Abstract
The lack of precision therapies combined with limited therapeutic access to intracranial tumor sites due to the presence of the blood-brain/blood-tumor barriers have contributed to glioblastoma (GBM) being one of the most difficult cancers to effectively treat. We have developed a novel precision medicine approach for GBM treatment that involves the use of brain-penetrant RNA interference (RNAi)-based spherical nucleic acids (SNAs). In this study, we used SNAs consisting of gold nanoparticle cores covalently conjugated with radially oriented and densely packed siRNA oligonucleotides. Based upon previous preclinical evaluation, we conducted toxicology and toxicokinetic studies in non-human primates, and a single-arm, open-label phase 0 first-in-human trial (NCT03020017) to determine safety, pharmacokinetics, intratumoral accumulation and gene-suppressive activity of systemically administered SNAs carrying siRNA specific for the GBM oncogene Bcl2Like12 (Bcl2L12). Patients with recurrent GBM were treated with intravenous administration of siBcl2L12-SNAs (drug moniker: NU-0129), at a dose corresponding to 1/50th of the no-observed-adverse-event-level (NOAEL), followed by tumor resection. Safety assessment revealed no significant treatment-related toxicities. Inductively coupled plasma mass spectrometry, X-ray fluorescence microscopy, and silver staining of resected GBM tissue demonstrated that intravenously administered SNAs reached patient tumors, with gold (Au) enrichment observed in the tumor-associated endothelium, macrophages and tumor cells. NU-0129 uptake into glioma cells correlated with significant reduction in tumor-associated Bcl2L12 protein expression, as indicated by comparison of NU-0129-treated recurrent vs. matched primary (i.e., untreated) tumor. Our results establish SNA nanoconjugates as a brain-penetrant precision medicine approach for the systemic treatment of GBM.
ONE SENTENCE SUMMARY.
This first-in human phase 0 study of gene-regulatory RNA interference-based Spherical Nucleic Acids demonstrates that SNA nanoconjugates represent a potentially safe and brain-penetrant precision medicine approach for the treatment of glioblastoma.
INTRODUCTION.
Current standard treatment for newly diagnosed glioblastoma (GBM) is surgical resection followed by radiotherapy with concurrent and adjuvant temozolomide (TMZ). This treatment approach prolongs patient median survival from 12.1 to 14.6 months and increases the five-year survival rate from 2% to 10% (1, 2). With the addition of tumor-treating fields (TTFs) to the standard-of-care regimen, median survival increases to 20.9 months, as indicated by the results of a large randomized control study (3). Treatment options for recurrent GBM include surgical resection, TTFs, re-irradiation, systemic administration of the DNA alkylator lomustine (4), surgical implantation of chemotherapy-bearing wafers (polifeprosan 20 with carmustine implant, Gliadel® Wafer) (5), and systemic administration of the anti-angiogenic monoclonal antibody bevacizumab to which a partial response rate of 20–26% has been reported (6, 7). Each of the above-mentioned therapies has shown some, albeit modest and inconsistent activity against recurrent tumor, and has been associated with significant treatment-related toxicities, including wound breakdown after surgical resection (8).
Genotoxic therapy continues to be the default approach to treating newly diagnosed GBM, in part due to the lack of success in developing effective precision GBM therapies. This is despite the results of high content, multiplatform oncogenomic characterization of GBM tumors that have revealed numerous gene alterations, which support unabated tumor cell proliferation, survival, invasion, angiogenesis, and therapy resistance (9). To date, studies applying a precision medicine strategy to the treatment of GBM have shown that these tumors compensate for approaches that target a single protein activity by activating related pathway components. Such pathway compensation, e.g., as that demonstrated for members of the receptor tyrosine kinase family (10, 11), suggests that co-inhibition strategies are required to improve treatment outcomes for patients with GBM. Due to drug-drug interactions and increased toxicities associated with combinatorial inhibitor and/or chemotherapy treatments, the development of multi-agent treatments is challenging.
RNA interference (RNAi)-mediated gene silencing to downregulate oncogene drivers has emerged as a powerful and novel concept of cancer therapy that can address many of the challenges of conventional drug development and application (12–15), in particular related to inhibiting oncogenic aberrations that encode ‘undruggable’ targets. However, the delivery of nucleic acid-based therapies to brain tumors is particularly challenging. Unmodified oligonucleotides are rapidly cleared, subject to degradation by serum nucleases, inefficiently taken up by cells and tissues, and may trigger immune responses (16). In addition, systemically administered therapies that are intended to act against brain tumors have to negotiate passage through the blood-brain barrier (BBB), the blood-cerebrospinal fluid barrier (BCSF), and the blood-tumor barrier (BTB), and they must withstand substantial dynamic forces in the brain interstitium that are caused by cerebrospinal fluid (CSF) flow, intratumoral edema, and tumor mass-related pressure effects (17).
While oligonucleotide carriers such as polymers, polyplexes, or lipids each display specific safety concerns and delivery limitations, several nanotechnological platforms have emerged as carriers for delivering oligonucleotide payloads to tumors. Seven different siRNA-based cancer nanotherapeutics have been or are currently under clinical investigations. These include lipid nanoconjugates, such as Atu027, which target protein kinase 3, a downstream effector of phosphatidylinositol 3 kinase signaling. Phase I/II clinical trials demonstrated that Atu027 was well tolerated in patients with advanced solid tumors and resulted in stable disease in 41% of patients (18, 19). Furthermore, systemic administration of CALAA-01, cyclodextrin-based polyplex, encapsulating siRNA targeted to ribonucleotide reductase subunit M2 (RRM2), and decorated with a transferrin receptor ligand for tumor cell targeting, resulted in preferential accumulation in human metastatic melanoma tumors in a dose-dependent manner, and inhibited melanoma-associated RRM2 expression (20). While the first RNAi-based nanotherapeutic has been FDA-approved for the treatment of transthyretin-related hereditary amyloidosis (21), FDA approval for an RNAi-based cancer therapeutic has not been reported yet.
To inhibit oncogene function in GBM, and to address the challenges of effective drug delivery to intracranial tumor, we have developed RNAi-based spherical nucleic acids (SNAs). Gene-regulatory SNAs are nanostructures that are densely functionalized at their nanoparticle surface with highly oriented siRNA oligonucleotides. In this study, we evaluated such SNAs composed of siRNA oligonucleotides assembled on gold (Au) nanoparticle cores that are backfilled with surface-passivating oligoethylene glycol or polyethylene glycol (OEG/PEG) to improve colloidal stability and circulation half-life (22). While SNAs are highly tunable structures that can be composed of a variety of different oligonucleotides and core materials, we have chosen an Au-based construct in previous preclinical proof-of-concept experiments (23, 24), and in the clinical studies outlined here. The use of Au as the SNA core allows for precise quantification of Au spatial distribution in cells and tumors by inductively coupled plasma mass spectrometry (ICP-MS), X-ray fluorescence microscopy (XFM), and silver histology staining. In preclinical proof-of-concept studies, we generated and characterized Au-based gene regulatory SNAs targeted to the GBM oncogene Bcl2Like12 (Bcl2L12), a proline-rich protein characterized by a C-terminal 14 amino acid sequence with significant homology to the BH (Bcl-2 Homology) 2 domain found in several members of the Bcl-2 family (25, 26). Bcl2L12 expression is increased in GBM through multiple mechanisms, including corresponding gene amplification [non-focal gain of chromosome 19(q)], transcriptional upregulation, and alterations in regulatory miRNA expression (24, 26). With regard to its tumor promoting mechanism, Bcl2L12 inhibits effector caspase-3 and caspase-7 activation through direct interaction with pro-caspase-7 (26) and upregulation of the small heat shock protein and pro-caspase-3-specific inhibitor, αB-crystallin (27). In addition, Bcl2L12 can bind wild-type p53 and, to a lesser extent, p53 conformational mutants, and in so doing decrease p53 protein stability as well as block p53-mediated transactivation by preventing p53 binding to target gene promoters (28). In view of the low-level expression of Bcl2L12 in the adult brain and its consistently elevated expression in GBM tumors, we consider Bcl2L12 a rational therapeutic target, and have extensively characterized siBcl2L12-carrying SNAs (23, 24). SNAs show robust uptake into patient-derived glioma-initiating cells (GICs), which results in reduced Bcl2L12 protein expression and enhanced effector caspase-3 and −7 activation, as well as reduced p53-associated transcriptional activity Using 5′ RNA ligase-mediated rapid amplification of cDNA ends (5-RLM-RACE), we demonstrated that SNA-driven downregulation of Bcl2L12 occurred through RNAi-mediated mRNA cleavage (23). Consistent with limited SNA off-target effects, as determined by genome-wide expression profiling (29), systemic SNA administration into Sprague-Dawley rats did not result in differences in body or organ weight, nor in inflammatory response within the brain or organs of the reticuloendothelial system (23). Importantly, SNAs cross the BBB/BTB in non-tumor-bearing mice, and to a greater extent in patient-derived xenograft (PDX)-bearing mice, respectively, as demonstrated by In Vivo Imaging System (IVIS) analysis, silver staining of tumor sections, and magnetic resonance imaging (MRI) of animal subjects injected with gadolinium (III)-conjugated SNAs (23). Accumulation and pervasive dissemination into extravascular tumor parenchyma resulted in intratumoral Bcl2L12 protein knockdown and impaired GBM cell tumorigenicity, as measured by bioluminescence imaging and as indicated by prolonged survival of PDX-bearing mice (23, 24, 30).
In this study, we have examined siBcl2L12-SNA (drug moniker: NU-0129) toxicology, toxicokinetics and no-observed-adverse-effect level (NOAEL) in non-human primates. Further, we have conducted a first-in-human phase 0 clinical trial involving the systemic administration of NU-0129 in patients with recurrent GBM, to assess safety, pharmacokinetics, intratumoral SNA nanoconjugate accumulation, and target knockdown (clinical trial identifier: NCT03020017). Our results provide the first evidence that systemic administration of a NU-0129 microdose is safe, results in nanoconjugate uptake by endothelial, immune and tumor cells, and correlates with reduced target protein expression in patient GBM tumor.
RESULTS.
Toxicology study in cynomolgus monkeys.
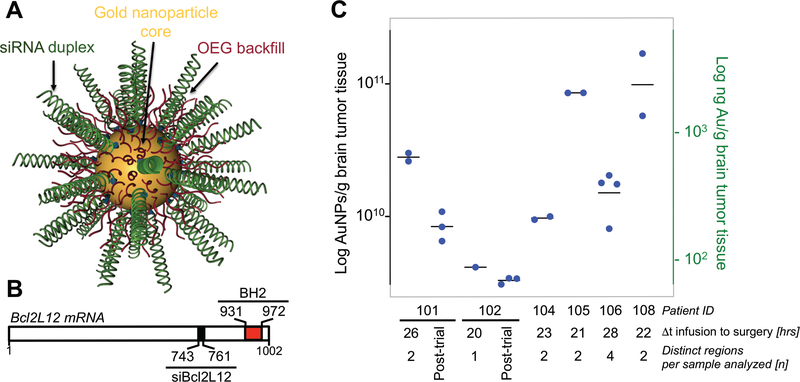
NU-0129 [Fig. 1A-B, showing a schematic of the SNA architecture, and the siRNA binding site within the Bcl2L12 mRNA (nucleotides 743–761), relative to nucleotides 931 to 972 encoding the C-terminal BH2 domain] was manufactured in compliance with current good manufacturing practice (cGMP) standards (see Materials and Methods for details), and assessed for compound stability, including siRNA duplex integrity and content, together with nanoconjugate size (table S1). To evaluate the potential toxicity of intravascular NU-0129 administration, we determined toxicity and toxicokinetic profiles of NU-0129 when administered via an intravenous bolus dose to cynomolgus monkeys, and evaluated the recovery, persistence, or progression of any effects following a 14-day recovery period (see Table 1A for summary of adverse effects in cynomolgus monkeys; see SI Materials and Methods for details on the study design). All animals survived to the scheduled necropsy. NU-0129 treatment did not affect body weight, hematology, coagulation, serum chemistry, urinalysis, or organ weight, and had no impact on heart rate, pulse pressure, body temperature, electrocardiography intervals (PR, QRS, RR, QT, or QTcB), or electrocardiography waveform morphology. NU-0129-related clinical observations of purple and/or blue discoloration of various body surfaces [facial area, gums, forelimb(s), hindlimb(s), oral cavity, and/or trunk (dorsal and ventral)] were noted in the 4 and 8 mg/kg toxicology group (both males and females) and in the 8 mg/kg cardiovascular group (both males and females) throughout the study, and in a single 1 mg/kg toxicology group (male) on study days 4 and 5. Similar to rodents systemically treated with NU-0129 (23), NU-0129-related macroscopic observations of blue discoloration due to Au accumulation were noted in many tissues [adrenals, gastrointestinal tissues (esophagus, stomach, duodenum, jejunum, ileum, cecum, colon, and rectum), epididymis, lymphoid tissues (spleen, lymph nodes, and thymus), skin, liver, tongue, urogenital tissues (seminal vesicles, testes, vagina, and urinary bladder)] of most males and females at 8 mg/kg on study days 2 and 14. In general, purple discoloration was noted only on the day of dose administration, while blue discoloration was observed for the remainder of the recovery period.
Fig. 1. ICP-MS analysis demonstrates Au accumulation in recurrent GBM patient tumors.
(A) Schematic of NU-0129. (B) Bcl2L12 mRNA with binding site for the siRNA oligonucleotide used to functionalize gold nanoparticle cores (nucleotide position 743–761; black box). Positions for nucleotides encoding the C-terminal BH2 are indicated (nucleotide position 931–972; red box). (C) ICP-MS analysis of bulk patient GBM tissue, including tumor recurrences post NU-0129 trial enrollment for patients 101 and 102; tumors recurred 159 and 174 days post NU-109 trial enrollment, respectively. n, number of tumor regions sampled. Dt, time from surgery to infusion. Shown is the median. Of note, tumor resections for patients 103 and 107 did not yield sufficient viable tumor tissue for ICP-MS.
In both males and females in the 8 mg/kg cardiovascular group, NU-0129-related lower systolic blood pressure was noted at 0–15 and 16–30 min post-dosing, and lower diastolic and mean arterial blood pressure values were noted at 0–15 min post-dosing (Table 1A). Injection site findings of acute perivascular hemorrhage were noted in all groups, including the control group, on study days 2 and 14. Procedure-related injection site findings of perivascular acute inflammation (neutrophils, edema, and fibrin exudation) was noted at all NU-0129 dose levels on study day 2, and subacute inflammation was noted in the 4 and 8 mg/kg group males on study day 14.
Exposure to NU-0129 increased with increasing drug dose level, as assessed by quantification of siRNA oligonucleotide content using Liquid Chromatography-High Performance Liquid Chromatography (LC-HPLC) using an atto-labeled peptide nucleic acid (PNA) probe (see fig. S1 for summary of toxicokinetic parameters). The increase in AUClast was greater than the proportional NU-0129 dose increase from 1 mg/kg to 8 mg/kg: an 8-fold increase in NU-0129 dose resulted in a nearly 40-fold increase in AUClast for both male and females. NU-0129 exposure, in terms of AUClast, was slightly higher in females than in males, but differences were less than two-fold. Mean clearance (CL), ranging from approximately 600 to 3000 mL/h/kg, was moderate to high [approximately 20% to 120% of hepatic blood flow in the monkey as reported (31)], and decreased as dose increased. Mean volume of distribution at steady state (VSS) was small, ranging from approximately 50 to 200 mL/kg [approximately 10% to 30% of total body water in the monkey as reported (31)], and also decreased with increasing dose. This resulted in a short terminal elimination half-life that ranged from approximately 2 to 9 min for individual animals. CL and VSS values were approximately 1.4- to 2-fold higher for males than females, but half-life values were similar. Based on the results of this study, a single intravenous (bolus) injection of NU-0129 to cynomolgus monkeys resulted in a no-observed-adverse-effect level (NOAEL) of 8 mg/kg for both systemic and local assessments. This dose corresponded to AUClast and clearance values of 9.82 μg•h/mL and 863 mL/h/kg for males, and 14.2 μg•h/mL and 570 mL/h/kg for females, respectively.
Patient enrollment and clinical workflow.
Based on our cumulative studies in cell-based assays and PDX mouse model in vivo (23), and non-human primate results described above, we conducted a single-arm, open-label, phase 0 first-in-human trial to determine the safety, pharmacokinetics, and biodistribution of intravenously administered NU-0129 in patients with recurrent GBM (NCT03020017). Patients were required to have a diagnosis of recurrent GBM or gliosarcoma, appropriate bone marrow [white blood cell count (WBC) ≥ 3,000/μL, absolute neutrophil count (ANC) ≥ 1,500/mm3, platelet count of ≥ 100,000/mm3, hemoglobin ≥8 g/dL], and organ function [Bilirubin ≤ 2 × upper limit normal (ULN), aspartate aminotransferase/alanine transaminase (AST/ALT) ≤ 2 × ULN, creatinine ≤ 1.5 × ULN] in order to qualify for enrollment in this study. Patients were also required to have failed prior front-line therapy with radiation and TMZ chemotherapy and were not to have had cancer therapy within 21 days of registration (biologic, cytotoxic, experimental therapies and nitrosoureas) with no limitations concerning the number of prior treatments, including with bevacizumab. All patients had a diagnosis of recurrent GBM, and therefore had received prior first line therapy, including radiation and TMZ chemotherapy (see table S2 for details on patient characteristics). A total of 8 patients [median age 55.5 (range 32–66)] consented and were registered on study, of which 4 were female. All 8 patients were evaluable for the primary outcome of patient safety and toxicity. Seven of the patients were White/Caucasian and one was Black/African American. At the time of study data freeze, 6 of the 8 patients were deceased from GBM tumor progression and not from treatment-related complications. Recurrent patient tumors, as well as their corresponding newly diagnosed pre-treatment tumors, were subjected to Next Generation Sequencing (NGS) to identify mutational status of key glioma oncogenes and tumor suppressors (see table S3 for patient characteristics and molecular features).
Based upon the NU-0129 NOAEL of 8 mg/kg siRNA dose, determined through toxicology studies in cynomolgus monkeys described above, we determined 1/50th the human equivalent dose to be 0.04 mg/kg. Patients were dosed via a single intravenous NU-0129 injection given 20–28 hrs prior to surgical start time. We have chosen this infusion schedule based upon preclinical studies in PDX-bearing mice that demonstrated maximum intratumoral target knockdown 24–48 hrs post intravenous SNA administration (23, 30). Tumor tissue obtained at surgery was collected and analyzed for Au content and target protein expression, as detailed below. Patients were monitored via telemetry, vital signs, and labs, before, during and after the NU-0129 infusion. After hospital discharge post-tumor resection, patients continued to be monitored for safety, toxicity, and adverse events through weekly outpatient monitoring visits for 3 weeks post-infusion. Patients were enrolled on study one-at-a-time and were closely observed for toxicity through the 21-day toxicity-monitoring period. Once this 21-day period was completed, safety data was given to an external and independent Data and Safety Monitoring Board, which reviewed safety data and once deemed appropriate, the study reopened for enrollment of the next patient.
NU-0129 safety assessment.
With regard to the primary outcome of the study, micro-dosing of the study drug NU-0129 was well-tolerated. No significant treatment-related toxicities, i.e., grade 4 or 5 clinically relevant adverse events were observed. Only two treatment-related severe adverse events (> grade 3) were noted, i.e., hypophosphatemia and lymphopenia, both of which were considered as ‘possibly’ related by the treating oncologists (see Table 1B for all treatment-related adverse events). The severity of both adverse events decreased by the end of the toxicity-monitoring period. One serious adverse event occurred, a cerebrospinal fluid leak, which was observed in one patient post-operatively (grade 3). It was deemed related to surgery and unrelated to NU-0129. Of note, skin discoloration and acute perivascular hemorrhage at the injection site that were observed in cynomolgus monkeys were not seen in human subjects enrolled on study.
NU-0129 pharmacokinetics.
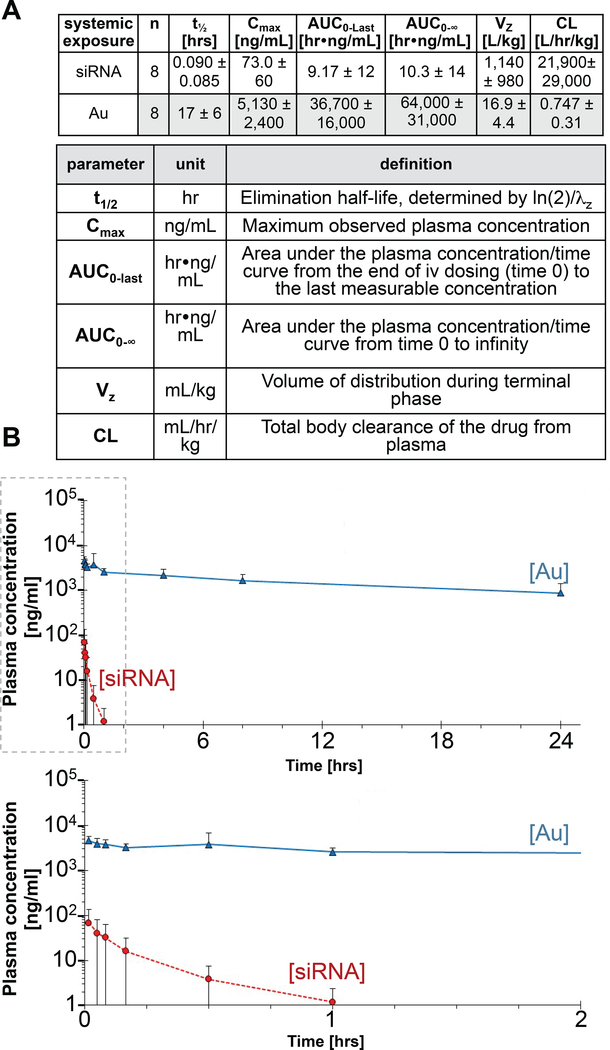
Mean plasma levels of NU-0129 were derived from time profiles for both siRNA and Au concentrations, with Au plasma level determined by ICP-MS, and siRNA concentration assessed by LC-HPLC using an atto dye-labeled PNA probe (see Materials and Methods). Fig. 2 summarizes the pharmacokinetic (PK) parameters based upon Au and siRNA oligonucleotide content. In general, inter-subject variability in systemic exposure was larger for siRNA than for Au. Upon intravenous NU-129 infusion at 0.04 mg/kg of body weight, siRNA was rapidly eliminated, with a mean t1/2 of 0.09 hrs (5.4 min; %CV 95; range 1 to 17 min, median 0.06, 95%Cl [0.02, 0.16] hrs). CL and Vz mean values for siRNA were 21,900 mL/hr/kg (%CV 133; median 7,470; 95%Cl [0, 46,400]) and 1,140 mL/kg (%CV 86; median 602; 95%Cl [322, 1,960]), respectively. CL for siRNA was very high when compared to hepatic blood flow (1,200 mL/hr/kg), while Vz was about twice the total body water volume in humans (600 mL/kg). The mean observed Cmax and AUC0−∞ were 73.0 ng/mL (%CV 82; median 62.1; 95%Cl [22.8, 123]), and 10.3 hr•ng/mL (%CV 133; median 5.38; 95%Cl [0, 21.7]), respectively. In contrast to siRNA, Au was slowly eliminated following IV infusion, with a mean t1/2 of 17 hrs (%CV 35; range 9 to 28 hrs; median 18; 95%Cl [12, 23]). CL and Vz mean values for Au were 0.747 mL/hr/kg (%CV 41; median 0.804; 95%Cl [0.489, 1.01]), and 16.9 mL/kg (%CV 26; median 15.9; 95%Cl [13.2, 20.6]), respectively. CL for Au was very low when compared to hepatic blood flow, while Vz was about 3% of the total body water volume in humans. The mean observed Cmax and AUC0−∞ were 5,130 ng/mL kg (%CV 47; median 4,290; 95%Cl [3,120, 7,140]), and 64,000 hr•ng/mL (%CV 49; median 50,000; 95%Cl [38,000, 90,000]), respectively.
Fig. 2. Average plasma concentration-time profiles of siRNA and Au following intravenous infusion in recurrent GBM patients.
(A) Key pharmacokinetic parameters of siRNA and Au in patient plasma. Standard deviations are indicated in brackets. (B) Plot of average plasma concentration time profiles of siRNA and Au following an IV Infusion with NU-0129; lower panel shows graph truncated at the 2 hrs time point post infusion. Data points are shown ± standard deviations. Blood was drawn at 0, 1, 3, 5, 10 min, 0.5, 1, 4, 8 and 24 hrs post NU-0129 infusion.
NU-0129 intratumoral accumulation.
For 6 of the 8 patients enrolled in the clinical trial, sufficient tumor tissue was available for comprehensive quantitative assessment of Au intratumoral accumulation by ICP-MS. The remaining two patients, i.e., patients 103 and 107, had smaller tumor resections, which resulted in insufficient amount of viable tumor tissue for ICP-MS analysis. Therefore, overall level and intratumoral distribution of Au were analyzed by ICP-MS for patients 101, 102, 104, 105, 106 and 108, and by XFM, and histological analysis of silver staining, for all patients. For ICP-MS, multiple areas of resected tumors were analyzed to quantify Au concentration, and to determine Au distribution in tumor specimens. Au was detectable in all tumor specimens analyzed, with variable intratumoral levels evident in each case (Fig. 1C). We also assessed long-term Au accumulation in 2 GBM tumors (patients 101 and 102) that recurred 159 and 174 days post NU-109 trial enrollment, respectively. As shown in Fig. 1C, Au was detected in both recurrences, with 41% and 81% of the initial Au level still present in tumors.
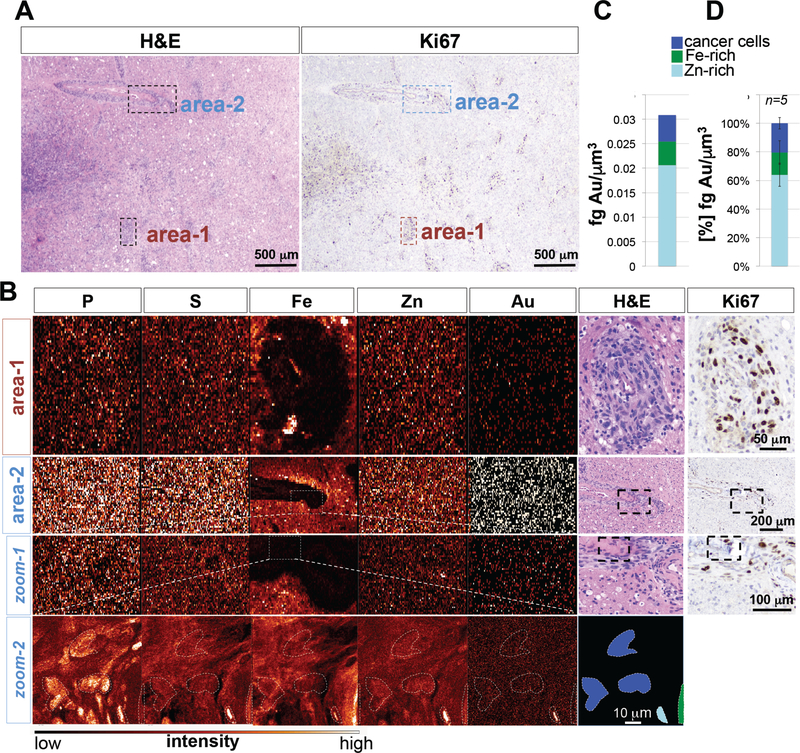
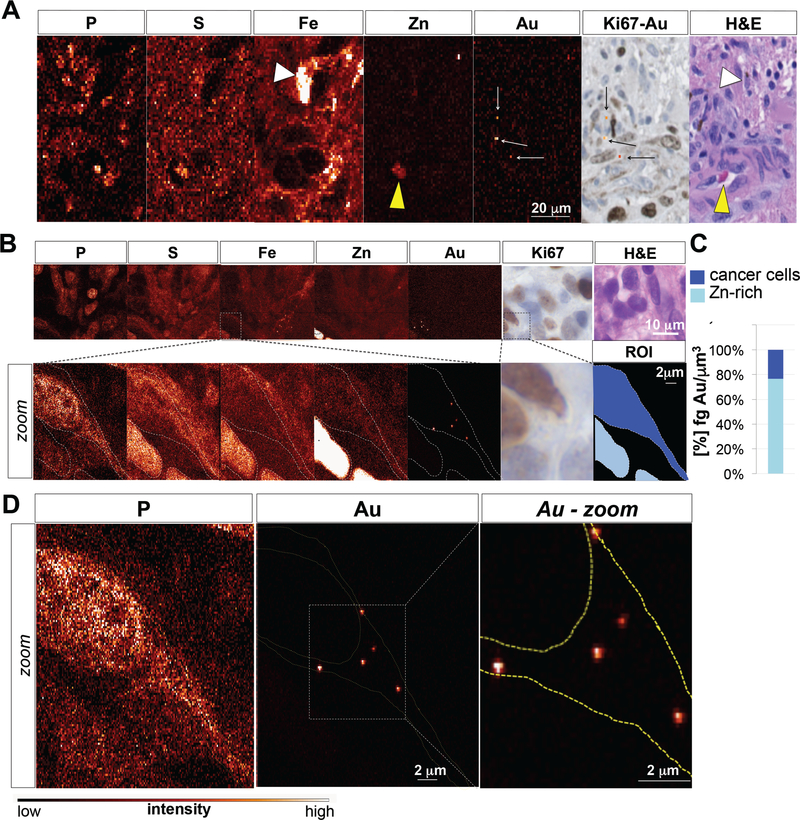
To analyze spatial distribution of Au within tumor tissue, synchrotron XFM elemental maps of GBM tissue slices were acquired at micron and sub-micron resolution and matched to adjacent H&E and Ki67-stained tumor sections. Large-step-size overview scans revealed Au distribution in both perivascular (Fig. 3A,B), and intraparenchymal tumor elements (fig S2A, B; fig. S3A, B). Subsequent analysis of small-step-size scans demonstrated Au accumulation in perivascular (Fig. 3B) and intraparenchymal Ki67-positive tumor cells (fig. S2B; fig. S3B). Quantification of elemental Au concentration revealed that the majority of Au was associated with Zn/Fe-rich vasculature, and that cancer cells contained on average 9.1×10−3 fg of Au per μm3 (2.7×10−3 to 4×10−2 fg/μm3) (Fig. 3C; fig. S2C; fig. S3C). XFM imaging and subsequent elemental quantification across multiple sections from different patient tumors confirmed Au accumulation in Ki67-positive glioma cells in both perivascular and intraparenchymal regions, with 20.5% (± 8.15%) of total Au found in cancer cells, and the majority of Au associated with the Zn-rich tumor vasculature (Fig. 3D). To gain subcellular information on Au distribution, we repeated XFM using the bionanoprobe (BNP) focused to an 80 nm X-ray beam in order to map Au distribution at high spatial resolution. This analysis confirmed Au accumulation in cancer cells (Fig. 4A-C), particularly within the cytoplasm (Fig. 4D). Au enrichment in Fe and Zn-rich areas additionally indicate Au presence associated with red blood cells (RBCs) inside the lumen of tumor-associated vessels (Fig. 4A, yellow arrowhead; fig. S4), and within tumor-associated marcophages (Fig. 4A, white arrowhead).
Fig. 3. XFM elemental maps of recurrent GBM tumor (X-ray beam spot 0.3 micron).
(A) Overview H&E and Ki67 staining of patient 106 tumor. Indicated are 2 perivascular tumor regions. (B) Phosphorus (P), sulfur (S), iron (Fe), zinc (Zn) and gold (Au) elemental maps of GBM tumor sample 106. Magnified areas from large step size scans are boxed. (C) Quantification of Au content in Zn-rich areas, Ki67-positive glioma cells, and Fe-rich areas. Color bar indicates pixel concentrations for each element ranging from black (no signal) to white (highest signal for a given element). (D) Cumulative quantification of Au distribution in the different cell populations across different sections and patient tumors. A total of 5 sections were analyzed. Shown is the median ± standard deviation.
Fig. 4. XFM-Bionanoprobe (X-ray beam spot 80 nanometers) assessment of patient tumor 107 reveals extranuclear/cytoplasmic distribution of Au in tumor cells.
(A) Phosphorus (P), sulfur (S), iron (Fe), zinc (Zn) and gold (Au) elemental maps of GBM tumor sample 106, using the XFM-Bionanoprobe (X-ray beam spot 80 nanometers), together with matching H&E and Ki67 stainings demonstrate Fe accumulation in tumor-associated macrophages (white arrow head), Zn enrichment in the lumen of blood vessels (yellow arrow head), and gold accumulation within perivascular Ki67-positive tumor cells. (B) Phosphorus (P), sulfur (S), iron (Fe), zinc (Zn) and gold (Au) elemental maps of tumor sample as assessed by XFM-BNP. Zoomed areas (0.1 micron step size) in overview scans (0.5 micron step size) are boxed. (C) High resolution image of a cancer cell demonstrates extranuclear/cytoplasmic localization of Au signal. Color bar indicates pixel concentrations for each element ranging from black (no signal) to white (highest signal for a given element). (D) Quantification of Au content in Zn-rich areas and Ki67-positive glioma cells. Color bar indicates pixel concentrations for each element ranging from black (no signal) to white (highest signal for a given element).
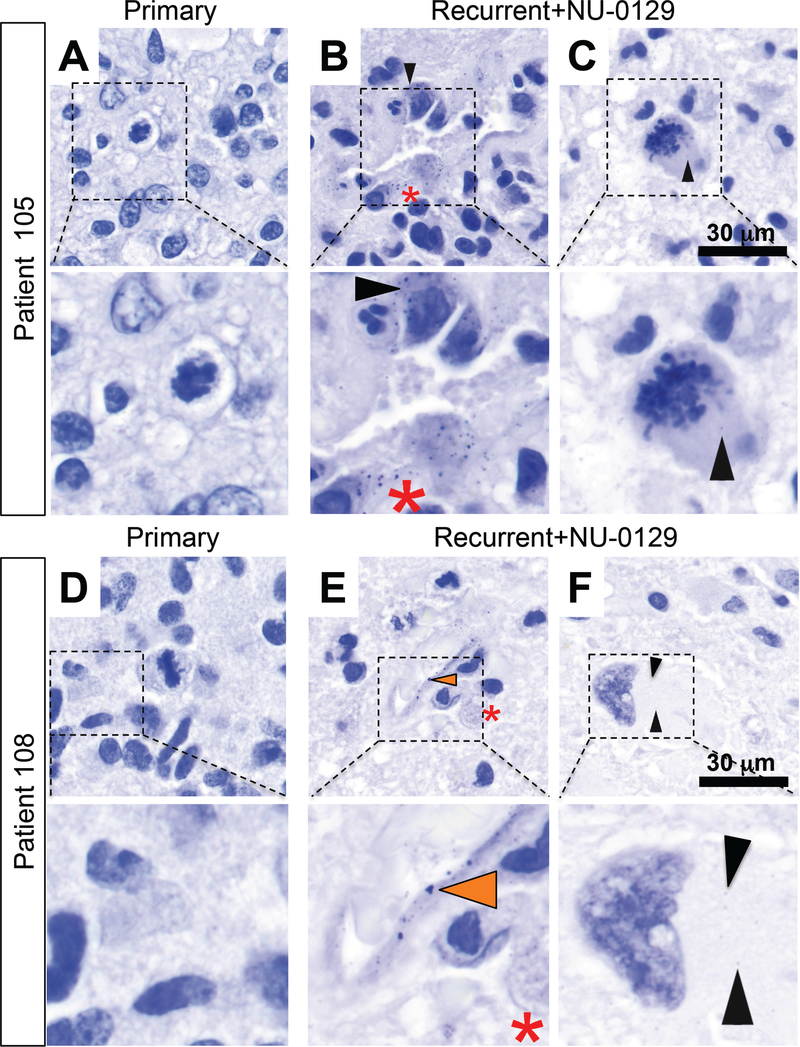
Cellular and subcellular accumulation of Au was confirmed by light microscopic analysis of silver-stained sections. Specificity of the silver histology stain was established by comparing staining intensities in matched primary versus NU-0129-treated recurrent patient tumors. This analysis confirmed Au signal in extranuclear areas of tumor cells (Fig. 5B, C, F; black arrowheads), in the tumor-associated endothelium (Fig. 5E, orange arrowhead), and within tumor-resident macrophages (Fig. 5B, E, red asterisk) in NU-0129-treated, but not in newly diagnosed (i.e., untreated) tumors (Fig. 5A, D). Taken together, our results show that Au accumulates in the tumor-associated endothelium and is taken in by tumor-associated macrophages and Ki67-positive tumor cells, in both perivascular and intraparenchymal tumor regions.
Fig. 5. Silver staining of pre- and post-treatment tumors 105 and 108.
(A, D) Silver staining of pre-treatment tumors in patients 105 and 108. (B-C; E-F) Silver staining of tumor sections post-NU-0129 treatment. In tumors resected after NU-0129 administration, Au was present within endothelial cells (E, orange arrowhead), tumor cells (B, C, F; black arrow head), and macrophages (B, E, red asterisk). Cell shown in panel F demonstrates therapy-related nuclear atypia typical for a post-therapy GBM tumor cell (panel F).
Intratumoral Bcl2L12 target expression in tumor samples pre and post NU-0129 treatment.
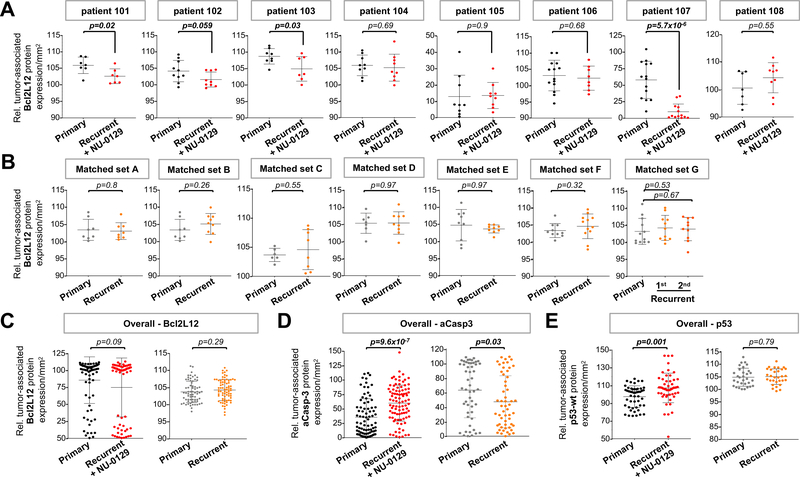
For quantification of Bcl2L12 protein in tumor cells from matched primary and NU0129-treated recurrent tumors, we analyzed immunohistochemistry (IHC) images using Histoquest 6.0.1.125 TissueGnostics software. A minimum of 8 regions of interest (ROIs) per sample was selected. Gating for cells within ROIs was set for relative mean Bcl2L12 staining intensity > 60, to quantify Bcl2L12 protein expression in cells with above-background Bcl2L12 signal only. Gating for tumor cells was set for large nuclei with nuclear areas > 50 μm2, and nuclear intensity > 67 (fig. S5A-C). To account for different tumor cellularity in different samples, we then calculated the fraction of IHC signal-positive tumor cells per total number of tumor cell nuclei per mm2. Anti-Bcl2L12 antibody specificity was verified by staining xenografted tumors expressing a vector control or Bcl2L12. As shown in fig. S5D, engrafted tumor with Bcl2L12 overexpression showed strong immunoreactivity in comparison to control tumors. Quantification of Bcl2L12 staining intensity in tumor cells revealed statistically significantly reduction in Bcl2L12 protein level in 4 out of 8 NU-0129 treated recurrent as compared to untreated primary tumors (Fig. 6A; see Fig. 6C for overall Bcl2L12 level in all matched tumor sets combined). To determine if reduced Bcl2L12 expression in NU-0129-treated recurrent tumors was due to downregulation of Bcl2L12 protein during recurrence, and not attributed to NU-0129 treatment, we quantified Bcl2L12 protein in 7 matched pairs of primary and recurrent tumor, using the same gating strategies described above, to identify Bcl2L12-positive tumor cells. As shown in Fig. 6B-C, Bcl2L12 protein level did not change significantly during tumor recurrence. In addition, we analyzed Bcl2L12 mRNA level in matched primary versus recurrent GBM tumors cataloged in the Glioma Longitudinal Analysis (GLASS) consortium (32) and GSE62153 datasets (33). This analysis showed that Bcl2L12 transcript levels between primary and recurrent tumors were not significantly different [fig. S6A-B; p=0.41 (GLASS); p=0.1 (GSE62153)]. To assess the effect of NU-0129 treatment on Bcl2L12 downstream signaling, protein abundance of active caspase-3 and wild-type p53 was quantified using IHC, and images were analyzed using Histoquest software. Our preclinical studies demonstrated enhanced caspase-3 activation upon Bcl2L12 knockdown in glioma cells in vitro and in murine orthotopic tumors (26, 27, 34, 35). Consistent with these results, NU-0129 treatment robustly increased levels of active caspase-3 in the majority of NU-0129-treated recurrent tumors, as compared to corresponding primary tumors (Fig. 6D, Fig. S7A). Consistent with the ability of Bcl2L12 to destabilize wild-type p53 protein (36), NU-0129-treated recurrent tumors harboring wild-type p53 alleles (patient tumors 103, 104, 105, 107; table S2) showed significantly elevated levels of wild-type p53 (Fig. 6E, Fig. S7C). Similar to Bcl2L12 protein and mRNA levels, caspase 3 and wild-type p53 transcript did not significantly change during GBM recurrence (fig. S6C, D). Of note, quantitative analysis of IHC stainings revealed that active caspase-3 level was significantly downregulated in trial-unrelated recurrent tumor, and p53 protein levels remained unchanged (Fig. 6D, E; Fig. S7B, D). Taken together, these results indicate that NU-0129 accumulation in recurrent GBM tumor cells correlates with significant downregulation of Bcl2L12 protein, elevated caspase-3 activation and increased level of wild-type p53.
Fig. 6. Protein expression of Bcl2L12, active caspase-3 and wild-type 53 in matched newly diagnosed and NU-0129-treated recurrent GBM.
(A-B) Histoquest quantification of IHC staining for Bcl2L12 in matched primary and NU-0129-treated recurrent GBM tumors, and in trial unrelated matched primary and recurrent GBM tumors. A minimum of 8 ROIs were selected per tumor samples. Shown is the mean ± standard deviations. (C-E) Histoquest quantification of overall IHC staining intensities for Bcl2L12, and the Bcl2L12 downstream effectors active caspase-3 and wild-type p53 (8–13 ROIs selected). Shown is the mean ± standard deviations. For all analyses, ROIs were randomly chosen. Cells within ROIs have above-background signal for IHC markers (mean intensity >60 for Bcl2L12; >15 for active caspase-3; >50 for p53). Tumor cells were identified based on the presence of larger nuclei with nuclear areas >50 μm2 and mean nuclear staining intensity of > 67. Shown is the fraction of marker positive tumor cells per total number of tumor cells per mm2. Outliers were identified by applying the interquartile range rule. p values were calculated using Student’s t-test. Patient tumors with significant differences in IHC marker expression level (p<0.05) are highlighted in bold.
DISCUSSION.
Several precision therapies are currently being evaluated for the treatment of GBM, including small molecule inhibitors that target growth factor receptors and their downstream signaling components, cell cycle and epigenetic regulators, and drivers of angiogenesis and antitumor immunity (37). While treatment responses have been observed in some patients, success in implementing targeted therapies as effective GBM treatments has yet to be realized. Here, in a non-human primate study and in a human phase 0 clinical trial, we provide first-time experimental evidence that gene-regulatory, siRNA-based SNA can be safely administered through microdose intravenous administration, cross the BBB/BTB, and show accumulation in GBM tumor cells, which correlated with reduced target protein level.
Consistent with results from our preclinical studies in rodents (23), systemic NU-0129 treatment in non-human primates and GBM patients was well-tolerated. Assessment of toxicity in non-human primates was based on body weights, blood chemistry, complete blood counts, and detailed histopathology, and in patients on the evaluation of vital signs and blood chemistry before, during, and after infusion. In total, these studies indicate the absence of acute or longer-term toxicity of systemic NU-0129 treatment.
Pharmacokinetic analysis revealed that plasma concentrations of the siRNA component of NU-0129 rapidly decline to undetectable by 1 hr post-infusion for all patients. Rates of siBcl2L12 oligonucleotide plasma clearance are consistent with results from other clinical studies evaluating pharmacokinetics of systemically administered nanoformulations carrying siRNA oligonucleotides, including siRNA targeted to the M2 subunit of ribonucleotide reductase (RRM2), delivered by a cyclodextrin-based polymeric nanostructure for the treatment of metastatic melanoma (38). Together, these observations suggest that the SNA structure influences siRNA plasma pharmacokinetics similar to other nanoparticle carriers, but that siRNA delivery and privileged access to intracranial tumor sites represent a unique enabling feature of the SNA architecture. Of note, consistent with pharmacokinetic studies in rodents (23), plasma Au showed much slower elimination compared to siRNA content, pointing to the loss of siRNA from Au over time, likely related to oligonucleotide degradation by plasma nucleases. RNase-mediated degradation of siRNA oligonucleotides conjugated to gold nanoparticle cores has been extensively studied. A previous study determined how siRNA immobilized on Au nanoparticle cores is recognized and degraded by serum nucleases (39). Results indicated that the degradation rate for siRNA on Au nanoparticle cores is rapid, and occurs via nuclease recognition and hydrolysis of specific sequence motifs (U–A and G–U, 5′–3′). One U–A motif is present in the siBcl2L12 sequences of NU-0129.
SNAs are modular structures, composed of densely functionalized and highly oriented nucleic acids that are spherically presented around a nanoparticle core. The type of oligonucleotide [e.g., DNA, siRNA, microRNA, PNA, or locked nucleic acid (LNA)] and nanoparticle core materials [e.g., Au (40), silver (Ag) (41), iron oxide (Fe3O4) (42, 43), quantum dots (CdSe, CdSe/ZnS) (43), platinum (43), silica (SiO2) (44), core-shell (Au@SiO2) (44), and liposomes (45)] provide numerous options for developing nucleic acid bearing nanoparticles to suit a specific diagnostic or therapeutic application. Importantly, SNA properties and capabilities stem from the densely functionalized and highly oriented nucleic acid shell, and not from the nanoparticle core (46–48). For our initial preclinical investigations (23), and the early phase clinical study described in this work, we have chosen an inorganic Au nanoparticle core that enables precise quantification and localization of intratumoral NU-0129, using ICP-MS and XFM. ICP-MS analysis revealed significant uptake of Au into bulk tumor tissue from patients that received infusion with a sub-therapeutic NU-0129 dose (0.04 mg/kg), with Au quantities ranging from 4×109 (patient 102) to 1.14×1011 particles per gram of tumor tissue (patient 108). The extent of intramural Au accumulation is likely driven by multiple factors, which may include timing of infusion, extent of necrosis, neovascularization, intratumoral edema, and tumor mass-related pressure effects. Larger clinical studies in the future will determine parameters that dictate accumulation within the patient tumor. In comparison, systemic administration of a therapeutic dosage of NU-0129 to immunocompromised mice bearing orthotopic GBM xenografts resulted in 9×1012 Au nanoparticles per gram of intracranial tumor (23). Taking into account differences in administration (infusion versus serial bolus injections), and dose (0.04 mg/kg in patients versus 10 mg/kg to orthotopic explant models), Au uptake in patient tumor is comparable to Au uptake observed in experimental murine tumors. We hypothesize that similar NU-0129 uptake in murine and human tumors may stem from similar cellular mechanisms of SNA uptake. Our previous studies have shown that SNA cellular uptake and BBB transcytosis, as indicated by use of an in vitro non-contact co-culture model, depends on the activity of scavenger receptors (SRs), especially class A scavenger receptor (SRA) (23). Rapid cellular entry via SRA-mediated endocytosis stems from the unique three-dimensional arrangement of oligonucleotides on the SNA surface, and involves a lipid raft-dependent, caveolae-mediated process (49). Structural insight into oligonucleotide recognition by SRs points to critical roles for a base-quartet-stabilized four-stranded helix. The nucleic acid quadruplexes form a negatively-charged surface, which recognizes the positively-charged surface of the collagenous SR ligand-binding domain (50). While the mechanisms of NU-0129 cellular uptake into GBM tumors in vivo has not been fully elucidated, it is likely facilitated by a functionally compromised BBB/BTB, and we speculate that SR expression on endothelial cells at the BBB promotes recognition of siRNA oligonucleotide corona, and subsequent robust penetration of SNAs into brain and tumor through transcytosis (51, 52).
Synchrotron XFM and silver staining results corroborated results from ICP-MS studies by enabling visualization and quantification of Au accumulation in different cell populations, and by providing high-resolution maps of cellular and subcellular Au distribution. Approximately 20% of intratumoral Au content localized to tumor cells, as indicated by localization of Au signal within cells characterized by large nuclei and Ki67 positivity. Quantification of Au content in tumors that recurred up to 174 days post NU-0129 trial enrollment indicated that >40% of the total Au content was still detectable, and future phase I clinical trials will have to assess the potential for adverse events associated with such long-term Au accumulation. The majority of Au was found associated with the Zn-rich vasculature (XFM maps of RBCs show particularly high Zn, although this element is also found in leukocytes and plasma), as well as in Fe-positive cells, in particular tumor-associated macrophages, which represent a critical source of Fe within the tumor microenvironment (53). Of note, mature red blood cells do not express scavenger receptors and lack intracellular organelles, such as endosomes, which are required for SNA uptake (22, 54). Therefore, we speculate that NU-0129 presence in blood is not due to internalization by mature red blood cells, but instead may be consequence of internalization by leukocytes and/or macrophages, surface adsorption by red blood cells, and/or enrichment in plasma.
Unbiased quantification of Bcl2L12 protein using IHC image analysis revealed that sub-therapeutic dose of NU-0129 resulted in moderate, but statistically significant downregulation of Bcl2L12 in tumor cells from 4 NU-0129-treated recurrent when compared to matched primary GBM patient tumors and triggered robust activation of caspase-3 and induction of wild-type p53 level. Analysis of Bcl2L12 mRNA and protein levels together with in silico analyses of publicly available datasets of matched primary and recurrent GBM tumors reveal that Bcl2L12 protein and transcript levels were not significantly changed during recurrence, suggesting that Bcl2L12 downregulation in NU-0129-treated recurrent patient tumors is likely due to suppression of Bcl2L12 expression by NU-0129, and is not a consequence of longitudinal changes in Bcl2L12 expression during disease recurrence. In further support of the NU-0129 gene silencing effect, and consistent with the role of Bcl2L12 in inducing caspase-3 activation and in destabilizing wild-type p53, protein levels active caspase-3 and wild-type 53 were significantly elevated in NU-0129-treated recurrent tumors when compared to matched primary tumors, while wild-type p53 protein levels remained unchanged, and levels of active caspase-3 decreased during tumor recurrence.
Limitations of our study relate to the lack of direct evidence for an RNAi-based mechanism of NU-0129 action, direct quantification of siRNA content in tumor and adjacent normal brain, and direct correlation between SNA level, Bcl2L12 protein knockdown and concomitant increases in active caspase-3 and wild-type p53 levels within the same set of tumor cells. In a first-in-human phase I clinical study of a systemically administered siRNA-carrying cyclodextrin-based polymeric nanostructure in treating metastatic melanoma patients (20), significant transcript and protein knockdown in 3 metastatic melanoma tumors was found, but mRNA cleavage by 5-RLM-RACE was only detected in one patient (20). This patient had stable disease, characterized by mostly quiescent tumor cells with very slow growth kinetics. These findings are consistent with previous reports suggesting that the robustness of RNAi effects triggered by siRNA delivered to cells in vitro and tumor tissue in vivo is dependent on the doubling time of the cells being analyzed (55). The duration of gene silencing is short for rapidly dividing cells, but longer for quiescent cells, suggesting that ‘siRNA dilution’ in rapidly expanding cells and tissue is a major factor governing RNAi efficacy and the ability to detect mRNA cleavage by 5′-RLM-RACE. Finally, due to rapid degradation of siRNA oligonucleotides in circulation, and the resulting difficulty in detecting siRNA in tumor or adjacent normal brain 22 to 28 hrs post infusion, we rely on Au quantification by ICP-MS, XFM and silver staining, and the assessment of intratumoral target knockdown as indicators for the presence of siRNA oligonucleotide in tumor. An intact siRNA oligonucleotide corona is required for SR-dependent cell entry of NU-0129, suggesting that the presence of Au is an adequate surrogate marker for siRNA oligonucleotide accumulation in GBM tissue. Finally, while the unbiased quantification of Bcl2L12, active caspase-3 and wild-type p53 protein levels in proximal, distinct tumor sections indicated NU-129 treatment-related effects on Bcl2L12 expression as well as the levels of downstream signaling surrogates, we were unable to establish a direct correlation between the amount of SNAs that accumulated in tumor cells, the extent of Bcl2L12 target knockdown and the ensuing increase in both active caspase-3 and wild-type p53 levels, using XFM and IHC analyses of the same set of tumor cells.
Several early phase clinical trials of siRNA-based nanostructures have been completed or are ongoing. These include evaluation of CALAA-01, a cyclodextrin-based polymeric nanostructure to deliver siRNA targeted to RRM2; the lipid-based nanoconstruct Atu27 for the delivery of protein kinase N3-specific siRNA; the lipid nanoparticles DCR-MYC to downregulate cancer-associated cMYC expression; ALN-RSV containing siRNA specific for VEGF and kinesin spindle protein (KSP); TKM-080301 with siRNA against polo-like kinase-1; liposomes for systemic siRNA delivery to inhibit Ephrin type-A receptor2; and the LODER® polymer matrix for silencing mutated K-RAS expression. Four of these constructs (CALAA-01, Atu27, siEphA2 and TKM-080301) have been designed for systemic intravenous injection/infusion for the treatment of advanced solid non-CNS cancers. NU-0129 is the first systemically administered siRNA-based nanostructure designed to target oncogene expression in malignant brain tumors.
The phase 0 clinical trial results presented here will motivate future clinical investigations to comprehensively assess NU-0129 safety, particularly related to adverse events associated with long-term Au accumulation and knockdown of Bcl2L12 in normal tissue. Of note, we have selected Bcl2L12 due its selective up-regulation in GBM tumors compared to normal tissue (23, 26, 27, 34–36). In addition, further clinical testing will assess NU-0129 dose-dependence of gene knockdown, as well as the impact of NU-0129 treatment on primary and recurrent GBM patient survival as a monotherapy and in combination with standard-of-care, in particular genotoxic therapy, such as RT, TMZ or CCNU. Future preclinical and clinical investigations will test the hypothesis that the combination of RT, RT+TMZ or CCNU in combination with NU-0129 will result in more robust p53 and effector caspase activation, more effective glioma cell apoptosis, and increased patient survival.
MATERIALS AND METHODS.
Study design of preclinical toxicokinetic and toxicology studies.
NU-0129 in vehicle (phosphate buffered saline, pH 7.4) was administered intravenously by bolus injection once to 3 toxicology groups (Groups 2–4) and 1 cardiovascular group (Group 4C) of cynomolgus monkeys. Dosage levels were 1, 4, and 8 mg/kg of siRNA per kg for Groups 2, 3, and 4/4C, respectively. Concurrent control groups (Groups 1 and 1C) received the vehicle on a comparable regimen. The dosage volume was 5 mL/kg for all groups. Each toxicology group consisted of 5 males and 5 females, and each cardiovascular group consisted of 2 males and 2 females. Two days following the single dose administration, 3 animals/sex/toxicology group were euthanized; the remaining 2 animals/sex/toxicology group were euthanized following a 14-day non-dosing (recovery) period. For toxicology assessment, animals were observed twice daily for mortality and moribundity. Clinical examinations were performed for at least 6 consecutive days prior to randomization, including the day of randomization, prior to dosing, and at 1–2 hrs following dose administration. During the recovery period, the animals were observed once daily. Detailed physical examinations were conducted on all animals at least weekly during acclimation, on the day of randomization, weekly (± 2 days) during the study period, and on the day of the scheduled necropsies. Individual body weights were recorded at least weekly during acclimation, on the day of randomization, on study day 0 (prior to dosing), weekly (± 2 days) during the study period, and on the day prior to the scheduled necropsies (non-fasted). Clinical pathology evaluations (hematology, coagulation, serum chemistry, and urinalysis) were performed once during acclimation (study day −28), and on the day of the primary (study days 2) and the recovery (study day 14) necropsies. Blood samples for toxicokinetic evaluation were collected from all animals prior to dose administration, and at approximately 3, 10, and 30 min and 2, 8, and 24 hrs after dose administration on study day 0. Complete necropsies were performed on all toxicology animals, and selected organs were weighed at the scheduled necropsies. Selected tissues were examined microscopically from all animals. For cardiovascular assessments, heart rate, arterial blood pressure (systolic, diastolic, and mean), pulse pressure, electrocardiogram (ECG) waveforms (from which the ECG intervals, PR, QRS, RR, QT, and QTcB were derived) were collected continuously for approximately 24 hrs during acclimation (study day −7), for approximately 1 h prior to dose administration and for approximately 24 hrs following dose administration.
Study design phase 0 clinical trial.
This study is a prospective, open-label, single-center, single-arm phase 0 first-in-human trial to determine the safety, pharmacokinetics, and biodistribution of intravenously administered NU-0129 in patients with recurrent GBM or gliosarcoma. This investigator-initiated clinical trial was conceived, developed, and performed at Northwestern University, received FDA approval and was registered appropriately in the clinicaltrials.gov database (NCT03020017). A total of 8 patients were enrolled onto study, each provided written informed consent in accordance with institutional guidelines. The study began in September of 2017 and ended in September of 2018.
a). Study Design/Study Endpoints.
The sample size n=8 was determined on the basis of feasibility of recruitment and conducting the study. This is the first study in humans, and both the primary and secondary outcomes were assessed with sufficient precision with such n, while few patients were put to risk.
b). Data analyses.
The primary objective of this clinical study was to evaluate the safety of NU-0129 infusion in patients with recurrent GBM or gliosarcoma. To this end, we counted and recorded the number of adverse events. The secondary objective was to evaluate biodistribution of NU-0109 in tumor tissue, using ICP-MS, XFM and silver staining methodologies, as well as the pharmacokinetics of NU-0129. For pharmacokinetic analysis, we collected blood samples at 1, 3, 5, 10, 30, 60 min and 4, 8, and 24 hrs post-dose, to determine appropriate pharmacokinetic curves and critical pharmacokinetic parameters. Exploratory objectives related to the assessment of Bcl2L12 expression levels, and the abundance of downstream markers for Bcl2L12 inactivation, including and especially active caspase-3 and p53-wt, by IHC and subsequent unbiased, automated IHC signal quantification.
NU-0129 manufacturing.
Three separate cGMP vendors participated in manufacturing. Nitto Denko Avecia, Inc. manufactured the Bcl2L12 siRNA duplex (active pharmaceutical ingredient, API) by solid-phase synthesis, and conjugated it to Au nanoparticle cores to form the intermediate drug substance. The drug substance was then transferred to Northwestern University, where it was stored in a controlled laboratory until it was transferred to University of Iowa Pharmaceuticals (UIP), which completed formulation, fill, and finish of the drug product under cGMP. The intermediate NU-0129 drug product consists of 25 mg of conjugated drug substance (0.987 mg of Bcl2L12 siRNA duplex) and 120 mg of D-mannitol (USP). The amount of mannitol contained in the formulated NU-0129 drug product is less than 10% (mg/kg) of that safely administered for the reduction of intracranial pressure and the treatment of cerebral edema. The drug product was then transferred in compliance with GMP to Northwestern University’s GMP Investigational Pharmacy, where it was stored at −20°C until prepared for clinical use. Final formulation (dilution) was completed at the Northwestern pharmacy, and the final drug product was released for clinical use. Long-term stability study of NU-0129 drug product was conducted on the intermediate NU-0129 drug product by UIP. No placebo has been manufactured for the Phase 0 study.
NU-0129 stability.
NU-0129 stability data were collected on both drug substance and drug product through 12 months of storage. The storage conditions for these studies were 25 °C and −20 °C for drug substance, and 5 °C and −20 °C for drug product. The testing performed included appearance, water Content by Karl Fischer, purity by AEX, purity by SEC, particle size by UV-Vis, siRNA content, bioburden, endotoxin content, and osmolality.
siRNA quantification in plasma.
siRNA quantification in plasma was performed by Wil Research (Ohio, USA; for non-human primate toxicokinetic studies) and Covance (Utah, USA; for pharmacokinetic studies in GBM patients), using LC-HPLC technology. For sample extraction, an adequate amount (~125 μL per sample) of lysis mixture was prepared by mixing 20 mg/mL Proteinase K and hybridization buffer. 100 μL of the above lysis mixture was added to each of the samples. Upon sealing the assay plate, sample plates were heated at 65°C for approximately 30 min in a thermocycler (Mastercycler Pro S, Eppendorf) to activate the proteinase K used for protein digestion. Plates were centrifuged at 500 rpm for approximately 2 min. Each well was supplemented with 40 μL of freshly prepared PNA-atto-probe solution [1 mL of 1 μM of PNA-atto-probe solution in 1 mL of annealing buffer (60 mM Tris-HCl pH=8)/MeCN]. The samples were immediately heated to 95°C for 15 min, and quickly cooled to approximately 20°C by placing it on a wet ice bath for approximately 5 min. Upon centrifugation at ~500 rpm for approximately 2 min, samples were transferred to a clean tapered plate stored at 1–8°C for subsequent analysis using fluorescence LC-HPLC. Standards with defined siRNA concentration (5 to 500 ng/mL of siRNA) were prepared for each run. Analyte chromatographic peak areas were integrated using Xcalibur software. The following equipment and conditions were used:
| Autosampler | LEAP (LEAP Technologies, Chapel Hill, NC) or Shimadzu SIL-20AC HT Prominence (Shimadzu, Columbia, MD) |
| HPLC pumps / Controller | Shimadzu CBM-20A or SCL-10A VP controller with LC-10/20AD pump (Shimadzu, Columbia, MD) and Eppendorf CH-30 column heater. |
| Autosampler Temp Settings | 5°C |
| Column Type | DNAPac-PA-200, 4x250 mm, Part#: 063000 Vendor: Dionex, Cedar Rapids, IA |
| Column Temp Settings | 60°C |
| Guard Column | Frit, change as needed |
| Flow Rate | 1 mL/min |
| Injection Volume | 30 μL |
| Mobile phase | A= 50:50 (1 mM EDTA in 25 mM Tis-HCl, pH 8.0): MeCN B= 50:50 (1 mM EDTA and 1.6 M NaCLO4 in 25 mM Tris-HCl, pH 8.0): MeCN (v/v) C= N/A |
| Needle Wash | 1 = 50:50 (0.2% TEA and 0.02% EDTA in water): DMPF (v/v) 2 = 0.1% FA in water |
| LC Program | Gradient |
| Typical Column Pressure | 160 bar |
| Detector | RF-20A xs fluorescence detector (Shimadzu, Columbia, MD) |
| Detector Temperature | 20 °C |
Au quantification in tumor tissue and plasma using ICP-MS.
High resolution (HR)-ICP-MS was performed using a ThermoElement2 HR-ICP-MS (Thermo Fisher Scientific, Waltham, MA, USA), controlled with the Thermo Element2 HR-ICP-MS 3.0 software package and equipped with an ESI PrepFAST M5 autosampler (Omaha, NE, USA). Internal standard was added inline using the prepFAST system and consisted of 1 ng/mL of a mixed element solution containing Bi, In, 6Li, Sc, Tb, Y (IV-ICPMS-71D from Inorganic Ventures, Christiansburg, VA, USA). Quantitative calibration and check standards were made using a 100 μg/mL Au standard from Inorganic Ventures (Christiansburg, VA, USA). Frozen tissue samples were thawed at room temperature for approximately 5 min and placed in a lyophilizer while still partly frozen. Sample were desiccated for at least 4 hrs or until dry and dissolved in 5 mL HNO3 and 5 mL of HCl. Each sample was acquired using 3 main runs (15 passes). The isotopes selected for analysis were 197Au and 209Bi (chosen as an internal standard for data interpolation and machine stability). Instrument performance was optimized daily through tuning followed by verification via a performance report (passing manufacturer specifications).
Pharmacokinetics.
Pharmacokinetic analyses were performed by IIT Research Institute. Individual human plasma siRNA and Au concentration data at scheduled (nominal) sampling times were analyzed using the non-compartmental model for constant infusion with Phoenix® WinNonlin® software (Version 8.1; Certara, Princeton, NJ). Values below the analytical method’s limit of quantitation (BQL) and suspected outliers were assigned a value of zero for PK calculations. Systemic exposure was calculated only for the post-infusion period since no plasma concentration data was available from during the infusion period. Area under the plasma concentration-time curve from time zero (defined as the time of termination of the IV infusion) to the last measured concentration (AUC0-last) was estimated by the log trapezoidal rule. Elimination rate constant values (λz) were calculated by log-linear regression on data points of the terminal phase (using Phoenix WinNonlin’s Best Fit Lambda Z Calculation Method option) when allowed by the data; the plasma elimination half-life (t1/2) was calculated as ln(2)/λz. Area under the plasma concentration-time curve extrapolated to infinity was also calculated when applicable, and was defined as AUC0−∞ = AUC0-last+Ct/λz, where Ct is the last measureable plasma concentration. PK parameter calculations included t1/2 (elimination half-life), Cmax (maximum observed concentration), AUC0−∞, AUC0-last, Vz (volume of distribution) and CL (total body clearance). Nominal doses and times were used in all PK analysis.
X-ray fluorescence microscopy.
XFM was performed on formalin-fixed, paraffin-embedded (FFPE) 5 micron thick sections of tumor samples from patients given NU-0129 treatment, using several instruments for hard X-ray fluorescence at the Advanced Photon Source (APS) at Argonne National Laboratory (ANL). Instruments included the following: large area microprobe at beamline 8BM-B, microprobes at beamlines 2ID-E and 2-IDD, and the Bionanoprobe instrument at the beamline 9ID-Bs. Samples were prepared by placing three consecutive FFPE tissue sections on glass or an Ultralene membrane, respectively. The first was placed onto regular glass microscopy slide for IHC, the second onto 4 micron thick Ultralene membrane (3525 - Ultralene® Window Film (Roll) from SPEX SamplePrep, Metuchen, NJ) held by a 3D-printed frame (LulzBot PolyLite PLA, 2.85 mm, Polymaker, Loveland, Colorado), and the third one again onto a glass slide. In each case, the first tissue section was stained by H&E, the second tissue section was used for XFM and left embedded in paraffin, while the third tissue section was stained for Ki67. This approach allowed us to co-register different tissue features and specific cell types, and align optical images with elemental maps produced by XFM, similarly to work done in the past (56–58). At the 8BM-B beamline, the beam was focused by KB-mirror system to a 30 micron spot suitable for low resolution tissue map overviews. Beam energy was tuned to 15 keV in order to excite Au fluorescence at the range of wavelengths suitable for best elemental detection accuracy. Spectra were collected with a SII Vortex ME4 4-element silicon drift detector (SII NanoTechnology USA, Northridge, CA). Tissue sections were raster-scanned with step sizes matching beam spot sizes and dwell times of 100–500 milliseconds. Elemental concentrations were calibrated using thin film AXO 10X standards (AXO Dresden GmbH, Germany), and data were calibrated and fitted using MAPS software (59). Per pixel counts were converted to elemental concentrations (μg/cm2).
Coarse elemental maps were used to select and extract a portion of a tissue section together with the supporting Ultralene membrane for high resolution scans. Subsequently, the subsection of the sample was remounted for more detailed imaging at the sector 2 beamlines, which use Fresnel zone plates for X-ray focusing. At the beamline 2-ID-E, the beam was focused to a 0.5 micron spot, while beam size at station 2-ID-D was 0.3 μm. Incident X-ray energy was tuned to 13.4 keV using a beam splitting Si(220) monochromator at 2ID-E and 12.0 keV by Kohzu monochromator at 2ID-D. Silicon drift energy dispersive detector SII Vortex ME4 positioned at 90° to the incident beam was used to collect the fluorescence signal from samples tilted 15° to detector. Areas of the sample several hundreds of microns wide were first scanned with a step size of 10 micron (overview scans) and then with the 1 micron step size and a per-pixel dwell time of 100 milliseconds. Per-pixel elemental concentration was obtained by comparison with the thin-film standards AXO (AXO Dresden GmbH) for 2ID-E data and NBS-1832 and NBS-1833 from the National Bureau of Standards (Gaithersburg, MD) for 2ID-D data. The analysis was performed using MAPS software (60). Finally, sample size was reduced again, and the samples mapped using Bionanoprobe instrument. In this case, the X-ray beam was focused to 80 nm spot size using Fresnel zone plate and small sample areas corresponding to few individual cells were imaged. Monochromatic 13.7 keV hard X-rays were used to excite fluorescence. The fluorescence spectra at each step of the scan were collected with a four-element silicon drift detector (Vortex ME-4, SII Nanotechnology). Data were fitted and quantified by comparison to a standard reference material (RF8–200-S2453, AXO Dresden GmbH) using the MAPS program (60).
Per pixel elemental data for chemical elements phosphorous (P), sulfur (S), calcium (Ca), iron (Fe), zinc (Zn) and gold (Au) were collected by focusing on the part of the elemental spectra with K alpha fluorescence signal for elements concluding with Zn; concentration of Au was evaluated by Au L lines fluorescence. Final measurement of elemental concentrations in cells and tissue segments was done as follows: For each selected region of interest (ROI), total elemental yield for the element of interest was obtained as a sum in femtograms, and this information was either used as-is or used to determine elemental content per unit volume of the tissue.
Silver staining.
Silver staining on FFPE BM tumor tissue was done using the BBI silver staining kit (BBI Solutions #16673) according to the manufacturer’s instructions. Briefly, tissue slides were deparaffinized and hydrated, and incubated with silver staining solution at RT for 10 min. After rinsing, slides were counterstained with hematoxylin for 1 min, dehydrated, and a coverslip was added with xylene-based mounting media.
Immunohistochemistry.
Slides were heated at 60°C for 1 hr, deparaffinized and hydrated using xylene (3 times for 5 min. each), 100% EtOH (2 times for 3 min each), and 95% EtOH for 3 min. Slides were washed with H2O, placed into 1x Retrieval solution, pH 6.0, and incubated in Biocare Medical Decloaking Chamber at 110°C for 10 min. Slides were allowed to cool to RT, washed 3 times with PBS, and incubated with peroxidase I for 10 min at RT. Upon rinsing with PBS, 2–5 drops protein block background with background sniper (Biocare Medical BS966H) were added, and slides were incubated for 15 min at RT. Slides were rinsed in PBS for 1–2 min, blocked with 10% normal goat serum for 60 min, rinsed with PBS, and incubated with primary antibody overnight at 4°C. The following antibodies were used: anti-p53 DO-7 (Cell Signaling 48818, 1:300), anti-Bcl2L12 (Thermo Fisher PA5–54267, 1:50), and anti-active caspase-3 (Cell Signaling 9664, 1:1000). Slides were subsequently rinsed with TBST/Tween 3 times for 1 min. Slides were incubated with secondary antibody (Biocare MACH2) for 30 min at RT, washed 3 times for 1 min with PBS, and incubated with DAB chromogen for up to 3 min. Slides were counterstained with hematoxylin for 1 min, washed in H2O until blue color faded, dehydrated with graded alcohol and xylene: 95% EtOH 3 min, 100% EtOH 2 times for 3 min, xylene (or Histo-Clear from national diagnostics) 3 times for 5 min, and a coverslip was added using a xylene-based mounting medium.
Quantification of IHC staining intensities.
Slides were imaged using the Hamamatsu Nanozoomer at the Northwestern Pathology core facility, and the images were analyzed using TissueGnostics Histoquest 6.0.1.125 software. Nuclear segmentation was done with nuclei size set at 15 micron, and IHC markers (Bcl2L12, p53 and active caspase-3) were considered present only when the staining occurred adjacent to or inside the nuclear mask. Positive cells were gated for a given IHC staining threshold with mean staining intensities >60 for Bcl2L12, >15 for active caspase-3, and >50 for p53, and for cell nuclei above 50 μm2 and nuclear intensity > 67. At least 8 regions of interest were selected for analysis in each case.
Statistical analysis.
Statistical analysis was conducted using Excel (Microsoft) and GraphPad Prism software (GraphPad Software Inc.). Statistical analysis of the toxicokinetic dataset was carried out by ordinary one-way ANOVA nonparametric test and Tukey’s multiple comparisons test to compare means. We used the limma package in R for differential gene expression analyses. Limma uses an empirical Bayes model to produce standard errors, t-statistics and raw p values. Statistical analyses of image quantitation were carried out by Student’s t test. P < 0.05 was considered significant. Data were calculated as the means ± SD. Outliers were identified by applying the interquartile range rule.
Supplementary Material
fig. S1. Toxicokinetic parameters of NU-0129 as assessed in cynomolgus monkeys.
fig. S2. XFM elemental maps of recurrent GBM tumor (patient ID 105).
fig. S3. XFM elemental maps of recurrent GBM tumor (patient ID 101).
fig. S4. XFM elemental maps of recurrent GBM tumors identify Fe and Zn as markers for RBCs in blood vessels.
fig. S5. ROI selection for IHC staining quantification.
fig. S6. mRNA expression of Bcl2L12, wild-type p53 and caspase-3 in primary versus recurrent GBM.
fig. S7. Protein expression of active caspase-3 and wild-type p53 in individual matched newly diagnosed and recurrent on trial and trial-unrelated GBM.
table S2. Demographics and molecular profiles of patients enrolled into the clinical trial.
table S3. Patient demographics and molecular profiles of matched primary and recurrent GBM used for IHC analysis of Bcl2L12.
table S1. NU-0129 stability and integrity.
Table 1.
Summary of NU-0129 treatment-related adverse events in cynomolgus monkeys (A) and GBM patients (B).
| A | |||||||
| Category | Term | animals affected | |||||
| Investigations | Blue discoloration of tissues | all | |||||
| Investigations | Injection site findings of acute perivascular hemorrhage | all | |||||
| Vascular disorders | lower diastolic and mean arterial pressure lower systolic blood pressure | 8 mg/kg group | |||||
| B | |||||||
| Grade | |||||||
| Category | Term | 1 | 2 | 3 | 4 | 5 | Patients affected |
| Blood and lymphatic system disorders | Anemia | 1 | 0 | 0 | 0 | 0 | 1 |
| Investigations | Alkaline phosphatase increased | 1 | 0 | 0 | 0 | 0 | 1 |
| Investigations | Lymphocyte count decreased | 0 | 1 | 1 | 0 | 0 | 2 |
| Investigations | White blood cell decreased | 1 | 0 | 0 | 0 | 0 | 1 |
| Metabolism and nutrition disorders | Hypophosphatemia | 0 | 0 | 1 | 1 | 0 | 2 |
| Vascular disorders | Hypertension | 0 | 1 | 0 | 0 | 0 | 1 |
| Total | 3 | 2 | 2 | 1 | 0 | 8 | |
ACKNOWLEDGMENTS.
We would like to acknowledge support provided by the following cores at the Northwestern University Feinberg School of Medicine and the Robert H. Lurie Comprehensive Cancer Center (RHLCC): Center for Advanced Microscopy/Nikon Imaging Center, NUSeq, Nervous System Tumor Bank, Pathology Core Facility and the Mouse Histology and Phenotyping Laboratory. Metal analysis was performed at the Northwestern University Quantitative Bio-element Imaging Center and was funded by the Chicago Biomedical Consortium with support from the Searle Funds at The Chicago Community Trust and supported by the Office of The Director, National Institutes of Health of the National Institutes of Health under Award Number S10OD020118. This research used resources of the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357. Implementation of the Bionanoprobe was supported by NIH ARRA grant SP0007167. We would like to thank Evan Maxey for expert advise and help with the execution of the XFM study, Dr. Roger Stupp for guidance on the clinical trial design and for critically reading the manuscript, and especially Dr. Chad Mirkin for spearheading pre-IND meeting with the FDA, for providing expert advice on study design, and the characterization of SNA biological effects, and for carefully reading the manuscript.
Funding: This research was supported by the Office of Research, Northwestern University, the Center for Cancer Nanotechnology Excellence Initiative of the NIH under award U54 CA151880 and 199091 (to T.P., C.D.J., G.E.W. and A.H.S.), R01CA208783 (to A.H.S), R01NS102669 (to C.H.), R01LM011297 (to R.V.D.), postdoctoral fellowship under award F32CA216996 to D.H., and a brain tumor SPORE grant P50CA221747 (to P.K., R.V.L, C.D.J., C.H., D.J.B. and A.H.S.). C.H.K. was in part supported by NewCures Accelerator funded by Office for Research at Northwestern University.
Footnotes
Competing interests: A.H.S is shareholder of Exicure Inc., which develops SNA therapeutic platforms. A.H.S and C.H.K. are inventors on patent US20150031745A1, which describes SNA nanoconjugates to cross the blood-brain barrier.
SUPPLEMENTAL MATERIALS AND METHODS.
Toxicokinetic and toxicology studies in cynomolgus monkeys.
Data and materials availability:
All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
REFERENCES and NOTES.
- 1.Stupp R et al. , Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. The New England journal of medicine 352, 987–996 (2005). [DOI] [PubMed] [Google Scholar]
- 2.Stupp R, Roila F, Group EGW, Malignant glioma: ESMO clinical recommendations for diagnosis, treatment and follow-up. Ann Oncol 20 Suppl 4, 126–128 (2009). [DOI] [PubMed] [Google Scholar]
- 3.Stupp R et al. , Effect of Tumor-Treating Fields Plus Maintenance Temozolomide vs Maintenance Temozolomide Alone on Survival in Patients With Glioblastoma: A Randomized Clinical Trial. JAMA 318, 2306–2316 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wick W et al. , Lomustine and Bevacizumab in Progressive Glioblastoma. The New England journal of medicine 377, 1954–1963 (2017). [DOI] [PubMed] [Google Scholar]
- 5.Brem H et al. , Placebo-controlled trial of safety and efficacy of intraoperative controlled delivery by biodegradable polymers of chemotherapy for recurrent gliomas. The Polymer-brain Tumor Treatment Group. Lancet 345, 1008–1012 (1995). [DOI] [PubMed] [Google Scholar]
- 6.Cohen MH, Shen YL, Keegan P, Pazdur R, FDA drug approval summary: bevacizumab (Avastin) as treatment of recurrent glioblastoma multiforme. Oncologist 14, 1131–1138 (2009). [DOI] [PubMed] [Google Scholar]
- 7.Friedman HS et al. , Bevacizumab alone and in combination with irinotecan in recurrent glioblastoma. Journal of clinical oncology : official journal of the American Society of Clinical Oncology 27, 4733–4740 (2009). [DOI] [PubMed] [Google Scholar]
- 8.Clark AJ et al. , Impact of bevacizumab chemotherapy on craniotomy wound healing. Journal of neurosurgery 114, 1609–1616 (2011). [DOI] [PubMed] [Google Scholar]
- 9.Brennan CW et al. , The somatic genomic landscape of glioblastoma. Cell 155, 462–477 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang PH et al. , Quantitative analysis of EGFRvIII cellular signaling networks reveals a combinatorial therapeutic strategy for glioblastoma. Proceedings of the National Academy of Sciences of the United States of America 104, 12867–12872 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stommel JM et al. , Coactivation of receptor tyrosine kinases affects the response of tumor cells to targeted therapies. Science 318, 287–290 (2007). [DOI] [PubMed] [Google Scholar]
- 12.Naghizadeh S, Mansoori B, Mohammadi A, Sakhinia E, Baradaran B, Gene silencing strategies in cancer therapy: An update for drug resistance. Curr Med Chem, (2018). [DOI] [PubMed] [Google Scholar]
- 13.Ren YJ, Zhang Y, An update on RNA interference-mediated gene silencing in cancer therapy. Expert Opin Biol Ther 14, 1581–1592 (2014). [DOI] [PubMed] [Google Scholar]
- 14.Yang WQ, Zhang Y, RNAi-mediated gene silencing in cancer therapy. Expert Opin Biol Ther 12, 1495–1504 (2012). [DOI] [PubMed] [Google Scholar]
- 15.Zuckerman JE, Davis ME, Clinical experiences with systemically administered siRNA-based therapeutics in cancer. Nat Rev Drug Discov 14, 843–856 (2015). [DOI] [PubMed] [Google Scholar]
- 16.Stegh AH, Toward personalized cancer nanomedicine - past, present, and future. Integrative biology : quantitative biosciences from nano to macro 5, 48–65 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abbott NJ, Evidence for bulk flow of brain interstitial fluid: significance for physiology and pathology. Neurochem Int 45, 545–552 (2004). [DOI] [PubMed] [Google Scholar]
- 18.Schultheis B et al. , First-in-human phase I study of the liposomal RNA interference therapeutic Atu027 in patients with advanced solid tumors. Journal of clinical oncology : official journal of the American Society of Clinical Oncology 32, 4141–4148 (2014). [DOI] [PubMed] [Google Scholar]
- 19.Strumberg D et al. , Phase I clinical development of Atu027, a siRNA formulation targeting PKN3 in patients with advanced solid tumors. Int J Clin Pharmacol Ther 50, 76–78 (2012). [DOI] [PubMed] [Google Scholar]
- 20.Davis ME et al. , Evidence of RNAi in humans from systemically administered siRNA via targeted nanoparticles. Nature 464, 1067–1070 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Coelho T et al. , A phase II, open-label, extension study of long-term patisiran treatment in patients with hereditary transthyretin-mediated (hATTR) amyloidosis. Orphanet J Rare Dis 15, 179 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cutler JI, Auyeung E, Mirkin CA, Spherical nucleic acids. Journal of the American Chemical Society 134, 1376–1391 (2012). [DOI] [PubMed] [Google Scholar]
- 23.Jensen SA et al. , Spherical Nucleic Acid Nanoparticle Conjugates as an RNAi-Based Therapy for Glioblastoma. Science translational medicine 5, 209ra152 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kouri FM et al. , miR-182 integrates apoptosis, growth, and differentiation programs in glioblastoma. Genes & development 29, 732–745 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scorilas A et al. , Molecular cloning, physical mapping, and expression analysis of a novel gene, BCL2L12, encoding a proline-rich protein with a highly conserved BH2 domain of the Bcl-2 family. Genomics 72, 217–221 (2001). [DOI] [PubMed] [Google Scholar]
- 26.Stegh AH et al. , Bcl2L12 inhibits post-mitochondrial apoptosis signaling in glioblastoma. Genes & development 21, 98–111 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stegh AH et al. , Bcl2L12-mediated inhibition of effector caspase-3 and caspase-7 via distinct mechanisms in glioblastoma. Proceedings of the National Academy of Sciences of the United States of America 105, 10703–10708 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stegh AH et al. , Glioma oncoprotein Bcl2L12 inhibits the p53 tumor suppressor. Genes Dev 24, 2194–2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Massich MD, Giljohann DA, Schmucker AL, Patel PC, Mirkin CA, Cellular response of polyvalent oligonucleotide-gold nanoparticle conjugates. ACS nano 4, 5641–5646 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sita TL et al. , Dual bioluminescence and near-infrared fluorescence monitoring to evaluate spherical nucleic acid nanoconjugate activity in vivo. Proceedings of the National Academy of Sciences of the United States of America 114, 4129–4134 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Davies B, Morris T, Physiological parameters in laboratory animals and humans. Pharmaceutical research 10, 1093–1095 (1993). [DOI] [PubMed] [Google Scholar]
- 32.Barthel FP et al. , Longitudinal molecular trajectories of diffuse glioma in adults. Nature 576, 112–120 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kwon SM et al. , Recurrent Glioblastomas Reveal Molecular Subtypes Associated with Mechanistic Implications of Drug-Resistance. PloS one 10, e0140528 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stegh AH, Chin L, Louis DN, DePinho RA, What drives intense apoptosis resistance and propensity for necrosis in glioblastoma? A role for Bcl2L12 as a multifunctional cell death regulator. Cell cycle 7, 2833–2839 (2008). [DOI] [PubMed] [Google Scholar]
- 35.Stegh AH, DePinho RA, Beyond effector caspase inhibition: Bcl2L12 neutralizes p53 signaling in glioblastoma. Cell cycle 10, 33–38 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stegh AH et al. , Glioma oncoprotein Bcl2L12 inhibits the p53 tumor suppressor. Genes & development 24, 2194–2204 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Touat M, Idbaih A, Sanson M, Ligon KL, Glioblastoma targeted therapy: updated approaches from recent biological insights. Ann Oncol 28, 1457–1472 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zuckerman JE et al. , Correlating animal and human phase Ia/Ib clinical data with CALAA-01, a targeted, polymer-based nanoparticle containing siRNA. Proceedings of the National Academy of Sciences of the United States of America 111, 11449–11454 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barnaby SN, Lee A, Mirkin CA, Probing the inherent stability of siRNA immobilized on nanoparticle constructs. Proceedings of the National Academy of Sciences of the United States of America 111, 9739–9744 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mirkin CA, Letsinger RL, Mucic RC, Storhoff JJ, A DNA-based method for rationally assembling nanoparticles into macroscopic materials. Nature 382, 607–609 (1996). [DOI] [PubMed] [Google Scholar]
- 41.Lee JS, Lytton-Jean AK, Hurst SJ, Mirkin CA, Silver nanoparticle-oligonucleotide conjugates based on DNA with triple cyclic disulfide moieties. Nano letters 7, 2112–2115 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cutler JI, Zheng D, Xu X, Giljohann DA, Mirkin CA, Polyvalent oligonucleotide iron oxide nanoparticle “click” conjugates. Nano letters 10, 1477–1480 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang C et al. , A general approach to DNA-programmable atom equivalents. Nature materials 12, 741–746 (2013). [DOI] [PubMed] [Google Scholar]
- 44.Young KL et al. , Hollow spherical nucleic acids for intracellular gene regulation based upon biocompatible silica shells. Nano letters 12, 3867–3871 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Banga RJ, Chernyak N, Narayan SP, Nguyen ST, Mirkin CA, Liposomal spherical nucleic acids. Journal of the American Chemical Society 136, 9866–9869 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Calabrese CM et al. , Biocompatible infinite-coordination-polymer nanoparticle-nucleic-acid conjugates for antisense gene regulation. Angewandte Chemie 54, 476–480 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cutler JI et al. , Polyvalent nucleic acid nanostructures. Journal of the American Chemical Society 133, 9254–9257 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Morris W, Briley WE, Auyeung E, Cabezas MD, Mirkin CA, Nucleic acid-metal organic framework (MOF) nanoparticle conjugates. Journal of the American Chemical Society 136, 7261–7264 (2014). [DOI] [PubMed] [Google Scholar]
- 49.Choi CH, Hao L, Narayan SP, Auyeung E, Mirkin CA, Mechanism for the endocytosis of spherical nucleic acid nanoparticle conjugates. Proceedings of the National Academy of Sciences of the United States of America 110, 7625–7630 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pearson AM, Rich A, Krieger M, Polynucleotide binding to macrophage scavenger receptors depends on the formation of base-quartet-stabilized four-stranded helices. The Journal of biological chemistry 268, 3546–3554 (1993). [PubMed] [Google Scholar]
- 51.Goti D et al. , Scavenger receptor class B, type I is expressed in porcine brain capillary endothelial cells and contributes to selective uptake of HDL-associated vitamin E. Journal of neurochemistry 76, 498–508 (2001). [DOI] [PubMed] [Google Scholar]
- 52.Mackic JB et al. , Human blood-brain barrier receptors for Alzheimer’s amyloid-beta 1–40. Asymmetrical binding, endocytosis, and transcytosis at the apical side of brain microvascular endothelial cell monolayer. The Journal of clinical investigation 102, 734–743 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jung M, Mertens C, Tomat E, Brune B, Iron as a Central Player and Promising Target in Cancer Progression. Int J Mol Sci 20, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rosi NL et al. , Oligonucleotide-modified gold nanoparticles for intracellular gene regulation. Science 312, 1027–1030 (2006). [DOI] [PubMed] [Google Scholar]
- 55.Bartlett DW, Davis ME, Insights into the kinetics of siRNA-mediated gene silencing from live-cell and live-animal bioluminescent imaging. Nucleic acids research 34, 322–333 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Deng J et al. (Natinal Cancer Institute, 2017), vol. 2018. [Google Scholar]
- 57.Paunesku T et al. , X-ray fluorescence microscopy for investigation of archival tissues. Health physics 103, 181–186 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Refaat T et al. , Distribution of Iron Oxide Core-Titanium Dioxide Shell Nanoparticles in VX2 Tumor Bearing Rabbits Introduced by Two Different Delivery Modalities. Nanomaterials 6, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nietzold T et al. , Quantifying X-Ray Fluorescence Data Using MAPS. J Vis Exp, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vogt S, MAPS: A set of software tools for analysis and visualization of 3D X-ray fluorescence data sets. J. Phys. IV 104, 635–638 (2003). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
fig. S1. Toxicokinetic parameters of NU-0129 as assessed in cynomolgus monkeys.
fig. S2. XFM elemental maps of recurrent GBM tumor (patient ID 105).
fig. S3. XFM elemental maps of recurrent GBM tumor (patient ID 101).
fig. S4. XFM elemental maps of recurrent GBM tumors identify Fe and Zn as markers for RBCs in blood vessels.
fig. S5. ROI selection for IHC staining quantification.
fig. S6. mRNA expression of Bcl2L12, wild-type p53 and caspase-3 in primary versus recurrent GBM.
fig. S7. Protein expression of active caspase-3 and wild-type p53 in individual matched newly diagnosed and recurrent on trial and trial-unrelated GBM.
table S2. Demographics and molecular profiles of patients enrolled into the clinical trial.
table S3. Patient demographics and molecular profiles of matched primary and recurrent GBM used for IHC analysis of Bcl2L12.
table S1. NU-0129 stability and integrity.
Data Availability Statement
All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.