Significance
Observations summarized herein contribute to an ongoing paradigm shift in microbial ecology, documenting an emergent property of ecosystem function that further challenges the perception that biogenic methane (CH4) production is strictly an anaerobic process. Relevant metabolites, a model bacterial isolate, gene, and enzyme are identified, and we show how this property can conceivably be broadly distributed in the biosphere and contribute to global CH4 emissions. Scientifically, this study will enable lines of investigation that will expand our understanding of CH4 synthesis and emission in nature and illustrates how CH4 synthesis may actually serve as a nexus for the C and N cycles in nature.
Keywords: methane, aerobic, methylamine, glycine betaine, bacteria
Abstract
Reports of biogenic methane (CH4) synthesis associated with a range of organisms have steadily accumulated in the literature. This has not happened without controversy and in most cases the process is poorly understood at the gene and enzyme levels. In marine and freshwater environments, CH4 supersaturation of oxic surface waters has been termed the “methane paradox” because biological CH4 synthesis is viewed to be a strictly anaerobic process carried out by O2-sensitive methanogens. Interest in this phenomenon has surged within the past decade because of the importance of understanding sources and sinks of this potent greenhouse gas. In our work on Yellowstone Lake in Yellowstone National Park, we demonstrate microbiological conversion of methylamine to CH4 and isolate and characterize an Acidovorax sp. capable of this activity. Furthermore, we identify and clone a gene critical to this process (encodes pyridoxylamine phosphate-dependent aspartate aminotransferase) and demonstrate that this property can be transferred to Escherichia coli with this gene and will occur as a purified enzyme. This previously unrecognized process sheds light on environmental cycling of CH4, suggesting that O2-insensitive, ecologically relevant aerobic CH4 synthesis is likely of widespread distribution in the environment and should be considered in CH4 modeling efforts.
The “methane paradox” is well documented and has drawn significant attention from a broad array of biologists, biogeochemists, biochemists, and physical scientists (see review in ref. 1). This supersaturation phenomenon is inconsistent with the extreme oxygen sensitivity of methanogens, a phylogenetically constrained group of archaea, which have long been viewed to be the sole source of biogenic CH4. Work in marine waters previously suggested this is due to methanogens active within anoxic microhabitats in suspended organic debris (2–5), digestive tracts, or fecal pellets of zooplankton (5–7) and/or of fish (8, 9). Recently, strong evidence for microbial dealkylation of methylphosphonate (referred to here as MPn) in marine (10, 11) and freshwater environments (12, 13) has been presented and thus provides at least a partial explanation for this phenomenon.
Pelagic methane enriched zones (PMEZ, ref. 12) are well-defined CH4 maxima occurring as a distinct region(s) of the water column in deep freshwater lakes (1, 12, 14). PMEZ offer a tractable environment in which to study the microbiology and biogeochemistry that underpins aerobic CH4 synthesis. Our prior efforts on Yellowstone Lake using 13C-labeled methanogen substrates, 16S ribosomal RNA (rRNA) gene based microbial community characterization, and methyl-coenzyme M reductase (mcrA) gene-targeted PCRs demonstrated the absence of recognizable methanogens in PMEZ waters (12). Instead, MPn metabolism was strongly associated with distinct populations of Pseudomonas sp. as being important contributors to PMEZ formation (12).
Efforts presented here summarize continuing studies that now identify aerobic metabolism of methylamine (MeA) as an important contributing metabolite to biogenic CH4. Methylotrophic methanogens anaerobically convert MeA to CH4 by disproportionation of MeA to carbon dioxide and CH4 (15, 16). Methylotrophic bacteria can metabolize MeA as a carbon source (17, 18), nonmethylotrophs for nitrogen (17, 19) (20), a carbon and N source (21), or methylovores can use it as an energy source (22). MeA utilization is viewed to occur as an oxidation via either MeA dehydrogenase, MeA oxidase, or involving methy group transfer to tetrahydrofolate involving the formation of formation of γ‐glutamyl‐methylamide (GMA) and N‐methylglutamate (NMG) (20, 23). In the current study, we report the discovery of an alternate, simpler route of MeA metabolism that yields CH4, involving a reaction catalyzed by a 5′pyridoxal-phosphate–dependent aspartate aminotransferase.
Results
Water Column Characterization.
Yellowstone Lake water columns sampled in 2016 shared similar EXO Sonde features (SI Appendix, Table S1 and Fig. S1). The PMEZs were located just below the approximate upper limit of the thermocline, although the July PMEZ was 0.5 m deeper (11.5 versus 11 m), and CH4 concentrations were approximately twofold greater (51 versus 26 nM) (Fig. 1A). Incubating July 29 PMEZ water samples (12 °C, initiated as aerobic) with 13C-labled MPn and 13C-labled substrates capable of supporting known methanogenesis pathways (acetate, formate, bicarbonate, H2 + bicarbonate) found no strong evidence of 13CH4 formation (SI Appendix, Fig. S2A). Samples spiked with 13C-MeA produced 13CH4 and to a much lesser extent 13C-methionine as well (Fig. 1B and SI Appendix, Fig. S2). Lack of MPn metabolism was not anticipated and thus to relocate the MPn active region, follow-up sampling (August 5) of the PMEZ as well as 1 m above and below were spiked with 13C-MeA, 13C-methionine, and 13C-MPn. On this second sampling date, 13C-MPn was the primary substrate leading to 13CH4 (Fig. 1C and SI Appendix, Fig. S2B) and thus consistent with our prior observations (12). The data showing that MeA was the sole contributing metabolite to PMEZ formation in July was striking. MeA is widespread in the environment (24), but much remains to be learned about its environmental fate, even though it is ubiquitous and viewed as an important nitrogen source in marine environments (19, 25). Subsequent work focused on its ecological relevance in this freshwater environment.
Fig. 1.
Characterization of the water column methane and MPn and MeA metabolism potential of the PMEZ microbial communities. (A) Methane profiles generated from equilibrium CH4 measurements. (B and C) 13CH4 generation from lake water samples spiked with 13C-labeled methylamine (MeA) or methylphosphonate (MPn). Data are mean ± range of n = 2 distinct water samples. On July 28, 2016, only water from 11.5 m was tested, whereas on August 5, 2016, water from three depths (as shown) was tested. VPDB, Vienna Pee Dee Belemnite; δ13CH4.
On July 24, 2017, the water column was sampled in higher resolution (0.5-m increments). As in 2016, the PMEZ was well defined (Fig. 2A) just below the upper boundary of the thermocline (8 m) in O2-saturated water (Fig. 2C). Also, a second more-robust PMEZ was present at 11.5 to 12.0 m depths (roughly 3,000% of saturation) (Fig. 2A) at near O2 saturation (98.6% of saturation, Fig. 2C). Neither PMEZ could be linked with the Chl a peak (14 to 16 m), and only the upper PMEZ was located within the photic zone (0 to 9 m). PCRs using degenerate universal mcrA primers (SI Appendix, Table S2) and total DNA failed to generate amplicons (SI Appendix, Fig. S3), indicating recognizable methanogens (26) were absent or below detection.
Fig. 2.
PMEZ characterization in 2017 for methane and relevant catabolic precursors. (A) Water column methane profile showing two concentration maxima at 8.0 and 11.5 to 12.0 m overlaid with relative abundance (%) of Burkholderiales. (B) Metabolite analysis showing change in concentrations of glycine betaine, trimethylamine, and methylamine. Open symbols indicate methylamine concentration below the detection limit. (C) Water column properties illustrating O2 saturation (or nearly so) at both methane peak depths. (D) Relative metabolic potentials of the microbial communities for converting 13C-MPn or 13C-MeA to 13C-CH4. Data in D denotes the mean ± range of two replicates except for MPn at 12 m, at which n = 1 due to broken serum bottle. VPDB, Vienna Pee Dee Belemnite; δ13CH4, ratio of stable isotopes 13C:12C in parts per thousand (per mil, ‰).
Targeted liquid chromatography–mass spectrometry (LC–MS) analyses of unamended PMEZ samples found MPn was below detection (1 μM in this lake water matrix; SI Appendix, Fig. S4). However, MeA was present (∼2.8 μM at 6 m) and decreased to or below detection at depths greater than 10 m (Fig. 2B). The lowest MeA levels coincided with the deepest and largest PMEZ maxima, suggesting MeA consumption may have contributed to its formation. Results of other parallel experiments examining microbial community metabolic potentials supported this hypothesis. 13C-MPn dealkylation activity indicated MPn could contribute to 13CH4 generation in water depths of 8, 11.5, and 12 m, whereas 13C-MeA exhibited the potential to contribute to 13CH4 synthesis only at 12 m (Fig. 2D).
Methylated amines are indicator metabolites of glycine betaine (GB) degradation and are commonly associated with algae (27–30). Microbial conversion of GB to trimethylamine (TMA) has been documented (30, 31), which can then be converted to trimethylamine oxide (TMAO) → dimethylamine (DMA) → MeA (17), and DMA can be converted directly to MeA (32). In this study, GB, TMA, and MeA profiles were correlated (R2 = 0.64 to 0.77, SI Appendix, Fig. S5), diminishing to or below the detection limits at depths that correspond to the deeper PMEZ associated with 13MeA → 13CH4 activity (Fig. 2 B and D). This suggests that the source, consumption, and metabolism of the three metabolites could be coordinated.
Microbial Community and Underlying Biology.
Analysis of Illumina 16S rRNA gene sequence libraries was used to determine if specific taxa exhibited increased abundance at the 12-m depth PMEZ and hence might indicate which organism(s) could be involved in 13MeA → 13CH4 activity. Taxa classified as belonging to the order Burkholderiales (annotated as Acid A-1 and Acid A-2 in the Freshwater Database) exhibited a sharp peak (13% abundance, Fig. 2A) coinciding specifically with the MeA metabolic potential at 12 m (Fig. 2D). However, in agreement with the apparent absence of amplifiable mcrA (SI Appendix, Table S2), recognizable methanogen signatures were absent in all libraries throughout the water column.
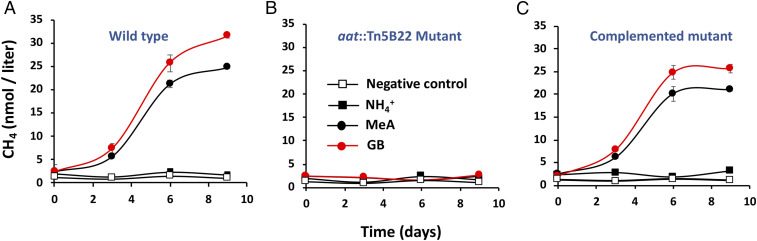
To more closely examine the biology underpinning the MeA→CH4 activity, microorganisms from PMEZ samples were isolated on SAR-MeA agar containing MeA as the sole nitrogen source (Methods) and then screened in liquid SAR-MeA for CH4 generation. We isolated several bacteria capable of converting MeA to CH4. Genus-level classification based on 16S rRNA gene sequence includes the following: Acidovorax sp. (MK896843.1), Pseudomonas sp. (MK896839.1), Caulobacter sp. (MK896844.1), Mesorhizobium sp. (MK896845.1), and Dietzia sp. (MK896847.1). For further characterization, we selected one of the Acidovorax sp. isolates obtained from the 12-m depth because this genus is a member of the Burkholderiales order that was significantly abundant at the depth corresponding to the MeA metabolic potential (Fig. 2 A and D) and because it was found to be genetically tractable. Batch cultures in liquid SAR-MeA in sealed serum bottles (maintained aerobic with sterile air injections at each gas sampling) demonstrated robust growth (SI Appendix, Fig. S6A) and CH4 generation from MeA (Fig. 3A). Gas chromatography–mass spectrometry (GC–MS) analysis of headspace samples of the Acidovorax sp. cultured with 13C-MeA confirmed 13CH4 generation (SI Appendix, Fig. S7), demonstrating transfer of the MeA carbon to CH4. GB was also investigated in the context of the above hypothesized catabolic pathway and also found to support growth (SI Appendix, Fig. S6A) and CH4 generation (Fig. 3A).
Fig. 3.
Conversion of glycine betaine and MeA to CH4 by pure culture Acidovorax sp. isolates. CH4 generation profiles are shown for the following: (A) wild-type Acidovorax sp. isolate; (B) the aat::Tn5B22 mutant 3-29; and (C) 3-29 mutant carrying the pCPP30::aat that complements the mutant back to wild type for growth on glycine betaine and MeA and for CH4 synthesis. All strains were cultured in SAR media containing NH4+, MeA, or glycine betaine as the sole nitrogen source (treatment symbols for all three panels are shown in B). Data points and error bars (where visible) are the average ± SD of three replicates.
Gene Identification and Mutational Analysis.
Transposon Tn5B22 mutagenesis of the Acidovorax sp. isolate yielded two mutants (among ∼8,000 transconjugates) capable of growth with ammonium chloride but not MeA nor GB as sole N source (SI Appendix, Fig. S6B) and were likewise defective for generating CH4 (Fig. 3B). Cultures initiated at cell densities equivalent to mid log phase under these growth conditions (starting optical density = 0.15; SI Appendix, Fig. S6, compare D and E) also failed to generate CH4, demonstrating lack of CH4 synthesis in the mutants is due to defective MeA and/or GB metabolism, not lack of biomass. The two Tn5B22 insertion sites differed, but the same gene was affected in both mutants (SI Appendix, Fig. S8), providing strong evidence that the mutated gene is essential to MeA/GB metabolism and CH4 synthesis in this organism. Basic local alignment search tool (BLAST)x analysis matched the gene to the family of pyridoxal 5′ phosphate-dependent aspartate aminotransferases (referred to herein as plp-aat). The specific plp-aat coding sequence without adjacent genomic DNA (GenBank accession no. MK170382) was PCR cloned from the Acidovorax isolate DNA into the broad host range plasmid pCPP30. Conjugation of the recombinant plasmid to the mutants reversed their negative growth and CH4 synthesis phenotypes (Fig. 3C and SI Appendix, Fig. S6C), confirming the importance of this gene.
Introducing plp-aat to Escherichia coli strain BL21(DE3) allowed this enteric bacterium to grow in M9 broth containing MeA as the N source (Fig. 4A) and to synthesize CH4 (Fig. 4B). However, growth in M9-MeA media required BL21 (pET28::aat) to first be grown in lysogeny broth (LB) broth with antibiotic selection; that is, washed cells failed to grow in M9-MeA. Consequently, subsamples of stationary phase cells were transferred directly (∼200-fold dilution) to the M9-MeA media (with isopropyl β-d-1-thiogalactopyranoside [IPTG] to induce aat transcription). The identity of the putative metabolite(s) apparently necessary for the recombinant E. coli to grow in M9-MeA is unknown at present, but it was not sufficient to allow the BL21 (pET28a+) negative control to grow in M9-MeA. Endpoint oxygen levels for the E. coli experiments were ∼43% of saturation at 37 °C (SI Appendix, Fig. S9), illustrating the cultures had not gone anaerobic. For both Acidovorax (Fig. 3 and SI Appendix, Fig. S6A) and E. coli (Fig. 4), CH4 synthesis was linked to growth on MeA, implying N acquisition and CH4 release are somehow metabolically coupled.
Fig. 4.
PLP-Aat enables E. coli to grow with MeA as a nitrogen source and synthesize CH4. (A) Growth profiles of E. coli BL21 with MeA or NH4+ and carrying the empty plasmid (control), cloned aat (pET28a::aat), or mutant aat wherein the catalytic lysine (K237) was replaced by alanine. (B) CH4 generation by BL21 with the same constructs. All data and error bars (where visible) are the mean ± SD of three replicate cultures.
PLP-Aat enzyme has been used as a model enzyme for understanding PLP-linked catalysis, and from these efforts, an essential catalytic lysine residue has been identified to be invariantly conserved (33, 34). Prior work with the E. coli PLP-Aat enzyme has shown that mutation of this conserved lysine to alanine results in an enzyme with only 0.5% native aspartate aminotransferase activity (34). In an analagous approach, we changed the catalytic lysine (K237) to alanine in the cloned plp-aat and repeated the bioassays. Growth and CH4 synthesis by BL21 carrying the K237A mutant clone was severely constrained in MeA media (Fig. 4).
Purified Enzyme Generates CH4 from MeA.
To more directly test the PLP-Aat protein for CH4 production, it was overexpressed and purified as a His-tagged protein (SI Appendix, Fig. S10). For assessing PLP-Aat activity, enzyme reactions were conducted in E. coli cytoplasm extract that was prefiltered through a 3 kDa molecular weight (MW) cutoff filter to remove proteins but retain all low molecular weight species (Methods) so as to include any other metabolite(s) that are unknown at present but which might be required for E. coli to host this activity. When purified PLP-Aat was added to the cytoplasmic extract along with α-ketoglutarate and PLP, CH4 was produced (Fig. 5). Control reactions using heat inactivated enzyme did not produce CH4. Likewise, negative were reactions where α-ketoglutarate or MeA were individually omitted. Methane was produced at a lower level in reactions where PLP was omitted. We attribute this to apo-form enzyme that purified with holo-form enzyme. This could be due to overproduction of the enzyme in the pET system and is consistent with the increased activity upon addition of PLP, which would generate more holo-form enzyme (PLP-Aat).
Fig. 5.
CH4 synthesis by purified Aat. CH4 synthesis by purified Aat in E. coli cytoplasmic extract. Complete reactions contained purified enzyme (typically ∼3 μg), MeA (4 mM), α-ketoglutarate (4 mM), and 5′pyridoxal phosphate (1 mM). Other reactions shown were conducted wherein a single substrate was omitted in order to illustrate the requirement of each for the generation of methane. All data are the mean ± SD of three replicate reactions.
Discussion
From experiments including analyses of microbial community composition and metabolic potential, targeted metabolomics, pure culture physiologic and genetic characterizations, and defined genetic transfer of this property, we put forth data that contributes to an ongoing paradigm shift for environmental CH4 synthesis that is distinct from classical methanogenesis or other presently known mechanisms of CH4 synthesis. We found MeA occurs at substantial concentrations in the pristine Yellowstone Lake environment in conjunction with GB and TMA (Fig. 2B and SI Appendix, Fig. S5), which are catabolic precursors of MeA. The extant microbial community exhibited the capacity to convert MeA to CH4 (Fig. 2D), and depending on as of yet undefined environmental conditions, MeA is an important contributor to PMEZ formation (Fig. 1, Fig. 2D) or indeed the only contributor (Fig. 1 A and B). The ability to convert MeA to CH4 is heritable by a single gene (Figs. 3C and 4) and importantly, MeA→CH4 conversion will occur under aerobic conditions (Figs. 3 and 4 and SI Appendix, Fig. S9). In sum, this study illustrates all the essential components of a relatively simple and ecologically relevant CH4 synthesis pathway; that is, aerobic bacterial CH4 synthesis.
Some have argued that lateral transport of CH4 derived from near shore–based methanogenesis to be the basis for metalimnetic CH4 (35–37). However, evidence supporting in situ CH4 synthesis in oxic lake waters is now clear and overwhelming (e.g., refs. 1, 12 to 14, 38, and 39). We suggest that defined CH4 maxima (PMEZs) that coincide with relevant microbes (ref. 12 and Figs. 2 and 3), substrates (ref. 12 and Fig. 2B), and functional gene occurrence (12) is not coincidental nor explainable by lateral transport. Furthermore, recent work directly comparing in situ oxic CH4 production versus lateral transport concluded that oxic biogeneic CH4 accounts for the majority of CH4 emissions for freshwater lakes with surface areas > 1 km2 (40). As such and given estimates that freshwater CH4 release accounts for roughly 16% of total annual global emissions (41), aerobic CH4 synthesis from GB and or MeA may contribute substantially to biosphere CH4 emissions.
While substrates supporting CH4 synthesis likely derive from photosynthetic primary producers in aqueous environments (42, 43), the Yellowstone Lake PMEZ is consistently (across four sampling seasons) not associated with a Chl a peak (e.g., Fig. 2A and SI Appendix, Fig. S1). Subsurface chlorophyll maxima result from phytoplankton sinking/migration behaviors associated with nutrient depletion or from photoadaption at deeper locations (44). Metabolite exchange between sinking phytoplankton and bacteria (45–47) may be an active (potentially mutualistic) process or derived from algal cell lysis caused by bacterial algaecides or virus lytic events (48–50). Increased water density at colder temperature transitions in the water column could result in transient accumulation of substrates (i.e., live functioning or lysed algae, particulate organic matter) (51) at specific depths. Enhanced residence time would facilitate CH4 synthesis and accumulation, resulting in PMEZ formation. CH4 concentration at these depths does not necessarily correlate with synthesis rate and may be more easily explained as synthesis exceeding consumption. The absence of dissolved MPn does not necessarily mean that alkylphosphonates are absent (detection limit 1 μM, SI Appendix, Fig. S4). Our assay methodology would not detect methylphosphonate (or any alkyphosphonates) covalently bound to organic matter in marine water (10). Indeed, the microbiome metabolic capacity to convert MPn to CH4 was present in both PMEZs and consistently at the more shallow PMEZ. Presumably, to some extent the occurrence and separation of MPn and MeA metabolic potentials in the water column is tied to bioavailability of relevant substrates. Methionine conversion to CH4 was limited relative to MPn and MeA but nevertheless detectable. Prior studies have documented methionine conversion to CH4, although under anaerobic conditions (52–54).
We predict compounds such as GB as well as choline (55–57) play an important role in generating methylated amines and thus are likely to contribute to this aerobic CH4 synthesis pathway, particularly in marine environments. GB is a ubiquitous osmolyte in bacteria, archaea, and eukaryotes (23, 58) and a precursor to the equally ubiquitous MeA, which is also recognized as an important N source (19, 23, 59) occurring at concentration ranges from nM to μM (60–64). Methylated amines are indicator metabolites of GB degradation (28), are common in algae (6, 32) or phototrophic mats (28, 29), and consistent with the documented biochemistry and genetics underlying GB conversion to TMA and TMA conversion to MeA in various organisms (17) (30, 31) (32). MeA metabolism has likewise been examined in some detail (23, 25), although this prior characterization work did not include synthesis of CH4 as an end product.
Ecological-based evidence in support of an aerobic GB → CH4 pathway includes research which infers CH4 production associated with methylamines derived from zooplankton grazing of phytoplankton (6) and recent studies showing TMA enhances CH4 synthesis in N-depleted incubations of Lake Stechlin oxic PMEZ water samples (65). Furthermore, the current study provides direct genetic evidence as illustrated by the inability of Acidovorax aat::Tn5B22 mutants to synthesize CH4 from GB nor use GB as an N source (Fig. 3B and SI Appendix, Fig. S6 B and E). By contrast, the mutants were reverted back to wild-type status when provided with the plp-aat gene (Fig. 3C). This indicates that, at least for this organism, acquiring N from GB must include the biochemical step catalyzed by the enzyme encoded by plp-aat described herein.
Several studies speak to the general importance of GB and MeA beyond simply their ubiquity and abundance in marine environments. Potential fates include assimilation as a carbon source, oxidized for energy, or used as a nitrogen source (19, 22, 25, 66–68). These examples represent metabolisms that would compete with a MeA → CH4 pathway, but it is still reasonable to consider its potential for CH4 synthesis. At the moment, information on marine MeA dynamics is limited, but MeA assimilation (0.005 to 54 nmol L−1 day−1), oxidation (0.09 to 3.43 nmol L−1 day−1) and turnover rates (40 to 57 nmol L−1 day−1) have been reported (60, 67), facilitating at least a preliminary estimate. Conservatively assuming a PLP-Aat catalyzed MeA turnover rate of 1 nmol L−1 day−1 in the top 100 m of the global oceans is associated with CH4 release, this would account for 11.6% of annual marine CH4 emission [2.1 Mt CH4 yr−1 (69); Methods].
This mode of CH4 synthesis could be highly relevant to any situation where GB and or MeA occur, including environments where CH4 synthesis has traditionally been interpreted to derive from methanogens. There is no obvious reason why this PLP-Aat reaction would be constrained by redox conditions per se because aminotransferase reactions are important for maintaining cellular amino acid equilibria in all self-replicating organisms. Indeed, numerous aerobic or anaerobic organisms may be involved with varying efficiency, and at present, we are unable to offer any specific criteria for excluding hundreds of more-remote aat homologs in metagenome libraries that may encode the same function, perhaps with greater efficiency. However, we draw attention to the fact that the PLP-Aat encoded in the genome of E. coli strain BL21(DE3) (GenBank genome accession no. AM946981, used herein for bioassays) shares only 21.6% identity with the Acidovorax enzyme and did not allow E. coli to grow with MeA nor make CH4 from MeA. Thus, apparently not all proteins annotated as PLP-dependent Aat enzymes are equal in regard to catalyzing the MeA → CH4 reaction. Characterizing homologs capable of this reaction will enhance our understanding of the universality of this process within and across environments. A cursory search identified thousands of homologs with a high degree of homology with the Acidovorax PLP-Aat, suggesting the possibility that MeA → CH4 activity occurs in other aqueous and terrestrial environments as well. Examples of highly homologous Aat proteins occurring in a variety of aquatic environments are provided in SI Appendix, Fig. S11.
The PLP-Aat catalytic mechanism resulting in CH4 release is unknown at present. In particular, the source of reductant represents a mechanistic enigma that further contributes to the methane paradox (70). Recreating the reaction milieu in E. coli cytoplasmic extract amended with MeA, α-ketoglutarate, PLP, and live purified PLP-Aat enzyme resulted in methane generation (Fig. 5). Methane was not generated when enzyme was heat inactivated before addition. Prefiltering the extract using a 3 kDa cutoff eliminates a direct contribution of other enzyme(s) but would not remove other metabolite(s), which may contribute or may be associated with the requirement for E. coli to be first grown in a complex medium. The functional importance of the catalytic lysine that is conserved across all PLP-Aat enzymes appears to be involved for the MeA → CH4 reaction and thus implies some basic similarities; that is, the Schiff base formation between the Aat catalytic lysine and the aldehyde carbon of the coenzyme pyridoxal 5′-phosphate (33, 34). However, beyond this initial step it is also not possible at this juncture to predict reaction mechanism, or efficiency, or any other metabolite/gene product apparently involved in the functioning cell, though it is relevant to point out that CH4 synthesis in both Acidovorax (Fig. 3) and E. coli (Fig. 4) appears connected to growth, implying a link between CH4 release and N acquisition to support growth. And furthermore, for at least organisms such as the Acidovorax sp. isolate characterized herein, growth with MeA was as robust as ammonium (SI Appendix, Fig. S6), arguing that MeA use as a nitrogen source does not represent a spurious metabolism. Indeed, aat::Tn5B22 mutants failed to grow with MeA, which argues that alternate MeA metabolic pathways (23) are missing in this organism, which is also consistent with the absence of genes annotated as encoding N-methylglutamate synthase, γ-glutamylmethylamide synthase, and N-methylglutamate dehydrogenase in the draft genome of this organism.
Establishing the essential components of a GB → MeA → CH4 pathway in a pure culture of an ecologically relevant microorganism and linking this to in situ metabolite analyses and community metabolic potentials reveals a route of CH4 synthesis occurring under fully oxygenated conditions. This also elucidates a nexus for C and N cycling in nature. Based on the characterization of the Acidovorax sp. isolate detailed herein, we view this organism as a robust, though almost certainly not unique, participant in PMEZ formation and useful as a first-generation model for illustrating gene–enzyme–organism–ecosystem linkages in this process.
Methods
Study Site, Sampling, and Initial Sample Analyses.
The Yellowstone Lake sampling location and YSI EXO1 multiparameter Sonde water column characterizations were as previously described (12). For 2016 samplings, PMEZ localizations were determined using a dissolved gas equilibration system also as described (12). In 2017, the more extensive water column sampling did not allow for the use of the more time-consuming flow-through gas equilibration device for PMEZ identification. Instead, the thermocline was identified and characterized using the EXO Sonde, which then allowed us to approximate the depth of the anticipated PMEZ (i.e., just below the upper limit of the thermocline) (12). Extensive sampling was then conducted above and below this depth. On the day of sampling in 2017, lake conditions were exceptionally placid, allowing for sampling in 0.5-m increments.
For in situ CH4 analysis, duplicate lake water samples for each depth were collected in 250 mL serum bottles, filling from the bottom and overflowing with at least one volume to expel bubbles. Vials were immediately sealed with gray chlorobutyl rubber stoppers and secured with aluminum crimps. Upon reaching shore, samples were killed by injecting 200 µL saturated HgCl2 solution and stored on ice for transport to the laboratory where they were stored at 4 °C. CH4 analysis was by GC of an introduced headspace (using ultra-high purity N2) on a Hewlett-Packard HP5890A (for 2016 work) or a Varian CP-3800 (for 2017 work) gas chromatograph, both with flame ionization detection. The original CH4 concentration in solution was calculated using Henry’s law and solubility equations (71). All other water samples were stored on ice in the dark for transport to the laboratory and then stored in a cold room (5 °C).
Upon identifying the PMEZs from the GC analysis (∼24 h post–lake acquisition), untreated cold room stored samples were then used to initiate enrichments with 13C-labeled substrates to identify and qualitatively assess potential methanogenesis substrates yielding 13CH4. For the 2016 samples, select water column samples were aseptically spiked to a final substrate concentration of 1 mM with filter sterilized stock solutions (25 mM, 99 atom%, Sigma-Aldrich) of 13C-labeled formate, acetate, NaHCO3, NaHCO3 + H2, MPn, MeA, or methionine. PMEZ samples taken in 2016 were incubated in the dark at 12 °C to mimic in situ conditions. 13C enrichments of samples taken in 2017 focused only on 13C-MPn and 13C-MeA. Also, by that time we had found all functionally relevant lake isolates to have mesophilic temperature optima and thus incubations were conducted at room temperature, which was more optimal for assessing the potential of lake microbial community samples to convert these substrates to 13CH4. The isotopic signature (δ13C) of the CH4 of the headspace was measured using a modification of the protocols described by Wang et al. (12) on a Picarro G2201-i cavity ring-down spectrometer (CRDS) equipped with a Small Sample Introduction Module 2 (SSIM2). Each vial was first pressurized with 20 mL of <0.2 µm filtered ambient air (δ13C = ∼−48‰ versus Vienna Pee Dee Belemnite) and then 20 mL was removed via gastight syringe and injected into the SSIM2. Factory calibration was used for the CRDS, and a tank of compressed ambient air was used as a reference material between injections to verify lack of instrument drift. For samples taken in 2017, the isotopic signature (δ13C) of the CH4 of the headspace was determined using GC–MS.
GC–MS Analysis.
For CH4 isotope analysis, 800 µL headspace gas was injected directly into an Agilent 7890 GC–MS equipped with Carboxen-1010 porous-layer open-tubular (PLOT) capillary GC column in splitless mode. The injection temperature was 200 °C, the ion source was set to 230 °C, helium was the carrier gas, and the column flow rate was 1 mL/min. The oven was held at 35 °C for 7.5 min and then heated at 25 °C/min to 250 °C and held at that temperature for 6.5 min. The mass spectrometer was operated in scan/selected ion monitoring (SIM) mode; scan range (10 to 100 m/z) for SIM m/z 14, 15, 16, 17 were used. The 13CH4 standard was purchased from Sigma-Aldrich, and for 12CH4, a standard high purity methane tank was purchased from American Welding and Gas.
For analysis of E. coli culture headspace oxygen, 250 µL headspace gas was injected directly into an Agilent 7890 GC–MS equipped with a Carboxen-1010 PLOT capillary GC column in splitless mode. The injector temperature was 200 °C, with the ion source at 230 °C and a helium flow rate of 1 mL/min. An isocratic GC program at 65 °C with the mass spectrometer in SIM mode for m/z 32 was used.
Detection of GB, TMA, and MeA.
Methylamine hydrochloride, trimethylamine hydrochloride, and glycine betaine hydrochloride standards were purchased from Sigma-Aldrich. For detection of MeA in lake waters, dansyl chloride (Dns-Cl, 1 -dimethylaminonaphthalene-5-sulfonyl chloride) was used to label the compounds of interest prior to analysis by LC–MS. The dansylation procedure was performed as described by Guo et al. (72). Briefly, a Hamilton gastight syringe was used to draw 50 μL water from the sample serum bottle, which was then placed into a 250 μL polypropylene analysis vial and pH adjusted to ∼9.5 with 2 μL 160 mM sodium hydroxide. Dns-Cl prepared in acetonitrile (20 mg/mL) was added to the sample in a volume of 46 μL. Samples were then incubated for 30 min at room temperature. After the incubation period, pH was adjusted to ∼4 with 2 μL 10% formic acid. At this point, the sample was ready for analysis. Chromatography experiments were done on an Agilent 6538 quadrupole time-of-flight (Q-TOF) mass spectrometer, positive mode, equipped with a reversed-phase Agilent Zorbax Eclepse Plus C18 column (2.1 × 150 mm). Solvent A was 0.1% formic acid in high-performance liquid chromatography (HPLC) water, and solvent B was 0.1% formic acid in acetonitrile. The 15 min binary gradient elution profile was as follows: t1) 1 min, 0% B; t2) 11 min, 55% B; t3) 14 min, 100% B; and t4) 15 min, 0% B. The wavelength was 320 nm, the flow rate was 600 µL/min, and the sample injection volume was 10 µL. Limit of detection was determined to be 20 nM.
For detection of trimethylamine, ethyl bromoacetate was used to label the compound of interest prior to analysis by LC–MS. The labeling procedure was performed as described by Johnson (73). A Hamilton gastight syringe was used to draw 90 μL water from the sample serum bottle and transferred to a 250 μL polypropylene analysis vial and then 10 μL (20mg/mL acetonitrile) ethyl bromoacetate was added. Samples were then incubated for 30 min at room temperature. At this point, the sample was ready for analysis. Chromatography was done on an Agilent 6538 Q-TOF mass spectrometer, positive mode, equipped with a normal-phase Waters ACQUITY BEH HILIC 1.7 μm column (2.1 × 100 mm). Solvent A was 0.1% formic acid in HPLC water, and solvent B was 0.1% formic acid in acetonitrile. The 4.7 min binary gradient elution profile was as follows: t1) 0 min, 90% B; t2) 2 min, 70% B; t3) 3.6 min, 60% B; and t4) 4.7 min, 90% B. The flow rate was 400 µL/min, and the sample injection volume was 5 µL. Limit of detection was determined to be 50 nM.
For detection of glycine betaine, a Hamilton gastight syringe was used to transfer 90 μL water from the sample serum bottle to a 250 μL polypropylene analysis vial. Methanol was added to give a 10% concentration, and the sample was then vortexed for 30 s. At this point, the sample was ready for analysis. Chromatography was done on an Agilent 6538 Q-TOF mass spectrometry, positive mode, equipped with a normal-phase Waters ACQUITY BEH HILIC 1.7 μm column (2.1 × 100 mm). Solvent A was 0.1% formic acid in HPLC-grade water, and solvent B was 0.1% formic acid in acetonitrile. The 4.7 min binary gradient elution profile was as follows: t1) 0 min, 90% B; t2) 2 min, 70% B; t3) 3.6 min, 60% B; and t4) 4.7 min, 90% B. The flow rate was 400 µL/min, and the sample injection volume was 10 µL. Limit of detection was determined to be 1 nM.
Isolate Enrichment, Cultivation, and Characterization.
All bacteria used in this study are listed in SI Appendix, Table S2. To isolate Acidovorax from the lake water, we used the same SAR media described previously as SAR-MPn (12), except modified such that 10 mM glucose replaced pyruvate, 1 mM MeA or GB was provided as the sole N source instead of NH4SO4, and inorganic phosphate was used instead of MPn as a P source (agar media referred to herein as SAR-MeA). Sample aliquots from the positive 13C-MeA → 13CH4 enrichments were plated directly onto SAR-MeA solidified with Agar Noble. Following a 2-wk incubation (room temperature), visually unique colonies were identified based on morphological differences and selected for several rounds of subculture on SAR-MeA agar to obtain pure cultures.
DNA from each culture was extracted using Wizard Genomic DNA Purification Kits (Promega) as per manufacturer’s instructions. The 16S rRNA genes were PCR cloned using 27F and 1492R PCR primers (SI Appendix, Table S2). Amplicons were individually cloned into pCR2.1 (Invitrogen); clone-bearing plasmids were purified via QIAprep Spin Miniprep Kit (Qiagen) and then sequenced by the Brigham Young University Central DNA sequencing facility. Resulting sequences were compared with GenBank sequences to identify closely related cultured organisms via BLASTn.
For culture characterizations, Acidovorax was grown in liquid SAR-MeA media containing glucose as the sole carbon source (4 g/L). Growth was tracked based on culture optical absorbance (Ab595) using a Molecular Dynamics microtiter plate reader. MeA → CH4 synthesis experiments were conducted in 70 mL sealed serum bottles in order to quantify CH4 synthesis, which was measured via GC as described in Study Site, Sampling, and Initial Sample Analyses.
DNA Isolation, PCR, and Sequencing.
DNA was extracted from all water column depths for use as PCR templates to determine whether the methanogen indicator gene, mcrA, was detectable. In each case, biomass from triplicate 1 L samples were separately collected on Sterivex-GV 0.22 μm filter cartridges (Millipore Sigma) and then DNA extracted using the PowerWater DNA isolation kit (MO BIO Laboratories, Inc.) following the manufacturer’s instructions.
PCR primers for near full-length amplification of the Acidovorax pure culture 16S rRNA gene used the primers describe by Lane (74), and for the mcrA gene, we used the universal primers described by Luton et al. (75) (SI Appendix, Table S2). For community 16S rRNA gene sequencing, DNA extracts were quantified using a SpectraMax Plus Microplate Reader (Molecular Devices) and PCR tested prior to submission for Illumina sequencing at the Institute for Genomics and Systems Biology Next Generation Sequencing Core at Argonne National Laboratory. Very briefly, PCR amplicons were generated using barcoded primers 515F and 806R targeting the V4 region of the 16S rRNA gene in the domains Bacteria and Archaea (76) and then sequenced using the Illumina MiSeq sequencing platform. Illumina sequence libraries were processed using the mothur software package [version 1.39, (77)]. Low-quality sequences were removed using minimum and maximum lengths of 100 and 500, respectively, and no ambiguous bases or mismatches were allowed in the primer sequence. Chimera sequences were removed using UCHIME (78), and the Silva v123 reference database was used to remove uninformative data (79). For each depth, the libraries were rarified to 18,800 quality and trimmed reads, and singletons were removed prior to calculating relative abundances. Operational taxonomic unit classification was based on 97% identity, and classification was based on using the “FreshTrain” database available at https://github.com/McMahonLab/TaxAss/tree/master/FreshTrain-files (80). Repeated resampling (n = 10) of the libraries generated the same relative abundances for the taxa described. Sequencing data can be found in GenBank as Bioproject ID PRJNA598368, Biosample accession no. SAMN13704654.
Transposon Mutagenesis, plp-aat Cloning, and Complementation.
Methods for transposon mutagenesis, genome walking, and cloning were as we previously described (81). Briefly, E. coli S17-1 was used to conjugate Tn5-B22 (82) to Acidovorax sp. GentR transconjugates were then screened for loss of growth on SAR-MeA agar, yielding two unique mutants. Cloning of the Tn5B22 and adjacent DNA, as well as additional primer walking, used the APAgene genome walking kit (Bio S&T Inc.) to obtain the full gene sequence. The specific aat sequence was then PCR cloned into pCR2.1 and then subcloned to the broad host range plasmid pCPP30, which was then transformed into E. coli S17-1 for conjugal transfer to each mutant for complementation experiments.
Catalytic Lysine Amino Acid Substitution Mutation.
The selected genes were refactored to match the codon usage of E. coli using an empirically derived codon usage table to remove NdeI and HindIII restriction sites and to overcome DNA synthesis constraints using Build-Optimization Software Tools (BOOST) (83). Synthetic DNA (Twist Biosciences) was inserted into the NcoI/XhoI site of pET28a+ by Gibson assembly. The E. coli Top10 transformants were plated on LB agar plates supplemented with kanamycin (50 µg/mL). All plasmids were sequence verified using the PacBio Sequel sequencing platform (Pacific Biosciences). Positive clones were DNA prepped and transformed into BL21 (DE3) cells (Novagen).
Enzyme Purification.
E. coli strain BSL21 carrying pET28a+::aat was used as the expression system for the His-tagged Aat. Briefly, cells were grown in LB media under kanamycin selection at 37 °C to an OD600 of 0.2 and then Aat protein production were induced by adding IPTG to a final concentration of 0.1 mM. Cells were incubated at 25 °C for 8 h, then spun down (3,000 rcf, 20 min, 4 °C). The resulting cell pellet was stored at −80 °C until purification. Cells were lysed using sonication in a lysis buffer of phosphate buffer (0.1 M sodium phosphate, pH 7.4), 10 mM imidazole, and cOmplete Mini EDTA-free Protease Inhibitor mixture (cOmplete, Sigma-Aldrich). Protein was purified using a step-wise imidazole gradient on a HisTrapFF affinity column (GE Healthcare). Following elution, the Aat protein fraction was dialyzed into phosphate buffer overnight using 10 k molecular weight cut off (MWCO) SnakeSkin pleated dialysis tubing (ThermoScientific). The Aat fraction was then concentrated using a Nanosep 30 k MWCO centrifugal device (PALL Life Sciences), and glycerol was added to a final concentration of 10%. Protein was stored at −80 °C until used in the enzymatic assays. Intact protein molecular weight determination was conducted using C4 reverse-phase LC–MS on Micro-TOF (Bruker Daltronics) coupled to a 1290 ultra-high performance liquid chromatography (Agilent Technologies). Intact mass analysis had an expected mass of 46,797 Da, with the observed MW of 46,794 Da equating to a 0.006% error. Digested gel samples were analyzed using a maXis Impact Ultra-High Resolution QTOF instrument (Bruker Daltonics) coupled to a Dionex 3000 nano-uHPLC (Thermo-Fisher). Data analysis was performed using the SearchGUI/PeptideShaker data analysis software (Compomics). Sequence coverage was obtained for residues 2 to 245 of Aat. Based on sodium dodecyl sulfate-polyacrylamide gel electrophoresis, Aat was estimated to be 90 to 95% of the total protein in the purified sample used for enzyme assays.
Enzyme Assays.
The standard protocol for generating the E. coli cytoploasmic extract was as follows. A 100 mL culture of E. coli BL21 was grown to stationary phase in M9 minimal media (84), collected by centrifugation, suspended in 10 mL phosphate buffer (0.1 M, pH 7.4), and then lysed by sonication on ice. The lysate was centrifuged (12,000 rpm, 20 min) to pellet unbroken cells and debris and then the supernatant was filtered through a 3 Kd MW filter (required multiple filters) to remove proteins but retain metabolites in a batch of extract, which was used for all assays. The standard assay volume was 1.0 mL containing 3 μg purified protein, 4 mM MeA, 4 mM α-ketoglutarate, and 1 mM 5′ pyridoxal phosphate and contained in 10 mL sealed serum bottles for tracking CH4 synthesis as described in Study Site, Sampling, and Initial Sample Analyses. Control reactions consisted of the same compositions except the Aat enzyme preparation was inactivated by boiling for 20 min.
Estimating Methane Release from Marine MeA.
MeA assimilation rates of 0.005 to 54 nmol L−1 day−1, oxidation rates of 0.09 to 3.43 nmol L−1 day−1, and overall turnover rates of 40 to 57 nmol L−1 day−1 have been reported (62, 67). Assuming MeA conversion to CH4 is restricted to the top 100 m of the marine water column where the majority of primary productivity occurs, and an average turnover rate of 1 nmol MeA L−1 day−1, the 3.6 × 1019 L of seawater in that volume would result in a MeA→ CH4 conversion rate of 1.31 × 1013 mol yr−1, equaling a potential release of 210 Mt CH4 yr−1 (MWCH4 = 16.043 g ⋅ mol−1). Assuming that 0.1, 1, or 10% of marine MeA turnover is due to PLP-Aat homologs capable of the MeA → CH4, 0.21, 2.1, or 21 Mt CH4 yr−1 would be released. This would account for 1.16, 11.6, and 116% of the global CH4 emissions from oceanic sources, which are estimated at 18 Mt yr−1 (69).
Supplementary Material
Acknowledgments
Primary project support was provided by the NSF EAR-1529461. Additional support to T.R.M. was from the Montana Agricultural Experiment Station (MAES Project 911310) and to T.R.M. and B.B. from NASA 80NSSC21K0487. Minor support to J.E.D. was received from the Montana Institute on Ecosystems NSF Established Program to Stimulate Competitive Research Program Grant EPS-1101342 and the NSF Systems and Synthetic Biology program (MCB-1817428) to R.H. Work for Award 504607 from the Department of Energy (DOE) Joint Genome Institute (JGI) Synthetic Biology Program was conducted by the US DOE JGI, a DOE Office of Science User Facility, supported under Contract DE-AC02-05CH11231. Funding for the Proteomics, Metabolomics and Mass Spectrometry Facility used in this study was made possible in part by the M.J. Murdock Charitable Trust and the National Institute of General Medical Sciences of the NIH under Award P20GM103474. Research on Yellowstone Lake was conducted under US Department of the Interior National Park Service Research Permit YELL-SCI-5700 to T.R.M. We thank Mary Ann Moran and Caroline Harwood for thoughtful comments prior to submission and Zackary Jay for helpful discussion on 16S rRNA gene microbial community analysis. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the author(s) and do not necessarily reflect the views of MAES nor the NSF.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2019229118/-/DCSupplemental.
Data Availability
In addition to the methods described herein, DNA sequence data have been deposited in GenBank (https://www.ncbi.nlm.nih.gov): 1) aat gene, MK170382 (85); 2) Illumina libraries, Biosample accession: SAMN13704654 (86). All other study data are included in the article and/or SI Appendix.
References
- 1.Tang K. W., et al., Methane production in oxic lake waters potentially increases aquatic methane flux to air. Environ. Sci. Technol. Lett. 3, 227–233 (2016). [Google Scholar]
- 2.Marty D., Nival P., Yoon W., Methanoarchaea associated with sinking particles and zooplankton collected in the Northeastern tropical Atlantic. Oceanogr. Lit. Rev. 6, 970 (1998). [Google Scholar]
- 3.Karl D. M., Tilbrook B. D., Production and transport of methane in oceanic particulate organic matter. Nature 368, 732–734 (1994). [Google Scholar]
- 4.Sasakawa M., et al., Carbon isotopic characterization for the origin of excess methane in subsurface seawater. J. Geophys. Res. Oceans 113, C03012 (2008). [Google Scholar]
- 5.Bianchi M., et al., Strictly aerobic and anaerobic bacteria associated with sinking particulate matter and zooplankton fecal pellets. Mar. Ecol. Prog. Ser. 88, 55–60 (1992). [Google Scholar]
- 6.de Angelis M. A., Lee C., Methane production during zooplankton grazing on marine phytoplankton. Limnol. Oceanogr. 39, 1298–1308 (1994). [Google Scholar]
- 7.Ditchfield A. K., et al., Identification of putative methylotrophic and hydrogenotrophic methanogens within sedimenting material and copepod faecal pellets. Aquat. Microb. Ecol. 67, 151–160 (2012). [Google Scholar]
- 8.Oremland R. S., Methanogenic activity in plankton samples and fish intestines A mechanism for in situ methanogenesis in oceanic surface waters. Limnol. Oceanogr. 24, 1136–1141 (1979). [Google Scholar]
- 9.van der Maarel M. J., Sprenger W., Haanstra R., Forney L. J., Detection of methanogenic archaea in seawater particles and the digestive tract of a marine fish species. FEMS Microbiol. Lett. 173, 189–194 (1999). [DOI] [PubMed] [Google Scholar]
- 10.Repeta D. J., et al., Marine methane paradox explained by bacterial degradation of dissolved organic matter. Nat. Geosci. 9, 884–887 (2016). [Google Scholar]
- 11.Sosa O. A., et al., Isolation and characterization of bacteria that degrade phosphonates in marine dissolved organic matter. Front. Microbiol. 8, 1786 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang Q., Dore J. E., McDermott T. R., Methylphosphonate metabolism by Pseudomonas sp. populations contributes to the methane oversaturation paradox in an oxic freshwater lake. Environ. Microbiol. 19, 2366–2378 (2017). [DOI] [PubMed] [Google Scholar]
- 13.Yao M., Henny C., Maresca J. A., Freshwater bacteria release methane as a byproduct of phosphorus acquisition. Appl. Environ. Microbiol. 82, 6994–7003 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grossart H.-P., Frindte K., Dziallas C., Eckert W., Tang K. W., Microbial methane production in oxygenated water column of an oligotrophic lake. Proc. Natl. Acad. Sci. U.S.A. 108, 19657–19661 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deppenmeier U., Lienard T., Gottschalk G., Novel reactions involved in energy conservation by methanogenic archaea. FEBS Lett. 457, 291–297 (1999). [DOI] [PubMed] [Google Scholar]
- 16.Welander P. V., Metcalf W. W., Loss of the mtr operon in Methanosarcina blocks growth on methanol, but not methanogenesis, and reveals an unknown methanogenic pathway. Proc. Natl. Acad. Sci. U.S.A. 102, 10664–10669 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen Y., Patel N. A., Crombie A., Scrivens J. H., Murrell J. C., Bacterial flavin-containing monooxygenase is trimethylamine monooxygenase. Proc. Natl. Acad. Sci. U.S.A. 108, 17791–17796 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nayak D. D., Marx C. J., Methylamine utilization via the N-methylglutamate pathway in Methylobacterium extorquens PA1 involves a novel flow of carbon through C1 assimilation and dissimilation pathways. J. Bacteriol. 196, 4130–4139 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Taubert M., et al., Methylamine as a nitrogen source for microorganisms from a coastal marine environment. Environ. Microbiol. 19, 2246–2257 (2017). [DOI] [PubMed] [Google Scholar]
- 20.Chen Y., McAleer K. L., Murrell J. C., Monomethylamine as a nitrogen source for a nonmethylotrophic bacterium, Agrobacterium tumefaciens. Appl. Environ. Microbiol. 76, 4102–4104 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen Y., et al., γ-Glutamylmethylamide is an essential intermediate in the metabolism of methylamine by Methylocella silvestris. Appl. Environ. Microbiol. 76, 4530–4537 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sun J., et al., One carbon metabolism in SAR11 pelagic marine bacteria. PLoS One 6, e23973 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chistoserdova L., Kalyuzhnaya M. G., Lidstrom M. E., The expanding world of methylotrophic metabolism. Annu. Rev. Microbiol. 63, 477–499 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Poste A. E., Grung M., Wright R. F., Amines and amine-related compounds in surface waters: A review of sources, concentrations and aquatic toxicity. Sci. Total Environ. 481, 274–279 (2014). [DOI] [PubMed] [Google Scholar]
- 25.Latypova E., et al., Genetics of the glutamate-mediated methylamine utilization pathway in the facultative methylotrophic beta-proteobacterium Methyloversatilis universalis FAM5. Mol. Microbiol. 75, 426–439 (2010). [DOI] [PubMed] [Google Scholar]
- 26.Du Z., et al., Landscape position influences microbial composition and function via redistribution of soil water across a watershed. Appl. Environ. Microbiol. 81, 8457–8468 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee C., “Amino acid and amine biogeochemistry in marine particulate material and sediments” in Nitrogen Cycling in Coastal Marine Environments, Blackburn T. H., Sørenson J., Eds. (SCOPE Series 33, Wiley & Sons, 1988), pp. 125–141. [Google Scholar]
- 28.Oren A., Formation and breakdown of glycine betaine and trimethylamine in hypersaline environments. Antonie van Leeuwenhoek 58, 291–298 (1990). [DOI] [PubMed] [Google Scholar]
- 29.King G. M., Methanogenesis from methylated amines in a hypersaline algal mat. Appl. Environ. Microbiol. 54, 130–136 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Müller C., et al., Seasonal variation of aliphatic amines in marine sub-micrometer particles at the Cape Verde islands. Atmos. Chem. Phys. 9, 9587–9597 (2009). [Google Scholar]
- 31.Möller B., et al., Sporomusa, a new genus of gram-negative anaerobic bacteria including Sporomusa sphaeroides spec. nov. and Sporomusa ovata spec. nov. Arch. Microbiol. 139, 388–396 (1984). [Google Scholar]
- 32.Lidbury I., Mausz M. A., Scanlan D. J., Chen Y., Identification of dimethylamine monooxygenase in marine bacteria reveals a metabolic bottleneck in the methylated amine degradation pathway. ISME J. 11, 1592–1601 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Toney M. D., Aspartate aminotransferase: An old dog teaches new tricks. Arch. Biochem. Biophys. 544, 119–127 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Malcolm B. A., Kirsch J. F., Site-directed mutagenesis of aspartate aminotransferase from E. coli. Biochem. Biophys. Res. Commun. 132, 915–921 (1985). [DOI] [PubMed] [Google Scholar]
- 35.DelSontro T., del Giorgio P. A., Prairie Y. T., No longer a paradox: The interaction between physical transport and biological processes explains the spatial distribution of surface water methane within and across lakes. Ecosystems (N. Y.) 21, 1073–1087 (2018). [Google Scholar]
- 36.Fernández J. E., Peeters F., Hofmann H., On the methane paradox: Transport from shallow water zones rather than in situ methanogenesis is the major source of CH4 in the open surface water of lakes. J. Geophys. Res. Biogeosci. 121, 2717–2726 (2016). [Google Scholar]
- 37.Peeters F., Encinas Fernandez J., Hofmann H., Sediment fluxes rather than oxic methanogenesis explain diffusive CH4 emissions from lakes and reservoirs. Sci. Rep. 9, 243 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bižić M., et al., Aquatic and terrestrial cyanobacteria produce methane. Sci. Adv. 6, eaax5343 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hartmann J. F., et al., High spatiotemporal dynamics of methane production and emission in oxic surface water. Environ. Sci. Technol. 54, 1451–1463 (2020). [DOI] [PubMed] [Google Scholar]
- 40.Günthel M., et al., Contribution of oxic methane production to surface methane emission in lakes and its global importance. Nat. Commun. 10, 5497 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bastviken D., Tranvik L. J., Downing J. A., Crill P. M., Enrich-Prast A., Freshwater methane emissions offset the continental carbon sink. Science 331, 50 (2011). [DOI] [PubMed] [Google Scholar]
- 42.Bogard M. J., et al., Oxic water column methanogenesis as a major component of aquatic CH4 fluxes. Nat. Commun. 5, 5350 (2014). [DOI] [PubMed] [Google Scholar]
- 43.Amin S. A., Parker M. S., Armbrust E. V., Interactions between diatoms and bacteria. Microbiol. Mol. Biol. Rev. 76, 667–684 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cullen J. J., Subsurface chlorophyll maximum layers: Enduring enigma or mystery solved? Annu. Rev. Mar. Sci. 7, 207–239 (2015). [DOI] [PubMed] [Google Scholar]
- 45.Amin S. A., et al., Interaction and signalling between a cosmopolitan phytoplankton and associated bacteria. Nature 522, 98–101 (2015). [DOI] [PubMed] [Google Scholar]
- 46.Durham B. P., et al., Recognition cascade and metabolite transfer in a marine bacteria-phytoplankton model system. Environ. Microbiol. 19, 3500–3513 (2017). [DOI] [PubMed] [Google Scholar]
- 47.Paul C., Mausz M. A., Pohnert G., A co-culturing/metabolomics approach to investigate chemically mediated interactions of planktonic organisms reveals influence of bacteria on diatom metabolism. Metabolomics 9, 349–359 (2013). [Google Scholar]
- 48.Mayali X., Azam F., Algicidal bacteria in the sea and their impact on algal blooms. J. Eukaryot. Microbiol. 51, 139–144 (2004). [DOI] [PubMed] [Google Scholar]
- 49.Seyedsayamdost M. R., Carr G., Kolter R., Clardy J., Roseobacticides: Small molecule modulators of an algal-bacterial symbiosis. J. Am. Chem. Soc. 133, 18343–18349 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mayers T. J., Bramucci A. R., Yakimovich K. M., Case R. J., A bacterial pathogen displaying temperature-enhanced virulence of the microalga Emiliania huxleyi. Front. Microbiol. 7, 892 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kindler K., Khalili A., Stocker R., Diffusion-limited retention of porous particles at density interfaces. Proc. Natl. Acad. Sci. U.S.A. 107, 22163–22168 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kiene R. P., Visscher P. T., Production and fate of methylated sulfur compounds from methionine and dimethylsulfoniopropionate in anoxic salt marsh sediments. Appl. Environ. Microbiol. 53, 2426–2434 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zinder S. H., Brock T. D., Methane, carbon dioxide, and hydrogen sulfide production from the terminal methiol group of methionine by anaerobic lake sediments. Appl. Environ. Microbiol. 35, 344–352 (1978). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rimbault A., Niel P., Virelizier H., Darbord J. C., Leluan G., l-Methionine, a precursor of trace methane in some proteolytic Clostridia. Appl. Environ. Microbiol. 54, 1581–1586 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.King G. M., Metabolism of trimethylamine, choline, and glycine betaine by sulfate-reducing and methanogenic bacteria in marine sediments. Appl. Environ. Microbiol. 48, 719–725 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jameson E., et al., Deltaproteobacteria (Pelobacter) and Methanococcoides are responsible for choline-dependent methanogenesis in a coastal saltmarsh sediment. ISME J. 13, 277–289 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lidbury I., Kimberley G., Scanlan D. J., Murrell J. C., Chen Y., Comparative genomics and mutagenesis analyses of choline metabolism in the marine Roseobacter clade. Environ. Microbiol. 17, 5048–5062 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Welsh D. T., Ecological significance of compatible solute accumulation by micro-organisms: From single cells to global climate. FEMS Microbiol. Rev. 24, 263–290 (2000). [DOI] [PubMed] [Google Scholar]
- 59.Gibb S. W., Hatton A. D., The occurrence and distribution of trimethylamine-N-oxide in Antarctic coastal waters. Mar. Chem. 91, 65–75 (2004). [Google Scholar]
- 60.Yang X.-H., Scranton M. I., Lee C., Seasonal variations in concentration and microbial uptake of methylamines in estuarine waters. Mar. Ecol. Prog. Ser. 108, 303 (1994). [Google Scholar]
- 61.King G. M., Klug M. J., Lovley D. R., Metabolism of acetate, methanol, and methylated amines in intertidal sediments of lowes cove, Maine. Appl. Environ. Microbiol. 45, 1848–1853 (1983). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fitzsimons M., Kahni-Danon B., Dawitt M., Distributions and adsorption of the methylamines in the inter-tidal sediments of an East Anglian Estuary. Environ. Exp. Bot. 46, 225–236 (2001). [Google Scholar]
- 63.Lee C., Olson B. L., Dissolved, exchangeable and bound aliphatic amines in marine sediments: Initial results. Org. Geochem. 6, 259–263 (1984). [Google Scholar]
- 64.Naqvi S. W. A., et al., Biogeochemical ocean-atmosphere transfers in the Arabian sea. Prog. Oceanogr. 65, 116–144 (2005). [Google Scholar]
- 65.Bižić-Ionescu M., et al., “Oxic methane cycling: New evidence for methane formation in oxic lake water” in Biogenesis of Hydrocarbons, Stams A. J. M., Souna D. Z., Eds. (Springer International Publishing, Basel, 2018), pp. 1–22. [Google Scholar]
- 66.Jones H. J., et al., A new family of uncultivated bacteria involved in methanogenesis from the ubiquitous osmolyte glycine betaine in coastal saltmarsh sediments. Microbiome 7, 120 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhuang G. C., Peña-Montenegro T. D., Montgomery A., Hunter K. S., Joye S. B., Microbial metabolism of methanol and methylamine in the Gulf of Mexico: Insight into marine carbon and nitrogen cycling. Environ. Microbiol. 20, 4543–4554 (2018). [DOI] [PubMed] [Google Scholar]
- 68.Deng W., Peng L., Jiao N., Zhang Y., Differential incorporation of one-carbon substrates among microbial populations identified by stable isotope probing from the estuary to South China Sea. Sci. Rep. 8, 15378 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Saunois M., et al., The global methane budget 2000–2012. Earth Syst. Sci. Data 8, 697–751 (2016). [Google Scholar]
- 70.Tang K. W., et al., Paradox reconsidered: Methane oversaturation in well‐oxygenated lake waters. Limnol. Oceanogr. 59, 275–284 (2014). [Google Scholar]
- 71.Wiesenburg D. A., N. L. Guinasso, Jr, Equilibrium solubilities of methane, carbon monoxide, and hydrogen in water and sea water. J. Chem. Eng. Data 24, 356–360 (1979). [Google Scholar]
- 72.Guo K., Li L., Differential 12C-/13C-isotope dansylation labeling and fast liquid chromatography/mass spectrometry for absolute and relative quantification of the metabolome. Anal. Chem. 81, 3919–3932 (2009). [DOI] [PubMed] [Google Scholar]
- 73.Johnson D. W., A flow injection electrospray ionization tandem mass spectrometric method for the simultaneous measurement of trimethylamine and trimethylamine N-oxide in urine. J. Mass Spectrom. 43, 495–499 (2008). [DOI] [PubMed] [Google Scholar]
- 74.Lane D. J., Nucleic Acid Techniques in Bacterial Systematics (Wiley, 1991), pp. 115–147. [Google Scholar]
- 75.Luton P. E., Wayne J. M., Sharp R. J., Riley P. W., The mcrA gene as an alternative to 16S rRNA in the phylogenetic analysis of methanogen populations in landfill. Microbiology (Reading) 148, 3521–3530 (2002). [DOI] [PubMed] [Google Scholar]
- 76.Caporaso J. G., et al., Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 6, 1621–1624 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Schloss P. D., et al., Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75, 7537–7541 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Edgar R. C., Haas B. J., Clemente J. C., Quince C., Knight R., UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27, 2194–2200 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Quast C., et al., The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Newton R. J., Jones S. E., Eiler A., McMahon K. D., Bertilsson S., A guide to the natural history of freshwater lake bacteria. Microbiol. Mol. Biol. Rev. 75, 14–49 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kashyap D. R., Botero L. M., Franck W. L., Hassett D. J., McDermott T. R., Complex regulation of arsenite oxidation in Agrobacterium tumefaciens. J. Bacteriol. 188, 1081–1088 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Simon R., Priefer U., Pühler A., A broad host range mobilization system for in vivo genetic engineering: Transposon mutagenesis in gram negative bacteria. Biotechnology 1, 784–791 (1983). [Google Scholar]
- 83.Oberortner E., Cheng J. F., Hillson N. J., Deutsch S., Streamlining the design-to-build transition with build-optimization software tools. ACS Synth. Biol. 6, 485–496 (2017). [DOI] [PubMed] [Google Scholar]
- 84.Miller J. H., M9 minimal medium (standard). Cold Spring Harb Protoc., 10.1101/pdb.rec12295 (2010). [DOI] [Google Scholar]
- 85.Wang Q., McDermott T. R., Acidovorax sp. strain MeA-13 asparatate aminotransferase (aat) gene, complete cds. Genbank. https://www.ncbi.nlm.nih.gov/nuccore/MK170382. Deposited 12 November 2018.
- 86.McDermott T., 2017 Yellowstone lake water 16S illumina. Raw sequence reads. Genbank. https://www.ncbi.nlm.nih.gov/biosample/?term=SAMN13704654. Deposited 31 December 2019.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
In addition to the methods described herein, DNA sequence data have been deposited in GenBank (https://www.ncbi.nlm.nih.gov): 1) aat gene, MK170382 (85); 2) Illumina libraries, Biosample accession: SAMN13704654 (86). All other study data are included in the article and/or SI Appendix.