Abstract
Objectives:
Gaucher disease (GD) is the most common autosomal recessive disorder of glycolipid storage. It results from mutations in the glucocerebrosidase (GBA) gene and leads to GBA deficiency. Different mutations are associated with different phenotypes in the three major types of GD.
Materials and Methods:
The spectrum of mutations in GBA gene in 26 unrelated patients with GD from different Iranian populations was determined by DNA sequencing, polymerase chain reaction (PCR)-restriction fragment length polymorphism (RFLP), and amplification-refractory mutation system (ARMS) methods. An in silico analysis was also performed for novel mutations.
Results:
Six new mutations were identified in this study. The newly detected mutations that could be theoretically harmful included p.I200T (c.599T>C), p.H312D (c.934C>G), p.L325S (c.974T>C), p.L393V (c.1177C>G), p.S439G (c.1315A>G), and p.M455R (c.1365G>A). Also, p.L483P, p.N409S, p.W420X, p.E379K, p.R398Q, p.N227S, p.R202Q, and p.D448H mutations were identified in the patients. Besides, two new complex mutations, namely, p.S439G/p.S439G+p.E379K/- and p.R202Q/p.R202Q+p.N227S/p.N227S, were detected. The most common GBA mutation in the population was p.L483P with an allele frequency of 32.7%, followed by p.N409S (19.2%).
Conclusion:
The present study detected six new mutations of GBA gene among GD patients. Two mutations (p.L483P and p.N409S) were especially common among Iranians; this finding can be used in implementing screening programs and understanding the molecular basis of GD.
Key Words: Gaucher Disease, Mutation, GBA, Sequencing
Introduction
Gaucher disease (GD) is an autosomal recessive disorder and the most common form of lysosomal storage disease (LSD). It results from deficiencies in acid β-glucosidase (glucocerebrosidase [GBA], E.C.3.2.1.45). In GD, the accumulation of GBA in macrophages leads to organ dysfunctions. These organ dysfunctions are generally characterized by anemia, thrombocytopenia, bone disease, and hepatosplenomegaly. However, GD is rarely associated with neurological symptoms (1-2). There are three clinical subtypes of GD according to the absence or presence of neurological complications. Type I GD (OMIM 230800) or non-neuronopathic form manifests without neurological involvement. Type II (OMIM 230900) or acute neuronopathic disease is a fatal neurodegenerative disorder of infancy, and finally, type III (OMIM 231000) is a chronic neuronopathic form of the disease (3, 4).
The GBA gene is located on chromosome 1q21. It has a sequence of 7.6 kb, comprising of 11 exons and ten introns (5). Determination of the GD mutation spectrum in different ethnic groups can be useful in genetic counseling, screening programs, and identification of the molecular basis of GD. The present study aimed to identify the spectrum of GBA gene mutations in Iranians with GD and to study the genotype/phenotype associations.
Materials and Methods
Study population
A total of 33 GD patients, including the patients’ siblings and other relatives, were identified in this study. Finally, 26 unrelated GD patients were recruited from different regions of Iran. The study sample consisted of 11 males and 15 females in the age range of 2.5 to 33 years. The patients’ characteristics, including major clinical features, are shown in Table 1. The parents were first cousins in 15 families, while in three families, they were distant relatives. Patients were recruited from specialized pediatric clinics of inherited metabolic diseases and digestive disorders at Mofid Children's Hospital and Ali-Asghar Children's Hospital (Tehran, Iran) over two years.
Table 1.
Primer sequences and product sizes for identification of specific GBA exons, exon–intron boundaries, and common mutations
| Exon or common mutation | Primer sequence (5’-3’) | Restriction enzyme | Product size (bp) | Annealing temperature (°C) |
|---|---|---|---|---|
| Exon 1 | F ATCCTCTGGGATTTAGGAGC R CTGGATTCAAAGAGAGTCTG |
- | 482 | 56 |
| Exon 2 | F GTCCTAATGAATGTGGGAGACC R AAGCTGAAGCAAGAGAATCG |
- | 478 | 60.5 |
| Exon 3-4 | F GTTCAGTCTCTCCTAGCAGATG R GCAGAGTGAGATTCTGCCTC |
- | 637 | 60.5 |
| Exon 5-6 | F GATAAGCAGAGTCCCATACTCTC R ACAGATCAGCATGGCTAAAT |
- | 644 | 56 |
| Exon 7 | F CTAATGGCTGAACCGGATG R ATAGTTGGGTAGAGAAATCG |
- | 1144 | 56 |
| Exon 8 | F CTAGTTGCATTCTTCCCGTC R GCTTCTGTCAGTCTTTGGTG |
- | 404 | 60.5 |
| Exon 9 | F TGTGCAAGGTCCAGGATCAG R GCTCCCTCGTGGTGTAGAGT |
- | 914 | 62 |
| Exon 10-11 | F ACTGGAACCTTGCCCTGAAC R CTCTTTAGTCACAGACAGCG |
- | 904 | 56 |
| N409S (c.1226A>G) | F GTCTCTTTGCCTTTGTCCTTACCCTCGA R ACTGTCGACAAAGTTACGCACCCAAT |
XhoI | 120 | 60.5 |
| L483P (c.1448T>C) | F ACTGGAACCTTGCCCTGAAC R CTCTTTAGTCACAGACAGCG |
NciI | 904 | 56 |
| S439G (c.1315A>G) |
F AGCTGCCTCTCCCACATGTGACCCTTAC (common) R GTCCTTGGTGATGTCTACAATGATGGAACT (wild) R GTCCTTGGTGATGTCTACAATGATGGAACC (mutant) |
- | 210 | 61 |
All patients were previously diagnosed with GD, according to a fluorometric assay of GBA activity or had Gaucher cells in the bone marrow. Additionally, a fluorometric assay was performed for measuring the GBA (6) and chitotriosidase (7) activities in patients. The high-density lipoprotein (HDL)-cholesterol level was also measured after precipitation of lipoproteins containing apo B with phosphotungstic acid, using an available kit (Pars Azmoon Co., Iran). Seventeen patients received enzyme replacement therapy (ERT). This study was approved by the Ethics Committee of Pasteur Institute of Iran (IR.PII.REC.1394.02). Written informed consent was also obtained from the controls, patients, or their parents.
Genotyping
Genomic DNA was extracted from peripheral blood leukocytes, based on the standard phenol-chloroform method using a commercial SinaClon kit (Tehran, Iran). First, the samples were screened for common p.N409S and p.L483P mutations by polymerase chain reaction (PCR)-restriction fragment length polymorphism (RFLP), using XhoI and NciI restriction enzymes, respectively. The presence of both mutations was confirmed by DNA sequencing.
For sequencing analysis, eight DNA fragments, which covered 11 exons and exon-intron boundaries of GBA gene (NM_001005742.3) and no GBA pseudogene, were amplified by PCR, using exon-specific primer sequences reported in previous studies (8, 9) (Table 1). On the other hand, the new GBA-specific PCR products were prepared by pairing different primers and comparing the sequencing results against both GBA gene and pseudogene sequences. Moreover, an ARMS PCR technique was designed for rapid detection of p.S439G mutation, as the third common mutation in our population. The primers were manually designed and evaluated (Table 1). Finally, all PCR products were sequenced at Macrogen Inc. (Seoul, South Korea).
For analyzing and predicting the potential pathogenicity of novel mutations in the GBA gene coding region, three web-based tools, that is, SNPs3D (http://www.snps3d.org), SIFT (http://sift.jcvi.org/), and PolyPhen (http://genetics.bwh.harvard.edu/pph2), were used (10-12). Finally, in four common new mutations (p.S439G, p.M455R, p.L393V, and p.H312D), mutant structures were constructed using the I-TASSER (13) server, according to the 1OGS native structure.
Results
A total of 49 GBA mutant alleles were identified in 52 chromosomes of 26 GD patients, using PCR-RFLP and direct sequencing of PCR products. No lesions were detected in 30 control alleles. Among mutant alleles, there were 49 single nucleotide missense substitutions (six new mutations and six previously described mutations), one nonsense, and two complex allele mutations.
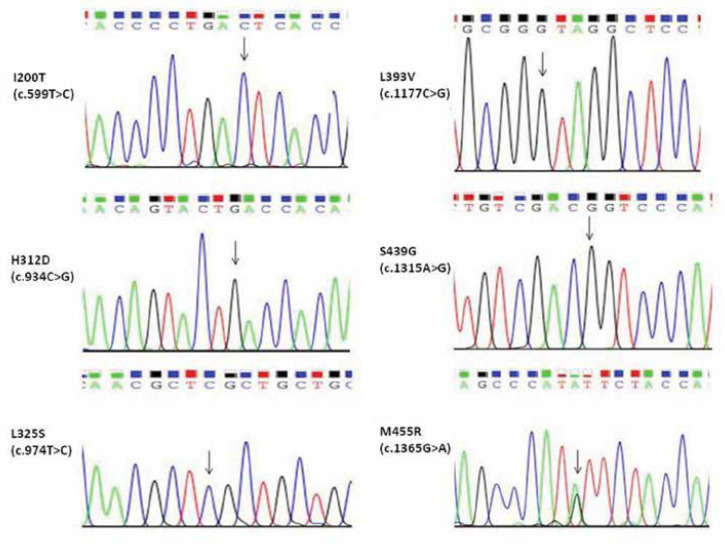
Mutations found in the present study are demonstrated in Table 2. The mutation results were also confirmed in the DNA samples of the patients’ parents if available. The most commonly observed clinical phenotypes were splenomegaly (84.6%), hepatomegaly (65.3%), anemia (61.5%), thrombocytopenia (61.5%), bone disease (26.9%), and failure to thrive (19.2%), respectively. The sequencing results of the newly detected mutations in the present study are shown in Figure 1. The mean activities of GBA and chitotriosidase were 0.64±0.41 µmol/L/h and 10477±11925 nmol/ml/h, respectively.
Table 2.
Demographic data, clinical characteristics, and geographic area of Iranians with GD
| No | Sex | Age (y) | Genotype | Phenotype | GD Type | Age at diagnosis (y) | CHIT1 | HDL2 | Cons3 | Ethnicity4 |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | M | 9.5 | L483P/L483P | At5, BD, FT, S, SNP, T | 3 | 1 | 2381 | 22 | + | Fars |
| 2 | M | 4 | L483P/L483P | A, H, Partial Sp | 1 | 3 | 11574 | 23 | + | Kurd |
| 3 | F | 7.5 | L483P/L483P | A, HS,OH, SS, St, T | 3 | 3 | 38131 | 32 | + | Fars |
| 4 | F | 3.5 | L483P/L483P | A, At, CI, DD, Hsz, St, S, SNP, T | 3 | 2 | 6193 | 25 | + | Fars |
| 5 | F | 8 | L483P/L483P | A, HS, T | 1 | 3 | 14250 | 16 | + | Fars |
| 6 | F | 7 | L483P/L483P | A, BD, FT | 1 | 2 | 1600 | 28 | - | Lor |
| 7 | M | 10 | L483P/L483P | A, HS, N, T | 1 | 1 | 3483 | 45 | - | Gilaki |
| 8 | M | 9 | L483P/H312D6 | A, HS | 1 | 8 | 38244 | 11 | - | Azari |
| 9 | F | 15 | L483P/N409S | A, BD, Sp | 1 | 8 | 1801 | 26 | - | Fars |
| 10 | M | 4 | L483P/M455R | HS, T | 1 | 3 | 3015 | 32 | - | Fars |
| 11 | F | 25 | N409S/N409S | H, Sp | 1 | 23 | 1450 | 35 | + | Fars |
| 12 | M | 33 | N409S/N409S | C, S, T | 1 | 31 | 3411 | 24 | + | Gilaki |
| 13 | F | 11 | N409S/N409S | HS | 1 | 6 | 4527 | 29 | + | Azari |
| 14 | M | 29 | N409S/N409S | A, HS, T | 1 | 14 | 5714 | 28 | - | Azari |
| 15 | F | 10 | N409S/W420X | HS | 1 | 7.5 | 6670 | 36 | - | Gilaki |
| 16 | M | 9 | S439G/S439G | A, BD, HS, T | 1 | 3 | 27406 | 12 | + | Kurd |
| 17 | F | 7 | [S439G/S439G + E379K] |
A, CI, DD, FT, HS | ? | 3 | 8004 | 18 | - | Kurd |
| 18 | F | 29 | M455R/? | A, BD, HS | 1 | 22 | 11577 | 19 | + | Azari |
| 19 | F | 9 | L393V/L393V | BD, S, T | 1 | 3 | 3491 | 10 | + | Azari |
| 20 | F | 19 | I200T/I200T | S, T | 1 | 15 | 101 | 33 | + | Kurd |
| 21 | M | 7.5 | [R202Q/R202Q +N227S/N227S] |
FT, S, T | 1 | 6.5 | 2947 | 30 | + | Azari |
| 22 | M | 2 | H312D/H312D | A, HS | 1 | 1.5 | 36731 | 18 | + | Lor |
| 23 | M | 3 | L325S/L325S | A, FT, HS, T | 1 | 0.5 | 0 | 19 | + | Gilaki |
| 24 | F | 2.5 | R398Q/R398Q | A, HS, T | 1 | 2 | 21647 | 19 | + | Fars |
| 25 | F | 18 | D448H/D448H | BD, OH, OMA, S, SS, T | 3 | 4 | 12390 | 24 | + | Fars |
| 26 | F | 20 | ?/? | A, C, HS, T | 1 | 19 | 5677 | 30 | + | Gilaki |
1CHIT: Chitotriosidase activity. 2HDL-C: HDL-cholesterol. 3Cons: Consanguinity. 4Main ethnicities in Iran include Fars, Azari, Kurd, Gilaki, and Lor, respectively.
5Abbreviations: F, female. M, male. y, year. A, anemia. At, ataxia. BD, bone disease. C, cirrhosis. CI, cognitive impairment. DD, development delay. FT, failure to thrive. H, hepatomegaly. Hsz, history of seizure. MC, microcephaly. N, nocturia. OH, ocular hypertension. OMA, oculomotor apraxia. S, splenomegaly. SNP, supranuclear gaze palsy. SS, slow speech. Sp, splenectomy. St, strabismus. T, thrombocytopenia
Figure 1.
The sequencing results for six new mutations
The type and frequency of the detected alleles are demonstrated in Table 3. The mutation nomenclature followed the Human Genome Variation Society’s mutation nomenclature recommendations (http://www.hgvs.org/mutnomen). Moreover, the traditional nomenclature, which subtracts the first 39 amino acids of the preprotein, was described (14). The p.L483P mutation was detected as the most common mutation (32.7%) in our patients. The p.N409S mutation was identified in ten alleles (19.2%). The p.S439G mutation was found to be the third most common mutation (7.6% of alleles). This mutation is the first common novel sequence alteration, caused by G to A variation at position 1315 of cDNA (exon 9). The previously described and new mutations of GBA gene, nucleotide alterations, amino acid changes, and their frequencies in our study are listed in Table 3.
Table 3.
Prevalence of various GBA mutant alleles, cDNA nucleotide substitutions, amino acid changes, and frequency of mutations
| Allele typea | Exon number | cDNAb | Protein | Frequency (%) |
|---|---|---|---|---|
| Missense L483P N409S S439G M455R L393V H312D I200T L325S R398Q D448H |
10 9 9 9 8 7 6 7 8 10 |
c.1448T>C c.1226A>G c.1315A>G c.1365G>A c.1177C>G c.934C>G c.599T>C c.974T>C c.1193G>A c.1342G>C |
p.Leu483Pro p.Asn409Ser p.Ser439Gly p.Met455Arg p.Leu383Val p.His312Asp p.Ile200Thr p.Leu325Ser p.Arg398Gln p.Asp448His |
17 (32.7) 10 (19.2) 3 (5.7) 2 (3.8) 2 (3.8) 3 (5.7) 2 (3.8) 2 (3.8) 2 (3.8) 2 (3.8) |
| Nonsense W420X |
9 | c.1259G>A | p.Trp420X | 1 (1.9) |
| Complex allele R202Q + N227S S439G + E379K |
6 & 6 9 & 8 |
c.605G>A c.680A>G c.1315A>G c.1135G>A |
p.Arg202Gln p.Asn225Ser p.Ser439Gly p.Glu379Lys |
2 (3.8) 1 (1.9) |
| Unidentified allele | - | - | - | 3 (5.7) |
| Total | 52 (100) |
a GBA mutations are named according to www.hgvs.org/mutnomen.
bNucleotides are numbered from A of the first ATG.
The new p.M455R mutation was found to be heterozygous in two patients. The second mutant allele was not identified in one of the two patients carrying the p.M455R mutation. In two patients, the combined alleles of R202Q+N227S and S439G+E379K were detected. Nevertheless, in another patient, no mutation in exons or exon-intron boundaries of GBA gene was found. The newly detected mutations of GBA gene were analyzed in silico, using web-based SNP3D, SIFT, and Polyphen2 tools to predict the possible damaging effects of new GBA mutations.
Discussion
The present study is the first report of GD mutations in Iranians with different ethnic backgrounds. The detection rate of mutations was 94.2%, and the p.L483P mutation was the most common mutation with a frequency of 32.7%. Our findings are consistent with several studies on Syrian, Western Indian, Korean, Filipino, and Chinese populations (15-19). However, among American, Brazilian, Venezuelan, European, and Jewish populations, the p.N409S mutation was found to be the most common mutation (20-24). The prevalence of p.L483P mutation clearly decreased from west to east, while the prevalence of p.N409S mutation increased.
Consistent with other reports (16), a similar phenotype-genotype correlation was found in GD patients with both GD type I and III carriers of p.L483P homozygous genotypes. Interestingly, the most severe involvement was reported in patient 4, who presented with ataxia, history of seizures, cognitive impairment, strabismus, supranuclear gaze palsy (frozen eyes), and other classic symptoms of GD. She also had a history of hypothyroidism and was unable to walk. The second most common mutation was p.N409S, which was found in six patients without any neurological symptoms, indicating the neuroprotective function of this mutation (25).
Moreover, the novel p.S439G mutation was found in the present study. Both patients with this mutation had GD type I with moderate-mild symptoms. However, patient 17 had an unknown type of GD, which co-occurred with beta-thalassemia minor and severe manifestations. She had a moderate cognitive impairment (according to the Mini Mental State Examination [MMSE]), slurred speech, failure to thrive, developmental delay, and learning disorders, but had no other neurological symptoms. Moreover, a patient with the p.L393V mutation was found, who had type I GD. This patient (No. 19) started ERT at the age of three, but suffered from bone pain and was at a high risk of bone fracture, according to the bone mineral density (BMD) test.
In our study, another interesting phenotype-genotype correlation was found in a patient with a homozygous p.D448H mutation. This young female patient had been diagnosed with type III GD at the age of four. She had been on ERT from the age of 14, with a good response to the treatment of anemia and organomegaly. However, at the age of 19, she suffered from oculomotor apraxia and poor eyelid function (decreased blink frequency), which led to a scar in the left eye with keratoconjunctivitis, ocular hypertension, and face asymmetry. She also had a history of kidney stone, migraine, slow speech, and calcification of heart valves, which have been previously reported for this genotype (26).
We identified homozygous p.I200T, p.H312D, and p.L325S mutations in three patients. All of these patients had GD type I with moderate complications and a good response to ERT. The results of in silico analysis revealed that all new mutations might be damaging. However, according to all three web-based tools, the p.H312D mutation might be a tolerable alteration in the GBA gene. Nonetheless, patient 22 only had this mutation in the GBA gene. After starting ERT, organomegaly and anemia improved in this patient after eight months of therapy. The p.I200T and p.M455R mutations were detected with different nucleotide changes in GD (27, 28). Clearly, these missense changes were pathogenic.
In the present study, the new homozygous p.R202Q+p.N227S mutation was detected in a patient with type I GD. This patient was diagnosed with severe thrombocytopenia and failure to thrive. In Korean patients, the p.N227S mutation (17) was associated with type III GD. In our study, the presence of p.N227S+p.R202Q led to type I GD, without any neurological symptoms.
Concerning the in silico results, change of charge in the residues of domain III in the presence of two new p.H312D and p.M455R (α8-Heilx) mutations could alter the spatial configuration of GBA molecule, possibly affecting its catalytic function. Histidine 312 and histidine 262 are on the surface of GBA opposite to loop 1 (29). Also, the presence of c.974T>C transition results in the substitution of leucine to serine (p.L325S), i.e., a non-polar amino acid replaced by a polar amino acid with a hydroxyl group in domain III. This domain has (β/α) an 8TIM barrel structure (located between α5 helix and β5 sheet); the mutation could change the polarity and affinity for H+ with severe phenotypic consequences (30). The p.S439G mutation could eventually disrupt the H-bond between D438 and S439 in loop 2 of GBA in both states of open and close conformations (31).
Moreover, the outputs of I-TASSER server were obtained, and all six new mutations were superposed by YASARA structure package (Figure 2). All mutant structures had a root mean square deviation (RMSD) of about 1.2-1.35 Å, which possibly destabilizes the GBA structure. Briefly, in the p.I200T mutation, an extra turn was formed in the α2-helical structure. In the p.H312D structure, a part of β5-sheet (near the catalytic site) altered to coil. In the p.L325S mutation, Asp322 to Arg324 residues in the α5-heilx forms a secondary structure turn. The L354V mutation moderately changed the catalytic site. The p.S439G mutation induced a small β-sheet transition to coil in domain I. Finally, the p.M455R mutation had the same effect on the β-sheet in domain II (Figure 2). The percentage of unknown alleles (5.7%) was similar to reports in European and Asian populations (32, 33).
Figure 2.
The representation of superposed native GBA structure (green) with different mutant structures (red).
Despite the availability of detailed and efficient methods for the detection of GBA gene mutations, many mutations are still unknown. In summary, the current study investigated the molecular basis of GD among Iranians. Six new mutations were detected among GD patients, and the presence of two common mutations, that is, p.L483P and p.N409S, was confirmed in the Iranian population. Our study provided valuable knowledge about the molecular basis of GD, genotype-phenotype correlations, and allelic heterogeneity of this disease.
Acknowledgments
This study was financially supported by a research grant from Iran National Science Foundation (grant No.: 92031115). We thank the staff members of Pasteur Institute of Iran, Tarbiat Modares University, Ali-Asghar Children's Hospital, and Mofid Children's Hospital of Tehran.
Authors’ contribution
Hadi Mozafari: Involved in design, performance of experiments and writing manuscript
Mohammad Taghikhani: Designed and advised the study
Asad Vaisi-Raygani: Involved in writing and editing of manuscript
Shahla Ansari: Involved in clinical data collection of patients
Shohreh Khatami: Main supervisor and designer of study
Reza Saghiri: Involved in advising of project and editing of manuscript
Rahimi Zohreh: Involved in writing and editing of manuscript
Mohammad Reza Alaei: Involved in clinical data collection of patients and editing of manuscript
Conflict of interest
All authors of this article have no financial or personal involvement or any conflicts of interest.
References
- 1.Shrestha B, Devgan A, Sharma M. Gaucher's disease: rare presentation of a rare disease. J Child Neurol. 2013;28:1296–8. doi: 10.1177/0883073812454940. [DOI] [PubMed] [Google Scholar]
- 2.Pacheco N, Uribe A. Enzymatic analysis of biomarkers for the monitoring of Gaucher patients in Colombia. Gene. 2013;521:129–35. doi: 10.1016/j.gene.2013.03.044. [DOI] [PubMed] [Google Scholar]
- 3.Tammachote R, Tongkobpetch S, Srichomthong C, Phipatthanananti K, Pungkanon S, Wattanasirichaigoon D, et al. A common and two novel GBA mutations in Thai patients with Gaucher disease. J Hum Genet. 2013;58:594–9. doi: 10.1038/jhg.2013.60. [DOI] [PubMed] [Google Scholar]
- 4.Jack A, Amato D, Morris G, Choy FY. Two novel mutations in glucocerebrosidase, C23W and IVS7-1 G>A, identified in Type 1 Gaucher patients heterozygous for N370S. Gene. 2014;538:84–7. doi: 10.1016/j.gene.2014.01.015. [DOI] [PubMed] [Google Scholar]
- 5.Hruska KS, LaMarca ME, Scott CR, Sidransky E. Gaucher disease: mutation and polymorphism spectrum in the glucocerebrosidase gene (GBA) Hum Mutat. 2008;29:567–83. doi: 10.1002/humu.20676. [DOI] [PubMed] [Google Scholar]
- 6.Olivova P, Cullen E, Titlow M, Kallwass H, Barranger J, Zhang K, et al. An improved high-throughput dried blood spot screening method for Gaucher disease. Clin Chim Acta. 2008;398:163–4. doi: 10.1016/j.cca.2008.08.024. [DOI] [PubMed] [Google Scholar]
- 7.Hollak CE, van Weely S, van Oers MH, Aerts JM. Marked elevation of plasma chitotriosidase activity. A novel hallmark of Gaucher disease. J Clin Invest. 1994;93:1288–92. doi: 10.1172/JCI117084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pomponio RJ, Cabrera-Salazar MA, Echeverri OY, Miller G, Barrera LA. Gaucher disease in Colombia: mutation identification and comparison to other Hispanic populations. Mol Genet Metab. 2005;86:466–72. doi: 10.1016/j.ymgme.2005.07.026. [DOI] [PubMed] [Google Scholar]
- 9.Siebert M, Bock H, Michelin-Tirelli K, Coelho JC, Giugliani R, Saraiva-Pereira ML. Novel mutations in the glucocerebrosidase gene of brazilian patients with Gaucher disease. JIMD Rep. 2013;9:7–16. doi: 10.1007/8904_2012_174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yue P, Moult J. Identification and Analysis of Deleterious Human SNPs. J Mol Biol. 2006;356:1263–74. doi: 10.1016/j.jmb.2005.12.025. [DOI] [PubMed] [Google Scholar]
- 11.Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4:1073–81. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 12.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–9. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang J, Yan R, Roy A, Xu D, Poisson J, Zhang Y. The I-TASSER Suite: protein structure and function prediction. Nat Methods. 2015;12:7–8. doi: 10.1038/nmeth.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miocić S, Filocamo M, Dominissini S, Montalvo AL, Vlahovicek K, Deganuto M, et al. Identification and functional characterization of five novel mutant alleles in 58 Italian patients with Gaucher disease type 1. Hum Mutat. 2005;25 doi: 10.1002/humu.9301. [DOI] [PubMed] [Google Scholar]
- 15.Alasmar D. Gaucher disease in Syrian children: common mutations identification, and clinical futures. Ann Saudi Med. 2015;35:127–32. doi: 10.5144/0256-4947.2015.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ankleshwaria C, Mistri M, Bavdekar A, Muranjan M, Dave U, Tamhankar P, et al. Novel mutations in the glucocerebrosidase gene of Indian patients with Gaucher disease. J Hum Genet. 2014;59:223–8. doi: 10.1038/jhg.2014.5. [DOI] [PubMed] [Google Scholar]
- 17.Jeong SY, Park SJ, Kim HJ. Clinical and genetic characteristics of Korean patients with Gaucher disease. Blood Cells Mol Dis. 2011;46:11–4. doi: 10.1016/j.bcmd.2010.07.010. [DOI] [PubMed] [Google Scholar]
- 18.Chiong MAD, Racoma MJC, Abacan MAR. Genetic and clinical characteristics of Filipino patients with Gaucher disease. Mol Genet Metab Rep. 2018;15:110–115. doi: 10.1016/j.ymgmr.2018.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Feng Y, Huang Y, Tang C, et al. Clinical and molecular characteristics of patients with Gaucher disease in Southern China. Blood Cells Mol Dis. 2018;68:30–34. doi: 10.1016/j.bcmd.2016.10.026. [DOI] [PubMed] [Google Scholar]
- 20.Orenstein M, Barbouth D, Bodamer OA, Weinreb NJ. Patients with type 1 Gaucher disease in South Florida, USA: demographics, genotypes, disease severity and treatment outcomes. Orphanet J Rare Dis. 2014;31:45. doi: 10.1186/1750-1172-9-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rozenberg R, Fox DC, Sobreira E, Pereira LV. Detection of 12 new mutations in Gaucher disease Brazilian patients. Blood Cells Mol Dis. 2006;37:204–9. doi: 10.1016/j.bcmd.2006.09.004. [DOI] [PubMed] [Google Scholar]
- 22.Gómez G, Arias S, Cárdenas L, Zoghbi D, Paradisi I. GBA mutations in Gaucher type I Venezuelan patients: ethnic origins and frequencies. J Genet. 2017;96:583–589. doi: 10.1007/s12041-017-0821-8. [DOI] [PubMed] [Google Scholar]
- 23.Mattošová S, Chandoga J, Hlavatá A, Saligová J, Maceková D. Spectrum of GBA mutations in patients with Gaucher disease from Slovakia: identification of five novel mutations. Isr Med Assoc J. 2015;17:166–70. [PubMed] [Google Scholar]
- 24.Duran R, McNeill A, Mehta A, Hughes D, Cox T, Deegan P, et al. Novel pathogenic mutations in the glucocerebrosidase locus. Mol Genet Metab. 2012;106:495–7. doi: 10.1016/j.ymgme.2012.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Perretti A, Parenti G, Balbi P, Titomanlio L, Marcantonio L, Iapoce M, et al. Study of multimodal evoked potentials in patients with type 1 Gaucher's disease. J Child Neurol. 2005;20:124–8. doi: 10.1177/08830738050200020801. [DOI] [PubMed] [Google Scholar]
- 26.Abrahamov A, Elstein D, Gross-Tsur V, Farber B, Glaser Y, Hadas-Halpern I, et al. Gaucher's disease variant characterised by progressive calcification of heart valves and unique genotype. Lancet. 1995;346:1000–3. doi: 10.1016/s0140-6736(95)91688-1. [DOI] [PubMed] [Google Scholar]
- 27.Liou B, Kazimierczuk A, Zhang M, Scott CR, Hegde RS, Grabowski GA. Analyses of variant acid beta-glucosidases: effects of Gaucher disease mutations. J Biol Chem. 2006;281:4242–53. doi: 10.1074/jbc.M511110200. [DOI] [PubMed] [Google Scholar]
- 28.Beutler E, Gelbart T, Scott CR. Hematologically important mutations: Gaucher disease. Blood Cells Mol Dis. 2005;35:355–64. doi: 10.1016/j.bcmd.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 29.Yap TL, Gruschus JM, Velayati A, Westbroek W, Goldin E, Moaven N, et al. Alpha-synuclein interacts with Glucocerebrosidase providing a molecular link between Parkinson and Gaucher diseases. J Biol Chem. 2011;286:28080–8. doi: 10.1074/jbc.M111.237859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dvir H, Harel M, McCarthy AA, Toker L, Silman I, Futerman AH, et al. X-ray structure of human acid-beta-glucosidase, the defective enzyme in Gaucher disease. EMBO Rep. 2003;4:704–9. doi: 10.1038/sj.embor.embor873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Premkumar L, Sawkar AR, Boldin-Adamsky S, Toker L, Silman I, Kelly JW, et al. X-ray structure of human acid-beta-glucosidase covalently bound to conduritol-B-epoxide. Implications for Gaucher disease. J Biol Chem. 2005;280:23815–9. doi: 10.1074/jbc.M502799200. [DOI] [PubMed] [Google Scholar]
- 32.Horowitz M, Tzuri G, Eyal N, Berebi A, Kolodny EH, Brady RO, et al. Prevalence of nine mutations among Jewish and non-Jewish Gaucher disease patients. Am J Hum Genet. 1993;53:921–30. [PMC free article] [PubMed] [Google Scholar]
- 33.Emre S, Gürakan F, Yüce A, Rolf A, Scott R, Ozen H. Molecular analysis of Turkish Gaucher disease patients: identification of novel mutations in glucocerebrosidase (GBA) gene. Eur J Med Genet. 2008;51:315–21. doi: 10.1016/j.ejmg.2008.02.004. [DOI] [PubMed] [Google Scholar]