Abstract
Presently, immunoinformatics and bioinformatics approaches are contributing actively to COVID-19 vaccine research. The first immunoinformatics-based vaccine construct against SARS-CoV-2 was published in February 2020. Following this, immunoinformatics and bioinformatics approaches have created a new direction in COVID-19 vaccine research. Several researchers have designed the next-generation COVID-19 vaccines using these approaches. Presently, immunoinformatics has accelerated immunology research immensely in the area of COVID-19. Hence, we have tried to depict the current scenario of immunoinformatics and bioinformatics in COVID-19 vaccine research.
Supplementary Information
The online version contains supplementary material available at 10.1007/s10989-021-10254-4.
Keywords: Immunoinformatics, Bioinformatics, COVID-19 vaccine research, Vaccinogenomics
The COVID-19 vaccines have rolled out worldwide, and the vaccination program has started in different countries. More than 13 approved vaccine candidates are being used throughout the world for the mass vaccination program. Among them, Pfizer (BioNTech mRNA vaccine: BNT162b2) and ModernaTX mRNA vaccine (mRNA-1273) are the first approved vaccines, which have shown excellent efficacy (95% and 94.1%, respectively) (Chakraborty et al. 2021a, b). These vaccines are capable of reducing COVID-19 infection. However, DNA-based (Ad5-nCoV) and peptide-based (EpiVacCorona) vaccines are also being used for vaccination (Table S1). Most of the vaccines are based on viral S (Spike) protein as the vital vaccine antigen. If we look back at the COVID-19 vaccine research scenario, the first vaccine research against SARS-CoV-2 was initiated using immunoinformatics.
The first vaccine construct of the SARS-CoV-2 was reported in the Journal of Medical Virology on 28 February 2020 online (Bhattacharya et al. 2020a). Chakraborty and his colleagues are the first group of researchers who have developed a next-generation epitope-based peptide vaccine construct, and the vaccine construct was generated through immunoinformatics. Moreover, Chakraborty and his colleagues analyzed this vaccine's stability, safety, and efficacy through immunoinformatics, showing that this next-generation vaccine candidate is safe and immunogenic (Bhattacharya et al. 2020b). Likewise, some vaccine development companies have used immunoinformatic techniques to search for the most antigenic epitope for the vaccine candidate development.
After the beginning of COVID-19 in December 2019 in China, WHO declared a health emergency on 30 January 2020. Since then, researchers have intensified the search for therapeutics against SARS-CoV-2 (Baden and Rubin 2020). Several clinical trials have been performed in this direction, where more than 100 countries have participated. A report shows that 3754 clinical trials had been completed for COVID-19. It has been noted that some of these clinical trial results had not been updated in trial repositories (Rodgers et al. 2021). Quite a few therapeutics have given better results in clinical trials for severe COVID-19 patients until today. Some therapeutic molecules have proven helpful for the treatment of COVID-19, which includes remdesivir (an antiviral molecule), baricitinib (an immunosuppressive molecule), dexamethasone (an immunosuppressive molecule), and some monoclonal antibodies (Collins 2021). At first, most researchers tried to search for therapeutics by repurposing existing drugs. However, selected drugs have not provided accurate and successful outcomes. Therefore, the only way to stop the pandemic is to vaccinate the people to develop immunity against COVID-19 by using approved vaccines. Presently new SARS-CoV-2 variant (VOC; variants of concern and VOI: variants of interest) are a concern for the whole world. The vaccine candidate using alternative multi-epitopes for Wuhan strain and significant variant can be a solution (Bhattacharya et al. 2021). Collectively, it has been well accepted that the vaccine is the only effective option to stop this pandemic situation.
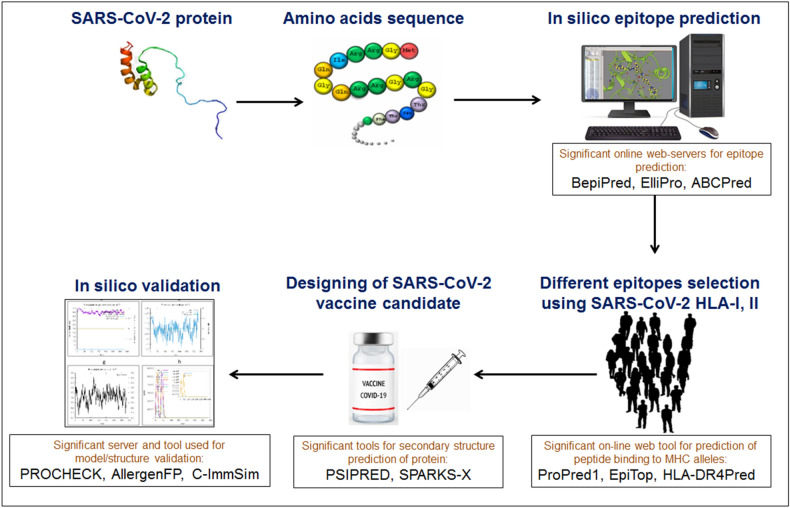
Immunoinformatics and bioinformatics have a significant role in COVID-19 vaccine research, especially in antigenic epitopes selection and vaccine construct development (Fig. 1). Bioinformatics, immunoinformatics, vaccinogenomics, structural biology, and molecular dynamics simulations have contributed significantly to COVID-19 vaccine research. It was observed that several vaccine constructs were developed using immunoinformatics and bioinformatics. We performed PubMed search and found that approximately 24 vaccine constructs have been developed through immunoinformatics and bioinformatics to date (Table 1). Simultaneously, several scientists identified T cell epitopes, B cell epitopes, and common T and B cell epitopes (Table 2) (Chakraborty et al. 2021c). The selected epitopes have suggested that the identified common epitopes can be used for vaccine construct development. However, the researchers did not further analyze the identified epitopes to develop vaccine constructs, having several essential parameters like allergenicity and immunogenicity, utilizing immunoinformatics and bioinformatics.
Fig. 1.
Schematic representation showing a flowchart of next-generation COVID-19 vaccine development through immunoinformatics. We have highlighted different tools, databases and servers which are using by the researchers for the vaccine development through immunoinformatics
Table 1.
Different immunoinformatics and bioinformatics research on next generation vaccine construct development against SARS-CoV-2
| Sl. no. | Researcher | Country | Nos. epitopes | Contributing viral proteins | Remarks | References |
|---|---|---|---|---|---|---|
| 1. | Bhattacharya M., et al., 2020 | India, South Korea | 19 epitopes | Spike glycoprotein | Peptide-based multi-epitopic vaccine contrast from S-protein | Bhattacharya et al. (2020a) |
| 2. | Kalita P., et al., 2020 | India, Japan | 33 epitopes | Nucleocapsid protein, membrane glycoprotein, surface spike glycoprotein | Multi-epitopic peptide-based subunit vaccine designed | Kalita et al. (2020) |
| 3. | Qamar M., et al., 2020 | China, Pakistan | 27 epitopes | Envelope protein, membrane glycoprotein, nucleocapsid protein | Designed a 505 amino acids containing effective multi-epitope vaccine | ul Qamar et al. (2020) |
| 4. | Saha R., et al., 2021 | India | 16 epitopes | Spike glycoprotein | B cell-derived T cell epitopes peptide based vaccine construct | Saha et al. (2021) |
| 5. | Yazdani Z., et al., 2020 | Iran | 6 epitopes | Spike glycoprotein, membrane glycoprotein, nucleocapsid phosphoprotein, envelope protein | Vaccine construct consists of immunodominant multi-epitopes from viral structural proteins | Yazdani et al. (2020) |
| 6. | Jain N., et al., 2020 | India | 29 epitopes | Nucleocapsid protein, surface glycoprotein, membrane protein, envelope protein | Multi-epitope peptide based vaccine candidate against SARS-CoV-2 | Jain et al. (2021) |
| 7. | Dong R., et al., 2020 | China | 44 epitopes | Nucleocapsid phosphoprotein, envelope protein, endoRNAse membrane glycoprotein | Multi-epitopic vaccine developed from T and B cell epitopes of S-protein | Dong et al. (2020) |
| 8. | Kumar A., et al., 2020 | India | 56 epitopes | Nucleocapsid protein, Envelope protein, spike glycoprotein | Prediction and selection of multi-epitope, and in silico cloning of vaccine construct | Kumar et al. (2020) |
| 9. | Khairkhah N., et al., 2020 | Iran | 46 epitopes | Spike glycoprotein, nucleocapsid protein, membrane protein | Three multi-epitope constructs for peptide based vaccine candidate | Khairkhah et al. (2020) |
| 10. | Samad A., et al., 2020 | Bangladesh, Saudi Arabia | 6 epitopes | Spike glycoprotein | Multi-epitopic subunit vaccine construction and structural evaluation | Samad et al. (2020) |
| 11. | Qamar M., et al., 2020 | China, Pakistan | 13 epitopes | Surface glycoprotein, envelope protein, and membrane glycoprotein | Multi-epitopic peptide vaccine construction and in silico cloning | Tahir ul Qamar et al. (2020) |
| 12. | Fatoba A., et al., 2021 | South Africa, Nigeria | 18 epitopes | Surface and membrane glycoproteins | Design of multi-epitope vaccine from surface and membrane glycoprotein | Fatoba et al. (2021) |
| 13. | Mahapatra S.R., et al., 2020 | India | 20 epitopes | Spike protein, envelope protein, membrane protein, nucleocapsid protein | Epitope selection from multiple glycoproteins and vaccine construction | Mahapatra et al. (2020) |
| 14. | Behmard E., et al., 2020 | Iran | 46 epitopes | Spike glycoprotein, envelope protein, membrane protein, and nucleocapsid phosphoprotein | Construction and molecular modeling of multi-epitopic peptide vaccine | Behmard et al. (2020) |
| 15. | Oladipo E.K., et al., 2021 | Nigeria | 15 epitopes | Surface glycoprotein | Conserved peptide-based antigenic, non-toxic and non-allergic subunit vaccine | Oladipo et al. (2021) |
| 16. | Srivastava S., et al., 2020 | India | 103 epitopes | ORF proteins | Multi-patch protein vaccine constructs | Srivastava et al. (2020) |
| 17. | Albagi S., et al., 2020 | Sudan, India, Turkey | 6 epitopes | Nucleocapsid phosphoprotein and spike glycoprotein | Peptides vaccine designed from the nucleocapsid phosphoprotein and S- protein | Abd Albagi et al. (2020) |
| 18. | Ghorbani A., et al., 2020 | Iran | 10 epitopes | Spike glycoprotein | Virus-like particle based vaccine developed from epitopes of S-protein | Ghorbani et al. (2020) |
| 19. | Waqas M., et al., 2020 | Pakistan | 28 epitopes | Main protease | Multi-epitopic peptide vaccine construct from SARS-CoV-2 | Waqas et al. (2021) |
| 20. | Abduljaleel Z., et al., 2020 | Saudi Arabia, Canada | 12 Epitopes | Spike protein, membrane glycoprotein, envelop protein and nucleocapsid protein | Vaccine construct developed by antigenic epitope peptides fragments | Abduljaleel et al. (2021) |
| 21. | Khan T., et al., 2021 | Bangladesh, USA | 26 epitopes | Nucleocapsid protein, membrane protein, envelope protein, spike, protein, ORF and non-structural proteins | Effective peptide-based multi-epitope vaccine | Khan et al. (2021b) |
| 22. | Lim H., et al., 2020 | Malaysia | 7 epitopes | Spike glycoprotein, nucleocapsid protein, membrane protein | Vaccine construct from conserved peptides epitopes | Lim et al. (2020) |
| 23. | Rahman N., et al., 2020 | Pakistan, Czech Republic | 4 epitopes | Surface glycoprotein | Peptide-based multi-epitope five vaccine constructs developed | Rahman et al. (2020) |
| 24. | Sanami S., et al., 2020 | Iran | 18 epitopes | Spike protein | Vaccine development from the T and B cell epitopes of S-protein | Sanami et al. (2020) |
| 25. | Bhattacharya M., et al., 2021 | India, South Korea | 23 epitopes | Spike protein | Multi-epitopic peptide vaccine construct against the Wuhan variant and all significant mutant variants of SARS-CoV-2 | Bhattacharya et al. (2021) |
| 26. | Khan et al., 2021 | China, Pakistan, Kuwait | 11 epitopes | Spike protein | Multi-epitopes subunit vaccine from the S- protein of the SARS-CoV-2 new variants | Khan et al. (2021a) |
Table 2.
Different immunoinformatics and bioinformatics approaches on epitopes identification towards SARS-CoV-2 vaccine research
| Sl. no. | Researcher | Country | Nos. epitopes | Contributing viral proteins | Remarks | References |
|---|---|---|---|---|---|---|
| 1. | Joshi A., et al., 2020 | India | 9 epitopes | Envelope protein, nulceocapsid phosphoprotein, membrane glycoprotein ORF-3a and ORF-7a | Putative epitope selection from SARS-CoV-2 against HLA allelic proteins | Joshi et al. (2020) |
| 2. | Singh J., et al., 2021 | India | 5 epitopes | Spike glycoprotein | Potential linear, structural B cell epitope and T cell epitopes were predicted from eight different SARS-COV-2 strain | Singh et al. (2021) |
| 3. | Kiyotani K., et al., 2020 | Japan | 3412 epitopes | Spike, envelope, membrane, and nucleocapsid proteins, nonstructural proteins (6 ORF) | Identified numbers of possible peptide epitopes from SARS-COV-2 structural and nonstructural proteins | Kiyotani et al. (2020) |
| 4. | Oliveira S C., et al., 2020 | Brazil, United States | 135 epitopes | Nucleocapsid Protein | Major B and T cell epitopes are predicted from the SARS-CoV-2 nucleocapsid protein | Oliveira et al. (2020) |
| 5. | Chen H., et al., 2020 | China | 63 epitopes | Spike protein, nucleocapsid protein | B cell epitopes and T cell epitopes were predicted from SARS-CoV-2 S-protein and N protein | Chen et al. (2020) |
| 6. | Wang D., et al., 2020 | China, USA | 71 epitopes | Spike protein | Potential B cell and T cell epitopes from S- protein were predicted for vaccine design | Wang et al. (2020) |
| 7. | Lin L., et al., 2020 | China | 30 epitopes | Surface glycoprotein, membrane glycoprotein and nucleocapsid protein | T cell epitopes and B cell epitopes identified from multiple protein segment of SARS-CoV-2 | Lin et al. (2020) |
| 8. | Rakib A., et al., 2020 | Bangladesh, Indonesia, Morocco, Saudi Arabia | 10 epitopes | Spike glycoprotein | Optimal epitopes were identified from S- protein of SARS-CoV-2 | Rakib et al. (2020) |
| 9. | Jakhar R., et al., 2020 | India | 10 epitopes | Envelope protein | Epitopes were identified from envelope protein of SARS-CoV-2 | Jakhar and Gakhar (2020) |
| 10. | Lizbeth R., et al., 2020 | México | 4 epitopes | Spike glycoprotein | Identified four epitopes from SARS-CoV-2 S-protein | Lizbeth et al. (2020) |
| 11. | Mukherjee S., et al., 2020 | Israel | 17 epitopes | Membrane glycoprotein, nucleocapsid phosphoprotein, spike glycoprotein | Epitopes were identified from whole genome and proteome of SARS-CoV-2 | Mukherjee et al. (2020) |
| 12. | Crooke S., et al., 2020 | USA | 47 epitopes | Spike glycoprotein, envelope protein, membrane protein | Identified T cell epitopes and B cell epitopes from structural, non-structural and accessory proteins of SARS-CoV-2 | Crooke et al. (2020) |
| 13. | Ranga V., et al., 2020 | Finland | 15 epitopes | RNA-dependent RNA polymerase, membrane glycoprotein, envelope protein, nucleocapsid phosphoprotein, 3C-like proteinase, surface glycoprotein, ORF and other non-structural protein | Epitopes were identified from 26 protein sequences encoded by the SARS-CoV-2 genomic sequence | Ranga et al. (2020) |
| 14. | Ashik A., et al., 2020 | Bangladesh | 3 epitopes | Spike glycoprotein | Altered epitopes were predicted from the S-protein of SARS-CoV-2 | Ashik et al. (2020) |
| 15. | Baruah V., et al., 2020 | India | 13 epitopes | Surface glycoprotein | Multiple, conserved epitopes were identified in the SARS-CoV-2 | Baruah and Bose (2020) |
| 16. | Bhattacharya M., et al., 2020 | India, South Korea | 4 epitopes | Spike glycoprotein | Common (B and T cell) epitopes were identified from the S-protein of SARS-CoV-2 | Bhattacharya et al. (2020c) |
| 17. | Tilocca B., et al., 2020 | Italy | 8 epitopes | Envelope protein | Epitopes having high antigenicity were mapped and characterized from SARS-CoV-2 | Tilocca et al. (2020) |
| 18. | Rencilin CF., et al., 2020 | India, USA | 18 epitopes | ORF, envelope protein, membrane glycoprotein, nucleocapsid Phosphoprotein | Conserved epitopes were identified from the complete proteome of SARS-CoV-2 | Rencilin et al. (2021) |
| 19. | Lon JR., et al., 2020 | China | 7 epitopes | spike protein, envelope protein and membrane protein | Seven epitopes were predicted from the nucleocapsid phosphoprotein of SARS-CoV-2 | Lon et al. (2020) |
| 20. | Ong E., et al., 2021 | USA | 301 epitopes | Spike protein | Numbers of T cell epitopes were identified from S-protein of SARS-CoV-2 | Ong et al. (2021) |
It was observed that only a few groups of scientists developed the vaccine construct against SARS-CoV-2 and performed docking with the Toll-like Receptor (TLR) group of molecules to understand the TLR based downstream regulation of the protective/adaptive immunity. Simultaneously, quite a few scientists have analyzed the complex stability with molecular dynamics simulation. Furthermore, we have found that a small number of scientist groups evaluated the vaccine construct's allergenicity and immunogenicity. Even few researchers have performed normal mode analysis (NMA) analyses, in-silico cloning of vaccine candidates, and analyzed the physicochemical properties using immunoinformatics and bioinformatics. Analysis of the physicochemical properties is necessary to understand the solubility, molecular weight, theoretical isoelectric point (pI), estimated half-life, instability index, aliphatic index, and grand average of hydropathicity (GRAVY) of the vaccine candidate. All these steps are very crucial for evaluating a successful vaccine construct while utilizing bioinformatics and immunoinformatics.
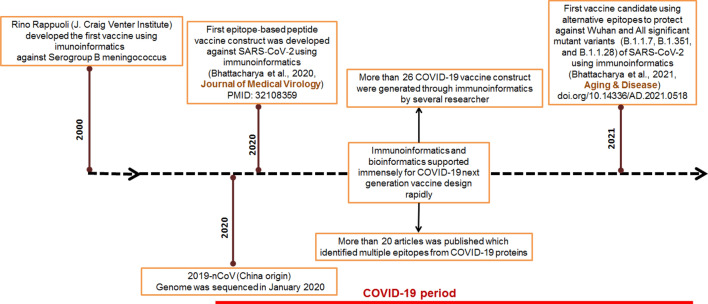
On 10 January 2020, the Chinese research group was the first to sequence the SARS-CoV-2 genome. Zhang and his colleagues sequenced the genome at Fudan University and made it publicly available in GenBank (Fan et al. 2020; Triggle et al. 2020). After the availability of the genome sequence in GenBank, several researchers started to identify the antigenic epitopes using the sequence through immunoinformatics and bioinformatics. Immunoinformatics approaches for COVID-19 vaccine research were triggered because of two reasons. Firstly, this approach can design the vaccine rapidly (Fig. 2). Secondly, there was an urgency for the COVID-19 vaccine throughout the globe. Most researchers targeted viral spike (S)-protein in their vaccine design analysis to identify the epitopes as it was found from the previous studies that S-protein has the maximum antigenic epitope regions (Dai and Gao 2020). In addition, the previous studies have also shown that S glycoprotein in the other coronaviruses (SARS-CoV-1, MERS-CoV-2) has the highest antigenic epitopes. So, this knowledge of the prior research helped the researchers to develop the COVID-19 vaccine candidates quickly. Alternatively, several researchers also tried to identify epitopic areas from other structural proteins (M protein, E protein, N protein)/ proteome along with S-protein.
Fig. 2.
Some important milestone of immunoinformatics and bioinformatics studies that stimulated the next-generation vaccine research against SARS-CoV-2
We have performed a comprehensive, advanced search on PubMed with the keywords "immunoinformatics" and "COVID-19" and found that 88 articles have been published so far on this topic (Fig. 3). Most of the article deals with the immunoinformatics-based vaccine development, the safety and efficacy analysis of vaccine construct, and different immunological component analyses related to SARS-CoV-2. The immunoinformatics approach has also been applied to find out different vaccine constructs for other coronaviruses (SARS-CoV-1, MERS-CoV-2). Few of them even have tried to develop a trivalent subunit vaccine construct for three emerging coronaviruses using immunoinformatics approaches. Several immunoinformatic databases have been developed to illustrate the immunogenicity and virulence of glycoproteins of coronaviruses and others. One such example of a database is DBCOVP which provides the information about conserved B cell, and T cell epitopes predicted from the protein (Sahoo et al. 2021).
Fig. 3.
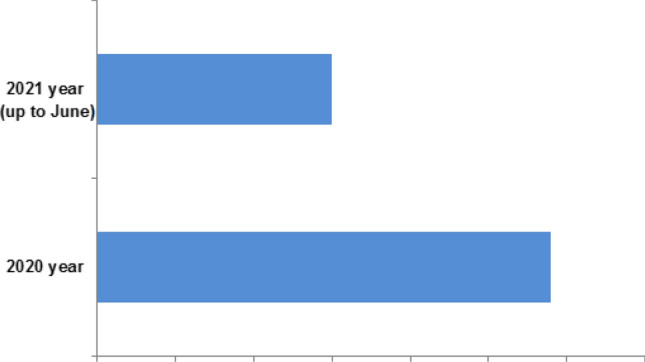
PubMed search using keywords “immunoinformatics” and “COVID-19” which illustrated the number of publications of immunoinformatics based COVID-19 research in the year of 2020 and 2021 (up to June)
Epitope-based COVID-19 vaccines are the next-generation COVID-19 vaccines, posing a highly antigenic part and an adjuvant. The antigenic component is also selected through the common epitopes (B and T cell) selection procedure. It can be more effective in generating adaptive immunity. Also, the vaccine can trigger innate immunity and stimulate the secretion of protective cytokines through interaction with TLRs. However, these vaccines have shown some limitations. One such limitation observed was blood clot formation after using the COVID-19 vaccine made by AstraZeneca (Wolf et al. 2021). Other types of vaccines (live attenuated COVID-19 vaccine) also have some limitations. For example, live attenuated vaccines may suffer secondary mutation, which can revive virulence from the attenuated microorganism and lead to the occurrence of disease.
Immunoinformatics is now at the forefront of the development of the next-generation COVID-19 vaccine. Recently, Ishack and Lipner have published a significant commentary that described the immense role of immunoinformatics and bioinformatics on COVID-19 vaccine development (Ishack and Lipner 2021). However, there are several challenges ahead for immunoinformatics in vaccine research that need to address instantly. Firstly, advancement in the development of algorithms for immunoinformatics and bioinformatics. These algorithms will help to perform a more accurate and faster calculation without any computational errors. Secondly, some algorithms are available to illustrate the adaptive and innate immunity scenario after vaccination; however, more research data (in vitro and in vivo) is required to validate their claim. Thirdly, consideration of several factors associated with effective multi-epitope vaccine construct activity, such as the combination of epitopes and peptide linkers. One such example is that the stability of the vaccine candidate depends on the linker peptide. Fourthly, no epitope-based vaccine has thrived against some diseases until today (e.g., HIV, malaria). For these diseases, the causative organism possesses several antigenic proteins. In these cases, epitopes of these proteins are not adequately mapped, and the highly potent antigenic protein is difficult to identify. Therefore more extensive researches are required in this direction. However, soon, immunoinformatics will address all the challenges for COVID-19 vaccine research and help to design next-generation vaccines for all the infectious diseases and neglected diseases in coming times.
Supplementary Information
Below is the link to the electronic supplementary material.
Author Contributions
Writing, review, and editing the final draft, CC, ARS; validation and analysis, MB; revising and supervision, CC, SSL. All authors have read and approved the final version of this manuscript.
Funding
None.
Data availability
All data includes within the manuscript.
Declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Chiranjib Chakraborty and Ashish Ranjan Sharma contributed equally to this work.
Contributor Information
Chiranjib Chakraborty, Email: drchiranjib@yahoo.com.
Sang-Soo Lee, Email: 123sslee@gmail.com.
References
- Abd Albagi SO, et al. A multiple peptides vaccine against COVID-19 designed from the nucleocapsid phosphoprotein (N) and Spike Glycoprotein (S) via the immunoinformatics approach. Inform Med Unlocked. 2020;21:100476. doi: 10.1016/j.imu.2020.100476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abduljaleel Z, Al-Allaf FA, Aziz SA. Peptides-based vaccine against SARS-n CoV-2 antigenic fragmented synthetic epitopes recognized by T cell and β-cell initiation of specific antibodies to fight the infection. Biodes Manuf. 2021;4:490–505. doi: 10.1007/s42242-020-00114-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashik AI, Hasan M, Tasnim AT, Chowdhury MB, Hossain T, Ahmed S. An immunoinformatics study on the spike protein of SARS-CoV-2 revealing potential epitopes as vaccine candidates. Heliyon. 2020;6:e04865. doi: 10.1016/j.heliyon.2020.e04865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baden LR, Rubin EJ. Covid-19—the search for effective therapy. N Engl J Med. 2020;382:1851–1852. doi: 10.1056/NEJMe2005477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baruah V, Bose S. Immunoinformatics-aided identification of T cell and B cell epitopes in the surface glycoprotein of 2019-nCoV. J Med Virol. 2020;92:495–500. doi: 10.1002/jmv.25698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behmard E, Soleymani B, Najafi A, Barzegari E. Immunoinformatic design of a COVID-19 subunit vaccine using entire structural immunogenic epitopes of SARS-CoV-2. Sci Rep. 2020;10:20864. doi: 10.1038/s41598-020-77547-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya M, et al. A SARS-CoV-2 vaccine candidate: in-silico cloning and validation. Inform Med Unlocked. 2020;20:100394. doi: 10.1016/j.imu.2020.100394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya M, et al. Development of epitope-based peptide vaccine against novel coronavirus 2019 (SARS-COV-2): immunoinformatics approach. J Med Virol. 2020;92:618–631. doi: 10.1002/jmv.25736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya M, Sharma AR, Mallick B, Sharma G, Lee S-S, Chakraborty C. Immunoinformatics approach to understand molecular interaction between multi-epitopic regions of SARS-CoV-2 spike-protein with TLR4/MD-2 complex. Infect Genet Evol. 2020;85:104587. doi: 10.1016/j.meegid.2020.104587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya M, Sharma AR, Ghosh P, Lee S-S, Chakraborty C. A next-generation vaccine candidate using alternative epitopes to protect against wuhan and all significant mutant variants of SARS-CoV-2: an immunoinformatics approach. Aging Dis. 2021 doi: 10.14336/AD.2021.0518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty C, Sharma AR, Bhattacharya M, Agoramoorthy G, Lee SS. Asian-origin approved COVID-19 vaccines and current status of COVID-19 vaccination program in Asia: a critical analysis. Vaccines. 2021;9:600. doi: 10.3390/vaccines9060600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty C, Sharma AR, Bhattacharya M, Sharma G, Lee S-S. Immunoinformatics approach for the identification and characterization of T cell and B cell epitopes towards the peptide-based vaccine against SARS-CoV-2. Arch Med Res. 2021;52:362–370. doi: 10.1016/j.arcmed.2021.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty C, Sharma AR, Bhattacharya M, Lee SS. From COVID-19 to cancer mRNA vaccines: moving from bench to clinic in the vaccine landscape. Front Immunol. 2021;12:2648. doi: 10.3389/fimmu.2021.679344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H-Z, Tang L-L, Yu X-L, Zhou J, Chang Y-F, Wu X. Bioinformatics analysis of epitope-based vaccine design against the novel SARS-CoV-2. Infect Dis Poverty. 2020;9:88. doi: 10.1186/s40249-020-00713-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins FS. COVID-19 lessons for research. American Science. 2021;371:1081. doi: 10.1126/science.abh3996. [DOI] [PubMed] [Google Scholar]
- Crooke SN, Ovsyannikova IG, Kennedy RB, Poland GA. Immunoinformatic identification of B cell and T cell epitopes in the SARS-CoV-2 proteome. Sci Rep. 2020;10:1–15. doi: 10.1038/s41598-020-70864-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai L, Gao GF. Viral targets for vaccines against COVID-19. Nat Rev Immunol. 2020;21:73–82. doi: 10.1038/s41577-020-00480-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong R, Chu Z, Yu F, Zha Y. Contriving multi-epitope subunit of vaccine for COVID-19: immunoinformatics approaches. Front Immunol. 2020;11:1784. doi: 10.3389/fimmu.2020.01784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan W, et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatoba AJ, Maharaj L, Adeleke VT, Okpeku M, Adeniyi AA, Adeleke MA. Immunoinformatics prediction of overlapping CD8+ T-cell, IFN-γ and IL-4 inducer CD4+ T-cell and linear B-cell epitopes based vaccines against COVID-19 (SARS-CoV-2) Vaccine. 2021;39:1111–1121. doi: 10.1016/j.vaccine.2021.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghorbani A, Zare F, Sazegari S, Afsharifar A, Eskandari M, Pormohammad A. Development of a novel platform of virus-like particle (VLP)-based vaccine against COVID-19 by exposing epitopes: an immunoinformatics approach. New Microbes New Infect. 2020;38:100786. doi: 10.1016/j.nmni.2020.100786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishack S, Lipner SR. Bioinformatics and immunoinformatics to support COVID-19 vaccine development. J Med Virol. 2021 doi: 10.1002/jmv.27017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain N, Shankar U, Majee P, Kumar A. Scrutinizing the SARS-CoV-2 protein information for designing an effective vaccine encompassing both the T-cell and B-cell epitopes. Infect Genet Evol. 2021;87:104648. doi: 10.1016/j.meegid.2020.104648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakhar R, Gakhar S. An Immunoinformatics study to predict epitopes in the envelope protein of SARS-CoV-2. Can J Infect Dis Med Microbiol. 2020;2020:7079356. doi: 10.1155/2020/7079356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi A, Joshi BC, Mannan MA, Kaushik V. Epitope based vaccine prediction for SARS-COV-2 by deploying immuno-informatics approach. Inform Med Unlocked. 2020;19:100338. doi: 10.1016/j.imu.2020.100338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalita P, Padhi AK, Zhang KY, Tripathi T. Design of a peptide-based subunit vaccine against novel coronavirus SARS-CoV-2. Microb Pathog. 2020;145:104236. doi: 10.1016/j.micpath.2020.104236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khairkhah N, Aghasadeghi MR, Namvar A, Bolhassani A. Design of novel multiepitope constructs-based peptide vaccine against the structural S, N and M proteins of human COVID-19 using immunoinformatics analysis. PLoS ONE. 2020;15:e0240577. doi: 10.1371/journal.pone.0240577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan A, et al. Immunogenomics guided design of immunomodulatory multi-epitope subunit vaccine against the SARS-CoV-2 new variants, and its validation through in silico cloning and immune simulation. Comput Biol Med. 2021;133:104420. doi: 10.1016/j.compbiomed.2021.104420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan MT, et al. Immunoinformatics and molecular modeling approach to design universal multi-epitope vaccine for SARS-CoV-2. Inform Med Unlocked. 2021;24:100578. doi: 10.1016/j.imu.2021.100578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyotani K, Toyoshima Y, Nemoto K, Nakamura Y. Bioinformatic prediction of potential T cell epitopes for SARS-Cov-2. J Hum Genet. 2020;65:569–575. doi: 10.1038/s10038-020-0771-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Kumar P, Saumya KU, Kapuganti SK, Bhardwaj T, Giri R. Exploring the SARS-CoV-2 structural proteins for multi-epitope vaccine development: an in-silico approach. Expert Rev Vaccines. 2020;19:887–898. doi: 10.1080/14760584.2020.1813576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim HX, Lim J, Jazayeri SD, Poppema S, Poh CL. Development of multi-epitope peptide-based vaccines against SARS-CoV-2. Biomed J. 2020;44:18–30. doi: 10.1016/j.bj.2020.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L, Ting S, Yufei H, Wendong L, Yubo F, Jing Z. Epitope-based peptide vaccines predicted against novel coronavirus disease caused by SARS-CoV-2. Virus Res. 2020;288:198082. doi: 10.1016/j.virusres.2020.198082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lizbeth R-SG, Jazmín G-M, José C-B, Marlet M-A. Immunoinformatics study to search epitopes of spike glycoprotein from SARS-CoV-2 as potential vaccine. J Biomol Struct Dyn. 2020;25:1–15. doi: 10.1080/07391102.2020.1780944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lon JR, Bai Y, Zhong B, Cai F, Du H. Prediction and evolution of B cell epitopes of surface protein in SARS-CoV-2. Virol J. 2020;17:165. doi: 10.1186/s12985-020-01437-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahapatra SR, Sahoo S, Dehury B, Raina V, Patro S, Misra N, Suar M. Designing an efficient multi-epitope vaccine displaying interactions with diverse HLA molecules for an efficient humoral and cellular immune response to prevent COVID-19 infection. Expert Rev Vaccines. 2020;19:871–885. doi: 10.1080/14760584.2020.1811091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee S, Tworowski D, Detroja R, Mukherjee SB, Frenkel-Morgenstern M. Immunoinformatics and structural analysis for identification of immunodominant epitopes in SARS-CoV-2 as potential vaccine targets. Vaccines. 2020;8:290. doi: 10.3390/vaccines8020290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oladipo EK, et al. Designing a conserved peptide-based subunit vaccine against SARS-CoV-2 using immunoinformatics approach. Silico Pharmacol. 2021;9:1–21. doi: 10.1007/s40203-020-00062-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira SC, de Magalhães MT, Homan EJ. Immunoinformatic analysis of SARS-CoV-2 nucleocapsid protein and identification of COVID-19 vaccine targets. Front Immunol. 2020;11:587615. doi: 10.3389/fimmu.2020.587615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong E, Huang X, Pearce R, Zhang Y, He Y. Computational design of SARS-CoV-2 spike glycoproteins to increase immunogenicity by T cell epitope engineering. Comput Struct Biotechnol J. 2021;19:518–529. doi: 10.1016/j.csbj.2020.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman N, et al. Vaccine design from the ensemble of surface glycoprotein epitopes of SARS-CoV-2: an immunoinformatics approach. Vaccines. 2020;8:423. doi: 10.3390/vaccines8030423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakib A, et al. Immunoinformatics-guided design of an epitope-based vaccine against severe acute respiratory syndrome coronavirus 2 spike glycoprotein. Comput Biol Med. 2020;124:103967. doi: 10.1016/j.compbiomed.2020.103967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranga V, Niemelä E, Tamirat MZ, Eriksson JE, Airenne TT, Johnson MS. Immunogenic SARS-CoV-2 epitopes: in silico study towards better understanding of COVID-19 disease—paving the way for vaccine development. Vaccines. 2020;8:408. doi: 10.3390/vaccines8030408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rencilin CF, Rosy JC, Mohan M, Coico R, Sundar K. Identification of SARS-CoV-2 CT L epitopes for development of a multivalent subunit vaccine for COVID-19. Infect Genet Evol. 2021;89:104712. doi: 10.1016/j.meegid.2021.104712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers F, Pepperrell T, Keestra S, Pilkington V. Missing clinical trial data: the evidence gap in primary data for potential COVID-19 drugs. Trials. 2021;22:59. doi: 10.1186/s13063-021-05024-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha R, Ghosh P, Burra VP. Designing a next generation multi-epitope based peptide vaccine candidate against SARS-CoV-2 using computational approaches. 3 Biotech. 2021;11:47. doi: 10.1007/s13205-020-02574-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahoo S, et al. DBCOVP: a database of coronavirus virulent glycoproteins. Comput Biol Med. 2021;129:104131. doi: 10.1016/j.compbiomed.2020.104131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samad A, Ahammad F, Nain Z, Alam R, Imon RR, Hasan M, Rahman MS. Designing a multi-epitope vaccine against SARS-CoV-2: an immunoinformatics approach. J Biomol Struct Dyn. 2020 doi: 10.1080/07391102.2020.1792347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanami S, et al. Design of a multi-epitope vaccine against SARS-CoV-2 using immunoinformatics approach. Int J Biol Macromol. 2020;164:871–883. doi: 10.1016/j.ijbiomac.2020.07.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh J, Malik D, Raina A. Immuno-informatics approach for B-cell and T-cell epitope based peptide vaccine design against novel COVID-19 virus. Vaccine. 2021;39:1087–1095. doi: 10.1016/j.vaccine.2021.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava S, et al. Computationally validated SARS-CoV-2 CTL and HTL Multi-Patch vaccines, designed by reverse epitomics approach, show potential to cover large ethnically distributed human population worldwide. J Biomol Struct Dyn. 2020 doi: 10.1080/07391102.2020.1838329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahir ul Qamar M, et al. Designing of a next generation multiepitope based vaccine (MEV) against SARS-COV-2: immunoinformatics and in silico approaches. PLoS ONE. 2020;15:e0244176. doi: 10.1371/journal.pone.0244176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilocca B, et al. Immunoinformatic analysis of the SARS-CoV-2 envelope protein as a strategy to assess cross-protection against COVID-19. Microbes Infect. 2020;22:182–187. doi: 10.1016/j.micinf.2020.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triggle CR, Bansal D, Abd Farag EAB, Ding H, Sultan AA. COVID-19: learning from lessons to guide treatment and prevention interventions. Msphere. 2020;5:e00317–e320. doi: 10.1128/mSphere.00317-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ul Qamar MT, et al. Reverse vaccinology assisted designing of multiepitope-based subunit vaccine against SARS-CoV-2. Infect Dis Poverty. 2020;9:132. doi: 10.1186/s40249-020-00752-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, et al. Immunoinformatic analysis of T-and B-cell epitopes for SARS-CoV-2 vaccine design. Vaccines. 2020;8:355. doi: 10.3390/vaccines8030355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waqas M, et al. Immunoinformatics and molecular docking studies predicted potential multiepitope-based peptide vaccine and novel compounds against novel SARS-CoV-2 through Virtual screening. Biomed Res Int. 2021;2021:1596834. doi: 10.1155/2021/1596834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf ME, Luz B, Niehaus L, Bhogal P, Bäzner H, Henkes H. Thrombocytopenia and intracranial venous sinus thrombosis after “COVID-19 vaccine AstraZeneca” exposure. J Clin Med. 2021;10:1599. doi: 10.3390/jcm10081599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yazdani Z, Rafiei A, Yazdani M, Valadan R. Design an efficient multi-epitope peptide vaccine candidate against SARS-CoV-2: an in silico analysis. Infect Drug Resist. 2020;13:3007. doi: 10.2147/IDR.S264573. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data includes within the manuscript.