cells, ubiquitinated glycoproteins were barely detectable in the absence of Btz. On the other hand, the level of ubiquitinated glycoproteins was significantly higher in NGLY1-KO cells when FBS2 was overexpressed, regardless of the presence of Btz (Fig. 5B Left, lanes 5 and 9). However, accumulated ubiquitinated glycoproteins were barely detectable in the absence of Btz in FBS2-overexpressing NGLY1;NRF1-dKO cells, as well as in wild-type cells (Fig. 5B Left, lanes 1 and 13), implying that NRF1 is the major substrate of SCFFBS2 in NGLY1-KO HeLa cells, presumably due to congestion of the ERAD pathway.
Finally, we investigated whether ubiquitinated NRF1 could be detected in Ngly1-KO mice embryos as well as NGLY1-KO cells. In the C57BL/6 background, Ngly1-deficient mice are embryonically lethal between E16.5 and birth (19). As shown in Fig. 5C and SI Appendix, Fig. S6C, the level of ubiquitinated NRF1 was specifically elevated in the torsos of Ngly1-KO day 14 or day 15 embryos. Notably, relative to Ngly1-KO embryos, ubiquitination of NRF1 was nearly completely eliminated in Fbs2−/−;Ngly1−/− mouse embryos (Fig. 5D), indicating that NRF1 ubiquitinated by SCFFbs2 accumulated in Ngly1-KO embryos. These results suggest that the accumulation of glycosylated NRF1 ubiquitinated by SCFFbs2 is critical for cytotoxicity in Ngly1-KO embryos.
Discussion
The findings presented herein show that the deletion of Fbs2 suppressed the embryonic lethality of Ngly1-KO mice. Fbs2;Ngly1 dKO mice exhibited no overt defects, even in old age, indicating that Fbs2 also induces the formation of postnatal detrimental phenotypes in Ngly1-KO mice (Fig. 1). Moreover, FBS2 overexpression resulted in growth arrest and cell death specifically in NGLY1-KO cells (Fig. 3D). Consistent with our observations, FBS2 expression is drastically down-regulated in lymphoblastoid cells derived from most patients with NGLY1 deficiency (“NGLY1 browser”; https://ngly.shiny.embl.de/). These results further support our conclusion that FBS2 is deleterious to NGLY1-KO cells and animals.
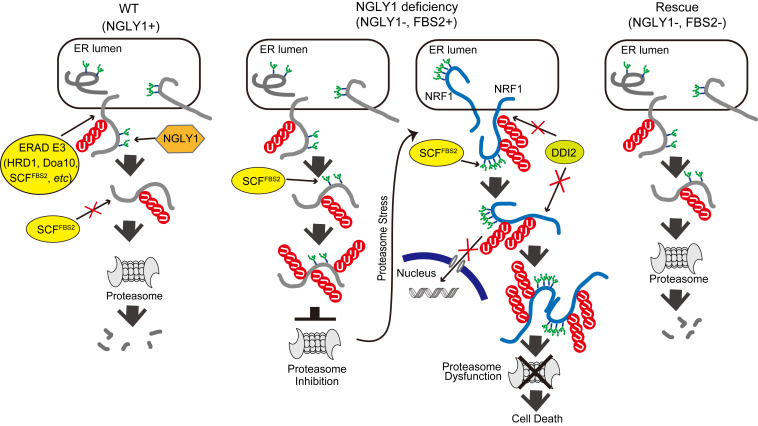
Collectively, our results are consistent with the following scenarios: in wild-type cells, if ERAD substrate glycoproteins are deglycosylated by NGLY1, they will no longer be recognized by FBS2 and degraded by proteasomes (Fig. 6A, Left). In the absence of NGLY1, however, glycoproteins will be constitutively ubiquitinated by SCFFBS2, which overloads proteasomes, thereby impairing them (Fig. 6A, Middle). Under conditions of proteasome inhibition or stress, N-glycosylated NRF1 ubiquitinated by SCFFBS2, which cannot be processed by DDI2 and is therefore retained in the cytosol, induces proteasome dysfunction in NGLY1-KO cells. Consistent with this view, the expression of “glycan-less” NRF1 or the additional deletion of NRF1 rescued FBS2-induced cell death in NGLY1-KO cells. In the absence of both NGLY1 and FBS2, most ERAD substrates are degraded normally, and abnormally processed NRF1 moves to the nucleus without accumulating in the cytosol; this may be because excessive ubiquitination of glycoproteins by SCFFBS2 is prevented, thereby suppressing the effects of NGLY1 deficiency (Fig. 6A, Right).
Fig. 6.
Schematic model of the molecular mechanism of induction of cell death by FBS2 in NGLY1-KO cells. (Left) In wild-type (WT) cells expressing NGLY1 with or without FBS2, glycoprotein ERAD substrates are ubiquitinated by multiple ERAD-E3s, such as HRD1, Doa10, or SCFFBS2, upon retrotranslocation to the cytosol. Most ubiquitinated glycoproteins are deglycosylated by NGLY1 prior to proteasomal degradation. (Middle) In NGLY1-KO cells expressing FBS2 (NGLY1 deficiency), glycoproteins that accumulate are ubiquitinated by SCFFBS2. Glycoproteins are constitutively ubiquitinated by SCFFBS2, which overloads the proteasome, thereby impairing it. The cells then sense proteasome stress, and transcriptional activation of proteasome subunits is induced by active NRF1. FBS2 expression causes inactivation of NRF1, which in turn suppresses NRF1-triggered expression of proteasome subunits. As with the ubiquitinated glycoprotein substrates, accumulation of NRF1 itself ubiquitinated by SCFFBS2 causes proteasome and ultimately induces cell death. (Right) In cells lacking both NGLY1 and FBS2, most ERAD substrates are degraded normally, and abnormally processed NRF1 moves to the nucleus without accumulating in the cytosol.
NGLY1 activity is important for the activation of NRF1 by sequence editing, which is critical for transcriptional activation of proteasome subunits upon inhibition of proteasome activity (12). However, the sequence-edited NRF1 9N/D mutant induced excessive expression of proteasome genes, even in the absence of a proteasome inhibitor (SI Appendix, Fig. S5 A, B, and E), suggesting that the deglycosylation of NRF1 fine-tunes NRF1 activity. Conversely, when NGLY1 activity is lost, NRF1, which is retained in the cytosol due to aberrant ubiquitination by SCFFBS2, causes proteasome dysfunction. Thus, NGLY1 regulates proteasome function in both NRF1’s transcriptional activity–dependent and –independent manners. We do not yet understand how exactly the abnormally ubiquitinated NRF1 impairs proteasome function. Although overexpression of FBS2 or FBS2/NRF1 decreased the in vitro chymotryptic activities of the proteasome in NGLY1-KO cells (Fig. 4D), proteasomes were not recovered from the detergent-insoluble fraction, and proteasome aggregates were not detected in NGLY1-KO cells expressing GFP-tagged proteasome subunits and FBS2. In addition, we detected no interaction between the proteasomes and NRF1 in immunoprecipitation analyses using cross-linked lysates of FBS2-overexpressing NGLY1-KO cells. Future studies should explore the precise mechanisms underlying the proteasomal dysfunction caused by abnormally ubiquitinated NRF1.
NGLY1 regulates cytosolic glycoproteostasis and is important for embryogenesis and survival in mice. About 30% of Ngly1−/− embryos were inviable at E17.5 to 18.5, and Ngly1−/− embryos at E16.5 developed a ventricular septal defect (VSD) and anemia (19). The expression of Fbs2 was up-regulated in both the heart and liver at later stage of embryonic development (SI Appendix, Fig. S1C), and even heterozygous Fbs2 mutation could attenuate the lethality of Ngly1−/− mice (Fig. 1D), suggesting that a high level of Fbs2 induces the VSD that causes lethality in Ngly1−/− mice. Although NRF1 is a crucial target for both NGLY1 and FBS2, it is possible that other ERAD substrate glycoproteins are also ubiquitinated by SCFFBS2 in the absence of NGLY1. These ubiquitinated glycoprotein substrates may not always impair proteasome function, and it remains unclear at this time whether cell type–specific proteotoxic substrates exist. NGLY1 also regulates NRF1-mediated mitochondrial function (37) and inflammation (36). Tissue-specific bone morphogenetic protein signaling in Drosophila and aquaporin transcription through Creb1 are also regulated by NGLY1 (38–40). The identification of ubiquitinated glycoproteins other than NRF1 in Ngly1-KO mouse/rat embryos would help reveal the precise cause of their lethality and defects. It is possible that the accumulation of specific NGLY1-dependent substrates is catastrophic for embryogenesis in Ngly1-KO mice. A wide spectrum of symptoms has been reported in patients with NGLY1 deficiency; however, the genotype–phenotype correlation was not so obvious (15, 16, 41). It will be necessary to determine whether there are single nucleotide polymorphsms (SNPs) in the FBS2 gene or differences in FBS2 expression levels in patients with NGLY1 deficiency to clarify whether FBS2 expression can indeed influence the severity of the symptoms of these patients.
Although there are several viable animal models for Ngly1-KO mice and rats (19, 20), Fbs2;Ngly1 dKO mice are by far the healthiest, indicating that Fbs2 contributes to most if not all of the phenotypic consequences of Ngly1-KO mice. Notably, the embryonic lethality of Ngly1-KO was also partially suppressed by heterozygous KO of Fbs2, and no dramatic defect in motor functions was observed in these animals. Furthermore, Fbs2-KO mice were completely normal, further indicating that development of an FBS2 inhibitor represents a promising strategy for treating NGLY1 deficiency.
Materials and Methods
Generation of Fbs2-KO Mice.
Genomic DNA containing the mouse MAD2L2-Fbs1 gene (SI Appendix, Fig. S1A) was isolated from a BAC (ATCC clone RP23-139J21) and a targeting vector was constructed by replacing a part of exon 1 and exons 2 to 4 of the Fbs2 gene with a loxP site and MC1-neo resistance gene. Another loxP site was also inserted into 3′ of the Fbs1 gene. The vector was transfected into ES cells (TT2) (42), and G418-resistant colonies were selected. A homologous recombinant ES clone containing the appropriately targeted allele was microinjected into eight-cell embryos of Crl:CD1 (ICR) mice. The resultant chimeric mice were backcrossed with C57BL/6 mice more than 12 times. Genotyping conditions are described in SI Appendix.
Phenotypic Analyses of Fbs2−/−; Ngly1−/− Mice.
Detailed methods about hindlimb clasping tests, Rotarod test, and histological analysis are described in SI Appendix.
Establishment of NGLY1-KO, FBS2;NGLY1-dKO, NRF1-KO, NGLY1;NRF1-dKO, and NGLY1-dKO;NRF1-low Cell Lines.
To generate KO cell lines, CRISPR target sites were designed using CHOPCHOP (https://chopchop.cbu.uib.no/) and cloned into a pX330-U6-Chimeric_BB-Cbh-hSpCas9 (Addgene plasmid #42230). Detailed methods for generation of KO cell lines are described in SI Appendix.
Transfections and Generation of Stable Cell Lines.
Detailed methods for plasmid construction are described in SI Appendix. For transient transfection of plasmids in Fig. 2C and SI Appendix, Fig. S2 A and B, Lipofectamine 2000 (Thermo Fisher Scientific) was used for HeLa cells, and FuGENE6 transfection reagent (Promega) was used for HCT116 cells. Stable cell lines were established by recombinant retrovirus infection as follows. Virus particles were produced in human embryonic kidney (HEK)293T cells by cotransfection with a retrovirus plasmid (expressing in pMXs-puro vector), Gag-Pol (Addgene plasmid #14887), and VSV-G (Addgene plasmid #8454) by Lipofectamine LTX with Plus reagent (Thermo Fisher). Detailed methods for generation of stable cell lines are described in SI Appendix.
Subcellular Protein Fractionation.
HeLa cells cultured for 4 d after infection with retrovirus expressing FLAG-FBS2 or FLAG- FBS2YW/AA were treated with or without 20 nM Btz for 5 h, harvested with trypsin-EDTA, and counted. Per sample, 1 × 106 cells were analyzed using the Subcellular Protein Fractionation Kit for Cultured Cells (Thermo Fisher Scientific). Aliquots of each fraction totaling 2 μl were used for immunoblotting.
Colony-Forming Unit Assay.
A total 20 h after infection with recombinant retrovirus generated by pMXs-puro vector transfection, cells were cultured in fresh medium for 16 h. Cells were harvested by trypsinization, counted, and plated in 6-well dishes at the same number of cells/well in the presence of 1 μg/mL puromycin. After culture for 11 to 14 d, cells were rinsed with phosphate-buffered saline (PBS) and stained with a mixture of 6% glutaraldehyde and 0.5% crystal violet for 30 min. The 6-well dishes were rinsed with water and dried, and colonies were counted.
qRT-PCR.
Cells were harvested 16 h after 20 nM bortezomib treatment. Total RNA was isolated using TRIzol Reagent (Thermo Fisher Scientific) and reverse-transcribed using the Transcriptor First Strand complementary DNA (cDNA) Synthesis Kit (Roche). qRT-PCR was performed with LightCycler 480 SYBR Green I Master (Nippon Genetics) on a LightCycler 480 (Roche). Primer sequences are provided in SI Appendix.
Proteasome Peptidase Activity Measurement.
Peptidase activity of the 26S proteasome in cell lysates was measured as described previously (43). Further details of the analysis are described in SI Appendix.
Statistical Analysis.
EXSUS version 8.0 (CAC Croit Corporation) was used for statistical analysis of the mouse experiments. Prism 7 (GraphPad Software) was used for statistical analysis of experiments using cell lines. Statistical tests are indicated in the corresponding figure legends. Numerical data are shown as means ± SD of the indicated biological replicates. P < 0.05 was considered statistically significant.
Additional information is provided in SI Appendix.
Supplementary Material
Acknowledgments
We thank Dr. Hamed Jafar-Nejad (Baylor College of Medicine) and Dr. Minoru Yoshida (RIKEN) for critical reading of the manuscript. We also thank Dr. Koji Yamano, Dr. Hikaru Tsuchiya, Dr. Yasushi Saeki, Dr. Haruhiko Fujihira, and Dr. Stuart Emmerson for discussion. This research was supported by Ministry of Education, Culture, Sports, Science, and Technology (MEXT)/Japan Society for the Promotion of Science (JSPS) KAKENHI Grants JP19H02926 (Y.Y.), JP18H02443 (N.M.), JP19H05712 (N.M.), JP19H00997 (K.T.), and JP18H03990 (T.S.); the RIKEN Pioneering Research Project “Glyco-lipidologue Initiative” (T.S.); Japan Science and Technology Agency-Core Research for Evolutional Science and Technology (JST-CREST) Grant JPMJCR16H3 (Y.Y.); Japan Agency for Medical Research and Development-Core Research for Evolutional Science and Technology (AMED-CREST) Grant JP20gm1410003 (T.S. and Y.Y.); and the Takeda Science Foundation (N.M. and K.T.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2102902118/-/DCSupplemental.
Data Availability.
All study data are included in the article and/or supporting information.
References
- 1.Collins G. A., Goldberg A. L., The logic of the 26S proteasome. Cell 169, 792–806 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hipp M. S., Kasturi P., Hartl F. U., The proteostasis network and its decline in ageing. Nat. Rev. Mol. Cell Biol. 20, 421–435 (2019). [DOI] [PubMed] [Google Scholar]
- 3.Pilla E., Schneider K., Bertolotti A., Coping with protein quality control failure. Annu. Rev. Cell Dev. Biol. 33, 439–465 (2017). [DOI] [PubMed] [Google Scholar]
- 4.Dantuma N. P., Bott L. C., The ubiquitin-proteasome system in neurodegenerative diseases: Precipitating factor, yet part of the solution. Front. Mol. Neurosci. 7, 70 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Steffen J., Seeger M., Koch A., Krüger E., Proteasomal degradation is transcriptionally controlled by TCF11 via an ERAD-dependent feedback loop. Mol. Cell 40, 147–158 (2010). [DOI] [PubMed] [Google Scholar]
- 6.Radhakrishnan S. K., et al., Transcription factor Nrf1 mediates the proteasome recovery pathway after proteasome inhibition in mammalian cells. Mol. Cell 38, 17–28 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim H. M., Han J. W., Chan J. Y., Nuclear factor erythroid-2 like 1 (NFE2L1): Structure, function and regulation. Gene 584, 17–25 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sommer T., Wolf D. H., Endoplasmic reticulum degradation: Reverse protein flow of no return. FASEB J. 11, 1227–1233 (1997). [DOI] [PubMed] [Google Scholar]
- 9.Vembar S. S., Brodsky J. L., One step at a time: Endoplasmic reticulum-associated degradation. Nat. Rev. Mol. Cell Biol. 9, 944–957 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koizumi S., et al., The aspartyl protease DDI2 activates Nrf1 to compensate for proteasome dysfunction. eLife 5, e18357 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lehrbach N. J., Ruvkun G., Proteasome dysfunction triggers activation of SKN-1A/Nrf1 by the aspartic protease DDI-1. eLife 5, e17721 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lehrbach N. J., Breen P. C., Ruvkun G., Protein sequence editing of SKN-1A/Nrf1 by peptide:N-glycanase controls proteasome gene expression. Cell 177, 737–750.e15 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tomlin F. M., et al., Inhibition of NGLY1 inactivates the transcription factor Nrf1 and potentiates proteasome inhibitor cytotoxicity. ACS Cent. Sci. 3, 1143–1155 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Suzuki T., Huang C., Fujihira H., The cytoplasmic peptide:N-glycanase (NGLY1)—Structure, expression and cellular functions. Gene 577, 1–7 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Caglayan A. O., et al., NGLY1 mutation causes neuromotor impairment, intellectual disability, and neuropathy. Eur. J. Med. Genet. 58, 39–43 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Enns G. M., et al., Mutations in NGLY1 cause an inherited disorder of the endoplasmic reticulum-associated degradation pathway. Genet. Med. 16, 751–758 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lam C., et al., Prospective phenotyping of NGLY1-CDDG, the first congenital disorder of deglycosylation. Genet. Med. 19, 160–168 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.He P., et al., A congenital disorder of deglycosylation: Biochemical characterization of N-glycanase 1 deficiency in patient fibroblasts. Glycobiology 25, 836–844 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fujihira H., et al., Lethality of mice bearing a knockout of the Ngly1-gene is partially rescued by the additional deletion of the Engase gene. PLoS Genet. 13, e1006696 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Asahina M., et al., Ngly1 -/- rats develop neurodegenerative phenotypes and pathological abnormalities in their peripheral and central nervous systems. Hum. Mol. Genet. 29, 1635–1647 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yoshida Y., et al., E3 ubiquitin ligase that recognizes sugar chains. Nature 418, 438–442 (2002). [DOI] [PubMed] [Google Scholar]
- 22.Yoshida Y., et al., Fbs2 is a new member of the E3 ubiquitin ligase family that recognizes sugar chains. J. Biol. Chem. 278, 43877–43884 (2003). [DOI] [PubMed] [Google Scholar]
- 23.Yoshida Y., et al., Ubiquitination of exposed glycoproteins by SCFFBXO27 directs damaged lysosomes for autophagy. Proc. Natl. Acad. Sci. U.S.A. 114, 8574–8579 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cardozo T., Pagano M., The SCF ubiquitin ligase: Insights into a molecular machine. Nat. Rev. Mol. Cell Biol. 5, 739–751 (2004). [DOI] [PubMed] [Google Scholar]
- 25.Mizushima T., et al., Structural basis of sugar-recognizing ubiquitin ligase. Nat. Struct. Mol. Biol. 11, 365–370 (2004). [DOI] [PubMed] [Google Scholar]
- 26.Mizushima T., et al., Structural basis for the selection of glycosylated substrates by SCF(Fbs1) ubiquitin ligase. Proc. Natl. Acad. Sci. U.S.A. 104, 5777–5781 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yamaguchi Y., et al., Fbs1 protects the malfolded glycoproteins from the attack of peptide:N-glycanase. Biochem. Biophys. Res. Commun. 362, 712–716 (2007). [DOI] [PubMed] [Google Scholar]
- 28.Koyano F., et al., Ubiquitin is phosphorylated by PINK1 to activate parkin. Nature 510, 162–166 (2014). [DOI] [PubMed] [Google Scholar]
- 29.Hart T., et al., High-resolution CRISPR screens reveal fitness genes and genotype-specific cancer liabilities. Cell 163, 1515–1526 (2015). [DOI] [PubMed] [Google Scholar]
- 30.Kulak N. A., Pichler G., Paron I., Nagaraj N., Mann M., Minimal, encapsulated proteomic-sample processing applied to copy-number estimation in eukaryotic cells. Nat. Methods 11, 319–324 (2014). [DOI] [PubMed] [Google Scholar]
- 31.Dantuma N. P., Lindsten K., Glas R., Jellne M., Masucci M. G., Short-lived green fluorescent proteins for quantifying ubiquitin/proteasome-dependent proteolysis in living cells. Nat. Biotechnol. 18, 538–543 (2000). [DOI] [PubMed] [Google Scholar]
- 32.Obeng E. A., et al., Proteasome inhibitors induce a terminal unfolded protein response in multiple myeloma cells. Blood 107, 4907–4916 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Suraweera A., Münch C., Hanssum A., Bertolotti A., Failure of amino acid homeostasis causes cell death following proteasome inhibition. Mol. Cell 48, 242–253 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Franken N. A., Rodermond H. M., Stap J., Haveman J., van Bree C., Clonogenic assay of cells in vitro. Nat. Protoc. 1, 2315–2319 (2006). [DOI] [PubMed] [Google Scholar]
- 35.Zhang Y., Ren Y., Li S., Hayes J. D., Transcription factor Nrf1 is topologically repartitioned across membranes to enable target gene transactivation through its acidic glucose-responsive domains. PLoS One 9, e93458 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang K., Huang R., Fujihira H., Suzuki T., Yan N., N-glycanase NGLY1 regulates mitochondrial homeostasis and inflammation through NRF1. J. Exp. Med. 215, 2600–2616 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kong J., et al., Mitochondrial function requires NGLY1. Mitochondrion 38, 6–16 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Galeone A., et al., Tissue-specific regulation of BMP signaling by Drosophila N-glycanase 1. eLife 6, e27612 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tambe M. A., Ng B. G., Freeze H. H., N-glycanase 1 transcriptionally regulates aquaporins independent of its enzymatic activity. Cell Rep. 29, 4620–4631.e4 (2019). [DOI] [PubMed] [Google Scholar]
- 40.Galeone A., et al., Regulation of BMP4/Dpp retrotranslocation and signaling by deglycosylation. eLife 9, e55596 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Heeley J., Shinawi M., Multi-systemic involvement in NGLY1-related disorder caused by two novel mutations. Am. J. Med. Genet. A. 167A, 816–820 (2015). [DOI] [PubMed] [Google Scholar]
- 42.Yagi T., et al., A novel ES cell line, TT2, with high germline-differentiating potency. Anal. Biochem. 214, 70–76 (1993). [DOI] [PubMed] [Google Scholar]
- 43.Sha Z., Goldberg A. L., Proteasome-mediated processing of Nrf1 is essential for coordinate induction of all proteasome subunits and p97. Curr. Biol. 24, 1573–1583 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information.