Abstract
Vaccination represents the best line of defense against infectious diseases and is crucial in curtailing pandemic spread of emerging pathogens to which a population has limited immunity. In recent years, mRNA vaccines have been proposed as the new frontier in vaccination, owing to their facile and rapid development while providing a safer alternative to traditional vaccine technologies such as live or attenuated viruses. Recent breakthroughs in mRNA vaccination have been through formulation with lipid nanoparticles (LNPs), which provide both protection and enhanced delivery of mRNA vaccines in vivo. In this review, current paradigms and state-of-the-art in mRNA-LNP vaccine development are explored through first highlighting advantages posed by mRNA vaccines, establishing LNPs as a biocompatible delivery system, and finally exploring the use of mRNA-LNP vaccines in vivo against infectious disease towards translation to the clinic. Furthermore, we highlight the progress of mRNA-LNP vaccine candidates against COVID-19 currently in clinical trials, with the current status and approval timelines, before discussing their future outlook and challenges that need to be overcome towards establishing mRNA-LNPs as next-generation vaccines.
Statement of significance
With the recent success of mRNA vaccines developed by Moderna and BioNTech/Pfizer against COVID-19, mRNA technology and lipid nanoparticles (LNP) have never received more attention. This manuscript timely reviews the most advanced mRNA-LNP vaccines that have just been approved for emergency use and are in clinical trials, with a focus on the remarkable development of several COVID-19 vaccines, faster than any other vaccine in history. We aim to give a comprehensive introduction of mRNA and LNP technology to the field of biomaterials science and increase accessibility to readers with a new interest in mRNA-LNP vaccines. We also highlight current limitations and future outlook of the mRNA vaccine technology that need further efforts of biomaterials scientists to address.
Keywords: Lipid nanoparticles, mRNA, Vaccines, COVID-19
Graphical abstract
1. Introduction
Deliberate priming of the immune system with a pathogen originated in primitive form in Medieval China [1]. Such inoculation was advanced by Edward Jenner on a farm in 1796, whose pioneering work on smallpox paved the way for its global eradication almost two centuries later [2]. A similar approach led to the development of a vaccine for cholera and rabies by Louis Pasteur in 1880, when he discovered that chickens exposed to bacterial culture of cholera didn't die from the disease [3]. These scientific breakthroughs are saving millions of lives and have had the greatest public health benefits aside from the provision of safe drinking water [4]. The lasting benefit of vaccines relies not only on the effectiveness of the technology, but also heavily on governmental vaccine policies. Vaccine campaigns, such as those launched by the World Health Organization (WHO), promote global protection, as high levels of population coverage are necessary for achieving herd immunity that disrupts chains of infection and stops or slows the spread of the disease [5]. While funding, government and regulatory agencies are key to the development and success of vaccines, this review will focus instead on the scientific aspects required for a successful vaccine technology with a specific focus on the development of lipid nanoparticle (LNP)-formulated messenger RNA (mRNA) vaccines.
mRNA, a single stranded RNA molecule encoding for a (viral) antigen of interest, is produced from DNA by an in vitro transcription process and subsequently injected in vivo where translation of the antigen occurs, towards which the body mounts an immune response. This is an attractive cell-free, rapid, scalable process, which is well suited to respond to pandemic outbreaks, such COVID-19. Major scientific advances in mRNA purification, sequence optimization and nucleoside chemistry have paved the way to tailoring the expression kinetics with potent immune responses. Several biotech companies have based their entire scientific approach and pipeline on one or a combination of these chemical RNA modifications, and claim an optimal activation of the innate immune system. Whilst the mRNA construct is key for successful in situ translation into a functional protein, it has become increasingly apparent that the delivery system is equally important to the design of an effective vaccine. Naked mRNA, modified or not, is prone to degradation in the systemic circulation resulting in degradation products that are small enough to be renally excreted. These molecular properties do not promote cellular uptake and exposure to organs of interest for antigen production and for subsequent immune response. The last decade has seen an avalanche of new nucleic acid nanoparticle delivery systems that aim at efficiently encapsulating mRNA, providing protection against serum nucleases, facilitating endocytosis, promoting endosomal escape, and eventually at eliciting an immune response. Two non-viral delivery systems currently in the spotlight due to their efficient delivery properties are lipoplexes and LNPs. Both comprising of similar lipids (cationic lipid, helper lipid, cholesterol), they mainly differ in size, heterogeneity, and the location of the nucleic acid in the particle; in the lipid bilayer (lipoplex) or in the particle core (LNPs) [6,7]. For vaccine purposes these nanoparticles aim at delivering mRNA to dendritic cells and in lymphoid compartments such as the spleen for optimal antigen presentation and immune response activation.
Here we review the state of the art in mRNA optimization and LNP design to tailor the immune response for vaccine applications. We explore the hurdles and successes to clinical translation of these technologies with a focus on the recent clinical development of mRNA-LNP vaccine candidates against COVID-19.
2. mRNA vaccines: an emerging vaccine technology
2.1. Nucleic acid vaccines: an overview
Vaccines represent the ultimate form of biomedical disease prevention. The introduction of an antigen into the body to stimulate an immune response is termed a ‘vaccination’, and leads to ‘immunization’ – the process of protecting individuals, but also the community (herd immunity) from disease by acquiring immunity [8]. Although the terms are often used interchangeably, passive immunization can be achieved by immunoglobulin administration for initial short-term protection, while vaccines promote T cell (cellular) and B cell (humoral) immune responses leading to adaptive immunity for long-lasting protection against diseases. There are four main subtypes of conventional vaccines: live-attenuated virus, inactivated virus, subunit and toxoid. Their production, mechanisms of action and advantages or disadvantages to their use are beyond the scope of this review, and are discussed in detail elsewhere [4,5,8].
The use of recombinant DNA technology in vaccine design has led to the evolution of nucleic acid vaccines. Historically, engineered vectors that contain the gene encoding for the subunit of the pathogen were inserted into yeast, bacteria, or replication defective viral expression systems that produce the required subunit antigen. These recombinant vector vaccines allow the safe and reproducible production of large quantities of purified antigen. Rather than inserting the DNA plasmid into a bacterial or mammalian cell to then purify the antigenic product, nucleic acid vaccines are instead administered directly into the patient where the antigen is produced within the patient's own cells (in situ) [9]. In a successful nucleic acid vaccine, the viral antigen, expressed by the host cells in situ, is recognized as non-self and prompts an immune response: an approach designed to generate immunity to the original virus without compromising the safety of the vaccine recipient. Plasmid DNA and messenger RNA (mRNA) vaccines have been used in pre-clinical models of viral infection and have shown promising results with good safety profiles, prompting a number of them to enter clinical trials in recent years [10], [11], [12]. With respect to the manufacturing process, nucleic acid vaccines are simpler to produce and to purify than recombinant protein antigens, a key consideration in reducing the response time during a pandemic outbreak, such as COVID-19. Once the sequence encoding an immunogen is known, clinical batches of nucleic acid vaccines can be produced within weeks. This can also be applied when a new viral strain has been prototyped (influenza), or mutations have occurred in a known virus. Due to being orders of magnitude smaller in size compared to protein-based therapeutics, multiple mRNAs encoding various antigens can be combined into a single immunization. This multi-antigenic approach could be a more robust solution for assembling multimeric protein complexes in situ (e.g. for neutralizing epitopes to be exposed) and thereby pave the way forward to a universal influenza vaccine.
The capacity for rapid, relatively cheap production and safe administration of nucleic acid vaccines renders them an attractive alternative to many conventional vaccines, in particular given the need for large scale deployment in response to emerging diseases such as COVID-19 [13]. Though both DNA and mRNA vaccines rely on the same basic principles of utilizing the cells’ ability to produce the antigen, there are advantages and drawbacks associated with both approaches, listed in Table 1 . Interest in developing mRNA therapeutics was rather scarce at the start of 2000s, mostly due to concerns of instability, poor in vivo delivery, and high innate immunogenicity. Hope, instead, was placed on DNA-based therapies and vaccines – especially after published reports, such as Ulmer et al. in 1993, demonstrated that mice injected with pDNA encoding a conserved influenza protein elicited protective antibodies against an influenza strain that was heterologous to the strain from which the DNA was cloned, thereby demonstrating cross-strain protective immunity [14]. There have been some promising results for DNA vaccines in animal models, leading to the development of animal health products such as a West Nile virus vaccine for horses [15] and a melanoma cancer vaccine for dogs [16]. Despite this, DNA vaccines have struggled to make it to the human market, as initial clinical studies have reported low and sporadic levels of both antibody and T cell responses. The reasons for these differences in immunogenicity for DNA vaccines are not clear, but are likely due, at least in part, to inefficient delivery of DNA into human cells and therefore inadequate stimulation of the human immune system.
Table 1.
Comparing DNA and mRNA vaccines.
| Advantages | ||
|---|---|---|
| mRNA | DNA | |
| Safety |
|
|
| Stability and Manufacturing |
|
|
| Clinical Application |
|
|
| Disadvantages | ||
| mRNA | DNA | |
| Safety |
|
|
| In vivodelivery |
|
|
| Clinical application |
|
|
2.2. mRNA: the cellular vaccine factory
Major technological advances in the fields of sequence optimization, nucleotide chemistry, and formulation design in the past decade have revolutionized the therapeutic application of mRNA. While the immunostimulatory effects of naked mRNA have been extensively documented in recent literature [24,[39], [40], [41]], it is worth pointing out that different approaches have emerged amongst research groups with respect to modifying the structural features of mRNA to manipulate immune responses. These bioengineering strategies for nucleic acid vaccines can be classified into four categories: nucleoside modifications, sequence optimization, HPLC purification, and self-amplifying mRNA (SAM) – with each having both advantages and drawbacks in relation to immune modulation. The main challenge of these bioengineering strategies is finding the right balance between adjuvancy and translation efficiency [40].
In vitro transcribed (IVT) mRNA was first reported to be effective in vivo following administration to mice in 1990 [42]. Within a few years, antigenic information was delivered via mRNA to antigen presenting cells (APCs) [43]. Since then, a plethora of technological advances and research have presented mRNA as a potential therapeutic tool for vaccine development [39] To produce IVT mRNA, a linear DNA template of the selected gene or antigen of interest is required. Bacteriophage-derived RNA polymerases such as T7, T4 or SP6 are then utilized for transcription of the DNA template to generate mRNA, which then undergoes further modification and purification prior to in vivo application. In general, IVT mRNA will contain sequences beyond the coding region of the related gene – these non-coding elements play a critical role in the pharmacology of the mRNA and can be optimized to suit their application. Modification of the 5′- and 3′-untranslated regions (UTRs), flanking the coding sequence, has been shown to dramatically impact the stability and translation efficiency of exogenous mRNA. A common approach is to select the UTRs from endogenously found proteins with long half-lives that lack active mRNA synthesis (e.g. α- and β-globins, which are functional proteins found abundantly in erythrocytes) [44,45]. Optimized motifs have been shown to generate a superior antigen-specific immune response by increasing the intensity and duration of gene expression in mice lymphoid tissues [46]. Other essential components to protect against premature degradation of the construct by exo-and endonucleases are the poly(A) tail and the 5′ cap. Slowing down deadenylation-dependent mRNA decay in P bodies (cytoplasmic ribonucleoprotein granules) within eukaryotic cells by increasing the length of the poly(A) tail between 120 and 150 nucleotides is now common practice [47]. Inclusion of segmented poly(A) tails utilizing a spacer element (typically short, around ten nucleotides) in the DNA template also leads to enhanced translational efficiency and half-life of mRNA [48]. Recent efforts towards rapid and large-scale production of mRNA therapeutics have led to development of a co-transcriptional 5′ capping strategy, called CleanCap®, proprietary to TriLink Biotechnologies. This method generates a natural Cap 1 structure which improves the translation of mRNA into a functional protein over the Cap 0 obtained with ARCA, which has shown to activate the pattern recognition receptors involved in breaking down non-self RNA [49], [50], [51]. The CleanCap technology also eliminates the cost and sample loss associated with conventional enzymatic capping methods, such as ARCA and mCap [52]. Alternatively, codons in the open reading frame (ORF) can also be optimized by replacing rare codons with more abundant, synonymous sequences for more efficient harnessing of cytosolic tRNAs during translation [53].
2.2.1. Sequence optimization
Bypassing codon degeneracy and enriching GC content is a form of sequence engineering, and has shown to increase protein levels in vivo [54]. CureVac, a German company at the forefront of prophylactic mRNA vaccine developments, has built an entire vaccine platform around this sequence engineering concept to achieve high levels of antigen expression [55]. Their RNAactive technology also includes a separate protamine-based adjuvant that acts via the TLR 7-RIG1 signaling pathway to stimulate the innate immune response. While their technology has demonstrated strong and durable immunogenicity in non-human primates (NHP) with the seasonal flu vaccine and in preclinical models of rabies, recent data from an open-label first-in-human phase 1 clinical trial (CV7201) indicated modest results. In the trial, vaccination by needle-syringe failed to induce an adequate level of neutralizing antibodies, independent of dose or route of administration [31]. Both Moderna's and BioNTech's COVID-19 vaccines encode for a pre-fusion stabilized conformation of the spike protein, made possible with by two proline substitutions at the apex of the central helix.
The utility of codon-optimization was interrogated by Mauro et al. [56] who highlighted complexities associated with mRNA secondary structure [55], altered post-transcriptional modifications, protein folding, protein function [57,58], and potential loss of naturally-occurring cryptic peptides, all contributing to a therapeutic immune response [56]. As a solution to this lack of immunogenicity, CureVac has coincidentally focused on the development of a RNA-based adjuvant (CV8102, RNAdjuvant®), a TLR 7/8 agonist and RIG I pathway activator [59]. In preclinical studies of influenza and rabies, CV8102 was combined with an existing, licensed vaccine and induced humoral immune responses [59]. Furthermore, combining CV8102 with antigen-derived T cell epitopes for therapeutic cancer vaccination also induced strong humoral and cellular immune responses [60]. The magnitude of these benefits were not observed in a clinical trial that investigated the combination of Rabipur, an inactivated licensed rabies vaccine, with CV8102 and additional severe reactogenicity was reported [61]. This adjuvant combination model has shown more promising results in cancer immunotherapy, and might be more suitable in therapeutic vaccines where higher reactogenicity is acceptable [62].
2.2.2. Nucleoside modification
A second approach to modulate the immunogenicity of mRNA is the use of modified nucleosides in the construct. This method has origins in the findings of Kariko et al., who demonstrated that incorporation of naturally occurring nucleoside modifications, such as (1-methyl)-pseudouridine, ablates activation of innate immune sensors and thereby reduces interferon alpha (IFNα) signaling which would otherwise lead to degradation of invading mRNA [63]. Additionally, these modifications were shown to reduce the activity of mRNA cleavage enzymes (RNAse L) and translation inhibition enzymes (protein kinase R) [64,65]. The vast majority of the animal studies with modified mRNA have been conducted in the context of enhancing the stability and efficiency of mRNA translation (in comparison to unmodified nucleoside), which are desirable features in protein replacement therapies [29,66]. While suppressing the intrinsic adjuvant activity of IVT mRNA may limit the design of potent vaccines, recent evidence suggests that improper or excessive activation of type I IFN can interfere with antigen expression and adaptive immunity, thereby impairing the efficacy of mRNA vaccines [67,68]. The biotech company Moderna (‘Mod-e-RNA’) has based its entire platform on base-modified mRNA, with a diverse pipeline ranging from gene therapies to prophylactic and cancer vaccines. Efforts around disease prevention have focused on vaccines against cytomegalovirus (CMV), Zika, respiratory syncytial virus (RSV), and influenza which are currently undergoing clinical trials. BioNTech is a German company that, amongst other platform technologies, incorporates a variety of modified nucleosides to suppress intrinsic immune activation by mRNA. This technology is used in a variety of applications where therapeutic proteins are translated by the mRNA, such as ‘RiboMab’ (mRNA-encoded antibody therapeutics), ‘RiboCytokine’ (mRNA-encoded cytokine therapeutics), and protein replacement therapies for rare diseases. Their (neo) antigen platform instead includes optimized unmodified mRNA, claiming that the natural adjuvancy of mRNA is a benefit when used for immunotherapies, such as the iNeST (Individualized Neoantigen Specific Immunotherapy) and FixVac (fixed vaccine combination of shared tumor-associated antigens against cancer) platforms.
2.2.3. Purification of mRNA by LC
Purification of mRNA via fast protein liquid chromatography (FPLC) or high-performance liquid chromatography (HPLC) is performed to remove any remaining reaction by-products and produce mRNA at a large scale in compliance with Good Manufacturing Practice (GMP) processes [69]. This is a third engineering strategy that both unmodified [54,70] and modified [69,71] nucleoside advocates have implemented to remove immunogenic contaminants (dsRNA) from IVT mRNA. Purified mRNA thereby avoids RNA sensor activation and increases translational levels of the encoded protein. This reduces the chances of toxicity upon repeat administration, which makes it more suitable for commercial in vivo therapies. Recently, a simple, highly scalable purification method has been developed by Baierdorfer et al. to remove these transcriptional by-products from IVT mRNA samples [72]. The purification is performed via selective adsorption of the dsRNA to cellulose in an ethanol-containing buffer and has shown equivalent efficacy to HPLC methodologies, facilitating large-scale manufacture of mRNA therapeutics.
2.2.4. Self-amplifying mRNA (SAM)
Lastly, SAM vaccines are an advanced strategy to enhance the immune response to mRNA vaccines. This vaccine format is based on the alphavirus genome, in which the viral RNA replication machinery is retained and the genes encoding for the structural proteins are replaced with the antigen of interest. The viral replicase drives cytoplasmic mRNA amplification through synthesis of the RNA-dependent RNA polymerase complex [73,74]. This leads to the production of multiple copies of the antigen-encoding mRNA, which has the potential to correlate with higher and sustained antigen expression levels compared to non-amplifying mRNA [75]. SAM vaccines have been shown to stimulate both T and B cell immune cascades [74,76]; however, the overactivation of Type I INF at the site of injection has shown to limit the initial antigen expression and promoted T cell exhaustion [77,78]. The SAM approach has the potential to lower the required dose of mRNA and thereby the cost associated with vaccine development and manufacture. BioNTech currently has an influenza (BNT161) and tuberculosis SAM vaccine in the preclinical development phase, as well as a COVID-19 SAM vaccine candidate (BNT162c2). However, degradation of SAM constructs, due to their inherently larger size, has the potential to be detrimental to their efficacy.
2.3. In vivo translation of the message
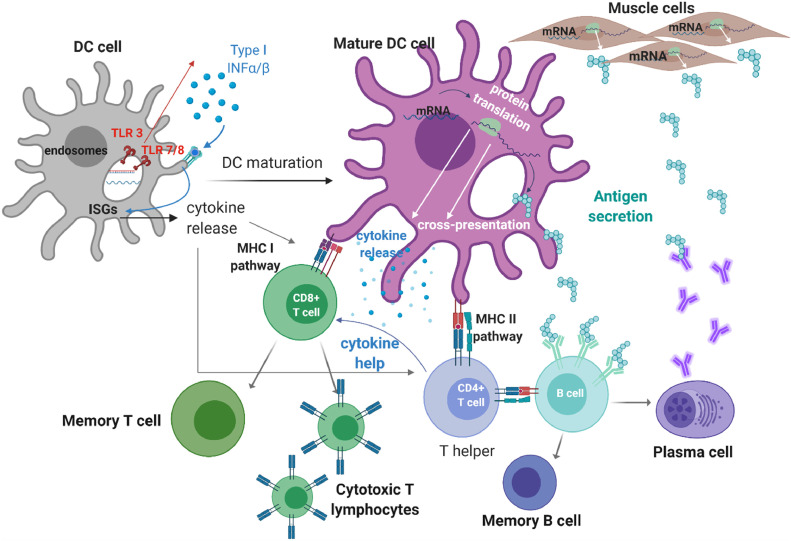
IVT mRNA pharmacodynamics depend on its delivery to the cytosol. Unlike IVT mRNA, endogenous mRNA travels from the nucleus to the cytoplasm through the nucleopore, while IVT mRNA travels into the cells from the extracellular space. Once in the cytoplasm, IVT mRNA mimics native mRNA translation [39]. IVT mRNA immunotherapies are delivered either ex vivo to dendritic cells (DCs) [85] or directly injected in vivo. The cascade from mRNA injection to immune response is depicted in Fig. 1 .
Fig. 1.
Scheme of induced immune response following mRNA vaccination. Dendritic cell (DC) maturation upon (ds) mRNA sensing by TLRs (adjuvant effect of mRNA) and subsequent Type I interferon (INF) production [[79], [80], [81]]. Type I INF initiates the transcription of interferon-stimulated genes (ISGs) involved in the DC maturation process, but also activates antiviral enzymes that promote mRNA degradation and inhibit antigen expression (innate immune response) [24]. The mature DC can express the antigenic proteins and present them on MHC class I and class II to CD8+ (cytotoxic) and CD4+(helper) T cells respectively (adaptive immune response) [82]. Secreted antigen can be presented through the MHC II pathway to T helper cells and B cells, to generate memory B cells and plasma cells that can engender antigen-specific immune defenses and antibody production resulting in a durable protection [83,84]. Figure created using BioRender.com.
In humans, DCs are present at sites with high antigen exposure such as body surfaces and lymphoid organs [86]. DCs are found in either a mature or immature state. The main role of immature DCs is to take up and process foreign antigens; as such, they are generally located in peripheral tissues. DCs can internalize extracellular antigens via phagocytosis, or endocytosis by receptor and non-receptor mediated mechanisms, which enables processing and presentation of antigens [87].
For ex vivo applications, immature DCs are isolated from the patient's blood following transfection with naked antigen-coding mRNA [88]. Alternatively, the antigen is transferred into immature cells in vitro [89] and subsequently cultured with a cocktail of inflammatory cytokines that are necessary for maturation. Antigen-loaded DCs are then administered to patients by injection. Re-administration of ex vivo modified DCs via intradermal (i.d.) and intranodal (i.n.) routes has been shown to be the most effective [86]. However, despite a large number of clinical trials towards anti-cancer applications utilizing the ex vivo DC approach, prophylactic vaccination is less favorable. For example, this approach was evaluated as a HIV immunotherapy but was unable to induce a significant humoral immune response [17].
Several problems remain with ex vivo DC vaccination. An insufficient number of DCs may reach the secondary lymphatic organs resulting in inadequate immune responses. Additionally, ex vivo DC vaccination is a costly, laborious method for infectious disease treatment, making the option unavailable to most patients [39,90].
Direct mRNA injection is more attractive. Direct polynucleotide transfer in vivo was first demonstrated following intramuscular (i.m.) administration of luciferase-encoding mRNA to mice in 1990 [42]. Naked, unprotected mRNA can produce luciferase expression equivalent to 74 pg 18 h post-injection. The effectiveness of naked mRNA delivery to the mice was confirmed following immunization with carcinoembryonic antigen (CEA) mRNA, which elicited anti-CEA responses to proto-oncogene and growth factor [91]. The first insights towards the mechanisms of mRNA transfection in vivo were reported in a mouse study a decade later [92,93]. Mechanisms governing micropinocytosis, uptake into clathrin-coated pits or caveolae, and cytosolic mobility were blocked with inhibitors to determine the active pathway that enables mRNA to reach the cytosol after intradermal injection. The uptake of mRNA in the dermis was determined to be mediated by a saturable mechanism that involves movement of vesicles. Further advances have demonstrated that the uptake of naked mRNA by DCs occurs via macropinocytosis, in which scavenger receptors actively take up mRNA via endocytosis and micropinocytosis inside the immature DC [20,94]. Both processes involve internalization of extracellular materials, outlining the importance of mRNA accumulation at the cellular interface.
In a first-in-human study of prophylactic naked mRNA vaccination delivered using needle-free administration as a sterile-lyophilizate, the candidate elicited a safe and tolerable profile but also insufficient antibody production [31]. Nucleoside base modification represents a fast approach to improve potency, while sequence optimization ensures robust protein expression and immunogenicity. Currently mRNA vaccine candidates comprise either unmodified sequence-optimized mRNA, nucleoside-modified mRNA or mRNA-protamine complexes [95]. Mechanical approaches, such as gene gun delivery and electroporation, have also been employed; however, due to the tissue damage induced, their use is limited [96].
As delivery, uptake and endosomal escape of naked nucleic acids is restricted, clinical translation of naked mRNA vaccines remains unaccomplished. Several obstacles constrain the potential of mRNA vaccines [17]. Firstly, the negatively charged phosphate backbone of mRNA prevents passive diffusion through a cell's phospholipid bilayer. Secondly, susceptibility to degradation by RNase and rapid clearance by the reticuloendothelial system ostensibly reduces in vivo availability to promote gene expression. Additionally, administration of RNA has the potential to be immunogenic, activating intracellular innate and extracellular immune responses which lead to degradation of mRNA molecules. Finally, escape of naked mRNA from the endosomal and lysosomal compartments is limited due to mRNA charge and its eventual degradation in the lysosomes. As a result of the above barriers, it is estimated that only 0.0001% of the administered dose is taken up by cells [96]. This highlights the need for suitable delivery systems that balance target distribution, endosomal escape and mRNA protection: lipid nanoparticles (LNPs) can overcome some of those barriers.
3. Lipid nanoparticles: a new frontier in RNA delivery
The utilization of lipids for the delivery of nucleic acids to cells is well established. Lipids present an ideal material for the transport of mRNA due to their fusogenic compatibility with lipid cell membranes, allowing ‘fusion’ of lipidic delivery agents with target cells and effective release of their cargo into the cell cytosol in an analogous fashion to certain viruses – considered nature's ‘perfect’ nucleic acid delivery vehicles. As early as the 1990s, liposomes were being utilized to deliver mRNA cargoes to mice in vivo; in the proceeding decades, lipid-based systems such as Lipofectamine (Invitrogen; a lipoplexing agent), Stemfect (Stemgent; a lipoplexing mix), and TransIT-mRNA (Mirus Bio LLC; a cationic polymer/lipid formulation) were designed and made commercially available to aid in vitro cell transfection of nucleic acids, including mRNA [98,99]. A number of other technologies, including ‘elastic’ liposomes, nanostructured lipid carriers, ‘lipobrids’ and lipid-polymer hybrid nanoparticles have been developed in the past decade and may be adapted for delivery of nucleic acids [[100], [101], [102], [103]]. However, these so-called ‘soft’ lipid nanostructures [104], i.e. micelles, liposomes and lipoplexes, with generally only one or two different lipid components in their formulation, can suffer from a lack of reproducibility in their production [105] and have low stability. Micellar particles can degrade if diluted below their critical micelle point, for example, and increasing the ratio of incorporated surfactant in order to promote particle stability can further result in increased cytotoxicity [104]. Lipoplexes have demonstrated little success in in vivo application, owing to particle instability and a broad size distribution, ranging even up to a few microns in diameter [106,107]. Liposomes, meanwhile, generally exhibit low encapsulation efficiency of oligonucleotides [108], making them impractical for development towards in vivo transport and delivery of nucleic acid vaccines.
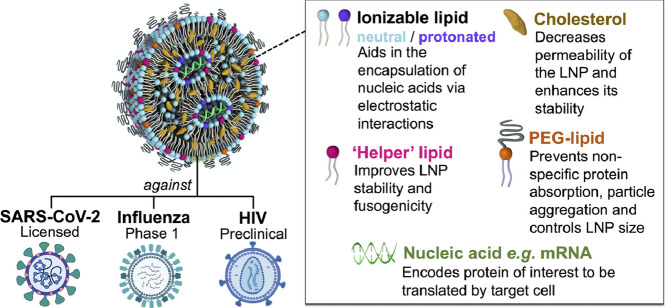
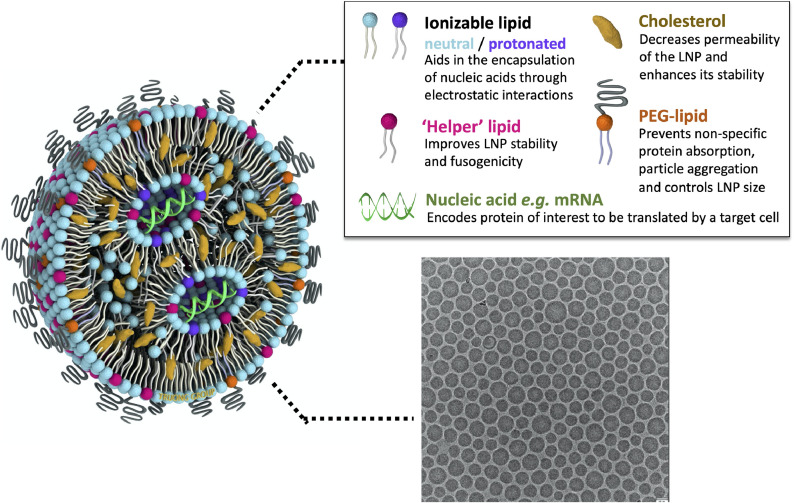
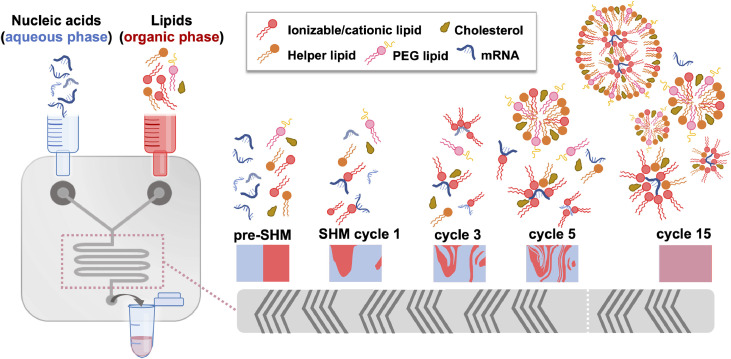
Lipid nanoparticles (LNPs), in their modern definition, are nano-sized (i.e. <1 μm) lipid systems made of two or more (generally four) lipids at varying ratios, and are differentiated from liposomes by lacking a hollow core: rather, they consist of a discontinuous mix of the different lipid components, and, depending on their formulation and cargo, may form a nanostructured core, a homogeneous core shell, multilamellar vesicular structures, or other self-assembled morphologies [[109], [110], [111]]. A scheme of classical LNP formulation components and morphology under cryogenic transmission electron microscopy (cryo-TEM) is shown in Fig. 2 . Though a nucleic acid cargo is incorporated into the particle in an ‘inverted micellar’ structure here – a prototypical visual representation of LNPs within the field – the state of the lipid-cargo complex within the particle interior remains unclear, with more research needed to confirm the morphology of mRNA-LNPs. Modern production of LNPs typically utilizes a microfluidic platform: one such version, depicted in Fig. 3 , incorporates a staggered herringbone micromixer (SHM), [112] while techniques such as two-phase and single-phase mixing may utilize a simpler ‘T’ platform [113]. Toroidal micromixing also represents an emerging technology that looks to improve on the current standard [114]. In addition to a low ‘dead’ volume, allowing economical utilization of the applied materials, these platforms can be facilely modulated to accommodate the requirements of certain formulation components, and/or streamline particle processing and purification [115,116]. Microfluidic formulation of LNPs allows controlled, rapid mixing of lipids (within an ethanolic or organic phase) with a nucleic acid cargo (typically in a low pH acetate buffer) to generate particles with high encapsulation rates (up to 100% efficiency), low polydispersity indices and reproducible physicochemical properties [108].
Fig. 2.
Structure of a typical lipid nanoparticle formulation. The cartoon scheme (left) highlights key components of a lipid nanoparticle with payload and how they contribute to its structure and function (above, right). A representative cryogenic transmission electron micrograph of LNPs with an mRNA cargo is shown on the bottom right, adapted with permission from Richner et al. 2017 [97].
Fig. 3.
Lipid nanoparticle formation utilizing a microfluidic platform with a staggered herringbone micromixer (SHM). Within the microfluidic channels, the SHM allows the aqueous phase (containing the nucleic acids, e.g. mRNA, under an acidic pH) and the water-miscible organic phase (containing the lipids and cholesterol) to proceed from laminar flow (pre-SHM) through several cycles of chaotic mixing until complete mixing of the phases has occurred (cycle 15). This process facilitates the complexation of cationic/ionizable lipids (dark pink) with the nucleic acids and the formation of micelles and early particle structures formed by the lipid mix, typically consisting of a cationic/ionizable lipid, a helper lipid (orange), cholesterol (brown), and a PEG-lipid (light pink), and eventually mRNA-encapsulated lipid nanoparticles.
LNPs have become one of the most common vehicles used for the delivery of mRNA, benefiting from substantial developments in LNP formulation towards in vivo small interfering RNA (siRNA) delivery. For example, Patisiran, an siRNA-LNP system for the therapeutic treatment of the genetic condition transthyretin amyloidosis, is the first RNA-LNP candidate to clear clinical trials and reach the market [117]. siRNA sequences are orders of magnitude shorter than mRNA, however – one study found that simply substituting an siRNA cargo for mRNA containing a >30-fold longer nucleotide sequence could increase LNP size by nearly 50% (size reported based on particle diameter) [109]. Furthermore, mRNA-LNP vaccines target different biological pathways to siRNA-LNP which are aimed at gene silencing [29]. Indeed, formulations optimized for siRNA delivery [118] or even pDNA delivery [119] have demonstrated poor performance when provided with an mRNA cargo, necessitating optimization of formulations towards effective encapsulation and delivery of antigen-encoding mRNA, towards establishing a new class of nanovaccines [120].
The most typical lipid composition utilized for mRNA-LNP systems consists of a cationic/ionizable lipid, a phospholipid ‘helper lipid’, cholesterol and/or a poly(ethylene glycol) (PEG) lipid. The ratios of these components are varied based on the desired target tissue; physicochemical attributes such as particle size, morphology, encapsulation efficiency, and outer surface charge within the LNP delivery system can be facilely modulated via tuning the lipid composition [121].
3.1. The cationic lipid
Cationic lipids are amphiphilic molecules formed of three parts: an amine group, which imparts a net positive charge, a hydrophobic chain, and a linkage group allowing attachment of the hydrophilic moiety to the hydrophobic chain. Due to their net positive charge, they readily complex with negatively charged nucleic acids, making them highly effective transfection agents. In the context of the LNP system, the cationic/ionizable lipid is a major component, present at the highest incorporated ratio. The positive charge in the head group serves two functions: first, improving the entrapment efficiency of LNPs through forming electrostatic interactions with the negatively charged mRNA molecules, and, second, increasing the cellular uptake and endosomal escape of the LNP system by disrupting the cell/endosomal membrane. An overview of the structures for a range of cationic lipids utilized for LNP formulations is provided in Fig. 4 . Generally, positively-charged LNPs are susceptible to rapid clearance from the blood circulation due to serum protein adsorption and uptake by the reticuloendothelial system (RES), thereby limiting their therapeutic value [122]. A positive charge also typically increases the cytotoxicity of a given system [123]. As such, initial development of LNP delivery systems harnessed electrostatic interactions of the cargo with neutral, zwitterionic lipids to mediate passive entrapment and ensure a more biocompatible delivery system. These efforts, however, had limited success, owing to low entrapment efficiency (< 40%) [124] and low transfection rates [125].
Fig. 4.
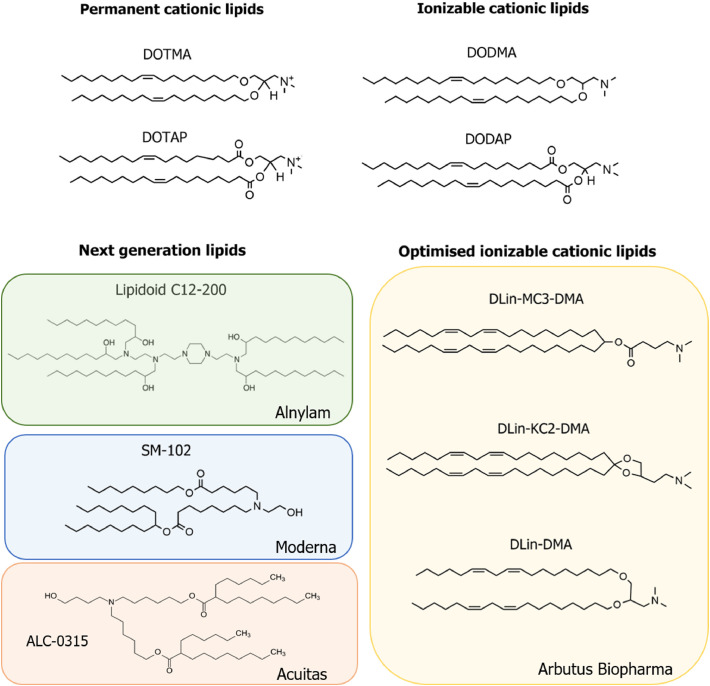
Chemical structures of the cationic lipid subtypes developed for utilization in lipid nanoparticle formulations for nucleic acid delivery. DOTMA, DOTAP, DODMA and DODAP represent the first generation of cationic and ionizable cationic lipids to be utilized in LNP formulations. Next generation lipids include the lipidoid C12-200 [136], invented by Drs. Anderson, Langer and colleagues and licensed by Alnylam, Moderna's SM-102, formerly known as 'Lipid H' [147], and ALC-0315 from Acuitas Therapeutics. Ionizable cationic lipids optimized for LNP formation and delivery include DLin-DMA [127], DLin-KC2-DMA [127] and DLin-MC3-DMA [126], all patented by Arbutus Biopharma. ALC-0315 from Acuitas and SM-102 from Moderna are the lipids utilized in the SARS-CoV-2 vaccines BNT162b2, from BioNTech and Pfizer, and mRNA-1273, from Moderna, respectively.
The utilization of ionizable cationic lipids as the entrapment vector for nucleic acid cargoes provides a solution to both the biocompatibility issues of cationic lipid systems and the low efficiency of neutral systems. Ionizable lipids are designed with a pKa below 7 to allow high encapsulation efficiencies of negatively nucleic acids at acidic pH (<6) while maintaining a neutral to mildly cationic charge at physiological pH (7.4), ultimately reducing toxicity associated with the positive charge and prolonging the circulation lifetime of the LNP system. Ionizable amino-lipids with an apparent pKa range between 6.2–6.5 display an ideal balance between maintaining a more biocompatible neutral charge during circulation and enough positive charge at acidic pH to ensure high encapsulation efficiency of nucleic acids [126].
The Cullis group have made considerable efforts in elucidating the relationship between the molecular structure of ionizable amino-lipids and the therapeutic activity of LNP–siRNA, developing new generations of pH-dependent ionizable lipids for optimized siRNA delivery to hepatocytes [126,127]. Decreasing the saturation of the lipid tail, switching from ester linkers between the lipid tail and amine group to other linkage groups, and modifying the distance between the head group and hydrophobic chain were found to affect the lipid pKa, thereby affecting the hepatic gene silencing potency of their LNP-siRNA system [[126], [127], [128]]. Their work identified two potent ionizable amino-lipids, DLin-KC2-DMA and DLin-MC3-DMA, with siRNA-LNPs incorporating these lipids shown to be 100-fold more effective in hepatocyte gene silencing compared to those utilizing other ionizable cationic lipids, such as DLin-DMA [126]. DLin-MC3-DMA, in particular, has become a gold standard as an ionizable cationic lipid in LNP formulations towards the development of mRNA-LNP candidates from established siRNA systems. LNPs formulated with DLin-MC3-DMA provided efficient delivery of frataxin (FXN) mRNA, successfully producing mFXN protein in hepatocytes after LNPs were administered intravenously (i.v.). Indeed, more than 50% mFXN protein was able to be detected a week post-administration of the FXN LNPs, indicating a half-life exceeding one week in vivo. This LNP formulation also demonstrated the ability to reach the dorsal root ganglia and produce mFXN protein after intrathecal (i.t.) administration [129].
Biodegradable DLin-MC3-DMA variants have also been developed to improve the safety of an applied mRNA-LNP system through adding an ester group to the hydrophobic chain. One variant, L319, enhanced rapid elimination of this lipid in the liver [130]. OF-Deg-Lin is a biodegradable lipid containing an ester group, developed from the nonbiodegradable, linoleic acid derived OF-02; mRNA-LNPs containing OF-Deg-Lin showed high expression in the spleen [131], whereas OF-02 systems accumulated and enhanced mRNA expression in the liver [118,131]. The utilization of unsaturated ionizable cationic lipids has additionally been identified as a factor to enhance intracellular delivery of mRNA. Heyes et al. demonstrated that increasing the double bonds from zero to two in the acyl chain structure of amino-lipids decreases their phase transition temperature; promoting the lipid transition from lamellar (Lα) to reversed hexagonal (HII) phase, and, consequently, increasing the fusogenicity of the LNP [128]. It is hypothesized that the polyunsaturated lipid chain can increase cell membrane fluidity, therefore enhancing LNP fusogenicity and resulting in improved intracellular uptake and endosomal escape. Sato et al. also recently reported that cationic lipids with long, linear hydrophobic scaffolds improved efficiency of nucleic acid delivery in vitro [132]. In addition to the lipid structure, the ratio of the ionizable lipid in the LNP lipid composition is critical for determining the encapsulation and the delivery efficiency of the LNP system. LNPs composed of ionizable cationic lipids, phospholipid, cholesterol, and PEG-lipids in a ratio of 20/25/45/10 mol%, respectively, exhibited high entrapment efficiencies, but were less efficacious in terms of gene silencing in hepatocytes [133]. Increasing the molar ratio of the ionizable lipid up to 40 mol% enhanced liver accumulation and gene silencing activity [127], with the lipid composition molar ratio of 50/10/38.5/1.5 mol% leading to a 6-fold improvement in hepatocyte gene silencing [126].
Concurrently, advances are being made in the development of 'next generation' lipids and lipid-like molecules for effective mRNA-LNP systems [146][147]. Lipidoids, ionizable lipid derivatives composed of tertiary-amines, have been primarily developed for LNP-mediated siRNA delivery, with formulations containing C12-200 and cKK-A12 demonstrating efficacy in a similar range to DLin-MC3-DMA for hepatocyte gene silencing [[134], [135], [136]]. Recent work has further demonstrated efficacy of lipidoids within mRNA-LNP systems. Anderson et al. optimized an LNP-mRNA formulation composed of C12-200 and DOPE, with the mRNA to C12-200 ratio adjusted to 1:10 (w/w) [118]. This formulation succeeded in raising production levels of translated protein comparative to DLin-MC3-DMA-containing LNPs, with both systems mainly distributed to the liver. Cationic lipid-modified aminoglycosides (CLAs), which are generated from natural existing aminoglycosides modified with alkyl epoxides and acrylates, have also been recently explored as candidates for the cationic component of mRNA-LNP systems. Yu and colleagues generated a library of CLAs paired with a DOPE/Cholesterol/DMG-PEG system to deliver EPO mRNA in mice, and demonstrated that EPO levels in the blood were over 3-fold higher 6 h after administration of LNPs containing GT-EP10, one of their candidates, comparative to the commercial MC3-based formulation [137]. Advances in ionizable lipid formulations, e.g. through achieving even tighter pKa windows, will ensure mRNA-LNP systems can maximize encapsulation efficiency without sacrificing biocompatibility.
3.2. Phospholipid ‘helper lipids’
More efficient entrapment and transfection potency of cationic lipid delivery systems has been found when a ‘helper lipid’ is mixed into the formulation. This was first demonstrated through combining cationic DOTMA and neutral DOPE to effectively complex DNA [138]. DOPE has a pronounced conical shape due to its unsaturated acyl chains and small head group, and tends to assemble in the hexagonal (HII) phase [139]. These properties allow DOPE to facilitate the formation of non-bilayer structures and disrupt the endosomal membrane to ensure endosomal escape of internalized LNPs, thus improving the transfection efficacy of the cationic lipid formulations [[139], [140], [141], [142]]. The use of DOPE is more commonly associated with LNP formulations for the delivery of large macromolecules such as plasmids and mRNA, however, and is less utilized for delivery systems for small molecules such as siRNA and oligonucleotides, as these cargoes require less electrostatic complexation. Cheng and Lee further suggested that DOPE could lead to a decrease in the colloidal stability of particles designed for the delivery of siRNAs [143].
As such, current LNP formulations for mRNA delivery generally utilize phosphatidylcholines (PCs) other than DOPE as the ‘helper’ lipid – particularly more saturated variants, such as 1,2-distearoyl-sn‑glycero-3-phosphocholine (DSPC) – which have a high melting temperature (Tm) and enhance the stability of LNP formulations through the formation of bilayer structures. In the commercial siRNA-LNP delivery system Patisiran, DSPC acts as the helper lipid with DLin-MC3-DMA as the ionizable cationic lipid. DLin-MC3-DMA has a polyunsaturated acyl chain, which supports the formation of the inverted micellar or non-bilayer structure at physiological temperature in the presence of a payload [144]. Replacing DOPE with DSPC in the LNP-siRNA formulation containing 40% ionizable lipid resulted in an increase of gene-silencing efficiency in vitro, likely a consequence of increased stability of the particle [145].
Though siRNA-containing LNP systems have required DSPC for efficient delivery, efficacy in mRNA-LNP formulations has been mixed, with reports of substituting DOPE with DSPC in LNP formulations for mRNA either improving [148] or decreasing the performance of the system [118,145,149]. One possible explanation for a more efficacious mRNA-LNP system formulated with DOPE compared to DSPC could be related to differences in the molecular interaction with RNA, which affects their packing ability. In a study exploring LNP morphology and encapsulation efficiency, Leung et al. demonstrated that DOPE, alongside cholesterol, is important for the formation of the inverted micellar structure and enhancing packing ability [145]. However, increasing the concentration of the ionizable cationic lipid and decreasing the amount of cholesterol in the particles decreased the encapsulation efficiency of the LNP. Kauffman et al. noted that, though their LNP formulations utilizing DSPC had superior mRNA encapsulation rates compared to those containing DOPE, the latter compositions induced better protein production [118,150]. While Oberli and colleagues reported mRNA-LNPs utilizing DSPC and another phospholipid (DOPS) demonstrated superior performance to DOPE-containing LNPs [148], the authors noted that, in 20% of mice, LNPs formulated with DSPC or DOPE triggered inflammation at the injection site 5–10 days post-administration.
Recent studies have also suggested that increasing the molar ratio of zwitterionic phospholipids to ionizable cationic lipids can result in improved delivery of mRNA [118,149,151], and a new generation of ionizable phospholipids has recently been synthesized to reduce the presence of cationic lipids while still assisting in mRNA capture [152]. Because mRNA is larger in size than siRNA, it is assumed that a large amount of the ionizable cationic lipid might tightly bind to mRNA and make its release from LNPs difficult. Therefore, the rationale for decreasing the amount of these lipids in a formulation is to reach an equilibrium where it is high enough to condense mRNA and form stable nanoparticles, yet low enough to allow the release of mRNA upon internalization into target cells. Finding the balance between maximizing both encapsulation and delivery of mRNA, while taking into account how outside variables (such as administration route) come into play, is a challenge in the selection and optimization of the ‘helper lipid’ in mRNA-LNP formulations that continues to be investigated.
3.3. Cholesterol
Cholesterol is a natural component of cell membranes [153,154], and plays an important role in lowering the transition temperature of LNPs, facilitating transition from the lamellar phase to hexagonal phase, and thereby aiding the release of mRNA from internalized LNPs into the cell cytosol [122,155]. There is still some ambiguity on the precise location of cholesterol within LNPs. A series of independent studies have demonstrated the presence of a crystalline cholesterol domain, either at the surface, within the lipid bilayer [156], or even complexed with the ionizable lipid in the core [144].
When cholesterol is mixed with unsaturated phospholipids in a lipid delivery system, it promotes the ability of the phospholipid membrane to form a bicontinuous cubic phase at physiological temperatures, further enhancing fusogenicity [157]. In addition, cholesterol improves lipid packing and modulates membrane fluidity and permeability of the bilayer membrane system through the ‘condensing effect’. For example, when cholesterol was incorporated into a mixture of dimyristoylphosphatidylcholine (DMPC), the complex shifted from a fluid state to tighter lipid packing through the reduction of the surface area per lipid [158]. Pozzi et al. demonstrated that increasing the molar fraction of cholesterol in DC-Chol-DOPE DNA lipoplexes was associated with increased transfection efficiency of the DNA cargo, owing to enhanced fusogenicity of higher cholesterol formulations [159]. Moreover, incorporation of 30 mol% cholesterol in pure DSPC liposomes led to improved pharmacokinetics and prolonged circulation half-life of this system from a few seconds to 5 h, though increasing the mol% of cholesterol beyond that did not further enhance the circulation time [160].
Recently, Patel and colleagues explored naturally occurring cholesterol analogs as potential alternatives to cholesterol for mRNA-LNP formulations, with a notable improvement in cell transfection reported in formulations containing β-sitosterol (Sito) compared to those with cholesterol [111]. Sito-formulated LNPs were observed to have a faceted morphology though cryo-EM whereas cholesterol-formulated LNPs are spherical with smooth curvature. This is likely a consequence of phase separation within the Sito-formulated LNPs [161], but also potentially the formation of two-dimensional lipid crystals at the LNP surface. In a follow-up study, the group further investigated the incorporation of C24 alkyl derivatives of cholesterol, known as phytosterols, in mRNA-LNP systems [110]. Formulations containing cholesterol derivates were all shown to be polymorphic, with lipid partitioning and phase separation observed to varying extents; for example, methyl and ethyl groups added to the C24 alkyl tail induced a 50% increase in multi-lamellarity, and introduction of a double bond increased lipid partitioning by 90%. Interestingly, higher gene transfection rates were observed in LNPs with multilamellar, faceted structures, in addition to those with a lamellar lipid phase. Cholesterol derivatives and, by extension, LNPs with varied morphologies may provide effective systems for mRNA vaccine delivery, though in vivo application is required to assess the benefit of replacing cholesterol as the gold standard in LNP formulations.
3.4. PEG-lipids
PEGylation of nanomaterials is a common strategy to provide them with steric stability, prevent opsonization and enhance their overall systemic circulation time [162]. The impact conferred by PEG-lipids on LNP circulation time is determined by the dissociation rate of the PEG-lipid from the surface of the LNP, which relates to the acyl chain length of the PEG-lipid: dissociation rate has been shown to decrease with chain length, with formulations comprised of longer chain PEG-lipids thus having a longer circulation time [163]. PEG-lipids are additionally one of the key factors controlling particle size: increasing the ratio of a PEG-lipid in LNP formulations has been hypothesized to increase compression in the lipid bilayer and enhance repulsive forces between particles, [164] consequently generating smaller particles as PEG-lipid ratio increases [165,166]. PEG-lipids further aid in the aqueous solubility of LNP systems and provide a ‘stealth’ effect to particles in vivo, through shielding the LNP surface to limit the adsorption of serum proteins, ensuring protection against mononuclear phagocyte systemic uptake and thus contributing to increased circulation time [162]. Generally, higher PEG density on the surface of lipid delivery systems generates better steric barriers through the formation of brush-like conformations; around 2% PEGylation of liposomes was found to be optimal to limit serum protein binding to the liposome surface on incubation with serum for 10 min [167]. Interestingly, though PEGylated lipids are generally incorporated at low molar ratios compared to other formulation components in mRNA-LNPs, Sago and colleagues reported mRNA-LNPs with 20% C14-PEG2000 content mediated effective delivery of their cargo [168].
Having a hydrophilic layer around LNPs can, however, interfere with cellular uptake and limit endosome destabilization, consequently decreasing transfection potency [135,169]. In this sense, a more rapid dissociation of PEG-lipid from the LNP surface, though decreasing the circulation time of the particle, can increase its fusogenicity, allowing the LNP to more effectively deliver its cargo to target cells. This is known as the “PEG dilemma”: improved pharmacokinetics of the LNP system mediated by PEG, at the cost of cellular uptake and a consequently reduced transfection efficiency [167]. Increasing the PEG content additionally affects particle size, and might therefore have a detrimental effect on the amount of nucleic acid that can be encapsulated and the transfection efficiency, which is especially relevant for pDNA. Optimizing the density and the chain length of the PEG-lipid component is therefore crucial for effective development of mRNA-LNP systems. ‘Diffusible’ PEG-lipid was developed to balance the transfection potency and stability of LNP systems. This consists of C14 acyl chain lipids which dissociate rapidly from the LNP surface after administration into blood circulation, allowing particle uptake by cells [163,170]. The use of diffusible PEG-lipid in siRNA-LNPs has shown enhanced accumulation and gene silencing in hepatocytes, through facilitating apolipoprotein E (ApoE) adsorption to the particle surface followed by rapid uptake via ApoE-dependent low-density lipoprotein receptors (LDLR) [171,172].
In order to target extrahepatic tissues for vaccination purposes, incorporation of ‘persistent’ PEG-lipids, i.e. composed of C18 acyl chains rather than C14, can be more favorable to prevent premature clearance of LNPs. Formulations incorporating C18 PEG-lipids at molar ratios of more than 1.5% lead to increased LNP circulation time, consequently enhancing the accumulation of the LNP system at target tissues without loss of transfection potency [172]. However, a key challenge related to the longer half-life of formulations with persistent or long-chain PEG-lipid is the potential for accelerated blood clearance (ABC) phenomenon, which has been observed for long-circulating PEGylated liposomes. In this phenomenon, the clearance of PEGylated nanoparticles is increased and half-life is reduced on repeated administration due to induction of anti-PEG antibodies [173,174]. This may result in clearance of the LNP before the cargo is effectively delivered. How anti-PEG immune responses impact mRNA-LNP systems in vivo needs to be explored to ensure it does not impact the efficacy of mRNA-LNP vaccine candidates. Recent research, however, indicates that stable LNP systems can be generated utilizing alternative materials to PEG-lipids, and may even provide a superior vaccine. For example, LNPs containing 3% Tween 20 in the place of a PEG lipid specifically delivered a DNA cargo to the draining lymph nodes of mice after i.m. injection, outperforming a standard formulation that utilized DSPE-PEG2000 for which expression was mainly seen at the injection site [175].
4. Tuning the vehicle: towards targeted vaccination of mRNA-LNPs
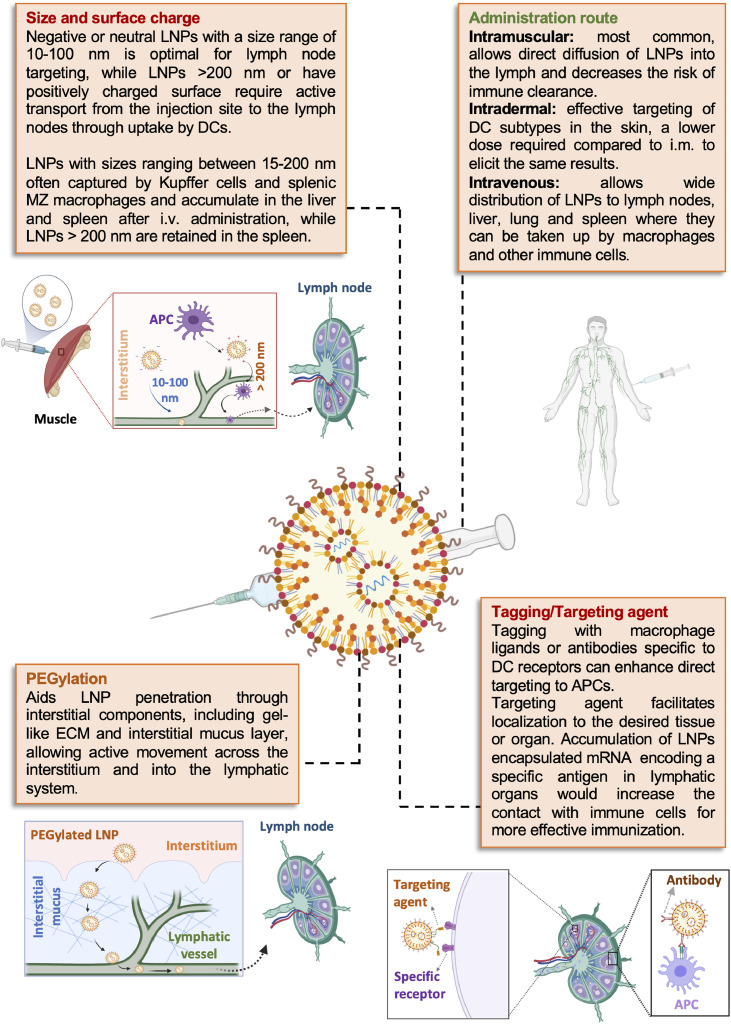
For successful delivery of the LNP vaccine payload, LNPs need to pass through several tissues and cell specific barriers to reach the target tissue, where they should display efficient cell internalization and nucleic acid release into the cytosol [176]. Tuning the formulation and physicochemical properties of an LNP system can thus direct particle fate in vivo; pertaining to LNP circulation half-life, macrophage uptake, tissue localization and extravasation through leaky vasculature. For example, nanoparticles with a hydrodynamic diameter under 5 nm generally clear rapidly through renal filtration after i.v. administration [177]; nanoparticles with sizes ranging between 15 and 200 nm often accumulate in the liver and spleen after being captured by Kupffer cells and splenic MZ macrophages, whereas particles larger than 200 nm are retained in the spleen [178,179]. The design of targeted mRNA-LNP vaccines can be categorized into three main strategies: targeting the lymphatic system, targeting key extralymphatic organs or tissues, and targeting APCs.
4.1. Targeting the lymph
Within the context of vaccination, delivery and retention of a vaccine candidate into the lymphatic system is generally desired for an effective adaptive immune response [[184], [185], [186]]. The lymphatic system consists of a series of vessels, nodes and lymphoid tissue where the majority of the immune cells within the body reside [187,188]. Entry into the lymphatic system typically occurs via small lymphatic capillaries within tissues, which flow into larger collecting lymphatic vessels and pass through one or more lymph nodes before joining the right or thoracic lymph duct, twhich empties into the systemic circulation at the subclavian veins [187,188]. An effective immune response typically requires that an antigen is presented via an APC (such as a DC) to T cells within the lymph node or lymphoid tissue. This results in activation and proliferation of T cells and can also lead to B cell activation and affinity maturation of antibodies. Many recent studies have demonstrated that enhancing the delivery of vaccine antigens and adjuvants to immune cells within the lymphatic system enhances efficacy [180,[189], [190], [191], [192], [193]].
Delivery of different lipid-based systems to the lymph has been reviewed elsewhere [104,194]. In general, enhanced delivery to the lymphatic system has been achieved via association of therapeutics or vaccines with exogenous or endogenous macromolecules or nano-sized carriers (such as nanoparticles, liposomes, polymers, proteins, lipoproteins etc.) that are too large to enter blood vessels from tissues and instead are transported via the lymphatics from tissues. The blood capillaries typically have gaps of approximately 5–10 nm in size between endothelial cells (depending on the tissue and blood vessel type) whereas lymphatic capillaries have open button-like junctions which enable the entry of larger molecules and particles [187,188,190,195]. The net charge and zeta potential of nanoparticles can also impact transport from the injection site into lymphatic vessels and, additionally, retention within the lymph node. The interstitium is composed of a complex of collagen fibers and glycosaminoglycans such as hyaluronic acid, which has a net negative charge. Positively charged LNPs may thus become trapped in the interstitium, whereas negatively-charged LNPs may more readily transfer through the interstitium to the lymphatics via charge repulsion [196].
The optimum size to enable lymphatic access after injection into tissues (i.e. intradermal, intramuscular or subcutaneous) is typically considered to be >20 kDa for macromolecules and 10–100 nm diameter for nanoparticles. This is because smaller particles are transported from injection sites via the blood capillaries as blood flows much faster (100–500 fold) than lymph fluid through capillaries. Particles larger than 100 nm in diameter, on the other hand, may become trapped in the interstitium at the injection site as the interstitium contains water channels that are ~100 nm in diameter and provide a route for interstitial transfer of nanoparticles. LNPs larger than 100 nm may thus have difficulty traversing the interstitium towards gaining access to the lymphatics [190]. Within the context of vaccine delivery, nanoparticles with a diameter less than 200 nm can have been found to drain to the lymph nodes within hours of intradermal injection. In contrast, larger particles (e.g. 200–500 nm) may be internalized by DCs at the injection site and subsequently trafficked through the lymphatic vessels to lymph nodes where the DCs present antigen to T cells. This mechanism of transport is typically slower and can require over 24 h to reach the lymph node [197]. Interestingly, PEGylation of lipid-based systems (generally liposomes) has shown the capacity to permit direct transport of particles > 100–200 nm to the lymph nodes [194]. Though this phenomenon may be at least partially attributed to the longer circulation time of PEGylated particles, there is evidence that PEGylation aids particle penetration through interstitial components, including gel-like ECM, in addition to the highly adhesive and viscoelastic interstitial mucus layer – a natural defense against invading pathogens – allowing active movement across the interstitium and into the lymphatic system [198,199].
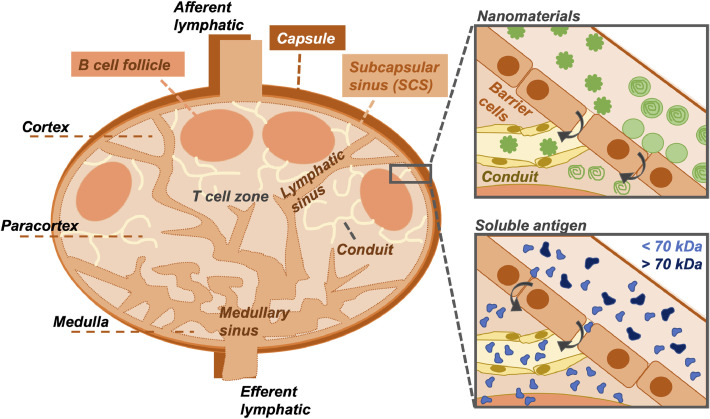
For vaccination, after entering the lymph node via the afferent lymphatic vessels draining the injection site, LNPs face further obstacles to reach the target immune cells within the interior of the lymph node [180]. On entering the lymph node (Fig. 5 ), lymph fluid passes around the subcapsular sinus, a channel that runs around the outside of the lymph node providing a rapid pathway for fluid to exit the node via the efferent lymphatic vessel. This sinus is lined by phagocytic cells including macrophages and DCs that remove ‘foreign’ materials such as LNPs from the inflowing lymph and may present these materials to underlying cells. The T cells and B cells within the lymph node are localized within the interior paracortical and cortical regions of the node. Access to these regions may occur via a reticular network that drains from the main lymphatic sinuses [180,181]. The reticular network is, however, very narrow with channels of 3–5 nm in diameter which have been shown to restrict entry to fluorescent macromolecules that are <70 kDa in size [180,181,183].
Fig. 5.
Structure of the lymph node and localization of foreign agents. Materials enter the lymph node after draining from the injection site into the afferent lymphatics and pass around the subcapsular sinus (SCS), which provides a rapid pathway for fluid to exit the lymph node via the efferent lymphatics. T cells and B cells are localized within the interior paracortical and cortical regions of the node; access to these regions by vaccine candidates is required to generate a potent immune response. Passage of materials across the SCS can occur via conduits in a reticular network that drains from the main lymphatic sinuses, though these channels are very narrow, spanning only 3–5 nm in diameter [180,181]. Certain nanomaterials may be able to enter the conduits through gaps between barrier cells; others, while trapped at the barrier, may be able to release a cargo (e.g. small molecule), which can freely diffuse across the SCS and into the cortex [182]. Soluble antigen <70 kDa may also access the lymph node interior, whereas larger macromolecules cannot [180,181,183].
An overview of design strategies for LNPs to target the lymphatics is provided in Fig. 6 . Though most studies have focused on exploring liposome formulations for effective lymph targeting, Nakamura and colleagues recently generated LNPs, neutrally charged at physiological pH, with different size distributions (30, 100 and 200 nm) and assessed their localization to the lymph after subcutaneous administration in mice [200]. Though low LN localization was observed with the larger particles, 12–16% of total cells in the LN internalized 30 nm LNPs, with up to 70% association seen with CD8α DCs. Further modification of lipid formulation at the 30 nm size point generated net positive and negatively-charged particles, which demonstrated 5% and 22% total LN cell association, respectively. Negatively-charged LNPs were shown to translocate from the SCS to the cortex and paracortex, demonstrating high association with B220+ B cells, whereas the neutral and positively charged particles remained at the barrier. However, these LNPs were ‘empty’, i.e. did not contain a cargo; how effective these systems are for facilitating the delivery and expression of mRNA in the lymph nodes is yet to be established.
Fig. 6.
Overview of lipid nanoparticle design and administration strategies for targeting the lymphatic system. Manipulating the physicochemical properties of LNPs, including size and surface charge, can either direct them to the lymphatics or promote their uptake by cells at the injection site, where i.m. is the most frequently used route of administration. Adjusting LNP formulation by tuning the level of PEGylation can promote access to the lymphatics; similarly, decoration of the particle surface with targeting agents can facilitate localization to antigen-presenting cells and/or specific cell types in the lymph node. Figure created using BioRender.com.
4.2. Targeting extralymphatic organs and tissues
Modifications to the surface charge of nanoparticles have been used to direct delivery of mRNA-LNPs to extralymphatic organs and tissues that have a key role the pathology of certain diseases. In influenza, for example, where the lungs are the main site of infection, the large surface area and numerous APCs present provide an attractive target for generating mucosal immunity. Several studies have demonstrated that a net cationic LNP system preferably accumulates in the lung after i.v. administration. Recent work has shown that mRNA-LNPs incorporating the cationic lipid 1,2-di-O-octadecenyl-3-trimethylammonium propane (DOTMA) effectively targeted the lung after i.v. administration, specifically transfecting endothelial cells, tissue-resident DCs and macrophages [40,201]. Selective organ targeting (SORT) lipid nanoparticles were designed to target specific tissues through incorporation of differentially charged phospholipids into the LNP formulation. Incorporation of 50% DOTAP into LNP formulations in order to generate positively charged particles resulted in localization to the lung, whereas negatively charged LNPs containing 30% DOPA trafficked mainly to the spleen [143]. Neutral SORT LNPs, in contrast, were shown to accumulate in the liver and spleen. Interestingly, other studies have illustrated that nanoparticles with a highly positive surface charge primarily accumulate in the lungs, whereas negatively charged nanoparticles have more propensity to accumulate in the spleen and liver [46,202]. In a follow-up to this study, the authors generated a library of 51 ionizable ‘iPhos’ phospholipids which were utilized to formulate mRNA-LNPs with similar size, zeta potential and pKa, yet displaying a range of in vivo efficacy and organ selectivity [152]. Post i.v. administration of a 0.1 mg/kg mRNA dose, iPhos lipid-containing LNPs consisting of a single tertiary amine and phosphate group with three alkyl tails demonstrated the highest efficacy, with alkyl chain length playing a key role in organ selectivity and functional mRNA delivery: at the amine side, 8–10 carbons facilitated high mRNA expression; at the phosphate side, translation was directed to the liver, where chains were under 12 carbons versus longer chains localized to the spleen. Ultimately, however, the discrete pathways by which tuning the physicochemical properties of formulation components directs LNPs to selectively traffic and express in certain organs and tissues require more detailed mechanistic studies to establish.
The physicochemical properties of the LNP system also impacts its interactions with serum proteins and consequent formation of a ‘protein corona’ which further determines particle fate in vivo. A protein corona composed of plasma opsonins, such as fibrinogen, IgG, and complement factors, for example, usually promotes rapid clearance of particles from the circulation [203]. In contrast, adsorption of plasma proteins like apolipoproteins elongate particle circulation half-lives and enhances interaction with LDL receptors, which facilitate accumulation of the particles in the liver and transportation across the blood-brain barrier [46,204]. Lipoproteins such as ApoE have been utilized for siRNA-LNP decoration due to their capacity to induce internalization into hepatocytes [171], with interactions between ApoE-bound LNPs and heparan sulfate proteoglycans shown to further promote rapid uptake [205], and albumin coating has recently been harnessed for mRNA-LNP delivery to the liver through apolipoprotein E (ApoE) independent cellular pathways [206]. Varying the proportion of charged and zwitterionic lipids at the surface of a lipid system has been reported to change the quantity and identity of the most abundant serum proteins [207]; as such, controlling protein corona formation could present a strategy for specific targeting in vivo.
LNPs may also be directly modified with a specific targeting agent in order to facilitate localization to the desired tissue or organ for more effective immunization. Li et al. recently designed LNPs targeted to plasmalemma vehicle-associated protein (PV1), known to be associated with lung endothelial caveolae, through conjugation of DLin-MC3-DMA LNPs (DSPC:Chol:DMG-PEG:DSPE-PEG maleimide, 50:10:38:1.75:0.25) to a Fab-C4 caveolae-targeting nanobody, and demonstrated significantly higher targeting specificity of PV1-targeted LNPs to mouse lungs compared to the control LNPs after i.v. administration [208]. Interestingly, increasing LNP size from 70 to 160 nm in this study further directed LNPs out of the lymphatics towards the lungs. The fluidic nature of the LNPs is advantageous, here: nanoparticle elasticity has been shown to be crucial for effective PV1-targeting when vehicle size is over 100 nm [209]. Modulation of the mRNA:lipid ratio also impacted localization of the LNPs: a mRNA:lipid (N/P) ratio of 3 trafficked predominantly to lungs, whereas a mRNA:lipid (N/P) ratio of 10 localized similarly between the lungs and the spleen, likely consistent with a higher proportion of cationic lipid in a given formulation mix (i.e., at a mRNA:lipid ratio of 3) appearing to favor lung targeting and expression.
4.3. Targeting APCs
Tagging with other ligands such as those that bind to macrophages (e.g. IgG and mannose [210]) or antibodies specific to DC receptors that favor less degradative intracellular trafficking pathways (e.g. DC-SIGN [211], DEC-205 [212] and langerin [213]) has been utilized to enhance targeting of nanostructured delivery systems to APCs, though mostly in the context of therapeutic vaccination rather than prophylactic [214]. Katakowski et al. showed effective targeting and knockdown of murine DCs with siRNA-LNPs coated with a nanobody targeting DEC-205 after i.v. administration, a targeting strategy that could be adapted for delivery of mRNA vaccines [215]. Mannosylated liposomes and lipoplexes have also provided successful delivery of mRNA to DCs [216,217]. Though mannose-based targeting approaches have been underutilized for mRNA-LNPs, Zhuang et al. recently demonstrated that intranasal (i.n.) administration of mannosylated LNPs can provide complete protection against influenza in mice challenged with a tenfold LD50 H1N1 influenza virus dose. The response to the mannosylated LNPs was also superior to cationic mRNA-LNPs lacking mannose [218]. Veiga and colleagues have also developed a monoclonal antibody-based targeting system in LNPs, dubbed ASSET, to direct RNA delivery to leukocytes after i.v. injection [219], and demonstrated that mRNA-LNPs could specifically target Ly6c+ inflammatory leukocytes and elicit a therapeutic effect in mice, though this system was not utilized in the context of infectious disease [220]. Direct targeting of APCs therefore represents a dual approach for effective vaccine delivery in lymph-directed mRNA-LNP vaccines, or an alternative pathway for candidates should their physicochemical properties prove unfavorable for uptake into the lymph.
5. mRNA-LNPs in vivo: routes of administration and practical considerations
5.1. Routes of administration
Once LNPs are ready for administration, routes of choice can vary depending on their specific application; for example, outside of an immunological context, delivery of drug-loaded lipid systems has been reported through oral [221,222], opthalmic [223,224] and even topical [225], [226], [227] means. For vaccination purposes, delivery methodologies that allow generation of mucosal immunity and subsequent IgA secretion in certain organs can be desirable for certain diseases, e.g. targeting the lungs through intranasal delivery for influenza [218]. Intestinal delivery [228] is beneficial in theory as particle size does not need to be controlled – LNPs pass into the lymph via diffusion – but vehicle design to resist degradation in the harsh environment of the gut remains a challenge. Similarly, invasive methods, such as direct lymph node injection, provide excellent immune responses but are relatively impractical for widespread use. Subcutaneous (s.c.) administration is a less practical route in humans compared to animals due to a far tighter connection between subcutaneous tissue and underlying bone and muscle tissues [229], and while used routinely in certain treatment regimens (e.g. insulin injection for diabetics), poor vascularity in subcutaneous fat resulting in slow mobilization and processing of the vaccine makes it less desirable for immunotherapeutic purposes [230,231].
A majority of LNPs purposed for siRNA delivery to the liver have been designed for i.v. administration, though this route has also been used for early screening of mRNA-LNPs. For example, Guimaraes and coworkers utilized barcoded mRNA to allow high-throughput screening of multiple different LNP formulations intravenously administered to mice in a single pool, quantifying tissue and organ localization of the different mRNA cargoes simultaneously via deep sequencing, with delivery efficacy then quantified for selected candidates utilizing a commercial luciferase-encoding mRNA [119]. A major pitfall of i.v. administration in the context of immunomodulatory agents, however, is the risk of systemic impact and cytokine storm, making this route less desirable to pursue for LNP-mRNA vaccine candidate development. Injection by i.v. also requires administration in a medical setting by a trained professional, and so is not as practical as other routes, e.g. s.c. and i.m., which can be self-administered by the patient.
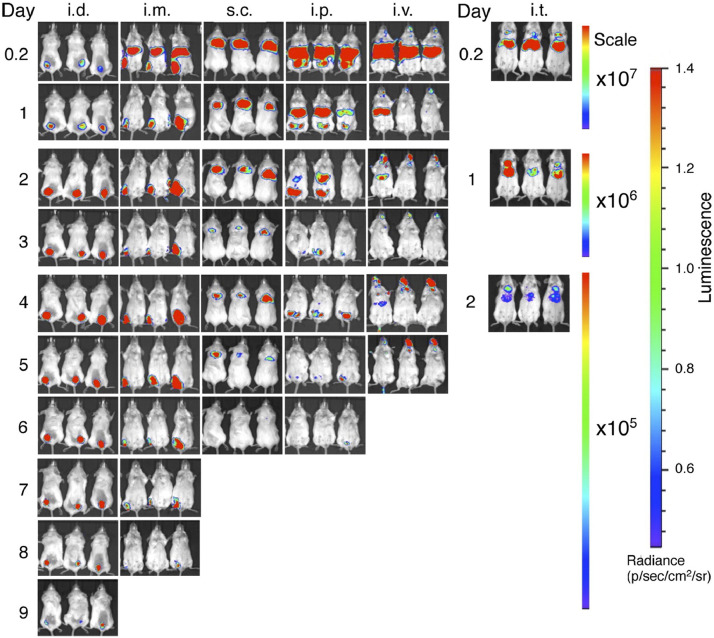
As such, for mRNA-LNP vaccines, typically i.m. administration [233,234] is the most common route of delivery, allowing diffusion of LNPs into the lymph rather than trapping them within the circulation, where they have less access to APCs and are at risk of systemic clearance. The i.d. route has also been highlighted as an advantageous route to explore for mRNA-LNP administration; due to effective targeting of DC subtypes in the skin, i.d. of a subunit vaccine was shown to require as little as one fifth of a standard i.m. dose to elicit the same results [235]. Pardi and colleagues used nucleoside-modified mRNA encoding firefly luciferase loaded into LNPs to investigate the impact of administration route on effective mRNA delivery and expression (Fig. 7 ) [232]. i.m. and i.d. administration were demonstrated to provide the highest levels and duration of luciferase expression in mouse models over the i.v., s.c., i.p. and i.t. routes, which was shown to peak at 4 h post-injection and maintained at the injection site for up to 8–10 days, depending on the dosage schedule. In rhesus macaques, both i.m. and i.d. administration of influenza H10-encapsulated LNPs provided protective titers, but response was shown to be more rapid in i.d. injected monkeys compared to those dosed via i.m. injection [236]. Further research in macaques of LNPs (~100 nm) carrying H10 mRNA demonstrated that two doses applied via i.m. or i.d. administration targeted APCs at both the site of administration and draining LNs, with vaccine-specific immunity ultimately developing in the draining LNs 9 days post immunization regardless of administration route. The H10 mRNA-LNP vaccines induced protective antibody titers for 25 weeks, effectively demonstrating protection over a given seasonal influenza transmission period without the need for an adjuvant [237]. As LNP systems have demonstrated self-adjuvating properties, and have even been utilized as adjuvants for an influenza subunit vaccine [238], this further confers their advantage as a vaccine platform comparative to other vaccine subtypes which require additional adjuvancy to generate a protective immune response. In a follow-up to the H10 mRNA-LNP study, Hassett and co-workers [147] screened ionizable lipids to determine an optimal formulation for i.m. delivery and observed that LNP formulations should be tailored towards administration routes; for example, lipids associated with negligible protein expression after i.v. administration demonstrated some of the highest levels of expression after i.m. administration. The pKa of effective lipids after i.m. administration was generally higher than those that showed efficacy after i.v. administration, with the optimal pKa for immunogenicity determined to be 6.6–6.8. As such, lipid pKa may play a role in how LNP formulations interact with the immune system independent of mRNA delivery, though the exact mechanism by which this occurs is yet to be elucidated.
Fig. 7.
Importance of administration route in tissue localization, translation efficacy and duration of mRNA-LNPs. Mice were injected with mRNA-LNPs expressing firefly luciferase by the intradermal (i.d.), intramuscular (i.m.), subcutaneous (s.c.), intravenous (i.v.), intraperitoneal (i.p.) or intratracheal (i.t.) routes. Administration by i.v. and i.p. resulted in a high level of localization to the liver, with some residual activity at the injection site (around the eyes and abdomen, respectively); similar distribution was observed with i.t. and i.m. injection, though with significant bioluminescent signal in the lungs and muscles; for i.m., this was detectable for up to eight days post-injection. LNPs injected via the s.c. and i.d. routes resulted in translation solely at the injection site, but bioluminescence could be detected up to six (s.c.) to ten (i.t.) days. Reproduced with permission from Pardi et al. 2015 [232].
5.2. Endosomal escape for functional delivery of mRNA
Beyond administration, mRNA-LNP delivery at a cellular level must also be taken into consideration. Less than 2% of siRNA and <1% of mRNA delivered to cells by LNPs can escape endosomes to reach the cytosol [239,240]. Endosomes have a high negative surface charge [241]; release of negatively-charged RNA can be complex and slow [242], especially given that different cell types have demonstrated variations in endosome acidification. Furthermore, lipids within the system that are positively charged at acidic pH may further limit mRNA release from endosomal compartments. Sago and colleagues developed a high throughput screening process to assess delivery of mRNA beyond biodistribution studies, which cannot distinguish between cell-associated or internalized LNPs unable to complete endosomal escape and those that successfully delivered their cargoes, i.e. functional delivery [168]. Through utilizing Cre mRNA as a reporter herein – which necessitates translation within the cytoplasm, translocation to the nucleus and successful editing of target DNA in order to generate a positive result – they identified two mRNA-LNP formulations, 7C2 (7C1:cholesterol:18:1 Lyso PC:C14-PEG2000; 50:23.5:20:6.5) and 7C3 (7C1:cholesterol:DOPE:C14-PEG2000; 60:10:5:25), that could achieve successful in vivo delivery to 22% and 43% of murine spleen endothelial cells, respectively. Though the authors focused on delivery to endothelial cells, they noted that this screening process is not specific to cell type, which would allow use of this system for assessing delivery of mRNA-LNP vaccines to APCs.
After endocytosis, however, delivered mRNA can be packaged into extracellular vesicles (endo-EVs) and secreted by the cell, which may be then taken up by neighboring cells, potentially conferring an additional delivery effect [240]. Recently, Maugeri et al. demonstrated that administration of DLin-MC3-DMA or DLin-DMA LNPs (50% ionizable lipid, 38.5% cholesterol, 10% DSPC, 1.5% DMPE-PEG) with an average size of ~82–90 nm resulted in detection of fractions of ionizable lipids and mRNA (1:1 molar ratio of mRNA nucleotides:ionizable lipids, compared to 3:1 ratio in LNPs) of endocytosed LNPs in endo-EVs at a low rate of 1 copy of mRNA per 10,000 EVs [240]. DLin-MC3-DMA-LNPs were also shown to be more effective as delivery vehicles for the mRNA of interest, generating 8x higher protein expression at the same dosage compared to the DLin-DMA LNPs. The authors noted that the stoichiometric ratio between the ionizable lipid in the LNP system and the mRNA cargo should be neutrally charged (1:1) to enable endosomal escape of encapsulated mRNA – however, doing so would greatly reduce efficient encapsulation of the negatively-charged mRNA compared to formulations with a higher ratio of ionizable or cationic lipids. As an emerging field, fundamental insight into mRNA-LNP activity in the biological interface is still in its nascent stages; elucidating these factors is critical in the effective development of mRNA-LNP formulation and delivery strategies towards generating optimal vaccine candidates.
5.3. Self-amplifying RNA (SAM)-LNPs for effective vaccination
LNP formulations incorporating SAM can potentially overcome challenges introduced by low encapsulation efficiency and poor delivery rates of mRNA. Geall et al. demonstrated that an LNP formulation optimized for siRNA could be effectively utilized for vaccination with SAM, with a 1 µg dose capable of eliciting a comparable immune response in rodent models to a single-cycle alphavirus vector [74]. The large size of SAM (over 9000 nucleotides) can, however, deem it impractical for encapsulation in LNPs. Blakney and coworkers pitched surface absorption, rather than encapsulation, as an effective method of transporting SAM within an LNP system, proposing that, lacking the need to optimize encapsulation, LNP vaccine candidates complexing a range of nucleic acid constructs can be rapidly prepared in the event of an outbreak [243]. Utilization of cationic lipids in the formulation was shown to protect surface-absorbed SAM against RNase-mediated degradation; in fact, surface-absorbed HIV-1 gp140 SAM generated a comparable immune response in vivo to LNP formulations with SAM encapsulated in the particle interior. The same group has recently developed a SAM-LNP vaccine candidate against COVID-19 [244], which has now entered Phase I clinical trials (see Table 2 ). The SARS-CoV-2 prefusion-stabilized spike protein encoded by SAM was found highly immunogenic in preclinical mouse studies, with protective antibody levels exceeding those reported previously for subunit vaccines for the SARS, MERS and SARS-2 coronaviruses [245]; their SAM-LNP formulation generated a potent immune response at doses as low as 0.01 µg, with neutralizing antibody titers greater than those in recovered COVID-19 patients. Furthermore, superior antibody titers, viral neutralization (IC50) and cellular responses were observed in SAM-LNPs comparative to electroporated control pDNA (even at a ten-fold lower dose), which the authors determine is a mediated by the LNP as the delivery system, as polyplexed SAM previously generated analogous immunogenicity to electroporated pDNA. Consequently, LNP systems delivering SAM cargoes may present an ideal system for effective vaccination against infectious disease.
Table 2.
mRNA-LNP vaccine candidates in clinical trials.
| Sponsoring Institution | Name | Infectious disease target [strain] | mRNA encoding [administration] | Trial numbers [phase] | Status |
|---|---|---|---|---|---|
| Arcturus Therapeutics | ARCT-021 | COVID-19 [SARS-CoV-2] | Spike protein [i.m.] |
NCT04480957 [I/II] NCT04668339 [II] NCT04728347 [II] |
Recruiting Ongoing Recruiting |
| BioNTech RNA Pharmaceuticals; Pfizer | BNT162a1; BNT162b1; BNT162b2 (branded as Comirnaty); BNT162b3; BNT162c2; BNT162-04 |
COVID-19 [SARS-CoV-2] | Spike protein; Spike receptor binding domain [i.m.] |
NCT04380701 [I/II] NCT04368728 [II/III] NCT04523571 [I] NCT04537949 [I/II] NCT04588480 [I/II] NCT04649021 [II] NCT04713553 [III] NCT04754594 [II/III] NCT04816643 [I] NCT04816669 [III] 2020-001038-36a [I/II] 2020-002641-42a [I/II/III] 2020-003267-26a [I/II] ChiCTR2000034825b [I] |
Recruiting Recruiting Ongoing Ongoing Ongoing Ongoing Recruiting Recruiting Recruiting Recruiting Ongoing Ongoing Ongoing Recruiting |
| Chulalongkorn University | ChulaCov19 | COVID-19 [SARS-CoV-2] | Spike protein [i.m.] | NCT04566276 [I] | Not yet recruiting |
| CureVac AG | CV702 | Rabies | Rabies virus glycoprotein [i.m.] | NCT03713086 [I] | Ongoing |
| CureVac AG; Coalition for Epidemic Preparedness Innovations (CEPI) | CVnCoV | COVID-19 [SARS-CoV-2] | Spike protein [i.m.] |
NCT04449276 [I] NCT04515147 [II] NCT04674189 [III] 2020-001286-36a [I] |
Ongoing Ongoing Recruiting Ongoing |
| Daiichi Sankyo | DS-5670a | COVID-19 [SARS-CoV-2] | Spike protein [i.m.] | NCT04821674 [I/II] | Recruiting |
| GlaxoSmithKline | CoV-2 SAM | COVID-19 [SARS-CoV-2] | Spike protein [i.m.] | NCT04758962 [I] | Recruiting |
| Imperial College London | LNP-nCoVsaRNA | COVID-19 [SARS-CoV-2] | Spike protein [i.m.] | 2020-001646-20a [I] | Ongoing |
| Moderna | mRNA-1647 mRNA-1443 |
Cytomegalovirus (CMV) | CMV glycoprotein H pentamer complex [i.m.] | NCT03382405 [I] NCT04232280 [II] | Completed Recruiting |
| mRNA-1653 | Combination human metapneumovirus (hMPV) and parainfluenza virus type 3 (PIV3) | hMPV and PIV3 membrane fusion protein [i.m.] |
NCT03392389 [I] NCT04144348 [I] |
Completed Recruiting |
|
| VAL-506440; mRNA-1440 | Influenza A virus [H10N8] | Membrane-bound hemagglutinin 10 (H10) protein mRNA; [i.m.] | NCT03076385 [I] | Completed | |
| VAL-339851; mRNA-1851 | Influenza A virus [H7N9] | Membrane-bound hemagglutinin 7 (H7) [i.m.] | NCT03345043 [I] | Completed | |
| mRNA-1345 | Respiratory syncytial virus (RSV) | RSV prefusion F protein [i.m.] | NCT04528719 [I] | Recruiting | |
| Moderna; Biomedical Advanced Research and Development Authority | VAL-181388; mRNA-1388; mRNA-1944 |
Chikungunya virus (CHIKV) | Antigenic proteins associated with CHIKV [i.m.] Anti-CHKV monoclonal antibody [i.m.] |
NCT03325075 [I] NCT03829384 [I] |
Completed Ongoing |
| mRNA-1325; mRNA-1893 |
Zika virus | prME structural protein [i.m.] |
NCT03014089 [I] NCT04064905 [I] |
Completed Ongoing |
|
| Moderna; Biomedical Advanced Research and Development Authority; National Institute of Allergy and Infectious Diseases |
mRNA-1273; mRNA-1283 |
COVID-19 [SARS-CoV-2] | Spike protein [i.m.] |
NCT04283461 [I] NCT04405076 [II] NCT04470427 [III] NCT04649151 [II/III] NCT04796896 [II/III] NCT04813796 [I] |
Ongoing Recruiting Ongoing Ongoing Recruiting Ongoing |
| National Institute of Allergy and Infectious Diseases | SAM-LNP-S | COVID-19 [SARS-CoV-2] | Spike protein [i.m.] | NCT04776317 [I] | Recruiting |
| Providence Therapeutics | PTX-COVID19-B | COVID-19 [SARS-CoV-2] | Spike protein [i.m.] | NCT04765436 [I] | Recruiting |
| Sanofi Pasteur | MRT5500 | COVID-19 [SARS-CoV-2] | Spike protein [i.m.] | NCT04798027 [I/II] | Recruiting |
| Shulan (Hangzhou) Hospital; Center for Disease Control and Prevention of Guangxi Zhuang Autonomous Region; Suzhou Abogen Biosciences; Yunnan Walvax Biotechnology | ARCoV | COVID-19 [SARS-CoV-2] | Spike receptor binding domain [i.m.] | ChiCTR2000034112b [I] | Recruiting |
EudraCT number.
Chinese Clinical Trial Registry.
However, challenges remain in the development of this emerging technology. Nucleoside-modified RNA can be incorporated in shorter RNA where stringency of the structure is not critical. The incorporation of nucleoside modified sequences can be difficult for large mRNA, and so has not been utilized for SAM. The difficulty appears in achieving full length RNA, but also the modified nucleosides can disrupt necessary secondary structures that are essential for SAM activity. Although partial incorporation may be possible, optimizing the level of this incorporation may be warranted for future work. The use of non-modified RNA in SAM may cause innate immune response activation. Furthermore, current SAM constructs are long sequences, which makes them fragile to processing and shearing thus limiting their long-term stability. To solve these issues, Beissert et al. explored the use of trans-amplifying RNA (taRNA), which comprises two RNA instead of a single long self-amplifying construct. The first RNA encodes the replication proteins and the second segment consists of the antigen of choice [246]. While this approach provides an advantage in expected RNA stability, the total dose of both transcripts remains high. As such, though SAM vaccines hold great promise to circumvent the prime-boost regimens with their efficacy at lower RNA doses, the fragile payload and current inability to modify nucleosides dictates that the true clinical benefit of these vaccines is yet to be discovered [95,247].
6. Clinical trials of mRNA-LNP systems
The first clinical trials utilizing direct application of mRNA therapeutics used a naked, unmodified mRNA vaccine against metastatic melanoma [248]. Though pronounced feasible and safe, it failed to show clinical efficacy and did not progress beyond Phase I clinical trials. Development of LNP-RNA systems focused initially on siRNA, and, until recently, the siRNA-LNP therapeutic Patisiran was the only LNP-RNA system to reach the market. Given that a recent analysis determined that only 13.8% of therapeutics are likely to pass clinical trials and reach the approval stage [249], Patisiran's success presented LNP systems as viable candidates for effective biological delivery of RNA therapeutics. Indeed, growing interest in mRNA therapies for infectious disease, coupled with this demonstrated efficacy of the LNP delivery system, have resulted in a number of mRNA-LNP candidates entering clinical trials in recent years – and, of particular interest at the current time, promising and proven vaccine candidates against COVID-19. mRNA-LNP systems developed by Moderna constitute the majority of vaccine candidates brought to trial, which are summarized in Table 2.
6.1. Non-COVID mRNA-LNP vaccine candidates in development
Current mRNA-LNP vaccines encode for structural or membrane proteins of the pathogen of interest which are intended to be secreted by and/or decorated on the surface of transfected cells. One candidate encodes for a monoclonal antibody. Though outcomes of several of these studies have been announced in media releases, including for Chikungunya virus [250], cytomegalovirus [251,252], and a hMPV and PIV3 combined vaccine [253], few peer-reviewed articles disseminating clinical trial results have been published. The first mRNA-LNP vaccine candidates against two highly virulent strains of influenza A, H10N8 and H7N9, were shown to be well-tolerated and immunogenic. After two i.m. vaccinations of the H10 mRNA-LNPs three weeks apart, all 31 patients developed HA-specific antibodies, which were further detected in the serum of 87% of patients. The reactogenicity profile of the H10N8 mRNA-LNP vaccine was comparable to the AS03-adjuvated H1N1 vaccine, and demonstrated similar efficacy in healthy adults (19–55 years) to the meningococcal conjugate vaccine [236]. Participants administered 25 μg of the H7N9 mRNA-LNP vaccine achieved HAI titers of over 1:40 in 96.3% of the cohort, and MN titers over 1:20 in 100 percent of patients administered [254].
Moderna's respiratory syncytial virus (RSV) candidate mRNA-1777 was generally well-tolerated in younger and older healthy adults, generating increased specific humoral and cellular immunity, but did not progress beyond Phase I [255]. However, mRNA-1345, Moderna's new RSV vaccine candidate, demonstrated a geometric mean fold rise (95% CI, relative to baseline) in RSV-A neutralizing antibody titer of nearly 8fold higher than mRNA-1777 at the same dosage level, per interim reporting in the first month post-administration in Phase I clinical trials [256]. Recently, Phase I interim data has been published regarding a rabies vaccine developed by CureVac, consisting of unmodified mRNA encoding the rabies virus glycoprotein (RABV-G) encapsulated in an LNP system [257]. Two 1 μg or 2 μg doses were well tolerated and elicited rabies neutralizing antibody responses analogous to the current licensed vaccine, Rabipur. Reactogenicity at the highest dose (5 μg), however, was not found acceptable for a prophylactic vaccine.
Vaccine candidates for a number of viruses, including Nipah, human immunodeficiency virus (HIV), Epstein-Barr virus, and seasonal influenza are currently in preclinical development, with many expected to progress to Phase I clinical trials during 2021.
6.2. mRNA-LNP vaccine candidates against SARS-CoV-2
A number of vaccine candidates against SARS-CoV-2, the causative agent of COVID-19, are currently progressing through clinical trials. The ease of synthesizing mRNA coupled with facile and streamlined encapsulation into an LNP system ensured development of mRNA-LNP vaccine candidates occurred rapidly upon the availability of the SARS-CoV-2 genome, resulting in the first clinical trial patient being dosed with an mRNA-LNP vaccine candidate just 63 days later: Moderna dosed their first patient in a US-based Phase I clinical trial of their candidate, mRNA-1273, in March. BioNTech, the first from Europe to put forth a COVID-19 vaccine candidate, entered a number of candidates (BNT162b1, 2 and 3) into Phase I trials in Germany in April and the US in May. It should be noted, however, that this rapid development was only made possible due to the availability of the crystal structure of the SARS-CoV-2 spike protein [258]. Candidates from both companies, in addition to CureVac's candidate CVnCoV, progressed to Phase III during 2020, representing the first mRNA-LNP therapeutics to advance this far in clinical trials. Authorization of BNT162b2 for emergency use in the UK established it as not only the first licensed mRNA-LNP vaccine, but also the world's first licensed vaccine against COVID-19 [259]. As millions of doses of mRNA-1273 and BNT162b2 are now being administered globally, other candidates currently progressing through Phase I or Phase I/II combined trials include ChulaCov19 (Chulalongkorn University), LNP-nCoVsaRNA (Imperial College London), ARCoV (Suzhou Abogen Biosciences, Yunnan Walvax Biotechnology), MRT5500 (Sanofi Pasteur), PTX-COVID19-B (Providence Therapeutics), DS-5670a (Daiichi Sankyo), CoV-2 SAM (GlaxoSmithKline), SAM-LNP-S (the US National Institute of Allergy and Infectious Diseases), and ARCT-021 (Arcturus Therapeutics).
Of the candidates now reporting clinical trial data, mRNA-LNP systems have been shown to perform well overall with regards to tolerability, immunogenicity and efficacy. Interim data for CureVac's CVnCoV showed strong binding and neutralizing antibody responses with good tolerance within the doses tested and the first indication of T cell activation [188]. Arcturus Therapeutics has reported 100% of Phase I trial participants showed immunogenicity after either a single (7.5 µg) or prime-boost (5 µg) dosing regimen of their candidate ARCT-021 [260]. As ARCT-021 utilizes a SAM cargo, this result in addition to preclinical data indicates this system may potentially provide one-shot protectivity. Interim analysis of BioNTech/Pfizer's BNT162b2 and Moderna's mRNA-1273 Phase III clinical trials reported 95% and 94.1% efficacy respectively and, importantly, achieved complete prevention of severe disease [261,262]. Overall, both mRNA-LNP systems were found to be generally well tolerated. Reactogenicity of the vaccines was lower for BioNTech/Pfizer's candidate [262] comparative to Moderna's [261], with 3.8% of participants reporting fatigue and 2% reporting headaches versus 9.7% and 4.5% respectively. In some participants mRNA-1273 also induced muscle (8.9%) and joint (5.2%) pain. Less than 2% of participants in both trials reported severe fever. As of January 27th, the US Vaccine Adverse Event Reporting System (VAERS) has reported around 3–5 cases of anaphylaxis per million doses of each candidate, mostly (>80%) in recipients who had a history of allergies; [263] clinical trials to assess allergic reactions to mRNA-1273 and BNT162b2 in atopic populations are ongoing [264].
Though more virulent strains of SARS-CoV-2 are now emerging, initial data indicates both mRNA-1273 and BNT162b2 are capable of protecting against certain variants. The sera of vaccinated Phase III trial participants was shown to effectively neutralize a number of variants including the B.1.1.7 (UK) strain in vitro, though reduced neutralization has been reported against the B.1.351 (South Africa) lineage [[265], [266], [267], [268], [269], [270], [271]]. Further investigation within populations vaccinated with BNT162b2 indicated increased incidence of breakthrough infections of B.1.1.7 in partially vaccinated individuals and of B.1.351 in fully vaccinated individuals in Israel [272]; however, estimated effectiveness against infections with these variants of concern in a fully vaccinated Qatari population was still high, at 89.5% for B.1.1.7 and 75% against B.1.351 [273]. Moderna has begun dosing previously vaccinated Phase II clinical trial patients in South Africa with mRNA-1273.351, a formulation designed to counter the B.1.351 variant, and mRNA-1273.211, a multivalent booster candidate incorporating a 50–50 mix of mRNA-1273 and mRNA-1273.351. Initial results demonstrate that a single 50 µg booster dose of mRNA-1273 or mRNA-1273.351 increased neutralizing antibody titer responses against both the wild type virus and B.1.351, in addition to the P.1 (Brazil) strain [274]. Incorporation into Phase III trials of BNT162b2SA, BioNTech/Pfizer's B.1.351-specific vaccine, is expected in the second quarter of 2021.
Further ‘second generation’ candidates are also tackling a major obstacle of global distribution of Moderna and BioNTech/Pfizer's vaccines: namely, the capacity to maintain cold and/or ultracold chain to ensure stability of mRNA-1273 (stable for up to a month at 4 °C and six months at −20 °C) [37] and BNT162b2 (stable for up to two weeks at −20 °C, storage required at −70 °C) [38]. [Moderna's next generation candidate, mRNA-1283, is being developed as a potentially refrigerator-stable vaccine, and is currently being assessed in Phase I clinical trials; BioNTech/Pfizer are also undertaking a Phase III clinical trial to test a lyophilized version of BNT162b2 [275]. A vaccine should ideally be able to be stored at room temperature for one to two months in order for distribution to regions without ready access to cold storage facilities, but even in countries where these are available, local facilities may still fail to meet the required standards [276]. Interestingly, preclinical studies of the candidate ARCoV indicated that it is stable for up to a week at room temperature [277] – further development of mRNA-LNP vaccine technologies may be able to increase this timeframe and thus ease the logistical barrier of delivering mRNA-LNP vaccines beyond the clinic.
Given the average time of vaccine development is around 11 years [278], with a 34% success rate upon reaching clinical trials [249], progression of a candidate from bench to clinic in under a year is a remarkable achievement. mRNA-LNP vaccines are proving their merit on the world stage as next generation vaccines, as validation of mRNA-LNP vaccine efficacy has now been shown in vaccinated populations: administration of BioNTech/Pfizer's BNT162b2 demonstrated >90% effectiveness against mild and severe infection across all strains in Israel [272], in addition to 89.5% against B.1.1.7. and 75% against the B.1.351 variants in Qatar, where, most notably, it was also observed to be 97.4% effective against severe, critical or fatal disease across all strains [273]. With a number of other COVID-19 vaccine technologies recently licensed or soon to complete Phase III efficacy testing, including adenovirus vector and protein-based nanoparticle candidates, how these vaccines will compare to mRNA technologies for safety, durability, efficacy and ease of production and distribution remains to be seen.
7. Conclusions and future outlook
Increased globalization has fostered the rapid spread of infectious disease, necessitating that any response to combat emerging threats, COVID-19 and beyond, must be swift. As such, mRNA has all elements to respond to a global pandemic in which a virus may mutate and evade the immune system: rapid synthesis, GMP manufacturing, and the capacity for dosage to be further reduced through utilizing SAM. Formulation of mRNA with LNPs has been key to the recent clinical success of mRNA vaccines after earlier failures, reducing mRNA degradation while providing enhanced delivery and the capacity to introduce specific targeting to cells, tissues and organs of interest for the generation of protective immunity. The vaccine candidates trialed by Moderna, BioNTech/Pfizer and others are validating the efficacy (up to 95%) and safety in humans of mRNA vaccines formulated with LNPs, with BioNTech's BNT162b2 becoming the first licensed mRNA-LNP vaccine in addition to the first licensed vaccine against SARS-CoV-2, and mRNA-1273 following soon after. Variant-specific and ‘second generation’ COVID-19 vaccines, including Moderna's mRNA-1283, are already being assessed in Phase I clinical trials – a testament to the capacity for rapid and dynamic development of mRNA-LNP vaccines to combat an ever-evolving global crisis.
Obstacles still remain ahead. PEG-associated immunogenicity due to prior exposure of patients to PEGylated products and the production of anti-PEG antibodies [174] still needs to be assessed as to how it may impact the safety and efficacy of mRNA-LNP vaccines. Furthermore, two doses are required for full efficacy of BNT162b2 and mRNA-1273, increasing the cost and likelihood of experiencing side effects, which are typically more frequent after the second dose. The World Health Organization highlights vaccine hesitancy as a global health issue; indeed, ensuring patient compliance to not only receive the first dose but return for the second, particularly if they have experienced side effects to the vaccine, may be a challenge faced when combating current and future pandemics. Design of a system that is efficacious after a single dose would thus be optimal, and incorporation of SAM may be a viable option to achieve this: the efficacy of a ‘single shot’ approach of such candidates, e.g. ARCT-021, will be determined if they advance to Phase III clinical trials. Additionally, given that a single mRNA construct can be designed to encode multiple antigens, there is potential for the development of ‘universal’ vaccines for diseases with seasonal outbreaks, such as influenza. Enhancing the stability of mRNA constructs and design of the LNP vehicle to protect them from degradation, however, is key for future development of mRNA-LNP vaccines.
A number of innovative nanovaccine candidates, such as polymer-based vaccines, continue to push the envelope with improved efficiency and vaccine stability [120]. LNP technology has ample room for improvement for vaccine applications, from elucidating direct targeting strategies – both for localizing to the lymph nodes, and for overcoming the barrier cells therein – to, at a cellular level, enhancing uptake, endosomal escape and ultimately functional delivery of mRNA. The design of novel ionizable lipids with narrower pKa windows may provide both effective encapsulation and endosomal escape of mRNA cargoes; introducing targeting ligands or antibodies to the LNP surface may provide an alternative to relying on lipid formulation for localization of LNPs in the desired organ or tissue, and in particular for enhancing delivery to the lymph. Balancing these factors to find an optimal solution for the most effective systems will require joint efforts of scientists across different fields, but will reap huge benefits when successful: given the versatility of these platforms, mRNA-LNPs can be designed for a myriad of applications across the scope of gene therapy, including both prophylactic vaccination in infectious disease but also therapeutic vaccination for various cancers. Ultimately, the coming years promise to be an exciting time for mRNA-LNP vaccines.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Boylston A. The origins of inoculation. J. R. Soc. Med. 2012;105:309–313. doi: 10.1258/jrsm.2012.12k044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baxby D. Edward Jenner’s inquiry after 200 years. BMJ. 1999;318:390. doi: 10.1136/bmj.318.7180.390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.L. Pasteur, Méthode pour prévenir la rage après morsure, 1885.
- 4.Pollard A.J. Childhood immunisation: what is the future? Arch. Dis. Child. 2007;92:426–433. doi: 10.1136/adc.2006.095760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Doherty M., Buchy P., Standaert B., Giaquinto C., Prado-Cohrs D. Vaccine impact: benefits for human health. Vaccine. 2016;34:6707–6714. doi: 10.1016/j.vaccine.2016.10.025. [DOI] [PubMed] [Google Scholar]
- 6.Granot Y., Peer D. Delivering the right message: challenges and opportunities in lipid nanoparticles-mediated modified mRNA therapeutics—an innate immune system standpoint. Semin. Immunol. 2017;34:68–77. doi: 10.1016/j.smim.2017.08.015. [DOI] [PubMed] [Google Scholar]
- 7.Felgner P.L., Barenholz Y., Behr J.P., Cheng S.H., Cullis P., Huang L., Jessee J.A., Seymour L., Szoka F., Thierry A.R., Wagner E., Wu G. Nomenclature for synthetic gene delivery systems. Hum. Gene Ther. 1997;8:511–512. doi: 10.1089/hum.1997.8.5-511. [DOI] [PubMed] [Google Scholar]
- 8.Greenwood B. The contribution of vaccination to global health: past, present and future. Philos. Trans. R. Soc. Lond., B, Biol. Sci. 2014;369 doi: 10.1098/rstb.2013.0433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.W. Jiskoot, G.F.A. Kersten, E. Mastrobattista, B. Slutter, Vaccines, in: D.J.A. Crommelin, R.D. Sindelar, B. Meibohm (Eds), Pharmaceutical Biotechnology, Springer Nature, 2016, pp. 381–304.
- 10.Bahl K., Senn J.J., Yuzhakov O., Bulychev A., Brito L.A., Hassett K.J., Laska M.E., Smith M., Almarsson O., Thompson J., Ribeiro A., Watson M., Zaks T., Ciaramella G. Preclinical and clinical demonstration of immunogenicity by mRNA vaccines against H10N8 and H7N9 Influenza viruses. Mol. Ther. 2017;25:1316–1327. doi: 10.1016/j.ymthe.2017.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leal L., Guardo A.C., Morón-López S., Salgado M., Mothe B., Heirman C., Pannus P., Vanham G., van den H.J., Ham R., Gruters A., Andeweg S.V., Meirvenne J., Pich J.A., Arnaiz J.M., Gatell C., Brander K. Thielemans, phase I clinical trial of an intranodally administered mRNA-based therapeutic vaccine against HIV-1 infection. AIDS. 2018;32:2533–2545. doi: 10.1097/QAD.0000000000002026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wilks A.B., Christian E.C., Seaman M.S., Sircar P., Carville A., Gomez C.E., Esteban M., Pantaleo G., Barouch D.H., Letvin N.L., Permar S.R. Robust vaccine-elicited cellular immune responses in breast milk following systemic simian immunodeficiency virus DNA prime and live virus vector boost vaccination of lactating rhesus monkeys. J. Immunol. 2010;185:7097–7106. doi: 10.4049/jimmunol.1002751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thi T.T.H., Suys E.J.A., Lee J.S., Nguyen D.H., Park K.D., Truong N.P. Lipid-based nanoparticles in the clinic and clinical trials: from cancer nanomedicine to COVID-19 vaccines. Vaccines. 2021;9:359. doi: 10.3390/vaccines9040359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ulmer J.B., Donnelly J.J., Parker S.E., Rhodes G.H., Felgner P.L., Dwarki V.J., Gromkowski S.H., Deck R.R., DeWitt C.M., Friedman A., Hawe L.A., Leander K.R., Martinez D., Perry H.C., Shiver J.W., Montgomery D.L., Liu M.A. Heterologous protection against influenza by injection of DNA encoding a viral protein. Science. 1993;259:1745–1749. doi: 10.1126/science.8456302. [DOI] [PubMed] [Google Scholar]
- 15.Jazayeri S.D., Poh C.L. Recent advances in delivery of veterinary DNA vaccines against avian pathogens. Vet. Res. 2019;50:78. doi: 10.1186/s13567-019-0698-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thalmensi J., Pliquet E., Liard C., Chamel G., Kreuz C., Bestetti T., Escande M., Kostrzak A., Pailhes-Jimenez A.-S., Bourges E., Julithe M., Bourre L., Keravel O., Clayette P., Huet T., Wain-Hobson S., Langlade-Demoyen P. A DNA telomerase vaccine for canine cancer immunotherapy. Oncotarget. 2019;10:364. doi: 10.18632/oncotarget.26927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gómez-Aguado I., Rodríguez-Castejón J., Vicente-Pascual M., Rodríguez-Gascón A., Solinís M., Del Pozo-Rodríguez A. Nanomedicines to deliver mRNA: state of the art and future perspectives. Nanomaterials (Basel) 2020;10:3361–3371. doi: 10.3390/nano10020364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shende P., Waghchaure M. Combined vaccines for prophylaxis of infectious conditions. Artif. Cells Nanomed. Biotechnol. 2019;47:695–704. doi: 10.1080/21691401.2019.1576709. [DOI] [PubMed] [Google Scholar]
- 19.Zhang C., Maruggi G., Shan H., Li J. Advances in mRNA vaccines for infectious diseases. Front. Immunol. 2019;10:594. doi: 10.3389/fimmu.2019.00594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Van Hoecke L., Roose K. How mRNA therapeutics are entering the monoclonal antibody field. J. Trans. Med. 2019;17:54. doi: 10.1186/s12967-019-1804-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rauch S., Jasny E., Schmidt K.E., Petsch B. New vaccine technologies to combat outbreak situations. Front. Immunol. 2018;9:1963. doi: 10.3389/fimmu.2018.01963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jahanafrooz Z., Baradaran B., Mosafer J., Hashemzaei M., Rezaei T., Mokhtarzadeh A., Hamblin M.R. Comparison of DNA and mRNA vaccines against cancer. Drug Discov. Today. 2020;25:552–560. doi: 10.1016/j.drudis.2019.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jones K.L., Drane D., Gowans E.J. Long-term storage of DNA-free RNA for use in vaccine studies. BioTechniques. 2007;43:675–681. doi: 10.2144/000112593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Linares-Fernández S., Lacroix C., Exposito J.-Y., Verrier B. Tailoring mRNA vaccine to balance innate/adaptive immune response. Trends Mol. Med. 2020;26:311–323. doi: 10.1016/j.molmed.2019.10.002. [DOI] [PubMed] [Google Scholar]
- 25.Zhang C., Maruggi G., Shan H., Li J. Advances in mRNA vaccines for infectious diseases. Front. Immunol. 2019;10:564. doi: 10.3389/fimmu.2019.00594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kreiter S., Diken M., Selmi A., Türeci Ö., Sahin U. Tumor vaccination using messenger RNA: prospects of a future therapy. Curr. Opin. Immunol. 2011;23:399–406. doi: 10.1016/j.coi.2011.03.007. [DOI] [PubMed] [Google Scholar]
- 27.Pollard C., Rejman J., De Haes W., Verrier B., Van Gulck E., Naessens T., De Smedt S., Bogaert P., Grooten J., Vanham G., De Koker S. Type I IFN counteracts the induction of antigen-specific immune responses by lipid-based delivery of mRNA vaccines. Mol. Ther. 2013;21:251–259. doi: 10.1038/mt.2012.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ulmer J.B., Mason P.W., Geall A., Mandl C.W. RNA-based vaccines. Vaccine. 2012;30:4414–4418. doi: 10.1016/j.vaccine.2012.04.060. [DOI] [PubMed] [Google Scholar]
- 29.Sahin U., Karikó K., Türeci Ö. mRNA-based therapeutics — developing a new class of drugs. Nat. Rev. Drug Discov. 2014;13:759–780. doi: 10.1038/nrd4278. [DOI] [PubMed] [Google Scholar]
- 30.Sahin U., Türeci Ö. Personalized vaccines for cancer immunotherapy. Science. 2018;359:1355–1360. doi: 10.1126/science.aar7112. [DOI] [PubMed] [Google Scholar]
- 31.Alberer M., Gnad-Vogt U., Hong H.S., Mehr K.T., Backert L., Finak G., Gottardo R., Bica M.A., Garofano A., Koch S.D., Fotin-Mleczek M., Hoerr I., Clemens R., von Sonnenburg F. Safety and immunogenicity of a mRNA rabies vaccine in healthy adults: an open-label, non-randomised, prospective, first-in-human phase 1 clinical trial. Lancet. 2017;390:1511–1520. doi: 10.1016/S0140-6736(17)31665-3. [DOI] [PubMed] [Google Scholar]
- 32.Valilou S.F., Keshavarz-Fathi M. In: Vaccines For Cancer Immunotherapy: An Evidence-Based Review on Current Status and Future Perspectives. Rezaei N., Keshavarz-Fathi M., editors. Elsevier; 2018. Vaccines for cancer immunotherapy; pp. 129–143. [Google Scholar]
- 33.Smith T.R.F., Patel A., Ramos S., Elwood D., Zhu X., Yan J., Gary E.N., Walker S.N., Schultheis K., Purwar M., Xu Z., Walters J., Bhojnagarwala P., Yang M., Chokkalingam N., Pezzoli P., Parzych E., Reuschel E.L., Doan A., Tursi N., Vasquez M., Choi J., Tello-Ruiz E., Maricic I., Bah M.A., Wu Y., Amante D., Park D.H., Dia Y., Ali A.R., Zaidi F.I., Generotti A., Kim K.Y., Herring T.A., Reeder S., Andrade V.M., Buttigieg K., Zhao G., Wu J.-M., Li D., Bao L., Liu J., Deng W., Qin C., Brown A.S., Khoshnejad M., Wang N., Chu J., Wrapp D., McLellan J.S., Muthumani K., Wang B., Carroll M.W., Kim J.J., Boyer J., Kulp D.W., Humeau L.M.P.F., Weiner D.B., Broderick K.E. Immunogenicity of a DNA vaccine candidate for COVID-19. Nat. Commun. 2020;11:2601. doi: 10.1038/s41467-020-16505-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu M.A. A comparison of plasmid DNA and mRNA as vaccine technologies. Vaccines. 2019;7:37. doi: 10.3390/vaccines7020037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lopes A., Vandermeulen G., Préat V. Cancer DNA vaccines: current preclinical and clinical developments and future perspectives. J. Exp. Clin. Cancer Res. 2019;38:146. doi: 10.1186/s13046-019-1154-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schlake T., Thran M., Fiedler K., Heidenreich R., Petsch B., Fotin-Mleczek M. mRNA: a novel avenue to antibody therapy? Mol. Ther. 2019;27:773–784. doi: 10.1016/j.ymthe.2019.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moderna Inc, Moderna announces longer shelf life for its COVID-19 vaccine candidate at refrigerated temperatures, https://investors.modernatx.com/news-releases/news-release-details/moderna-announces-longer-shelf-life-its-covid-19-vaccine, 2020 (accessed 13 May 2021).
- 38.BioNTech SE, Pfizer Inc, Pfizer and Biontech Conclude Phase 3 Study of Covid-19 Vaccine Candidate, Meeting All Primary Efficacy Endpoints, https://www.pfizer.com/news/press-release/press-release-detail/pfizer-and-biontech-conclude-phase-3-study-covid-19-vaccine, 2020 (Accessed 13 May 2021).
- 39.Pardi N., Hogan M.J., Porter F.W., Weissman D. mRNA vaccines — a new era in vaccinology. Nat. Rev. Drug Discov. 2018;17:261–279. doi: 10.1038/nrd.2017.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Verbeke R., Lentacker I., De Smedt S.C., Dewitte H. Three decades of messenger RNA vaccine development. Nano Today. 2019;28 [Google Scholar]
- 41.Pastor F., Berraondo P., Etxeberria I., Frederick J., Sahin U., Gilboa E., Melero I. An RNA toolbox for cancer immunotherapy. Nat. Rev. Drug Discov. 2018;17:751–767. doi: 10.1038/nrd.2018.132. [DOI] [PubMed] [Google Scholar]
- 42.Wolff J.A., Malone R.W., Williams P., Chong W., Acsadi G., Jani A., Felgner P.L. Direct gene transfer into mouse muscle in vivo. Science. 1990;247:1465–1468. doi: 10.1126/science.1690918. [DOI] [PubMed] [Google Scholar]
- 43.Martinon F., Krishnan S., Lenzen G., Magné R., Gomard E., Guillet J.-G., Lévy J.-P., Meulien P. Induction of virus-specific cytotoxic T lymphocytes in vivo by liposome-entrapped mRNA. Eur. J. Immunol. 1993;23:1719–1722. doi: 10.1002/eji.1830230749. [DOI] [PubMed] [Google Scholar]
- 44.Ross J., Sullivan T.D. Half-lives of beta and gamma globin messenger RNAs and of protein synthetic capacity in cultured human reticulocytes. Blood. 1985;66:1149–1154. [PubMed] [Google Scholar]
- 45.Aviv H., Voloch Z., Bastos R., Levy S. Biosynthesis and stability of globin mRNA in cultured erythroleukemic friend cells. Cell. 1976;8:495–503. doi: 10.1016/0092-8674(76)90217-8. [DOI] [PubMed] [Google Scholar]
- 46.Kranz L.M., Diken M., Haas H., Kreiter S., Loquai C., Reuter K.C., Meng M., Fritz D., Vascotto F., Hefesha H., Grunwitz C., Vormehr M., Hüsemann Y., Selmi A., Kuhn A.N., Buck J., Derhovanessian E., Rae R., Attig S., Diekmann J., Jabulowsky R.A., Heesch S., Hassel J., Langguth P., Grabbe S., Huber C., Türeci Ö., Sahin U. Systemic RNA delivery to dendritic cells exploits antiviral defence for cancer immunotherapy. Nature. 2016;534:396–401. doi: 10.1038/nature18300. [DOI] [PubMed] [Google Scholar]
- 47.Luo Y., Na Z., Slavoff S.A. P-bodies: composition, properties, and functions. Biochemistry. 2018;57:2424–2431. doi: 10.1021/acs.biochem.7b01162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Trepotec Z., Geiger J., Plank C., Aneja M.K., Rudolph C. Segmented poly(A) tails significantly reduce recombination of plasmid DNA without affecting mRNA translation efficiency or half-life. RNA. 2019;25:507–518. doi: 10.1261/rna.069286.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Abbas Y.M., Laudenbach B.T., Martínez-Montero S., Cencic R., Habjan M., Pichlmair A., Damha M.J., Pelletier J., Nagar B. Structure of human IFIT1 with capped RNA reveals adaptable mRNA binding and mechanisms for sensing N1 and N2 ribose 2′-O methylations. Proc. Natl. Acad. Sci. U.S.A., 2017;114:E2106–E2115. doi: 10.1073/pnas.1612444114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Katibah G.E., Qin Y., Sidote D.J., Yao J., Lambowitz A.M., Collins K. Broad and adaptable RNA structure recognition by the human interferon-induced tetratricopeptide repeat protein IFIT5. Proc. Natl. Acad. Sci. U.S.A., 2014;111:12025–12030. doi: 10.1073/pnas.1412842111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schuberth-Wagner C., Ludwig J., Bruder A.K., Herzner A.-M., Zillinger T., Goldeck M., Schmidt T., Schmid-Burgk J.L., Kerber R., Wolter S., Stümpel J.-P., Roth A., Bartok E., Drosten C., Coch C., Hornung V., Barchet W., Kümmerer B.M., Hartmann G., Schlee M. A conserved histidine in the RNA sensor RIG-I controls immune tolerance to N1-20O-methylated Self RNA. Immunity. 2015;43:41–51. doi: 10.1016/j.immuni.2015.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vaidyanathan S., Azizian K.T., Haque A., Henderson J.M., Hendel A., Shore S., Antony J.S., Hogrefe R.I., Kormann M.S.D., Porteus M.H., McCaffrey A.P. Uridine depletion and chemical modification increase Cas9 mRNA activity and reduce immunogenicity without HPLC purification. Mol. Ther. Nucleic Acids. 2018;12:530–542. doi: 10.1016/j.omtn.2018.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hanson G., Coller J. Codon optimality, bias and usage in translation and mRNA decay. Nat. Rev. Mol. Cell Biol. 2018;19:20–30. doi: 10.1038/nrm.2017.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Thess A., Grund S., Mui B.L., Hope M.J., Baumhof P., Fotin-Mleczek M., Schlake T. Sequence-engineered mRNA without chemical nucleoside modifications enables an effective protein therapy in large animals. Mol. Ther. 2015;23:1456–1464. doi: 10.1038/mt.2015.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kudla G., Murray A.W., Tollervey D., Plotkin J.B. Coding-sequence determinants of gene expression in Escherichia coli. Science. 2009;324:255–258. doi: 10.1126/science.1170160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mauro V.P., Chappell S.A. A critical analysis of codon optimization in human therapeutics. Trends Mol. Med. 2014;20:604–613. doi: 10.1016/j.molmed.2014.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Agashe D., Martinez-Gomez N.C., Drummond D.A., Marx C.J. Good codons, bad transcript: large reductions in gene expression and fitness arising from synonymous mutations in a key enzyme. Mol. Biol. Evol. 2013;30:549–560. doi: 10.1093/molbev/mss273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Spencer P.S., Siller E., Anderson J.F., Barral J.M. Silent substitutions predictably alter translation elongation rates and protein folding efficiencies. J. Mol. Biol. 2012;422:328–335. doi: 10.1016/j.jmb.2012.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ziegler A., Soldner C., Lienenklaus S., Spanier J., Trittel S., Riese P., Kramps T., Weiss S., Heidenreich R., Jasny E., Guzmán C.A., Kallen K.-J., Fotin-Mleczek M., Kalinke U. A new RNA-based adjuvant enhances virus-specific vaccine responses by locally triggering TLR- and RLH-dependent effects. J. Immunol. 2017;198:1595. doi: 10.4049/jimmunol.1601129. [DOI] [PubMed] [Google Scholar]
- 60.Heidenreich R., Jasny E., Kowalczyk A., Lutz J., Probst J., Baumhof P., Scheel B., Voss S., Kallen K.-J., Fotin-Mleczek M. A novel RNA-based adjuvant combines strong immunostimulatory capacities with a favorable safety profile. Int. J. Cancer. 2015;137:372–384. doi: 10.1002/ijc.29402. [DOI] [PubMed] [Google Scholar]
- 61.Doener F., Hong H.S., Meyer I., Tadjalli-Mehr K., Daehling A., Heidenreich R., Koch S.D., Fotin-Mleczek M., Gnad-Vogt U. RNA-based adjuvant CV8102 enhances the immunogenicity of a licensed rabies vaccine in a first-in-human trial. Vaccine. 2019;37:1819–1826. doi: 10.1016/j.vaccine.2019.02.024. [DOI] [PubMed] [Google Scholar]
- 62.Sebastian M., Schröder A., Scheel B., Hong H.S., Muth A., von Boehmer L., Zippelius A., Mayer F., Reck M., Atanackovic D., Thomas M., Schneller F., Stöhlmacher J., Bernhard H., Gröschel A., Lander T., Probst J., Strack T., Wiegand V., Gnad-Vogt U., Kallen K.J., Hoerr I., von der Muelbe F., Fotin-Mleczek M., Knuth A., Koch S.D. A phase I/IIa study of the mRNA-based cancer immunotherapy CV9201 in patients with stage IIIB/IV non-small cell lung cancer. Cancer Immunol. Immunother. 2019;68:799–812. doi: 10.1007/s00262-019-02315-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Karikó K., Buckstein M., Ni H., Weissman D. Suppression of RNA recognition by toll-like receptors: the impact of nucleoside modification and the evolutionary origin of RNA. Immunity. 2005;23:165–175. doi: 10.1016/j.immuni.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 64.Nallagatla S.R., Bevilacqua P.C. Nucleoside modifications modulate activation of the protein kinase PKR in an RNA structure-specific manner. RNA. 2008;14:1201–1213. doi: 10.1261/rna.1007408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Anderson B.R., Muramatsu H., Nallagatla S.R., Bevilacqua P.C., Sansing L.H., Weissman D., Karikó K. Incorporation of pseudouridine into mRNA enhances translation by diminishing PKR activation. Nucleic. Acids. Res. 2010;38:5884–5892. doi: 10.1093/nar/gkq347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Karikó K., Muramatsu H., Keller J.M., Weissman D. Increased erythropoiesis in mice injected with submicrogram quantities of pseudouridine-containing mRNA encoding erythropoietin. Mol. Ther. 2012;20:948–953. doi: 10.1038/mt.2012.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.De Beuckelaer A., Pollard C., Van Lint S., Roose K., Van Hoecke L., Naessens T., Udhayakumar V.K., Smet M., Sanders N., Lienenklaus S., Saelens X., Weiss S., Vanham G., Grooten J., De Koker S. Type I interferons interfere with the capacity of mRNA lipoplex vaccines to elicit cytolytic T cell responses. Mol. Ther. 2016;24:2012–2020. doi: 10.1038/mt.2016.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Murira A., Lamarre A. Type-I Interferon Responses: from Friend to Foe in the Battle against Chronic Viral Infection. Front. Immunol. 2016;7:609. doi: 10.3389/fimmu.2016.00609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Karikó K., Muramatsu H., Ludwig J., Weissman D. Generating the optimal mRNA for therapy: HPLC purification eliminates immune activation and improves translation of nucleoside-modified, protein-encoding mRNA. Nucleic. Acids. Res. 2011;39:e142. doi: 10.1093/nar/gkr695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kauffman K.J., Mir F.F., Jhunjhunwala S., Kaczmarek J.C., Hurtado J.E., Yang J.H., Webber M.J., Kowalski P.S., Heartlein M.W., DeRosa F., Anderson D.G. Efficacy and immunogenicity of unmodified and pseudouridine-modified mRNA delivered systemically with lipid nanoparticles in vivo. Biomaterials. 2016;109:78–87. doi: 10.1016/j.biomaterials.2016.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pardi N., Parkhouse K., Kirkpatrick E., McMahon M., Zost S., Mui B., Tam Y., Kariko K., Barbosa C., Madden T. Nucleoside-modified mRNA immunization elicits influenza virus hemagglutinin stalk-specific antibodies. Nat. Commun. 2018;9:3361. doi: 10.1038/s41467-018-05482-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Baiersdörfer M., Boros G., Muramatsu H., Mahiny A., Vlatkovic I., Sahin U., Karikó K. A facile method for the removal of dsRNA contaminant from in vitro-transcribed mRNA. Mol. Ther. Nucleic Acids. 2019;15:26–35. doi: 10.1016/j.omtn.2019.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Vogel A.B., Lambert L., Kinnear E., Busse D., Erbar S., Reuter K.C., Wicke L., Perkovic M., Beissert T., Haas H., Reece S.T., Sahin U., Tregoning J.S. Self-amplifying RNA vaccines give equivalent protection against influenza to mRNA vaccines but at much lower doses. Mol. Ther. 2018;26:446–455. doi: 10.1016/j.ymthe.2017.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Geall A.J., Verma A., Otten G.R., Shaw C.A., Hekele A., Banerjee K., Cu Y., Beard C.W., Brito L.A., Krucker T., O’Hagan D.T., Singh M., Mason P.W., Valiante N.M., Dormitzer P.R., Barnett S.W., Rappuoli R., Ulmer J.B., Mandl C.W. Nonviral delivery of self-amplifying RNA vaccines. Proc. Natl. Acad. Sci. U.S.A., 2012;109:14604–14609. doi: 10.1073/pnas.1209367109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Johanning F., Conry R., LoBuglio A., Wright M., Sumerel L., Pike M., Curiel D. A Sindbis virus mRNA polynucleotide vector achieves prolonged and high level heterologous gene expression in vivo. Nucleic. Acids. Res. 1995;23:1495–1501. doi: 10.1093/nar/23.9.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Brazzoli M., Magini D., Bonci A., Buccato S., Giovani C., Kratzer R., Zurli V., Mangiavacchi S., Casini D., Brito L.M. Induction of broad-based immunity and protective efficacy by self-amplifying mRNA vaccines encoding influenza virus hemagglutinin. J. Virol. 2016;90:332–344. doi: 10.1128/JVI.01786-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pepini T., Pulichino A.-M., Carsillo T., Carlson A.L., Sari-Sarraf F., Ramsauer K., Debasitis J.C., Maruggi G., Otten G.R., Geall A.J., Yu D., Ulmer J.B., Iavarone C. Induction of an IFN-mediated antiviral response by a self-amplifying RNA vaccine: implications for vaccine design. J. Immunol. 2017;198:4012. doi: 10.4049/jimmunol.1601877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.De Beuckelaer A., Grooten J., De Koker S. Type I interferons modulate CD8(+) T cell immunity to mRNA vaccines. Trends Mol. Med. 2017;23:216–226. doi: 10.1016/j.molmed.2017.01.006. [DOI] [PubMed] [Google Scholar]
- 79.Karikó K., Muramatsu H., Ludwig J., Weissman D. Generating the optimal mRNA for therapy: HPLC purification eliminates immune activation and improves translation of nucleoside-modified, protein-encoding mRNA. Nucleic. Acids. Res. 2011;39:e142. doi: 10.1093/nar/gkr695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tojo S., Zhang Z., Matsui H., Tahara M., Ikeguchi M., Kochi M., Kamada M., Shigematsu H., Tsutsumi A., Adachi N., Shibata T., Yamamoto M., Kikkawa M., Senda T., Isobe Y., Ohto U., Shimizu T. Structural analysis reveals TLR7 dynamics underlying antagonism. Nat. Commun. 2020;11:5204. doi: 10.1038/s41467-020-19025-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tanji H., Ohto U., Shibata T., Taoka M., Yamauchi Y., Isobe T., Miyake K., Shimizu T. Toll-like receptor 8 senses degradation products of single-stranded RNA. Nat. Struct. Mol. Biol. 2015;22:109–115. doi: 10.1038/nsmb.2943. [DOI] [PubMed] [Google Scholar]
- 82.Wykes M., Pombo A., Jenkins C., MacPherson G.G. Dendritic cells interact directly with naive B lymphocytes to transfer antigen and initiate class switching in a primary T-dependent response. J. Immunol. 1998;161:1313–1319. [PubMed] [Google Scholar]
- 83.Tam H.H., Melo M.B., Kang M., Pelet J.M., Ruda V.M., Foley M.H., Hu J.K., Kumari S., Crampton J., Baldeon A.D., Sanders R.W., Moore J.P., Crotty S., Langer R., Anderson D.G., Chakraborty A.K., Irvine D.J. Sustained antigen availability during germinal center initiation enhances antibody responses to vaccination. Proc. Natl. Acad. Sci. U.S.A. 2016;113:E6639–E6648. doi: 10.1073/pnas.1606050113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Vu M.N., Kelly H.G., Tan H.-X., Juno J.A., Esterbauer R., Davis T.P., Truong N.P., Wheatley A.K., Kent S.J. Hemagglutinin functionalized liposomal vaccines enhance germinal center and follicular helper T cell immunity. Adv. Healthcare Mater. 2021;10 doi: 10.1002/adhm.202002142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Garg N.K., Dwivedi P., Prabha P., Tyagi R.K. RNA pulsed dendritic cells: an approach for cancer immunotherapy. Vaccine. 2013;31:1141–1156. doi: 10.1016/j.vaccine.2012.12.027. [DOI] [PubMed] [Google Scholar]
- 86.Benteyn D., Heirman C., Bonehill A., Thielemans K., Breckpot K. mRNA-based dendritic cell vaccines. Expert Rev. Vaccines. 2015;14:161–176. doi: 10.1586/14760584.2014.957684. [DOI] [PubMed] [Google Scholar]
- 87.Liu Z., Roche P.A. Macropinocytosis in phagocytes: regulation of MHC class-II-restricted antigen presentation in dendritic cells. Front. Physiol. 2015;6:1. doi: 10.3389/fphys.2015.00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Heiser A., Coleman D., Dannull J., Yancey D., Maurice M.A., Lallas C.D., Dahm P., Niedzwiecki D., Gilboa E., Vieweg J. Autologous dendritic cells transfected with prostate-specific antigen RNA stimulate CTL responses against metastatic prostate tumors. J. Clin. Invest. 2002;109:409–417. doi: 10.1172/JCI14364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Van Lint S., Heirman C., Thielemans K., Breckpot K. mRNA: from a chemical blueprint for protein production to an off-the-shelf therapeutic. Hum. Vaccin. Immunother. 2013;9:265–274. doi: 10.4161/hv.22661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Freitas-Silva R., Brelaz-de-Castro M.C., Pereira V.R. Dendritic cell-based approaches in the fight against diseases. Front. Immunol. 2014;5:78. doi: 10.3389/fimmu.2014.00078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Conry R.M., LoBuglio A.F., Wright M., Sumerel L., Pike M.J., Johanning F., Benjamin R., Lu D., Curiel D.T. Characterization of a messenger RNA polynucleotide vaccine vector. Cancer Res. 1995;55:1397–1400. [PubMed] [Google Scholar]
- 92.Schlake T., Thess A., Fotin-Mleczek M., Kallen K.-J. Developing mRNA-vaccine technologies. RNA Biol. 2012;9:1319–1330. doi: 10.4161/rna.22269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Probst J., Weide B., Scheel B., Pichler B.J., Hoerr I., Rammensee H.G., Pascolo S. Spontaneous cellular uptake of exogenous messenger RNA in vivo is nucleic acid-specific, saturable and ion dependent. Gene Ther. 2007;14:1175–1180. doi: 10.1038/sj.gt.3302964. [DOI] [PubMed] [Google Scholar]
- 94.Wadhwa A., Aljabbari A., Lokras A., Foged C., Thakur A. Opportunities and challenges in the delivery of mRNA-based vaccines. Pharmaceutics. 2020;12:102. doi: 10.3390/pharmaceutics12020102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Maruggi G., Zhang C., Li J., Ulmer J.B., Yu D. mRNA as a transformative technology for vaccine development to control infectious diseases. Mol. Ther. 2019;27:757–772. doi: 10.1016/j.ymthe.2019.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Pardi N., Hogan M.J., Weissman D. Recent advances in mRNA vaccine technology. Curr. Opin. Immunol. 2020;65:14–20. doi: 10.1016/j.coi.2020.01.008. [DOI] [PubMed] [Google Scholar]
- 97.Richner J.M., Himansu S., Dowd K.A., Butler S.L., Salazar V., Fox J.M., Julander J.G., Tang W.W., Shresta S., Pierson T.C., Ciaramella G. Modified mRNA vaccines protect against Zika virus infection. Cell. 2017;168:1114–1125. doi: 10.1016/j.cell.2017.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Gonzalez G., Pfannes L., Striker R. Selection of an optimal RNA transfection reagent and comparison to electroporation for the delivery of viral RNA. J. Virol. Methods. 2007;145:14–21. doi: 10.1016/j.jviromet.2007.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zhang H., De Smedt S.C., Remaut K. Fluorescence correlation spectroscopy to find the critical balance between extracellular association and intracellular dissociation of mRNA complexes. Acta Biomater. 2018;75:358–370. doi: 10.1016/j.actbio.2018.05.016. [DOI] [PubMed] [Google Scholar]
- 100.Garg N.K., Tandel N., Jadon R.S., Tyagi R.K., Katare O.P. Lipid–polymer hybrid nanocarrier-mediated cancer therapeutics: current status and future directions. Drug Discov. Today. 2018;23:1610–1621. doi: 10.1016/j.drudis.2018.05.033. [DOI] [PubMed] [Google Scholar]
- 101.Chaudhari R., Tandel N., Sahu K., Negi S., Bashir H., Rupareliya A., Mishra R.P.N., Dalai S.K., Tyagi R.K. Transdermal immunization of elastic liposome-laden recombinant chimeric fusion protein of P. falciparum (PfMSP-Fu24) mounts protective immune response. Nanomaterials (Basel) 2021;11:406. doi: 10.3390/nano11020406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Garg N.K., Singh B., Kushwah V., Tyagi R.K., Sharma R., Jain S., Katare O.P. The ligand(s) anchored lipobrid nanoconstruct mediated delivery of methotrexate: an effective approach in breast cancer therapeutics. Nanomedicine. 2016;12:2043–2060. doi: 10.1016/j.nano.2016.05.008. [DOI] [PubMed] [Google Scholar]
- 103.Garg N.K., Tyagi R.K., Singh B., Sharma G., Nirbhavane P., Kushwah V., Jain S., Katare O.P. Nanostructured lipid carrier mediates effective delivery of methotrexate to induce apoptosis of rheumatoid arthritis via NF-κB and FOXO1. Int. J. Pharm. 2016;499:301–320. doi: 10.1016/j.ijpharm.2015.12.061. [DOI] [PubMed] [Google Scholar]
- 104.Jeong S.-H., Jang J.-H., Cho H.-Y., Lee Y.-B. Soft- and hard-lipid nanoparticles: a novel approach to lymphatic drug delivery. Arch. Pharm. Res. 2018;41:797–814. doi: 10.1007/s12272-018-1060-0. [DOI] [PubMed] [Google Scholar]
- 105.Ribeiro L.N.D.M., Couto V.M., Fraceto L.F., de Paula E. Use of nanoparticle concentration as a tool to understand the structural properties of colloids. Sci. Rep. 2018;8:982. doi: 10.1038/s41598-017-18573-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Filion M.C., Phillips N.C. Toxicity and immunomodulatory activity of liposomal vectors formulated with cationic lipids toward immune effector cells. BBA Biomembranes. 1997;1329:345–356. doi: 10.1016/s0005-2736(97)00126-0. [DOI] [PubMed] [Google Scholar]
- 107.Lappalainen K., Jääskeläinen I., Syrjänen K., Urtti A., Syrjänen S. Comparison of cell proliferation and toxicity assays using two cationic liposomes. Pharm. Res. 1994;11:1127–1131. doi: 10.1023/a:1018932714745. [DOI] [PubMed] [Google Scholar]
- 108.Evers M.J.W., Kulkarni J.A., van der Meel R., Cullis P.R., Vader P., Schiffelers R.M. State-of-the-art design and rapid-mixing production techniques of lipid nanoparticles for nucleic acid delivery. Small Methods. 2018;2 [Google Scholar]
- 109.Viger-Gravel J., Schantz A., Pinon A.C., Rossini A.J., Schantz S., Emsley L. Structure of lipid nanoparticles containing siRNA or mRNA by dynamic nuclear polarization-enhanced NMR spectroscopy. J. Phys. Chem. B. 2018;122:2073–2081. doi: 10.1021/acs.jpcb.7b10795. [DOI] [PubMed] [Google Scholar]
- 110.Eygeris Y., Patel S., Jozic A., Sahay G. Deconvoluting lipid nanoparticle structure for messenger RNA delivery. Nano Lett. 2020;20:4543–4549. doi: 10.1021/acs.nanolett.0c01386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Patel S., Ashwanikumar N., Robinson E., Xia Y., Mihai C., Griffith J.P., Hou S., Esposito A.A., Ketova T., Welsher K., Joyal J.L., Almarsson O., Sahay G. Naturally-occurring cholesterol analogues in lipid nanoparticles induce polymorphic shape and enhance intracellular delivery of mRNA. Nat. Commun. 2020;11:983. doi: 10.1038/s41467-020-14527-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Stroock A.D., Dertinger S.K.W., Ajdari A., Mezic I., Stone H.A., Whitesides G.M. Chaotic mixer for microchannels. Science. 2002;295:647–651. doi: 10.1126/science.1066238. [DOI] [PubMed] [Google Scholar]
- 113.Kulkarni J.A., Thomson S.B., Zaifman J., Leung J., Wagner P.K., Hill A., Tam Y.Y.C., Cullis P.R., Petkau T.L., Leavitt B.R. Spontaneous, solvent-free entrapment of siRNA within lipid nanoparticles. Nanoscale. 2020;12:23959–23966. doi: 10.1039/d0nr06816k. [DOI] [PubMed] [Google Scholar]
- 114.Roces C.B., Lou G., Jain N., Abraham S., Thomas A., Halbert G.W., Perrie Y. Manufacturing considerations for the development of lipid nanoparticles using microfluidics. Pharmaceutics. 2020;12:1095. doi: 10.3390/pharmaceutics12111095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.He Z., Hu Y., Nie T., Tang H., Zhu J., Chen K., Liu L., Leong K.M., Chen Y., Mao H.-Q. Size-controlled lipid nanoparticle production using turbulent mixing to enhance oral DNA delivery. Acta Biomater. 2018;81:195–207. doi: 10.1016/j.actbio.2018.09.047. [DOI] [PubMed] [Google Scholar]
- 116.Arduino I., Liu Z.H., Rahikkala A., Figueiredo P., Correia A., Cutrignelli A., Denora N., Santos H.A. Preparation of cetyl palmitate-based PEGylated solid lipid nanoparticles by microfluidic technique. Acta Biomater. 2020;121:566–578. doi: 10.1016/j.actbio.2020.12.024. [DOI] [PubMed] [Google Scholar]
- 117.Patisiran, an RNAi Therapeutic, for hereditary transthyretin amyloidosis. N. Engl. J. Med. 2018;379:11–21. doi: 10.1056/NEJMoa1716153. [DOI] [PubMed] [Google Scholar]
- 118.Kauffman K.J., Dorkin J.R., Yang J.H., Heartlein M.W., DeRosa F., Mir F.F., Fenton O.S., Anderson D.G. Optimization of lipid nanoparticle formulations for mrna delivery in vivo with fractional factorial and definitive screening designs. Nano Lett. 2015;15:7300–7306. doi: 10.1021/acs.nanolett.5b02497. [DOI] [PubMed] [Google Scholar]
- 119.Guimaraes P.P.G., Zhang R., Spektor R., Tan M., Chung A., Billingsley M.M., El-Mayta R., Riley R.S., Wang L., Wilson J.M., Mitchell M.J. Ionizable lipid nanoparticles encapsulating barcoded mRNA for accelerated in vivo delivery screening. J. Control. Release. 2019;316:404–417. doi: 10.1016/j.jconrel.2019.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Bhardwaj V., Bhatia E., Sharma S., Ahamad N., Banerjee R. Advancements in prophylactic and therapeutic nanovaccines. Acta Biomater. 2020;108:1–21. doi: 10.1016/j.actbio.2020.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Vu M.N., Kelly H.G., Wheatley A.K., Peng S., Pilkington E.H., Veldhuis N.A., Davis T.P., Kent S.J., Truong N.P. Cellular interactions of liposomes and PISA nanoparticles during human blood flow in a microvascular network. Small. 2020;16 doi: 10.1002/smll.202002861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Allen T.M., Cullis P.R. Liposomal drug delivery systems: from concept to clinical applications. Adv. Drug. Deliv. Rev. 2013;65:36–48. doi: 10.1016/j.addr.2012.09.037. [DOI] [PubMed] [Google Scholar]
- 123.Hafez I.M., Maurer N., Cullis P.R. On the mechanism whereby cationic lipids promote intracellular delivery of polynucleic acids. Gene Ther. 2001;8:1188–1196. doi: 10.1038/sj.gt.3301506. [DOI] [PubMed] [Google Scholar]
- 124.Fraley R., Subramani S., Papahadjopoulos D. Introduction of liposome-encapsulated SV40 DNA into cells. J. Biol. Chem. 1980;255:10431–10435. [PubMed] [Google Scholar]
- 125.Fraley R., Straubinger R.M., Rule G., Springer E.L., Papahadjopoulos D. Liposome-mediated delivery of deoxyribonucleic acid to cells: enhanced efficiency of delivery by changes in lipid composition and incubation conditions. Biochemistry. 1981;20:6978–6987. doi: 10.1021/bi00527a031. [DOI] [PubMed] [Google Scholar]
- 126.Jayaraman M., Ansell S.M., Mui B.L., Tam Y.K., Chen J., Du X., Butler D., Eltepu L., Matsuda S., Narayanannair J.K., Rajeev K.G., Hafez I.M., Akinc A., Maier M.A., Tracy M.A., Cullis P.R., Madden T.D., Manoharan M., Hope M.J. Maximizing the potency of siRNA lipid nanoparticles for hepatic gene silencing in vivo. Angew. Chem. Int. Ed. 2012;51:8529–8533. doi: 10.1002/anie.201203263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Semple S.C., Akinc A., Chen J., Sandhu A.P., Mui B.L., Cho C.K., Sah D.W., Stebbing D., Crosley E.J., Yaworski E., Hafez I.M., Dorkin J.R., Qin J., Lam K., Rajeev K.G., Wong K.F., Jeffs L.B., Nechev L., Eisenhardt M.L., Jayaraman M., Kazem M., Maier M.A., Srinivasulu M., Weinstein M.J., Chen Q., Alvarez R., Barros S.A., De S., Klimuk S.K., Borland T., Kosovrasti V., Cantley W.L., Tam Y.K., Manoharan M., Ciufolini M.A., Tracy M.A., de Fougerolles A., MacLachlan I., Cullis P.R., Madden T.D., Hope M.J. Rational design of cationic lipids for siRNA delivery. Nat. Biotechnol. 2010;28:172–176. doi: 10.1038/nbt.1602. [DOI] [PubMed] [Google Scholar]
- 128.Heyes J., Palmer L., Bremner K., MacLachlan I. Cationic lipid saturation influences intracellular delivery of encapsulated nucleic acids. J. Control. Release. 2005;107:276–287. doi: 10.1016/j.jconrel.2005.06.014. [DOI] [PubMed] [Google Scholar]
- 129.Nabhan J.F., Wood K.M., Rao V.P., Morin J., Bhamidipaty S., LaBranche T.P., Gooch R.L., Bozal F., Bulawa C.E., Guild B.C. Intrathecal delivery of frataxin mRNA encapsulated in lipid nanoparticles to dorsal root ganglia as a potential therapeutic for Friedreich’s ataxia. Sci. Rep. 2016;6:20019. doi: 10.1038/srep20019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Maier M.A., Jayaraman M., Matsuda S., Liu J., Barros S., Querbes W., Tam Y.K., Ansell S.M., Kumar V., Qin J., Zhang X., Wang Q., Panesar S., Hutabarat R., Carioto M., Hettinger J., Kandasamy P., Butler D., Rajeev K.G., Pang B., Charisse K., Fitzgerald K., Mui B.L., Du X., Cullis P., Madden T.D., Hope M.J., Manoharan M., Akinc A. Biodegradable lipids enabling rapidly eliminated lipid nanoparticles for systemic delivery of RNAi therapeutics. Mol. Ther. 2013;21:1570–1578. doi: 10.1038/mt.2013.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Fenton O.S., Kauffman K.J., McClellan R.L., Appel E.A., Dorkin J.R., Tibbitt M.W., Heartlein M.W., DeRosa F., Langer R., Anderson D.G. Bioinspired alkenyl amino alcohol ionizable lipid materials for highly potent in vivo mRNA delivery. Adv. Mater. 2016;28:2939–2943. doi: 10.1002/adma.201505822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Sato Y., Okabe N., Note Y., Hashiba K., Maeki M., Tokeshi M., Harashima H. Hydrophobic scaffolds of pH-sensitive cationic lipids contribute to miscibility with phospholipids and improve the efficiency of delivering short interfering RNA by small-sized lipid nanoparticles. Acta Biomater. 2020;102:341–350. doi: 10.1016/j.actbio.2019.11.022. [DOI] [PubMed] [Google Scholar]
- 133.Semple S.C., Klimuk S.K., Harasym T.O., Dos Santos N., Ansell S.M., Wong K.F., Maurer N., Stark H., Cullis P.R., Hope M.J., Scherrer P. Efficient encapsulation of antisense oligonucleotides in lipid vesicles using ionizable aminolipids: formation of novel small multilamellar vesicle structures. Biochim. Biophys. Acta. 2001;1510:152–166. doi: 10.1016/s0005-2736(00)00343-6. [DOI] [PubMed] [Google Scholar]
- 134.Whitehead K.A., Sahay G., Li G.Z., Love K.T., Alabi C.A., Ma M., Zurenko C., Querbes W., Langer R.S., Anderson D.G. Synergistic silencing: combinations of lipid-like materials for efficacious siRNA delivery. Mol. Ther. 2011;19:1688–1694. doi: 10.1038/mt.2011.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Akinc A., Zumbuehl A., Goldberg M., Leshchiner E.S., Busini V., Hossain N., Bacallado S.A., Nguyen D.N., Fuller J., Alvarez R., Borodovsky A., Borland T., Constien R., de Fougerolles A., Dorkin J.R., Narayanannair Jayaprakash K., Jayaraman M., John M., Koteliansky V., Manoharan M., Nechev L., Qin J., Racie T., Raitcheva D., Rajeev K.G., Sah D.W.Y., Soutschek J., Toudjarska I., Vornlocher H.-P., Zimmermann T.S., Langer R., Anderson D.G. A combinatorial library of lipid-like materials for delivery of RNAi therapeutics. Nat. Biotechnol. 2008;26:561–569. doi: 10.1038/nbt1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Love K.T., Mahon K.P., Levins C.G., Whitehead K.A., Querbes W., Dorkin J.R., Qin J., Cantley W.L., Qin L.L., Racie T., Frank-Kamenetsky M., Yip K.N., Alvarez R., Sah D.W.Y., de Fougerolles A., Fitzgerald K., Koteliansky V., Akinc A., Langer R., Anderson D.G. Lipid-like materials for low-dose, in vivo gene silencing. Proc. Natl. Acad. Sci. U.S.A. 2010;107:1864–1869. doi: 10.1073/pnas.0910603106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Yu X., Liu S., Cheng Q., Wei T., Lee S., Zhang D., Siegwart D.J. Lipid-modified aminoglycosides for mRNA delivery to the liver. Adv. Healthcare Mater. 2020;9 doi: 10.1002/adhm.201901487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Felgner P.L., Gadek T.R., Holm M., Roman R., Chan H.W., Wenz M., Northrop J.P., Ringold G.M., Danielsen M. Lipofection: a highly efficient, lipid-mediated DNA-transfection procedure. Proc. Natl. Acad. Sci. U.S.A. 1987;84:7413–7417. doi: 10.1073/pnas.84.21.7413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Hafez I.M., Cullis P.R. Roles of lipid polymorphism in intracellular delivery. Adv. Drug. Deliv. Rev. 2001;47:139–148. doi: 10.1016/s0169-409x(01)00103-x. [DOI] [PubMed] [Google Scholar]
- 140.Farhood H., Serbina N., Huang L. The role of dioleoyl phosphatidylethanolamine in cationic liposome mediated gene transfer. BBA Biomembranes. 1995;1235:289–295. doi: 10.1016/0005-2736(95)80016-9. [DOI] [PubMed] [Google Scholar]
- 141.Mok K.W., Lam A.M., Cullis P.R. Stabilized plasmid-lipid particles: factors influencing plasmid entrapment and transfection properties. Biochim. Biophys. Acta. 1999;1419:137–150. doi: 10.1016/s0005-2736(99)00059-0. [DOI] [PubMed] [Google Scholar]
- 142.Xiao K., Li Y., Luo J., Lee J.S., Xiao W., Gonik A.M., Agarwal R.G., Lam K.S. The effect of surface charge on in vivo biodistribution of PEG-oligocholic acid based micellar nanoparticles. Biomaterials. 2011;32:3435–3446. doi: 10.1016/j.biomaterials.2011.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Cheng Q., Wei T., Farbiak L., Johnson L.T., Dilliard S.A., Siegwart D.J. Selective organ targeting (SORT) nanoparticles for tissue-specific mRNA delivery and CRISPR–Cas gene editing. Nat. Nanotechnol. 2020;15:313–320. doi: 10.1038/s41565-020-0669-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Kulkarni J.A., Darjuan M.M., Mercer J.E., Chen S., van der Meel R., Thewalt J.L., Tam Y.Y.C., Cullis P.R. On the formation and morphology of lipid nanoparticles containing ionizable cationic lipids and siRNA. ACS Nano. 2018;12:4787–4795. doi: 10.1021/acsnano.8b01516. [DOI] [PubMed] [Google Scholar]
- 145.Leung A.K., Tam Y.Y., Chen S., Hafez I.M., Cullis P.R. Microfluidic mixing: a general method for encapsulating macromolecules in lipid nanoparticle systems. J. Phys. Chem. B. 2015;119:8698–8706. doi: 10.1021/acs.jpcb.5b02891. [DOI] [PubMed] [Google Scholar]
- 146.Sabnis S., Kumarasinghe E.S., Salerno T., Mihai C., Ketova T., Senn J.J., Lynn A., Bulychev A., McFadyen I., Chan J., Almarsson O., Stanton M.G., Benenato K.E. A Novel amino lipid series for mRNA delivery: improved endosomal escape and sustained pharmacology and safety in non-human primates. Mol. Ther. 2018;26:1509–1519. doi: 10.1016/j.ymthe.2018.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Hassett K.J., Benenato K.E., Jacquinet E., Lee A., Woods A., Yuzhakov O., Himansu S., Deterling J., Geilich B.M., Ketova T., Mihai C., Lynn A., McFayden I., Moore M.J., Senn J.J., Stanton M.G., Almarsson O., Ciaramella G., Brito L.A. Optimization of lipid nanoparticles for intramuscular administration of mRNA vaccines. Mol. Ther. Nucleic Acids. 2019;15:1–11. doi: 10.1016/j.omtn.2019.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Oberli M.A., Reichmuth A.M., Dorkin J.R., Mitchell M.J., Fenton O.S., Jaklenec A., Anderson D.G., Langer R., Blankschtein D. Lipid nanoparticle assisted mRNA delivery for potent cancer immunotherapy. Nano Lett. 2017;17:1326–1335. doi: 10.1021/acs.nanolett.6b03329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Cheng Q., Wei T., Jia Y., Farbiak L., Zhou K., Zhang S., Wei Y., Zhu H., Siegwart D.J. Dendrimer-based lipid nanoparticles deliver therapeutic FAH mRNA to normalize liver function and extend survival in a mouse model of hepatorenal tyrosinemia type I. Adv. Mater. 2018;30 doi: 10.1002/adma.201805308. [DOI] [PubMed] [Google Scholar]
- 150.Kulkarni J.A., Witzigmann D., Leung J., Tam Y.Y.C., Cullis P.R. On the role of helper lipids in lipid nanoparticle formulations of siRNA. Nanoscale. 2019;11:21733–21739. doi: 10.1039/c9nr09347h. [DOI] [PubMed] [Google Scholar]
- 151.Miller J.B., Zhang S., Kos P., Xiong H., Zhou K., Perelman S.S., Zhu H., Siegwart D.J. Non-viral CRISPR/Cas gene editing in vitro and in vivo enabled by synthetic nanoparticle co-delivery of Cas9 mRNA and sgRNA. Angew. Chem. Int. Ed. Engl. 2017;56:1059–1063. doi: 10.1002/anie.201610209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Liu S., Cheng Q., Wei T., Yu X., Johnson L.T., Farbiak L., Siegwart D.J. Membrane-destabilizing ionizable phospholipids for organ-selective mRNA delivery and CRISPR–Cas gene editing. Nat. Mater. 2021;20:701–710. doi: 10.1038/s41563-020-00886-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Demel R.A., De Kruyff B. The function of sterols in membranes. Biochim. Biophys. Acta. 1976;457:109–132. doi: 10.1016/0304-4157(76)90008-3. [DOI] [PubMed] [Google Scholar]
- 154.Ercole F., Whittaker M.R., Quinn J.F., Davis T.P. Cholesterol modified self-assemblies and their application to nanomedicine. Biomacromolecules. 2015:1886–1914. doi: 10.1021/acs.biomac.5b00550. 2015. [DOI] [PubMed] [Google Scholar]
- 155.Takahashi H., Sinoda K., Hatta I. Effects of cholesterol on the lamellar and the inverted hexagonal phases of dielaidoylphosphatidylethanolamine. BBA Gen. Subj. 1996;1289:209–216. doi: 10.1016/0304-4165(95)00170-0. [DOI] [PubMed] [Google Scholar]
- 156.Yanez Arteta M., Kjellman T., Bartesaghi S., Wallin S., Wu X., Kvist A.J., Dabkowska A., Székely N., Radulescu A., Bergenholtz J., Lindfors L. Successful reprogramming of cellular protein production through mRNA delivered by functionalized lipid nanoparticles. Proc. Natl Acad. Sci. 2018;115:E3351–E3360. doi: 10.1073/pnas.1720542115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Tenchov B.G., MacDonald R.C., Siegel D.P. Cubic phases in phosphatidylcholine-cholesterol mixtures: cholesterol as membrane “fusogen”. Biophys. J. 2006;91:2508–2516. doi: 10.1529/biophysj.106.083766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Hung W.-C., Lee M.-T., Chen F.-Y., Huang H.W. The condensing effect of cholesterol in lipid bilayers. Biophys. J. 2007;92:3960–3967. doi: 10.1529/biophysj.106.099234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Pozzi D., Marchini C., Cardarelli F., Amenitsch H., Garulli C., Bifone A., Caracciolo G. Transfection efficiency boost of cholesterol-containing lipoplexes. Biochim. Biophys. Acta. 2012;1818:2335–2343. doi: 10.1016/j.bbamem.2012.05.017. [DOI] [PubMed] [Google Scholar]
- 160.Semple S.C., Chonn A., Cullis P.R. Influence of cholesterol on the association of plasma proteins with liposomes. Biochemistry. 1996;35:2521–2525. doi: 10.1021/bi950414i. [DOI] [PubMed] [Google Scholar]
- 161.Imam Z.I., Kenyon L.E., Ashby G., Nagib F., Mendicino M., Zhao C., Gadok A.K., Stachowiak J.C. Phase-separated liposomes enhance the efficiency of macromolecular delivery to the cellular cytoplasm. Cell. Mol. Bioeng. 2017;10:387–403. doi: 10.1007/s12195-017-0489-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Suk J.S., Xu Q., Kim N., Hanes J., Ensign L.M. PEGylation as a strategy for improving nanoparticle-based drug and gene delivery. Adv. Drug. Deliv. Rev. 2016;99:28–51. doi: 10.1016/j.addr.2015.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Ambegia E., Ansell S., Cullis P., Heyes J., Palmer L., MacLachlan I. Stabilized plasmid-lipid particles containing PEG-diacylglycerols exhibit extended circulation lifetimes and tumor selective gene expression. Biochim. Biophys. Acta. 2005;1669:155–163. doi: 10.1016/j.bbamem.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 164.Garbuzenko O., Barenholz Y., Priev A. Effect of grafted PEG on liposome size and on compressibility and packing of lipid bilayer. Chem. Phys. Lipids. 2005;135:117–129. doi: 10.1016/j.chemphyslip.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 165.Belliveau N.M., Huft J., Lin P.J., Chen S., Leung A.K., Leaver T.J., Wild A.W., Lee J.B., Taylor R.J., Tam Y.K., Hansen C.L., Cullis P.R. Microfluidic synthesis of highly potent limit-size lipid nanoparticles for in vivo delivery of siRNA. Mol. Ther. Nucleic Acids. 2012;1:e37. doi: 10.1038/mtna.2012.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Leung A.K.K., Hafez I.M., Baoukina S., Belliveau N.M., Zhigaltsev I.V., Afshinmanesh E., Tieleman D.P., Hansen C.L., Hope M.J., Cullis P.R. Lipid nanoparticles containing siRNA synthesized by microfluidic mixing exhibit an electron-dense nanostructured core. J. Phys. Chem. C. 2012;116:18440–18450. doi: 10.1021/jp303267y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Dos Santos N., Allen C., Doppen A.M., Anantha M., Cox K.A., Gallagher R.C., Karlsson G., Edwards K., Kenner G., Samuels L., Webb M.S., Bally M.B. Influence of poly(ethylene glycol) grafting density and polymer length on liposomes: relating plasma circulation lifetimes to protein binding. Biochim. Biophys. Acta. 2007;1768:1367–1377. doi: 10.1016/j.bbamem.2006.12.013. [DOI] [PubMed] [Google Scholar]
- 168.Sago C.D., Lokugamage M.P., Paunovska K., Vanover D.A., Monaco C.M., Shah N.N., Castro M.G., Anderson S.E., Rudoltz T.G., Lando G.N., Tiwari P.M., Kirschman J.L., Willett N., Jang Y.C., Santangelo P.J., Bryksin A.V., Dahlman J.E. High-throughput in vivo screen of functional mRNA delivery identifies nanoparticles for endothelial cell gene editing. Proc. Natl. Acad. Sci. U.S.A. 2018;115:E9944–E9952. doi: 10.1073/pnas.1811276115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Bao Y., Jin Y., Chivukula P., Zhang J., Liu Y., Liu J., Clamme J.P., Mahato R.I., Ng D., Ying W., Wang Y., Yu L. Effect of PEGylation on biodistribution and gene silencing of siRNA/lipid nanoparticle complexes. Pharm. Res. 2013;30:342–351. doi: 10.1007/s11095-012-0874-6. [DOI] [PubMed] [Google Scholar]
- 170.Chen S., Tam Y.Y.C., Lin P.J.C., Sung M.M.H., Tam Y.K., Cullis P.R. Influence of particle size on the in vivo potency of lipid nanoparticle formulations of siRNA. J. Control. Release. 2016;235:236–244. doi: 10.1016/j.jconrel.2016.05.059. [DOI] [PubMed] [Google Scholar]
- 171.Akinc A., Querbes W., De S., Qin J., Frank-Kamenetsky M., Jayaprakash K.N., Jayaraman M., Rajeev K.G., Cantley W.L., Dorkin J.R., Butler J.S., Qin L., Racie T., Sprague A., Fava E., Zeigerer A., Hope M.J., Zerial M., Sah D.W.Y., Fitzgerald K., Tracy M.A., Manoharan M., Koteliansky V., de Fougerolles A., Maier M.A. Targeted delivery of RNAi therapeutics with endogenous and exogenous ligand-based mechanisms. Mol. Ther. 2010;18:1357–1364. doi: 10.1038/mt.2010.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Allison S.J., Milner J. RNA interference by single- and double-stranded siRNA with a DNA extension containing a 3′ nuclease-resistant mini-hairpin structure. Mol. Ther. Nucleic Acids. 2014;2:e141. doi: 10.1038/mtna.2013.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Abu Lila A.S., Kiwada H., Ishida T. The accelerated blood clearance (ABC) phenomenon: clinical challenge and approaches to manage. J. Control. Release. 2013;172:38–47. doi: 10.1016/j.jconrel.2013.07.026. [DOI] [PubMed] [Google Scholar]
- 174.Thi T.T.H., Pilkington E.H., Nguyen D.H., Lee J.S., Park K.D., Truong N.P. The importance of poly(ethylene glycol) alternatives for overcoming PEG immunogenicity in drug delivery and bioconjugation. Polymers (Basel) 2020;12:298. doi: 10.3390/polym12020298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Zukancic D., Suys E.J.A., Pilkington E.H., Algarni A., Al-Wassiti H., Truong N.P. The importance of poly(ethylene glycol) and lipid structure in targeted gene delivery to lymph nodes by lipid nanoparticles. Pharmaceutics. 2020;12:1068. doi: 10.3390/pharmaceutics12111068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Bertrand N., Wu J., Xu X., Kamaly N., Farokhzad O.C. Cancer nanotechnology: the impact of passive and active targeting in the era of modern cancer biology. Adv. Drug. Deliv. Rev. 2014;66:2–25. doi: 10.1016/j.addr.2013.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Choi H.S., Liu W., Misra P., Tanaka E., Zimmer J.P., Itty Ipe B., Bawendi M.G., Frangioni J.V. Renal clearance of quantum dots. Nat. Biotechnol. 2007;25:1165–1170. doi: 10.1038/nbt1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Braet F., Wisse E., Bomans P., Frederik P., Geerts W., Koster A., Soon L., Ringer S. Contribution of high-resolution correlative imaging techniques in the study of the liver sieve in three-dimensions. Microsc. Res. Tech. 2007;70:230–242. doi: 10.1002/jemt.20408. [DOI] [PubMed] [Google Scholar]
- 179.Moghimi S.M., Hunter A.C., Andresen T.L. Factors controlling nanoparticle pharmacokinetics: an integrated analysis and perspective. Annu. Rev. Pharmacol. Toxicol. 2012;52:481–503. doi: 10.1146/annurev-pharmtox-010611-134623. [DOI] [PubMed] [Google Scholar]
- 180.Schudel A., Francis D.M., Thomas S.N. Material design for lymph node drug delivery. Nat. Rev. Mater. 2019;4:415–428. doi: 10.1038/s41578-019-0110-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Porter C.J.H., Trevaskis N.L. Targeting immune cells within lymph nodes. Nat. Nanotechnol. 2020;15:423–425. doi: 10.1038/s41565-020-0663-z. [DOI] [PubMed] [Google Scholar]
- 182.Schudel A., Chapman A.P., Yau M.-K., Higginson C.J., Francis D.M., Manspeaker M.P., Avecilla A.R.C., Rohner N.A., Finn M.G., Thomas S.N. Programmable multistage drug delivery to lymph nodes. Nat. Nanotechnol. 2020;15:491–499. doi: 10.1038/s41565-020-0679-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Gretz J.E., Norbury C.C., Anderson A.O., Proudfoot A.E.I., Shaw S. Lymph-borne chemokines and other low molecular weight molecules reach high endothelial venules via specialized conduits while a functional barrier limits access to the lymphocyte microenvironments in lymph node cortex. J. Exp. Med. 2000;192:1425–1440. doi: 10.1084/jem.192.10.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Swartz M.A. The physiology of the lymphatic system. Adv. Drug. Deliv. Rev. 2001;50:3–20. doi: 10.1016/s0169-409x(01)00150-8. [DOI] [PubMed] [Google Scholar]
- 185.Irvine D.J., Aung A., Silva M. Controlling timing and location in vaccines. Adv. Drug. Deliv. Rev. 2020;158:91–115. doi: 10.1016/j.addr.2020.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Thomas S.N., Rohner N.A., Edwards E.E. Implications of lymphatic transport to lymph nodes in immunity and immunotherapy. Annu. Rev. Biomed. Eng. 2016;18:207–233. doi: 10.1146/annurev-bioeng-101515-014413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Oliver G., Kipnis J., Randolph G.J., Harvey N.L. The lymphatic vasculature in the 21st century: novel functional roles in homeostasis and disease. Cell. 2020;182:270–296. doi: 10.1016/j.cell.2020.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Petrova T.V., Koh G.Y. Biological functions of lymphatic vessels. Science. 2020;369:eaax4063. doi: 10.1126/science.aax4063. [DOI] [PubMed] [Google Scholar]
- 189.Sixt M., Kanazawa N., Selg M., Samson T., Roos G., Reinhardt D.P., Pabst R., Lutz M.B., Sorokin L. The conduit system transports soluble antigens from the afferent lymph to resident dendritic cells in the T cell area of the lymph node. Immunity. 2005;22:19–29. doi: 10.1016/j.immuni.2004.11.013. [DOI] [PubMed] [Google Scholar]
- 190.Trevaskis N.L., Kaminskas L.M., Porter C.J.H. From sewer to saviour — targeting the lymphatic system to promote drug exposure and activity. Nat. Rev. Drug Discov. 2015;14:781–803. doi: 10.1038/nrd4608. [DOI] [PubMed] [Google Scholar]
- 191.Reddy S.T., Rehor A., Schmoekel H.G., Hubbell J.A., Swartz M.A. In vivo targeting of dendritic cells in lymph nodes with poly(propylene sulfide) nanoparticles. J. Control. Release. 2006;112:26–34. doi: 10.1016/j.jconrel.2006.01.006. [DOI] [PubMed] [Google Scholar]
- 192.Liu H., Moynihan K.D., Zheng Y., Szeto G.L., Li A.V., Huang B., Van Egeren D.S., Park C., Irvine D.J. Structure-based programming of lymph-node targeting in molecular vaccines. Nature. 2014;507:519–522. doi: 10.1038/nature12978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Rincon-Restrepo M., Mayer A., Hauert S., Bonner D.K., Phelps E.A., Hubbell J.A., Swartz M.A., Hirosue S. Vaccine nanocarriers: coupling intracellular pathways and cellular biodistribution to control CD4 vs CD8 T cell responses. Biomaterials. 2017;132:48–58. doi: 10.1016/j.biomaterials.2017.03.047. [DOI] [PubMed] [Google Scholar]
- 194.Nakamura A., Harashima H. Dawn of lipid nanoparticles in lymph node targeting: potential in cancer immunotherapy. Adv. Drug. Deliv. Rev. 2020;167:78–88. doi: 10.1016/j.addr.2020.06.003. [DOI] [PubMed] [Google Scholar]
- 195.Feeney O.M., Crum M.F., McEvoy C.L., Trevaskis N.L., Williams H.D., Pouton C.W., Charman W.N., Bergström C.A.S., Porter C.J.H. 50 years of oral lipid-based formulations: provenance, progress and future perspectives. Adv. Drug. Deliv. Rev. 2016;101:167–194. doi: 10.1016/j.addr.2016.04.007. [DOI] [PubMed] [Google Scholar]
- 196.Rao D.A., Forrest M.L., Alani A.W.G., Kwon G.S., Robinson J.R. Biodegradable PLGA based nanoparticles for sustained regional lymphatic drug delivery. J. Pharm. Sci. 2010;99:2018–2031. doi: 10.1002/jps.21970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Manolova V., Flace A., Bauer M., Schwarz K., Saudan P., Bachmann M.F. Nanoparticles target distinct dendritic cell populations according to their size. Eur. J. Immunol. 2008;38:1404–1413. doi: 10.1002/eji.200737984. [DOI] [PubMed] [Google Scholar]
- 198.Wang Y.Y., Lai S.K., Suk J.S., Pace A., Cone R., Hanes J. Addressing the PEG mucoadhesivity paradox to engineer nanoparticles that “slip” through the human mucus barrier. Adv. Drug. Deliv. Rev. 2008;47:9726–9729. doi: 10.1002/anie.200803526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Cu Y., Saltzman W.M. Stealth particles give mucus the slip. Nat. Mater. 2009;8:11–13. doi: 10.1038/nmat2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Nakamura T., Kawai M., Sato Y., Maeki M., Tokeshi M., Harashima H. The effect of size and charge of lipid nanoparticles prepared by microfluidic mixing on their lymph node transitivity and distribution. Mol. Pharm. 2020;17:944–953. doi: 10.1021/acs.molpharmaceut.9b01182. [DOI] [PubMed] [Google Scholar]
- 201.Rosigkeit S., Meng M., Grunwitz C., Gomes P., Kreft A., Hayduk N., Heck R., Pickert G., Ziegler K., Abassi Y., Röder J., Kaps L., Vascotto F., Beissert T., Witzel S., Kuhn A., Diken M., Schuppan D., Sahin U., Haas H., Bockamp E. Monitoring translation activity of mRNA-loaded nanoparticles in mice. Mol. Pharm. 2018;15:3909–3919. doi: 10.1021/acs.molpharmaceut.8b00370. [DOI] [PubMed] [Google Scholar]
- 202.Levchenko T.S., Rammohan R., Lukyanov A.N., Whiteman K.R., Torchilin V.P. Liposome clearance in mice: the effect of a separate and combined presence of surface charge and polymer coating. Int. J. Pharm. 2002;240:95–102. doi: 10.1016/s0378-5173(02)00129-1. [DOI] [PubMed] [Google Scholar]
- 203.Owens D.E., Peppas N.A. Opsonization, biodistribution, and pharmacokinetics of polymeric nanoparticles. Int. J. Pharm. 2006;307:93–102. doi: 10.1016/j.ijpharm.2005.10.010. [DOI] [PubMed] [Google Scholar]
- 204.Monopoli M.P., Walczyk D., Campbell A., Elia G., Lynch I., Baldelli Bombelli F., Dawson K.A. Physical−chemical aspects of protein corona: relevance to in vitro and in vivo biological impacts of nanoparticles. J. Am. Chem. Soc. 2011;133:2525–2534. doi: 10.1021/ja107583h. [DOI] [PubMed] [Google Scholar]
- 205.Sato Y., Kinami Y., Hashiba K., Harashima H. Different kinetics for the hepatic uptake of lipid nanoparticles between the apolipoprotein E/low density lipoprotein receptor and the N-acetyl-d-galactosamine/asialoglycoprotein receptor pathway. J. Control. Release. 2020;322:217–226. doi: 10.1016/j.jconrel.2020.03.006. [DOI] [PubMed] [Google Scholar]
- 206.Miao L., Lin J., Huang Y., Li L., Delcassian D., Ge Y., Shi Y., Anderson D.G. Synergistic lipid compositions for albumin receptor mediated delivery of mRNA to the liver. Nat. Commun. 2020;11:2424. doi: 10.1038/s41467-020-16248-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Yang K., Mesquita B., Horvatovich P., Salvati A. Tuning liposome composition to modulate corona formation in human serum and cellular uptake. Acta Biomater. 2020;106:314–327. doi: 10.1016/j.actbio.2020.02.018. [DOI] [PubMed] [Google Scholar]
- 208.Li Q., Chan C., Peterson N., Hanna R.N., Alfaro A., Allen K.L., Wu H., Dall’Acqua W.F., Borrok M.J., Santos J.L. Engineering Caveolae-targeted lipid nanoparticles to deliver mRNA to the lungs. ACS Chem. Biol. 2020;15:830–836. doi: 10.1021/acschembio.0c00003. [DOI] [PubMed] [Google Scholar]
- 209.Myerson J.W., Braender B., Mcpherson O., Glassman P.M., Kiseleva R.Y., Shuvaev V.V., Marcos-Contreras O., Grady M.E., Lee H.-S., Geineder C.F., Stan R.V., Composto R.J., Eckmann D.M., Muzykantov V.R. Flexible nanoparticles reach sterically obscured endothelial targets inaccessible to rigid nanoparticles. Adv. Mater. 2018;30 doi: 10.1002/adma.201802373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Martinez-Pomares L. The mannose receptor. J. Leukoc. Biol. 2012;92:1177–1186. doi: 10.1189/jlb.0512231. [DOI] [PubMed] [Google Scholar]
- 211.Geijtenbeek T.B.H., Torensma R., van Vliet S.J., Dujinhoven G.C.F., Adema G.J., van Kooyk Y., Figdor C.G. Identification of DC-SIGN, a novel dendritic cell–specific ICAM-3 receptor that supports primary immune responses. Cell. 2000;100:575–585. doi: 10.1016/s0092-8674(00)80693-5. [DOI] [PubMed] [Google Scholar]
- 212.Jiang W., Swiggard W.J., Heufler C., Peng M., Mirza A., Steinman R.M., Nussenzweig M.C. The receptor DEC-205 expressed by dendritic cells and thymic epithelial cells is involved in antigen processing. Nature. 1995;375:151–155. doi: 10.1038/375151a0. [DOI] [PubMed] [Google Scholar]
- 213.Valladeau J., Ravel O., Dezutter-Dambuyant C., Moore K., Kleijmeer M., Liu Y., Duvert-Frances V., Vincent C., Schmitt D., Davoust J., Caux C., Lebecque S., Saeland S. Langerin, a novel C-type lectin specific to langerhans cells, is an endocytic receptor that induces the formation of birbeck granules. Immunity. 2000;12:71–81. doi: 10.1016/s1074-7613(00)80160-0. [DOI] [PubMed] [Google Scholar]
- 214.Phua K.K.L. Towards targeted delivery systems: ligand conjugation strategies for mrna nanoparticle tumor vaccines. J. Immunol. Res. 2015 doi: 10.1155/2015/680620. 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Katakowski J.A., Mukherjee G., Wilner S.E., Maier K.E., Harrison M.T., DiLorenzo T.P., Levy M., Palliser D. Delivery of siRNAs to dendritic cells using DEC205-targeted lipid nanoparticles to inhibit immune responses. Mol. Ther. 2016;24:146–155. doi: 10.1038/mt.2015.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Wang F., Xiao W., Elbahnasawy M.A., Bao X., Zheng Q., Gong L., Zhou Y., Yang S., Fang A., Farag M.M.S., Wu J., Song X. Optimization of the linker length of mannose-cholesterol conjugates for enhanced mRNA delivery to dendritic cells by liposomes. Front. Pharmacol. 2018;9:980. doi: 10.3389/fphar.2018.00980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Perche F., Benvegnu T., Berchel M., Lebegue L., Pichon C., Jaffrès P.A., Midoux P. Enhancement of dendritic cells transfection in vivo and of vaccination against B16F10 melanoma with mannosylated histidylated lipopolyplexes loaded with tumor antigen messenger RNA. Nanomedicine. 2011;7:445–453. doi: 10.1016/j.nano.2010.12.010. [DOI] [PubMed] [Google Scholar]
- 218.Zhuang X., Qi Y., Wang M., Yu N., Nan F., Zhang H., Tian M., Li C., Lu H., Jin N. mRNA vaccines encoding the HA protein of Influenza A H1N1 virus delivered by cationic lipid nanoparticles induce protective immune responses in mice. Vaccines. 2020;8:123. doi: 10.3390/vaccines8010123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Kedmi R., Veiga N., Ramishetti S., Goldsmith M., Rosenblum D., Dammes N., Hazan-Halevy I., Nahary L., Leviatan-Ben-Arye S., Harlev M., Behlke M., Benhar I., Lieberman J., Peer D. A modular platform for targeted RNAi therapeutics. Nat. Nanotechnol. 2018;13:214–219. doi: 10.1038/s41565-017-0043-5. [DOI] [PubMed] [Google Scholar]
- 220.Veiga N., Goldsmith G., Granot Y., Rosenblum D., Dammes N., Kedmi R., Ramishetti S., Peer D. Cell specific delivery of modified mRNA expressing therapeutic proteins to leukocytes. Nat. Commun. 2018;9:4493. doi: 10.1038/s41467-018-06936-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Yang S., Zhu J., Lu Y., Liang B., Yang C. Body distribution of camptothecin solid lipid nanoparticles after oral administration. Pharm. Res. 1999;16:751–757. doi: 10.1023/a:1018888927852. [DOI] [PubMed] [Google Scholar]
- 222.Pandey R., Sharma S., Khuller G.K. Oral solid lipid nanoparticle-based antitubercular chemotherapy. Tuberculosis. 2005;85:415–420. doi: 10.1016/j.tube.2005.08.009. [DOI] [PubMed] [Google Scholar]
- 223.Cavalli R., Gasco M.R., Chetoni P., Burgalassi S., Saettone M.F. Solid lipid nanoparticles (SLN) as ocular delivery system for tobramycin. Int. J. Pharm. 2002;238:241–245. doi: 10.1016/s0378-5173(02)00080-7. [DOI] [PubMed] [Google Scholar]
- 224.Friedrich I., Reichl S., Müller-Goymann C.C. Drug release and permeation studies of nanosuspensions based on solidified reverse micellar solutions (SRMS) Int. J. Pharm. 2005;305:167–175. doi: 10.1016/j.ijpharm.2005.09.007. [DOI] [PubMed] [Google Scholar]
- 225.de Vringer T., de Tonde H.A.G. Preparation and structure of a water-in-oil cream containing lipid nanoparticles. J. Pharm. Sci. 1994;84:466–472. doi: 10.1002/jps.2600840415. [DOI] [PubMed] [Google Scholar]
- 226.Dingler A., Blum R.P., Niehus H., Muller R.H., Gohla S. Solid lipid nanoparticles (SLNTM/LipopearlsTM) a pharmaceutical and cosmetic carrier for the application of vitamin E in dermal products. J. Microencapsul. 1999;16:751–767. doi: 10.1080/026520499288690. [DOI] [PubMed] [Google Scholar]
- 227.Lippacher A., Müller R.H., Mäder K. Semisolid SLNTM dispersions for topical application: influence of formulation and production parameters on viscoelastic properties. Eur. J. Pharm. Biopharm. 2002;53:155–160. doi: 10.1016/s0939-6411(01)00233-8. [DOI] [PubMed] [Google Scholar]
- 228.Sznitowska M., Gajewska M., Janicki S., Radwanska A., Lukowski G. Bioavailability of diazepam from aqueous-organic solution, submicron emulsion and solid lipid nanoparticles after rectal administration in rabbits. Eur. J. Pharm. Biopharm. 2001;52:159–163. doi: 10.1016/s0939-6411(01)00157-6. [DOI] [PubMed] [Google Scholar]
- 229.Chen S., Zaifman J., Kulkarni J.A., Zhigaltsev I.V., Tam Y.K., Ciufolini M.A., Tam Y.Y.C., Cullis P.R. Dexamethasone prodrugs as potent suppressors of the immunostimulatory effects of lipid nanoparticle formulations of nucleic acids. J. Control. Release. 2018;286:46–54. doi: 10.1016/j.jconrel.2018.07.026. [DOI] [PubMed] [Google Scholar]
- 230.Zuckerman J.N. The importance of injecting vaccines into muscle. BMJ. 2000;321:1237–1238. doi: 10.1136/bmj.321.7271.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Davies N., Hovdal D., Edmunds N., Nordberg P., Dahlén A., Dabkowska A., Arteta M.Y., Radulescu A., Kjellman T., Höijer A., Seeliger F., Holmedal E., Andihn E., Bergenhem N., Sandinge A.-S., Johansson C., Hultin L., Johansson M., Lindqvist J., Björsson L., Jing Y., Bartesaghi S., Lindfors L., Andersson S. Functionalized lipid nanoparticles for subcutaneous administration of mRNA to achieve systemic exposures of a therapeutic protein. Mol. Ther. Nucleic Acids. 2021;24:369–384. doi: 10.1016/j.omtn.2021.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Pardi N., Tuyishime S., Muramatsu H., Kariko K., Mui B.L., Tam Y.K., Madden T.D., Hope M.J., Weissman D. Expression kinetics of nucleoside-modified mRNA delivered in lipid nanoparticles to mice by various routes. J. Control. Release. 2015;217:345–351. doi: 10.1016/j.jconrel.2015.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Yang S.C., Lu L.F., Cai Y., Zhu J.B., Liang B.W., Yang C.Z. Body distribution in mice of intravenously injected camptothecin solid lipid nanoparticles and targeting effect on brain. J. Control. Release. 1999;59:299–307. doi: 10.1016/s0168-3659(99)00007-3. [DOI] [PubMed] [Google Scholar]
- 234.Wissing S.A., Kayser O., Müller R.H. Solid lipid nanoparticles for parenteral drug delivery. Adv. Drug. Deliv. Rev. 2004;56:1257–1272. doi: 10.1016/j.addr.2003.12.002. [DOI] [PubMed] [Google Scholar]
- 235.Kenney R.T., Frech S.A., Muenz L.R., Villar C.P., Glenn G.M. Dose sparing with intradermal injection of influenza vaccine. N. Engl. J. Med. 2004;351:2295–2301. doi: 10.1056/NEJMoa043540. [DOI] [PubMed] [Google Scholar]
- 236.Lindgren G., Ols S., Liang F., Thompson E.A., Lin A., Hellgren F., Bahl K., John S., Yuzhakov O., Hassett K.J., Brito L.A., Salter H., Ciaramella G., Loré K. Induction of robust B cell responses after Influenza mRNA vaccination is accompanied by circulating hemagglutinin-specific ICOS+ PD-1+ CXCR3+ T follicular helper cells. Front. Immunol. 2017;8:1539. doi: 10.3389/fimmu.2017.01539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Liang F., Lindgren G., Lin A., Thompson E.A., Ols S., Röhss J., John S., Hassett K.J., Olga Y., Bahl K., Brito L.A., Salter H., Ciaramella G., Loré K. Efficient targeting and activation of antigen-presenting cells in vivo after modified mRNA vaccine administration in rhesus macaques. Mol. Ther. 2017;25:2635–2647. doi: 10.1016/j.ymthe.2017.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Swaminathan G., Thoryk E.A., Cox K.S., Meschino S., Cubey S.A., Vora K.A., Celano R., Gindy M., Casimiro D.R., Bett A.J. A novel lipid nanoparticle adjuvant significantly enhances B cell and T cell responses to sub-unit vaccine antigens. Vaccine. 2016;34:110–119. doi: 10.1016/j.vaccine.2015.10.132. [DOI] [PubMed] [Google Scholar]
- 239.Gilleron J., Querbes W., Zeigerer A., Borodovsky A., Marsico G., Schubert U., Manygoats K., Seifert S., Andree C., Stöter M., Epstein-Barash H., Zhang L., Koteliansky V., Fitzgerald K., Fava E., Bickle M., Kalaidzidis Y., Akinc A., Maier M.A., Zerial M. Image-based analysis of lipid nanoparticle–mediated siRNA delivery, intracellular trafficking and endosomal escape. Nat. Biotechnol. 2013;31:638–646. doi: 10.1038/nbt.2612. [DOI] [PubMed] [Google Scholar]
- 240.Maugeri M., Nawaz M., Papadimitriou A., Angerfors A., Camponeschi A., Na M., Hölttä M., Skantze P., Johansson S., Sundqvist M., Lindquist J., Kjellman T., Martensson I.L., Jin T., Sunnerhagen P., Östman S., Lindfors L., Valadi H. Linkage between endosomal escape of LNP-mRNA and loading into EVs for transport to other cells. Nat. Commun. 2019;10:4333. doi: 10.1038/s41467-019-12275-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Huotari J., Helenius A. Endosome maturation. EMBO J. 2011;30:3481–3500. doi: 10.1038/emboj.2011.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Juliano R.L. The delivery of therapeutic oligonucleotides. Nucleic. Acids. Res. 2016;44:6518–6548. doi: 10.1093/nar/gkw236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Blakney A.K., McKay P.F., Yus B.I., Aldon Y., Shattock R.J. Inside out: optimization of lipid nanoparticle formulations for exterior complexation and in vivo delivery of saRNA. Gene Ther. 2019;26:363–372. doi: 10.1038/s41434-019-0095-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.McKay P.F., Hu K., Blakney A.K., Samnuan K., Brown J.C., Penn R., Zhou J., Bouton C.R., Rogers P., Polra K., Lin P.J.C., Barbosa C., Tam Y.K., Barclay W.S., Shattock R.J. Self-amplifying RNA SARS-CoV-2 lipid nanoparticle vaccine candidate induces high neutralizing antibody titers in mice. Nat. Commun. 2020;11:3523. doi: 10.1038/s41467-020-17409-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Kim E., Erdos G., Huang S., Kenniston T.W., Balmert S.C., Carey C.D., Raj V.S., Epperly M.W., Klimstra W.B., Haagmans B.L., Korkmaz E., Falo L.D., Gambotto A. Microneedle array delivered recombinant coronavirus vaccines: immunogenicity and rapid translational development. EBioMedicine. 2020;55 doi: 10.1016/j.ebiom.2020.102743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Beissert T., Perkovic M., Vogel A., Erbar S., Walzer K.C., H. T., Brill S., Haefner E., Becker R., Türeci Ö., Sahin U. A Trans-amplifying RNA vaccine strategy for induction of potent protective immunity. Mol. Ther. 2020;28:119–128. doi: 10.1016/j.ymthe.2019.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Bloom K., van den Berg F., Arbuthnot P. Self-amplifying RNA vaccines for infectious diseases. Gene Ther. 2020;28:117–129. doi: 10.1038/s41434-020-00204-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Weide B., Carralot J.-P., Reese A., Scheel B., Eigentler T.K., Ingmar H., Rammensee H.-G., Garbe C., Pascolo S. Results of the first phase I/II clinical vaccination trial with direct injection of mRNA. J. Immunother. 2008;31:180–188. doi: 10.1097/CJI.0b013e31815ce501. [DOI] [PubMed] [Google Scholar]
- 249.Wong C.H., Siah K.W., Lo A.W. Estimation of clinical trial success rates and related parameters. Biostatistics. 2019;20:273–286. doi: 10.1093/biostatistics/kxx069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Moderna Inc, Moderna announces positive phase 1 results for the first systemic messenger RNA therapeutic encoding a secreted protein (mRNA-1944), https://investors.modernatx.com/news-releases/news-release-details/moderna-announces-positive-phase-1-results-first-systemic, 2019 (Accessed 13 May 2021).
- 251.Moderna Inc, Moderna announces positive interim results from phase 1 cytomegalovirus (CMV) vaccine (mRNA-1647) study and progress toward phase 2 and pivotal trials, https://investors.modernatx.com/news-releases/news-release-details/moderna-announces-positive-interim-results-phase-1, 2019 (Accessed 13 May 2021).
- 252.Moderna Inc, Moderna announces additional positive phase 1 data from cytomegalovirus (CMV) vaccine (mRNA-1647) and first participant dosed in phase 2 study, https://investors.modernatx.com/news-releases/news-release-details/moderna-announces-additional-positive-phase-1-data, 2020 (accessed 19 February 2021).
- 253.Moderna Inc, Moderna announces positive interim phase 1 data for first combination vaccine against the respiratory viruses hMPV and PIV3, https://investors.modernatx.com/news-releases/news-release-details/moderna-announces-positive-interim-phase-1-data-first, 2019 (Accessed 13 May 2021).
- 254.Feldman R.A., Fuhr R., Smolenov I., Ribeiro A., Panther L., Watson M., Senn J.J., Smith M., Almarsson O., Pujar H.S., Laska M.E., Thompson J., Zaks T., Ciaramella G. mRNA vaccines against H10N8 and H7N9 influenza viruses of pandemic potential are immunogenic and well tolerated in healthy adults in phase 1 randomized clinical trials. Vaccine. 2019;37:3326–3334. doi: 10.1016/j.vaccine.2019.04.074. [DOI] [PubMed] [Google Scholar]
- 255.Aliprantis A.O., Shaw C.A., Griffin P., Farinola N., Railkar R.A., Cao X., Liu W., Sachs J.R., Swenson C.J., Lee H., Cox K.S., Spellman D.S., Winstead C.J., Smolenov I., Lai E., Zaks T., Espeseth A.S., Panther L. A phase 1, randomized, placebo-controlled study to evaluate the safety and immunogenicity of an mRNA-based RSV prefusion F protein vaccine in healthy younger and older adults. Hum. Vaccines Immunother. 2021;17:1248–1261. doi: 10.1080/21645515.2020.1829899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Moderna Inc, Moderna announces clinical progress from its industry-leading mRNA vaccine franchise and continues investments to accelerate pipeline development, https://investors.modernatx.com/news-releases/news-release-details/moderna-announces-clinical-progress-its-industry-leading-mrna, 2021 (Accessed 13 May 2021).
- 257.Aldrich C., Leroux-Roels I., Huang K.B., Bica M.A., Loeliger E., Schoenborn-Kellenberger O., Walz L., Leroux-Roels G., von Sonnenburg F., Oostvogels L. Proof-of-concept of a low-dose unmodified mRNA-based rabies vaccine formulated with lipid nanoparticles in human volunteers: a phase 1 trial. Vaccine. 2021;39:1310–1318. doi: 10.1016/j.vaccine.2020.12.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.-L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.BioNTech SE, Pfizer Inc, Pfizer and BioNTech achieve first authorization in the world for a vaccine to combat COVID-19, in, https://investors.biontech.de/news-releases/news-release-details/pfizer-and-biontech-achieve-first-authorization-world-vaccine, 2020 (Accessed 13 May 2021).
- 260.Arcturus Therapeutics, Arcturus therapeutics received approval from Singapore Health Sciences Authority to proceed with phase 2 study of ARCT-021 (LUNAR-COV19) vaccine candidate and provides new and updated clinical and preclinical data, https://www.businesswire.com/news/home/20201228005277/en/, 2020 (Accessed 13 May 2021).
- 261.Baden L.R., El Sahly H.M., Essink B., Kotloff K., Frey S., Novak R., Diemert D., Spector S.A., Rouphael N., Creech C.B., McGettigan J., Khetan S., Segall N., Solis J., Brosz A., Fierro C., Schwartz H., Neuzil K., Corey L., Gilbert P., Janes H., Follmann D., Marovich M., Mascola J., Polakowski L., Ledgerwood J., Graham B.S., Bennett H., Pajon R., Knightly C., Leav B., Deng W., Zhou H., Han S., Ivarsson M., Miller J., Zaks T. Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. N. Engl. J. Med. 2020;384:403–416. doi: 10.1056/NEJMoa2035389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Polack F.P., Thomas S.J., Kitchin N., Absalon J., Gurtman A., Lockhart S., Perez J.L., Marc G.P., Moreira E.D., Zerbini C., Bailey R., Swanson K.A., Roychoudhury S., Koury K., Li P., Kalina W.V., Cooper D., Frenck R.W., Hammitt L.L., Türeci O., Nell H., Schaefer A., Ünal S., Tresnan D.B., Mather S., Dormitzer P.R., Şahin U., Jansen K.U., Gruber W.C. Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. N. Engl. J. Med. 2020;383:2603–2615. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.National Center for Immunization & Respiratory Diseases, COVID-19 vaccine safety update, https://www.cdc.gov/vaccines/acip/meetings/downloads/slides-2021-01/06-COVID-Shimabukuro.pdf, 2021 (Accessed 13 May 2021).
- 264.National Institutes of Health, NIH begins study of allergic reactions to Moderna, Pfizer-BioNTech COVID-19 vaccines, https://www.nih.gov/news-events/news-releases/nih-begins-study-allergic-reactions-moderna-pfizer-biontech-covid-19-vaccines, 2021 (Accessed 13 May 2021).
- 265.A. Muik, A.-K. Wallisch, B. Sänger, K.A. Swanson, J. Mühl, W. Chen, H. Cai, R. Sarkar, O. Türeci, P.R. Dormitzer, U. Sahin, Neutralization of SARS-CoV-2 lineage B.1.1.7 pseudovirus by BNT162b2 vaccine-elicited human sera, bioRxiv, (2021) doi: 10.1101/2021.01.18.426984. [DOI] [PMC free article] [PubMed]
- 266.X. Xie, J. Zou, C.R. Fontes-Garfias, H. Xia, K.A. Swanson, M. Cutler, D. Cooper, V.D. Menachery, S. Weaver, P.R. Dormitzer, P.-Y. Shi, Neutralization of N501Y mutant SARS-CoV-2 by BNT162b2 vaccine-elicited sera, bioRxiv, (2021) doi: 10.1101/2021.01.18.426984. [DOI] [PubMed]
- 267.K. Wu, A.P. Werner, J.I. Moliva, M. Koch, A. Choi, G.B.E. Stewart-Jones, H. Bennett, S. Boyoglu-Barnum, W. Shi, B.S. Graham, A. Carfi, K.S. Corbett, R.A. Seder, D.K. Edwards, mRNA-1273 vaccine induces neutralizing antibodies against spike mutants from global SARS-CoV-2 variants, bioRxiv, (2021) doi: 10.1101/2021.01.25.427948. [DOI]
- 268.T. Tada, B.M. Dcosta, M. Samanovic-Golden, R.S. Herati, A. Cornelius, M.J. Mulligan, N.R. Landau, Neutralization of viruses with European, South African, and United States SARS-CoV-2 variant spike proteins by convalescent sera and BNT162b2 mRNA vaccine-elicited antibodies, bioRxiv, (2021) doi: 10.1101/2021.02.05.430003. [DOI]
- 269.Kuzmina A., Khalaila Y., Keren-Naus A., Boehm-Cohen L., Raviv Y., Shemer-Avni Y., Rosenberg E., Taube R. SARS-CoV-2 spike variants exhibit differential infectivity and neutralization resistance to convalescent or post-vaccination sera. Cell. Host. Microbe. 2021;29:522–528. doi: 10.1016/j.chom.2021.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Xie X., Liu Y., Liu J., Zhang X., Zou J., Fontes-Garfias C.R., Xia H., Swanson K.A., Cutler M., Cooper D., Menachery V.D., Weaver S.C., Dormitzer P.R., Shi P.-Y. Neutralization of SARS-CoV-2 spike 69/70 deletion, E484K and N501Y variants by BNT162b2 vaccine-elicited sera. Nat. Med. 2021;27:620–621. doi: 10.1038/s41591-021-01270-4. [DOI] [PubMed] [Google Scholar]
- 271.Collier D.A., De Marco A., Ferreira I.A.T.M., Meng B., Datir R.P., Walls A.C., Kemp S.A., Bassi J., Pinto D., Silacci-Fregni C., Bianchi S., Tortorici M.A., Bowen J., Culap K., Jaconi S., Cameroni E., Snell G., Pizzuto M.S., Pellanda A.F., Garzoni C., Riva A. The CITIID-NIHR BioResource COVID-19 Collaboration. Nature. 2021;593:136–141. Sensitivity of SARS-CoV-2 B1.1.7 to mRNA vaccine-elicited antibodies. [Google Scholar]
- 272.T. Kustin, N. Harel, U. Finkel, S. Perchik, S. Harari, M. Tahor, I. Caspi, R. Levy, M. Leschinsky, S.K. Dror, G. Bergerzon, H. Gadban, F. Gadban, E. Eliassian, O. Shimron, L. Saleh, H. Ben-Zvi, D. Amichay, A. Ben-Dor, D. Sagas, M. Strauss, Y.S. Avni, A. Huppert, E. Kepten, R.D. Balicer, D. Nezer, S. Ben-Shachar, Evidence for increased breakthrough rates of SARS-CoV-2 variants of concern in BNT162b2 mRNA vaccinated individuals, medRxiv, (2021) doi: 10.1101/2021.04.06.21254882. [DOI] [PMC free article] [PubMed]
- 273.Abu-Raddad L.J., Chemaitelly H., Butt A.A. Effectiveness of the BNT162b2 Covid-19 vaccine against the B.1.1.7 and B.1.351 variants. New Engl. J. Med. 2021 doi: 10.1056/NEJMc2104974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Moderna Inc, Moderna announces positive initial booster data against SARS-CoV-2 variants of concern, https://investors.modernatx.com/news-releases/news-release-details/moderna-announces-positive-initial-booster-data-against-sars-cov, 2021 (Accessed 13 May 2021).
- 275.Moderna Inc, First participants dosed in phase 1 study evaluating mRNA-1283, Moderna's next generation COVID-19 vaccine, https://investors.modernatx.com/news-releases/news-release-details/first-participants-dosed-phase-1-study-evaluating-mrna-1283, 2021 (Accessed 13 May 2021).
- 276.Thielmann A., Puth M.-T., Weltermann B. Visual inspection of vaccine storage conditions in general practices: a study of 75 vaccine refrigerators. PLoS ONE. 2019;14 doi: 10.1371/journal.pone.0225764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Zhang N.-N., Li X.-F., Deng Y.-Q., Zhao H., Huang Y.-J., Yang G., Huang W.-J., Gao P., Zhou C., Zhang R.-R., Guo Y., Sun S.-H., Hang F., Zu S.-L., Chen Q., He Q., Cao T.-S., Huang X.-Y., Qiu H.-Y., Nie J.-H., Jiang Y., Yan H.-Y., Ye Q., Zhong X., Xue X.-L., Zha Z.-Y., Zhou D., Yang X., Wang Y.-C., Ying B., Qin C.-F. A thermostable mRNA vaccine against COVID-19. Cell. 2020;182:1271–1283. doi: 10.1016/j.cell.2020.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Pronker E.S., Weenen T.C., Commandeur H., Claassen E.H.J.H.M., Osterhaus A.D.M.E. Risk in vaccine research and development quantified. PLoS ONE. 2013;8:e57755. doi: 10.1371/journal.pone.0057755. [DOI] [PMC free article] [PubMed] [Google Scholar]