Significance
Disruption of circadian rhythms enhances cancer risk, but the underlying mechanisms are largely unknown. The circadian repressors CRY1 and CRY2 evolved from light-activated DNA-repair enzymes, suggesting that they may be involved. Here, we demonstrate that missense mutations in CRY2 reported in The Cancer Genome Atlas suppress P53 target-gene expression and enhance the growth of MYC-transformed fibroblasts. Our identification of point mutations in CRY2 in human tumors that influence P53 activity and cell growth provides evidence for a clinically relevant molecular connection between CRYs and P53. Furthermore, these mutants will enable new strategies to investigate underlying molecular mechanisms and inform a growing effort to identify selective chemical modulators of CRY1 and/or CRY2.
Keywords: cryptochromes, circadian clocks, tumor suppressor protein P53
Abstract
Disruption of circadian rhythms increases the risk of several types of cancer. Mammalian cryptochromes (CRY1 and CRY2) are circadian transcriptional repressors that are related to DNA-repair enzymes. While CRYs lack DNA-repair activity, they modulate the transcriptional response to DNA damage, and CRY2 can promote SKP1 cullin 1–F-box (SCF)FBXL3-mediated ubiquitination of c-MYC and other targets. Here, we characterize five mutations in CRY2 observed in human cancers in The Cancer Genome Atlas. We demonstrate that two orthologous mutations of mouse CRY2 (D325H and S510L) accelerate the growth of primary mouse fibroblasts expressing high levels of c-MYC. Neither mutant affects steady-state levels of overexpressed c-MYC, and they have divergent impacts on circadian rhythms and on the ability of CRY2 to interact with SCFFBXL3. Unexpectedly, stable expression of either CRY2 D325H or of CRY2 S510L robustly suppresses P53 target-gene expression, suggesting that this may be a primary mechanism by which they influence cell growth.
Circadian rhythms entrain many aspects of physiology to the daily solar cycle (1). Mammalian circadian rhythms are generated by a cell-autonomous molecular clock based on a transcription–translation feedback loop (TTFL): a heterodimer of circadian locomotor output cycles kaput (CLOCK) and brain and muscle ARNT-like protein 1 (BMAL1) drives transcription of their own repressors, period (PER1, PER2, and PER3), and cryptochrome (CRY1 and CRY2). The F-box and leucine-rich repeat proteins 3 (FBXL3) and 21 (FBXL21) are substrate adaptors for S phase kinase–associated protein 1 (SKP1)–cullin 1 (CUL1)–F-box protein (SCF) E3 ubiquitin ligases that stimulate CRY ubiquitination and degradation and contribute to circadian rhythms (2–8).
In addition to driving circadian rhythms, clock components transmit temporal information to other physiological processes (1, 9–11). For example, CRY1/2 suppress the activity of several noncircadian transcription factors (11–17), thereby influencing their susceptibility to activation in a time-of-day–dependent manner. In the case of c-MYC and Early 2 factor (E2F) family members, this suppression is driven by stimulated proteolysis (12, 16), in which CRY2 acts as a cofactor to recruit c-MYC or E2F family members to the SCFFBXL3 complex to promote ubiquitination and subsequent degradation.
Mammalian CRYs evolved from bacterial light-activated DNA-repair enzymes also known as photolyases but lack DNA-repair activity and light sensitivity (18). Several studies have identified molecular links between CRYs and cancer-related pathways (19). The CRY photolyase homology region (PHR) comprises most of the protein, excluding the disordered carboxyl-terminal tail (CTT). The PHR includes a flavin adenine dinucleotide (FAD) binding pocket, a secondary pocket [analogous to the antenna chromophore-binding pocket of photolyases (20)], and a coiled coil (CC) just upstream of the CTT. FBXL3 and FBXL21 interact with the FAD binding pocket of CRYs; a carboxyl-terminal tryptophan in each of these F-box proteins occupies the FAD binding site (2–8). Similarly, a tryptophan in the disordered loop that connects helices H and I (i.e., the HI loop) of CLOCK interacts with the secondary pocket of CRY1 (21). Sequence differences between CRY1 and CRY2 surrounding the secondary pocket have a major influence on their differential binding affinity for CLOCK:BMAL1 (22, 23) and their divergent influence on circadian period (23). The CC helix of CRY1 interacts with both PER2 and with the transcriptional activation domain (TAD) of BMAL1 and likely reduces the association of BMAL1 with coactivators (24–26). The CTTs in CRY1 and CRY2 are highly divergent both from the carboxyl termini of CRYs in other organisms and from each other. The CTTs contain many phosphorylation sites, which modulate stability (27–30), are regulated by DNA damage signaling (29, 30), and influence circadian timing (31).
The circadian and cell cycles influence each other (9, 32–35), and several studies have demonstrated that environmental or genetic disruption of circadian rhythms alters cancer development (36). Regarding the repressive arm of the TTFL and the period and cryptochrome proteins, previous studies have reported mixed results regarding how their disruption impacts cancer progression. In p53-deficient mice, loss of both Cry1 and Cry2 reduces tumor formation and increases survival (37). By contrast, in wild-type (WT) mice, loss of both Cry1 and Cry2 increases the development of liver tumors spontaneously and in response to either whole-body radiation or treatment with diethylnitrosamine (38–40). Furthermore, contradictory findings appear with irradiated Per2-deficient mice; while several studies have reported enhanced tumor formation upon deletion of mutation of PER2 (39–45), another study found that irradiated Per2-deficient mice are not tumor prone (46). Further investigation is needed to understand the influence of genetic backgrounds, age, and tumor-driving stimuli on these outcomes.
We previously demonstrated that genetic deletion of Cry2 renders cells more susceptible to transformation by diverse oncogenic insults (16), and we and others found that Cry2-deficient cells accumulate more DNA damage (30, 47), which could impact cell growth. Here, we investigate five missense mutations in CRY2 that have each been observed in multiple human tumors, selected based on their predicted impacts on the interactions between CRY2, FBXL3, and c-MYC. Expression of WT CRY2, but not of two of the mutant mouse orthologs studied here, suppresses the growth of c-MYC–transformed Cry2−/− fibroblasts (16). One of the mutants, human CRY2 S532L (mouse CRY2 S510L), exhibits greatly reduced association with FBXL3 as predicted. In contrast, the human CRY2 D347H mutation (mouse CRY2 D325H) does not impact the association of CRY2 with FBXL3 or c-MYC. Instead, we found that it reduces the ability of CRY2 to interact with and repress the CLOCK:BMAL1 heterodimer, and, therefore, it cannot support circadian rhythms in fibroblasts. To gain insight into common effects of the mouse CRY2 D325H and S510L mutations, we globally sequenced RNA prepared from cells expressing WT or mutant CRY2 in combination with c-MYC. This unbiased analysis revealed that both CRY2 missense mutations that accelerate transformation by c-MYC robustly reduce the expression of P53 target genes, suggesting that these mutations influence cell growth by suppressing P53.
Results
Missense Mutation of CRY2 Alters Cell Growth in a Context-Dependent Manner.
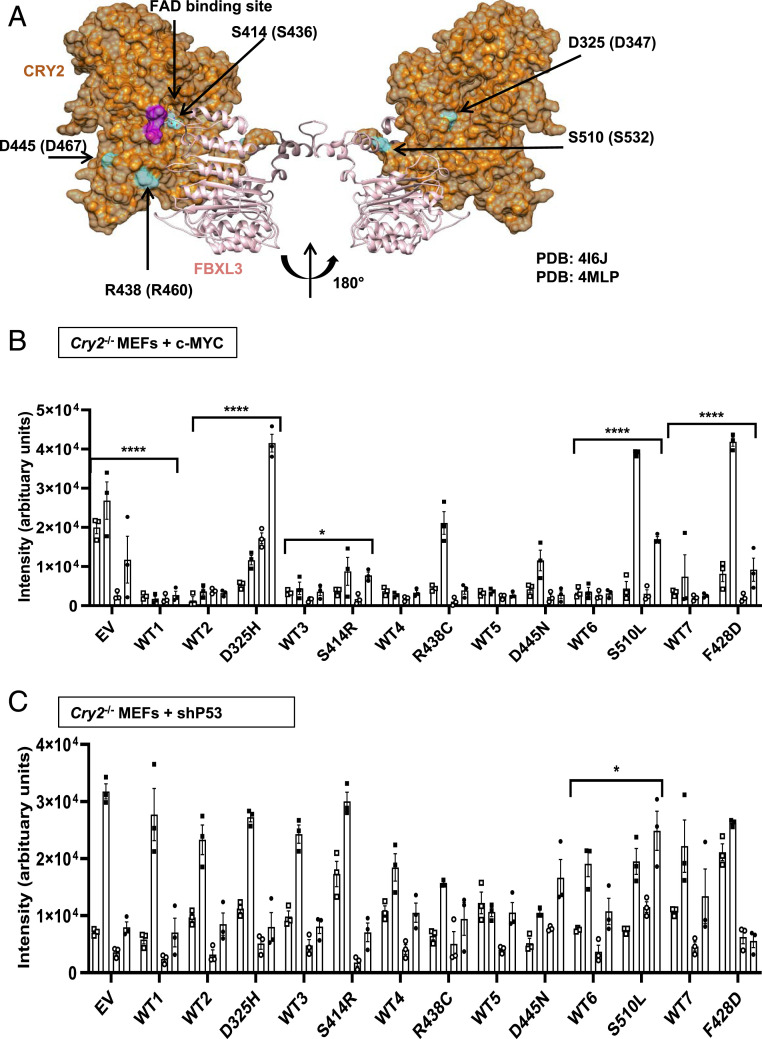
We used cBioPortal (48, 49) (last accessed November 13, 2020) to identify recurrent missense mutations in CRY2 reported in human tumor samples in The Cancer Genome Atlas (TCGA) Pan-Cancer Atlas studies (Fig. 1 and SI Appendix, Fig. S1A). We selected five mutations to investigate based on 1) their frequency of observation in diverse tumor types (when cBioPortal was first accessed in August 2016) and 2) their surface-exposed locations on the three-dimensional structure of CRY2 in close proximity to the CRY2:FBXL3 interface and the phosphate-binding loop where c-MYC interacts with CRY2 (Fig. 1A), suggesting that these mutations may affect the interactions between CRY2, FBXL3, and c-MYC.
Fig. 1.
Missense mutations in CRY2 alter cell growth. (A) Three-dimensional structure of mouse CRY2 (orange) and FBXL3 (light pink) (Protein Data Bank ID: 4I6J). The location and numbering of amino acids orthologous to missense mutations found in TCGA are depicted in blue, with the relevant mutations in human CRY2 indicated in parentheses. Magenta shading on CRY2 indicates the phosphate binding loop that interacts with c-MYC. (B and C) Quantitation of crystal violet staining of colonies formed by Cry2−/− MEFs stably expressing c-MYC (B) or shRNA targeting P53 (C) after plating at low density. Bars represent mean ± SEM for quantification of staining from four biological replicates each analyzed in triplicate, with individual measurements for each replicate depicted by a unique symbol type: open or filled square, open or filled circle. Each condition was compared with controls that were plated in wells on the same plates (see also SI Appendix, Figs. S3 and S4). *P ≤ 0.05, ****P ≤ 0.0001 by two-way ANOVA with Tukey’s multiple comparison test.
Malignant cells must replicate indefinitely (50). To investigate the potential for mutant CRY2 to influence cell proliferation, we monitored cell numbers over 10 d after sparsely plating Cry2−/− mouse embryonic fibroblasts (MEFs) expressing either c-MYC or short hairpin RNA (shRNA) targeting P53 in combination with WT or mutant CRY2. Cells expressing elevated c-MYC proliferated more quickly, consistent with the well-established role of MYC in promoting entry into S phase (51). Stable expression of WT CRY2 slowed proliferation of Cry2−/− fibroblasts expressing c-MYC and tended to slow the growth of P53-depleted cells (Fig. 1 and SI Appendix, Fig. S1 B and C). While the impact of CRY2 expression on proliferation was reduced by several of the mutations examined, the high variability and small effect size made it difficult to measure significant differences in this assay. Intriguingly, cells expressing CRY2 S414R seemed to proliferate more slowly in the context of P53 depletion.
Two-dimensional (2D) colony formation tests cells’ ability to survive and grow under limited paracrine-signaling conditions. We previously demonstrated that Cry2−/− fibroblasts form colonies much more efficiently than matched WT cells in the context of either c-MYC overexpression or depletion of p53. Notably, this phenomenon was most robust in primary fibroblasts that were maintained in culture only for very few passages (16), perhaps due to accelerated accumulation of DNA damage in Cry2−/− cells (30). To study the impact of CRY2 missense mutations, we generated Cry2−/− fibroblasts stably expressing either c-MYC or shRNA targeting P53 and then further manipulated those lines to stably express WT or mutant CRY2. The process of generating the cell lines and growing a sufficient number to plate multiple replicates required performing colony formation assays after 10 to 12 passages. In this context, we found that WT CRY2 consistently suppresses colony growth in c-MYC–expressing Cry2−/− MEFs (Fig. 1B), but the tendency for CRY2 to suppress colony formation in P53-depleted cells was less robust (Fig. 1C). Consistent with our previous studies (16), a mutant that abolishes the interaction between CRY2 and FBXL3 (F428D) (2) fails to suppress colony growth of c-MYC–expressing cells (Fig. 1B). Each of the cancer-associated missense mutants of CRY2 that we examined tended to be somewhat less effective at suppressing colony formation in c-MYC–expressing cells compared with WT CRY2, but most had a mild impact. Strikingly, mouse CRY2 D325H and S510L consistently failed to suppress colony growth in c-MYC–expressing cells (Fig. 1B and SI Appendix, Fig. S1D). In the context of P53 depletion, expression of CRY2 S510L slightly enhanced colony formation, but neither WT nor mutant CRY2 had a major impact on colony formation of P53-depleted cells under the conditions tested (Fig. 1C and SI Appendix, Fig. S1E). Protein levels of the CRY2 missense mutants varied across biological replicates, likely due to heterogeneity of genome-integration sites within the stable-cell population, and did not have consistent effects on c-MYC protein accumulation in the Cry2−/− c-MYC–expressing cells (SI Appendix, Fig. S2).
CRY2 Missense Mutations Alter Protein–Protein Interactions.
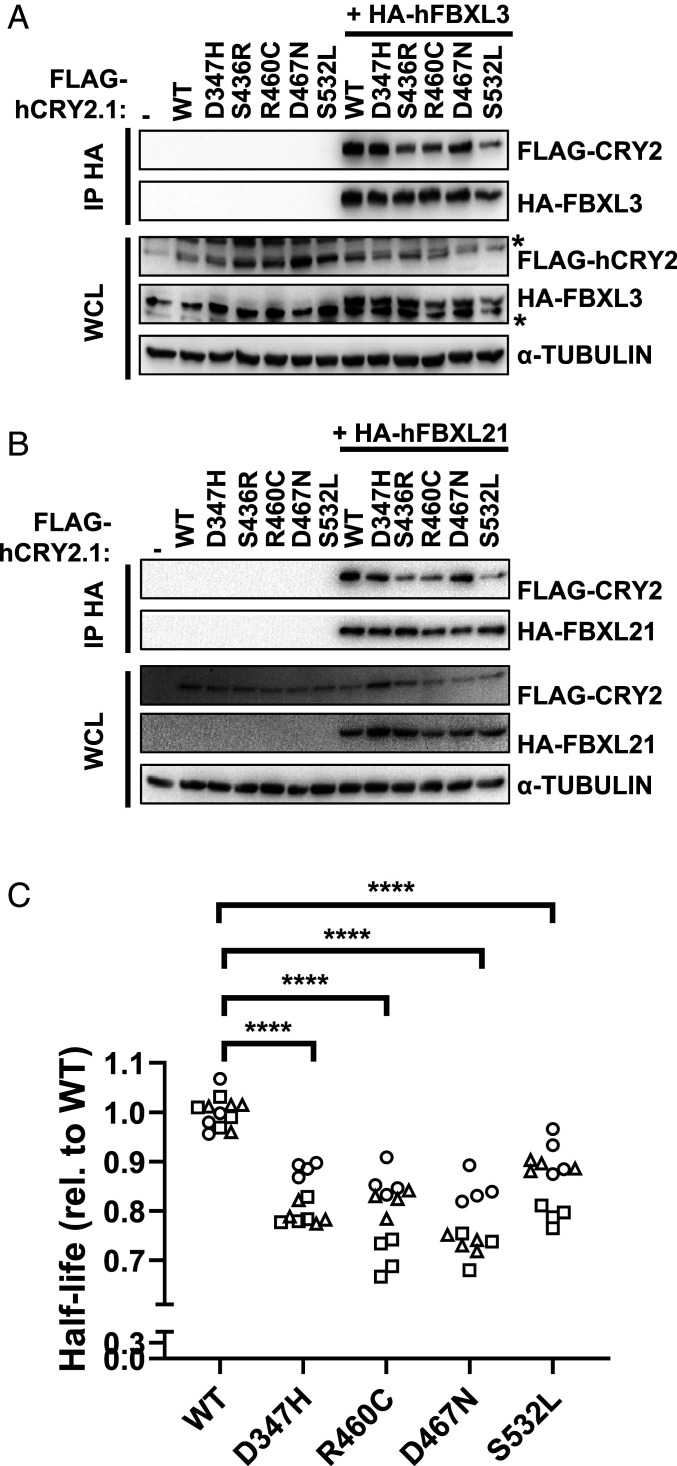
To determine whether the observed impact of CRY2 missense mutations on cell growth could be explained by altered interaction with SCF complexes and/or with c-MYC, we examined the interactions of WT and mutant CRY2 with FBXL3, FBXL21, and c-MYC. Human CRY2 S436R, R460C, and S532L each exhibit decreased interactions with FBXL3 and FBXL21 compared with WT CRY2 (Fig. 2 A and B), though their impact is more subtle than that of the CRY2 F428D mutant, which was established from structural studies to critically disrupt interaction with FBXL3 (2) (SI Appendix, Fig. S3 A and B). None of the mutants consistently affected interactions with c-MYC (SI Appendix, Fig. S3C).
Fig. 2.
CRY2 S436R, R460C, and S532L exhibit reduced interaction with F-box proteins. (A and B) Proteins detected by immunoblot (IB) following HA immunoprecipitation (IP) or in whole-cell lysates (WCL) from HEK293T cells expressing the indicated plasmids with the indicated tags. * denotes nonspecific band. (C) Half-life of CRY2-Luciferase fusions proteins containing the indicated missense mutations in CRY2 expressed in AD293 cells. Each symbol represents the half-life calculated from fitting luminescence data recorded continuously after addition of cycloheximide to an exponential decay function. Data represent three independent experimental replicates indicated by different symbols, each performed in quadruplicate. ****P ≤ 0.0001 by two-way ANOVA with Tukey’s multiple comparison test.
Since FBXL3 and FBXL21 are primary determinants of CRY stability (2–8), we hypothesized that the half-lives of CRY2 S436R, R460C, and S532L would be increased due to their reduced interaction with FBXL3 and FBXL21. We generated tools to express fusion proteins in which luciferase is appended to the carboxyl terminus of WT or mutant CRY2 (CRY2::LUC) in AD293 cells to measure the impact of each mutation on CRY2 stability. Contrary to expectations, each of the missense mutations studied reduces the half-life of the CRY2::LUC fusion protein (Fig. 2C and SI Appendix, Fig. S3 F–H). This assay may not accurately reflect CRY2 half-life under physiological conditions, given that the fusion protein may behave differently than CRY2 alone as well as the probability that expression of the fusion protein alters the stoichiometry of CRY2, endogenous F-box proteins, and/or other components that regulate CRY2 stability in vivo. Nevertheless, we did not observe consistent alterations in the steady-state accumulation of either CRY2 or c-MYC under the conditions in which cell-growth properties were impacted by mutant CRY2 (SI Appendix, Fig. S2).
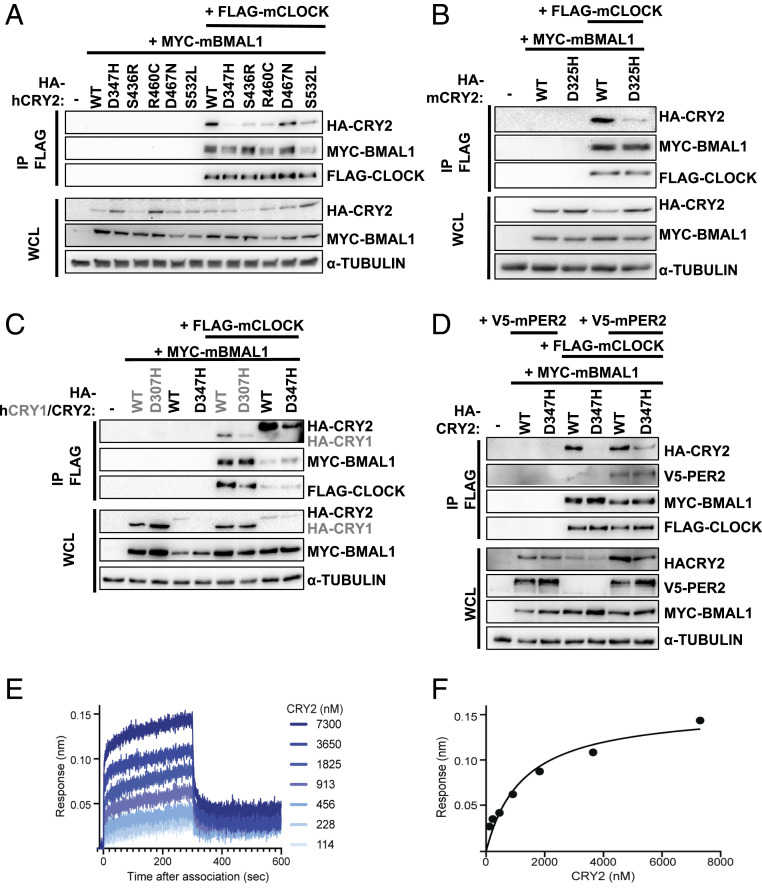
Within the core circadian-clock mechanism, CRYs directly repress CLOCK and BMAL1 in the presence or absence of PERs (52, 53). Several studies have suggested that disruption of circadian rhythms may contribute to tumor development (36). So, we examined whether the five selected tumor-associated CRY2 mutants exhibit altered interaction with core clock components. Indeed, several of the missense mutations under investigation reduced the recruitment of CRY2 to the CLOCK:BMAL1 heterodimer. Most strikingly, the interaction of human CRY2 D347H with CLOCK in the presence of BMAL1 was almost undetectable (Fig. 3A). The ability of overexpressed CRY2 to interact with endogenous BMAL1 recapitulated the pattern of interaction with overexpressed CLOCK in the presence of overexpressed BMAL1, suggesting that these mutants may influence the interaction of CRY2 with the CLOCK:BMAL1 heterodimer under physiological conditions (Fig. 3 and SI Appendix, Fig. S4A). Importantly, none of the mutations examined had any effect on the interactions of CRY2 with overexpressed PER1 or PER2 (Fig. 3 and SI Appendix, Fig. S4 B and C), suggesting that the decreased interaction of CRY2 D347H with CLOCK in the presence of BMAL1 is due to local disruptions in the structure rather than a global alteration of protein fold. The analogous mutations in human CRY1 (D307H) and mouse CRY2 (D325H) similarly impacted their interactions with CLOCK and BMAL1 (Fig. 3 B and C).
Fig. 3.
CRY2 D347H, S436R, R460C, and S532L exhibit reduced interaction with CLOCK. (A–D) Proteins detected by IB following IP of the FLAG tag or in WCL from HEK293T cells expressing the indicated plasmids. (E) Representative BLI binding data of mouse CRY2 D325H PHR domain binding to the CLOCK:BMAL1 PAS domain core (PAS-AB domain heterodimer) immobilized on streptavidin tips. (F) Equilibrium analysis of the BLI data in E fit to a one-site binding model. KD = 1.8 ± 0.4 µM from two independent BLI experiments.
The interaction of PER2 with CRY2 was previously shown to enhance its ability to coimmunoprecipitate with CLOCK:BMAL1 (22, 23), resulting from an approximately twofold decrease in the dissociation constant (KD) of the CRY2 PHR domain for the PER-ARNT-SIM (PAS) domain core of CLOCK:BMAL1 in vitro (22). Consistent with this, coexpression of PER2 enables detection of CRY2 D347H in complex with CLOCK:BMAL1, although the mutant binds less robustly than WT CRY2 (Fig. 3D). Given that relatively modest changes in affinity impact the ability of CRY2 to coimmunoprecipitate with CLOCK:BMAL1 from cells, we used biolayer interferometry (BLI) to determine how the CRY2 D325H mutation influences the affinity of the mouse CRY2 PHR domain for the PAS domain core of CLOCK:BMAL1 in vitro (Fig. 3E). The D325H mutation decreased the affinity 1.5-fold compared with WT CRY2 (22) for a KD of 1.8 ± 0.4 µM (Fig. 3F), helping to explain the reduced interaction of these proteins in cellular extracts.
CRY2 D347H Does Not Repress CLOCK:BMAL1.
Given that CRY2 D347H exhibits reduced interaction with CLOCK in the presence of BMAL1, we predicted that it would be a less effective repressor of CLOCK:BMAL1 transcriptional activity. In U2OS cells, transient overexpression of CLOCK and BMAL1 enhances the expression of luciferase driven by the Per2 promoter (Fig. 4 A and B). CRY2 dose-dependently decreases the bioluminescent signal, indicating repression of the CLOCK:BMAL1 heterodimer by CRY2 as expected. A previous study demonstrated that CRY2 G351D is unable to repress CLOCK:BMAL1 (54); we have recapitulated that finding and use CRY2 G351D as a control in our assay. Four CRY2 missense mutants (S436R, R460C, D467N, and S532L) examined behave indistinguishably from WT CRY2 in this assay. However, consistent with its impact on the interaction of CRY2 with CLOCK and BMAL1 in a coimmunoprecipitation assay, CRY2 D347H does not repress CLOCK:BMAL1 when all three are overexpressed in U2OS cells (Fig. 4B).
Fig. 4.
CRY2 D347H neither represses CLOCK/BMAL1 nor supports circadian rhythms. (A) Schematic diagram of the luciferase assay performed in B. (B) Luminescence detected from U2OS cells expressing Per2-Luciferase and the indicated additional plasmids (mCherry, mCh). Data are represented as mean ± SEM and are normalized to the gray bar and CLOCK-BMAL1 alone. Triangles on the x-axis indicate increasing amounts of CRY WT or mutant transfected, 2 or 5 ng. ****P ≤ 0.0001 by Student’s t test versus CLOCK-BMAL1 alone in GraphPad Prism 9. (C) Schematic diagram of rhythmicity rescue assay in Cry1−/−;Cry2−/− MEFs. (D) Periods of Cry1, Cry2 WT, or mutant or control (no Cry transfected) across three independent experiments (open circles, triangles, or squares) with six technical replicates in each. Periods calculated using the Lumicycle analysis program using running-average background subtraction and fitting to a damped sine wave by least mean squares method. Replicates for which the best-fit damped sine had period >40 h and/or goodness of fit <80% were considered arrhythmic and are indicated as 0 on the graph. (E) Periods of the mutants that showed rhythmicity (R438C and S510L). Data represent mean ± SEM of two independent experiments with two to six replicates each as indicated by filled circles in the graph. ***P ≤ 0.001, ****P ≤ 0.0001 by one-way ANOVA using Dunnett’s multiple comparisons in GraphPad Prism 9. (F) Background-subtracted data generated from continuous recording of luciferase activity from Cry1−/−;Cry2−/− fibroblasts transfected with Per2-Luciferase and WT Cry1 (gray), or WT (black) or mutant (as indicated) Cry2. Note that, because the background signal decreases exponentially over time, the background-subtracted signal falls below zero. Data represent mean bioluminescence for two to six replicates from a representative of at least two experiments.
To examine whether cancer-associated missense mutations in CRY2 alter the ability of CRY2 to sustain endogenous circadian rhythms, we used the rhythmicity rescue assay developed in ref. 55. In this assay, arrhythmic Cry1−/−;Cry2−/− MEFs (56) are transiently transfected with a plasmid encoding destabilized luciferase under the control of the Per2 promoter, in combination with CRY1 or CRY2 under the control of elements derived from the endogenous Cry1 promoter and first intron. Expression of WT CRY1 (57) or CRY2 (23) under these conditions enables rhythmic expression of the luciferase reporter with long or short periodicity characteristic of Cry2−/− or Cry1−/− MEFs, respectively. Continuous recording of luciferase activity enables precise measurement of the period of the resulting rhythmic luciferase expression, which allows sensitive detection of perturbations of those rhythms.
Using this system, we found that all of the cancer-associated missense mutations tested impair the ability of CRY2 to sustain short-period circadian rhythms of luciferase expression driven by the Per2 promoter (Fig. 4 D–F and SI Appendix, Fig. S5). Consistent with its inability to repress CLOCK:BMAL1 transcriptional activity, orthologous mouse CRY2 D325H was unable to rescue rhythms. Unexpectedly, CRY2 D445N also failed to support circadian expression of Per2-luciferase despite having minimal impact on the interaction of CRY2 with either FBXL3/21 or CLOCK:BMAL1. Expression of CRY2 R438C or S510L resulted in longer periods than those observed with expression of WT CRY2. CRY2 S414R sustained detectable rhythms in only one-third of replicates, and those rhythms also had a longer period than those observed with WT CRY2, similar to those supported by CRY2 R438C and S510L.
CRY2 Mutants That Fail to Suppress Colony Formation Repress P53 Target-Gene Expression.
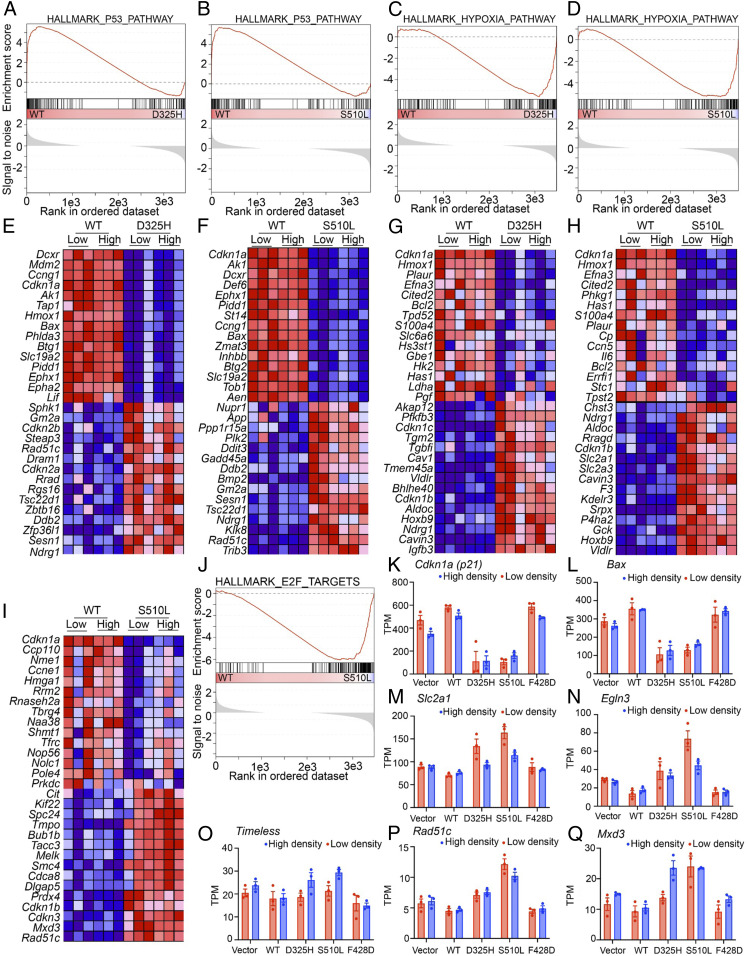
Because neither their impact on protein–protein interactions nor their ability to rescue circadian rhythms distinguished the two CRY2 mutants that fail to suppress 2D colony growth in c-MYC–expressing fibroblasts (D325H and S510L) (Fig. 5 and SI Appendix, Table S1), we took an unbiased approach to examine their influence on global gene expression in those cells. We sequenced RNA from Cry2−/− MEFs overexpressing c-MYC and either an empty vector (EV, Control), or WT or mutant CRY2 plated at a low or high density. Gene set enrichment analysis (GSEA) (57) revealed dramatic suppression of a hallmark set of P53 target genes (58) in cells expressing either CRY2 D325H (false discovery rate, FDR = 0.004) or S510L (FDR = 0.01) compared with those expressing WT CRY2 (Fig. 5 A, B, E, F, K, and L and SI Appendix, Fig. S6). Expression of the transcript encoding P53 (Trp53) is decreased in cells expressing CRY2 D325H or S510L (SI Appendix, Fig. S7A), but P53 protein levels do not seem to be affected (SI Appendix, Fig. S7B). This suggests that the decreased expression of the Trp53 transcript in D325H and S510L does not explain the reduced expression of P53 target genes. Instead, the reduction in Trp53 mRNA is likely a consequence of decreased P53 activity, since it has been reported to regulate its own expression (59).
Fig. 5.
mCRY2 D325H and mCRY2 S510L suppress transcription in the P53 pathway. (A–J) GSEA enrichment plots (A–D, and J) and heat maps representing the top 15 up- or down-regulated transcripts of the leading edge that were driving the enrichment score (ES) (E–I) for the hallmark gene sets representing the P53 pathway (A, B, E, and F), hypoxia pathway (C, D, G, and H), or E2F target genes (I and J). In A–D and J, the top portion shows the running ES for the gene set as the analysis walks down the ranked list. The middle portion shows where the members of the gene set appear in the ranked list of genes. The bottom portion shows the value of the ranking metric as you move down the list of ranked genes. The ranking metric measures a gene’s correlation with a phenotype. Here, the phenotypes are defined as the cell lines stably expressing WT or mutant (D325H or S510L). In E–I, red, high expression; blue, low expression. (K–Q) Extracted gene-expression profiles of the indicated genes from the RNA-seq data. Light red, low-density plating; light blue, high-density plating.
Given that P53 is a key mediator of growth suppression, reduced expression of P53 target genes in cells expressing CRY2 D325H or S510L with concomitant overexpression of c-MYC suggests that reduced P53 activity may be a critical effector of the CRY2 mutants’ impact on the growth of c-MYC–expressing cells. To understand whether regulation of P53 is required for the influence of CRY2 on colony formation in c-MYC–overexpressing cells, we used shRNA to deplete P53 or Luciferase (control) in cells that also express c-MYC and WT or mutant CRY2 (SI Appendix, Fig. S8). As expected, depleting P53 robustly increases colony formation in c-MYC–expressing cells (SI Appendix, Fig. S8A). Consistent with the idea that CRY2 suppression of colony formation in cells overexpressing c-MYC involves regulation of P53, expression of WT CRY2 is less effective at suppressing colony formation in P53-depleted cells. However, in contrast to Cry2−/− cells in which P53 is depleted by shRNA without overexpression of c-MYC (Fig. 1C), expression of WT CRY2 consistently suppresses colony formation in these cells in which c-MYC overexpression is combined with shRNA-mediated depletion of P53 (SI Appendix, Fig. S8A). Intriguingly, reduced P53 expression does not significantly impact colony formation in Cry2-deficient cells or in those expressing CRY2 S510L when compared with shLuc (SI Appendix, Fig. S8B), suggesting that P53 activity may be already suppressed in those cells even when P53 protein is present.
While the effect was less robust than the observed suppression of the P53 pathway, we also measured significantly increased expression of the hallmark hypoxia gene network (FDR = 0.016 or 0.05, Fig. 5 C, D, G, H, M, and N and SI Appendix, Fig. S6). Finally, consistent with our demonstration that CRYs cooperate with SCFFBXL3 to regulate E2F protein accumulation (12), E2F target-gene expression was elevated in cells expressing CRY2 S510L (which exhibits decreased interaction with FBXL3) when plated at high density (FDR = 0.21 including only high-density samples, Fig. 5 I, J, O, P, and Q and SI Appendix, Fig. S6). Taken together, these data suggest that CRY2 D325H and S510L are unable to suppress colony formation of Cry2−/− MEFs overexpressing c-MYC due to overlapping functional impairments including suppression of the P53 transcriptional network.
Discussion
Most missense mutations reported in TCGA have an unknown functional impact on the cancers in which they are found. Although missense mutations in CRY2 are rare in TCGA, human genome-sequencing data spanning six global and eight subcontinental ancestries (60) demonstrate that core clock genes, including CRY2, are resistant to mutation in healthy somatic tissues, suggesting that recurrent cancer-associated mutations in these genes may be functionally important. Several lines of evidence indicate that CRY2 may play a role in preventing tumor development. In a variety of cancers, CRY2 expression is significantly reduced compared with matched normal tissues (16, 61). Furthermore, inactivation or reduced expression of CRY2 is strongly associated with altered activity of established oncogenic or tumor suppressive pathways (61). Here, we demonstrate that two cancer-associated mutations in CRY2 (D347H and S532L, using numbering for human CRY2) prevent CRY2-mediated suppression of colony growth in the context of c-MYC overexpression in Cry2−/− fibroblasts.
Several molecular mechanisms have been proposed to explain the apparent inhibition of tumor development by CRY2 (19). For example, CRY2 can cooperate with SCFFBXL3 to stimulate ubiquitination and subsequent degradation of c-MYC (16), the DNA damage–activated protein kinase TLK2 (62), E2F family members (12), and likely other proteins. CRY2 S532L exhibits decreased interaction with FBXL3 and FBXL21, and this likely contributes to the elevated expression of E2F target genes in cells expressing CRY2 S532L. However, disrupted interaction with FBXL3/21 may not be sufficient to prevent CRY2 from suppressing colony formation because the S436R and R460C mutations, which had a similar impact on the ability of CRY2 to interact with FBXL3/21, can suppress colony formation. We cannot exclude the possibility that quantitative differences in the impacts of these mutations on CRY2-FBXL3/21 affinity underlie their diverse functional impacts on cell growth. Conversely, CRY2 D347H fails to suppress colony formation but does not prevent CRY2 from interacting with its F-box–protein partners. Thus, disruption of the CRY2-FBXL3 interaction is not required for loss of CRY2 suppression of colony formation in the context of c-MYC overexpression.
CRY2 D347H exhibits reduced interaction with and an inability to repress CLOCK:BMAL1 and cannot sustain circadian rhythms of Per2-luciferase expression in Cry1−/−;Cry2−/− fibroblasts.
The secondary pocket of CRYs provides an extensive interaction surface for the CLOCK PAS-B domain that plays a critical role in the stable recruitment of cryptochromes to CLOCK:BMAL1 (22, 23). The electrostatic surface of the CRY secondary pocket is mostly negatively charged, while the HI loop of the CLOCK PAS-B domain that docks into this pocket is positively charged (21), suggesting that electrostatic attraction could help to support the CRY–CLOCK interaction. Notably, D347 is located on the edge of the secondary pocket of CRY2, providing a rationale for how the mutation from aspartic acid (D) to histidine (H) could contribute to reduced affinity of the CRY2 mutant for CLOCK:BMAL1. Given that differences in the affinity of CRY1 and CRY2 define their ability to stably interact with CLOCK:BMAL1 and repress its activity (22, 23), the D437H mutation represents another example of how a modest change in binding at this critical interface can manifest as a substantial change in CRY function in the clock.
Disrupted circadian rhythms increase cancer risk in humans and increase tumor formation in a variety of genetically engineered mouse models of cancer (36). The inability of the mouse ortholog of CRY2 D347H to suppress colony formation in c-MYC–expressing fibroblasts supports the contention that circadian rhythms play a protective role against tumorigenesis. However, as for the role of CRY2 in promoting SCFFBXL3/21–mediated protein turnover, loss of the ability to sustain circadian rhythms is not sufficient to prevent suppression of colony formation since the CRY2 D467N ortholog (D445N) suppresses colony formation but cannot support cell-autonomous rhythms. The inability of CRY2 D445N to sustain circadian rhythms despite its seemingly normal interaction with and repression of CLOCK:BMAL1 suggests that it must disrupt some other aspect of the concerted and periodic interaction with other endogenous proteins (63), appropriate degradation (3, 5, 7), or other posttranslational regulation within the clock mechanism. We cannot exclude the possibility that the impact of the CRY2 mutants examined could differ in the presence of WT CRY1 and/or CRY2, both of which are likely present in tumor samples in which the mutations occur in a single copy. Taken together, the impacts of the five missense mutations in CRY2 on cell growth, circadian rhythms, and interaction with FBXL3/21 suggest that disruption of core clock function or circadian proteolysis may contribute to enhanced cell growth, but neither is sufficient nor required for loss of growth suppression by CRY2. We measured global RNA-expression patterns to identify functional impacts shared by the two mutations with the greatest impact on cell growth. This analysis revealed a striking suppression of a P53 transcriptional network and concomitant elevation of hypoxia target genes in cells expressing mutant CRY2. We do not exclude the possibility that multiple mechanisms contribute to the failure of CRY2 D325H and S510L to suppress colony growth.
We previously demonstrated that CRY1 is stabilized and CRY2 is destabilized in response to DNA damage (30). Furthermore, transcriptional activation of at least some P53 target genes is blunted in Cry2−/− cells and sustained in Cry1−/− cells following DNA damage (30). PER2, which cooperates with CRYs to repress transcriptional activation of the CLOCK:BMAL1 heterodimer, stabilizes P53 by inhibiting its MDM2-mediated ubiquitination and degradation (64–66). These mechanisms could be at play, but further investigation will be required to understand how CRY2 D325H and CRY2 S510L suppress P53 target-gene expression.
Elevated expression of hypoxia-regulated transcripts can enhance cell survival under stressful conditions and contribute to tumorigenesis (67–69). We and others recently demonstrated that CRY1 and CRY2 suppress both the protein accumulation and transcriptional activation of HIF1-α and HIF2-α (70, 71). HIFs are closely related to CLOCK and BMAL1 (72), and we and others have shown that BMAL1 dimerizes with HIF1-α (70, 73, 74). Given that the CRY2-D325H mutation reduces its association with and repression of CLOCK:BMAL1, it may also influence repression of HIF1-α/BMAL1 by altering protein–protein interactions, although it is unclear whether the interaction of CRYs with HIFs involves the secondary pocket. In contrast, the CRY2 S510L mutation may enhance hypoxia target-gene expression by preventing CRY-mediated suppression of HIF1-α protein accumulation.
E2Fs are a family of transcription factors that regulate G1- to S-phase progression in the cell cycle (75). The expression of E2F target genes is elevated in cells expressing CRY2 S510L but not in those expressing CRY2 D325H. This distinction likely reflects reduced SCFFBXL3/21–mediated turnover of E2F family members (12) in the presence of CRY2 S510L, which exhibits reduced association with FBXL3 and FBXL21. The additional elevation of E2F target genes in combination with suppression of P53 likely contributes to the greater impact of CRY2 S510L in cell-growth and colony-formation assays.
Over the past decade, several small molecules that target CRYs have been identified, and some are under development for therapeutic use in metabolic disease and cancer (76–80). Intriguingly, one of these (TH301) forms a hydrogen bond with the serine in CRY1 analogous to CRY2 S414 and selectively increases the half-life of CRY2 (and not of CRY1), presumably by reducing its interaction with FBXL3 (80). The disordered CTT of CRY2 was required for the impact of TH301 on CRY2 stability, suggesting an intramolecular interaction between the CTT and the globular PHR, which contains the FAD binding pocket in which S414 is located. The apparently greater impact of the CRY2 mutant (D325H) to decrease interaction with CLOCK:BMAL1 in cells compared with the modest difference in affinity between the WT and D325H mutant CRY2 PHR for the BMAL1:CLOCK PAS domain core in vitro may also be explained by participation of the CTT. Improved understanding of the molecular mechanisms connecting circadian clocks, and CRYs in particular, to cancer-driving networks will inform efforts for further development of CRY-modulating compounds and other strategies to combat the increased incidence of metabolic disease, cancer, and other pathologies associated with chronic circadian disruption.
Materials and Methods
Cell Culture.
Cry2−/− MEFs were generated as described in ref. 16 with minor modifications (SI Appendix, Materials and Methods). by Andrew Liu, University of Florida, Gainesville, FL, and Hiroki Ueda, University of Tokyo, Tokyo, Japan and the RIKEN Quantitative Biology Center, Osaka, Japan (55) and were gifted to us. HEK 293T and U2OS cells were purchased from the American Type Culture Collection. AD293 cells were gifted to us by Steve Kay, University of Southern California, Los Angeles, CA. Cell culture methods were as in ref. 12. Fetal bovine serum was purchased from Thermo Fisher (catalog no. 16000044). Transfections were carried out using standard protocols and various plasmids described in SI Appendix, Materials and Methods. Stable cells were generated using various lentiviruses or retroviruses as detailed in SI Appendix, Materials and Methods.
Proliferation and 2D Colony-Formation Assays.
For proliferation assays, cells were plated at 5,000 cells per well in triplicate in a 6-well plate and counted every 2 d for 10 d via hemocytometer. Two independent experiments were performed. For 2D colony-formation assays, cells were plated 150 cells per well in a 6-well plate in triplicate and grown for 11 to 18 d prior to fixing and staining with crystal violet. Three independent experiments were performed.
Real-Time Bioluminescence Rhythmicity Rescue Assays.
Cry1−/−;Cry2−/− cells (from Hiroki Ueda, University of Tokyo and the RIKEN Quantitative Biology Center) were plated at 4 × 105 per 3.5-cm dish and transfected the same day with FuGene 6 (Promega, catalog no. E2311) according to the manufacturer’s protocol. Assays were performed as in ref. 23 with minor modifications (SI Appendix, Materials and Methods).
Protein Degradation and Luciferase Assays.
The protein degradation assay was performed as in ref. 76 with slight modifications (SI Appendix, Materials and Methods). Luciferase assays were performed with U2OS cells as in ref. 13.
Mice.
Cry1−/−;Cry2−/− mice from which Cry2−/− primary MEFs were derived were from Aziz Sancar, University of North Carolina, Chapel Hill, NC (81). All animal care and treatments were in accordance with Scripps Research guidelines and regulations for the care and use of animals. All procedures involving experimental animals were approved by the Scripps Research Institutional Animal Care and Use Committee under protocol no. 10–0019.
RNA Sequencing and Analysis.
Cry2−/− primary MEFs stably overexpressing c-MYC and EV, WT, D325H, S510L, or F428D were plated at two plating densities. Extracted total RNA samples were sent to BGI Group, Beijing, China for library preparation and sequencing. Reads (single-end 50-bp at a sequencing depth of 20 million reads per sample) were generated by BGISEQ-500. Kallisto (https://pachterlab.github.io/kallisto/) was used to align to the reference transcriptome (ftp://ftp.ensembl.org/pub/current_fasta/mus_musculus/cdna/) and estimate transcript abundance. GenePattern (https://www.genepattern.org/use-genepattern), GSEA (57), and differential expression analysis (using R [https://www.r-project.org/]) were used to analyze the RNA sequencing (RNA-seq) data for differentially expressed genes and to find suppressed or enriched gene sets across samples.
BLI.
Polyhistidine (His)-tagged mouse CRY2 D325H PHR domain was expressed in Sf9-suspension insect cells (Expression Systems) and was purified as described in detail in SI Appendix, Materials and Methods. The CLOCK:BMAL1 PAS domain core was purified and biotinylated as reported in ref. 22. BLI experiments were performed using an eight-channel Octect-RED96e (ForteBio) as detailed in SI Appendix, Materials and Methods.
Supplementary Material
Acknowledgments
This work was supported by NIH Grant CA211187 and by a gift from the Brown Foundation for Cancer Research to K.A.L. and NIH Grant GM107069 to C.L.P. G.C.G.P. was supported by an HHMI Gilliam Fellowship with support from the University of California, Santa Cruz (UCSC) graduate division. J.L.F. was supported by a University of California Office of the President (UCOP) and UCSC Chancellor’s Postdoctoral Fellowship. We thank Drs. Hiroki Ueda, Carla Green, Tsuyoshi Hirota, Clark Rosensweig, Anne-Laure Huber, Megan Vaughan, Anna Kriebs, and Marie Pariollaud, and Ms. Rebecca Mello for helpful discussions and/or gifting us critical reagents. The results published here are, in whole or in part, based on data generated by the TCGA Research Network. We thank Toni Thomas, Judy Valecko, and Yolanda Slivers for administrative assistance.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2101416118/-/DCSupplemental.
Data Availability
RNA-seq data have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus (accession no. GSE165647).
References
- 1.Bass J., Circadian topology of metabolism. Nature 491, 348–356 (2012). [DOI] [PubMed] [Google Scholar]
- 2.Xing W., et al., SCF(FBXL3) ubiquitin ligase targets cryptochromes at their cofactor pocket. Nature 496, 64–68 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Godinho S. I., et al., The after-hours mutant reveals a role for Fbxl3 in determining mammalian circadian period. Science 316, 897–900 (2007). [DOI] [PubMed] [Google Scholar]
- 4.Busino L., et al., SCFFbxl3 controls the oscillation of the circadian clock by directing the degradation of cryptochrome proteins. Science 316, 900–904 (2007). [DOI] [PubMed] [Google Scholar]
- 5.Siepka S. M., et al., Circadian mutant overtime reveals F-box protein FBXL3 regulation of cryptochrome and period gene expression. Cell 129, 1011–1023 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dardente H., Mendoza J., Fustin J. M., Challet E., Hazlerigg D. G., Implication of the F-box protein FBXL21 in circadian pacemaker function in mammals. PLoS One 3, e3530 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hirano A., et al., FBXL21 regulates oscillation of the circadian clock through ubiquitination and stabilization of cryptochromes. Cell 152, 1106–1118 (2013). [DOI] [PubMed] [Google Scholar]
- 8.Yoo S. H., et al., Competing E3 ubiquitin ligases govern circadian periodicity by degradation of CRY in nucleus and cytoplasm. Cell 152, 1091–1105 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kowalska E., et al., NONO couples the circadian clock to the cell cycle. Proc. Natl. Acad. Sci. U.S.A. 110, 1592–1599 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hunt T., Sassone-Corsi P., Riding tandem: Circadian clocks and the cell cycle. Cell 129, 461–464 (2007). [DOI] [PubMed] [Google Scholar]
- 11.Koike N., et al., Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science 338, 349–354 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chan A. B., Huber A. L., Lamia K. A., Cryptochromes modulate E2F family transcription factors. Sci. Rep. 10, 4077 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kriebs A., et al., Circadian repressors CRY1 and CRY2 broadly interact with nuclear receptors and modulate transcriptional activity. Proc. Natl. Acad. Sci. U.S.A. 114, 8776–8781 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lamia K. A., Cryptochromes mediate rhythmic repression of the glucocorticoid receptor. Nature 480, 552–556 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jang H., et al., SREBP1c-CRY1 signalling represses hepatic glucose production by promoting FOXO1 degradation during refeeding. Nat. Commun. 7, 12180 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huber A. L., et al., CRY2 and FBXL3 cooperatively degrade c-MYC. Mol. Cell 64, 774–789 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jordan S. D., et al., CRY1/2 selectively repress PPARδ and limit exercise capacity. Cell Metab. 26, 243–255.e6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Partch C. L., Sancar A., Photochemistry and photobiology of cryptochrome blue-light photopigments: The search for a photocycle. Photochem. Photobiol. 81, 1291–1304 (2005). [DOI] [PubMed] [Google Scholar]
- 19.Chan A. B., Lamia K. A., Cancer, hear my battle CRY. J. Pineal Res. 69, e12658 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sancar A., Structure and function of DNA photolyase and cryptochrome blue-light photoreceptors. Chem. Rev. 103, 2203–2237 (2003). [DOI] [PubMed] [Google Scholar]
- 21.Michael A. K., et al., Formation of a repressive complex in the mammalian circadian clock is mediated by the secondary pocket of CRY1. Proc. Natl. Acad. Sci. U.S.A. 114, 1560–1565 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fribourgh J. L., et al., Dynamics at the serine loop underlie differential affinity of cryptochromes for CLOCK:BMAL1 to control circadian timing. eLife 9, e55275 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rosensweig C., et al., An evolutionary hotspot defines functional differences between CRYPTOCHROMES. Nat. Commun. 9, 1138 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schmalen I., et al., Interaction of circadian clock proteins CRY1 and PER2 is modulated by zinc binding and disulfide bond formation. Cell 157, 1203–1215 (2014). [DOI] [PubMed] [Google Scholar]
- 25.Nangle S. N., et al., Molecular assembly of the period-cryptochrome circadian transcriptional repressor complex. eLife 3, e03674 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu H., et al., Cryptochrome 1 regulates the circadian clock through dynamic interactions with the BMAL1 C terminus. Nat. Struct. Mol. Biol. 22, 476–484 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harada Y., Sakai M., Kurabayashi N., Hirota T., Fukada Y., Ser-557-phosphorylated mCRY2 is degraded upon synergistic phosphorylation by glycogen synthase kinase-3 beta. J. Biol. Chem. 280, 31714–31721 (2005). [DOI] [PubMed] [Google Scholar]
- 28.Kurabayashi N., Hirota T., Sakai M., Sanada K., Fukada Y., DYRK1A and glycogen synthase kinase 3beta, a dual-kinase mechanism directing proteasomal degradation of CRY2 for circadian timekeeping. Mol. Cell. Biol. 30, 1757–1768 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gao P., et al., Phosphorylation of the cryptochrome 1 C-terminal tail regulates circadian period length. J. Biol. Chem. 288, 35277–35286 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Papp S. J., et al., DNA damage shifts circadian clock time via Hausp-dependent Cry1 stabilization. eLife 4, e04883 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parico G. C. G., et al., The human CRY1 tail controls circadian timing by regulating its association with CLOCK:BMAL1. Proc. Natl. Acad. Sci. U.S.A. 117, 27971–27979 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matsuo T., et al., Control mechanism of the circadian clock for timing of cell division in vivo. Science 302, 255–259 (2003). [DOI] [PubMed] [Google Scholar]
- 33.Geyfman M., et al., Brain and muscle Arnt-like protein-1 (BMAL1) controls circadian cell proliferation and susceptibility to UVB-induced DNA damage in the epidermis. Proc. Natl. Acad. Sci. U.S.A. 109, 11758–11763 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hong C. I., et al., Circadian rhythms synchronize mitosis in Neurospora crassa. Proc. Natl. Acad. Sci. U.S.A. 111, 1397–1402 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bieler J., et al., Robust synchronization of coupled circadian and cell cycle oscillators in single mammalian cells. Mol. Syst. Biol. 10, 739 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pariollaud M., Lamia K. A., Cancer in the fourth dimension: What is the impact of circadian disruption? Cancer Discov. 10, 1455–1464 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ozturk N., Lee J. H., Gaddameedhi S., Sancar A., Loss of cryptochrome reduces cancer risk in p53 mutant mice. Proc. Natl. Acad. Sci. U.S.A. 106, 2841–2846 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mteyrek A., et al., Critical cholangiocarcinogenesis control by cryptochrome clock genes. Int. J. Cancer 140, 2473–2483 (2017). [DOI] [PubMed] [Google Scholar]
- 39.Kettner N. M., et al., Circadian homeostasis of liver metabolism suppresses hepatocarcinogenesis. Cancer Cell 30, 909–924 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee S., Donehower L. A., Herron A. J., Moore D. D., Fu L., Disrupting circadian homeostasis of sympathetic signaling promotes tumor development in mice. PLoS One 5, e10995 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fu L., Pelicano H., Liu J., Huang P., Lee C., The circadian gene Period2 plays an important role in tumor suppression and DNA damage response in vivo. Cell 111, 41–50 (2002). [DOI] [PubMed] [Google Scholar]
- 42.Hwang-Verslues W. W., et al., Loss of corepressor PER2 under hypoxia up-regulates OCT1-mediated EMT gene expression and enhances tumor malignancy. Proc. Natl. Acad. Sci. U.S.A. 110, 12331–12336 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sun C. M., et al., Per2 inhibits k562 leukemia cell growth in vitro and in vivo through cell cycle arrest and apoptosis induction. Pathol. Oncol. Res. 16, 403–411 (2010). [DOI] [PubMed] [Google Scholar]
- 44.Gu X., et al., The circadian mutation PER2(S662G) is linked to cell cycle progression and tumorigenesis. Cell Death Differ. 19, 397–405 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mteyrek A., Filipski E., Guettier C., Okyar A., Lévi F., Clock gene Per2 as a controller of liver carcinogenesis. Oncotarget 7, 85832–85847 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Antoch M. P., Toshkov I., Kuropatwinski K. K., Jackson M., Deficiency in PER proteins has no effect on the rate of spontaneous and radiation-induced carcinogenesis. Cell Cycle 12, 3673–3680 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hoffman A. E., et al., Phenotypic effects of the circadian gene cryptochrome 2 on cancer-related pathways. BMC Cancer 10, 110 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gao J., et al., Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 6, pl1 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cerami E., et al., The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2, 401–404 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hanahan D., Weinberg R. A., Hallmarks of cancer: The next generation. Cell 144, 646–674 (2011). [DOI] [PubMed] [Google Scholar]
- 51.Kaczmarek L., Hyland J. K., Watt R., Rosenberg M., Baserga R., Microinjected c-myc as a competence factor. Science 228, 1313–1315 (1985). [DOI] [PubMed] [Google Scholar]
- 52.Gustafson C. L., Partch C. L., Emerging models for the molecular basis of mammalian circadian timing. Biochemistry 54, 134–149 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ye R., et al., Dual modes of CLOCK:BMAL1 inhibition mediated by cryptochrome and period proteins in the mammalian circadian clock. Genes Dev. 28, 1989–1998 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McCarthy E. V., Baggs J. E., Geskes J. M., Hogenesch J. B., Green C. B., Generation of a novel allelic series of cryptochrome mutants via mutagenesis reveals residues involved in protein-protein interaction and CRY2-specific repression. Mol. Cell. Biol. 29, 5465–5476 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ukai-Tadenuma M., et al., Delay in feedback repression by cryptochrome 1 is required for circadian clock function. Cell 144, 268–281 (2011). [DOI] [PubMed] [Google Scholar]
- 56.van der Horst G. T., et al., Mammalian Cry1 and Cry2 are essential for maintenance of circadian rhythms. Nature 398, 627–630 (1999). [DOI] [PubMed] [Google Scholar]
- 57.Subramanian A., Kuehn H., Gould J., Tamayo P., Mesirov J. P., GSEA-P: A desktop application for gene set enrichment analysis. Bioinformatics 23, 3251–3253 (2007). [DOI] [PubMed] [Google Scholar]
- 58.Liberzon A., et al., The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 1, 417–425 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Deffie A., Wu H., Reinke V., Lozano G., The tumor suppressor p53 regulates its own transcription. Mol. Cell. Biol. 13, 3415–3423 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Karczewski K. J.et al.; Genome Aggregation Database Consortium , The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 581, 434–443 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ye Y., et al., The genomic landscape and pharmacogenomic interactions of clock genes in cancer chronotherapy. Cell Syst. 6, 314–328.e2 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Correia S. P., et al., The circadian E3 ligase complex SCFFBXL3+CRY targets TLK2. Sci. Rep. 9, 198 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Aryal R. P., et al., Macromolecular assemblies of the mammalian circadian clock. Mol. Cell 67, 770–782.e6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gotoh T., et al., Model-driven experimental approach reveals the complex regulatory distribution of p53 by the circadian factor Period 2. Proc. Natl. Acad. Sci. U.S.A. 113, 13516–13521 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gotoh T., Vila-Caballer M., Liu J., Schiffhauer S., Finkielstein C. V., Association of the circadian factor Period 2 to p53 influences p53's function in DNA-damage signaling. Mol. Biol. Cell 26, 359–372 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gotoh T., et al., The circadian factor Period 2 modulates p53 stability and transcriptional activity in unstressed cells. Mol. Biol. Cell 25, 3081–3093 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Choudhry H., Harris A. L., Advances in hypoxia-inducible factor biology. Cell Metab. 27, 281–298 (2018). [DOI] [PubMed] [Google Scholar]
- 68.Semenza G. L., Hypoxia-inducible factors: Coupling glucose metabolism and redox regulation with induction of the breast cancer stem cell phenotype. EMBO J. 36, 252–259 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Semenza G. L., HIF-1 mediates metabolic responses to intratumoral hypoxia and oncogenic mutations. J. Clin. Invest. 123, 3664–3671 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Vaughan M. E., et al., Cryptochromes suppress HIF1a in muscles. iScience 23, 101338 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dimova E. Y., et al., The circadian clock protein CRY1 is a negative regulator of HIF-1α. iScience 13, 284–304 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fribourgh J. L., Partch C. L., Assembly and function of bHLH-PAS complexes. Proc. Natl. Acad. Sci. U.S.A. 114, 5330–5332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wu Y., et al., Reciprocal regulation between the circadian clock and hypoxia signaling at the genome level in mammals. Cell Metab. 25, 73–85 (2017). [DOI] [PubMed] [Google Scholar]
- 74.Hogenesch J. B., Gu Y. Z., Jain S., Bradfield C. A., The basic-helix-loop-helix-PAS orphan MOP3 forms transcriptionally active complexes with circadian and hypoxia factors. Proc. Natl. Acad. Sci. U.S.A. 95, 5474–5479 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Attwooll C., Lazzerini Denchi E., Helin K., The E2F family: Specific functions and overlapping interests. EMBO J. 23, 4709–4716 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hirota T., et al., Identification of small molecule activators of cryptochrome. Science 337, 1094–1097 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Oshima T., et al., C-H activation generates period-shortening molecules that target cryptochrome in the mammalian circadian clock. Angew. Chem. Int. Ed. Engl. 54, 7193–7197 (2015). [DOI] [PubMed] [Google Scholar]
- 78.Lee J. W., et al., Development of small-molecule cryptochrome stabilizer derivatives as modulators of the circadian clock. ChemMedChem 10, 1489–1497 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Dong Z., et al., Targeting glioblastoma stem cells through disruption of the circadian clock. Cancer Discov. 9, 1556–1573 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Miller S., et al., Isoform-selective regulation of mammalian cryptochromes. Nat. Chem. Biol. 16, 676–685 (2020). [DOI] [PubMed] [Google Scholar]
- 81.Thresher R. J., et al., Role of mouse cryptochrome blue-light photoreceptor in circadian photoresponses. Science 282, 1490–1494 (1998). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq data have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus (accession no. GSE165647).