Significance
Antibiotic resistance among clinically relevant bacteria presents an increasing threat and raises the urgent need for new compounds. Antimicrobial peptides and peptide antibiotics are potent membrane-active molecules and valuable prototypes for drug development. In gram-negative infections, killing of bacteria by antimicrobials is accompanied by the release of lipopolysaccharide (LPS), an endotoxin that causes severe hyperinflammation and pathology. We demonstrate how two medically relevant peptides, the cathelicidin LL-32 and polymyxin B, disarm endotoxins by peptide–LPS interaction. Furthermore, our studies reveal a mechanism of peptide-mediated immune control by acting on signaling domains of the immune cell membrane. Our results significantly enhance our understanding of how peptide antibiotics can regulate inflammation and will be important for their development and therapeutic use.
Keywords: antimicrobial peptides, membrane domains, hyperinflammation, immune regulation, endotoxin
Abstract
Antimicrobial peptides (AMPs) contribute to an effective protection against infections. The antibacterial function of AMPs depends on their interactions with microbial membranes and lipids, such as lipopolysaccharide (LPS; endotoxin). Hyperinflammation induced by endotoxin is a key factor in bacterial sepsis and many other human diseases. Here, we provide a comprehensive profile of peptide-mediated LPS neutralization by systematic analysis of the effects of a set of AMPs and the peptide antibiotic polymyxin B (PMB) on the physicochemistry of endotoxin, macrophage activation, and lethality in mice. Mechanistic studies revealed that the host defense peptide LL-32 and PMB each reduce LPS-mediated activation also via a direct interaction of the peptides with the host cell. As a biophysical basis, we demonstrate modifications of the structure of cholesterol-rich membrane domains and the association of glycosylphosphatidylinositol (GPI)-anchored proteins. Our discovery of a host cell–directed mechanism of immune control contributes an important aspect in the development and therapeutic use of AMPs.
Antimicrobial peptides (AMPs), a central part of the innate immune system, represent a phylogenetically conserved mechanism of immune defense. In humans, AMPs are found at all body interfaces, including the skin and mucosal surfaces of the lung, intestine, and urogenital tract where they provide efficient first-line protection against environmental pathogens and naturally acquired microbiota. The broad contributions of AMPs to human immune defenses against infections are exemplified by cathelicidin (1–3).
The coincidence of stagnating efforts toward new antibiotic development for more than a decade (4) and the emergence of antibiotic-resistant strains of pathogens have led to a lack of effective antibacterial treatment options. Particularly, multidrug-resistant pathogens represent a major challenge for clinicians and hospitals (5). Notably, AMPs have long been used and optimized evolutionarily for efficiency and applicability in the human body. The success of this evolutionary process is demonstrated by the low emergence of microbial resistance to this class of defense molecules. Accordingly, AMPs represent an ideal alternative to conventional antibiotics in the development of new therapeutics against infectious diseases (6, 7).
AMPs mainly exert direct antibacterial activity, although the increasing recognition of other biological activities (8, 9) has led to the description of these molecules as host defense peptides. Particularly, AMPs play key roles in wound healing and repair by modulating immune responses and angiogenesis. The ability to modulate inflammation in the context of infection represents a central step in the avoidance of excessive immune-mediated damage and devastating consequences such as sepsis-related multiorgan failure, shock, and death (10). Dysregulated AMP expression has been linked to several diseases associated with high morbidity (11), including Crohn’s disease (CD) (12), cystic fibrosis (CF) (13), chronic obstructive pulmonary disease (COPD), and asthma (14, 15).
AMPs are small (length: 20 to 40 amino acids) and structurally diverse (α-helical, β-sheet, circular) peptides that share a common structural motif with a strong amphiphilic nature and a net cationic charge. Serum concentrations can be in the micromolar range. This particular structure forms the basis of their antimicrobial activity, which targets bacterial membranes and induces dysfunction via pore formation, membrane thinning, or lipid segregation (16). The simple and highly efficient lytic specificity of AMPs depends on charge selectivity for anionic lipids in the microbial membrane, such that the neutral surfaces of host cells are largely unaffected (17, 18). Some antibiotic peptides, such as colistin and other clinically used polymyxins, efficiently also exploit this bactericidal mechanism. However, the therapeutic potential of the immune-modulating activity of the peptides is hindered by our limited understanding of the molecular mechanism.
The effects of AMPs on host cells have been attributed to the binding of AMPs to cellular receptors or intracellular targets (19–21), however, the detailed mechanisms remain unclear. Consequently, we analyzed a panel of AMPs from different molecular classes: the cathelicidin LL-37 (8) and short-variant LL-32; hBD-3-l, a variant of the β-defensin hBD-3; NK-2, a derivative of the lymphocytic effector protein NK-lysin; Pep19-2.5 (Aspidasept), a de novo–designed peptide (22); and polymyxin B (PMB), a peptide-based antibiotic which binds with high specificity to LPS and leads to its aggregation (23), and that has been used widely in studies of LPS bioactivity neutralization. We analyzed the effects of individually administered LL-32 or PMB on the host cell response to lipopolysaccharide (LPS; endotoxin), the main molecular trigger for the immune detection of gram-negative infection. LPS induces severe hyper-inflammatory responses and is one of the most potent inducers of sepsis and septic shock (10, 24). LPS activates immune cells via a complex interplay of transport and receptor proteins. Specifically, its recognition by the TLR4/MD-2 receptor complex on the cytoplasmic membrane (25–28) initiates several intracellular signaling cascades, leading to the production of proinflammatory mediators such as TNF-α, IL-6, and IL-8 (29). This recognition is strongly enhanced by the transport of LPS via LPS-binding protein (LBP) and soluble CD14 to the TLR4/MD-2 receptor complex, which enables the recognition of picogram amounts of LPS by monocytes and macrophages (30–32).
Our analyses of the effects of LL-32 and of PMB on different stages of LPS-induced cell activation revealed that the AMPs or PMB interact with LPS and induce structural and biophysical changes that reduce the bioactivity of this endotoxin. Changes in the aggregate structure of LPS by cationic and amphiphilic molecules has been discussed for polymers and peptides (33, 34). Moreover, we discovered specific interactions of cathelicidin AMPs and PMB with host cell cytoplasmic membranes, as well as effects of the peptides on signaling domain membrane organization. Our findings indicate a host cell–directed mechanism by which antibiotic peptides restrain proinflammatory immune responses.
Results
LL-32 and PMB Reduce LPS-Induced Inflammation In Vitro and In Vivo.
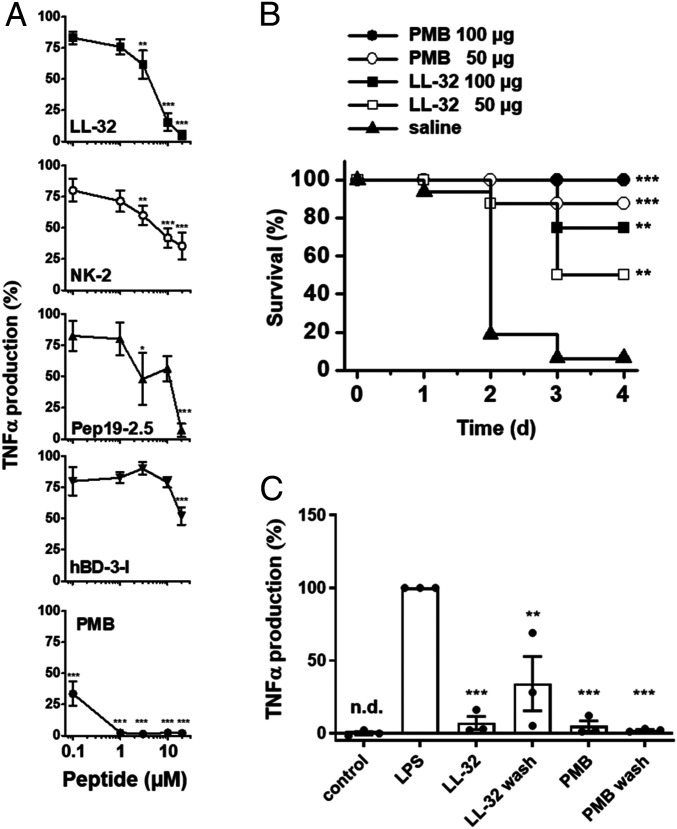
Initially, we analyzed the abilities of various AMPs and of PMB to reduce the proinflammatory responses of human macrophages to LPS. The preincubation of macrophages with the peptides at concentrations of 1 to 20 µM for 30 min reduced or even abolished the LPS-induced production of TNF-α (Fig. 1A). Specifically, LPS-mediated cell activation was abrogated fully by LL-32 at 10 µM and Pep19-2.5 at 20 µM and inhibited by 58% and 21% in response to 10 µM NK-2 or hBD-3-l, respectively and by 65% and 48% in response to 20 µM concentrations of the latter peptides, respectively. Notably, PMB exhibited the most potent activity and abrogated TNF-α production at 1 µM. Consequently, we focused on LL-32 and PMB as the most potent peptides in subsequent analyses.
Fig. 1.
AMPs and PMB inhibit LPS-mediated inflammation in vitro and in vivo. (A) Human macrophages were incubated in medium alone or in the presence of 0.1, 1, 3, 10, and 20 µM concentrations of the peptides LL-32, NK-2, Pep19-2.5, hBD-3-l, or PMB for 30 min at 37 °C. The macrophages were subsequently stimulated with 5 nM LPS for 4 h at 37 °C. The concentrations of TNF-α were determined in supernatants. TNF-α values of samples stimulated with LPS in the absence of peptide were set 100% and all other values were calculated accordingly. The data are shown as the means ± SEM of n = 8 (LL-32), n = 4 to 5 (NK-2, Pep19-2.5, hBD-3-l), or n = 5 to 9 (PMB) independent experiments using cells from different donors. (B) Galactosamine-sensitized mice were injected intraperitoneally with LPS (100 ng/mouse; equivalent to 5 µg/kg) and subsequently with 50 or 100 µg/mouse of LL-32 or PMB or saline (n = 8 mice per group) at a different injection site. The survival of the mice was monitored daily. (C) Macrophages were incubated with 10 µM LL-32 or PMB for 30 min at 37 °C and subsequently washed three times with serum-free Roswell Park Memorial Institute (RPMI) 1640 medium to remove unbound peptide or left untreated, followed by stimulation with 5 nM LPS for 4 h. The concentrations of TNF-α in the supernatants were determined. TNF-α values of samples stimulated with LPS in the absence of peptide were set 100%, and all other values were calculated accordingly. The data are shown as the means ± SEM of n = 3 independent experiments using cells from different donors. Black dots represent the individual data points. Control, unstimulated cells; n.d., not detectable. Statistical analyses were performed via one-way ANOVA and Dunnett’s post test; *P ≤ 0.05, **P ≤ 0.01, and ***P ≤ 0.001 (peptide groups versus LPS control).
We next induced a mouse model of endotoxin shock via the intravenous injection of LPS into the bloodstream. In the saline control group, 88% of the animals died within 4 d after LPS injection. The administration of LL-32 or PMB increased the survival rate to 75% after treatment with LL-32 (100 µg/mouse) and was maintained at 100% after treatment with PMB (100 µg/mouse) (Fig. 1B). These results demonstrate the effective ability of these peptides to reduce the exaggerated immune response to endotoxin.
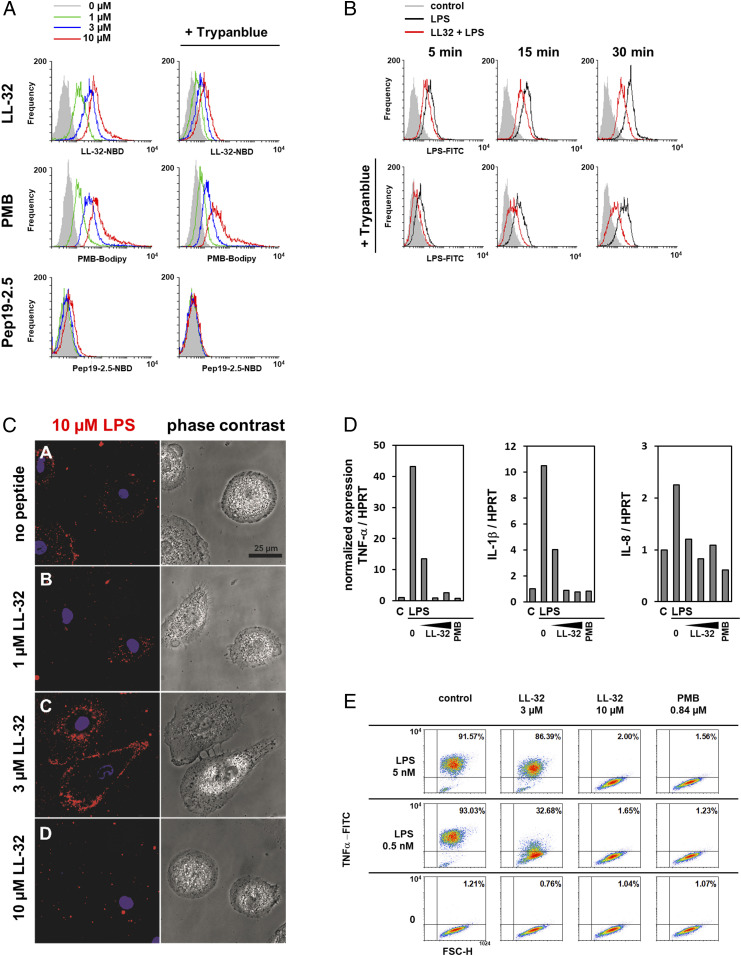
The LPS-neutralizing activities of these cationic peptides have been attributed to strong interactions with the negatively charged LPS molecule and subsequent physicochemical changes in the LPS structure (35). To differentiate whether the observed immunomodulatory functions of AMPs are based on the neutralization of LPS or on the modulation of host cell functions, we performed washing experiments in which human macrophages were incubated with the peptides for 30 min and subsequently washed to remove free peptide prior to LPS stimulation. We observed that the preincubation of macrophages with the individual peptides leads to a significant reduction (LL-32 **P ≤ 0.01, PMB ***P ≤ 0.001 peptide versus LPS control) in LPS-mediated TNF-α production, even if the cells were washed intensively before stimulation with LPS (Fig. 1C). These data suggest that the anti-inflammatory effects of the peptides are not solely dependent on LPS neutralization via direct peptide binding but also rely on interactions between peptides and the host cell. Accordingly, our flow cytometry data reveal the dose-dependent binding of fluorophore-conjugated LL-32 and PMB to human macrophages (Fig. 2A; reference SI Appendix, Fig. S1 for the gating strategy). The results of fluorescence quenching assays with trypan blue demonstrated that considerable proportions of LL-32 and PMB were not internalized but remained exposed on the cell surface. In contrast, Pep19-2.5 exhibited very low binding to macrophage membranes.
Fig. 2.
LL-32 and PMB binding modifies cellular LPS processing and inhibits inflammatory responses in human macrophages. (A) Macrophages were incubated with LL-32-NBD, Pep19-2.5-NBD, or PMB-BODIPY for 5 min at 37 °C, then washed and fixed. Subsequently, the samples were split and analyzed by flow cytometry to determine the total bound peptide directly and after quenching with 0.2% trypan blue to determine the amount of intracellular peptide. (B) Macrophages were incubated with 3 µM LPS-FITC in the presence of 3 µM LL-32 for 5, 15, or 30 min at 37 °C. The samples were analyzed by flow cytometry. (C) Macrophages cultured on µ-slides were stimulated with rhodamine-labeled LPS in the presence of 1, 3, and 10 µM LL-32 for 5 min at room temperature. Cell nuclei were counterstained with Hoechst. (Scale bar, 25 µM). (D) Macrophages were preincubated for 30 min with LL-32 (1, 3, and 10 µM) or PMB (1 µg/mL, 0.84 µM) and stimulated with 5 nM LPS for 1 h at 37 °C. Gene expression was analyzed by qRT-PCR. Data are relative expression ratios of the target to reference gene (HPRT), normalized to the untreated control. (E) Macrophages were preincubated with LL-32 (3 and 10 µM), PMB (1 µg/mL, 0.84 µM), or buffer (control) for 30 min at 37 °C and subsequently stimulated with LPS for 4 h in the presence of 10 µg/mL bafilomycin. Intracellular TNF-α was stained with a specific antibody, and the cells were analyzed by flow cytometry. Numbers in the upper right quadrants of plots indicate the percentages of gated macrophages positive for TNF-α. Data are representative of n = 3 (A, C–E) and n = 5 (B) independent experiments.
We next characterized the effects of the peptides on different stages of host cell activation. Both LL-32 (Fig. 2B) and PMB (SI Appendix, Fig. S2) reduce but do not completely abrogate the binding and internalization of LPS by human macrophages. Confocal microscopy analysis further demonstrated a condensing effect of LL-32 on the intracellular LPS pool, an observation that could be relevant for the activation of intracellular LPS-receptors such as caspase-4, -5, and -11 (36). As shown in Fig. 2C, larger intracellular LPS aggregates are visible at LL-32 concentrations of 3 and 10 µM. LL-32 attenuated the transcriptional activation of the proinflammatory cytokines TNF-α, IL-1β, and IL-8 at 1 µM and suppressed this expression at 3 and 10 µM. PMB yielded similar results (Fig. 2D). A similar dose-dependent effect of AMPs on intracellular TNF-α was observed (Fig. 2E), demonstrating that the anti-inflammatory effects of these peptides are exerted at an early stage of activation. We did not observe any cytotoxic effects of the peptides on human macrophages or HEK293-TLR4/MD-2 cells at the experimental concentrations; however, LL-32 exerted a low level of hemolytic activity against human erythrocytes (SI Appendix, Fig. S3).
LL-32 and PMB Modulate the Three-Dimensional Structure, Surface Charge, and Transport of LPS.
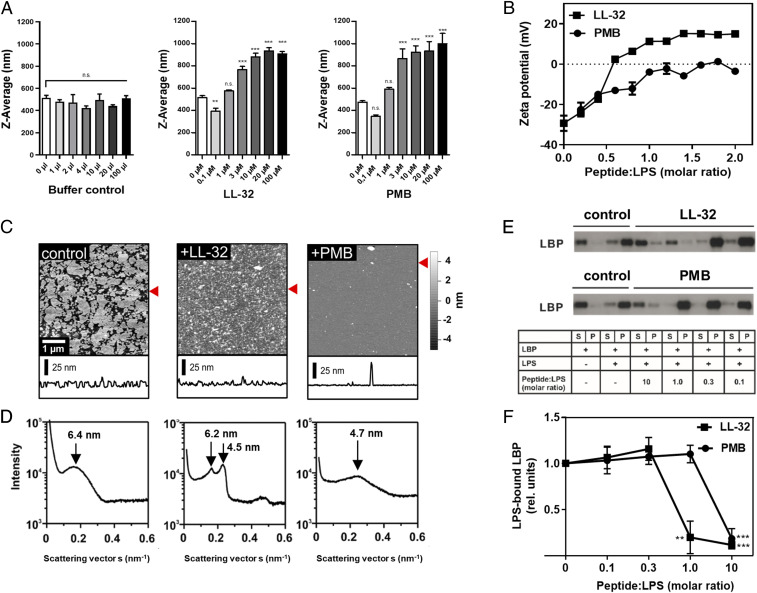
The biological activity of LPS is dependent on the aggregation state, the presentation of the negatively charged phosphate groups on the backbone, and the overall three-dimensional (3D) structure (37), and this activity is modified by the binding of cationic peptides and proteins. The titration of LL-32 or PMB to LPS aggregates in solution led to a significant increase of the size of aggregates (Fig. 3A) and neutralized the negative surface charge of the LPS aggregates (zeta potential: −27 ± 4.8 mV) to varying degrees. The addition of PMB almost fully neutralized the surface charge (zeta potential: −5.2 ± 1.7 mV), whereas the addition of LL-32 induced charge overcompensation (zeta potential: +17 ± 2.2 mV; Fig. 3B). Accordingly, the addition of LL-32 or PMB to LPS coated on mica induced strong changes in the lateral membrane organization, as visualized by atomic force microscopy (AFM; Fig. 3C). PMB treatment yielded a smooth bilayer surface suggestive of deeper penetration of this peptide into the LPS bilayer core, whereas LL-32 accumulated on the membrane surface and formed larger irregular domains (Fig. 3C). These findings are consistent with our small-angle X-ray scattering (SAXS) data revealing the supramolecular membrane structure (Fig. 3D). Specifically, pure LPS yielded a diffuse symmetric scattering curve characteristic of the formation of unilamellar aggregates caused by the negative surface charge, which led to a net electrostatic repulsion of the LPS bilayers (38). In the presence of LL-32, the appearance of Bragg peaks in the SAXS profile clearly indicates the formation of strongly correlated LPS bilayers, presumably due to the shielding of negative charges by the bound peptide and a consequent drastic change in the aggregation structure. In contrast, the addition of PMB did not significantly alter the shape of the scattering profile relative to pure LPS but significantly shifted the maximum to higher angles, indicating a thinning of the LPS bilayer. This observation may be explained by partial intercalation of the peptide into the hydrophobic core, consistent with the AFM measurements (Fig. 3C).
Fig. 3.
LL-32 and PMB affect the supramolecular organization and binding interactions of LPS. (A) The size of LPS aggregates (1 µM) was determined by dynamic light-scattering experiments at 37 °C. Buffer (volume control) or peptides were added at the indicated final concentrations. The data represent the means ± SEM of n = 4 (buffer control), n = 7 (LL-32), and n = 5 (PMB) independent experiments. Data were analyzed as peptide versus control by one-way ANOVA with Dunnett’s post test. **P ≤ 0.01 and ***P ≤ 0.001; n.s., not significant. (B) Zeta potentials of LPS aggregates in solution. LL-32 and PMB were titrated to LPS at the indicated molar ratios. The data represent the means ± SEM of two independent experiments with three technical replicates. (C) AFM images and height profiles of solid-supported layers of LPS WBB01. LPS was immobilized as the control or preincubated with peptide at a ratio of 2:1 (by weight). Data are representative of n = 3 independent experiments. The red arrows label the line of the respective height profile. All AFM images were only flattened and not further processed. (D) SAXS diffractograms of pure LPS aggregates (Left) and aggregates prepared in the presence of LL-32 (Middle) or PMB (Right) at a LPS:peptide ratio of 2:1 (by weight). The diffractograms are representative of n = 3 independent experiments. The respective repeating distances are indicated by arrows. (E) LPS (4.55 µM) and LBP (500 ng) were coincubated with peptide for 30 min at room temperature and subsequently sedimented via ultracentrifugation. Supernatant (S) and pellet (P) fractions were separated by SDS-PAGE and immunoblotted to detect LBP. Representative images of the blots of LL-32 (Upper) and PMB (Lower) from n = 4 independent experiments each are shown. (F) Quantification of the band intensities of the pellet fractions. The data were quantified using Image J software, and the pixel intensities of the pellet fractions were normalized to the control sample (LBP + LPS). The statistical analyses by one-way ANOVA and Dunnett’s post test. The graph presents the means ± SEM of n = 4 independent experiments; **P ≤ 0.01 and ***P ≤ 0.001 (LPS only versus LPS + peptide).
LPS-induced cell activation is enhanced greatly by LBP activity in the serum, which enables cells to respond sensitively to minute amounts of LPS (30). To determine the effects of AMPs on the LBP–LPS interaction, we incubated LPS aggregates with LBP in the presence of LL-32 or PMB and sedimented the aggregates by centrifugation before subjecting the supernatant and pellet fractions to Western blotting for LBP. Notably, LBP was detectable in the supernatant fraction (S) in the absence of LPS but sedimented into the pellet fraction (P) upon incubation with LPS aggregates, thus demonstrating the binding of LBP to LPS aggregates. The addition of LL-32 or PMB to the LPS aggregates strongly reduced the amount of LPS-bound LBP (Fig. 3E). Densitometric analysis confirmed that LL-32 and PMB significantly reduced the binding of LBP to LPS aggregates (Fig. 3F). These data are consistent with our observation that both peptides reduce the binding of LPS to cells (Fig. 2B and SI Appendix, Fig. S2) and demonstrate their effects on LPS transport.
Inhibitory Effects of LL-32 and PMB Specifically Impair Cell Activation by LPS.
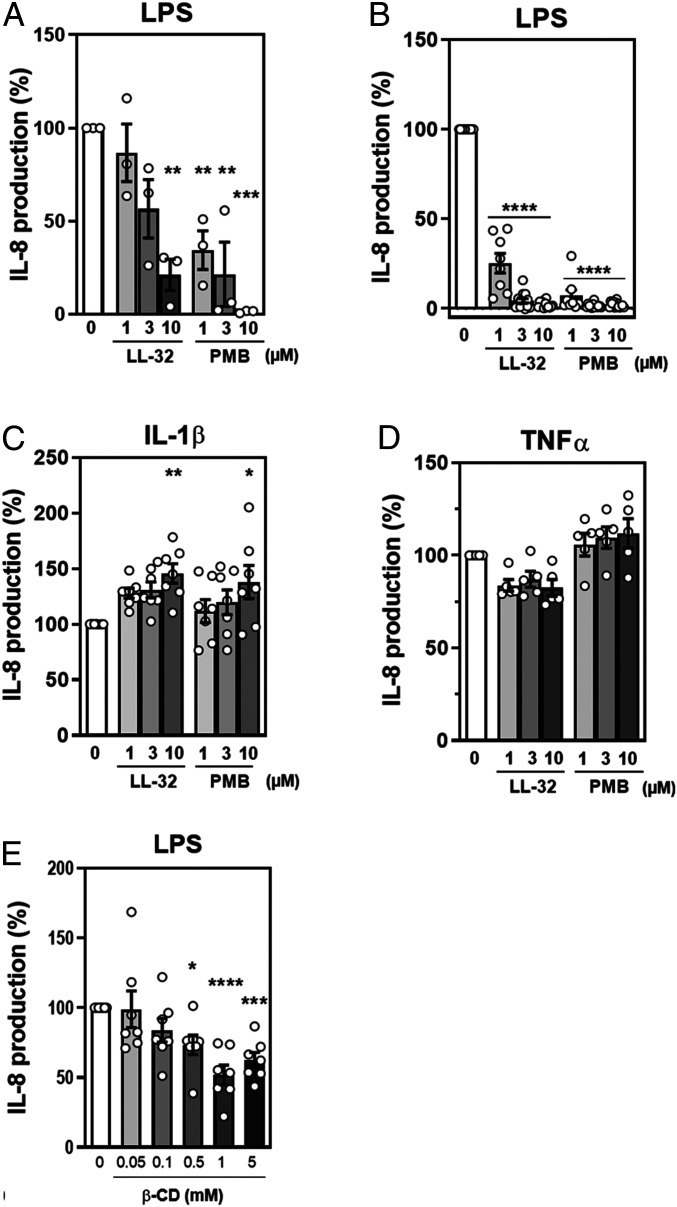
Primary cells, such as human macrophages, exhibit strongly variable donor-dependent responses. Therefore, we subjected human embryonic kidney (HEK)293 cells expressing the TLR4/MD-2 receptor to washing experiments. Cells washed after peptide exposure exhibited a significantly reduced ability to respond to LPS (**P ≤ 0.01, ***P ≤ 0.001) when compared with unexposed cells (Fig. 4A), whereas unwashed cells exhibited enhanced peptide-mediated inhibitory activity (****P ≤ 0.0001; Fig. 4B). As shown in SI Appendix, Fig. S4, this phenomenon was not restricted to LL-32 or PMB, as other cathelicidin peptide family members, including the full-length peptide LL-37 (***P ≤ 0.001), rabbit fragment rCAP18 (****P ≤ 0.0001), murine CRAMP (****P ≤ 0.0001), and bovine BMAP-27 and BMAP-28 (****P ≤ 0.0001), strongly reduced LPS-induced proinflammatory activity in washing experiments (SI Appendix, Fig. S4). In contrast, the NK-lysin derivative NK-2, an unrelated peptide, lacks this capacity, as demonstrated by a complete loss of its endotoxin-inhibitory potential after cell washing (SI Appendix, Fig. S5 A and B). This observation demonstrates that NK-2 cannot exploit the inhibitory mechanism used by cathelicidins, probably due to differences in the interactions of these two types of compounds with cells and/or LPS.
Fig. 4.
The anti-inflammatory effects of LL-32 and PMB are mediated by peptide–cell interactions. (A) HEK293-TLR4/MD-2 cells in DMEM containing 10% FCS were seeded into plates at a density of 5 × 105/well and treated with LL-32 or PMB at the indicated concentrations for 30 min at 37 °C. The cells were washed three times in DMEM to remove peptides from the medium and were subsequently stimulated with 10 nM LPS for 24 h. (B) HEK293-TLR4/MD-2 cells were treated with LL-32 or PMB for 30 min at 37 °C, followed directly by stimulation with 10 nM LPS for 24 h. (C) HEK293-TLR4/MD-2 cells were treated with LL-32 or PMB for 30 min at 37 °C, followed directly by stimulation with 100 nM IL-1β or (D) 50 nM TNF-α. The concentrations of secreted IL-8 in the supernatants were determined by enzyme-linked immunosorbent assay (ELISA). IL-8 values of cells stimulated with LPS in the absence of peptide were set 100% and all other values were calculated accordingly. The data are reported as the means ± SEM of n = 3 (A), n = 8 to 13 (B), n = 7 (C), and n = 5 (D) independent experiments. White dots represent the individual data points. (E) Cholesterol-dependent down-regulation of the LPS response. HEK293-TLR4/MD-2 cells in DMEM containing 1% FCS were treated with 0.05 to 5 mM β-methyl-cyclodextrin (β-CD) for 60 min at 37 °C, washed with DMEM containing 10% FCS, and stimulated with 50 nM LPS for 24 h. Data are shown as the means ± SEM of n = 7 independent experiments. White dots represent the individual data points. The statistical analyses (A–E) were performed using one-way ANOVA and Dunnett’s post test; *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, and ****P ≤ 0.0001 (peptide groups versus LPS control).
To determine the ability of LL-32 or PMB to inhibit proinflammatory signaling cascades not triggered by LPS via the TLR4/MD-2 pathway, we analyzed the effects of both peptides on cell activation mediated by IL-1β (Fig. 4C) and TNF-α (Fig. 4D). The inability of either peptide to prevent cytokine-induced cell activation suggests that the immunomodulatory mechanism specifically targets the TLR4/MD-2 signaling cascade. Notably, IL-8 production in response to IL-1β–mediated cell activation was even enhanced by LL-32 and PMB, and these peptides had only minor inhibitory effects on intracellular TNF-α levels (SI Appendix, Fig. S6A) or TNF-α secretion (SI Appendix, Fig. S6B). This observation is particularly important, as the IL-1 and TLR4 receptors share a conserved cytoplasmic domain, the Toll/IL-1 receptor homologous region, which recruits the intracellular signaling adaptor MyD88 upon receptor activation.
To differentiate the direct LPS-neutralizing and immunomodulatory effects of these peptides, we compared the biological responses of cells preincubated with peptides and those treated with LPS+peptides. Both LL-32 and PMB inhibited cell activation under both conditions (SI Appendix, Fig. S5 C and D). In contrast, the peptides NK-2, hBD-3-l, and Pep19-2.5 exhibited the most pronounced inhibitory effects only when preincubated with LPS (SI Appendix, Fig. S5C). A biophysical analysis of all investigated peptides revealed clear changes in the 3D structure of LPS as determined by the SAXS analysis of diffraction and AFM analysis of membrane organization (Fig. 3 C and D and SI Appendix, Fig. S2 B and C). These data demonstrate differences in the interactions of the peptides with LPS and with the host cell. Therefore, we suggest that LL-32 and PMB exhibit an anti-inflammatory effect via interactions with the host cell membrane, whereas other peptides neutralize LPS primarily via direct interactions.
LL-32 and PMB Interact with and Modify the Organization of Signaling Domains in the Host Cell Membrane.
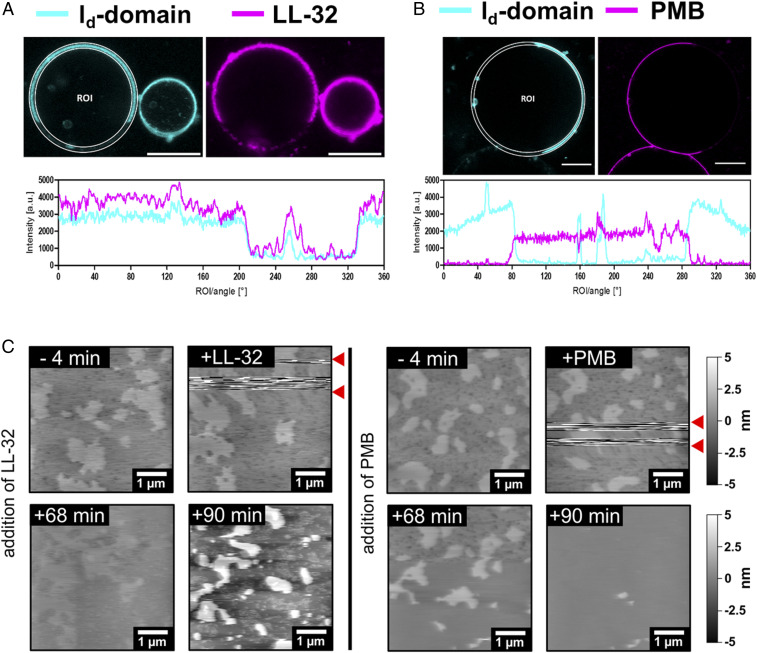
TLR4/MD-2 pathway signaling relies on the recruitment of the receptors into cholesterol-containing membrane domains (39, 40). We observed that the β-cyclodextrin–mediated depletion of cholesterol from HEK293-TLR4/MD-2 cells reduced the LPS-induced production of IL-8 (Fig. 4E). These data, together with data from the washing and preincubation experiments, led us to investigate the effects of LL-32 and PMB on cholesterol-containing cytoplasmic membrane domains, using the well-established dioleoylphosphatidylcholine (DOPC):sphingomyelin (SM):cholesterol (Chol) (2:2:1 M) vesicle model. We implemented giant unilamellar vesicles (GUVs) exhibiting phase separation to study the lateral distributions of the peptides. The low-cholesterol areas (i.e., liquid-disordered, ld domain) of the vesicles were highlighted by a fluorescent lipid–dye conjugate, whereas the cholesterol-rich domains (liquid-ordered, lo domain) appeared black due to exclusion of the dye. The addition of fluorescence-labeled peptides to the vesicles revealed that the peptides interact with the phospholipid membrane with different specificities; LL-32 (ld domains) and PMB (lo domains) favor opposite sites of interaction on the membrane (Fig. 5 A and B). The biological activity of the fluorescent peptides was only marginally reduced compared to the unconjugated peptides (SI Appendix, Fig. S6C). An evaluation of the fluorescence intensities of the fluid domain marker and peptides indicated positive and negative correlations of both signals for LL-32 and PMB, respectively.
Fig. 5.
LL-32 and PMB interact via opposing interaction sites on cholesterol-rich model membranes. (A and B) False-color presentation of (A) LL-32-Rho and (B) PMB-BODIPY on giant vesicles reconstituted from DOPC:SM:Chol (2:2:1 M). The ld domain (cyan) of the membrane was labeled using the lipid-dye conjugate PC-BODIPY or Atto633-DOPE and cholesterol-rich (lo-) domains appear black. Peptides were added at 4.5 μM LL-32-Rho or 22.5 μM PMB-BODIPY (magenta). (Scale bars, 10 μm.) The data are representative of n = 3 independent experiments. (C) AFM images of solid-supported bilayers of the macrophage mimetic lipid mixture PLMAK:SM:Chol (2:0.5:0.2 M). Bilayers were immobilized on mica and peptides were added to a final concentration of 25 µM. The peptides were added in between the two red arrows and were responsible for the visible disturbances. All AFM images were only flattened and not further processed. The presented images were obtained before and after the addition of peptide or buffer at the indicated times. The data are representative of n = 2 (LL-32) and n = 6 (PMB) independent experiments.
The effects of peptides on the lateral organization of membrane domains on solid-supported bilayers composed of DOPC:SM:Chol (9:9:2 M) was investigated by AFM. The image of pure membranes demonstrates that the lo domains are 0.74 ± 0.39 nm higher than the ld domains. LL-32 reduced the lo domain size, as indicated by the change in the ld/lo ratio from <1 to >1 after peptide addition (height histogram, SI Appendix, Fig. S7D) and by an increase in the interdomain height difference to 3.55 ± 0.85 nm. PMB only induced slight changes in the lo domain sizes and a marginal change in the domain height (0.82 ± 0.43 nm), indicating that the two peptides interact differently with the membrane. X-ray reflectivity (XRR) experiments on solid-supported membrane multilayers provide high resolution data on the membrane organization. XRR data confirm a thickening of ordered membrane domains from 4.68 ± 0.6 nm to 11.62 ± 0.61 nm (distance headgroup to headgroup) in the presence of LL-32 (SI Appendix, Fig. S7A) and show a stabilization of the domain structure by LL-32 even at a higher temperature of 40 °C (SI Appendix, Fig. S7B). The XRR data do not indicate changes in the membrane thickness in the presence of PMB, but they demonstrate effects of PMB on the domain structure, with more variability in membrane phases indicated by the broadening of the reflection peaks observed at 40 °C (SI Appendix, Fig. S7 A and B). Thermodynamic analyses of DOPC:SM:Chol membranes did not indicate that the interaction of PMB with the lipid system induces any relevant change in the phase transition enthalpy nor a change in the broadness of the phase transition (SI Appendix, Fig. S7C).
To account for the much more complex composition of biological membranes, we next investigated a lipid mixture closer resembling the lipid composition of macrophage membranes. AFM imaging of PLMAK:SM:Chol (2:0.5:0.2 M) membranes corroborated the results obtained for LL-32. This lipid mixture is much more fluid, less structured, and the cholesterol-containing domains are smaller. LL-32 first bound to the ld domains (Fig. 5C, 60 and 68 min) and then induced a dramatic change in the domain structure leading to smaller domains with an interdomain height difference of about 5.8 nm (Fig. 5C, 90 min). Furthermore, a clear interaction of PMB with cholesterol-containing domains could be observed with this lipid mixture leading to a time-dependent height increase of about 1 nm (Fig. 5C, 68 min) and a following change of the domain structure to smaller domains or complete disintegration (Fig. 5C, 90 min).
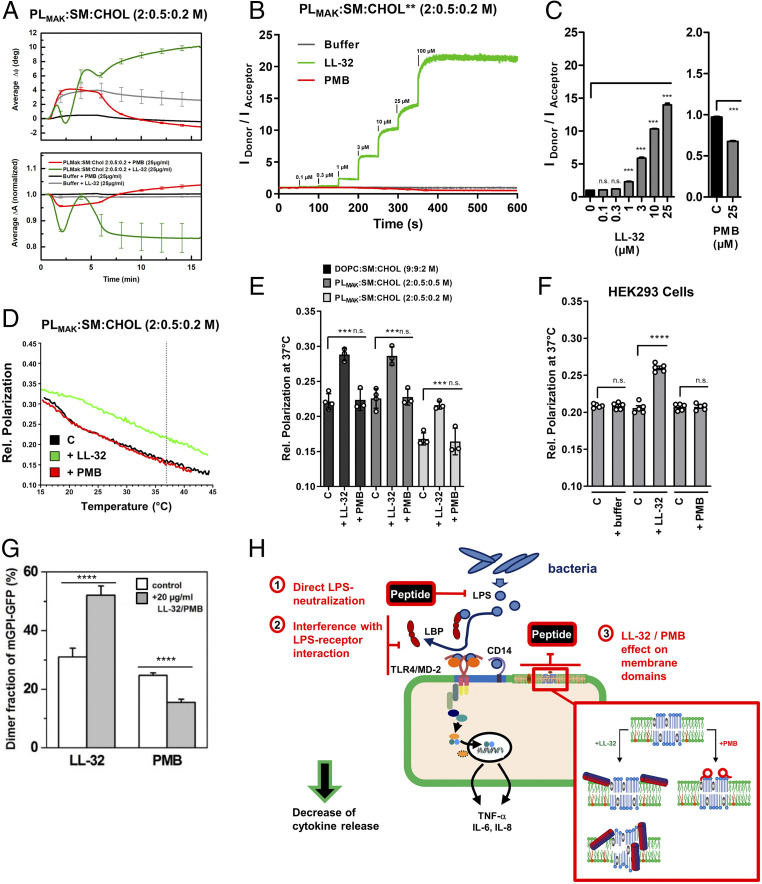
Consequently, we characterized the effects of LL-32 and PMB on eukaryotic membranes in more detail, now with a focus on the macrophage mimetic PLMAK membranes. Whereas LL-32 and PMB both demonstrate binding to PLMAK:SM:Chol membranes (Fig. 6A), the mode of interaction is different for the peptides. PMB dissociated from the membrane when the peptide loading was terminated at t = 6 min, whereas LL-32 remained membrane bound. Probing the membrane surface area by a Förster resonance energy transfer–based assay showed a dose-dependent increase of the membrane surface area consistent with membrane intercalation for LL-32, whereas no such effect was observed for PMB (Fig. 6B). Analysis of several titration experiments shows a clear increase of the membrane area for LL-32 at biologically relevant doses starting at 1 µM concentration. For PMB, a significant reduction in membrane surface area is observed, supporting the conclusion that PMB binds but does not intercalate into the membrane leaflets (Fig. 6C). In line with these results, LL-32 induced a rigidification of the lipid acyl chains over a broad range of temperatures in DOPC:SM:Chol membranes and in PLMAK membranes (Fig. 6D), with a significant reduction of membrane fluidity at the physiological temperature 37 °C (Fig. 6E). In contrast, PMB did not affect the membrane fluidity in any of the lipid systems. Investigation of biological membranes using the HEK293 cell line confirmed these results also in living cells (Fig. 6F).
Fig. 6.
LL-32 and PMB differentially interact with complex eukaryotic membranes and influence the homo-association of GPI-anchored proteins in the exoplasmic leaflet of the plasma membrane. (A) Binding of peptides to immobilized PLMAK:SM:Chol (2:0.5:0.2 M) membranes was determined by surface acoustic wave measurements. The graph presents the means ± SEM of n = 3 independent experiments. (B) Changes in the membrane surface area were analyzed by a Förster resonance energy transfer–based assay. PLMAK:SM:Chol (2:0.5:0.2 M) liposomes containing *NBD-PE (donor) and *Rh-DHPE (acceptor) were diluted to 10 µM. Peptides were added at the indicated final concentrations and signals recorded for 50 s after each titration step. Ratios IDonor/IAcceptor were calculated. Data represent the means of n = 3 independent measurements. (C) Ratios IDonor/IAcceptor after peptide titration from experiments displayed in B on PLMAK:SM:Chol (2:0.5:0.2 M) liposomes. Data are values at 50 s after addition of peptides and display means ± SEM of n = 3 independent experiments. Data were analyzed by two-tailed t test of paired samples (peptide versus control); ***P ≤ 0.001, n.s. not significant. (D) PLMAK:SM:Chol (2:0,5:0,2 M) liposomes were labeled with DPH, and membrane fluidity was determined as relative polarization of the fluorescence emission of DPH during temperature scans in the presence or absence of peptides at 25 µM final concentration. Data are representative of n = 3 independent experiments. (E) Relative polarization data of DOPC:SM:Chol (9:9:2 M) or PLMAK:SM:Chol (2:0.5:0.5 and 2:0,5:0,2 M) membranes at 37 °C from experiments performed as outlined in D. Data are mean ± SEM of n = 3 to 4 independent experiments. Data were analyzed by two-tailed t test of unpaired samples (peptide or buffer versus control); ***P ≤ 0.001, n.s. not significant. (F) HEK293 wild-type cells were labeled with DPH and measured at 0.2*106 cells/mL at 37 °C. Baseline signal was recorded for 50 s, then peptides were added to a final concentration of 25 µM and signals recorded until 300 s. Data are means ± SEM at t = 50 s (C) and t = 300 s (+ buffer/peptide) of n = 5 independent experiments. Data were analyzed by two-tailed t test of paired samples (peptide or buffer versus control); ****P ≤ 0.0001, n.s. not significant. (G) Single-molecule TOCCSL (“Thinning Out Clusters while Conserving Stoichiometry of Labeling”) experiments were performed to determine the degree of mGFP-GPI homo-association in the plasma membranes of living CHO cells. The addition of both the LL-32 and PMB peptides caused a substantial change in mGFP-GPI homo-association, as demonstrated by an increase (LL-32, n = 10 cells) and decrease (PMB, n = 8 cells) in the mGFP-GPI dimer fraction. Data are shown as the mean ± SEM. Statistical analysis was performed using a two-sample Kolmogorov–Smirnov test; ****P ≤ 0.0001 (peptide versus control). (H) Model of the modes of LL-32 and PMB function. The peptides have different levels of interaction that lead to the neutralization of inflammatory cell activation by LPS: 1) direct interaction with LPS, leading to biophysical changes and a biologically less active LPS structure; 2) interference with the interaction between LPS and transport proteins in serum (i.e., LBP, soluble CD14) or cellular LPS receptor proteins (membrane-bound CD14, TLR4/MD-2); and 3) reorganization of cholesterol-containing membrane domains. Enlargement box of the host cell cytoplasmic membrane depicts the differential mechanisms of membrane interaction as observed for LL-32 (Left; step I model is based on the experimental data, step II model is the suggested most likely biophysical model to explain the data) and PMB (Right) membrane interaction.
To obtain evidence for the consequences of LL-32 and PMB membrane interaction on the organization of cholesterol-rich domains in living cells, we used a monomeric green-fluorescent protein (mGFP) attached to the outer leaflet of the plasma membrane of CHO cells via a glycosylphosphatidylinositol (GPI) anchor as a model system for cholesterol-dependent protein interactions (41). The fraction of mGFP-GPI homo-associates (α2) was changed highly significant after peptide addition, showing an increase after the addition of LL-32 (from 31 ± 3% to 53 ± 3%) and a decrease after addition of PMB (from 25 ± 1% to 16 ± 1%) (Fig. 6G), consistent with the data from the reconstituted membrane systems. Thus, we conclude that LL-32 binds to the ld domains and induces a domain-condensing effect, while PMB binds to the lo domains and induces the spreading of molecules associated with cholesterol-rich domains.
Discussion
Severe inflammatory diseases such as sepsis, acute respiratory distress syndrome, CD, CF, COPD, and asthma demonstrate the impact of immune dysregulation on disease development and progression. In gram-negative infectious diseases, the neutralization of LPS and regulation of the anti-inflammatory immune response are as important as the cytotoxic effect on bacteria. Considering this, the combined antibacterial and anti-inflammatory effects of AMPs emerge as an example of the perfect adaptation of innate immunity to these requirements.
Here, we present a mechanism of immune modulation by members of the cathelicidin family, namely LL-32, LL-37, CAP18, CRAMP, BMAP-27/28, and the peptide antibiotic PMB, in human macrophages and human HEK293 cells. Our findings strongly support a multifaceted nature of the anti-inflammatory activities of these peptides (Fig. 6H). Our cellular and biophysical investigations of the mechanism underlying this host cell–based immunomodulatory response identified the cytoplasmic membrane as a target of the peptides. Our DOPC:SM:Chol and macrophage-mimicking PLMAK model membrane experiments revealed the modes of interaction of LL-32 and PMB with the lipid matrix of the cytoplasmic membrane. In-depth biophysical analysis of the structural and biophysical effects of the peptides on membrane domains revealed differential mechanisms of interaction leading to subsequent changes in the membrane domain organization: LL-32 preferentially interacts with liquid-disordered domains and induces a subsequent reduction of overall membrane fluidity as well as condensation and thickening of cholesterol-containing membrane domains. We assume that this latter step is accompanied by diffusion of LL-32 into the cholesterol-containing domains, since this is currently the best model to explain the observed membrane thickening. PMB binds preferentially to the headgroup region of the liquid-ordered membrane domains. The biophysical effects are less obvious but clearly include conversion of ordered membrane domains to smaller size as observed for the PLMAK membranes, but without affecting the overall membrane fluidity. The concentrations applied were above those of endogenous AMPs in serum and below any cytotoxic concentrations. With that, they are in the therapeutic concentration range.
The homo-association of GPI-anchored protein is a hallmark of confinement within cholesterol-dependent nanodomains (41). The LL-32–mediated increase in mGFP-GPI homo-association in the plasma membranes of live cells is consistent with the condensing effect of this peptide on cholesterol-containing domains in phase-separated model membranes and reflects the direct dependence of protein organization on the lipid environment. Vice versa, the direct interaction of PMB with cholesterol-dependent membrane domains reduced the homo-association of mGFP-GPI in live cells. Thus, despite differences in domain specificity, we identified a common mechanism by which both peptides mediated changes in protein organization and association in cholesterol-dependent domains.
LPS-signaling complex activation is closely associated with dynamic TLR4 recruitment to cholesterol-containing domains upon LPS-binding (39, 40). This process leads to TLR4/MD-2 homodimer formation (42, 43) and active signal transduction (29, 44–46). Based on our findings, we propose the following mechanistic scenarios: The peptides 1) may hinder recruitment of the TLR4 receptor to the condensed cholesterol-containing domains in which GPI-anchored proteins are present at a higher degree of association or 2) may hinder signaling complex assembly by reducing the homo-association of GPI-anchored LPS receptor protein CD14 or its association with TLR4, leading to an increased activation threshold. Neither mechanism relies on a particular peptide receptor, as was demonstrated previously for some biological functions of LL-37 (47). The LPS receptor system is among the most stringently regulated innate immune receptors and employs several sensitization and deactivation circuits. The membrane-based regulation of LPS activation by cathelicidin AMPs and PMB described in this work is an additional anti-inflammatory mechanism.
In this study, the inability of LL-32 and PMB to inhibit IL-1β– and TNF-α–induced cell activation demonstrates the strong specificity of this mechanism for LPS signal transduction. Although IL-1β and TNF-α receptor activation has been linked to lipid raft domains, this process does not involve the degree of dynamic receptor protein association as described above for TLR4 receptor activation. This difference may explain the lesser effects of membrane disturbances on IL-1β and TNF-α receptor signaling. IL-1 receptor type 1 (48) and TNF receptor type 1 (TNFR1) are constitutively present in lipid rafts, and a study of murine macrophages revealed that TNFR1 NF-κB–signaling is not sensitive to lipid raft manipulation (49). Moreover, our data, in which LL-32 and PMB only moderately interfere with cytokine-induced cell activation, demonstrate that peptide-mediated immunomodulation does not result from a general suppression of the host immune cell response. Previous observations regarding IL-1β– and TNF-α–signaling in human peripheral blood mononuclear cells (PBMCs) support our findings (50, 51). Immunosuppression during the later phases of hyper-inflammatory diseases, such as systemic infection and sepsis, is a critical driver of immune pathology (52, 53). The signaling specificity reported in our work is important to the consideration of AMPs and PMB as potential antibacterial or anti-inflammatory therapeutic agents.
Previous studies of the interactions of AMPs with lipid bilayers have identified membrane lesions or pore formation as the basis for antimicrobial activity (16). The negative aspect of this mode of action is reflected by the cytotoxic effects of numerous AMPs at higher concentrations. Notably, the peptides used in this study exhibited low or no cytotoxic effects on human macrophages, HEK293, and red blood cells in vitro. We thus conclude that host cell–directed membrane interaction represents a relevant biological function as opposed to the harmful effects of cytotoxicity at higher concentrations of peptides. Our data also demonstrate that the peptides exert varied effects on different domain-associated signaling cascades. We therefore assume the existence of various types of membrane domains.
The further development of AMPs or polymyxin-based compounds as drugs for clinical use will require more detailed knowledge about the modes of action of these peptides (54). Their potential uses as antimicrobial agents are highly apparent, as demonstrated by the recently described new class of PMB-derived peptidomimetics that exhibit high potential for the treatment of resistant gram-negative pathogens of the ESKAPE group (55). Our discovery of a peptide-mediated mechanism of immune control adds an important aspect for the development and clinical use of antibiotic peptides.
In conclusion, our findings reveal that the interactions of cathelicidin AMPs and PMB with lipid bilayers not only provide the basis for the antimicrobial activities of these peptides against bacterial membranes but also support the host-directed modulation of the inflammatory responses of immune cells. This latter function may be important in the context of acute hyper-inflammatory responses, such as bacterial sepsis, and may also be applicable to chronic hyper-inflammatory diseases induced by recurrent infections, such as COPD or CF. We have demonstrated that both LL-32 and PMB confer LPS neutralization via three actions: 1) modification of the agonistic LPS conformation to an antagonistic conformation, 2) detection of LPS in serum by LBP, and 3) modification of the receptor domain. This multitargeted function likely explains the observed high level of activity and broad-spectrum, LPS-neutralizing activity observed in vivo. Further studies of the host-directed functions are needed to elucidate fully the physiological impacts and therapeutic potential of AMPs and peptide antibiotics as anti-inflammatory immune response modifiers.
Materials and Methods
Reagents.
Peptides were synthesized and purified by high-performance liquid chromatography. The purity of each peptide was >95%. Peptide identity was confirmed by mass spectrometry. The peptide sequences are presented in SI Appendix, Table S1. For fluorophore-labeling of peptides, lipids and dyes, refer to SI Appendix.
LPS Aggregate Preparation.
Deep-rough type LPS Re was extracted from Escherichia coli strain WBB01 grown at 37 °C (56). After extraction via the phenol/chloroform/petrol ether method, LPS was purified and lyophilized (57). LPS aggregate dispersions were prepared in 20 mM Hepes, 150 mM NaCl, pH 7.4, or water by applying a pulsed ultrasound (Ultrasonic-Homogenizer HTU Soni130, 1 min, pulse on/off: 2 s, amplitude 30%) followed by three rounds of thermocycling between 4 °C and 56 °C for 30 min each and stored overnight at 4 °C before use.
Macrophage Model Membranes.
The lipid mixture resembling the composition of macrophage membranes (PLMAK) was prepared to a final molar ratio [PC:PS:PE]:SM = 1:0.4:0.7:0.5 M + cholesterol 0.5 M or 0.2 M. DOPC:SM:Chol model membranes contained molar ratios of 9:9:2 M or 2:2:1 M. Details of the liposome preparation are given in SI Appendix.
Animal Model of Endotoxicity.
Seven-week-old female C57BL/6 mice (Harlan Interfauna Iberica S.A.) were distributed randomly in experimental groups (n = 8 per group). Endotoxemia was induced by intraperitoneal coadministration of LPS and D-galactosamine (18 mg/mouse), a compound that sensitizes animals to LPS (58). LPS was prepared as described above, dissolved in endotoxin-free saline and injected at 100 ng/mouse (LD90). Immediately after the LPS injection, animals received an injection of 100 or 50 µg peptide in 150 µl of pyrogen-free saline (control group) at a different peritoneal site. Survival was monitored at daily intervals for 96 h. Survival plots were compared statistically using the log-rank test, whereas intersecting plots were compared using the Breslow–Gehan–Wilcoxon test. All P values represent comparisons of mortality data from the same experiment (treated versus untreated mice). All mouse experiments were approved by the University of Navarra Animal Research Committee (Permission 069/09). Animal experiments were assessed without blinding of the treatment group identity.
Stimulation of Human Macrophages by LPS.
Human mononuclear cells (MNC) from healthy donors were isolated from heparinized peripheral blood. The experimental use of MNC was approved by the Ethical Commission of the University of Lübeck (12-202A). All volunteer donors provided informed consent prior to the procedure. MNCs were harvested, and the monocytes were differentiated to macrophages. For stimulation experiments, macrophages were seeded, incubated with peptides for 30 min at 37 °C, and were subsequently washed three times to remove non–cell-bound peptide or stimulated directly with LPS for 4 h at 37 °C. Cell-free supernatants were analyzed for TNF-α. For further experimental details and the method for detection of intracellular TNF-α protein, refer to SI Appendix.
Cell Lines.
Wild-type HEK293 cells were maintained in Dulbecco's Modified Eagle Medium (DMEM) containing 10% low–endotoxin-grade fetal calf serum (FCS, Linaris), 100 U/mL penicillin, and 100 μg/mL streptomycin. The HEK293-TLR4/MD-2 cell line was described earlier (59). For experiments, HEK293-TLR4/MD-2 were seeded into 96-well plates. The peptides were added to the wells at the indicated concentrations. After a 30-min incubation at 37 °C, the cells were washed three times to remove free peptide or stimulated directly with LPS, IL-1β, or TNF-α (PeproTec) for 24 h at 37 °C. Cell-free supernatants were analyzed for IL-8. For experimental details and cholesterol-depletion experiments, refer to SI Appendix.
CHO cells (ATTC #CCL-61) that had been stably transfected with mGFP-GPI (41) were grown in DMEM/F12 medium (PAA Laboratories) supplemented with 10% FCS (PAA Laboratories) and 400 µg/mL G418 (PAA Laboratories).
qRT-PCR.
Human macrophages were seeded in 96-well plates at a density of 5 × 105 cells/well, incubated with peptides or control medium for 30 min at 37 °C, and subsequently stimulated with LPS. After 1 h of stimulation, the cells were harvested, RNA extracted, and analyzed by qrt-PCR as outlined in SI Appendix.
Studies of Peptide and LPS-Binding to Human Macrophages.
Human macrophages were seeded in flow cytometry tubes at a density of 105 cells/tube. After adding peptides in phosphate buffered saline (PBS) containing 2% FCS at the indicated concentrations, the cells were incubated at 4 °C or 37 °C for 5, 15, or 30 min. Subsequently, the cells were washed in ice-cold PBS with 2% FCS and azide (azide-PBS, 2% FCS), fixed in 2% paraformaldehyde, washed, and resuspended in 1 mL of azide-PBS and 2% FCS. For the fluorophore-quenching analysis, the samples were split, one aliquot was pelleted by centrifugation and resuspended in 0.2% trypan blue in 0.75% NaCl directly prior to measurement. To study the effects of peptides on LPS-binding, the macrophages were incubated with FITC-conjugated LPS in the presence or absence of peptides for 5, 15, or 30 min and subsequently washed and fixed. All samples were analyzed on a flow cytometer.
Confocal Microscopic Analysis of Human Macrophages.
Human macrophages were seeded in µ-Slides VI (Ibidi) at a density of 2 × 104 cells/well and allowed to adhere for 24 h in an atmosphere of 37 °C and 5% CO2. LPS was labeled 10:1 M with rhodamine-DHPE (Invitrogen) in chloroform/methanol and prepared as described above. The cells were incubated with rhodamine-labeled LPS aggregates in the presence of 1, 3, or 10 µM LL32 in PBS for 5 min at room temperature. Subsequently, the cells were washed with PBS, fixed with 4% paraformaldehyde, and the nuclei were counterstained with Hoechst (Invitrogen). The samples were analyzed using a Leica TCS SP5 confocal laser-scanning microscope (Leica Microsystems), and all images were acquired using Leica LAS AF software with identical settings.
SAXS.
X-ray scattering analyses of LPS in the presence and absence of peptides were performed using a SAXS camera equipped with a linear position-sensitive detector (HECUS X-ray Systems). The camera was mounted on a sealed-tube X-ray generator (Seifert), which was operated at 2 kW. CuKα radiation (λ = 1.542 Å) was selected using a Ni filter and a pulse height discriminator. Silver stearate was used to perform the angular calibration of the scattered intensities. LPS dispersions (50 mg/mL) or LPS:peptide mixtures (2:1 by weight) were prepared in 20 mM Hepes (pH 7.0) as described for the preparation of LPS aggregates. The samples were measured using a thin-walled quartz capillary (diameter, 1 mm) in a steel cuvette (Anton Paar) that had been inserted into a brass block. Temperature control was provided by a programmable Peltier unit. After a 10-min equilibration period, scattering data for the small-angle region were recorded for each sample with an exposure time of 1 h.
Aggregate Size and Zeta Potential Measurements.
The size of LPS aggregates was measured by dynamic light scattering using a ZetaSizer Nano device (Malvern Instruments). LPS aggregates at 1 µM in 20 mM Hepes, 150 mM NaCl, and pH 7.4 were equilibrated for 3 min to 37 °C. Peptides were added consecutively to final concentrations of 1 to 100 µM, and the aggregate size was determined by triplicate measurements. The data represent the means and ± SEM of ≥4 independent experiments. The Zeta potentials of LPS aggregates at 2 µM in 20 mM Hepes (pH 7.0) were measured at 25 °C. Peptides were added to the indicated LPS to peptide molar ratio. The velocity (v) of the LPS aggregates in a driving electric field with an effective voltage of 152 V was measured via dynamic light scattering, and the corresponding electrophoretic mobilities (v/E) were calculated. The associated Zeta potentials were calculated using the Smulochowski approximation.
Analysis of the LBP–LPS Interaction by Ultracentrifugation.
The LBP–LPS interaction was studied using samples of LPS aggregates that had been incubated with LBP in the absence or presence of peptides. LPS (4.55 µM) was incubated with recombinant human LBP (XOMA) at a molar ratio of 100:1 for 30 min at room temperature in tubes that had been previously blocked for 1 h at 37 °C with 10% bovine serum albumin (BSA) (wt/vol) in 20 mM Hepes. Peptides were added to the LPS aggregates at the indicated molar ratios before the addition of LBP. LPS aggregates were sedimented by ultracentrifugation at 117,000 g for 1 h at 4 °C. Supernatant and pellet fractions were collected, separated by 12% sodium dodecylsulfate–polyacrylamide gel electrophoresis (SDS-PAGE), and transferred to nitrocellulose membranes. The membranes were incubated with an anti-LBP antibody (biG 42, 1:4,000, Biometec) and goat anti-mouse IgG-HRP (1:10.000, Jackson ImmunoResearch). Immunolabeled proteins were visualized by ECL Plus Western blotting detection system (GE Healthcare). Band intensities were quantified using ImageJ 1.45S analysis software (NIH).
Confocal Microscopic Analysis of GUVs.
GUVs were prepared from DOPC:SM:Chol 2:2:1 M by electroformation as described (60). For microscopy analysis, GUVs were immobilized to microscopy slides and fluorophore-labeled peptides added in solution (for experimental details, refer to SI Appendix). Fluorescence dye distribution was detected using a Leica TCS SP5 confocal laser-scanning microscope (Leica Microsystems).
AFM.
The deposition of peptides on membranes was investigated using an MPF3D atomic force microscope (Asylum Research). LPS, DOPC:SM:Chol, and PLMAK membranes were prepared by allowing vesicles to spread on mica plates (1 cm2) and were imaged in 2 to 3 mL of buffer at 23 °C. The final LPS or lipid concentration was 25 µg/mL or 25 µM, respectively. The buffer, including any unbound LPS/lipids, was replaced prior to the addition of peptides at final concentrations of 25 µM during imaging. RC800PSA cantilevers (Olympus; typical spring constant: k ∼0.1 N·m−1) or qp-BioAC (Nanosensors; typical spring constant: k ∼0.1 N·m−1) were used in the oscillating mode. Images were processed in MFP-3D using IGOR Pro.
Surface Acoustic Wave Biosensor.
Measurements were performed at 22 °C using functionalized gold-coated chips (S-sens K5 Biosensor Quartz Chips, SAW [Surface Acoustic Wave] Instruments GmbH). Biomolecular interaction processes on the surface of the sensor chip can affect phase and amplitude of the surface-guided acoustic wave. Following the immobilization of liposomes (500 μM) on the sensor chip surface, 100 µl of 25 µM solution of LL-32 or PMB were injected, and phase and amplitude were recorded over time.
Fluorescence Polarization Experiments.
DOPC:SM:Chol (9:9:2 M) or PLMAK:SM:Chol (2:0.5:0.5 M and 2:0.5:0.2 M) liposomes at 1 mM were labeled at 0.5% (vol/vol) with 2 mM 1,6-diphenyl 1,3,5 hexatriene (DPH, Fluka). Temperature scans were performed at a heating rate of 1 °C/min in a stirred cuvette in a Fluorolog SPEX (Jobin Yvon Incorporated). Excitation light was polarized and emission analyzed parallel and perpendicular to the excitation light. Relative polarization of DPH emission was calculated according to the equation P = (I| - I⊥)/(I| + I⊥). For analysis of biological membranes, wild-type HEK293 cells were labeled with DPH at 0.05% (vol/vol). Experiments were performed at 37 °C as time scans with 50 s of background measurement and subsequent addition of buffer or peptides.
Förster Resonance Energy Transfer Assay.
PLMAK:SM:Chol (2:0.5:0.2 M) liposomes labeled with *NBD-PE (donor) and *Rh-DHPE (acceptor) at 100:1:1 M were diluted to 10 µM in 20 mM Hepes, 150 mM NaCl, and pH 7.4. Measurements were performed at 37 °C on a Fluorolog-3 (Jobin Yvon Incorporated). The fluorescence intensities IDonor and IAccceptor were adjusted to equal intensities (ratio = 1) before the measurement and recorded for 50 s to obtain the baseline signal. Peptides were added at the indicated final concentrations and signals recorded for 50 s after each titration step. The ratios IDonor/IAcceptor were calculated, with a ratio >1 indicating an increase and a ratio <1 indicating a decrease in membrane surface area.
Single-Molecule Fluorescence Microscopy.
TOCCSL (“Thinning Out Clusters while Conserving Stoichiometry of Labeling”), a single-molecule fluorescence modality (41, 61), was used to evaluate the mGFP-GPI homo-association on the plasma membranes of living CHO cells. Briefly, an Axiovert 200 microscope equipped with a 100x Plan-Apochromat objective (NA = 1.46; Zeiss) was used to illuminate samples in an objective-based total internal reflection configuration via the epiport. After recording a prebleach image at a power density of 2 kW/cm2 and an illumination time of 1 ms, the samples were bleached. After a recovery period of 600 to 2,400 ms, sequences of up to 10 images at a delay of 20 ms were recorded. The first image after recovery was used to analyze the brightness of individual mGFP-GPI homo-associates, while the last image of the sequence was used to determine the reference brightness of a single mGFP molecule. Because only a small area of the cell was photobleached, multiple bleach and recovery runs could be performed on a single cell. All experiments were performed at 37 °C. For further experimental details and data analysis, refer to SI Appendix.
Quantification and Statistical Analysis.
The statistical analysis was performed using GraphPad Prism, versions 5 and 9 or Matlab. Details of the analyses are provided in the respective figure legends. A P value ≤0.05 (*) was considered significant. No specific randomization method was used when handling samples or during experiments.
Supplementary Material
Acknowledgments
We are grateful for the excellent technical assistance provided by Sabrina Groth, Irina von Cube, Christine Hamann, Kerstin Stephan, and our technician trainees. We thank the Bioanalytical Chemistry Group for mass spectrometry of synthetic peptides; confocal microscopy and flow cytometry were performed on instruments of the Flow Cytometry Core Unit. SAXS measurements were performed at the Institute of Molecular Biosciences, University of Graz, and the European Molecular Biology Laboratory (EMBL) beamlines P08 and P12 at the Deutsches Elektronen-Synchrotron, Hamburg (beamtime granted to A.B.S., K.B., and T.G). This project was supported by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation), Cluster of Excellence 306/2 “Inflammation at Interfaces” (Excellence Initiative, Germany, since 2006, DFG-Project No. 49701054), Grant Exc306 RA3 to A.B.S. and T.G. G.M.T. was funded by grants from Proyectos de Investigación Universidad de Navarra (PIUNA-P2011-17 and P2015-14) and by a grant from Ministerio de Sanidad y Consumo (FIS-PI050768), Spain.
Footnotes
Competing interest statement: K.B. holds a patent for Aspidasept® and is the Chief Executive Officer of Brandenburg Antiinfectiva GmbH.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2101721118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix.
References
- 1.Kovach M. A., et al., Cathelicidin-related antimicrobial peptide is required for effective lung mucosal immunity in Gram-negative bacterial pneumonia. J. Immunol. 189, 304–311 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McHugh B. J., et al., Cathelicidin is a “fire alarm”, generating protective NLRP3-dependent airway epithelial cell inflammatory responses during infection with Pseudomonas aeruginosa. PLoS Pathog. 15, e1007694 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang L. J., et al., Age-related loss of innate immune antimicrobial function of dermal fat is mediated by transforming growth factor beta. Immunity 50, 121–136.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.WHO , Antimicrobial Resistance: Global Report on Surveillance (World Health Organization, 2014), pp. 1–232. [Google Scholar]
- 5.U.S. Department of Health and Human Services , Antibiotic Resistance Threats in the United States, 2013 (U.S. Department of Health and Human Services, Centers for Disease Control and Prevention, 2013), pp. 1–114. [Google Scholar]
- 6.Lazzaro B. P., Zasloff M., Rolff J., Antimicrobial peptides: Application informed by evolution. Science 368, eaau5480 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hancock R. E., Nijnik A., Philpott D. J., Modulating immunity as a therapy for bacterial infections. Nat. Rev. Microbiol. 10, 243–254 (2012). [DOI] [PubMed] [Google Scholar]
- 8.Mansour S. C., Pena O. M., Hancock R. E., Host defense peptides: Front-line immunomodulators. Trends Immunol. 35, 443–450 (2014). [DOI] [PubMed] [Google Scholar]
- 9.Hancock R. E., Haney E. F., Gill E. E., The immunology of host defence peptides: Beyond antimicrobial activity. Nat. Rev. Immunol. 16, 321–334 (2016). [DOI] [PubMed] [Google Scholar]
- 10.Cohen J., The immunopathogenesis of sepsis. Nature 420, 885–891 (2002). [DOI] [PubMed] [Google Scholar]
- 11.Haney E. F., Straus S. K., Hancock R. E. W., Reassessing the host defense peptide landscape. Front Chem. 7, 43 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wehkamp J., et al., Reduced Paneth cell alpha-defensins in ileal Crohn’s disease. Proc. Natl. Acad. Sci. U.S.A. 102, 18129–18134 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Turner J., Cho Y., Dinh N. N., Waring A. J., Lehrer R. I., Activities of LL-37, a cathelin-associated antimicrobial peptide of human neutrophils. Antimicrob. Agents Chemother. 42, 2206–2214 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beisswenger C., et al., Allergic airway inflammation inhibits pulmonary antibacterial host defense. J. Immunol. 177, 1833–1837 (2006). [DOI] [PubMed] [Google Scholar]
- 15.Hiemstra P. S., Amatngalim G. D., van der Does A. M., Taube C., Antimicrobial peptides and innate lung defenses: Role in infectious and noninfectious lung diseases and therapeutic applications. Chest 149, 545–551 (2016). [DOI] [PubMed] [Google Scholar]
- 16.Lohner K., Membrane-active antimicrobial peptides as template structures for novel antibiotic agents. Curr. Top. Med. Chem. 17, 508–519 (2017). [PubMed] [Google Scholar]
- 17.Lohner K., Blondelle S. E., Molecular mechanisms of membrane perturbation by antimicrobial peptides and the use of biophysical studies in the design of novel peptide antibiotics. Comb. Chem. High Throughput Screen. 8, 241–256 (2005). [DOI] [PubMed] [Google Scholar]
- 18.Hancock R. E., Sahl H. G., Antimicrobial and host-defense peptides as new anti-infective therapeutic strategies. Nat. Biotechnol. 24, 1551–1557 (2006). [DOI] [PubMed] [Google Scholar]
- 19.Yang De, et al., LL-37, the neutrophil granule- and epithelial cell-derived cathelicidin, utilizes formyl peptide receptor-like 1 (FPRL1) as a receptor to chemoattract human peripheral blood neutrophils, monocytes, and T cells. J. Exp. Med. 192, 1069–1074 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bowdish D. M. E., Davidson D. J., Speert D. P., Hancock R. E. W., The human cationic peptide LL-37 induces activation of the extracellular signal-regulated kinase and p38 kinase pathways in primary human monocytes. J. Immunol. 172, 3758–3765 (2004). [DOI] [PubMed] [Google Scholar]
- 21.Mookherjee N., et al., Intracellular receptor for human host defense peptide LL-37 in monocytes. J. Immunol. 183, 2688–2696 (2009). [DOI] [PubMed] [Google Scholar]
- 22.Heinbockel L., et al., Preclinical investigations reveal the broad-spectrum neutralizing activity of peptide Pep19-2.5 on bacterial pathogenicity factors. Antimicrob. Agents Chemother. 57, 1480–1487 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Domingues M. M., et al., Biophysical characterization of polymyxin B interaction with LPS aggregates and membrane model systems. Biopolymers 98, 338–344 (2012). [DOI] [PubMed] [Google Scholar]
- 24.Beutler B., Rietschel E. T., Innate immune sensing and its roots: The story of endotoxin. Nat. Rev. Immunol. 3, 169–176 (2003). [DOI] [PubMed] [Google Scholar]
- 25.Poltorak A., et al., Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: Mutations in Tlr4 gene. Science 282, 2085–2088 (1998). [DOI] [PubMed] [Google Scholar]
- 26.Hoshino K., et al., Cutting edge: Toll-like receptor 4 (TLR4)-deficient mice are hyporesponsive to lipopolysaccharide: Evidence for TLR4 as the Lps gene product. J. Immunol. 162, 3749–3752 (1999). [PubMed] [Google Scholar]
- 27.Shimazu R., et al., MD-2, a molecule that confers lipopolysaccharide responsiveness on Toll-like receptor 4. J. Exp. Med. 189, 1777–1782 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schromm A. B., et al., Molecular genetic analysis of an endotoxin nonresponder mutant cell line: A point mutation in a conserved region of MD-2 abolishes endotoxin-induced signaling. J. Exp. Med. 194, 79–88 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gay N. J., Symmons M. F., Gangloff M., Bryant C. E., Assembly and localization of Toll-like receptor signalling complexes. Nat. Rev. Immunol. 14, 546–558 (2014). [DOI] [PubMed] [Google Scholar]
- 30.Schumann R. R., et al., Structure and function of lipopolysaccharide binding protein. Science 249, 1429–1431 (1990). [DOI] [PubMed] [Google Scholar]
- 31.Wright S. D., Ramos R. A., Tobias P. S., Ulevitch R. J., Mathison J. C., CD14, a receptor for complexes of lipopolysaccharide (LPS) and LPS binding protein. Science 249, 1431–1433 (1990). [DOI] [PubMed] [Google Scholar]
- 32.Gioannini T. L., et al., Isolation of an endotoxin-MD-2 complex that produces Toll-like receptor 4-dependent cell activation at picomolar concentrations. Proc. Natl. Acad. Sci. U.S.A. 101, 4186–4191 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gutsmann T., et al., New antiseptic peptides to protect against endotoxin-mediated shock. Antimicrob. Agents Chemother. 54, 3817–3824 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Uppu D. S., Haldar J., Lipopolysaccharide neutralization by cationic-amphiphilic polymers through pseudoaggregate formation. Biomacromolecules 17, 862–873 (2016). [DOI] [PubMed] [Google Scholar]
- 35.Andrä J., Gutsmann T., Garidel P., Brandenburg K., Mechanisms of endotoxin neutralization by synthetic cationic compounds. J. Endotoxin Res. 12, 261–277 (2006). [DOI] [PubMed] [Google Scholar]
- 36.Rathinam V. A. K., Zhao Y., Shao F., Innate immunity to intracellular LPS. Nat. Immunol. 20, 527–533 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schromm A. B., et al., The charge of endotoxin molecules influences their conformation and IL-6-inducing capacity. J. Immunol. 161, 5464–5471 (1998). [PubMed] [Google Scholar]
- 38.Garidel P., et al., Divalent cations affect chain mobility and aggregate structure of lipopolysaccharide from Salmonella minnesota reflected in a decrease of its biological activity. Biochim. Biophys. Acta 1715, 122–131 (2005). [DOI] [PubMed] [Google Scholar]
- 39.Triantafilou M., Miyake K., Golenbock D. T., Triantafilou K., Mediators of innate immune recognition of bacteria concentrate in lipid rafts and facilitate lipopolysaccharide-induced cell activation. J. Cell Sci. 115, 2603–2611 (2002). [DOI] [PubMed] [Google Scholar]
- 40.Triantafilou M., Morath S., Mackie A., Hartung T., Triantafilou K., Lateral diffusion of Toll-like receptors reveals that they are transiently confined within lipid rafts on the plasma membrane. J. Cell Sci. 117, 4007–4014 (2004). [DOI] [PubMed] [Google Scholar]
- 41.Brameshuber M., et al., Imaging of mobile long-lived nanoplatforms in the live cell plasma membrane. J. Biol. Chem. 285, 41765–41771 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Saitoh S., et al., Lipid A antagonist, lipid IVa, is distinct from lipid A in interaction with Toll-like receptor 4 (TLR4)-MD-2 and ligand-induced TLR4 oligomerization. Int. Immunol. 16, 961–969 (2004). [DOI] [PubMed] [Google Scholar]
- 43.Park B. S., et al., The structural basis of lipopolysaccharide recognition by the TLR4-MD-2 complex. Nature 458, 1191–1195 (2009). [DOI] [PubMed] [Google Scholar]
- 44.Motshwene P. G., et al., An oligomeric signaling platform formed by the Toll-like receptor signal transducers MyD88 and IRAK-4. J. Biol. Chem. 284, 25404–25411 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sun Y., et al., Free cholesterol accumulation in macrophage membranes activates Toll-like receptors and p38 mitogen-activated protein kinase and induces cathepsin K. Circ. Res. 104, 455–465 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fessler M. B., Parks J. S., Intracellular lipid flux and membrane microdomains as organizing principles in inflammatory cell signaling. J. Immunol. 187, 1529–1535 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hilchie A. L., Wuerth K., Hancock R. E., Immune modulation by multifaceted cationic host defense (antimicrobial) peptides. Nat. Chem. Biol. 9, 761–768 (2013). [DOI] [PubMed] [Google Scholar]
- 48.Oakley F. D., Smith R. L., Engelhardt J. F., Lipid rafts and caveolin-1 coordinate interleukin-1beta (IL-1beta)-dependent activation of NFkappaB by controlling endocytosis of Nox2 and IL-1beta receptor 1 from the plasma membrane. J. Biol. Chem. 284, 33255–33264 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Doan J. E. S., Windmiller D. A., Riches D. W. H., Differential regulation of TNF-R1 signaling: Lipid raft dependency of p42mapk/erk2 activation, but not NF-kappaB activation. J. Immunol. 172, 7654–7660 (2004). [DOI] [PubMed] [Google Scholar]
- 50.Mookherjee N., et al., Modulation of the TLR-mediated inflammatory response by the endogenous human host defense peptide LL-37. J. Immunol. 176, 2455–2464 (2006). [DOI] [PubMed] [Google Scholar]
- 51.Yu J., et al., Host defense peptide LL-37, in synergy with inflammatory mediator IL-1beta, augments immune responses by multiple pathways. J. Immunol. 179, 7684–7691 (2007). [DOI] [PubMed] [Google Scholar]
- 52.López-Collazo E., del Fresno C., Pathophysiology of endotoxin tolerance: Mechanisms and clinical consequences. Crit. Care 17, 242 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.van der Poll T., van de Veerdonk F. L., Scicluna B. P., Netea M. G., The immunopathology of sepsis and potential therapeutic targets. Nat. Rev. Immunol. 17, 407–420 (2017). [DOI] [PubMed] [Google Scholar]
- 54.Boto A., Pérez de la Lastra J. M., González C. C., The road from host-defense peptides to a new generation of antimicrobial drugs. Molecules 23, 311 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Luther A., et al., Chimeric peptidomimetic antibiotics against Gram-negative bacteria. Nature 576, 452–458 (2019). [DOI] [PubMed] [Google Scholar]
- 56.Brabetz W., Müller-Loennies S., Holst O., Brade H., Deletion of the heptosyltransferase genes rfaC and rfaF in Escherichia coli K-12 results in an Re-type lipopolysaccharide with a high degree of 2-aminoethanol phosphate substitution. Eur. J. Biochem. 247, 716–724 (1997). [DOI] [PubMed] [Google Scholar]
- 57.Galanos C., Lüderitz O., Westphal O., A new method for the extraction of R lipopolysaccharides. Eur. J. Biochem. 9, 245–249 (1969). [DOI] [PubMed] [Google Scholar]
- 58.Galanos C., Freudenberg M. A., Reutter W., Galactosamine-induced sensitization to the lethal effects of endotoxin. Proc. Natl. Acad. Sci. U.S.A. 76, 5939–5943 (1979).293694 [Google Scholar]
- 59.Keese S. P., Brandenburg K., Roessle M., Schromm A. B., Pulmonary surfactant protein A-induced changes in the molecular conformation of bacterial deep-rough LPS lead to reduced activity on human macrophages. Innate Immun. 20, 787–798 (2014). [DOI] [PubMed] [Google Scholar]
- 60.Veatch S. L., Electro-formation and fluorescence microscopy of giant vesicles with coexisting liquid phases. Methods Mol. Biol. 398, 59–72 (2007). [DOI] [PubMed] [Google Scholar]
- 61.Moertelmaier M., Brameshuber M., Linimeier M., Schütz G. J., Stockinger H., Thinning out clusters while conserving stoichiometry of labeling. Appl. Phys. Lett. 87, 263903 (2005). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.